- 1Dryland Agricultural Research Institute, Agricultural Research, Education and Extension Organization (ARREO), Maragheh, Iran
- 2Department of Biotechnology, College of Agriculture, Isfahan University of Technology, Isfahan, Iran
- 3Department of Plant Pathology, Faculty of Agricultural Sciences and Engineering, College of Agriculture and Natural Resources, University of Tehran, Karaj, Iran
- 4Department of Plant Pathology, Faculty of Agriculture, Tarbiat Modares University, Tehran, Iran
- 5Wageningen University and Research, Wageningen Plant Research, Wageningen, Netherlands
Zymoseptoria tritici is one of the most economically destructive wheat diseases all over the world and is a model fungal plant pathogen within the ascomycetes. In this study, the instrumental role of the ZtRlm1 gene encoding a MADS-box transcription factor (TF) in the infection process of Z. tritici was functionally characterized as these proteins play critical roles in the global gene regulation required for various developmental and physiological processes. Our infection assays showed that ZtRlm1 mutants were attenuated in disease development as a 30 and 90% reduction in chloro-necrotic lesions and pycnidia formation, respectively, were observed in plants inoculated with ZtRlm1 mutant strains demonstrating that ZtRlm1 is a crucial factor playing a significant role in the late stage of infection corresponding with pycnidial formation. Our expression analysis demonstrated that the transcript level of ZtRlm1 is induced at 2 and 20 days post-inoculation, coinciding with pycnidial sporulation. In addition, microscopic analyses showed that branch intensity and biomass production were significantly reduced, indicating that impaired pycnidia formation is a result of impaired differentiation and biomass production in the ZtRlm1 mutants. Furthermore, melanization, a phenomenon required for fruiting body formation, was significantly hampered in ZtRlm1 mutants as they were not melanized under all tested temperature and media conditions. Overall, our data showed that impaired disease development of the ZtRlm1 mutants is mainly due to the significant impact of ZtRlm1 in different cellular processes, including differentiation, branching, fungal biomass production, and melanization, in which identification of downstream genes are of interest to increase our understanding of this pathosystem.
Introduction
Zymoseptoria tritici (Desm.) Quaedvlieg & Crous (Quaedvlieg et al., 2011) (formerly known as Mycosphaerella graminicola), causes septoria tritici blotch (STB), which is one of the most economically destructive wheat diseases all over the worlds. STB results in imposing remarkable yield losses annually in wheat-growing regions with high rainfall and moderate temperature during the growing season (Eyal and Levy, 1987; Fones and Gurr, 2015). Currently, the application of resistant cultivars is the most cost-effective and environmentally friendly approach to manage STB disease (Brown et al., 2015). However, only a limited number of resistant cultivars are currently commercially applied to manage this damaging disease (Fraaije et al., 2005; Orton et al., 2011). The importance of STB is increasing, particularly after the introduction of high yielding cultivars that are specifically improved for resistance to other biotic and abiotic stresses. In addition, the rapid emergence of fungicide resistant isolates in natural populations of this fungus has brought a strong demand for wheat protection against STB.
Z. tritici-wheat pathosystem has attracted considerable attention from many researchers as this fungus has a unique lifestyle, active sexual cycle, genomic feature, and high genome plasticity (Dean et al., 2012). Z. tritici is considered as a hemibiotrophic pathogen with a symptomless biotrophic stage of around 10-days, followed by a rapid switch to the necrotrophic stage. The fungus directly penetrates the leaves through natural opening (stomata) by forming appressorium-like swellings at the hyphal tips and afterward intercellularly colonizes the apoplastic space, without establishing particular feeding structures such as haustoria. During the biotrophy, the fungal biomass is low and increases exponentially after switching to necrotrophy in which the typical symptoms of the disease including irregular chlorotic lesions develops leading to the formation of necrotic blotches that eventually bear pycnidia and pycnidiospores in sub-stomatal cavities (Eyal et al., 1987; Kema et al., 1996; Mehrabi et al., 2006a). Sexual reproduction of this fungus can be completed several times during growing seasons resulting in developing natural populations with high genetic diversity (Chen and McDonald, 1996; Hunter et al., 1999; Morais et al., 2019).
To initiate infection, plant pathogenic fungi are able to perceive host signals followed by proper molecular and phenotypic responses, including penetration and colonization. During co-evolution, both pathogens and their hosts have achieved elaborated tactics leading to either compatible or incompatible interactions (Dodds and Rathjen, 2010). For example, fungal pathogens use specific virulence factors to suppress or manipulate host defense mechanisms, whereas host plants have developed molecular components to recognize and activate other defense processes against invading pathogens (De Wit et al., 2009). Plant-pathogenic fungi are equipped with numerous genes to effectively regulate the distinctive developmental and differentiation processes to attack host cells. Additionally, it is well documented that dynamic and complex interactions presented in the battlefield between a host and a fungal pathogen would determine the outcome of the interactions (compatibility/incompatibility). Targeted gene replacement strategies were employed to functionally characterize some of these genes (Mehrabi and Kema, 2006; Mehrabi et al., 2006a, b, 2007, 2009; Orton et al., 2011). Among the genes involved in the infection process, transcription factors (TFs) play pivotal roles in the global regulation of other genes required for various developmental processes as well as a successful infection. For example Mirzadi Gohari et al. (2014) demonstrated that ZtWor1 was implicated in the infection process of Z. tritici since deletion mutant failed to cause disease and complementation of the mutant by ZtWor1 gene restored the wild-type phenotypes. In another study Mohammadi et al. (2017) showed that ZtVf1 was involved in different developmental stages, including melanization, hyphal branching, pycnidia differentiation and biomass production and acts as a functional pathogenicity factor. The MADS-box proteins belong to the family of TFs that directly bind to specific motifs in the promoter region of the targeted genes and participate in a diverse range of biological activities (Messenguy and Dubois, 2003). Typically, filamentous ascomycetes have only two MADS-box transcription factor proteins, serum response factor (SRF-type or type I) and myocyte enhancer factor (MEF-type or type II). Unlike many fungi, Saccharomyces cerevisiae has four MADS-box transcription factors; Mcm1 and Arg80 proteins are associated with the SRF-type while Rlm1 and Smp1 belong to the MEF-type (de Nadal et al., 2003). In S. cerevisiae, Rlm1 is involved in the expression of genes that are essential for cell wall integrity, whereas Smp1 is associated with regulating genes that are responsible for osmotic stress response (de Nadal et al., 2003). Furthermore, Mcm1 and Arg80 regulate arginine metabolism in yeast cells (Messenguy and Dubois, 2003). Targeted gene deletion of MIG1 (MADS-box protein responsible for infectious growth 1 gene) as homolog of ZtRlm1 in the fungal plant pathogen Magnaporthe oryzae revealed their involvement in plant infection (Mehrabi et al., 2008). In mig1 mutant strains, infectious structures, including appressoria, penetration pegs, and primary infectious hyphae were generated. However, these mutants were unable to create secondary infectious hyphae in living cells. Additionally, mutant strains of mig1 were able to penetrate and develop infectious hypha-like structures in plant cells killed by heating or cellophane membranes, indicating that this gene might be acting as a downstream of Magnaporthe grisea MAP kinase (MPS1) to regulate genes required for suppressing plant defense responses (Mehrabi et al., 2008).
Although few reports showed the instrumental roles of MADS-box TFs in the virulence of various fungal pathogens (Mehrabi et al., 2008; Lin et al., 2018), no functional study has been performed to unveil the functional role of these TFs in Z. tritici. Therefore, we functionally characterized the biological function of the ZtRlm1, homolog of MIG1, in Z. tritici, and our results confirmed that this gene is implicated in several developmental processes to successfully establish tissue colonization.
Materials and Methods
Biological Materials
We applied Z. tritici IPO323, which is highly pathogenic on the wheat cultivar Obelisk (Mehrabi et al., 2006a) in our assays. This strain was propagated on PDA media (Sigma-Aldrich Chemie, Steinheim, Germany) or Yeast Glucose Broth (YGB) medium (yeast extract 10 g/L and glucose 20 g/L) to generate abundant yeast-like cells at 18°C for 5–7 days. Eventually, the cells generated on the cultured plates were harvested by softly scratching cultures, and kept at –80°C (Kema and van Silfhout, 1997).
Constructs Generation
The gene deletion construct designated as pZtRlm1KO was generated by using the USER-friendly cloning approach to delete the ZtRlm1 (Frandsen et al., 2008). Firstly, primer pairs, including ZtRlm1-PRF-F1, R1, and ZtRlm1-PRF-F2, R2 to amplify 1.8 kb bp upstream and downstream regions of the targeted genes through PfuTurbo® CxHotstart DNA polymerase were applied (Stratagene, Cedar Creek, TX, United States). Additionally, the pRF-HU2 vector contained the hygromycin phosphotransferase (hph) gene as a selectable marker was digested via enzymes named PacI and Nt.BbvCI, to create an overhang ends that are compatible with the PCR products. Afterward, the resultant materials were combined and treated with the USER enzyme mix (New England Biolabs, Ipswich, MA, United States) and placed at 37°C for 30 min followed by another incubation period at 25°C for 30 min. Eventually, the developed reaction was directly transformed into Escherichia coli strain DH5α using an electroporation machine. Subsequently, the treated materials were cultured on a medium possessing the antibiotic kanamycin. The right bacteria harboring the interesting fragment with the expected insertions at the right places were evaluated using colony PCR technique via hph-R2/ZtRlm1-PRF-F1 and hph-F2/ZtRlm1-PRF-R2 primer pairs (Table 1 and Figure 2A). The correct construct amplified 2065 bp bands with both primer pairs, indicating that the upstream and downstream of ZtRlm1 placed in the right position in the pRF-HU2 vector leading to the development of pZtRlm1KO. All primers are listed in Table 1.
Fungal Transformation
We cloned the pZtRlm1KO into Agrobacterium tumefaciens strain LBA1100 through electroporation technique. A. tumefaciens-mediated transformation (ATMT) method was performed to delete ZtRlm1 in the Z. tritici IPO323, as previously described (Mehrabi et al., 2006a). Following 20 days, separate Z. tritici transformants were picked up and transferred to a PDA medium supplemented with 100 μg/mL hygromycin plus 200 μg/mL cefatoxime. We subsequently extracted the genomic DNAs of each transformant based on the protocol depicted by the Dellaporta et al. (1983). They extracted DNA were applied in a PCR-based screening approach to test the presence of expected fragments. Our PCR-based screening showed that the band of 743 bp using the pair primer (ZtRlm1-F1 and ZtRlm1-R1) was amplified in the WT and ectopic transformants, whereas this band was not generated in the deleted mutant for ZtRlm1 (Figure 2B), implying that this gene was eliminated from these independent transformants. All the transformants including ectopic and ZtRlm1 mutants amplified a band of 764 bp, when hph specific primers (hph-F1 and hph-R1) were used in PCR amplification (Figure 2C). Additionally, elimination of ZtRlm1 occurred in the chromosomal region was investigated by a primer pair named hph-F2 and ZtRlm1 -R2 demonstrating the anticipated band of 2065 bp were exclusively amplified in the deleted mutant for ZtRlm1 (Figure 2D).
Infection Assay
The wheat cv. Obelisk was grown in a glasshouse by the time that the first leaves have appeared. Inoculum of the tested strains was generated in the YGB at 18°C for 7 days, and yeat-like cells were collected by centrifuging at 3000 rpm for 3 min followed by two washing steps by distilled water. Subsequently, spore concentrations were adjusted to 107∘ml–1 spores by a hemocytometer instrument, and the 0.15% Tween 20® was added to the spore suspensions to enhance spore adhering to leaf surfaces. Infection assay was performed as described in detail previously (Mohammadi et al., 2017) in three replicates and each replicate contained a pot of 5 seedlings. The infected plants were kept under black plastic bags for 48 h to increase the humidity and then transferred to a greenhouse compartment at 22°C with a relative humidity > 90% and 16 h light and 8-h darkness. Disease development was evaluated daily and recorded following disease symptoms expression around 8 dpi, which is coincident with the transition phase. We documented the percentage of chloro-necrotic regions along with the chloronecrotic lesions covered by asexual fruiting bodies (pycnidia). At 21 dpi, 10 leaves infected by all examined strains were harvested, and the formed pycnidia in the distance down to 1 cm from the leaf tips were calculated and analyzed using SPSS software package (IBM SPSS Statistics 19, United States).
RNA Isolation and Quantitative RT-PCR
Transcript abundance of ZtRlm1 at various time courses was conducted through a quantitative real-time PCR (q-RT-PCR) technique. For evaluating the expression level of ZtRlm1 in planta, we infected the wheat cv. Obelisk by Z. tritici IPO323 and, subsequently, the inoculated leaves were harvested in three biological replicates (four leaves per each biological replicates) (Mehrabi et al., 2006a). The samples were immediately frozen by placing them in the liquid nitrogen and, then, ground in this liquid via a mortar and pestle. Total RNA of the inoculated leaves and fungal biomass generated in YGB was extracted through the RNeasy plant mini kit (Qiagen, location, United States). We applied the DNA-free kit (Ambion, Cambridgeshire, United Kingdom) to eliminate DNA contamination, and afterward synthesized the first-strand cDNA from around two μg of total RNA primed with oligo(dT) via the SuperScript III enzyme based on the instructions provided by the manufacturer. One μl of the synthesized cDNA was employed in a 25 μl PCR reaction via a QuantiTect SYBR Green PCR Kit (Applied Biosystems, Warrington, United Kingdom), and run and analyzed using an ABI 7500 Real-Time PCR machine. Three technical replicates for each biological replicates were used, allowing us to perform the statistical analysis through the SPSS software package (IBM SPSS Statistics 19, United States). We initially normalized the relative expression of ZtRlm1 with the constitutively expressed Z. tritici beta-tubulin gene (Keon et al., 2007; Motteram et al., 2009) and, eventually, estimated according to the comparative C (t) method defined previously (Schmittgen and Livak, 2008).
In vitro Phenotyping
We used two nutritionally diverse solid cultures, including YMDA (4 g yeast, 4 g malt, 10 g dextrose, and 15 g agar in 1 L distilled water) and PDA along with an application of three various temperatures (16, 20, and 28°C) to characterize morphologically the WT and mutant strains. We spotted ~1 μl of spore suspension (107 spores∘ml–1) on the above-cultures, and we monitored and recorded their morphological features for 10 days.
We harvested spores of the WT and mutant strains of ZtRlm1 produced in the YGB medium at 18°C for 5 days. Afterward, we washed spore suspensions by the distilled water to remove remaining media and, subsequently, adjusted the spores to 105, spores ml–1. Next, we placed 12μl of the adjusted spore suspensions on a piece of the Water Agar (WA) or PDA fragment placed on a microscope slide covered by a coverslip. Eventually, we incubated the prepared materials at 20°C under dark conditions, and at least two spores of each sample were analyzed using an Olympus BX51 microscope equipped with Olympus DP72 digital camera every 12 h until 48 h. Pictures were proceeded with Adobe Photoshop version 15.2.2. After 48 h, for comparison of spores biomass and spore branches, randomly 10 spores of mutants and control strains cultivated on the PDA and WA media, were selected and analyzed using Digimizer version 4.1.1.0 (MedCalc, 2009) and SPSS softwares package (IBM SPSS Statistics 19, United States).
In vivo Histopathological Assay
We employed two methodologies to determine the germination and penetration patterns, plus the colonization of Z. tritici in the attacked leaves. In the first method, we collected the inoculated leaves at various time courses, including 8, 12, 16, and 20 dpi and then we instantly immersed them in 15 ml of 0.05% trypan blue dissolved in lactophenol-ethanol (1:2, v/v), and boiled for 10 min. We subsequently destained samples in a saturated chloral hydrate solution (5:2, w/v) for at least 10 h and then stored in 87% (v/v) glycerol by the time of analysis. In another method, we cleared the harvested samples with a mixture of glacial acetic acid: absolute ethanol (1:3, v/v) followed by transferring onto filter paper saturated with lactoglycerol (1:1:1, lactic acid: glycerol: water, v/v/v) until analysis by a cytological observation (Shetty et al., 2003).
Phylogenetic Tree Construction
We employed the MEGA software package version 5.05 (Tamura et al., 2011) to construct and analyze the phylogenetic tree. We applied the unweighted pair group approach with the arithmetic average (UPGMA) algorithm to build up the phylogenetic tree, and we evaluated the accuracy of the constructed phylogenetic tree by running the bootstrap analysis of 1000 repetitions.
Results
Identification and Characterization of ZtRlm1
To identify the homolog of Rlm1 in the fully sequenced genome of Z. tritici, a BLASTp search of Z. tritici genome using S. cerevisiae Rlm1 (GenBank number: BAA09658.1) as the query was performed resulting in the retrieval of the homolog of Rlm1 protein (protein ID = 71585). This protein was designated as ZtRlm1 and subsequently subjected to further analyses. ZtRlm1 has an open reading frame of 1903 bp, which is interrupted by three introns, and it is located on chromosome 4 encoding a protein of 597 amino acids. Interestingly, the overall similarity and identity between ZtRlm1 and yeast Rlm1 is about 34%, but they are over 90% identical in the MADS-box domain. Phylogenetic analysis of ZtRlm1 and related MADS-box proteins from other selected fungi indicated ZtRlm1 grouped with MEF-type MADS-box proteins (Figure 1). The specific motif, including the 58-amino-acid MADS-box region and 75-amino-acid MEF2 region determining the features of TFs are located at the N terminus region of ZtRlm1. Alignment analysis showed that that the N terminus region of ZtRlm1 is more conserved than the other part of protein sequences.
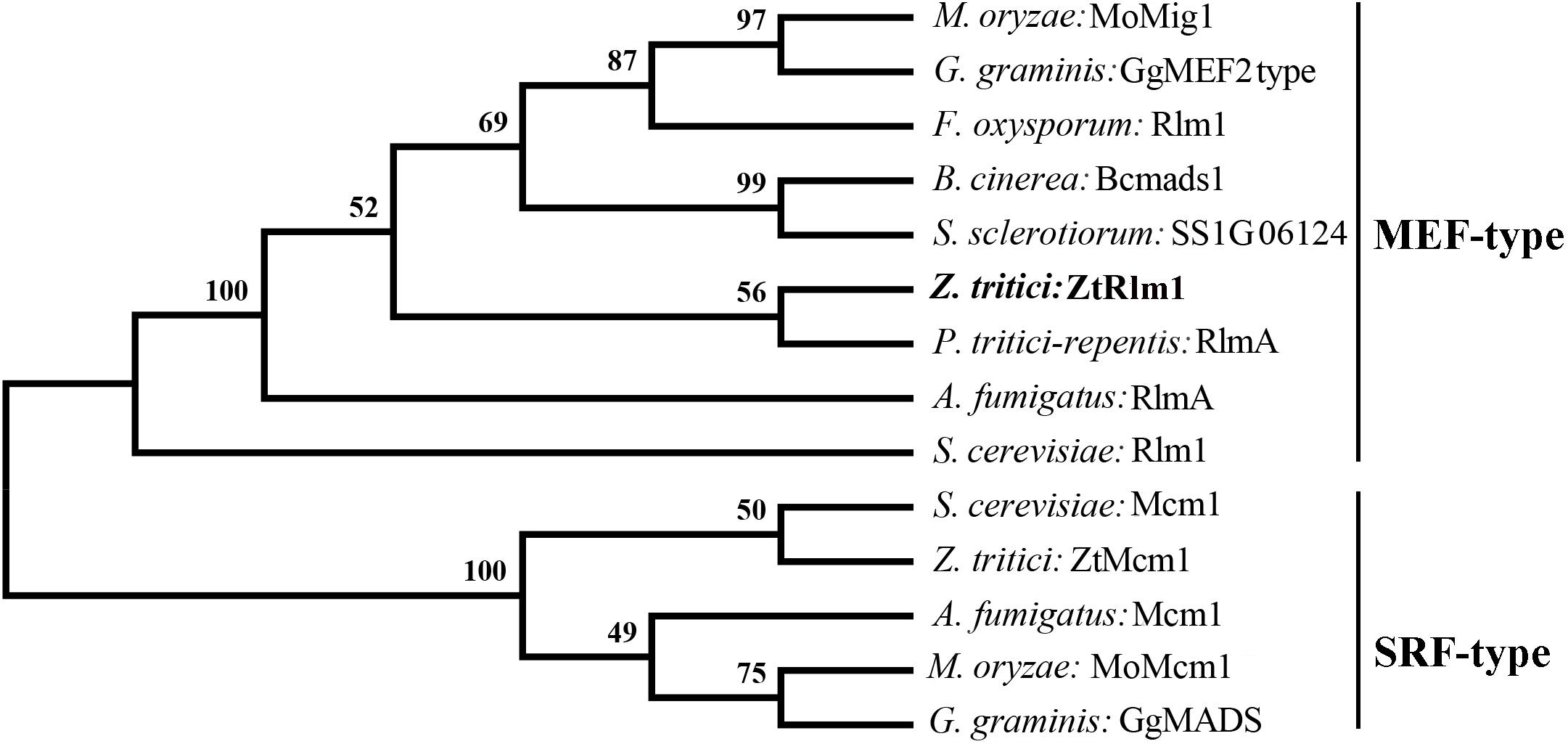
Figure 1. Phylogenetic comparison of Zymoseptoria tritici Rlm1 (ZtRlm1) with members of the MADS-box family based on amino acid sequence alignments. The two subfamilies of MADS-box family: SRF-like/Type I and MEF2-like/type II are indicated. The accession number of the proteins are included as ENH69552.1 (Rlm1) (Fusarium oxysporum), XP_001559829.1 (Bcmads1) (Botrytis cinerea), XP_001593202.1 (SS1G06124) (Sclerotinia sclerotiorum), XP_001938412.1 (RlmA) (Pyrenophora tritici-repentis), NP_013757.1 (Mcm1) and NP_009741.3 (Rlm1) (Saccharomyces cervisiae), XP_003852780.1 (ZtMcm1) and XP_003853236.1 (ZtRlm1) (Zymoseporia ritici), XP_747866.1 (Mcm1) and XP_754763.1 (RlmA) (Aspergillus fumigatus), XP_003720973.1 (MoMcm1) and XP_003714096.1 (MoMig1) (Magnaporthe oryzae), XP_009220065.1 (GgMADS) and XP_009220065.1 (GgMEF2 type) (Geaumaomyces graminis). The phylogenetic tree was constructed using MEGA 5 software. The bootstrap values (1000 replicates) are shown above the branches.
Deletion of ZtRlm1
To study the biological role of ZtRlm1 during the infection process of Z. tritici, the deletion construct based on the USER-friendly method as described previously (Frandsen et al., 2008) was made. As shown in Figure 2A, about 1.8 kb of upstream and downstream of the ZtRlm1 were used to generate the deletion construct that eventually was used to knock-out the Z. tritici ZtRlm1 in IPO323 strain through Agrobacterium tumefaciens–mediated transformation (ATMT). After transformation, the ZtRlm1 mutants were found using the ZtRlm1 gene-specific primers, ZtRlm1-F1, and ZtRlm1 -R1 (Figures 2A,B). After performing many transformations and screening more than 600 transformants, two independent mutants were identified and designated as ΔZtRlm1#1 and ΔZtRlm1#2. ZtRlm1 gene-specific primers confirmed that only the WT and ectopic transformants could produce a band of 743 bp while this PCR band was absent in the ZtRlm1 mutants showing that the ZtRlm1 was deleted in these mutants (Figure 2B). In addition, the ZtRlm1 mutants and ectopic transformants amplified the expected hph band (764 bp) whereas this band was absent in the WT showing that the construct was inserted in the genome of transformants (Figure 2C). The exact position of homologous recombination in the ZtRlm1 chromosomal region was also confirmed using the hph-F2 primer located at 3’ end of the hph marker gene along with the ZtRlm1 -R2 primer located 211 bp at the right border of ZtRlm1 downstream (Figure 2D). The results revealed that only ZtRlm1 mutants could amplify the expected band of 2065 bp, showing that the ZtRlm1 was exclusively deleted in the mutant strains (Figure 2D). Eventually, it worth mentioning that our laboring effort to generate a complemented strain of Rlm1 deletion mutant was unsuccessful. After transformation, we have screened more than 400 transformants, but none of them were positive.
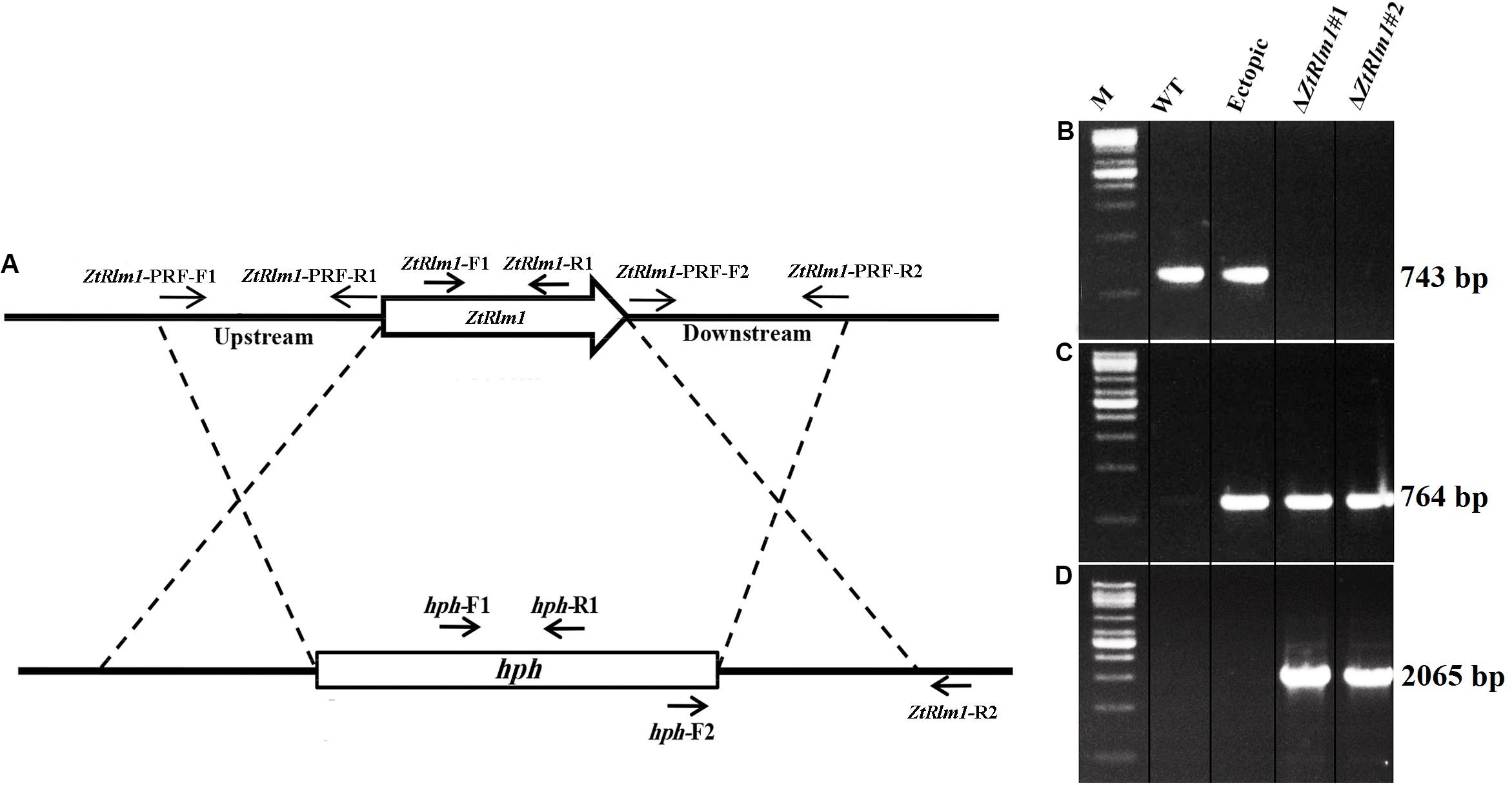
Figure 2. Generation of the ZtRlm1 gene replacement mutant. Diagram showing the replacement of ZtRlm1 by hygromycin phosphotransferase resistance cassette (hph) through homologous recombination. The dotted lines depict the flanking regions where homologous recombination occurred (A). Identification of replacement mutants by PCR approach. The WT and ectopic transformants generated the expected PCR band (743 bp) using primers ZtRlm -F1 and ZtRlm1 -R1, while this band was not amplified in the ZtRlm1 mutants indicating that the ZtRlm1 gene was deleted from these independent transformants (B). The ZtRlm1 mutants and ectopic transformants amplified the expected hph band (764 bp) using hph-F1/hph-R1 primers, whereas this band was absent in the WT (C). A primer pair (hph-F2 and ZtRlm1-R2) was used to confirm targeted gene deletion that occurred in the ZtRlm1 chromosomal region. Only the mutant strains were able to produce the expected amplicons (2065 bp) (D).
ZtRlm1 Delayed Disease Development
Infection assay was performed by the inoculation of susceptible wheat cv. Obelisk using the mutant strains (ΔZtRlm1#1 and ΔZtRlm1#2) and the control strains (IPO323 wild type (WT) and ectopic transformants) and disease development was monitored every 48 h. Both WT and ectopic strains caused chlorotic flecks at 9 days post-inoculation (dpi), especially at the leaf tips, which generated extended chlorosis at 12 dpi, and eventually expanded into large necrotic areas containing abundant mature pycnidia at 16–18 dpi (Figure 3). In contrast, ZtRlm1 mutants were not able to generate disease symptoms until 13 dpi when a few scattered chlorotic and necrotic lesions were observed that slowly merged into larger lesions along with the expansion of chlorotic areas becoming necrotic at 20 dpi but did not sporulate pycnidia even at 30 dpi. The results showed that both independent mutants were unable to produce pycnidia and significantly reduced the percentage of chloro-necrotic lesions (Figure 4 and Supplementary Figure 1). In order to determine the effect of ZtRlm1 deletion on chlorosis and pycnidia formation, two disease indices (CN and P) were used. In contrast to control strains, the ZtRlm1 mutant strains did not generate symptoms until 13 dpi when limited chlorotic areas developed that progressively merged into necrotic at 20 dpi without mature and normal pycnidial sporulation (Figures 3, 4). CN and P analysis revealed a significant reduction in CN (~30%) and P (~90%) in plants inoculated with ZtRlm1 mutant strains compared to that of the WT strain, indicating that ZtRlm1 is vital for the pycnidial formation (Figure 4 and Supplementary Tables 1–3).

Figure 3. Determination of the effect of ZtRlm1 deletion on disease development of Zymoseptoria tritici on the susceptible wheat cv. Obelisk at 20 dpi. Note that the WT and ectopic strains caused extensive necrosis bearing abundant pycnidia, while the ZtRlm1 mutant strains generated significantly reduced necrotic regions without pycnidia.
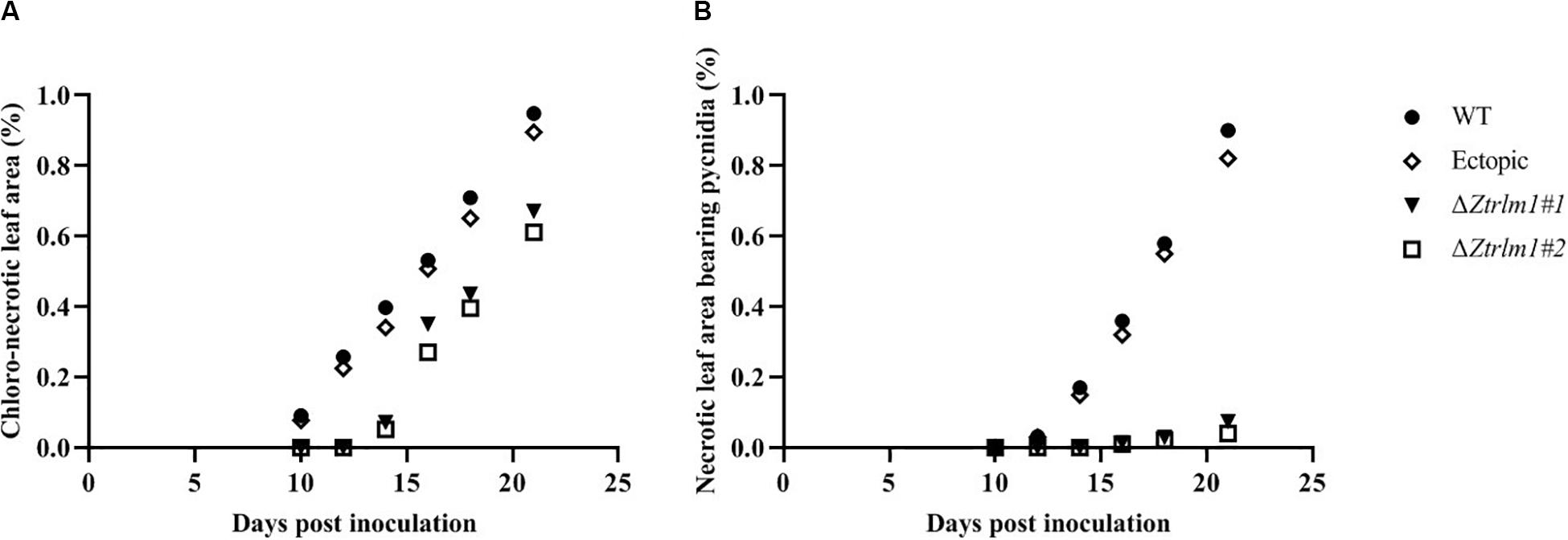
Figure 4. Disease development of Zymoseptoria tritici ZtRlm1 mutant strains compared with that of the control strains over a period of 10 days (from 10 to 20 days post-inoculation). The average of chloro-necrotic leaf area (%) (A). The average of necrotic leaf area bearing pycnidia (B). Note that the disease development of the ZtRlm1 mutant strains was significantly reduced compared with that of the control strains.
ZtRlm1 Plays in Early and Late Phases of Infection
As infection assay indicated that ZtRlm1 mutants were significantly impaired in virulence, we performed a histopathological study to determine and monitor the behavior of ΔZtRlm1 strain during infection stages in further detail. The WT and ectopic strains were able to penetrate stomata by infectious hypha, whereas ΔZtRlm1 strain demonstrated a 30% reduction in penetration frequency compared with that of control strains (Supplementary Table 1). Additionally, the ZtRlm1 mutant strain, in contrast with WT, could not colonize mesophyll cells completely (Figure 5), suggesting that ZtRlm1 has a pivotal role in the penetration strength as well as the initial establishment of Z. tritici colonization. At 12 dpi, infectious hypha of the WT strain totally colonized substomatal cavities of penetrated stomata which was significantly different from the ZtRlm1 mutant where ∼65% of the penetrated stomata was colonized (Supplementary Table 1). Additionally, the biomass of infectious hypha was much lesser compared to the WT strain. This finding might be the reason for no disease symptom expression until 13 dpi. Finally, the inoculated plants by the Z. tritici IPO323 resulted in the pycnidial sporulation at 16 and 20 dpi while that of the ZtRlm1 deletion mutant strain was failed to produce mature pycnidia at the mentioned time points and we rarely noticed few immature pycnidia in the infected plants by ZtRlm1 mutant strain in microscopic analysis (Figure 5). These findings indicated that ZtRlm1 plays a significant function in both of pycnidial production and differentiation (Supplementary Table 1 and Supplementary Figure 1).
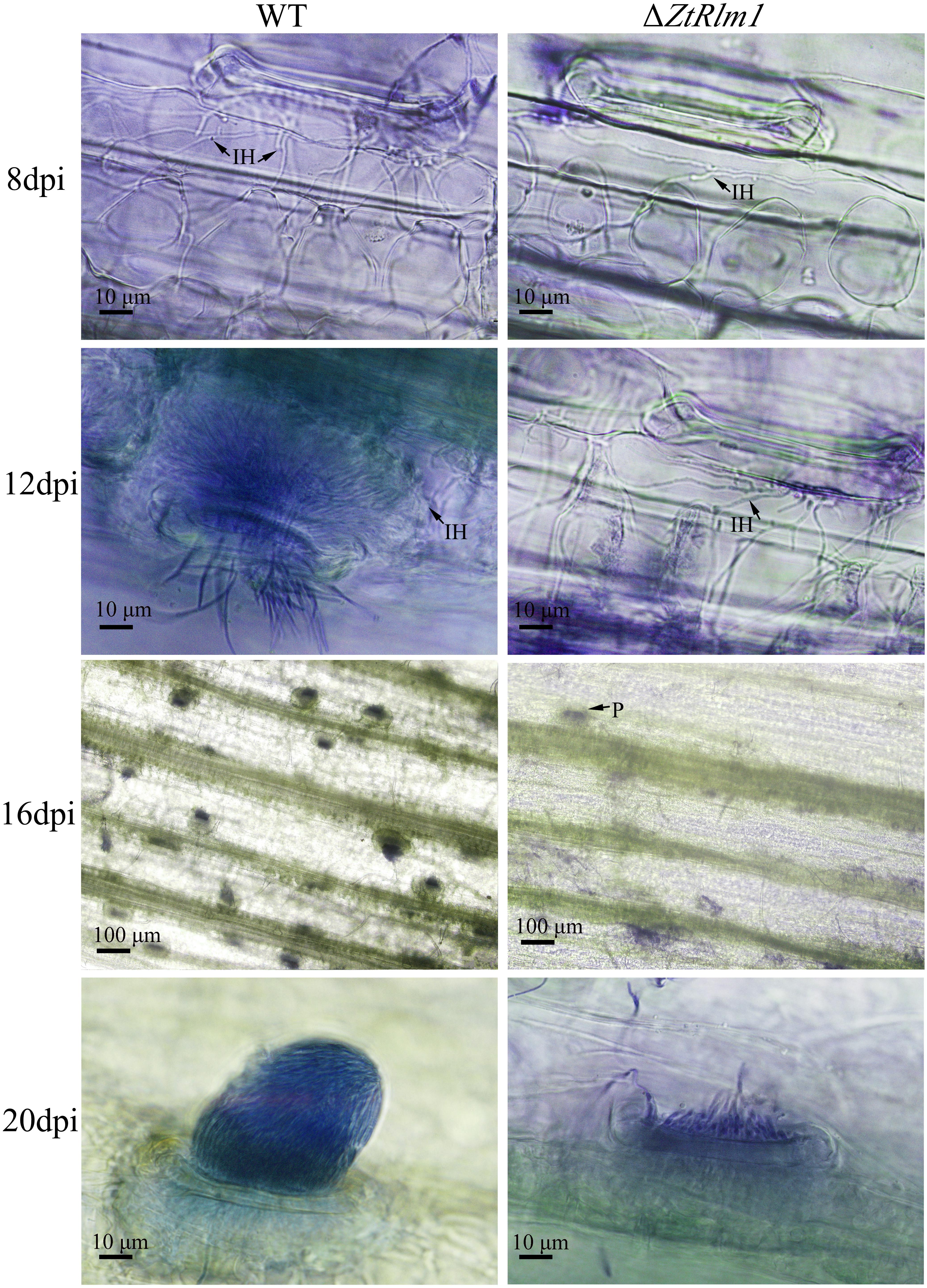
Figure 5. Comparative histopathological analysis of Zymoseptoria tritici WT and ZtRlm1 mutant strain on the susceptible wheat cv. Obelisk at 8, 12, 16, and 20 days post-inoculation (dpi). At 8 dpi, the colonization of mesophyll cells by infectious hypha (IH) of the ZtRlm1 mutant strain was significantly less than the WT strain. At 12 dpi, the colonization intensity of stomatal cavities in the ZtRlm1 mutant was significantly lower than that of the WT strain. At 16 and 20 dpi, the infected stomata by the WT resulted in mature pycnidia (P), while in the ZtRlm1 mutant strain, the majority of infected stomata produced immature pycnidia.
ZtRlm1 Is Induced at Early and Late Stages of Infection
As our result showed that ZtRlm1 acts in the infection process of Z. tritici, we analyzed the transcript abundance of ZtRlm1 in vitro and in planta using a quantitative RT-PCR approach. ZtRlm1 expression is induced during the early stage of infection (2 dpi), and subsequently, its expression was decreased sharply by 8 dpi. Again, relative expression of ZtRlm1 remarkably was triggered by 20 dpi, the stage of infection corresponding to the pycnidial formation. The transcript accumulation of ZtRlm1 in mycelial condition was similar to the in planta expression at 2 dpi, whereas that of ZtRlm1 in yeast-like cells was similar to the in planta expression at 12 dpi coinciding with the asexual reproduction (Figure 6).
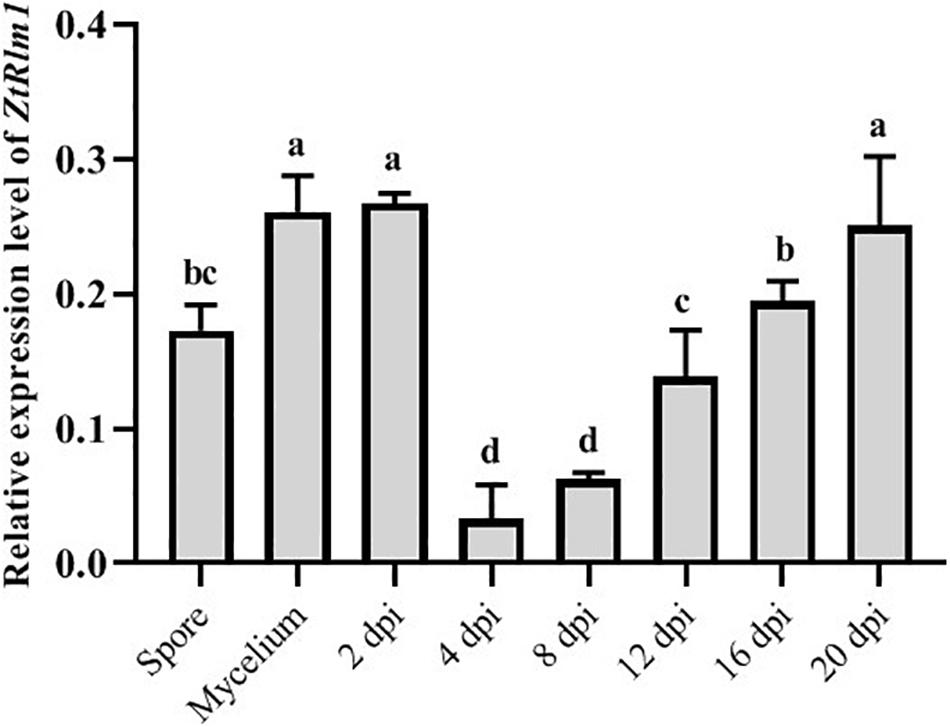
Figure 6. | In vitro and in planta expression levels of Zymoseptoria tritici Rlm1. In vitro conditions (18 and 25°C to induce yeast-like cells and mycelia formation, respectively) were selected to compare the expression levels of ZtRlm1 in mycelial and conidial cultures with in planta conditions. The susceptible cv. Obelisk was infected with the Z. tritici WT strain and inoculated leaves were collected 2, 4, 8, 12, 16, and 20 days after inoculation followed by RNA isolation and cDNA synthesis. The expression level of ZtRlm1 was normalized with the constitutively expressed Z. tritici beta-tubulin gene. Results display the mean ± SD of three biological reps with three technical replicates for each biological replicates. Bars with different alphabets are significantly different (p < 0.05).
ZtRlm1 Impacts Germination Pattern, Hyphal Branching, and Biomass Production
A successful infection requires proper differentiation and the establishment of a parasitic relationship between the pathogen and the host. Differentiation starts from the development of germ tubes that eventually develop into infectious hyphae to penetrate host tissue. We evaluate the impact of ZtRlm1 in germination, growth pattern, hyphal branching, and biomass production, by applying two solid media, including water agar (WA) and potato dextrose agar (PDA) and monitor the germination pattern of ZtRlm1 spores every 12 h (Figure 7). WA is expected to mimic the leaf surface conditions as this induces Z. tritici to establish germ tubes as noticed on leaves surface of wheat before penetration (Kema et al., 1996; Mehrabi and Kema, 2006). On WA, we observed that spores of the control strains germinated from both apical cells and formed primary germ tubes within 12 h. The secondary germ tubes established from the same or other cells of the spore after 24 h, which was followed by the expansion of tertiary hyphal as detected within 36 h. Eventually, the tertiary hyphal developed from the primary and secondary germ tubes after 48 h and established the web of compressed filaments (Figure 7). This showed that the poor medium promotes filamentation growth. Furthermore, we did not detect significantly changed germination patterns for the ΔZtRlm1 strains compared with that of the control strains after 36 h. Nevertheless, the branch intensity and biomass formation were remarkably decreased to 85 and 60% compared with that of the WT control, respectively (Supplementary Tables 2, 3), culminating in less dense filamentation growth compared with that of the WT strain.
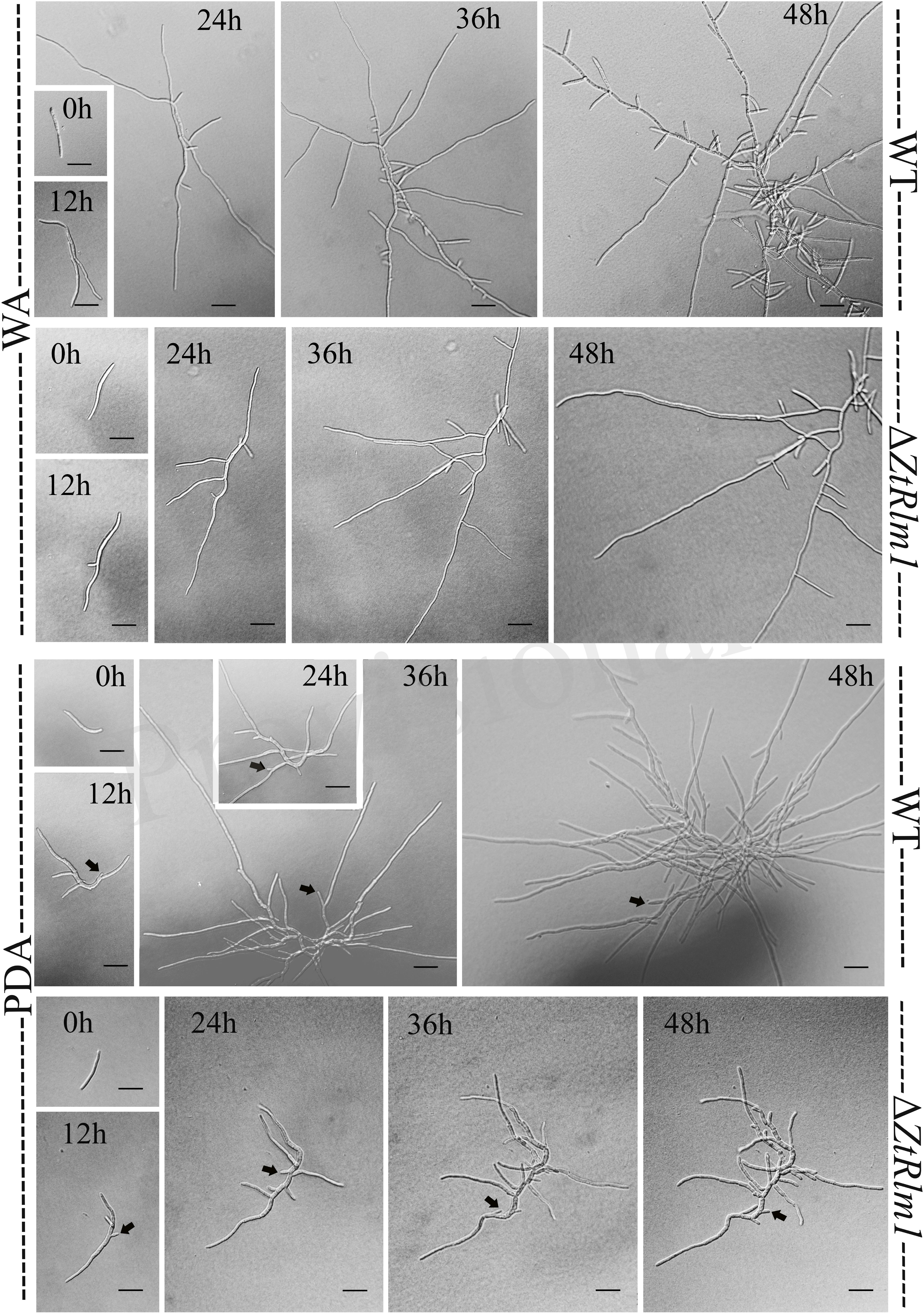
Figure 7. Comparative analysis of the germination pattern of Zymoseptoria tritici ZtRlm1 mutant compared with that of the WT strain over 48 h. Strains were grown on WA (upper panel) and PDA (lower panel) at 20°C under dark conditions. The branch intensity, as well as filamentation, were reduced in the ZtRlm1 mutant resulting in less dense filaments compared to the WT strain. The black arrows indicate the budding cells. Bars represent 20 μm.
The early pattern of colony development on PDA was significantly different in a comparison between the WT and the ZtRlm1 mutant strain. Following germination event, germ tubes did not extend longitudinally but rather develop simultaneously budding cells on intermediate cells of the spore, and on secondary filaments hyphae during the first 24 h continued by producing intensely extra budding cells partially extended in comparatively short filamentous hyphae at 36 h and stored in the center of the dense colony at 48 h. The germination pattern of ZtRlm1 mutant strains was remarkably dissimilar from the WT strain. Furthermore, the filamentation event was much limited and delayed filamentous growth in the ZtRlm1 mutant strain was observed. The hyphae produced by the ZtRlm1 mutant strain was thicker and high bulbous than those formed WT, and germ tubes turned into unusual hyphal swellings after 48 (Figure 7). We confirmed that the branch intensity and biomass production were significantly and statistically reduced by 83 and 89% in comparison with that of the WT control, respectively (Figure 8 and Supplementary Tables 2, 3).
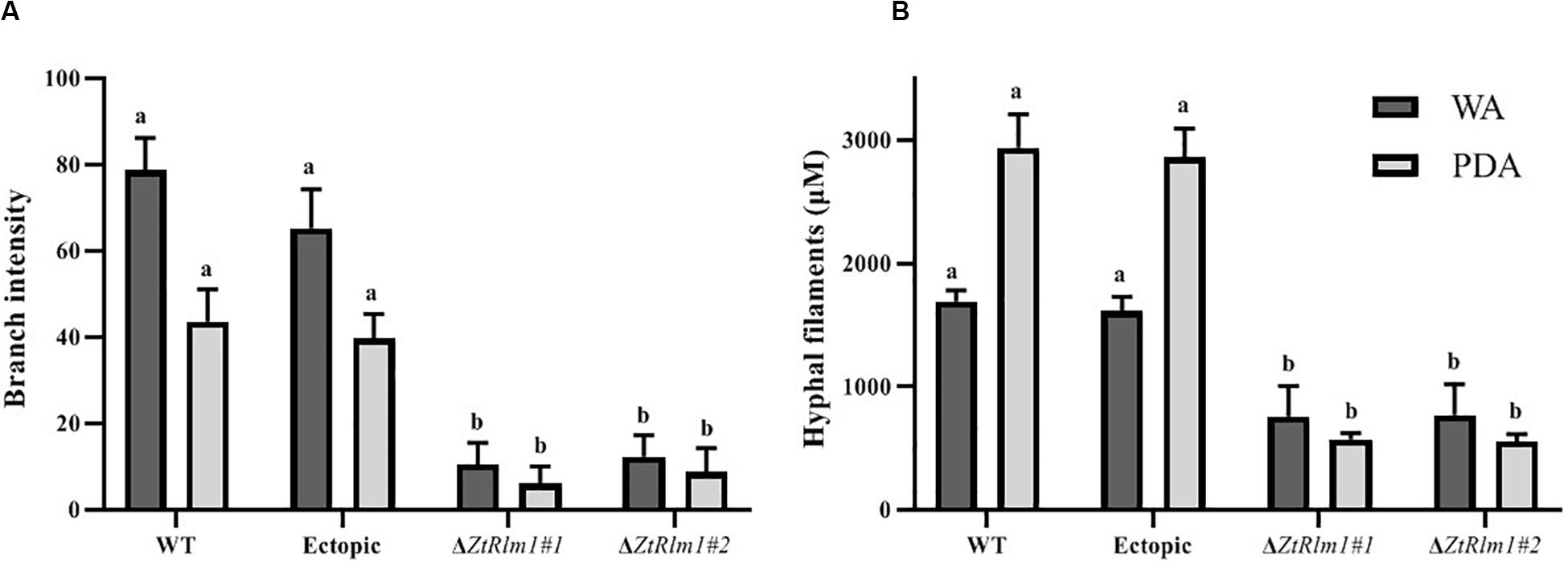
Figure 8. The ZtRlm1 affects growth and development in Zymoseptoria tritici. The WT, ectopic and ZtRlm1 mutant strains were grown on WA and PDA at 20°C in darkness, and the data was recorded after 48 h. The average branch intensity (A) and hyphal filaments (B) of ZtRlm1 mutants were significantly reduced compared with that of the control strains. a,bStatistically significant differences.
ZtRlm1 Affects Melanization Phenomenon
We inoculated the WT and ectopic strains along with the ΔZtRlm1 strains on two diverse solid media (PDA and YMDA) under three temperature conditions for 10 days to investigate the instrumental role of ZtRlm1 in melanization event (16, 20, and 28°C). Colony morphology and pattern growth of the ΔZtRlm1 strains on applied media at three different temperatures did not show significant differences and were similar to that of the control strains. Interestingly, melanization did not occur under all conditions tested, even after extending an incubation period to 1 month (Figure 9).
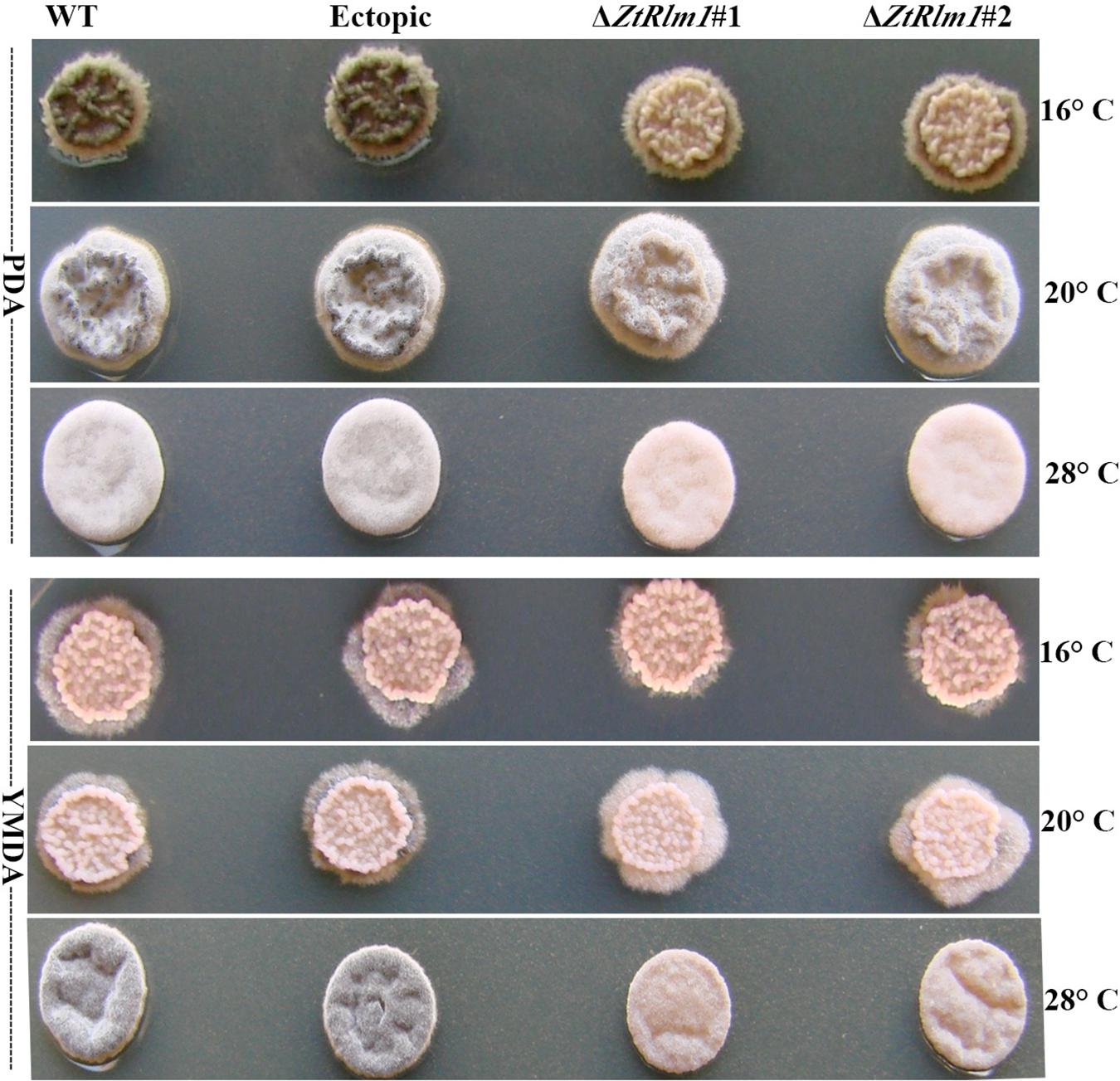
Figure 9. Macroscopic colony morphology of Zymoseptoria tritici WT, ectopic, ΔZtRlm1 #1, ΔZtRlm1 #2 strains under different growing conditions. Approximately 2 μl of spore suspension (107 spores/mL) was spotted on PDA (top panel) or YMDA (bottom panel) and grown for 10 days at 16, 20, and 28°C.
Discussion
To successfully establish a parasitic relationship with its host plant, Z. tritici utilizes diverse tactics to colonize its host efficiently. Up to now, some virulence factors like ZtWor1, and ZtVf1, (Mirzadi Gohari et al., 2014; Mohammadi et al., 2017) have been functionally characterized and to some extent, their contributions to the infection process of Z. tritici were investigated to understand how this fungus evade defense mechanisms and establishes STB. Despite this fact that several pathogenicity factors of Z. tritici being crucial during different stages of the infection process are functionally characterized (Orton et al., 2011; Mirzadi Gohari et al., 2014), molecular strategies underlying Z. tritici infection process remains weakly understood.
Here, we functionally analyzed the biological role of ZtRlm1, and our results revealed that this protein is required for successful infection and an asexual fructification in planta. To fully understand the instrumental role of ZtRlm1 in other developmental processes such as penetration, colonization, and pycnidial formation, a cytological analysis was carried out. Eventually, we demonstrated that ZtRlm1 is involved in penetration, colonization, pycnidial formation, and differentiation (Figure 5).
The members of MADS-box TFs contain a preserved motif within the DNA binding domains of these proteins (Shore and Sharrocks, 1995), and it was demonstrated several proteins belonging to this family have substantial functional roles in a variety of organisms (Mehrabi et al., 2008; Delgado-Silva et al., 2014). Like other filamentous ascomycetes, Z. tritici has only two MADS-box transcription factors (XP_003852780.1 and XP_003853236.1) that we named them as ZtRlm1 and ZtMcm1.
MADS-box TFs possess a wide array of biological functions, and their contributions to diverse developmental processes as well as virulence in fungal pathogens have been functionally investigated. It is shown that Rlm1 homologs in Aspergillus niger, A. nidulans, and Candida albicans play critical roles in maintaining the cell wall integrity (CWI) in reacting to cell wall- perturbing agents through transcriptional regulation of the genes encoding proteins associated with cell wall (Fujioka et al., 2007; Delgado-Silva et al., 2014; Rocha et al., 2016). A. fumigatus RlmA is required for reinforcing the CWI and virulence since mutant strains deleted for RlmA showed a changed cell wall arrangement and tolerance to cell wall stress agents. Additionally, the mutant strains had weakened virulence in a neutropenic murine model of invasive pulmonary aspergillosis (Rocha et al., 2016). Furthermore, it was demonstrated that M. oryzae Mig1, an ortholog of S. cerevisiae Rlm1, is involved in the development of the secondary infectious hyphae inside the living cells of the plant as the deleted strains were blocked in this stage, thereby they were failed to infect rice leaves. Finally, Bcmads1encoding MEF-type MADS-box TF was known to play an instrumental role in the pathogenicity of Botrytis cinerea through its impact on the protein secretion process.
Here, we demonstrated that ZtRlm1 play a significant role in diverse developmental process, and pathogenicity of Z. tritici. Our in vitro phenotyping assays suggested that ZtRlm1 is responsible for hyphal branching growth since lowly branched hypha in ΔZtRlm1 under both WA and PDA media was observed, indicating that this gene is a positive regulator of both hyphal growth and branching. This might be a reasonable explanation to interpret the results of infection assay, and significantly reducing the number of pycnidial formation on plants inoculated by control strains and ΔZtRlm1.
It is generally hypothesized that controlled branching promotes fungal invasion as previously demonstrated that hyphal branching probably increases surface area for colonization, which is a critical issue for successful infection (Harris, 2008). Additionally, hyphal branching might enable the fusion of hyphae, thereby facilitating the process of exchanging genetic materials between diverse hyphae of the same or distinctive fungi. Additionally, in Z. tritici, pycnidial formation is initiated in substomatal cavities by enormous growth, branching and a wide fusion of hyphae deriving originally from 1 or 2 hyphae resulting in exponential improve in fungal biomass generation coinciding with disease symptoms expression (Hilu and Bever, 1957; Kema et al., 1996; Shetty et al., 2003). Several studies have identified TFs such as Tup1 and Rbp1 that are key regulators of filamentious growth (Braun et al., 2000). A mutant strain deleted for tup1 showed uncontrolled filamentous growth under all tested conditions, suggesting a key role in the filamentation process (Braun and Johnson, 1997; Celera and Claderone, 2001). These studies proved that the successful colonization of plants by invading fungal pathogens necessitates efficient and normal hyphal branching, hyphal fusion and colonization, and defect in one of these processes culminated in reduced virulence in host plants.
We demonstrated that ZtRlm1 is a crucial factor playing a major role in the late stage of infection coinciding with pycnidial formation. This data is in agreement with that of in vivo expression analysis when ZtRlm1 expression was highly induced at 20 dpi corresponding to the pycnidia sporulation. Furthermore, in vitro expression of ZtRlm1 at mycelial form is comparable with that of in planta at 2 dpi when yeast-like cells of Z. tritici germinated to produce infectious hyphae just before penetration. Interestingly, we proved that ZtRlm1 is a pivotal regulator of melanization event in the Z. tritici as the examined mutant strains were not melanized under all tested temperature and media conditions. The melanization in Z. tritici is a complex process, and several studies previously indicated that different genetic factors partly have roles in melanin biosynthesis (Mehrabi et al., 2006a, b, Mehrabi and Kema, 2006; Cousin et al., 2006; Lendenmann et al., 2014; Derbyshire et al., 2018). Although our observations demonstrated that ZtRlm1 plays an important function in the melanization phenomenon of the Z. tritici, molecular mechanisms underlying this phenotypic observation is unknown. The future in-depth studies are required to further explore the association of the ZtRlm1 with genes involved in the melanin biosynthesis pathway.
Our previous functional analysis of ZtPKS genes encoding polyketide synthases demonstrated that melanization process is not involved in the pathogenicity of Z. tritici in contrast to other phytopathogens such as Magnaporthe oryzae that has been demonstrated that an appressorium melanization is a central event in the infection process (Casadevall et al., 2000; Jacobson, 2000; Hamilton and Gomez, 2002; Eisenman and Casadevall, 2012). Nevertheless, the melanin is a multifunctional agent, providing defense against environmental stresses but a major role of melanin in plant-pathogenic fungi is the pigment’s contribution to virulence by diminishing the susceptibility of fungi to host defense strategies.
It has been previously shown that the pycnidia of Z. tritici are heavily melanized during the infection process (Kema et al., 1996; Duncan and Howard, 2000; Mehrabi and Kema, 2006), suggesting that melanization event is required for asexual fructification. Thus, this may explain the failure of the ZtRlm1 mutants to produce pycnidia in vitro.
To sum up, we conclude ZtRlm1 is a putative transcriptional regulator in Z. tritici playing a central function in the diverse developmental process, including differentiation, asexual fructification, and pathogenicity. As this TF have combinatorial interactions with other regulatory proteins involved in the signaling cascade, it is required to discover the downstream components of ZtRlm1 that lead to the identification of further pathogenicity factors regulating morpho-pathogenic behavior of Z. tritici. This will contribute to the further understanding of the Z. tritici – wheat pathosystem.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
RM designed the study. AM performed the generation of the required constructs and fungal transformation. NM conducted other described assays in this study and wrote the manuscript with substantial input from RM, AM, MR, and EM. RM and GK coordinated the project. All authors contributed to the article and approved the submitted version.
Funding
This study was partly supported by the Iranian National Scientific Foundation (Project No. 92043155) provided to RM, Tarbiat Modares University, Wageningen University and University of Tehran.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Department of Plant Pathology, Faculty of Agriculture, Tarbiat Modares University to financially support this project, and provide the required facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01976/full#supplementary-material
References
Braun, B. R., Head, W. S., Wang, M. X., and Johnson, A. D. (2000). Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156, 31–44.
Braun, B. R., and Johnson, A. D. (1997). Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277, 105–109. doi: 10.1126/science.277.5322.105
Brown, J. K., Chartrain, L., Lasserre-Zuber, P., and Saintenac, C. (2015). Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genet. Biol. 79, 33–41. doi: 10.1016/j.fgb.2015.04.017
Casadevall, A., Rosas, A. L., and Nosanchuk, J. D. (2000). Melanin and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 3, 354–358. doi: 10.1016/s1369-5274(00)00103-x
Celera, J. A., and Claderone, R. (2001). Signalling and the Biology of Human Fungal Pathogens. Fungal Pathogenesis: Principles and Clinical Applications. New York, NY: Marcel Dekker, 115–137.
Chen, R., and McDonald, B. A. (1996). Sexual reproduction plays a major role in the genetic structure of populations of the fungus Mycosphaerella graminicola. Genetic 142, 1119–1127.
Cousin, A., Mehrabi, R., Guilleroux, M., Dufresne, M., Van Der Lee, T., Waalwijk, C., et al. (2006). The MAP kinase-encoding gene MgFus3 of the non appressorium phytopathogen Mycosphaerella graminicola is required for penetration and in vitro pycnidia formation. Mol. Plant Pathol. 7, 269–278. doi: 10.1111/j.1364-3703.2006.00337.x
de Nadal, E., Casadomé, L., and Posas, F. (2003). Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol. Cell Biol. 23, 229–237. doi: 10.1128/mcb.23.1.229-237.2003
De Wit, P. J. G. M., Mehrabi, R., Van Den Burg, H. A., and Stergiopoulos, I. (2009). Fungal effector proteins: past, present and future. Mol. Plant Pathol. 10, 735–747. doi: 10.1111/j.1364-3703.2009.00591.x
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Delgado-Silva, Y., Vaz, C., Carvalho-Pereira, J., Carneiro, C., Nogueira, E., Correia, A., et al. (2014). Participation of Candida albicans transcription factor RLM1 in cell wall biogenesis and virulence. PLoS One 9:e86270. doi: 10.1371/journal.pone.0086270
Dellaporta, S. L., Wood, J., and Hicks, J. B. (1983). A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1, 19–21. doi: 10.1007/bf02712670
Derbyshire, M. C., Gohari, A. M., Mehrabi, R., Kilaru, S., Steinberg, G., Ali, S., et al. (2018). Phosphopantetheinyl transferase (Ppt)-mediated biosynthesis of lysine, but not siderophores or DHN melanin, is required for virulence of Zymoseptoria tritici on wheat. Sci. Rep. 8:17069.
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Duncan, K. E., and Howard, R. J. (2000). Cytological analysis of wheat infection by the leaf blotch pathogen Mycosphaerella graminicola. Mycol. Res. 104, 1074–1082. doi: 10.1017/s0953756299002294
Eisenman, H. C., and Casadevall, A. (2012). Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93, 931–940. doi: 10.1007/s00253-011-3777-2
Eyal, Z., and Levy, E. (1987). Variations in pathogenicity patterns of Mycosphaerella graminicola within Triticum Spp. in Israel. Euphytica 36, 237–250. doi: 10.1007/bf00730670
Eyal, Z., Scharen, A. L., Prescott, J. M., and van Ginkel, M. (1987). The Septoria Diseases of Wheat: Concepts and Methods of Disease Management. Mexico: CIMMYT.
Fones, H., and Gurr, S. (2015). The impact of Septoria tritici Blotch disease on wheat: an EU perspective. Fungal Genet. Biol. 79, 3–7. doi: 10.1016/j.fgb.2015.04.004
Fraaije, B. A., Cools, H. J., Fountaine, J., Lovell, D. J., Motteram, J., West, J. S., et al. (2005). Role of ascospores in further spread of QoI-resistant cytochrome b alleles (G143A) in field populations of Mycosphaerella graminicola. Phytopathology 95, 933–941. doi: 10.1094/phyto-95-0933
Frandsen, R., Andersson, J., Kristensen, M., and Giese, H. (2008). Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 9:70. doi: 10.1186/1471-2199-9-70
Fujioka, T., Mizutani, O., Furukawa, K., Sato, N., Yoshimi, A., Yamagata, Y., et al. (2007). MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 6, 1497–1510. doi: 10.1128/EC.00281-06
Hamilton, A. J., and Gomez, B. L. (2002). Melanins in fungal pathogens. J. Med. Microbiol. 51, 189–191. doi: 10.1099/0022-1317-51-3-189
Harris, S. D. (2008). Branching of fungal hyphae: regulation, mechanisms and comparison with other branching systems. Mycologia 100, 823–832. doi: 10.3852/08-177
Hilu, H. M., and Bever, W. M. (1957). Inoculation, oversummering, and suscept—pathogen relationship of Septoria tritici on Triticum species. Phytopathology 47, 474–480.
Hunter, T., Coker, R. R., and Royle, D. J. (1999). The teleomorph stage, Mycosphaerella graminicola, in epidemics of septoria tritici blotch on winter wheat in the UK. Plant Pathol. 48, 51–57. doi: 10.1046/j.1365-3059.1999.00310.x
Jacobson, E. S. (2000). Pathogenic roles for fungal melanins. Clin. Microbiol. Rev. 13, 708–717. doi: 10.1128/cmr.13.4.708
Kema, G. H. J., and van Silfhout, C. H. (1997). Genetic variation for virulence and resistance in the wheat-Mycosphaerella graminicola pathosystem. III. Comparative seedling and adult plant experiments. Phytopathology 87, 266–272. doi: 10.1094/phyto.1997.87.3.266
Kema, G. H. J., Yu, D., Rijkenberg, F. H. J., Shaw, M. W., and Baayen, R. P. (1996). Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology 86, 777–786.
Keon, J., Antoniw, J., Carzaniga, R., Deller, S., Ward, J. L., Baker, J. M., et al. (2007). Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant Microbe Interact. 20, 178–193. doi: 10.1094/mpmi-20-2-0178
Lendenmann, M. H., Croll, D., Stewart, E. L., and McDonald, B. A. (2014). Quantitative trait locus mapping of melanization in the plant pathogenic fungus Zymoseptoria tritici. G3 Genes Genomes Genetics 4, 2519–2533. doi: 10.1534/g3.114.015289
Lin, L., Ye, W., Wu, J., Xuan, M., Li, Y., Gao, J., et al. (2018). The MADS-box transcription factor PsMAD1 is involved in zoosporogenesis and pathogenesis of Phytophthora sojae. Front. Microbiol. 9:2259. doi: 10.3389/fmicb.2018.02259
Mehrabi, R., Ben, M., Barek, S., Van der Lee, T. A. J., Waalwijk, C., de Wit, P. J. G. M., et al. (2009). G alpha and G beta proteins regulate the cyclic amp pathway that is required for development and pathogenicity of the phytopathogen Mycosphaerella graminicola. Eukaryot. Cell 8, 1001–1013. doi: 10.1128/ec.00258-08
Mehrabi, R., Ding, S., and Xu, J. R. (2008). MADS-box transcription factor Mig1 is required for infectious growth in Magnaporthe oryzae. Eukaryot. Cell 7, 791–799. doi: 10.1128/ec.00009-08
Mehrabi, R., and Kema, G. H. J. (2006). Protein kinase a subunits of the ascomycete pathogen Mycosphaerella graminicola regulate asexual fructification, filamentation, melanization and osmosensing. Mol. Plant Pathol. 7, 565–577. doi: 10.1111/j.1364-3703.2006.00361.x
Mehrabi, R., Taga, M., and Kema, G. H. (2007). Electrophoretic and cytological karyotyping of the foliar wheat pathogen Mycosphaerella graminicola reveals many chromosomes with a large size range. Mycologia 99, 868–876. doi: 10.3852/mycologia.99.6.868
Mehrabi, R., Van der Lee, T., Waalwijk, C., and Kema, G. H. J. (2006a). MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant Microbe Interact. 19, 389–398. doi: 10.1094/mpmi-19-0389
Mehrabi, R., Zwiers, L. H., de Waard, M. A., and Kema, G. H. J. (2006b). MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 19, 1262–1269. doi: 10.1094/mpmi-19-1262
Messenguy, F., and Dubois, E. (2003). Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316, 1–21. doi: 10.1016/s0378-1119(03)00747-9
Mirzadi Gohari, A., Mehrabi, R., Robert, O., Ince, I. A., Boeren, S., Schuster, M., et al. (2014). Molecular characterization and functional analyses of ZtWor1, a transcriptional regulator of the fungal wheat pathogen Zymoseptoria tritici. Mol. Plant Pathol. 15, 394–405. doi: 10.1111/mpp.12102
Mohammadi, N., Mehrabi, R., Gohari, A. M., Goltapeh, E. M., Safaie, N., and Kema, G. H. J. (2017). The ZtVf1 transcription factor regulates development and virulence in the foliar wheat pathogen Zymoseptoria tritici. Fungal Genet. Biol. 109, 26–35. doi: 10.1016/j.fgb.2017.10.003
Morais, D., Duplaix, C., Sache, I., Laval, V., Suffert, F., and Walker, A. S. (2019). Overall stability in the genetic structure of a Zymoseptoria tritici population from epidemic to interepidemic stages at a small spatial scale. Eur. J. Plant Pathol. 154, 423–436. doi: 10.1007/s10658-018-01666-y
Motteram, J., Küfner, I., Deller, S., Brunner, F., Hammond-Kosack, K. E., Nürnberger, T., et al. (2009). Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain-containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 22, 790–799. doi: 10.1094/mpmi-22-7-0790
Orton, E. S., Deller, S., and Brown, J. K. M. (2011). Mycosphaerella graminicola: from genomics to disease control. Mol. Plant Pathol. 12, 413–424. doi: 10.1111/j.1364-3703.2010.00688.x
Quaedvlieg, W., Kema, G. H. J., Groenewald, J. Z., Verkley, G. J. M., Seifbarghi, S., Razavi, M., et al. (2011). Zymoseptoria gen. nov.: a new genus to accommodate Septoria-like species occurring on graminicolous hosts. Persoonia 26, 57–69. doi: 10.3767/003158511x571841
Rocha, M. C., Fabri, J. H. T. M., de Godoy, K. F., de Castro, P. A., Hori, J. I., da Cunha, A. F., et al. (2016). Aspergillus fumigatus MADS-Box transcription factor rlmA is required for regulation of the cell wall integrity and virulence. G3 Genes Genomes Genetics 6, 2983–3002.
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shetty, N. P., Kristensen, B. K., Newman, M. A., Møller, K., Gregensen, P. L., and Jørgensen, H. J. L. (2003). Association of hydrogen peroxide with restriction of Septoria tritici in wheat. Physiol. Mol. Plant Pathol. 62, 333–346. doi: 10.1016/s0885-5765(03)00079-1
Shore, P., and Sharrocks, A. D. (1995). The MADS−box family of transcription factors. Eur. J. Biochem. 229, 1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x
Keywords: Zymoseptoria tritici, gene deletion, pathogenicity assay, phenotyping, Agrobacterium tumefaciens-mediated transformation
Citation: Mohammadi N, Mehrabi R, Mirzadi Gohari A, Roostaei M, Mohammadi Goltapeh E, Safaie N and Kema GHJ (2020) MADS-Box Transcription Factor ZtRlm1 Is Responsible for Virulence and Development of the Fungal Wheat Pathogen Zymoseptoria tritici. Front. Microbiol. 11:1976. doi: 10.3389/fmicb.2020.01976
Received: 10 March 2020; Accepted: 27 July 2020;
Published: 18 August 2020.
Edited by:
Maria Rosa Simon, National University of La Plata, ArgentinaReviewed by:
Javier Palma-Guerrero, ETH Zürich, SwitzerlandFIona M. Doohan, University College Dublin, Ireland
Copyright © 2020 Mohammadi, Mehrabi, Mirzadi Gohari, Roostaei, Mohammadi Goltapeh, Safaie and Kema. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rahim Mehrabi, bWVocmFiaUBpdXQuYWMuaXI=; cmFoaW0ubWVocmFiaUBnbWFpbC5jb20=
 Naser Mohammadi
Naser Mohammadi Rahim Mehrabi
Rahim Mehrabi Amir Mirzadi Gohari3
Amir Mirzadi Gohari3