- 1Laboratorio Controllo Alimenti, Istituto Zooprofilattico Sperimentale dell’Umbria e delle Marche, Ancona, Italy
- 2Facoltà di Medicina Veterinaria, Università degli Studi di Teramo, Teramo, Italy
- 3Facoltà di Biologia, Università degli Studi di Trieste, Trieste, Italy
- 4Istituto Nazionale di Oceanografia e di Geofisica Sperimentale, Trieste, Italy
This research aimed to study the abundance and molecular diversity of Vibrio parahaemolyticus-specific Halobacteriovorax strains isolated from seawater of the Adriatic Sea and the relationship between predator and prey abundances. Moreover, predator efficiency of the Halobacteriovorax isolates toward V. parahaemolyticus and Vibrio cholerae non-O1/O139 strains was tested. V. parahaemolyticus NCTC 10885 was used as primary host for the isolation of Halobacteriovorax from seawater by the plaque assay. Molecular identification was performed by PCR detection of a fragment of the 16S rRNA gene of the Halobacteriovoraceae family members. Moreover, 700 bp PCR products were sequenced and compared between them and to clones described for other sampling sites. Vibrio counts were performed on TCBS agar from 100 ml of filtered water samples and presumptive colonies were confirmed by standard methods. Predatory efficiency of Halobacteriovorax isolates was tested by monitoring abilities of 3-day enrichments to form clear lytic halos on a lawn of Vibrio preys, by the plaque assay. Out of 12 seawater samples monthly collected from June 2017 to May 2018, 10 were positive for V. parahaemolyticus specific Halobacteriovorax with counts ranging from 4 to 1.4 × 103 PFU per 7.5 ml. No significant relationship was found between Halobacteriovorax and Vibrio abundances. The 16SrRNA sequences of our Halobacteriovorax strains, one for each positive sample, were divided into three lineages. Within the lineages, some sequences had 100% similarity. Sequence similarity between lineages was always <94.5% suggesting that they may therefore well belong to three different species. All Halobacteriovorax isolates had the ability to prey all tested Vibrio strains. Additional research is necessary to assess whether stable strains of Halobacteriovorax are present in the Adriatic Sea and to understand the mechanisms by which Halobacteriovorax may modulate the abundance of V. parahaemolyticus and other vibrios in a complex marine ecosystem.
Introduction
In many ecological communities, predation has a key role in regulating community structure or function. In the marine environment, predation has been extensively explored in animals, microbial eukaryotes and viruses (Baum and Worm, 2009) while predation by bacteria is less well understood (Williams et al., 2015). The genus Halobacteriovorax, in the class Deltaproteobacteria, family Bacteriovoracaceae, consists of small, Gram-negative, flagellated, marine predator bacteria that are members of a broader group of predatory bacteria known as Bdellovibrio and like organisms (BALOs), also including non-marine (terrestrial and freshwater) forms (Williams and Baer, 2005). BALOs enter into a susceptible Gram-negative prey bacterium and reside within the periplasmic space where they use the cytoplasmic nutrients of the prey to support growth and replication. The replicative form, known as bdelloplast, extends within the prey and divides into progeny cells that are released as soon as the host is lysed and are able to attack other preys (Williams and Baer, 2005). The genus Halobacteriovorax, according to the results of the analysis of the 16S rRNA gene sequence, includes two species Halobacillus litoralis and Halobacteriovorax marinus for which the similarity between the type strains is 92.61% (Koval et al., 2015). Recently, the new species Halobacteriovorax vibrionivorans has been also proposed (Ye et al., 2019). Furthermore, analysis of the 16S rRNA gene sequence from Halobacteriovorax saltwater strains identified multiple distinct phylogenetic clusters in different marine habitats, grouped based on a sequence identity with values >96.5% (Pineiro et al., 2007). Many of these Halobacteriovorax could represent potential new species. Halobacteriovorax favors predation on Vibrio species and other saltwater prey (Rice et al., 1998; Chen et al., 2012; Koval et al., 2015; Ye et al., 2019). Different lineages within this genus favor estuarine or marine waters and some isolates have been found in salt lakes (Koval et al., 2015). There are no known freshwater isolates (Koval et al., 2015). Recent studies reported that Halobacteriovorax were capable of containing Vibrio parahaemolyticus levels in seawater and oysters, at laboratory scale (Chen et al., 2011; Li et al., 2011; Richards et al., 2012, 2016; Williams et al., 2015; Ottaviani et al., 2018a). Furthermore, there is evidence that Halobacteriovorax contributes to Vibrio vulnificus mortality in a simulated natural seawater system with greater efficiency than other natural predators, such as bacteriophages (Chen et al., 2018). A previous study demonstrated that Halobacteriovorax was part of the coral microbiome (Welsh et al., 2016). In light of this evidence to understand if Halobacteriovorax has a role in modulating the natural levels of V. parahaemolyticus and other vibrios in the marine environment it is essential to know its natural abundance and understand in details how the predator infects and kills the prey. Quantitative PCR has been used to detect total Halobacteriovorax abundances in the marine environment, but it does not identify strains capable of targeting specific host bacteria (Zheng et al., 2008). On the other hand, Pineiro et al. (2007) isolated and characterized selected Halobacteriovorax phylotypes against V. parahaemolyticus in seawater, but results were not quantitative. To date, only two previous (plaque assay-based) studies have documented the abundance of V. parahaemolyticus specific Halobacteriovorax in seawater (Richards et al., 2013; Ottaviani et al., 2018a). However, as far as we know, there are no data on correlations between levels of Halobacteriovorax, V. parahaemolyticus and total vibrios in the marine environment. V. parahaemolyticus strains producing thermostable direct haemolysin (TDH) and/or TDH-related haemolysin (TRH) are recognized as a cause of diarrhoeal diseases worldwide, with bivalves, eaten raw or undercooked being the most frequent sources of infection (Potasman et al., 2002; Letchumanan et al., 2014). In Italy Halobacteriovorax and pathogenic vibrios, including V. parahaemolyticus and non-O1/O139 Vibrio cholerae, have been isolated in seawater and mussels coming from the Adriatic Sea (Masini et al., 2007; Ottaviani et al., 2018a). Moreover, in the last years, illness due to V. parahaemolyticus and non-O1/O139 V. cholerae, with mussels or seawater of Adriatic Sea as the source of infection, has been reported (Ottaviani et al., 2010, 2013, 2018b). In this work we studied the abundance and molecular diversity of V. parahaemolyticus-specific Halobacteriovorax strains isolated from seawater of a mussels growing area at the Conero Riviera of the Adriatic Sea. From the same samples V. parahaemolyticus and total vibrios were also counted and the relationship between these and Halobacteriovorax was studied. Finally, the predatory efficiency of Halobacteriovorax isolates toward V. parahaemolyticus and V. cholerae non-O1/O139 strains from clinical and environmental sources, the majority linked to the Adriatic sea of Italy, was tested.
Materials and Methods
Sampling Site
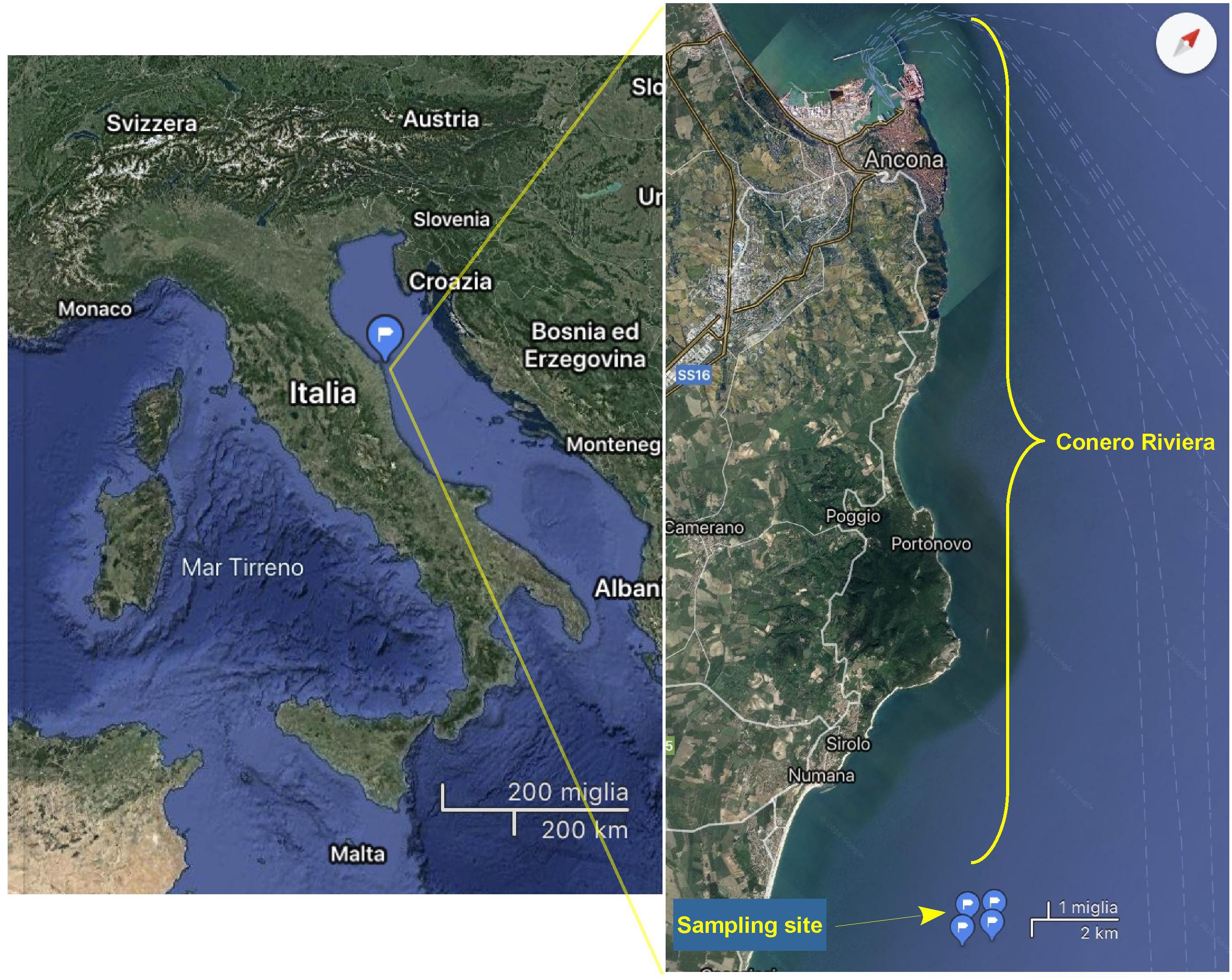
The Conero Riviera, located halfway of the Italian Adriatic coast, is a coastal marine ecosystem with an extension of 20 km and a low anthropic impact. This riviera includes several important sea towns, such as Ancona, Portonovo, Sirolo, and Numana, and lies in a protected area representing the most important marine reserve of the Marches where there are also many natural banks of mussels (Mytilus galloprovincialis) (Figure 1). Subsurface seawater samples were monthly collected from a site of approved mussels growing area in the Conero Riviera (43°30′12″N–13°41′12″E) from June 2017 to May 2018 (Figure 1). Two liters of seawater for each sample were collected using sterile polypropylene bottles. Water temperature and salinity were determined in situ with a multiparametric probe (Handy Gamma, Oxyguard, Denmark). All samples were transported to the laboratory on ice for analysis. Salinity remained steady between 38 and 39 ppt. Seawater was immediately transported to the laboratory in an insulated cooler and analyzed within 4 h. The same seawater samples that were used for Halobacteriovorax analyses were also used for V. parahaemolyticus and total Vibrio analyses.
Prey Strains
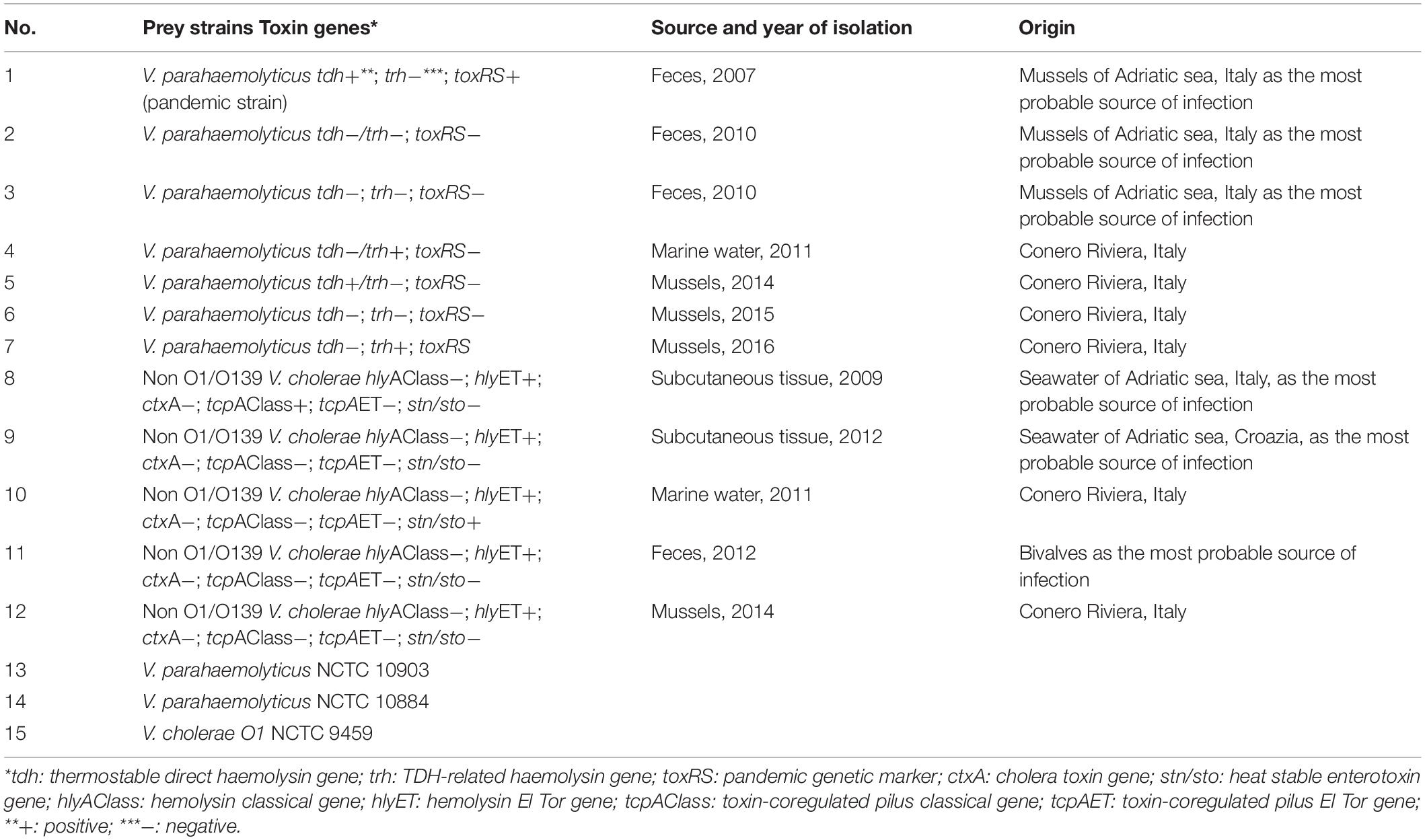
Vibrio parahaemolyticus NCTC 10885 strain was used as prey. Prey specificity and predatory efficiency of Halobacteriovorax isolates were tested on V. parahaemolyticus and V. cholerae strains of environmental and clinical origin, the majority directly or indirectly linked to the Adriatic sea of Italy (Table 1). For all Vibrio strains, fresh enrichments were prepared from a stock culture grown on 3% NaCl Luria-Bertani broth until prey reached an OD600 of 0.20 (∼1.8 × 108 CFU/ml).
Halobacteriovorax Enumeration
It was performed by a double layer agar plating technique (Richards et al., 2012). The prey was grown in LB–3% NaCl broth until it reached an OD600 of 0.20 (∼1.8 × 108 CFU/ml). For the analysis, 500 ml of test seawater was first filtered through a 0.45-μm, 500-ml filter to remove particulates and bacteria. Then, the filtered seawater was serially diluted 10-fold in sterilized artificial seawater (30 ppt). For each assay, 25 ml of bottom-layer Pp 20 agar (polypeptone peptone supplemented with Bacto agar) were dispensed into a 100-mm Petri dish and allowed to harden. Top agar was pipetted into sterile glass tubes (7.5 ml/tube) while the agar was hot and allowed to cool to 48°C in a water bath. The plaque assay was conducted by combining 1 ml of host V. parahaemolyticus culture, at an OD600 of 0.20 and 7.5 ml of undiluted and diluted test seawater to 7.5 ml of molten (48°C) Pp 20 agar in tubes. The tubes were inverted 3 times to mix and poured on top of the existing bottom layer. Counts of Halobacteriovorax were performed after incubation at 26°C for 7 days. The number of viable Halobacteriovorax isolates was estimated as PFU per 7.5 ml seawater.
Halobacteriovorax Molecular Identification and Sequencing Analysis
For each presumptively positive sample, five plaques appearing on plates of the highest dilution were picked up for molecular identification. The templates used for PCR were individual plaques re-suspended in 100 ml of sterile double-distilled water and vortexed at a high speed. The liquid phase was transferred to a new tube and subjected to heating in boiling water for 3 min (Jurkevitch et al., 2000). 16S rRNA PCR analysis was performed on a fragment of the 16S rRNA gene of the Halobacteriovoraceae family members using the primers Bac676F and Bac1442R, as previously described (Davidov et al., 2006; Richards et al., 2013). Samples showing a band of 700 bp were considered Halobacteriovorax. For each positive sample, sequencing analysis was performed on PCR product of 700 bp from a unique plaque. PCR products were purified with the High Pure PCR Product Purification kit Roche Diagnostics (GmbH, Mannheim, Germany). Sequencing analysis was performed using the reverse primer BAC1442R and ABI Prism® BigDye® Terminator v1.1 Cycle Sequencing kit (Applied BiosystemsTM, Life Technologies, United States), according to the manufacturer’s instructions. Sequenced products were analyzed in an automated capillary sequencer ABI Prism® 310 Genetic Analyzer (Applied BiosystemsTM, United States). Nucleotide sequences were manually edited, aligned and analyzed using CLC genomics workbench V.12 (Qiagen Bioinformatics). Phylogenetic trees of the 16S rRNA gene sequences from the isolates generated in this study were reconstructed using the maximum likelihood (ML) method. These sequences were also compared to those of the following type strains of the Halobacteriovorax species: H. marinus SJ (GenBank accession number 102485), H. litoralis JS5 (GenBank accession number 028724) and H. vibrionivorans BL9 (GenBank accession number MH150810) Initially the 16SrRNA sequences were aligned with a progressive alignment tool (Feng and Doolittle, 1987) within the CLC genomics workbench 20 environment. The best substitution model fitting the alignment was the General Time Reversible (GTR), gamma distribution 0 and transition/transversion rate ratio 2. The tree topology was tested with 1000 bootstrap replicates.
Total Vibrio spp. and V. parahaemolyticus Enumeration
Each water sample was mixed and 100 ml of undiluted and 1:10, 1:100, 1:1000 diluted sample was filtered using a 0.45-mm-pore membrane filter (Millipore, Bedford, MA, United States); the filter was placed on thiosulfatecitrate-bile-salts-sucrose-agar (TCBS, Difco Laboratories, Detroit, MI, United States) and incubated at 37°C for 24 h. For each presumptively positive sample, five colonies appearing on plates of the highest dilution were selected and subcultured on trypticase soy agar with 2% NaCl (TSAs, Oxoid). Presumptive Vibrio spp. were biochemically identified at genus level by a standardized protocol (Ottaviani et al., 2003). Presumptive V. parahaemolyticus strains were confirmed by a standard method (ISO/TS, 2017). The number of viable V. parahaemolyticus and total Vibrio isolates was estimated as CFU per 100 ml seawater.
Prey Specificity and Predatory Efficiency of Halobacteriovorax Isolates
Three-day enrichments of each Halobacteriovorax isolate (approximately 1 × 106 PFU ml–1) were filtered through a 0.45- m-pore-size Millex HV syringe filter (Millipore Corp., Billerica, MA, United States) to remove the primary prey. Prey specificity and predator efficiency of Halobacteriovorax strains were determined by monitoring their abilities to form clear lytic halos on a lawn of the reference V. parahaemolyticus and V. cholerae preys listed in Table 1, using a double layer agar plating technique. Briefly, 1 μl of filtered predator undiluted enrichment, 1 ml of prey culture at an OD600 of 0.2 for Vibrio preys (approximately 108 PFU ml–1), were added to 7.5 ml of sterilized artificial seawater (30 ppt salinity) and 7.5 ml of Pp 20 top agar (Richards et al., 2012, 2016). For each prey specificity assay a positive control, represented by V. parahaemolyticus NCTC 10885 and the Halobacteriovorax strain, and a negative control, which consisted of the prey alone, were performed.
Statistical Analysis
On each 2 l seawater sample the analyses were performed in six independent replicates for Halobacteriovorax and Vibrio (n = 6). Results of microbiological analyses were reported as mean values (log-transformed) ± standard deviation. Correlation coefficients were determined and associated p-values < 0.05 were considered significant.
Results
Halobacteriovorax Enumeration
Out of 12 seawater samples collected, 10 (83.3%) were positive for presumptive V. parahaemolyticus specific Halobacteriovorax, with counts ranging from 4 PFU per 7.5 ml (November 2017) to 1.4 × 103 PFU per 7.5 ml (January 2018) (Figure 2). Plaques on primary prey became visible after 72 h. After that, sizes expanded over time, reaching the maximum diameter of 9 mm after 5 days of incubation at 26°C. A lack of plaques was observed on all replicates of February and May 2018 samples even when the incubation was prolonged up to 10 days. No significant relationship was found between Halobacteriovorax levels and seawater temperature (Figure 2).
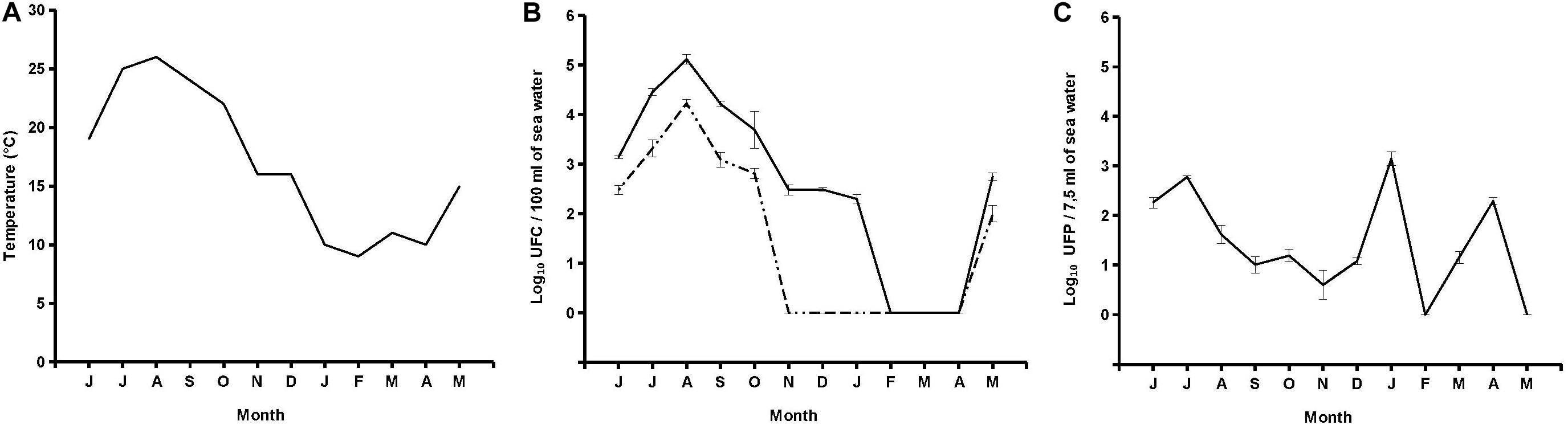
Figure 2. Trend of temperature (A), V. parahaemolyticus and total vibrio (B), and Halobacteriovorax (C).
Halobacteriovorax Molecular Identification and Sequencing Analysis
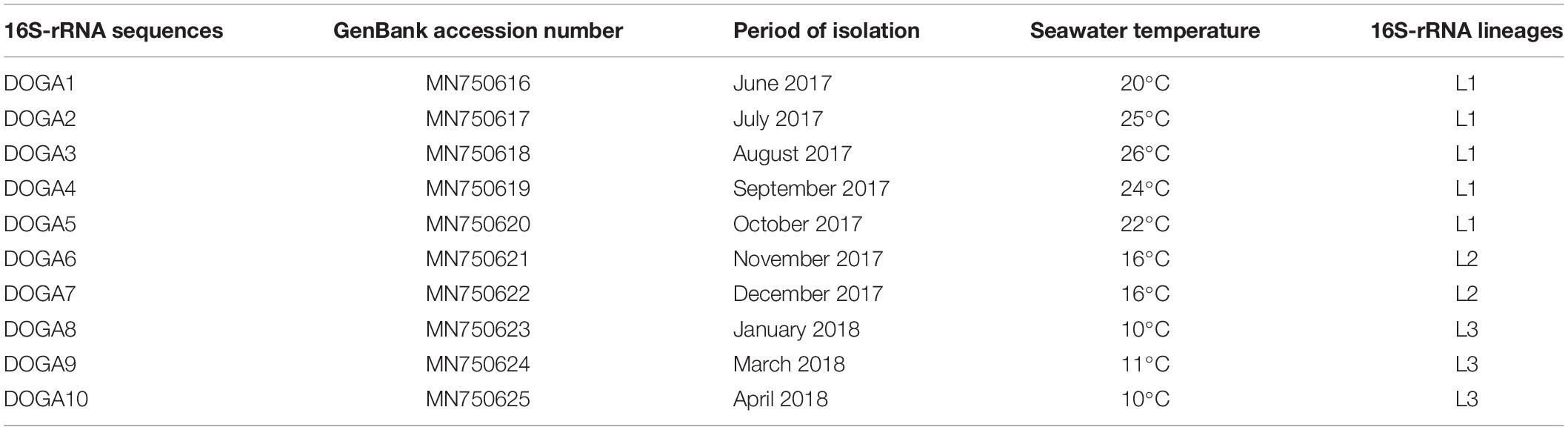
All plaques for each seawater sample were confirmed by molecular methods as Halobacteriovorax. The sequences obtained by analysis of the 10 16SrRNA 700pb PCR fragments, one for each positive sample, were named DOGA1-DOGA10. The partial 16SrRNA gene sequences were deposited in GenBank under accession numbers MN750616–MN750625 (Table 2). Sequences from DOGA1 to DOGA7 were obtained from the strains isolated in 2017 while sequences from DOGA8 to DOGA10 were from the strains isolated in 2018 (Table 2). Overall, the divergence between our sequences was always <10% (Supplementary Figure 1). A similarity >99% was between the sequences DOGA1-DOGA5, DOGA6-DOGA7 and DOGA8-DOGA10. Between the three groups, the sequence similarity ranged from 90.63% (between DOGA 1 and DOGA 8) to 94.51% (between DOGA 6 and DOGA 9–10) (Supplementary Figure 1). These three distinct lineages were named L1, L2, L3, respectively (Table 2). DOGA2, DOGA3, and DOGA5 within L1 showed 100% sequence identity (Figure 3 and Table 2). Also DOGA9 and DOGA10 within L3 showed 100% sequence identity (Figure 3 and Table 2). Sequences within L3 shared 98.54–99.19% similarity with H. vibrionivorans BL9, 94.83–95.32% with H. litoralis JS5 and 91.75–92.23%. with H. marinus SJ (Figure 3 and Supplementary Figure 1). Sequences within L2 shared 94.18–94.51% similarity with H. vibrionivorans BL9, 95.64–95.96% with H. litoralis JS5 and 93.38–93.70%. with H. marinus SJ (Figure 3 and Supplementary Figure 1). Sequences within L1 shared 91.44–92.57% similarity with H. vibrionivorans BL9, 91.11–92.25% with H. litoralis JS5 and 92.41–93.21%. with H. marinus SJ (Figure 3 and Supplementary Figure 1).
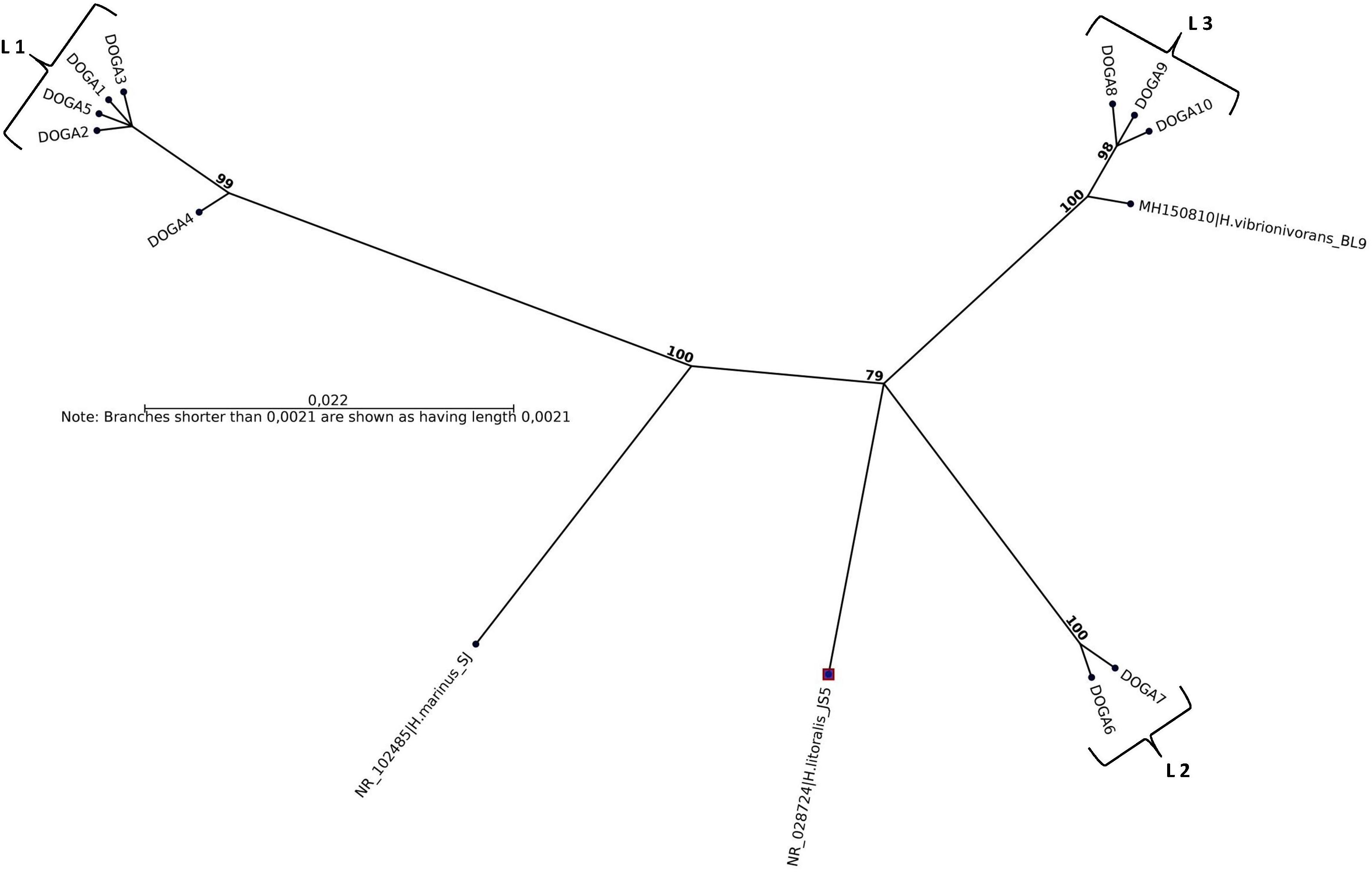
Figure 3. Maximum-likelihood phylogenetic tree, based on 16S rRNA gene sequence comparisons, showing the position of strains DOGA1-10 and related type strains. Numbers at branch nodes are bootstrap values (per 1000 trials).
Total Vibrio spp. and V. parahaemolyticus Enumeration
Six samples (50%) were positive for V. parahaemolyticus with counts ranging from 1.0 × 102 CFU per 100 ml (May 2018) to 1.7 × 104 CFU per 100 ml (August 2017). A lack of detection was reported from November to December 2017 and from January to April 2018. Nine samples (75%) were positive for total vibrios with counts ranging from 2.0 × 102 CFU per 100 ml (January 2018) to 1.3 × 105 CFU per 100 ml (August 2017). A lack of detection was reported from February to April 2018. No significant relationship was found between levels of Halobacteriovorax and those of total Vibrio and V. parahaemolyticus (Figure 2).
Prey Specificity and Predatory Efficiency of Halobacteriovorax Isolates
All Halobacteriovorax isolates were able to lyse all tested V. parahaemolyticus and V. cholerae reference strains. Plaques of lysis became visible after 72 h and then, their sizes expanded over time, reaching the maximum after 5 days of incubation at 26°C. At the end of the incubation, the diameter of lysis plaques on vibrios was similar to those on primary prey, ranging between 7 and 9 mm diameter.
Discussion
Halobacteriovorax Enumeration
Indigenous V. parahaemolyticus-specific Halobacteriovorax is present in seawater of a mussel growing area of the Conero Riviera at levels higher than those reported for Atlantic and Pacific Oceans which never exceeded values of 102 PFU per mL (Richards et al., 2013). Moreover, the Halobacteriovorax counts in this study should be considered minimum counts since some portion of the predators were undoubtedly removed by filtration (Richards et al., 2013). Surprisingly, we found the Halobacteriovorax higher abundances on January 2018 while Halobacteriovorax in Atlantic and Pacific Oceans were mainly concentrated in the warmer months of the year (Richards et al., 2013). It is known that Halobacteriovorax against V. parahaemolyticus is able to proliferate in the temperature range between 10 and 30°C (Williams et al., 2015). In the marine areas investigated by Richards et al. (2013) temperatures over a 12-month period ranged from lows of 5°C in winter and highs of 30°C in summer. Halobacteriovorax replication was therefore completely inhibited in the winter months. In the Conero Riviera temperatures over a 12-month period ranged from lows of 10°C in winter and highs of 25°C in summer. Therefore, throughout the year, the temperatures were suitable for a good replication of Halobacteriovorax.
Halobacteriovorax Molecular Identification and Sequencing Analysis
In BALOs, as in other bacteria, a 90% sequence similarity means grouping at the genus level while the species level is identified by a >98% similarity (Koval et al., 2015). In this study the sequence similarity of Halobacteriovorax strains inter-lineages was always <94.5% suggesting that they may therefore well belong to three different species. Moreover, sequences in L1 and L2 formed independent branches separated from the type strains of Halobacteriovorax. Instead the sequences in L3 shared >99% similarity with H. vibrionivorans BL9. For this reason these Halobacteriovorax strains would clustered with the proposed new species H. vibrionivorans. In this period there is a great interest from the scientific world in the taxonomic and phylogenetic study of this genus. Shortly we will perform the genome sequencing of our Halobacteriovorax strains in order to obtain more detailed information on their taxonomic position and understand if they could belong to new species within the genus. Our results show that different strains of Halobacteriovorax circulated in the Conero Riviera in the 2017–2018 period. In this study we have limited ourselves to the molecular characterization of the dominant population of Halobacteriovorax. For this reason, the plaques for each isolate were enumerated and treated as if they were a single clonal population. Moreover, in this study we have used a unique V. parahaemolyticus strain as prey to recover Halobacteriovorax. These working methods may have limited the Halobacteriovorax diversity detected in the marine area investigated. Previous studies associated different clusters of Halobacteriovorax to marine waters with different salinity (Pineiro et al., 2007, 2013; Crossman et al., 2013; Richards et al., 2013). In disagreement, our strains belonging to different lineages, were isolated from a marine area with a constant level of salinity equal to 38–39 ppt throughout the year. An association was found among the lineages of our Halobacteriovorax strains and the temperature of seawater. The strains in L1 were isolated in the period June–October 2017 when the temperature ranged between 20 and 25°C. The strains in L2 were isolated in the period November–December 2017 when the temperature was 16°C. Finally, the strains in L3 were isolated in the period January–April 2018 when the temperature ranged between 10 and 11°C. In a future investigation we will extend the period and marine area of the study, by using different Vibrio parahemolyticus prey strains and sequencing 16SrRNA fragments from more plaques for each sample. The scope will be to assess if, in the Adriatic Sea, diversities of Halobacteriovorax may exist in respect of seasonality and prey specificity and whether particular strains can be present here stably.
Total Vibrio spp. and V. parahaemolyticus Enumeration
No significant correlations between Halobacteriovorax, total Vibrio spp. and V. parahaemolyticus abundance was detected in the marine area investigated in this study. A previous study confined to microcosms in which Halobacteriovorax/V. parahaemolyticus at a ratio of 105 PFU/105 CFU per ml were added, produced a different effect, where Halobacteriovorax increased as V. parahaemolyticus decreased (Ottaviani et al., 2018a). The fact that no relationship between the abundance of Halobacteriovorax and V. parahaemolyticus was observed in the present study could be related to the very low abundance of both genera in seawater which was about 10–100 cells per ml. This may have reduced the chance for high predation and consequently a significant increase and decrease in the abundance of Halobacteriovorax and V. parahaemolyticus, respectively. Moreover, it is known that some Halobacteriovorax has broader host specificity than others and the mechanisms that drive Halobacteriovorax host specificity within a complex bacterial community are not known. In this regard, our Halobacteriovorax strains demonstrated predatory activity in vitro toward all V. parahaemolyticus and V. cholerae reference strains tested, many of them isolated from the same marine area. Halobacteriovorax may well target one microorganism over another within the same species and also between different species of marine bacteria. The extent to which this occurs remains unresolved. In the next future it would be essential to perform microcosm-studies to investigate the unwanted effect of Halobacteriovorax isolates toward the vital core-microbiome bacteria in marine seawater. Moreover, Halobacteriovorax and Vibrio abundances may be subject to control by bacteriophages, other predators, environmental conditions, nutrient levels, the effects of competing microbes, and the development of host resistance. Finally, the lack of correlation between predator and prey levels could be linked to the different speed of Halobacteriovorax and Vibrio replication, so that they cannot compete with maximum efficiency between them.
Prey Specificity and Predatory Efficiency of Halobacteriovorax Isolates
Our Halobacteriovorax isolates, although captured using a single V. parahaemolyticus strain as primary prey, were able to lyse a wide range of V. parahaemolyticus and V. cholerae strains, including those toxigenic and/or involved in human infections. Previous scientific information reported that V. parahaemolyticus is the most efficient prey known for Halobacteriovorax recovery, at least for the strains against Vibrio (Pineiro et al., 2007). The broad spectrum of action of our isolates would seem to confirm this hypothesis. Most of the vibrio prey came from the same marine area from which Halobacteriovorax strains were also isolated. Some had been isolated from mussels, others from seawater in the Conero Riviera. We believe that Halobacteriovorax similarly to other bacteria through the filtration from seawater passes inside the mollusk where continues to parasitize vibrios and other potential pathogens. A previous study investigated the Halobacteriovorax predation in coral-associated microbiome (Welsh et al., 2016). The authors speculated that coral microbiome could allow the predator to remain alive and vital while Halobacteriovorax could regulate and maintain the microbiome structure in balance and at the same time protect the host by consuming potential pathogens. Similar symbiotic relationships may also exist between Halobacteriovorax and the microbiome associated with other aquatic organisms, such as bivalves. Laboratory-scale studies are currently underway to evaluate the effects of Halobacteriovorax in the reduction of V. parahaemolyticus in bivalves during the purification phase and to understand the potential unwanted effects of the predator on the mussel microbiome.
Conclusion
In conclusion, different V. parahaemolyticus-specific Halobacteriovorax strains are present in seawater of a mussel growing area on the Conero Riviera where they, probably, play a physiological role as natural modulators versus V. parahaemolyticus and other vibrios populations. These predatory bacteria could have a primary role in regulating and structuring marine bacterial communities and the nutrient cycle but have not received the attention they deserve to date. Additional research is necessary to better understand the mechanisms by which Halobacteriovorax may modulate V. parahaemolyticus and other vibrios in a complex marine ecosystem, and the overall effect they exert on the structure of seawater microbial communities and marine host-associated microbiomes.
Data Availability Statement
The datasets generated for this study can be found in the GenBank under the following accession numbers: SUB6636935 DOGA1 MN750616, SUB6636935 DOGA2 MN750617, SUB6636935 DOGA3 MN750618, SUB6636935 DOGA4 MN750619, SUB6636935 DOGA5 MN750620, SUB6636935 DOGA6 MN750621, SUB6636935 DOGA7 MN750622, SUB6636935 DOGA8 MN750623, SUB6636935 DOGA9 MN750624, and SUB6636935 DOGA10 MN750625.
Author Contributions
GA, SP, and FM isolated bacteria and performed the laboratory measurements. DO, FL, ER, ML, and PT made substantial contributions to conception and design. AP aligned and compared 16SrRNA sequences. DO wrote and revised the manuscript. DO and GA drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by the Health Ministry financed projects RF-2013-023355019 and RC-2017-D33C17000030001.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01575/full#supplementary-material
References
Baum, J. K., and Worm, B. (2009). Cascading top-down effects of changing oceanic predator abundances. J. Anim. Ecol. 78, 699–714. doi: 10.1111/j.1365-2656.2009.01531.x
Chen, H., Athar, R., Zheng, G., and Williams, H. N. (2011). Prey bacteria shape community structure of their predators. ISME J. 5, 1314–1342.
Chen, H., Laws, E. A., Martin, J. L., Berhane, T.-K., Gulig, P. A., and Williams, H. N. (2018). Relative contributions of Halobacteriovorax and bacteriophage to bacterial cell death under various environmental conditions. mBio 9:e01202-18. doi: 10.1128/mBio.01202-18
Chen, H., Young, S., Berhane, T.-K., and Williams, H. N. (2012). Predatory Bacteriovorax communities ordered by various prey species. PLoS One 7:e34174. doi: 10.1371/journal.pone.0034174
Crossman, L. C., Chen, H., Cerdeño-Tárraga, A. M., Brooks, K., Quail, M. A., Piñeiro, S. A., et al. (2013). A small predatory core genome in the divergent marine Bacteriovorax marinus SJ and the terrestrial Bdellovibrio bacteriovorus. ISME J. 7, 148–160. doi: 10.1038/ismej.2012.90
Davidov, Y., Friedjung, A., and Jurkevitch, E. (2006). Structure analysis of a soil community of predatory bacteria using culture dependent and culture-independent methods reveals a hitherto undetected diversity of Bdellovibrio-and like organisms. Environ. Microbiol. 8, 1667–1673. doi: 10.1111/j.1462-2920.2006.01052.x
Feng, D. F., and Doolittle, R. F. (1987). Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evol. 25, 351–360. doi: 10.1007/bf02603120
ISO/TS (2017). Microbiology of Food and Animal Feeding Stuffs — Horizontal Method for the Detection of Potentially Enteropathogenic Vibrio spp. — Part 1: Detection of Vibrio parahaemolyticus and Vibrio cholerae International Organization for Standardization (ISO) 1, Chemin de la Voie Creuse, case postale 56. Geneva: ISO.
Jurkevitch, E., Minz, D., Ramati, B., and Barel, G. (2000). Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl. Environ. Microbiol. 66, 2365–2371. doi: 10.1128/aem.66.6.2365-2371.2000
Koval, S. F., Williams, H. N., and Stine, O. C. (2015). Reclassification of Bacteriovorax marinus as Halobacteriovorax marinus gen. nov., comb. nov. and Bacteriovorax litoralis as Halobacteriovorax litoralis comb. nov.; description of Halobacteriovoraceae fam. nov. in the class Deltaproteobacteria. Int. J. Syst. Evol. Microbiol. 65, 593–597. doi: 10.1099/ijs.0.070201-0
Letchumanan, V., Chan, K. G., and Lee, L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Li, H., Liu, C., Chen, L., Zhang, X., and Cai, J. (2011). Biological characterization of two marine Bdellovibrio-and-like organisms isolated from Daya bay of Shenzhen, China and their application in the elimination of Vibrio parahaemolyticus in oyster. Int. J. Food Microbiol. 151, 36–43. doi: 10.1016/j.ijfoodmicro.2011.07.036
Masini, L., De Grandis, G., Principi, F., Mengarelli, C., and Ottaviani, D. (2007). Research and characterization of pathogenic vibrios from bathing water along the Conero Riviera (Central Italy). Water Res. 41, 4031–4040. doi: 10.1016/j.watres.2007.05.003
Ottaviani, D., Leoni, F., Rocchegiani, E., Canonico, C., Potenziani, S., Santarelli, S., et al. (2010). Vibrio parahaemolyticus-associated gastroenteritis in Italy: persistent occurence of O3:K6 pandemic clone and emergence of O1:KUT serotype. Diagn. Microbiol. Infect. Dis. 66, 452–455. doi: 10.1016/j.diagmicrobio.2009.11.015
Ottaviani, D., Leoni, F., Rocchegiani, E., Mioni, R., Costa, A., Virgilio, S., et al. (2013). An extensive investigation into the prevalence and the genetic and serological diversity of toxigenic Vibrio parahaemolyticus in Italian marine coastal waters. Environ. Microbiol. 15, 1377–1386. doi: 10.1111/j.1462-2920.2012.02839.x
Ottaviani, D., Masini, L., and Bacchiocchi, S. (2003). A biochemical protocol for the isolation and identification of current species of Vibrio in seafood. J. Appl. Microbiol. 95, 1277–1284. doi: 10.1046/j.1365-2672.2003.02105.x
Ottaviani, D., Chierichetti, S., Angelico, G., Forte, C., Rocchegiani, E., Manuali, E., et al. (2018a). Halobacteriovorax isolated from marine water of the Adriatic sea, Italy, as an effective predator of Vibrio parahaemolyticus, non-O1/O139 V. cholerae, V. vulnificus. J. Appl. Microbiol. 4, 1199–1207. doi: 10.1111/jam.14027
Ottaviani, D., Medici, L., Talevi, G., Napoleoni, M., Serratore, P., Zavatta, E., et al. (2018b). Molecular characterization and drug susceptibility of non-O1/O139 V. cholerae strains of seafood, environmental and clinical origin, Italy. Food Microbiol. 72, 82–88. doi: 10.1016/j.fm.2017.11.011
Pineiro, S., Chauhan, A., Berhane, T. K., Athar, R., Zheng, G., Wang, C., et al. (2013). Niche partition of Bacteriovorax operational taxonomic units along salinity and temporal gradients in the Chesapeake Bay reveals distinct estuarine strains. Microb. Ecol. 65, 652–660. doi: 10.1007/s00248-013-0186-3
Pineiro, S. A., Stine, O. C., Chauhan, A., Steyert, S. R., Smith, R., and Williams, H. N. (2007). Global survey of diversity among environmental saltwater Bacteriovoracaceae. Environ. Microbiol. 9, 2441–2450. doi: 10.1111/j.1462-2920.2007.01362.x
Potasman, I., Paz, A., and Odeh, M. (2002). Infectious outbreaks associated with bivalve shellfish consumption: a worldwide perspective. Clin. Infect.Dis. 35, 921–928. doi: 10.1086/342330
Rice, T. D., Williams, H. N., and Turng, B. F. (1998). Susceptibility of bacteria in estuarine environments to authchthonous Bdellovibrios. Microb. Ecol. 35, 256–264. doi: 10.1007/s002489900081
Richards, G. P., Fay, J. P., Dickens, K. A., Parent, M. A., Soroka, D. S., and Boyd, E. F. (2012). Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Appl. Environ. Microbiol. 78, 7455–7466. doi: 10.1128/aem.01594-12
Richards, G. P., Fay, J. P., Uknalis, J., Olanya, O. M., and Watson, M. A. (2016). Purification and host specificity of predatory Halobacteriovorax isolates from seawater. Appl. Environ. Microbiol. 82, 922–927. doi: 10.1128/aem.03136-15
Richards, G. P., Watson, M. A., Boyd, E. F., Burkhardt, W. I. I. I., Lau, R., Uknalis, J., et al. (2013). Seasonal levels of the Vibrio predator Bacteriovorax in Atlantic, Pacific and Gulf Coast seawater. Int. J. Microbiol. 2013:375371. doi: 10.1155/2013/375371
Welsh, R. M., Zaneveld, J. R., Rosales, S. M., Payet, J. P., Burkepile, D. E., and Thurber, R. V. (2016). Bacterial predation in a marine host-associated microbiome. ISME J. 10, 1540–1544. doi: 10.1038/ismej.2015.219
Williams, H. N., and Baer, M. L. (2005). “Genus II Bacteriovorax,” in Bergey’s Manual of Systematic Bacteriology: The Proteobacteria, Vol. 2, eds G. M. Garrity, D. J. Brenner, N. R. Krieg, and J. T. Staley (New York, NY: Springer), 1053–1057.
Williams, H. N., Lymperopoulou, D. S., Athar, R., Chauhan, A., Dickerson, T. L., Chen, H., et al. (2015). Halobacteriovorax, an underestimated predator on bacteria: potential impact relative to viruses on bacterial mortality. ISME J. 10, 491–499. doi: 10.1038/ismej.2015.129
Ye, X. S., Chen, M. X., Li, H. Y., He, X. Y., and Zhao, Y. (2019). Halobacteriovorax vibrionivorans sp. nov., a novel prokaryotic predator isolated from coastal seawater of China. Int. J. Syst. Evol. Microbiol. 69, 3917–3923. doi: 10.1099/ijsem.0.003703
Keywords: V. parahaemolyticus-specific Halobacteriovorax, Vibrio spp., V. parahaemolyticus, V. cholerae non-O1/O139, Adriatic Sea
Citation: Ottaviani D, Pieralisi S, Rocchegiani E, Latini M, Leoni F, Mosca F, Pallavicini A, Tiscar PG and Angelico G (2020) Vibrio parahaemolyticus-specific Halobacteriovorax From Seawater of a Mussel Harvesting Area in the Adriatic Sea: Abundance, Diversity, Efficiency and Relationship With the Prey Natural Level. Front. Microbiol. 11:1575. doi: 10.3389/fmicb.2020.01575
Received: 03 January 2020; Accepted: 17 June 2020;
Published: 08 July 2020.
Edited by:
Luigi Vezzulli, University of Genoa, ItalyReviewed by:
Sanin Musovic, Danish Technological Institute (DTI), DenmarkTaviani Elisa, Universidade Eduardo Mondlane, Mozambique
Copyright © 2020 Ottaviani, Pieralisi, Rocchegiani, Latini, Leoni, Mosca, Pallavicini, Tiscar and Angelico. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donatella Ottaviani, d.ottaviani@izsum.it
 Donatella Ottaviani
Donatella Ottaviani Silvia Pieralisi1
Silvia Pieralisi1 Francesca Leoni
Francesca Leoni Francesco Mosca
Francesco Mosca Alberto Pallavicini
Alberto Pallavicini Pietro Giorgio Tiscar
Pietro Giorgio Tiscar Gabriele Angelico
Gabriele Angelico