- 1Department of Biomedical Sciences and Pathobiology, Virginia-Maryland College of Veterinary Medicine, Virginia Tech, Blacksburg, VA, United States
- 2Biocomplexity Institute and Initiative, University of Virginia, Virginia Tech, Charlottesville, VA, United States
- 3Department of Large Animal Clinical Sciences, Virginia-Maryland College of Veterinary Medicine, Blacksburg, VA, United States
- 4Department of Veterinary Biomedical Sciences, College of Veterinary Medicine, Long Island University, Brookville, NY, United States
Histophilus somni and Pasteurella multocida are two of multiple agents responsible for bovine respiratory disease (BRD) in cattle. Following respiratory infection of calves with H. somni, P. multocida may also be isolated from the lower respiratory tract. Because H. somni may form a biofilm during BRD, we sought to determine if P. multocida can co-exist with H. somni in a polymicrobial biofilm in vitro and in vivo. Interactions between the two species in the biofilm were characterized and quantified by fluorescence in situ hybridization (FISH). The biofilm matrix of each species was examined using fluorescently tagged lectins (FTL) specific for the exopolysaccharide (EPS) using confocal laser scanning microscopy. Bacterial interactions were determined by auto-aggregation and biofilm morphology. Pasteurella multocida and H. somni were evenly distributed in the in vitro biofilm, and both species contributed to the polymicrobial biofilm matrix. The average biomass and biofilm thickness, and the total carbohydrate and protein content of the biofilm, were greatest when both species were present. Polymicrobial bacterial suspensions auto-aggregated faster than single species suspensions, suggesting physical interactions between the two species. Almost 300 P. multocida genes were significantly differentially regulated when the bacteria were in a polymicrobial biofilm compared to a mono-species biofilm, as determined by RNA-sequencing. As expected, host genes associated with inflammation and immune response were significantly upregulated at the infection site following H. somni challenge. Encapsulated P. multocida isolates not capable of forming a substantial biofilm enhanced an in vitro polymicrobial biofilm with H. somni, indicating they contributed to the polymicrobial biofilm matrix. Indirect evidence indicated that encapsulated P. multocida also contributed to a polymicrobial biofilm in vivo. Only the EPS of H. somni could be detected by FTL staining of bovine tissues following challenge with H. somni. However, both species were isolated and an immune response to the biofilm matrix of both species was greater than the response to planktonic cells, suggesting encapsulated P. multocida may take advantage of the H. somni biofilm to persist in the host during chronic BRD. These results may have important implications for the management and prevention of BRD.
Introduction
Bovine respiratory disease (BRD) complex causes significant economic losses to the beef industry through increased treatment costs, reduced carcass value due to treatment and prevention measures, morbidity, and mortality (Griffin, 1997; Stovall et al., 2000). BRD is a collective term describing respiratory infections from causative agents including the predominant bacterial species Histophilus somni, Pasteurella multocida, Mannheimia haemolytica, and Mycoplasma bovis, as well as several viruses (Griffin et al., 2010). It is common to detect more than one causative agent during an outbreak (Gershwin et al., 2005; Booker et al., 2008; Agnes et al., 2013), suggesting BRD is often polymicrobial.
A definitive diagnosis of BRD can only be made after post-mortem examination, resulting in broad, poorly defined diagnostic criteria (Taylor et al., 2010). Signs of disease can go undetected, and include a lack of appetite, nasal discharge, coughing, rapid breathing, fever, and diarrhea. Polymicrobial infections can display a more complex pathology than single species infections (Sibley et al., 2008; Burmølle et al., 2014; Murray et al., 2014), and are more resistant to antibiotics (Perez et al., 2014). Cattle are predisposed to disease after experiencing stress, including situations such as weaning, excessive handling, a change in diet, and transportation or exposure to new locations and/or herds (Snowder et al., 2006; Holman et al., 2017). Genetic factors may also predispose cattle to disease (Snowder et al., 2006). It is generally accepted that transmission occurs via contaminated aerosols in locations with limited ventilation such as during transportation, in auction houses or crowded barns, or after a change in climate or location.
All of the primary bacterial agents responsible for BRD have been demonstrated to produce biofilms (Sandal et al., 2007; Simmons and Dybvig, 2009; Boukahil and Czuprynski, 2015; Petruzzi et al., 2017), which may be associated with their pathogenic role during BRD. H. somni biofilm formation has been directly associated with BRD (Sandal et al., 2009). Bacterial populations within a biofilm are pleural in respect to their phenotype and genotype (Ehrlich et al., 2005), and therefore the study of bacteria in a monophasic system (e.g., a planktonic log phase culture) may be highly irrelevant to their status in vivo. Prior experimental bovine respiratory challenge showed that during chronic infection H. somni exists as a biofilm in the cardiopulmonary tissue of calves raised from birth and in isolation (Sandal et al., 2009). However, P. multocida was also isolated from these biofilm samples, as well as from other H. somni respiratory challenge experiments (Gogolewski et al., 1987a, a; Elswaifi et al., 2012; Murray et al., 2017). BRD research over the past 4 decades has aided in the reduction of disease incidence, provided a better understanding of disease pathogenesis and the immune response, aided in the detection of new infectious agents, and has promoted the implementation of preventative techniques by veterinarians. However, despite such progress, BRD is still the largest cause of morbidity and mortality in the beef industry and current vaccines offer little relief, indicating the need for further research (Griffin, 1997; Fulton, 2009; Taylor et al., 2010).
The work described in this report aims to characterize the interactive polymicrobial relationship between the BRD pathogens H. somni and P. multocida during biofilm formation. Confocal laser scanning microscopy (CLSM) of fluorescently labeled components enabled each species to be observed simultaneously within the biofilm matrix, providing a detailed understanding of their polymicrobial relationship. Additionally, data obtained from an experimental infection of calves supported the co-habitation of H. somni and P. multocida in polymicrobial biofilms in vivo. The information presented here further supports the role of biofilm as an important phase of BRD, provides evidence that BRD can be a complex polymicrobial biofilm disease, and supports the need for treatment and preventative measures targeting biofilm formation.
Materials and Methods
Bacterial Growth
Histophilus somni strain 2336, Pasteurella multocida strain C0513, and capsule-deficient variant C0513-P5 (isolated after 5 serial passages of a single colony of C0513 to obtain a rough colony variant) were used (Petruzzi et al., 2017). H. somni was grown in Brain Heart Infusion (BHI) broth (BD Scientific; Irvine, CA, United States) containing 0.5% yeast extract (BD Scientific; Irvine, CA, United States), 0.1% Trizma base, 0.01% thiamine monophosphate (TMP) (Sigma-Aldrich, St. Louis, MO, United States), and 1% bovine serum at 37°C with rapid shaking until growth reached 109 colony forming units (CFU)/ml (determined by spectrophotometric reading and confirmed by viable plate count). P. multocida strains C0513 and C0513-P5 were grown in BHI broth alone, incubated and shaken as above to the same density. Biofilms were grown in 50-mL conical tubes using the same medium at 37°C with rotation at 50 rpm or less. H. somni biofilms were grown for approximately 3 days (Sandal et al., 2011) before the addition of P. multocida to obtain polymicrobial biofilm growth. After the addition of P. multocida, biofilm incubations were continued for an additional 2 days to enable the P. multocida biofilm to reach maturity (Petruzzi et al., 2017). Single species biofilms were grown for 5 days for H. somni, and 2 days for P. multocida due to the difference in growth rate of the two species.
Fluorescence in situ Hybridization
Fluorescence in situ hybridization (F.I.S.H.) was performed using the following 16S rRNA probes: ′/5FluorT/TT AAG AGA TTA ATT GAT TGA′ to detect H. somni, and ′/5Cy5/CT ATT TAA CAA CAT CCC TTC′ to detect P. multocida. DNA-specific probes were purchased from Integrated DNA Technologies Inc. (Coralville, IA, United States), and confirmed to be species-specific using a BLAST search and through experimental hybridization with homologous and heterologous bacterial species. F.I.S.H. was carried out as described (Thurnheer et al., 2004), with the following modifications. The hybridization buffer determined to optimize attachment of both probes contained 25% formamide. Biofilms were grown on glass coverslips and fixed with 4% paraformaldehyde for at least 60 min. Biofilms were incubated with hybridization buffer for at least 15 min at 37°C. After pre-incubation, 0.2 μg of each probe was added to the fixed biofilm and incubated overnight at 37°C. Biofilms were then washed 1× with wash buffer, then rinsed thoroughly with water. Glass coverslips and slides were embedded biofilm-side down with 20 μl of polyvinyl alcohol (Mowiol® 4–88; Millipore Sigma-Aldrich, St. Louis, MO, United States) onto glass slides, and stored in the dark. CLSM was performed on a Zeiss 880 confocal laser scanning microscope (Zeiss; Oberkochen, Germany) at 40× magnification using Cy5 (deep red, 660–710 nm) and fluorescein (green, 500–540 nm) filters.
Fluorescence Lectin Staining of the Biofilm Exopolysaccharide
Fluorescein isothiocyanate (FITC)-conjugated Griffonia simplicifolia lectin (GS-II) (EY laboratories; San Mateo, CA, United States) was used to detect glycogen EPS (Hennigar et al., 1986, 1987) produced by P. multocida (Petruzzi et al., 2017). Tetramethylrhodamine (TRITC)-conjugated Moringa M Lectin (MNA-M) (EY laboratories) was used to detect the H. somni galactomannan EPS (Sandal et al., 2009). Lectins were suspended in 10 mM phosphate buffer, pH 7.5, at a concentration of 10 μg/ml. Fifty microliters of lectin solution was applied to the biofilms and incubated 30–60 min in the dark at room temperature, then washed with phosphate buffer four times. Coverslips were fixed onto slides with 20 μl of polyvinyl alcohol and stored in the dark for at least 6 h before imaging. CSLM was performed on a Zeiss LSM 880 CSLM (Zeiss) at 40× magnification using TRITC (red, 532 nm) and fluorescein (green, 500–540 nm) filters.
Polymerase Chain Reaction
The primers specific to the P. multocida and H. somni 16S rRNA gene that were used in this study are described elsewhere (Angen et al., 1998; Mbuthia et al., 2001). Isolated colonies were suspended in sterile water and boiled for 10 min. Unbroken cells were removed by centrifugation at 12000 rpm for 5 min, and the supernatant used as a DNA template for PCR amplification. DNA was extracted from sections of bovine lung tissue using the Blood and Tissue DNeasy Kit (Qiagen) according to manufacturer’s instructions. PCR was performed on extracted DNA material using OneTaq Kit (NEB), according to manufacturer’s instructions.
Enzyme-Linked Immunosorbent Assay
Enzyme-linked immunosorbent assays (ELISA) were performed to determine the antibody response to H. somni whole cells, P. multocida whole cells, or crude single species biofilm material. Biofilms were grown for 5 days, most of the fluid aspirated, the matrix material suspended in distilled water and lyophilized, and each sample resuspended in sterile saline to 1 mg/ml. Twenty micrograms of this biofilm material per well were used to coat the surface of Immulon 4HBX plates (Thermo Fisher Scientific). Bacteria were grown to 109 CFU/ml (mid-log phase), then resuspended to the same concentration in carbonate coating buffer, pH 9.6 (15 mM Na2CO3, 35 mM NaHCO3, 3 mM NaN3). Wells were coated with 108 CFU of whole cells. Phosphate buffered saline (PBS) supplemented with 0.05% Tween 20 (Sigma) and 2% non-fat dry milk (Kroger) was used as a blocking agent. All washes between each step (5× each) were carried out with PBS supplemented with 0.05% Tween-20. The primary antibody used was calf serum that was separated from whole blood at the time of euthanasia. The secondary antibody used was rabbit IgG to bovine heavy & light chain conjugated to horseradish peroxidase (Jackson laboratory; Bar Harbor, ME, United States). TMB substrate (Pierce Biotechnology; Rockford, IL, United States) was used for color development, per the manufacturer’s instructions. Control wells included all components except the initial biofilm antigen.
Biofilm Protein and Carbohydrate Concentrations
The protein and carbohydrate content of the biofilms was determined using the BCA assay (Thermo Fisher Scientific) and the Anthrone assay (Yemm and Willis, 1954; Laurentin and Edwards, 2003), respectively. Equal concentrations of biofilm material were suspended in 1 ml of PBS, and vortexed rapidly for several seconds to break up the biofilm matrix prior to assay.
Auto-Aggregation of Single Species and Polymicrobial Suspensions
Auto-aggregation was performed as previously described (Sorroche et al., 2012) with modifications. Bacterial cells from P. multocida C0513, C0513-P5, H. somni 2336, or in combination were suspended in sterile PBS with or without supplementation with 100 μg/ml of H. somni crude biofilm material. OD562 measurements were taken prior to the start of the experiment, after 1 h, and after 24 h. The initial OD562 was between 0.8 and 1.0. The auto-aggregation percentage at each time period was calculated from [1 - (ODfinal/ODinitial)] × 100.
Determination of Polymicrobial Biofilm Formation in vivo
Two, eight-week old, male Holstein calves (#65 and 66) were challenged with 109 CFU of H. somni strain H.S.25 in 10 ml sterile PBS containing 0.15 mM CaCl2 and 0.5 mM MgCl2 (PCM) intratracheally using a syringe (6–12 cc) and a Tom Cat/Small Animal Catheter (3.5 Fr, 5.5 inches long). Calves were monitored daily for signs of respiratory infection. Twenty-two days after challenge, calves were humanely euthanized, and the lungs were harvested for further analysis. Bacterial load within the pulmonary tissue was determined by manually breaking the lung tissue in sterile PCM buffer. The resulting digest was serially diluted. Dilutions were spread onto Columbia blood agar and incubated for 72 h at 37°C in 6% CO2. The presence of H. somni and P. multocida was determined by colony morphology upon isolation and subsequent PCR of isolated colonies and lung tissue. Tissue sections were embedded in paraffin and cut into 5 μm sections on glass slides. The slides were deparaffinated and stained by F.I.S.H. and fluorescently tagged lectins as described above. Histology was carried out on sections of paraffin-fixed tissue that were stained with hematoxylin and eosin.
RNA-Sequencing (RNA-Seq)
RNA was isolated from bacterial biofilms in triplicate on 3 separate occasions using Qiagen RNA Protect bacterial reagent, Qiagen QiaShredder, and Qiagen RNeasy kits (Qiagen; Hilden, Germany) according to manufacturer’s instructions for prokaryotic RNA. The isolated RNA was immediately transcribed into cDNA using the Quanta qScript kit (Quanta Biosciences; Gaithersburg, MD, United States) following the manufacturer’s instructions.
Lung samples collected at the time of euthanasia were stored in RNAlater RNA storage reagent (Qiagen; Hilden, Germany) for extraction of host RNA. Lung samples collected at the time of euthanasia were also stored in both liquid nitrogen and 10% buffered formalin for further analysis. Twenty 50-mg sections of lung were homogenized in 900 μl TriZol (Thermo Fisher Scientific; Waltham, MA, United States), and stored at room temperature for several minutes. Chloroform (180 μl) was then added to the homogenate, and vortexed vigorously. After 3 min at room temperature, the homogenate solutions were centrifuged at 12,000 × g for 15 min at 4°C. The aqueous phase was transferred to a clean tube containing an equal volume of 70% ethanol, and the RNA extracted with the Qiagen RNeasy kit (Qiagen; Hilden, Germany). The purity of the RNA was determined by the A260/280 and A260/230 ratios using nanodrop (NanoDrop; Wilmington, DE, United States). Fifty ng of pure RNA was transcribed into cDNA using the Quanta Biosciences qScriptTM XLT cDNA Supermix kit (Quanta Biosciences; Gaithersburg, MD, United States) according to the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR) of the 16S rRNA gene was used as a control for RNA-Seq. qRT-PCR was performed using the Quanta Biosciences PerfeCta® SYBR® Green FastMix, ROXTM (Quanta Biosciences; Gaithersburg, MD, United States) according to the manufacturer’s instructions on an Applied Biosciences 7300 Real-Time PCR system (Applied Biosciences; Foster City, CA, United States). Two oligonucleotide probes specific for the 16S rRNA gene of H. somni (5′-TTCGGGCACCAAGT(A/G) TTCA-3′) (Angen et al., 1998) and P. multocida (5′-ACGAGACTCTAGACTCCC-3′) (Mbuthia et al., 2001), respectively, were obtained from MWG-BIOTECH AG, Ebensburg, Germany and were labeled with the isothiocyanate derivative Cy3. Quantitative RT-PCR was performed in 20 μl reactions using 50 ng cDNA template and 300 mM of each primer. Genetic data were annotated using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.81.
The original RNA-Seq data presented in this study are publicly available. These data can be found at the following NCBI SRA site2.
Statistical Analysis
Analysis of polymicrobial biofilms was performed on z-stack images obtained by CLSM. z-stacks were analyzed using COMSTAT coded files (Heydorn et al., 2000) through MATLAB software (MathWorks; Natick, MA, United States). All measured experiments were performed in triplicate in three different experiments on different days (9 datum points total). Statistical analysis was performed using GraphPad Prism software version 6.01 (GraphPad; La Jolla, CA, United States). Statistical significance was considered if p-values were ≤0.05.
Results
Fluorescence in situ Hybridization (F.I.S.H.) of Polymicrobial Biofilms Using DNA-Specific Probes
Fluorescent DNA probes specific for the 16S rRNA region of H. somni or P. multocida DNA were used to determine the spatial arrangement of P. multocida within an established H. somni biofilm. After 48 h, P. multocida integrated evenly throughout the H. somni biofilm (Figures 1A–C). Large micro-colonies of P. multocida were not visible, which has been reported for other polymicrobial bacterial biofilms (Zainal-Abidin et al., 2012; Zhu et al., 2013). The H. somni biofilm continued to mature after the addition of P. multocida over the monitored 48-h period, indicated by increased green fluorescence between time points (Table 1, Figure 2A). COMSTAT analysis was used to determine the three-dimensional spatial occupation (biomass) of the DNA from each species, as well as the two dimensional thickness of the DNA occupied by each species (Table 1). Thickness was used as a determination of the depth to which P. multocida had integrated into the established biofilm matrix.
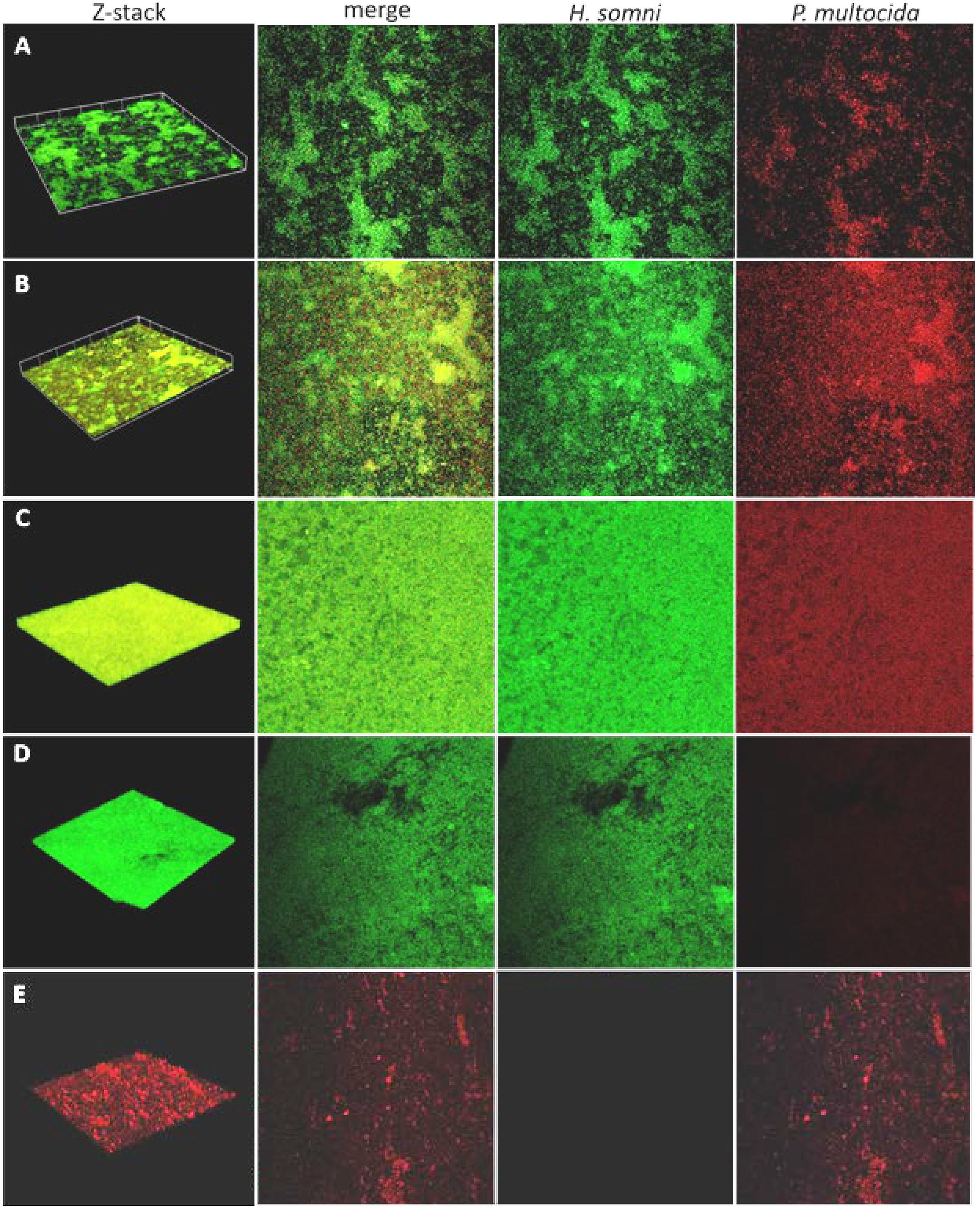
Figure 1. Fluorescence in situ hybridization (FISH) of polymicrobial and single species biofilms. Biofilms were imaged over a 48-h period by CLSM after FISH using probes labeled with fluorescein for H. somni or Cy5 for P. multocida. (A–C), 6-h, 12-h, and 48-h time points, respectively, for biofilms containing H. somni strain 2336 and P. multocida strain C0513-P5. Each column shows the 3-D z-stack, a merged view of images with each probe, the probe for only H. somni strain 2336, and the probe for only P. multocida strain C0513-P5. (D) Single species biofilm of H. somni strain 2336 after 48 h showing in each column the 3-D z-stack, the merged view of images with each probe, the probe for only H. somni strain 2336, and the probe for only P. multocida strain C0513-P5. (E) A single species biofilm of P. multocida strain C0513 after 48 h showing in each column the 3D z-stack, the merged view of images with each probe, the probe for only H. somni strain 2336, and the probe for only P. multocida strain C0513-P5.
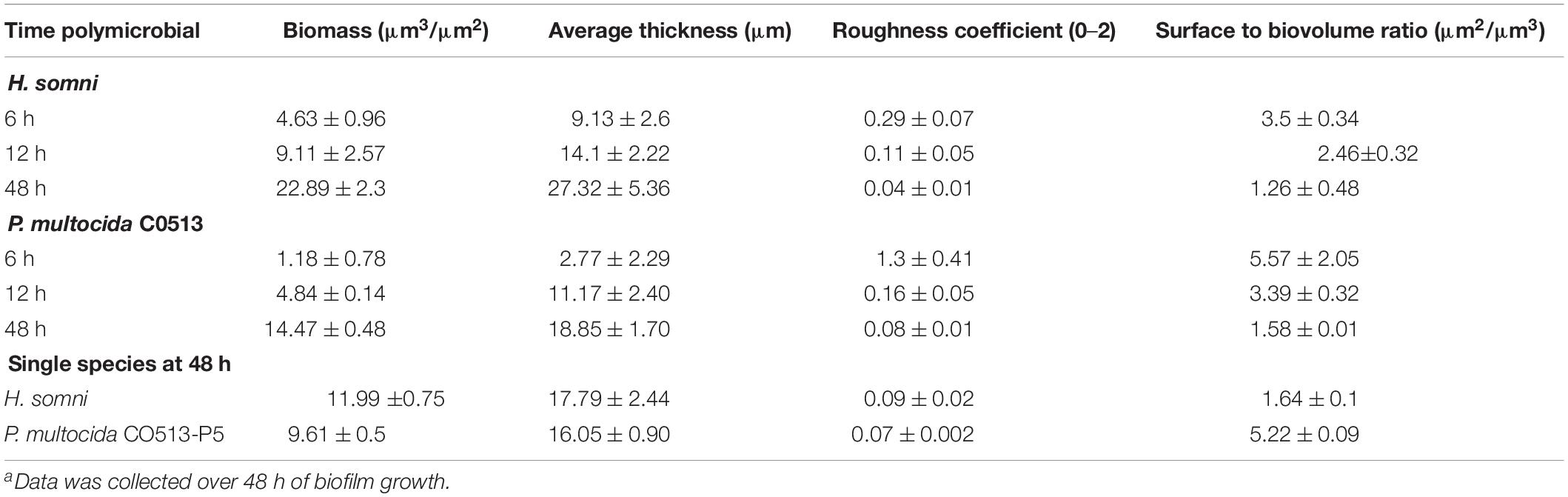
Table 1. COMSTAT analysis of fluorescence in situ hybridization of polymicrobial and single species biofilms.a
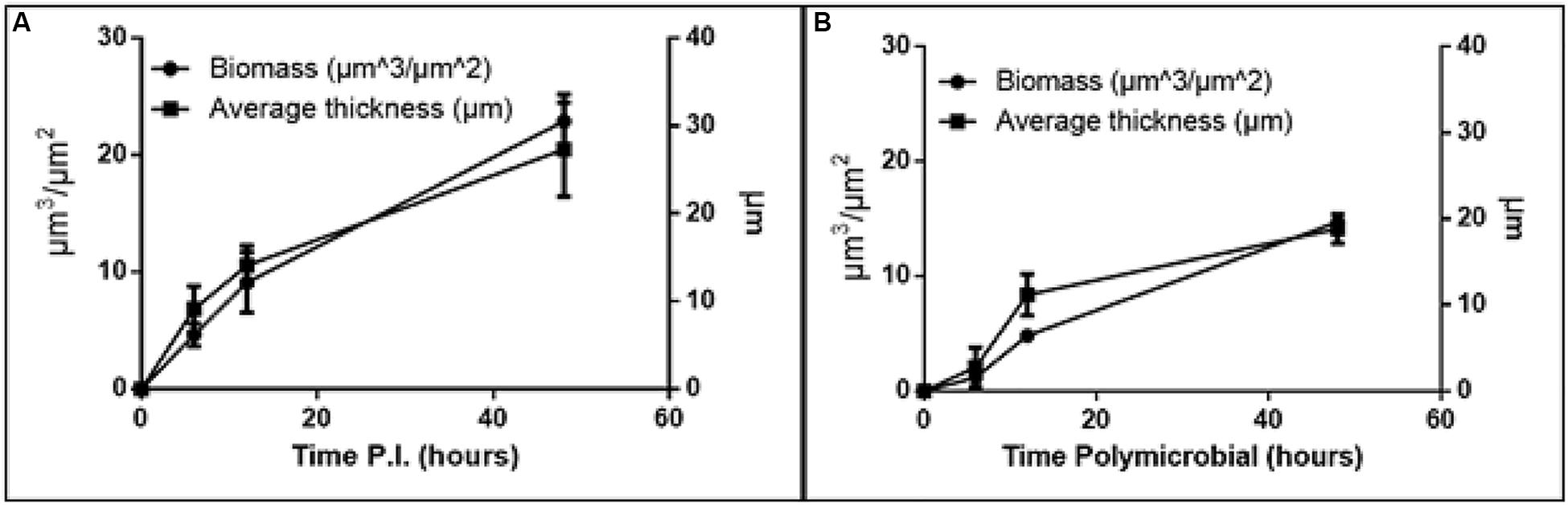
Figure 2. Increase in biomass and thickness within the polymicrobial biofilm. (A) H. somni strain 2336 biomass and thickness within the polymicrobial biofilm. (B) P. multocida strain C0513-P5 biomass and thickness within the polymicrobial biofilm. All experiments were performed in triplicate in three different experiments (9 datum points total).
The H. somni biomass increased from 4 ± 1 μm3/μm2 at 6 h to 22 ± 2 μm3/μm2 at 48 h (Table 1, Figure 2A). The average thickness of the H. somni biomass also increased over this 48 h period. The roughness coefficient is an arbitrary value used to determine the topography of the biofilm surface, which remained relatively constant throughout the experiment (Table 1). Therefore, the addition of P. multocida did not significantly alter biofilm architecture. The surface to bio-volume ratio decreased over the 48 h period, indicating biofilm growth increased (Table 1). The values obtained using the H. somni DNA probe were used to determine the total thickness of the biofilm, while the total biomass was determined by the addition of the H. somni and P. multocida biomass values.
Six hours after its addition to the established H. somni biofilm, P. multocida attached to the H. somni biofilm, as an apparent monolayer (Figure 1A). This is indicated by a P. multocida thickness measuring 2.77 ± 2.29 μm. The P. multocida biomass was determined to be 1.18 ± 0.78 μm3/μm2 at 6 h. However, at the 48 h polymicrobial time point, P. multocida had expanded to a thickness of 18.85 ± 1.7 μm and a biomass of 14.76 ± 0.48 μm3/μm2 (Table 1, Figures 1C, 2B), indicating that P. multocida had incorporated into 69% of the total H. somni biofilm matrix and contributed to 39% of the total biomass. Over the 48 h period, the roughness coefficient of the P. multocida biofilm contribution decreased to values similar to the H. somni roughness coefficient (Table 1), indicating that P. multocida conformed to the H. somni biofilm structure. Despite their apparent cooperation, the polymicrobial biofilm did not appear to provide a growth advantage to either individual species. The biomass and thickness values for the polymicrobial biofilm (indicated by the values provided for H. somni within the polymicrobial biofilm) were not significantly different than the addition of the values for each single species biofilm (Table 1, Figures 1D,E).
Fluorescent-Tagged Lectin Staining of Biofilm EPS
Fluorescent lectins were used to determine the presence of EPS from each species in the in vitro polymicrobial biofilm. Galactomannan EPS from H. somni and glycogen EPS from P. multocida was detected in the polymicrobial biofilm with FITC-conjugated GS-II and TRITC-conjugated MNA-M, respectively. Glycogen EPS was detected in the polymicrobial biofilm for both P. multocida strains tested: biofilm-deficient strain C0513 and biofilm-proficient passed variant C0513-P5 (Figures 3C,D).
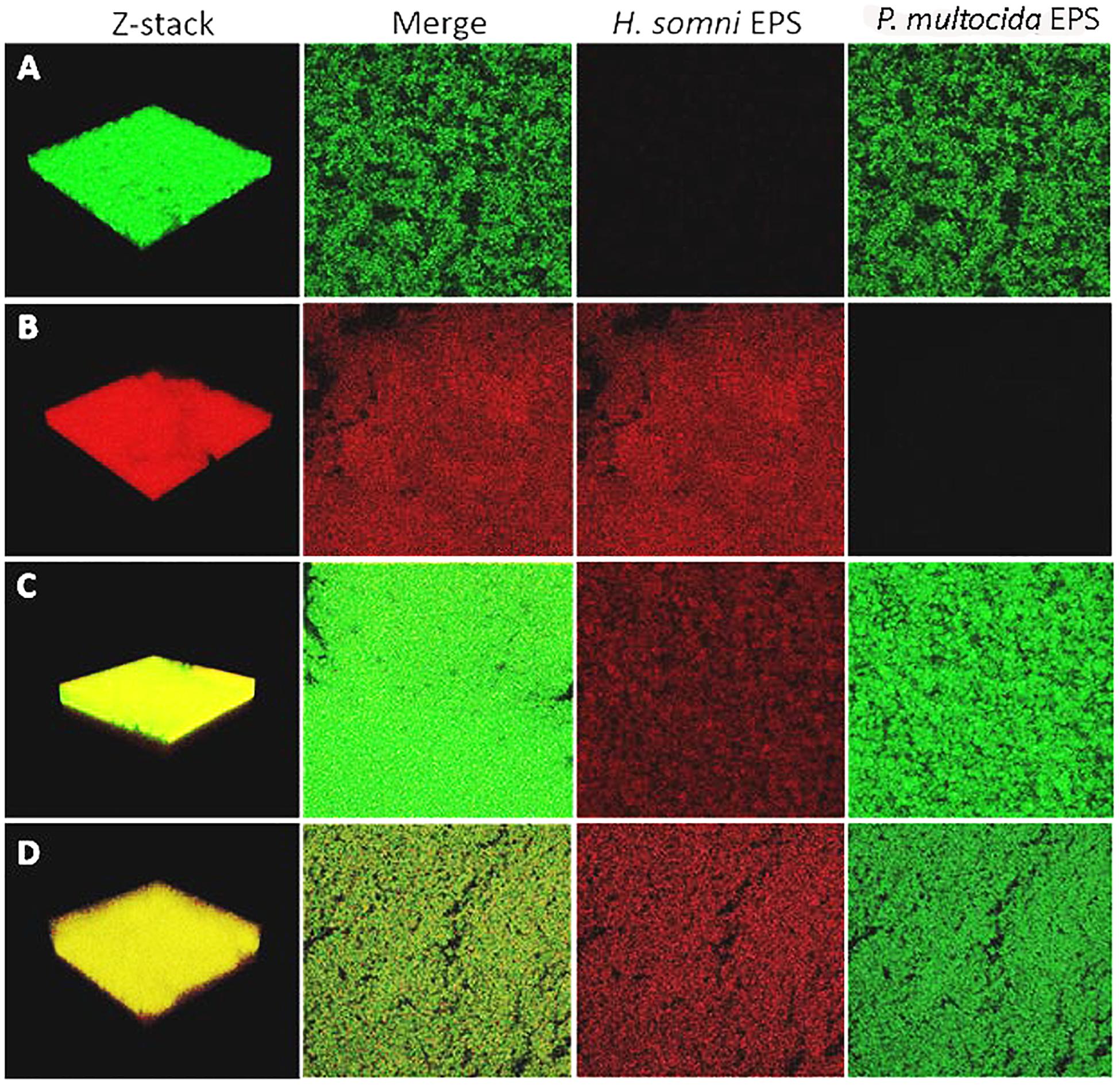
Figure 3. Fluorescently tagged lectin staining of in vitro biofilm EPS. Moringa M labeled with TRITC was used as the specific lectin for the galactomannan EPS from H. somni, and Griffonia simplicifolia labeled with FITC was used as the specific lectin for the glycogen EPS of P. multocida. (A) P. multocida strain C0513-P5 single species biofilm Z-stack, the merged view of images with each lectin, H. somni strain 2336 EPS, P. multocida strain C0513-P5 EPS. (B) H. somni strain 2336 single species biofilm Z-stack, the merged view of images with each lectin, H. somni strain 2336 EPS, P. multocida strain C0513-P5 EPS. (C) Polymicrobial biofilm with P. multocida C0513-P5 Z-stack, the merged view of images with each lectin, H. somni strain 2336 EPS, P. multocida strain C0513-P5 EPS. (D) Polymicrobial biofilm with P. multocida C0513 Z-stack, the merged view of images with each lectin, H. somni strain 2336 EPS, P. multocida strain C0513 EPS.
COMSTAT analysis of polymicrobial biofilms indicated that 48 h after P. multocida C0513 incorporated into an established H. somni biofilm, the average thickness of EPS produced by C0513 in the polymicrobial biofilm was 20.73 ± 17.03 μm (Table 2). The average thickness of EPS produced by H. somni in the polymicrobial biofilm was 20.58 ± 28.07 μm, indicating variability between the spatial arrangement of EPS within the biofilm. The average biomass and roughness coefficient displayed similar variability. The surface to bio-volume ratios for EPS produced by C0513 and H. somni in the polymicrobial biofilm were 5.47 ± 3.16 and 7.21 ± 1.92, respectively. Therefore, although P. multocida strain C0513 did not produce substantial biofilm or EPS independently, this strain did produce some EPS and contribution to the polymicrobial biofilm when incorporated into the H. somni biofilm.
Polymicrobial biofilms in which passed variant strain C0513-P5 integrated into an established H. somni biofilm contained more EPS overall than what was observed for polymicrobial biofilms grown with P. multocida strain C0513 (Table 2). COMSTAT analysis 48 h after P. multocida strain C0513-P5 was incorporated into the H. somni biofilm indicated an average thickness of 31.98 ± 19.84 μm for strain C0513-P5 EPS, and an average thickness of 25.06 ± 18.54 μm for H. somni EPS. The average biomass values were also larger when strain C0513-P5 was integrated into the biofilm, and the roughness coefficient was closer to 0 than was observed for polymicrobial biofilms incorporating strain C0513.
Single species biofilms of C0513 and C0513-P5 contained less EPS than what was present in polymicrobial biofilms. The average EPS thickness and biomass for a single species C0513 biofilm was 2.31 ± 0.12 μm and 1.65 ± 0.09 μm3/μm2, respectively. The average EPS thickness and biomass for a single species C0513-P5 biofilm was 24.12 ± 0.14 μm and 19.23 ± 0.68 μm3/μm2, respectively (Figure 3A). However, the H. somni single species biofilm contained more H. somni EPS than polymicrobial biofilms (Figure 3B). The average EPS thickness and biomass for the H. somni single species biofilm was 26.2 ± 1.99 μm and 19.69 ± 1.16 μm3/μm2, respectively.
Concentrations of Protein and Carbohydrate in Polymicrobial Biofilms
Carbohydrate concentrations within the polymicrobial biofilm were determined daily over a period of 6 days (Figure 4). The carbohydrate concentration of the established single species H. somni biofilm increased slightly over time, with the greatest increase occurring between days 3 and 4 (Figure 4A). The average carbohydrate concentration of the P. multocida single species biofilm remained relatively constant throughout the duration of biofilm growth (Figure 4B). The carbohydrate concentration of the polymicrobial biofilm remained constant until day 3, when P. multocida was added to the established H. somni biofilm. At this point an increase in the amount of biofilm carbohydrate was measured, and this increase continued until the end of the experiment on day 6 (Figure 4C).
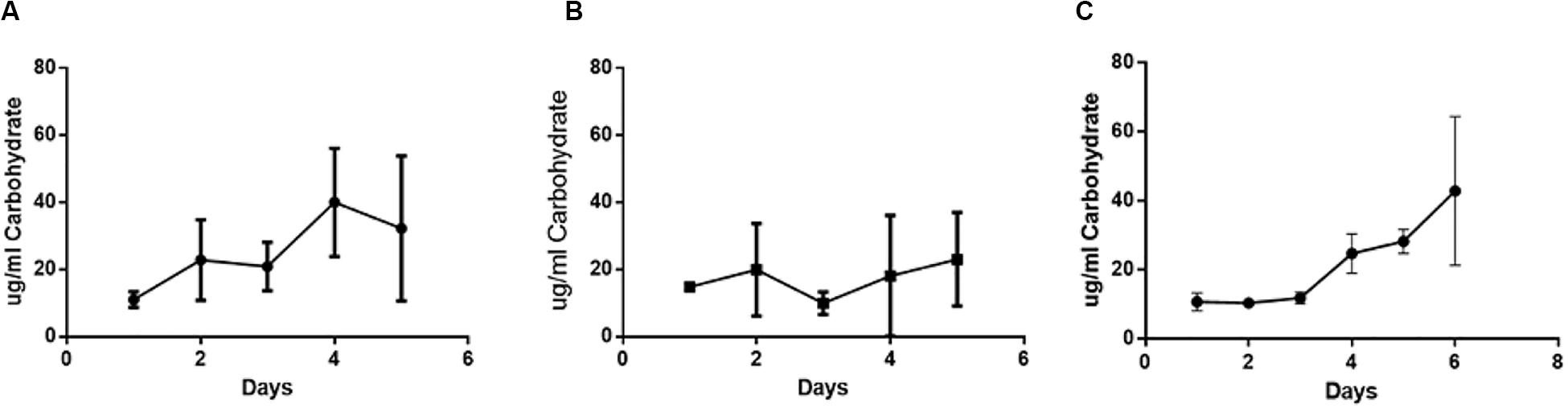
Figure 4. Changes in carbohydrate content during polymicrobial biofilm growth. (A) Single species H. somni biofilm grown for 5 days. (B) Single species P. multocida strain C0513-P5 biofilm grown for 5 days. (C) Polymicrobial biofilm of H. somni grown for 3 days, P. multocida added, and growth continued for an additional 3 days. The carbohydrate content of the biofilm was measured by Anthrone assay. All experiments were performed in triplicate in three different experiments (9 datum points total).
Protein concentration was measured daily throughout the biofilm growth period (Figure 5). During single species biofilm growth of H. somni, protein concentration increased over time until the end of the study at day 5 (Figure 5A). Protein concentration did not increase over time during single species P. multocida biofilm growth, and remained at concentrations seen during early stages of biofilm maturation (Figure 5B). Similar to the carbohydrate concentration of the polymicrobial biofilm, the protein concentration of the polymicrobial biofilm increased over time, with the most dramatic increase occurring between days 3 and 4, which coincided with the addition of P. multocida to the established H. somni biofilm on day 3 (Figure 5C).
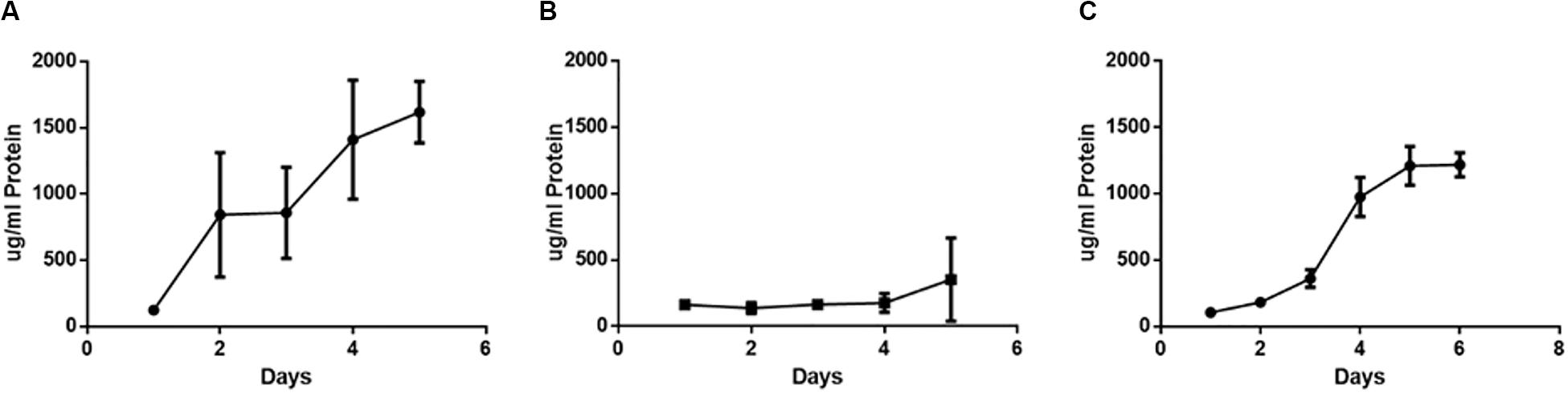
Figure 5. Changes in protein content during polymicrobial biofilm growth. (A) Single species H. somni strain 2336 biofilm grown for 5 days. (B) Single species P. multocida strain C0513-P5 biofilm grown for 5 days. (C) Polymicrobial biofilm of H. somni grown for 3 days, P. multocida added, and growth continued for an additional 3 days. The protein content of the biofilms was determined by BCA assay. All experiments were performed in triplicate in three different experiments (9 datum points total).
Auto-Aggregation of Polymicrobial Cultures
Adhesion is the first stage of biofilm formation, and auto-aggregation can be used as a measure of cellular adhesion (Felek et al., 2008; Sorroche et al., 2012; Arenas et al., 2015). In this study, auto-aggregation of polymicrobial bacterial suspensions was used to determine if interactions between the two species occurred. H. somni auto-aggregated 80% within 24 h in a single species suspension (Figure 6A), while P. multocida strains C0513-P5 and C0513 auto-aggregated significantly less: 29 and 18% within 24 h (Figures 6C,D, respectively), and was confirmed by viable plate count (data not shown). No additional auto-aggregation was observed for P. multocida or H. somni after 24 h incubation (data not shown). Polymicrobial bacterial suspensions auto-aggregated 50% when either the encapsulated (strain C0513) or capsule-deficient strain P. multocida (C0513-P5) was used (Figure 6B), suggesting that interaction occurred between H. somni and P. multocida when in a polymicrobial environment. Polymicrobial auto-aggregation experiments carried out in the presence of crudely extracted H. somni biofilm material did not display aggregation (data not shown), indicating that cells must be present to observe this effect.
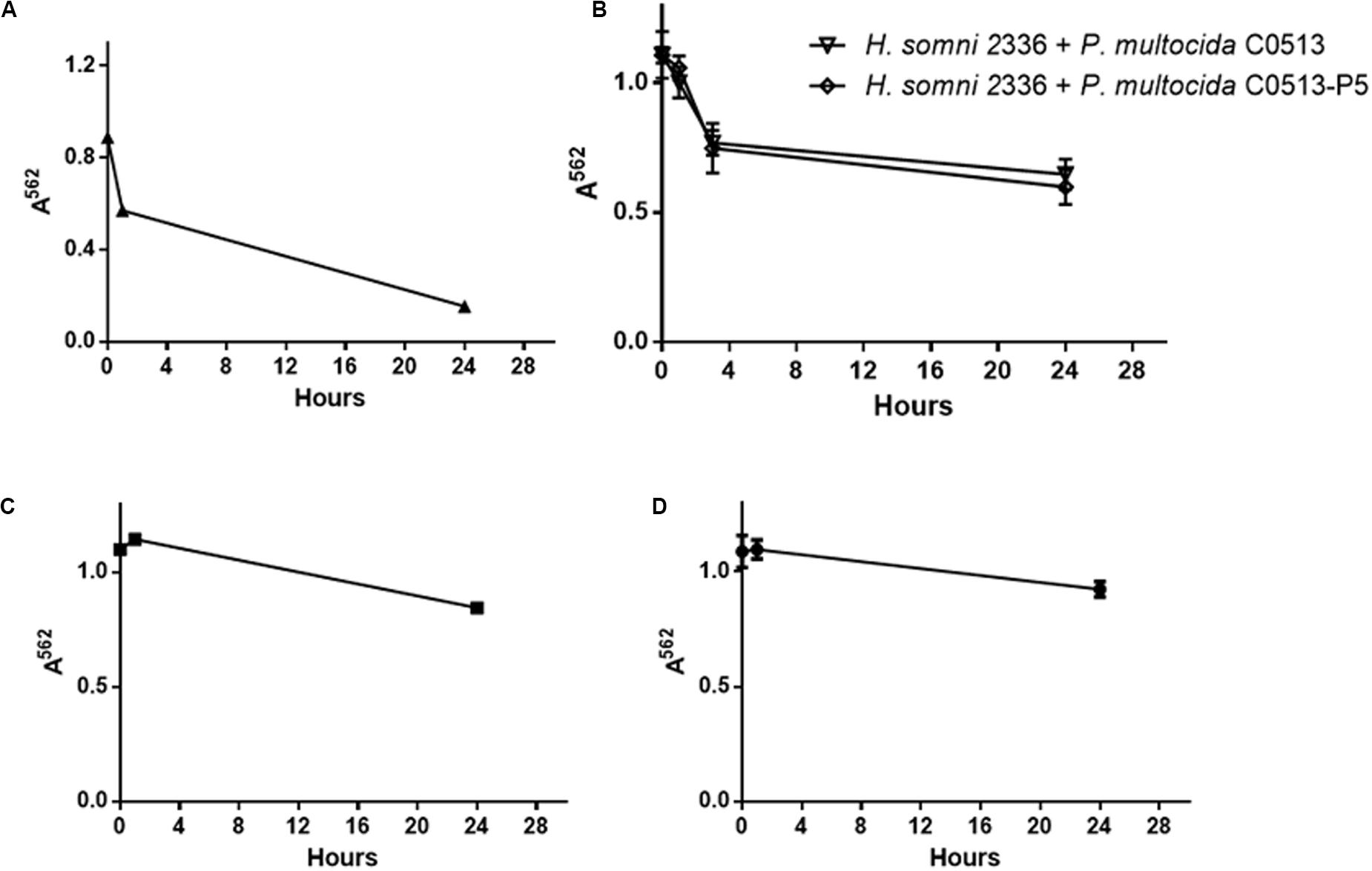
Figure 6. Auto-aggregation of polymicrobial suspensions over 24 h. (A) Auto-aggregation of H. somni strain 2336 as a single species. (B) Auto-aggregation between H. somni strain 2336 and P. multocida strain C0513 or strain C0513-P5. (C) Auto-aggregation of P. multocida strain C0513-P5. (D) Auto-aggregation of P. multocida strain C0513. All experiments were performed in triplicate in three different experiments (9 datum points total).
Polymicrobial Biofilm Formation in vivo
Lung tissue from calves experimentally infected with H. somni was analyzed for polymicrobial biofilm formation by histopathology. Previous experimental infections have shown that P. multocida can be isolated from the lower respiratory tract of calves in conjunction with H. somni following experimental challenge with H. somni alone (Gogolewski et al., 1987a, b; Elswaifi et al., 2012; Murray et al., 2017). Both H. somni and P. multocida were isolated from calf 65, but not calf 66. However, PCR revealed P. multocida and H. somni DNA present in the lungs of both calves 65 and 66. H. somni and P. multocida could not be detected by F.I.S.H. in vivo. However, fluorescent lectin staining did demonstrate the presence of biofilm matrix-specific EPS produced by H. somni in calf 65, but not P. multocida EPS (Figure 7).
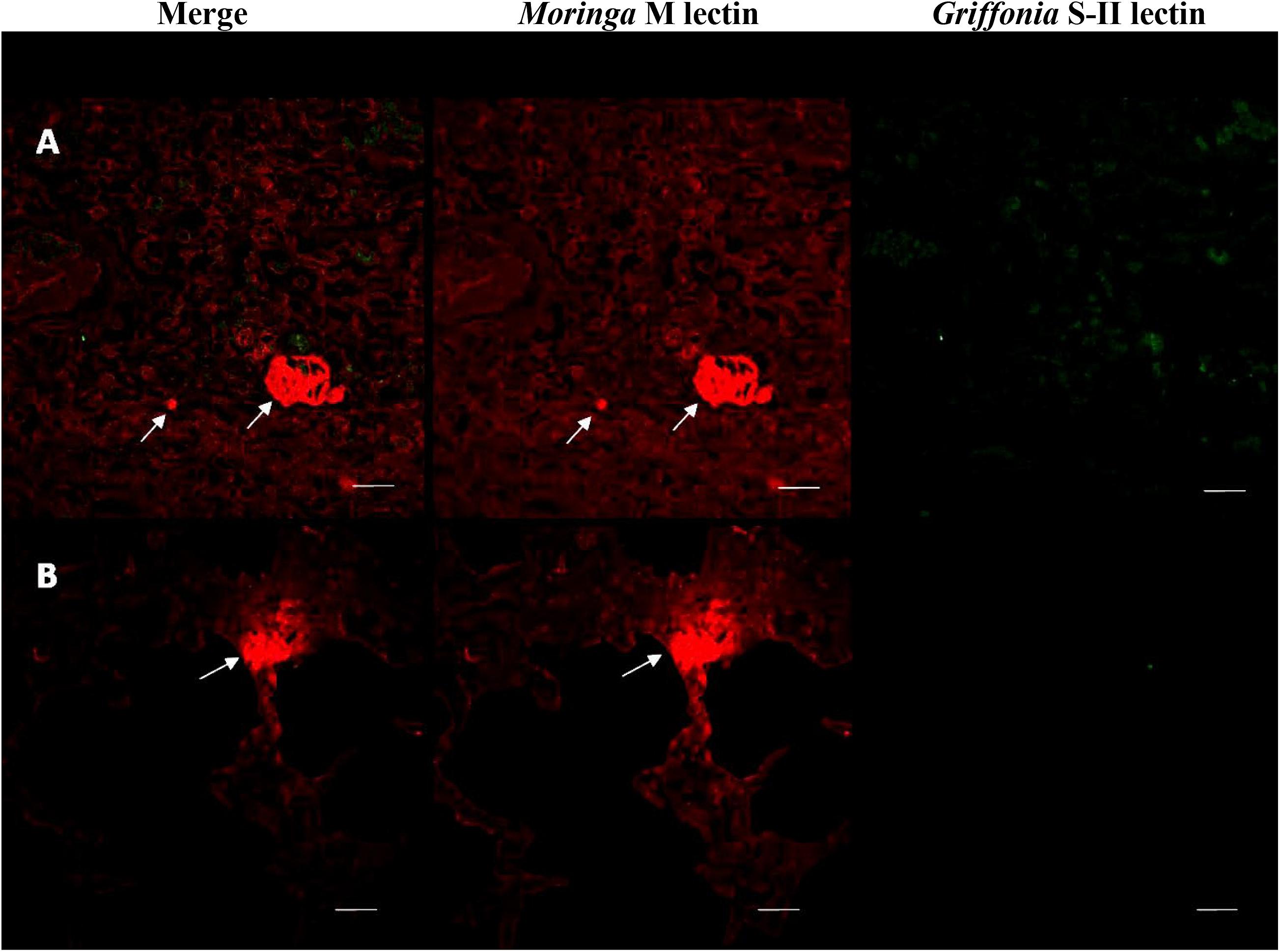
Figure 7. Fluorescently tagged lectin staining of in vivo biofilm EPS. Lung tissue sections from calf 65 stained with fluorescently tagged lectins. (A,B) Two separate samples showing merged images of images using each lectin, section stained with Moringa M lectin labeled with TRITC for EPS from H. somni only, section stained with Griffonia simplicifolia labeled with FITC for EPS from P. multocida only. The H. somni EPS is only made during biofilm formation indicating the presence of a biofilm in these sections. Scale bar is equal to 20 um.
Histological Analysis of Polymicrobial Respiratory Disease
Tissue sections from calf 65 and 66 were observed for histological analysis. In both calves the lungs appeared slightly inflamed and partially collapsed. Sections of lung contained pleural fibrosis. Neutrophils and macrophages were seen in lung airways, and lymphocytes were present at the lung periphery. Acute pulmonary edema was present in the interlobular septa, and bronchioles displayed minor hemorrhaging (data not shown).
Enzyme-Linked Immunosorbent Assay
Calves 65 and 66 produced an antibody titer to H. somni planktonic cells of 1:20. However, both calves produced an antibody titer of 1:800 to the H. somni biofilm. Both calves produced an antibody titer to P. multocida planktonic cells of 1:1600, and a titer of 1:3200 to the P. multocida biofilm. There was no antibody titer to P. multocida or H. somni detected prior to challenge with H. somni.
RNA-Seq Analysis
Previous RNA-Seq analysis of H. somni indicated that up to half of the bacterial genes are significantly differentially expressed when the bacteria are grown in a biofilm, compared to planktonic growth (Petruzzi et al., 2015). Such differential regulation was also observed with P. multocida when grown as a polymicrobial biofilm compared to a single species biofilm. A total of 282 genes were differentially regulated when P. multocida was grown as a biofilm together with H. somni compared to growth as a mono-species biofilm. Examples of some genes that were significantly upregulated are shown in Table 3. Many of these genes encode for proteins involved in iron acquisition or outer membrane composition, including lipopolysaccharide (LPS).
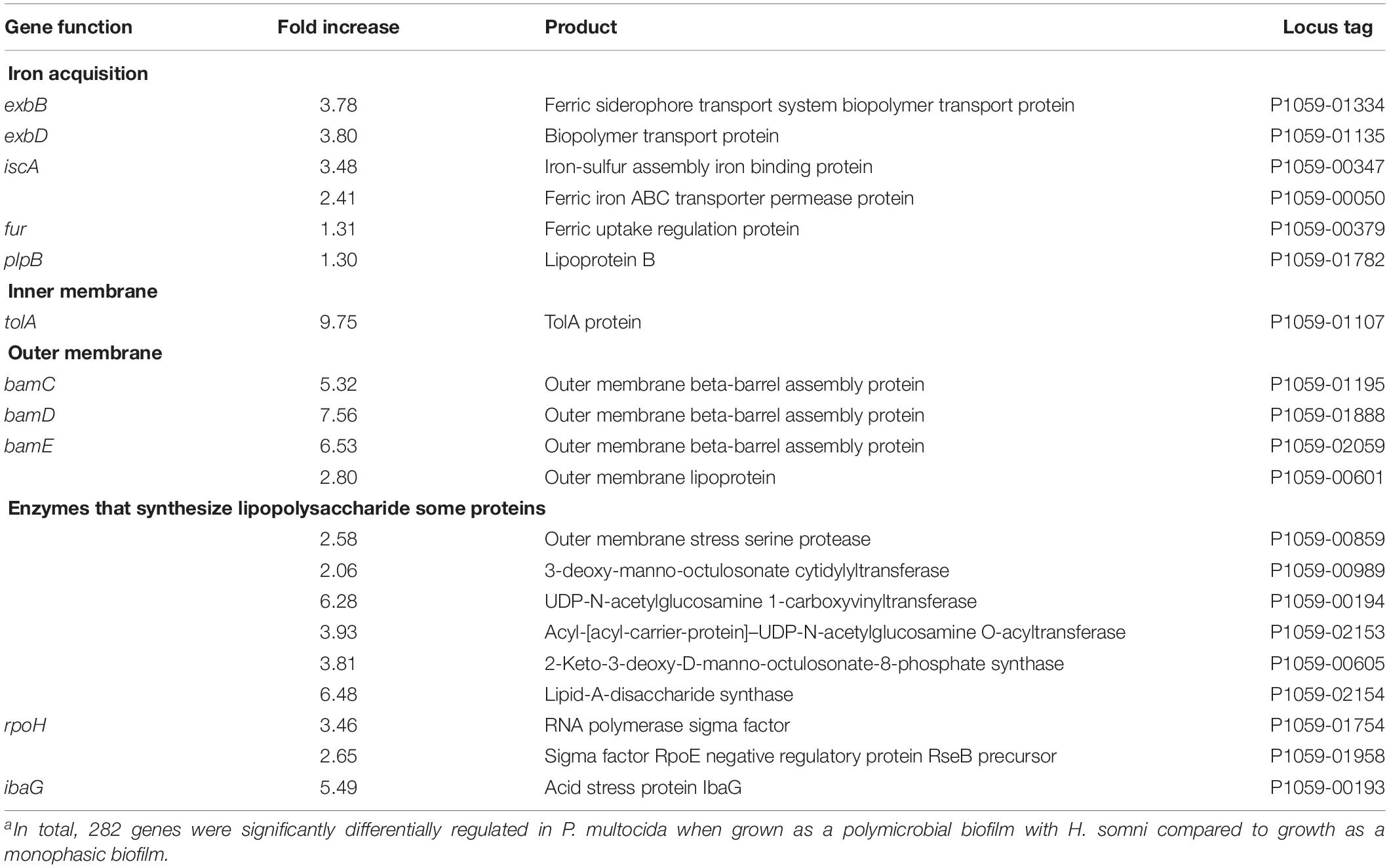
Table 3. Examples of genes significantly differentially regulated when P. multocida was grown in a poly-microbial biofilm with H. somni compared to a monophasic biofilm.a
The expression of host genes at the infected site were compared to the same genes at normal tissue sites of the same lung. Ninety-three genes were expressed greater than 2.5-fold at the infected site, most of which are involved in inflammation and the host immune response. A partial list of the gene ontology (GO) terms and these genes is listed in Table 4. Genes that were upregulated most significantly were those encoding for cytokines and chemokines that contribute to inflammation and the host immune response, chemotaxis and differentiation of phagocytic cells, and other related cellular catabolic processes.

Table 4. Gene Ontology (GO) terms enriched among 93 genes expressed >2.5 fold higher in infected site.a
Discussion
Despite available vaccines, BRD is still the leading cause of morbidity and mortality in the North American beef and dairy industries. Research over the past 40 years has aided our understanding of the transmission and pathogenesis of the causative agents of BRD, but the disease and improved vaccines and treatments remain in need (Griffin, 1997; Stovall et al., 2000). BRD is a multifactorial disease, and diagnosis is often complicated by the presence of several potential etiologic agents, many of whom are opportunistic pathogens. The 4 most common bacterial agents of BRD (H. somni, P. multocida, M. haemolytica, and M. bovis) have been shown to be capable of forming biofilms (Sandal et al., 2007; Simmons and Dybvig, 2009; Boukahil and Czuprynski, 2015; Petruzzi et al., 2017). Some species, such as H. somni, have been documented to also form biofilms in cardiopulmonary tissue during BRD (Sandal et al., 2009; O’Toole and Sondgeroth, 2016; O’Toole et al., 2017). In order to further understand the potential role of biofilms in BRD, we have characterized the polymicrobial relationship between two of these important BRD agents: H. somni and P. multocida.
The adherence of bacteria to a framework or scaffolding, such as a biofilm matrix that is assembled by different bacterial species or by the host, is known as coadhesion (Bos et al., 1994; Rickard et al., 2003). Preliminary studies in our lab indicated that the ideal conditions for polymicrobial biofilm formation between H. somni and P. multocida were created by allowing H. somni to establish a preliminary biofilm before the addition of the secondary colonizer, P. multocida. This is consistent with experimental results obtained with other bacterial agents that formed coadhesive polymicrobial biofilms (Bos et al., 1994; Rickard et al., 2003; Castonguay et al., 2006; Karched et al., 2015). P. multocida was not inhibitory to H. somni; but grew much faster and if growth of the two species was initiated at the same time, P. multocida simply over-grew H. somni.
Using labeled DNA probes the spatial arrangement of the bacterial species within the polymicrobial and single species biofilms could be observed, and allowed the monitoring of bacterial growth over time. Antibodies raised against H. somni or P. multocida surface antigens displayed cross reactivity, limiting the microscopic and fluorescent techniques available for discernable observation (Pan et al., 2014). The use of specific DNA probes allowed for the distinct identification of DNA (and hence the cells) from each species simultaneously (Thurnheer et al., 2004). Although we cannot negate the possibility that 16S rRNA DNA was present in the extracellular matrix, we presume that the DNA probes detected predominately, or only cellular-associated, DNA (Okshevsky and Meyer, 2015). Over a 48-h period of polymicrobial growth, the DNA present from both species increased and did not appear to be separated into large micro-colonies or separate biofilms, indicating an integrated polymicrobial biofilm had formed.
The polymicrobial relationship of the two species was further confirmed using fluorescently tagged lectins specific for the EPS produced by H. somni or P. multocida. Interestingly, the use of lectins provided insight into the glycogen synthesis of a P. multocida capsule-producing bovine-associated strain: C0513. This strain did not produce a substantial single-species biofilm. However, once incorporated into the H. somni biofilm matrix in vitro, it began producing EPS and contributing to the overall biofilm matrix. These results indicate that encapsulated P. multocida that cannot efficiently produce a biofilm on its own can contribute to biofilm formation in the H. somni biofilm, which may be advantageous to both species. The capsule-deficient passed variant of this strain, C0513-P5 did produce a single species biofilm, as previously reported (Petruzzi et al., 2017), and produced visibly more EPS when incorporated into the H. somni biofilm matrix than in a single species biofilm. However, the amount of EPS produced by H. somni in the polymicrobial biofilm, compared to when grown as a single species, decreased. The reason for this was not determined, but may be due to a synergistic relationship with P. multocida, resulting in a decreased need for H. somni EPS (Castonguay et al., 2006).
The overall biofilm matrix carbohydrate content was determined to increase over time, further supporting that the incorporation of P. multocida into the H. somni biofilm had an additive effect on the matrix. However, there was little protein content in the single species P. multocida biofilm, and no additive protein to the polymicrobial biofilm, suggesting that, unlike most biofilms, protein may not be a major component of the P. multocida biofilm. This observation needs to be confirmed. While the majority of H. somni cells auto-aggregated within 24 h, only the minority of P. multocida cells did. Encapsulated cells auto-aggregated less than capsule-deficient cells, indicating that the capsule (likely the electrostatic negative charge of the capsule) interfered with auto-aggregation. We presume that although capsule-deficient, strain C0513-P5 also had a substantially more negative surface charge than H. somni. Polymicrobial aggregation experiments to determine interactions between H. somni and P. multocida cell surfaces showed that aggregation of the two species together was greater than the aggregation of P. multocida alone, suggesting that H. somni interacted with P. multocida to enhance the aggregation of P. multocida. However, there was no effect if only H. somni biofilm material was added, indicating cell-cell interaction was occurring. Further experimentation on species interactions is warranted.
Calves challenged with H. somni formed a polymicrobial respiratory infection with both H. somni and P. multocida, even though the calves were raised in isolation from the day of birth and challenged with only H. somni. This polymicrobial interaction has been reported in previous experimental infections (Gogolewski et al., 1987a, b; Elswaifi et al., 2012; Murray et al., 2017). Although the calves presented with only mild symptoms of respiratory disease, at necropsy histological analysis revealed that both calves had a respiratory infection consistent with experimental BRD. The lungs appeared inflamed and slightly collapsed, and contained pleural fibrosis. However, lesions were only visible in one lobe of both calves.
Lung sections taken from visible lesions were microscopically observed by the same techniques used for in vitro polymicrobial biofilms. DNA from neither H. somni nor P. multocida were detected in the lungs of calves using F.I.S.H., but DNA from both species was detected in the lungs of calves by PCR. H. somni biofilm EPS was detected in the lungs of one calf, and EPS is produced by H. somni only during biofilm formation (Sandal et al., 2007). However, EPS produced by P. multocida was not detected in the lungs of either calf. P. multocida EPS may have been masked due to the presence of capsular polysaccharide, which is required for full virulence in vivo. However, both calves produced an antibody response to crude P. multocida biofilm material, indicating the calves were exposed to biofilm matrix material produced by P. multocida during infection. These results support the in vitro results that showed encapsulated P. multocida could generate a biofilm when in the H. somni biofilm. Additionally, calves produced a weaker antibody response to P. multocida whole planktonic cells, a much weaker response to H. somni whole planktonic cells, and the most robust response to the P. multocida and H. somni biofilm matrixes. These results support that both P. multocida and H. somni were present predominately as a biofilm during chronic BRD infection.
Bacterial gene expression is highly regulated and heavily influenced by environmental factors (Balleza et al., 2009). As described previously and reported here, not only does biofilm formation alter gene expression, in comparison to planktonic growth, but growth in a polymicrobial biofilm further alters gene expression, in comparison to a mono-species biofilm. Likely as a result of competition for resources it is not surprising that many genes, such as those involved in regulation and iron acquisition, were significantly upregulated when P. multocida was grown in a biofilm with H. somni. Furthermore, outer membrane proteins play a major role in transport of molecules across the outer membrane (Koebnik et al., 2000), and the LPS plays an important role in outer membrane permeability and integrity (Bertani and Ruiz, 2018). Therefore, while not inhibiting H. somni, it appeared that P. multocida altered gene expression to obtain a competitive advantage, and to persist and contribute to the developing H. somni biofilm.
The host response at the site of infection, compared to healthy tissue, was enhanced for the expression of genes associated with inflammation and the immune response, including multiple cytokines and chemokines. Of interest was that while there was an enhanced response by TLR2, there was not a measured differential response by TLR4, indicating the response to bacterial proteins was greater than to LPS. However, because we could not control the polymicrobial nature of the biofilm in vivo we could not determine if the host response to the polymicrobial infection was different than it would have been to a mono-species infection. Biofilms associated with infections of animals are a significant problem that have been poorly studied (Clutterbuck et al., 2007). Reports of polymicrobial biofilms of veterinary importance are even less common. However, the relevance of polymicrobial biofilms to human medicine has become well established (Brogden et al., 2005; Peters et al., 2012). This is the first report of a coadhesive polymicrobial biofilm relationship associated with BRD, leading us to suspect polymicrobial biofilm interactions between other bacterial agents of BRD may also occur. Polymicrobial BRD has been reported, but the presence of biofilm was not considered (Booker et al., 2008; Hotchkiss et al., 2010; Holman et al., 2017; Murray et al., 2017). While further research to reduce losses caused by BRD has been recommended (Griffin, 1997; Fulton, 2009; Taylor et al., 2010), much of this new work should be focused on means to treat or prevent biofilm infections.
Data Availability Statement
The databases presented in this study can be found in the NCBI SRA online repository https://www.ncbi.nlm.nih.gov/sra/?term=SRR11948555.
Ethics Statement
The animal study was reviewed and approved by the Virginia Tech Institutional Animal Care and Use Committee (Approval number 16-128 CVM).
Author Contributions
BP carried out most of the experimental work and wrote and edited the manuscript. KL harvested the tissues and performed gross and histopathologic pathology of the calves. AD carried out bioinformatic analyses of the RNA-Seq results, and contributed data to the manuscript. WS challenged, monitored, and collected samples from the calves. TI designed the experiments, contributed to writing the manuscript, and did the final editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the United States Department of Agriculture-National Institute of Food and Agriculture grant 2013-67015-21314 to TI, and by the Virginia Tech College of Veterinary Medicine.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Shuo Huang for excellent technical assistance during these studies, Dr. Virginia Buechner-Maxwell (Department of Large Animal Clinical Sciences, Virginia Tech) for assistance with challenge of the calves, Sai Karyala and the Virginia Tech Biocomplexity Institute for RNA-Seq results, and Dr. Dianjun Cao for RNA-Seq repository deposition.
Footnotes
References
Agnes, J. T., Zekarias, B., Shao, M., Anderson, M. L., Gershwin, L. J., and Corbeil, L. B. (2013). Bovine respiratory syncytial virus and Histophilus somni interaction at the alveolar barrier. Infect. Immun. 81, 2592–2597. doi: 10.1128/iai.00108-13
Angen, O., Ahrens, P., and Tegtmeier, C. (1998). Development of a PCR test for identification of Haemophilus somnus in pure and mixed cultures. Vet. Microbiol. 63, 39–48. doi: 10.1016/s0378-1135(98)00222-3
Arenas, J., Cano, S., Nijland, R., Dongen, V., Rutten, L., Ende, A., et al. (2015). The meningococcal autotransporter AutA is implicated in autoaggregation and biofilm formation. Environ. Microbiol. 17, 1321–1337. doi: 10.1111/1462-2920.12581
Balleza, E., Lopez-Bojorquez, L. N., Martinez-Antonio, A., Resendis-Antonio, O., Lozada-Chavez, I., Balderas-Martinez, Y. I., et al. (2009). Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol. Rev. 33, 133–151. doi: 10.1111/j.1574-6976.2008.00145.x
Bertani, B., and Ruiz, N. (2018). Function and biogenesis of lipopolysaccharides. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0001-2018
Booker, C. W., Abutarbush, S. M., Morley, P. S., Jim, G. K., Pittman, T. J., Schunicht, O. C., et al. (2008). Microbiological and histopathological findings in cases of fatal bovine respiratory disease of feedlot cattle in Western Canada. Can. Vet. J. 49, 473–481.
Bos, R., van der Mei, H. C., Meinders, J. M., and Busscher, H. J. (1994). A quantitative method to study co-adhesion of microorganisms in a parallel plate flow chamber: basic principles of the analysis. J. Microbiol. Meth. 20, 289–305. doi: 10.1016/0167-7012(94)90053-1
Boukahil, I., and Czuprynski, C. J. (2015). Characterization of Mannheimia haemolytica biofilm formation in vitro. Vet. Microbiol. 175, 114–122. doi: 10.1016/j.vetmic.2014.11.012
Brogden, K. A., Guthmiller, J. M., and Taylor, C. E. (2005). Human polymicrobial infections. Lancet 365, 253–255. doi: 10.1016/s0140-6736(05)70155-0
Burmølle, M., Ren, D., Bjarnsholt, T., and Sørensen, S. J. (2014). Interactions in multispecies biofilms: do they actually matter? Trends Microbiol. 22, 84–91. doi: 10.1016/j.tim.2013.12.004
Castonguay, M.-H., Van der Schaaf, S., Koester, W., Krooneman, J., Van der Meer, W., Harmsen, H., et al. (2006). Biofilm formation by Escherichia coli is stimulated by synergistic interactions and co-adhesion mechanisms with adherence-proficient bacteria. Res. Microbiol. 157, 471–478. doi: 10.1016/j.resmic.2005.10.003
Clutterbuck, A., Woods, E., Knottenbelt, D., Clegg, P., Cochrane, C., and Percival, S. (2007). Biofilms and their relevance to veterinary medicine. Vet. Microbiol. 121, 1–17. doi: 10.1016/j.vetmic.2006.12.029
Ehrlich, G. D., Hu, F. Z., Shen, K., Stoodley, P., and Post, J. C. (2005). Bacterial plurality as a general mechanism driving persistence in chronic infections. Clin. Orthopaed. Related Res. 437, 20–24. doi: 10.1097/00003086-200508000-00005
Elswaifi, S. F., Scarratt, W. K., and Inzana, T. J. (2012). The role of lipooligosaccharide phosphorylcholine in colonization and pathogenesis of Histophilus somni in cattle. Vet. Res. 43:49. doi: 10.1186/1297-9716-43-49
Felek, S., Lawrenz, M. B., and Krukonis, E. S. (2008). The Yersinia pestis autotransporter YapC mediates host cell binding, autoaggregation and biofilm formation. Microbiology 154, 1802–1812. doi: 10.1099/mic.0.2007/010918-0
Fulton, R. W. (2009). Bovine respiratory disease research (1983–2009). Anim. Health Res. Rev. 10, 131–139. doi: 10.1017/s146625230999017x
Gershwin, L. J., Berghaus, L. J., Arnold, K., Anderson, M. L., and Corbeil, L. B. (2005). Immune mechanisms of pathogenetic synergy in concurrent bovine pulmonary infection with Haemophilus somnus and bovine respiratory syncytial virus. Vet. Immunol. Immunopathol. 107, 119–130. doi: 10.1016/j.vetimm.2005.04.004
Gogolewski, R. P., Kania, S. A., Inzana, T. J., Widders, P. R., Liggitt, H. D., and Corbeil, L. B. (1987a). Protective ability and specificity of convalescent serum from calves with Haemophilus somnus pneumonia. Infect. Immun. 55, 1403–1411. doi: 10.1128/iai.55.6.1403-1411.1987
Gogolewski, R. P., Leathers, C. W., Liggitt, H. D., and Corbeil, L. B. (1987b). Experimental Haemophilus somnus pneumonia in calves and immunoperoxidase localization of bacteria. Vet. Pathol. 24, 250–256. doi: 10.1177/030098588702400309
Griffin, D. (1997). Economic impact associated with respiratory disease in beef cattle. Vet. Clin. North Am. Food Anim. Pract. 13, 367–377. doi: 10.1016/s0749-0720(15)30302-9
Griffin, D., Chengappa, M., Kuszak, J., and McVey, D. S. (2010). Bacterial pathogens of the bovine respiratory disease complex. Vet. Clin. North Am. Food Anim. Pract. 26, 381–394.
Hennigar, R., Mayfield, R., Harvey, J., Ge, Z.-H., and Sens, D. (1987). Lectin detection of renal glycogen in rats with short-term streptozotocin-diabetes. Diabetologia 30, 804–811.
Hennigar, R. A., Schulte, B. A., and Spicer, S. S. (1986). Histochemical detection of glycogen using Griffonia simplicifolia agglutinin II. Histochem. J. 18, 589–596. doi: 10.1007/bf01675294
Heydorn, A., Nielsen, A. T., Hentzer, M., Sternberg, C., Givskov, M., Ersbøll, B. K., et al. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395–2407. doi: 10.1099/00221287-146-10-2395
Holman, D. B., Timsit, E., Amat, S., Abbott, D. W., Buret, A. G., and Alexander, T. W. (2017). The nasopharyngeal microbiota of beef cattle before and after transport to a feedlot. BMC Microbiol. 17:70. doi: 10.1186/s12866-017-0978-6
Hotchkiss, E. J., Dagleish, M. P., Willoughby, K., McKendrick, I. J., Finlayson, J., Zadoks, R. N., et al. (2010). Prevalence of Pasteurella multocida and other respiratory pathogens in the nasal tract of Scottish calves. Vet. Rec. 167, 555–560. doi: 10.1136/vr.c4827
Karched, M., Bhardwaj, R. G., and Asikainen, S. E. (2015). Coaggregation and biofilm growth of Granulicatella spp. with Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans. BMC Microbiol. 15:114. doi: 10.1186/s12866-015-0439-z
Koebnik, R., Locher, K. P., and Van Gelder, P. (2000). Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37, 239–253. doi: 10.1046/j.1365-2958.2000.01983.x
Laurentin, A., and Edwards, C. A. (2003). A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal. Biochem. 315, 143–145. doi: 10.1016/s0003-2697(02)00704-2
Mbuthia, P. G., Christensen, H., Boye, M., Petersen, K. M., Bisgaard, M., Nyaga, P. N., et al. (2001). Specific detection of Pasteurella multocida in chickens with fowl cholera and in pig lung tissues using fluorescent rRNA in situ hybridization. J. Clin. Microbiol. 39, 2627–2633. doi: 10.1128/jcm.39.7.2627-2633.2001
Murray, G. M., More, S. J., Sammin, D., Casey, M. J., McElroy, M. C., O’Neill, R. G., et al. (2017). Pathogens, patterns of pneumonia, and epidemiologic risk factors associated with respiratory disease in recently weaned cattle in Ireland. J. Vet. Diagn. Invest. 29, 20–34. doi: 10.1177/1040638716674757
Murray, J. L., Connell, J. L., Stacy, A., Turner, K. H., and Whiteley, M. (2014). Mechanisms of synergy in polymicrobial infections. J. Microbiol. 52, 188–199. doi: 10.1007/s12275-014-4067-3
Okshevsky, M., and Meyer, R. L. (2015). The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 41, 341–352. doi: 10.3109/1040841x.2013.841639
O’Toole, D., Hunter, R., Allen, T., Zekarias, B., Lehmann, J., Kim, K. S., et al. (2017). Effect of Histophilus somni on heart and brain microvascular endothelial cells. Vet. Pathol. 54, 629–639.
O’Toole, D., and Sondgeroth, K. S. (2016). Histophilosis as a natural disease. Curr. Top. Microbiol. Immunol. 396, 15–48. doi: 10.1007/82_2015_5008
Pan, Y., Fisher, T., Olk, C., and Inzana, T. J. (2014). Detection of antibodies to the biofilm exopolysaccharide of Histophilus somni following infection in cattle by enzyme-linked immunosorbent assay. Clin. Vaccine Immunol. 21, 1463–1467. doi: 10.1128/cvi.00384-14
Perez, A. C., Pang, B., King, L. B., Tan, L., Murrah, K. A., Reimche, J. L., et al. (2014). Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog. Dis. 70, 280–288. doi: 10.1111/2049-632x.12129
Peters, B. M., Jabra-Rizk, M. A., Graeme, A., Costerton, J. W., and Shirtliff, M. E. (2012). Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25, 193–213. doi: 10.1128/cmr.00013-11
Petruzzi, B., Briggs, R. E., Tatum, F. M., Swords, W. E., De Castro, C., Molinaro, A., et al. (2017). Capsular polysaccharide interferes with biofilm formation by Pasteurella multocida serogroup A. mBio 8:e01843-17. doi: 10.1128/mBio.01843-17
Petruzzi, B., Scarratt, W. K., Buechner-Maxwell, V., and Inzana, T. J. (2015). “Inhibition of Pasteurella multocida biofilm formation by capsular polysaccharide and interaction with Histophilus somni in a polymicrobial biofilm,” in Proceedings of #151 Conference Research Workers Animal Disease, Chicago, IL.
Rickard, A. H., Gilbert, P., High, N. J., Kolenbrander, P. E., and Handley, P. S. (2003). Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11, 94–100. doi: 10.1016/s0966-842x(02)00034-3
Sandal, I., Hong, W., Swords, W. E., and Inzana, T. J. (2007). Characterization and comparison of biofilm development by pathogenic and commensal isolates of Histophilus somni. J. Bacteriol. 189, 8179–8185. doi: 10.1128/jb.00479-07
Sandal, I., Inzana, T. J., Molinaro, A., De Castro, C., Shao, J. Q., Apicella, M. A., et al. (2011). Identification, structure, and characterization of an exopolysaccharide produced by Histophilus somni during biofilm formation. BMC Microbiol. 11:186. doi: 10.1186/1471-2180-11-186
Sandal, I., Shao, J. Q., Apicella, M. A., Boye, M., Saunders, G. K., and Inzana, T. J. (2009). Histophilus somni biofilm formation in cardiopulmonary tissue of the bovine host following respiratory challenge. Microbes Infect. 11, 254–263. doi: 10.1016/j.micinf.2008.11.011
Sibley, C. D., Duan, K., Fischer, C., Parkins, M. D., Storey, D. G., Rabin, H. R., et al. (2008). Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4:e1000184. doi: 10.1371/journal.ppat.1000184
Simmons, W. L., and Dybvig, K. (2009). Mycoplasma biofilms ex vivo and in vivo. FEMS Microbiol. Lett. 295, 77–81.
Snowder, G. D., Van Vleck, L. D., Cundiff, L. V., and Bennett, G. L. (2006). Bovine respiratory disease in feedlot cattle: environmental, genetic, and economic factors. J. Anim. Sci. 84, 1999–2008. doi: 10.2527/jas.2006-046
Sorroche, F. G., Spesia, M. B., Zorreguieta, Á, and Giordano, W. (2012). A positive correlation between bacterial autoaggregation and biofilm formation in native Sinorhizobium meliloti isolates from Argentina. Appl. Environ. Microbiol. 78, 4092–4101. doi: 10.1128/aem.07826-11
Stovall, T., Gill, D., Smith, R., and Ball, R. (2000). “Impact of bovine respiratory disease during the receiving period on feedlot performance and carcass traits,” in Animal Science Research Report, (Stillwater, OK: Oklahoma State University).
Taylor, J. D., Fulton, R. W., Lehenbauer, T. W., Step, D. L., and Confer, A. W. (2010). The epidemiology of bovine respiratory disease: what is the evidence for preventive measures. Can. Vet. J. 51, 1351–1359.
Thurnheer, T., Gmür, R., and Guggenheim, B. (2004). Multiplex FISH analysis of a six-species bacterial biofilm. J. Microbiol. Meth. 56, 37–47. doi: 10.1016/j.mimet.2003.09.003
Yemm, E., and Willis, A. (1954). The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 57:508. doi: 10.1042/bj0570508
Zainal-Abidin, Z., Veith, P. D., Dashper, S. G., Zhu, Y., Catmull, D. V., Chen, Y.-Y., et al. (2012). Differential proteomic analysis of a polymicrobial biofilm. J. Proteome Res. 11, 4449–4464. doi: 10.1021/pr300201c
Keywords: biofilm, polymicrobial, bovine respiratory disease, Histophilus somni, Pasteurella multocida
Citation: Petruzzi B, Dickerman A, Lahmers K, Scarratt WK and Inzana TJ (2020) Polymicrobial Biofilm Interaction Between Histophilus somni and Pasteurella multocida. Front. Microbiol. 11:1561. doi: 10.3389/fmicb.2020.01561
Received: 27 April 2020; Accepted: 16 June 2020;
Published: 10 July 2020.
Edited by:
David William Waite, Ministry for Primary Industries, New ZealandReviewed by:
Chuck Czuprynski, University of Wisconsin-Madison, United StatesAnthony W. Confer, Oklahoma State University, United States
Copyright © 2020 Petruzzi, Dickerman, Lahmers, Scarratt and Inzana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas J. Inzana, Thomas.Inzana@liu.edu
 Briana Petruzzi1
Briana Petruzzi1 Kevin Lahmers
Kevin Lahmers William K. Scarratt
William K. Scarratt Thomas J. Inzana
Thomas J. Inzana