- Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), Gif-sur-Yvette, France
Cadmium, although not redox active is highly toxic. Yet, the underlying mechanisms driving toxicity are still to be characterized. In this study, we took advantage of the purple bacterium Rubrivivax gelatinosus strain with defective Cd2 +-efflux system to identify targets of this metal. Exposure of the ΔcadA strain to Cd2 + causes a decrease in the photosystem amount and in the activity of respiratory complexes. As in case of Cu+ toxicity, the data indicated that Cd2 + targets the porphyrin biosynthesis pathway at the level of HemN, a S-adenosylmethionine and CxxxCxxC coordinated [4Fe-4S] containing enzyme. Cd2 + exposure therefore results in a deficiency in heme and chlorophyll dependent proteins and metabolic pathways. Given the importance of porphyrin biosynthesis, HemN represents a key metal target to account for toxicity. In the environment, microorganisms are exposed to mixture of metals. Nevertheless, the biological effects of such mixtures, and the toxicity mechanisms remain poorly addressed. To highlight a potential cross-talk between Cd2 + and Cu+ -efflux systems, we show (i) that Cd2 + induces the expression of the Cd2 +-efflux pump CadA and the Cu+ detoxification system CopA and CopI; and (ii) that Cu+ ions improve tolerance towards Cd2 +, demonstrating thus that metal mixtures could also represent a selective advantage in the environment.
Introduction
Metal accumulation, through environmental contamination by anthropogenic release, results in toxicity leading to impaired growth of microorganisms (Nunes et al., 2016). Indeed, excess metal can affect and disrupt different cellular metabolic pathways. Because of their thiophilicity, metals can compete or displace each other in the binding sites of metalloproteins. For instance, copper (Cu+), zinc (Zn2 +) or cadmium (Cd2 +) represent a standing threat for Fe-S proteins in bacteria. They can either directly damage exposed [4Fe-4S] clusters (Macomber and Imlay, 2009) or inhibit components of the iron-sulfur (Fe-S) biogenesis machinery (Tan et al., 2017; Roy et al., 2018; Li et al., 2019). In Escherichia coli, Cu+, Zn2 +or Cd2 + exert their toxic effects by disrupting the solvent exposed [4Fe-4S] clusters of dehydratases (Macomber et al., 2007; Macomber and Imlay, 2009; Xu and Imlay, 2012). These cations can also compete and prevent iron or iron-sulfur cluster binding in IscA, IscU or ferredoxin in the iron-sulfur cluster biogenesis system (Tan et al., 2017; Roy et al., 2018; Li et al., 2019).
In the human pathogen Neisseria gonorrhoeae, and the photosynthetic bacterium Rubrivivax (R.) gelatinosus, accumulation of Cu+ in the cytoplasm affect heme biosynthesis (Azzouzi et al., 2013; Djoko and McEwan, 2013). A hypothesis put forward is that Cu+ excess targets HemN, the anaerobic coproporphyrinogen III Oxidase. This enzyme has a conserved cysteine-rich motif, CxxxCxxC, which coordinates a solvent [4Fe–4S] cluster with an iron that anchors a S-adenosylmethionine (SAM) moiety (Layer et al., 2003). It was suggested that Cu+ targets this solvent exposed SAM-[4Fe–4S] cluster, thereby affecting the enzyme activity (Azzouzi et al., 2013; Djoko and McEwan, 2013).
Given the importance of Fe-S clusters and heme metabolism in bacteria but also in eukaryotes, the increase of metal concentration in the environment is therefore challenging to all living organisms. Homeostasis maintenance and detoxification of metal excess are therefore crucial to enable bacterial survival in contaminated environments or in macrophages for pathogenic bacteria (Arguello et al., 2007; von Rozycki and Nies, 2009; Capdevila et al., 2017; Chandrangsu et al., 2017).
The intracellular concentration of metals is tightly regulated at the uptake, storage and/or excretion level (Arguello et al., 2007; von Rozycki and Nies, 2009; Capdevila et al., 2017; Chandrangsu et al., 2017). For metals such as copper, cadmium or silver, efflux pumps are effective detoxification systems that allow bacteria to deal with excess metals in their immediate environment (Arguello et al., 2007; von Rozycki and Nies, 2009; Capdevila et al., 2017; Chandrangsu et al., 2017). Indeed, most free-living bacteria possess efflux system to remove metal excess. Members of P1B-type ATPases family of heavy metal transporters are universally present in bacteria. They are part of the large superfamily of ATP-driven pumps involved in metal transport across bacterial inner membrane (Arguello et al., 2007). These transporters extrude excess or toxic metal ions such as Cu+, Zn2 +, Cd2 +, or Ag+ from the cytoplasm to the periplasm in which metal is handled by other detoxifying proteins (Arguello et al., 2007). Cu+, Zn2 +, or Cd2 + efflux ATPase mutants accumulate these cations in their cytoplasm (Legatzki et al., 2003; Gonzalez-Guerrero et al., 2010; Djoko and McEwan, 2013; Lu et al., 2016) and very often display growth inhibition phenotype.
A strain of R. gelatinosus (TN414) efficient at removing Cd2 + and Zn2 + from cadmium and zinc contaminated soil has been isolated from paddy fields (Sakpirom et al., 2017). R. gelatinosus S1 can also grow in medium containing up to 3 mM of cadmium. The mechanisms governing Cd2 + tolerance are however, not yet known. In this study, we first identified the Cd2 +-efflux system to ask which pathways Cd2 +, a divalent non-redox active cation, impacts in the absence of an efficient efflux system. Interestingly, it is also shown that Cd2 + induces Cu+ homeostasis system and that Cu+ alleviates Cd2 + toxicity in R. gelatinosus, a phenomenon that could represent a selective advantage for bacteria growing in multiple metal-polluted environment.
Materials and Methods
Bacterial Strains and Growth
Escherichia coli was grown at 37°C in LB medium. R. gelatinosus (S1) was grown at 30°C, in the dark aerobically (high oxygenation: 250 ml flasks containing 20 ml medium) and microaerobically (low oxygenation: 50 ml flasks filled with 50 ml medium) or in light (photosynthetically in filled tubes with residual oxygen in the medium) in malate growth medium. Antibiotics were used at the following concentrations: kanamycin (Km), ampicillin and trimethoprim (Tp) at 50 μg/ml, tetracycline at 2 μg/ml. Bacterial strains and plasmids are listed in Supplementary Table SI. Growth inhibition curves were monitored at OD680nm with measurements taken every 15 min for 24 h using a Tecan Infinite M200 luminometer (Tecan, Mannerdorf, Switzerland) for aerobic condition. For photosynthesis conditions, strains were grown as described above and OD was measured after 24 h using the Tecan luminometer. Inhibition growth experiments were done in triplicates starting from a three diluted culture (OD680 at 0.02).
Gene Cloning and Plasmid Constructions for Allele Replacement
Standard methods were performed according to Sambrook et al. (1989) unless indicated otherwise. To inactivate cadA, a 1.8 kb fragment was amplified using the primers cadAF1 and cadAR1 (Supplementary Table SII) and cloned into the PCR cloning vector pGEM-T to give pGcadA. cadA gene was inactivated by deletion of 0.8 kb fragment and the insertion of the Tp cassette at the NarI sites within the cadA coding sequence. The resulting recombinant plasmid was designated pGcadA:Tp. Furthermore, a plasmid (pB106) containing the full cadA and cadR genes was also isolated from the DNA library. A 6.6 kb fragment containing cadA was subcloned from pB106 in pBBR1MCS-2 at the KpnI-SacI sites. The resulting plasmid was designated pBKCadA. cadR was PCR cloned using primers cadRF1 and cadRR1 in pGemT plasmid. To inactivate cadR, the 1.2 kb Km cassette was inserted in the StuI site within cadR in the pGcadR plasmid. To generate HemN-H6 fusion, hemN was amplified from wild-type genomic DNA using primers hemN-NdeI and hemN-XhoI and cloned in the corresponding sites within pET-28b.
Gene Transfer and Strain Selection
Transformation of R. gelatinosus cells was performed by electroporation (Ouchane et al., 1996). The plasmid pGcadA:Tp was used to transform the wild-type, copA–, ΔcopI, or copA-H6 strains. Transformants were selected on malate plates supplemented with appropriate antibiotics under aerobic conditions. Following transformant selection, template genomic DNA was prepared from the ampicillin sensitive transformants and confirmation of the antibiotic resistance marker’s presence at the desired locus was performed by PCR. Finally, selected colonies were spotted on metal containing plates to confirm their metal sensitive phenotype. Plasmid phemN-H6 was used to transform the wild-type strain to generate the HemN-H6 strain. Clones were selected on kanamycin plates.
Membrane and Soluble Protein Preparation
The membranes or solubles fractions were prepared by cell disruption with a French press in 0.1 M sodium phosphate buffer (pH 7.4) containing 1 mM phenylmethylsulfonyl fluoride, followed by differential centrifugation. Samples were subjected to a low speed centrifugation step (25, 000 g, 30 min, 4°C) and supernatants were subjected to ultracentrifugation (200, 000 g, 1h30, 4°C) to purify the soluble proteins and collect the membrane fraction in the pellets.
The membranes were then resuspended in the same buffer. Protein concentration was estimated using the bicinchoninic acid assay (Sigma) with bovine serum albumin as a standard. Periplasmic fractions were purified as previously described by Durand et al. (2015).
Spectrophotometric Measurements
Absorption spectroscopy was performed with a Cary 500 spectrophotometer. For difference spectra (reduced minus oxidized), total pigments were extracted from cell pellets or membranes with acetone-methanol (7/2 [vol/vol]). For each sample, the spectrum was collected on oxidized sample upon addition of 50 μM K6Fe(CN)3. The sample was reduced by addition of dithionite (few crystals), and the spectrum was collected to generate the reduced minus oxidized spectrum.
Blue-Native Gel Electrophoresis
To assay cbb3 and succinate dehydrogenase activities, wild-type and ΔcadA strains were grown microaerobically. Membranes were prepared as previously described (Khalfaoui Hassani et al., 2010). Blue-native polyacrylamide gel (BN-PAGE) electrophoresis and in gel-Cox activity assays (DAB staining) were performed as described in (Khalfaoui Hassani et al., 2010) and succinate dehydrogenase activity was assayed using succinate and NBT (Nitroblue tetrazolium) as described for the succinate-NBT reductase assay in reference Wittig et al. (2007).
Western Blot and Immunodetection
Equal amount of proteins or disrupted cells (OD680nm = 1) were separated by SDS–PAGE (15% polyacrylamide) and further transferred to a Hybond ECL PVDF membrane, (GE Healthcare). Coomassie blue of the SDS gels are shown in Supplementary Material. Membranes were then probed with the HisProbe-HRP (from Pierce) according to the manufacturer instruction and positive bands were detected using a chemiluminescent HRP substrate according to the method of Haan and Behrmann (2007). Image capture was performed with a ChemiDoc camera system (Biorad).
Results
The P-type ATPase CadA Is the Primary Cadmium Tolerance Element in R. gelatinosus
Cadmium tolerance in bacteria usually involves the metal efflux pump P1B-type ATPase CadA (ZntA) protein that translocates cadmium from the cytoplasm to the periplasm (Rensing et al., 1998). A gene (hereafter referred to as cadA) encoding a putative ATPase involved in Zn2 + or Cd2 + efflux was identified in R. gelatinosus genome. Sequence analyses showed high similarities with putative heavy metal translocating P1B-type ATPases from β-proteobacteria (Supplementary Figure S1). In addition to conserved structural features and motifs in the ATPase sequence, an N-terminal 30 amino acids stretch consisting of 14 histidines and 7 acidic residues, flanked by two CxxxC motifs were found in R. gelatinosus CadA (Supplementary Figure S1). This N-terminal cytoplasmic stretch could play a role in metal binding and transport regulation as reported for other metal transporting ATPases (Mana-Capelli et al., 2003; Baekgaard et al., 2010). A gene (referred to as cadR) encoding a MerR regulator protein (CadR) was found upstream cadA. To ascertain the role of CadA in Cd2 + tolerance, the ΔcadA deletion strain was generated and its ability to grow on solid medium in presence of CdCl2 was tested in comparison with wild-type strain under aerobic respiration and photosynthesis conditions (Figure 1). When challenged with Cd2 +, the growth of ΔcadA strain is inhibited with 100 or 250 μM CdCl2 under aerobic or photosynthesis conditions, respectively. In contrast, the wild-type strain was still able to grow in presence of 1.5 or 3 mM CdCl2 (Figure 1B). The tolerance towards Cd2 + was restored by the expression of the wild-type copy of cadA gene in trans in ΔcadA strain (Supplementary Figure S2). Growth inhibition by Cd2 + of these strains was also assessed in liquid medium in the presence of increasing CdCl2 concentration. Whereas growth of the wild-type strain remained unaffected under both conditions, growth of ΔcadA mutant was reduced starting at 150 μM or 50 μM CdCl2 under photosynthesis (Figure 1C) and respiration (Figure 1D) conditions, respectively. ΔcadA mutant was also sensitive to excess Zn2 + as shown by growth inhibition on plates of the wild-type and ΔcadA grown in the presence of CdCl2, ZnSO4 or CuSO4 (Supplementary Figure S3A). Similarly, dose response and growth curves in liquid media (Supplementary Figures S3B–D) confirmed that cadA was also required for full zinc resistance. We should note that ΔcadA was more tolerant to Zn2 + than Cd2 + probably because under Zn2 + excess, Zn2 + uptake (ZnuABC transporter) could be limited by Zur regulator and other Zn2 +-efflux systems like CzcCBA could mediate Zn2 + efflux (Arguello et al., 2007; von Rozycki and Nies, 2009; Capdevila et al., 2017; Chandrangsu et al., 2017). Genes encoding these proteins are present in R. gelatinosus genome, but their role was not investigated yet. These data confirmed that cadA gene encodes for the Cd2 +/Zn2 +-efflux ATPase CadA, required for Cd2 + (and involved in Zn2 +) tolerance in R. gelatinosus, and that the ΔcadA mutant is more sensitive to cadmium under respiratory conditions.
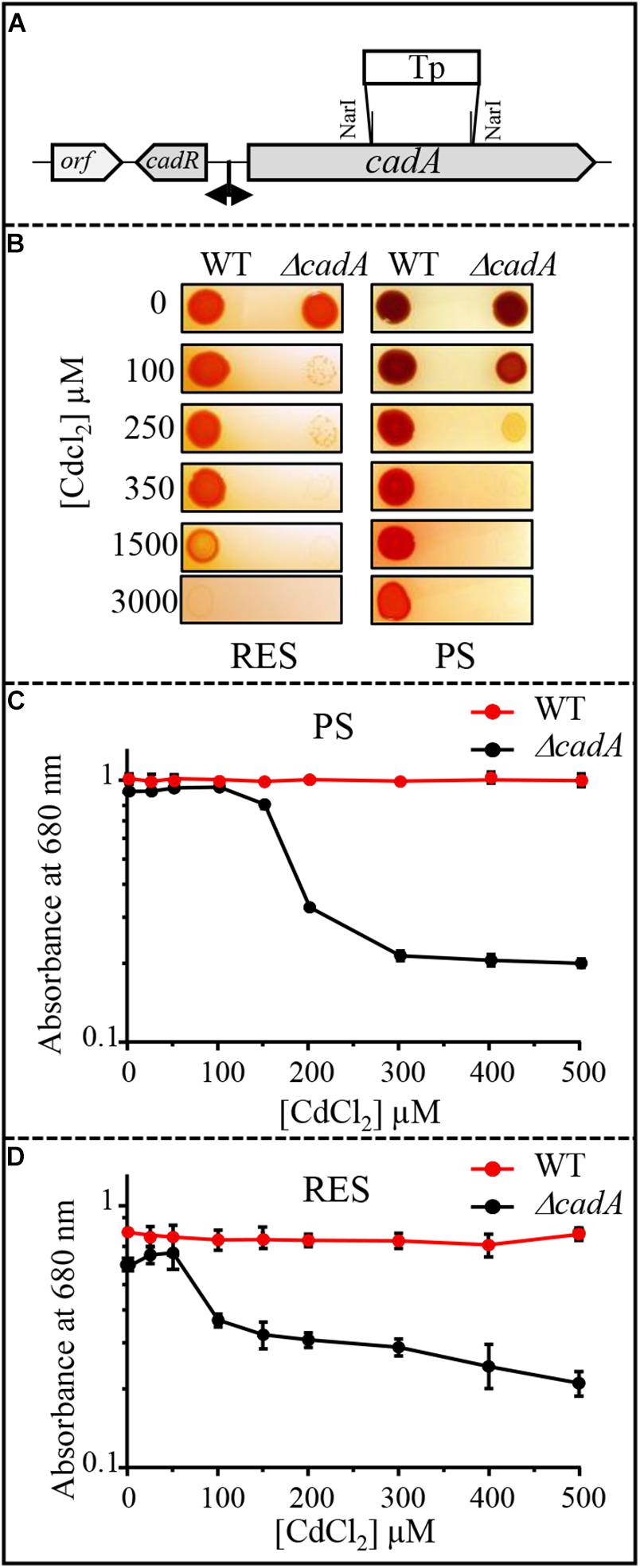
Figure 1. (A) Genetic organization of the cadAR locus in R. gelatinosus. To inactivate cadA, the NarI fragment was replaced by antibiotic resistance Tp cassette. (B) Growth phenotype of the wild-type (WT) and ΔcadA on solid medium supplemented with CdCl2. Plates were incubated under aerobic respiration (RES) in the dark or under photosynthesis (PS) for 24h at 30°C prior to photography. (C,D) Growth inhibition of the WT and ΔcadA mutant challenged with increasing CdCl2 concentrations under photosynthesis (PS) (C) and respiration (RES) (D) conditions. Cells were grown for 24 h at 30°C before OD680nm measurement. The error bars represent the standard deviation of the mean of 3 independent experiments.
Induction of CadA in Response to Excess Cd2 + Involves the MerR-Type Transcriptional Regulator CadR
To cope with excess toxic metal exposure, bacteria usually induce the expression of metal detoxification proteins including the efflux ATPases. To gain better insight into the expression profile of CadA in response to excess Cd2 +, we analyzed the expression of CadA by Western blot in the wild-type and ΔcadA strains grown under photosynthesis condition in presence of increasing CdCl2 concentration. The presence of the histidine stretch within CadA N-terminus sequence allowed the detection of CadA on Western blot using the horseradish peroxidase (HRP)-conjugated HisProbe. As shown in Figure 2A, addition of 150 μM or 1 mM CdCl2 to the medium during wild-type growth resulted in a significant increase in the amount of an 80 kDa protein, likely corresponding to CadA. This band was not detected in the ΔcadA cells grown without or with the addition of 150 μM CdCl2. To confirm that this band was indeed CadA, the ΔcadA complemented strain was also subjected to Cd2 + stress and Western blot showed the expression and induction of the 80 kDa band encoded by cadA gene on the replicative plasmid (Supplementary Figure S2B). These findings demonstrated that CadA is induced by cadmium Unexpectedly, in addition to CadA induction, Cd2 + also induced an increase in the amount of the periplasmic Cu+ tolerance protein CopI (Durand et al., 2015) in both wild-type and ΔcadA cells suggesting a putative role of CopI in Cd2 + tolerance and very likely a cross talk between Cu+ and Cd2+ (see below). To unequivocally show that the Cd2 +-induced band correspond to CopI, we generated a mutant (ΔcadA-ΔcopI) in which cadA and copI were deleted, and probed the presence of CopI in the periplasmic fractions. As shown in Figure 2B, while CopI was induced in ΔcadA under Cd2 + excess, the band was absent in the ΔcadA_ΔcopI mutant showing that the cadmium-induced band correspond to CopI.
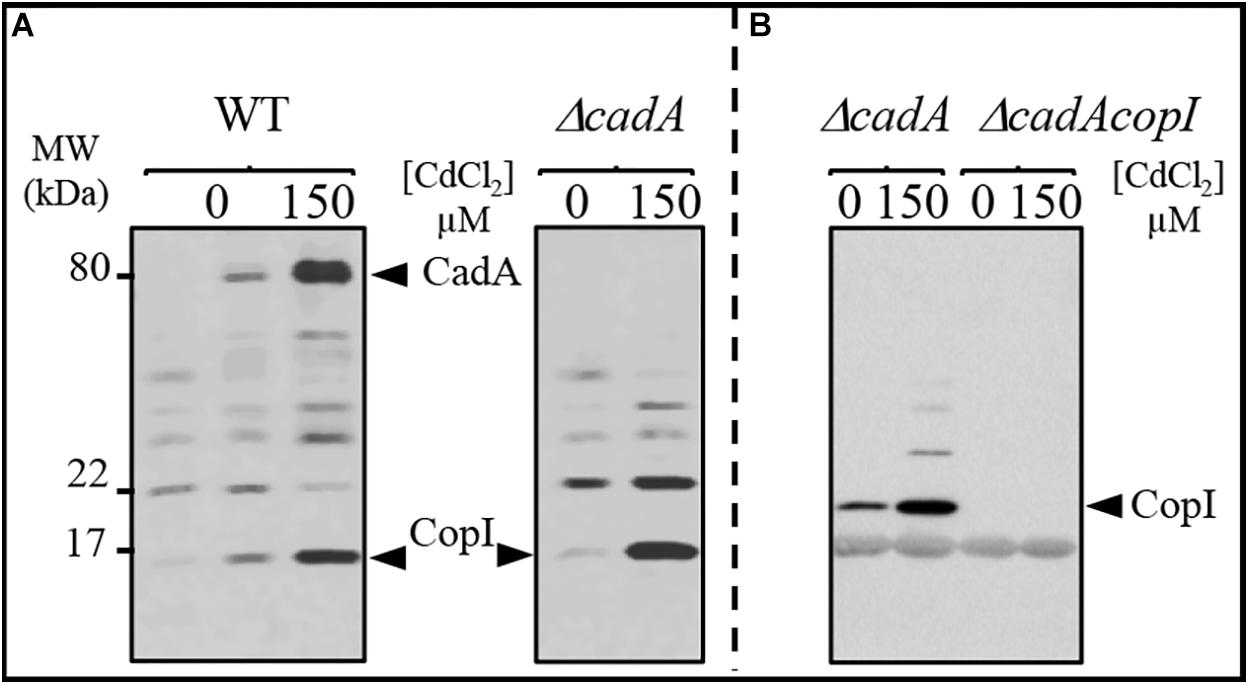
Figure 2. Expression profile of CadA and CopI in R. gelatinosus wild-type (WT) and ΔcadA cells challenged with Cd2 +. Cells were grown overnight (18 h) under photosynthesis and total protein extract from the same amount of cells (OD680nm = 0.1) were separated on a 15% Tris-glycine SDS-PAGE (A). Periplasmic fractions were purified from ΔcadA and ΔcadA_ΔcopI cells challenged or not with Cd2 + and separated on a 15% Tris-glycine SDS-PAGE (B). Proteins were visualized after Western blotting using the HRP-HisProbe (Pierce).
It has been reported that the transcription regulator CadR/ZntR recognizes a palindromic sequence in the promoter region of cadA/zntA genes (Singh et al., 1999). In the intergenic sequence (54 bp) between cadA and cadR (Figure 1), an inverted repeat sequence (–27-ACCCGCTACAGGGT-–13) might allow the binding of regulatory proteins was identified. To further highlight the role of CadR, the gene was inactivated and the resulting cadR– strain was shown to be sensitive to cadmium (Supplementary Figure S4) suggesting its involvement in cadA expression. To unambiguously show the role of cadR in CadA expression, we checked the expression level of CadA in the wild-type and the cadR– strains challenged with excess Cd2 +. For this purpose, cells were grown overnight under photosynthesis in the presence of increasing CdCl2 concentration and CadA expression profile was assessed by Western blot. In contrast to the wild-type, overnight growth of cadR– cells was inhibited beyond 600 μM CdCl2. Western blot analysis confirmed that CadA is induced with increasing concentration of CdCl2 (Figure 3) in the wild-type and this induction is CadR-dependent since CadA was not detected in cadR– cells. In conclusion, these data showed that the CadA efflux ATPase is required for cadmium detoxification and its expression is induced by CdCl2 under the control of CadR. Note that the cadR– mutant tolerates a higher cadmium concentration than the ΔcadA mutant, but less than the wild-type (Supplementary Figure S4). This difference could be due to a low expression of CadA (undetectable on Western blot) in cadR–, but sufficient enough to warrant tolerance toward low cadmium concentrations. The Western blot also confirmed the induction of CopI expression in response to Cd2 + stress and showed that induction of CopI by Cd2 + occurred, even in the absence of CadR.
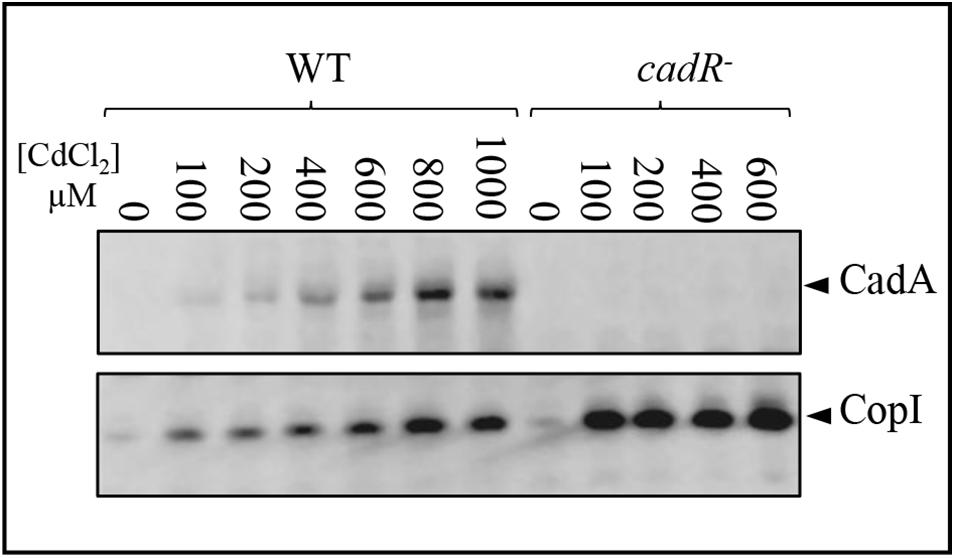
Figure 3. Expression level of CadA and CopI in R. gelatinosus wild-type (WT) and cadR- mutant cells challenged with increasing concentration of CdCl2. Cells were grown overnight (18 h) under photosynthetic condition. Total protein extract from the same amount of cells (OD680nm = 0.1) were separated on a 15% Tris-glycine SDS-PAGE. Proteins were visualized after Western blotting using the HRP-HisProbe.
Cadmium Effect on Photosynthesis and Respiratory Membrane Complexes
The toxic effect of Cu+ on photosynthesis and respiration in R. gelatinosus was previously assessed in the Cu+-P1B-type ATPase efflux mutant copA– (Azzouzi et al., 2013; Liotenberg et al., 2015). Because photosynthetic organisms can also be exposed to Cd2 + in their environment, we took advantage of the ΔcadA strain that should likely accumulate Cd2 + in the cytoplasm, to ask whether Cd2 + affects protein complexes and cytochromes involved in photosynthesis and microaerobic respiration. Membranes from the wild-type and ΔcadA cells grown microaerobically in medium supplemented or not with 100 μM Cd2 + were enriched and the amount of photosynthetic complexes (RC-LH 860 nm and 800 nm) was assessed with UV-visible absorbance spectra. In contrast to the wild-type, the amount of the photosynthetic complexes was shown to significantly decrease in the membrane of ΔcadA cells exposed to Cd2 + (Figure 4A). We also performed redox spectral analyses (reduced minus oxidized spectra) to checked the effect of Cd2 + on cytochromes c (peak at 552 nm) and b (peak at 560 nm) in these membranes by comparing the total cytochrome content in the wild-type and ΔcadA membranes (Figure 4B). Comparable spectra were recorded for Cd2 + treated and untreated wild-type membrane. Unlike the wild-type, in the ΔcadA cells, Cd2 + addition resulted in a drastic decrease of cytochrome c and b content within the membranes.
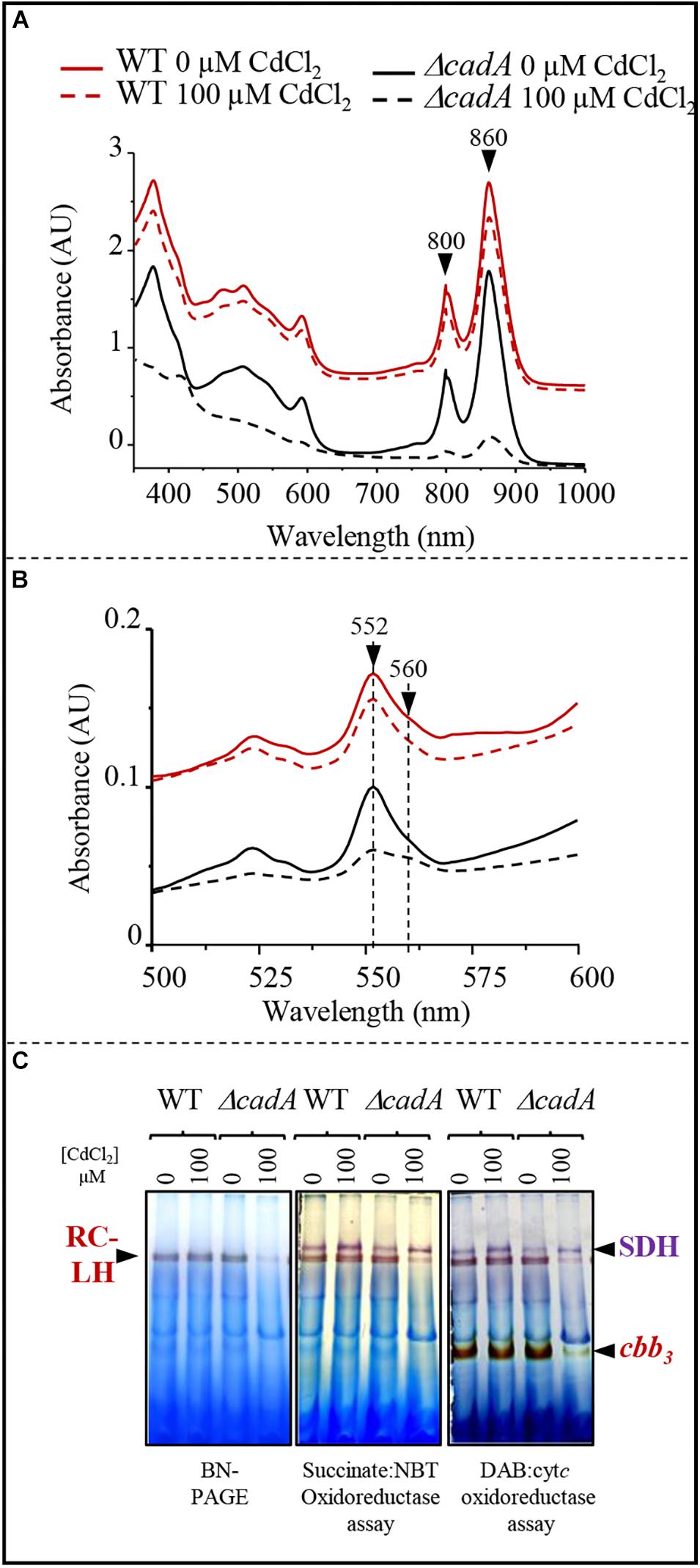
Figure 4. Cadmium effect on photosynthesis and respiratory membrane complexes in the wild-type (WT) and ΔcadA mutant grown overnight (18 h) under microaerobic condition. (A) Effect of CdCl2 on photosystem (860–800 nm) amount in the membranes. (B) Difference (reduced minus oxidized) pyridine haemochrome spectra of WT (red) and ΔcadA (black) membranes with 100 μM (dashed lines) or without cadmium (lines). (C) cbb3 cytochrome c oxidase and succinate dehydrogenase (SDH) in-gel activity assays. Equal amount of DDM-solubilized membrane proteins from wild-type (WT) and ΔcadA cells grown without or in the presence of 150 μM CdCl2 were separated on 3–12% acrylamide-bisacrylamide gradient BN-PAGE. Gels were first assayed for cbb3 cytochrome c oxidase activity (DAB staining) and subsequently assayed for SDH activity (Succinate/NBT). The photosynthetic RC-LH complexes are also visible on the gel thanks to their photopigments.
BN-PAGE analysis confirmed the decreased amount of reaction centre and light harvesting antenna (RC-LH) in the Cd2 + stressed ΔcadA mutant membranes (Figure 4C). Furthermore, cytochrome c oxidase in-gel assay revealed comparable amount of active cytochrome c oxidase cbb3 in the wild-type membrane from cells grown with or without Cd2 +; whereas in ΔcadA membrane, the cbb3 active band decreased significantly in the Cd2 + stressed mutant. Concomitantly, succinate-NBT reductase in-gel assay revealed an active band, likely corresponding to SDH, in the wild-type and ΔcadA membranes. This band increased slightly in the presence of Cd2 + in both strains. However, the activity was also slightly reduced in ΔcadA membranes compared to the wild-type membranes (Figure 4C). The decrease in active cytochrome c oxidase cbb3 was more pronounced than the SDH. The SDH complex requires only one heme per complex while the cbb3 oxidase requires five hemes for its assembly and activity.
Photosynthetic and respiratory complexes require chlorophylls and heme cofactors respectively for their assembly and activity. The decreased level of these complexes in presence of excess Cd2 + could be attributed to a decreased amount of cofactors under Cd2 + stress condition in the ΔcadA mutant. The total heme and chlorophylls were extracted from cells exposed or not to Cd2 + and dithionite-reduced and ferricyanide-oxidized pyridine hemochrome spectra were recorded. The reduced minus oxidized spectra showed a comparable amount of heme (peaks α and β at 550 nm and 520 nm respectively) and bacteriochlorophylls (756 nm) for photosynthetic grown wild-type strain in presence/absence of Cd2 + in these samples (Figure 5A). The ΔcadA mutant cells stressed by 100 μM of Cd2 + presented a strong decrease of the heme and bacteriochlorophylls content compared to the unstressed mutant strain (Figure 5B). Heme and chlorophyll share the same biosynthesis pathway down to protoporphyrin IX. An effect of cadmium on the porphyrin biosynthesis pathway upstream protoporphyrin IX would then affect concomitantly heme and chlorophyll biosynthesis.
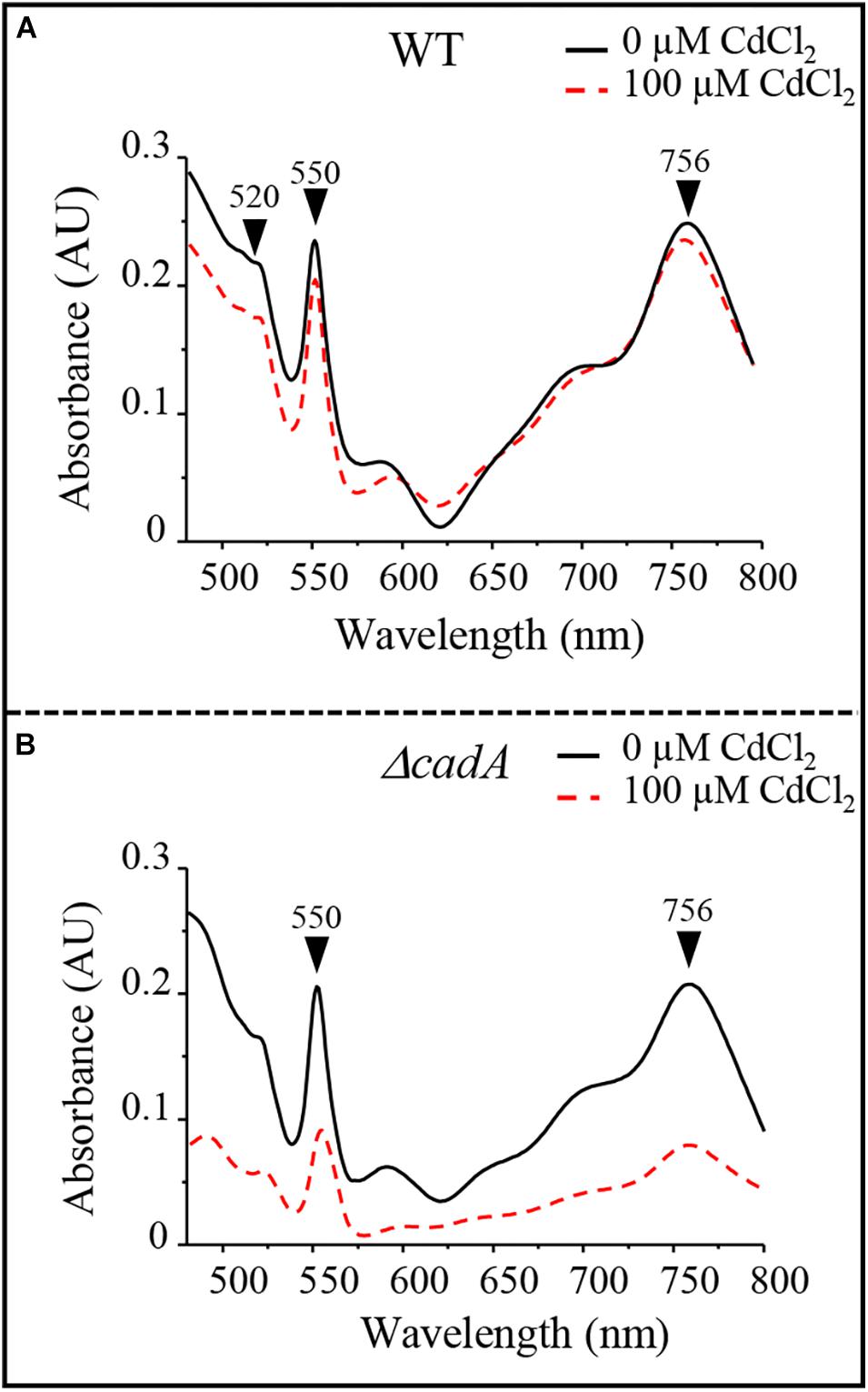
Figure 5. Cadmium effect on total heme (550 nm) and bacteriochlorophyll (756 nm) content in the wild-type (WT) (A) and ΔcadA strain (B) grown by photosynthesis for 18 h in the presence of CdCl2.
Cadmium Excess Is Associated With an Accumulation of Coproporphyrin III
The wild-type strain and the ΔcadA mutant were subjected to increasing sub-lethal concentrations of CdCl2 (0–150 μM CdCl2) under photosynthetic growth conditions. No effect of excess CdCl2 was observed on the wild-type cells (not shown). Conversely, for the ΔcadA cells, growth was affected by increasing CdCl2 concentration and a UV-fluorescent pigment was released in the spent medium as a CdCl2 concentration-dependent manner (Figure 6A). This phenotype also seen under microaerobic conditions, is presumably due to the excretion of porphyrin intermediate and is reminiscent of the copA– strain one when exposed to Cu+ (Azzouzi et al., 2013). The spent medium of the ΔcadA strain was therefore compared to that of wild-type and copA– strains using UV-Vis absorbance spectra (Figure 6B). In view of these spectra, the ΔcadA strain extrudes the same pigment as the copA– strain exposed to Cu+. Two absorbance peaks in the Soret region at 371 and 395 nm with minor peaks at 501, 537, and 553 nm were observed. These peaks are characteristic of coproporphyrin III (oxidized coproporphinogen III), a protoporphyrin IX precursor. The presence of coproporphyrin III in the medium may correlate with a decreased amount of chlorophylls and heme in the ΔcadA cells exposed to Cd2 +. To ascertain this assumption, UV-vis spectra were recorded on total pigments extracted from wild-type and ΔcadA strains grown with 100 μM CdCl2 or from copA– strain grown with 50 μM CuSO4 under the same conditions. In both ΔcadA and copA– strains, the addition of CdCl2 or CuSO4 respectively to the growth medium correlated with a decrease in the bacteriochlorophyll a content (peak at 770 nm) and an increase of the 395 nm peak (Figure 6C). In contrast, in the wild-type strain grown in the presence of 100 μM CdCl2, the higher amount of bacteriochlorophyll a correlated with the absence of the 395 nm peak in the extract (Figure 6C). These data confirm the accumulation of coproporphyrin III and reflects the decrease of bacteriochlorophyll a and heme under photosynthetic and microaerobic conditions in ΔcadA cells exposed to Cd2 + as previously reported for the copA– mutant exposed to excess CuSO4. As for Cu+ toxicity, accumulation of coproporphyrin III in the ΔcadA strain when stressed with Cd2 + suggested that excess Cd2 + affected the enzyme for which coproporphyrinogen III is a substrate. We therefore assumed that Cd2 + targets the coproporphyrinogen III oxidase HemN in the porphyrin biosynthesis pathway. This is the first in vivo demonstration that, as oxygen and Cu+, Cd2 + affects the porphyrin biosynthesis pathway presumably at the level of the SAM and [4Fe-4S] containing HemN enzyme.
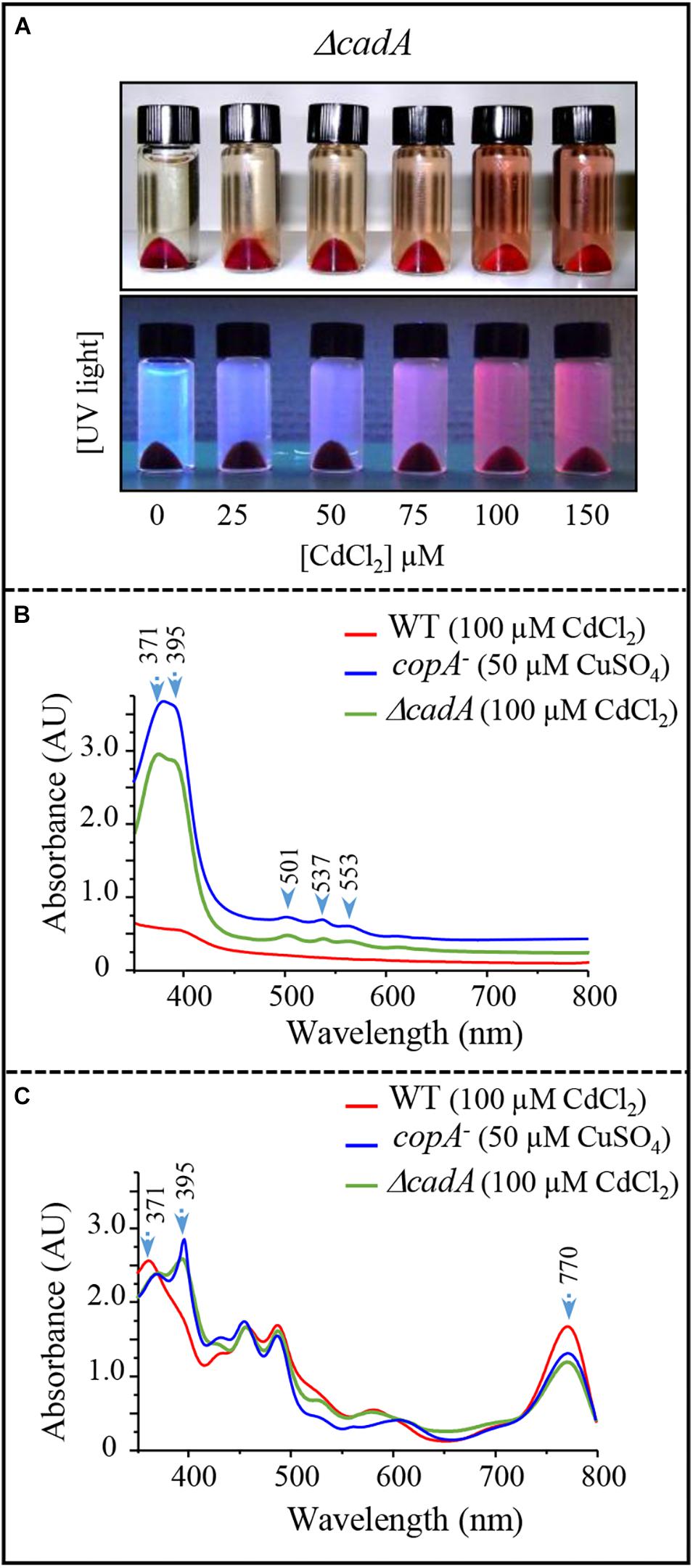
Figure 6. Effect of cadmium on photosynthetic growth in the ΔcadA strain and characterization of the pigment extruded in the culture medium. (A) Phenotype of the ΔcadA mutant grown in the presence of increasing CdCl2 concentration. Appearance of coproporphyrin III in the spent medium under visible and UV light. (B) UV-visible absorption spectra of the spent medium from the wild-type and ΔcadA strain grown with 100 μM CdCl2, in comparison with the spent medium of copA– strain that accumulates coproporphyrin III (395–371 nm) when grown in the presence of 50 μM CuSO4. (C) Absorption spectra of total pigment extract from membranes of the wild-type, ΔcadA cells grown with 100 μM CdCl2 and copA– cells grown with 50 μM CuSO4. The spectra show the decrease in the amount of bacteriochlorophyll a (770 nm) in ΔcadA and copA– mutants and reveal the presence of coproporphyrin III in these cells.
HemN Is a Target of Cd2 + and Cu+ Toxicity in R. gelatinosus
Accumulation of coproporphyrin III in the ΔcadA strain when exposed to excess cadmium, or in the CopA defective strains when challenged with excess copper both in R. gelatinosus and N. gonorrhoea, strongly suggests that HemN activity is affected by excess metal (Azzouzi et al., 2013; Djoko and McEwan, 2013). Inactivating the [4Fe-4S] of HemN by Cd2 + or Cu+ would be consistent with the published evidences showing that Cd2 + or Cu+ damage exposed [4Fe-4S] in dehydratases (Macomber and Imlay, 2009; Xu and Imlay, 2012). Attempt to express and purify HemN of R. gelatinosus and N. gonorrhoea (Djoko and McEwan, 2013) in E. coli to assess the effect of metal on the enzyme activity were unfortunately unsuccessful. Therefore, to provide direct evidence that Cd2 + or Cu+ damage HemN, we hypothesized that the amount of the enzyme would decrease if the [4Fe-4S] cluster is damaged by excess Cu+ or Cd2 +. To test this assumption, we generated a strain in which the wild-type hemN gene was substituted by a histidine tagged copy on the chromosome. In this strain, the hemN-H6 gene is under its own FNR-regulated promoter and should therefore be inducible under micro-aerobic and photosynthesis conditions. This strain was subjected to elevated concentrations of CdCl2 or CuSO4 and the amount of HemN-H6 in the soluble fraction was assessed on Western blot.
A band of 54 kDa likely corresponding to HemN-H6 was detected on Western blot in the soluble fraction of HemN-H6 strain but not in the wild-type soluble fraction (Figure 7). This band was however significantly decreased in the soluble fractions purified from HemN-H6 strain grown in the medium supplemented either with 1000 μM CdCl2 or with 500 μM CuSO4 (Figure 7). In contrast to the decrease of HemN-H6, CopI protein expression was increased in presence of Cd2 + and highly increased by Cu+ as expected in these samples (Figure 7), confirming the Cd2 + and Cu+ stress status encountered by these cells. These results, clearly demonstrated that in vivo, HemN stability is affected by excess Cd2 + and Cu+, thus elucidating the effect of metal excess on heme biosynthesis and the cause of coproporphyrin III accumulation.
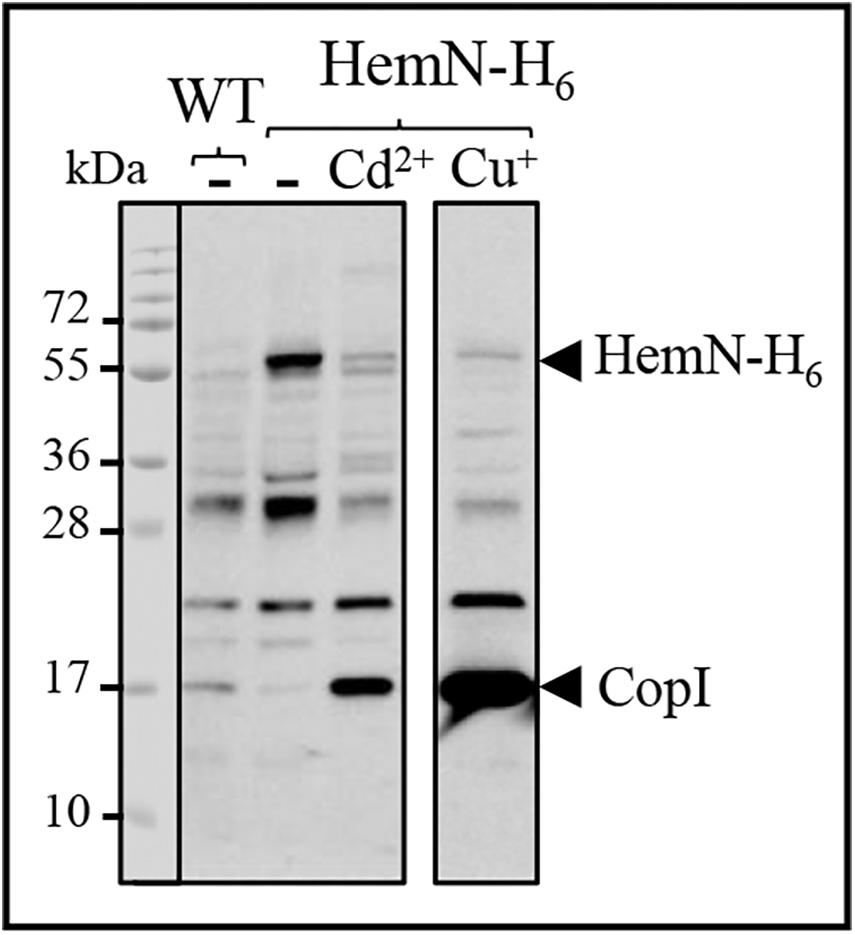
Figure 7. Expression level of HemN-H6 in R. gelatinosus WT and HemN-H6 strain in malate medium (-) or challenged with 1000 μM CdCl2 (Cd2 +) or 500 μM CuSO4 (Cu+). Cells were grown overnight (18 h) under photosynthetic condition. Soluble protein fractions (20 μg) were separated on a 15% Tris-glycine SDS-PAGE. Proteins were visualized after Western blotting using the HRP-HisProbe.
Cadmium and Copper Cross-Tolerance: CopI Is Involved in Cadmium Tolerance
On Western Blot, we observed an increase of the periplasmic copper tolerance protein CopI with the addition of CdCl2 in both wild-type and ΔcadA cells (Figure 2). This CopI induction, in presence of 150 μM CdCl2, is higher in the ΔcadA mutant than in the wild-type, very likely resulting from Cd2 + accumulation in the cytoplasm of the ΔcadA strain. CopI induction by Cd2 + suggested a cross-talk between the Cd2 + and Cu+ response systems and a role of CopI and/or the copper detoxification system in Cd2 + response. A band at about 22 kDa apparent molecular weight was also noticeably induced in the ΔcadA mutant. This band corresponds to the cytosolic superoxide dismutase SodB (Steunou et al., 2020a). Similarly, in the cadR– mutant, CopI is also more induced compared to the wild-type strain. Once again, this difference in expression could arise from the accumulation of Cd2 + in the cytoplasm of cadR– cells devoid of the Cd2 +-efflux ATPase. The data suggested that either CopR can interact with Cd2 + to induce CopI or that other regulators can induce CopI expression in the cadR– strain.
Given the induction of CopI, the periplasmic copper tolerance protein, by excess Cd2 + (Figures 2, 3), we wondered whether CopI was required for Cd2 + resistance. The ΔcopI mutant was sensitive only to high Cd2 + concentration. However, the presence of CadA in the ΔcopI mutant made the conclusion elusive. To confirm its requirement for Cd2 + tolerance, we question whether inactivation of copI in the ΔcadA background could give rise to a higher Cd2 + sensitivity in such a strain. The double mutant ΔcadA_ΔcopI was constructed and its tolerance towards Cd2 + was compared to that of ΔcadA strain on plates (Figure 8A) and liquid (Figure 8B and Supplementary Figure S5) under photosynthesis conditions. The double mutant ΔcadA_ΔcopI was indeed more sensitive to Cd2 + than the ΔcadA mutant when exposed to 150 μM and 200 μM CdCl2 on plates, and to 75 μM CdCl2 in liquid, thus confirming that CopI is somehow involved in Cd2 + tolerance. The underlying mechanism is not yet known, however, CopI was shown to be periplasmic and displays conserved His and Met residues that could interact with cations (Durand et al., 2015). It is then tempting to speculate that CopI may handle Cd2 + within the periplasm.
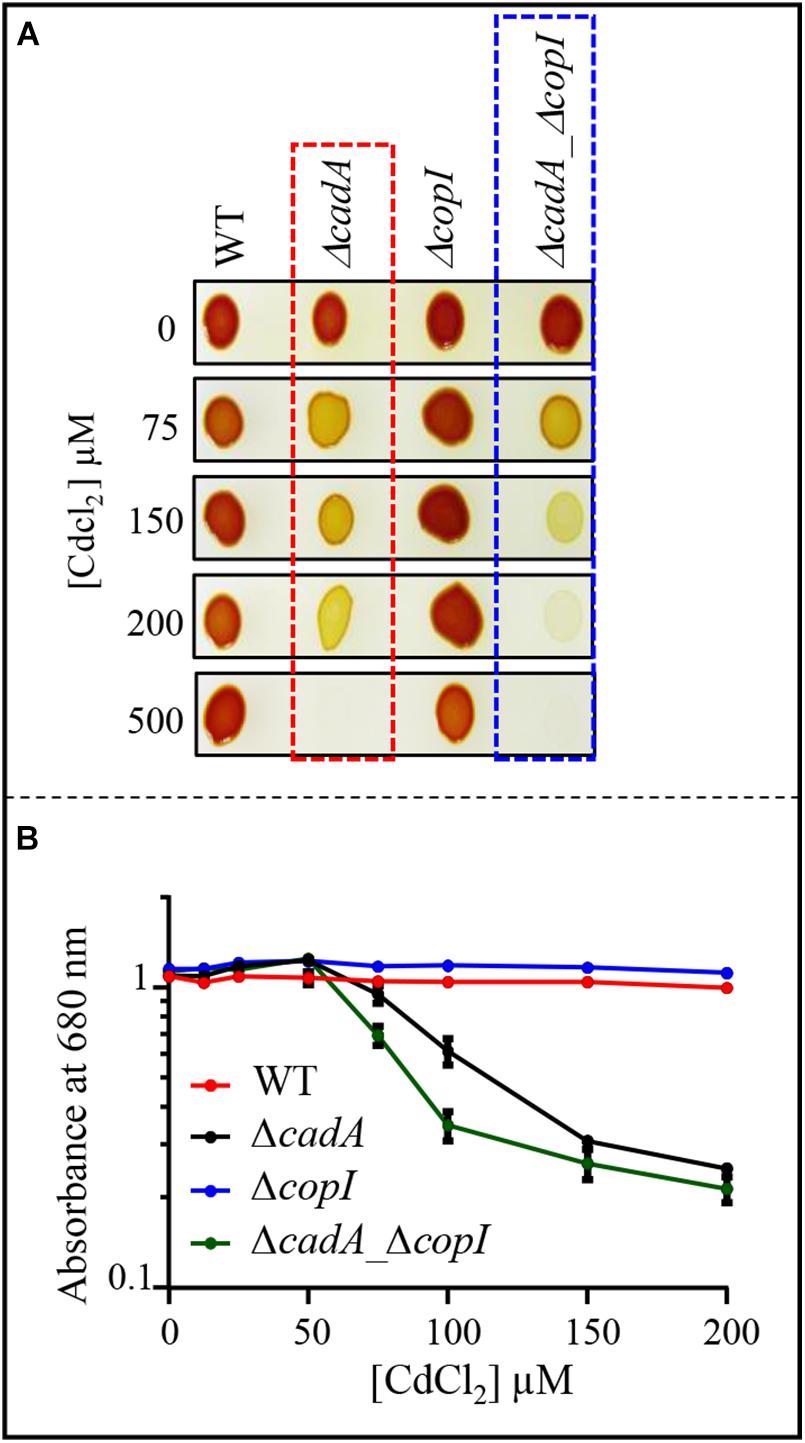
Figure 8. (A) Growth phenotype of the wild-type (WT), ΔcadA, ΔcopI, and ΔcadA-ΔcopI mutants in the presence of increasing CdCl2 concentrations on solid plates. Cells were grown by photosynthesis for 48 h at 30°C prior to photography. (B) Growth inhibition of the wild-type, ΔcadA, ΔcopI, and ΔcadA_ΔcopI mutants challenged with increasing CdCl2 concentration. Cells were grown in liquid under photosynthetic condition for 21 h at 30°C before OD680nm measurement. The error bars represent the standard deviation of the mean of 3 independent experiments.
The Cu+-ATPase CopA Is Induced by Cd2 + and Cu+ Enhances Cadmium Tolerance in the ΔcadA Strain
Induction of CopI expression in response to both Cu+ and Cd2 +, suggests a cross-tolerance towards these metals and raised the question on the ability of Cu+ to enhance the resistance of ΔcadA strain to increasing concentration of Cd2 +. To answer this question, we assessed the growth inhibition of ΔcadA strain grown under photosynthesis condition in Cd2 + containing medium supplemented or not with 200 μM CuSO4. No significant difference in growth of the wild-type strain was observed between the two conditions, both on solid (Figure 9A) and liquid media (Figure 9B). Interestingly, for the ΔcadA strain, the addition of CuSO4 in the medium significantly improved growth of the mutant, suggesting that, Cu+ alleviates Cd2 + stress. Similar results were obtained under aerobic respiration (Supplementary Figure S6).
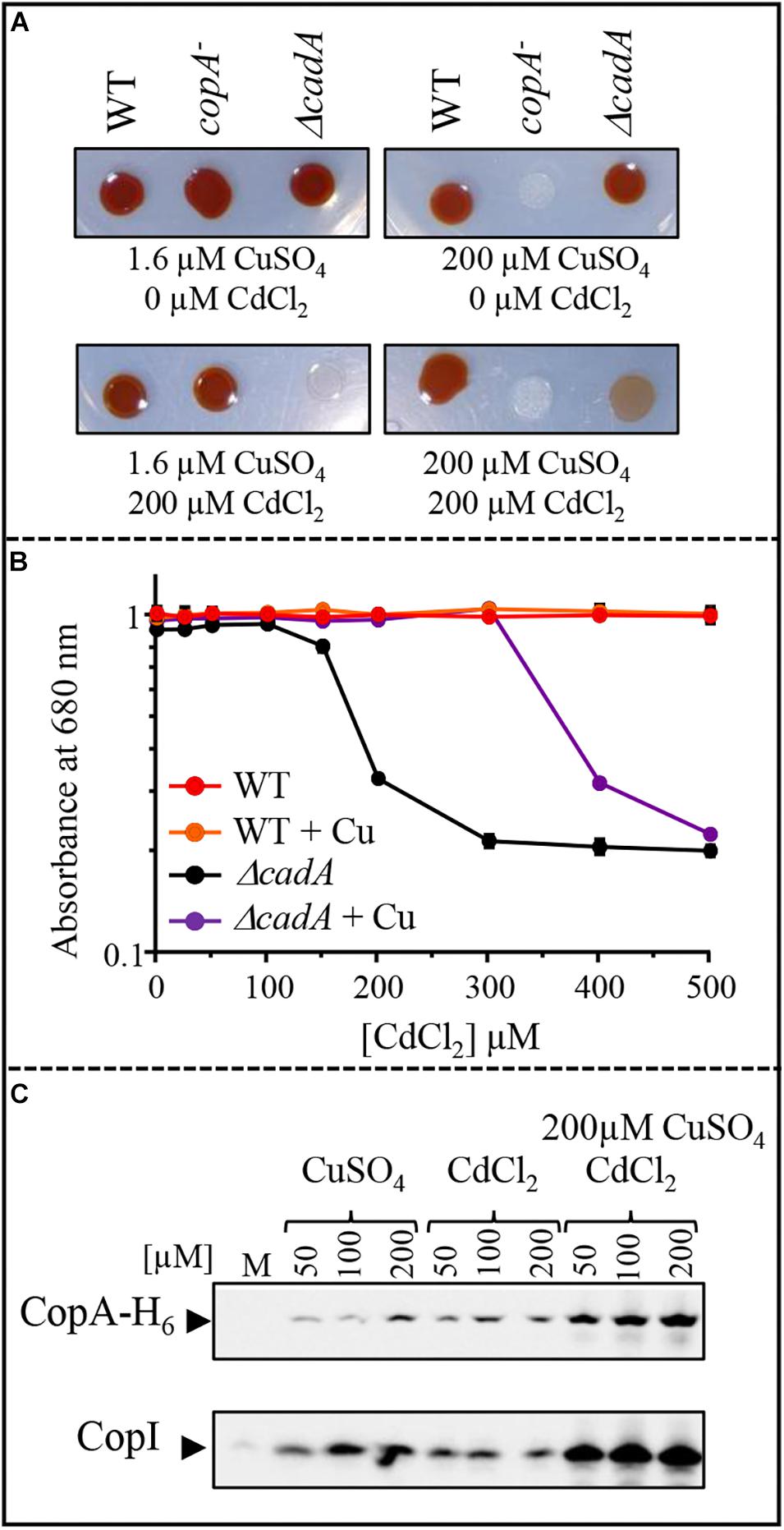
Figure 9. Effect of CuSO4 on growth in the presence of increasing concentration of CdCl2. (A) Growth phenotype of the wild-type (WT), ΔcadA and copA– strains in presence of increasing CdCl2 concentration with or without addition of 200 μM CuSO4 on solid plates. (B) Growth inhibition of the wild-type and ΔcadA mutant challenged with increasing CdCl2 concentration in the medium supplemented or not with 200 μM CuSO4. Cells were grown under photosynthetic condition for 18 h at 30°C before OD680nm measurement. The error bars represent the standard deviation of the mean of 3 independent experiments. (C) Expression level of CopA-H6 and CopI in R. gelatinosus ΔcadA-copA-H6 mutant challenged with increasing concentration of CuSO4, CdCl2, or CdCl2 + CuSO4. Cells were grown in the malate (M) medium supplemented or not with metals, under photosynthetic condition. Total protein extract from the same amount of cells (OD680nm = 0.1) were separated on a 15% tris-glycine SDS-PAGE. Proteins were visualized after Western blotting using the HRP-HisProbe.
Given that exposure to Cu+ caused an induction of the CopA-CopI system (Azzouzi et al., 2013; Durand et al., 2015), the enhanced growth observed in ΔcadA strain could be the result of an up-regulation of CopA and CopI expression by the simultaneous effect of Cu+ and Cd2 + in the cell. To check the expression of CopA in response to both Cd2 + and Cu+ in the ΔcadA background, we constructed a ΔcadA strain bearing a copA histidine fusion gene (copA-H6) integrated to the copA locus on the chromosome of R. gelatinosus to allow detection on Western blot. In this ΔcadA_copA-H6 strain, the transcription of copA-H6 is under the control of its endogenous Cu+-regulated promoter. The strain was grown under photosynthesis condition with increasing CuSO4 and/or CdCl2 concentrations and whole cell extracts were probed for the presence of CopA-H6. The presence of both metals resulted in a significant increase in the amount of CopA-H6 and CopI within the cells (Figure 8C). These findings clearly showed that as for CopI and CadA, Cd2 + induced the Cu+-ATPase CopA. Although transport of Cd2 + by CopA is not yet shown, the data suggest a role of CopA and CopI in Cd2 + tolerance. In agreement with this assumption, we assessed growth on Cd2 + alone or on Cd2 + and Cu+ of a ΔcadA_copA– double mutant devoid of both CadA and CopA ATPases. In contrast to ΔcadA single mutant, no difference in growth was observed between the two conditions for the double mutant (Supplementary Figure S7). Together with the involvement of Cd2 + in Cu+-homeostasis system induction, these new findings suggested a cross-talk and -tolerance between the Cd2 + and Cu+ efflux system in R. gelatinosus.
Discussion
Heavy metal contamination represents a serious threat to the environment and all living organisms because of their acute toxicity. Understanding toxicity is an intricate issue because of the presence of efficient metal detoxification systems; however, the availability of strains defective in a metal homeostasis machinery allowed the identification of new metal targets and shed light on the toxicity mechanisms. Successful efforts have been made to understand the mechanisms by which bacteria respond and deal with single metal excess (Arguello et al., 2007; Barwinska-Sendra and Waldron, 2017; Capdevila et al., 2017; Chandrangsu et al., 2017). In the present study, the Cd2 +-efflux ATPase defective mutant of R. gelatinosus provides a useful tool to identify targets of Cd2 + within the cells. Exposure of ΔcadA to excess Cd2 + resulted in a decreased amount of the light harvesting and reaction center (LH-RC) photosynthetic complexes and a decreased amount of the respiratory complexes succinate dehydrogenase (SDH) and cbb3 cytochrome c oxidase. Both cbb3 and SDH are hemoproteins, while the LH-RC complex assembly and activity are bacteriochlorophyll dependent. This decrease is concomitant to the decrease in heme and bacteriochlorophyll production. Moreover, noticeably, under microaerobic respiration and photosynthesis growth conditions, the ΔcadA mutant extruded coproporphyrin III. These data showed that when challenged with excess Cd2 +, the tetrapyrrole biosynthesis pathway was affected at the level of the anaerobic coproporphyrinogen III oxidase HemN. HemN is an [4Fe-4S] cluster-dependent enzyme, with a cluster coordinated only by three cysteines and the methionine group of S-adenosylmethionine. This cluster is located within the active site pocket that accommodates two SAM molecules and the coproporphyrinogen III (Layer et al., 2003, 2004). This pocket is likely solvent accessible, making the [4Fe-4S] cluster vulnerable to molecular oxygen but also very likely towards metal ions (Azzouzi et al., 2013; Djoko and McEwan, 2013). The Cd2 +-related porphyrin phenotype was observed only under microaerobic respiration or photosynthesis conditions in which HemN is required for the tetrapyrrole biosynthesis pathway, but not under high aeration when the aerobic coproporphyrinogen III oxidase HemF performs the oxidation of coproporphyrinogen III (Breckau et al., 2003). We therefore suggest that Cd2 +, like oxygen an Cu+, affects tetrapyrrole biosynthesis presumably at the level of the SAM and [4Fe-4S] containing HemN enzyme.
Under aerobic respiration condition, ΔcadA growth is even more affected by Cd2 + than under photosynthesis condition, this might be related to the synergetic toxic effects of Cd2 + and oxygen. Yet, the Cd2 + targets remain to be identified in R. gelatinosus. Given that Cd2 + targets [4Fe-4S] clusters under microaerobic respiration and photosynthesis conditions, it is tempting to suggest that other oxygen sensitive [4Fe-4S] dehydratases may be poisoned by the simultaneous presence of oxygen and Cd2 +, as shown in E. coli (Xu and Imlay, 2012) and in R. gelatinosus (Liotenberg et al., 2015).
In their environment, bacteria are exposed to a mixture of metals (Ye et al., 2017). This can cause interactions between metals, enhanced toxicity or on the contrary, generate cross-resistance towards different metals. Interestingly, we identified CopI, known to be involved in Cu+ tolerance (Durand et al., 2015), as also involved in Cd2 + tolerance since deletion of copI in ΔcadA mutant resulted in an increased sensitivity to Cd2 +. The mechanism by which CopI interferes with Cd2 + tolerance is unknown, however, since CopI interacts with Cu+, Cd2 + may also interact with the apo-CopI protein in the absence of Cu+. In P. aeruginosa, the pcoAB genes are induced by Cu+ and encode periplasmic Cu+ binding proteins. The pcoAB genes are also suggested to be involved in Zn2 + and probably Cd2 + tolerance since the pcoAB mutants are more sensitive to Zn2 + (Teitzel et al., 2006).
Interaction of Cd2 + with cupro-protein, albeit with low affinity, have been reported in the case of the Cu+ chaperone CopZ involved in copper efflux in Bacillus (B.) subtilis and other bacteria (Kihlken et al., 2008). As for copI, the copZ deletion in B. subtilis resulted in an increased sensitivity to both Cu+ and Cd2 + and expression of the copZA operon was induced in response to elevated concentrations of both metals (Moore et al., 2005; Steunou et al., 2020a). Similarly, in Pseudomonas (P.) aeruginosa Cu+ induces the activation of the czcCBA operon involved in the efflux of Zn2 +, Cd2 +, and Co2 + (Caille et al., 2007). In our study, Cd2 + is shown to induce the Cd2 +-efflux ATPase CadA and the Cu+-efflux ATPase CopA in R. gelatinosus. In addition, Cu+ improves Cd2 + tolerance in the CadA deficient mutant, suggesting that CopA and CopI are somehow involved in Cd2 + detoxification. The involvement of CopA in Cd2 + or Zn2 + efflux has never been demonstrated and seems very unlikely. Nevertheless, the cross-talk between Cu+ and or Zn2 +/Cd2 + efflux systems, at least at the expression level, was previously reported in other bacteria. In P. aeruginosa, both PA3920 gene encoding CopA and PA3690 encoding ZntA/CadA homologue are induced by Cu+ stress (Teitzel et al., 2006). We should stress out that in this study, P. aeruginosa wild type cells were exposed to a very high concentration of CuSO4 (10 mM). In a more recent study, Arguello’s group used RNA-Seq to characterize the response of P. aeruginosa to 0.5 mM CuSO4 to avoid pleiotropic effects that could be related to elevated concentration of CuSO4 (Quintana et al., 2017). PA3690 (ZntA/CadA) was not induced in the wild type, nevertheless, in the ΔcueR and ΔcopR mutant, a 3.6 and 2 fold change in the expression of PA3690 was observed in these mutants (Quintana et al., 2017). Likewise, in the filamentous cyanobacterium Oscillatoria brevis, expression of the ATPase Bxa1 is also induced by both Cd2 + and Cu+ cations (Liu et al., 2004). Cross-tolerance towards Cu+ and Zn2 + or Cd2 + involving these ATPases, was not investigated in these bacteria.
Although the mechanism and physiological significance behind Cu+/Cd2 + growth improvement reported in this work remain unclear and require further investigation, the cross-talk and cross-tolerance reported in all these studies emphasize on the importance of interactions and toxicity that could arise from metal mixtures and show that research should further consider mixtures effects.
Metal ability to undergo redox reactions is important for its function as protein cofactor but also for its intrinsic toxicity. Cd2 + is not redox active and cannot directly trigger ROS formation, yet, it is very toxic. It was suggested that Cd2 + could indirectly promote ROS production by displacing and releasing redox active ions such as iron or copper from proteins (Xu and Imlay, 2012; Barwinska-Sendra and Waldron, 2017). Induction of SodB expression in the ΔcadA strain suggests an increase in ROS production or a dysregulation of iron homeostasis since SodB is known to be regulated by Fur regulator in bacteria. Together with the degradation of solvent exposed [4Fe-4S] cluster from key metabolic enzymes, iron homeostasis dysregulation and ROS should be considered to account for the Cd2 + toxicity in the ΔcadA strain (Steunou et al., 2020b).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
AS, AD, MB, SL, and SO designed the research. AS, AD, M-LB, RT, and SO performed the research. AS, AD, MB, SL, M-LB, and SO analyzed the data. AS and SO wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the support of the CNRS and the Microbiology department of I2BC.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00893/full#supplementary-material
Abbreviations
BN-PAGE, blue native polyacrylamide gel electrophoresis; DAB, 3,3’-diaminobenzidine tetrahydrochloride; DDM, n-dodecyl - β -D-maltopyranoside; HRP, horseradish peroxidase; LH-RC, light harvesting-reaction center; PVDF, polyvinylidene difluoride.
References
Arguello, J. M., Eren, E., and Gonzalez-Guerrero, M. (2007). The structure and function of heavy metal transport P1B-ATPases. Biometals 20, 233–248. doi: 10.1007/s10534-006-9055-6
Azzouzi, A., Steunou, A. S., Durand, A., Khalfaoui-Hassani, B., Bourbon, M. L., Astier, C., et al. (2013). Coproporphyrin III excretion identifies the anaerobic coproporphyrinogen III oxidase HemN as a copper target in the Cu+-ATPase mutant copA– of Rubrivivax gelatinosus. Mol. Microbiol. 88, 339–351. doi: 10.1111/mmi.12188
Baekgaard, L., Mikkelsen, M. D., Sorensen, D. M., Hegelund, J. N., Persson, D. P., Mills, R. F., et al. (2010). A combined zinc/cadmium sensor and zinc/cadmium export regulator in a heavy metal pump. J. Biol. Chem. 285, 31243–31252. doi: 10.1074/jbc.m110.111260
Barwinska-Sendra, A., and Waldron, K. J. (2017). The Role of Intermetal Competition and Mis-Metalation in Metal Toxicity. Adv. Microb. Physiol. 70, 315–379. doi: 10.1016/bs.ampbs.2017.01.003
Breckau, D., Mahlitz, E., Sauerwald, A., Layer, G., and Jahn, D. (2003). Oxygen-dependent coproporphyrinogen III oxidase (HemF) from Escherichia coli is stimulated by manganese. J. Biol. Chem. 278, 46625–46631. doi: 10.1074/jbc.m308553200
Caille, O., Rossier, C., and Perron, K. (2007). A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 189, 4561–4568. doi: 10.1128/jb.00095-07
Capdevila, D. A., Edmonds, K. A., and Giedroc, D. P. (2017). Metallochaperones and metalloregulation in bacteria. Essays Biochem. 61, 177–200. doi: 10.1042/ebc20160076
Chandrangsu, P., Rensing, C., and Helmann, J. D. (2017). Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350. doi: 10.1038/nrmicro.2017.15
Djoko, K. Y., and McEwan, A. G. (2013). Antimicrobial action of copper is amplified via inhibition of heme biosynthesis. ACS Chem. Biol. 8, 2217–2223. doi: 10.1021/cb4002443
Durand, A., Azzouzi, A., Bourbon, M. L., Steunou, A. S., Liotenberg, S., Maeshima, A., et al. (2015). c-type cytochrome assembly is a key target of copper toxicity within the bacterial periplasm. mBio 6, e01007–e01015.
Gonzalez-Guerrero, M., Raimunda, D., Cheng, X., and Arguello, J. M. (2010). Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 78, 1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x
Haan, C., and Behrmann, I. (2007). A cost effective non-commercial ECL-solution for Western blot detections yielding strong signals and low background. J. Immunol. Methods 318, 11–19. doi: 10.1016/j.jim.2006.07.027
Khalfaoui Hassani, B., Steunou, A. S., Liotenberg, S., Reiss-Husson, F., Astier, C., and Ouchane, S. (2010). Adaptation to oxygen: Role of terminal oxidases in photosynthesis initiation in the purple photosynthetic bacterium. Rubrivivax gelatinosus. J. Biol. Chem. 285, 19891–19899. doi: 10.1074/jbc.m109.086066
Kihlken, M. A., Singleton, C., and Le Brun, N. E. (2008). Distinct characteristics of Ag+ and Cd2+ binding to CopZ from Bacillus subtilis. J. Biol. Inorg. Chem. 13, 1011–1023. doi: 10.1007/s00775-008-0388-1
Layer, G., Heinz, D. W., Jahn, D., and Schubert, W. D. (2004). Structure and function of radical SAM enzymes. Curr. Opin. Chem. Biol. 8, 468–476. doi: 10.1016/j.cbpa.2004.08.001
Layer, G., Moser, J., Heinz, D. W., Jahn, D., and Schubert, W. D. (2003). Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes. EMBO J. 22, 6214–6224. doi: 10.1093/emboj/cdg598
Legatzki, A., Grass, G., Anton, A., Rensing, C., and Nies, D. H. (2003). Interplay of the Czc system and two P-type ATPases in conferring metal resistance to Ralstonia metallidurans. J. Bacteriol. 185, 4354–4361. doi: 10.1128/jb.185.15.4354-4361.2003
Li, J., Ren, X., Fan, B., Huang, Z., Wang, W., Zhou, H., et al. (2019). Zinc Toxicity and Iron-Sulfur Cluster Biogenesis in Escherichia coli. Appl. Environ. Microbiol. 85, e1967–e1918.
Liotenberg, A., Steunou, A. S., Durand, A., Bourbon, M. L., Bollivar, D. W., Hansson, M., et al. (2015). Oxygen-dependent copper toxicity: targets in the chlorophyll biosynthesis pathway identified in the copper efflux ATPase CopA deficient mutant. Environ. Microbiol, 17, 1963–1976. doi: 10.1111/1462-2920.12733
Liu, T., Nakashima, S., Hirose, K., Shibasaka, M., Katsuhara, M., Ezaki, B., et al. (2004). A novel cyanobacterial SmtB/ArsR family repressor regulates the expression of a CPx-ATPase and a metallothionein in response to both Cu(I)/Ag(I) and Zn(II)/Cd(II). J. Biol. Chem. 279, 17810–17818. doi: 10.1074/jbc.m310560200
Lu, M., Li, Z., Liang, J., Wei, Y., Rensing, C., and Wei, G. (2016). Zinc Resistance Mechanisms of P1B-type ATPases in Sinorhizobium meliloti CCNWSX0020. Sci. Rep. 6:29355.
Macomber, L., and Imlay, J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349. doi: 10.1073/pnas.0812808106
Macomber, L., Rensing, C., and Imlay, J. A. (2007). Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189, 1616–1626. doi: 10.1128/jb.01357-06
Mana-Capelli, S., Mandal, A. K., and Arguello, J. M. (2003). Archaeoglobus fulgidus CopB is a thermophilic Cu2+-ATPase: functional role of its histidine-rich-N-terminal metal binding domain. J. Biol. Chem. 278, 40534–40541. doi: 10.1074/jbc.m306907200
Moore, C. M., Gaballa, A., Hui, M., Ye, R. W., and Helmann, J. D. (2005). Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol. Microbiol. 57, 27–40. doi: 10.1111/j.1365-2958.2005.04642.x
Nunes, I., Jacquiod, S., Brejnrod, A., Holm, P. E., Johansen, A., Brandt, K. K., et al. (2016). Coping with copper: legacy effect of copper on potential activity of soil bacteria following a century of exposure. FEMS Microbiol. Ecol. 92:fiw175. doi: 10.1093/femsec/fiw175
Ouchane, S., Picaud, M., Reiss-Husson, F., Vernotte, C., and Astier, C. (1996). Development of gene transfer methods for Rubrivivax gelatinosus S1: construction, characterization and complementation of a puf operon deletion strain. Mol. Gen. Genet. 252, 379–385. doi: 10.1007/bf02173002
Quintana, J., Novoa-Aponte, L., and Arguello, J. M. (2017). Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. J. Biol. Chem. 292, 15691–15704. doi: 10.1074/jbc.m117.804492
Rensing, C., Sun, Y., Mitra, B., and Rosen, B. P. (1998). Pb(II)-translocating P-type ATPases. J. Biol. Chem. 273, 32614–32617. doi: 10.1074/jbc.273.49.32614
Roy, P., Bauman, M. A., Almutairi, H. H., Jayawardhana, W. G., Johnson, N. M., and Torelli, A. T. (2018). Comparison of the response of bacterial IscU and SufU to Zn(2+) and select transition-metal ions. ACS Chem. Biol. 13, 591–599. doi: 10.1021/acschembio.7b00442
Sakpirom, J., Kantachote, D., Nunkaew, T., and Khan, E. (2017). Characterizations of purple non-sulfur bacteria isolated from paddy fields, and identification of strains with potential for plant growth-promotion, greenhouse gas mitigation and heavy metal bioremediation. Res. Microbiol. 168, 266–275. doi: 10.1016/j.resmic.2016.12.001
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning, A Laboratory Manual, 2nd Edn. New York, NY: Cold Spring Harbor.
Singh, V. K., Xiong, A., Usgaard, T. R., Chakrabarti, S., Deora, R., Misra, T. K., et al. (1999). ZntR is an autoregulatory protein and negatively regulates the chromosomal zinc resistance operon znt of Staphylococcus aureus. Mol. Microbiol. 33, 200–207. doi: 10.1046/j.1365-2958.1999.01466.x
Solovieva, I. M., and Entian, K. D. (2004). Metalloregulation in Bacillus subtilis: the copZ chromosomal gene is involved in cadmium resistance. FEMS Microbiol. Lett. 236, 115–122. doi: 10.1111/j.1574-6968.2004.tb09636.x
Steunou, A. S., Babot, M., Bourbon, M. L., Tambosi, R., Durand, A., Liotenberg, S., et al. (1999). Additive effects of metal excess and superoxide, a highly toxic mixture in bacteria. Microb. Biotechnol. (In press)Google Scholar
Steunou, A. S., Babot, M., Bourbon, M. L., Tambosi, R., Durand, A., Liotenberg, S., et al. (2020a). Additive effects of metal excess and superoxide, a highly toxic mixture in bacteria. Microb. Biotechnol. (in press).
Steunou, A. S., Bourbon, M. L., Babot, M, Durand, A., Liotenberg, S., Yamaichi, Y., et al. (2020b). Increasing the copper sensitivity of microorganisms by restricting iron supply, a strategy for bio-management practices. Microb. Biotechnol. (in press).
Tan, G., Yang, J., Li, T., Zhao, J., Sun, S., Li, X., et al. (2017). Anaerobic copper toxicity and iron-sulfur cluster biogenesis in Escherichia coli. Appl. Environ. Microbiol. 83:e00867-17.
Teitzel, G. M., Geddie, A., De Long, S. K., Kirisits, M. J., Whiteley, M., and Parsek, M. R. (2006). Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 188, 7242–7256. doi: 10.1128/jb.00837-06
von Rozycki, T., and Nies, D. H. (2009). Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie Van Leeuwenhoek 96, 115–139. doi: 10.1007/s10482-008-9284-5
Wittig, I., Karas, M., and Schagger, H. (2007). High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell Proteomics 6, 1215–1225. doi: 10.1074/mcp.m700076-mcp200
Xu, F. F., and Imlay, J. A. (2012). Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl. Environ. Microbiol. 78, 3614–3621. doi: 10.1128/aem.07368-11
Keywords: CadA/ZntA, cadmium/copper, metal homeostasis, metal toxicity, cross-talk, [4Fe-4S], porphyrin biosynthesis
Citation: Steunou AS, Durand A, Bourbon M-L, Babot M, Tambosi R, Liotenberg S and Ouchane S (2020) Cadmium and Copper Cross-Tolerance. Cu+ Alleviates Cd2 + Toxicity, and Both Cations Target Heme and Chlorophyll Biosynthesis Pathway in Rubrivivax gelatinosus. Front. Microbiol. 11:893. doi: 10.3389/fmicb.2020.00893
Received: 28 January 2020; Accepted: 16 April 2020;
Published: 03 June 2020.
Edited by:
Davide Zannoni, University of Bologna, ItalyReviewed by:
Daniel Raimunda, Medical Research Institute Mercedes and Martín Ferreyra (INIMEC), ArgentinaManuel González-Guerrero, Polytechnic University of Madrid, Spain
Copyright © 2020 Steunou, Durand, Bourbon, Babot, Tambosi, Liotenberg and Ouchane. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soufian Ouchane, soufian.ouchane@i2bc.paris-saclay.fr
 Anne Soisig Steunou
Anne Soisig Steunou Anne Durand
Anne Durand Reem Tambosi
Reem Tambosi Soufian Ouchane
Soufian Ouchane