- 1Li Dak Sum Yip Yio Chin Kenneth Li Marine Biopharmaceutical Research Center, College of Food and Pharmaceutical Sciences, Ningbo University, Ningbo, China
- 2Institute of Drug Discovery Technology, Ningbo University, Ningbo, China
- 3Zhejiang Provincial Key Laboratory of Pathophysiology, School of Medicine, Ningbo University, Ningbo, China
- 4Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA, United States
- 5National Institute of Molecular Biology and Biotechnology, University of the Philippines Diliman, Quezon, Philippines
Six new dihydroisocoumarins, aspergimarins A−F (1−6), were discovered together with five known analogs (7−11) from a monoculture of the sponge-derived fungus Aspergillus sp. NBUF87. The structures of these compounds were elucidated through comprehensive spectroscopic methods, and absolute configurations were assigned after X-ray crystallography, use of the modified Mosher’s method, and comparison of electronic circular dichroism (ECD) data with literature values for previously reported analogs. Compounds 1−11 were evaluated in a variety of bioassays, and at 100 μM, both 1 and 5 showed significant inhibitory effects on the lateral root growth of Arabidopsis thaliana Columbia-0 (Col-0). Moreover, at 100 μM, 5 also possessed notable inhibition against the primary root growth of Col-0. Meanwhile, 1−11 were all found to be inactive in vitro against acetylcholinesterase (AChE) (IC50 > 100 μM), four different types of human-derived cancer cell lines (IC50 > 50 μM), as well as methicillin-resistant Staphylococcus aureus and Escherichia coli (MIC > 50 μg/mL), and Plasmodium falciparum W2 (EC50 > 100 μg/mL), in phenotypic tests.
Introduction
Sponges are among the most primitive multicellular invertebrates and harbor vast microbial populations, owing largely to the unique filter-feeding physiology that is full of pores and channels (Hentschel et al., 2006; Webster and Taylor, 2012). As a result of the long-standing interaction and coevolution with sponges, marine symbiotic microorganisms have differentiated from those of terrestrial origins in terms of their biosynthetic pathways that lead to the production of structurally interesting and biologically active compounds (Thomas et al., 2010; Fan et al., 2012). Therefore, sponge-associated microbes have become an exciting area of drug discovery research (Thomas et al., 2010; Pita et al., 2016). Endophytic fungi that are associated with sponges, especially members of the genus Aspergillus, have been recognized as a source of structurally diverse natural products with biological activities that provide value for drug discovery (Blunt et al., 2018; Zhang et al., 2018). In the past decade, the secondary metabolites from sponge-derived Aspergillus fungi have been reported from many classes, including polyketides (Wang et al., 2014; Kong et al., 2015), terpenoids (Liu et al., 2009; Li D. et al., 2012), alkaloids (Zhou et al., 2013, 2014), diketopiperazines (Ahmed et al., 2017), and peptides (Lee et al., 2011). Many of these metabolites have been shown to exhibit strong antitumor, antibacterial, antiviral, and other bioactivities.
Isocoumarins and 3,4-dihydroisocoumarins, subclasses of polyketide compounds, are also lactone-containing natural products that are abundantly produced among fungi, bacteria, liverworts, lichens, as well as some higher plants (Elsebai and Ghabbour, 2016; Saeed, 2016; Hussain and Green, 2017; Chen M. et al., 2019). Moreover, these compounds have been isolated from marine sponges, insect pheromones, and venoms (Saeed, 2016). Almost 400 isocoumarins and dihydroisocoumarins have been reported to date, and these compounds have been found to be of broad interest across many pharmacological applications (Saeed, 2016; Chen M. et al., 2019). For example, isocoumarin derivatives from some marine-derived fungi are found to possess a wide range of biological properties including enzyme inhibitory (Kim et al., 2015; Chen S. et al., 2016; Wiese et al., 2016; Cai et al., 2018), cytotoxic (Wang et al., 2019; Wu et al., 2019), antibacterial (Li S. et al., 2012; Lei et al., 2017; Chen Y. et al., 2018; Wang et al., 2019), antiproliferative (Tsukada et al., 2011), anti-food allergic (Niu et al., 2018), as well as anti-inflammatory (Kim et al., 2015; Chen Y. et al., 2018; Liu et al., 2018) activities.
As part of a continuing research program investigating the biologically active secondary metabolites from sponge-derived fungi (Ding et al., 2018; Huang et al., 2019; Li W. et al., 2019), a detailed chemical investigation was initiated on the culture of fungus Aspergillus sp. NBUF87. The fungus was isolated from a South China Sea marine sponge of the genus Hymeniacidon. The EtOAc extract of the culture of fungus Aspergillus sp. NBUF87 exhibited inhibitory effects on the root growth of Arabidopsis thaliana Columbia-0 (Col-0), a typical model organism for studying plant growth and development. The separation and purification of the bioactive extract led to the discovery of six new dihydroisocoumarin compounds (1−6) and five known analogs (7−11) (Figure 1). Herein, the detailed isolation and structure elucidation of these dihydroisocoumarin derivatives, together with the evaluation of their inhibitory effects against the root growth of Col-0 and a preliminary broader biological activity screening are described.
Materials and Methods
General Experimental Procedures
Optical rotation measurements were conducted with a JASCO P-2000 digital polarimeter. IR and UV spectra were obtained with a Thermo Scientific Nicolet iS5 FT-IR spectrometer and a Thermo Scientific Evolution 201 spectrophotometer, respectively. Electronic circular dichroism (ECD) spectra were collected on a JASCO J-1500 spectrophotometer. 1D and 2D NMR spectra were recorded in DMSO-d6 (or CDCl3) with a Palo Alto Varian 600 MHz spectrometer, using standard pulse sequences. HRESIMS data were collected on an Agilent Technologies 6224 TOF MS. X-ray single-crystal diffraction data were acquired using an Agilent Gemini Ultra diffractometer with Cu Kα radiation (λ = 1.54178 Å). Medium-pressure liquid chromatography (MPLC) was performed using a Bonna-Agela FLEXA purification instrument. Column chromatography (CC) was carried out with silica gel (200−300 mesh, Qingdao) and Amersham Biosciences Sephadex LH-20. Reversed-phase HPLC (RP-HPLC) was conducted using a Waters 1525 binary HPLC pump equipped with a Waters 2996 photodiode array detector and a YMC-Pack C18 column (YMC, 20 × 250 mm, 5 μm).
Fungal Material
The fungus Aspergillus sp. NBUF87 was isolated from the sponge Hymeniacidon sp. obtained from the Paracel Islands in the South China Sea, and was determined as being Aspergillus sp. by its morphology and gene sequence (ITS rDNA region) analyses (GenBank accession no. MH595747.1). The strain specimen was deposited in PDB medium to the repository conserved at the College of Food and Pharmaceutical Sciences, Ningbo University, China.
Fermentation, Extraction, and Isolation
Spores of Aspergillus sp. NBUF87 were initially inoculated into Erlenmeyer flasks (1 L) containing 400 mL of the seed medium (potato dextrose broth powder 26 g/L and sea salt 35 g/L dissolved in distilled water) and were grown on a shaker (150 r/min) for 100 h at 28°C. The subsequent amplified fermentation was conducted in 105 × 1 L Erlenmeyer flasks, each containing solid rice medium (rice 120 g, sea salt 6.3 g, purified H2O 180 mL), followed by inoculation with 30 mL of the seed culture. After fermentation at room temperature under static conditions for 6 weeks, the fermented substrate was repeatedly extracted with EtOAc. Removal of EtOAc under reduced pressure yielded 54 g of crude extract, which was subjected to vacuum liquid chromatography (VLC) on silica gel column eluting with a petroleum ether/EtOAc stepwise gradient system (from 1:0 to 0:1) to generate seven fractions (Fr. 1−7).
Fraction 4 (3.9 g) was subjected to Sephadex LH-20 gel filtration chromatography, eluted with isocratic CH2Cl2-CH3OH (1:1, v/v) to furnish five sub-fractions (Fr. 4A−4F). The further separation of Fr. 4E (1.8 g) was conducted by reversed-phase ODS MPLC with a gradient elution of CH3OH/H2O (from 25 to 100% CH3OH, flow rate 20 mL/min, 180 min, UV detection at 210 nm) to afford 60 test tube sub-fractions. Subsequently, aspergimarin A (1, 15.3 mg, tR 33.4 min) was purified from Fr. 4E-20 by semi-preparative RP-HPLC [YMC-Pack C18 column (YMC, 20 × 250 mm, 5 μm), UV detection at 210 and 246 nm] with 28% CH3CN/H2O at 2 mL/min. Aspergimarin E (5, 3.6 mg, tR 40.2 min), compounds 10 (4.3 mg, tR 72.3 min) and 11 (4.7 mg, tR 58.2 min) were purified from Fr. 4E-19 by semi-preparative RP-HPLC with 24% CH3CN/H2O at 2 mL/min. Aspergimarin F (6, 3.4 mg, tR 57.3 min) was purified from Fr. 4E-18 by semi-preparative RP-HPLC with 24% CH3CN/H2O at 2 mL/min. Fr. 4B (1.0 g) was conducted by MPLC (30−90% CH3OH/H2O, flow rate 20 mL/min, 120 min, UV detection at 210 nm) to produce 10 sub-fractions (Fr. 4B-1−4B-10). Fr. 4B-4 was further purified by semi-preparative RP-HPLC with 42% CH3OH/H2O at 2 mL/min to yield compound 9 (43.1 mg, tR 45.0 min). Fr. 4B-8 was further purified by semi-preparative RP-HPLC with 60% CH3OH/H2O at 2 mL/min to afford compound 7 (56.0 mg, tR 26.7 min) (Supplementary Figures S1–S10, S41–S60).
Fraction 5 (4.8 g) was separated by Sephadex LH-20 gel filtration chromatography utilizing an isocratic elution gradient of CH2Cl2−CH3OH (1:1, v/v) to afford three sub-fractions (Fr. 5A−5C). The further separation of Fr. 5B (2.5 g) was conducted by RP-MPLC (ODS, 15−100% CH3OH/H2O, flow rate 20 mL/min, 140 min, UV detection at 210 nm) to give 35 tubes. Aspergimarin B (2, 2.5 mg, tR 31.4 min) and aspergimarin C (3, 3.8 mg, tR 22.9 min) were further purified from Fr.5B-23 by semi-preparative RP-HPLC with 30% CH3CN/H2O at 4 mL/min. Fr. 5A (0.9 g) was separated by MPLC (ODS, 25−100% CH3OH/H2O, flow rate 20 mL/min, 150 min, UV detection at 210 nm) to obtain 38 tubes. Fr.5A-15 was further purified by semi-preparative RP-HPLC with 25% CH3CN/H2O at 4 mL/min to afford aspergimarin D (4, 6.3 mg, tR 31.0 min). Fr. 5A-14 was purified by semi-preparative RP-HPLC with 22% CH3CN/H2O at 4 mL/min to afford compound 8 (26.1 mg, tR 31.6 min) (Supplementary Figures S11–S40).
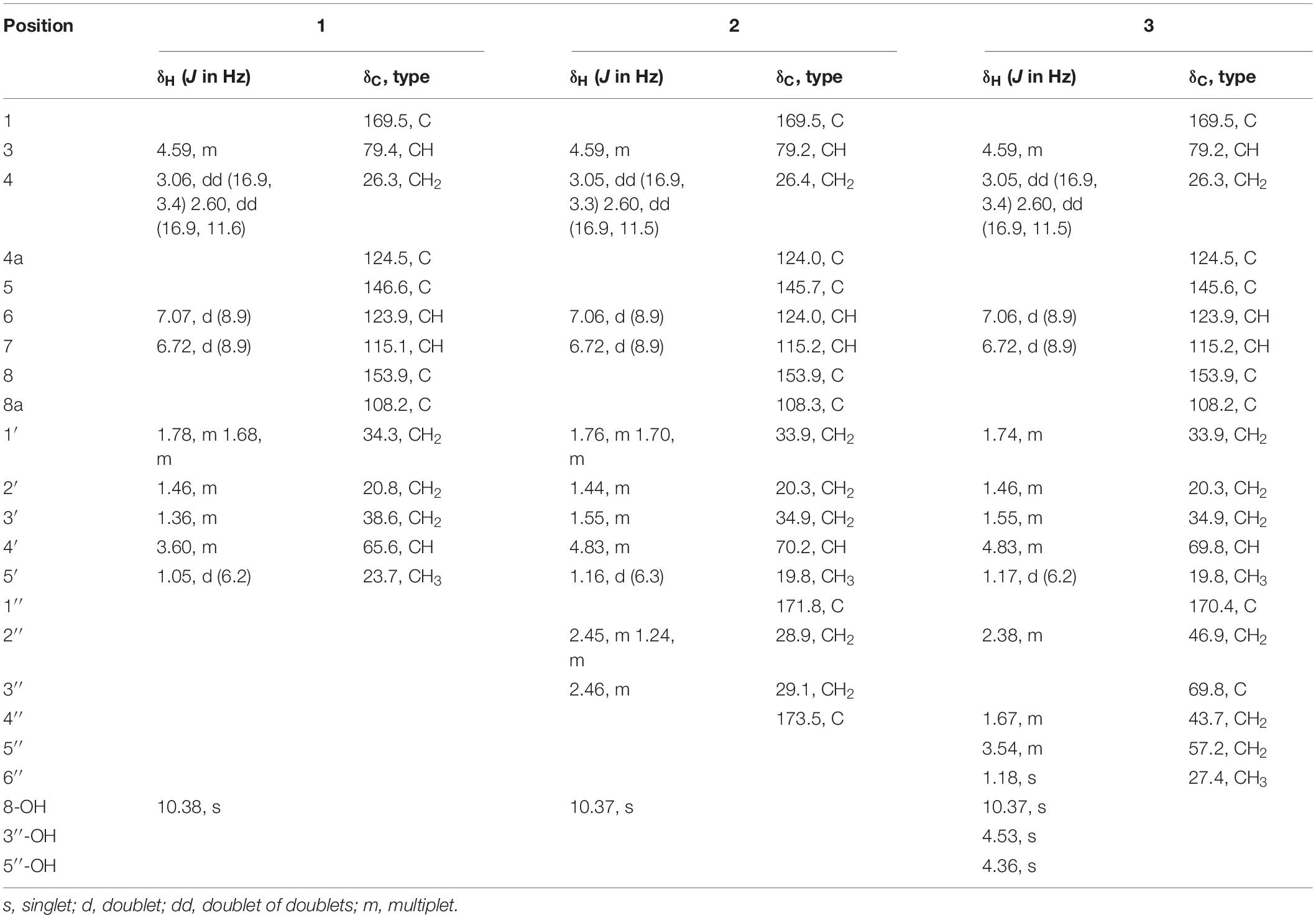
Aspergimarin A (1): white, crystals; mp 177.9−178.8 °C; [α]20 D −34.0 (c 0.2, CHCl3); UV (MeOH) λmax (log ε): 219 (4.14), 248 (3.77), 346 (3.58) nm; CD (c = 0.68 mM, MeOH) λmax (Δε): 223 (+ 2.15), 237 (−1.93), 245 (−1.10), 259 (−6.32) nm; IR (KBr) νmax: 3205, 1647, 1588, 1483, 1354, 1281, 820, 795, 700 cm–1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 265.1082 [M − H] – (calcd for C14H17O5, 265.1081).
Aspergimarin B (2): brown, oil; [α]24 D −10.6 (c 0.2, MeOH); UV (MeOH) λmax (log ε): 211 (4.24), 345 (3.66) nm; CD (c = 0.33 mM, MeOH) λmax (Δε): 225 (+ 0.88), 235 (−1.54), 243 (−0.81), 257 (−5.77) nm; IR (KBr) νmax: 3226, 2929, 1723, 1675, 1469, 1379, 1207, 829 cm–1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 365.1232 [M − H] – (calcd for C18H21O8, 365.1242).
Aspergimarin C (3): brown, oil; [α]24 D −10.8 (c 0.3, MeOH); UV (MeOH) λmax (log ε): 219 (4.27), 344 (3.63) nm; CD (c = 0.45 mM, MeOH) λmax (Δε): 229 (+ 0.86), 238 (−1.44), 243 (−0.32), 257 (−5.37) nm; IR (KBr) νmax: 3368, 2921, 1671, 1469, 1378, 1287, 1206, 1123 cm–1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 435.1416 [M + K]+ (calcd for C20H28KO8, 435.1415).
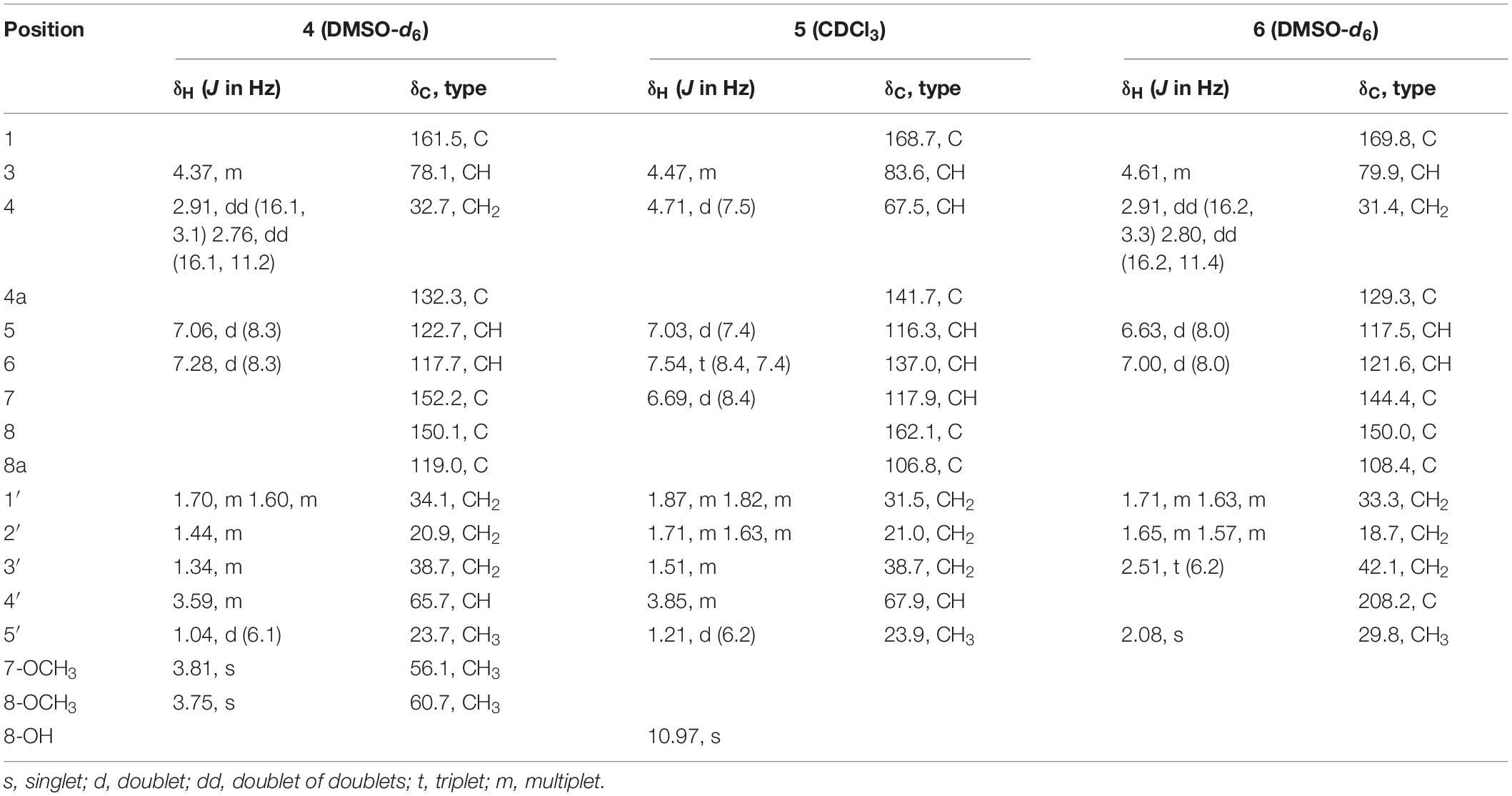
Aspergimarin D (4): yellow, oil; [α]24 D −82.2 (c 0.5, MeOH); UV (MeOH) λmax (log ε): 214 (4.16), 312 (3.24) nm; CD (c = 0.68 mM, MeOH) λmax (Δε): 221 (−6.98), 241 (−0.28), 256 (−2.47), 284 (−0.48), 311 (−1.61) nm; IR (KBr) νmax: 3402, 2935, 1717, 1489, 1455, 1418, 1259, 1131, 1052, 963 cm–1; 1H and 13C NMR data, see Table 2; HRESIMS m/z 295.1537 [M + H]+ (calcd for C16H23O5, 295.1540).
Aspergimarin E (5): colorless, oil; [α]24 D + 17.7 (c 0.3, MeOH); UV (MeOH) λmax (log ε): 209 (4.17), 246 (3.52), 312 (3.42) nm; CD (c = 0.56 mM, MeOH) λmax (Δε): 206 (−8.37), 218 (−0.47), 224 (−1.04), 243 (+ 3.53), 264 (−0.71), 280 (−0.01), 311 (−0.53) nm; IR (KBr) νmax: 3367, 2920, 1672, 1461, 1230, 1117, 823 cm–1; 1H and 13C NMR data, see Table 2; HRESIMS m/z 289.1034 [M + Na]+ (calcd for C14H18NaO5, 289.1046).
Aspergimarin F (6): yellow, amorphous powder; [α]24 D −7.5 (c 0.2, MeOH); UV (MeOH) λmax (log ε): 221 (4.14), 256 (3.71), 332 (3.44) nm; CD (c = 0.57 mM, MeOH) λmax (Δε): 208 (−8.75), 240 (+ 2.67), 266 (−1.47) nm; IR (KBr) νmax: 3360, 2920, 2849, 2361, 1707, 1664, 1452, 1272, 1135, 668 cm–1; 1H and 13C NMR data, see Table 2; HRESIMS m/z 287.0879 [M + Na]+ (calcd for C14H16NaO5, 287.0890).
X-Ray Crystal Structure Analysis of 1
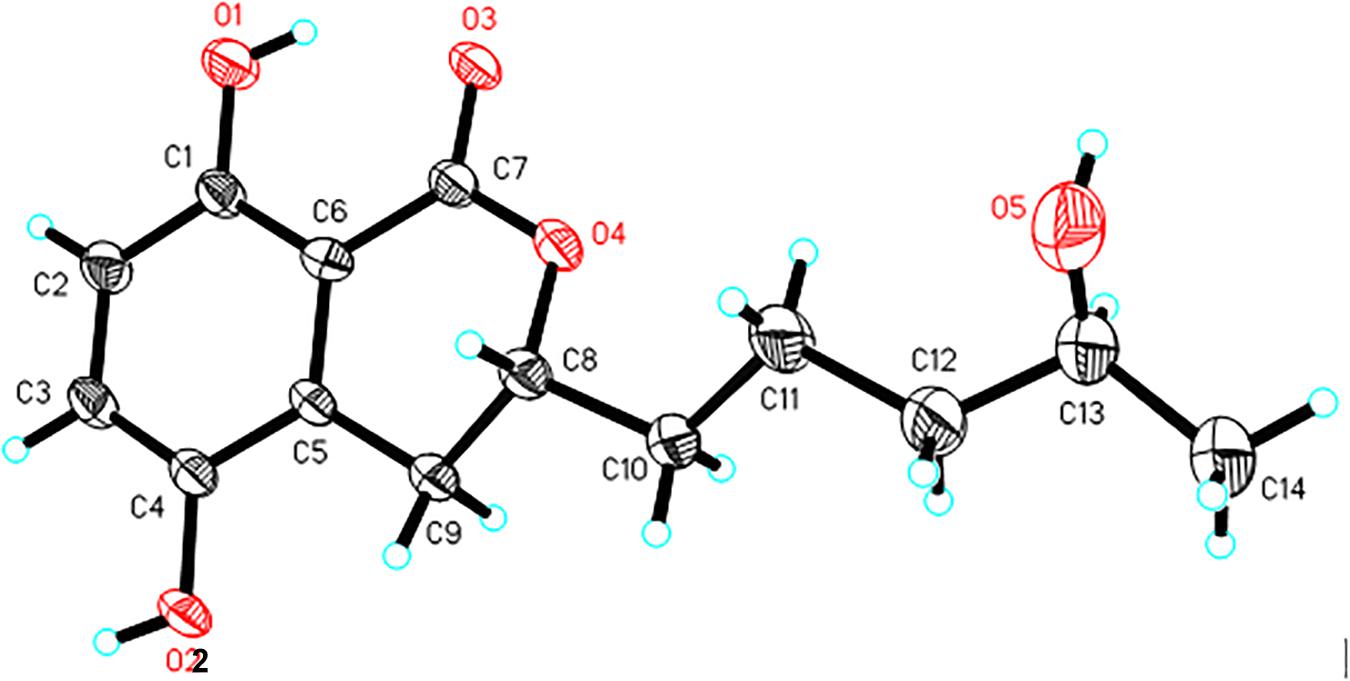
A single crystal of 1 was obtained from 90% CH3OH/H2O. Crystal X-ray diffraction data was collected on an Agilent Gemini Ultra diffractometer with Cu Kα radiation (λ = 1.54178 Å). The structure was solved by direct methods (SHELXS−97) and refined with full-matrix least-squares difference Fourier techniques. All non-hydrogen atoms were refined anisotropically, and hydrogen atoms were placed in the idealized geometrical positions and refined isotropically with a riding model. Crystallographic data for 1 have been deposited in the Cambridge Crystallographic Data Center as supplementary publication no. CCDC 1919904. Copies of the data can be obtained, free of charge, on application to the Director, CCDC, 12 Union Road, Cambridge CB21EZ, United Kingdom (fax: + 44-(0)1223-336033, or e-mail: deposit@ccdc.cam.ac.uk).
Crystal data of aspergimarin A (1): C14H18O5, MR = 266.28, monoclinic, a = 4.9195(3) Å, b = 24.7434(17) Å, c = 5.6478(4) Å, α = γ = 90°, β = 101.812(2)°, V = 672.92(8) Å3, space group P21, Z = 2, Dc = 1.314 mg/m3, μ = 0.829 mm–1, and F(000) = 284. Crystal size: 0.200 × 0.170 × 0.130 mm3. Independent reflections: 2294 (Rint = 0.0377). Final R indices [I > 2 sigma (I)], R1 = 0.0717, wR2 = 0.2076. Goodness of fit on F2 was 1.109.
Preparation of MTPA Esters of 3−5 for Modified Mosher’s Analysis
Under an atmosphere of nitrogen, pyridine-d5 (500 μL) and (R)-MTPA-Cl (8 μL) was sequentially added to an EP tube containing compounds 3, 4, or 5 (1.0 mg), separately. The mixture was shaken at 28 °C for 12 h and then purified by RP-HPLC to obtain the (S)-MTPA esters 3a, 4a, and 5a. By the same procedure, the (R)-MTPA esters 3b, 4b, and 5b were obtained using (S)-MTPA-Cl as a reagent. Key 1H NMR signals used for configurational assignments were determined by respective 1H−1H COSY correlations and the already completed full assignments of 1H NMR data for 3, 4, and 5 (see Supplementary Figures S61–S72).
The C-3′′ absolute configuration of 3 were established as R on the basis of the Δδ values (ΔδH–2^″: −0.01; ΔδH–4^″: + 0.03; ΔδH–5^″: + 0.01) of the (S)- and (R)-MTPA esters (3a and 3b). 3a: H-2′′ (δH 2.37), H-4′′ (δH 1.89), H-5′′ (δH 4.45); 3b: H-2′′ (δH 2.38), H-4′′ (δH 1.86), H-5′′ (δH 4.44).
The C-4′ absolute configuration of 4 was established as S on the basis of the Δδ values (ΔδH–3′: + 0.05, + 0.05; ΔδH–5−0.10) of the (S)– and (R)–MTPA esters (4a and 4b). 4a: H–3′ (δH 1.64, 1.27), H-5′ (δH 1.21); 4b: H-3′ (δH 1.59, 1.22), H-5′ (δH 1.31).
Both C-4 and C-4′ absolute configuration of 5 were established as S on the basis of the Δδ values (ΔδH–3: + 0.17; ΔδH–5: −0.19; ΔδH–6: −0.02; ΔδH–1ΔδH–2ΔδH–3ΔδH–5−0.07) of the (S)– and (R)–MTPA esters (5a and 5b). 5a: H–3 (δH 4.65), H-5 (δH 7.27), H-6 (δH 7.63), H-1′ (δH 1.66, 1.53), H-2′ (δH 1.49), H-3′ (δH 1.60, 1.32), H-5′ (δH 1.25); 5b: H-3 (δH 4.48), H-5 (δH 7.46), H-6 (δH 7.65), H-1′ (δH 1.58, 1.40), H-2′ (δH 1.46), H-3′ (δH 1.53, 1.24), H-5′ (δH 1.32).
Plant Growth Response Assays
Arabidopsis thaliana Col-0, a model organism for plant growth and development, was used to test each isolated compound according to a previously described protocol (Li X. et al., 2018; Huang et al., 2019). Plants grown in 2% (v/v) DMSO were used as the negative control. Seeds of Col-0 incubated in 1 μM of 6-benzylaminopurine (BAP) were selected as the positive control. Test samples were dissolved in 2% (v/v) DMSO at various test concentrations for the experiment.
In vitro AChE Activity Assays
For compounds 1−11, the in vitro acetylcholinesterase (AChE) activity was assessed by the colorimetric method in 96-well plates according to a previously reported method (Santos et al., 2012). Donepezil was selected as the positive control with IC50 value of 11.9 nM (Chen H. et al., 2018).
In vitro Cancer Cell Cytotoxicity Assays
The in vitro cytotoxic activities of all isolated compounds against four human cancer cell lines (CCRF-CEM, MDA-MB-231, HCT-116, and AGS) were evaluated by the MTT method as previously described (Huang et al., 2019; Li W. et al., 2019). 7-Ethyl-10-hydroxycamptothecin (1.3, 10.8, 9.9, and 4.2 nM, respectively) was used as the positive control against four above-mentioned human cancer cell lines.
Antibacterial Activity Assays
Compounds 1−11 were evaluated for their antibacterial activities against methicillin-resistant Staphylococcus aureus ATCC43300 and Escherichia coli ATCC25922 in 96-well plates according to the method described by Gu et al. (2018). Ciprofloxacin was selected as a positive control against the above-mentioned bacteria with MIC values of 0.5 μg/mL.
In vitro Antimalarial Activity Assays
The in vitro antimalarial activity of the compounds was evaluated against the parasite (Plasmodium falciparum W2), which was cultured continuously according to a previously described method (Lazaro et al., 2006). Chloroquine, atovaquone, and artemisinin were used as positive controls against the above parasite with EC50 values of 112, 2.5, and 160 nM, respectively.
Results and Discussion
Structure Elucidation
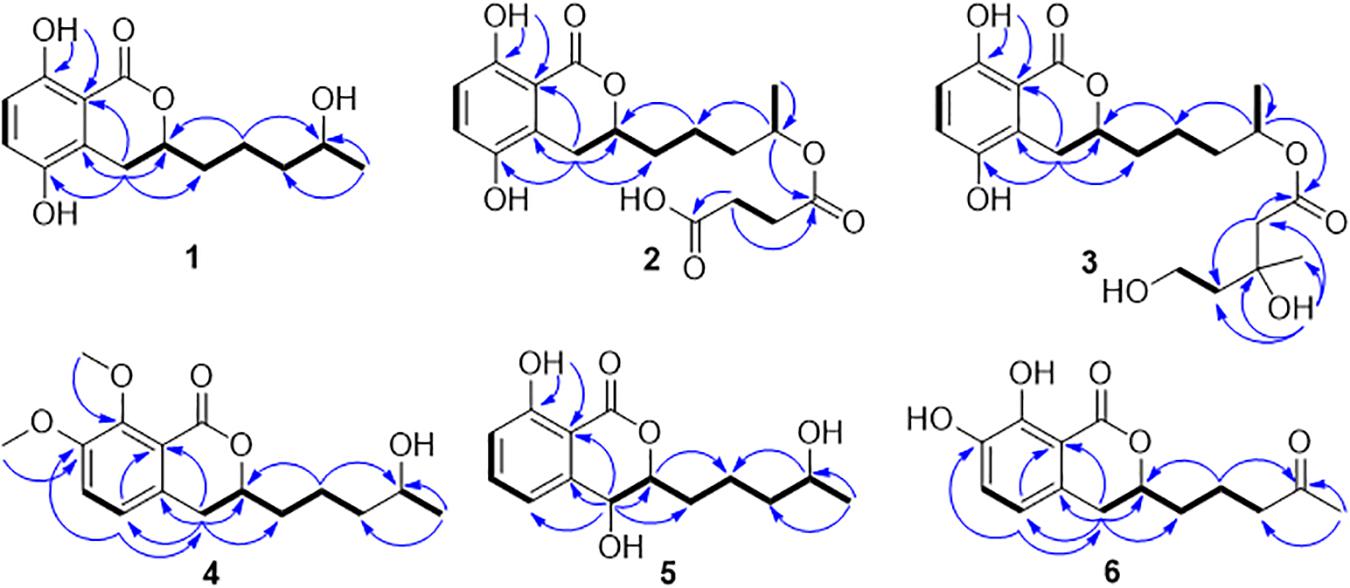
Compound 1 was isolated as white crystals. The molecular formula of 1 was determined to be C14H18O5 based on an ion peak observed in HRESIMS spectrum (m/z [M - H]– 265.1082, calcd for C14H17O5, 265.1081), and this implied the compound has six degrees of unsaturation. The 1H NMR data of 1 (Table 1) demonstrated two aromatic signals [(δH 7.07, d, J = 8.9 Hz, H-6) and (δH 6.72, d, J = 8.9 Hz, H-7)] that were suggestive of two ortho protons. In addition, one phenolic hydroxy group at δH 10.38 (1H, s, 8-OH), two oxygenated methine groups at δH 4.59 (1H, m, H-3) and 3.60 (1H, m, H-4′), four methylene groups at δH 3.06 (1H, dd, H-4α), 2.60 (1H, dd, H-4β), 1.78 (1H, m, H-1′α), 1.68 (1H, m, H-1′β), 1.46 (2H, m, H-2′), and 1.36 (2H, m, H-3′), and one methyl group at δH 1.05 (3H, d, H-5′) were observed in the 1H NMR spectrum of 1. The 13C NMR and DEPT spectra together for 1 indicated 14 carbon signals (Table 1), including a carbonyl at δC 169.5 (C-1), six aromatics at δC 153.9 (C-8), 146.6 (C-5), 124.5 (C-4a), 123.9 (C-6), 115.1 (C-7), and 108.2 (C-8a), and seven alkyl carbons that were two oxygenated methines at δC 79.4 (C-3) and 65.6 (C-4′), four hydrocarbon methylenes at δC 38.6 (C-3′), 34.3 (C-1′), 26.3 (C-4), and 20.8 (C-2′), and a methyl at δC 23.7 (C-5′). In total, the NMR data suggested the presence of a dihydroisocoumarin skeleton contained in 1. The 1H−1H COSY correlations of H-5′/H-4′/H-3′/H-2′/H-1′ further indicated a continuous spin system in the molecule, identified as -CH2-CH2-CH2-CH(OH)-CH3 (Figure 2). Furthermore, according to the key HMBC correlations presented in Figure 2, including from H-2′ to C-3, from H-4 to C-1′, C-3, C-5, C-4a, and C-8a, as well as from 8-OH to C-7, C-8, and C-8a, the planar structure of 1 was established as shown. Finally, on the basis of X-ray single-crystal diffraction analysis (Figure 3), the absolute configuration of 1 was established as 3R,4′S, and this new molecule was given the trivial name aspergimarin A.
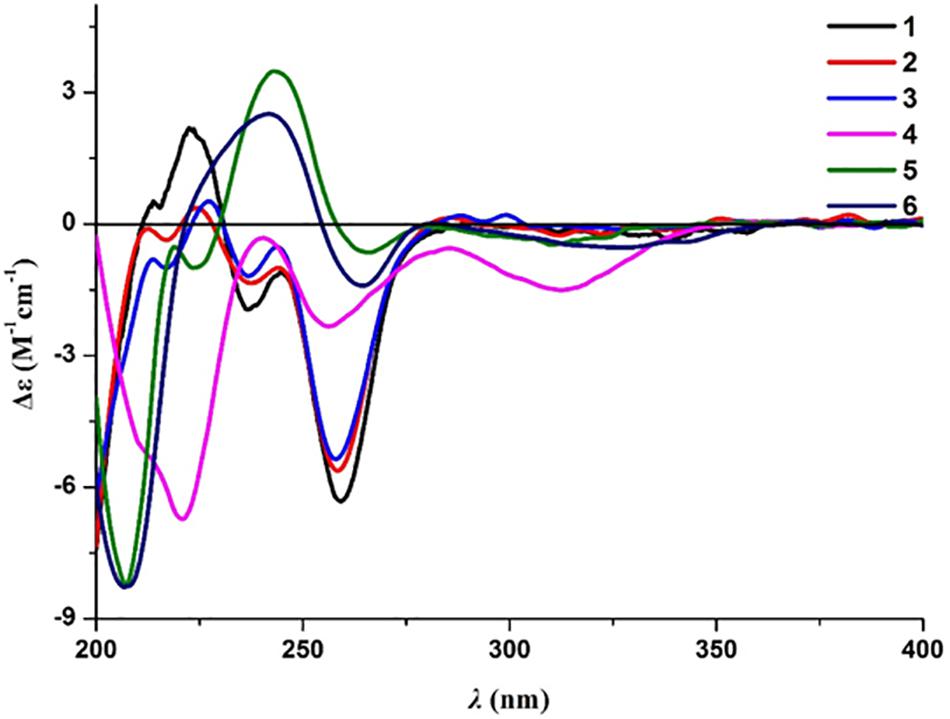
Compound 2 was obtained as a brown oil, and the molecular formula of C18H22O8 was assigned to this molecule by the anion HRESIMS peak at m/z 365.1232 [M − H]– (calcd for C18H21O8, 365.1242). The 1H and 13C NMR spectroscopic data of 2 (Table 1) resembled those of 1, and it was determined that the core structure of an oxygenated hydrocarbon-extended dihydroisocoumarin was shared between these molecules. The 1H−1H COSY correlation between H-2′′ and H-3′′, along with key HMBC correlations from H-3′′ (δH 2.46) to C-1′′ (δC 171.8) and C-4′′ (δC 173.5) (Figure 2), indicated the existence of the linear chain -OCO-CH2-CH2-CO2H in the structure of 2. Furthermore, according to the same biosynthetic pathway with 1 based on and the key HMBC correlation observed from H-4′ (δH 4.83) to C-1′′, the planar structure of 2 was established. The absolute configuration for 2 was suggested as being 3R,4′S based on the biosynthetic logic that it would match that of 1. The CD spectra of 1 and 2 (Figure 4) are able to be overlapped, with the same Cotton effects observed, which further support the configurational assignment. Accordingly, the resolved structure of compound 2 was afforded the trivial name aspergimarin B.
Compound 3 was also obtained as a brown oil, and its molecular formula of C20H28O8 was determined by the potassium cation adduct peak in the HRESIMS spectrum at m/z 435.1416 [M + K]+ (calcd for C20H28KO8, 435.1416). The CD spectrum of 3 (Figure 4) and the 1H and 13C NMR spectroscopic data (Table 1) were similar to those of both 1 and 2, indicating that this molecule is another oxygenated hydrocarbon-extended dihydroisocoumarin analog with the same absolute configuration at C-3 and C-4′. It was determined from a 1H−1H COSY correlation of H-4′′/H-5′′ and the key HMBC correlations from H-2′′ (δH 2.38) to C-1′′ (δC 170.4) and C-4′′ (δC 43.7), as well as from 3′′-OH (δH 4.53) to C-2′′ (δC 46.9), C-3′′ (δC 69.8) and C-6′′ (δC 27.4) (Figure 2), that there is a different secondary carbon side chain [-OCO-CH2-C(CH3)(OH)-CH2-CH2OH] in the molecule of 3 as compared to 2. Furthermore, the key HMBC correlation observed from H-4′ (δH 4.83) to C-1′′ allowed for the planar structure of 3 to be completed. Since the biosynthetic logic of 3 with relation to 1 and 2, together with the matching CD data of these molecules allowed the partial absolute configuration to be assigned as 3R,4′S, only one stereocenter remained uncertain. Using the modified Mosher’s method (Figure 5) (Gu et al., 2018) the absolute configuration of C-3′′ of 3 was established as being R. Therefore, the absolute configuration of 3 was determined to be 3R,4′S,3′′R.
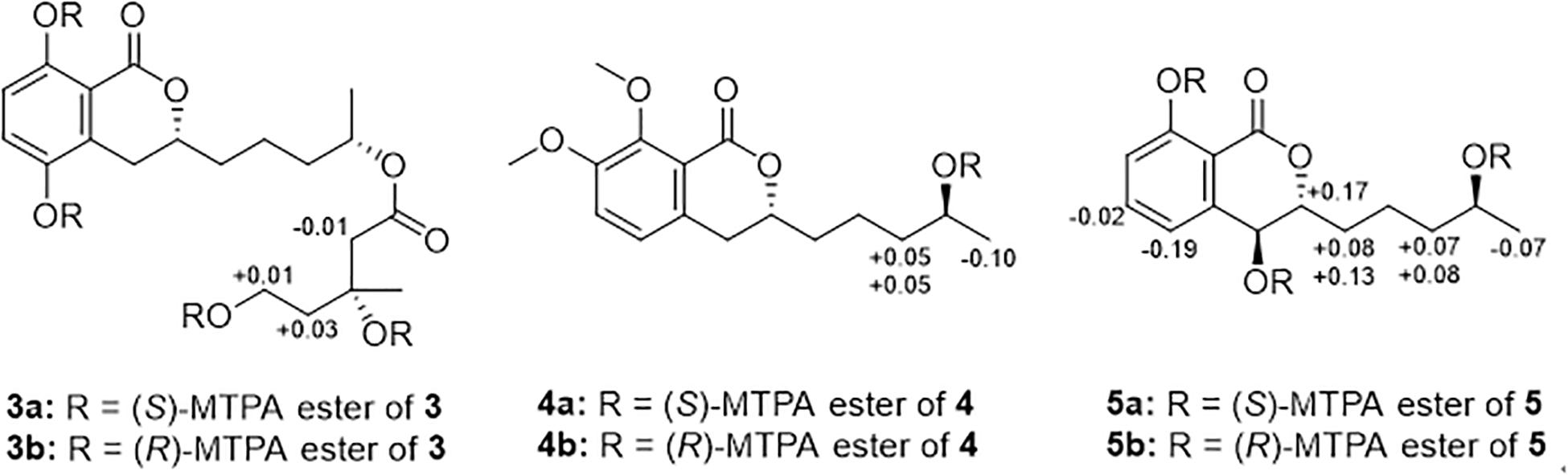
Figure 5. Modified Mosher’s analysis for 3−5. Values shown denote Δδ (δS −δR) (ppm) for the MTPA esters of 3−5.
Compound 4 was obtained as a yellow oil, and the molecular formula of this molecule was determined to be C16H22O5 was based on a peak observed in the HRESIMS spectrum at m/z 295.1537 [M + H]+ (calcd for C16H23O5, 295.1540). The data of 4 from spectroscopic analysis, including UV, IR, and NMR, were extremely similar to those reported for the known compound, penicimarin C (9) (Qi et al., 2013). The exceptions noted were determined to be due to the substitution of one methoxy group for a proton at C-7. The placement of the additional methoxy group at C-7 was determined from key HMBC correlations observed from H-5 (δH 7.06) to C-4 (δC 32.7), C-7 (δC 152.2), and C-8a (δC 119.0), and from 7-O-CH3 (δH 3.81) to C-7, together with the 1H−1H COSY correlation of H-5/H-6 (δH 7.28) (Figure 2). Accordingly, the planar structure of 4 was unambiguously established as shown. Compared to the CD data of 1 and some previously described values for dihydroisocoumarins (Choukchou-Braham et al., 1994), the observed CD spectrum of 4 (Figure 4) indicated the R configuration at C-3. The absolute configuration at the side chain was suggested as 4′S to match that of 1 according to biosynthetic logic, and this was confirmed by use of the modified Mosher’s method (Figure 5). Thus, the absolute configuration of 4 was determined to be 3R,4′S.
Compound 5 was isolated as a colorless oil, and its molecular formula was established as being C14H18O5 according to the associated sodiated molecular ion peak in the HRESIMS spectrum at m/z 289.1034 [M + Na]+ (calcd for C14H18NaO5, 289.1046). This formula for 5 corresponds to an additional OH with respect to 1, and the 1H and 13C NMR data (Table 2) suggested shared carbon skeletons with the substitution of an additional hydroxy group. From the chemical shift differences calculated between 5 and 1, the additional hydroxy group of 5 was suggested to be at C-4. This assignment was further supported by key HMBC correlations from H-4 (δH 4.71) to C-5 (δC 116.3) and from H-5 (δH 7.03) to C-4 (δC 67.5), together with the 1H−1H COSY correlation of H-4/H-3 (δH 4.47) (Figure 2). The absolute configuration at C-3 was determined to be R by the comparison of CD data with 1 and 4 (Figure 4). The absolute configuration of chiral centers at both C-4 and C-4′ in 5 was established as being S by use of the modified Mosher’s method (Figure 5). Therefore, the absolute configuration of 5 was determined to be 3R,4S,4′S.
Compound 6 was isolated as a yellow amorphous powder, and its molecular formula was determined to be C14H16O5 by an associated sodiated molecular ion peak in the HRESIMS spectrum at m/z 287.0879 [M + Na]+ (calcd for C14H16NaO5+, 287.0890). The spectroscopic data of 6, including UV, IR, and NMR, resembled those previously reported for the known molecule penicilloxalone B (10) (Ren et al., 2019). From the 1H and 13C NMR data, it was obvious that the aromatic substitution patterns of 6 and 10 differed, with 6 bearing protons at C-5 and C-6 while 10 has protons at C-6 and C-7. Furthermore, the key HMBC correlations observed from H-5 (δH 6.63) to C-4 (δC 31.4), C-7 (δC 144.4), and C-8a (δC 108.4) together with the 1H−1H COSY correlation of H-5/H-6 (δH 7.00) (Figure 2) corroborated that the two phenolic hydroxy groups of 6 were situated at C-7 and C-8 (δC 150.0). The absolute configuration of 6 was determined to be 3R, the same as for 10, by comparison of the observed CD spectrum for this molecule with reported data for 10 (Figure 4).
Five additional compounds isolated from Aspergillus sp. NBUF87 in the course of this study were determined, by comparison of the spectroscopic and spectrometric data of each with reported values, to be the known isocoumarin derivatives aspergillumarin B (7) (Li S. et al., 2012), penicimarin B (8) (Qi et al., 2013), penicimarin C (9) (Qi et al., 2013), penicilloxalone B (10) (Ren et al., 2019), and (R)-3-(3-hydroxypropyl)-8-hydroxy-3,4-dihydroisocoumarin (11) (Sun et al., 2017). The absolute configurations for 7-9 were established to be 3R, 4′S because the CD spectra (see Supplementary Figures S61, S73), and optical rotation data matched literature values (Li S. et al., 2012; Qi et al., 2013). The absolute configuration of 10 and 11 was also determined to be 3R because the optical rotation data matched literature reported values for these molecules (Sun et al., 2017; Ren et al., 2019).
Effects of Compounds 1−11 on Plant Growth of Arabidopsis thaliana Columbia-0
The isolated compounds 1−11 were subjected to bioassays for testing plant growth response using A. thaliana Col-0, a model plant growth organism. At 100 μM, for both 1 and 5, root growth inhibitory activity was observed against Col-0. Interestingly, while 1 showed only significant inhibitory effect on the lateral root growth of Col-0, 5 caused notable inhibition of both lateral root and primary root growth, as shown in Figure 6. Compounds 2-4 and 6-11 did not show any obvious activity in the same plant growth response assay at 100 μM.
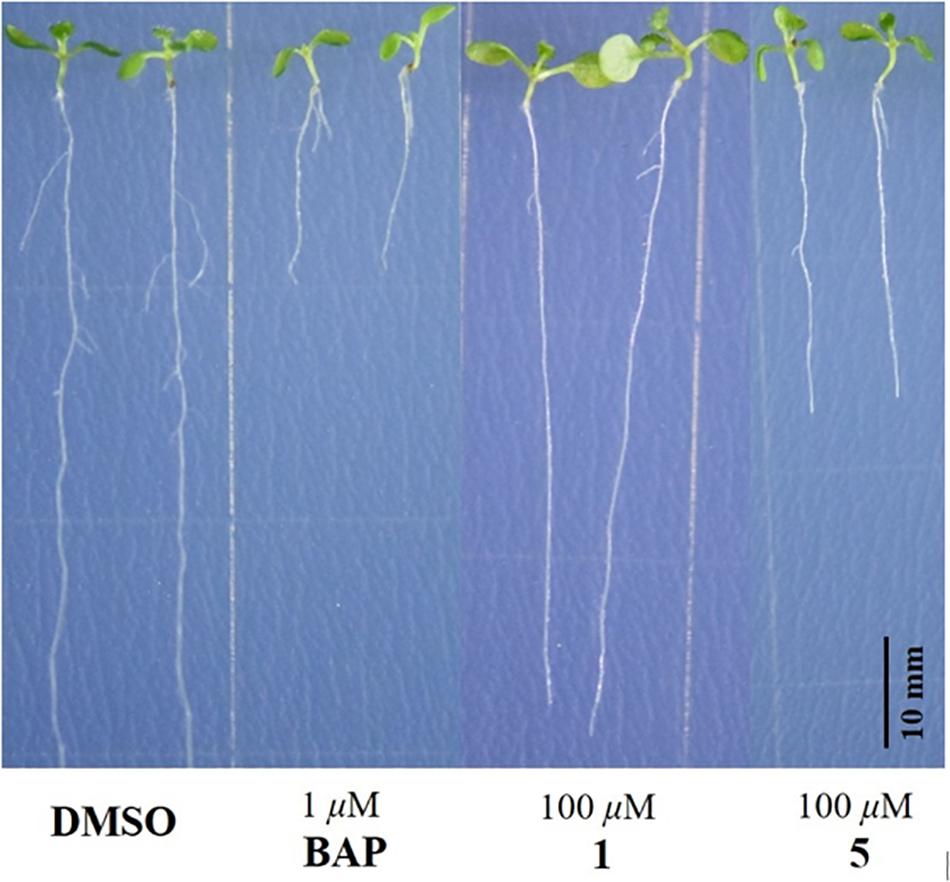
Figure 6. Responses of 100 μM addition of 1 and 5 on the root growth of Arabidopsis thaliana Columbia-0 at 10 days after seed germination. DMSO: negative control; BAP: positive control.
Results of Compounds 1−11 Against Four Additional Bioassays
All compounds isolated in this study (1−11) were also tested for their in vitro inhibitory activity of AChE in a biochemical assay, and phenotypic tests for cytotoxicity against four human-derived cancer cell lines, namely, CCRF-CEM (acute lymphoblastic leukemia T lymphocyte), MDA-MB-231 (breast cancer), HCT-116 (colon cancer), and AGS (gastric adenocarcinoma), and antibacterial activity toward methicillin-resistant S. aureus ATCC43300 and E. coli ATCC25922, and antimalarial activity against P. falciparum W2. None of these compounds exhibited inhibition of AChE (IC50 > 100 μM), cytotoxic activities against any of the cell lines tested (IC50 > 50 μM), antibacterial activities toward the two bacteria (MIC > 50 μg/mL), or antimalarial activity against the parasite (EC50 > 100 μg/mL).
Conclusion
In summary, six new dihydroisocoumarin derivatives, aspergimarins A−F (1−6) were obtained along with five known analogs (7−11) from the fermentation of a fungus Aspergillus sp. NBUF87, isolated from the sponge Hymeniacidon sp. collected from the Paracel Islands in the South China Sea. Structurally, compounds 2 and 3 are esters of the C-4′ hydroxy group in the C-3 side chain of 1, representing relatively rare isocoumarin derivatives according to previous literature reports (Saeed, 2016). In a plant growth response assay using A. thaliana Col-0 as a model organism, only 1 and 5 showed inhibitory activity against root growth, while the others were inactive at up to 100 μM. This finding indicates that the substitution pattern of hydroxy groups in these dihydroisocoumarins may play an important role in root growth inhibitory activity, and further studies remain necessary to interrogate this phenomenon. Since none of the isolated compounds, including 1 and 5, were found to be broadly AChE inhibitors, anticancer agents, antibacterial agents, or antimalarial agents, it is proposed that these root growth inhibitors act through more elaborate signaling pathways. Moreover, 1 and 5 were here as the root growth inhibitors of Col-0, suggesting that they have an important impact in agricultural production.
Data Availability Statement
The datasets generated for this study can be found in the Cambridge Structural Database https://www.ccdc.cam.ac.uk/structures/accession1919904.
Author Contributions
All authors conceived the research, analyzed the data, contributed to the study, and approved the final version of the manuscript. LH, LD, XL, NW, WC, XW, and JL performed the experiments. LH wrote the manuscript. LD, CN, JL, XY, and SH read and revised the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2018YFC0310900), the National Natural Science Foundation of China (41776168, 41706167, and 81850410553), Ningbo Public Service Platform for High-Value Utilization of Marine Biological Resources (NBHY-2017-P2), Zhejiang Provincial Public Welfare Technology Program (LGC19B020002), the Natural Science Foundation of Ningbo (2018A610303 and 2018A610320), Ningbo Science and Technology Projects for Common Wealth (2017C10016), the National 111 Project of China (D16013), the Li Dak Sum Yip Yio Chin Kenneth Li Marine Biopharmaceutical Development Fund, and the K. C. Wong Magna Fund in Ningbo University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02846/full#supplementary-material
References
Ahmed, E. F., Rateb, M. E., El-Kassem, L. A., and Hawas, U. W. (2017). Anti-HCV protease of diketopiperazines produced by the Red Sea sponge-associated fungus Aspergillus versicolor. Appl. Biochem. Microbiol. 53, 101–106. doi: 10.1134/s0003683817010021
Blunt, J. W., Carroll, A. R., Copp, B. R., Davis, R. A., Keyzers, R. A., and Prinsep, M. R. (2018). Marine natural products. Nat. Prod. Rep. 35, 8–53. doi: 10.1039/c7np00052a
Cai, R., Wu, Y., Chen, S., Cui, H., Liu, Z., Li, C., et al. (2018). Peniisocoumarins A–J: isocoumarins from Penicillium commune QQF-3, an endophytic fungus of the mangrove plant Kandelia candel. J. Nat. Prod. 81, 1376–1383. doi: 10.1021/acs.jnatprod.7b01018
Chen, H., Xiang, S., Huang, L., Lin, J., Hu, S., Mak, S. H., et al. (2018). Tacrine (10)-hupyridone, a dual-binding acetylcholinesterase inhibitor, potently attenuates scopolamine-induced impairments of cognition in mice. Metab. Brain Dis. 33, 1131–1139. doi: 10.1007/s11011-018-0221-7
Chen, Y., Liu, Z., Liu, H., Pan, Y., Li, J., Liu, L., et al. (2018). Dichloroisocoumarins with potential anti-inflammatory activity from the mangrove endophytic fungus Ascomycota sp. CYSK-4. Mar. Drugs 16, 54–63. doi: 10.3390/md16020054
Chen, M., Wang, R., Zhao, W., Yu, L., Zhang, C., Chang, S., et al. (2019). Isocoumarindole A, a chlorinated isocoumarin and indole alkaloid hybrid metabolite from an endolichenic fungus Aspergillus sp. Org. Lett. 21, 1530–1533. doi: 10.1021/acs.orglett.9b00385
Chen, S., Liu, Y., Liu, Z., Cai, R., Lu, Y., Huang, X., et al. (2016). Isocoumarins and benzofurans from the mangrove endophytic fungus Talaromyces amestolkiae possess α-glucosidase inhibitory and antibacterial activities. Rsc Adv. 6, 26412–26420. doi: 10.1039/c6ra02566h
Choukchou-Braham, N., Asakawa, Y., and Lepoittevin, J. P. (1994). Isolation, structure determination and synthesis of new dihydroisocoumarins from Ginkgo biloba L. Tetrahedron Lett. 35, 3949–3952. doi: 10.1016/s0040-4039(00)76710-4
Ding, L., Li, T., Liao, X., He, S., and Xu, S. (2018). Asperitaconic acids A–C, antibacterial itaconic acid derivatives produced by a marine-derived fungus of the genus Aspergillus. J. Antibiot. 71, 902–904. doi: 10.1038/s41429-018-0079-2
Elsebai, M. F., and Ghabbour, H. A. (2016). Isocoumarin derivatives from the marine-derived fungus Phoma sp. 135. Tetrahedron Lett. 57, 354–356. doi: 10.1016/j.tetlet.2015.12.024
Fan, L., Reynolds, D., Liu, M., Stark, M., Kjelleberg, S., Webster, N. S., et al. (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. RSC Adv. 109, E1878–E1887. doi: 10.1073/pnas.1203287109
Gu, B. B., Jiao, F. R., Wu, W., Jiao, W. H., Li, L., Sun, F., et al. (2018). Preussins with inhibition of IL-6 expression from Aspergillus flocculosus 16D-1, a fungus isolated from the marine sponge Phakellia fusca. J. Nat. Prod. 81, 2275–2281. doi: 10.1021/acs.jnatprod.8b00662
Hentschel, U., Usher, K. M., and Taylor, M. W. (2006). Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55, 167–177. doi: 10.1111/j.1574-6941.2005.00046.x
Huang, L., Ding, L., Li, X., Wang, N., Yan, Y., Yang, M., et al. (2019). A new lateral root growth inhibitor from the sponge-derived fungus Aspergillus sp. LS45. Bioorg. Med. Chem. Lett. 29, 1593–1596. doi: 10.1016/j.bmcl.2019.04.051
Hussain, H., and Green, I. R. (2017). A patent review of two fruitful decades (1997-2016) of isocoumarin research. Expert Opin. Ther. Pat. 27, 1267–1275. doi: 10.1080/13543776.2017.1344220
Kim, D. C., Quang, T. H., Ngan, N. T. T., Yoon, C. S., Sohn, J. H., Yim, J. H., et al. (2015). Dihydroisocoumarin derivatives from marine-derived fungal isolates and their anti-inflammatory effects in lipopolysaccharide-induced BV2 microglia. J. Nat. Prod. 78, 2948–2955. doi: 10.1021/acs.jnatprod.5b00614
Kong, F., Zhao, C., Hao, J., Wang, C., Wang, W., Huang, X., et al. (2015). New α-glucosidase inhibitors from a marine sponge-derived fungus, Aspergillus sp. OUCMDZ-1583. RSC Adv. 5, 68852–68863. doi: 10.1016/j.bmcl.2017.12.049
Lazaro, J. E. H., Nitcheu, J., Mahmoudi, N., Ibana, J. A., Mangalindan, G. C., Black, G. P., et al. (2006). Antimalarial activity of crambescidin 800 and synthetic analogues against liver and blood stage of Plasmodium sp. J. Antibiot. 59, 583–590. doi: 10.1038/ja.2006.78
Lee, Y. M., Li, J., Zhang, P., Hong, J. K., Lee, C. O., and Jung, J. H. (2011). A cytotoxic fellutamide analogue from the sponge-derived fungus Aspergillus versicolor. B Korean Chem. Soc. 32, 3817–3820. doi: 10.5012/bkcs.2011.32.10.3817
Lei, H., Lin, X., Han, L., Ma, J., Ma, Q., Zhong, J., et al. (2017). New metabolites and bioactive chlorinated benzophenone derivatives produced by a marine-derived fungus Pestalotiopsis heterocornis. Mar. Drugs 15, 69–78.
Li, D., Xu, Y., Shao, C. L., Yang, R. Y., Zheng, C. J., Chen, Y. Y., et al. (2012). Antibacterial bisabolane-type sesquiterpenoids from the sponge-derived fungus Aspergillus sp. Mar. Drugs 10, 234–241. doi: 10.3390/md10010234
Li, S., Wei, M., Chen, G., and Lin, Y. (2012). Two new dihydroisocoumarins from the endophytic fungus Aspergillus sp. collected from the south china sea. Chem. Nat. Compd. 48, 371–373. doi: 10.1007/s10600-012-0254-9
Li, W., Ding, L., Wang, N., Xu, J., Zhang, W., Zhang, B., et al. (2019). Isolation and characterization of two new metabolites from the sponge-derived fungus Aspergillus sp. LS34 by OSMAC approach. Mar. Drugs 17, 283–291. doi: 10.3390/md17050283
Li, X., Yang, R., and Chen, H. (2018). The Arabidopsis thaliana mediator subunit MED8 regulates plant immunity to Botrytis Cinerea through interacting with the basic helix-loop-helix (bHLH) transcription factor FAMA. PLoS One 13:e0193458. doi: 10.1371/journal.pone.0193458
Liu, H., Edrada-Ebel, R., Ebel, R., Wang, Y., Schulz, B., Draeger, S., et al. (2009). Drimane sesquiterpenoids from the fungus Aspergillus ustus isolated from the marine sponge Suberites domuncula. J. Nat. Prod. 72, 1585–1588. doi: 10.1021/np900220r
Liu, J. T., Wu, W., Cao, M. J., Yang, F., and Lin, H. W. (2018). Trienic α-pyrone and ochratoxin derivatives from a sponge-derived fungus Aspergillus ochraceopetaliformis. Nat. Prod. Res. 32, 1791–1797. doi: 10.1080/14786419.2017.1402325
Niu, S., Liu, Q., Xia, J. M., Xie, C. L., Luo, Z. H., Shao, Z., et al. (2018). Polyketides from the deep-sea-derived fungus Graphostroma sp. MCCC 3A00421 showed potent antifood allergic activities. J. Agric. Food Chem. 66, 1369–1376. doi: 10.1021/acs.jafc.7b04383
Pita, L., Fraune, S., and Hentschel, U. (2016). Emerging sponge models of animal-microbe symbioses. Front. Microbiol. 7:2102. doi: 10.3389/fmicb.2016.02102
Qi, J., Shao, C. L., Li, Z. Y., Gan, L. S., Fu, X. M., Bian, W. T., et al. (2013). Isocoumarin derivatives and benzofurans from a sponge-derived Penicillium sp. fungus. J. Nat. Prod. 76, 571–579. doi: 10.1021/np3007556
Ren, Y., Chao, L. H., Sun, J., Chen, X. N., Yao, H. N., Zhu, Z. X., et al. (2019). Two new polyketides from the fungus Penicillium oxalicum MHZ153. Nat. Prod. Res. 33, 347–353. doi: 10.1080/14786419.2018.1452001
Saeed, A. (2016). Isocoumarins, miraculous natural products blessed with diverse pharmacological activities. Eur. J. Med. Chem. 116, 290–317. doi: 10.1016/j.ejmech.2016.03.025
Santos, W. P., da Silva Carvalho, A. C., dos Santos, Estevam, C., Santana, A. E., and Marçal, R. M. (2012). In vitro and ex vivo anticholinesterase activities of erythrina velutina leaf extracts. Pharm. Biol. 50, 919–924. doi: 10.3109/13880209.2011.649429
Sun, J., Zhu, Z. X., Song, Y. L., Ren, Y., Dong, D., Zheng, J., et al. (2017). nti-neuroinflammatory constituents from the fungus Penicillium purpurogenum MHZ 111. Nat. Prod. Res. 31, 562–567. doi: 10.1080/14786419.2016.1207075
Thomas, T. R. A., Kavlekar, D. P., and LokaBharathi, P. A. (2010). Marine drugs from sponge-microbe association-A review. Mar. Drugs 8, 1417–1468. doi: 10.3390/md8041417
Tsukada, M., Fukai, M., Miki, K., Shiraishi, T., Suzuki, T., Nishio, K., et al. (2011). Chemical constituents of a marine fungus, Arthrinium sacchari. J. Nat. Prod. 74, 1645–1649. doi: 10.1021/np200108h
Wang, W., Liao, Y., Zhang, B., Gao, M., Ke, W., Li, F., et al. (2019). Citrinin monomer and dimer derivatives with antibacterial and cytotoxic activities isolated from the deep sea-derived fungus Penicillium citrinum NLG-S01-P1. Mar. Drugs 17, 46–54. doi: 10.3390/md17010046
Wang, X., Mou, Y., Hu, J., Wang, N., Zhao, L., Liu, L., et al. (2014). Cytotoxic polyphenols from a sponge-associated fungus Aspergillus versicolor Hmp-48. Chem. Biodivers. 11, 133–139. doi: 10.1002/cbdv.201300115
Webster, N. S., and Taylor, M. W. (2012). Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14, 335–346. doi: 10.1111/j.1462-2920.2011.02460.x
Wiese, J., Imhoff, J., Gulder, T., Labes, A., and Schmaljohann, R. (2016). Marine fungi as producers of benzocoumarins, a new class of inhibitors of glycogen-synthase-kinase 3β. Mar. Drugs 14, 200–208.
Wu, Y., Chen, S., Liu, H., Huang, X., Liu, Y., Tao, Y., et al. (2019). Cytotoxic isocoumarin derivatives from the mangrove endophytic fungus Aspergillus sp. HN15-5D. Arch. Pharm. Res. 42, 326–331. doi: 10.1007/s12272-018-1019-1
Zhang, X., Li, Z., and Gao, J. (2018). Chemistry and biology of secondary metabolites from Aspergillus Genus. Nat. Prod. J. 8, 275–304. doi: 10.2174/2210315508666180501154759
Zhou, Y., Debbab, A., Mándi, A., Wray, V., Schulz, B., Müller, W. E., et al. (2013). Alkaloids from the sponge-associated fungus Aspergillus sp. Eur. J. Org. Chem. 2013, 894–906.
Keywords: dihydroisocoumarin, root growth inhibitor, sponge-derived fungus, Aspergillus sp., electronic circular dichroism
Citation: Huang L, Ding L, Li X, Wang N, Cui W, Wang X, Naman CB, Lazaro JEH, Yan X and He S (2019) New Dihydroisocoumarin Root Growth Inhibitors From the Sponge-Derived Fungus Aspergillus sp. NBUF87. Front. Microbiol. 10:2846. doi: 10.3389/fmicb.2019.02846
Received: 06 September 2019; Accepted: 25 November 2019;
Published: 10 December 2019.
Edited by:
László Galgóczy, Hungarian Academy of Sciences, HungaryReviewed by:
Yongbo Xue, Huazhong University of Science and Technology, ChinaAjmal Khan, University of Nizwa, Oman
Mahmoud Fahmi Elsebai, Mansoura University, Egypt
Copyright © 2019 Huang, Ding, Li, Wang, Cui, Wang, Naman, Lazaro, Yan and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijian Ding, dinglijian@nbu.edu.cn; Shan He, heshan@nbu.edu.cn
 Liming Huang1
Liming Huang1 Lijian Ding
Lijian Ding C. Benjamin Naman
C. Benjamin Naman J. Enrico H. Lazaro
J. Enrico H. Lazaro Shan He
Shan He