- 1Department of Molecular Biology, Ariel University, Ariel, Israel
- 2Koret School of Veterinary Medicine, The Robert H. Smith Faculty of Agriculture, Food and Environment, The Hebrew University of Jerusalem, Rehovot, Israel
- 3Government Central Laboratories, Ministry of Health, Jerusalem, Israel
- 4The Dr. Miriam and Sheldon G. Adelson School of Medicine, Ariel University, Ariel, Israel
Salmonella enterica is a major causative pathogen of human and animal gastroenteritis. Antibiotic resistant strains have emerged due to the production of extended-spectrum β-lactamases (ESBLs) posing a major health concern. With the increasing reports on ESBL-producing Enterobacterales that colonize companion animals, we aimed to investigate ESBL dissemination among ESBL-producing Salmonella enterica (ESBL-S) in hospitalized horses. We prospectively collected ESBL-S isolates from hospitalized horses in a Veterinary-Teaching Hospital during Dec 2015–Dec 2017. Selection criteria for ESBL-S were white colonies on CHROMagarESBL plates and an ESBL phenotypic confirmation. Salmonella enterica serovars were determined using the Kaufmann-White-Le-Minor serological scheme. ESBL-encoding plasmids were purified, transformed and compared using restriction fragment length polymorphism (RFLP). Whole genome sequencing (Illumina and MinION platforms) were performed for detailed phylogenetic and plasmid analyses. Twelve ESBL-S were included in this study. Molecular investigation and Sequence Read Archive (SRA) meta-analysis revealed the presence of three unique Salmonella enterica serovars, Cerro, Havana and Liverpool, all reported for the first time in horses. PFGE revealed the clonal spread of S. Cerro between seven horses. All twelve isolates carried blaCTX–M–3 and showed an identical multidrug resistance profile with co-resistance to trimethoprim/sulfamethoxazole and to aminoglycosides. Plasmid RFLP proved the inter-serovar horizontal spread of a single blaCTX–M–3-encoding plasmid. Complete sequence of a representative plasmid (S. Havana strain 373.3.1), designated pSEIL-3 was a -86.4 Kb IncM2 plasmid, that encoded nine antibiotic resistance genes. pSEIL-3 was virtually identical to pCTX-M3 from Citrobacter freundii, and showed high identity (>95%) to six other blaCTX–M–3 or blaNDM–1 IncM2 broad host range plasmids from various Enterobacterales of human origin. Using a specific six gene-based multiplex PCR, we detected pSEIL-3 in various Enterobacterales species that co-colonized the horses’ gut. Together, our findings show the alarming emergence of ESBL-S in hospitalized horses associated with gut shedding and foal morbidity and mortality. We demonstrated the dissemination of CTX-M-3 ESBL among different Salmonella enterica serovars due to transmission of a broad host range plasmid. This report highlights horses as a zoonotic reservoir for ESBL-S, including highly transmissible plasmids that may represent a ‘One-Health’ hazard. This risk calls for the implementation of infection control measures to monitor and control the spread of ESBL-S in hospitalized horses.
Introduction
Salmonella enterica is the major causative pathogen of human and animal Salmonellosis (Scallan et al., 2011; Omer et al., 2018). Human Salmonellosis recently poses a major health concern due to the dissemination of multidrug resistant (MDR) strains that produce extended-spectrum β-lactamases (ESBLs) that limit the appropriate treatment options (Antonelli et al., 2019; Jajere, 2019). ESBL-producing Salmonella enterica (ESBL-S) are increasingly reported from livestock animals (European Centre for Disease Prevention and Control., 2019). Shared ESBL-S serovars among livestock and humans suggest that food animals are possible zoonotic reservoir for this human-associated pathogen (Sjölund-Karlsson et al., 2013).
In the last decade, along with food animals, ESBL-producing Enterobacterales colonization in companion animals is steadily increasing (Doi et al., 2017). Although the zoonotic potential of these bacteria is still enigmatic, there is a consensus regarding their role as being a reservoir for antibiotic resistance, and as a possible hazard to human health due to the close physical contact between companion animals and humans (Madec et al., 2017).
Horses in specific are in close interaction with humans and children in various interfaces including private use, sport events and as therapeutic animals. As such, they may serve as a zoonotic source for antibiotic resistant pathogens. Horses have been shown previously to be colonized and infected with various clinically important pathogens including methicillin-resistant Staphylococcus aureus (MRSA) (Tirosh-Levy et al., 2015), Acinetobacter baumannii, and various ESBL-producing enteric pathogens (Walther et al., 2018; Shnaiderman-Torban et al., 2019). As for the genus Salmonella, horses may be sub-clinically infected with the bacterium or suffer from clinical signs which may vary from mild disease as fever and dehydration, to diarrhea, colic and manifestations of septicemia (Hernandez et al., 2014; Cummings et al., 2016). However, reports on ESBL-S strains in horses are still rare. A report from Germany described an SHV-12-producing S. Newport causing an outbreak in an equine hospital, which led to a three-month facility closure (Rankin et al., 2005). Another report from the United States described 11 ESBL-S clinical isolates from an equine referral hospital that belonged to various serovars including Braenderup, Anatum, Agona, Rubislaw, and Newport (Leon et al., 2018).
In a previous study, we investigated the shedding rate of ESBL-producing Enterobacterales in farm horses versus hospitalized horses and observed a significant increase in ESBL shedding rate among hospitalized horses together with first isolation of three colonizing isolates that we identified as ESBL-S isolates (Shnaiderman-Torban et al., 2020). The present study investigated and characterized the molecular epidemiology of ESBL-S isolates that were isolated during our surveys, together with ESBL-S isolates recovered from clinical infections from hospitalized horses during the study period. We aimed to describe the emergence of ESBL-S in hospitalized horses and to explore the dissemination of ESBL in this important pathogen.
Materials and Methods
Isolation of ESBL-Producing Enterobacterales From Hospitalized Horses
During a prospective surveillance study of ESBL-producing Enterobacterales (ESBL-E) gut colonization in hospitalized horses that we performed in the Koret School of Veterinary Medicine-Veterinary Teaching Hospital (KSVM-VTH) in Israel (Dec 2015–Dec 2017), rectal swabs were collected from horses on admission and after 72 h of hospitalization. The study protocol was approved by the Internal Research Review Institution Committee (Protocol number: KSVM-VTH/15_2015). Isolation of ESBL-E from swabs was performed after swab enrichment in Tryptic Soy Broth supplemented with Ampicillin (100 mg/L), and an overnight incubation at 37°C to increase sensitivity of detection (Jazmati et al., 2016). After incubation, samples were plated onto CHROMagarESBL plates (HyLabs, Rehovot, Israel). In addition, Salmonella enterica clinical isolates recovered from horses during the study period that were processed at the Clinical Microbiology Lab at the Meir Medical Center, Kfar Saba, Israel, were collected and stored for retrospective molecular characterization.
Isolation of ESBL-Producing Salmonella enterica and the Identification of ESBL Genes
Following the former described procedure, all the white colonies that were obtained on the CHROMagarESBL plates, suspected as ESBL-producing Salmonella enterica (ESBL-S) were further isolated onto selective Salmonella/Shigella-agar plates (HyLabs) following verification using the slide agglutination polyvalent serum assay (Remel Inc., United States). All ESBL-S isolates (both fecal and clinical isolates) were identified by the VITEK 2 automated system (Biomerieux, United States) together with antibiotic susceptibility testing using AST-N270 and AST-GN65 cards. Susceptibility results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. All isolates were confirmed for a positive ESBL production phenotype using the cephalosporin/clavulanic-acid combination disk assay (Oxoid, United Kingdom). The blaCTX–M genes were identified by multiplex-PCR (Woodford et al., 2006) and Sanger sequencing (Macrogen, Netherlands). Sequences were analyzed (Snap-Gene) and compared with NCBI database to identify the specific ESBL gene allele.
Salmonella enterica Serovar Identification and Pulsed Field Gel Electrophoresis (PFGE)
Salmonella enterica serotyping was performed using the Kauffmann-White-Le Minor scheme (Le Minor et al., 1982). XbaI-restricted (New England BioLabs) PFGE was performed according to the PulseNet International Standard Protocol (Ribot et al., 2006) with S. Braenderup H9812 as a reference strain. The PFGE fingerprinting patterns were analyzed with BioNumerics software (version 7.6.3, Applied Maths, Sint-Martens-Latem, Belgium). The unweighted-pair group method using average linkages (UPGMA) clustering method and Dice similarity coefficients were used (1% optimization and 1% tolerance). Isolates were defined as genetically related if they presented ≥ 98% PFGE similarity.
Meta Data of the NCBI Sequence Read Archive (SRA) for Statistical Analysis
We explored the global occurrences and the isolation sources of the Salmonella enterica serovars using the NCBI SRA data. In order to perform the mata-analysis, we retrieved the SRA accession numbers and meta-data for all the publically available isolates using the NCBI E-Utilities. Statistical correlations between the serovar type and the isolation source were calculated using Phi coefficients with p-values < 0.01. P-values were corrected for multiple tests in step-down method using Bonferroni adjustments (alpha = 0.01). All statistical analyses were performed using Python statistics modules.
Salmonella enterica Whole Genome Sequencing (WGS) and Data Analysis
Total DNA was isolated using Blood and Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. WGS was performed by Illumina MiSeq platform using 2 × 250 paired-end libraries prepared with the NEBNext Ultra II FS DNA Library Prep Kit. Assembly was performed using SPAdes-3.11.1. Plasmid replicon types and antibiotic resistance genes (ARGs) were identified using the Center for Genomic Epidemiology (CGE) pipeline.
Whole Genome Multi Locus Sequence Typing (wgMLST) Phylogenetic Analysis
The EnteroBase database was searched for sequences predicted as S. Cerro, S. Havana, or S. Liverpool, according to the SISTR1 and SeqSero2 algorithms. Strains with source country metadata were selected for a phylogenetic analysis and for comparison with the Israeli sequences. A GrapeTree depiction of a NINJA NJ tree based on the wgMLST allelic distances was generated for each serovar population.
Purification and Characterization of Salmonella enterica ESBL-Encoding Plasmids
ESBL-encoding plasmid DNA was extracted using the Plasmid Midi Kit (Qiagen) following the manufacturer’s instructions. Plasmids were transformed into electro-competent Escherichia coli DH10B and transformants were selected on ampicillin containing LB plates (100 mg/L), followed by blaCTX–M PCR screening (Woodford et al., 2006). A second transformation and plasmid purification was performed to ensure plasmid purity. ESBL-encoding plasmids purified from all the 12 ESBL-S isolates were compared using RFLP analysis following restriction with SacI, EcoRI and HindIII (New England BioLabs) and electrophoresis.
Complete Sequencing and Annotation of blaCTX–M–3-Encoding Plasmid pSEIL-3
Since ten of 12 isolates harbored CTX-M-3-encoding plasmids with identical RFLP patterns, one representative plasmid (pSEIL-3 from S. Havana strain 373.3.1) was sequenced using MinION device (Oxford Nanopore Technologies, ONT, Oxford, United Kingdom) following hybrid assembly, resulting in a complete plasmid sequence (Wick et al., 2017). Plasmid DNA (200 ng) fragment library was prepared (SQK-RBK004 ONT Rapid barcoding sequencing kit) according to the manufacturer’s instructions, and loaded onto the MinION flow cell FLO-MIN106. The hybrid read set (WGS Illumina and Nanopore reads) was assembled using Unicycler (v0.4.0) to yield a single circular plasmid designated pSEIL-3, annotated by RAST (Aziz et al., 2008). Replicon type assignment, ARG content, virulence genes and IS elements identification were performed using the CGE pipeline, and by ISfinder (Siguier et al., 2012). Homologous plasmids were identified from the NCBI Nucleotide (nt/nr) database using BLASTn search. Linear plasmid maps were generated using Easyfig-2.2.3.
GenBank Submission
WGS Illumina reads of the three S. serovars were deposited in the NCBI Sequence Read Archive database under project number PRJNA559324 (Table 2). The complete pSEIL-3 sequence isolated from S. Havana strain 373.3.1 (BioSample SAMN12532154) was submitted to the NCBI Nucleotide database under the accession number MN380440.
Conjugation Experiments of pSEIL-3
Conjugation experiments were performed with S. Cerro strain 339.3.3 and S. Havana strain 373.3.1 as the donor strains and Klebsiella pneumoniae B199 (resistant to nalidixic-acid) and E. coli J53 (rifampicin resistant) as the recipient strains. Filter-mating was performed (donor and recipient, 1:1 ratio) on LB plates followed by selection of transconjugant colonies on LB agar plates containing ceftriaxone (2 mg/L) and either nalidixic acid (64 mg/L) or rifampicin (300 mg/L). Transconjugants were verified by the colony color obtained on CHROMagarESBL plates and by PCR detection of blaCTX–M–3, and then were subjected to the VITEK 2 for antibiotic susceptibility testing.
Molecular Screening for the Presence of pSEIL-3
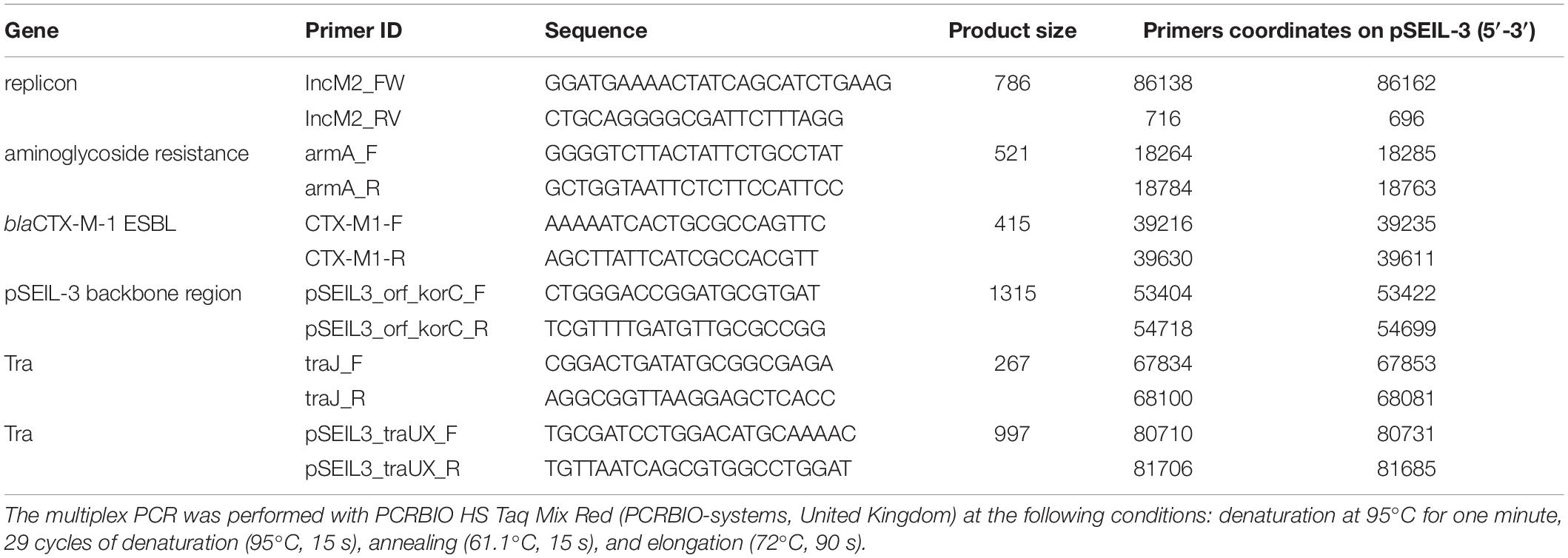
A novel six-gene multiplex PCR scheme for the molecular screening of pSEIL-3 was developed. The primers were designed (Table 3) based on the sequences of six genes whose combination was unique according to the NCBI Nucleotide database search. The multiplex PCR was performed with PCRBIO HS Taq Mix Red (PCRBIO-systems, United Kingdom) at the following conditions: denaturation at 95°C for one minute, 29 cycles of denaturation (95°C, 15 s), annealing (61.1°C, 15 s) and elongation (72°C, 90 s).
Results
ESBL-Producing Salmonella enterica Isolates Recovered From Hospitalized Horses
Overall, 12 ESBL-S isolates were recovered from 12 hospitalized horses during the study period. All these strains were isolated > 72 h after admission and therefore were defined as nosocomial (Table 1). Ten out of the 12 horses were also sampled on admission, as part of an ESBL surveillance study, and two of them were found to be colonized with an ESBL-producing Enterobacterales (ESBL-E) strain. Nine out of the 12 horses (75%) that were colonized with an ESBL-S were also found to be colonized with different ESBL-E species. Diverse ESBL-E colonizing species were found: E. coli (n = 7), K. pneumoniae (n = 4), Klebsiella oxytoca (n = 2), and Citrobacter freundii (n = 1) (Table 1). Ten of the ESBL-S isolates originated from rectal swabs, of which, four were sampled from asymptomatic horses, and six from horses with clinical signs of gastroenteritis. Two additional ESBL-S isolates were clinical isolates that caused joint and umbilicus infections. The majority of the horses from which ESBL-S was recovered were neonates (8/12, 67%, Table 1), which were all diagnosed with sepsis (Wong et al., 2018), presenting various clinical signs. These foals were all treated with ampicillin and amikacin, and if they suffered from diarrhea, metronidazole therapy was added. Five out of eight (62.5%) died or were euthanized during hospitalization.
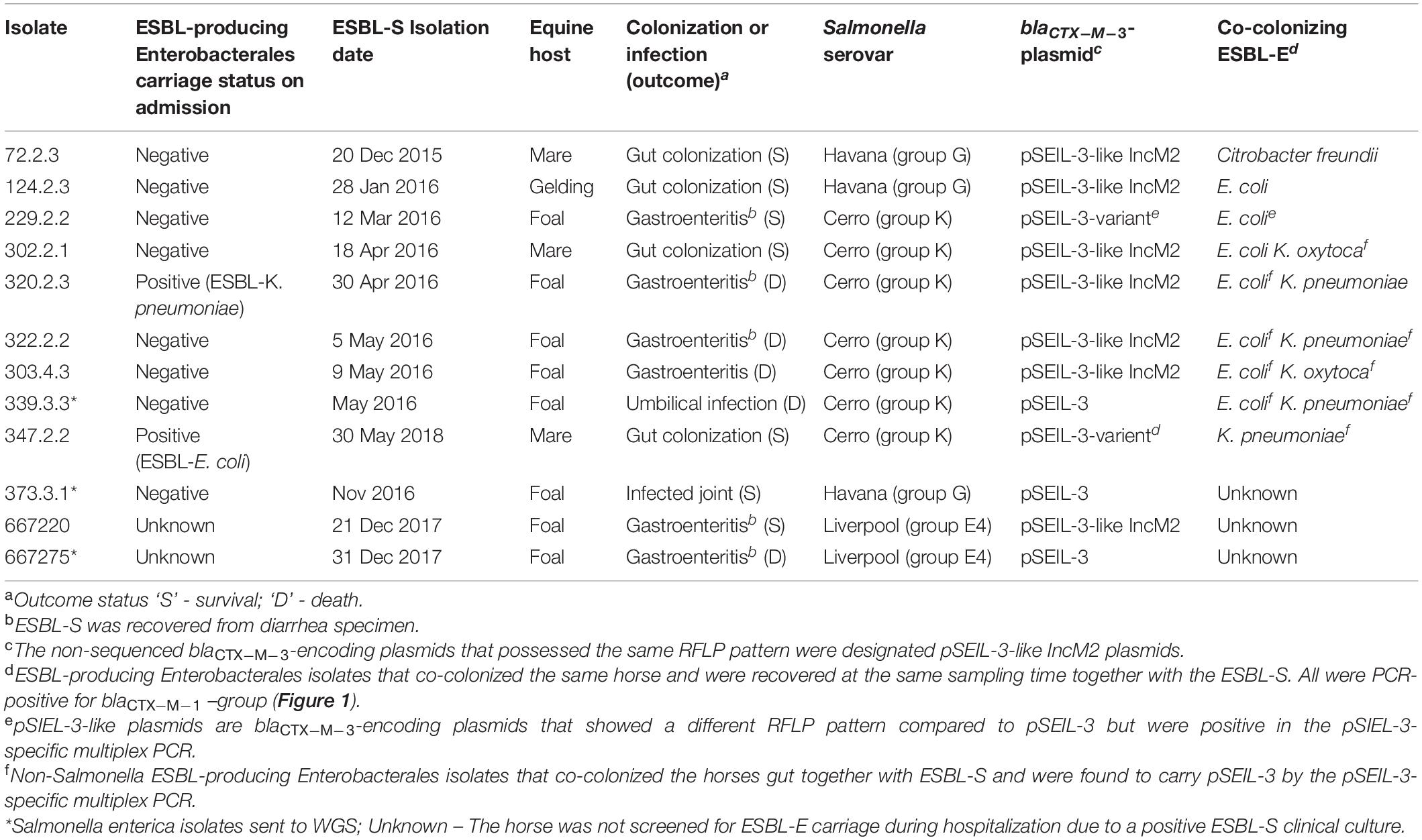
Table 1. Molecular and epidemiological characteristics of the 12 ESBL-producing Salmonella enterica (ESBL-S) isolates included in this study and their equine host.
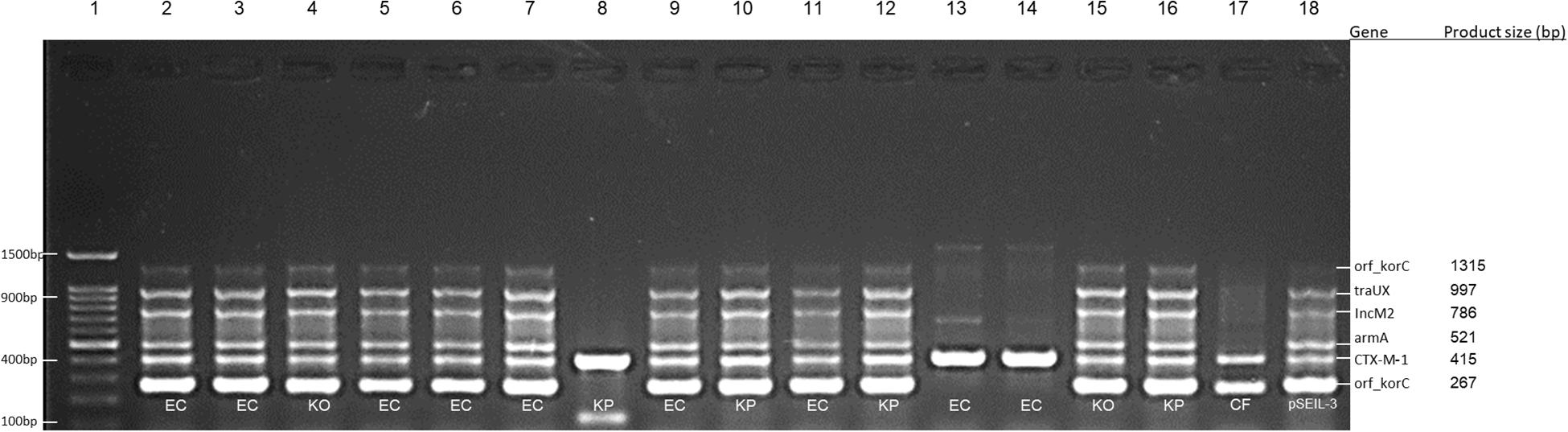
Figure 1. Multiplex PCR for screening of pSEIL-3 in non-Salmonella CTX-M-1-producing Enterobacteriales species co-colonizing eight horses. Six-gene-multiplex PCR amplification for the detection of pSIEL-3 was performed on 16 CTX-M-1 positive ESBL-E isolates co-colonizing (together with ESBL-S) eight horses. foal 229, lane 2; foal 303, lanes 3–6; foal 320, lanes 7–8; foal 322, lanes 9–10; foal 339, lanes 11–12; gelding 124, lane 13; mare 302, lanes 14–15; mare 347, lane 16; mare 72, lane 17; pSIEL-3, lane 18; DNA 100 bp ladder, lane 1. EC - E. coli; KP - K. pneumoniae; KO - K. oxytoca; CF - Citrobacter freundii.
Serovars, Genotyping and Antibiotic Susceptibility Profiles
ESBL-S isolates belonged to three different serovars – Cerro (n = 7), Havana (n = 3), and Liverpool (n = 2), with Cerro being the major serovar, representing more than 50% of the isolates (Table 1). All the seven ESBL-producing S. Cerro isolates clustered in time (a two-month period) and PFGE genotyping suggested the clonal expansion of this serovar (87.8–100% isolate identity, Supplementary Figure 1).
All the 12 ESBL-S isolates carried blaCTX–M–3 and showed an identical MDR profile independent with their serovar antibiotic susceptibility testing showed resistance to ceftriaxone, aminoglycosides, amikacin, tobramycin, gentamicin, and trimethoprim-sulfamethoxazole. Isolates were susceptible to carbapenems, quinolones and fosfomycin (Supplementary Table 1).
WGS of ESBL-S Serovars and Identification of Plasmid Replicons and Resistome
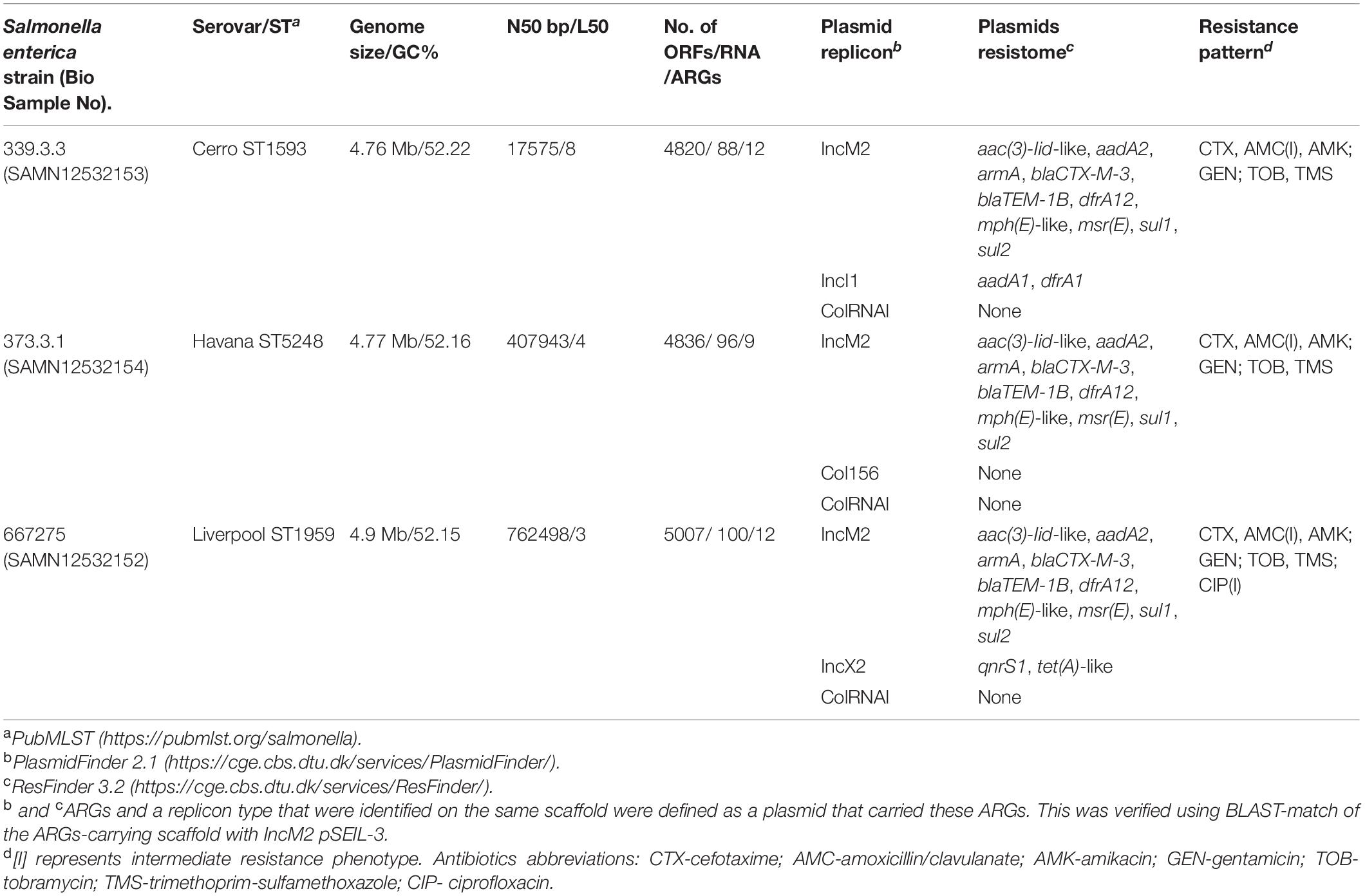
To further explore the three ESBL-S serovars identified in the equine population, we performed WGS of three representative isolates, one of each serovar (data was deposited under project number PRJNA559324 in the GenBank). The WGS data is summarized in Table 2. Sequence types were identified in silico, and plasmid replicon analysis revealed that all three serovars harbored common IncM2 and ColRNAI plasmids, accompanied by other plasmids, that were unique for each isolate. Alongside with blaCTX–M–3, they all encoded a wide resistome encompassing nine to 12 ARGs that correlated with their susceptibility profiles (Table 2).
Local and Global Occurrences and Comparative Genomics of S. enterica Serovars Cerro, Havana, and Liverpool
In order to assess the origin of the Salmonella enterica serovars identified in this study, we analyzed the Salmonella national database that consists data on all human and non-human Salmonella isolates recovered in Israel (the reference Salmonella laboratory, the Ministry of Health, Israel). The data indicated that during the study period (2015–2017), the annual prevalence of Cerro, Havana and Liverpool serovars was relatively low; In human infections it ranged from 0.1–0.2% (out of an average of 3,952 Salmonella isolates/year). In non-human sources, the prevalence increased throughout these years but was also low (0, 0.2% and 1% for Cerro, Havana and Liverpool in 2015, to - 0.6%, 0.4%, and 3.3%, in 2017, respectively).
In order to evaluate the global abundance of these Salmonella serovars and to hypothesize about their main reservoirs we performed a meta-analysis on a global dataset of SRA Salmonella enterica isolates belonging to the respected serovars (Supplementary Table 2, n = 1394). The meta-analysis indicated that these serovars were recovered previously from various human, animal and food sources, with Cerro being the most prevalent serovar. This meta-analysis confirmed that these three serovars are reported herein for the first time in horses.
In order to study the relations with globally reported lineages and clusters, we compared our Israeli genomic sequences with all global genomes of S. Cerro, S. Havana, and S. Liverpool with geographical source, available in Enterobase database. A minimum spanning tree representing all wgMLST profiles for each serovar is shown in Figure 2. The analysis of the global population highlights closely clustered genotypes that originate from specific geographical locations. The Israeli genotypes did not significantly cluster with strains from other countries, and the minimum allelic distances from the nearest neighbors were 40 for S. Cerro, 779 for S. Havana, and 39 for S. Liverpool (Figure 2). The analysis indicated that the antibiotic resistance phenotypes of the Israeli strains are linked to genomic profiles unique to Israel.
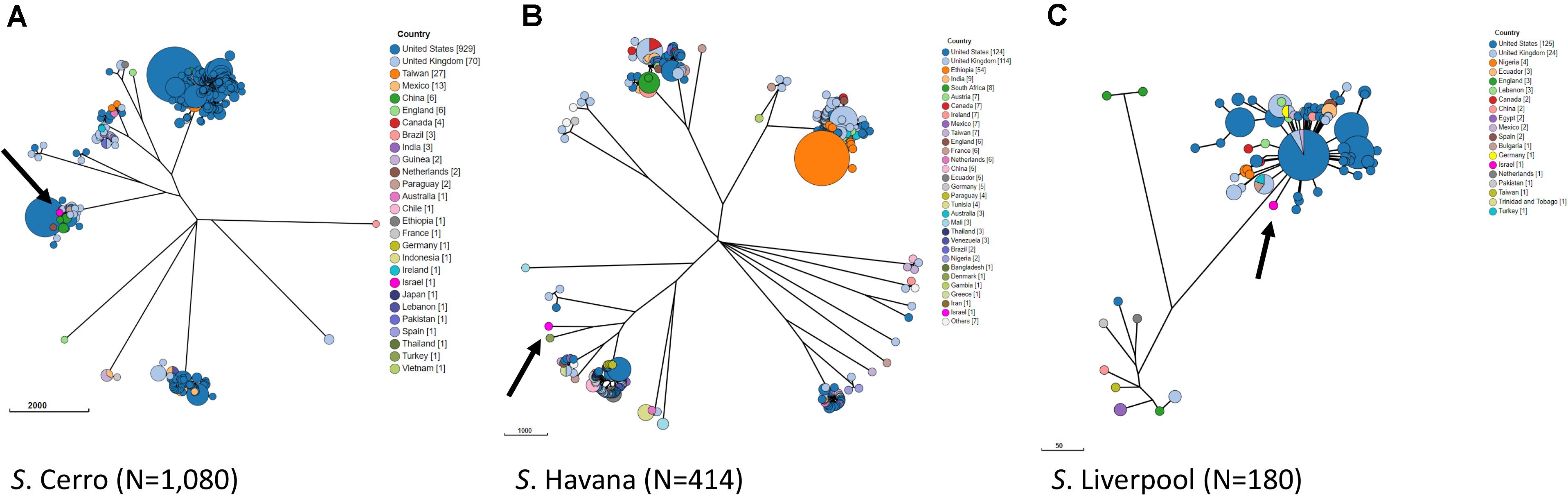
Figure 2. Phylogenetic analysis by wgMLST of global populations of S. Cerro (A), S. Havana (B), and S. Liverpool (C). GrapeTree visualization of a NINJA NJ tree based on wgMLST allelic distances. Strains with allelic distances of ≤10 alleles were clustered into a single node. The three Israeli strains of each serovar are shown in pink and are indicated by an arrow.
Characterization of the ESBL-Encoding Salmonella Plasmids
The WGS data revealed similar plasmid content between the serovars with a common IncM blaCTX–M–3-encoding plasmid (Table 2). The blaCTX–M–3-encoding plasmids of all twelve isolates were successfully transformed into E. coli DH10B The blaCTX–M–3–positive transformants possessed exactly the same antibiotic susceptibility profile showing resistance to all cephalosporins except for ceftazidime and co-resistance to trimethoprim/sulfamethoxazole and aminoglycosides (Supplementary Table 1).
To examine and support the possible inter-serovar plasmid transmission we compared all 12 blaCTX–M–3-encoding plasmids by plasmid-RFLP. Ten out of 12 plasmids (83%) showed an identical RFLP pattern, suggesting an inter-serovar horizontal plasmid transfer. We designated this plasmid as pSEIL-3 (plasmid of S. enterica from Israel encoding blaCTX–M–3). In silico analysis of the WGS data confirmed the presence of pSEIL-3 in all three serovars. A representative RFLP analysis of pSEIL-3 is presented in Figure 3. Two out of the 12 blaCTX–M–3-carrying ESBL-S isolates (229.2.2 and 347.2.2, Table 1) were IncM2 plasmids with a different RFLP pattern (Results are not presented) suggesting the presence of a variant of this plasmid (Table 1).
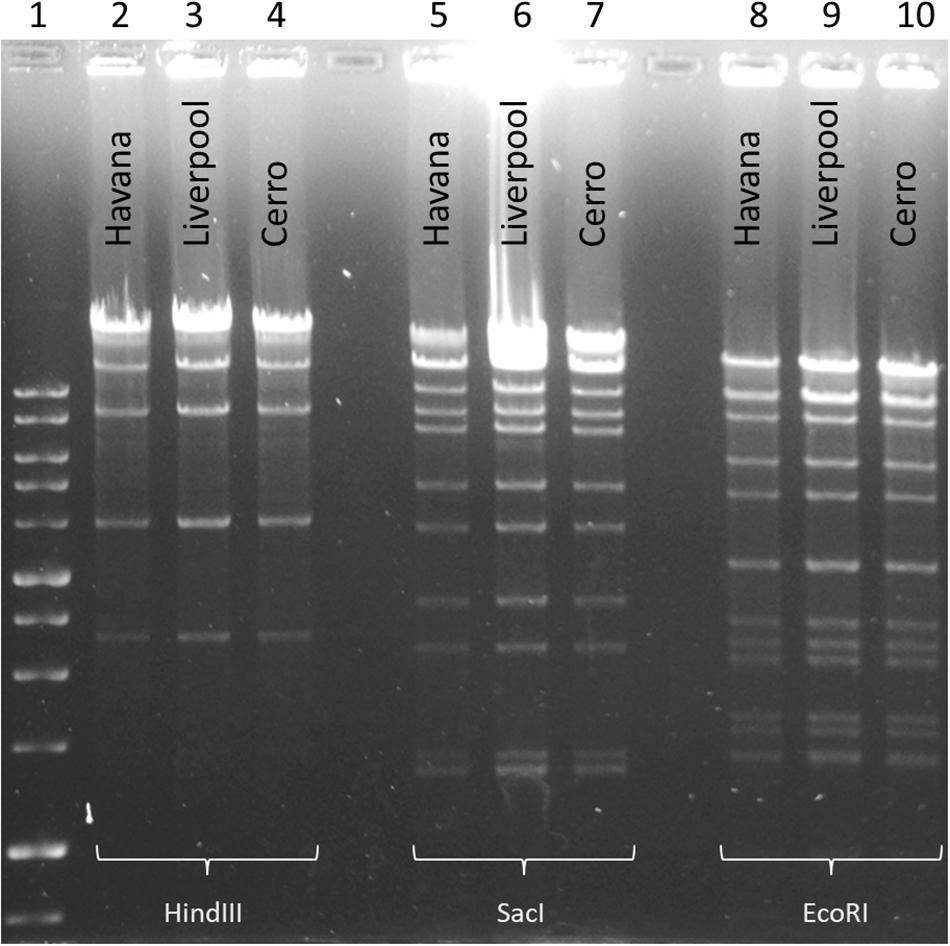
Figure 3. Plasmid RFLP mapping of the CTX-M-3-encoding plasmid DNA from Salmonella serovars. RFLP analysis of plasmid DNA (1 μg) isolated from three Salmonella isolates representing each of the serovars (S. Liverpool 667220; S. Havana 373.3.1; and S. Cerro 303.4.3) was restricted with three restriction enzymes, SacI (Lanes 2–4), EcoRI (Lanes 5–7), and HindIII (Lanes 8–10). 1 kb DNA size ladder (Lane 1). Plasmid RFLP analysis revealed identical restriction patterns demonstrating the presence of an identical plasmid common to all the three serovars.
Complete Sequence of pSEIL-3
To deepen our understanding on the transferability of pSEIL-3 we sequenced the purified plasmid (Havana 373.3.1) and performed long-read MinION sequencing. Using a hybrid assembly, we generated the complete sequence of the circular 86207-bp plasmid. pSEIL-3 was an IncM2 plasmid (Carattoli et al., 2015) and encoded 118 ORFs, 24 conjugation genes and a single toxin-antitoxin pair pemIK. The pSEIL-3 resistome encompassed nine ARGs conferring broad resistance to cephalosporins (blaTEM–1B and blaCTX–M–3), aminoglycosides (the modifying enzymes, aac2 and aadA2 and the 16S rRNA methyl transferase, armA), trimethoprim (dfrA12), sulfonamide (sul1) and to macrolides (msrE and mphE) (Figure 4).
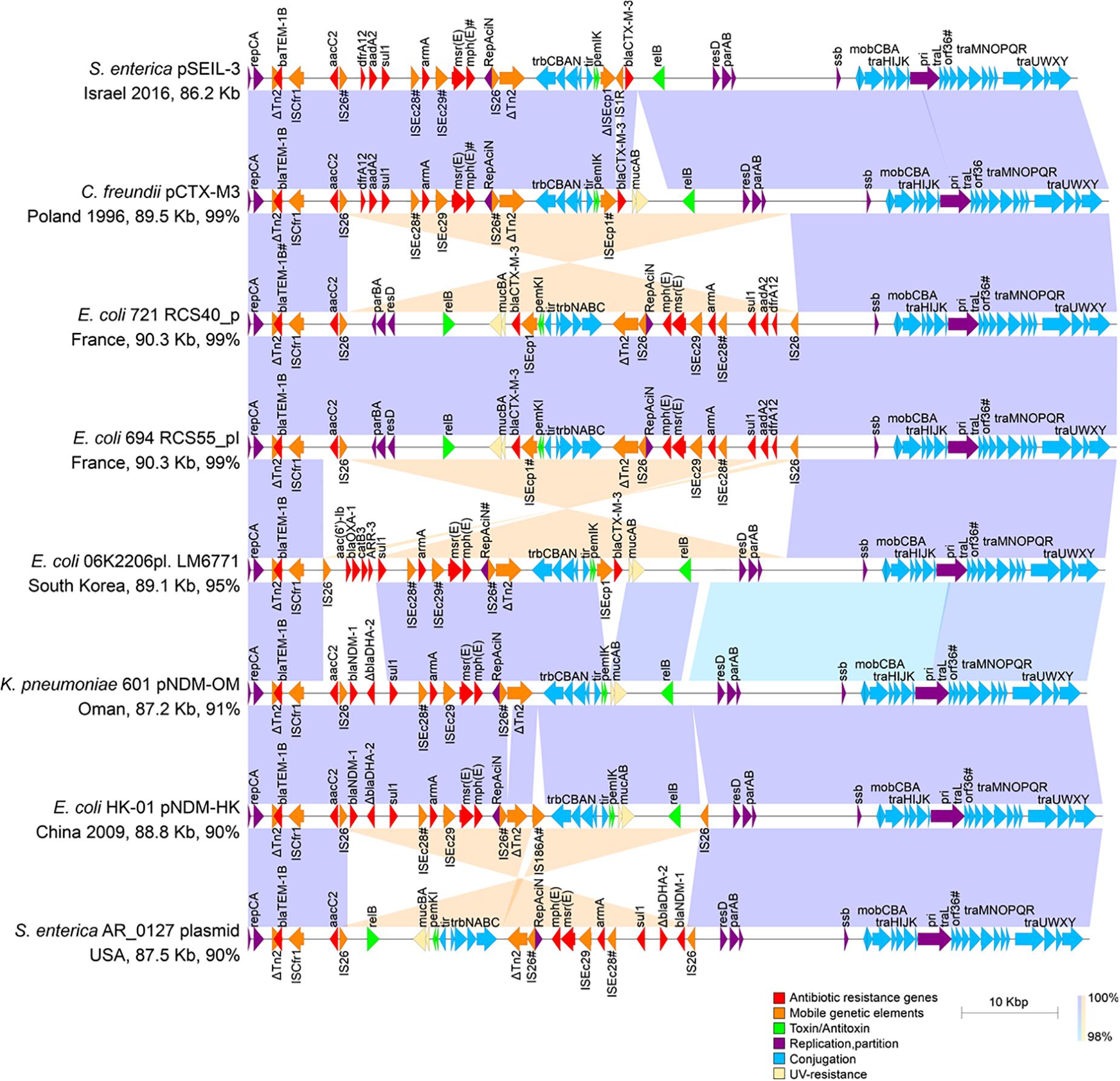
Figure 4. Linear plasmid maps of pSEIL-3 and seven highly related IncM2 related plasmids. Linear schematic maps of pSEIL-3 and IncM2-related plasmids retrieved from the GenBank (aligning for >90%). Plasmids’ GenBank accession numbers: pCTX-M3 - AF550415; RCS40_p - LT985241; RCS55_pI - LT985387; LM6771 - KX009507; pNDM-OM - JX988621; pNDM-HK - HQ451074; AR_0127 - CP032193. Arrows (colored by function) represent the major annotated gene groups and IS elements. The alignment between related plasmids is shown (purple, same direction alignment; orange, and opposite direction). The year of plasmid isolation is indicated when available. Truncated genes are labeled with ‘Δ’ and mutated genes (identity > 99%) are labeled with #.
Blast-based search of plasmids related to pSEIL-3 revealed that our sequenced plasmid was virtually identical to pCTX-M-3 from Citrobacter freundii except for a 3902 bp region that encodes mucAB of the umuDC-like gene family that is involved in UV-resistance, and four additional ORFs that encode hypothetical proteins in pCTX-M-3. The conjugation genes in both plasmids were identical except for a truncation in orf36 that was shown previously to be involved in plasmid mobilization efficiency (Dmowski et al., 2018). In addition, pSEIL-3 resembled six other blaCTX–M–3 and blaNDM-encoding plasmids aligning to >90% of its sequence (Figure 4). These plasmids were isolated from various human Enterobacterales strains (E. coli - 4; C. freundii - 1; K. pneumoniae - 1; S. enterica - 1) isolated from different countries and years, demonstrating the broad-host-range and high stability nature of these plasmids. Plasmid alignment revealed several DNA rearrangements that seemed to be host-dependent and presumably were linked to the presence of IS26 (Figure 4).
Transferability of pSEIL-3
Plasmid pSEIL-3 proved to be self-conjugable and was transferrable into both E. coli and K. pneumoniae. Acquisition of pSEIL-3 resulted in the same antibiogram as the donor ESBL-S strains (Supplementary Table 1).
In order to examine the in situ transferability of pSEIL-3 and pSEIL-3-like IncM2 plasmids in the horses’ gut, we screened 16 non-Salmonella isolates that co-colonized the horses, and that were PCR positive for blaCTX–M–1 –group, for the presence of pSEIL-3 using a specific six-gene multiplex PCR we have developed (Table 3). We identified pSEIL-3 and pSEIL-3-like plasmids in 12/16 (75%) of the ESBL-E isolates tested. These isolates belonged to various Enterobacterales species, including E. coli, K. pneumoniae, K. oxytoca, and C. freundii. Therefore, we defined it as a broad host range plasmid (Figure 1).
Discussion
In this study, we report for the first time the emergence of three MDR CTX-M-3-producing S. enterica serovars - Cerro, Havana and Liverpool, which colonize and cause severe infections in hospitalized horses. Based on WGS and molecular studies we elucidated the route of ESBL spread in S. enterica and discovered an inter-serovar horizontal transfer of an IncM2 broad host range plasmid, pSEIL-3. Furthermore, we identified the clonal expansion of blaCTX–M–3-producing S. Cerro that was responsible for more than half of the cases.
Global phylogenetic serovar analysis indicated the genetic uniqueness of our strains, and the metadata analysis revealed that these three serovars have not been described before in horses. Previously, S. Cerro was mainly reported in cattle in the United States (Tewari et al., 2012; Webb et al., 2017), and as the main causative Salmonellosis pathogen in dairy farms (Van Kessel et al., 2007; Kovac et al., 2017). In the United States and the Far East, S. Cerro has also been reported in poultry (Roy et al., 2002; Murase et al., 2004). The second serovar we found, S. Havana was reported both in humans (Backer, 2000; Bekal et al., 2013) and in poultry (Clemente et al., 2013), and less frequently in wild birds (Reche et al., 2003) and in environmental setting (Magwedere et al., 2015). It was also identified previously as an ESBL-producer, carrying various blaCTX–M alleles (Bekal et al., 2013; Clemente et al., 2013). S. Liverpool is a more rarely reported serovar, with a single report that describes its origin from cattle feces in an EU registered slaughterhouse (Madden et al., 2007).
The source of the S. enterica serovars that we identified is unknown. In spite a large nation-wide survey of poultry-associated Salmonella enterica was recently reported from Israel (Cohen et al., 2020), data on the serovars that are circulating in the community or in hospital equine populations is still lacking. The large animal department in the KSVM-VTH serves equine patients from diverse farms that occasionally may be housed together with different farm animals. In addition, various animals, often rescued from rural areas, are sporadically admitted for intensive care to the same department. These farm animals may be the source for these Salmonella serovars however, a solid support for this is lacking.
Interestingly, the majority of the horses included in our study were not detected as positive ESBL carriers on admission to the hospital, suggesting the nosocomial acquisition of the ESBL-producing strains or the ESBL genetic elements (the blaCTX–M–3 gene or its encoding plasmid). In the United States, studies that describe asymptomatic community carriage of Salmonella in horses report the prevalence of 0.8% without information on the existing serovars (Traub-Dargatz et al., 2000). Other studies in horses that describe the prevalence of clinical Salmonella isolates indicate that the main serovars are Typhimurium, Newport, Agona, Javiana, Anatum, Infantis, and Braenderup (Hernandez et al., 2014; Martelli et al., 2019). Nevertheless, the serovars that we describe herein are unique and are mentioned for the first time in the context of equine population.
Dissemination of ESBL among the hospitalized horses showed a complex epidemiology that included the clonal expansion of S. Cerro between seven horses alongside with an in-hospital spread of pSEIL-3 that horizontally transferred to all three Salmonella serovars. Acquisition of this single plasmid with its wide resistome was responsible for the dissemination of multidrug resistance. Complete plasmid sequencing of pSEIL-3 indicated that it is merely identical to the previously reported wide-host-range pCTX-M-3 plasmid from Citrobacter freundii (Golebiewski et al., 2007) and to other MDR plasmids, that encode various carbapenemases, all from human origin. The findings of pSEIL-3-like IncM2 plasmids in other non-Salmonella ESBL-E species that colonized the horses’ gut is alarming, and proves their high inter-species transmissibility. The presence of pSEIL-3 in horses, and previously in humans, highlights the risk of horizontal transmission of MDR plasmids between human, animals and environmental pools.
The potential transmission of pSEIL-3-like plasmids is disturbing not only due to their broad host range, but also due to their wide resistome, which confers resistance to all aminoglycosides and to trimethoprim/sulfamethoxazole. Considering the massive use of aminoglycosides antibiotics, often combined with β-lactamase inhibitors, for treating ESBL-producing pathogens in humans, food and companion animals, emphasizes the risk of this plasmid as it may lead to limited treatment options. Additional reports regarding this clinically important ARGs combination in S. enterica are infrequent, with one recent study that described a similar MDR pattern of S. Virchow from food animals in South Korea (Na et al., 2020), and another study describing shedding of quinolone resistant and ESBL-producing S. enterica serovars in swine population in the United States (Elnekave et al., 2019).
The clinical impact of ESBL-S and specifically pSEIL-3-like plasmids in a ‘One-Health’ perspective is vast. The clonal expansion of the S. Cerro underlines the lack of current infection-control measures for detecting and controlling Salmonella infections in the veterinary hospital, and calls for the implementation of control measures to prevent further spread. The existence of highly transmissible plasmids such as pSEIL-3 and its spread into three uncommon S. enterica serovars highlights the importance of detailed molecular analyses for elucidation of these transmission paths. The developed multiplex PCR in this study enables the tracking of pSEIL-3 in future studies and in active surveillance actions.
This study describes horse-to-horse spread of a zoonotic pathogen harboring a wide-host-range MDR plasmid, which was reported previously in human pathogens, representing a major public health concern. Although the source of this highly transferable plasmid in the veterinary hospital and its circulating routes remains unclear, its disseminative nature is alarming.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, SAMN12532153; https://www.ncbi.nlm.nih.gov/genbank/, SAMN12532154; https://www.ncbi.nlm.nih.gov/genbank/, SAMN12532152.
Author Contributions
ZD and AS-T collected the specimens. ZD performed all the microbiological and molecular analyses. KK assisted in the bioinformatics analysis. MD-C and AR performed the serovar typing, the PFGE and the wgMLST analyses. AS was involved in the study design. SN-V was responsible for the design of the study and data analyses. ZD and SN-V wrote the manuscript. All authors read and approved the submitted manuscript.
Funding
This study was supported by a personal grant on ‘Antibiotic Resistance in Companion Animals’ provided by Ariel University to SN-V. MSc studies scholarship was awarded to ZD by Ariel University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Dr. Paitan Y, the director of the Clinical Microbiology and Virology Laboratories, Meir Medical Center, Kfar Saba, Israel for performing the VITEK 2 antibiotic susceptibility analysis. We thank Prof. Ohad Galmore from the Infectious Diseases Research Laboratory at the Sheba Medical Center, for providing us with the E. coli RifR strain for the conjugation experiments. This work was performed in partial fulfillment of Ziv Dor MSc degree requirements at the Department of Molecular Biology, Faculty of Life Sciences at Ariel University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.616032/full#supplementary-material
References
Antonelli, P., Belluco, S., Mancin, M., Losasso, C., and Ricci, A. (2019). Genes conferring resistance to critically important antimicrobials in Salmonella enterica isolated from animals and food: a systematic review of the literature, 2013–2017. Res. Vet. Sci. 126, 59–67. doi: 10.1016/j.rvsc.2019.08.022
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genom. 9:75. doi: 10.1186/1471-2164-9-75
Backer, H. D. (2000). High incidence of extra-intestinal infections in a Salmonella Havana outbreak associated with alfalfa sprouts. Public Health Rep. 115, 339–344. doi: 10.1093/phr/115.4.339
Bekal, S., Lefebvre, B., Bergevin, M., and Tremblay, C. (2013). CTX-M-15 type ESBL-producing Salmonella havana associated with international adoption in canada. Can. J. Microbiol. 59, 57–57. doi: 10.1139/cjm-2012-0667
Carattoli, A., Seiffert, S. N., Schwendener, S., Perreten, V., and Endimiani, A. (2015). Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS ONE 10:e0123063. doi: 10.1371/journal.pone.0123063
Clemente, L., Manageiro, V., Ferreira, E., Jones-Dias, D., Correia, I., Themudo, P., et al. (2013). Occurrence of extended-spectrum β-lactamases among isolates of Salmonella enterica subsp. enterica from food-producing animals and food products, in Portugal. Int. J. Food Microbiol. 167, 221–228. doi: 10.1016/j.ijfoodmicro.2013.08.009
Cohen, E., Davidovich, M., Rokney, A., Valinsky, L., Rahav, G., and Gal-Mor, O. (2020). Emergence of new variants of antibiotic resistance genomic islands among multidrug-resistant Salmonella enterica in poultry. Environ. Microbiol. 22, 413–432. doi: 10.1111/1462-2920.14858
Cummings, K. J., Perkins, G. A., Khatibzadeh, S. M., Warnick, L. D., Aprea, V. A., and Altier, C. (2016). Antimicrobial resistance trends among Salmonella isolates obtained from horses in the northeastern United States (2001–2013). Am. J. Vet. Res. 77, 505–513. doi: 10.2460/ajvr.77.5.505
Dmowski, M., Gołêbiewski, M., and Kern-Zdanowicz, I. (2018). Characteristics of the conjugative transfer system of the IncM Plasmid pCTX-M3 and identification of its putative regulators. J. Bacteriol. 200, e218–e234. doi: 10.1128/JB.00234-18
Doi, Y., Iovleva, A., and Bonomo, R. A. (2017). The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J. Travel Med. 24, S44–S51. doi: 10.1093/jtm/taw102
European Centre for Disease Prevention and Control. (2019). The european union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 17:e06007. doi: 10.2903/j.efsa.2019.5598
Elnekave, E., Hong, S. L., Lim, S., Hayer, S. S., Boxrud, D., Taylor, A. J., et al. (2019). Circulation of plasmids harboring resistance genes to quinolones and/or extended-spectrum cephalosporins in multiple Salmonella enterica serotypes from swine in the united states. Antimicrob. Agents Chemother. 63, e2602–e2618. doi: 10.1128/AAC.02602-18
Golebiewski, M., Kern-Zdanowicz, I., Zienkiewicz, M., Adamczyk, M., Zylinska, J., Baraniak, A., et al. (2007). Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum -lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51, 3789–3795. doi: 10.1128/AAC.00457-07
Hernandez, J. A., Long, M. T., Traub-Dargatz, J. L., and Besser, T. E. (2014). “Salmonellosis,” in Equine Infectious Diseases. Amsterdam: Elsevier, 321–333. doi: 10.1016/B978-1-4557-0891-8.00035-X
Jajere, S. M. (2019). A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World 12, 504–521. doi: 10.14202/vetworld.2019.504-521
Jazmati, N., Hein, R., and Hamprecht, A. (2016). Use of an enrichment broth improves detection of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in clinical stool samples. J. Clin. Microbiol. 54, 467–470. doi: 10.1128/JCM.02926-15
Kovac, J., Cummings, K. J., Rodriguez-Rivera, L. D., Carroll, L. M., Thachil, A., and Wiedmann, M. (2017). Temporal genomic phylogeny reconstruction indicates a geospatial transmission path of Salmonella cerro in the united states and a clade-specific loss of hydrogen sulfide production. Front. Microbiol. 8:737. doi: 10.3389/fmicb.2017.00737
Le Minor, L., Véron, M., and Popoff, M. (1982). [The taxonomy of Salmonella]. Ann. Microbiol. 133, 223–243.
Leon, I. M., Lawhon, S. D., Norman, K. N., Threadgill, D. S., Ohta, N., Vinasco, J., et al. (2018). Serotype diversity and antimicrobial resistance among Salmonella enterica isolates from patients at an equine referral hospital. Appl. Environ. Microbiol. 84, e2817–e2829. doi: 10.1128/AEM.02829-17
Madden, R. H., Murray, K. A., and Gilmour, A. (2007). Carriage of four bacterial pathogens by beef cattle in northern ireland at time of slaughter. Lett. Appl. Microbiol. 44, 115–119. doi: 10.1111/j.1472-765X.2006.02064.x
Madec, J.-Y., Haenni, M., Nordmann, P., and Poirel, L. (2017). Extended-spectrum β-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: a threat for humans? Clin. Microbiol. Infec. 23, 826–833. doi: 10.1016/j.cmi.2017.01.013
Magwedere, K., Rauff, D., De Klerk, G., Keddy, K. H., and Dziva, F. (2015). Incidence of nontyphoidal Salmonella in food-producing animals. animal feed, and the associated environment in south africa, 2012–2014. Clin. Infect. Dis. 61, S283–S289. doi: 10.1093/cid/civ663
Martelli, F., Kidd, S., and Lawes, J. (2019). Surveillance for Salmonella in horses in great britain. Vet. Rec. 184, 56–58. doi: 10.1136/vr.l149
Murase, T., Nagato, M., Shirota, K., Katoh, H., and Otsuki, K. (2004). Pulsed-field gel electrophoresis-based subtyping of DNA degradation-sensitive Salmonella enterica subsp. Enterica serovar livingstone and serovar cerro isolates obtained from a chicken layer farm. Vet. Microbiol. 99, 139–143. doi: 10.1016/j.vetmic.2003.11.014
Na, S. H., Moon, D. C., Kang, H. Y., Song, H.-J., Kim, S.-J., Choi, J.-H., et al. (2020). Molecular characteristics of extended-spectrum β-lactamase/AmpC-producing Salmonella enterica serovar virchow isolated from food-producing animals during 2010–2017 in south korea. Int. J. Food Microbiol. 322:108572. doi: 10.1016/j.ijfoodmicro.2020.108572
Omer, M. K., Álvarez-Ordoñez, A., Prieto, M., Skjerve, E., Asehun, T., and Alvseike, O. A. (2018). A systematic review of bacterial foodborne outbreaks related to red meat and meat products. Foodborne Pathog. Dis. 15, 598–611. doi: 10.1089/fpd.2017.2393
Rankin, S. C., Whichard, J. M., Joyce, K., Stephens, L., O’Shea, K., Aceto, H., et al. (2005). Detection of a blaSHV Extended-spectrum β-Lactamase in Salmonella enterica serovar newport MDR-AmpC. J. Clin. Microbiol. 43, 5792–5793. doi: 10.1128/JCM.43.11.5792-5793.2005
Reche, M. P., Jiménez, P. A., Alvarez, F., García de los Rios, J. E., Rojas, A. M., and de Pedro, P. (2003). Incidence of Salmonellae in captive and wild free-living raptorial birds in central spain. J. Vet. Med. B Infect. Dis. Vet. Public Health 50, 42–44. doi: 10.1046/j.1439-0450.2003.00623.x
Ribot, E. M., Fair, M. A., Gautom, R., Cameron, D. N., Hunter, S. B., Swaminathan, B., et al. (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7. salmonella, and shigella for PulseNet. Foodborne Pathog. Dis. 3, 59–67. doi: 10.1089/fpd.2006.3.59
Roy, P., Dhillon, A. S., Lauerman, L. H., Schaberg, D. M., Bandli, D., and Johnson, S. (2002). Results of Salmonella isolation from poultry products, poultry, poultry environment, and other characteristics. Avian Dis. 46, 17–24.
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the united states—major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Shnaiderman-Torban, A., Navon-Venezia, S., Dor, Z., Paitan, Y., Arielly, H., Abu Ahmad, W., et al. (2020). Extended-Spectrum β-lactamase-producing Enterobacteriaceae shedding in farm horses versus hospitalized horses: prevalence and risk factors. Animals 10:282. doi: 10.3390/ani10020282
Shnaiderman-Torban, A., Paitan, Y., Arielly, H., Kondratyeva, K., Tirosh-Levy, S., Abells-Sutton, G., et al. (2019). Extended-spectrum β-lactamase-producing Enterobacteriaceae in hospitalized neonatal foals: prevalence, risk factors for shedding and association with infection. Animals 9:600. doi: 10.3390/ani9090600
Siguier, P., Varani, A., Perochon, J., and Chandler, M. (2012). “Exploring Bacterial Insertion Sequences with ISfinder: Objectives, Uses, and Future Developments,” in Mobile Genetic Elements, ed. Y. Bigot (Totowa, NJ: Humana Press), 91–103.
Sjölund-Karlsson, M., Howie, R. L., Blickenstaff, K., Boerlin, P., Ball, T., Chalmers, G., et al. (2013). Occurrence of β-lactamase genes among non-Typhi Salmonella enterica isolated from humans, food animals, and retail meats in the United States and Canada. Microb. Drug Resist. 19, 191–197. doi: 10.1089/mdr.2012.0178
Tewari, D., Sandt, C. H., Miller, D. M., Jayarao, B. M., and M’ikanatha, N. M. (2012). Prevalence of Salmonella Cerro in laboratory-based submissions of cattle and comparison with human infections in pennsylvania, 2005–2010. Foodborne Pathog. Dis. 9, 928–933. doi: 10.1089/fpd.2012.1142
Tirosh-Levy, S., Steinman, A., Carmeli, Y., Klement, E., and Navon-Venezia, S. (2015). Prevalence and risk factors for colonization with methicillin resistant Staphylococcus aureus and other Staphylococci species in hospitalized and farm horses in Israel. Prev. Vet. Med. 122, 135–144. doi: 10.1016/j.prevetmed.2015.09.007
Traub-Dargatz, J. L., Garber, L. P., Fedorka-Cray, P. J., Ladely, S., and Ferris, K. E. (2000). Fecal shedding of Salmonella spp by horses in the United States during 1998 and 1999 and detection of Salmonella spp in grain and concentrate sources on equine operations. J. Am. Vet. Med. Assoc. 217, 226–230. doi: 10.2460/javma.2000.217.226
Van Kessel, J. S., Karns, J. S., Wolfgang, D. R., Hovingh, E., and Schukken, Y. H. (2007). Longitudinal study of a clonal. subclinical outbreak of salmonella enterica subsp. enterica serovar cerro in a u.s. dairy herd. Foodborne Pathog. Dis. 4, 449–461. doi: 10.1089/fpd.2007.0033
Walther, B., Klein, K.-S., Barton, A.-K., Semmler, T., Huber, C., Wolf, S. A., et al. (2018). Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Acinetobacter baumannii among horses entering a veterinary teaching hospital: the contemporary “Trojan Horse.”. PLoS One 13:0191873. doi: 10.1371/journal.pone.0191873
Webb, H. E., Brichta-Harhay, D. M., Brashears, M. M., Nightingale, K. K., Arthur, T. M., Bosilevac, J. M., et al. (2017). Salmonella in peripheral lymph nodes of healthy cattle at slaughter. Front. Microbiol. 8:2214. doi: 10.3389/fmicb.2017.02214
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 3:e000132. doi: 10.1099/mgen.0.000132
Wong, D. M., Ruby, R. E., Dembek, K. A., Barr, B. S., Reuss, S. M., Magdesian, K. G., et al. (2018). Evaluation of updated sepsis scoring systems and systemic inflammatory response syndrome criteria and their association with sepsis in equine neonates. J. Vet. Intern. Med. 32, 1185–1193. doi: 10.1111/jvim.15087
Keywords: Salmonella enterica, WGS, ESBL, serovars, IncM2, blaCTX–M–3, MDR plasmid, horizontal transfer
Citation: Dor Z, Shnaiderman-Torban A, Kondratyeva K, Davidovich-Cohen M, Rokney A, Steinman A and Navon-Venezia S (2020) Emergence and Spread of Different ESBL-Producing Salmonella enterica Serovars in Hospitalized Horses Sharing a Highly Transferable IncM2 CTX-M-3-Encoding Plasmid. Front. Microbiol. 11:616032. doi: 10.3389/fmicb.2020.616032
Received: 10 October 2020; Accepted: 27 November 2020;
Published: 17 December 2020.
Edited by:
Jian-Hua Liu, South China Agricultural University, ChinaReviewed by:
Yvonne Pfeifer, Robert Koch Institute (RKI), GermanyIzabela Kern-Zdanowicz, Institute of Biochemistry and Biophysics Polish Academy of Sciences, Poland
Copyright © 2020 Dor, Shnaiderman-Torban, Kondratyeva, Davidovich-Cohen, Rokney, Steinman and Navon-Venezia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiri Navon-Venezia, shirinv@ariel.ac.il
 Ziv Dor1
Ziv Dor1 Anat Shnaiderman-Torban
Anat Shnaiderman-Torban Kira Kondratyeva
Kira Kondratyeva Maya Davidovich-Cohen
Maya Davidovich-Cohen Assaf Rokney
Assaf Rokney Amir Steinman
Amir Steinman Shiri Navon-Venezia
Shiri Navon-Venezia