- 1Tianjin Key Laboratory of Food Science and Health, School of Medicine, Nankai University, Tianjin, China
- 2Institute of Radiation Medicine, Chinese Academy of Medical Sciences and Peking Union Medical Collage, Tianjin, China
- 3Institute of Infection and Immunity, Taihe Hospital, Hubei University of Medicine, Shiyan, China
Virulence traits and antibiotic resistance are frequently provided by genes located on plasmids. However, experimental verification of the functions of these genes is often lacking due to a lack of related experimental technology. In the present study, an integrated suicide vector was used to efficiently and specifically delete a bacterial endogenous plasmid in Cronobacter sakazakii. The pESA3 plasmid was removed from C. sakazakii BAA-894, and we confirmed that this plasmid contributes to the invasion and virulence of this strain. In addition, the pGW1 plasmid was expunged from C. sakazakii GZcsf-1, and we confirmed that this plasmid confers multidrug resistance. We further screened plasmid-curing agents and found that p-coumaric acid had a remarkable effect on the curing of pESA3 and pGW1 at sub-inhibitory concentrations. Our study investigated the contribution of endogenous plasmids pESA3 and pGW1 by constructing plasmid-cured strains using suicide vectors and suggested that p-coumaric acid can be a safe and effective plasmid-curing agent for C. sakazakii.
Introduction
Previously developed strategies for the treatment of systemic infections caused by pathogenic bacteria still rely on the application of various antibiotics. However, the frequent and extensive utilization of antibiotics has dramatically increased the possibility of severe infections related to bacteria with multidrug resistance, posing a more aggressive threat to public health and necessitating the development of novel antimicrobials and bacterial vaccines. Moreover, prospective research on the underlying mechanism of multidrug resistance could facilitate the modification of currently employed antibiotics and the development of novel reagents. To date, multidrug resistance has been observed in a variety of pathogenic bacteria, including Neisseria gonorrhoeae (Yuan et al., 2019; Chen et al., 2020; Seong et al., 2020), Pseudomonas aeruginosa (Horcajada et al., 2019; Kumarage et al., 2019; Tada et al., 2019), and Acinetobacter baumannii (da Silva et al., 2018; Salehi et al., 2018).
Related studies have unveiled the inseparable correlation between bacterial antibiotic resistance and plasmids, which could be verified by the frequent distribution of virulence determinants and antibiotic resistance genes in plasmids (Stratev and Odeyemi, 2016; Kopotsa et al., 2019). Importantly, the abuse of antibiotics promoted the spread of antibiotic-resistant plasmids that have the potential to increase the occurrence of virulence or drug resistance genes, posing an increased threat to society (Fraise, 2002; Zhang et al., 2011; Mangat et al., 2017).
Bacterial plasmids can significantly vary in size from 2 to 2,000 kbp (Canteros, 1990). Large plasmids can potentially harbor more transcriptomic information and perform relatively important functions that contribute to host survival.
Recently, Cronobacter sakazakii, an emerging pathogenic bacterium belonging to the family Enterobacteriaceae, has attracted a great deal of attention due to its multidrug resistance and ability to cause disease with a high mortality rate (Parra-Flores et al., 2018; Odeyemi and Abdullah Sani, 2019). C. sakazakii is a foodborne pathogen associated with life-threatening sepsis (Hassan and Naser, 2018), meningitis (Chaves et al., 2018; Zeng et al., 2018), and necrotizing enterocolitis (Fan et al., 2019) in premature and full-term infants. C. sakazakii has been widely isolated and identified from powder infant formula (PIF) (Li et al., 2019), fruit powders, vegetables, tea leaves, herbs, cereals, and spices (Shi et al., 2016).
The pathogenicity and multidrug resistance of C. sakazakii are believed to be associated with two large plasmids that it harbors, pESA3 and pGW1, with sizes of 134 and 340 kbp, respectively. A variety of virulence-related genes have been identified in pESA3 (Aly et al., 2019), and pGW1 has been reported to carry a number of drug resistance genes (Zeng et al., 2018).
The highly conserved replicon of RepFIB in pESA3 and its frequent identification in almost 97% of Cronobacter isolates has been previously described (Singh et al., 2017). pESA3 encodes two iron uptake systems (lucABCD/lutA and EitCBAD) that may facilitate host survival and pathogenesis. In 2018, one multidrug-resistant strain of C. sakazakii, GZcsf-1, was reported to be responsible for a case of meningitis in a neonate in China (Zeng et al., 2018). Two internalized plasmids, namely, pGW1 (340 kbp) and pGW2 (135 kbp), were identified, with 17 predicted drug resistance genes encoded by pGW1 and a high similarity observed for pGW2 to pESA3 (Zeng et al., 2018). Moreover, an mcr-1-harboring plasmid identified in C. sakazakii was confirmed to be correlated with colistin resistance (Liu et al., 2017; Nang et al., 2019), which is widely used as an antibiotic of last-resort against bacterial infections. Considering the ubiquitous presence of C. sakazakii in nature, as well as its excellent capacity for plasmid internalization, it may be associated with an increased risk of potential infections and an increased threat to public health, especially in infants.
Given the frequent observation of plasmid-mediated bacterial drug resistance, functional characterization of these plasmids is still needed. Currently, such functional studies primarily rely on comparative genomic analysis without further experimental verification, which may be attributed to the difficulty of transforming plasmids into Escherichia coli to further study their functions, especially large plasmids (Tu et al., 2016; Chung et al., 2017). Moreover, the common presence of multiple replicons in large plasmids impedes the elucidation of their function through replicon incompatibility-mediated plasmid curing (Medaney et al., 2016; Douarre et al., 2020).
In this study, we used an integrated suicide vector to efficiently and specifically delete an endogenous plasmid of choice in C. sakazakii and analyzed the contributions of endogenous plasmids pESA3 and pGW1. The plasmid-cured strains were used to screen plasmid-curing agents and showed that p-coumaric acid had a remarkable effect on the plasmid curing, suggesting that p-coumaric acid can be a safe and effective plasmid-curing agent for C. sakazakii.
Results
Strategy of Plasmid Curing in C. sakazakii
A strategy based on suicide vector pCVD442 was developed for plasmid curing in the current study. Generally, the plasmid-specific fragment of the suicide vector was taken from an endogenous plasmid (Figure 1), integration of the hybrid plasmid could then be specifically selected using the antibiotic corresponding to the resistance marker on the suicide vector. Then, the suicide plasmid was spontaneously removed in culture without antibiotic stress, leaving a suspension of two types of bacteria with or without the hybrid plasmid. Finally, based on the sensitivity to sucrose, the bacteria with the hybrid plasmid were eliminated by sucrose supplementation. The loss of the plasmid was verified by plasmid-specific PCR. 58 out of randomly selected 60 colonies growing on LB plates containing sucrose in pESA3 and pGW1 curing assays were verified to be plasmid curing strains, indicating a high frequency of loss of the plasmid. One hundred single colonies were selected in the sucrose selection step and verified to be plasmid-cured isolates. Compared to the traditional molecular method based on plasmid incompatibility, where 92% (92/100) of C. sakazakii transformant colonies retained pESA3 after 5 rounds of culturing in the presence of ampicillin, the method developed in the present study resulted in 100% (100/100) of transformant colonies being verified as pESA3-cured isolates (Figure 2). These results clearly showed that this method is efficient for expunging plasmids in C. sakazakii.
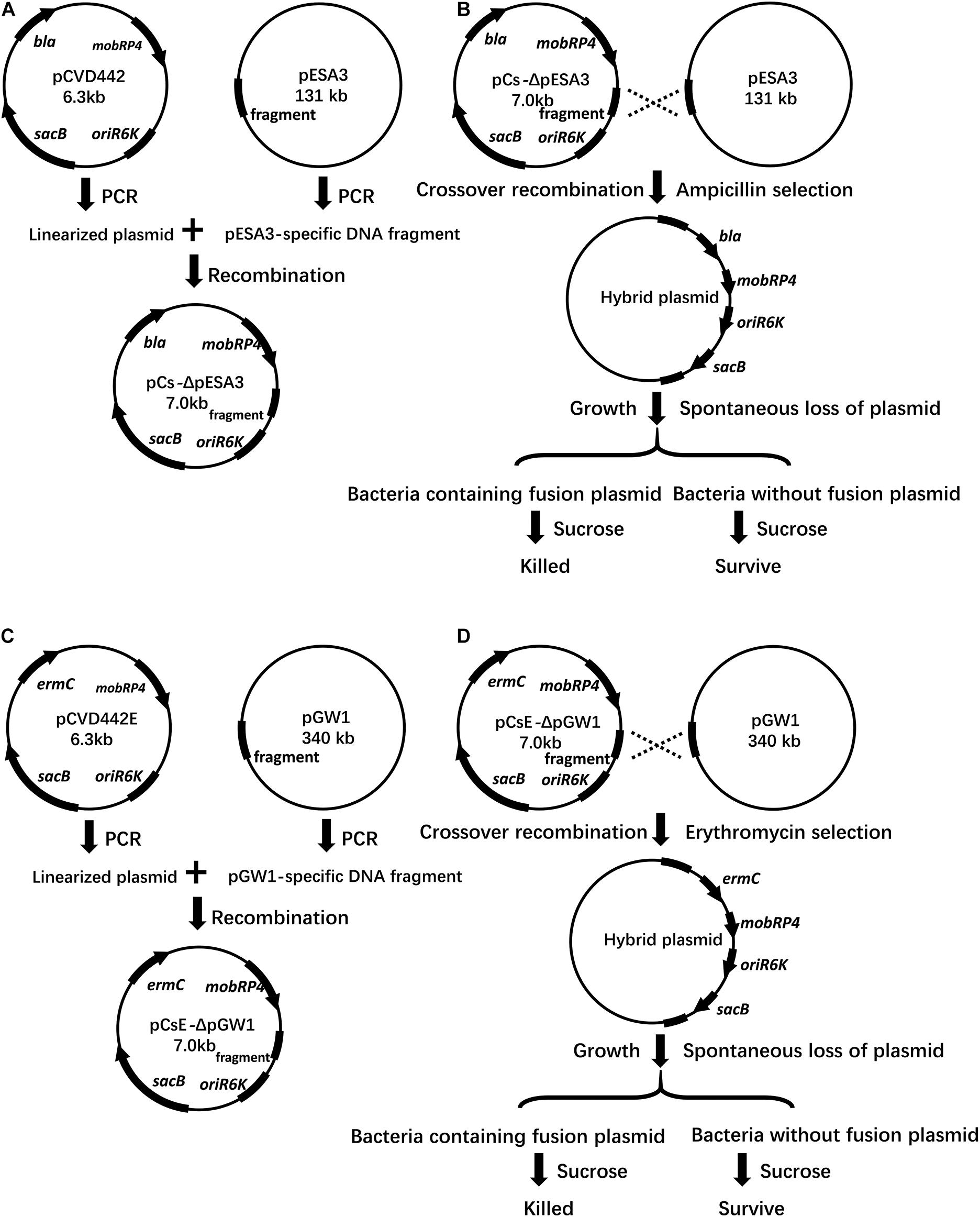
Figure 1. Schematic representation of vectors, crossover recombination, plasmid hybrid, and sucrose selection. (A) The vector pCs-ΔpESA3 is shown with the pESA3-specific hybrid fragment and gene orientation of the ampicillin resistance (bla), pir replication initiator (oriR6K), sucrose sensitivity marker (sacB) and broad-host-range mobilizable region (mobRP4). pCVD442 was linearized by PCR and ligated with a pESA3-specific DNA fragment, resulting in the plasmid pCs-ΔpESA3. (B) The plasmid pCs-ΔpESA3 was extracted from S17–1 lambda pir E. coli and transformed into C. sakazakii BAA-894. The integration of pCs-ΔpESA3 into the endogenous plasmid pESA3 of C. sakazakii BAA-894 by crossover recombination resulted in the strain C. sakazakii BAA-894 containing the hybrid plasmid. Growth of the plasmid crossover recombinants allowed for the spontaneous loss of the hybrid plasmid in a small fraction of the bacteria. Exposure to sucrose resulted in selective killing of the bacteria containing the hybrid plasmid that retained the sucrose sensitivity marker sacB, and the surviving bacteria cured of the plasmid were identified by testing for ampicillin susceptibility and via plasmid-specific PCR. (C) The vector pCs-ΔpGW1 was similar to pCsE-ΔpESA3, but ermC was used instead of bla due to the existence of the cephalosporin resistance gene blaDHA–1 in pESA3, and the fragment was amplified from pGW1. (D) The procedure was the same as that described in (B), except the endogenous plasmid to be deleted was pGW1, and the integrated suicide plasmid was pCs-ΔpGW1. (B,D) The hybrid plasmid was used for plasmid complementation in the plasmid-cured strain.
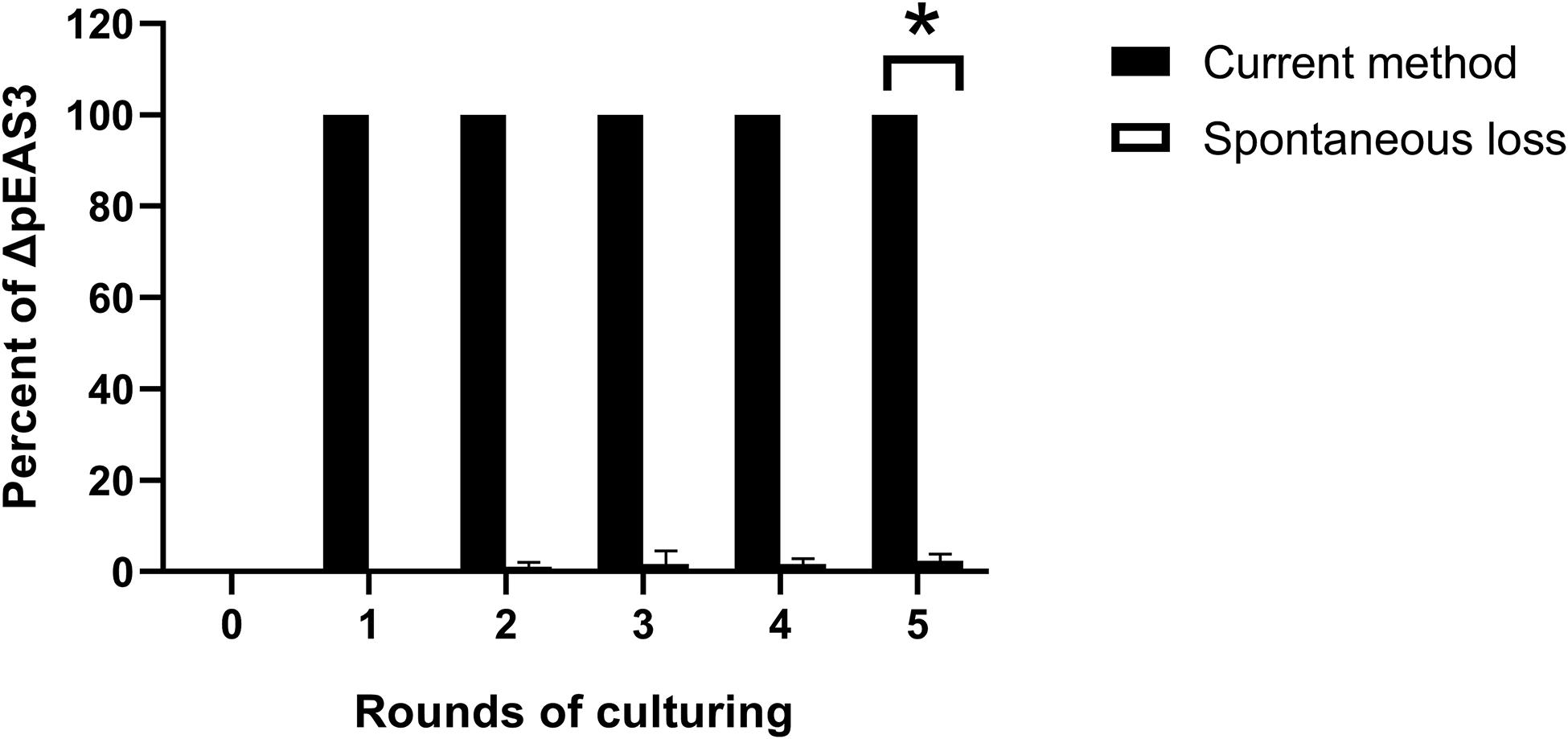
Figure 2. Percentage of pESA3 plasmid-cured isolates after continuous culturing. The method described in the present study represents a strategy for plasmid curing in C. sakazakii, and the spontaneous loss of an endogenous plasmid represents the traditional molecular method based on plasmid incompatibility using another plasmid with the same replicon. The loss of the plasmid was verified by plasmid-specific PCR using primers listed in Table 3. At the beginning of the experiment, 105 CFU/mL bacteria harboring the integrated plasmid pCs-ΔpESA3 (Figure 1B) and bacteria harboring the incompatible plasmid pUC57-RepFIB were prepared and subsequently continuously cultured in LB containing sucrose and ampicillin, respectively. The cultures were shaken overnight at 37°C and then inoculated into 20 mL of fresh sterile LB medium at a 1:100 dilution the next day. After each round of culturing, bacterial samples were serially diluted and plated on agar to obtain signal colonies. Finally, 100 single colonies were selected, and their genotypes were determined by PCR using plasmid-specific primers. Each data point represents the average and standard deviation of three biological repeats. Student’s t-test; *P < 0.05.
pESA3 Contributes to Invasion and Virulence
Many of the genes distributed on the pESA3 plasmid are predicted to be involved in the adhesion and infection of the host pathogens (Aly et al., 2019). To verify the role of pESA3 in pathogenicity, the C. sakazakii strain BAA-894 encapsulated with or without pESA3 was used to assess the contribution of pESA3 to the adhesion, invasion and virulence of the host. In addition, the hybrid plasmid was used for complementation pESA3 in the pESA3-cured strain (Figures 1C,D). In vitro analysis indicated that pESA3-cured bacteria were significantly attenuated in their ability to invade Caco-2 cells than the wild-type strain (Figure 3B), but no difference was observed with respect to adhesion (Figure 3A). In addition, the results of an in vivo study revealed that C. sakazakii BAA-894 lacking pESA3 had notably lower lethality than wild-type bacteria after intragastric administration to 3-day-old rats (Figure 3C). The survival rate of rats infected by wild-type bacteria was 40% at 120 h postinfection, while 80% survival was observed for those infected with pESA3-cured bacteria. Furthermore, the organs of young rats exhibited a significantly lower distribution of pESA3-cured bacteria 24 h postinfection compared to those infected with the wild-type or complemented strains (Figures 3D–F).
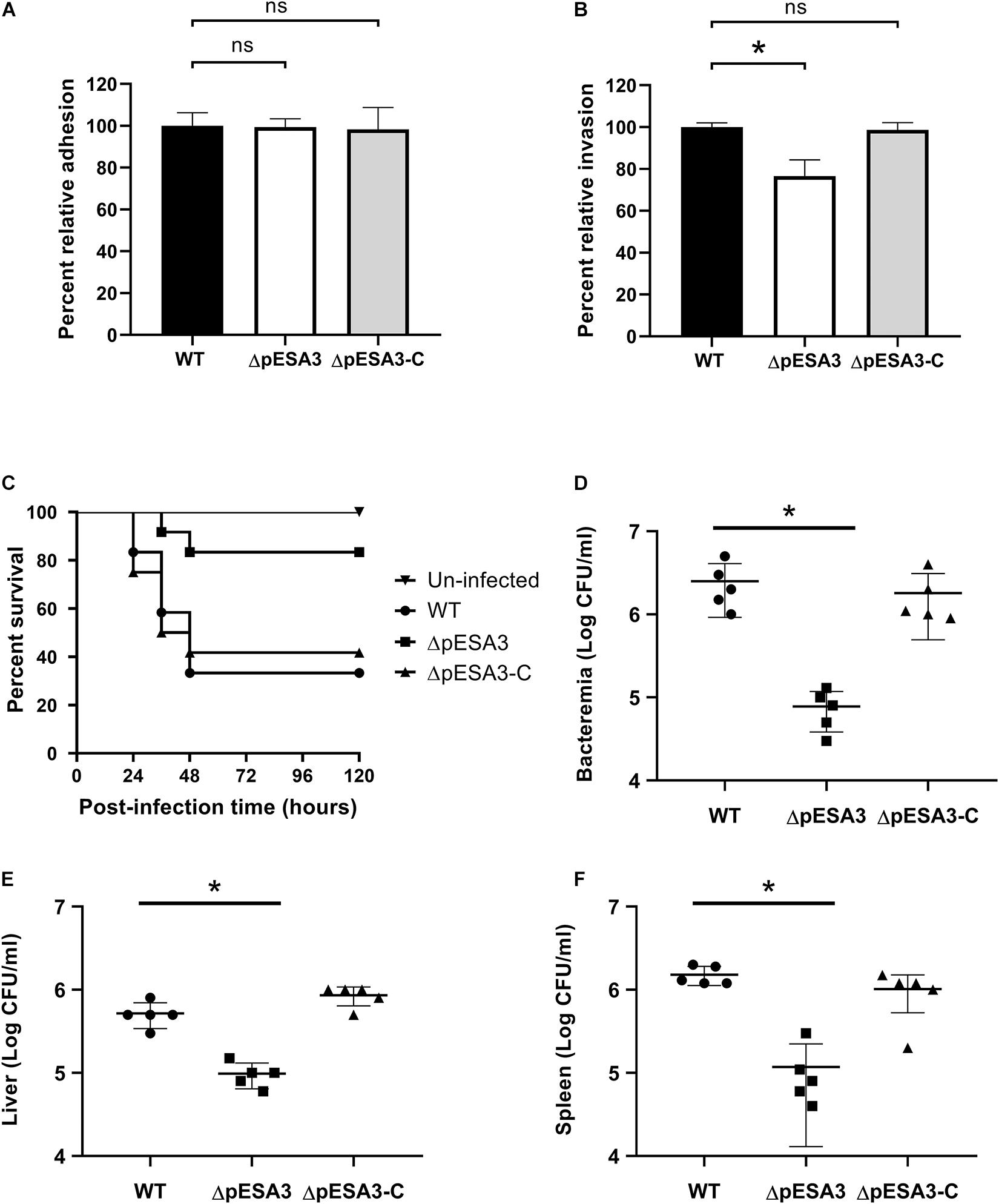
Figure 3. The endogenous plasmid pESA3 contributes to the virulence of Cronobacter sakazakii BAA-894. Adhesion (A) and invasion (B) of Caco-2 cells by WT, ΔpESA3 and ΔpESA3 complementation (ΔpESA3-C) C. sakazakii strains (n = 3). (C) Survival curves of rat pups 120 h after oral infection. (D–F) Colony-forming unit (CFU) counts at 24 h postinfection from the blood, liver, and spleen of orally infected rats. Blood samples were collected from facial veins, liver and spleen, homogenized and serially diluted. The diluents were plated on LB agar for colony enumeration. The data are presented as the means and standard deviation of five biological repeats (Student’s t-test; ns, no significant difference, *P < 0.05).
pGW1 Is Involved in Multidrug Resistance
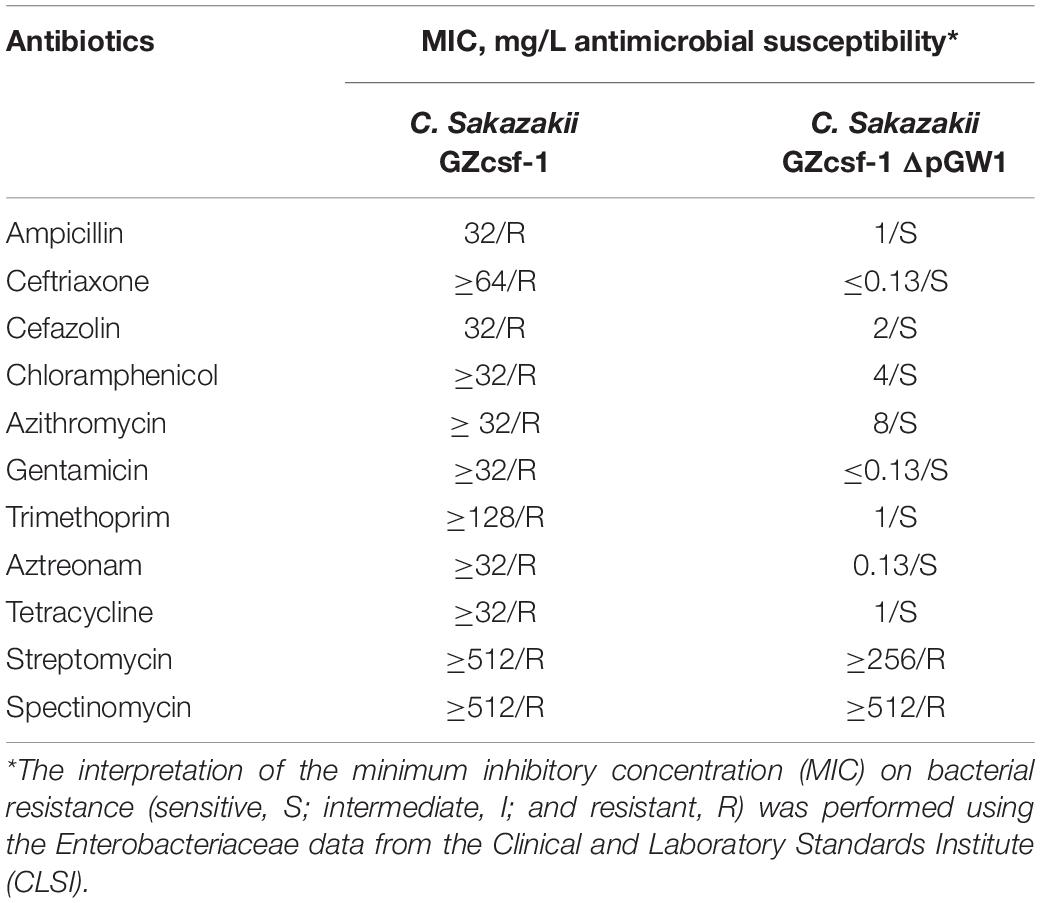
C. sakazakii GZcsf-1 was initially isolated from the brain abscess fluid of an infant meningitis patient, and multidrug resistance to various antibiotics has been reported to be potentially linked with the endogenous plasmid pGW1 (Zeng et al., 2018). However, the dominant role of pGW1 in multidrug resistance remains obscure and needs to be investigated. Therefore, in the present study, the antibiotic sensitivity of wild-type C. sakazakii GZcsf-1 and C. sakazakii GZcsf-1 without pGW1 was determined (Table 1). The results of a minimum inhibitory concentration (MIC) analysis demonstrated the resistance of wild-type C. sakazakii GZcsf-1 to multiple antibiotics, namely, ampicillin, ceftriaxone, cefazolin, chloramphenicol, azithromycin, gentamicin, trimethoprim, aztreonam, and tetracycline, while the cured pGW1 variant was susceptible to all these antibiotics. In addition, both C. sakazakii GZcsf-1 strains harboring or lacking pGW1 exhibited resistance to spectinomycin and streptomycin. These results clearly demonstrated the association of pGW1 with multidrug resistance in C. sakazakii GZcsf-1, although not against spectinomycin and streptomycin, which is probably dependent on genes present in the genome. Indeed, the presence of aminoglycoside adenyltransferase (aadA1), a gene that confers antibiotic resistance to spectinomycin and streptomycin, has been confirmed in the genome of C. sakazakii GZcsf-1 (Zeng et al., 2018), which could potentially explain the obtained results.
Plasmid Curing of C. sakazakii With p-Coumaric Acid
It has been found that plant-derived chemicals have the effect of plasmid elimination (39). Considering that polyphenols are naturally existing substances in food and are relatively safe, we next selected plasmid elimination drugs from polyphenols. The plasmid knockout strains obtained in our project were used to screen plasmid elimination drugs by comparing the growth rates of plasmid knockout strains and wild-type strains at sub-inhibitory concentrations of drugs. The minimal inhibitory concentration (MIC) of 12 polyphenols, including daidzin, trans-chalcone, apigenin, quercetin, flavanone, polydatin, trihydroxyflavone, caffeic acid, 5,7-Dihydroxyflavone, p-coumaric acid, isoferulic acid and phenethyl cinnamate, were determined using broth dilution methods. However, the MICs of 13 of these polyphenols were above 1,000 mg/L, and only p-coumaric acid had a MIC of 500 mg/L for both C. sakazakii GZcsf-1 and C. sakazakii BAA-894, irrespective of bacterial antibiotic resistance behavior. Thus, the plasmid-curing effects of these polyphenols at 250 mg/L were tested by comparing the growth rates between plasmid knockout strains and wild-type strains. In LB, wild-type C. sakazakii BAA-894 showed a similar growth rate as C. sakazakii BAA-894 ΔpESA3, and wild-type C. sakazakii GZcsf-1 showed a similar growth rate as C. sakazakii GZcsf-1 ΔpGW1 (Figure 4A). However, in LB complemented with 250 mg/L p-coumaric acid, wild-type C. sakazakii BAA-894 showed a significant reduction in optical density at 600 nm (OD600) compared to that of the C. sakazakii BAA-894 ΔpESA3 strain (Figure 4B). Similar results were also observed in C. sakazakii GZcsf-1 and its pGW1 curing mutant, indicating an in vitro fitness cost of the endogenous plasmids imposed on C. sakazakii with the existence of p-coumaric acid. Subsequently, the plasmid curing effect of p-coumaric acid was directly examined by continuous culture of wild-type C. sakazakii in LB or LB with p-coumaric acid. We found that with the increase of culture cycles, the proportion of plasmid-eliminating bacteria of C. sakazakii in the medium supplemented with p-coumaric acid increased gradually, while no plasmid-cured bacteria were detected in the medium without p-coumaric acid (Figure 4C). These results revealed that p-coumaric acid exhibits plasmid-curing effect on C. sakazakii strains.
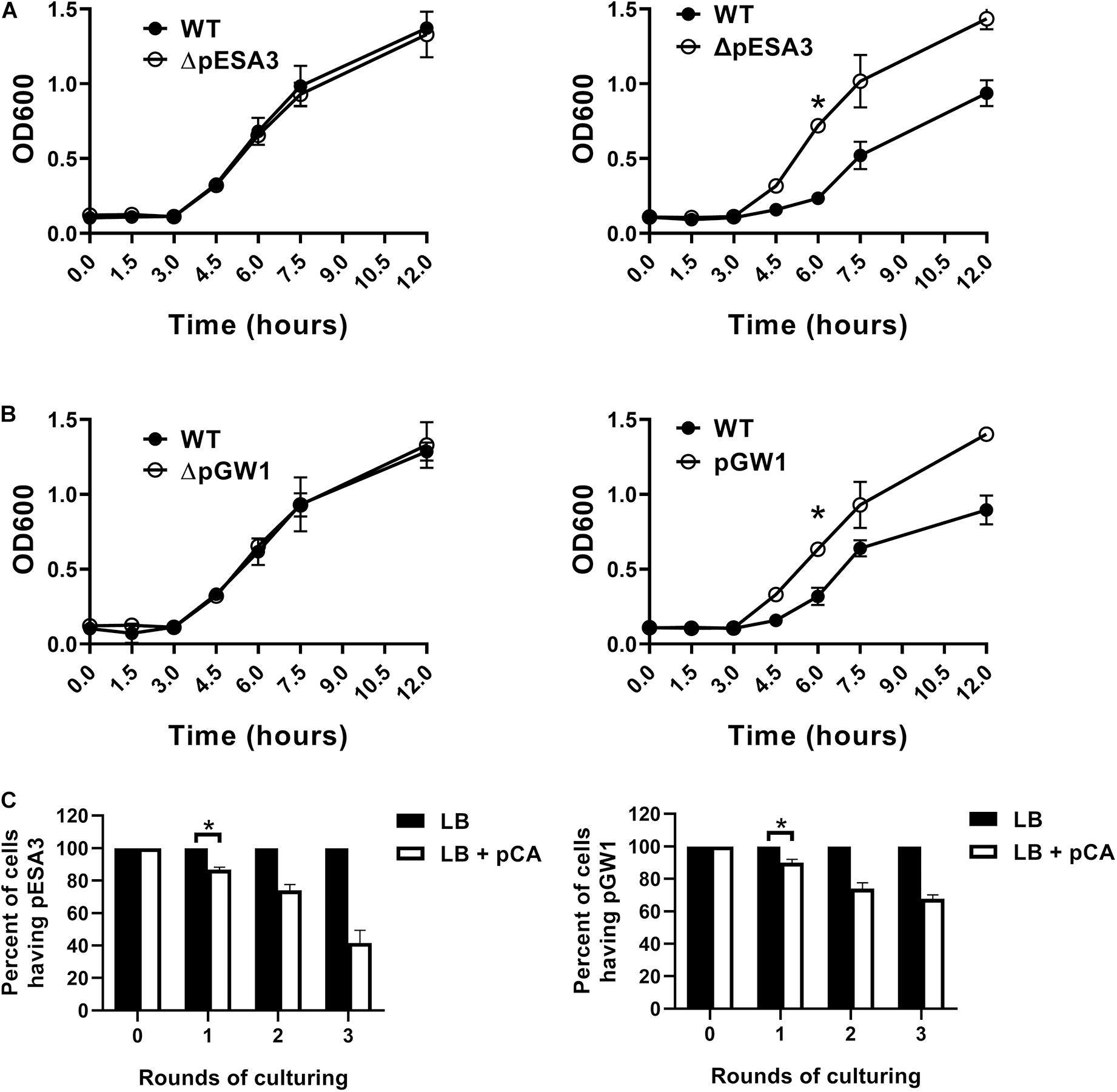
Figure 4. Plasmid-curing ability of p-coumaric acid in Cronobacter sakazakii. (A) Growth of wild-type C. sakazakii BAA-894 and its ΔpESA3 derivative in LB (left) and in LB containing 250 mg/L p-coumaric acid (right). (B) Growth of wild-type C. sakazakii GZcsf-1 and its ΔpGW1 derivative in LB (left) and in LB containing 250 mg/L p-coumaric acid (right). (C) Percent of plasmid-cured isolates of each culture round in LB or in LB containing 250 mg/L p-coumaric acid (pCA). Left: initial strain was wild-type C. sakazakii BAA-894, right: initial strain was wild-type C. sakazakii GZcsf-1. At the beginning of the experiment, a total of 105 CFU of wild-type bacteria was added to 10 mL of LB or LB containing pCA. The cultures were shaken overnight at 37°C and then inoculated into 10 mL of fresh sterile LB medium or LB containing pCA at a 1:100 dilution the next day, respectively. After each round of culturing, bacterial samples were serially diluted and plated on agar to obtain signal colonies. Finally, 100 single colonies were selected, and their genotypes were determined by PCR using plasmid-specific primers. All data represent the average and standard deviation of three biological repeats (Student’s t-test; *P < 0.05).
Discussion
C. sakazakii is an opportunistic pathogen that is widely distributed in foods (Zeng et al., 2020), the gut (Chandrasekaran et al., 2018) and various environments, increasing its chance of coming into contact with a variety of antibiotics, which may account for the accumulation of drug resistance genes in this bacterium, especially multidrug resistance genes in plasmids or the genome. In particular, the ability of C. sakazakii to harbor plasmids larger than 300 kbp would further increase its propensity to acquire resistance (Zeng et al., 2018). The resistance plasmids harbored by C. sakazakii could potentially serve as reservoirs for and as the origin for resistance in other strains through conjugation, transduction and transformation. The investigated fragment in pGW1 from C. sakazakii is also conserved in plasmids from S. enterica, S. flexneri, C. freundii, and K. pneumoniae, which could explain the dissemination of drug resistance plasmids in different bacteria.
The conventional strategy used to study the function of plasmids primarily depends on the removal of endogenous plasmids or their exogenous transformation into E. coli for functionality analysis. The exogenous transformation of plasmids of interest can promote a better understanding of their relationship to resistance to specific antibiotics but cannot provide sufficient insight into their roles in bacterial pathogenicity. In addition, the lower transformation efficiency of large plasmids or their restricted replication in E. coli can further restrict the application of the traditional strategy used to evaluate plasmids. The typical approaches used to cure endogenous plasmids primarily include chemical-based methods and molecular biological methods. Common chemical methods depend on the use of a high concentration of SDS, EDTA or ethidium bromide to interfere with plasmid replication, which has certain disadvantages, including non-specific removal, increased tendency for genomic mutation, and high toxicity (Buckner et al., 2018). Alternatively, molecular biology methods based on plasmid incompatibility involve the construction of plasmids containing replicons to cure endogenous plasmids, which primarily rely on the accurate cloning of the replicon from endogenous plasmids (Buckner et al., 2018). However, this method is subject to the prediction of plasmid incompatibility, and the multiple replicons that are frequently present in endogenous plasmids can further restrict the applicability of the plasmid incompatibility-based method (Citterio et al., 2020). Another traditional approach uses transposons harboring the sacB or rpsL genes to cure various plasmids that were established decades ago. However, transposons have insertion sites, and the frequency of the occurrence of hybridization of the plasmid to be cured and transposon is low, resulting in a time-consuming colony screening process (Hynes et al., 1989; Stojiljković et al., 1991; Brom et al., 1992). A method based on the CRISPR/Cas9 system and homing endonuclease shows the capability of plasmid curing in certain species (Cao et al., 2017; Lauritsen et al., 2017; Tang et al., 2018; Wang et al., 2019). However, the off-target effect of the system leads to the concern over unwanted cleavages and mutations in chromosomal DNA. Furthermore, intracellular DNA ligase could repair the CRISPR/Cas9 system/endonuclease-induced DNA double-strand breaks, and an additional procedure to inhibit activity of DNA ligase has been proposed (Weber et al., 2015), casting a shadow over the application of this plasmid-curing method. Recently, suicide vectors were used in specific plasmid curing in Agrobacterium and Salmonella strains (Yamamoto et al., 2018; Yin et al., 2018), suggesting that this method has the advantages of plasmid specificity and convenience in plasmid elimination, but its broad-spectrum application needs more verification.
The current research described the integrated suicide vector pCVD442 to efficiently and specifically delete an endogenous plasmid of choice in C. sakazakii and analyzed the contribution of endogenous plasmids pESA3 and pGW1. The plasmid elimination method provided in the current study could facilitate the generation of plasmid knockout strains and could be applied for evaluation of selected plasmid elimination drugs by comparisons of their growth rates with those of wild-type strains under fixed pressure. Furthermore, the lower toxicity of the plasmid knockout strains could contribute to the development of attenuated vaccines after detailed investigation.
Plant-derived chemicals have many functions in the field of microorganisms, including antibacterial activity, bactericidal activity, regulation of quorum sensing and so on. The plasmid-cured strains obtained in our study were used to screen plasmid-curing agents from plant polyphenols and found that p-coumaric acid had a remarkable effect on the plasmid curing. Previous studies have found that p-coumaric acid can compete with ethidium bromide in binding to DNA (Lou et al., 2012), suggesting that p-coumaric acid can intercalate into the DNA base pairs. In fact, many DNA intercalating agents including ethidium bromide, such as methyl orange and acriflavine, have been found to eliminate plasmids from various strains (Buckner et al., 2018). Therefore, the plasmid-curing activity of p-coumaric acid in C. sakazakii may due to intercalation of p-coumaric into plasmid DNA, blocking the replication of plasmid. Previous studies have also found some polyphenols with plasmid elimination activity for specific bacteria. It seems that the plasmid elimination activity of plant polyphenols is not broad-spectrum, which may be due to the different tolerances of different bacteria to polyphenols. Our study found that p-coumaric acid inhibited the growth of C. sakazakii at high concentrations and had plasmid elimination effects at low concentrations. This is the first report on the antibacterial effect of p-coumaric acid on C. sakazakii. Considering the safety of p-coumaric acid derived from food, it may be a potential food additive for the inhibition of C. sakazakii. In addition, this is the first report in which p-coumaric acid can inhibit the plasmid-carrying capacity of C. sakazakii at sub-inhibitory concentrations. Most importantly, we found that the antibiotic resistance of C. sakazakii was not related to the elimination and growth inhibition of its plasmid by p-coumaric acid. Therefore, p-coumaric acid may solve the multiple drug resistance problem of C. sakazakii.
Overall, the current research analyzed the contributions of the endogenous plasmids pESA3 and pGW1 by constructing plasmid-cured strains using an integrated suicide vector. The plasmid-cured strains were used to screen plasmid-curing agents and showed that p-coumaric acid derived from plants had a remarkable effect on the plasmid curing, suggesting that p-coumaric acid can be a safe and effective plasmid-curing agent for C. sakazakii.
Materials and Methods
Strains and Plasmids
All strains and plasmids used in the present study are listed in Table 1, and primers are listed in Table 2. Bacteria were stored in LB medium (Solarbio, China) containing 15% glycerol (Solarbio, China) at –80°C. To initiate all experiments, strains were cultured overnight in LB. If necessary, antibiotics (ampicillin or kanamycin) were added at a final concentration of 100 μg/mL (Solarbio, China), and E. coli S17–1 lambda pir (Weidi, China) was used to prepare the pCVD442 suicide vector (Miaoling, China). The gene cloning and transformation of C. sakazakii were performed using standard techniques.
Curing of Endogenous Plasmids
To remove pESA3 from the C. sakazakii strain BAA-894, the specific fragment amplified from the plasmid to be cured using the primer pair ΔpESA3-F and ΔpESA3-R was mixed with the linearized plasmid pCVD442 (Miaoling, China) using the primer pair pCVD442-F and pCVD442-R, and plasmid cyclization was achieved by homogenous recombination using a recombination kit according with protocols recommended by the manufacturer (Vazyme, China). The hybrid plasmid was selected by transformation into S17–1 lambda pir competent cells (Weidi, China) and selective medium. Single colonies growing on LB agar were selected. Subsequently, the plasmid was extracted from the prepared S17–1 lambda pir-Δplasmid, and C. sakazakii BAA-894 was transformed by electroporation as previously described (Kim and Loessner, 2008). A colony was cultured in LB overnight and then transferred to LB agar containing 20% sucrose. Strains harboring the integrated suicide plasmid will be killed by sucrose due to harboring sacB in the plasmid, while strains that lost the plasmid will survive. The genotype of colonies growing on sucrose LB agar was further verified by PCR and sequencing using dual pESA3 plasmid-specific primer pairs (see primer pairs, Table 3).
To cure pGW1 from the strain C. sakazakii GZcsf-1, the specific fragment that was amplified from the plasmid to be removed using the primer pair ΔpGW1-F and ΔpGW1-R was mixed with plasmid pCVD442 that was linearized using the primer pair pCVD442-F1 and pCVD442-R1 and the ermC fragment amplified from pUC57-ermC (Ji et al., 2019) using the primer pair ermC-F and ermC-R. The following procedure was identical as the protocols described for the removal of pESA3, but 300 mg/L erythromycin was used to select strains harboring the hybrid plasmid. The genotype of colonies growing on sucrose agar was further verified by PCR and sequencing using dual pGW1 plasmid-specific primer pairs (see primer pairs, Table 3).
Generation of the Plasmid Complemented Strains
Hybrid plasmid was used for plasmid complementation in the corresponding plasmid-cured strain (Figures 1C,D). Specifically, the hybrid plasmid was extracted from S17–1 lambda pir-Δplasmid, and plasmid-cured C. sakazakii BAA-894 was transformed by electroporation as previously described (Kim and Loessner, 2008). The plasmid complemented strains were selected by plating the bacteria on LB agar containing corresponding antibiotics. The genotype of colonies growing on selective medium was further verified by PCR and sequencing using dual plasmid-specific primer pairs (see primer pairs, Table 3).
Susceptibility Assays
The antimicrobial sensitivity of C. sakazakii strains was tested by the agar dilution method according to Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th Edition1. All antibiotics were purchased from Beijing Solarbio Science and Technology Co., Ltd., China. All polyphenols were purchased from Shanghai yuanye Bio-Technology Co., Ltd., China. Briefly, bacteria cultured overnight were diluted in LB media to 107 CFU/mL, and 10 μL of liquid was aliquoted on LB agar plates containing a two-fold dilution series of antimicrobials. The MICs were determined as the lowest concentration at which growth could not be observed when the plates were incubated at 37°C for 24–48 h.
Adhesion Assay
Bacterial adhesion assays were performed as described previously (Choi et al., 2015). Briefly, Caco-2 cells were washed in PBS (Solarbio, China) and then resuspended in fresh RPMI 1640 medium. C. sakazakii was prepared and added to a Caco-2 cell monolayer at a multiplicity of infection (MOI) of 100. After incubating for 45 min, the cells were washed with PBS three times and lysed in 1% Triton X-100. Then, the suspension was serially diluted and plated on LB agar to enumerate the CFU.
Invasion Assays
Bacterial invasion assays were performed as described previously (Choi et al., 2015). In short, Caco-2 cells were washed in PBS, and fresh RPMI 1640 media was added. Then, C. sakazakii cells were prepared and added to a Caco-2 cell monolayer at an MOI of 100. After incubating for 90 min, the cells were washed with PBS three times and then incubated with 100 μg/mL gentamycin for 1 h to kill extracellular bacteria. Subsequently, the wells were washed with PBS three times and lysed in 1% Triton X-100. Then, the suspension was serially diluted and plated on LB agar to enumerate the CFU.
In vivo Rat Pup Virulence Assay
Bacterial cells were washed and resuspended in PBS (Solarbio, China), and a mixed inoculum of 5 × 109 CFU of bacteria was orally administered to 3-days-old female Sprague-Dawley rat pups (4 rats/group). Where appropriate, chemicals were orally administered 1 h after bacterial administration. To analyze the colonization of bacteria in organs, after 24 h of infection, the bacterial load in the blood was determined by diluting facial vein blood and plating the diluents on LB agar. Then, the rats were sacrificed, and the liver and spleen were aseptically removed. The organs were homogenized in ice-cold PBS and serially diluted. The diluent was plated on LB agar to determine the bacterial loads.
Statistical Analysis
Statistical significance was analyzed using the GraphPad Prism (version 8.4) with the unpaired t-test. The data are represented as the mean and standard deviation. A P-value of < 0.05 was considered to indicate a significant difference.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the Nankai University.
Author Contributions
XJ and PL conceived, designed the research, conducted the experiments, and analyzed the data. YH and XJ contributed new reagents or analytical tools. JW, BZ, YZ, LD, and HL provided useful suggestions. SW systematically reviewed the literature and wrote the manuscript. All authors contributed to the editing of the manuscript.
Funding
This work was funded by grants from the National Key R&D Program of China (2019YFC1605005).
Conflict of Interest
The authors have filed a China patent application (No. 202010973714.6) based on the results reported in this manuscript.
Acknowledgments
We are grateful to Ping Li (Tianjin University of Science and Technology, China) for her technical assistance.
Footnotes
References
Aly, M. A., Domig, K. J., Kneifel, W., and Reimhult, E. (2019). Whole genome sequencing-based comparison of food isolates of Cronobacter sakazakii. Front. Microbiol. 10:1464. doi: 10.3389/fmicb.2019.01464
Brom, S., De Los Santos, A. G., Stepkowsky, T., Flores, M., Dávila, G., and Romero, D., et al. (1992). Different plasmids of Rhizobium leguminosarum bv. phaseoli are required for optimal symbiotic performance. J. Bacteriol. 174, 5183–5189. doi: 10.1128/jb.174.16.5183-5189.1992
Buckner, M. M. C., Ciusa, M. L., and Piddock, L. J. V. (2018). Strategies to combat antimicrobial resistance: anti-plasmid and plasmid curing. FEMS Microbiol. Rev. 42, 781–804. doi: 10.1093/femsre/fuy031
Canteros, B. I. (1990). Diversity of Plasmids and Plasmid-Encoded Phenotypic Traits in Xanthomonas campestris pv. vesicatoria. Ph.D thesis. Gainesville, FL: University of Florida.
Cao, Q.-H., Shao, H.-H., Qiu, H., Li, T., Zhang, Y.-Z., and Tan, X.-M. (2017). Using the CRISPR/Cas9 system to eliminate native plasmids of Zymomonas mobilis ZM4. Biosci. Biotechnol. Biochem. 81, 453–459. doi: 10.1080/09168451.2016.1189312
Chandrasekaran, S., Burnham, C. D., Warner, B. B., Tarr, P. I., and Wylie, T. N. (2018). Carriage of Cronobacter sakazakii in the very preterm infant gut. Clin. Infect. Dis. 67, 269–274. doi: 10.1093/cid/ciy062
Chaves, C. E. V., Brandão, M. L. L., Lacerda, M. L. G. G., and Rocha, C. A. B. C. (2018). Fatal Cronobacter sakazakii sequence type 494 meningitis in a newborn, Brazil. Emerg. Infect. Dis. 24:1948. doi: 10.3201/eid2410.180373
Chen, S.-C., Hu, L.-H., Zhu, X.-Y., and Yin, Y.-P. (2020). Gonococcal urethritis caused by a multidrug resistant Neisseria gonorrhoeae strain with high-level resistance to spectinomycin in China. Emerg. Microbes Infect. 9, 517–519. doi: 10.1080/22221751.2020.1732836
Choi, Y., Kim, S., Hwang, H., Kim, K. P., Kang, D. H., and Ryu, S. (2015). Plasmid-encoded MCP is involved in virulence, motility, and biofilm formation of Cronobacter sakazakii ATCC 29544. Infect. Immun 83, 197–204. doi: 10.1128/iai.02633-14
Chung, M. E. I, Yeh, H., Sung, L. Y., Wu, M. Y., Chao, Y. P. I, and Ng, S., et al. (2017). Enhanced integration of large DNA into E. coli chromosome by CRISPR/Cas9. Biotechnol. Bioeng. 114, 172–183. doi: 10.1002/bit.26056
Citterio, B., Andreoni, F., Simoni, S., Carloni, E., Magnani, M., and Mangiaterra, G., et al. (2020). Plasmid replicon typing of antibiotic-resistant Escherichia coli from clams and marine sediments. Front. Microbiol. 11:1101. doi: 10.3389/fmicb.2020.01101
da Silva, K. E., Maciel, W. G., Croda, J., Cayô, R., Ramos, A. C., and de Sales, R. O., et al. (2018). A high mortality rate associated with multidrug-resistant Acinetobacter baumannii ST79 and ST25 carrying OXA-23 in a Brazilian intensive care unit. PLoS One 13:e0209367. doi: 10.1371/journal.pone.0209367
de Lorenzo, V., Eltis, L., Kessler, B., and Timmis, K. N. (1993). Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123, 17–24. doi: 10.1016/0378-1119(93)90533-9
Donnenberg, M. S., and Kaper, J. B. (1991). Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59, 4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991
Douarre, P.-E., Mallet, L., Radomski, N., Felten, A., and Mistou, M.-Y. (2020). Analysis of COMPASS, a new comprehensive plasmid database revealed prevalence of multireplicon and extensive diversity of IncF plasmids. Front. Microbiol. 11:483. doi: 10.3389/fmicb.2020.00483
Fan, H., Chen, Z., Lin, R., Liu, Y., Wu, X., and Puthiyakunnon, S., et al. (2019). Bacteroides fragilis strain ZY-312 defense against Cronobacter sakazakii-induced necrotizing enterocolitis in vitro and in a neonatal rat model. Msystems 4, e00305–e00319.
Fraise, A. P. (2002). Biocide abuse and antimicrobial resistance—a cause for concern? J. Antimicrob. Chemother. 49, 11–12. doi: 10.1093/jac/49.1.11
Hassan, J. S., and Naser, W. E. (2018). Incidence of Cronobacter sakazakii in Iraqi Infants with neonatal sepsis. Indian J. Public Health Res. Dev. 9, 908–913. doi: 10.5958/0976-5506.2018.01256.1
Horcajada, J. P., Montero, M., Oliver, A., Sorlí, L., Luque, S., and ómez-Zorrilla, S. G., et al. (2019). Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32:e00031-19.
Hynes, M. F., Quandt, J., O’Connell, M. P., and Pühler, A. (1989). Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene 78, 111–120. doi: 10.1016/0378-1119(89)90319-3
Ji, X., Lu, P., and van der Veen, S. (2019). Development of a dual-antimicrobial counterselection method for markerless genetic engineering of bacterial genomes. Appl. Microbiol. Biotechnol. 103, 1465–1474. doi: 10.1007/s00253-018-9565-5
Kim, K.-P., and Loessner, M. J. (2008). Enterobacter sakazakii invasion in human intestinal Caco-2 cells requires the host cell cytoskeleton and is enhanced by disruption of tight junction. Infect. Immun. 76, 562–570. doi: 10.1128/iai.00937-07
Kopotsa, K., Osei Sekyere, J., and Mbelle, N. M. (2019). Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann. N. Y. Acad. Sci. 1457, 61–91.
Kumarage, J., Khonyongwa, K., Khan, A., Desai, N., Hoffman, P., and Taori, S. K. (2019). Transmission of multi-drug resistant Pseudomonas aeruginosa between two flexible ureteroscopes and an outbreak of urinary tract infection: the fragility of endoscope decontamination. J. Hosp. Infect. 102, 89–94. doi: 10.1016/j.jhin.2019.02.015
Lauritsen, I., Porse, A., Sommer, M. O. A., and Nørholm, M. H. H. (2017). A versatile one-step CRISPR-Cas9 based approach to plasmid-curing. Microb. Cell Fact. 16:135.
Li, C., Zeng, H., Zhang, J., He, W., Ling, N., and Chen, M., et al. (2019). Prevalence, antibiotic susceptibility, and molecular characterization of Cronobacter spp. isolated from edible mushrooms in China. Front. Microbiol. 10:283. doi: 10.3389/fmicb.2019.00283
Liu, B.-T., Song, F.-J., Zou, M., Hao, Z.-H., and Shan, H. (2017). Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and in Escherichia coli from the same animal. Antimicrob. Agents Chemother. 61:e01444-16.
Lou, Z., Wang, H., Rao, S., Sun, J., Ma, C., and Li, J. (2012). p-coumaric acid kills bacteria through dual damage mechanisms. Food Control 25, 550–554. doi: 10.1016/j.foodcont.2011.11.022
Mangat, C. S., Bekal, S., Irwin, R. J., and Mulvey, M. R. (2017). A novel hybrid plasmid carrying multiple antimicrobial resistance and virulence genes in Salmonella enterica serovar Dublin. Antimicrob. Agents Chemother. 61, e02601–e02616.
Medaney, F., Ellis, R. J., and Raymond, B. (2016). Ecological and genetic determinants of plasmid distribution in Escherichia coli. Environ. Microbiol. 18, 4230–4239. doi: 10.1111/1462-2920.13552
Nang, S. C., Li, J., and Velkov, T. (2019). The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841x.2018.1492902
Odeyemi, O. A., and Abdullah Sani, N. (2019). Antibiotic resistance, putative virulence factors and curli fimbrination among Cronobacter species. Microb Pathog 136:103665. doi: 10.1016/j.micpath.2019.103665
Parra-Flores, J., Aguirre, J., Juneja, V., Jackson, E. E., Cruz-Córdova, A., and Silva-Sanchez, J., et al. (2018). Virulence and antibiotic resistance profiles of Cronobacter sakazakii and Enterobacter spp. involved in the diarrheic hemorrhagic outbreak in Mexico. Front. Microbiol. 9:2206. doi: 10.3389/fmicb.2018.02206
Salehi, B., Goudarzi, H., Nikmanesh, B., Houri, H., Alavi-Moghaddam, M., and Ghalavand, Z. (2018). Emergence and characterization of nosocomial multidrug-resistant and extensively drug-resistant Acinetobacter baumannii isolates in Tehran, Iran. J. Infect. Chemother. 24, 515–523. doi: 10.1016/j.jiac.2018.02.009
Seong, Y. J., Alhashimi, M., Mayhoub, A., Mohammad, H., and Seleem, M. N. (2020). Repurposing fenamic acid drugs to combat multidrug-resistant Neisseria gonorrhoeae. Antimicrob Agents Chemother. 64, e02206–e02219.
Shi, C., Sun, Y., Zhang, X., Zheng, Z., Yang, M., and Ben, H., et al. (2016). Antimicrobial effect of lipoic acid against Cronobacter sakazakii. Food Control 59, 352–358. doi: 10.1016/j.foodcont.2015.05.041
Singh, N., Raghav, M., Narula, S., Tandon, S., and Goel, G. (2017). Profiling of virulence determinants in Cronobacter sakazakii isolates from different plant and environmental commodities. Curr. Microbiol. 74, 560–565. doi: 10.1007/s00284-017-1219-9
Stojiljković, I., Trgovčević, Z. e, and Salaj-S̆mic, E. (1991). Tn5-rpsL: a new derivative of transposon Tn5 useful in plasmid curing. Gene 99, 101–104. doi: 10.1016/0378-1119(91)90039-e
Stratev, D., and Odeyemi, O. A. (2016). Antimicrobial resistance of Aeromonas hydrophila isolated from different food sources: a mini-review. J. Infect. Public Health 9, 535–544. doi: 10.1016/j.jiph.2015.10.006
Tada, T., Hishinuma, T., Watanabe, S., Uchida, H., Tohya, M., and Kuwahara-Arai, K., et al. (2019). Molecular characterization of multidrug-resistant Pseudomonas aeruginosa isolates in hospitals in Myanmar. Antimicrob. Agents Chemother. 63:e02397-18.
Tang, Q., Lu, T., and Liu, S.-J. (2018). Engineering the bacterium Comamonas testosteroni CNB-1: plasmid curing and genetic manipulation. Biochem. Eng. J. 133, 74–82. doi: 10.1016/j.bej.2018.01.030
Tu, Q., Yin, J., Fu, J., Herrmann, J., Li, Y., and Yin, Y., et al. (2016). Room temperature electrocompetent bacterial cells improve DNA transformation and recombineering efficiency. Sci. Rep. 6:24648.
Wang, P., He, D., Li, B., Guo, Y., Wang, W., and Luo, X., et al. (2019). Eliminating mcr-1-harbouring plasmids in clinical isolates using the CRISPR/Cas9 system. J. Antimicrob. Chemother. 74, 2559–2565. doi: 10.1093/jac/dkz246
Weber, T., Wefers, B., Wurst, W., Sander, S., Rajewsky, K., and Kühn, R. (2015). Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548. doi: 10.1038/nbt.3198
Yamamoto, S., Sakai, A., Agustina, V., Moriguchi, K., and Suzuki, K. (2018). Effective removal of a range of Ti/Ri plasmids using a pBBR1-type vector having a repABC operon and a lux reporter system. Appl. Microbiol. Biotechnol. 102, 1823–1836. doi: 10.1007/s00253-017-8721-7
Yin, C., Xu, L., Li, Y., Liu, Z., Gu, D., and Li, Q., et al. (2018). Construction of pSPI12-cured Salmonella enterica serovar pullorum and identification of IpaJ as an immune response modulator. Avian Pathol. 47, 410–417. doi: 10.1080/03079457.2018.1471195
Yuan, Q., Li, Y., Xiu, L., Zhang, C., Fu, Y., and Jiang, C., et al. (2019). Identification of multidrug-resistant Neisseria gonorrhoeae isolates with combined resistance to both ceftriaxone and azithromycin, China, 2017–2018. Emerg. Microbes Infect. 8, 1546–1549. doi: 10.1080/22221751.2019.1681242
Zeng, H., Lei, T., He, W., Zhang, J., Liang, B., and Li, C., et al. (2018). Novel multidrug-resistant Cronobacter sakazakii causing meningitis in Neonate, China, 2015. Emerg. Infect. Dis. 24, 2121–2124.
Zeng, H., Li, C., Ling, N., Zhang, J., Chen, M., and Lei, T., et al. (2020). Prevalence, genetic analysis and CRISPR typing of Cronobacter spp. isolated from meat and meat products in China. Int. J. Food Microbiol. 321:108549. doi: 10.1016/j.ijfoodmicro.2020.108549
Keywords: plasmid curing, virulence, antibiotic resistance, Cronobacter sakazakii, p-coumaric acid
Citation: Ji X, Lu P, Hu Y, Xue J, Wu J, Zhang B, Zhang Y, Dong L, Lv H and Wang S (2021) Function Characterization of Endogenous Plasmids in Cronobacter sakazakii and Identification of p-Coumaric Acid as Plasmid-Curing Agent. Front. Microbiol. 12:687243. doi: 10.3389/fmicb.2021.687243
Received: 29 March 2021; Accepted: 25 May 2021;
Published: 25 June 2021.
Edited by:
Gloria Soberón-Chávez, National Autonomous University of Mexico, MexicoReviewed by:
Angelika Lehner, University of Zurich, SwitzerlandDavid Romero, National Autonomous University of Mexico, Mexico
Copyright © 2021 Ji, Lu, Hu, Xue, Wu, Zhang, Zhang, Dong, Lv and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuo Wang, wangshuo@nankai.edu.cn
†These authors have contributed equally to this work
 Xuemeng Ji1†
Xuemeng Ji1† Yaozhong Hu
Yaozhong Hu Juan Xue
Juan Xue Bowei Zhang
Bowei Zhang Shuo Wang
Shuo Wang