- 1School of Bioengineering, Dalian University of Technology, Dalian, China
- 2College of Life Science, Dalian Minzu University, Dalian, China
- 3Key Laboratory of Biotechnology and Bioresources Utilization, Ministry of Education, Dalian, China
Fresh foods are vulnerable to foodborne pathogens which cause foodborne illness and endanger people’s life and safety. The rapid detection of foodborne pathogens is crucial for food safety surveillance. An in situ-synthesized gene chip for the detection of foodborne pathogens on fresh-cut fruits and vegetables was developed. The target genes were identified and screened by comparing the specific sequences of Salmonella Typhimurium, Vibrio parahemolyticus, Staphylococcus aureus, Listeria monocytogenes, and Escherichia coli O157:H7 from the National Center for Biotechnology Information database. Tiling array probes were designed to target selected genes in an optimized hybridization system. A total of 141 specific probes were selected from 3,227 hybridization probes, comprising 26 L. monocytogenes, 24 S. aureus, 25 E. coli O157:H7, 20 Salmonella Typhimurium, and 46 V. parahemolyticus probes that are unique to this study. The optimized assay had strong amplification signals and high accuracy. The detection limit for the five target pathogens on fresh-cut cantaloupe and lettuce was approximately 3 log cfu/g without culturing and with a detection time of 24 h. The detection technology established in this study can rapidly detect and monitor the foodborne pathogens on fresh-cut fruits and vegetables throughout the logistical distribution chain, i.e., processing, cleaning, fresh-cutting, packaging, storage, transport, and sale, and represents a valuable technology that support the safety of fresh agricultural products.
Introduction
The incidence of foodborne diseases resulting from the consumption of food contaminated with pathogens has increased significantly in many countries over the last few decades (Fusco and Quero, 2014). In 2011, 69,553 cases of food poisoning resulted in 7,125 hospitalizations and 93 deaths in Europe (European Food Safety Autority [EFSA] and European Centre for Disease Control, and Prevention [ECDC], 2013). The total cases from foodborne diseases reach 48,000,000 individuals, with 128,000 patients in hospitals, and cause 3,000 deaths each year in the United States according to the report from the Centers for Disease Control and Prevention (CDC) (Center for Disease Control and Prevention [CDC], 2011; Scallan et al., 2011). Food poisoning from fresh agricultural products contaminated with pathogens also occurs with surprising frequency. In the United States, the outbreak of foodborne illness was related to cantaloupe, which infected 1,751 individuals and caused 34 deaths (Michelle et al., 2014; Feng et al., 2016). With the increase in the demands for nutritious, healthy, and convenient foods, the consumption of fresh-cut produce involving fresh agricultural products has increased substantially worldwide (Oliveira et al., 2015; Ma et al., 2017). However, the processing of fresh-cut produce by cutting, shucking, carving, slicing, and peeling is highly susceptible to contamination by microorganisms and may lead to leakage or loss of nutrients (Gleeson and Beirne, 2005).
A potential safety risk from the contamination of foodborne pathogens from seed, soil, irrigation water, and organic fertilizers exists in fresh-cut fruit and vegetable products (Hoagland et al., 2018). Some studies have shown that some foodborne pathogens can survive and reach a high density (∼6 log cfu/g) on fresh-cut melon, papaya, celery, onions, and tomatoes (Penteado and Leitão, 2004; Feng et al., 2015; Salazar et al., 2017; Jayeol et al., 2019). Disease outbreaks were associated with foodborne pathogens on fresh-cut produce (Alegre et al., 2010; Center for Disease Control and Prevention [CDC], 2013). For example, fresh-cut cantaloupe, watermelon, and honeydew contaminated with Salmonella Carrau and Salmonella Adelaide resulted in the hospitalization of 38 and 36 people in 2018 and 2019, respectively (Center for Disease Control and Prevention [CDC], 2018, 2019).
The traditional pathogen detection methods are excessively time-consuming, which takes up to 8 days to obtain the results, making these methods inadequate for the rapid identification of foodborne pathogens (Lazcka et al., 2007). Considering that the shelf-life of fresh-cut fruits and vegetables ranges from 3 to 5 days, more rapid pathogen detection methods are required for these products. Detection methods employing non-culture-based identification and quantification technologies, such as real-time PCR and quantitative reverse-transcription PCR, are less time-consuming than the traditional methods but are susceptible to amplification biases and errors (Wolffs et al., 2005). For example, false-positive amplification can occur among genetically related microorganisms in the environment, and false-negative amplification can result from sequence variation in the primer binding sites of the target diagnostic region or from low reaction sensitivity (Davison, 1999).
To overcome these limitations, high-throughput DNA microarrays that can rapidly identify foodborne pathogens with a high degree of specificity have been developed (Kostić and Sessitsch, 2012). The DNA microarrays comprise of hundreds of oligonucleotide probes to positively detect a single target pathogen. Arraying many specific probes and setting a sufficiently high threshold for the positive identification of the presence of a target pathogen substantially avoid the false-positives that result from cross-contamination between foodborne pathogens (Wilson et al., 2002). The traditional gene chip arrays contain some probes that target the coding sequence of the virulence gene of the target pathogens (Mockler and Ecker, 2005; Bertone et al., 2005). The identification for foodborne pathogens is no longer considered reliable when only one area of the genome is targeted. The strategy of tiling probe arrays as applied on the gene chip can target the contiguous genome regions of the target foodborne pathogen and detect the base of the target gene sequences (Selinger et al., 2000; Johnson et al., 2005). For the development of a microarray detection technology, no such chip is commercially available, and the tiling arrays must be designed and synthesized for specific purposes. The customization of arrays permits total control over the probes on the chip, allowing researchers to select specific probes for the detection of the pathogen and to control the distribution of probes over the array. The in situ-synthesized gene chips are high-density arrays that have high specificity and can rapidly screen and identify the sources of contamination and the foodborne pathogens during food poisoning outbreaks.
The purpose of this research was to develop a rapid detection technique based on an in situ-synthesized gene chip comprising of virulence genes for detecting Salmonella Typhimurium (ST), Listeria monocytogenes (LM), Staphylococcus aureus (SA), Vibrio parahemolyticus (VP), and Escherichia coli O157:H7 (EC O157:H7), which are commonly associated with fresh-cut fruits and vegetables. The pathogens targeted by the chip (LM, ST, SA, EC O157:H7, and VP) are clinically relevant to food safety. The tiling probes of the whole target gene of the pathogens on the microarrays were used to enhance the detection accuracy of the chip. The in situ-synthesized virulence gene array can be used as a diagnostic tool in food poisoning outbreaks related to fresh-cut fruits and vegetables contaminated with foodborne pathogens.
Materials and Methods
Fresh-Cut Cantaloupe and Lettuce
Fresh cantaloupes and lettuces were purchased from the New-Mart supermarket in Dalian City, China. They were chosen based on uniformity of maturation stage, size, and absence of defects or injuries. The samples were stored at 4°C for approximately 1 h (transport time) prior to processing. The fresh cantaloupe and lettuce were cleaned twice with 75% (v/v) distilled water, sterile water, and ethyl alcohol (Tianjin Kemiou Chemical Reagent Co., Ltd., Tianjin, China), respectively. The samples were dried in a biosafety cabinet (1300 Series A2 Class II, Type A2, Thermo Fisher Scientific, Shanghai, China) prior to cutting. The cantaloupe was peeled and cut into 1-cm3 cubes without a rough outside surface. The lettuce was cut into 1-cm2 pieces using a sterile knife.
Bacterial Strains
The bacterial strains including ST (CICC 21484), LM (CICC 21633), SA (CICC 21600), VP (CICC 21617), and EC O157:H7 (CICC 21530) were purchased from the China Center of Industrial Culture Collection (CICC, Beijing, China). The LM was preserved in trypticase soy–yeast extract broth (Qingdao Hopebio-Technology Co., Ltd., Qingdao, Shandong, China) with 80% glycerol (Tianjin Kemiou Chemical Reagent Co.) at −20°C. The SA, EC O157:H7, and ST were preserved in TSB (Qingdao Hopebio-Technology Co.) with 80% glycerol at −20°C. The VP was stored at −20°C in 3% NaCl alkaline peptone water (Qingdao Hopebio-Technology Co.) with 80% glycerol. All bacterial strains were revived from frozen stocks before use by cultivation in a culture medium at 37°C for 24 h. The bacterial suspensions were made using 0.1% (w/v) peptone water (Aobox Biotechnology, Beijing, China) to obtain the proper concentration. The LM was enumerated on trypticase soy agar (TSA) with 0.6% (w/v) yeast extract (Qingdao Hopebio-Technology Co.). The ST, SA, and EC O157:H7 were enumerated on TSA (Qingdao Hopebio-Technology Co., Ltd.). The VP was enumerated on 3% NaCl TSA (Qingdao Hopebio-Technology Co., Ltd.). All bacteria were cultured for 24 h at 37°C, and the colonies were measured with an aCOLyte colony counter (Acolyte Technologies Corp., London, United Kingdom). The microbial counts are expressed as log cfu/ml.
DNA Extraction
For DNA extraction, the bacterial suspensions (1 ml of 108 cfu/ml) were centrifuged at 12,000 × g for 2 min. The sediment was suspended and washed twice using 0.1% peptone water. Then, the samples were centrifuged again at 12,000 × g for 2 min. The genomic DNA was obtained from foodborne pathogens according to the following procedure of DNA extraction: add 200 μl of cetyltriethylammnonium bromide extraction buffer (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) to mix and thoroughly vortex the sediment of 1 ml of suspension, transfer the homogenate to a 65°C bath for 30 min, place the sample at −80°C for 15 min, and put it at 65°C for 30 min again; repeat this process once for freezing and thawing, add 200 μl of chloroform/isoamyl alcohol (24:1), vortex for 5 s, and then centrifuge the sample at 12,000 × g for 15 min to separate the phases; transfer the upper aqueous phase to a new tube containing 80 μl magnetic beads (DNA Binding Beads, Thermo Fisher Scientific, MA, United States), incubate the sample at room temperature (∼25°C) for 5 min to allow the DNA to bind to the beads, and place the sample on the magnetic separator (24-well magnetic separator, Thermo Fisher Scientific) for 5 min. Without removing the plate from the magnetic separator, carefully remove and discard the supernatant, subsequently wash it with 80% ethanol twice, remove the residual ethanol by drying in a biosafety cabinet, and then dissolve DNA in 20 μl TE buffer (TE Buffer, Thermo Fisher Scientific). The genomic DNA concentration was determined using a Multiskan FC Microplate Photometer (Thermo Fisher Scientific), and the purity was calculated by the ratio between the OD of the DNA at 260 nm and at 280 nm.
Target Gene Screening
The entire genome sequences of the five foodborne pathogens were obtained from the GeneBank sequence database produced by the National Center for Biotechnology Information (NCBI). The number of genome sequences for the LM, ST, SA, VP, and EC O157:H7 was 31, 153, 93, 19, and 8, respectively, in all genomes (Supplementary Table S1). The sequences were initially screened using the NCBI BLAST to identify those with less than 80% sequence identity among the target strains and which were longer than 500 bp. Pairwise comparisons of the selected genome sequences were performed to identify similar sequences between any two genomes. For each bacterial genome, an inverse complement of the sequence operation was performed according to the sequence intervals of the obtained similarity comparison results to obtain the sequence intervals specific to the bacterial species. These sequences were compared against the NCBI database to screen out specific gene sequences and then aligned within the species to identify the conserved regions. Based on the specific sequences above, the target specific gene was screened for the second time according to the principle of sequence identity >95% intraspecific and <75% interspecific. The primers were designed based on the screening target gene of the LM, SA, EC O157:H7, ST, and VP (Table 1).
Multiplex PCR
The optimized multiplex PCR condition contained 0.8 ng/μl of genomic DNA, 1 × ex Taq Buffer plus Mg2+, 0.2 mM of dNTPs, 2.5 U of ex Taq DNA polymerase, and 1 nM of Cy3-dCTP RNase-free water to a final volume of 50 μl. The primer concentrations were 0.46 μM for LM, SA, and VP, 0.44 μM for ST, and 0.17 μM for EC O157:H7. All primers and reagents in the multiplex PCR assay were obtained from Takara Bio. Inc. (Dalian, Liaoning, China). The thermal profile for the assay includes incubating for 2 min at 95°C for pre-degeneration, subsequently setting 35 cycles involving 30 s at 95°C for denaturing, 30 s at 55°C for annealing, and 30 s at 72°C for extension, followed by 10 min at 72°C for the final extension. The amplicon of multiplex PCR was separated by electrophoresis on 3% agarose gels supplemented with Gelred dye (Biotium, Inc., Hayward, CA, United States) and visualized on a UVP BioSpectrum Imaging System (UVP, LLC, CA, United States). The multiplex PCR products were purified with LCS beads (LC science, Houston, TX, United States) to remove the redundant primers and the impurities.
Design of Tiling Probe Arrays
A microfluidic chip made by in situ-synthesis was manufactured by LC Sciences (Houston, TX, United States). The chip contains 3,968 probe sites (128 rows × 31 lines) in 1.4 cm2. The tiling array probes (25 bp in length) were designed according to the lengths and the region of the PCR amplicons derived from LM, SA, EC O157:H7, ST, and VP. The adjacent probe sequences were separated by a single base until the entire target sequence of each of the five target strains was covered. A total of 3,227 probes were obtained: 958 from LM, 547 from SA, 350 from EC O157:H7, 932 from ST, and 440 from VP. The layout of the tiling array probes is shown in Supplementary Table S2.
Microarray Hybridization, Washing, and Scanning
The preparation of chips for hybridization, target hybridization, washing, and scanning were performed by the procedure described as follows (LC Science, Houston, TX, United States). Briefly, the mixture of 25 μl of PCR amplification products (200 ng/μl) and 25 μl of hybridization buffer was added on the in situ-synthesis gene chip. Subsequently, hybridization for 18 h at 40°C, washing at 40°C, and staining were performed in a fluidics station at LC Science. The hybridization buffer (1 ml) was flushed through the chip at 500 μl/min for 20 min. The array chips were scanned using a GenPix 4000B scanner (Molecular Devices/Axon, Sunnyvale, CA, United States). The scanning pixel size was 10 mm. The Cy3 signals were collected using 532-nm channels (Zhu et al., 2007).
Internal Verification and Optimized Probe Specificity
To test the specificity among the target pathogens, the genomic DNA of each target pathogen (1 ml of 108 cfu/ml) was extracted, and 0.8 ng/μl of genomic DNA was amplified in the PCR system (50 μl) as described above. The mixture (50 μl), with an equal ratio of PCR amplicons (200 ng/μl) and hybridization buffer, was hybridized to the constructed gene chip (as described above). The 100 probes with the strongest hybridization signals were selected for further analysis, and the probes that detect hybridization signals from the non-target bacteria were excluded. The array layout was repeated ten times to verify the accuracy of hybridization (Supplementary Table S3).
Pathogens in Artificially Contaminated Fresh-Cut Cantaloupe and Lettuce
Fresh-cut cantaloupes (10 g) and lettuce (10 g) were placed on petri dishes and inoculated with 0.5 ml of the mixture of LM, SA, EC O157:H7, ST, and VP (each 0.1 ml of 3–4 log cfu/ml) using a sterile micropipette. Each sample was dried in a biosafety cabinet for 1 h and then put into a sterile stomacher bag (Interscience, Saint Nom la Breteche, France) containing 90 ml of 0.1% peptone water. The sterile stomacher bag was blended in a stomacher at high-speed setting for 1 min. The sample residues were removed using a nylon membrane with 15-μm pore in the filtration apparatus (Hangzhou Hengqing Technology Co., Ltd., Hangzhou, Zhejiang, China). The filtrate was collected again using a polyethersulfone filtration membrane with 0.22-μm pore. The filtration membrane enriching the foodborne pathogens was transferred to centrifuge tubes containing 10 ml of phosphate buffer (0.2 mM) and vortexed for 15 min. The genomic DNA of the bacterial suspensions was extracted as described above (“DNA Extraction”). The number of ST, LM, EC O157:H7, SA, and VP on fresh-cut cantaloupes was respectively, measured on Salmonella spp. chromogenic media and Oxford agar base (both from Qingdao Hopebio-Technology), EC O157:H7 chromogenic media, SA chromogenic media, and VP chromogenic media (from Shanghai Central Bio-Engineering Ltd., Co., Shanghai, China). The array layout was repeated 12 times on the in situ-synthesized gene chip (Supplementary Table S4). The multiplex PCR amplification products from the genomic DNA of the target pathogens hybridized with probes and hybridization signals on the in situ-synthesized gene chip were detected as described above (“Microarray Hybridization, Washing, and Scanning”).
Data Analysis
The fluorescence signal values were extracted from the images using Array-Pro Analyzer software (Media Cybernetics Inc., Rockville, MD, United States), which yielded values between 0 and 65,535 arbitrary units. The positive results were defined as the difference value between the medians of the feature pixel intensities, and the background pixel intensities were above three times than the standard deviation of the background pixel intensities at 532 nm. The subsequent data were measured using Microsoft Excel (Microsoft Corporation, Redmond, WA, United States).
Results
Probe Screening for Detecting Pathogenic Bacteria
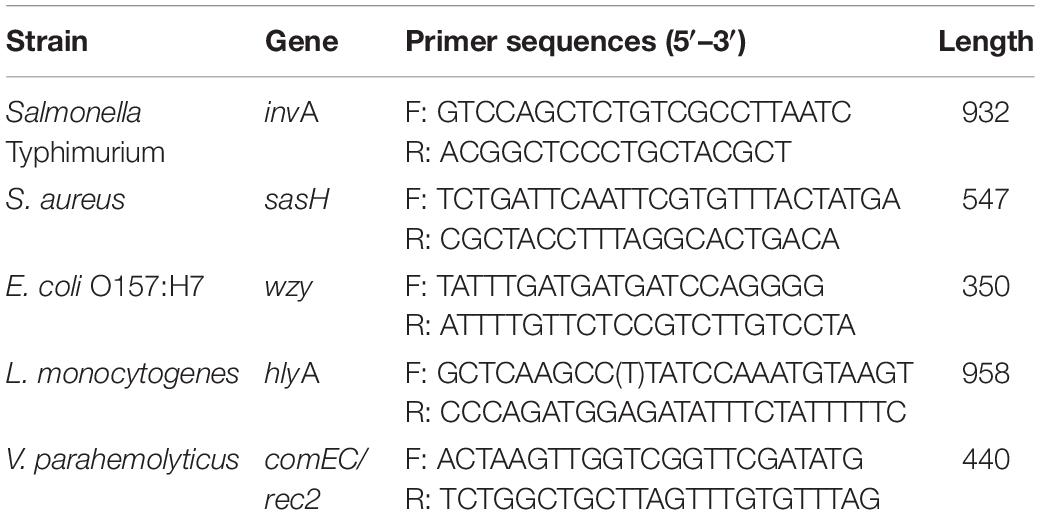
The PCR amplification product of each pathogen was respectively, hybridized with the in situ-synthesized gene chip containing the tiling array probes. The performance of the in situ-synthesized gene chip for the sequence-specific detection of amplification products of PCR was assessed (Figure 1). The fluorescence signal values were observed on the site which synthesized probes for detecting the target pathogen. The results of hybridization for LM, ST, SA, VP, and EC O157:H7 are shown in Figures 1A–E. The 100 probes with the strongest signal values were selected from the positive result of each target foodborne pathogen (Supplementary Tables S5–S9). However, some signal values were observed on the in situ-synthesized gene chip with non-target pathogen. It is obvious that the signal values were observed on the positive probe site of the LM from the in situ-synthesized gene chip with SA (Figure 1C). There are still some signal values on the other in situ-synthesized gene chips with non-target pathogen (Figures 1A,B,D,E). Therefore, the differences between the hybridization signal values of the top 100 probes and the non-target pathogenic bacteria was detected by Array-Pro software, and the probes with signal values similar to the negative probes were selected as final probe. A total of 141 specific probes were screened: 26 from LM, 20 from ST, 24 from SA, 46 from VP, and 25 from EC O157:H7 (Table 2).
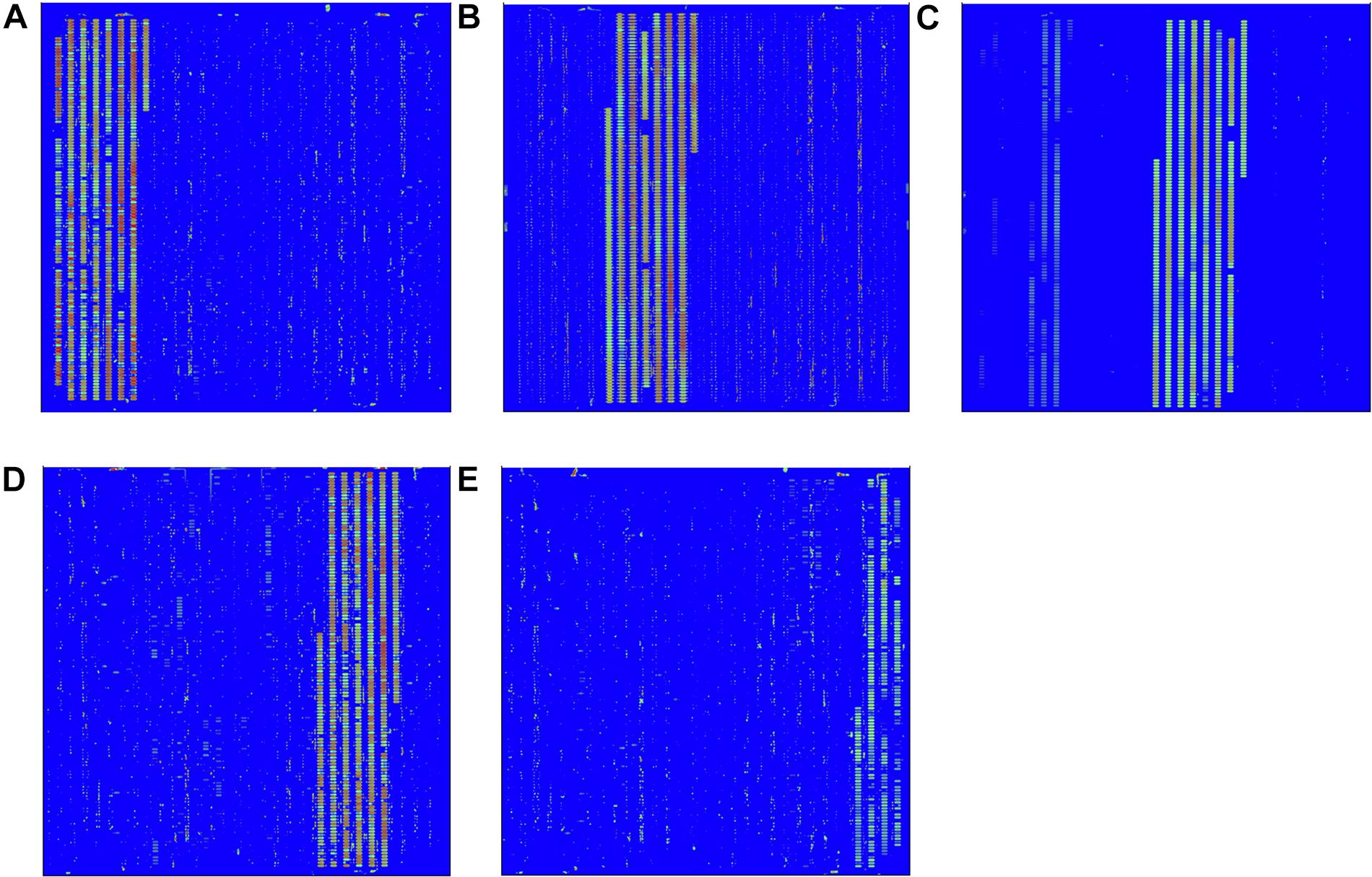
Figure 1. Probe screening on in situ-synthesized gene chip for detecting foodborne pathogens. (A) Listeria monocytogenes, (B) Salmonella Typhimurium, (C) Staphylococcus aureus, (D) Vibrio parahemolyticus, (E) Escherichia coli O157:H7. The array layout is presented as a list in Supplementary Table S2.

Table 2. The specific probe for detection Listeria monocytogenes, Salmonella Typhimurium, Staphylococcus aureus, Escherichia coli O157:H7, and Vibrio parahemolyticus.
Verification of Specific Probes Among Pathogenic Bacteria
The hybridization specificity was evaluated between the screening probes and the target and non-target pathogen (Figure 2). The hybridization results for each pathogen including EC O157:H7, LM, VP, SA, and ST are shown in Figures 2A–E, respectively. Strong signal values were present on sites between the specific screening probes and the target pathogen. No hybridization signals were found between the screening probes and the non-target pathogens. To determine the stability of the in situ-synthesized gene chip for detecting the pathogenic bacteria, the corresponding specific probes were repeatedly synthesized ten times. The signal values remained consistent with each repeated experiment. It demonstrated that the in situ-synthesized gene chip including screening probes has a high specificity for detecting EC O157:H7, LM, VP, SA and ST.
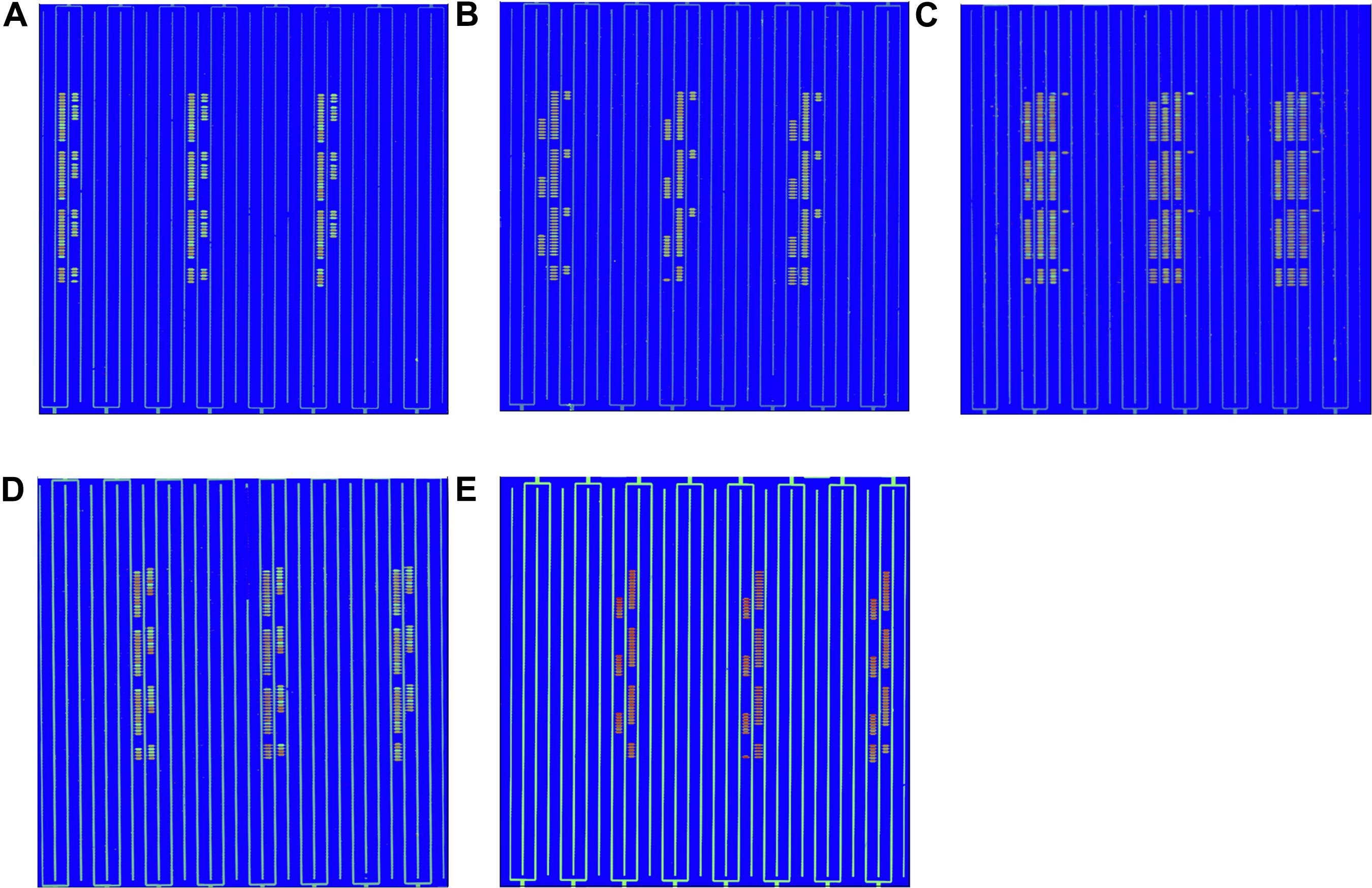
Figure 2. Specific detection by in situ-synthesized gene chips of five types of foodborne pathogens. (A) Escherichia coli O157:H7, (B) Listeria monocytogenes, (C) Vibrio parahemolyticus, (D) Staphylococcus aureus, (E) Salmonella Typhimurium. The array layout is presented as a list in Supplementary Table S3.
Identification of FoodBorne Pathogens on Fresh-Cut Cantaloupe and Lettuce
The detection of pathogenic bacteria inoculated into fresh-cut cantaloupe and fresh-cut lettuce using the in situ-synthesized gene chip is shown in Figure 3. The numbers of EC O157:H7, LM, SA, ST, and VP on fresh-cut cantaloupe were 3.05, 2.19, 2.02, 3.12, and 3.35 log cfu/g, respectively, as determined by colony counting, and strong signal values were found on the regions with probes specific to EC O157:H7, ST, and VP (Figure 3A). Contrarily, low signal values were present on the regions containing probes specific to LM and SA. This discrepancy might be due to cell lysis efficiency during the DNA extraction for the Gram-positive strains (LM and SA). The low abundance of LM and SA might also be the main factor for the low signal values on the fresh-cut cantaloupe. Therefore, the sensitivity of the in situ-synthesized gene chip for detecting foodborne pathogens inoculated into fresh-cut cantaloupe was ∼ 3 log cfu/g.
Figure 3. Detection by in situ-synthesized gene chips of five types of foodborne pathogens on fresh-cut fruits and vegetables. (A) Fresh-cut cantaloupe. (B) Fresh-cut lettuce. The array layout is presented as a list in Supplementary Table S4.
The numbers of EC O157:H7, LM, SA, ST, and VP on fresh-cut lettuce were 3.75, 2.77, 3.34, 3.27, and 3.35 log cfu/g, respectively, as measured according to colony counting. Strong, regular, and obvious signals were observed at specific probes from all five pathogenic bacteria on fresh-cut lettuce (Figure 3B). The hybridization signal from 141 specific probes was found on the in situ-synthesized gene chips. Especially the signal values of the probe site on the in situ-synthesized gene chip detecting LM and SA on fresh-cut lettuce were higher than those on fresh-cut cantaloupe. It might be due to the fact that the population of LM and SA on fresh-cut lettuce was higher than that on fresh-cut cantaloupe. The 141 specific probes were repeatedly synthesized 12 times on the in situ-synthesized gene chips. The signal value remained consistent with each repeated experiment, demonstrating that the probes designed and screened in this study can effectively improve the detection efficiency of these foodborne pathogens on fresh-cut cantaloupe and lettuce.
Discussion
The foodborne pathogens, primarily LM, ST, SA, VP, and EC O157:H7, are major causes of foodborne diseases worldwide. These pathogens, in particular, are the main reasons responsible for enteritidis, meningitis, and even death (Scallan et al., 2011). The rapid and accurate detection of pathogens on food products and in environmental samples are important for the prevention of outbreaks of foodborne diseases and of spreading of foodborne pathogens (Law et al., 2015). The traditional culture methods have several drawbacks mainly related to the time and labor required, which delay the feedback of information about the pathogens from suspected food (Andrews and Ryan, 2015). The detection techniques for foodborne pathogens have evolved significantly to overcome the limitations of conventional detection in the recent years. Generally, new strategies and methods based on nucleic acid amplification, the recognition of antigen and antibody in immunology, and signal identification with biosensor have been developed in the recent years (Zhao et al., 2014). The DNA microarrays are a comprehensive platform that combine nucleic-acid- and biosensor-based approaches and are a powerful tool for the detection and identification of pathogens on food matrices. The DNA microarrays are useful in the survey of outbreak of foodborne diseases, especially in the screening of foodborne pathogen in a large number of samples (Herrera-Rodriguez et al., 2013). The DNA microarray technology can be used to detect all potential pathogenic bacteria in a sample in a single assay. The microarrays can be designed to detect a broad spectrum of foodborne pathogens without compromising the sensitivity or increasing the time required for detection (Yu et al., 2016). For example, the DNA microarray was found to be more precise compared with the conventional culture for detecting Campylobacter jejuni and Campylobacter coli (Keramas et al., 2004). Similarly, a mixed genomic microarray was designed and could discriminate between the closely related L. monocytogenes isolates within a serotype obtained from similar geographic and epidemiologic sources (Borucki et al., 2003). Another microarray designed for the multi-pathogen identification of 18 pathogenic bacteria was found to be highly specific and sensitive. This array, containing 53,660 probes, could discriminate the amplification products and the false-positive amplification products for positive identifications (Wilson et al., 2002).
However, the microarray technology developed to date faces several challenges for the detection of pathogens as part of food surveillance. The current microarray methods generally require some cultivation of bacteria, DNA preparation, and hybridization during the detection process (Kim et al., 2012; Yu et al., 2016). The detection efficiency may be affected by low amounts of pathogenic bacteria, false-positive results, and inadequate sensitivity (Tarca et al., 2006). It is possible to overcome these challenges and achieve reasonable detection levels by optimizing the sample preparation methodology and by appropriate screening of hybridization probes.
Compared to other nucleic-acid-based platforms, the in situ-synthesized gene chips provide greater flexibility in design. The efficacy of this methodology is independent of the efficiency of the target gene amplification, the oligonucleotide length can be easily controlled, and the hybridization probe sequence can be screened from the different virulence gene regions for specific hybridization. Designing and screening specific probes is key to the establishment of in situ-synthesis chip technology. However, probe selection is often complicated by the complexity of the target genomes and the differences in sequence composition, which need to be accounted for during probe design. A large number of repeated experiments were performed to verify and screen thousands of effective probes (Gard et al., 2009). High-density in situ-synthesis microarrays were used to screen the whole genome of the target pathogens and yielded effective probe hybridization. The tiling probe design was used to screen and verify a single experiment, which saves time and enhances the efficiency of the in situ-synthesized microarrays in this study. The high resolution of the tiling arrays was also exploited in this study to differentiate between foodborne pathogens, which is particularly challenging for other traditional methods. The method of tiling probe array on the gene chip expanded the resequencing technology, and the main characteristic was the higher efficiency to identify each base of the oligonucleotide sequence for the detection of base mutation sites (Chee et al., 1996; Winzeler et al., 1998; Jackson et al., 2007). Although the approach of tiling probe array has been developed for decades, there is surprisingly little information about the application of this approach to food safety. In the current study, the probes were arranged as a tiling array on an in situ-synthesis microarray platform. The tiling array platform consisted of approximately 3,227 oligonucleotides with 1-bp intervals between the probes. This interval provided redundancy that allowed the detection of specific genes from the foodborne pathogens and also exposed the sites of possible genetic changes that may have altered the genomic sequence of the pathogens. The approach of tiling probe array has the advantage for discovering specific probes to identify virulence genes from the target bacteria. In the tiling array, a specific gene sequence in the genome is identified by comparing the hybridization signal of a target foodborne pathogen to that of a reference strain by matching the probe sequences, and 141 specific probes were obtained from the five foodborne pathogens in this study. This method enabled the detection of multiple target pathogens using customized tiling arrays in contrast to most pathogen detection methods that focus on the detection of pathogens individually. Other research developed a DNA microarray containing the random genomic DNA fragments of L. monocytogenes or four virulence genes of E. coli O157:H7, which can only detect L. monocytogenes or E. coli O157:H7 and distinguish it from other foodborne pathogens (Bang et al., 2013; Call et al., 2001). It is obvious that the in situ-synthesized gene chip with a tiling probe array contains a more specific probe which hybridizes with the multiplex PCR amplicon of the foodborne pathogen in this study. It can enhance the accuracy and the efficacy of detecting the target foodborne pathogens.
In our arrays, the species of a foodborne pathogen could be identified by the presence of virulence genes related to pathogenicity. The primers and probes designed to target the bacterial virulence genes were able to enhance the specificity for detection. The virulence genes screened in this research have shown a strong specificity toward LM, ST, SA, VP, and EC O157:H7. The gene targets were obtained by multiple comparisons and were based on the whole genome sequences of the five target foodborne pathogens. The five target genes, hlyA from LM, invA from ST, sash from SA, comEC/rec2 from VP, and wzy from EC O157:H7, were selected for the design of the primers and probes. The amplification results of this detection system had a strong specificity compared with those of other studies based on random amplification (Poretsky et al., 2014). At the species level, the 16S rRNA gene, as the target gene, has been used to detect the pathogens, however, the detection accuracy of 16s rRNA was limited for discriminating at or below the species level due to the low base substitution mutation rates (Ruan et al., 2017; Liu et al., 2019). The in situ-synthesis microarray detection platform for the foodborne pathogens is based on nucleic acid hybridization technology rather than on a single marker gene. Multiplex PCR was used to prepare the amplification products for probe hybridization. The reliability and the accuracy of the combined microarray chip and multiplex PCR detection system were further enhanced by the simultaneous identification of the specific regions of the multiple pathogens (Wilson et al., 2002). However, there may be variations in the genes of the different pathogenic bacteria, resulting in mismatches between some probes and the products of PCR. Therefore, the multiple probes were screened for each strain to ensure a specific identification of the target strain. Ultimately, one signal between the probe and the PCR product could be utilized to confirm the presence of the target strain.
In addition, the ingredient in fruits and vegetables also negatively impact the detection efficiency of the in situ-synthesized gene chip. The interference from inhibitors, such as humic acid, has been noted as a critical issue by researchers in identifying pathogens in the samples using other detection platforms, such as PCR (Loge et al., 2002). Such inhibitors can often be carried over during DNA extraction, which is the first step in many analytical protocols. However, the purity and quality of DNA is an important factor for DNA microarray analysis (Vital et al., 2016). In fact, the interference from element in food matrices to DNA has not been avoided during some DNA extraction methods, such as column-based extraction, magnetic bead-based extraction, and even automated extraction. In our detection platform, the filtration system effectively removed the micro-particles and the flocculent precipitates after the homogenization of fruits and vegetables, thus reducing the interference of fruit and vegetable DNA to subsequent microbial detection. Our extraction protocol can be used as a sort of a bacterial enrichment device to maximize the enrichment of pathogenic foodborne microorganisms in fruits and vegetables and improve the efficiency and sensitivity of positive detection. This effectively shortens the culture time required for microbial growth.
The application of DNA microarray technology for detecting foodborne pathogen on food matrix still has some difficulties to be resolved. The false-positive result is the major problem while identifying the multiple foodborne pathogens by DNA hybridization. In the current study, the combination of tiling probe array and in situ-synthesized gene chip enhanced the accuracy and efficiency. There are also other methods, like digital PCR technology, nanotechnology platform, and biosensor chip technology, that may contribute to further improvements in the detection of DNA from food samples combined with microarrays in the future (Avarre et al., 2007). The in situ-synthesized gene chip method provided a comprehensive, highly efficient, and rapid detection for foodborne pathogen. In the case of food poison outbreak associated with foodborne pathogen, the detection of in situ-synthesized gene chip enables to provide comprehensive data and strategy, thereby performing a suitable treatment for patients and encouraging a rapid recall of the contaminated food.
CONCLUSION
This study developed an in situ-synthesized gene chip for detecting foodborne pathogens on fresh-cut fruits and vegetables. This assay showed a strong amplification signal and high accuracy. We obtained 141 specific probes by screening 3,227 potential hybridization probes. The detection limit was approximately 3 log cfu/g without culturing, and the detection time was 24 h. The foodborne pathogen detection technology established in this study can effectively detect and monitor the foodborne pathogens on fresh-cut cantaloupe and lettuce during processing, cleaning, fresh-cutting, packaging, storage, logistics, and sale, thereby improving the quality and safety controls of fresh-cut fruits and vegetables.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: The accession number have been included in the Supplementary Table S1.
Author Contributions
S, KF, and AJ performed the development of molecular biology method, microarray detection, and sequence screening and drafted the manuscript. WH and ZX conceived and designed the experiments and helped to polish the language. YLa, YLi, and YLo participated in sequence alignment and microbial experiments.
Funding
This research was supported by the “Thirteenth Five-Year Plan” for National Key Research and Development Program (grant no. 2016YFD0400903), National Natural Science Foundation of China (grant no. 31471923), and “Twelfth Five-Year Plan” for National Science and Technology Support Program (grant no. 2012BAD38B05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03089/full#supplementary-material
References
Alegre, I., Abadias, M., Anguera, M., Usall, J., and Viñas, I. (2010). Fate of Escherichia coli O157:H7, Salmonella and Listeria innocua on minimally-processed peaches under different storage conditions. Food Microbiol. 27, 862–868. doi: 10.1016/j.fm.2010.05.008
Andrews, J. R., and Ryan, E. T. (2015). Diagnostics for invasive Salmonella infections: current challenges and future directions. Vaccine 33, C8–C15. doi: 10.1016/j.vaccine.2015.02.030
Avarre, J., de Lajudie, P., and Béna, G. (2007). Hybridization of genomic DNA to microarrays: a challenge for the analysis of environmental samples. J. Microbiol. Meth. 69, 242–248. doi: 10.1016/j.mimet.2006.11.007
Bang, J. Y., Beuchat, L. R., Song, H., Gu, M. B., Chang, H., Kim, H. S., et al. (2013). Development of a random genomic DNA microarray for the detection and identification of Listeria monocytogenes in milk. Int. J. Food Microbiol. 161, 134–141. doi: 10.1016/j.ijfoodmicro.2012.11.023
Bertone, P., Gerstein, M., and Snyder, M. (2005). Applications of DNA tiling arrays to experimental genome annotation and regulatory pathway discovery. Chromosome Res. 13, 259–274. doi: 10.1007/s10577-005-2165-0
Borucki, M. K., Krug, M. J., Muraoka, W. T., and Call, D. R. (2003). Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92, 351–362. doi: 10.1016/S0378-1135(02)00423-6
Call, D. R., Brockman, F. J., and Chandler, D. P. (2001). Detecting and genotyping Escherichia coli O157:H7 using multiplexed PCR and nucleic acid microarrays. Int. J. Food Microbiol. 67, 71–80. doi: 10.1016/S0168-1605(01)00437-8
Center for Disease Control and Prevention [CDC] (2013). Outbreak of staphylococcal food poisoning from a military unit lunch party-United States, July 2012. Morb. Mortal. Week. Rep. 62, 1026–1028.
Center for Disease Control and Prevention [CDC] (2011). Estimates of Foodborne Illness in the United States. Factsheet Printable and Downloadable From the Following. Available at: https://www.cdc.gov/foodborneburden/2011-foodborne-estimates.html (accessed November 8, 2013).
Center for Disease Control and Prevention [CDC] (2018). Multistate Outbreak of Salmonella Adelaide Infections Linked to Pre-Cut Melon. Available at: https://www.cdc.gov/salmonella/adelaide-06-18/index.html (accessed July 26, 2018).
Center for Disease Control and Prevention [CDC] (2019). Outbreak of Salmonella Infections Linked to Pre-Cut Melons. Available at: https://www.cdc.gov/salmonella/carrau-04-19/index.html (accessed May 24, 2019).
Chee, M., Yang, R., Hubbell, E., Berno, A., Huang, X. C., Stern, D., et al. (1996). Accessing genetic information with high-density DNA arrays. Science 274, 610–614. doi: 10.1126/science.274.5287.610
Davison, J. (1999). Genetic exchange between bacteria in the environment. Plasmid 42, 73–91. doi: 10.1006/plas.1999.1421
European Food Safety Autority [EFSA] and European Centre for Disease Control, and Prevention [ECDC], (2013). Scientific report of EFSA and ECDC. The European Union summary report on trends and sources of zoonoses, zoonotic agents and foodborne outbreaks in 2011. EFSA J. 11, 3129–3379.
Feng, K., Hu, W., Jiang, A., Sarengaowa, Xu, Y., Zou, Y., et al. (2016). A dual filtration-based multiplex PCR method for simultaneous detection of viable Escherichia coli O157:H7, Listeria monocytogenes, and Staphylococcus aureus on fresh-cut cantaloupe. PLoS One 11:e0166874. doi: 10.1371/journal.pone.0166874
Feng, K., Hu, W. Z., Jiang, A. L., Xu, Y. P., Sarengaowa, Li, X. B., et al. (2015). Growth potential of Listeria monocytogenes and Staphylococcus aureus on fresh-cut tropical fruits. J. Food Sci. 80, M2548–M2554. doi: 10.1111/1750-3841.13089
Fusco, V., and Quero, G. M. (2014). Culture-dependent and culture-independent nucleic-acid-based methods used in the microbial safety assessment of milk and dairy products. Compr. Rev. Food Sci. F. 13, 493–537. doi: 10.1111/1541-4337.12074
Gard, O. S., Alexander, D. R., Karin, L., Lindvall, J. M., and Rognes, T. (2009). Custom design and analysis of high-density oligonucleotide bacterial tiling microarrays. PLoS One 4:e5943. doi: 10.1371/journal.pone.0005943
Gleeson, E., and Beirne, D. O. (2005). Effects of process severity on survival and growth of Escherichia coli and Listeria innocua on minimally processed vegetables. Food Control 16, 677–685. doi: 10.1016/j.foodcont.2004.06.004
Herrera-Rodriguez, S. E., Elizondo-Quiroga, D., and Alvarez-Maya, I. (2013). Infectious diseases detection by microarray: an overview of clinical relevant infections. J. Biomed. Sci. Eng. 6, 1006–1013. doi: 10.4236/jbise.2013.610125
Hoagland, L., Ximenes, E., Ku, S., and Ladishch, M. (2018). Foodborne pathogens in horticultural production systems: ecology and mitigation. Sci. Hortic. 236, 192–206. doi: 10.1016/j.scienta.2018.03.040
Jackson, S. A., Mammel, M. K., Patel, I. R., Mays, T., Albert, T. J., LeClerc, J. E., et al. (2007). Interrogating genomic diversity of E. coli O157:H7 using DNA tiling arrays. Forensic Sci. Int. 168, 183–199. doi: 10.1016/j.forsciint.2006.06.079
Jayeol, V., Jeong, S., Almenar, E., Marks, B. P., Vorst, K. L., Brown, J. W., et al. (2019). Predicting the growth of Listeria monocytogenes and Salmonella typhimurium in diced celery, onions, and tomatoes during simulated commercial transport, retail storage, and display. J. Food Protect. 82, 287–300. doi: 10.4315/0362-028X.JFP-18-277
Johnson, J. M., Edwards, S., Shoemaker, D., and Schadt, E. E. (2005). Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 21, 93–102. doi: 10.1016/j.tig.2004.12.009
Keramas, G., Bang, D. D., and Lund, M. (2004). Use of culture, PCR analysis, and DNA microarrays for detection of Campylobacter jejuni and Campylobacter coli from chicken feces. J. Clin. Microbiol. 42, 3985–3991. doi: 10.1128/JCM.42.9.3985-3991.2004
Kim, J. M., Kim, S. Y., Park, Y. B., Kim, H. J., Min, B. S., Cho, J. C., et al. (2012). Simultaneous detection of major enteric viruses using a combimatrix microarray. J. Microbiol. 50, 970–977. doi: 10.1007/s12275-012-2228-9
Kostić, T., and Sessitsch, A. (2012). Microbial diagnostic microarrays for the detection and typing of food- and water-borne (bacterial) pathogens. Microarrays 1, 3–24. doi: 10.3390/microarrays1010003
Law, J. W., Mutalib, N. A., Chan, K., and Lee, L. (2015). Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi: 10.3389/fmicb.2014.00770
Lazcka, O., Campo, F. J., and Muñoz, F. X. (2007). Pathogen detection: a perspective of traditional methods and biosensors. Biosens. Bioelectron. 22, 1205–1217. doi: 10.1016/j.bios.2006.06.036
Liu, Y., Cao, Y., Wang, T., Dong, Q., Li, J., and Niu, C. (2019). Detection of 12 common food-borne bacterial pathogens by TaqMan real-time PCR using a single set of reaction conditions. Front. Microbiol. 10:222. doi: 10.3389/fmicb.2019.00222
Loge, F. J., Thompson, D. E., and Call, D. R. (2002). PCR detection of specific pathogens in water: a risk-based analysis. Environ. Sci. Technol. 36, 2754–2759. doi: 10.1021/es015777m
Ma, L., Zhang, M., Bhandari, B., and Gao, Z. (2017). Recent developments in novel shelf life extension technologies of fresh-cut fruits and vegetables. Trends Food Sci. Tech. 64, 23–38. doi: 10.1016/j.tifs.2017.03.005
Michelle, D. D., Loretta, M. F., and Donald, W. S. (2014). Modeling the growth of Listeria monocytogenes on cut cantaloupe, honeydew and watermelon. Food Microbiol. 38, 52–55. doi: 10.1016/j.fm.2013.08.001
Mockler, T. C., and Ecker, J. R. (2005). Applications of DNA tiling arrays for whole-genome analysis. Genomics 85, 1–15. doi: 10.1016/j.ygeno.2004.10.005
Oliveira, M., Abadias, M., Usall, J., Torres, R., TeixidÓ, N., and Viñas, I. (2015). Application of modified atmosphere packaging as a safety approach to fresh-cut fruits and vegetables—a review. Trends Food Sci. Tech. 46, 13–26. doi: 10.1016/j.tifs.2015.07.017
Penteado, A. L., and Leitão, M. F. (2004). Growth of Listeria monocytogenes in melon, watermelon and papaya pulps. Int. J. Food Microbiol. 92, 89–94. doi: 10.1016/j.ijfoodmicro.2003.08.020
Poretsky, R., Rodriguez-R, L. M., Luo, C., Tsementzi, D., and Konstantinidis, K. T. (2014). Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One 9:e93827. doi: 10.1371/journal.pone.0093827
Ruan, J. H., Wang, W. J., Zhang, T. Y., Bai, Q. Y., Zheng, T., Zhang, Z. D., et al. (2017). Rapid detection of serratia fonticola by TaqMan quantitative real-time PCR using primers targeting the gyrB Gene. Curr. Microbiol. 74, 1343–1348. doi: 10.1007/s00284-017-1323-x
Salazar, J. K., Sahu, S. N., Hildebrandt, I. M., Zhang, L. J., Qi, Y., Liggans, G., et al. (2017). Growth kinetics of Listeria monocytogenes in cut produce. J. Food Protect. 80, 1328–1336. doi: 10.4315/0362-028X.JFP-16-516
Scallan, E., Griffin, P. M., Angulo, F. J., Tauxe, R. V., and Hoekstra, R. M. (2011). Foodborne illness acquired in the United States—unspecified agents. Emerg. Infect. Dis. 17, 16–22. doi: 10.3201/eid1701.P21101
Selinger, D. W., Cheung, K. J., Mei, R., Johansson, E. M., Richmond, C. S., Blattner, F. R., et al. (2000). RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18, 1262–1268. doi: 10.1038/82367
Tarca, A. L., Romero, R., and Draghici, S. (2006). Analysis of microarray experiments of gene expression profiling. Am. J. Obstet. Gynecol. 195, 373–388. doi: 10.1016/j.ajog.2006.07.001
Vital, P. G., Van Ha, N. T., Tuyet, L. T., and Widmer, K. W. (2016). Application of quantitative real-time PCR compared to filtration methods for the enumeration of Escherichia coli in surface waters within Vietnam. J. Water Health 15, 155–162. doi: 10.2166/wh.2016.173
Wilson, W. J., Strout, C. L., DeSantis, T. Z., Stilwell, J. L., Carrano, A. V., and Andersen, G. L. (2002). Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probe. 16, 119–127. doi: 10.1006/mcpr.2001.0397
Winzeler, E. A., Richards, D. R., Conway, A. R., Goldstein, A. L., Kalman, S., McCullough, M. J., et al. (1998). Direct allelic variation scanning of the yeast genome. Science 281, 1194–1197. doi: 10.1126/science.281.5380.1194
Wolffs, P., Norling, B., and Rådström, P. (2005). Risk assessment of false-positive quantitative real-time PCR results in food, due to detection of DNA originating from dead cells. J. Microbiol. Meth. 60, 315–323. doi: 10.1016/j.mimet.2004.10.003
Yu, C., Wales, S. Q., Mammel, M. K., Hida, K., and Kulka, M. (2016). Optimizing a custom tiling microarray for low input detection and identification of unamplified virus targets. J. Virol. Methods 234, 54–64. doi: 10.1016/j.jviromet.2016.03.013
Zhao, X., Lin, C. W., Wang, J., and Oh, D. H. (2014). Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechn. 24, 297–312. doi: 10.4014/jmb.1310.10013
Keywords: Salmonella Typhimurium, Vibrio parahemolyticus, Staphylococcus aureus, Listeria monocytogenes, Escherichia coli O157:H7, in situ-synthesized gene chip, tiling array, fresh-cut fruits and vegetables
Citation: Sarengaowa, Hu W, Feng K, Jiang A, Xiu Z, Lao Y, Li Y and Long Y (2020) An in situ-Synthesized Gene Chip for the Detection of Food-Borne Pathogens on Fresh-Cut Cantaloupe and Lettuce. Front. Microbiol. 10:3089. doi: 10.3389/fmicb.2019.03089
Received: 05 August 2019; Accepted: 20 December 2019;
Published: 05 February 2020.
Edited by:
Lin Lin, Jiangsu University, ChinaReviewed by:
Haiying Cui, Jiangsu University, ChinaQing Fan, Northwestern University, United States
Zaixiang Lou, Jiangnan University, China
Copyright © 2020 Sarengaowa, Hu, Feng, Jiang, Xiu, Lao, Li and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenzhong Hu, wenzhongh@sina.com
 Sarengaowa1,2,3
Sarengaowa1,2,3 Wenzhong Hu
Wenzhong Hu Zhilong Xiu
Zhilong Xiu