- Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, China
Few bacteriocins with antibacterial activity against Shigella flexneri have been reported. Here, a novel bacteriocin (LFX01) produced by Lactiplantibacillus plantarum strain LF-8 from the intestine of tilapia was purified and extensively characterized. LFX01 possesses a molecular weight of 1049.56 Da and an amino acid sequence of I-T-G-G-P-A-V-V-H-Q-A. LFX01 significantly inhibited S. flexneri strain 14 (S. flexneri_14) growth. Moreover, it exhibited excellent stability under heat and acid-base stress, and presented sensitivity to a variety of proteases, such as proteinase K, pepsin, and trypsin. The minimum inhibitory concentration (MIC) of LFX01 against S. flexneri_14 was 12.65 μg/mL, which was smaller than that of most of the previously found bacteriocins. Furthermore, LFX01 significantly inhibited (p < 0.05) S. flexneri_14 cells and decreased their cell viability. In addition, LFX01 could significantly (p < 0.05) inhibit biofilm formation of S. flexneri_14. Scanning electron microscopy analysis presented that the cell membrane permeability of S. flexneri_14 was demolished by LFX01, leading to cytoplasmic contents leakage and cell rupture death. In summary, a novel bacteriocin of lactic acid bacteria (LAB) was found, which could effectively control S. flexneri in both planktonic and biofilm states.
Introduction
Shigella flexneri is a Gram-negative, non-spore-bearing facultative anaerobic bacterium that can cause serious gastrointestinal infections (food poisoning), especially bacillary dysentery (Cui et al., 2015; Jabbar and Al-azawi, 2020). It is also widely known as the “first killer” in the foodborne pathogenic bacteria in children in developing countries (Cui et al., 2015; Nisa et al., 2020). Previous studies reported an estimated 165 million cases of shigellosis worldwide each year, with approximately 1.1 million deaths accounting for 69% of the cases in the immunocompromised 5-year-olds (Mardaneh et al., 2013; Saima et al., 2018). Additionally, S. flexneri infecting an animal body could induce microfold cell transportation in animal intestinal mucosa by entering the lymphatic tissues through endocytosis (Ranganathan et al., 2019; Rey et al., 2020). Although macrophages can swallow S. flexneri, it can escape by binding S. flexneri IpaB with caspase-1 and activate caspase pathways in the host intestinal cells through lysis (Ranganathan et al., 2019; Li L.H. et al., 2020). Thus, preventing and controlling the S. flexneri that induced foodborne pathogen infection has become the primary concern of the researchers. S. flexneri strains primarily contaminate many high nutritional foods such as fish and poultry, which affect food quality, resulting in human food safety problems (Paul et al., 2019). At present, the primary way to control S. flexneri in food is via the use of unhealthful chemical preservatives. Therefore, exploring new methods to control foodborne S. flexneri is necessary.
Biofilm is a community of bacteria attached to surfaces of biotic (foods) or abiotic (processing equipment) (Lindsay and Von, 2006; Shahin et al., 2019). Biofilm of foodborne pathogenic bacteria possesses resistance to disinfectants and antibacterial agents, with high resistance to heat and drying. Furthermore, high-density structure of the biofilm protects pathogenic bacteria from acidic and antibiotic treatments (Tsukatani et al., 2020). In particular, cells of pathogenic bacteria in biofilms are 10–1,000 times less sensitive to disinfectants than planktonic cells (Li et al., 2021; Xiang et al., 2021). Moreover, biofilm is one of the primary causes of equipment damage, thus leading to increased energy cost, as well as is involved in the occurrence of food spoilage and foodborne diseases (Nové et al., 2020; Tsukatani et al., 2020). Currently, biofilm formed by pathogenic bacteria and biofilm mediated foodborne disease prevalence has become one of the major challenges for food safety. To date, several studies have explored the biofilm formation ability and mechanism of S. flexneri in foods, and employed high-concentration antibiotics and bacteriostatic synthetic compounds to control S. flexneri biofilm (Kang et al., 2020; Kaoukab et al., 2020). However, no such effective biological control approaches to inhibit planktonic cells and biofilm of S. flexneri have been reported so far.
Bacteriocin from bacteria is a class of natural macromolecular protein or small molecule with short peptide (Li et al., 2017; Enriqueta et al., 2020; Peng et al., 2021). It is also known to be a class of natural antimicrobial agents that can effectively inhibit or inactivate many foodborne pathogenic bacteria (Guo et al., 2020; Darvishi et al., 2021). In particular, many lactic acid bacteria (LAB) bacteriocins have been demonstrated with high safety and tolerance to heat, acids, and bases (Guo et al., 2020). For instance, LAB bacteriocins derived from Lactococcus lactis 2Mt and Lactobacillus salivarius CGMCC20700 were found to effectively inhibit foodborne Staphylococcus aureus under a wide range of acid-base and temperature (Pmka et al., 2019; Li et al., 2021). Although many LAB bacteriocins have been identified so far, their effects on biofilm formation of foodborne S. flexneri are still unexplored.
Lactiplantibacillus plantarum (previously named as Lactobacillus plantarum; Zheng et al., 2020) is a potential probiotic additive (Bu et al., 2021) and is abundant in animal gastrointestinal tracts and feces (Lei et al., 2020), such as bacteriocins produced by swun5815 strain from yak intestine (Zhou et al., 2018), bacteriocin ZJ316 from healthy infant feces (Chen et al., 2018), and bacteriocin zrx03 from newborn children feces (Lei et al., 2020). L. plantarum bacteriocins and other LAB bacteriocins are mostly isolated from the mammalian intestine, but bacteriocins isolated from other animal groups, such as fish are extremely rare. Therefore, the limited bacteriocin resources have also limited the application of LAB bacteriocins.
As the second most commonly cultured freshwater fish worldwide, tilapia aquaculture has been extensively conducted in more than 100 countries, including China, Indonesia, and Egypt (Subasinghe, 2017). Previous studies have isolated and identified bacteriocin-producing LAB strains from the intestine of Oreochromis niloticus in Cameroon, Africa (Pmka et al., 2019). However, these studies only obtained crude LAB bacteriocins from tilapia by chloroform/methanol method, and its molecular weight was roughly checked by tricine-SDS-PAGE (Pmka et al., 2019). This evidence implied that intestine of tilapia is an important source of LAB bacteriocin resources, and it likely contains LAB bacteriocins with antibacterial activity against S. flexneri. In addition, further purification and characterization (e.g., molecular weight, amino acid sequence composition, antibacterial, and antibiofilm activity) of LAB bacteriocins from tilapia is highly necessitated.
The present study sought to screen the L. plantarum strains with high antibacterial activity against S. flexneri from tilapia intestines. Subsequently, the L. plantarum-producing bacteriocin was purified and its molecular weight and amino acid sequence were determined. Furthermore, the minimum inhibitory concentration (MIC), time-kill kinetics, acid-base, thermal stability, and enzyme sensitivity of bacteriocin were assessed. The inhibitory effects of LFX01 on S. flexneris biofilm were investigated.
Materials and Methods
Isolation and Culture of Strain
Healthy juvenile tilapia (Oreochromis niloticus) samples were purchased from the food market located in Chenggong District, Kunming, Yunnan, China, and sent to the laboratory immediately within 3–5 h. Later, six individuals (mean body weight: 12 ± 1.5 g, mean body length: 7 ± 1.2 cm) were anesthetized with ethyl 3-aminobenzoate methane sulfonate (concentration: 15 mg/L, MS-222, Sigma, MO, United States). Then, the tilapia individuals were washed twice with 75% ethanol and 5 times with sterile water, and the intestines of tilapia were aseptically cut on a super-clean worktable (Optec, Chongqing, China) and placed in a 5.0-mL sterile Eppendorf tube with 2.0-mL sterile water, and thoroughly ground with a grinding rod. Afterward, 100 μL dilutions of four concentrations (i.e., 10–1, 10–3, 10–5, and 10–7) obtained by gradient dilution with sterile water were dispersed on the surface of MRS Culture Medium (Solarbio, Beijing, China) (37°C, 24 h). Furthermore, single colonies were picked out by sterile inoculation loop. Finally, these single colonies were preliminarily identified and stored at 4°C according to their phenotypic characteristics.
Screening and Identification of Bacteria With Antibacterial Activity
S. flexneri strain BDS14 (referred to as S. flexneri_14) isolated from chicken was obtained in our previous studies (He et al., 2021), and it is being stored at Faculty of Life Science and Technology, Kunming University of Science and Technology. Species of LAB strains were confirmed by morphological and microscopic observation under a light microscopy (400 × magnification) (Zeiss Primo Star, Jena, Germany) according to the “manual for systematic identification of common bacteria” (Goodfellow et al., 2012). LAB strains possess several morphological characteristics such as rod-shaped appearance, gram-positive stain, no flagella, and no capsule.
The candidate LAB strains were cultured in MRS liquid medium (37°C, 24 h) and the culture solution was centrifuged (7,104 × g, 10 min) to discard the precipitate. Subsequently, a 0.22-μm filter membrane was employed to obtain a cell-free suspension. The obtained cell-free supernatant was adjusted to neutral pH (6.5) to exclude the interference of organic acids (including lactic acid). Screening of LAB strains against S. flexneri_14 was performed by the Oxford cup (diameter 8.0 mm, sample volume 200 μL) double-layer plate method (Voulgari et al., 2010). In brief, 100 μL of a suspension of S. flexneri_14 (107 CFU/mL) cells in exponential phase was spread on the surface of LB semisolid medium (Solarbio). The Oxford cup was placed onto the surface of the LB semisolid medium and 200 μL of the cell-free supernatant was added, cultivated at 37°C for 24 h. Finally, a vernier caliper was used to determine the clearing zone around the well which was a result of the antibacterial activity of the LAB supernatant. Sterile water was used as control and three replicates were performed.
Based on the above obtained inhibitory zone, the LAB strain showing the best antibacterial effects against S. flexneri_14 was selected for the downstream experiments. The target strain was added to MRS liquid medium and then incubated at 37°C for 24 h. The taxonomic status of the target strain was verified using molecular markers. Briefly, DNA extraction of the target LAB strain was performed according to the directions of DNA Purification Kit (Tiangen, Beijing, China). PCR reaction system: 2 × Mix 12.5 μL, DNA template 1 μL, each universal primer (8F and 1492R for the bacterial 16S rRNA gene) 0.5 μL. PCR reaction conditions: 94°C for 3 min, 94°C for 32 s, 52°C for 32 s, 72°C for 50 s, final extension at 72°C for 5 min after 30 cycles. The PCR amplicons were detected by 1.5% agarose electrophoresis, and Sanger sequencing was performed by Geneseed Biotech Co., Ltd. (Guangzhou, China). The obtained 16S DNA sequences were aligned into the NCBI database using the online BLAST. MEGA6 software was used to reconstruct a phylogenetic tree (Tamura et al., 2013).
Purification and Determination of Bacteriocin
The target strain (L. plantarum) against S. flexneri_14 was added to MRS liquid medium (37°C, 24 h), and then centrifuged (7,104 × g, 10 min) to remove cell precipitate. The obtained cell-free supernatant was purified by an ÄKTA purifier automatic chromatograph in tandem coupled with a Superdex™ 30 Increase 10/300 GL Exclusion Chromatography columns (GE Healthcare, Marlborough, United States), as previously described (Li Z.R. et al., 2020; Li et al., 2021). The corresponding substances under each eluted peak were separately collected into a 25-mL centrifuge tube, and their antibacterial activity was determined by the Oxford cup double-plate method. The collection that showed the highest antibacterial activity was then loaded and purified again to obtain a purified antibacterial active substance (named as LFX01). Finally, the LFX01 fraction was preserved by freeze-drying in a FD-2A freeze dryer (Biocoll, Beijing, China), for downstream experiments.
The molecular weight and amino acid sequence of the purified LFX01 were determined by a Nano LC-MS/MS system, as previously described (Li Z.R. et al., 2020; Li et al., 2021). Briefly, LFX01 were eluted with 0.1% formic acid (Macklin, Shanghai, China), 100% water and 0.1% formic acid, and 100% acetonitrile. The LC linear gradient elution was implemented: from 6 to 9% B for 6 min; from 9 to 50% B for 30 min; and from 50 to 95% B for 3 min and 95–95% B for 5 min, with a flow rate at 0.3 μL/min. The primary mass spectrum was obtained by a single full scan (MS), and the secondary mass spectrum data is obtained by step-normalized collision energy. The obtained mass spectra signals by MS scan were further assessed for protein identification in Peaks Studio X (Bioinformatics, Waterloo, Canada). Antimicrobial peptide databases (i.e., NCBI, UniProt, and Swiss-Prot) were searched amino acid sequence of bacteriocin using the Blast online server. Finally, the antibacterial effects of LFX01 were verified by the Oxford cup double-plate method.
Determination of Bacteriocin Concentration
Bacteriocin concentration was estimated by the Pierce® BCA Protein quantification Kit (Thermo Fisher Scientific, Waltham, United States), according to user instruction. Briefly, bovine serum albumin (BSA) standards (2.0 mg/mL) were diluted in ampoules to prepare the required concentration of the standard solution. Subsequently, the standard solution (25 μL) and the equivalent amount of LFX01 were pipetted into a 96-well plate. A total of 200 μL BCA working reagent was added to each well, the content was thoroughly mixed for 1 min, incubated room temperature for 25 min. Absorbance of the solution was measured at 565 nm at room temperature using a microplate reader (MR-96A, Mindray, Shenzhen, China). Free-BSA samples were set as control. The concentration of LFX01 in the samples was measured by plotting a standard curve according to y = 1.058x+0.1164, R2 = 0.9962.
Enzyme Sensitivity Test
LFX01 (1.0 mg/mL) was added separately to catalase and different protease solutions [i.e., pepsin (20 mM in a in PBS, pH 2), trypsin, proteinase K, papain (20 mM in PBS, pH 7.5), and alkaline protease (20 mM in PBS, pH 9.0)] with a final concentration of 1.0 mg/mL (Solarbio). The mixture of LFX01 and enzyme was treated (37°C, 2 h), followed by deactivation (80°C, 5 min). Enzyme without LFX01 was set as control. The effect of each different enzyme on the antibacterial activity of LFX01 was confirmed by the Oxford cup double-plate method. Each experimental sample was independently tested in triplicate.
Acid-Base and Thermal Stability Test
The following tests were performed to detect the acid-base and thermal stability of LFX01 (1.0 mg/mL), as previously described (Yi et al., 2018; Zhang et al., 2019): (1) treatment at 37, 60, 80, 100, and 121°C for 30 min; (2) pH of LFX01 was adjusted to 2.0, 4.0, 6.0, 8.0, 10.0, and 12.0 by 1.0 moL/L HCl or NaOH solution and then treated at 37°C for 2 h. Finally, pH of LFX01 solution was adjusted back to neutral pH (6.5). The antibacterial activity of LFX01 against S. flexneri_14 under different pH values and temperature was separately assessed by the Oxford cup double-plate method. LFX01 under room temperature (25°C) and neutral pH value was set as control. Each experimental sample was independently tested in triplicate.
Minimum Inhibitory Concentration and Time-Kill Kinetics
Determination of the MIC values of LFX01 against S. flexneri_14 was conducted, as previously described (Zhang et al., 2019; Li et al., 2021). Briefly, S. flexneri_14 was precultured in LB liquid medium by incubation (37°C for 12 h). Then, LFX01 (1.0 mg/mL) was serially diluted into PBS. These dilutions (10 μL) were added separately into 90 μL of LB liquid medium with S. flexneri_14 (final concentration: 107 CFU/mL), incubated at 37°C in a 96-well plate for 24 h. Growth of S. flexneri_14 was assessed in triplicate using a microplate reader at 595 nm. MIC was the lowest concentration of antibacterial substances that completely inhibited growth of S. flexneri_14, as previously described (Yi et al., 2018; Li et al., 2021). 2 × MIC was used as the effective concentration for bacteriostatic treatment, as previously reported (Yi et al., 2018; Lanhua et al., 2020).
The time-killing kinetics test was conducted to assess the dynamic changes in killing effects of LFX01 against the indicator strain across various treatment times. Namely, LFX01 was mixed with S. flexneri_14 (107 CFU/mL) to reach a final concentration (2 × MIC) of LFX01 and then incubated for 0, 0.5, 1, 1.5, and 2 h. Subsequently, 100 μL of LFX01 at each incubation time points were spread on LB solid medium by 10 fold serial dilution method. After incubating at 37°C for 24 h, 30∼300 single colonies of S. flexneri_14 were selected for counting. LFX01-free samples were set as control groups. Each experimental sample was independently tested in triplicate.
Cell Proliferation and Viability Assay
The metabolic activity of S. flexneri_14 cells was determined by the XTT (i.e., 2,3-BIS (2-methoxy-4-nitro-5-sulfonyl)-2H-tetrazo-5-carboxyl) assay kit (Abcam, Cambridge, United Kingdom), according to use instruction strictly. Briefly, planktonic S. flexneri_14 samples and 10 μL of LFX01 (final concentrations: 2 × MIC) were added into a 96-well plate containing 80 μL of LB liquid medium, followed by the mixing and incubated (37°C, 1 h). Subsequently, 10 μL of XTT solution was added, mixed for 30 s and incubated (37°C, 2 h). Following a gentle homogenization of the microplate, absorbance of samples was measured using a microplate reader at 450 nm. Each experimental sample was independently tested in triplicate.
The survival rate of planktonic S. flexneri_14 cells after exposure to LFX01 was assessed by the Cell-Check™ viability/cytotoxicity kit (ABP Biosciences, Rockville, United States). Briefly, planktonic S. flexneri_14 cells were treated with LFX01 (2 × MIC) at 37°C for 10 min. Later, 3 μL of mixture composed of fluorescent dyes NucView Green and propidium iodide (PI) was added to the bacterial suspension, followed by incubated at 37°C for 10 min. LFX01-free planktonic S. flexneri_14 cells were set as control. The experimental samples were detected on the slides using fluorescence microscopy (Boschida, Shenzhen, China) (200 magnification). Each experimental sample was independently tested in triplicate.
SEM Analysis
Three milliliters of S. flexneri_14 (107 CFU/mL) were centrifuged (7,104 × g, 5 min) to collect the precipitated bacterial cells. The bacterial cells were washed thrice with PBS and incubated (37°C, 2 h) with 1 mL of LFX01 (2 × MIC). Subsequently, the LFX01 treated bacterial cells were then fixed with 2.5% glutaraldehyde at 4°C for 8 h and dehydrated in the concentration gradient of ethanol (20, 40, 60, 80, and 95%) for 30 min each. Afterward, all samples were transferred to polished silicon wafers (10 × 10 mm) and dried at room temperature (25°C). The samples covered by gold powder were imaged with an SEM (S-3000N, Hitachi, Tokyo, Japan). LFX01-free S. flexneri_14 was set as control, and each treatment was performed in three replicates.
Determination of Antibiofilm Activity
LFX01 was mixed with LB liquid medium and transferred to a 96-well plate to assess the effects of LFX01 exposure on S. flexneri_14 biofilm formation at final concentrations of: LFX01-free controls, 1/2 × MIC, 1 × MIC, 2 × MIC. Later, 100 μL of cell suspension of S. flexneri_14 was added to these LFX01 dilutions at different concentrations. After incubation at 37°C for 24 h (the optimal temperature and time for biofilm formation of S. flexneri_14), the liquid medium was discarded to remove planktonic S. flexneri_14 cells and expose the biofilm. The plates washing wells using PBS buffer, a MycoLight™ Live Bacteria Fluorescence Imaging Kit (AAT Bioquest, CA, United States) was employed to stain the exposed biofilm following the product manuals. Excessive dye was removed using PBS buffer. The formed biofilms were detected with fluorescence microscopy (100 magnification). PBS buffer was added to each well followed by homogenization. Absorbance (OD595) of the mixture was determined in a microplate reader. Samples not treated with LFX01 were used as control.
Data Analysis
Data analysis was conducted, as previously described (Yi et al., 2018; Li et al., 2021). In brief, results were exhibited as mean ± standard deviation (SD) of independent replicates. IBM® SPSS® Statistics version 22 was used to calculate the significance between two groups employing a non-paired two-tailed t-test. One-way ANOVA test was conducted to evaluate significance levels among groups. P-values < 0.05 indicated significant statistical difference.
Results
Screening of the Target Bacteriocin-Producing Lactic Acid Bacteria
Among pure cultures of LAB strains isolated from the tilapia intestine, the inhibitory zone diameter of the LF-8 strain against S. flexneri_14 was the largest (up to 26.69 ± 0.32 mm). Thus, this strain was selected as the object for further study. Colony morphology and micro-structure of LF-8 revealed white globular, smooth surface, short chain globular (Figures 1A,B). Furthermore, 16S rRNA amplicon of LF-8 with a nucleotide sequence of 1,465 bp (Genbank accession No. MW979597.1) was obtained. BLAST alignment results elucidated a 100.0% similarity of the sequence with L. plantarum species (Genbank No. NR117813.1), and the phylogenetic tree presented a cluster of LF-8 strain with L. plantarum (Figure 1C). Based on the above-obtained phenotypic and molecular evidence, LF-8 was identified as L. plantarum.
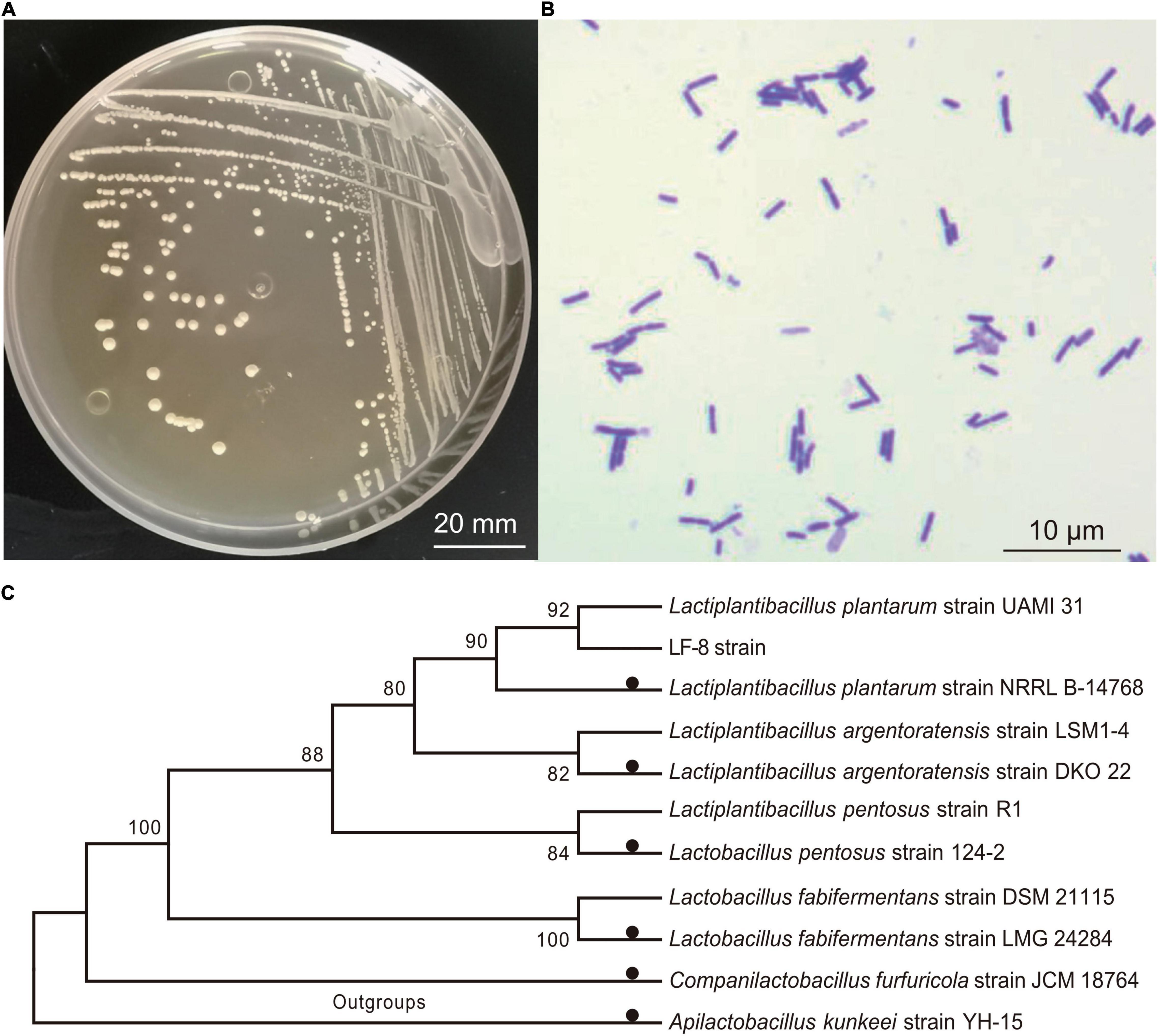
Figure 1. Identification of Lactiplantibacillus plantarum. Colony morphology (A) and microscopic characteristics; (B) following Gram staining (C); phylogenetic analysis of L. plantarum LF-8 from 16 rRNA nucleotide sequences. All sequences originated from Lactobacillaceae family and other non-Lactobacillus species were used as outgroups. Type strains were marked by solid black circles, and each species of Lactiplantibacillus/Lactobacillus included an additional strain.
Molecular Weight and Amino Acid Sequence of LFX01
Among the four peaks identified in the chromatogram of the preliminary purified extract, peak A4 showed the greatest antibacterial activity (Figure 2A). The corresponding product in B1 peak was collected after purifying A4 and the antibacterial activity of B1 substances (LFX01) against S. flexneri_14 was confirmed by the formation of an inhibitory zone (25.12 ± 0.16 mm) (Figure 2B).
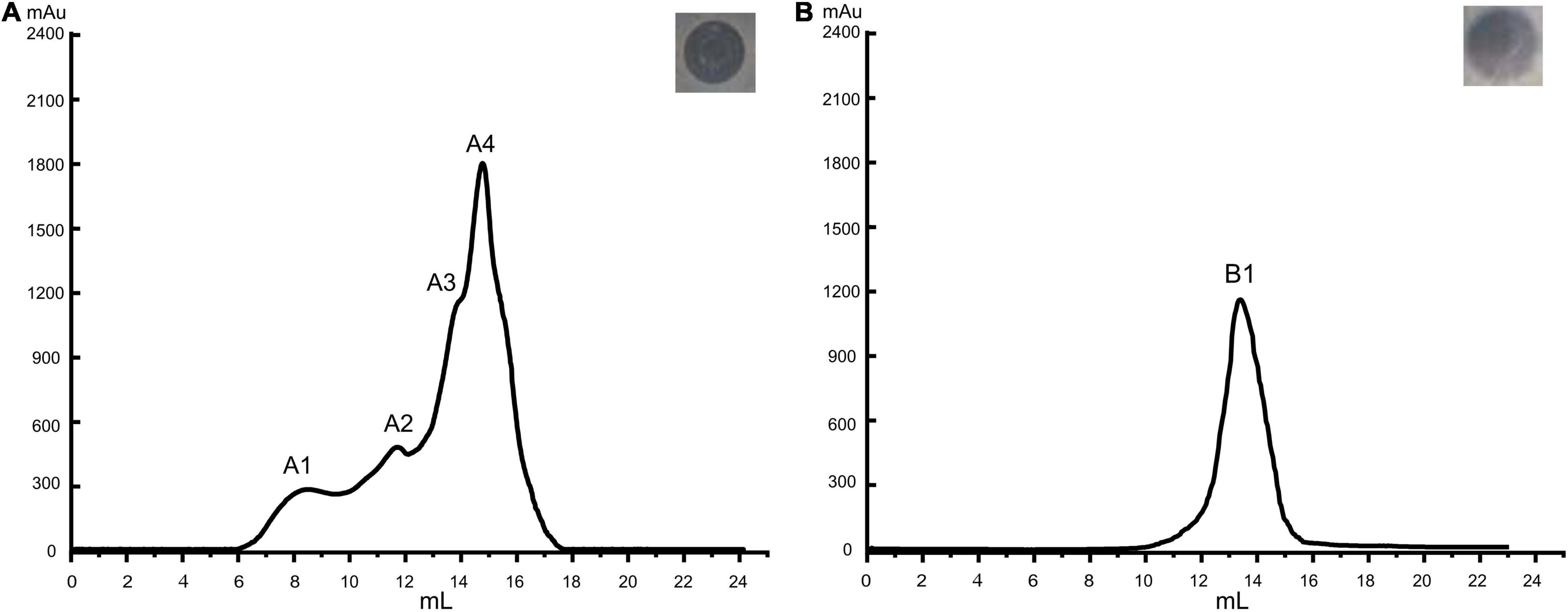
Figure 2. Purification of antibacterial substances produced by L. plantarum LF-8. Base peak chromatograms of (A) initially purification and (B) further purified bacteriocin extract accompanied by the inhibition zones of the corresponding peak against S. flexneri_14 as assessed by the Oxford cup double-plate method.
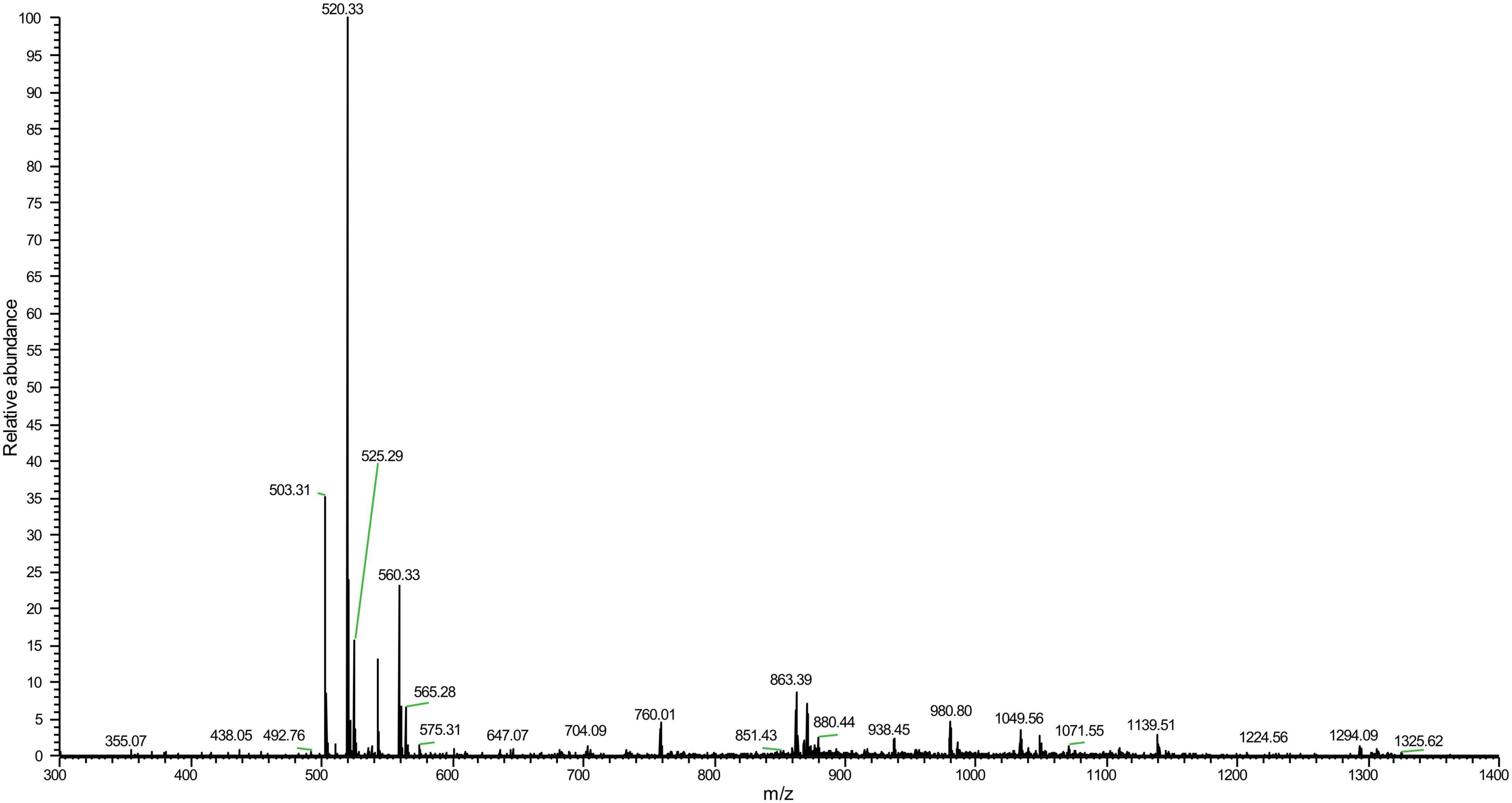
Mass spectrometry analysis determined a molecular weight of 1,049.56 Da (Figure 3) and amino acid composition of I-T-G-G-P-A-V-V-H-Q-A of LFX01 (Supplementary Figure 1). BLAST alignment did not search any similar protein sequence with LFX01 in protein storage databases.
Enzyme Sensitivity, Acid-Base, and Thermal Stability
LFX01 treated with catalase retained 97.02% activity compared to the controls (no significance, p > 0.05). This result excluded hydrogen peroxide-based antibacterial activity of LFX01. After treated with different proteases, the antibacterial activity of LFX01 significantly reduced (p < 0.05), particularly proteinase K, trypsin, and pepsin (reduced by more than 80%), indicating the sensitivity of LFX01 to various proteases (Figure 4). Additionally, the antibacterial activity of LFX01 gradually decreased with increase in pH values (Figure 5A). The antibacterial activity of LFX01 reached the lowest at pH 12.0 (reduced by 73.74%). The fastest decrease of antibacterial activity of LFX01 occurred at pH 8∼12 (reduced from 79.16 to 26.26%), and this change in antibacterial activity was significant compared with the controls (p < 0.05). Furthermore, the antibacterial activity of LFX01 gradually decreased with the increase of temperature (37∼121°C) (Figure 5B). Compared with the control, LFX01 retained 56.06% antibacterial activity at 121°C. The fastest decline in antibacterial activity of LFX01 occurred at 80∼121°C (reduced from 81.83 to 56.06%).
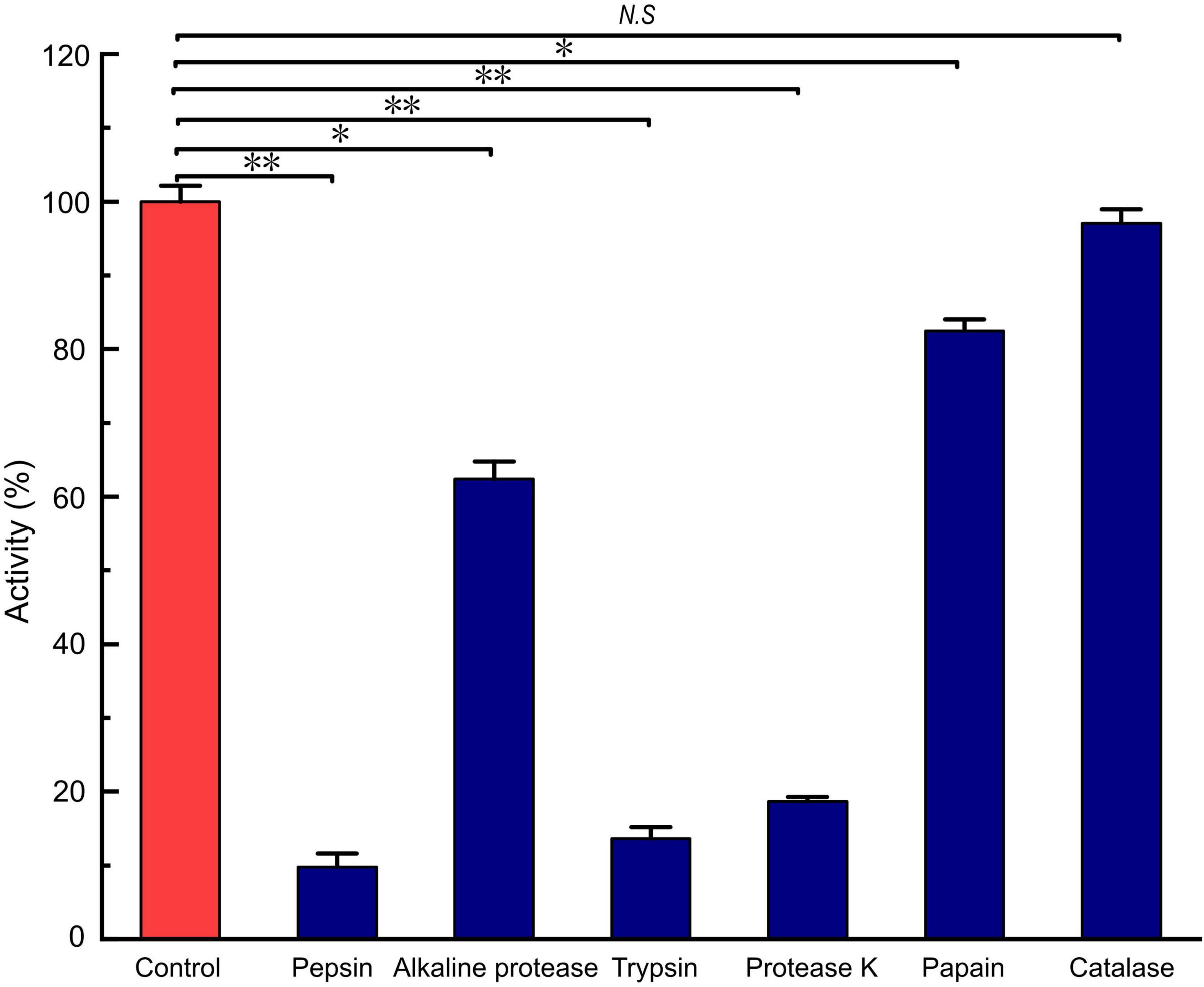
Figure 4. Analysis of enzymes on the stability of LFX01. *p < 0.05, **p < 0.01; N.S: no significant difference.
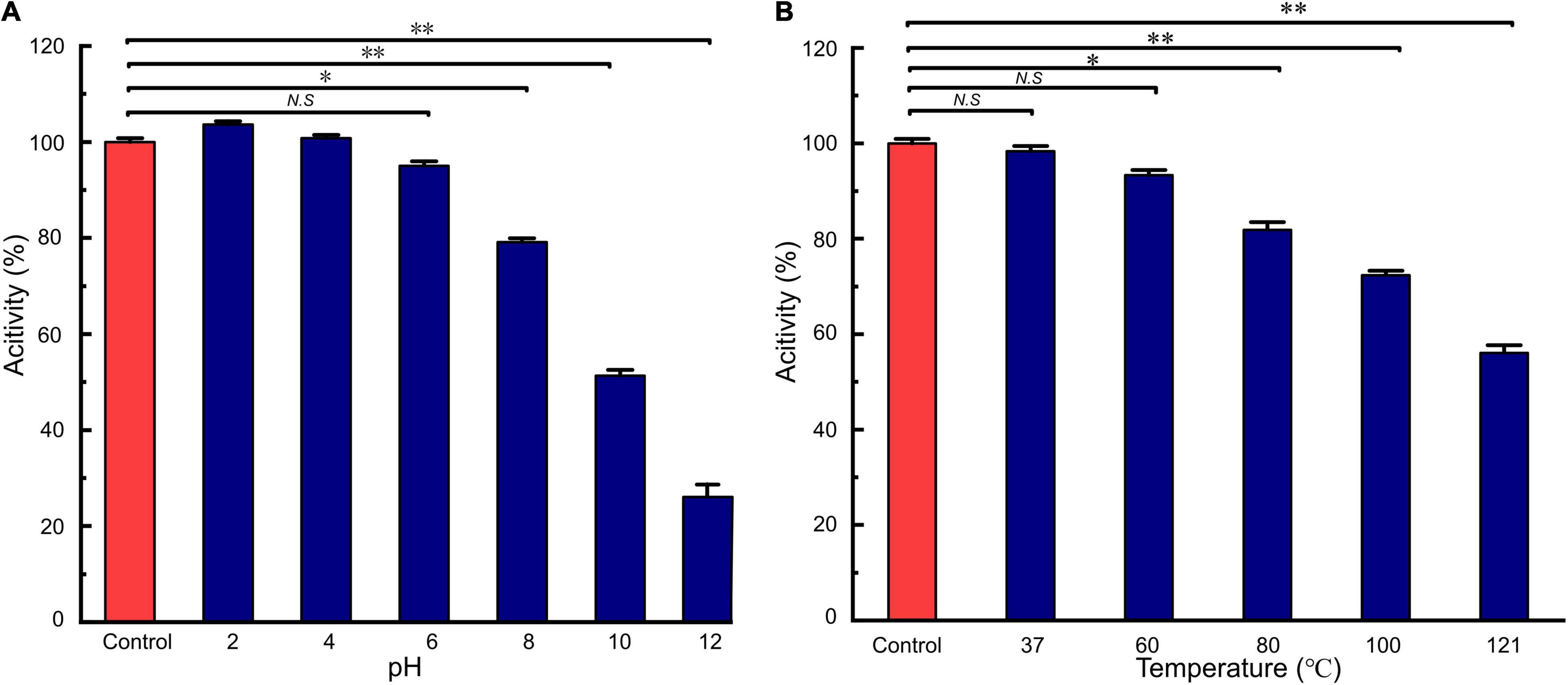
Figure 5. Analysis of acid-base (A) and heat (B) on the stability of LFX01. *p < 0.05, **p < 0.01; N.S: no significant difference.
Minimum Inhibitory Concentration and Time-Kill Kinetics
MIC of LFX01 against S. flexneri_14 was 12.65 μg/mL. The colony number of S. flexneri_14 gradually decreased with the treatment time and significantly decreased after 0.5 h (p < 0.05) when treated with 2 × MIC for 0∼2 h (Figure 6). The colony number of S. flexneri_14 reaches to the lowest value (lg4.65 CFU/mL) at 2 h, confirming the effectiveness of LFX01 to terminate planktonic S. flexneri_14 survival.
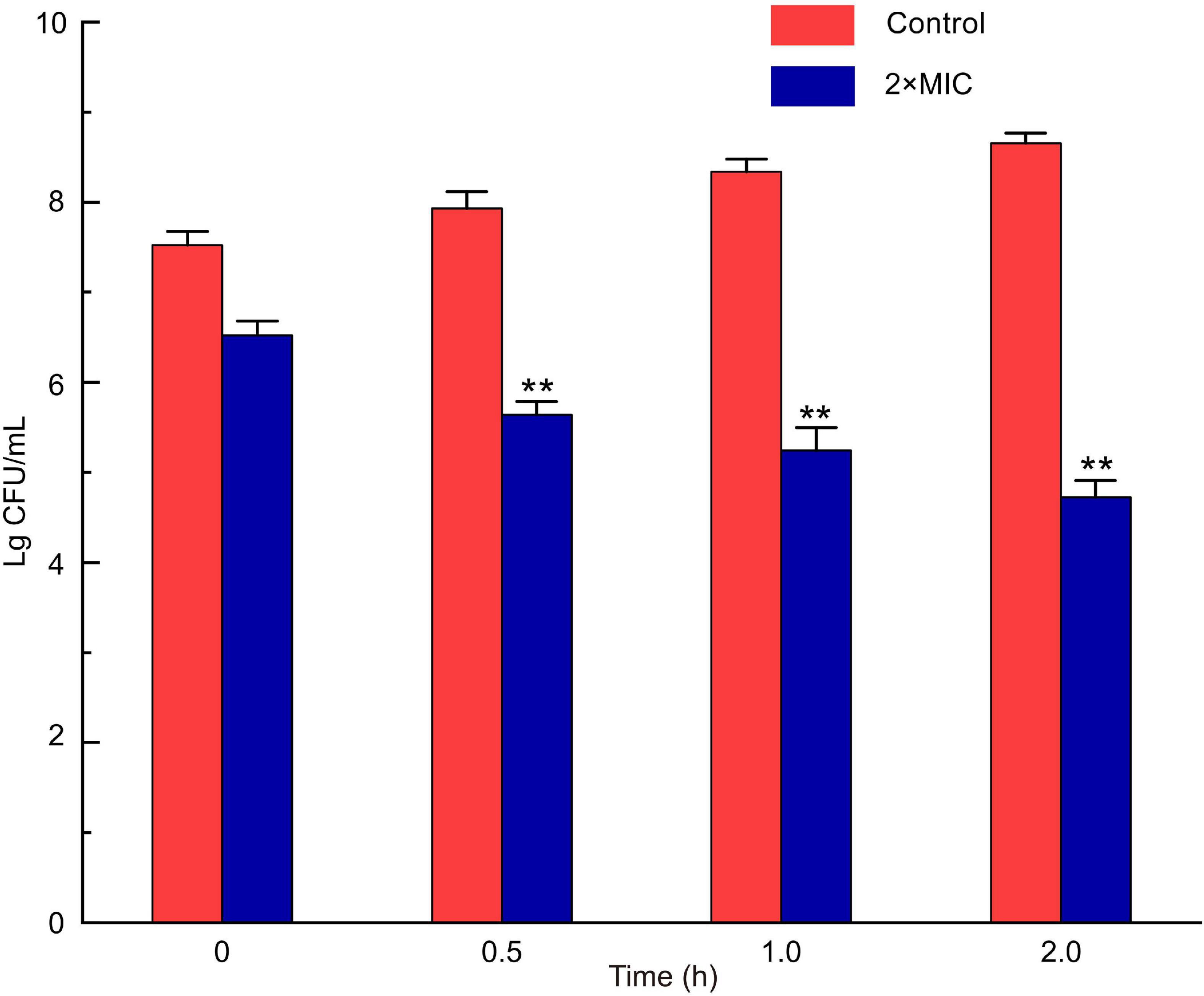
Figure 6. Analysis of time-kill kinetics of S. flexneri_14 cells treated with LFX01. **p < 0.01; N.S: no significant difference.
Proliferation and Cell Viability of Planktonic S. flexneri_14 Cells
The XTT assays showed that absorbance values of planktonic S. flexneri_14 cells was decreased to 45.32% upon exposure to LFX01 (2 × MIC) for 2 h (p < 0.01) (Figure 7A). Compared with the untreated controls (live cells are stained green) (Figure 7B), S. flexneri_14 cells died after treatment with LFX01 (2 × MIC) (red-stained) (Figure 7C). These results further confirmed the effective killing of LFX01 against planktonic S. flexneri_14 cells.
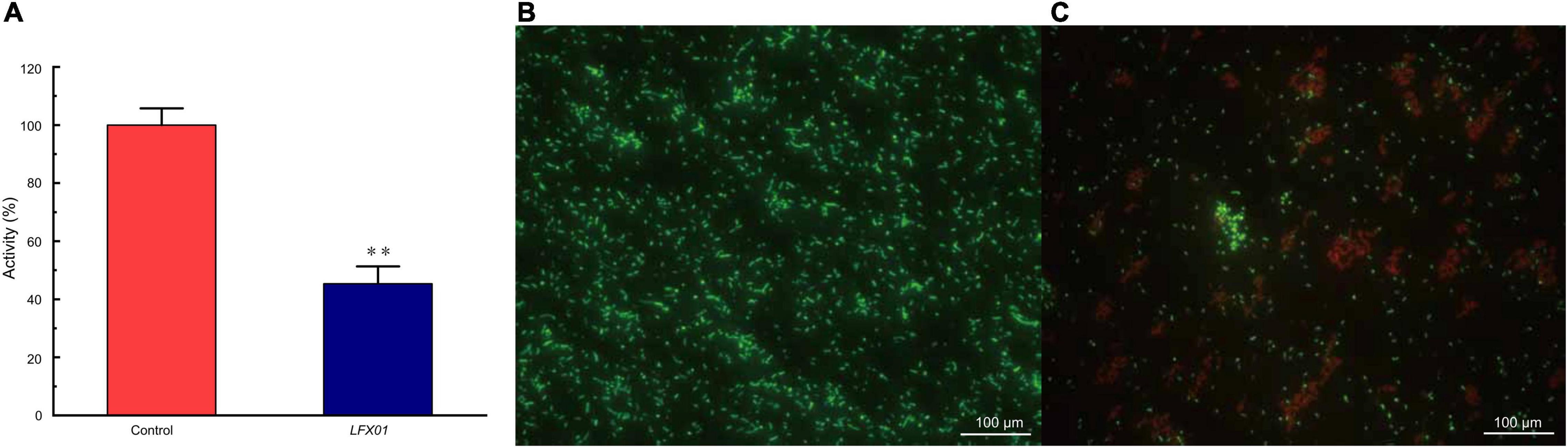
Figure 7. Cell viability of planktonic S. flexneri_14 cells after exposure to LFX01 compared with the untreated control (A). **p < 0.01. Images of planktonic S. flexneri_14 cells detected by fluorescence microscopy. (B) Control sample and (C) LFX01-treated S. flexneri_14 cells. Live and dead bacteria present green and fluorescence, respectively.
Antibiofilm Activity of LFX01
Biofilm density of S. flexneri_14 appeared to be significantly reduced (p < 0.05) after treating with LFX01 at concentrations of 1/2 × MIC, 1 × MIC, and 2 × MIC. Furthermore, quantitative analysis determined that the biofilm exposed to the control (0 × MIC; Figure 8A), 1/2 × MIC (Figure 8B), 1 × MIC (Figure 8C), and 2 × MIC (Figure 8D) of LFX01 yielded OD595 values of 1.35 ± 0.15, 0.96 ± 0.09, 0.56 ± 0.05, and 0.29 ± 0.03, respectively. P-values of all comparison pairs between controls and each experimental group were less than 0.05. The SEM analysis further presented LFX01-free S. flexneri_14 cells (control) with uniform rod-shaped, smooth surface, neat edge, clear outline, and complete cell structure (Figures 9A,B). However, compared with the control, S. flexneri_14 cells incubated with LFX01 presented uneven shape, wrinkled surface, disruptive cells, content leakage, and cell surface perforations (Figures 9C,D).
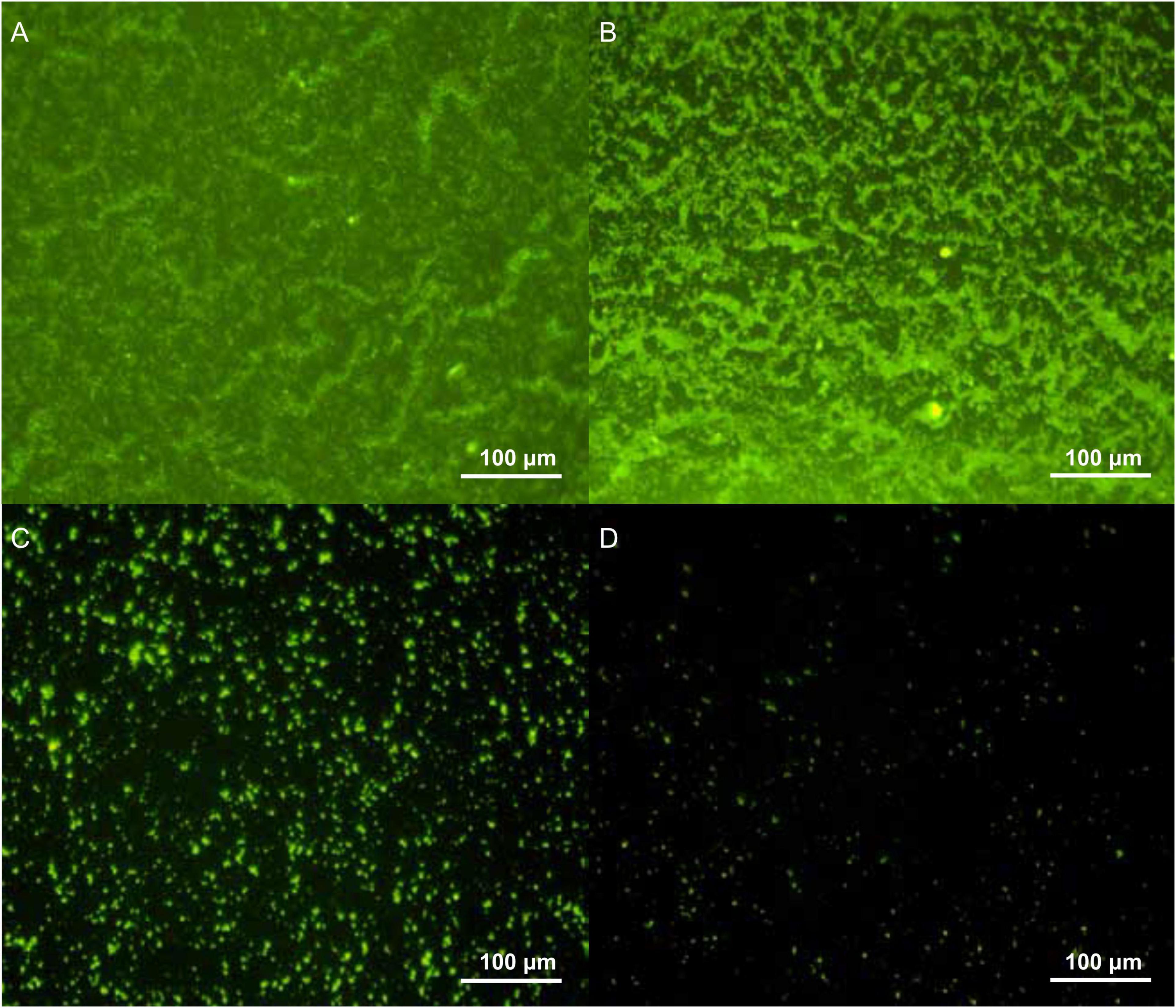
Figure 8. Effect of different concentrations of LFX01 (A): Control; (B): 1/2 × MIC; (C): 1 × MIC; (D): 2 × MIC) on the biofilm-forming ability of S. flexneri_14, as detected by fluorescence microscopy.
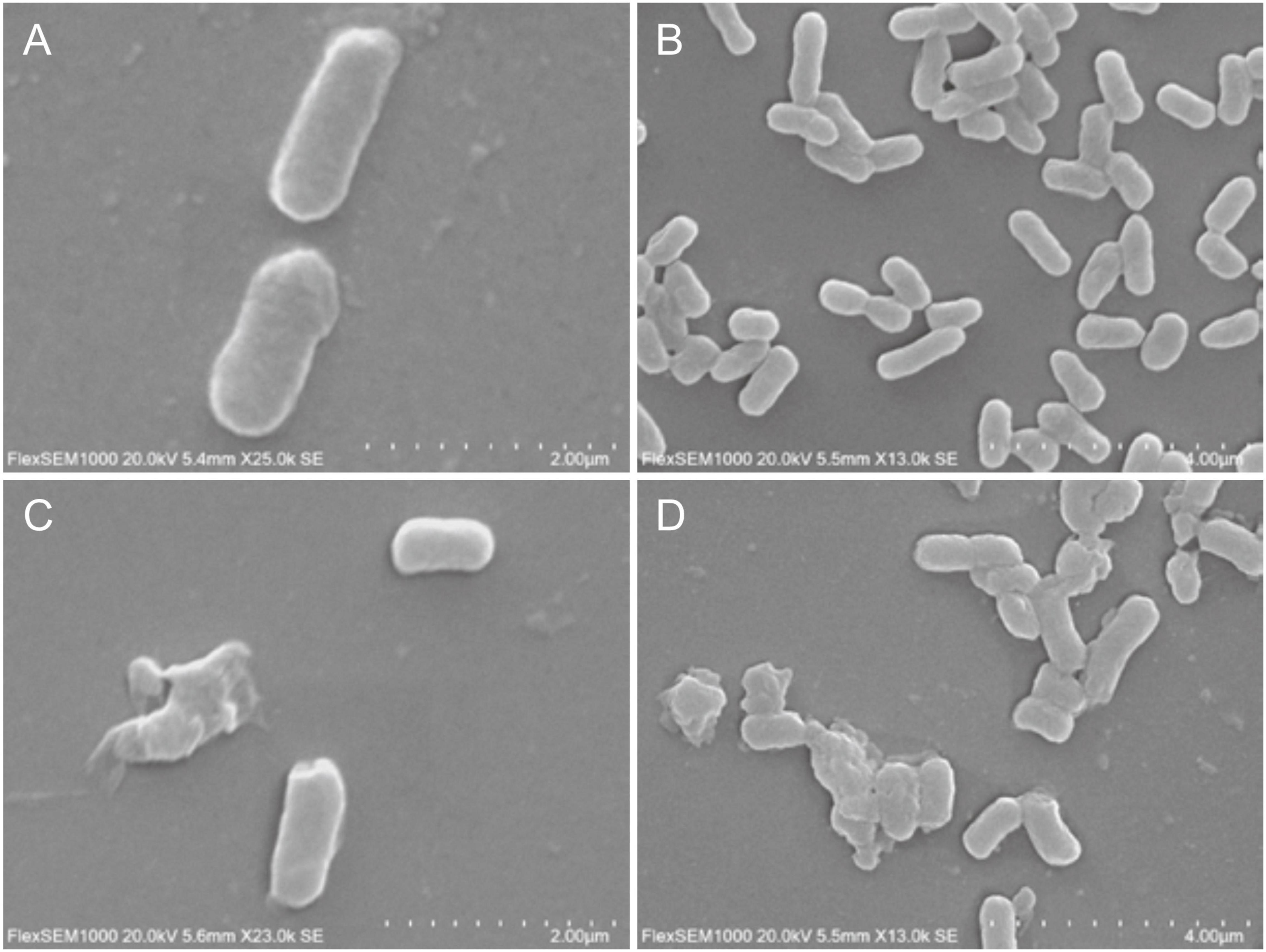
Figure 9. Effect of LFX01 on biofilm formation of S. flexneri_14, detected by SEM. (A,B) indicate the control, (C,D) indicate treatment.
Discussion
To the best of our knowledge, LFX01 is the first bacteriocin against S. flexneri_14 found in fish intestine. Compared with the molecular weight of other LAB bacteriocins reported in previous studies, LFX01 belongs to a small molecular bacteriocin, as reported in (5592.22 Da from Lactobacillus pentosus 31-1) (Zhang et al., 2009), (5383.2 Da from Lactobacillus salivarius SMXD51) (Messaoudi et al., 2012), and (1221.074 Da from L. salivarius SPW1) (Wayah and Philip, 2018) (665.30 Da from L. salivarius CGMCC2070) (Li et al., 2021). Generally, a spatial structure of bacteriocins with smaller molecular weight is more stable under environmental stressors (e.g., temperature, pH, and humidity) (Messaoudi et al., 2012; Wayah and Philip, 2018), implying that LFX01 can be widely applied to diverse food processing conditions. Additionally, LFX01 exhibits no homology with other previously reported bacteriocins, indicating that LFX01 is a novel.
Antibacterial characteristics of bacteriocins are essential parameters of the food industry. The enzyme sensitivity tests excluded the effects of hydrogen peroxide produced by LAB on S. flexneri_14 in this study. Meanwhile, LFX01 sensitivity to proteases indicated that LFX01 was a bacteriocin possessing protein-like properties (Sung and Jo, 2019). In addition, the antibacterial activity of LFX01 remained between pH 2 and pH 6, and then significantly decreased by approximately 20, 45, and 74% at pH 8, pH 10, and pH 12, respectively. Previous studies have reported that the antibacterial activity of LAB bacteriocins (produced by L. salivarius SPW1, Lactobacillus fermentum BZ532, and Lactobacillus paracasei HD1-7) significantly reduced by at least 90% when pH exceeded 8 for 2 h (Ge et al., 2016; Baranzan and Koshy, 2018; Rasheed et al., 2020). Accumulating studies have reported that the antibacterial activity of LAB bacteriocins significantly decreased over alkaline conditions (Am et al., 2019; Rasheed et al., 2020). For instance, LAB bacteriocin produced by Lactococcus lactis could be completely inactivated by alkali treatment at pH 12 for 2 h (Pmka et al., 2019). In this study, the antibacterial activity of LFX01 retained 26.26% of the control under pH 12 for 2 h. Additionally, the antibacterial activity of Lactobacillus rhamnosus zrx01 bacteriocin and L. plantarum B21 bacteriocin significantly reduced by at least 50% when heat treatment exceeded 100°C for 30 min (Golneshin et al., 2020; Zhao et al., 2020). Lactobacillus acidophilus NX2-6 bacteriocin exerted 20.00% antibacterial activity of the control under 121°C for 1 h (Meng et al., 2020). In this study, the antibacterial activity of LFX01 retained approximately 72.32–56.06 % of the control after treatment at 100 and 121°C for 30 min, respectively. Certainly, several previously reported LAB bacteriocins exhibited excellent thermal stability similar to that of LFX01 (Ge et al., 2016; Pmka et al., 2019; Li et al., 2021). These results suggested that LFX01 possesses excellent acid-base and temperature tolerance effects. Diversity in the thermal stability and acid-base tolerance of LAB bacteriocins confirms the significant potentiality of LAB bacteriocins as an antibacterial agent in a wide range of temperature and acid-base treatments of food by their combination.
The MIC of LFX01 against S. flexneri_14 assessed in this study was close to or lower than the previously purified LAB bacteriocins (Yi et al., 2018; Wang et al., 2019). For instance, MIC of XSJ01 against S. aureus was 9.85 μg/mL (Li et al., 2021), MIC of bacteriocin BMP11 against various foodborne pathogenic bacteria (e.g., S. flexneri, L. monocytogenes, Cronobacter sakazakii, and S. aureus) was 0.3–38.4 μg/mL (Yi et al., 2018), MIC of bacteriocin BM1157 against S. aureus was 25.0 μg/mL (Yi et al., 2020). This mostly attributed to good killing effects of LFX01 on planktonic S. flexneri_14 cells. Especially, the cell viability of S. flexneri_14 significantly reduced after treatment with LFX01 in the XTT experiments. However, planktonic Enterococcus faecalis cells exhibited no reduction with Pediococcus acidilactici bacteriocin during XTT assay (Yoon and Kang, 2020). P. acidilactici bacteriocin hardly retained the cell viability of Pseudomonas aeruginosa cells (Lee et al., 2020). Notably, XTT assay revealed that the cell viability of S. aureus cells was reduced by 47% after treating with LAB bacteriocins (produced by L. salivarius) (Li et al., 2021). Therefore, LAB bacteriocins presented better reduction effects on cell viability of planktonic foodborne pathogens than non-LAB bacteriocins.
In this study, different MICs of LFX01 were used to inhibit the formation of S. flexneri_14 biofilm. The quantitative analysis showed that LFX01 could significantly inhibit S. flexneri_14 biofilm formation. Further observation revealed that with the increase of MIC concentration, cell density of S. flexneri_14 biofilm gradually decreased. Notably, the biofilm formation was effectively reduced at the concentration of 1/2 × MIC, and the biofilm was hardly observed at the concentration of 2 × MIC, indicating an effective concentration of LFX01 under different antibiofilm requirements. These results confirmed LFX01 as an effective inhibitor of S. flexneri_14 biofilm formation. Therefore, LFX01 could be further developed to LAB bacteriocin products for inhibiting S. flexneri_14 biofilm. Despite these findings, the mechanisms underlying the antibacterial effects of bacteriocins on foodborne pathogens remain indefinite. The SEM observation revealed uneven shape, wrinkled surface, disruptive cells, content leakage, and perforations on the cell surface, which was consistent with the previous study results (Sun et al., 2018; Hossain et al., 2020; Lanhua et al., 2020). Previous studies have concluded that bacteriocins exposure could interfere with DNA metabolism, resulting in cytoplasmic content leakage and decreased activity of key kinases, eventually causing cell death of pathogenic bacteria (Yi et al., 2018; Li et al., 2021; Zhu et al., 2021). Therefore, antibacterial effects of LFX01 on biofilm formation and activity of planktonic S. flexneri_14 cells are primarily attributed to the reduced metabolic activity and perforated cell membrane.
Conclusion
In summary, a novel bacteriocin LFX01 produced by L. plantarum was purified and characterized for antibacterial activities. LFX01 exhibited excellent thermal stability and acid-base tolerance. Moreover, the antibacterial activity of LFX01 against S. flexneri_14 showed a reduced rate after treatment with proteases. Furthermore, LFX01 significantly decreased the cell viability of S. flexneri_14 and terminated planktonic bacterial cells. LFX01 significantly inhibited the S. flexneri_14 biofilm formation at low concentrations. Therefore, the antibacterial effects of LFX01 against S. flexneri_14 were attributed to the decreased bactericidal activity and the S. flexneri_14 cell membrane injury. This study suggests that LFX01 could be a promising alternative agent to control S. flexneri_14 in foods.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The experimental protocol was approved by the Ethical Committee of Research of Kunming University of Science and Technology.
Author Contributions
Q-LZ, X-YD, and L-BL conceived and designed the research. Y-HJ and W-GX conducted the experiments. Q-LZ and X-YD contributed new reagents and analytical tools. Y-HJ, W-GX, and X-YD analyzed the data and wrote the manuscript. X-YD guided the experiments. All authors read and approved the manuscript.
Funding
This study was supported by Yunnan Fundamental Research Projects (Grant No. 202101BE070001-046).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.779315/full#supplementary-material
Supplementary Figure 1 | Analysis of the amino acid composition of LFX01.
References
Am, A., Gbb, C., Pm, D., Akr, B., Gc, E., and Kg, A. (2019). Antibacterial substances produced by pathogen inhibitory gut bacteria in labeo rohita: physico-chemical characterization, purification and identification through MALDI-TOF mass spectrometry-sciencedirect. Microb. Pathog. 130, 146–155. doi: 10.1016/j.micpath.2019.02.028
Baranzan, W. S., and Koshy, P. (2018). Purification, characterization, mode of action and enhanced production of Salivaricin mmaye1, a novel bacteriocin from Lactobacillus salivarius SPW1 of human gut origin. Electron. J. Biotechnol. 35, 39–47. doi: 10.1016/j.ejbt.2018.08.003
Bu, Y. S., Liu, Y. X., Li, J. X., Liu, T. J., Gong, P., Zhang, L. W., et al. (2021). Analyses of plantaricin Q7 synthesis by Lactobacillus plantarum Q7 based on comparative transcriptomics. Food Control. 124:107909. doi: 10.1016/j.foodcont.2021.107909
Chen, L., Gu, Q., Li, P., Li, Y. J., Song, D. F., and Yang, J. (2018). Purification and characterization of plantaricin ZJ316, a novel bacteriocin against Listeria monocytogenes from Lactobacillus plantarum ZJ316. J. Food Prot. 81, 1929–1935. doi: 10.4315/0362-028X.JFP-18-306
Cui, X. Y., Jian, W., Chao, J. Y., Bei, B. L., Qiu, X. M., Sheng, J. Y., et al. (2015). Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China. Front. Microbiol. 6:00435. doi: 10.3389/fmicb.2015.00435
Darvishi, N., Fard, N. A., and Sadrnia, M. (2021). Genomic and proteomic comparisons of bacteriocins in probiotic species Lactobacillus and Bifidobacterium and inhibitory ability of Escherichia coli MG 1655. Biotechnol. Rep. 31:e00654. doi: 10.1016/j.btre.2021.e00654
Enriqueta, G. G., Paula, M. O., Ian, J. C., Natalia, M. V., Juan, M. R., Melinda, J., et al. (2020). Production of multiple bacteriocins, including the novel bacteriocin gassericin M, by Lactobacillus gasseri LM19, a strain isolated from human milk. Appl. Microbiol. Biotechnol. 104, 3869–3884. doi: 10.1007/s00253-020-10493-3
Ge, J., Sun, Y., Xin, X., Wang, Y., and Ping, W. (2016). Purification and partial characterization of a novel bacteriocin synthesized by Lactobacillus paracasei HD1-7 isolated from Chinese Sauerkraut Juice. Sci. Rep. 6:19366. doi: 10.1038/srep19366
Golneshin, A., Gor, M. C., Vezina, B., Williamson, N., and Smith, A. T. (2020). Discovery and characterisation of novel circular bacteriocin plantacyclin B21AG from Lactobacillus plantarum B21. Heliyon 6:e04715. doi: 10.1016/j.heliyon.2020.e04715
Goodfellow, M. K., Buss, H. J., Trujillo, M. E., Suzuki, K., Ludwig, W., and Whitman, W. B. (2012). “Begey’s manual of systematic bacteriology,” in Vol 5 The Actinobateria, Part A, B, 2nd Edn, eds W. B. Whitman, M. Goodfellow, P. Kämpfer, H. J. Busse, M. E. Trujillo, W. Ludwig, et al. (New York, NY: Springer), doi: 10.1007/978-0-387-21609-6
Guo, X., Chen, J., Sun, H., Luo, L., Gu, Y., and Yi, Y. (2020). Mining, heterologous expression, purification and characterization of 14 novel bacteriocins from Lactobacillus rhamnosus LS-8. Int. J. Biol. Macromol. 164, 2162–2176. doi: 10.1016/j.ijbiomac.2020.08.067
He, X., Deng, Z. Y., Wang, F., Zhang, Q. L., Lin, L. B., and Deng, X. Y. (2021). Isolation and biological characterization of a bacteriophage infecting Shigella flexneri. Microbiol. China 48, 3165–3175. doi: 10.13344/j.microbiol.china.201179
Hossain, M. L., Mizan, M. R., Ashrafudoulla, M., Nahar, S., Joo, H. J., Jahid, I., et al. (2020). Inhibitory effects of probiotic potential lactic acid bacteria isolated from kimchi against Listeria monocytogenes biofilm on lettuce, stainless-steel surfaces, and MBEC™ biofilm device. LWT Food Sci. Technol. 118:108864. doi: 10.1016/j.lwt.2019.108864
Jabbar, A. H., and Al-azawi, I. (2020). Molecular detection and genetic characterization of resistance genes in Shigella spp isolates in Al-Diwaniyah city. Int J Psychosoc Rehabil. 24, 1998–2006.
Kang, J., Liu, L., Liu, Y., and Wang, X. (2020). Ferulic acid inactivates Shigella flexneri through cell membrane destructieon, biofilm retardation, and altered gene expression. J. Agric. Food Chem. 68, 7121–7131. doi: 10.1021/acs.jafc.0c01901
Kaoukab, R. A., Latéfa, B., and Allaoui, A. (2020). Inactivation of the sfgtr4 gene of Shigella flexneri induces biofilm formation and affects bacterial pathogenicity. Microorg. 8:841. doi: 10.3390/microorganisms8060841
Lanhua, Y., Luo, L., Chen, J., Sun, H., Xin, W., Yi, Y., et al. (2020). Cell wall and DNA damage of Staphylococcus aureus by bacteriocin BM1157. LWT Food Sci. Technol. 134, 109842. doi: 10.1016/j.lwt.2020.109842
Lee, D. H., Kim, B. S., and Kang, S. S. (2020). Bacteriocin of Pediococcus acidilactici HW01 inhibits biofilm formation and virulence factor production by Pseudomonas aeruginosa. Probiotics Antimicrob. Proteins. 12, 73–81. doi: 10.1007/s12602-019-09623-9
Lei, S., Zhao, R., Sun, J., Ran, J., and Zhu, Y. (2020). Partial purification and characterization of a broad: pectrum bacteriocin produced by a Lactobacillus plantarum zrx03 isolated from infant’s feces. Food Sci. Nutr. 8, 2214–2222. doi: 10.1002/fsn3.1428
Li, H. W., Xiang, Y. Z., Zhang, M., Jiang, Y. H., Zhang, Y., Liu, Y. Y., et al. (2021). A novel bacteriocin from Lactobacillus salivarius against Staphylococcus aureus: isolation, purification, identification, antibacterial and antibiofilm activity. LWT Food Sci. Technol. 140:110826. doi: 10.1016/j.lwt.2020.110826
Li, L. H., Chen, T. L., Chiu, H. W., Hsu, C. H., Wang, C. C., Tai, T. T., et al. (2020). Critical role for the NLRP3 inflammasome in mediating IL-1β production in Shigella sonnei-infected macrophages. Front. Immunol. 11:01115. doi: 10.3389/fimmu.2020.01115
Li, S. W., Chen, Y. S., Lee, Y. S., Yang, C. H., Srionnual, S., Wu, H. C., et al. (2017). Comparative genomic analysis of bacteriocin-producing Weissella cibaria 110. Appl. Microbiol. Biotechnol. 101, 1227–1237. doi: 10.1007/s00253-016-8073-8
Li, Z. R., Han, J. C., Liu, R. X., Liu, D. Y., and Liang, J. F. (2020). Purification and characterization of bacteriocin produced by Lactobacillus pentosus LS1. Food Sci. 141, 112–118. doi: 10.7506/spkx1002-6630-20190826-278
Lindsay, D., and Von, H. A. (2006). What food safety professionals should know about bacterial biofilms. Br. Food J. 108, 27–37. doi: 10.1108/00070700610637616
Mardaneh, J., Poor, S. A., and Afrugh, P. (2013). Prevalence of Shigella species and antimicrobial resistance patterns of isolated strains from infected pediatrics in Tehran. Int. J. Enteric. Pathog. 1, 28–31. doi: 10.17795/ijep10705
Meng, F., Zhu, X., Zhao, H., Nie, T., and Lu, Y. (2020). A class III bacteriocin with broad-spectrum antibacterial activity from Lactobacillus acidophilus NX2-6 and its preservation in milk and che Saimese. Food Control. 121:107597. doi: 10.1016/j.foodcont.2020.107597
Messaoudi, S., Kergourlay, G., Dalgalarrondo, M., Choiset, Y., Ferchichi, M., and Pr’evost, H. (2012). Purification and characterization of a new bacteriocin active against Campylobacter produced by Lactobacillus salivarius SMXD51. Food Microbiol. 32, 129–134. doi: 10.1016/j.fm.2012.05.002
Nisa, I., Qasim, M. Q., Driessen, A., Nijland, J., Bari, F., Haroon, M., et al. (2020). Molecular epidemiology of Shigella flexneri isolated from pediatrics in a diarrhea-endemic area of Khyber Pakhtunkhwa, Pakistan. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1–15. doi: 10.1007/s10096-020-03811-0
Nové, M., Kincses, A., Szalontai, B., Rácz, B., Blair, J., González, P. A., et al. (2020). Biofilm eradication by symmetrical selenoesters for food-borne pathogens. Microorganisam 8:566. doi: 10.3390/microorganisms8040566
Paul, A. N., Fokam, T. M., Olivier, F., Mukam, J., Wambe, H., and Kamgang, R. (2019). Effects of Crinum jagus water/ethanol extract on Shigella flexneri-induced Diarrhea in Rats. Evid Based Compl Alt. 2:9537603. doi: 10.1155/2019/9537603
Peng, S. D., Song, J. J., Zeng, W. Y., Wang, H. W., Zhang, Y., Xin, J. W., et al. (2021). A broad-spectrum novel bacteriocin produced by Lactobacillus plantarum SHY 21–2 from yak yogurt: purification, antimicrobial characteristics and antibacterial mechanism. LWT Food Sci. Technol. 142:110955. doi: 10.1016/j.lwt.2021.110955
Pmka, C., Emfk, A., Mltt, A., Jbt, A., Cw, B., and Fzn, A. (2019). Nisin-producing Lactococcus lactis subsp. Lactis 2MT isolated from freshwater Nile tilapia in Cameroon: bacteriocin screening, characterization, and optimization in a low-cost medium. LWT Food Sci. Technol. 107, 272–279. doi: 10.1016/j.lwt.2019.03.007
Ranganathan, S., Doucet, M., Grassel, C. L., Delaine-Elias, B. O., and Barry, E. M. (2019). Evaluating S. flexneri pathogenesis in the human enteroid model. Infect. Immun. 87:e00740. doi: 10.1128/IAI.00740-18
Rasheed, H. A., Tuoheti, T., Zhang, Y., Fidelis, A., and Dong, M. (2020). Purification and partial characterization of a novel bacteriocin produced by bacteriocinogenic Lactobacillus fermentum BZ532 isolated from Chinese fermented cereal beverage (Bozai). LWT Food Sci. Technol. 124:109113. doi: 10.1016/j.lwt.2020.109113
Rey, C., Chang, Y. Y., Latour-Lambert, P., Varet, H., Proux, C., Legendre, R., et al. (2020). Transcytosis subversion by M cell-to-enterocyte spread promotes Shigella flexneri and Listeria monocytogenes intracellular bacterial dissemination. PLoS Pathog. 16:e1008446. doi: 10.1371/journal.ppat.1008446
Saima, S. A., Abbas, F., and Roomeela, S. S. (2018). Isolation & Identification of Shigella species from food and water samples of Quetta, Pakistan. Pure Appl. Biol. 7, 227–235. doi: 10.19045/bspab.2018.70027
Shahin, K., Bouzari, M., and Wang, R. (2019). Complete genome sequence analysis of a lytic Shigella flexneri vB-SflS-ISF001 bacteriophage. Turk. J. Biol. 43, 99–112. doi: 10.3906/biy-1808-97
Subasinghe, R. (2017). FAO Fisheries and Aquaculture Circular FIAA/C1140 (En) World Aquaculture 2015: A Brief Overview. Rome: FAO.
Sun, Z., Wang, X., Zhang, Wu, H., Zou, Y., Li, P., et al. (2018). Class III bacteriocin Helveticin-M causes sublethal damage on target cells through impairment of cell wall and membrane. J. Ind. Microbiol. Biotechnol. 45, 213–227. doi: 10.1007/s10295-018-2008-6
Sung, H. S., and Jo, Y. L. (2019). Purification and characterization of an antibacterial substance from Aerococcus urinaeequi strain HS36. J. Microbiol. Biotechnol. 30, 93–100. doi: 10.4014/jmb.1910.10015
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tsukatani, T., Sakata, F., Kuroda, R., and Akao, T. (2020). Biofilm eradication activity of herb and spice extracts alone and in combination against oral and food-borne pathogenic bacteria. Curr. Microbiol. 77, 2486–2495. doi: 10.1007/s00284-020-02017-z
Voulgari, K., Hatzikamari, M., Delepoglou, A., Georgakopoulos, P., Tzanetaki, L. E., and Tzanetakis, N. (2010). Antifungal activity of non-starter lactic acid bacteria isolates from dairy products. Food Control. 21, 136–142. doi: 10.1016/j.foodcont.2009.04.007
Wang, Y., Qin, Y., Zhang, Y., Wu, R., and Li, P. (2019). Antibacterial mechanism of plantaricin LPL-1, a novel class IIa bacteriocin against Listeria monocytogenes. Food Control. 97, 87–93. doi: 10.1016/j.foodcont.2018.10.025
Wayah, S. B., and Philip, K. (2018). Pentocin MQ1: a novel, broad-spectrum, poreforming bacteriocin from Lactobacillus pentosus CS2 with quorum sensing regulatory mechanism and biopreservative potential. Front. Microbiol. 9:564. doi: 10.3389/fmicb.2018.00564
Xiang, Y. Z., Zhang, Y. M., Liu, Y. Y., Zhang, M., Lin, L. B., and Zhang, Q. L. (2021). Purification, characterization, and antibacterial and antibiofilm activity of a novel bacteriocin against Salmonella Enteritidis. Food Control. 127:108110. doi: 10.1016/j.foodcont.2021.108110
Yi, L., Li, X., Luo, L., Lu, Y., Yan, H., Qiao, Z., et al. (2018). A novel bacteriocin BMP11 and its antibacterial mechanism on cell envelope of Listeria monocytogenes and Cronobacter sakazakii. Food Control. 91, 160–169. doi: 10.1016/j.foodcont.2018.03.038
Yi, L., Luo, L., Chen, J., Sun, H., Wang, X., and Yi, Y. (2020). Cell wall and DNA damage of Staphylococcus aureus by bacteriocin BM1157. LWTFood Sci. Technol. 134:109842. doi: 10.1016/J.LWT.2020.109842
Yoon, J., and Kang, S. S. (2020). In vitro antibiofilm and anti-inflammatory properties of bacteriocins produced by Pediococcus acidilactici against Enterococcus faecalis. Front. Microbiol. 17:764–771. doi: 10.1089/fpd.2020.2804
Zhang, J., Liu, G., Shang, N., Chen, W., Chen, S., and Li, P. (2009). Purification and partial amino acid sequence of pentocin 31-1, an anti-Listeria bacteriocin produced by Lactobacillus pentosus 31-1. J. Food Prot. 72:2524. doi: 10.4315/0362-028x-72.12.2524
Zhang, Y., Yang, J., Liu, Y., Wu, Y., Fang, Z., Wang, Y., et al. (2019). A novel bacteriocin PE-ZYB1 produced by Pediococcus pentosaceus zy-B isolated from intestine of Mimachlamys nobilis: purification, identification and its anti-listerial action. LWT Food Sci. Technol. 118:108760. doi: 10.1016/j.lwt.2019.108760
Zhao, R., Lu, R., Ran, J., Li, G., Lei, G., Zhu, Y., et al. (2020). Purification and characterization of bacteriocin produced by Lactobacillus rhamnosus zrx01. Food Biosci. 38:100754. doi: 10.1016/j.fbio.2020.100754
Zheng, J. S., Wittouck, S., Salvetti, E., Franz, C., Harris, H. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Zhou, Y., Kong, X., Mei, W., and Tang, C. (2018). Probiotic characteristics of a bacteriocin-producing Lactobacillus plantarum strain from yak. Food Sci. 39, 132–137. doi: 10.7506/spkx1002-6630-201814020
Keywords: tilapia, Lactiplantibacillus plantarum, bacteriocin, Shigella flexneri, antibiofilm properties
Citation: Jiang Y-H, Xin W-G, Zhang Q-L, Lin L-B and Deng X-Y (2022) A Novel Bacteriocin Against Shigella flexneri From Lactiplantibacillus plantarum Isolated From Tilapia Intestine: Purification, Antibacterial Properties and Antibiofilm Activity. Front. Microbiol. 12:779315. doi: 10.3389/fmicb.2021.779315
Received: 18 September 2021; Accepted: 03 December 2021;
Published: 05 January 2022.
Edited by:
Mostafa Ghanem, University of Maryland, College Park, United StatesReviewed by:
Huaxi Yi, Ocean University of China, ChinaKoshy Philip, University of Malaya, Malaysia
Copyright © 2022 Jiang, Xin, Zhang, Lin and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian-Yu Deng, dengxy1008@126.com
†These authors have contributed equally to this work
 Yu-Hang Jiang
Yu-Hang Jiang Wei-Gang Xin
Wei-Gang Xin Qi-Lin Zhang
Qi-Lin Zhang Lian-Bing Lin
Lian-Bing Lin