- 1CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Shenzhen, China
Heavy metal contamination is an environmental issue on a global scale. Particularly, cadmium poses substantial threats to crop and human health. Saccharomyces cerevisiae is one of the model organisms to study cadmium toxicity and was recently engineered as a cadmium hyperaccumulator. Therefore, it is desirable to overcome the cadmium sensitivity of S. cerevisiae via genetic engineering for bioremediation applications. Here we performed genome-scale overexpression screening for gene targets conferring cadmium resistance in CEN.PK2-1c, an industrial S. cerevisiae strain. Seven targets were identified, including CAD1 and CUP1 that are known to improve cadmium tolerance, as well as CRS5, NRG1, PPH21, BMH1, and QCR6 that are less studied. In the wild-type strain, cadmium exposure activated gene transcription of CAD1, CRS5, CUP1, and NRG1 and repressed PPH21, as revealed by real-time quantitative PCR analyses. Furthermore, yeast strains that contained two overexpression mutations out of the seven gene targets were constructed. Synergistic improvement in cadmium tolerance was observed with episomal co-expression of CRS5 and CUP1. In the presence of 200 μM cadmium, the most resistant strain overexpressing both CAD1 and NRG1 exhibited a 3.6-fold improvement in biomass accumulation relative to wild type. This work provided a new approach to discover and optimize genetic engineering targets for increasing cadmium resistance in yeast.
Introduction
Heavy metal contamination is a severe environmental problem (Vareda et al., 2019; Hu et al., 2020). Cadmium is one of the primary risks to human health due to the pollution of food crops and drinking water (Nordberg et al., 2018). Saccharomyces cerevisiae is a model organism to study cadmium toxicity, and related mechanisms include glutathione biosynthesis, stress response, vacuole transportation and sorting, metal ion homeostasis, and chromatin remodeling (Wysocki and Tamas, 2010). Notably, cadmium tolerance mechanisms are conserved between S. cerevisiae and plants, including gene overexpression of glutathione reductase (Kim et al., 2012), metallothionein (MT) (Wei et al., 2016; Ansarypour and Shahpiri, 2017), and phytochelatin (Cahoon et al., 2015; Yang et al., 2017).
Physiochemical and biological remediation are common strategies to treat heavy metal contamination. For example, chemical precipitation is frequently utilized, but the main limitations include secondary waste generation and complex infrastructure requirements (Fu and Wang, 2011; Li et al., 2019; Vardhan et al., 2019). Phytoremediation by “hyperaccumulator” plants is a promising alternative, but it suffers from the long life cycle and engineering challenges of plants (Rascio and Navari-Izzo, 2011; Reeves et al., 2018). On the other hand, microbial bioremediation may provide a scalable and cost-effective solution. In particular, S. cerevisiae has been recently engineered as a heavy-metal hyperaccumulator when equipped with selected membrane transporters and enhanced vacuolar compartmentalization (Sun et al., 2019, 2020). Furthermore, S. cerevisiae can be utilized to ferment the biomass of phytoremediation plants (Jing et al., 2020). However, S. cerevisiae is relatively sensitive to cadmium, and it is desirable to improve its cadmium robustness of S. cerevisiae as a future bioremediation agent.
Chemical tolerance is a complex phenotype with hundreds of genetic determinants (Almeida et al., 2007). Due to the lack of mechanistic understanding, it often requires genome-wide screening to identify mutations that confer resistance (Warner et al., 2010; Si et al., 2015b; Chen et al., 2020; Liu et al., 2020). For such endeavors, gene-deletion strain libraries are often utilized in S. cerevisiae, but most identified mutations lead to cadmium sensitivity (Jin et al., 2008; Ruotolo et al., 2008; Serero et al., 2008; Thorsen et al., 2009). Genetic overexpression libraries are less studied, and hence beneficial mutations are relatively scarce (Hwang et al., 2009; Fang et al., 2016). Notably, existing screening campaigns for cadmium resistance are confined to certain laboratory strains derived from the S288c background. Moreover, only single genetic mutations are investigated, whereas combinatorial optimization of multiplex gene targets is often needed for resistance engineering (Si et al., 2015a; Jiang et al., 2020).
Here we performed genome-wide screening in S. cerevisiae using a comprehensive cDNA plasmid library that was previously constructed (Si et al., 2017). We utilized a widely used industrial strain, CEN.PK2-1c, whose cadmium resistance has not been studied. Both known and new gene targets were identified to confer increased resistance. Further improvement in cadmium tolerance was achieved via the combinatorial introduction of individual mutations. Dosage-dependence and genetic interactions were also observed among different gene targets.
Materials and Methods
Strains, Media, and Cultivation Conditions
A previously reported derivative (CAD) of CEN.PK2-1c (MATa ura3-52 trp1-289 leu2-3,112 his3D1 MAL2-8C SUC2) was utilized in this study as the wild-type (WT) background, harboring an integrated RNA-interference pathway (Si et al., 2015a). Overexpression cassettes were either integrated into the genomic LEU2 locus or cloned in episomal plasmids (Supplementary Tables 1, 2). Saccharomyces cerevisiae strains were cultivated in either synthetic dropout medium (0.17% Difco yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate and 0.083% amino acid dropout mix, 0.01% adenine hemisulfate, and 2% glucose) or YPAD medium (1% yeast extract, 2% peptone, 0.01% adenine hemisulfate, and 2% glucose). For cadmium stresses, 100 mM Cd(NO3)2 solution stock was added to the above medium to indicated final concentrations. All media were adjusted to pH = 4.5. Saccharomyces cerevisiae strains were cultured at 30°C and with 250 r.p.m. agitation in culture tubes or 96-well microplates. Cadmium nitrate tetrahydrate was purchased from Aladdin (C118495). Other chemicals were purchased through Sigma-Aldrich.
Genome-Scale Overexpression Screening
The overexpression library was constructed by cloning the normalized cDNA library into a single-copy plasmid pRS416, which contains a PTEF1 promoter and a TPGK1 terminator (Si et al., 2017). The library was transformed into CAD strain by the standard LiAc/ssDNA/PEG protocol (Gietz and Schiestl, 2007) with an optimized condition, where 15 μg plasmid DNA was used to transform 30 OD600(optical density at 600 nm) unit competent yeast cells by heat shock at 42°C for 1 h. Cell transformant suspension was diluted 10-fold and spread onto SC-Ura agar plates containing 200 μM Cd(NO3)2. Approximately 1.5 × 106 independent yeast clones were obtained. The WT strain containing an empty pRS416 vector was used as a control. Clones with substantially larger colony sizes relative to WT were selected for further characterization. The overexpression cassettes of isolated plasmids were subjected to Sanger sequencing and Basic Local Alignment Search Tool (BLAST) analysis for target identification. Plasmid retransformation into a fresh WT background was performed to confirm positive targets.
Cadmium Tolerance Assays
Yeast strain cultures transformed with the empty vector or gene-overexpressing plasmids were inoculated and cultivated to the early stationary phase. OD600 values were measured using a Ultrospec 10 cell density meter (Biochrom, 80-2116-30). For spot assay, one OD600 unit of yeast cells was serially diluted at tenfold, spotted (3 μL) on SC-Ura agar supplemented with 200 μM Cd(NO3)2, and incubated at 30°C for 2 to 3 days. For microculture growth assay, growth phase-synchronized cultures were diluted to an initial OD600 of 0.2 in 200 μL of SC-Ura, SC-Ura/Leu or SC-Ura/Leu/Trp medium supplemented with indicated Cd(NO3)2 concentrations and cultivated at 30°C in 96-well, round-bottom plates (Corning). A BioTek Epoch2 microplate reader was utilized to monitor OD600 at 30 min intervals. The OD600 values at 48 h from three independent experiments were used to calculate relative cadmium tolerance [Equation (1), see below].
Real-Time, Quantitative PCR (RT-qPCR) Analyses
The mid-log phase cell cultures of the WT strain were treated with 50, 100, or 200 μM Cd(NO3)2 for 2 h. Total RNA was isolated using a Bacteria RNA Extraction Kit (Vazyme, R403-01), and 1 μg total RNA was reverse transcribed using HiScript III RT SuperMix for qPCR with gDNA wiper (Vazyme, R323-01). RT-qPCR reaction and data analysis were performed in an Analytik Jena qTOWER3 Real-Time PCR Thermal Cycler using ChamQ Universal SYBR qPCR Master Mix (Vzyme, Q711-02) following the manufacturer’s instructions. Primers for qPCR reactions were listed in Supplementary Table 3.
Dual-Mutation Strain Construction
The identified overexpression cassettes of cadmium-tolerant genes were PCR amplified with the TEF1p and PGK1t primers and cloned into the pRS415 vector using the Gibson Assembly Cloning Kit (New England Biolabs) (Supplementary Table 2). The primers used are listed in Supplementary Table 3. For genomic integration, the expression cassettes and LEU2 marker gene were PCR amplified with primers fusing with 40 bp homology arms and integrated into the LEU2 locus via homologous recombination.
Calculation of Relative Cadmium Tolerance and Combinatorial Effect Scores
The final OD600 at 48 h was used to calculate relative cadmium tolerance using Equation (1):
where G indicates a gene-overexpression target and V means the empty-vector control. T indicates cadmium treatment and C means the cadmium-free control. A value larger than 1 indicates improved cadmium tolerance relative to WT. The combinatorial effects of cadmium-tolerant genes were calculated using Equation (2) (Anastassiou, 2007):
where CTG1,G2 indicates cadmium tolerance of the strain co-expressing genes 1 and 2, whereby CTG1 and CTG2 means cadmium tolerance of the strain overexpressing only gene 1 or gene 2, respectively. A positive value of CEG1,G2 indicates synergistic effect.
Results
Genome-Wide Overexpression Screening for Enhanced Cadmium Resistance
We first studied the cadmium sensitivity of an industrial yeast strain CEN.PK2-1c using a previously constructed derivative (Si et al., 2015a). The WT strain transformed with the empty plasmid (pRS416) was cultivated in the synthetic dropout medium containing Cd(NO3)2 as a source of cadmium. We observed growth inhibition in a cadmium concentration-dependent manner (Figure 1A), and a 50% decrease in total biomass accumulation was elicited by approximately 70 μM cadmium (Figure 1B). Nearly complete growth inhibition was observed at 200 μM cadmium, which was selected to screen for resistant strains (Figures 1A,B). Similarly, the lethal dose of cadmium for BY4741 yeast strain, an S288c derivative, was 150 μM (Kuang et al., 2015).
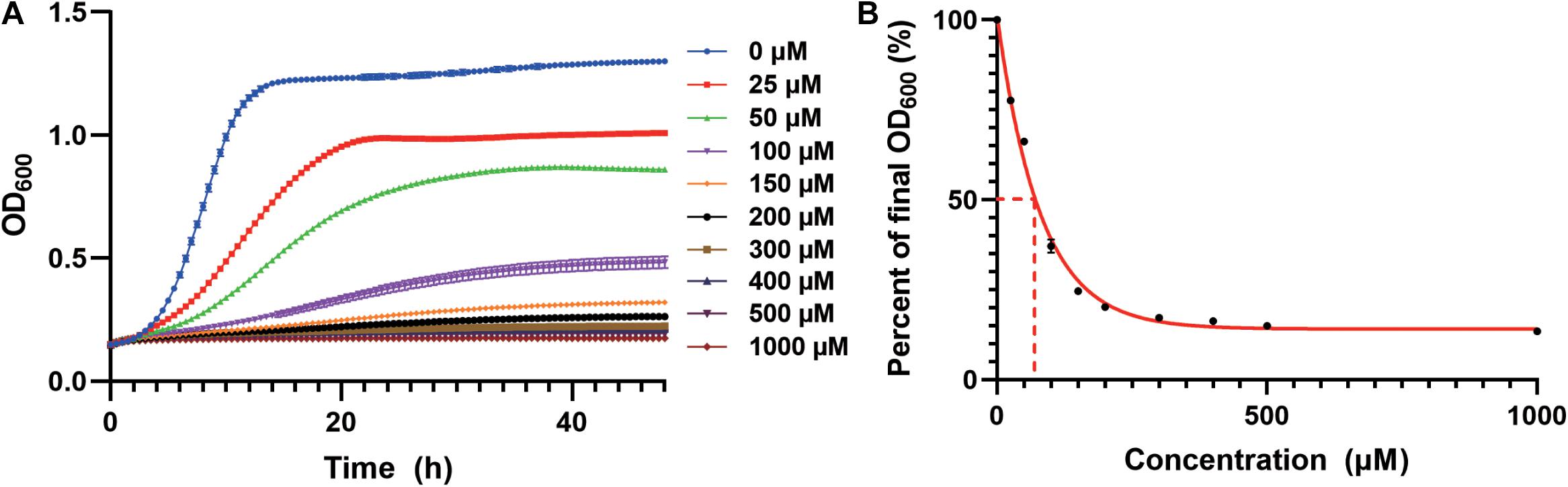
Figure 1. Growth inhibition by cadmium in the WT strain. (A) Cd(NO3)2 was added to the SC-Ura medium at indicated concentrations. Cellular growth was monitored every 30 min for 48 h as optical density at 600 nm (OD600). (B) Cadmium concentration-dependent reduction in the normalized final OD600 at 48 h. The curve was best fitted using a one-phase exponential decay model. Dash lines indicated the cadmium concentration for 50% growth inhibition.
We transformed the WT strain with a genome-scale library on the pRS416 vector, which was previously constructed based on a normalized, full-length enriched cDNA library of S. cerevisiae (Si et al., 2017). Over 106 independent transformants were obtained to ensure sufficient coverage of >92% of all yeast genes (Si et al., 2017). Resistant clones were selected based on colony sizes (Figure 2A). Plasmid DNA sequencing of the 160 largest colonies revealed 95 (59.4%), 27 (16.9%), and 5 (3.13%) clones contained the upregulation cassettes of CUP1, CAD1, and PPH21 genes, respectively. After retransformation into a fresh WT background, seven gene targets were confirmed with resistant phenotypes to 100 μM cadmium in liquid media, including CAD1, CRS5, CUP1, NRG1, PPH21, BMH1, and QCR6 (Figure 2C). Except for BMH1 and QCR6, gene overexpression of all other targets also conferred enhanced growth on agar media containing 200 μM cadmium (Figure 2B; Supplementary Figure 1). Gene Ontology (GO) analysis suggested that these genes are involved in stress response to chemical, oxidative, osmotic and DNA damage, regulation of transcription, translation, protein modification and cell cycle, and organization of organelle, cell wall and vacuole (Supplementary Table 4), which are all relevant mechanisms to cadmium toxicity.
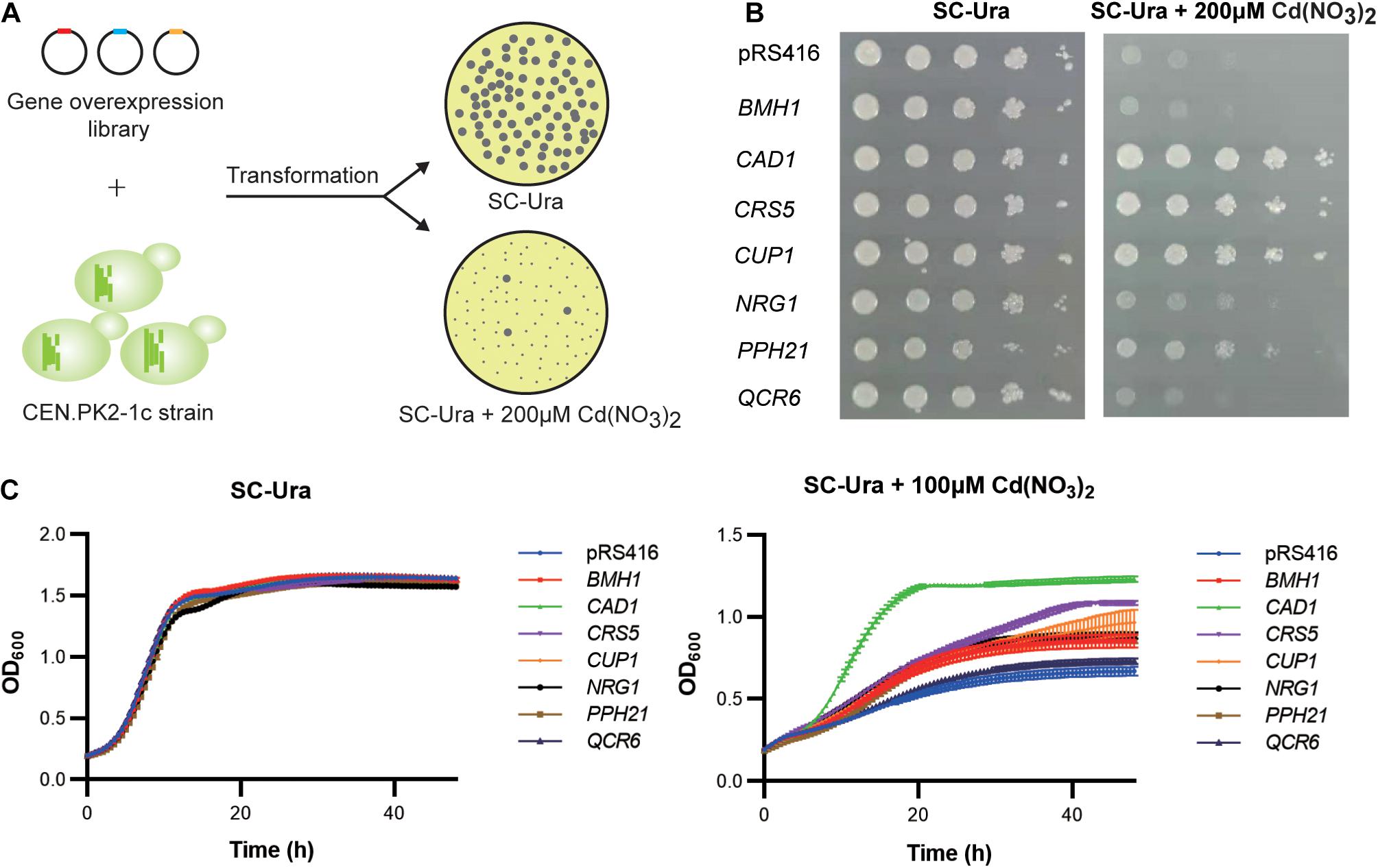
Figure 2. Genome-scale screening for cadmium-tolerant genes. (A) Scheme of genome-wide overexpression screening for cadmium resistant clones. (B) Spot assay on agar media. Yeast cultures were serially diluted, spotted, and cultivated for 2–3 days on agar plates with or without 200 μM cadmium. (C) Time-course of cellular growth in liquid media. The time-course of OD600 was collected at 30 min intervals for 48 h with or without 100 μM cadmium. Error bars indicate standard deviations of three biological replicates.
Gene Regulation of Identified Targets in Response to Cadmium Stress
To examine whether gene expression of identified targets is regulated by cadmium stress, we challenged the WT strain at the exponential growth phase with different cadmium concentrations. qPCR results indicated that the mRNA levels of CAD1, CRS5, CUP1, and NRG1 were significantly upregulated by cadmium in a concentration-dependent manner (Figure 3). Genetic upregulation of CRS5, CUP1, and NRG1 by cadmium was consistent with previous transcriptomic studies in the S288c background (Jeyaprakash et al., 1991; Fauchon et al., 2002; Jin et al., 2008). On the other hand, cadmium treatment suppressed PPH21 expression, and no significant changes in mRNA levels were observed for BMH1 and QCR6 (Figure 3). Consistently, previous studies did not report substantial transcriptional alterations in PPH21, BMH1, or QCR6 upon cadmium treatment (Jeyaprakash et al., 1991; Fauchon et al., 2002; Jin et al., 2008).
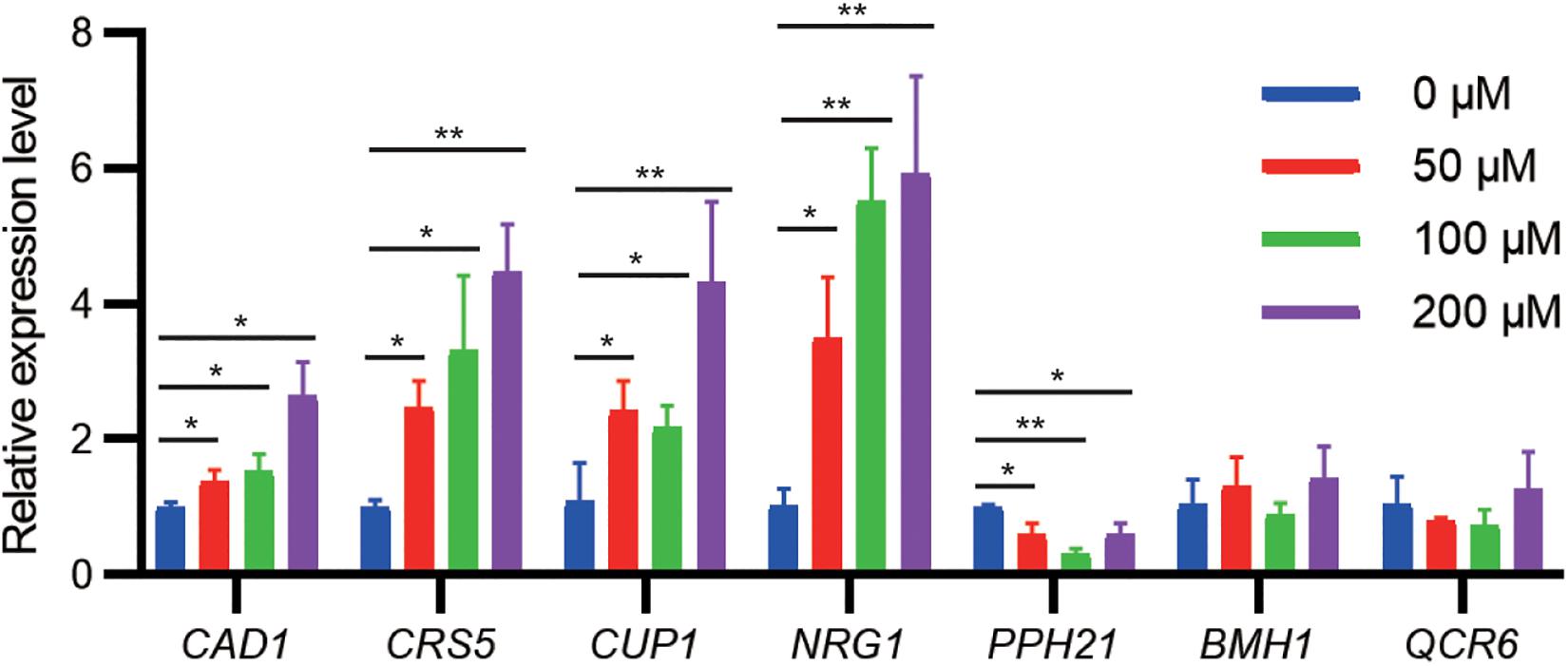
Figure 3. qPCR analysis of cadmium-tolerant genes upon cadmium stress. During the exponential growth phase, the WT strain was treated with Cd(NO3)2 at indicated concentrations for 2 h. Error bars indicate standard derivations of three biological replicates. *P < 0.05; **P < 0.01.
Combinatorial Optimization of Cadmium-Tolerant Genes
We then constructed mutant strains harboring two overexpression targets to improve cadmium tolerance (Figure 4A; Supplementary Figure 2). First, each identified cassette was separately integrated at the LEU2 locus of the yeast genome. In the presence of 100 μM cadmium, genomic and episomal overexpression of CAD1, NRG1, BMH1, or QCR6 conferred comparative improvement in cadmium tolerance relative to WT (Supplementary Figure 3). On the contrary, the strains integrated with CRS5, CUP1, or PPH21 cassettes showed substantially weaker cadmium resistance improvement relative to their plasmid-overexpression counterparts (Supplementary Figure 3). Differences in gene expression levels were previously noted between genomic integration and episomal expression (Crook et al., 2014; Si et al., 2015a). Therefore, genomic integration (1 per cell) of the CUP1 cassette may result in a lower overexpression level compared with pRS416 (3-4 copies per cell), so that a minimum threshold of CUP1 overexpression required for cadmium resistance was not achieved (Jeyaprakash et al., 1991; Adamo et al., 2012). To further confirm dosage effects, we constructed yeast strains overexpressing a single-gene target on pRS426, which is a 2 μ multicopy plasmid (20–80 copies per cell). Indeed, cadmium tolerance of CUP1-overexpressing yeast strains showed a pRS426 > pRS416 > integration pattern. No substantial dosage effects were observed for other targets except for CRS5 (Supplementary Figures 3, 4). Notably, both CUP1 and CRS5 encode yeast metallothionein, which suggested a possibly shared mechanism.
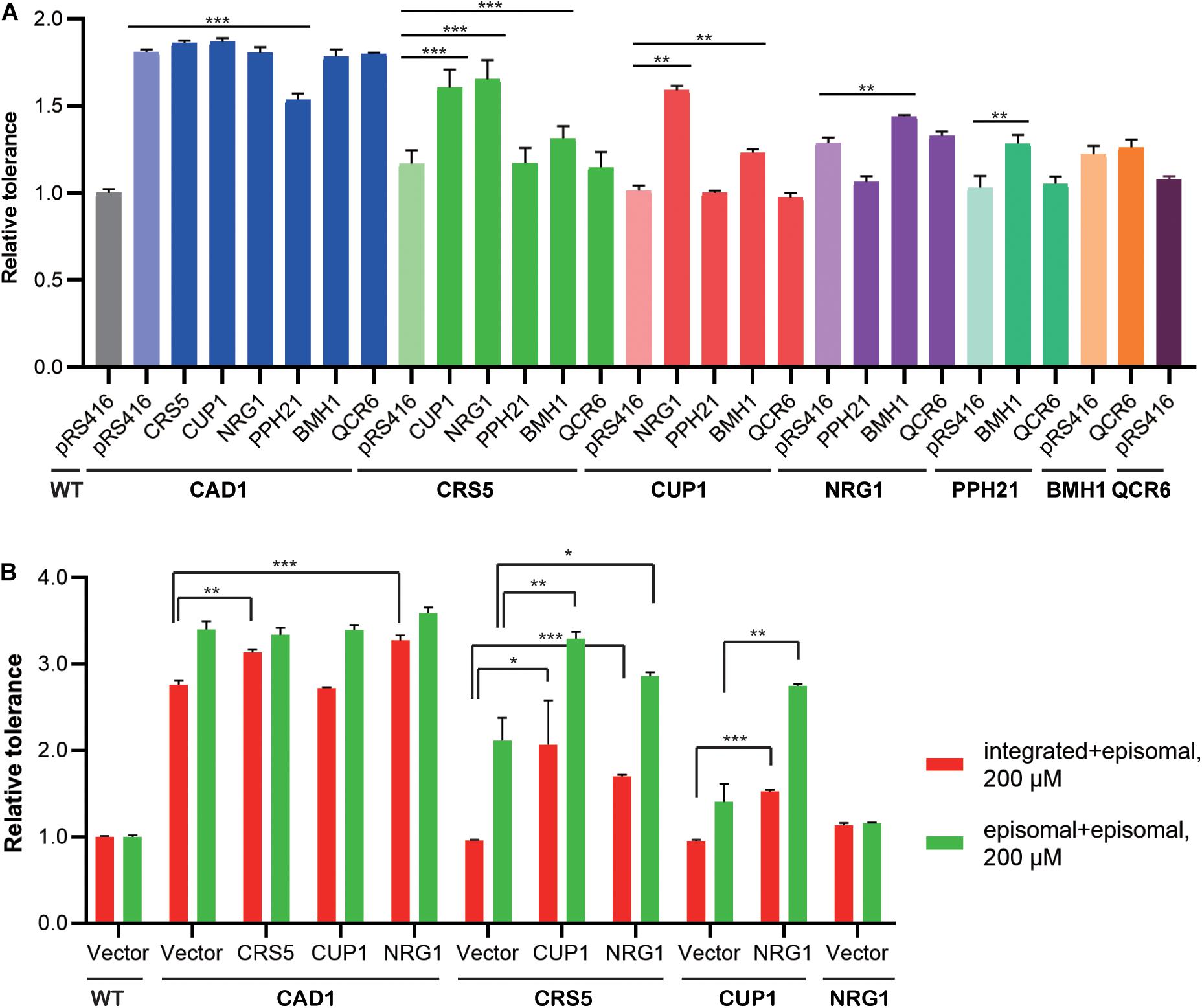
Figure 4. Combinatorial effects of cadmium-tolerant genes. Relative cadmium tolerance of the engineered strains with episomal plasmids in the presence of 100 μM (A) or 200 μM (B) cadmium was calculated as described above in Materials and Methods. (A) Genomic and episomal expression are labeled as bold and italic, respectively. (B) Red bars indicate overexpression from genomic (under the line) and episomal (above the line) vectors, and green bars indicate episomal expression for both targets. Values are means and standard derivations (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001.
Dual overexpression was then achieved by transforming the above-mentioned LEU2-integrated strains (Figure 4A, bold) with overexpression plasmids (Figure 4A, italic). Only selected combinations were constructed to save experimental efforts, whereby a more potent target was integrated into the yeast genome, and a weaker target was placed on the episomal plasmid. In the presence of 100 μM cadmium, CAD1-integrated strains generally exhibited high levels of resistance, except when transformed with a PPH21-overexpressing plasmid. Mutant strains harboring CRS5-CUP1, CRS5-NRG1, and CUP1-NRG1 combinations also showed robust resistance. Therefore, CAD1, CRS5, CUP1, and NRG1 were selected for further optimization, and new strains with two targets both expressed from episomal plasmids (pRS415 and pRS416) were constructed. Mutant strains containing integration-plasmid and plasmid-plasmid combinations were evaluated for the time-course of cellular growth under 200 μM cadmium (Figure 4B; Supplementary Figure 5). The highest tolerance was achieved by CAD1 and NRG1 co-expression both from plasmids, and the corresponding strain exhibited a 3.6-fold improvement in the final OD600 relative to WT. Unfortunately, no further improvement was achieved with triple overexpression of CAD1-NRG1-CRS5 or CAD1-NRG1-CUP1 from episomal plasmids (pRS414, pRS415, and pRS416) (Supplementary Figure 6).
Also, we examined the combinatorial effects among identified gene targets. Genetic interactions were evaluated using Equation (2) as previously reported, based on relative tolerance values calculated by Equation (1) (see Materials and Methods). In the presence of 100 μM cadmium, synergistic effects were observed between genome-integrated CRS5 and plasmid-borne NRG1, as well as between genome-integrated CUP1 and plasmid-borne NRG1 (Figure 4A). In the presence of 200 μM cadmium, synergistic interaction was noted between CRS5 and CUP1 when both expressed episomally (Figure 4B). Synergistic improvement confirmed the necessity for multiplex, combinatorial optimization of individual engineering targets (Wang et al., 2009; Si et al., 2015a, 2017; Cao et al., 2020; Chen et al., 2020).
Discussion
Here we performed the first genome-wide overexpression screening to engineer cadmium resistance in an industrial CEN.PK-derived S. cerevisiae strain. Possible mechanisms underlying identified targets may be deduced from GO analysis (Supplementary Table 4) and known functions reported in literature (Supplementary Table 5). Particularly, identifying well-established targets such as CAD1 and CUP1 indicated our approach was effective. These results also suggested that the corresponding mechanisms are conserved among different S. cerevisiae backgrounds for cadmium tolerance. For example, CAD1 encodes an AP-1-like basic leucine zipper (bZIP) transcriptional activator, which increased cadmium resistance when expressed on a multicopy plasmid (Wu et al., 1993; Hirata et al., 1994). Cad1p was imported to the nucleus from cytoplasm upon cadmium stress, upregulating FRM2 for oxidative stress attenuation, as well as activating gene expression of SLT2, RLM1, and CHS1 for cell-wall maintenance (Fernandes et al., 1997; Bilsland et al., 2004; Azevedo et al., 2007; Mazzola et al., 2015). In addition to nuclear import, our qPCR results implied that upregulation of CAD1 induced by cadmium may also participate in these processes (Figure 3). Moreover, we confirmed another known target, CUP1, which was reported to increase copper and cadmium resistance when constitutively expressed on high-copy episomal plasmids (Ecker et al., 1986).
Several less studied overexpression targets were also isolated. For example, Nrg1p is a Rim101-mediated transcriptional repressor. NRG1 overexpression enhanced transcriptional repression of TAT1, whose deletion also increased cadmium tolerance (Ruotolo et al., 2008) (Supplementary Table 5). Moreover, PPH21 encodes a catalytic subunit of protein phosphatase 2A (PP2A), which mediates cadmium-induced repression of class I transcription by targeting the formation/dissociation of the Pol I-Rxn3 complex in yeast (Zhou et al., 2013). It was reported that PP2A homolog prevented cadmium-induced cell death via inactivation of Erk1/2 and JNK and participated in MT expression regulation through dephosphorylation of MTF-1 in mammalian cells (Chen et al., 2008, 2014) (Supplementary Table 5). Also, BMH1 encodes a member of 14-3-3 proteins that function in various biological processes such as stress response (Kumar, 2017), and yeast cells expressing the human 14-3-3β/α showed enhanced cadmium resistance (Clapp et al., 2012) (Supplementary Table 5).
Notably, two new overexpression targets were identified in this study. We demonstrated plasmid-overexpression of CRS5 increased cadmium tolerance in S. cerevisiae for the first time (Figures 2B,C). CRS5 and CUP1 both encode MT in S. cerevisiae (Chatterjee et al., 2020), and Crs5p possesses a Cd(II)-binding ability (Pagani et al., 2007) (Supplementary Table 5). Gene deletion of crs5 did not lead to cadmium sensitivity (Culotta et al., 1994). Like CUP1, genomic overexpression of CRS5 was less effective than episomal expression (Supplementary Figures 3, 4). In addition, both CUP1 and CRS5 transcription levels were upregulated upon cadmium exposure (Figure 3), suggesting dosage effects of MT may be common when regarding cadmium tolerance (Jeyaprakash et al., 1991; Adamo et al., 2012). Furthermore, QCR6 encodes the subunit 6 of the ubiquinol cytochrome-c reductase complex (complex III). Complex III is a component of the mitochondrial inner membrane electron transport chain, which is suggested as a substantial source of reactive oxygen species (ROS) induced by cadmium in mammalian tissues (Wang et al., 2004) (Supplementary Table 5). Here we showed QCR6 overexpression weakly increased cadmium tolerance in yeast (Figures 2B,C).
In addition to single targets, we also explored multiplex overexpression optimization for improving cadmium resistance in S. cerevisiae. The most resistant strain overexpressing both CAD1 and NRG1 on plasmids exhibited a 3.6-fold improvement in biomass accumulation compared with WT under 200 μM cadmium, and 1.1-fold and 3.1-fold improvement relative to the mutant strains containing individual overexpression of CAD1 and NRG1, respectively. Furthermore, additive and synergistic effects were observed among overexpression targets. Additive effects for NRG1 and BMH1 were observed in CRS5-, CUP1-, NRG1-, or PPH21-integrated strains (Figure 4), possibly mediated via the cooperation of different tolerant mechanisms. For example, the gene products of CRS5, CUP1, NRG1, and PPH21 are metal-binding proteins, whereby NRG1 and BMH1 both encode transcription factors (Supplementary Table 4). Synergistic tolerance improvement was achieved by CRS5-NRG1, CUP1-NRG1, and CRS5-CUP1 co-expression (Figure 4), suggesting the presence of genetic interactions that warrant future mechanistic investigation.
Certain discrepancies were observed between this study and previous literature. For example, overexpression of PPH21 improved cadmium tolerance (Figure 2), but it was down regulated in response to cadmium stress (Figure 3). It is not surprising, however, because the genes with substantially altered transcription and translation revealed by transcriptomic and proteomic profiling do not necessarily represent effective targets for genetic engineering (Momose, 2001; Vido et al., 2001; Fauchon et al., 2002; Jin et al., 2008; Ruotolo et al., 2008; Serero et al., 2008; Thorsen et al., 2009; Huang et al., 2016; Marmiroli et al., 2016). Indeed, little overlap was observed between transcriptomics and genome-deletion screening results for cadmium sensitivity (Serero et al., 2008). Moreover, different sets of gene targets were often identified during different genome-wide screening campaigns targeting cadmium resistance (Thorsen et al., 2009), and the main reasons may include variations in strain background, screening conditions, and the formats of genetic mutations. For example, we observed different effects of the same gene target between genomic and episomal expression (Supplementary Figure 3), as well as between liquid and agar media (Figure 2). Therefore, it is necessary to perform genome-wide screening and multiplex optimization in a particular target strain under conditions that best reflect the real-world application scenario. Considering a range of 1–100 mg/L (∼8.9–890 μM) cadmium in wastewater (Huang et al., 2020), existing efforts on engineering cadmium-resistant yeast for bioremediation has only achieved moderate success. For example, upon heterologous introduction of TaPCS1 that encodes a phytochelatin synthase, S. cerevisiae was engineered as a plant-like cadmium hyperaccumulator in the presence of around 100 μM cadmium (Sun et al., 2019). In this study, we engineered a yeast strain that can tolerate up to 400 μM cadmium by co-overexpression of CAD1 and NRG1 (Supplementary Figure 6). But further endeavor is warranted to further improve the resistance to cadmium and other inhibitors in waste water for real-world applications using S. cerevisiae.
Another limitation of the current approach is the inability to explore functional potential within the S. cerevisiae pan-genome due to the use of a cDNA-derived library of a select yeast strain. For example, PCA1 encodes a cadmium efflux transporter. Whereas some natural S. cerevisiae isolates harbor a functional PCA1 gene that confers cadmium resistance, most laboratory S. cerevisiae strains including CEN.PK2-1c used in this study contain a nonfunctional Pca1p due to a G970R missense mutation (Adle et al., 2007; Wei et al., 2014; Wong et al., 2021). Therefore, it is unlikely to identify PCA1 as a positive hit via genome-scale screening using a CEN.PK cDNA-derived overexpression library. Moreover, the cDNA-templated library can only cover endogenous genes. To engineer CEN.PK yeast strains as bioremediation agents, it is necessary to screen heterologous genes to further improve its cadmium tolerance in future studies.
Conclusion
Using a genome-wide approach, we identified new and known targets and their dual combinations that substantially improved cadmium resistance of an industrial S. cerevisiae strain. Genome-wide search and optimization of more than two targets are desirable and underway for achieving superior cadmium robustness via iterations of the current workflow either manually (Si et al., 2015a) or using an automated biofoundries (Si et al., 2017). This workflow can also be extended to engineer robustness toward other chemical inhibitors in S. cerevisiae.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YC, TS, and BW conceived this study. YC designed and performed genome-scale screening, gene expression analysis, combinatorial optimization, and drafted the manuscript. JL measured growth inhibition of the WT strain by cadmium. ZC assisted with mutant screening and confirmation. TS and BW supervised the research. All authors proofread the manuscript before submission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge financial support from the National Key R&D Program of China (2020YFA0908501), Shenzhen Institute of Synthetic Biology Scientific Research Program (JCHZ20190003), and National Natural Science Foundation of China (32071428). BW thanks the funding support from the National Natural Science Foundation of China (22008252).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.662512/full#supplementary-material
References
Adamo, G. M., Lotti, M., Tamas, M. J., and Brocca, S. (2012). Amplification of the CUP1 gene is associated with evolution of copper tolerance in Saccharomyces cerevisiae. Microbiology 158, 2325–2335. doi: 10.1099/mic.0.058024-0
Adle, D. J., Sinani, D., Kim, H., and Lee, J. (2007). A cadmium-transporting P1B-type ATPase in yeast Saccharomyces cerevisiae. J. Biol. Chem. 282, 947–955. doi: 10.1074/jbc.M609535200
Almeida, J. R. M., Modig, T., Petersson, A., Hahn-Hagerdal, B., Liden, G., and Gorwa-Grauslund, M. F. (2007). Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 82, 340–349. doi: 10.1002/jctb.1676
Anastassiou, D. (2007). Computational analysis of the synergy among multiple interacting genes. Mol. Syst. Biol. 3:83. doi: 10.1038/msb4100124
Ansarypour, Z., and Shahpiri, A. (2017). Heterologous expression of a rice metallothionein isoform (OsMTI-1b) in Saccharomyces cerevisiae enhances cadmium, hydrogen peroxide and ethanol tolerance. Braz. J. Microbiol. 48, 537–543. doi: 10.1016/j.bjm.2016.10.024
Azevedo, D., Nascimento, L., Labarre, J., Toledano, M. B., and Rodrigues-Pousada, C. (2007). The S. cerevisiae Yap1 and Yap2 transcription factors share a common cadmium-sensing domain. FEBS Lett. 581, 187–195. doi: 10.1016/j.febslet.2006.11.083
Bilsland, E., Molin, C., Swaminathan, S., Ramne, A., and Sunnerhagen, P. (2004). Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol. Microbiol. 53, 1743–1756. doi: 10.1111/j.1365-2958.2004.04238.x
Cahoon, R. E., Lutke, W. K., Cameron, J. C., Chen, S., Lee, S. G., Rivard, R. S., et al. (2015). Adaptive engineering of phytochelatin-based heavy metal tolerance. J. Biol. Chem. 290, 17321–17330. doi: 10.1074/jbc.M115.652123
Cao, M. F., Tran, V. G., and Zhao, H. M. (2020). Unlocking nature’s biosynthetic potential by directed genome evolution. Curr. Opin. Biotechnol. 66, 95–104. doi: 10.1016/j.copbio.2020.06.012
Chatterjee, S., Kumari, S., Rath, S., Priyadarshanee, M., and Das, S. (2020). Diversity, structure and regulation of microbial metallothionein: metal resistance and possible applications in sequestration of toxic metals. Metallomics 12, 1637–1655. doi: 10.1039/d0mt00140f
Chen, L., Liu, L., and Huang, S. L. (2008). Cadmium activates the mitogen-activated protein kinase (MAPK) pathway via induction of reactive oxygen species and inhibition of protein phosphatases 2A and 5. Free Radic. Biol. Med. 45, 1035–1044. doi: 10.1016/j.freeradbiomed.2008.07.011
Chen, L. P., Ma, L., Bai, Q., Zhu, X. N., Zhang, J. M., Wei, Q., et al. (2014). Heavy metal-induced metallothionein expression is regulated by specific protein phosphatase 2A complexes. J. Biol. Chem. 289, 22413–22426. doi: 10.1074/jbc.M114.548677
Chen, Y., Guo, E., Zhang, J., and Si, T. (2020). Advances in RNAi-assisted strain engineering in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 8:731. doi: 10.3389/fbioe.2020.00731
Clapp, C., Portt, L., Khoury, C., Sheibani, S., Norman, G., Ebner, P., et al. (2012). 14-3-3 Protects against stress-induced apoptosis. Cell Death Dis. 3:e348. doi: 10.1038/cddis.2012.90
Crook, N. C., Schmitz, A. C., and Alper, H. S. (2014). Optimization of a yeast RNA interference system for controlling gene expression and enabling rapid metabolic engineering. ACS Synth. Biol. 3, 307–313. doi: 10.1021/sb4001432
Culotta, V. C., Howard, W. R., and Liu, X. F. (1994). CRS5 encodes a metallothionein-like protein in Saccharomyces cerevisiae. J. Biol. Chem. 269, 25295–25302. doi: 10.1016/S0021-9258(18)47246-8
Ecker, D. J., Butt, T. R., Sternberg, E. J., Neeper, M. P., Debouck, C., Gorman, J. A., et al. (1986). Yeast metallothionein function in metal ion detoxification. J. Biol. Chem. 261, 16895–16900.
Fang, Z., Chen, Z., Wang, S., Shi, P., Shen, Y., Zhang, Y., et al. (2016). Overexpression of OLE1 enhances cytoplasmic membrane stability and confers resistance to cadmium in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 83, e02319–16. doi: 10.1128/aem.02319-16
Fauchon, M., Lagniel, G., Aude, J. C., Lombardia, L., Soularue, P., Petat, C., et al. (2002). Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9, 713–723. doi: 10.1016/s1097-2765(02)00500-2
Fernandes, L., Rodrigues-Pousada, C., and Struhl, K. (1997). Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell Biol. 17, 6982–6993. doi: 10.1128/mcb.17.12.6982
Fu, F. L., and Wang, Q. (2011). Removal of heavy metal ions from wastewaters: a review. J. Environ. Manage. 92, 407–418. doi: 10.1016/j.jenvman.2010.11.011
Gietz, R. D., and Schiestl, R. H. (2007). Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 38–41. doi: 10.1038/nprot.2007.15
Hirata, D., Yano, K., and Miyakawa, T. (1994). Stress-induced transcriptional activation mediated by YAP1 and YAP2 genes that encode the Jun family of transcriptional activators in Saccharomyces cerevisiae. Mol. Gen. Genet. 242, 250–256. doi: 10.1007/BF00280413
Hu, B. F., Shao, S., Ni, H., Fu, Z. Y., Hu, L. S., Zhou, Y., et al. (2020). Current status, spatial features, health risks, and potential driving factors of soil heavy metal pollution in China at province level. Environ. Pollut. 266:114961. doi: 10.1016/j.envpol.2020.114961
Huang, H., Jia, Q., Jing, W., Dahms, H. U., and Wang, L. (2020). Screening strains for microbial biosorption technology of cadmium. Chemosphere 251:126428. doi: 10.1016/j.chemosphere.2020.126428
Huang, X., Li, Y., Pan, J., Li, M., Lai, Y., Gao, J., et al. (2016). RNA-Seq identifies redox balance related gene expression alterations under acute cadmium exposure in yeast. Environ. Microbiol. Rep. 8, 1038–1047. doi: 10.1111/1758-2229.12484
Hwang, G. W., Sasaki, K., Takahashi, T., Yamamoto, R., and Naganuma, A. (2009). Overexpression of Ycg1 or Ydr520c confers resistance to cadmium in Saccharomyces cerevisiae. J. Toxicol. Sci. 34, 441–443. doi: 10.2131/jts.34.441
Jeyaprakash, A., Welch, J. W., and Fogel, S. (1991). Multicopy CUP1 plasmids enhance cadmium and copper resistance levels in yeast. Mol. Gen. Genet. 225, 363–368. doi: 10.1007/BF00261675
Jiang, S. Y., Si, T., and Dai, J. B. (2020). Whole-genome regulation for yeast metabolic engineering. Small Methods 4:1900640. doi: 10.1002/smtd.201900640
Jin, Y. H., Dunlap, P. E., McBride, S. J., Al-Refai, H., Bushel, P. R., and Freedman, J. H. (2008). Global transcriptome and deletome profiles of yeast exposed to transition metals. PloS Genet. 4:e1000053. doi: 10.1371/journal.pgen.1000053
Jing, L. H., Zhang, X. H., Ali, I., Chen, X. M., Wang, L., Chen, H., et al. (2020). Usage of microbial combination degradation technology for the remediation of uranium contaminated ryegrass. Environ. Int. 144:106051. doi: 10.1016/j.envint.2020.106051
Kim, I. S., Kim, Y. S., and Yoon, H. S. (2012). Glutathione reductase from Oryza sativa increases acquired tolerance to abiotic stresses in a genetically modified Saccharomyces cerevisiae strain. J. Microbiol. Biotechnol. 22, 1557–1567. doi: 10.4014/jmb.1202.02028
Kuang, X., Fang, Z. J., Wang, S., Shi, P., and Huang, Z. W. (2015). Effects of cadmium on intracellular cation homoeostasis in the yeast Saccharomyces cerevisiae. Toxicol. Environ. Chem. 97, 922–930. doi: 10.1080/02772248.2015.1074689
Kumar, R. (2017). An account of fungal 14-3-3 proteins. Eur. J. Cell Biol. 96, 206–217. doi: 10.1016/j.ejcb.2017.02.006
Li, C. F., Zhou, K. H., Qin, W. Q., Tian, C. J., Qi, M., Yan, X. M., et al. (2019). A review on heavy metals contamination in soil: effects, sources, and remediation techniques. Soil Sediment Contam. 28, 380–394. doi: 10.1080/15320383.2019.1592108
Liu, R. M., Liang, L. Y., Freed, E. F., Choudhury, A., Eckert, C. A., and Gill, R. T. (2020). Engineering regulatory networks for complex phenotypes in E. coli. Nat. Commun. 11:4050. doi: 10.1038/s41467-020-17721-4
Marmiroli, M., Pagano, L., Pasquali, F., Zappettini, A., Tosato, V., Bruschi, C. V., et al. (2016). A genome-wide nanotoxicology screen of Saccharomyces cerevisiae mutants reveals the basis for cadmium sulphide quantum dot tolerance and sensitivity. Nanotoxicology 10, 84–93. doi: 10.3109/17435390.2015.1019586
Mazzola, D., Pimentel, C., Caetano, S., Amaral, C., Menezes, R., Santos, C. N., et al. (2015). Inhibition of Yap2 activity by MAPKAP kinase Rck1 affects yeast tolerance to cadmium. FEBS Lett. 589, 2841–2849. doi: 10.1016/j.febslet.2015.07.049
Momose, Y. I. H. (2001). Bioassay of cadmium using a DNA mciroarray: genome-wide expression patterns of Saccharomyces cerevisiae response to cadmium. Environ. Toxicol. Chem. 20, 2353–2360. doi: 10.1897/1551-50282001020<2353:bocuad>2.0.co;2
Nordberg, G. F., Bernard, A., Diamond, G. L., Duffus, J. H., Illing, P., Nordberg, M., et al. (2018). Risk assessment of effects of cadmium on human health (IUPAC Technical Report). Pure Appl. Chem. 90, 755–808. doi: 10.1515/pac-2016-0910
Pagani, A., Villarreal, L., Capdevila, M., and Atrian, S. (2007). The Saccharomyces cerevisiae Crs5 metallothionein metal-binding abilities and its role in the response to zinc overload. Mol. Microbiol. 63, 256–269. doi: 10.1111/j.1365-2958.2006.05510.x
Rascio, N., and Navari-Izzo, F. (2011). Heavy metal hyperaccumulating plants: how and why do they do it? And what makes them so interesting? Plant Sci. 180, 169–181. doi: 10.1016/j.plantsci.2010.08.016
Reeves, R. D., Baker, A. J. M., Jaffre, T., Erskine, P. D., Echevarria, G., and van der Ent, A. (2018). A global database for plants that hyperaccumulate metal and metalloid trace elements. New Phytol. 218, 407–411. doi: 10.1111/nph.14907
Ruotolo, R., Marchini, G., and Ottonello, S. (2008). Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast. Genome Biol. 9:R67. doi: 10.1186/gb-2008-9-4-r67
Serero, A., Lopes, J., Nicolas, A., and Boiteux, S. (2008). Yeast genes involved in cadmium tolerance: identification of DNA replication as a target of cadmium toxicity. DNA Repair (Amst) 7, 1262–1275. doi: 10.1016/j.dnarep.2008.04.005
Si, T., Chao, R., Min, Y. H., Wu, Y. Y., Ren, W., and Zhao, H. M. (2017). Automated multiplex genome-scale engineering in yeast. Nat. Commun. 8:15187. doi: 10.1038/ncomms15187
Si, T., Luo, Y., Bao, Z., and Zhao, H. (2015a). RNAi-assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering. ACS Synth. Biol. 4, 283–291. doi: 10.1021/sb500074a
Si, T., Xiao, H., and Zhao, H. M. (2015b). Rapid prototyping of microbial cell factories via genome-scale engineering. Biotechnol. Adv. 33, 1420–1432. doi: 10.1016/j.biotechadv.2014.11.007
Sun, G. L., Reynolds, E. E., and Belcher, A. M. (2019). Designing yeast as plant-like hyperaccumulators for heavy metals. Nat. Commun. 10:5080. doi: 10.1038/s41467-019-13093-6
Sun, G. L., Reynolds, E. E., and Belcher, A. M. (2020). Using yeast to sustainably remediate and extract heavy metals from waste waters. Nat. Sustain. 3:311. doi: 10.1038/s41893-020-0478-9
Thorsen, M., Perrone, G. G., Kristiansson, E., Traini, M., Ye, T., Dawes, I. W., et al. (2009). Genetic basis of arsenite and cadmium tolerance in Saccharomyces cerevisiae. BMC Genomics 10:105. doi: 10.1186/1471-2164-10-105
Vardhan, K. H., Kumar, P. S., and Panda, R. C. (2019). A review on heavy metal pollution, toxicity and remedial measures: current trends and future perspectives. J. Mol. Liq. 290:111197. doi: 10.1016/j.molliq.2019.111197
Vareda, J. P., Valente, A. J. M., and Duraes, L. (2019). Assessment of heavy metal pollution from anthropogenic activities and remediation strategies: a review. J. Environ. Manage. 246, 101–118. doi: 10.1016/j.jenvman.2019.05.126
Vido, K., Spector, D., Lagniel, G., Lopez, S., Toledano, M. B., and Labarre, J. (2001). A proteome analysis of the cadmium response in Saccharomyces cerevisiae. J. Biol. Chem. 276, 8469–8474. doi: 10.1074/jbc.M008708200
Wang, H. H., Isaacs, F. J., Carr, P. A., Sun, Z. Z., Xu, G., Forest, C. R., et al. (2009). Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898. doi: 10.1038/nature08187
Wang, Y., Fang, J., Leonard, S. S., and Rao, K. M. (2004). Cadmium inhibits the electron transfer chain and induces reactive oxygen species. Free Radic. Biol. Med. 36, 1434–1443. doi: 10.1016/j.freeradbiomed.2004.03.010
Warner, J. R., Reeder, P. J., Karimpour-Fard, A., Woodruff, L. B. A., and Gill, R. T. (2010). Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat. Biotechnol. 28, 856–862. doi: 10.1038/nbt.1653
Wei, Q., Zhang, H., Guo, D., and Ma, S. (2016). Cell surface display of four types of solanum nigrum metallothionein on Saccharomyces cerevisiae for biosorption of cadmium. J. Microbiol. Biotechnol. 26, 846–853. doi: 10.4014/jmb.1512.12041
Wei, W., Smith, N., Wu, X., Kim, H., Seravalli, J., Khalimonchuk, O., et al. (2014). YCF1-mediated cadmium resistance in yeast is dependent on copper metabolism and antioxidant enzymes. Antioxid. Redox Signal. 21, 1475–1489. doi: 10.1089/ars.2013.5436
Wong, A., Lam, E. M., Pai, C., Gunderson, A., Carter, T. E., and Kebaara, B. W. (2021). Variation of the response to metal ions and nonsense-mediated mRNA decay across different Saccharomyces cerevisiae genetic backgrounds. Yeast 1–14. doi: 10.1002/yea.3565
Wu, A. L., Wemmie, J. A., Edgington, N. P., Goebl, M., Guevara, J. L., and Moyerowley, W. S. (1993). Yeast Bzip proteins mediate pleiotropic drug and metal resistance. J. Biol. Chem. 268, 18850–18858. doi: 10.1016/S0021-9258(17)46705-6
Wysocki, R., and Tamas, M. J. (2010). How Saccharomyces cerevisiae copes with toxic metals and metalloids. FEMS Microbiol. Rev. 34, 925–951. doi: 10.1111/j.1574-6976.2010.00217.x
Yang, C. E., Chu, I. M., Wei, Y. H., and Tsai, S. L. (2017). Surface display of synthetic phytochelatins on Saccharomyces cerevisiae for enhanced ethanol production in heavy metal-contaminated substrates. Bioresour. Technol. 245, 1455–1460. doi: 10.1016/j.biortech.2017.05.127
Keywords: gene overexpression, combinatorial optimization, genome-scale engineering, Saccharomyces cerevisiae, cadmium tolerance
Citation: Chen Y, Liang J, Chen Z, Wang B and Si T (2021) Genome-Scale Screening and Combinatorial Optimization of Gene Overexpression Targets to Improve Cadmium Tolerance in Saccharomyces cerevisiae. Front. Microbiol. 12:662512. doi: 10.3389/fmicb.2021.662512
Received: 01 February 2021; Accepted: 18 June 2021;
Published: 14 July 2021.
Edited by:
Yizhi (Patrick) Cai, The University of Manchester, United KingdomReviewed by:
Ahmed Gomaa, National Research Centre, EgyptSoo Rin Kim, Kyungpook National University, South Korea
Rongming Liu, University of Colorado, Boulder, United States
Zhen Kang, Jiangnan University, China
Copyright © 2021 Chen, Liang, Chen, Wang and Si. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tong Si, tong.si@siat.ac.cn; Bo Wang, bo.wang@siat.ac.cn
 Yongcan Chen
Yongcan Chen Jun Liang1
Jun Liang1 Bo Wang
Bo Wang Tong Si
Tong Si