- 1Austrian Agency for Health and Food Safety (AGES), Institute for Medical Microbiology and Hygiene, Vienna, Austria
- 2Research Division of Biochemical Technology, Institute of Chemical, Environmental and BioScience Engineering, Technische Universität Wien, Vienna, Austria
- 3French Agency for Food, Environmental and Occupational Health & Safety (ANSES), Ploufragan-Plouzané-Niort Laboratory, Ploufragan, France
- 4French Agency for Food, Environmental and Occupational Health & Safety (ANSES), Fougeres Laboratory, Fougeres, France
- 5Institute for Hygiene and Applied Immunology - Water Microbiology, Medical University Vienna, Vienna, Austria
- 6Division Water Quality and Health, Karl Landsteiner University of Health Sciences, Krems an der Donau, Austria
- 7Interuniversity Cooperation Centre for Water and Health, Vienna, Austria
Vibrio cholerae belonging to serogroups other than O1 and O139 are opportunistic pathogens which cause infections with a variety of clinical symptoms. Due to the increasing number of V. cholerae non-O1/non-O139 infections in association with recreational waters in the past two decades, they have received increasing attention in recent literature and by public health authorities. Since the treatment of choice is the administration of antibiotics, we investigated the distribution of antimicrobial resistance properties in a V. cholerae non-O1/non-O139 population in a large Austrian lake intensively used for recreation and in epidemiologically linked clinical isolates. In total, 82 environmental isolates - selected on the basis of comprehensive phylogenetic information - and nine clinical isolates were analyzed for their phenotypic antimicrobial susceptibility. The genomes of 46 environmental and eight clinical strains were screened for known genetic antimicrobial resistance traits in CARD and ResFinder databases. In general, antimicrobial susceptibility of the investigated V. cholerae population was high. The environmental strains were susceptible against most of the 16 tested antibiotics, except sulfonamides (97.5% resistant strains), streptomycin (39% resistant) and ampicillin (20.7% resistant). Clinical isolates partly showed additional resistance to amoxicillin-clavulanic acid. Genome analysis showed that crp, a regulator of multidrug efflux genes, and the bicyclomycin/multidrug efflux system of V. cholerae were present in all isolates. Nine isolates additionally carried variants of blaCARB–7 and blaCARB–9, determinants of beta-lactam resistance and six isolates carried catB9, a determinant of phenicol resistance. Three isolates had both blaCARB–7 and catB9. In 27 isolates, five out of six subfamilies of the MATE-family were present. For all isolates no genes conferring resistance to aminoglycosides, macrolides and sulfonamides were detected. The apparent lack of either known antimicrobial resistance traits or mobile genetic elements indicates that in cholera non-epidemic regions of the world, V. cholerae non-O1/non-O139 play a minor role as a reservoir of resistance in the environment. The discrepancies between the phenotypic and genome-based antimicrobial resistance assessment show that for V. cholerae non-O1/non-O139, resistance databases are currently inappropriate for an assessment of antimicrobial resistance. Continuous collection of both data over time may solve such discrepancies between genotype and phenotype in the future.
Introduction
Antibiotics are supposed to be one of the most effective drugs to treat infectious diseases transmitted by bacteria (Martinez, 2009). Nevertheless, antimicrobial resistant bacteria pose one of the most significant healthcare challenges (WHO, 2017). Emerging antibiotic resistance is not only restricted to healthcare associated settings, but also appears in environmental settings, such as natural aquatic ecosystems (Taylor et al., 2011). The Gram-negative, potentially pathogenic bacterium Vibrio cholerae is a natural inhabitant of aquatic environments from freshwater to seawater. It can thrive in a free floating manner in the water body (Worden et al., 2006; Kirschner et al., 2008) but it has also been found on surfaces of crustaceans, algae, plants, and insects (Colwell, 1996; Halpern et al., 2004; Banerjee et al., 2014). Currently, more than 200 different serogroups have been described and only serogroups O1 and O139 are the agents of epidemic cholera. The remaining non-epidemic serogroups are therefore referred as V. cholerae non-O1/non-O139. They are opportunistic pathogens associated with small outbreaks of diarrheal disease (Deshayes et al., 2015; Hasan et al., 2015; Crowe et al., 2016). They can also cause severe wound infections and acute sepsis especially in immunocompromised patients (Sack et al., 2004; Huhulescu et al., 2007; Hirk et al., 2016; Siriphap et al., 2017). In recent years, infections due to V. cholerae non-O1/non-O139 have been increasingly reported for countries of the Northern hemisphere (Newton et al., 2012; Baker-Austin et al., 2013), primarily acquired through contact by swimming in seawater (Baker-Austin et al., 2016b; Marinello et al., 2017) or freshwater lakes (Dalsgaard et al., 2000a; Ninin et al., 2000; Andersson and Ekdahl, 2006; Ottaviani et al., 2011; Dobrovic et al., 2016; Hirk et al., 2016; Kechker et al., 2017). This significant increase in the number of infections has been linked to global warming and associated increase in water temperatures (Baker-Austin et al., 2016a, b; ECDC, 2019). Infections caused by V. cholerae non-O1/non-O139 have also been reported for the large subsaline lake Neusiedler See in Austria (Huhulescu et al., 2007). This lake is part of the largest bird sanctuary in Central Europe (National Park Neusiedler See - Seewinkel) and it is intensively used for recreational purposes throughout the year (swimming, surfing, sailing, fishing etc.). Recently, it was demonstrated, that the lake harbors an abundant and genetically diverse endemic V. cholerae community (Schauer et al., 2015; Pretzer et al., 2017). In the latter study (Pretzer et al., 2017), it was also shown that many V. cholerae isolates from other European countries were genetically related to the strains present in the lake, most probably due to the long distance import of strains (via birds or humans) from these countries. Thus, the Neusiedler See may serve as a bioreactor for the appearance of new strains with new (pathogenic) properties, including antibiotic resistance.
Despite the increasing clinical importance of environmental V. cholerae, only very limited information is available on antimicrobial susceptibility of V. cholerae non-O1/non-O139 populations in aquatic ecosystems of the Northern hemisphere (Aberkane et al., 2015; Bier et al., 2015; Baron et al., 2017). Drug resistance of V. cholerae is mediated by efflux pumps, by chromosomal mutations and by acquisition of conjugative plasmids, transposons, integrative and conjugative elements (ICE) such as SXT elements carrying antibiotic resistance genes (Kitaoka et al., 2011; Das et al., 2019). Aquatic ecosystems provide a suitable milieu for spreading antimicrobial resistant traits through horizontal gene transfer among bacterial populations (Baquero et al., 2008). Recent publications reported a limited capacity of V. cholerae non-O1/non-O139 strains to acquire resistance. Apparently, most of the investigated collections from Western European waters were susceptible to most antibiotic tested (Bier et al., 2015; Baron et al., 2017). However, multidrug resistant V. cholerae have been increasingly reported worldwide, mainly in clinical O1 and O139 strains (Faruque et al., 2007; Mandal et al., 2011; Verma et al., 2019), but also in environmental non-O1/non-O139 isolates in cholera epidemic areas (Kitaoka et al., 2011). First carbapenemase producing V. cholerae non-O1/non-O139 strains were recently described in Western Europe (Aberkane et al., 2015; Bier et al., 2015). Therefore, monitoring antimicrobial resistance of environmental populations of V. cholerae non-O1/non-O139 is highly relevant for public health.
The aim of this study was to determine the phenotypic and genotypic antimicrobial resistance properties of 91 V. cholerae non-O1/non-O139 strains (82 representative isolates from Neusiedler See and 9 Austrian clinical, epidemiologically linked isolates), by combining in vitro antimicrobial susceptibility testing and in silico detection of antimicrobial resistance traits from whole genome sequencing (WGS) data. By this a comprehensive assessment of the role of the V. cholerae population in Neusiedler See as a potential reservoir of environmental antimicrobial resistance was achieved. Since this lake may serve as model bioreactor for the appearance of new pathogenic/resistant Vibrio strains our findings are also relevant for other subsaline/hyposaline aquatic environments that are used for recreation.
MATERIALS AND METHODS
Description of the Study Area and Origin of Isolates
Neusiedler See is a large (320 km2) subsaline, alkaline lake in Eastern Austria, intensively used for recreational purposes with several hundred thousand visitors throughout the year. The lake is located in the temperate climate zone and has experienced an increase in the average water temperature during the past 30 years of around 1.5°C (Dokulil et al., 2010). Details on the lake can be found in earlier publications (Kirschner et al., 2008; Schauer et al., 2015).
Eighty-two environmental V. cholerae non-O1/non-O139 isolates for the analysis of antibiotic resistance properties were selected from an existing strain collection consisting of 472 isolates (Pretzer et al., 2017). The majority of these 472 isolates belong to 24 phylogenetic clades, based on multi-locus sequence analysis (MLSA) (Pretzer et al., 2017). Depending on the size of the clades (i.e., the number of isolates belonging to the clades), between one and six representatives of each clade (n = 46) and additional singletons (n = 36) were randomly selected, in order to cover the bulk of the genetic diversity of V. cholerae in the lake (Supplementary Figure S1). All selected V. cholerae environmental isolates were collected at Neusiedler See between May 2011 and October 2012 (Supplementary Figure S1).
For comparison, nine V. cholerae non-O1/non-O139 strains from human infections which had a reported epidemiological link to Neusiedler See, collected between 2000 and 2015, were also included in this study (Supplementary Table S1). All of these strains had been sent to the national reference laboratory for Vibrio at the Austrian Agency for Health and Food Safety (AGES). Six of the strains had caused otitis externa, one of them sepsis, another one prostatitis and for one strain no clinical symptoms had been reported.
Phenotypic Susceptibility Testing
The susceptibility of the 91 isolates was determined by disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines for rapidly growing pathogens (CLSI, 2018b) including the following 16 antimicrobial agents: ampicillin, amoxicillin-clavulanic acid, cefotaxime, imipenem, chloramphenicol, nalidixic acid, ciprofloxacin, norfloxacin, amikacin, gentamicin, streptomycin, tetracycline, doxycycline, sulfonamides (sulfamethoxazole, sulfisoxazole, sulfadiazine), trimethoprim-sulfamethoxazole, and erythromycin. The CLSI interpretative criteria for disk diffusion susceptibility testing of Vibrio spp. (CLSI, 2015, 2018a) were used (Supplementary Table S2), except for erythromycin for which no breakpoint was available. In Baron et al. (2017), the disc diffusion zones of inhibition of the wild type population were distributed between 17 and 24 mm. Therefore, any erythromycin inhibition zone diameter measured larger or equal to 17 mm was interpreted as susceptible. Non-susceptible categories (intermediate and resistant) were interpreted as resistant for this study. The term “multidrug resistant” was used for isolates being non-susceptible to at least one agent of three or more different antimicrobial classes as previously defined by ECDC and CDC experts (Magiorakos et al., 2012).
Whole-Genome Sequencing and Antimicrobial Resistance Gene Detection
From the 82 phenotypically tested environmental isolates, 46 isolates were selected for WGS from all but three identified phylogenetic clades (n = 36) and a few singletons (n = 10), in order to cover the bulk of the genetic diversity of V. cholerae in Neusiedler See (Supplementary Figure S1). In addition, eight of the nine clinical isolates were also sequenced. Bacterial total DNA was isolated from overnight cultures using the MagAttract HMW DNA Kit (Qiagen, Hilden, Germany) and DNA concentration was determined with a Trinean DropSense16 (Unchained Labs, CA, United States). Libraries for WGS were prepared using the Nextera XT DNA Library Preparation Kit (Illumina, CA, United States) according to the manufacturer’s instructions. All isolates were paired-end sequenced using MiSeq Reagent Kit v3 with a read length of 2 × 300 base pairs on a MiSeq instrument (Illumina, CA, United States). Velvet version 1.1.04 (Zerbino and Birney, 2008) was used for de novo assembly of raw reads and further WGS data interpretation was carried out in the analysis software SeqSphere + (Ridom, Münster, Germany).
The presence of antibiotic resistance genes was assessed with the Comprehensive Antibiotic Resistance Database (CARD) (Jia et al., 2017) using default settings including “Perfect” and “Strict” hits only. In addition, the ResFinder database was used with default settings. Allele libraries for aminoglycosides (167 targets), macrolides (132 targets) and sulfonamides (49 targets) resistance were downloaded from the ResFinder database (11/2017) available from the Center for Genomic Epidemiology1 and integrated in the SeqSphere + analysis software. In silico target scan procedure settings in SeqSphere + for reference sequences were defined with 90% required identity to reference sequence and 99% aligned to reference sequence. The following reference sequences were included: species-specific gene (ompW; accession no. FJ462451), SXT element integrase gene (int; AF099172), class 1 integron (intI1; EU436855), dihydrofolate reductase (dfrA18; AY034138), quinolone resistance protein (qnrVC1; EU436855), bicyclomycin/multidrug efflux system from V. cholerae O1 biovar El Tor str. N16961 (locus VC1634) and VcmA, VcmB, VcmD, VcmH, VcmN, VcrM belonging to the MATE family multidrug efflux pumps (Multidrug And Toxic Compound Extrusion) (Huda et al., 2001, 2003; Begum et al., 2005; Kuroda and Tsuchiya, 2009).
Nucleotide Sequence Accession Numbers
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession PRJNA5519292. For details see Supplementary Table S3. The version described in this paper is the first version.
Results
Phenotypic Susceptibility
Environmental Isolates
All isolates were susceptible to amoxicillin-clavulanic acid, cefotaxime, imipenem, chloramphenicol, ciprofloxacin, norfloxacin, amikacin, gentamicin, tetracycline, trimethoprim, trimethoprim-sulfamethoxazole (SXT) and erythromycin. All but one of 82 isolates were resistant to at least one of the tested antibiotics (Figure 1). Eighty (97.5%) were resistant to sulfonamides, 54 of them were only detected resistant to this antibiotic (44%). Twenty-five isolates (30%) were resistant to sulfonamides and streptomycin, twelve (15%) to sulfonamides and ampicillin and one isolate (1.1%) was only resistant to ampicillin. Seven isolates (9%) were multidrug resistant against sulfonamides, streptomycin and ampicillin.
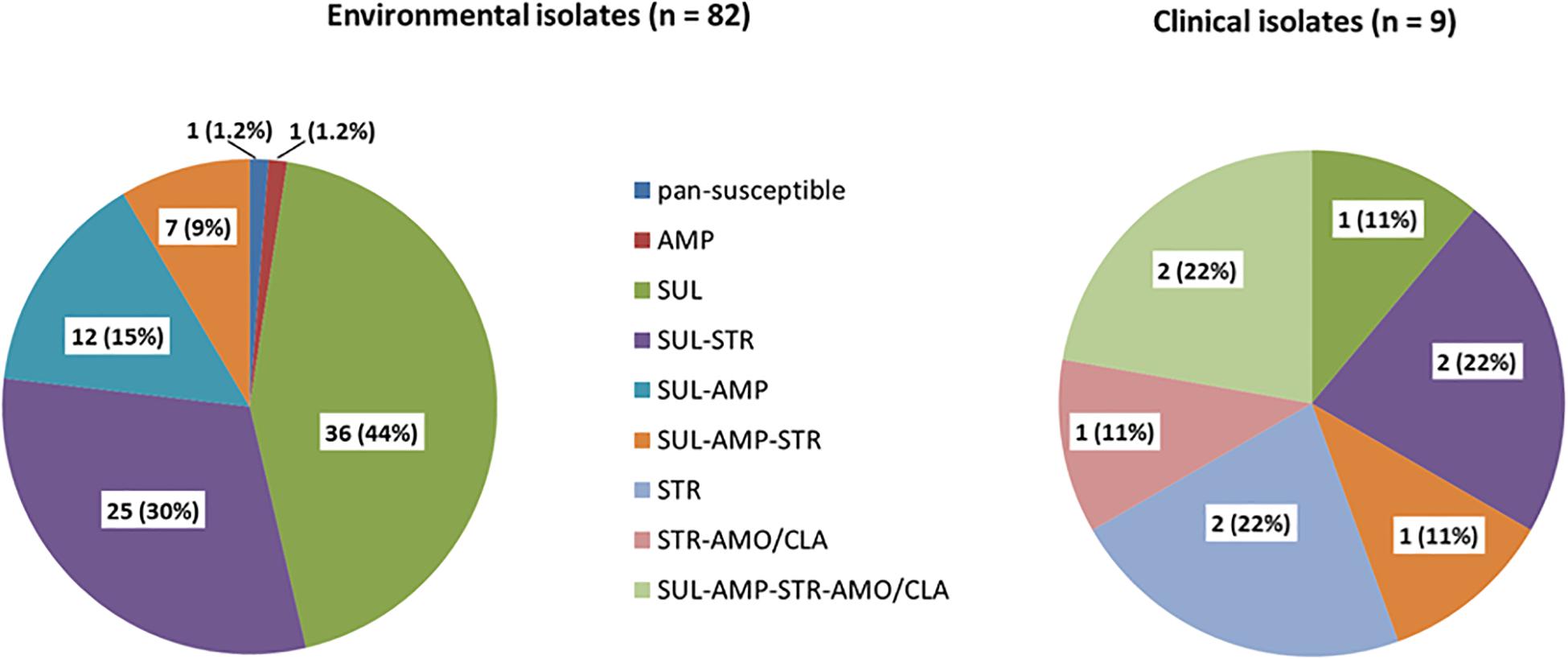
Figure 1. Percentage of phenotypic antibiotic resistance of 82 environmental and nine clinical isolates. SUL (sulfonamides), STR (streptomycin), AMP (ampicillin), NAL (nalidixic acid), AMO/CLA (amoxicillin-clavulanic acid).
Clinical Isolates
Overall, antibiotic resistance profiles were very similar in human and environmental isolates. Resistances against sulfonamides, streptomycin and ampicillin were the most frequently detected (Figure 1). The main difference observed was the occurrence of amoxicillin/clavulanic acid resistance in three clinical isolates. Three out of the nine isolates were multidrug resistant. None was pan-susceptible.
The full detailed list of all isolates tested and the results of each phenotypic antimicrobial resistance test can be found in the Supplementary Table S4.
Identification of Antibiotic Resistance Genes
The coverage obtained for the 54 whole genome sequenced isolates (46 environmental and eight clinical) was 68.2-fold and the species-specific gene ompW for V. cholerae was present in all investigated isolates. Analysis via CARD identified gene tet(34), an antibiotic inactivation enzyme and determinant of tetracycline resistance, and gene CRP, a regulator of multidrug efflux genes, to be present in all 54 investigated isolates (Supplementary Table S4).
Twelve isolates additionally carried variants of the antibiotic inactivation enzymes blaCARB–7 (n = 10) or blaCARB–9 (n = 2), determinants of beta-lactam resistance, perfectly matching the observed phenotypic resistance against the beta-lactam antibiotic ampicillin. Six isolates additionally carried the inactivation enzyme catB9, a determinant of phenicol resistance, despite the fact that no chloramphenicol resistance was observed for these isolates. Three isolates carried both blaCARB–7 and catB9. For all 54 isolates, no genes conferring resistance to aminoglycosides, macrolides nor sulfonamides were detected. The integron 1 integrase gene intI1, the two SXT integron-related resistance factors dfrA18, qnrVC1 and diverse SXT elements were absent in all investigated isolates. The bicyclomycin/multidrug efflux system locus of V. cholerae strain VC1634, which is involved in sulfonamide and bicyclomycin resistance, was present in all isolates. In 27 of 54 isolates five (VcmA, VcmB, VcmD, VcmH, VcrM) out of six subfamilies of the MATE (Multidrug and toxic compound extrusion)-family were present (Table 1 and Supplementary Table S4). VcmN was never detected.
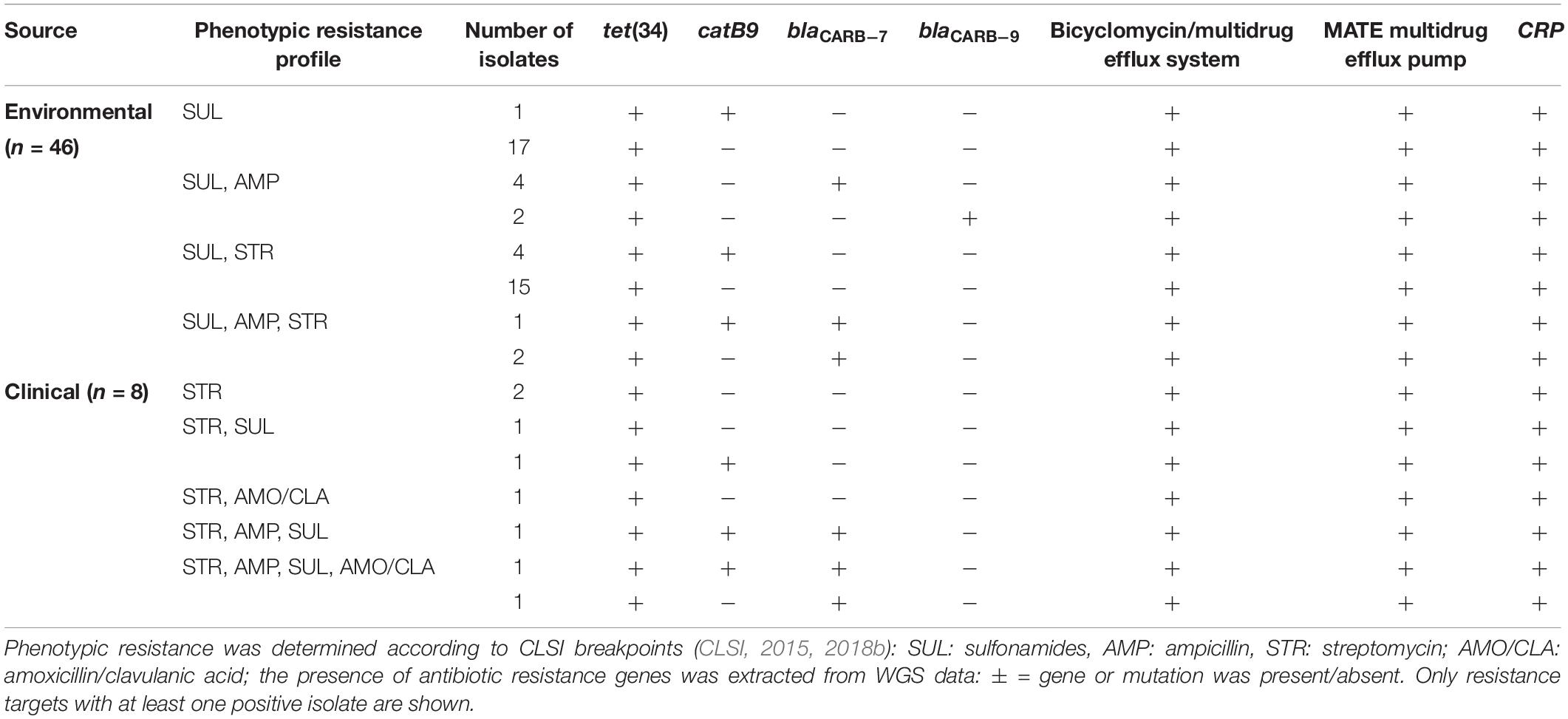
Table 1. Phenotypic and genotypic resistance profiles of the 54 whole genome sequenced V. cholerae isolates.
Discussion
All Strains Are Susceptible to First-Line Treatment Options and Carbapenems
All environmental and clinical isolates were susceptible to ciprofloxacin, norfloxacin, doxycycline and extended spectrum cephalosporins, the recommended treatment options for V. cholerae non-O1/non-O139 infections (Daniels and Shafaie, 2000). Thus, a loss of effectiveness of first-line treatments for V. cholerae infections related to Neusiedler See is not observed so far. Moreover, in agreement with the susceptibility to extended spectrum cephalosporins and carbapenems, no extended spectrum β-lactamase or carbapenemase genes were identified. Carbapenemase genes in Vibrio cholerae non-O1/non-O139 populations are uncommon in Europe, carbapenemase genes were so far only found in avian swab samples in France (Aberkane et al., 2015), a United Kingdom patient flying back from India (Darley et al., 2012) and the presence of undefined carbapenemases was reported for four Baltic Sea isolates (Bier et al., 2015).
Wide-Spread Resistance Against “Old Antibiotics”
Among the 82 environmental isolates, phenotypic resistances were only observed against the “old antibiotics” such as sulfonamides, streptomycin and ampicillin which had been widely used before the 1970’s, 9% of the isolates were multidrug resistant. Widespread resistance of V. cholerae non-O1/non-O139 strains against these substances has also been observed in wastewater and cockles of a Western European estuary (Baron et al., 2017). In other aquatic ecosystems from cholera free regions of the temperate climate zone, resistance to streptomycin and ampicillin was reported as well in the Baltic Sea and the North Sea (Bier et al., 2015) and the Chesapeake Bay (Ceccarelli et al., 2015). However, resistance to sulfonamides alone was not tested in those studies but only in combination with trimethoprim. Overall, the percentage of resistance against these three antibiotics was higher in Neusiedler See in comparison to the other investigations. In our study, 98% of the strains were resistant to sulfonamides, while in the Western European estuary it was 45%. Concerning ampicillin, 24% of our isolates showed resistance in comparison to 9% (Baron et al., 2017), 10% (Bier et al., 2015) and 7% (Ceccarelli et al., 2015); concerning streptomycin it was 39% resistant isolates from Neusiedler See in comparison to 22% (Baron et al., 2017), 25% (Bier et al., 2015) and 8% (Ceccarelli et al., 2015). The high rates of acquired resistances against sulfonamides and streptomycin in the isolates of Neusiedler See indicate a continuous selective pressure from these antibiotic classes. So far, there is no evidence showing the presence of those antibiotics in the lake triggering such resistance and monitoring of these antibiotics in the lake would be necessary. Currently, the presence of antibiotics in water by constant monitoring is not regulated in Austria and other European countries, which makes it difficult to assess the actual cause for the observed antibiotic resistance of V. cholerae non-O1/non-O139 strains (Inreiter et al., 2016). Streptomycin is widely used in plant agriculture, predominantly for combating fire blight and it has been permitted on an emergency use basis, subject to annual review and under tightly restricted conditions, in Austria (Stockwell and Duffy, 2012). High acquired resistance rates to sulfonamides have been observed in V. cholerae O1 (Eibach et al., 2016) but investigations on V. cholerae non-O1/non-O139 are rare. Concerning resistance to beta-lactams, the first strain of V. cholerae with acquired resistance to ampicillin due to a plasmid located beta-lactamase has been detected in 1977 (Hedges et al., 1977). From the 1990s onward several studies describe the prevalence of ampicillin resistance observed in environmental strains of V. cholerae non-O1/non-O139 in different parts of the world (Choury et al., 1999; Dalsgaard et al., 2000b; Melano et al., 2002; Petroni et al., 2004). The ampicillin resistance genes are carried by class 1 integrons. At least three carbenicillin-hydrolyzing beta-lactamases (CARB-6, CARB-7, and CARB-9) have been described in V. cholerae (Choury et al., 1999; Melano et al., 2002; Petroni et al., 2004). In our study, twenty ampicillin resistant environmental isolates were identified. From the nine sequenced isolates, a perfect match between the presence beta-lactamase genes and ampicillin resistance phenotype was observed.
The clinical isolates showed similar resistance patterns as the environmental isolates. However, three of them (33%) were additionally resistant to the combination of amoxicillin and clavulanic acid. No such resistance was found for patient and environmental isolates from the North and Baltic Sea region (Bier et al., 2015), but 19 of 42 environmental isolates from the Adriatic Sea (45%) and 3 of 9 Italian clinical isolates (33%) were resistant to the combination of penicillins and beta-lactamase inhibitors (Ottaviani et al., 2018). Nevertheless, prudent interpretation of the present data is required, as the three strains categorized as resistant to amoxicillin – clavulanic acid, displayed an inhibition zone diameter just below the susceptibility breakpoint. This breakpoint is supposed to be used “for Vibrio spp. other than V. cholerae” (CLSI, 2015). Due to limitations of the method, it can be hypothesized that these three strains have probably been falsely categorized as resistant.
Divergent Phenotypic and Genotypic Antimicrobial Profiles
Several divergent phenotypic and genotypic antimicrobial findings were obtained. All sequenced isolates harbored the tet(34) gene, encoding for a tetracycline inactivation enzyme (Nonaka and Suzuki, 2002), while all isolates were susceptible to tetracyclines. It should be noticed that the tet(34) gene was originally cloned from a Vibrio spp. and oxytetracycline resistance was only expressed in presence of MgCl2 (Nonaka and Suzuki, 2002). It is possible that under in vitro conditions of our susceptibility tests, gene tet(34) was not expressed. Moreover, to date the clinical relevance of gene tet(34) is still unknown (Grossman, 2016). In six out of 46 environmental and three out of eight fully sequenced clinical isolates, the catB9 gene was present. This gene, a chromosome-encoded variant of the cat gene found in V. cholerae encodes for a chloramphenicol acetyltransferase and confers resistance to chloramphenicol (Rowe-Magnus et al., 2002). In contrast, none of the isolates was resistant to chloramphenicol. No gene conferring sulfonamide resistance (e.g., sul2) and streptomycin (e.g., strB) for V. cholerae was found using either CARD or ResFinder databases, although nearly all of our strains were tested phenotypically resistant to these two antibiotics. These findings indicate that current databases are partly incomplete for assessing antimicrobial resistance properties of V. cholerae non-O1/non-O139 strains on genotypic characterization alone. Despite the fact that WGS allows detection of a much higher number of resistance markers, the clinical relevance of most of them still needs to be determined. Clinicians should thus still rely on phenotypic characterization of antimicrobial resistance properties of V. cholerae strains instead of depending on in silico assessment alone (Nahar and Bin Rashid, 2017). Vice-versa, when a resistance mechanism is detected in the genome, while the isolate is phenotypically susceptible, the interpretation criteria of the antibiogram may also be questioned. In fact, the literature comparing phenotypic and genotypic antimicrobial resistance in V. cholerae non-O1/non-O139 is still poor compared to either E. coli, K. pneumoniae or S. aureus. Continuous collection of both data over time may solve such discrepancies between genotype and phenotype in the future.
Both Clinical and Environmental Isolates Do Not Harbor Integrons
Despite the fact that V. cholerae is a highly diverse species that is able to easily take up extracellular DNA (Matthey and Blokesch, 2016), no common integrons and other integrative and conjugative elements able to carry antimicrobial resistance determinants have been identified. All sequenced clinical and environmental strains were negative for SXT elements or class I integron. This finding is in agreement with the other studies (Bier et al., 2015; Ceccarelli et al., 2015; Baron et al., 2017) and suggests that in non-cholera epidemic regions, the dissemination of antibiotic resistance genes via mobile genetic elements through V. cholerae colonization of the aquatic environment is probably of very limited impact.
Ampicillin Resistance Is Concentrated in Specific Phylogenetic Lineages
Finally, we tried to elucidate whether the observed phenotypic and genotypic profiles of the environmental strains were reflected by their phylogeny (see Supplementary Figure S1; Pretzer et al., 2017). Only two resistance traits were suitable for such inter-comparison - ampicillin resistance (n = 20) and the presence of the catB9 gene (n = 6), as all other traits were too abundant. From the identified 150 sequence types (STs) (Pretzer et al., 2017), the 20 ampicillin resistant isolates belonged to 15 different STs with eleven of the isolates belonging to only two out of 24 identified phylogenetic clades (Supplementary Table S5). Four isolates belonged to ST 75 (clade III) and three to ST 59 (clade I). In addition, one ST 62 and one ST 65 isolate clustered with ST 75 isolates in clade III and one ST 64 and one ST 76 isolate clustered with ST 59 isolates in clade I. These data show that ampicillin resistance, and consequently the CARB-7 and CARB-9 encoding genes are present only in specific phylogenetic lineages and not evenly distributed among the V. cholerae population in the lake. No such phylogenic relationship was observed for the catB9 gene.
Conclusion
The large Austrian lake Neusiedler See is an important bathing water with several hundred thousand visitors each year. As it is harboring an abundant and diverse natural V. cholerae non-O1/non-O139 community associated with several cases of human infection each year, the comprehensive assessment of the antimicrobial resistance properties of this pathogen is of public health concern. For this purpose, we combined phenotypic antibiotic susceptibility tests with genotypic characterization of environmental and clinical whole-genome-sequenced isolates, covering most of phylogenetic V. cholerae diversity in the lake. The specific model characteristics of the lake as a bioreactor for the appearance of new pathogenic/resistant Vibrio strains render our findings relevant also for other subsaline/hyposaline aquatic environments that are used for recreational purposes.
Our results clearly showed that all isolates were susceptible to all important first-line and last-line treatment options. Widespread resistance was only observed against old antibiotics such as sulfonamides, streptomycin and ampicillin. The apparent lack of either known antimicrobial resistance traits or mobile genetic elements indicates that in cholera non-epidemic regions of the world, V. cholerae non-O1/non-O139 play only a minor role as reservoir of antibiotic resistance genes. Nonetheless, the observed discrepancies between the phenotypic and genotypic resistance data highlight the need for improvement of the resistance databases for V. cholerae non-O1/non-O139 to better understand how these pathogens acquire and disseminate resistance traits in the aquatic environment.
Data Availability Statement
The whole genome sequencing datasets generated for this study can be found in the DDBJ/EMBL/GenBank; accession PRJNA551929.
Author Contributions
AK, AI, SP, SB, RM, and WR: conceptualization. CP, EL, SB, SL, and SG: methodology. SL, WR, CP, SB, and SG: software analysis. AK, AI, SP, and SB: validation. CP, EL, SB, and SL: investigation. AK, AI, and SB: resources. AK and WR: data curation. SL, SP, SB, AK, CP, EL, SG, and WR: writing–original draft preparation. AK, SB, AI, AF, SP, SG, WR, and RM: writing–review and editing. AK: project administration. AK, AF, AI, and SB: funding acquisition.
Funding
This work was funded by the Austrian Science Fund (FWF, project nr P21625-B20) and the Province of Burgenland.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Stefan Jakwerth for his help in isolating environmental V. cholerae strains from Neusiedler See.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02600/full#supplementary-material
Footnotes
References
Aberkane, S., Compain, F., Barraud, O., Ouedraogo, A. S., Bouzinbi, N., Vittecoq, M., et al. (2015). Non-O1/Non-O139 Vibrio cholerae avian isolate from france cocarrying the bla(VIM-1) and bla(VIM-4) genes. Antimicrob. Agents Chemother. 59, 6594–6596. doi: 10.1128/AAC.00400-15
Andersson, Y., and Ekdahl, K. (2006). Wound infections due to Vibrio cholerae in Sweden after swimming in the Baltic Sea, summer 2006. Eur. Surveill. 11:E060803060802.
Baker-Austin, C., Trinanes, J., Gonzalez-Escalona, N., and Martinez-Urtaza, J. (2016a). Non-cholera Vibrios: the microbial barometer of climate change. Trends Microbiol. 11, 30140–30148. doi: 10.1016/j.tim.2016.09.008
Baker-Austin, C., Trinanes, J. A., Salmenlinna, S., Lofdahl, M., Siitonen, A., Taylor, N. G., et al. (2016b). Heat wave-associated vibriosis, Sweden and Finland, 2014. Emerg. Infect. Dis. 22, 1216–1220. doi: 10.3201/eid2207.151996
Baker-Austin, C., Trinanes, J. A., Taylor, N. G. H., Hartnell, R., Siitonen, A., and Martinez-Urtaza, J. (2013). Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Change 3, 73–77. doi: 10.1038/nclimate1628
Banerjee, R., Das, B., Balakrish Nair, G., and Basak, S. (2014). Dynamics in genome evolution of Vibrio cholerae. Infect. Genet. Evol. 23, 32–41. doi: 10.1016/j.meegid.2014.01.006
Baquero, F., Martínez, J. L., and Cantón, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Baron, S., Larvor, E., Chevalier, S., Jouy, E., Kempf, I., Granier, S. A., et al. (2017). Antimicrobial susceptibility among urban wastewater and wild shellfish isolates of non-O1/Non-O139 Vibrio cholerae from La Rance Estuary (Brittany, France). Front. Microbiol. 8:1637. doi: 10.3389/fmicb.2017.01637
Begum, A., Rahman, M. M., Ogawa, W., Mizushima, T., Kuroda, T., and Tsuchiya, T. (2005). Gene cloning and characterization of four MATE family multidrug efflux pumps from Vibrio cholerae non-O1. Microbiol. Immunol. 49, 949–957. doi: 10.1111/j.1348-0421.2005.tb03690.x
Bier, N., Schwartz, K., Guerra, B., and Strauch, E. (2015). Survey on antimicrobial resistance patterns in Vibrio vulnificus and Vibrio cholerae non-O1/non-O139 in Germany reveals carbapenemase-producing Vibrio cholerae in coastal waters. Front. Microbiol. 6:1179. doi: 10.3389/fmicb.2015.01179
Ceccarelli, D., Chen, A., Hasan, N. A., Rashed, S. M., Huq, A., and Colwell, R. R. (2015). Non-O1/non-O139 Vibrio cholerae carrying multiple virulence factors and V. cholerae O1 in the Chesapeake Bay, Maryland. Appl. Environ. Microbiol. 81, 1909–1918. doi: 10.1128/AEM.03540-14
Choury, D., Aubert, G., Szajnert, M. F., Azibi, K., Delpech, M., and Paul, G. (1999). Characterization and nucleotide sequence of CARB-6, a new carbenicillin-hydrolyzing beta-lactamase from Vibrio cholerae. Antimicrob. Agents Chemother. 43, 297–301. doi: 10.1128/aac.43.2.297
CLSI, (2015). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria CLSI guideline, 3rd Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI, (2018a). Performance Standards for Antimicrobial Disk Suseptibility Tests; 13th edition; Standard M02. Wayne, PA: CLSI.
CLSI, (2018b). Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI Supplement M100. Wayne, P.A: Clinical and Laboratory Standards Institute.
Colwell, R. R. (1996). Global climate and infectious disease: the cholera paradigm. Science 274, 2025–2031. doi: 10.1126/science.274.5295.2025
Crowe, S. J., Newton, A. E., Gould, L. H., Parsons, M. B., Stroika, S., Bopp, C. A., et al. (2016). Vibriosis, not cholera: toxigenic Vibrio cholerae non-O1, non-O139 infections in the United States, 1984-2014. Epidemiol. Infect. 144, 3335–3341. doi: 10.1017/s0950268816001783
Dalsgaard, A., Forslund, A., Hesselbjerg, A., and Bruun, B. (2000a). Clinical manifestations and characterization of extra-intestinal Vibrio cholerae non-O1, non-O139 infections in Denmark. Clin. Microbiol. Infect. 6, 625–627. doi: 10.1046/j.1469-0691.2000.00174.x
Dalsgaard, A., Forslund, A., Serichantalergs, O., and Sandvang, D. (2000b). Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob. Agents Chemother. 44, 1315–1321. doi: 10.1128/aac.44.5.1315-1321.2000
Daniels, N. A., and Shafaie, A. (2000). A review of pathogenic Vibrio infections for clinicians. Infect. Med. 17, 665–685.
Darley, E., Weeks, J., Jones, L., Daniels, V., Wootton, M., Macgowan, A., et al. (2012). NDM-1 polymicrobial infections including Vibrio cholerae. Lancet 380:1358. doi: 10.1016/s0140-6736(12)60911-8
Das, B., Verma, J., Kumar, P., Ghosh, A., and Ramamurthy, T. (2019). Antibiotic resistance in Vibrio cholerae: understanding the ecology of resistance genes and mechanisms. Vaccine doi: 10.1016/j.vaccine.2019.06.031 [Epub ahead of print].
Deshayes, S., Daurel, C., Cattoir, V., Parienti, J. J., Quilici, M. L., and De La Blanchardiere, A. (2015). Non-O1, non-O139 Vibrio cholerae bacteraemia: case report and literature review. Springerplus 4:575. doi: 10.1186/s40064-015-1346-3
Dobrovic, K., Rudman, F., Ottaviani, D., Sestan Crnek, S., Leoni, F., and Skrlin, J. (2016). A rare case of necrotizing fasciitis caused by Vibrio cholerae O8 in an immunocompetent patient. Wien Klin. Wochenschr. 128, 728–730. doi: 10.1007/s00508-016-1060-3
Dokulil, M. T., Teubner, K., Jagsch, A., Nickus, U., Adrian, R., Straile, D., et al. (2010). “The impact of climate change on lakes in Central Europe,” in Aquatic Ecology Series 4: The Impact of Climate Change on European Lakes, ed. G. D. George, (Dordrecht: Springer), 387–409. doi: 10.1007/978-90-481-2945-4_20
ECDC. (2019). “Monitoring environmental suitability of Vibrio growth in the Baltic Sea, Summer 2019,” in Communicable Disease Threats Report Week 27, 30 June-6 July, 2019, (Solna Municipality: ECDC).
Eibach, D., Herrera-Leon, S., Gil, H., Hogan, B., Ehlkes, L., Adjabeng, M., et al. (2016). Molecular epidemiology and antibiotic susceptibility of Vibrio cholerae associated with a large cholera outbreak in Ghana in 2014. PLoS Negl. Trop. Dis. 10:e0004751. doi: 10.1371/journal.pntd.0004751
Faruque, A. S., Alam, K., Malek, M. A., Khan, M. G., Ahmed, S., Saha, D., et al. (2007). Emergence of multidrug-resistant strain of Vibrio cholerae O1 in Bangladesh and reversal of their susceptibility to tetracycline after two years. J. Health Popul. Nutr. 25, 241–243.
Grossman, T. H. (2016). Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 6:a025387. doi: 10.1101/cshperspect.a025387
Halpern, M., Broza, Y. B., Mittler, S., Arakawa, E., and Broza, M. (2004). Chironomid egg masses as a natural reservoir of Vibrio cholerae non-O1 and non-O139 in freshwater habitats. Microb. Ecol. 47, 341–349.
Hasan, N. A., Rezayat, T., Blatz, P. J., Choi, S. Y., Griffitt, K. J., Rashed, S. M., et al. (2015). Nontoxigenic Vibrio cholerae non-O1/O139 isolate from a case of human gastroenteritis in the U.S. Gulf Coast. J. Clin. Microbiol. 53, 9–14. doi: 10.1128/JCM.02187-14
Hedges, R. W., Vialard, J. L., Pearson, N. J., and O’grady, F. (1977). R plasmids from Asian strains of Vibrio cholerae. Antimicrob. Agents Chemother. 11, 585–588. doi: 10.1128/aac.11.4.585
Hirk, S., Huhulescu, S., Allerberger, F., Lepuschitz, S., Rehak, S., Weil, S., et al. (2016). Necrotizing fasciitis due to Vibrio cholerae non-O1/non-O139 after exposure to Austrian bathing sites. Wien Klin. Wochenschr. 128, 141–145. doi: 10.1007/s00508-015-0944-y
Huda, M. N., Chen, J., Morita, Y., Kuroda, T., Mizushima, T., and Tsuchiya, T. (2003). Gene cloning and characterization of VcrM, a Na+-coupled multidrug efflux pump, from Vibrio cholerae non-O1. Microbiol. Immunol. 47, 419–427. doi: 10.1111/j.1348-0421.2003.tb03379.x
Huda, M. N., Morita, Y., Kuroda, T., Mizushima, T., and Tsuchiya, T. (2001). Na+-driven multidrug efflux pump VcmA from Vibrio cholerae non-O1, a non-halophilic bacterium. FEMS Microbiol. Lett. 203, 235–239. doi: 10.1016/s0378-1097(01)00363-9
Huhulescu, S., Indra, A., Feierl, G., Stoeger, A., Ruppitsch, W., Sarkar, B., et al. (2007). Occurrence of Vibrio cholerae serogroups other than O1 and O139 in Austria. Wien Klin. Wochenschr. 119, 235–241. doi: 10.1007/s00508-006-0747-2
Inreiter, N., Huemer, B., Springer, B., Humer, F., and Allerberger, F. (2016). Antibiotics in Austrian drinking water resources, survey 2014. Die Bodenkultur 67, 35–43. doi: 10.1515/boku-2016-0004
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Kechker, P., Senderovich, Y., Ken-Dror, S., Laviad-Shitrit, S., Arakawa, E., and Halpern, M. (2017). Otitis media caused by V. cholerae O100: a case report and review of the literature. Front Microbiol 8:1619. doi: 10.3389/fmicb.2017.01619
Kirschner, A. K. T., Schlesinger, J., Farnleitner, A. H., Hornek, R., Süß, B., Golda, B., et al. (2008). Rapid growth of planktonic Vibrio cholerae non-O1/non-O139 strains in a large alkaline lake in Austria: dependence on temperature and dissolved organic carbon quality. Appl. Environ. Microbiol. 74, 2004–2015. doi: 10.1128/AEM.01739-07
Kitaoka, M., Miyata, S. T., Unterweger, D., and Pukatzki, S. (2011). Antibiotic resistance mechanisms of Vibrio cholerae. J. Med. Microbiol. 60, 397–407. doi: 10.1099/jmm.0.023051-0
Kuroda, T., and Tsuchiya, T. (2009). Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 1794, 763–768. doi: 10.1016/j.bbapap.2008.11.01
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mandal, S., Mandal, M. D., and Pal, N. K. (2011). Cholera: a great global concern. Asian Pac. J. Trop. Med. 4, 573–580. doi: 10.1016/S1995-7645(11)60149-1
Marinello, S., Marini, G., Parisi, G., Gottardello, L., Rossi, L., Besutti, V., et al. (2017). Vibrio cholerae non-O1, non-O139 bacteraemia associated with pneumonia. Italy 2016. Infection 45, 237–240. doi: 10.1007/s15010-016-0961-4
Martinez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902. doi: 10.1016/j.envpol.2009.05.051
Matthey, N., and Blokesch, M. (2016). The DNA-uptake process of naturally competent Vibrio cholerae. Trends Microbiol. 24, 98–110. doi: 10.1016/j.tim.2015.10.008
Melano, R., Petroni, A., Garutti, A., Saka, H. A., Mange, L., Pasteran, F., et al. (2002). New carbenicillin-hydrolyzing beta-lactamase (CARB-7) from Vibrio cholerae non-O1, non-O139 strains encoded by the VCR region of the V. cholerae genome. Antimicrob. Agents Chemother. 46, 2162–2168. doi: 10.1128/aac.46.7.2162-2168.2002
Nahar, N., and Bin Rashid, R. (2017). In silico assessment of the genotypic distribution of virulence and antibiotic resistance genes in Vibrio cholerae. Dhaka Univ. J Pharm. Sci. 16, 77–85. doi: 10.3329/dujps.v16i1.33385
Newton, A., Kendall, M., Vugia, D. J., Henao, O. L., and Mahon, B. E. (2012). Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin. Infect. Dis. 54(Suppl. 5), S391–S395. doi: 10.1093/cid/cis243
Ninin, E., Caroff, N., El Kouri, D., Espaze, E., Richet, H., Quilici, M. L., et al. (2000). Nontoxigenic Vibrio cholerae O1 bacteremia: case report and review. Eur. J. Clin. Microbiol. Infect. Dis. 19, 489–491. doi: 10.1007/s100960000296
Nonaka, L., and Suzuki, S. (2002). New Mg2+-dependent oxytetracycline resistance determinant tet 34 in Vibrio isolates from marine fish intestinal contents. Antimicrob. Agents Chemother. 46, 1550–1552. doi: 10.1128/aac.46.5.1550-1552.2002
Ottaviani, D., Leoni, F., Rocchegiani, E., Canonico, C., Masini, L., Pianetti, A., et al. (2011). Unusual case of necrotizing fasciitis caused by Vibrio cholerae O137. J. Clin. Microbiol. 49, 757–759. doi: 10.1128/JCM.02257-10
Ottaviani, D., Medici, L., Talevi, G., Napoleoni, M., Serratore, P., Zavatta, E., et al. (2018). Molecular characterization and drug susceptibility of non-O1/O139 V. cholerae strains of seafood, environmental and clinical origin. Italy. Food Microbiol. 72, 82–88. doi: 10.1016/j.fm.2017.11.011
Petroni, A., Melano, R. G., Saka, H. A., Garutti, A., Mange, L., Pasteran, F., et al. (2004). CARB-9, a carbenicillinase encoded in the VCR region of Vibrio cholerae non-O1, non-O139 belongs to a family of cassette-encoded beta-lactamases. Antimicrob. Agents Chemother. 48, 4042–4046. doi: 10.1128/aac.48.10.4042-4046.2004
Pretzer, C., Druzhinina, I. S., Amaro, C., Benediktsdóttir, E., Hedenström, I., Hervio-Heath, D., et al. (2017). High genetic diversity of Vibrio cholerae in the European lake Neusiedler see is associated with intensive recombination in the reed habitat and the long-distance transfer of strains. Environ. Microbiol. 19, 328–344. doi: 10.1111/1462-2920.13612
Rowe-Magnus, D. A., Guerout, A. M., and Mazel, D. (2002). Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 43, 1657–1669. doi: 10.1046/j.1365-2958.2002.02861.x
Schauer, S., Jakwerth, S., Bliem, R., Baudart, J., Lebaron, P., Huhulescu, S., et al. (2015). Dynamics of Vibrio cholerae abundance in Austrian saline lakes, assessed with quantitative solid-phase cytometry. Environ. Microbiol. 17, 4366–4378. doi: 10.1111/1462-2920.12861
Siriphap, A., Leekitcharoenphon, P., Kaas, R. S., Theethakaew, C., Aarestrup, F. M., Sutheinkul, O., et al. (2017). Characterization and genetic variation of Vibrio cholerae isolated from clinical and environmental sources in Thailand. PLoS One 12:e0169324. doi: 10.1371/journal.pone.0169324
Stockwell, V. O., and Duffy, B. (2012). Use of antibiotics in plant agriculture. Rev. Sci. Tech. 31, 199–210.
Taylor, N. G., Verner-Jeffreys, D. W., and Baker-Austin, C. (2011). Aquatic systems: maintaining, mixing and mobilising antimicrobial resistance? Trends Ecol. Evol. 26, 278–284. doi: 10.1016/j.tree.2011.03.004
Verma, J., Bag, S., Saha, B., Kumar, P., Ghosh, T. S., Dayal, M., et al. (2019). Genomic plasticity associated with antimicrobial resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 116, 6226–6231. doi: 10.1073/pnas.1900141116
WHO (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization.
Worden, A. Z., Seidel, M., Smriga, S., Wick, A., Malfatti, F., Bartlett, D., et al. (2006). Trophic regulation of Vibrio cholerae in coastal marine waters. Environ. Microbiol. 8, 21–29. doi: 10.1111/j.1462-2920.2005.00863.x
Keywords: Vibrio cholerae, antibiotic resistance, bathing water, climate change, non-O1/non-O139
Citation: Lepuschitz S, Baron S, Larvor E, Granier SA, Pretzer C, Mach RL, Farnleitner AH, Ruppitsch W, Pleininger S, Indra A and Kirschner AKT (2019) Phenotypic and Genotypic Antimicrobial Resistance Traits of Vibrio cholerae Non-O1/Non-O139 Isolated From a Large Austrian Lake Frequently Associated With Cases of Human Infection. Front. Microbiol. 10:2600. doi: 10.3389/fmicb.2019.02600
Received: 14 August 2019; Accepted: 25 October 2019;
Published: 08 November 2019.
Edited by:
Jens Andre Hammerl, Federal Institute for Risk Assessment (BfR), GermanyReviewed by:
Christopher John Grim, United States Food & Drug Administration, United StatesEiji Arakawa, National Institute of Infectious Diseases (NIID), Japan
Copyright © 2019 Lepuschitz, Baron, Larvor, Granier, Pretzer, Mach, Farnleitner, Ruppitsch, Pleininger, Indra and Kirschner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander K. T. Kirschner, alexander.kirschner@meduniwien.ac.at
†These authors have contributed equally to this work
 Sarah Lepuschitz1,2†
Sarah Lepuschitz1,2† Sandrine Baron
Sandrine Baron Sophie A. Granier
Sophie A. Granier Carina Pretzer
Carina Pretzer Robert L. Mach
Robert L. Mach Alexander K. T. Kirschner
Alexander K. T. Kirschner