- 1Department of Medical Microbiology, Section Experimental Bacteriology, Leiden University Medical Center, Leiden, Netherlands
- 2Centre for Microbial Cell Biology, Leiden, Netherlands
- 3Institute of Biotechnology, Technische Universität Berlin, Berlin, Germany
Faithful DNA replication is crucial for viability of cells across all kingdoms. Targeting DNA replication is a viable strategy for inhibition of bacterial pathogens. Clostridioides difficile is an important enteropathogen that causes potentially fatal intestinal inflammation. Knowledge about DNA replication in this organism is limited and no data is available on the very first steps of DNA replication. Here, we use a combination of in silico predictions and in vitro experiments to demonstrate that C. difficile employs a bipartite origin of replication that shows DnaA-dependent melting at oriC2, located in the dnaA-dnaN intergenic region. Analysis of putative origins of replication in different clostridia suggests that the main features of the origin architecture are conserved. This study is the first to characterize aspects of the origin region of C. difficile and contributes to our understanding of the initiation of DNA replication in clostridia.
Introduction
Clostridioides difficile (formerly Clostridium difficile) (Lawson et al., 2016) is a Gram-positive anaerobic bacterium. C. difficile infections (CDI) can occur in individuals with a disturbed microbiota and is one of the main causes of hospital associated diarrhea, but can also be found in the environment (Smits et al., 2016). The incidence of CDI has increased worldwide since the beginning of the century (Smits et al., 2016; Warriner et al., 2017). Consequently, the interest in the physiology of the bacterium has increased as a way to understand its interaction with the host and the environment and to explore news pathways for intervention (van Eijk et al., 2017; Crobach et al., 2018).
One such pathway is the replication of the chromosome. Overall, DNA replication is a highly conserved process across different kingdoms (O’Donnell et al., 2013; Bleichert et al., 2017). In all bacteria, DNA replication is a tightly regulated process that occurs with high fidelity and efficiency, and is essential for cell survival. The process involves many different proteins that are required for the replication process itself, or to regulate and aid replisome assembly and activity (Katayama et al., 2010; Murray and Koh, 2014; Chodavarapu and Kaguni, 2016; Jameson and Wilkinson, 2017; Schenk et al., 2017). Replication initiation and its regulation arguably are candidates for the search for novel therapeutic targets (Fossum et al., 2008; Grimwade and Leonard, 2017; van Eijk et al., 2017).
In most bacteria, replication of the chromosome starts with the assembly of the replisome at the origin of replication (oriC) and proceeds bidirectionally (Chodavarapu and Kaguni, 2016). In the majority of bacteria replication is initiated by the DnaA protein, an ATPase Associated with diverse cellular Activities (AAA+ protein) that binds specific sequences in the oriC region. The binding of DnaA induces DNA duplex unwinding, which subsequently drives the recruitment of other proteins, such as the replicative helicase, primase and DNA polymerase III proteins (Chodavarapu and Kaguni, 2016). Termination of replication eventually leads to disassembly of the replication complexes (Chodavarapu and Kaguni, 2016).
In C. difficile, knowledge on DNA replication is limited. Though many proteins appear to be conserved between well-characterized species and C. difficile, only certain replication proteins have been experimentally characterized for C. difficile (Torti et al., 2011; Briggs et al., 2012; van Eijk et al., 2016). DNA polymerase C (PolC, CD1305) of C. difficile has been studied in the context of drug-discovery and appears to have a conserved primary structure similar to other low-[G + C] gram-positive organisms (Torti et al., 2011). It is inhibited in vitro and in vivo by compounds that compete for binding with dGTP (van Eijk et al., 2019; Xu et al., 2019). Helicase (CD3657), essential for DNA duplex unwinding, was found to interact in an ATP-dependent manner with a helicase loader (CD3654) and loading was proposed to occur through a ring-maker mechanism (Davey and O’Donnell, 2003; van Eijk et al., 2016). However, in contrast to helicase of the Firmicute Bacillus subtilis, C. difficile helicase activity is dependent on activation by the primase protein (CD1454), as has also been described for Helicobacter pylori (Bazin et al., 2015; van Eijk et al., 2016). C. difficile helicase stimulates primase activity at the trinucleotide 5′d(CTA), but not at the preferred trinucleotide 5′-d(CCC) (van Eijk et al., 2016).
DnaA of C. difficile has not been studied to date. Although no full-length structure has been determined for DnaA, individual domains of the DnaA protein from different organisms have been characterized (Majka et al., 1997; Zawilak et al., 2003; Erzberger et al., 2006; Zawilak-Pawlik et al., 2017). DnaA proteins generally comprise four domains (Zawilak-Pawlik et al., 2017). Domain I is involved in protein-protein interactions and is responsible for DnaA oligomerization (Weigel et al., 1999; Abe et al., 2007; Natrajan et al., 2009; Jameson et al., 2014; Kim et al., 2017; Zawilak-Pawlik et al., 2017; Martin et al., 2018; Matthews and Simmons, 2019; Nowaczyk-Cieszewska et al., 2020). Little is known about a specific function of Domain II and this domain may even be absent (Erzberger et al., 2002). It is thought to be a flexible linker that promotes the proper conformation of the other DnaA domains (Abe et al., 2007; Nozaki and Ogawa, 2008). Domain III and Domain IV are responsible for the DNA binding. Domain III contains the AAA+ motif and is responsible for binding ATP, ADP and single-stranded DNA, as well as certain regulatory proteins (Kawakami et al., 2005; Cho et al., 2008; Ozaki et al., 2008; Ozaki and Katayama, 2012). Recent studies have also revealed the importance of this domain for binding phospholipids present in the bacterial membrane (Saxena et al., 2013). The C-terminal Domain IV contains a helix-turn-helix motif (HTH) and is responsible for the specific binding of DnaA to so called DnaA boxes (Blaesing et al., 2000; Erzberger et al., 2002; Fujikawa et al., 2003).
DnaA boxes are typically 9-mer non-palindromic DNA sequences, and the Escherichia coli DnaA box consensus sequence is TTWTNCACA (Schaper and Messer, 1995; Wolanski et al., 2014). The boxes can differ in their affinity for DnaA, and even demonstrate different dependencies on the ATP co-factor (Speck et al., 1999; Patel et al., 2017). Binding of Domain IV to the DnaA boxes promotes higher-order oligomerization of DnaA, forming a filament that wraps around DNA (Erzberger et al., 2006; Ozaki et al., 2012; Scholefield and Murray, 2013). It is thought that the interaction of the DnaA filament with the DNA helix introduces a bend in the DNA (Erzberger et al., 2006; Patel et al., 2017). The resulting superhelical torsion facilitates the melting of the adjacent [A + T]-rich DNA Unwinding Element (DUE) (Kowalski and Eddy, 1989; Erzberger et al., 2006; Zorman et al., 2012). Upon melting, the DUE provides the entry site for the replisomal proteins. Another conserved structural motif, a triplet repeat called DnaA-trio, is involved in the stabilization of the unwound region (Richardson et al., 2016, 2019).
The oriC region has been characterized for several bacterial species. These analyses show that oriC regions are quite diverse in sequence, length and even chromosomal location, all of which contribute to species-specific replication initiation requirements (Zawilak-Pawlik et al., 2005; Ekundayo and Bleichert, 2019). In Firmicutes, including C. difficile, the genomic context of the origin regions appears to be conserved and encompasses the rnpA-rpmH-dnaA-dnaN genes (Ogasawara and Yoshikawa, 1992; Briggs et al., 2012).
The oriC region can be continuous (i.e., located at a single chromosomal locus) or bipartite (Wolanski et al., 2014). Bipartite origins were initially identified in B. subtilis (Moriya et al., 1988) but more recently also in H. pylori (Donczew et al., 2012). The separated subregions of the bipartite origin, oriC1 and oriC2, are usually separated by the dnaA gene. Both oriC1 and oriC2 contain clusters of DnaA boxes, and one of the regions contains the DUE region. The DnaA protein binds to both subregions and places them in close proximity to each other, consequently looping out the dnaA gene (Krause et al., 1997; Donczew et al., 2012). In H. pylori, DnaA Domain I and II are important for maintaining the interactions between both oriC regions (Nowaczyk-Cieszewska et al., 2020).
In this study, we identified the putative oriC of C. difficile through in silico analysis and demonstrate DnaA-dependent unwinding of the oriC2 region in vitro. A clear conservation of the origin of replication organization is observed throughout the clostridia. The present study contributes to our understanding of clostridial DNA replication initiation in general, and replication initiation of C. difficile specifically.
Materials and Methods
Sequence Alignments and Structure Modeling
Multiple sequence alignment of amino acid sequences was performed with Protein BLAST (blastP suite)1 for individual alignment scores and the PRALINE program2 (Bawono and Heringa, 2014) for multiple sequence alignment. Sequences were retrieved from the NCBI Reference Sequences. DnaA protein sequences from C. difficile 630Δerm (CEJ96502.1), C. acetobutylicum DSM 1731 (AEI33799.1), Bacillus subtilis 168 (NP_387882.1), Escherichia coli K-12 (AMH32311.1), Streptomyces coelicolor A3(2) (TYP16779.1), Mycobacterium tuberculosis RGTB327 (AFE14996.1), Helicobacter pylori J99 (Q9ZJ96.1) and Aquifex aeolicus (WP_010880157.1) were selected for alignment. Alignment was visualized in Jalview version 2.11, with coloring by percentage identity.
Secondary structure prediction and homology modeling were performed using Phyre23 (Kelley et al., 2015) using the intensive default settings. Phyre2 modeling of C. difficile 630Δerm DnaA (CEJ96502.1) was performed with 3 templates from A. aeolicus (PDB 2HCB, chain C), B. subtilis (PDB 4TPS, chain D) and E. coli (PDB 2E0G, chain A) and 21 residues were modeled ab initio. 95% of the residues were modeled with >90% confidence. Graphical representation was performed with the PyMOL Molecular Graphics System, Version 1.76.6. (Schrödinger, LLC).
Prediction of the C. difficile oriC
To identify the oriC region of C. difficile the genome sequence of C. difficile 630Δerm (GenBank accession no. LN614756.1) was analyzed through different software in a stepwise procedure (Mackiewicz et al., 2004).
The GenSkew Java Application4 was used with default settings for the analysis of the normal and the cumulative skew of two selectable nucleotides of the genomic nucleotide sequence [(G−C)/(G + C)]. Calculations where performed with a window size of 4293 bp and a step size of 4293 bp. The inflection values of the cumulative GC skew plot are indicative of the chromosomal origin (oriC) and terminus of replication (ter).
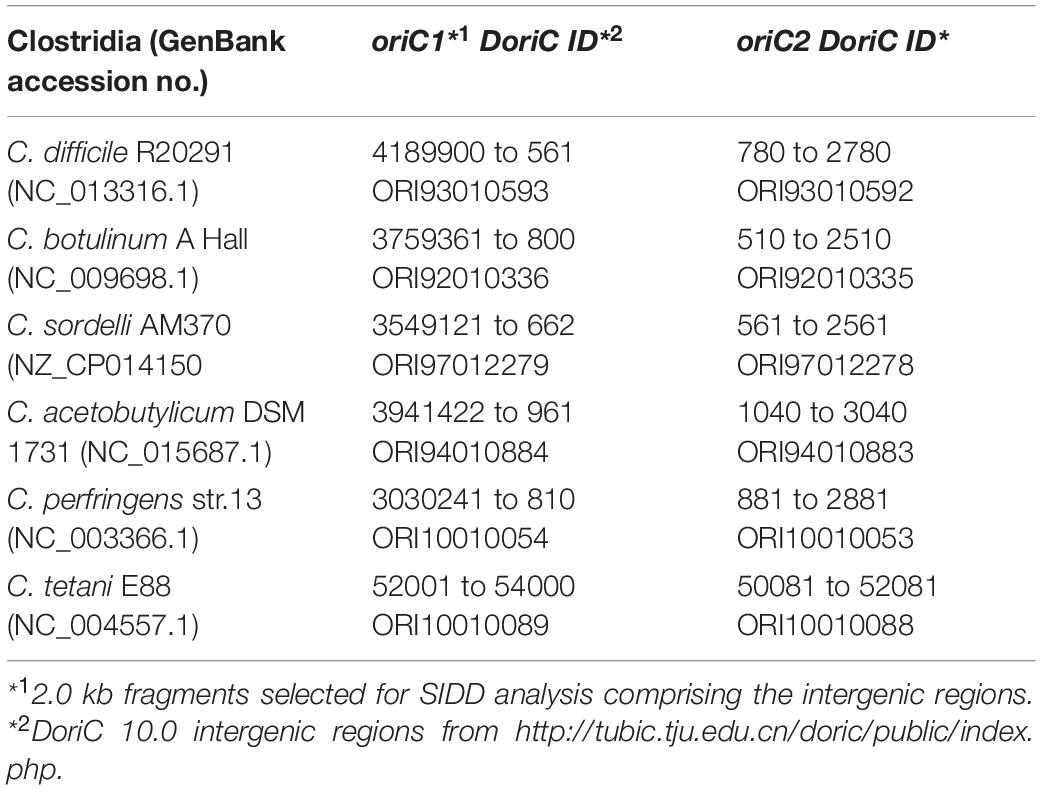
Prediction of superhelicity-dependent helically unstable DNA stretches (SIDDs) was performed in the vicinity of the inflection point of the GC-skew plot, in 2.0 kb fragments comprising intergenic regions from nucleotide position 4291795 to 745 (oriC1) and 466 to 2465 (oriC2) of the C. difficile 630Δerm chromosome. Prediction of the SIDDs in the different clostridia (Table 1) was performed in the vicinity of the inflection points of the GC-plot retrieved from DoriC 10.0 database5 (Luo and Gao, 2019), in 2.0 kb fragments comprising intergenic regions summarized in Table 1. The SIST program6 (Zhabinskaya et al., 2015) was used to predicted free energies G(x) by running the melting transition algorithm only (SIDD) with default values (copolymeric energetics; default: σ = −0.06; T = 37°C; x = 0.01 M) and with superhelical density σ = −0.04.
We performed the identification of the DnaA box clusters by search of the motif TTWTNCACA with one mismatch (Supplementary Material) in the leading strand on a 4432 bp sequence between the nucleotide position 4291488 to 2870 of the C. difficile 630Δerm chromosome, using Pattern Locator7 (Mrazek and Xie, 2006). Identification of the DnaA boxes in the different clostridia was performed with the same pattern motif in the leading strand of the intergenic regions summarized in Table 1.
DnaA-trio sequences and ribosomal binding sites where manually predicted based on Richardson et al. (2016) and Vellanoweth and Rabinowitz (1992), respectively.
All output data was obtained as raw text files and further processed with Prism 8.3.1 (GraphPad, Inc., La Jolla, CA, United States) and CorelDRAW X7 (Corel).
Strains and Growth Conditions
Escherichia coli strains were grown aerobically at 37°C in lysogeny broth (LB, Affymetrix) supplemented with 15 μg/mL chloramphenicol or 50 μg/mL kanamycin when required. E. coli strains DH5α and MC1061 (Table 2) were used to maintain dnaA- and oriC-containing plasmids, respectively. E. coli strain MS3898, kindly provided by Alan Grossman (MIT, Cambridge, United States) (Table 2) was used for recombinant DnaA expression. E. coli transformation was performed using standard procedures (Sambrook et al., 1989). The growth was followed by monitoring the optical density at 600 nm (OD600).
Construction of the Plasmids
For overexpression of DnaA, the dnaA nucleotide sequence (CEJ96502.1) from C. difficile 630Δerm (GenBank accession no. LN614756.1) was amplified by PCR from C. difficile 630Δerm genomic DNA using primers oEVE-7 and oEVE-21 (Table 3). The PCR product was subsequently digested with NcoI and BglII. The vector pAV13 (Smits et al., 2011; Table 4), containing B. subtilis dnaA cloned in pQE60 (Qiagen) was kindly provided by Alan Grossman (MIT, Cambridge, MA, United States) and was digested with the same enzymes and ligated to the digested fragment to yield vector pEVE40 (Table 4).
To construct a plasmid carrying the complete predicted oriC, the predicted oriC region (nucleotide 4292150 to 1593 from C. difficile 630 GenBank accession no. LN614756.1) was amplified by PCR from C. difficile 630Δerm genomic DNA using primers oAP40 and oAP41 (Table 3). The PCR product was subsequently digested with EcoRI and PstI and ligated into pori1ori2 (Table 4), kindly provided by Anna Zawilak-Pawlik (Hirszfeld Institute of Immunology and Experimental Therapy, PAS, Wrocław, Poland), that was digested with the same enzymes, to yield vector pAP205 (Table 4).
For the cloning of the predicted oriC1 region (nucleotide 4292150 to 24 of C. difficile 630Δerm genomic DNA) the primer set oAP30/oAP31 (Table 3) was used. The amplified fragment was digested with EcoRI and PstI and inserted onto pori1ori2 digested with same enzymes, yielding vector pAP83 (Table 4). For the cloning of the predicted oriC2 region (nucleotide 1291 to the 1593 of C. difficile 630Δerm genomic DNA) the primer set oAP32/oAP33 (Table 3) was used. The amplified fragment was digested with EcoRI and PstI and inserted onto pori1ori2 (Table 4) digested with same enzymes, yielding vector pAP76 (Table 4).
All DNA sequences introduced into the cloning vectors were verified by Sanger sequencing. For oriC containing vectors primers oAP56 and oAP57 (Table 3) were used for sequencing.
Overproduction and Purification of DnaA-6xHis
Overexpression of DnaA-6xHis was carried out in E. coli strain CYB1002 (Table 2), harboring the expression plasmid pEVE40 (Table 4). Cells were grown in 800 mL LB and induced with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at an OD600 of 0.6 for 3 h. The cells were collected by centrifugation at 4°C and stored at −80°C. Cells were resuspended in Binding buffer (1X Phosphate buffer pH7.4, 10 mM Imidazole, 10% glycerol) lysed by French Press and collected in phenylmethylsulfonyl fluoride (PMSF) at 0.1 mM (end concentration). Separation of the soluble fraction was performed by centrifugation at 13000 × g at 4°C for 20 min. Purification of the protein from the soluble fraction was done in Binding buffer on a 1 mL HisTrap Column (GE Healthcare) according to manufacturer’s instructions. Elution was performed with Binding buffer in stepwise increasing concentrations of imidazole (20, 60, 100, 300, and 500 mM). DnaA-6xHis was mainly eluted at a concentration of imidazole equal to or greater than 300 mM.
Fractions containing the DnaA-6xHis protein were pooled together and applied to Amicon Ultra Centrifugal Filters with 30 kDa cutoff (Millipore). Buffer was exchanged to Buffer A (25 mM HEPES-KOH pH 7.5, 100 mM K-glutamate, 5 mM Mg-acetate, 10% glycerol). The concentrated DnaA-6xHis protein was subjected to size exclusion chromatography on an Äkta pure instrument (GE Healthcare). 200 μL of concentrated DnaA-6xHis was applied to a Superdex 200 Increase 10/30 column (GE Healthcare) in buffer A at a flow rate of 0.5 ml min–1. UV detection was done at 280 nm. The column was calibrated with a mixture of proteins of known molecular weights (Mw): thyroglobulin (669 kDa), Apoferritin (443 kDa), β-amylase (200 kDa), Albumin (66 kDa), and Carbonic anhydrase (29 kDa). Eluted fractions containing DnaA-6xHis of the expected molecular weight (51 kDa) were quantified and visualized by Coomassie. Pure fractions were aliquoted and stored at −80°C for further experiments.
Immunoblotting and Detection
For immunoblotting, proteins were separated on a 12% SDS-PAGE gel and transferred onto nitrocellulose membranes (Amersham), according to the manufacturer’s instructions. The membranes were probed in PBST (PBS pH 7.4, 0.05% (v/v) Tween-20) with a mouse anti-his antibody (1:3000, Invitrogen) and a secondary goat anti-mouse-HRP antibody (1:3000, DAKO) was used. The membranes were visualized using the chemiluminescence detection kit Clarity ECL Western Blotting Substrates (Bio-Rad) in an Alliance Q9 Advanced machine (Uvitec).
P1 Nuclease Assay
For the P1 nuclease assay, 100 ng pAP205 plasmid was incubated with increasing concentrations of DnaA-6xHis (0.14, 0.54, 1, and 6.3 μM), when required, in P1 buffer (25 mM Hepes-KOH (pH 7.6), 12% (v/v) glycerol, 1 mM CaCl2, 0.2 mM EDTA, 5 mM ATP, 0.1 mg/ml BSA), at 30°C for 12 min. 0.75 unit of P1 nuclease (Sigma), resuspended in 0.01 M sodium acetate (pH 7.6) was added to the reaction and incubated at 30°C for 5 min. 220 μl of buffer PB (Qiagen) was added and the fragments purified with the minElute PCR Purification Kit (Qiagen), according to manufacturer’s instructions. Digestion with BglII, NotI or ScaI (NEB) of the purified fragments was performed according to manufacturer’s instructions for 1 h at 37°C. Digested samples were resolved on 1% agarose gels in 0.5xTAE (40 mM Tris, 20 mM CH-COOH, 1 mM EDTA PH 8.0) and stained with 0.01 mg/mL ethidium bromide solution afterward. Visualization of the gels was performed on the Alliance Q9 Advanced machine (Uvitec). Images were processed in CorelDraw X7 software. For all experiments at least three independent replicates were performed with various concentrations of DnaA. To quantify the results, background-corrected band intensities were determined using ImageJ, values were normalized against the total signal in a lane in MS Excel, and plotted using GraphPad.
Results
C. difficile DnaA Protein
Clostridioides difficile 630Δerm encodes a homolog of the bacterial replication initiator protein DnaA (GenBank: CEJ96502.1; CD630DERM_00010). Alignment of the full-length C. difficile DnaA amino acid sequence with selected DnaA homologs from other organisms demonstrates a sequence identity of 35 to 67%, with an even higher similarity (57 to 83%, Figure 1A). C. difficile DnaA displays a greater sequence identity between the low-[G + C] Firmicutes (>60%). When compared with the extensively studied DnaA proteins from E. coli and B. subtilis, the full-length protein has 43 and 62% identity, and a similarity of 63 and 78%, respectively (Figure 1A).
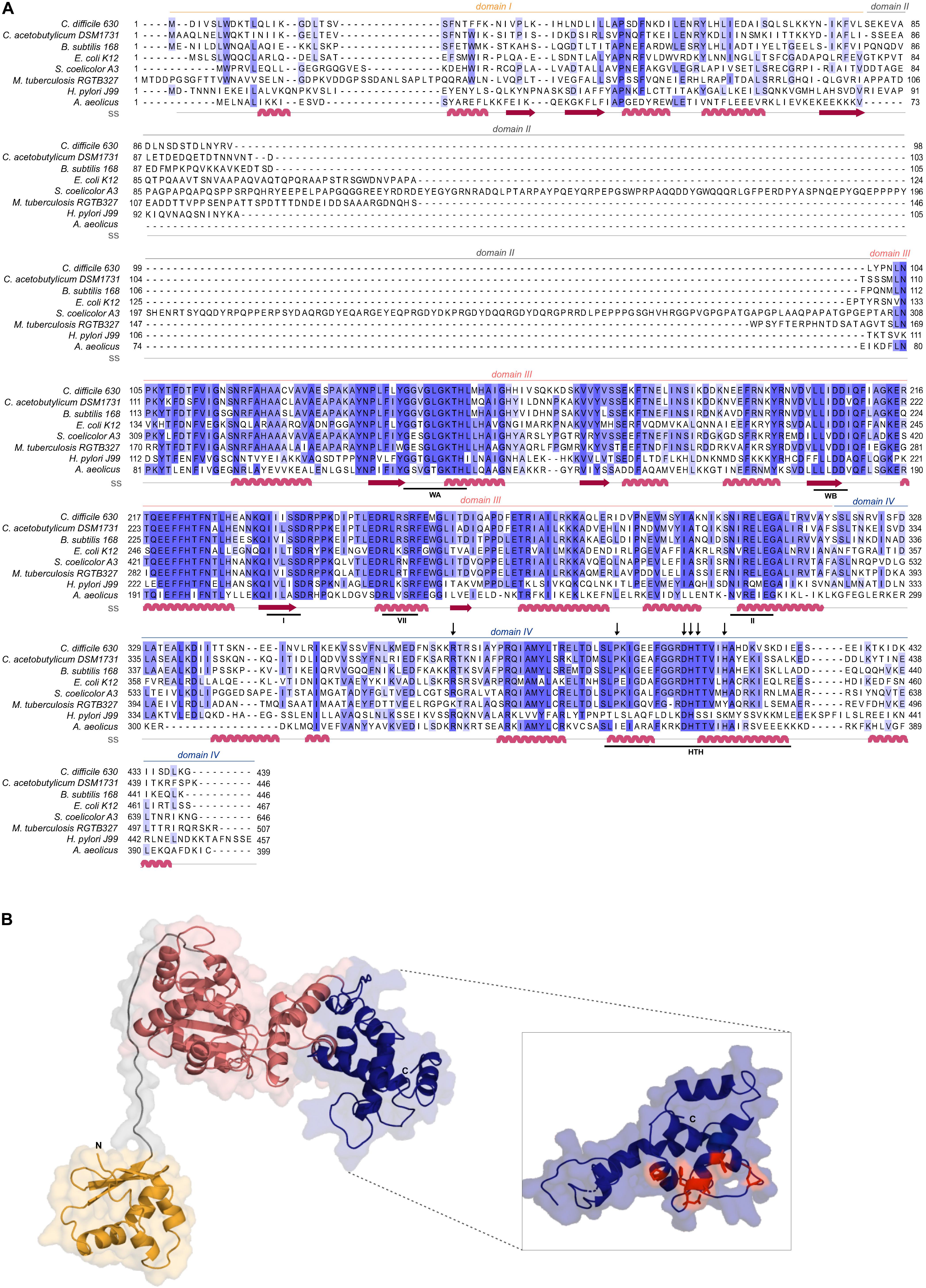
Figure 1. C. difficile DnaA DNA-binding domain is conserved. (A) Multiple sequence alignment (PRALINE) of C. difficile DnaA with homologous proteins retrieved from GenBank. The amino acid sequences from C. difficile 630Δerm (CEJ96502.1), C. acetobutylicum DSM 1731 (AEI33799.1), B. subtilis 168 (NP_387882.1), E. coli K-12 (AMH32311.1), S. coelicolor A3(2) (TYP16779.1), M. tuberculosis RGTB327 (AFE14996.1), H. pylori J99 (Q9ZJ96.1) and Aquifex aeolicus (WP_010880157.1) were used. Residues are colored according to sequence identity conservation using blue shading (dark blue more conserved), as analyzed in Jalview. Secondary structure prediction (ss) is indicated, according to Phyre2 modeled structure. DnaA domains are represented, with the conserved AAA+ ATPase fold motifs Walker A, Walker B, VII box, sensor I and sensor II highlighted (WA, WB, I, VII, and II motifs), as well as the Domain IV helix-turn-helix (HTH). Residues involved in the base-specific recognition of the 9-mer DnaA box sequence are identified with an arrow. (B) Structural model of C. difficile DnaA determined by Phyre2. Domains are colored as in alignment. Both the N-terminus and the C-terminus are indicated in the figure. The DnaA Domain IV is enhanced (inset) with the DnaA-box binding specific residues represented in red sticks.
To assess the structural properties of C. difficile DnaA, we predicted the secondary structure and generated a model of the protein using Phyre2 (Kelley et al., 2015; Figure 1B). The predicted DnaA model is based on three DnaA structures from different organisms: A. aeolicus (residues 101 to 318 and 334 to 437) (Erzberger et al., 2006) for Domain III and IV, and B. subtilis (residues 2 to 79) (Jameson et al., 2014) and E. coli (residues 5 to 97) (Abe et al., 2007) for Domain I and II.
Domain I of DnaA mediates interactions with a diverse set of regulators, and is involved in DnaA oligomerization (Zawilak-Pawlik et al., 2017; Nowaczyk-Cieszewska et al., 2020). We observe limited homology of C. difficile DnaA Domain I with the equivalent domain of the selected organisms (Figure 1A), although the overall fold is clearly conserved (Figure 1B). Nevertheless, some residues (P45, F48) appear to be conserved in most of the selected organisms (Figure 1A), though no functional role for these residues is known. Potentially, these residues might be involved in protein-protein interactions or DnaA oligomerization, as these functions have been mapped to Domain I of DnaA (Weigel et al., 1999; Abe et al., 2007; Natrajan et al., 2009; Jameson et al., 2014; Kim et al., 2017; Zawilak-Pawlik et al., 2017; Martin et al., 2018; Matthews and Simmons, 2019; Nowaczyk-Cieszewska et al., 2020).
Domain II is a flexible linker that is possibly involved in aiding the proper conformation of the DnaA domains, and thus requires a minimal length for DnaA function in vivo (Nozaki and Ogawa, 2008). No clear sequence similarity is observed on Domain II and modeling of the C. difficile DnaA protein suggests a putative disordered nature of this domain (Figure 1).
Domain III is responsible for binding to the co-factors ATP and ADP, and is in conjunction with Domain IV essential for DNA binding (Kawakami et al., 2005; Ozaki et al., 2008; Ozaki and Katayama, 2012). Within Domain III we readily identified the Walker A and Walker B motifs (WA and WB in Figure 1A) of the AAA+ fold (residues 135–317), crucial for binding and hydrolyzing ATP. This domain is highly conserved among all the selected organisms (Figure 1A) and comprises a structural center of β-sheets (Figure 1B, pink domain). Other features of the AAA+ ATPase fold are present and conserved between the organisms, such as the sensor I and sensor II motifs required for the nucleotide binding (I and II, Figure 1A). The arginine finger motif (the equivalent of R285 of E. coli DnaA in the VII box), important for the ATP dependent activation of DnaA (Kawakami et al., 2005), is conserved in C. difficile DnaA as well (R256 in motif box VII; Figure 1A).
The C-terminal Domain IV of the DnaA protein (residues 317–439, Figure 1A), contains the HTH motif required for the specific binding to DnaA-boxes (Erzberger et al., 2002; Zawilak et al., 2003). Previous studies identified several residues involved in specific interactions with the DnaA boxes, that bind through hydrogen bonds and van der Waals contacts with thymines present in the DNA sequence (Blaesing et al., 2000; Fujikawa et al., 2003; Tsodikov and Biswas, 2011). The residues are conserved among all Firmicutes and E. coli, including the residues R371 (position R399 in E. coli), P395 (P423), D405 (D433), H406 (H434), T407 (T435), and H411 (H439), (Figure 1B inset, red residues) (Fujikawa et al., 2003). Structural modeling of C. difficile DnaA predicts these residues to be exposed, providing an interface for DNA binding (Figure 1B). Residues involved in base-specific recognition of the DnaA box sequence are conserved between the Firmicutes and E. coli (Figure 1A), suggesting that C. difficile DnaA is likely to recognize the consensus DnaA box TTWTNCACA (Schaper and Messer, 1995). Notably, with the exception of a single arginine, these residues are not conserved between C. difficile and Thermotoga maritima DnaA (Supplementary Figure S1). As the latter recognizes an extended 12-bp motif (Ozaki et al., 2006; Richardson et al., 2019), this provides additional support for the notion that C. difficile DnaA recognizes a classical 9-bp DnaA box. In addition, residues found to be involved in non-specific interactions with the phosphate backbone of the DNA (some of which contribute to sequence specificity) (Fujikawa et al., 2003; Tsodikov and Biswas, 2011) appear less conserved between the selected organisms (Figure 1A).
Expression and Purification of DnaA-6xHis
To allow for in vitro characterization of DnaA activity, we recombinantly expressed the C. difficile DnaA with a C-terminal 6xHis-tag in E. coli cells. To prevent the co-purification of C. difficile DnaA with host DnaA protein, E. coli strain CYB1002 was used (a kind gift of A. D. Grossman). This strain is a derivative of E. coli MS3898, that lacks the dnaA gene and replicates in a DnaA-independent fashion (Sutton and Kaguni, 1997). Induction of the DnaA-6xHis protein was confirmed by Coomassie staining and immunoblotting with anti-his antibody at the expected molecular weight of 51 kDa (Supplementary Figure S2A, red arrow). Upon overexpression of DnaA-6xHis, smaller fragments were observed, which accumulated with a prolonged time of expression (Supplementary Figure S2A), most likely corresponding to proteolytic fragments of the DnaA-6xHis protein.
Purification of the recombinant DnaA-6xHis showed a clear band at the expected size when eluted at 300 mM imidazole concentration, but several lower molecular size bands were observed (Supplementary Figure S2B). Therefore, the eluted fractions where further purified with size exclusion chromatography (SEC). This yielded a single product at the expected molecular weight of DnaA-6xHis, and its identity was confirmed by western-blot with anti-his antibody (Supplementary Figure S2C, red arrow). A minor band of lower molecular weight (approximately 38 kDa, < 1% of total protein) was observed (Supplementary Figure S2C, green asterisk), which may reflect some instability of the N-terminus of the DnaA-6xHis protein, as it appears to have retained the C-terminal 6xHis tag.
In silico Prediction of the oriC Region
To identify the oriC region and the elements that are part of it (DUE, DnaA-trio and DnaA boxes) we performed different prediction approaches in a stepwise procedure, as initially described (Mackiewicz et al., 2004).
We first analyzed the DNA asymmetry of the genome of C. difficile 630Δerm (GenBank accession no. LN614756.1) (van Eijk et al., 2015), by plotting the normalized difference of the complementary nucleotides (GC-skew plot) (Necsulea and Lobry, 2007). C. difficile 630Δerm has a circular genome of 4293049 bp and an average [G + C] content of 29.1%. We used the GenSkew Java Application8 for determining the chromosomal asymmetry. Asymmetry changes in a GC-skew plot can be used to predict the origin of replication region and the terminus region of bacterial genomes. Based on this analysis, the origin is predicted at approximately position 1 of the chromosome. The terminus location is predicted at approximately 2.18 Mbp from the origin region (Figure 2A). These results were confirmed when artificially reassigning the starting position of the chromosomal assembly (data not shown). The gene organization in the putative origin region is rnpA-rpmH-dnaA-dnaN (position 4291488 to 2870, Figure 2B), identical to the origin of B. subtilis (Ogasawara et al., 1985; Briggs et al., 2012), and therefore encompasses the dnaA gene (CD630DERM_00010).
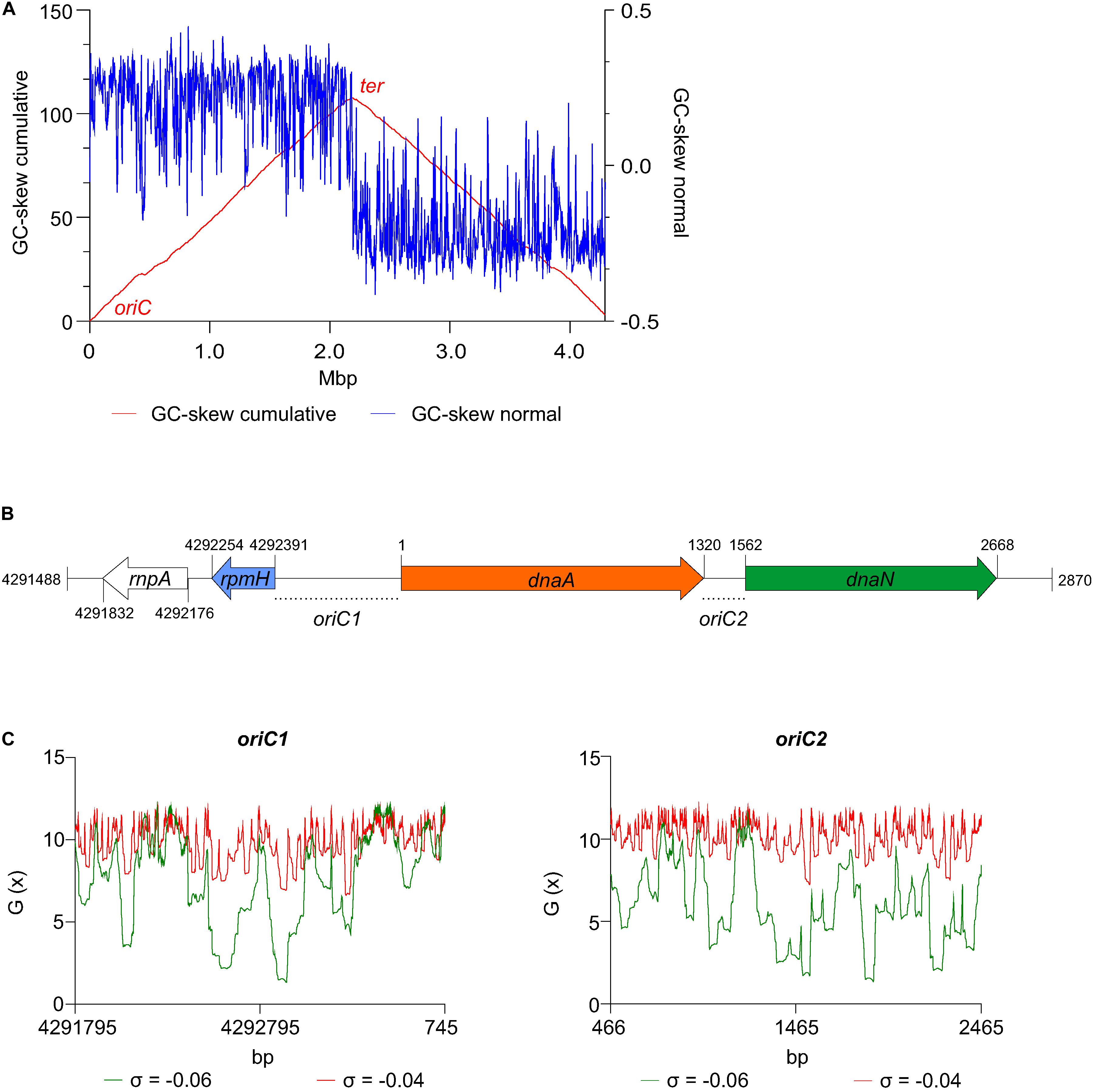
Figure 2. Prediction of the C. difficile origin of replication. (A) GC skew analysis of the C. difficile 630Δerm (LN614756.1) genome sequence. Normal GC skew analysis [(G – C)/(G + C)] performed on leading strand (blue line) and respective cumulative GC skew plot (red line). Calculations where performed with a window size of 4293 bp and a step size of 4293 bp. The origin (oriC) and terminus (ter) regions are indicated. (B) Representation of the predicted origin region and genomic context (from residues at position 4291488 to 2870 of the C. difficile 630Δerm chromosome). The rmpA, rpmH (blue arrow), dnaA (orange arrow) and dnaN (green arrow) genes are indicated. Putative origins in intergenic regions are represented oriC1 (rpmH-dnaA) and oriC2 (dnaA-dnaN). (C) SIDD analysis of 2.0 kb fragments comprising oriC1 (nucleotide 4291795 to 745) and oriC2 (nucleotide 466 to 2465). Predicted free energies G(x) for duplex destabilization at a superhelical density of σ = –0.06 (green) or σ = –0.04 (red).
We next used the SIST program (Zhabinskaya et al., 2015) to localize putative DUEs in the intergenic regions in the chromosomal region predicted to contain the oriC. Hereafter we refer to these regions as oriC1 (in the intergenic region of rpmH-dnaA) and oriC2 (in the intergenic region dnaA-dnaN), in line with nomenclature in other organisms (Ogasawara et al., 1985; Donczew et al., 2012; Figure 3B). SIST identifies helically unstable AT-rich DNA stretches (Stress-Induced Duplex Destabilization regions; SIDDs) (Donczew et al., 2012; Zhabinskaya et al., 2015). In regions with a lower free energy (G(x) < y kcal/mol) the double-stranded helix has a high probability to become single-stranded DNA. With increasing negative superhelicity (σ = −0.06, Figure 2C, green line) regions of both oriC1 and oriC2 become single stranded DNA (G(x) < 2 kcal/mol). At low negative superhelicity (σ = −0.04, Figure 3C, red line) short stretches of DNA of approximately 27 bp were identified with a significantly lower free energy. These regions with lower free energy at a negative superhelicity of −0.04 and −0.06 are potential DUE sites. The nucleotide sequence of the possible unwinding elements identified are represented in detail in Figure 3 (gray boxes).

Figure 3. Identification of the C. difficile oriC region. Nucleotide sequence of the oriC1 region (nucleotide 4292328 to 48 of the C. difficile 630Δerm LN614756.1 genome sequence) and oriC2 region (nucleotide 1274 to 1587). Identification of the possible unwinding AT-rich regions previously identified in the SIDD analysis (gray boxes). The putative DnaA boxes found are represented (pink boxes) and orientation in the leading (right) and lagging strand (left) are shown. Possible DnaA-trio sequence are denoted (light blue boxes). Coding sequence of the genes rpmH (blue arrow), dnaA (orange arrow) and dnaN (green arrow) and respective putative ribosome binding sites (dashed line) are indicated. Pattern identification is described in section “Materials and Methods.”
We then performed the identification of DnaA box clusters through a search of the consensus DnaA box TTWTNCACA containing up to one mismatch, using Pattern Locator (Mrazek and Xie, 2006). 22 putative DnaA boxes where identified in both the leading and lagging strand in the predicted C. difficile oriC regions (Figure 3, pink boxes), 14 in the oriC1 region and 8 in the oriC2 region. Both the consensus DnaA box TTWTNCACA and variant boxes are found. A cluster of DnaA boxes was proposed to contain at least three boxes with an average distance lower than 100 bp in between (Mackiewicz et al., 2004). At least one such cluster can be found in each origin region (Figure 3).
Though these are not crucial to origin function, we also manually identified the putative ribosomal binding sites for the annotated genes (Figure 3, dashed line) based on previously identified characteristics (Vellanoweth and Rabinowitz, 1992).
Finally, we manually predicted DnaA-trio sequences (3′-[G/A]A[T/A]n>3-5′ preceded by a GC-cluster) in the predicted oriC regions, as this motif is required for successful replication in both E. coli and B. subtilis (Richardson et al., 2016) and can also be identified in E. coli (Katayama et al., 2017), though a role in binding of DnaA to ssDNA has yet to be experimentally demonstrated in this organism. We identified a clear DnaA-trio in the lagging strand upstream of a predicted DUE region in the oriC2 region, with the nucleotide sequence 5′-CACCTACTACTATTACTACTATGA-3′ (Figure 3, light blue box), but no clear DnaA-trio was identified in the oriC1 region.
From all the observations, we anticipate that a bipartite origin is located in the dnaA chromosomal region of C. difficile with unwinding occurring downstream of dnaA, at the oriC2 region.
DnaA-Dependent Unwinding
To analyze DnaA-dependent unwinding of oriC, we used the purified C. difficile DnaA-6xHis protein and the predicted oriC sequence, to perform P1 nuclease assays as previously described (Sekimizu et al., 1988; Donczew et al., 2012). Localized melting resulting from DnaA activity exposes ssDNA to the action of the ssDNA-specific P1 nuclease. After incubation of a vector containing the oriC fragment with DnaA protein and cleavage by the P1 nuclease, the vector is purified and digested with different endonucleases to map the location of the unwound region.
We constructed vectors, based on pori1ori2 (Donczew et al., 2012), harboring C. difficile oriC1 (pAP76) or oriC2 (pAP83) individually (Supplementary Figure S3A), as well as the complete oriC region (pAP205) (Figure 4A). For a more accurate determination of the unwound region, the vectors were subjected to digestion by two different restriction enzymes (BglII and NotI), resulting in different restriction patterns. Limited spontaneous unwinding of the plasmid was observed in the C. difficile oriC-containing vectors (Figure 4B and Supplementary Figure S3B). No DnaA-dependent change in restriction pattern was observed when using the single oriC regions (Supplementary Figure S3B), suggesting oriC1 and oriC2 individually lack the requirements for DnaA-dependent unwinding.
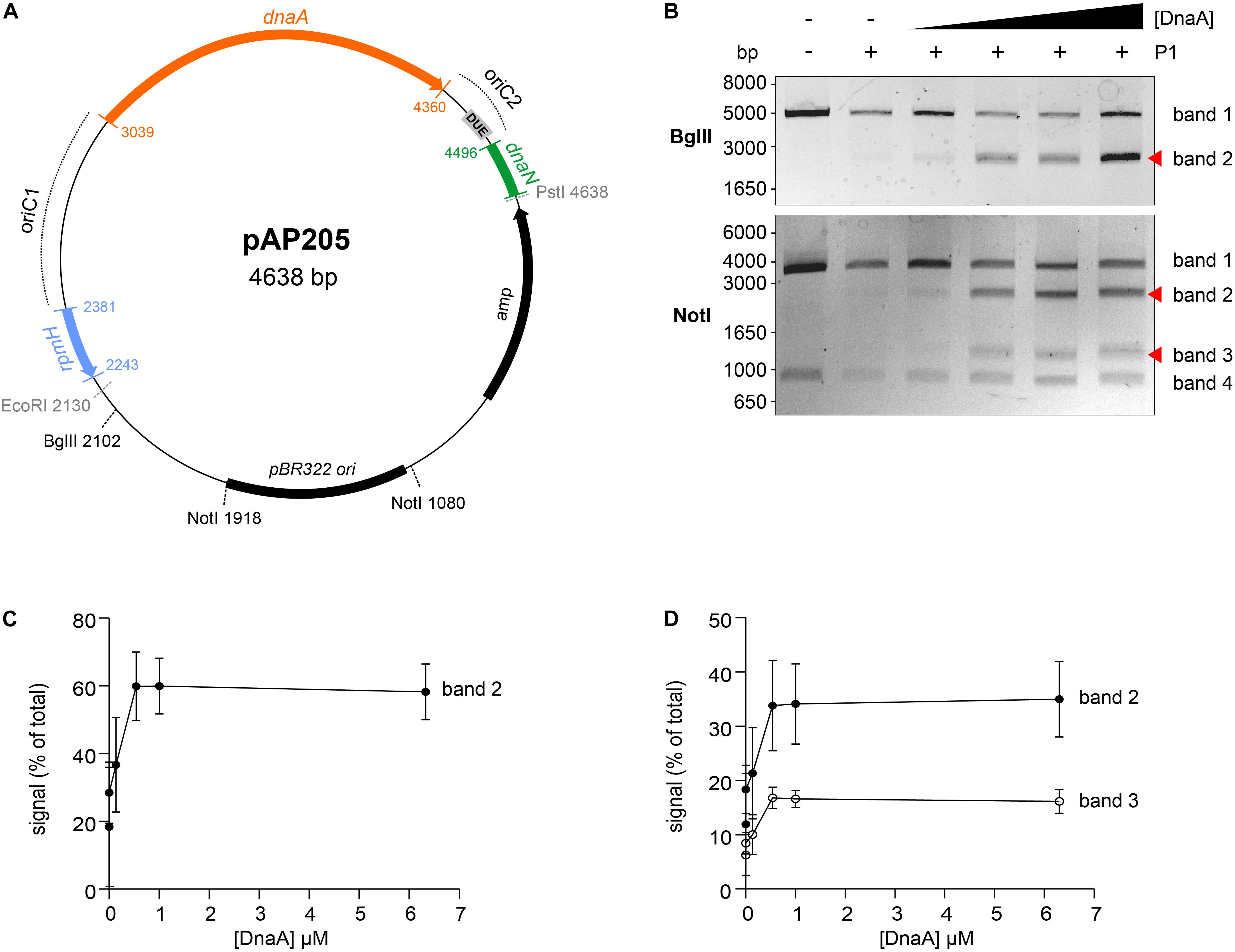
Figure 4. Identification of the unwinding region in C. difficile oriC. (A) Representation of the oriC1oriC2 containing vector pAP205 used in the P1 nuclease assay. The predicted oriC1 and oriC2 regions (dotted lines) and included genes are represented, rpmH (blue), dnaA (orange), and dnaN (green). The bla gene, the pBR322 plasmid origin of replication and the positions of used restriction sites are marked. The unwinding region (DUE) is denoted in a gray circle. (B) P1 nuclease assay of the oriC1oriC2-containing vector pAP205. Digestion of the vector (lane 1) with different restriction enzymes BglII (upper panel) or NotI (lower panel). Treatment of the fragments with P1 nuclease only (lane 2) and incubated with increasing amounts of C. difficile DnaA protein (lanes 3–6). The DNA fragments were separated in a 1% agarose gel and analyzed after ethidium bromide staining. Fragments resulting from DnaA-dependent unwinding are indicated with a red arrow (see Results for details). A typical result is shown. (C). Quantification of band 2 (black circles) of the P1/BglII digested vector. (D). Quantification of bands 2 (black circles) and 3 (open circles) of the P1/NotI digested vector. For panels (C,D), error bars indicate the standard deviation of the mean of n = 3 independent experiments.
We did observe a DnaA-dependent change in digestion patterns for the oriC1oriC2-containing vector pAP205 (Figure 4). Digestion of this vector with BglII in the absence of DnaA-6xHis and P1 nuclease resulted in a linear DNA fragment (4638 bp) due to the presence of a unique BglII restriction site (Figure 4B, upper panel, first lane). The addition of P1 nuclease leads to the appearance of a faint band between 1650 and 3000 bp (Figure 4B, upper panel, second lane), consistent with previous observations that the presence of a plasmid DUE can result in low-level spontaneous unwinding due to the inherent instability of these AT-rich regions (Jaworski et al., 2016). Upon the addition of the DnaA-6xHis protein the observed band becomes more intense, suggesting a strong increase in unwinding (Figure 4B, upper panel, red arrow).
Digestion of pAP205 with NotI in the absence of DnaA-6xHis and P1 nuclease results in fragments of 3804 and 842 bp, due to two NotI recognition sites in the vector (Figure 4B, lower panel, first lane). In the presence of just P1 nuclease, a similar low level of spontaneous unwinding is observed, resulting in the appearance of two additional faint bands, one between 1650 and 3000 bp and other between 1000 and 1650 bp (Figure 4B, lower panel, second lane). The addition of DnaA-6xHis results in an increase in intensity of both these bands in a dose dependent manner (Figure 4B, lower panel, red arrows).
We quantified the intensity of the bands from three independent P1 nuclease assays in order to determine the reproducibility of the assay (Figures 4C,D and Supplementary Figure S4). For the BglII-digested vector, we observed a DnaA-dependent increase of 20 to 60% of the total signal for the band between 1650 and 3000 bp (Figure 4C, band 2). For the NotI-digested vector, the signals of the second and third band increase from approximately 10% of the total signal to approximately 35% (1650–3000 bp, band 2) and 20% (1000–1650 bp, band 3) of total signal in the lane (Figure 4D). The observed increase was highly consistent, and appeared to saturate around 0.54–1 uM of DnaA (Figures 4C,D). The quantification also revealed a concomitant decrease in the signal for the upper bands in the gels of the BglII and NotI digests (Supplementary Figure S4, band 1).
The DnaA-dependent appearance of the ∼2000 bp band in the BglII digest, and the ∼1200 and ∼2200 bp bands in the NotI digest localize the DnaA-dependent unwinding of the C. difficile oriC in the oriC2 region (Figure 4A, gray rectangle, DUE). Moreover, these results suggest that C. difficile has a bipartite origin of replication, as successful DnaA-dependent unwinding of C. difficile in the oriC2 region requires both oriC regions (oriC1 and oriC2).
Conservation of the Origin Organization in Related Clostridia
Our results suggest that the origin organization of C. difficile resembles that of a more distantly related Firmicute, B. subtilis. To extend our observations, we evaluated the genomic organization of the oriC region in different organisms phylogenetically related to C. difficile. We followed a similar approach as described above for C. difficile 630Δerm, taking advantage of the DoriC 10.0 database (Luo and Gao, 2019). Importantly, our results with respect to the C. difficile origin of replication described above were largely congruent with the DoriC 10.0 database despite being based on different methods (a notable exception is the prediction for C. difficile strain 630; data not shown). We retrieved the predicted oriC regions from the DoriC 10.0 database and performed an in-depth analysis of these regions for the closely related C. difficile strain R20291 (NC_013316.1), as well as the more distantly related C. botulinum A Hall (NC_009698.1), C. sordelli AM370 (NZ_CP014150), C. acetobutylicum DSM 1731 (NC_015687.1), C. perfringens str.13 (NC_003366.1), and C. tetani E88 (NC_004557.1) (Table 1).
Similar to C. difficile 630Δerm, the genomic context of the origin contains the rpmH-dnaA-dnaN region for most of the clostridia selected and mirrors that of B. subtilis (Figure 5). The only exception is C. tetani E88 where the uncharacterized CLOTE0041 gene lies upstream of the dnaA-dnaN cluster (Figure 5).
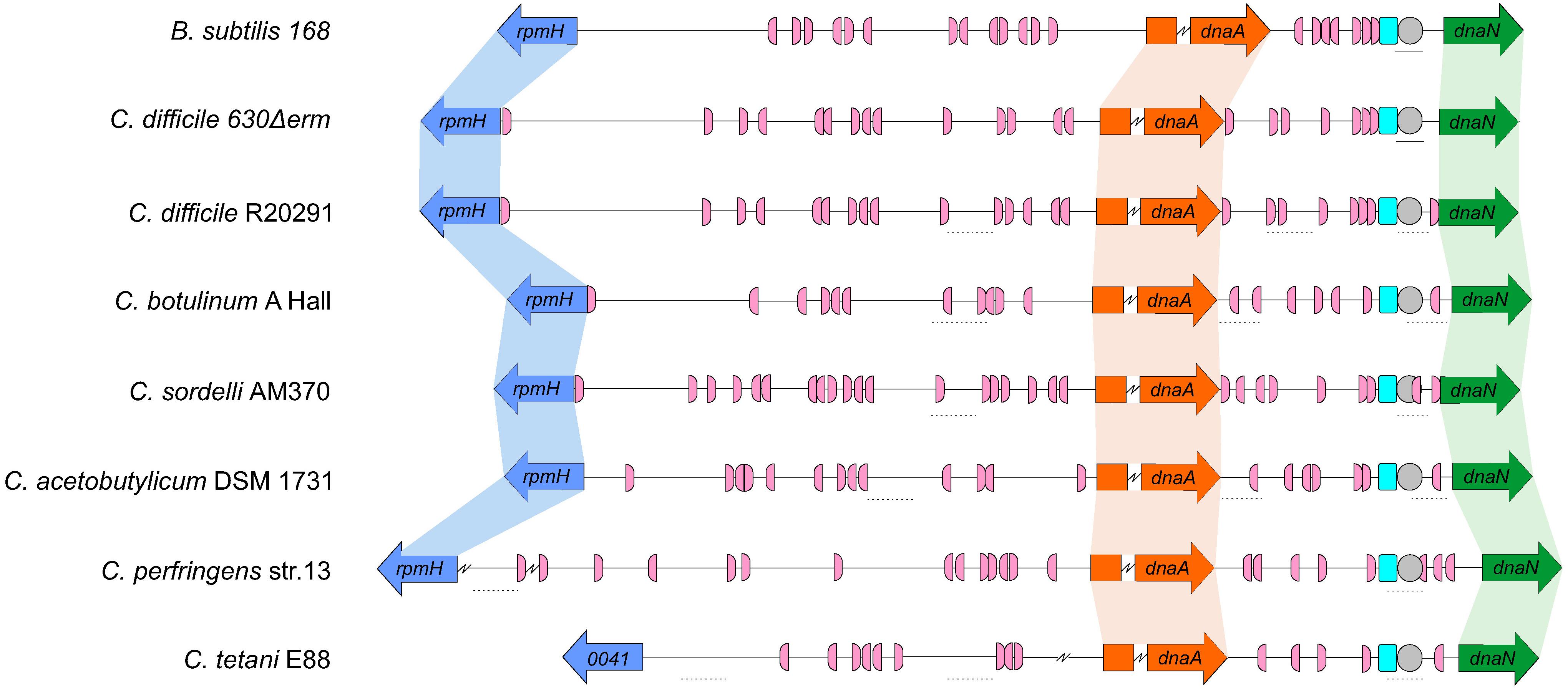
Figure 5. Comparison of the clostridial oriC regions. Representation of the origin region and genomic context of B. subtilis, C. difficile 630Δerm chromosome and the predicted regions for C. difficile R20291, C. botulinum A Hall, C. sordelli AM370, C. acetobutylicum DSM 1731, C. perfringens str.13, C. tetani E88 (see Table 1). The rpmH (blue arrow), dnaA (orange arrow) and dnaN (green arrow) genes are indicated. Predicted DnaA-boxes are indicated by pink boxes and orientation on the leading (right) and lagging strand (left) are shown. Identification of the experimentally identified unwinding AT-rich regions (lines) and the SIDD-predicted helical instability are shown (dashed lines). The putative DUE is denoted (gray circle). Possible DnaA-trio sequences are shown in light blue boxes. See section “Materials and Methods” for detailed information. Alignment of the represented chromosomal regions is based on the location of the DnaA-trio.
We also identified the possible DnaA boxes for the selected clostridia (Figure 5, pink semi-circle). Across the analyzed clostridia, oriC1 region presented more variability in the number of putative DnaA boxes, from 9 to 19, whereas oriC2 contained 5 to 9 DnaA boxes, with C. tetani E88 with the lowest number of possible DnaA boxes, both at the oriC1 (9 boxes) and oriC2 (5 boxes) regions (Figure 5, pink semi-circle). In all the organisms we observe at least 1 DnaA cluster in each origin region, as also observed for C. difficile 630Δerm.
Prediction of DUEs using the SIST program (Zhabinskaya et al., 2015) identified several helically unstable regions that are candidate sites for unwinding (Figure 5, dashed lines, and Supplementary Figure S5). Notably, in all cases one such region in oriC2 (Figure 5, gray circle) is preceded immediately by the manually identified DnaA-trio (Figure 5, light blue circle). Based on our experimental data for C. difficile 630Δerm, we suggest that in all analyzed clostridia, DnaA-dependent unwinding occurs at a conserved DUE downstream of the DnaA-trio in the oriC2 region (Figure 5).
Discussion
Chromosomal replication is an essential process for the survival of the cell. In most bacteria DnaA protein is the initiator protein for replication and through a cascade of events leads to the successful loading of the replication complex onto the origin of replication (Duderstadt et al., 2011; Chodavarapu and Kaguni, 2016).
Initial characterization of bacterial replication has been assessed in the model organisms E. coli and B. subtilis (Jameson and Wilkinson, 2017). Despite the similarities (location in an intergenic region, presence of a DUE, several DnaA boxes in both orientations) the structure of the replication origins and the regulation mechanisms are variable among bacteria (Wolanski et al., 2014). In contrast to E. coli, the B. subtilis origin region is bipartite, with two intergenic regions upstream and downstream of the dnaA gene. In C. difficile the genomic organization in the predicted cluster rnpA-rpmH-dnaA-dnaN, and the presence of [A + T]-rich sequences in the intergenic regions is consistent with a bipartite origin, as in B. subtilis (Figure 3).
The origin region contains several DnaA-boxes with different properties that are recognized by the DnaA protein. The specific binding of DnaA to the DnaA-boxes is mediated mainly through Domain IV of the DnaA protein. From DNA bound structures of DnaA it was possible to identify several residues involved in the contact with the DnaA boxes, some of which confer specificity (Blaesing et al., 2000; Fujikawa et al., 2003; Tsodikov and Biswas, 2011). Analysis of the of C. difficile DnaA homology in Domain IV did not show any difference in the residues involved on the DnaA-box specificity (Figure 1, vertical arrows), suggesting the same consensus motif conservation as the DnaA-box TTWTNCACA for E. coli (Schaper and Messer, 1995). The conserved DnaA-box motif allowed us to identify several DnaA boxes along the intergenic regions of the oriC. Like in the bipartite origin of B. subtilis, we identified at least one cluster of DnaA-boxes in the C. difficile oriC1 and oriC2 regions (Figures 3, 5). In the case of B. subtilis, it has been shown that different DnaA boxes fulfill different roles in replication initiation: two out of three DnaA boxes immediately upstream of the DnaA-trio are part of the basal unwinding system (i.e., required for DnaA-dependent strand separation), whereas other DnaA affect coordination and regulation of DNA replication (Richardson et al., 2019). For C. difficile, we also find three DnaA boxes immediately upstream of the DnaA trio (Figure 3 and Supplementary Figure S6), but the role of these boxes has not been experimentally verified to date.
The P1 nuclease assays place a region in which DnaA-dependent unwinding occurs in the oriC2 region of C. difficile, supported by the presence of the several features on the oriC2, such as the identified DUE and DnaA-trio, both required for unwinding (Kowalski and Eddy, 1989; Richardson et al., 2016). The presence of both oriC regions (oriC1 and oriC2) is required for melting in vitro, as observed for other bipartite origins (Wolanski et al., 2014). In contrast to the bipartite origin identified in H. pylori (Donczew et al., 2012), we did not observe unwinding of the oriC2 region alone. Though this may be a specific aspect of C. difficile oriC2, we cannot exclude that differences in the experimental setup (e.g. DnaA protein purification) could affect these observations. Nevertheless, our data are consistent with DnaA binding the DnaA-box clusters in both oriC regions, leading to potential DnaA oligomerization, loop formation, and unwinding at the [A + T]-rich DUE site.
When analyzing the origin region between different clostridia, features similar to those of C. difficile are observed, such as conservation of DnaA-box clusters within both oriC regions in the vicinity of the dnaA gene. Similar to C. difficile and B. subtilis, a putative DUE element, preceded by the DnaA-trio, was also located within the oriC2 region (Figures 4, 6). Thus, the overall origin organization and mechanism of DNA replication initiation is likely to be conserved within the Firmicutes (Briggs et al., 2012). As spacing of the DnaA-boxes are determinants for the species-specific effective replication (Zawilak et al., 2003; Zawilak-Pawlik et al., 2005), these similarities do no exclude the possibilities that subtle differences in replication initiation exist, and further studies are required. For instance, our work does not address which DnaA boxes in either oriC1 or oriC2 are important for unwinding, and whether the requirement is due to DnaA-dependent changes in structure of origin DNA (as has been shown for B. subtilis) (Richardson et al., 2019), or as a cis-acting regulatory element like DARS/DatA (Katayama et al., 2010, 2017). Further experiments could provide insights into the DnaA-box conservation and affinities and establish which DnaA boxes are crucial for origin firing and/or transcriptional regulation.
Several proteins can interact with the oriC region or DnaA, including YabA, Rok, DnaD/DnaB, Soj and HU (Briggs et al., 2012; Jameson and Wilkinson, 2017). In doing so they shape the origin conformation and/or stabilize the DnaA filament or the unwound region, consequently affecting replication initiation.
YabA or Rok affect B. subtilis replication initiation (Goranov et al., 2009; Schenk et al., 2017; Seid et al., 2017), but no homologs of these proteins have been identified in C. difficile (van Eijk et al., 2017). Similarly, no homologs are identified of other well-characterized DnaA-interacting proteins from gram-negative bacteria (van Eijk et al., 2017), such as Hda, DiaA/HobA (Zawilak-Pawlik et al., 2017) or HdaB (Frandi and Collier, 2020); it is unknown how C. difficile regulates DnaA activity.
In B. subtilis, DnaD, DnaB, and DnaI helicase loader proteins associate sequentially with the origin region resulting in the recruitment of the DnaC helicase protein (Marsin et al., 2001; Velten et al., 2003; Smits et al., 2010; Jameson and Wilkinson, 2017). In B. subtilis, DnaD binds to DnaA and it is postulated that this affects the stability of the DnaA filament and consequently the unwinding of the oriC (Ishigo-Oka et al., 2001; Martin et al., 2018; Matthews and Simmons, 2019). B. subtilis DnaB protein also affects the DNA topology and has been shown to be important for recruiting oriC to the membrane (Rokop et al., 2004; Zhang et al., 2005). C. difficile lacks a homolog for the DnaB protein, although the closest homolog of the DnaD protein (CD3653) (van Eijk et al., 2017) may perform similar functions in the origin remodeling (van Eijk et al., 2016). Direct interaction of DnaA-DnaD through the DnaA Domain I was structurally determined and the residues present at the interface were solved (Martin et al., 2018). Despite high variability of this domain between organisms, half of the identified contacts for the DnaA-DnaD interaction are conserved within C. difficile, the S22 (S23 in B. subtilis DnaA), T25 (T26), F48 (F49), D51 (D52) and L68 (L69) (Figure 1; Martin et al., 2018; Matthews and Simmons, 2019). This might suggest a similar interaction surface for CD3653 on C. difficile DnaA. A characterization of the putative interaction between CD3653 and DnaA, and the resulting effect on DnaA oligomerization and origin melting awaits purification and functional characterization of CD3653.
The Soj protein, also involved in chromosome segregation, has been shown to interact with DnaA via Domain III, regulating DnaA-filament formation (Scholefield et al., 2012) and the C. difficile encodes at least one uncharacterized Soj homolog, but a role in DNA replication has not been experimentally demonstrated.
Bacterial histone-like proteins (such as HU and HBsu) can modulate DNA topology and might therefore influence oriC unwinding and replication initiation. However, the importance of HU for replication initiation has only been clearly demonstrated for E. coli (Krause et al., 1997; Chodavarapu et al., 2008). Several studies have shown HU independent origin unwinding even in gram-negative bacteria (Donczew et al., 2012; Makowski et al., 2016; Jaworski et al., 2018; Plachetka et al., 2019), suggesting that HU-dependence of origin unwinding may be limited to a narrow phylogenetic group. C. difficile encodes a homolog of HU, HupA (Oliveira Paiva et al., 2019) but whether this protein plays a role in DNA replication initiation remains to be established.
Finally, Spo0A, the master regulator of sporulation, binds to several Spo0A-boxes present in this the oriC region in B. subtilis (Boonstra et al., 2013). Some of the Spo0A-boxes partially overlap with DnaA-boxes and binding of Spo0A can prevent the DnaA-mediated unwinding, thus playing a significant role on the coordination of between cell replication and sporulation (Boonstra et al., 2013). In C. difficile, Spo0A-binding has previously been investigated (Rosenbusch et al., 2012), but a role in DNA replication has not been assessed.
For all the regulators with a C. difficile homolog discussed above (i.e. CD3653, Soj, HupA, and Spo0A), further studies can be envisioned employing the P1 nuclease assays described here to assess the effects on DnaA-mediated unwinding of the origin. Our experiments show, however, they are not strictly required for origin unwinding (Figure 4).
In summary, through a combination of different in silico predictions and in vitro studies, we have shown the DnaA-dependent unwinding in the dnaA-dnaN intergenic region in the bipartite C. difficile origin of replication. We have analyzed the putative origin of replication in different clostridia and a conserved organization is observed throughout the Firmicutes, although different mechanisms and regulation could be behind the initiation of replication. The present study is the first to characterize the origin region of C. difficile and forms the start to further unravel the mechanism behind the DnaA-dependent regulation of C. difficile initiation of replication.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AO and WS designed the experiments, analyzed the data and wrote the manuscript. AO and CW performed the in silico analyses. AO, EE, and AF performed the experiments. All authors read and approved the final version for submission.
Funding
This work in the group of WS was supported by a Vidi Fellowship (Grant No: 864.13.003) of the Netherlands Organization for Scientific Research (NWO) and a Gisela Thier Fellowship from the Leiden University Medical Center.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Alan Grossman for kindly providing the pAV13 vector and E. coli strain CYB1002. We also thank Anna Zawilak-Pawlik for kindly providing the pori1ori2 vector and expert help in setting up the P1 assays. We would like to thank Luís Sousa for their help with the SIDD and Pattern Locator coding files.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.581401/full#supplementary-material
Footnotes
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://www.ibi.vu.nl/programs/pralinewww/
- ^ http://www.sbg.bio.ic.ac.uk/phyre2
- ^ http://genskew.csb.univie.ac.at/
- ^ http://tubic.tju.edu.cn/doric/public/index.php
- ^ https://bitbucket.org/benhamlab/sist_codes/src/master/
- ^ https://www.cmbl.uga.edu//downloads/programs/Pattern_Locator/patloc.c
- ^ http://genskew.csb.univie.ac.at/
References
Abe, Y., Jo, T., Matsuda, Y., Matsunaga, C., Katayama, T., and Ueda, T. (2007). Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 282, 17816–17827. doi: 10.1074/jbc.M701841200
Bawono, P., and Heringa, J. (2014). PRALINE: a versatile multiple sequence alignment toolkit. Methods Mol. Biol. 1079, 245–262. doi: 10.1007/978-1-62703-646-7_16
Bazin, A., Cherrier, M. V., Gutsche, I., Timmins, J., and Terradot, L. (2015). Structure and primase-mediated activation of a bacterial dodecameric replicative helicase. Nucleic Acids Res. 43, 8564–8576. doi: 10.1093/nar/gkv792
Blaesing, F., Weigel, C., Welzeck, M., and Messer, W. (2000). Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol. Microbiol. 36, 557–569. doi: 10.1046/j.1365-2958.2000.01881.x
Bleichert, F., Botchan, M. R., and Berger, J. M. (2017). Mechanisms for initiating cellular DNA replication. Science 355:eaah6317. doi: 10.1126/science.aah6317
Boonstra, M., de Jong, I. G., Scholefield, G., Murray, H., Kuipers, O. P., and Veening, J. W. (2013). Spo0A regulates chromosome copy number during sporulation by directly binding to the origin of replication in Bacillus subtilis. Mol. Microbiol. 87, 925–938. doi: 10.1111/mmi.12141
Briggs, G. S., Smits, W. K., and Soultanas, P. (2012). Chromosomal replication initiation machinery of low-G+C-content Firmicutes. J. Bacteriol. 194, 5162–5170.
Cho, E., Ogasawara, N., and Ishikawa, S. (2008). The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtils. Genes Genet. Syst. 83, 111–125. doi: 10.1266/ggs.83.111
Chodavarapu, S., Felczak, M. M., Yaniv, J. R., and Kaguni, J. M. (2008). Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol. Microbiol. 67, 781–792. doi: 10.1111/j.1365-2958.2007.06094.x
Chodavarapu, S., and Kaguni, J. M. (2016). Replication Initiation in Bacteria. Enzymes 39, 1–30. doi: 10.1016/bs.enz.2016.03.001
Crobach, M. J. T., Vernon, J. J., Loo, V. G., Kong, L. Y., Pechine, S., Wilcox, M. H., et al. (2018). Understanding Clostridium difficile Colonization. Clin. Microbiol. Rev. 31:e00021-17. doi: 10.1128/CMR.00021-17
Davey, M. J., and O’Donnell, M. (2003). Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 13, R594–R596. doi: 10.1016/s0960-9822(03)00523-2
Donczew, R., Weigel, C., Lurz, R., Zakrzewska-Czerwinska, J., and Zawilak-Pawlik, A. (2012). Helicobacter pylori oriC–the first bipartite origin of chromosome replication in Gram-negative bacteria. Nucleic Acids Res. 40, 9647–9660. doi: 10.1093/nar/gks742
Duderstadt, K. E., Chuang, K., and Berger, J. M. (2011). DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213. doi: 10.1038/nature10455
Ekundayo, B., and Bleichert, F. (2019). Origins of DNA replication. PLoS Genet. 15:e1008320. doi: 10.1371/journal.pgen.1008320
Erzberger, J. P., Mott, M. L., and Berger, J. M. (2006). Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13, 676–683. doi: 10.1038/nsmb1115
Erzberger, J. P., Pirruccello, M. M., and Berger, J. M. (2002). The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 21, 4763–4773.
Fossum, S., De Pascale, G., Weigel, C., Messer, W., Donadio, S., and Skarstad, K. (2008). A robust screen for novel antibiotics: specific knockout of the initiator of bacterial DNA replication. FEMS Microbiol. Lett. 281, 210–214. doi: 10.1111/j.1574-6968.2008.01103.x
Frandi, A., and Collier, J. (2020). HdaB: a novel and conserved DnaA-related protein that targets the RIDA process to stimulate replication initiation. Nucleic Acids Res. 48, 2412–2423. doi: 10.1093/nar/gkz1193
Fujikawa, N., Kurumizaka, H., Nureki, O., Terada, T., Shirouzu, M., Katayama, T., et al. (2003). Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086.
Goranov, A. I., Breier, A. M., Merrikh, H., and Grossman, A. D. (2009). YabA of Bacillus subtilis controls DnaA-mediated replication initiation but not the transcriptional response to replication stress. Mol. Microbiol. 74, 454–466. doi: 10.1111/j.1365-2958.2009.06876.x
Grimwade, J. E., and Leonard, A. C. (2017). Targeting the bacterial orisome in the search for new antibiotics. Front. Microbiol. 8:2352. doi: 10.3389/fmicb.2017.02352
Ishigo-Oka, D., Ogasawara, N., and Moriya, S. (2001). DnaD protein of Bacillus subtilis interacts with DnaA, the initiator protein of replication. J. Bacteriol. 183, 2148–2150. doi: 10.1128/JB.183.6.2148-2150.2001
Jameson, K. H., Rostami, N., Fogg, M. J., Turkenburg, J. P., Grahl, A., Murray, H., et al. (2014). Structure and interactions of the Bacillus subtilis sporulation inhibitor of DNA replication, SirA, with domain I of DnaA. Mol. Microbiol. 93, 975–991. doi: 10.1111/mmi.12713
Jameson, K. H., and Wilkinson, A. J. (2017). Control of initiation of DNA Replication in Bacillus subtilis and Escherichia coli. Genes 8:22. doi: 10.3390/genes8010022
Jaworski, P., Donczew, R., Mielke, T., Thiel, M., Oldziej, S., Weigel, C., et al. (2016). Unique and universal features of epsilonproteobacterial origins of chromosome replication and DnaA-DnaA Box Interactions. Front. Microbiol. 7:1555. doi: 10.3389/fmicb.2016.01555
Jaworski, P., Donczew, R., Mielke, T., Weigel, C., Stingl, K., and Zawilak-Pawlik, A. (2018). Structure and function of the campylobacter jejuni chromosome replication origin. Front. Microbiol. 9:1533. doi: 10.3389/fmicb.2018.01533
Katayama, T., Kasho, K., and Kawakami, H. (2017). The DnaA Cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front. Microbiol. 8:2496. doi: 10.3389/fmicb.2017.02496
Katayama, T., Ozaki, S., Keyamura, K., and Fujimitsu, K. (2010). Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170. doi: 10.1038/nrmicro2314
Kawakami, H., Keyamura, K., and Katayama, T. (2005). Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 280, 27420–27430. doi: 10.1074/jbc.M502764200
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kim, J. S., Nanfara, M. T., Chodavarapu, S., Jin, K. S., Babu, V. M. P., Ghazy, M. A., et al. (2017). Dynamic assembly of Hda and the sliding clamp in the regulation of replication licensing. Nucleic Acids Res. 45, 3888–3905. doi: 10.1093/nar/gkx081
Kowalski, D., and Eddy, M. J. (1989). The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 8, 4335–4344.
Krause, M., Ruckert, B., Lurz, R., and Messer, W. (1997). Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 274, 365–380. doi: 10.1006/jmbi.1997.1404
Lawson, P. A., Citron, D. M., Tyrrell, K. L., and Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prevot 1938. Anaerobe 40, 95–99. doi: 10.1016/j.anaerobe.2016.06.008
Luo, H., and Gao, F. (2019). DoriC 10.0: an updated database of replication origins in prokaryotic genomes including chromosomes and plasmids. Nucleic Acids Res. 47, D74–D77. doi: 10.1093/nar/gky1014
Mackiewicz, P., Zakrzewska-Czerwinska, J., Zawilak, A., Dudek, M. R., and Cebrat, S. (2004). Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 32, 3781–3791. doi: 10.1093/nar/gkh699
Majka, J., Messer, W., Schrempf, H., and Zakrzewska-Czerwinska, J. (1997). Purification and characterization of the Streptomyces lividans initiator protein DnaA. J. Bacteriol. 179, 2426–2432.
Makowski, L., Donczew, R., Weigel, C., Zawilak-Pawlik, A., and Zakrzewska-Czerwinska, J. (2016). Initiation of Chromosomal Replication in Predatory Bacterium Bdellovibrio bacteriovorus. Front. Microbiol. 7:1898. doi: 10.3389/fmicb.2016.01898
Marsin, S., McGovern, S., Ehrlich, S. D., Bruand, C., and Polard, P. (2001). Early steps of Bacillus subtilis primosome assembly. J. Biol. Chem. 276, 45818–45825. doi: 10.1074/jbc.M101996200
Martin, E., Williams, H. E. L., Pitoulias, M., Stevens, D., Winterhalter, C., Craggs, T. D., et al. (2018). DNA replication initiation in Bacillus subtilis: structural and functional characterization of the essential DnaA-DnaD interaction. Nucleic Acids Res. 47, 2101–2112. doi: 10.1093/nar/gky1220
Matthews, L. A., and Simmons, L. A. (2019). Cryptic protein interactions regulate DNA replication initiation. Mol. Microbiol. 111, 118–130. doi: 10.1111/mmi.14142
Moriya, S., Fukuoka, T., Ogasawara, N., and Yoshikawa, H. (1988). Regulation of initiation of the chromosomal replication by DnaA-boxes in the origin region of the Bacillus subtilis chromosome. EMBO J. 7, 2911–2917.
Mrazek, J., and Xie, S. (2006). Pattern locator: a new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics 22, 3099–3100. doi: 10.1093/bioinformatics/btl551
Murray, H., and Koh, A. (2014). Multiple regulatory systems coordinate DNA replication with cell growth in Bacillus subtilis. PLoS Genet. 10:e1004731. doi: 10.1371/journal.pgen.1004731
Natrajan, G., Noirot-Gros, M. F., Zawilak-Pawlik, A., Kapp, U., and Terradot, L. (2009). The structure of a DnaA/HobA complex from Helicobacter pylori provides insight into regulation of DNA replication in bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 21115–21120. doi: 10.1073/pnas.0908966106
Necsulea, A., and Lobry, J. R. (2007). A new method for assessing the effect of replication on DNA base composition asymmetry. Mol. Biol. Evol. 24, 2169–2179. doi: 10.1093/molbev/msm148
Nowaczyk-Cieszewska, M., Zyla-Uklejewicz, D., Noszka, M., Jaworski, P., Mielke, T., and Zawilak-Pawlik, A. M. (2020). The role of Helicobacter pylori DnaA domain I in orisome assembly on a bipartite origin of chromosome replication. Mol. Microbiol. 113, 338–355. doi: 10.1111/mmi.14423
Nozaki, S., and Ogawa, T. (2008). Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 154(Pt 11), 3379–3384. doi: 10.1099/mic.0.2008/019745-0
O’Donnell, M., Langston, L., and Stillman, B. (2013). Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 5:a010108. doi: 10.1101/cshperspect.a010108
Ogasawara, N., Moriya, S., von Meyenburg, K., Hansen, F. G., and Yoshikawa, H. (1985). Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J. 4, 3345–3350.
Ogasawara, N., and Yoshikawa, H. (1992). Genes and their organization in the replication origin region of the bacterial chromosome. Mol. Microbiol. 6, 629–634. doi: 10.1111/j.1365-2958.1992.tb01510.x
Oliveira Paiva, A. M., Friggen, A. H., Qin, L., Douwes, R., Dame, R. T., and Smits, W. K. (2019). The Bacterial Chromatin Protein HupA Can Remodel DNA and Associates with the Nucleoid in Clostridium difficile. J. Mol. Biol. 431, 653–672. doi: 10.1016/j.jmb.2019.01.001
Ozaki, S., Fujimitsu, K., Kurumizaka, H., and Katayama, T. (2006). The DnaA homolog of the hyperthermophilic eubacterium Thermotoga maritima forms an open complex with a minimal 149-bp origin region in an ATP-dependent manner. Genes Cells 11, 425–438. doi: 10.1111/j.1365-2443.2006.00950.x
Ozaki, S., and Katayama, T. (2012). Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665. doi: 10.1093/nar/gkr832
Ozaki, S., Kawakami, H., Nakamura, K., Fujikawa, N., Kagawa, W., Park, S. Y., et al. (2008). A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283, 8351–8362. doi: 10.1074/jbc.M708684200
Ozaki, S., Noguchi, Y., Hayashi, Y., Miyazaki, E., and Katayama, T. (2012). Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 287, 37458–37471. doi: 10.1074/jbc.M112.372052
Patel, M. J., Bhatia, L., Yilmaz, G., Biswas-Fiss, E. E., and Biswas, S. B. (2017). Multiple conformational states of DnaA protein regulate its interaction with DnaA boxes in the initiation of DNA replication. Biochim. Biophys. Acta 1861, 2165–2174. doi: 10.1016/j.bbagen.2017.06.013
Plachetka, M., Zyla-Uklejewicz, D., Weigel, C., Donczew, R., Donczew, M., Jakimowicz, D., et al. (2019). Streptomycete origin of chromosomal replication with two putative unwinding elements. Microbiology 165, 1365–1375. doi: 10.1099/mic.0.000859
Richardson, T. T., Harran, O., and Murray, H. (2016). The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature 534, 412–416. doi: 10.1038/nature17962
Richardson, T. T., Stevens, D., Pelliciari, S., Harran, O., Sperlea, T., and Murray, H. (2019). Identification of a basal system for unwinding a bacterial chromosome origin. EMBO J. 38:e101649. doi: 10.15252/embj.2019101649
Rokop, M. E., Auchtung, J. M., and Grossman, A. D. (2004). Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol. Microbiol. 52, 1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x
Rosenbusch, K. E., Bakker, D., Kuijper, E. J., and Smits, W. K. (2012). C. difficile 630Derm Spo0A Regulates Sporulation, but Does Not Contribute to Toxin Production, by Direct High-Affinity Binding to Target DNA. PLoS One 7:e48608. doi: 10.1371/journal.pone.0048608
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning : A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Saxena, R., Fingland, N., Patil, D., Sharma, A. K., and Crooke, E. (2013). Crosstalk between DnaA protein, the initiator of Escherichia coli chromosomal replication, and acidic phospholipids present in bacterial membranes. Int. J. Mol. Sci. 14, 8517–8537. doi: 10.3390/ijms14048517
Schaper, S., and Messer, W. (1995). Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 270, 17622–17626. doi: 10.1074/jbc.270.29.17622
Schenk, K., Hervas, A. B., Rosch, T. C., Eisemann, M., Schmitt, B. A., Dahlke, S., et al. (2017). Rapid turnover of DnaA at replication origin regions contributes to initiation control of DNA replication. PLoS Genet. 13:e1006561. doi: 10.1371/journal.pgen.1006561
Scholefield, G., Errington, J., and Murray, H. (2012). Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 31, 1542–1555. doi: 10.1038/emboj.2012.6
Scholefield, G., and Murray, H. (2013). YabA and DnaD inhibit helix assembly of the DNA replication initiation protein DnaA. Mol. Microbiol. 90, 147–159. doi: 10.1111/mmi.12353
Seid, C. A., Smith, J. L., and Grossman, A. D. (2017). Genetic and biochemical interactions between the bacterial replication initiator DnaA and the nucleoid-associated protein Rok in Bacillus subtilis. Mol. Microbiol. 103, 798–817. doi: 10.1111/mmi.13590
Sekimizu, K., Bramhill, D., and Kornberg, A. (1988). Sequential early stages in the in vitro initiation of replication at the origin of the Escherichia coli chromosome. J. Biol. Chem. 263, 7124–7130.
Smits, W. K., Goranov, A. I., and Grossman, A. D. (2010). Ordered association of helicase loader proteins with the Bacillus subtilis origin of replication in vivo. Mol. Microbiol. 75, 452–461. doi: 10.1111/j.1365-2958.2009.06999.x
Smits, W. K., Lyras, D., Lacy, D. B., Wilcox, M. H., and Kuijper, E. J. (2016). Clostridium difficile infection. Nat. Rev. Dis. Prim. 2:16020. doi: 10.1038/nrdp.2016.20
Smits, W. K., Merrikh, H., Bonilla, C. Y., and Grossman, A. D. (2011). Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. J. Bacteriol. 193, 640–648. doi: 10.1128/JB.01253-1210
Speck, C., Weigel, C., and Messer, W. (1999). ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 18, 6169–6176. doi: 10.1093/emboj/18.21.6169
Sutton, M. D., and Kaguni, J. M. (1997). Novel alleles of the Escherichia coli dnaA gene. J. Mol. Biol. 271, 693–703. doi: 10.1006/jmbi.1997.1209
Torti, A., Lossani, A., Savi, L., Focher, F., Wright, G. E., Brown, N. C., et al. (2011). Clostridium difficile DNA polymerase IIIC: basis for activity of antibacterial compounds. Curr. Enzym. Inhib. 7, 147–153.
Tsodikov, O. V., and Biswas, T. (2011). Structural and thermodynamic signatures of DNA recognition by Mycobacterium tuberculosis DnaA. J. Mol. Biol. 410, 461–476. doi: 10.1016/j.jmb.2011.05.007
van Eijk, E., Anvar, S. Y., Browne, H. P., Leung, W. Y., Frank, J., Schmitz, A. M., et al. (2015). Complete genome sequence of the Clostridium difficile laboratory strain 630Deltaerm reveals differences from strain 630, including translocation of the mobile element CTn5. BMC Genomics 16:31. doi: 10.1186/s12864-015-1252-7
van Eijk, E., Boekhoud, I. M., Kuijper, E. J., Bos-Sanders, I., Wright, G., and Smits, W. K. (2019). Genome location dictates the transcriptional response to PolC inhibition in Clostridium difficile. Antimicrob. Agents Chemother. 63:e01363-18. doi: 10.1128/AAC.01363-18
van Eijk, E., Paschalis, V., Green, M., Friggen, A. H., Larson, M. A., Spriggs, K., et al. (2016). Primase is required for helicase activity and helicase alters the specificity of primase in the enteropathogen Clostridium difficile. Open Biol. 6:160272. doi: 10.1098/rsob.160272
van Eijk, E., Wittekoek, B., Kuijper, E. J., and Smits, W. K. (2017). DNA replication proteins as potential targets for antimicrobials in drug-resistant bacterial pathogens. J. Antimicrob. Chemother. 72, 1275–1284. doi: 10.1093/jac/dkw548
Vellanoweth, R. L., and Rabinowitz, J. C. (1992). The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 6, 1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x
Velten, M., McGovern, S., Marsin, S., Ehrlich, S. D., Noirot, P., and Polard, P. (2003). A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell 11, 1009–1020. doi: 10.1016/s1097-2765(03)00130-8
Warriner, K., Xu, C., Habash, M., Sultan, S., and Weese, S. J. (2017). Dissemination of Clostridium difficile in food and the environment: Significant sources of C. difficile community-acquired infection? J. Appl. Microbiol. 122, 542–553. doi: 10.1111/jam.13338
Weigel, C., Schmidt, A., Seitz, H., Tungler, D., Welzeck, M., and Messer, W. (1999). The N-terminus promotes oligomerization of the Escherichia coli initiator protein DnaA. Mol. Microbiol. 34, 53–66. doi: 10.1046/j.1365-2958.1999.01568.x
Wolanski, M., Donczew, R., Zawilak-Pawlik, A., and Zakrzewska-Czerwinska, J. (2014). oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol 5:735. doi: 10.3389/fmicb.2014.00735
Xu, W. C., Silverman, M. H., Yu, X. Y., Wright, G., and Brown, N. (2019). Discovery and development of DNA polymerase IIIC inhibitors to treat Gram-positive infections. Bioorg. Med. Chem. 27, 3209–3217. doi: 10.1016/j.bmc.2019.06.017
Zawilak, A., Durrant, M. C., Jakimowicz, P., Backert, S., and Zakrzewska-Czerwinska, J. (2003). DNA binding specificity of the replication initiator protein, DnaA from Helicobacter pylori. J. Mol. Biol. 334, 933–947.
Zawilak-Pawlik, A., Kois, A., Majka, J., Jakimowicz, D., Smulczyk-Krawczyszyn, A., Messer, W., et al. (2005). Architecture of bacterial replication initiation complexes: orisomes from four unrelated bacteria. Biochem. J. 389(Pt 2), 471–481. doi: 10.1042/BJ20050143
Zawilak-Pawlik, A., Nowaczyk, M., and Zakrzewska-Czerwinska, J. (2017). The role of the N-terminal domains of bacterial initiator DnaA in the assembly and regulation of the bacterial replication initiation complex. Genes 8:136. doi: 10.3390/genes8050136
Zhabinskaya, D., Madden, S., and Benham, C. J. (2015). SIST: stress-induced structural transitions in superhelical DNA. Bioinformatics 31, 421–422. doi: 10.1093/bioinformatics/btu657
Zhang, W., Carneiro, M. J., Turner, I. J., Allen, S., Roberts, C. J., and Soultanas, P. (2005). The Bacillus subtilis DnaD and DnaB proteins exhibit different DNA remodelling activities. J. Mol. Biol. 351, 66–75. doi: 10.1016/j.jmb.2005.05.065
Keywords: oriC, Clostridioides difficile, P1 nuclease, unwinding, DnaA
Citation: Oliveira Paiva AM, van Eijk E, Friggen AH, Weigel C and Smits WK (2020) Identification of the Unwinding Region in the Clostridioides difficile Chromosomal Origin of Replication. Front. Microbiol. 11:581401. doi: 10.3389/fmicb.2020.581401
Received: 08 July 2020; Accepted: 31 August 2020;
Published: 02 October 2020.
Edited by:
Monika Glinkowska, University of Gdañsk, PolandReviewed by:
Julia Grimwade, Florida Institute of Technology, United StatesShogo Ozaki, Kyushu University, Japan
Copyright © 2020 Oliveira Paiva, van Eijk, Friggen, Weigel and Smits. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wiep Klaas Smits, w.k.smits@lumc.nl
†ORCID: Ana M. Oliveira Paiva, orcid.org/0000-0002-6122-832X; Erika van Eijk, orcid.org/0000-0003-3003-9822; Annemieke H. Friggen, orcid.org/0000-0001-5780-2053; Wiep Klaas Smits, orcid.org/0000-0002-7409-2847
 Ana M. Oliveira Paiva1,2†
Ana M. Oliveira Paiva1,2† Wiep Klaas Smits
Wiep Klaas Smits