- State Key Laboratory of Crop Stress Biology for Arid Areas, Shaanxi Key Laboratory of Agricultural and Environmental Microbiology, College of Life Sciences, Northwest A&F University, Yangling, China
HpaR, a MarR family transcriptional regulator, was first identified in Escherichia coli W for its regulation of the hpa-meta operon. Little else is known regarding its functionality. Here, we report that in Yersinia pseudotuberculosis, HpaR negatively regulates the hpa-meta operon similar to in E. coli W. To investigate additional functions of HpaR, RNA sequencing was performed for both the wild-type and the ΔhpaR mutant, which revealed that the type VI secretion system (T6SS) was positively regulated by HpaR. T6SS4 is important for bacteria resisting environmental stress, especially oxidative stress. We demonstrate that HpaR facilitates bacteria resist oxidative stress by upregulating the expression of T6SS4 in Y. pseudotuberculosis. HpaR is also involved in biofilm formation, antibiotic resistance, adhesion to eukaryotic cells, and virulence in mice. These results greatly expand our knowledge of the functionality of HpaR and reveal a new pathway that regulates T6SS4.
Introduction
HpaR has been characterized as a repressor of the hpa-meta operon (Galan et al., 2003). It has been shown that Escherichia coli strains B, C, and W can use 4-hydroxyphenylacetic acid (4-HPA) as a carbon source via the hpa-meta pathway, while E. coli K-12 does not have this ability (Cooper and Skinner, 1980). Studies of the hpa-meta cluster have focused mainly on E. coli W (Galan et al., 2001). The hpa-meta cluster of E. coli W is composed of 11 genes in two putative operons (Prieto et al., 1996; Galan et al., 2003). The upper operon consists of hpaBC, which encodes the two-component 4-HPA monooxygenase, which converts 4-HPA to 3,4-dihydroxyphenylacetic acid (3,4-HPA) (Prieto and Garcia, 1994; Galan et al., 2000). Transcription of the hpaBC operon is controlled by HpaA, which belongs to the AraC/XylS family and functions as an activator to activate the expression of hpaBC, except for the meta operon in the presence of 4-HPA or 3-HPA (Prieto and Garcia, 1997). The meta operon is composed of hpaGEDFHI, which encodes enzymes that catalyze 3,4-HPA in the Krebs cycle (Roper et al., 1993; Prieto et al., 1996). The expression of both the meta operon and hpaR are regulated by HpaR (Galan et al., 2003). HpaR was first identified as a repressor in E. coli W and is identical to HpcR in E. coli C (Roper et al., 1993; Galan et al., 2003). The hpa-meta cluster has also been found in other bacteria. In Pseudomonas putida U, the hpa pathway is coupled with the tyn pathway to degrade tyramine and dopamine (Barbour and Bayly, 1981; Arcos et al., 2010). Together these form the 3,4-HPA catabolon, in which 3,4-HPA is the central intermediate. The hpa-meta pathway in P. putida U is composed of the genes hpaRBCIHXFDEG2G1AY. HpaR of P. putida U is similar to that in E. coli in that it represses the meta operon (Arcos et al., 2010). Interestingly, a second hpaR (hpaY) was found in P. putida U with similar functionality, ensuring stronger control of the meta operon (Arcos et al., 2010). In Burkholderia xenovorans LB400, the hpa-meta pathway is made up of hpaG1G2EDFHI and hpaBC, while hpaA and hpaX are absent and hpaBC is not adjacent to the meta operon (Mendez et al., 2011). HpaR of B. xenovorans LB400 also regulates the meta operon as a repressor (Mendez et al., 2011). However, previous research has defined HpaR as a repressor that negatively controls expression of the meta operon, but not hpaBC. According to sequence comparison and a three-dimensional model, HpaR is classified into the MarR transcriptional regulator family, which is involved in various physiological processes (Alekshun et al., 2001; Galan et al., 2003; Grove, 2013). However, whether HpaR might exert other functions in addition to acting as a repressor of the hpa-meta operon remains unknown.
Yersinia pseudotuberculosis is a Gram-negative enteric pathogen of animals and humans that causes a variety of diseases such as acute ileitis, mesenteric lymphadenitis, and septicemia (Brubaker, 1991; Smego et al., 1999; Fallman and Gustavsson, 2005). During infection, environmental stress and host immunity reactions can cause an increase in reactive oxygen species (ROS) levels of Y. pseudotuberculosis (Green et al., 2016). Elevated cellular ROS levels lead to oxidative stress, which induces oxidative damage to macromolecules such as proteins, lipids, and DNA (D’Autreaux and Toledano, 2007). Protection against the adverse effects of ROS is vital—bacteria have developed a wide range of systems including antioxidant enzymes, i.e., peroxidase, superoxide dismutase, glutaredoxin, and thioredoxin; low molecular weight antioxidants, i.e., the tripeptide glutathione (GSH) and β-carotene; and vitamins, i.e., vitamins C and E (D’Autreaux and Toledano, 2007; Si et al., 2015, 2017a; Staerck et al., 2017). Recently, we found that the type VI secretion system (T6SS) in Y. pseudotuberculosis was also involved in resistance to oxidative stress; it secretes a zinc-binding protein that imports zinc to mitigate ROS (Wang et al., 2015).
T6SS is a versatile transmembrane machine used by many Gram-negative bacteria to inject effector proteins into cells, either prokaryotic or eukaryotic, or the extracellular milieu (Durand et al., 2014; Russell et al., 2014; Basler, 2015). Although traditionally T6SS is recognized as a contact-dependent bacterial weapon for interspecies competition, some T6SSs from diverse species are also found to play roles in bacterial pathogenesis, biofilm formation, and stress response (Durand et al., 2014; Russell et al., 2014; Yang et al., 2018). For example, a Vibrio anguillarum T6SS regulated by the general stress response regulator RpoS is involved in resistance to hydrogen peroxide, ethanol, and low pH (Weber et al., 2009). Enterohemorrhagic E. coli (EHEC) uses its T6SS to deliver KatN, an Mn-containing catalase, into the host cytosol, resulting in reduced levels of intracellular ROS and greater survival of the pathogen (Wan et al., 2017). In B. thailandensis, T6SS4 exports TseZ and TseM into the extracellular medium to acquire the antioxidant metal ions Zn2+ and Mn2+, respectively, to combat oxidative stress by reducing intracellular ROS levels (Si et al., 2017b,c). The avian pathogenic E. coli (APEC) strain TW-XM harbors two functional T6SSs. The first, T6SS1, plays versatile roles in adherence to host cells, biofilm formation, and bacterial competition; the second, T6SS2, is responsible only for cerebral infection (Ma et al., 2014; Navarro-Garcia et al., 2019).
Despite being reported to control expression of the hpa-meta operon, knowledge of HpaR in Y. pseudotuberculosis is lacking. Unexpectedly, in this study, we found that HpaR not only acts as a repressor of the hpa-meta operon in a similar manner to that of E. coli but also acts as an activator to upregulate the expression of T6SS4 to acquire Zn2+ and resist oxidative stress. HpaR is also involved in biofilm formation, antibiotic resistance, adhesion to eukaryotic cells, and virulence in mice. These results greatly expand our current knowledge of the functions of HpaR.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Escherichia coli strains were cultured at 37°C in Lysogeny Broth (LB) or LB plates. Y. pseudotuberculosis strains were grown in Yersinia-Lysogeny-Broth (YLB) broth (tryptone 1%, yeast extract 0.5%, NaCl 0.5%) or YLB plates at 30 or 26°C. The Y. pseudotuberculosis strain YPIII was the parent of all derivatives used in this study. To construct the hpaR in-frame deletion mutant, the wild-type Y. pseudotuberculosis was mated with E. coli S17-1λpir carrying pDM4-hpaR and chromosomal integration was selected by plating on YLB agar plates supplemented with nalidixic acid and chloramphenicol. The hpaR deletion mutant was subsequently screened on YLB agar plates with 20% sucrose and confirmed by polymerase chain reaction (PCR) and DNA sequencing. Appropriate antibiotics were included in growth medium at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 20 μg/ml; chloramphenicol, 20 μg/ml.
Plasmid Construction
Primers used in this study are listed in Supplementary Table S2. The lacZ fusion reporter vector pDM4-T6SS4p:lacZ was made in previous study (Zhang et al., 2013). To obtain the expression plasmid, hpaR-F-BamHI/hpaR-R-SalI primer pair was used to amplify hpaR gene fragment from Y. pseudotuberculosis genomic DNA by PCR. The hpaR gene fragment were digested with BamHI/SalI and then inserted into similar digested pET28a yielding the pET28a-hpaR. The suicide plasmid pDM4-hpaR used to construct the hpaR mutant was prepared by overlap PCR. Briefly, hpaR-UF-XbaI/hpaR-UR and hpaR-DF/hpaR-DR-SpeI primers were used to amplify the 1000 bp upstream fragment and 1000 bp downstream fragment of hpaR, respectively. Then, the amplified fragments were fused together by overlap PCR with hpaR-UF-XbaI/hpaR-DR-SpeI. The fused PCR products were digested with XbaI/SpeI and cloned into similar digested pDM4 resulting pDM4-hpaR. In order to construct the complementary plasmid pKT100-hpaR, hpaR-F-BamHI/hpaR-R-SalI primers were used to amplify hpaR gene from template. The PCR product was digested with BamHI/SalI and inserted into similarly digested pKT100 to produce pKT100-hpaR, which is subsequently electroporated into ΔhpaR to construct the complementation strains. To construct the hpaG1 promoter reporter vector, hpaG1p-F-SalI/hpaG1p-R-XbaI primer pair was used to amplify the 387 bp promoter fragment from Y. pseudotuberculosis. The product of PCR was digested with SalI/XbaI and cloned into similar digested pDM4-T6SS4p:lacZ to produce pDM4-hpaG1p:lacZ. To construct the T6SS4 promoter fragments with HpaR binding site mutations, overlap PCR was performed to replace the HpaR binding site with identical amount of irrelevant base pairs. T6SS4pMHpaR-R and T6SS4pMHpaR-F were designed to contain 22 bp overlapping DNA fragment (ATTTGTTAGATTCCGAACCGTC) used to replace the HpaR binding site. T6SS4p-F-SalI/T6SS4pMHpaR-R and T6SS4pMHpaR-F/T6SS4p-R-XbaI primers used to amplify the up-fragment and down-fragment of T6SS4p promoter, respectively. The PCR products were ligated with T6SS4p-F-SalI/T6SS4p-R-XbaI by overlap PCR to produce the mutant promoter fragment T6SS4pMHpaR. The T6SS4pMHpaR fragment were digested with SalI/XbaI and ligated into similarly digested pDM4-T6SS4p:lacZ to produce pDM4-T6SS4pMHpaR:lacZ. The validity of all the plasmids constructed above was confirmed by DNA sequencing.
Sequence Data Analysis
Sequence alignment was performed as previously described (Shen et al., 2005; Huang et al., 2008). The sequence of hpaRGEDFHIXABC (Accession No. Z37980.2) of E. coli W were retrieved from GenBank. Sequence comparisons were carried out using BLAST program at National Center for Biotechnology Information website1. Multiple protein sequence alignment was made by CLUSTAL W. The sequences used in alignment have been deposited in the GenBank database [Accession No. EcC E. coli C (S56952.1), EcW E. coli W (Z37980.2), Yptb Y. pseudotuberculosis YPIII (ACA68739.1), PpU P. putida U (FJ904934.1), BxLB400 B. xenovorans LB400 (ABE33958.1)]. The result was exported by ESPript2.
Construction of Chromosomal Fusion Reporter Strains and β-Galactosidase Activity Assay
The lacZ fusion reporter vector pDM4-T6SS4p:lacZ, pDM4-T6SS4pMHpaR:lacZ and pDM4-hpaG1p:lacZ was transformed into E. coli S17-1λpir and then introduced into Y. pseudotuberculosis by conjugation as described (Xu et al., 2014). All constructed lacZ fusion reporter strains were grown in YLB broth at 26°C and o-nitrophenyl-β-galactoside (ONPG) as substrate for measuring β-galactosidase activities as described by Miller (1992). The β-galactosidase results shown represent the mean of one representative assay performed in triplicate, and error bars represent standard deviation. Statistical analysis was carried out with Student’s t-test.
Protein Expression and Purification
To express and purify His6- tagged HpaR, the constructed expression vector pET28a-hpaR were transformed into BL21(DE3). Single colony was cultured in 5 ml LB broth at 37°C overnight and diluted 100-fold into 500 ml LB. Until the OD600 = 0.5, the culture was shifted to 22°C, induced with 0.3 mM IPTG and further cultivated for 12 h to express the recombinant proteins. Cell pellet was collected by centrifugation, washed and resuspended in His binding buffer, and lysed by sonication. The recombinant proteins were purified with the His⋅Bind Ni-NTA resin (Novagen) according to the manufacturer’s instructions. Eluted recombinant proteins were dialyzed against the appropriate buffer at 4°C for 4 h and then stored at −80°C until used.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed as previously described (Zhang et al., 2013; Wang et al., 2015). Bio-T6SS4pHpaR-F/Bio-T6SS4pHpaR-R primers were used to amplify the biotin 5′-end-labeled promoter probes (Bio-T6SS4pHpaR) from Y. pseudotuberculosis genomic DNA. The unlabeled T6SS4pHpaR was amplified with T6SS4pHpaR-F/T6SS4pHpaR-R from template, which used as a competitor. In addition, the unrelated protein BSA was used as negative control. All promoter probes were purified by EasyPure Quick Gel Extraction Kit (TransGen Biotech). According to the manufacturer’s protocol (LightShift Chemiluminescent EMSA Kit; Thermo Fisher Scientific), each 20 μl EMSA reaction solution was prepared as follows: 1 × binding buffer, 50 ng poly (dI-dC), 2.5% glycerol, 0.05% Nonidet P-40, 5 mM MgCl2, 3 ng Bio-T6SS4pHpaR DNA, 1 ng T6SS4pHpaR DNA as competitor, and different concentration of protein (0, 0.6, 0.8, 0.8 μg). Reaction solutions were incubated for 20 min at room temperature. The samples were loaded onto a 6% polyacrylamide native gel and transferred to a Biodyne B nylon membrane (Thermo Fisher Scientific). The biotin-labeled DNA bands were visualized by chemiluminescent substrate according to the manufacturer’s protocol. hpaG1p-F/hpaG1p-R primers were used to amplify the hpaG1p probes from template. An unrelated DNA (URD) in the similar length or bovine serum albumin (BSA) was included in the binding assay system to serve as negative controls. Increasing concentrations of purified His6-HpaR (0, 0.4, 0.8, 1.2, 1.2 μg) were incubated with 20 ng hpaG1p probes in EMSA buffer (20 mM Tris-HCl [pH 7.4], 4 mM MgCl2, 100 mM NaCl, 1 mM dithiothreitol, 10% glycerol). After incubation for 20 min at 26°C, the sample was subjected to electrophoresis on a 6% polyacrylamide native gel. Then the DNA probe was detected using SYBR green.
Stress Survival Assay
Yersinia pseudotuberculosis strains grown in YLB broth until mid-exponential are diluted 50-fold into M9 medium (Na2HPO4, 6 g/L; KH2PO4, 3 g/L; NaCl, 0.5 g/L; NH4Cl, 1 g/L; MgSO4, 1 mM; CaCl2, 0.1 mM; glucose 0.2%) with or without H2O2 (1.5 mM), cumene hydroperoxide (CHP, 0.5 mM), ampicillin (0.5 μg/ml), gentamicin (0.2 μg/ml) at 26°C for 1 h at 100 rpm. After treatment, the cultures were diluted 1000-fold and plated onto YLB agar plates with nalidixic acid. Colonies were counted after 24 h at 26°C. The survival percentage of Y. pseudotuberculosis is calculated by dividing the number of CFU of stressed cells by the number of CFU of cells without stress. All these assays were performed in triplicate at least three times.
Fluorescence Dye-Based Intracellular ROS Detection
The intracellular ROS levels were detected by the fluorescent reporter dye 5-(and-6)-chloromethyl-2′,7′-dichlorodihydro fluorescein diacetate, acetylester (CM-H2DCFDA, Invitrogen) as previously reported (Si et al., 2017c). Briefly, 1 ml culture were collected after treatment, washed with M9 medium and resuspended in 1 ml of M9 medium containing 10 μM CM-H2DCFDA. Samples were incubated at 26°C in the dark for 20 min. Then the cells were pelleted, washed with M9 medium and resuspended in 1 ml of PBS. Then 200 μl samples were transferred to a dark 96-well plate. Fluorescence signals were measured using a SpectraMax M2 Plate Reader (Molecular Devices) with excitation/emission wavelengths of 495/520 nm. The results shown represented the mean of one representative assay performed in triplicate, and error bars represent the standard deviation (SD).
Protein Secretion Assay
Secretion of YezP (encoded by ypk_3459) was detected as previously described (Wang et al., 2015). Briefly, all strains were grown in 180 ml YLB broth at 26°C on a rotary shaker (220 rpm) until OD600 = 1.6. A 2 ml culture was centrifuged and the cell pellet was resuspended in 100 μl SDS-loading buffer to serve as the total cell pellet sample. A total of 150 ml culture was centrifuged at low speed (5000 rpm) for 10 min to remove most cell pellets, and the supernatant was further centrifuged at high speed (8000 rpm) for 15 min to eliminate bacteria. To remove the remaining bacteria, the supernatant was filtered through a 0.22-μm filter (Millipore, Billerica, MA, United States). The supernatant was filtered three times through a nitrocellulose filter (BA85) (Whatman) to collect all secretion proteins. The filter was cut into pieces in 1.5 ml tubes and resuspended in 100 μl SDS-loading buffer for 20 min at 65°C to recover the proteins. All samples were normalized to the OD600 of the culture and volume used in preparation.
Western Blot Analysis
Western blot analysis was performed as described previously (Shen et al., 2009; Lin et al., 2017). Samples were resuspended in SDS-Loading buffer and separated in 15% polyacrylamide gel, then transferred onto PVDF membranes (Millipore). The membrane was blocked in 5% (w/v) BSA for 4 h at room temperature, and incubated with primary antibody at 4°C overnight. The primary antibodies were anti-VSVG (sc-365019, Santa Cruz biotechnology, United States), 1:1000 and anti-ICDH, 1:6000. The ICDH antisera were made in our previous study (Xu et al., 2010). The membrane was washed five times with TBST buffer (50 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 7.4), and incubated with 1:5,000 dilution of horseradish peroxidase conjugated secondary antibody (Shanghai Genomics) for 4 h at 4°C. Signals were detected using the ECL plus kit (GE Healthcare, Piscataway, NJ, United States) according to the manufacturer’s specified protocol.
DNase I Footprinting Assay
DNase I footprinting assay was performed as described preciously (Zhao et al., 2017). Briefly, hpaG1p and T6SS4p were amplified by PCR with hpaG1p-FP-F/hpaG1p-FP-R and T6SS4p-FP-F/T6SS4p-FP-R primers, respectively. The PCR products were cloned into the pMD-18T vector (TaKaRa) as template to further amplify the fluorescent FAM labeled probes with primers M13R(FAM-labeled) and M13F(-47). The FAM-labeled probes were purified by the Wizard SV Gel and PCRClean-Up System (Promega) and quantified with NanoDrop 2000C (Thermo). For DNase I footprinting assay, 400 ng probes were incubated with various amount of His6-HpaR in 40 μl of each sample. After binding at 30°C for 30 min, 10 μl solution containing about 0.010 unit DNase I (Promega) and 100 nmol CaCl2 were added and further incubated for 1 min at 25°C. 140 μl DNase I stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA and 0.15% SDS) was added into samples to stop the reaction. Samples were extracted by phenol/chloroform to remove proteins, precipitated with ethanol, washed and dissolved in 35 μl MiniQ water. The preparation of the DNA ladder, electrophoresis, and data analysis were similar to described previously (Wang et al., 2012).
RNA-seq Experiments
Yersinia pseudotuberculosis and the ΔhpaR mutant were used for RNA-Seq transcriptomics analysis (Wang et al., 2017). Two strains (three biological replicates) were grown in YLB medium at 26°C to OD600 = 1.6. Total RNA was extracted from each sample for cDNA library construction by using bacteria total RNA isolation kit (Tiangen) and analyzed with the Bioanalyzer 2100 system (Agilent Technologies). Library sequencing was completed at the Beijing Genomic Institute (BGI-Shenzhen). The result of sequencing was aligned with the reference genome of Y. pseudotuberculosis and RPKM (Reads per kilobase transcriptome per million mapped reads) was used to normalize the expression level of genes. The differential expressed genes were shown as fold change calculated by log2 (RPKM of ΔhpaR/WT).
Quantitative Real-Time PCR
All strains were cultured at mid-exponential, harvested, washed with PBS. Total RNA was isolated from each sample using the RNAprep Pure Cell/Bacteria Kit, treated with RNase-free DNase (Tiangen) and measured the purity and concentration by gel electrophoresis and spectrophotometer (NanoDrop, Thermo Scientific). 1.5 μg of total RNA was used to synthesis the first-strand cDNA by reverse transcriptase (TransGen Biotech). Quantitative real-time PCR (qRT-PCR) was performed in CFX96 Real-Time PCR Detection System (Bio-Rad) with TransStart Green qPCR SuperMix (TransGen Biotech). The qRT-PCR primers were listed in Supplementary Table S2 and qRT-PCR parameter was set as follows: 95°C for 30 s followed by 40 cycles of 94°C for 15 s, 52°C for 30 s. The relative abundance of 16S rRNA was used as the internal standard for standardization of results.
Determination of Intracellular Ion Content
Intracellular ion content was determined as described previously (Wang et al., 2015). Briefly, Y. pseudotuberculosis wild-type, ΔhpaR and ΔhpaR (hpaR) strains were grown in YLB medium to mid-exponential. 20 ml culture were collected, washed with PBS, resuspended in 20 ml PBS buffer containing 0.4% glucose, 1.5 mM H2O2 and 1 μM Zn2+, and then incubated further for 20 min at 26°C. After centrifugation at low speed (4000 rpm) for 10 min, the supernatant was removed and the cell pellet weight was measured. Then the pellet was resuspended and chemically lysed on a rotating mixer at a slow setting for 20 min by using Bugbuster (Novagen, Madison, WI, United States) according to the manufacturer’s instructions. The protein concentration was analyzed with NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) according to the manufacturer’s instructions. The lysis solution was diluted 100-fold in 2% molecular grade nitric acid to 10 ml. These samples were analyzed by Inductively Coupled Plasma Mass Spectrometry (ICP-MS) (Varian 802-MS), and the results were corrected using the appropriate buffers for reference and dilution factors. Triplicate cultures of each strain were analyzed during a single experiment and the experiment was repeated at least three times.
Biofilm Formation Assay
Biofilm formation was measured using the test tube, performed as previously described (Guan et al., 2015). Y. pseudotuberculosis strains were grown in YLB broth, and then transferred into 3 ml of M9 medium containing 0.4% glucose on a rotary shaker (220 rpm) at 26°C. After 24 h, the test tube was washed gently three times with PBS and stained with 1% crystal violet for 15 min, then washed three times with PBS again to remove redundant crystal violet. The remaining crystal violet was resolved with 95% ethanol and 200 μl of samples were measured the absorbance at 595 nm by a microplate reader (BioTek Instruments, Inc.).
Cell Adhesion Assay
The adhesion ability of bacteria was measured as previously reported (Tan et al., 2017). HeLa cells were seeded into 12-well plate at a concentration of 1 × 105 cells/ml before infection. Y. pseudotuberculosis wild-type, ΔhpaR and ΔhpaR (pKT100-hpaR) strains were grown at 26°C till OD600 = 1.0 and then collected, washed and resuspended in DMEM. The bacterial suspensions were serial diluted and plated onto YLB agar plates to count the actual number of the bacteria. HeLa cells were infected with bacterial strains at a multiplicity of infection (MOI) of 100 and then centrifuged for 5 min at 1500 rpm to promote contact between bacteria and HeLa cells. After 2 h, HeLa cells were washed for three times using prewarmed PBS. Cell-associated bacteria were liberated by treatment with 0.1% Triton X-100 in sterile PBS for 5 min. The number of cell-associated bacteria was divided by the total number of bacteria in a well to calculate the adhesion percentage.
Mouse Infection
All mice were maintained and handled in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People’s Republic of China. The protocol was approved by the Animal Welfare and Research Ethics Committee of Northwest A&F University (protocol number: NWAFUSM2018001). All strains growing in YLB broth at 26°C until mid-exponential were harvested, washed, resuspended in sterilized PBS to a final concentration of 3 × 109 bacteria for survival assays. The 6–8 weeks old female C57BL/6 mice were orogastric infected with Y. pseudotuberculosis using a ball-tipped feeding needle. The survival rate of the mice was determined by monitoring the survival everyday for 24 days.
Statistical Analysis
Statistical analyses of gene expression data, LacZ activity, survival assay, ROS determination, intracellular ion content determination, and cell adhesion assay were performed using paired two-tailed Student’s t-test. Statistical analyses were performed using GraphPad Prism software (GraphPad Software).
Results
Regulation of the hpa Gene Cluster in Y. pseudotuberculosis
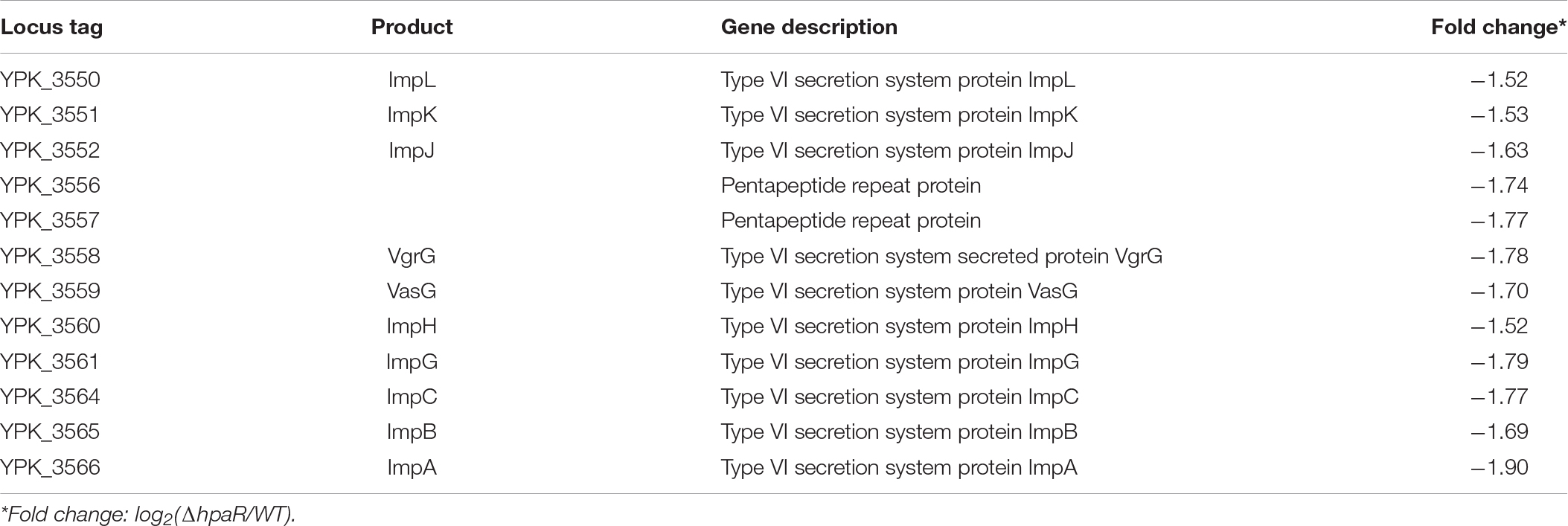
The 4-hydroxyphenylacetic acid (4-HPA) catabolic pathway was determined by alignment with E. coli W (ATCC 11105) while searching for the complete genome sequence of Y. pseudotuberculosis (Figure 1A). Analyses of the open reading frames of the gene cluster suggested that the 4-HPA metabolic cluster is composed of 11 genes in Y. pseudotuberculosis, similar to that of E. coli W (Roper et al., 1993; Prieto and Garcia, 1994). These genes include the genes of 4-HPA monooxygenase (hpaBC), the genes of homoprotocatechuate (HPC) meta cleavage enzymes (hpaG1G2EDFHI), the HPA transport protein gene (hpaX), and one regulatory gene (hpaR). All genes (hpaG1G2EDFHIXBC) lay in a continuous row in the same direction as in E. coli. The translation products of these genes were identified as the homologs of the 4-HPA hydroxylases in E. coli W with high identities (Figure 1A). HpaBC, the HPA monooxygenase, oxidizes 4-HPA or 3-HPA to HPC. HpaD (HPC 2,3-dioxygenase), HpaE (CHMS dehydrogenase), HpaF (CHM isomerase), HpaG (OPET decarboxylase), HpaH (hydratase), and HpaI (HHED aldolase) form the HPA catabolic pathway, which degrades HPC in the pyruvic acid or succinic semialdehyde pathways. The HpaR protein shares 66, 66, 48, and 34% amino acid sequence identities with identified HpaR in E. coli C, E. coli W, P. putida U, and B. xenovorans LB400, respectively (Supplementary Figure S1). Thus, we propose the gene cluster from ypk_2452 to ypk_2462 as an hpa-meta gene cluster in Y. pseudotuberculosis.
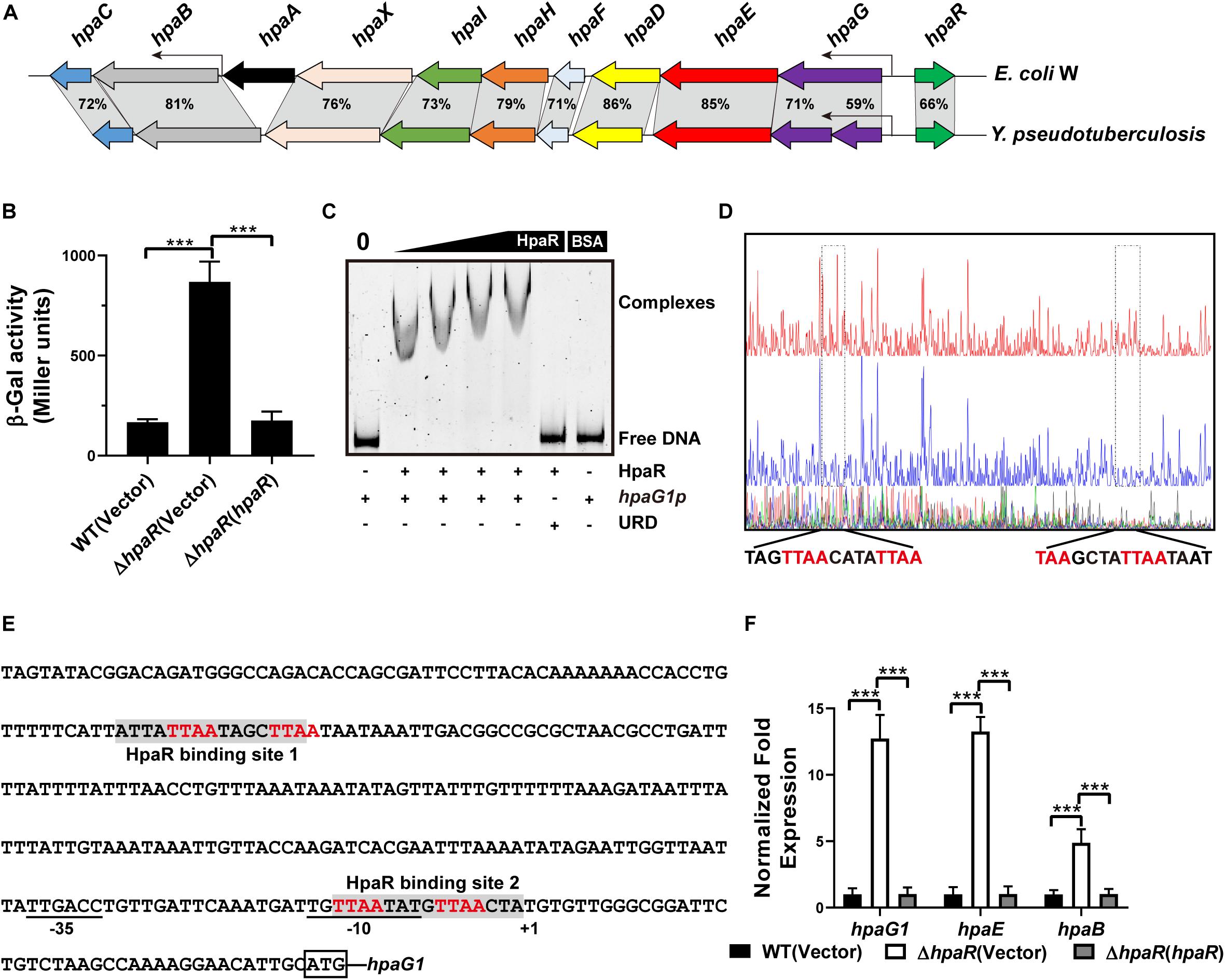
Figure 1. HpaR negatively regulates hpa-meta operon in Yersinia pseudotuberculosis. (A) Gene organization of hpa-meta cluster and identities of protein sequence in Escherichia coli W and Y. pseudotuberculosis. (B) β-Galactosidase activity of the hpaG1 promoter in Y. pseudotuberculosis Wild-type, ΔhpaR mutant, and its complementary strain ΔhpaR(hpaR). Data shown are the average of three independent experiments. The error bars indicate standard deviations of the average. ***P < 0.001. (C) EMSA was performed to analyze the interactions between His6-HpaR and the hpaG1 promoter. URD, unrelated DNA fragment. (D) Identification of the HpaR-binding site within the hpaG1 promoter using the DNase I footprinting assay. Letters in red denote inverted repeat sequences. (E) The HpaR binding sites detected by DNase I footprinting assay were shown in the hpaG1 promoter region by shading. Letters in red denote inverted repeat sequences. (F) Relative expression levels of hpaG1, hpaE, and hpaB were measured by quantitative RT-PCR in wild-type, ΔhpaR mutant, and complementary strain ΔhpaR(hpaR).
In E. coli W, the meta cleavage enzymes (HpaGEDFHI) that catabolize 4-HPA are repressed by HpaR, and the HPA monooxygenase (HpaBC) is positively regulated by HpaA (Prieto and Garcia, 1997). To verify the role of HpaR in expression of the hpa gene cluster in Y. pseudotuberculosis, a single-copy hpaG1p:lacZ fusion reporter was introduced into the chromosomes of wild-type, ΔhpaR mutant, and complemented ΔhpaR(hpaR) strains. The β-galactosidase activity of the three strains was quantitatively measured (Figure 1B). The deletion of hpaR significantly increased the activity of the hpa promoter, which could be fully restored by introducing a complementary plasmid expressing HpaR (Figure 1B). Next, we analyzed the interaction between HpaR and the putative operator with an electrophoresis mobility shift assay (EMSA). Incubation of HpaR with a probe containing the promoter of the hpa-meta operon (hpaG1p) led to the formation of DNA–protein complexes (Figure 1C), which suggests direct interaction of HpaR and the hpaG1p promoter. DNase I footprinting analysis further revealed that the putative HpaR binding site was protected from digestion in DNA–HpaR complexes. Two HpaR-protected sites upstream of the transcriptional start site in the hpaG1p promoter were identified (Figures 1D,E). Thus, HpaR specifically recognizes an operator within the hpa-meta promoter, likely influencing its expression. Indeed, negative regulation of the hpa-meta operon by HpaR was confirmed by qRT-PCR analyses. The data indicated that the expression of hpaG1, hpaE, and hpaB was upregulated in ΔhpaR, and such an increase could be restored to wild-type levels by complementation with the expression plasmid pKT100-hpaR (Figure 1F). Overall, in Y. pseudotuberculosis, HpaR functions as a repressor and negatively regulates the hpa-meta gene cluster in a similar manner to E. coli W.
Genome-Wide Analysis of the Genes Regulated by HpaR
The multiple antibiotic resistance regulator (MarR) family transcriptional regulator has a variety of biological functions, including resistance to multiple antibiotics and other toxic chemicals such as organic solvents, household disinfectants, and oxidative stress agents (Alekshun and Levy, 1999; Wei et al., 2007). As a MarR family regulator, we speculate that HpaR may play other regulatory roles besides aromatic degradation. Thus, RNA sequencing (RNA-seq) was employed to detect the genes regulated by HpaR, except the hpa-meta gene cluster.
To identify the HpaR-dependent expression genes in Y. pseudotuberculosis, RNA was extracted for RNA-seq from both wild-type and ΔhpaR mutant strains in mid-exponential phase (SRA accession: PRJNA612340). After RNA-seq, quality control and gene expression level analysis were performed by fragments per kilobase of transcript per million mapped reads (FPKM). A total of 106 gene candidates that exhibited differential expression were found between wild-type and ΔhpaR mutant strains, including 57 upregulated and 49 downregulated genes (Supplementary Table S3). To verify the RNA-seq data, qRT-PCR analysis of 11 representative genes was conducted. The log2-transformed values of each gene were in good agreement with the RNA-seq data (Figure 2A), which confirmed the credibility of the RNA-seq data. Subsequently, the functions of differentially expressed genes were identified by KEGG pathway analysis. As shown in Figure 2B, 17 different pathways were found in the DGE (Different Gene Expression), in which eight pathways were up-regulated and nine were down-regulated. These included degradation of aromatic compounds, metabolism, microbial metabolism in a diverse environment, purine metabolism, bacterial secretion system, and tyrosine metabolism. In the RNA-seq data, two gene clusters were found to be fully regulated by HpaR. One cluster was the hpa-meta operon, which was 2.3–4.56-fold upregulated in the ΔhpaR mutant (Table 1). The other was the T6SS4 gene cluster in Y. pseudotuberculosis. T6SS is a newly described secretion system in many Gram-negative bacteria (Bingle et al., 2008; Jani and Cotter, 2010). The RNA-seq data revealed that the genes in the T6SS4 operon were 0.54–0.96-fold downregulated in the ΔhpaR mutant (Table 2). Hemolysin co-regulated protein (Hcp), which forms the hexameric rings for the T6SS syringe needle, was 0.68-fold downregulated. Valine-glycine repeat protein G (VgrG) was 0.54-fold downregulated. The periplasmic domain protein IcmF was 0.62-fold downregulated. ImpA/B (VipA/VipB), which serve as tail sheath proteins in T6SS, were 0.73/0.94-fold downregulated. The same was true with other structural genes. These data suggest that the hpa-meta operon was negatively regulated by HpaR, consistent with the transcription of chromosomal hpaG1p:lacZ fusions. The data also suggest that T6SS4 is positively regulated by the transcriptional regulator HpaR.
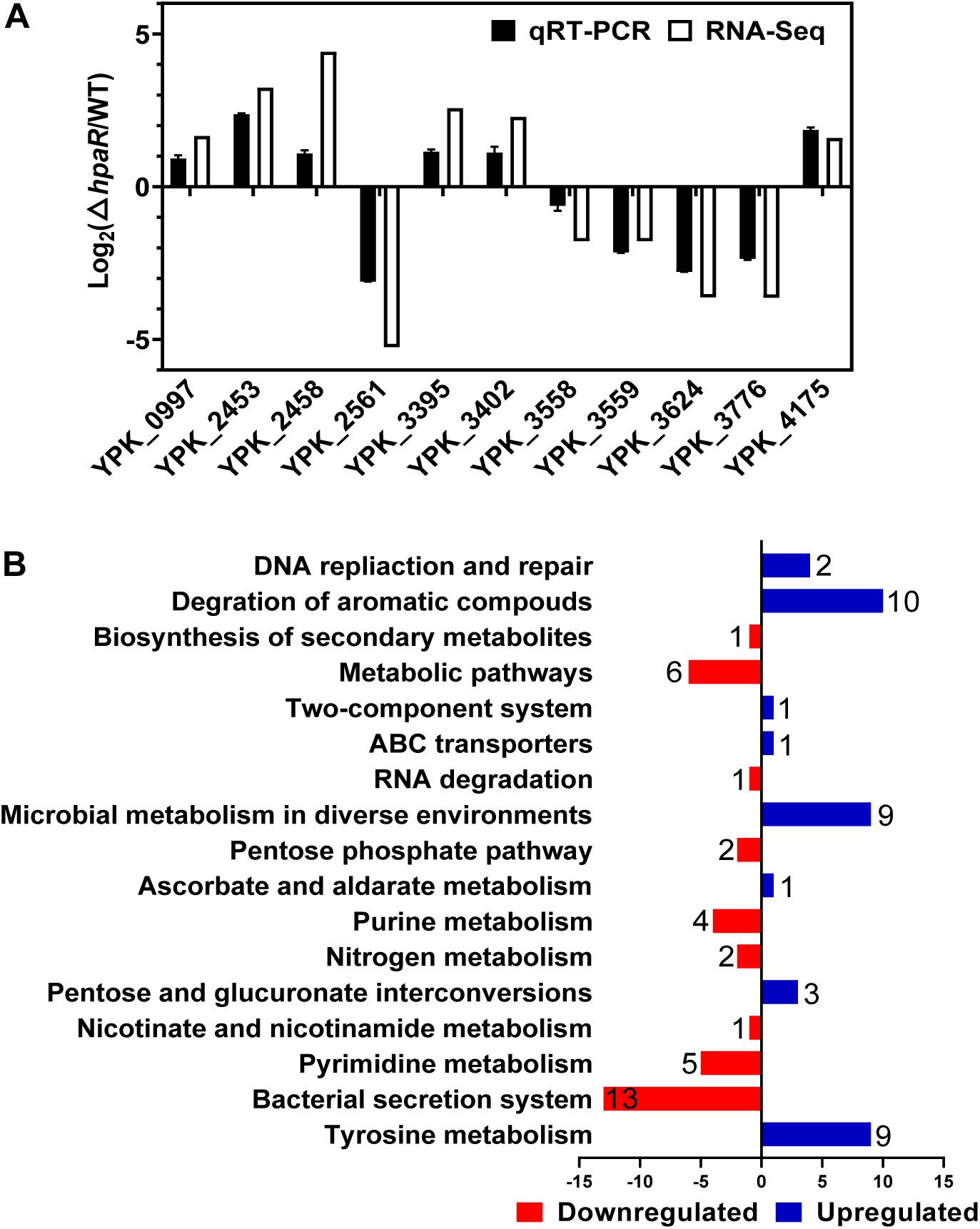
Figure 2. Transcriptomics analysis of HpaR regulated genes in Y. pseudotuberculosis. (A) Validation of RNA-Seq data using qRT-PCR. Eleven representative genes were evaluated for validation of the RNA-seq data using qRT-PCR. (B) KEGG pathway analysis of differentially expressed genes (hpaR mutant VS wild-type). The red and blue bars represent up- and down-regulated genes, respectively, and the numeric labels represent the number of genes with corresponding function.
HpaR Positively Regulates T6SS4 Expression by Directly Binding to Its Promoter
Based on the RNA-seq data, we found HpaR could positively regulate the expression of T6SS4. To further determine the role of HpaR in the expression of T6SS4, a single copy T6SS4p:lacZ fusion was introduced into the chromosomes of wild-type, hpaR mutant, and complemented ΔhpaR(hpaR) strains and LacZ activity of the resulting strains was quantitatively measured (Figure 3A). Compared to the wild-type strain, the T6SS4p:lacZ fusion was decreased significantly in ΔhpaR; this decrease could be restored by the complementary plasmid (pKT100-hpaR). This suggests that HpaR positively regulates T6SS4 expression. To test whether the T6SS4 gene clusters were regulated by HpaR directly, the interaction between HpaR and the T6SS4 promoter was examined by EMSA. Incubation of a probe containing the T6SS4 promoter with HpaR led to the formation of DNA–protein complexes (Figure 3C). DNase I footprinting analysis revealed that the binding site was protected from digestion in DNA–HpaR complexes, further indicating the recognition of this DNA element by HpaR (Figure 3D). An HpaR-protected site (CCTCTTATTTTGGCTATTCATCCACGTCATCGTGCTA) upstream of the -35 and -10 elements was identified (Figure 3E).
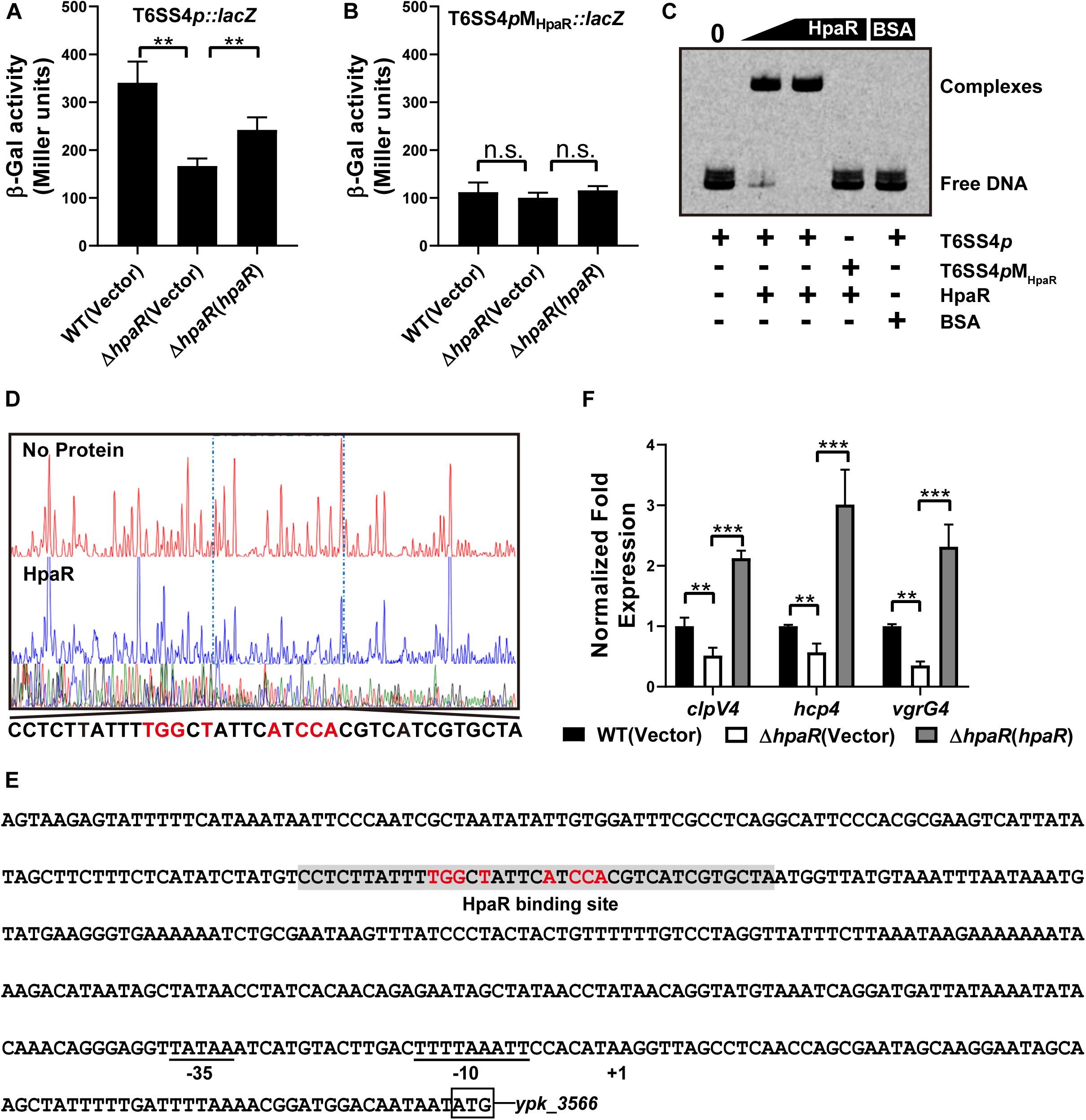
Figure 3. HpaR positively regulates the T6SS4 expression in Y. pseudotuberculosis. β-galactosidase activity of the T6SS4 promoter (A) and T6SS4 promoter with mutations in the HpaR binding site (B) in Y. pseudotuberculosis Wild-type, ΔhpaR mutant, and its complementary strain ΔhpaR(hpaR). Data shown are the average of three independent experiments. The error bars indicate standard deviations of the average. **P < 0.01; n.s., not significant. (C) EMSA was performed to analyze the interactions between His6-HpaR and the T6SS4 promoter (T6SS4p) or the promoter mutant identified His6-HpaR binding region (T6SS4pMHpaR). (D) Identification of the HpaR-binding site within the T6SS4 promoter using the DNase I footprinting assay. Letters in red denote inverted repeat sequences. (E) The HpaR binding site detected by DNase I footprinting assay were shown in the T6SS4 promoter region by shading. Letters in red denote inverted repeat sequences. (F) Relative expression levels of clpV4, hcp4, and vgrG4 were measured by qRT-PCR. The error bars indicate the standard deviation from three independent experiments. ***P < 0.001; **P < 0.01.
The interactions between HpaR and the T6SS4 promoter appeared to be site-specific, as mutation of the HpaR binding site in the T6SS4 promoter prevented formation of the protein–DNA complex (Figure 3C) and abolished HpaR dependent T6SS4 regulation (Figure 3B). The positive regulation of T6SS4 by HpaR was further confirmed by qRT-PCR analysis (Figure 3F). The data indicated that the expression of T6SS4 components clpV4, hcp4, and vgrG4 were reduced in ΔhpaR, which could be restored by plasmid (pKT100-hpaR). Thus, HpaR recognizes and binds to the promoter of T6SS4 specifically to activate expression.
HpaR Affects the Antioxidant Activity of Y. pseudotuberculosis by Regulating T6SS4
In our previous study, T6SS4 of Y. pseudotuberculosis was discovered to play important roles in multiple stress defenses, especially oxidative stress (Wang et al., 2015). The regulation of T6SS4 by HpaR prompted us to examine whether HpaR plays a role in oxidative stress resistance via T6SS4. To test the hypothesis that HpaR exerts antioxidant activity by regulating T6SS4, the survival rate and ROS level in cells treated with cumene hydroperoxide (CHP) or hydrogen peroxide (H2O2) were measured. After incubation with CHP, approximately 60% of wild-type cells survived. The survival rate of the mutant lacking hpaR decreased to 16%, and a 13% survival rate was observed for the double mutant ΔhpaRΔclpV4. Expressing HpaR could fully rescue such a decrease in the ΔhpaR, but only increased the survival rate slightly in the double mutant ΔhpaRΔclpV4 (Figure 4A). The ROS level under oxidative stress was also examined using CM-H2DCFDA fluorescent dye. The data showed that the ROS level in ΔhpaR and ΔhpaRΔclpV4 was higher than the wild-type strain. Complementation with the plasmid pKT100-hpaR could reduce the ROS level in ΔhpaR, but not in the ΔhpaRΔclpv4 double mutant (Figure 4B). Similar results were obtained with H2O2 treatment (Figures 4C,D). T6SS4 could secrete the zinc-binding protein YezP for zinc import in Y. pseudotuberculosis (Wang et al., 2015). Thus, YezP-VSVG was expressed in relevant Y. pseudotuberculosis strains to test the secretion of YezP. Significant amounts of YezP-VSVG could be readily detected in culture supernatant from wild-type bacteria. The secretion of YezP decreased by 32% in the ΔhpaR mutant and was completely restored in a mutant complemented with HpaR (Figure 4E). The zinc contents in bacteria challenged with H2O2 were measured using inductively coupled plasma mass spectrometry (ICP-MS). The results revealed that the deletion of hpaR significantly lowered the intracellular Zn2+ level and expression of HpaR restored the defects (Figure 4F). These results established that HpaR benefited Y. pseudotuberculosis resisting oxidative stress by upregulating T6SS4.
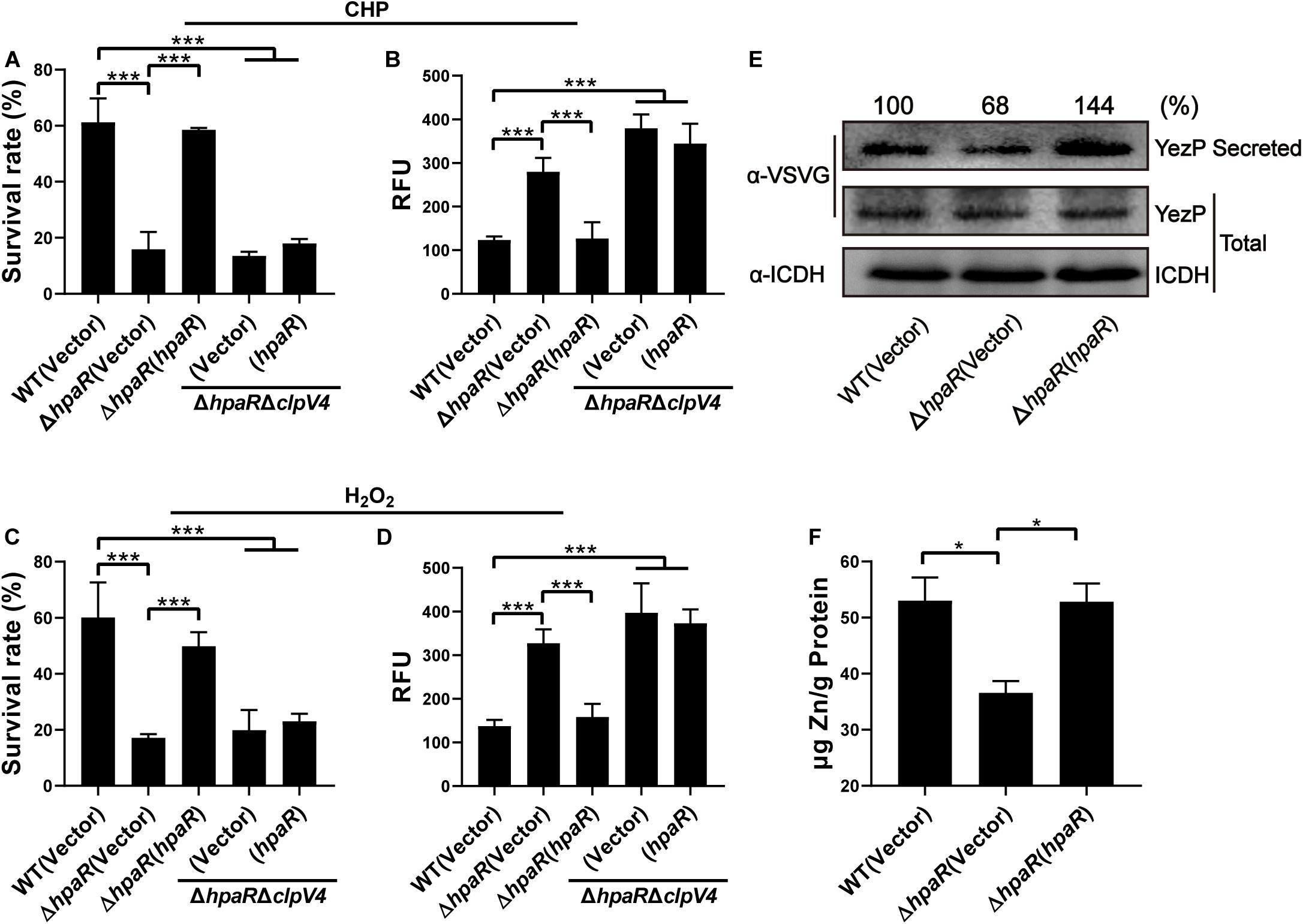
Figure 4. HpaR plays a role in resisting oxidative stress via T6SS4. Survival rate of indicated Y. pseudotuberculosis strains were determined after treatment with CHP (A) or H2O2 (C) for 1 h. (B,D) Intracellular ROS of indicated strains were stained with H2DCFDA dye (Invitrogen) and fluorescence signals were measured using a SpectraMax M2 Plate Reader (Molecular Devices) with excitation/emission wavelengths of 495/520. (E) Western blot detection of YezP secretion in Y. pseudotuberculosis wild-type, ΔhpaR mutant, and its complementary strain ΔhpaR(hpaR). Proteins in culture supernatant of relevant Y. pseudotuberculosis strains expressing YezP-VSVG were probed for VSVG (upper) by immunoblotting. (F) Mid-exponential phase of Y. pseudotuberculosis strains were exposed to 1.5 mM H2O2 for 30 min in PBS containing 1 μM ZnCl2. Zn2+ associated with bacterial cells was measured by inductively coupled plasma mass spectrometry (ICP-MS). Data shown are the average of three independent experiments. The error bars indicate the standard deviation from three independent experiments. ***P < 0.001; **P < 0.01; *P < 0.05.
HpaR Contributes to Biofilm Formation and Adhesion to Epithelial Cells
Similar to other Gram-negative bacteria, Y. pseudotuberculosis is capable of forming biofilms on abiotic surfaces. The biofilm formation ability of wild-type, ΔhpaR mutant, and complemented ΔhpaR(hpaR) strains were observed in a test tube by staining with crystal violet (Figure 5A) and quantified by resolving with 95% ethanol and measuring the absorbance at 595 nm (Figure 5B). The ΔhpaR mutant showed attenuated biofilm formation when compared to wild-type and the decrease could be restored by expressing hpaR. Note that there’s no significant difference between the growth activities of these strains (Supplementary Figure S2). Biofilm formation plays an important role in the pathogen lifecycle. It is imperative in chronic infection, antibiotic resistance, and adhesion to host cells. Thus, ampicillin and gentamicin, two commonly used antibiotics in clinics, were used to verify viability of the hpaR mutant in antibiotic resistance. The survival rates of the ΔhpaR strain were only 34 and 18% following gentamicin and ampicillin treatment, respectively, while approximately 72 and 51% of wild-type cells survived under the same conditions (Figure 5C). This suggests that the ΔhpaR mutant was more sensitive to antibiotics than the wild-type. Adhesion to host cells is a crucial step in the infection process and host colonization. Thus, the adherence of wild-type, hpaR mutant, and complemented strains to HeLa cells was evaluated. As shown in Figure 5D, the ΔhpaR strain had fewer adherences to HeLa cells than wild-type. Overall, hpaR may assist in the maintenance of biofilm formation activity, antibiotic resistance, and adhesion to eukaryotic cells.
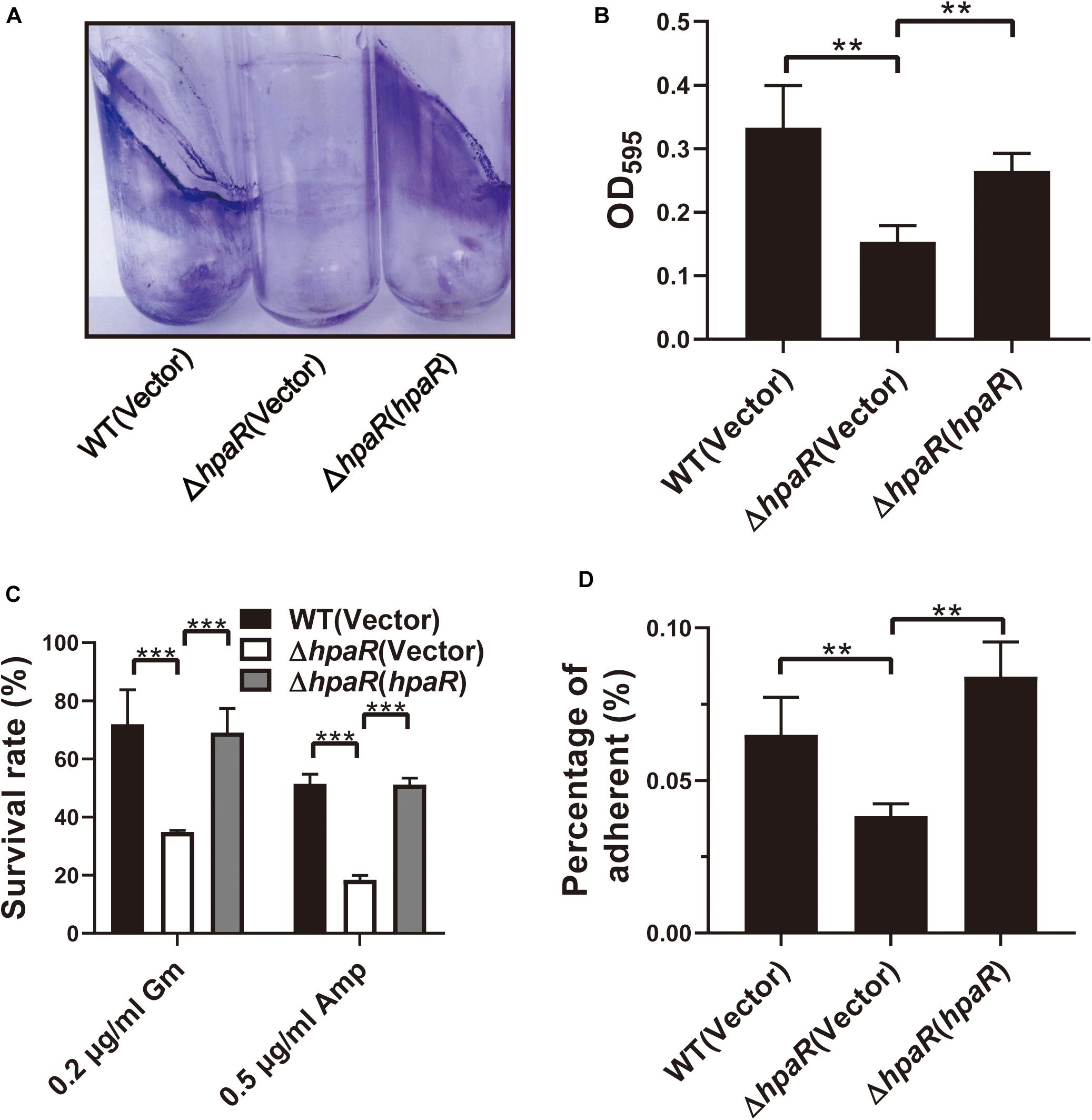
Figure 5. HpaR is involved in biofilm formation, antibiotic resistance and adhesion to eukaryotic cells. (A) Direct observation of Y. pseudotuberculosis biofilm formation by Wild-type, ΔhpaR mutant, and its complementary strain ΔhpaR (hpaR). The results shown the biofilm biomass stained with crystal violet in glass test tubes. (B) Quantitative measurement of biofilm formation by resolving in 95% ethanol and measuring the absorbance at OD595. (C) The survival rate of Wild-type, ΔhpaR mutant, and its complementary strain ΔhpaR (hpaR) under 0.2 μg/ml gentamicin (Gm) or 0.5 μg/ml ampicillin (Amp) treatment. (D) Adhesion of the strains to HeLa cells. Adherence level is expressed as the percentage of adherent bacteria relative to the inoculum count. Data shown are the average of three independent experiments; the error bars indicate the standard deviation from three independent experiments. ***P < 0.001; **P < 0.01; *P < 0.05; n.s., not significant.
Y. pseudotuberculosis Mutant Lacking hpaR Is Defective in Virulence in Mice
Biofilm formation is helpful for the survival and colonization of Y. pseudotuberculosis in hosts. In addition, oxidative burst is an important microbial-killing mechanism in phagocytes. The findings that HpaR affects the antioxidant activity, biofilm formation and the ability of adhesion to eukaryotic cells point to its role in the virulence of Y. pseudotuberculosis. To examine this, C57BL/6 mice were inoculated orogastrically with relevant bacterial strains. Wild-type bacteria caused more than 90% lethality within 3 weeks of inoculation. By contrast, consistent with the hypothesis, mice infected with mutants lacking hpaR or T6SS4 had better survival rates than the wild-type (Figure 6). The data indicate that hpaR affects the virulence of Y. pseudotuberculosis in the infection of mammalian hosts.
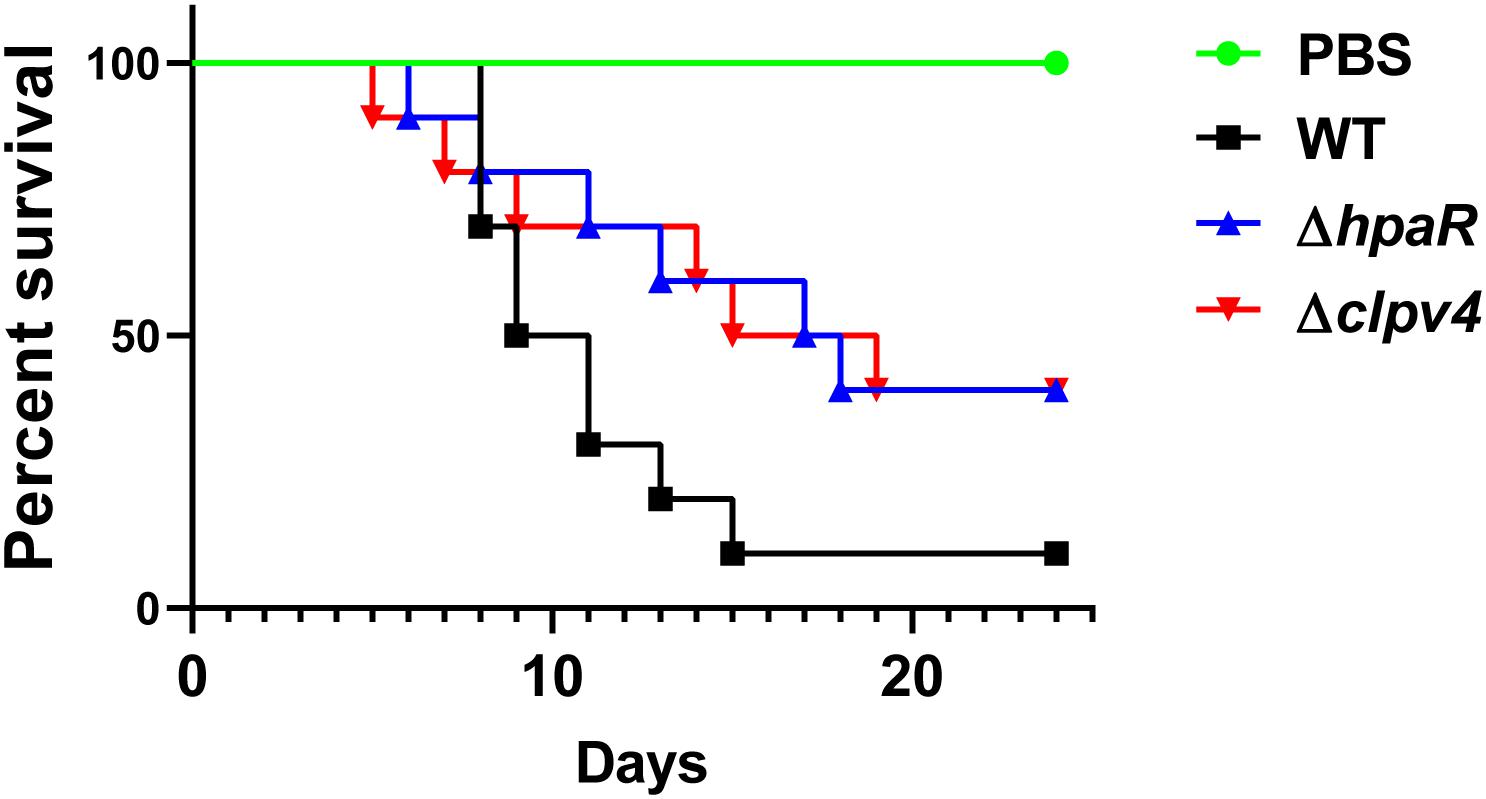
Figure 6. Yptb mutants lacking hpaR or T6SS4 are defective in virulence against mice. Bacterial strains grown in YLB were washed twice in sterilized PBS and used for orogastric infection of 6–8 weeks old female C57BL/6 mice using a ball-tipped feeding needle. For survival assays 3 × 109 bacteria of each strain were applied to different groups of mice (n = 10/strain), and the survival rate of the mice was determined by monitoring the survival daily for 24 days.
Discussion
MarR family transcriptional regulators are widely distributed in bacteria and archaea (Perez-Rueda and Collado-Vides, 2001; Perez-Rueda et al., 2004). They play crucial roles in the bacterial lifecycle as they control various genes involved in antibiotic resistance, aromatic compound catabolism, oxidative stress responses, and produce of virulence factors (Alekshun and Levy, 1999; Fiorentino et al., 2007; Deochand and Grove, 2017). Some of these regulators are involved in the degradation of specific aromatic compounds (Diaz and Prieto, 2000). For example, BadR in Rhodopseudomonas palustris regulates anaerobic benzoate degradation in cooperation with AadR (Egland and Harwood, 1999). In Comamonas testosteroni BR60, CbaR controls the cbaABC operon for converting 3-chlorobenzoate to protocatechuate (PCA) and 5-Cl-PCA (Providenti and Wyndham, 2001). HpaR was first identified in E. coli W (Roper et al., 1993). As a repressor, HpaR regulates the hpa-meta operon in 4-HPA catabolism (Prieto et al., 1996; Galan et al., 2003). However, whether HpaR has other functions remains unclear. Here, we report that in Y. pseudotuberculosis, HpaR acts as a dual-functional regulator. It not only negatively regulates the hpa-meta cluster but also positively controls T6SS4 expression to resist oxidative stress, which maintains cellular ROS levels.
The hpa-meta cluster in E. coli W is composed of 11 genes within two operons, hpaBC and hpaGEDFHI, which are controlled by HpaA and HpaR, respectively (Galan et al., 2001). Based on sequence alignment with E. coli W, we identified the hpa-meta cluster in Y. pseudotuberculosis. An HpaA homolog was not identified in Y. pseudotuberculosis. The hpa-meta cluster of one operon consisted of 11 genes, hpaG1G2EDFHIXBC, and one regulator, HpaR (Figure 1A). Using a β-galactosidase activity assay and qRT-PCR, we revealed that HpaR represses expression of the hpaG1G2EDFHIXBC operon in Y. pseudotuberculosis, similar to E. coli W. HpaR binding sites 1 and 2, both composed of a 15 bp region, were identified on the hpaG1 promoter containing a palindromic sequence of 4 bp (TTAA-XXXX-TTAA) on each side separated by 4 bp similar to E. coli W (Figure 1E). Site 1 was located -189 bp relative to the transcription start site + 1 of the hpaG1 promote. Site 2 was centered at position -8 of the hpaG1 promoter, overlapping the -10 element, which implies that HpaR inhibits the binding of RNA polymerase on the promoter to repress transcription of the hpa-meta cluster.
So far HpaR was specifically recognized as a repressor of the 4-HPA catabolic pathway (Galan et al., 2003; Arcos et al., 2010; Mendez et al., 2011). Unexpectedly, in the RNA-seq based transcriptomics analysis we found that in addition to the hpa-meta cluster genes, genes of the T6SS4 cluster were also found to be regulated by HpaR. Based on the β-galactosidase activity assay and qRT-PCR results, we confirmed that HpaR acts as an activator rather than a repressor to regulate T6SS4 (Figure 3A). We also identified the HpaR binding site on the T6SS4 promoter located -241 bp relative to the transcription start site + 1 of the T6SS4 promote, containing an imperfect invert sequence TGGXT-XXXX-AXCCA on each side separated by 4 bp (Figure 3E). The HpaR binding site of hpaG1 and T6SS4 promoter both contain a 4–5 bp palindromic sequence on each side separated by 4 bp but the site on T6SS4 promoter is imperfect. The binding site of T6SS4 promoter is far from the transcription start site. Besides, the GC content of binding site on T6SS4 promoter is higher than that on hpaG1 promoter. We presume that HpaR may bind to the stable site upstream of the -35 and -10 elements recruiting RNA polymerase to activate the transcription of T6SS4.
T6SS has been recognized as an anti-bacterial weapon to attack target cells in a contact-dependent manner (Durand et al., 2014; Ho et al., 2014; Russell et al., 2014). However, our previous report showed that the T6SS4 in Y. pseudotuberculosis is related to resistance to multiple environmental stresses, including high osmolarity, low pH, and oxidative stress (Song et al., 2015; Wang et al., 2015). We therefore speculated that HpaR might protect against oxidative stress via T6SS4. And we confirmed this by observing the survival rate and determining the intracellular ROS levels of wild-type and mutant strains under oxidative stress (Figures 4A–D). Y. pseudotuberculosis could import Zn2+ via secretion of YezP through T6SS4 to maintain intracellular ROS levels (Wang et al., 2015). Our results showed that the levels of YezP secreted by T6SS4 and the intracellular Zn2+ levels of ΔhpaR were also decreased under H2O2 stress (Figures 4E,F), indicating that HpaR regulates the expression of T6SS4 to resist oxidative stress.
Biofilm formation was also affected by HpaR (Figures 5A,B). However, no other biofilm-related gene was found to be differentially expressed in the RNA-seq results. Biofilm formation is complex—it may be indirectly regulated by other mechanisms that will require further investigation. Biofilm formation is beneficial to bacterial survival in a harmful environment and is associated with adhesion to eukaryotic cells (Flemming and Wingender, 2010; Hall and Mah, 2017). Our results also showed that HpaR was involved in antibiotic resistance and adhered to HeLa cells (Figures 5C,D). Y. pseudotuberculosis is an enteric pathogen of animals and humans (Smego et al., 1999). In host cells, the oxidative burst is an important method to kill bacteria and biofilm formation is essential for bacterial infection (Flemming and Wingender, 2010; Slauch, 2011). Besides, the adhesion to host cell is also crucial for bacterial colonization. It has been shown that T6SS4 is required for Y. pseudotuberculosis virulence in a mouse model (Wang et al., 2015). This work revealed that HpaR also plays important roles in virulence and biofilm formation, potentially also via regulation of T6SS4.
In summary, we revealed that HpaR is a dual-functional regulator in Y. pseudotuberculosis. HpaR not only represses expression of genes of the 4-HPA catabolic pathway but also activates the transcription of T6SS4 to acquire Zn2+ to maintain ROS levels in response to oxidative stress. Biofilm formation, antibiotic resistance, eukaryotic cell adhesion, and bacterial virulence were also affected by hpaR (Figure 7). This study has greatly expanded the function of HpaR beyond the regulation of aromatic compound catabolism and revealed a new pathway to regulate T6SS4 in response to environmental stress.
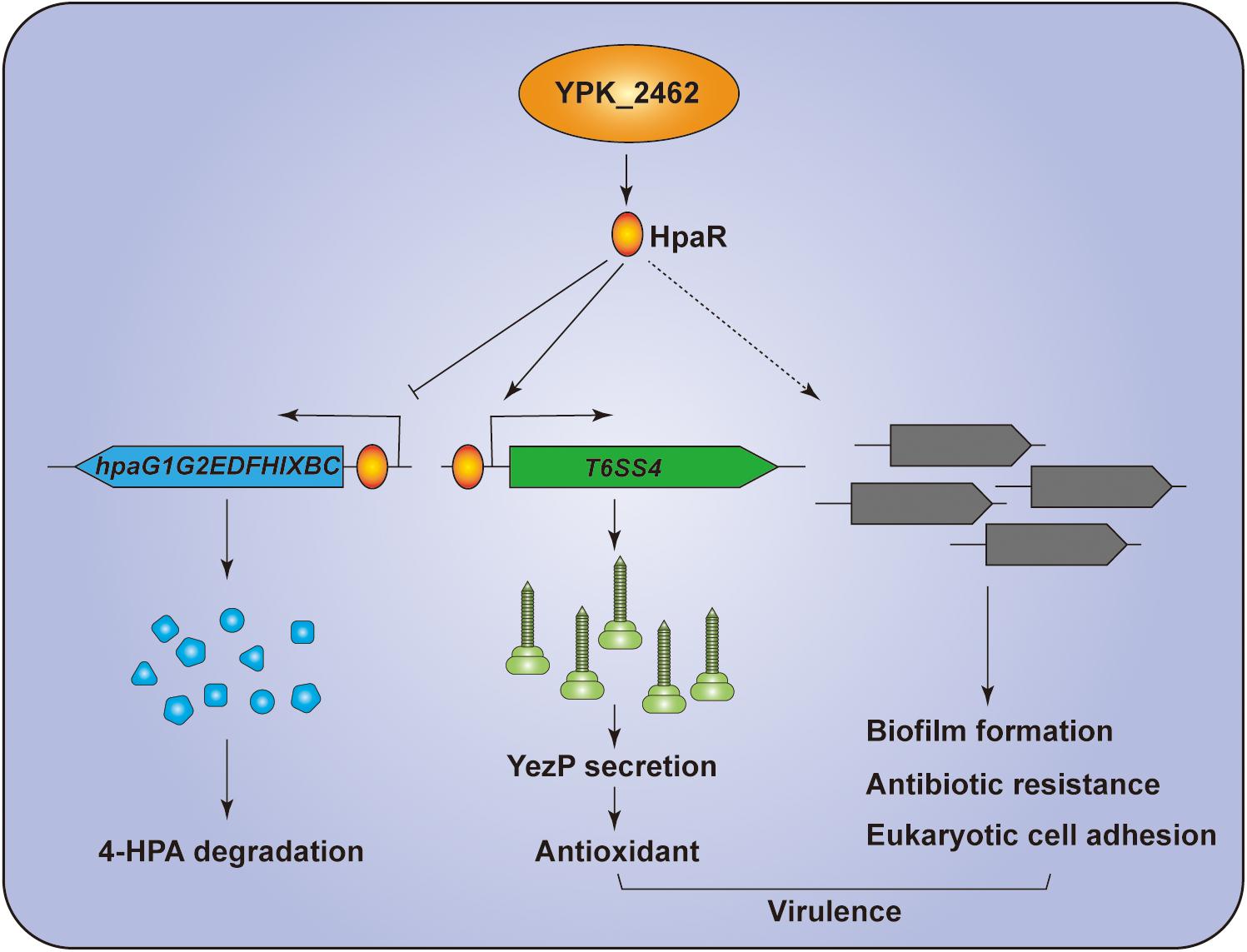
Figure 7. Hypothetical model for the function of HpaR in Y. pseudotuberculosis. HpaR represses the 4-HPA catabolic pathway but activates the transcription of T6SS4 to maintain ROS levels in response to oxidative stress. HpaR also affects the biofilm formation, eukaryotic cell adhension, antibiotic resistance, and virulence of Y. pseudotuberculosis.
Data Availability Statement
The datasets generated for this study can be found in the RNA-seq data was deposited in SRA (SRA accession: PRJNA612340).
Ethics Statement
All mice were maintained and handled in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People’s Republic of China. The protocol was approved by the Animal Welfare and Research Ethics Committee of Northwest A&F University (protocol number: NWAFUSM2018001).
Author Contributions
ZW, TW, YY, and XS conceived the study, designed the experiments, analyzed the data, and wrote the manuscript. ZW, TW, RC, ZZ, KC, ML, YH, HG, LX, and YW performed the experimental work.
Funding
This work was supported by the grant of National Key R&D Program of China (2018YFA0901200 to XS), the National Natural Science Foundation of China (31670053 and 31725003 to XS and, 31970114 and 31671292 to YW), the Open Project Program of the State Key Laboratory of Pathogen and Biosecurity (SKLPBS1825 to XS). LX was supported by China Postdoctoral Science Foundation (2018M631201) and Shaanxi Postdoctoral Science Foundation (2018BSHTDZZ20).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Teaching and Research Core Facility at College of Life Sciences, NWAFU for their support in this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00705/full#supplementary-material
FIGURE S1 | Protein sequence alignment of HpaR homologs by CLUSTAL W. The sequences used in alignment have been deposited in the GenBank database [Accession No. EcC Escherichia coli C (S56952.1), EcW Escherichia coli W (Z37980.2), Yptb Yersinia pseudotuberculosis YPIII (ACA68739.1), PpU Pseudomonas putida U (FJ904934.1), BxLB400 Burkholderia xenovorans LB400 (ABE33958.1)]. The result was exported by ESPript (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
FIGURE S2 | Growth curves of the wild-type (WT), ΔhpaR mutant, and the complemented strain ΔhpaR (hpaR) under normal condition. The growth of the indicated strains in YLB medium was monitored by measuring OD600 at indicated time points under 26°C.
TABLE S1 | The strains and plasmids used in this study.
TABLE S2 | Primers used in this study.
TABLE S3 | Genes differentially transcribed in ΔhpaR mutant compared to the Y. pseudotuberculosis wild-type detected by RNA-seq.
Footnotes
References
Alekshun, M. N., and Levy, S. B. (1999). The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7, 410–413. doi: 10.1016/s0966-842x(99)01589-1589
Alekshun, M. N., Levy, S. B., Mealy, T. R., Seaton, B. A., and Head, J. F. (2001). The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat. Struct. Biol. 8, 710–714. doi: 10.1038/90429
Arcos, M., Olivera, E. R., Arias, S., Naharro, G., and Luengo, J. M. (2010). The 3,4-dihydroxyphenylacetic acid catabolon, a catabolic unit for degradation of biogenic amines tyramine and dopamine in Pseudomonas putida U. Environ. Microbiol. 12, 1684–1704. doi: 10.1111/j.1462-2920.2010.02233.x
Barbour, M. G., and Bayly, R. C. (1981). Control of meta-cleavage degradation of 4-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 147, 844–850. doi: 10.1128/jb.147.3.844-850.1981
Basler, M. (2015). Type VI secretion system: secretion by a contractile nanomachine. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 1679. doi: 10.1098/rstb.2015.0021
Bingle, L. E., Bailey, C. M., and Pallen, M. J. (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol. 11, 3–8. doi: 10.1016/j.mib.2008.01.006
Brubaker, R. R. (1991). Factors promoting acute and chronic diseases caused by yersiniae. Clin. Microbiol. Rev. 4, 309–324. doi: 10.1128/cmr.4.3.309
Cooper, R. A., and Skinner, M. A. (1980). Catabolism of 3- and 4-hydroxyphenylacetate by the 3,4-dihydroxyphenylacetate pathway in Escherichia coli. J. Bacteriol. 143, 302–306. doi: 10.1128/jb.143.1.302-306.1980
D’Autreaux, B., and Toledano, M. B. (2007). ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824. doi: 10.1038/nrm2256
Deochand, D. K., and Grove, A. (2017). MarR family transcription factors: dynamic variations on a common scaffold. Crit. Rev. Biochem. Mol. Biol 52, 595–613. doi: 10.1080/10409238.2017.1344612
Diaz, E., and Prieto, M. A. (2000). Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 11, 467–475. doi: 10.1016/s0958-1669(00)00126-129
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
Egland, P. G., and Harwood, C. S. (1999). BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodo Pseudomonas palustris in concert with AadR, an Fnr family member. J. Bacteriol. 181, 2102–2109. doi: 10.1128/jb.181.7.2102-2109.1999
Fallman, M., and Gustavsson, A. (2005). Cellular mechanisms of bacterial internalization counteracted by Yersinia. Int. Rev. Cytol. 246, 135–188. doi: 10.1016/S0074-7696(05)46004-46000
Fiorentino, G., Ronca, R., Cannio, R., Rossi, M., and Bartolucci, S. (2007). MarR-like transcriptional regulator involved in detoxification of aromatic compounds in Sulfolobus solfataricus. J. Bacteriol. 189, 7351–7360. doi: 10.1128/JB.00885-887
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Galan, B., Diaz, E., Prieto, M. A., and Garcia, J. L. (2000). Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new Flavin:NAD(P)H reductase subfamily. J. Bacteriol. 182, 627–636. doi: 10.1128/jb.182.3.627-636.2000
Galan, B., Kolb, A., Garcia, J. L., and Prieto, M. A. (2001). Superimposed levels of regulation of the 4-hydroxyphenylacetate catabolic pathway in Escherichia coli. J. Biol. Chem. 276, 37060–37068. doi: 10.1074/jbc.M103033200
Galan, B., Kolb, A., Sanz, J. M., Garcia, J. L., and Prieto, M. A. (2003). Molecular determinants of the hpa regulatory system of Escherichia coli: the HpaR repressor. Nucleic Acids Res. 31, 6598–6609. doi: 10.1093/nar/gkg851
Green, E. R., Clark, S., Crimmins, G. T., Mack, M., Kumamoto, C. A., and Mecsas, J. (2016). Fis is essential for yersinia pseudotuberculosis virulence and protects against reactive oxygen species produced by phagocytic cells during infection. PLoS Pathog. 12:e1005898. doi: 10.1371/journal.ppat.1005898
Grove, A. (2013). MarR family transcription factors. Curr. Biol. 23, R142–R143. doi: 10.1016/j.cub.2013.01.013
Guan, J., Xiao, X., Xu, S., Gao, F., Wang, J., Wang, T., et al. (2015). Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J. Microbiol. 53, 633–642. doi: 10.1007/s12275-015-0099-96
Hall, C. W., and Mah, T. F. (2017). Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 41, 276–301. doi: 10.1093/femsre/fux010
Ho, B. T., Dong, T. G., and Mekalanos, J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microb. 15, 9–21. doi: 10.1016/j.chom.2013.11.008
Huang, Y., Zhao, K. X., Shen, X. H., Jiang, C. Y., and Liu, S. J. (2008). Genetic and biochemical characterization of a 4-hydroxybenzoate hydroxylase from Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 78, 75–83. doi: 10.1007/s00253-007-1286-1280
Jani, A. J., and Cotter, P. A. (2010). Type VI secretion: not just for pathogenesis anymore. Cell Host Microb. 8, 2–6. doi: 10.1016/j.chom.2010.06.012
Lin, J., Zhang, W., Cheng, J., Yang, X., Zhu, K., Wang, Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. doi: 10.1038/ncomms14888
Ma, J., Bao, Y., Sun, M., Dong, W., Pan, Z., Zhang, W., et al. (2014). Two functional type VI secretion systems in avian pathogenic Escherichia coli are involved in different pathogenic pathways. Infect. Immun. 82, 3867–3879. doi: 10.1128/IAI.01769-1714
Mendez, V., Agullo, L., Gonzalez, M., and Seeger, M. (2011). The homogentisate and homoprotocatechuate central pathways are involved in 3- and 4-hydroxyphenylacetate degradation by Burkholderia xenovorans LB400. PLoS One 6:e17583. doi: 10.1371/journal.pone.0017583
Miller, J. H. (1992). A Short Course in Bacterial Genetics - A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Navarro-Garcia, F., Ruiz-Perez, F., Cataldi, A., and Larzabal, M. (2019). Type VI secretion system in pathogenic Escherichia coli: structure, role in virulence, and acquisition. Front. Microbiol. 10:1965. doi: 10.3389/fmicb.2019.01965
Perez-Rueda, E., and Collado-Vides, J. (2001). Common history at the origin of the position-function correlation in transcriptional regulators in archaea and bacteria. J. Mol. Evol. 53, 172–179. doi: 10.1007/s002390010207
Perez-Rueda, E., Collado-Vides, J., and Segovia, L. (2004). Phylogenetic distribution of DNA-binding transcription factors in bacteria and archaea. Comput. Biol. Chem. 28, 341–350. doi: 10.1016/j.compbiolchem.2004.09.004
Prieto, M. A., Diaz, E., and Garcia, J. L. (1996). Molecular characterization of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli W: engineering a mobile aromatic degradative cluster. J. Bacteriol. 178, 111–120. doi: 10.1128/jb.178.1.111-120.1996
Prieto, M. A., and Garcia, J. L. (1994). Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. A two-protein component enzyme. J. Biol. Chem. 269, 22823–22829.
Prieto, M. A., and Garcia, J. L. (1997). Identification of a novel positive regulator of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli. Biochem. Biophys. Res. Commun. 232, 759–765. doi: 10.1006/bbrc.1997.6368
Providenti, M. A., and Wyndham, R. C. (2001). Identification and functional characterization of CbaR, a MarR-like modulator of the cbaABC-encoded chlorobenzoate catabolism pathway. Appl. Environ. Microbiol. 67, 3530–3541. doi: 10.1128/AEM.67.8.3530-3541.2001
Roper, D. I., Fawcett, T., and Cooper, R. A. (1993). The Escherichia coli C homoprotocatechuate degradative operon: hpc gene order, direction of transcription and control of expression. Mol. Gen. Genet. 237, 241–250. doi: 10.1007/bf00282806
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Shen, X., Banga, S., Liu, Y., Xu, L., Gao, P., Shamovsky, I., et al. (2009). Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 11, 911–926. doi: 10.1111/j.1462-5822.2009.01301.x
Shen, X. H., Jiang, C. Y., Huang, Y., Liu, Z. P., and Liu, S. J. (2005). Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 71, 3442–3452. doi: 10.1128/AEM.71.7.3442-3452.2005
Si, M., Wang, T., Pan, J., Lin, J., Chen, C., Wei, Y., et al. (2017a). Graded response of the multifunctional 2-cysteine peroxiredoxin, CgPrx, to increasing levels of hydrogen peroxide in Corynebacterium glutamicum. Antioxid. Redox. Signal. 26, 1–14. doi: 10.1089/ars.2016.6650
Si, M., Wang, Y., Zhang, B., Zhao, C., Kang, Y., Bai, H., et al. (2017b). The type VI secretion system engages a redox-regulated dual-functional heme transporter for zinc acquisition. Cell Rep. 20, 949–959. doi: 10.1016/j.celrep.2017.06.081
Si, M., Zhao, C., Burkinshaw, B., Zhang, B., Wei, D., Wang, Y., et al. (2017c). Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. U.S.A. 114, E2233–E2242. doi: 10.1073/pnas.1614902114
Si, M., Zhang, L., Chaudhry, M. T., Ding, W., Xu, Y., Chen, C., et al. (2015). Corynebacterium glutamicum methionine sulfoxide reductase A uses both mycoredoxin and thioredoxin for regeneration and oxidative stress resistance. Appl. Environ. Microbiol. 81, 2781–2796. doi: 10.1128/AEM.04221-4214
Slauch, J. M. (2011). How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol. Microbiol. 80, 580–583. doi: 10.1111/j.1365-2958.2011.07612.x
Smego, R. A., Frean, J., and Koornhof, H. J. (1999). Yersiniosis I: microbiological and clinicoepidemiological aspects of plague and non-plague Yersinia infections. Eur. J. Clin. Microbiol. Infect. Dis. 18, 1–15. doi: 10.1007/s100960050219
Song, Y., Xiao, X., Li, C., Wang, T., Zhao, R., Zhang, W., et al. (2015). The dual transcriptional regulator RovM regulates the expression of AR3- and T6SS4-dependent acid survival systems in response to nutritional status in Yersinia pseudotuberculosis. Environ. Microbiol. 17, 4631–4645. doi: 10.1111/1462-2920.12996
Staerck, C., Gastebois, A., Vandeputte, P., Calenda, A., Larcher, G., Gillmann, L., et al. (2017). Microbial antioxidant defense enzymes. Microb. Pathog. 110, 56–65. doi: 10.1016/j.micpath.2017.06.015
Tan, Y., Liu, W., Zhang, Q., Cao, S., Zhao, H., Wang, T., et al. (2017). Yersinia pestis YopK inhibits bacterial adhesion to host cells by binding to the extracellular matrix adaptor protein matrilin-2. Infect. Immun. 85:e01069-16. doi: 10.1128/IAI.01069-1016
Wan, B., Zhang, Q., Ni, J., Li, S., Wen, D., Li, J., et al. (2017). Type VI secretion system contributes to enterohemorrhagic Escherichia coli virulence by secreting catalase against host reactive oxygen species (ROS). PLoS Pathog 13:e1006246. doi: 10.1371/journal.ppat.1006246
Wang, T., Chen, K., Gao, F., Kang, Y., Chaudhry, M. T., Wang, Z., et al. (2017). ZntR positively regulates T6SS4 expression in Yersinia pseudotuberculosis. J. Microbiol. 55, 448–456. doi: 10.1007/s12275-017-6540-6542
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Wang, Y., Cen, X. F., Zhao, G. P., and Wang, J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. doi: 10.1128/JB.00989-912
Weber, B., Hasic, M., Chen, C., Wai, S. N., and Milton, D. L. (2009). Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 11, 3018–3028. doi: 10.1111/j.1462-2920.2009.02005.x
Wei, K., Tang, D. J., He, Y. Q., Feng, J. X., Jiang, B. L., Lu, G. T., et al. (2007). hpaR, a putative marR family transcriptional regulator, is positively controlled by HrpG and HrpX and involved in the pathogenesis, hypersensitive response, and extracellular protease production of Xanthomonas campestris pathovar campestris. J. Bacteriol. 189, 2055–2062. doi: 10.1128/JB.01331-1336
Xu, L., Shen, X., Bryan, A., Banga, S., Swanson, M. S., and Luo, Z. Q. (2010). Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 6:e1000822. doi: 10.1371/journal.ppat.1000822
Xu, S., Peng, Z., Cui, B., Wang, T., Song, Y., Zhang, L., et al. (2014). FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ. Microbiol. 16, 1090–1104. doi: 10.1111/1462-2920.12222
Yang, X., Pan, J., Wang, Y., and Shen, X. (2018). Type VI secretion systems present new insights on pathogenic yersinia. Front. Cell Infect. Microbiol. 8:260. doi: 10.3389/fcimb.2018.00260
Zhang, W., Wang, Y., Song, Y., Wang, T., Xu, S., Peng, Z., et al. (2013). A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ. Microbiol. 15, 557–569. doi: 10.1111/1462-2920.12005
Keywords: HpaR, type VI secretion system, Yersinia pseudotuberculosis, oxidative stress, biofilm, aromatic compounds degradation
Citation: Wang Z, Wang T, Cui R, Zhang Z, Chen K, Li M, Hua Y, Gu H, Xu L, Wang Y, Yang Y and Shen X (2020) HpaR, the Repressor of Aromatic Compound Metabolism, Positively Regulates the Expression of T6SS4 to Resist Oxidative Stress in Yersinia pseudotuberculosis. Front. Microbiol. 11:705. doi: 10.3389/fmicb.2020.00705
Received: 23 December 2019; Accepted: 26 March 2020;
Published: 17 April 2020.
Edited by:
Eric Cascales, Aix-Marseille Université, FranceReviewed by:
Gabriella Fiorentino, University of Naples Federico II, ItalySébastien Bontemps-Gallo, Institut Pasteur de Lille, France
Copyright © 2020 Wang, Wang, Cui, Zhang, Chen, Li, Hua, Gu, Xu, Wang, Yang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yantao Yang, yyt811113@nwsuaf.edu.cn; Xihui Shen, xihuishen@nwsuaf.edu.cn
†These authors have contributed equally to this work
 Zhuo Wang
Zhuo Wang Tietao Wang
Tietao Wang Rui Cui
Rui Cui Huawei Gu
Huawei Gu Lei Xu
Lei Xu Yao Wang
Yao Wang Yantao Yang
Yantao Yang Xihui Shen
Xihui Shen