- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Ministry of Agriculture (MOA) Laboratory of Quality and Safety Risk Assessment for Agro-Products (Hangzhou), Institute of Agro-Product Safety and Nutrition, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2Key Laboratory of Food Safety Risk Assessment, National Health Commission of the People’s Republic of China, China National Center for Food Safety Risk Assessment, Beijing, China
- 3College of Food and Pharmaceutical Sciences, Ningbo University, Ningbo, China
- 4College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 5Department of Biological and Agricultural Engineering, University of Arkansas, Fayetteville, AR, United States
Chlorine disinfectants have been widely used in the poultry supply chain but this exposure can also result in the development of bacterial tolerance to chlorine and this is often linked to antibiotic cross-resistance. The objectives of this study were to investigate sodium hypochlorite (NaClO) tolerance of Salmonella isolated from poultry supply chains and evaluate cross-resistance. We collected 172 Salmonella isolates from poultry farms, slaughter houses and retail markets in China during 2019–2020. We found that S. Enteritidis, S. Kentucky, and S. Typhimurium constituted > 80% of our Salmonella isolates. Overall, 68% of Salmonella isolates were resistant to > 3 antibiotics and S. Kentucky displayed a significantly (p > 0.05) higher frequency (93.2%) of multidrug resistance than the other serovars. Tolerance to chlorine at MIC > 256 mg/L was detected in 93.6% of isolates (161/172) and tolerant isolates displayed higher decimal reduction times (D value) and less ultrastructural damage than did the suspectable strains under chlorine stress. Spearman analysis indicated significant positive correlations between chlorine tolerance (evaluated by the OD method) and antibiotic resistance (p < 0.05) to ceftiofur, tetracycline, ciprofloxacin and florfenicol and this was most likely due to efflux pump over-expression. The most frequently detected chlorine resistance gene was qacEΔ1 (83.1%, n = 143) and we found a positive correlation between its presence and MIC levels (r = 0.66, p < 0.0001). Besides, we found weak correlations between chlorine-tolerance and antibiotic resistance genes. Our study indicated that chlorine disinfectants most likely played an important role in the emergence of chlorine tolerance and spread of antibiotic resistance and therefore does not completely control the risk of food-borne disease. The issue of disinfectant resistance should be examined in more detail at the level of the poultry production chain.
Introduction
Salmonella is a frequent cause of gastroenteritis in humans and is an important public health concern worldwide and in China causes 9.87 M gastroenteritis cases annually (Xiao et al., 2019; Yang et al., 2020a). More than 2600 Salmonella enterica serovars have been identified and S. Enteritidis, S. Typhimurium, S. Kentucky, and S. Indiana are the most frequent causes of human salmonellosis in China (Andoh et al., 2016; Elnekave et al., 2018; Zeng et al., 2021). Poultry products are the most common vehicles for Salmonella transmission and a recent study in China found that 37.5% of poultry samples were contaminated with Salmonella (Yang et al., 2020b). Contamination sources include the animals when introduced into the poultry house environment and direct or cross-contamination between poultry carcasses. All these events can result in foodborne salmonellosis when improperly cooked or handled products are consumed (Xiao et al., 2019).
Chlorine disinfection is one of the most common disinfection technologies for controlling the risks of microorganisms in industrial processing (Jia et al., 2020). In this process, bacteria can survive and even reproduce in residual chlorine and are designated chlorine tolerant (Langsrud et al., 2003; Luo et al., 2020). In particular, Salmonella possesses a high level of regeneration capacity in reclaimed water after chlorine disinfection (Li et al., 2013). Recent studies have focused on chlorine tolerance of bacteria in water while few studies addressed contamination on the processed poultry products (Roy and Ghosh, 2017; Bhojani et al., 2018; Wang et al., 2019; Luo et al., 2020). In China, 50–100 mg/L of sodium hypochlorite (NaClO) is commonly used in poultry processing and environmental disinfection while the inactivation of Salmonella was limited (< 1 Log) and could be due to chlorine tolerance (Jun et al., 2013; Lee et al., 2014; Xiao et al., 2019).
Current studies have also demonstrated that long-term antibiotic usage during animal breeding has led to a marked increase in the levels of antibiotic resistance (Zeng et al., 2021). Antibiotic susceptibility testing of the 318 poultry-associated Salmonella revealed that only 5 (1.6%) were susceptible to all 22 tested antibiotics, while 191 (60.1%) exhibited multidrug resistance. Poultry frequently harbor antibiotic-resistant Salmonella isolates that can be transmitted to humans via the food chain (Yang et al., 2020a). The emergence of quinolone, tetracycline and extended-spectrum β-lactam-resistant Salmonella is a serious public health concern (Mechesso et al., 2020). Cross-resistance between disinfectants and antibiotics is also becoming widespread for bacterial pathogens (Carey and McNamara, 2015; Puangseree et al., 2021). For example, a higher level of chlorine tolerance in antibiotic-resistant Escherichia coli (E. coli) was found when compared to antibiotic-susceptible strains (Templeton et al., 2013). Bacterial exposure to chlorine increases the expression of efflux pumps and activates qac genes that also export chloramphenicols, sulfonamide and β-lactams (Karumathil et al., 2014). However, chlorine tolerance and cross-resistance of Salmonella that are present in the poultry supply chain have not been fully investigated.
The focus of the current study was to investigate (i) chlorine tolerance and antibiotic resistance in Salmonella spp. isolated from poultry supply chains and (ii) to determine the level of cross-resistance in these isolates. This study examined on a broad scale the important role played by chlorine disinfectants as a non-antibiotic selective pressure for the emergence and spread of antibiotic resistance.
Materials and Methods
Salmonella Isolates and Serovar Identification
We isolated 172 Salmonella spp. from poultry supply chains as follows: poultry farms (n = 10), slaughter houses (n = 102) and retail markets (n = 60) in Guangzhou, Shandong and Zhejiang provinces during 2019–2020. Salmonella isolates were subcultured and serotyped by slide agglutination using commercial O and H antisera to distinguish Salmonella serovars (SSI-Diagnostica, Copenhagen, Denmark; Tianrun Bio-Pharmaceutical, Ningbo, China) in accordance with the White-Kauffmann-Le Minor scheme (Grimont and Weill, 2007; Slotved et al., 2016). All isolates were stored in brain heart infusion (BHI) broth (Becton Dickinson, Franklin Lakes, NJ, United States) containing 20% glycerol at −80°C until use.
NaClO Tolerance Determinations
Determination of Minimum Inhibitory Concentrations and Optical Density
The MIC values for NaClO in the Salmonella isolates were measured using broth microdilution. The E. coli ATCC 29522 and S. Enteritidis CVCC 1806 were included as controls for susceptibility testing. Bacterial suspensions were prepared by suspending 3–5 individual overnight colonies from trypticase soy (TSA) agar (Becton Dickinson) plates into 3 mL of 0.9% saline, equivalent to the turbidity of a 0.5 McFarland standard. The 0.5 McFarland inoculum suspensions were further diluted at 1:100 in Mueller-Hinton (MH) broth (Becton Dickinson). NaClO stock solutions containing 56.8 mg/mL chlorine was purchased from Sangon Biotech (Shanghai, China) and were prepared by dilution in sterile Milli-Q water (PALL, Buckinghamshire, United Kingdom). Chlorine concentrations were determined using a Palintest ChlorSense meter (Gateshead, United Kingdom). Preliminary tests indicated the MIC of S. Enteritidis CVCC 1806 to NaClO was 256 mg/L and tolerance to NaClO was defined as > 256 mg/L. These MIC assays for chlorine tolerance were performed in 96-well microtiter test plates containing 100 μL NaClO solution and were inoculated with 100 μL of suspended bacterial cultures to a final inoculum density of 5 Log CFU/mL per well. The plates were sealed using a perforated plate seal and incubated at 37°C for 24 h. The MIC values of NaClO were recorded as the lowest concentration of NaClO where no visible growth was observed. Control wells containing bacteria and lacking chlorine and wells with MH broth were used as positive and negative controls, respectively. Each assay was repeated three times on different days. Cell growth in the plates was also quantified by measuring the optical density at 600 nm (OD600) (Bernardez and de Andrade Lima, 2015; Yang et al., 2020a) using a Polarstar spectrophotometer (Omega, Ortenberg, Germany).
Inactivation Kinetics
Chlorine susceptible and tolerant isolates were selected based on the MIC and OD600 tests and then separately incubated in BHI at 37°C for 24 h and cultured to approximately 9 Log CFU/mL. The initial inoculum level was 5 ± 0.2 Log CFU/mL. Cell suspensions were treated with 100 mg/L of NaClO for 0, 30, 60, 90, and 120 min. Then the suspension was put into a sterile centrifuge tube containing sodium thiosulfate (Na2S2O3) to instantaneously quench the residual disinfectant and 10-fold diluted in buffered peptone water (Becton Dickenson) and a 50 μL portion of appropriate dilutions were plated in duplicate onto TSA agar plates using a spiral plater (WASP 2, Don Whitley Scientific, Shipley, United Kingdom). The plates were incubated at 37°C for 18 h. Colonies on TSA agar plates were enumerated by a ProtoCOL 3 automated colony counter (Synbiosis, Cambridge, United Kingdom). The limit of detection was one colony per 50 μL sample (1.3 Log CFU/mL). Each treatment was repeated three times on different days and duplicate plates were used in microbial tests for each sample.
The decimal reduction time (D value) was used as an index to evaluate the bacterial inactivation rate vs. NaClO treatment as previously described (Luo et al., 2020). D values were calculated at the initial linear portion of survivor plots assuming the logarithmic numbers of bacterial growth were a linear function of the isothermal treatment time (Ahn et al., 2007). The log-linear model was used to investigate the pattern of pathogen inactivation after NaClO treatment as follows:
where Nt (CFU/mL) is the bacterial population at time t (s), N0 (CFU/mL) is the initial bacterial population, and D is the decimal reduction time (min) at a specific treatment condition.
Membrane Damage
Bacterial viability and cell properties between NaClO tolerant and susceptible bacteria were measured using flow cytometry (FCM) to quantify viable, dead and injured cells after treatment. Salmonella cell suspensions (8 Log CFU/mL) were treated with NaClO at 100 mg/L for 0, 10, and 30 min. A commercial live/dead staining kit (BacLight, L7012, Molecular Probes, Carlsbad, CA, United States) was used to distinguish living, dead, and damaged states of bacteria. In brief, 1 mL of bacteria solution was mixed with 1.5 μL SYTO 9 and 1.5 μL propidium iodide (PI) dyes. The mixture was kept in darkness at room temperature for 15 min. FCM tests were performed using a Accuri C6 flow cytometer (Becton Dickinson) equipped with lasers emitting at 488 and 640 nm. Some influences of impurities (such as large particles) on bacterial detection were excluded according to the difference in forward and side light scatter and 10,000 events were collected. The non-treated stained and non-stained cells served as control samples to adjust the detectors (Barros et al., 2021).
Cell structure damage under NaClO stress was observed using scanning electron (SEM, Hitachi 8010, Ibaraki, Japan) and transmission electron (TEM, Hitachi 7650, Ibaraki, Japan) microscopy. Salmonella cell suspensions (8 Log CFU/mL) were harvested by centrifugation following NaClO treatments (see below) and fixed with a 2.5% glutaraldehyde (Sangon Biotech, Shanghai, China) solution overnight at 4°C. The cells were centrifuged and the pellets were washed three times with 0.1 M sodium phosphate buffer solution. Each resuspension was serially dehydrated with 25, 50, 75, 90, and 100% ethanol, respectively, and examined using SEM and TEM as previously described (Tyagi and Malik, 2012).
Antibiotic Resistance Determination
Bacterial suspensions were prepared by suspending 3–5 individual colonies grown at 37°C for 18 h on TSA into 3 mL 0.9% saline, equivalent to the turbidity of a 0.5 McFarland standard. The 0.5 McFarland inoculum suspensions were further diluted at 1:100 in MH. A panel of antibiotic agents was reconstituted by adding 200 μL/well of the inoculum and incubated at 37°C for 18 h. The E. coli ATCC 25922 was used as the reference strain. Antibiotic susceptibility testing was performed using the broth microdilution method with the commercial Gram-negative antibiotic panel (Biofosun, Fosun Diagnostics, Shanghai, China) consisting of ampicillin (resistant, AMP ≥ 32 μg/mL), amoxicillin/clavulanate (resistant, AMC ≥ 32 μg/mL), cefotaxime (resistant, CTX ≥ 4 μg/mL), meropenem (resistant, MEM ≥ 4 μg/mL), amikacin (resistant, AMK ≥ 64 μg/mL), gentamicin (resistant, GEN ≥ 16 μg/mL), colistin (resistant, CS ≥ 2 μg/mL), ceftiofur (resistant, CEF ≥ 8 μg/mL), ciprofloxacin (resistant, CIP ≥ 1 μg/mL), sulfamethoxazole (resistant, T/S ≥ 4/76 μg/mL), tetracycline (resistant, TET ≥ 16 μg/mL), tigecycline (resistant, TIG ≥ 8 μg/mL), and florfenicol (resistant, FFC ≥ 16 μg/mL). The breakpoints for each antibiotic agent were set by the 2019 Clinical and Laboratory Standards Institute (CLSI) and the 2017 European Committee on Antimicrobial Susceptibility Testing (EUCAST). Bacteria that were susceptible to all 13 antibiotics were classified as drug-suspectable and bacteria resistant to 1 or 2 classes of antibiotics were termed drug resistant. Multidrug resistance was defined as resistance to 3 or more different classes of antibiotics.
Chlorine and Antibiotic Resistance Genes in Salmonella Isolates
DNA Extraction
DNA was extracted from pure Salmonella colonies using a QIAamp1 Mini DNA Extraction Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
PCR Detection of Chlorine and Antibiotic Resistance Genes
Antibiotic resistance and chorine resistance genes were selected based on previous studies (Hu et al., 2018, 2021; Chen et al., 2021) and the use of the ResFinder database.1 We screened for the presence of -eight chlorine resistance genes (exbD, motB, qacE, qacEΔ1, qacF, sugE, ybhR, and yedQ) and ten antibiotic resistance genes (blaOXA, blaCTX, blaTEM, qnrB6, qnrS7, tetG, tetA, floR, cat86, and cmx) using PCR assays (Table 1) as previously described (Wu et al., 2015). Amplicons were electrophoresed in 1% gel agarose gels containing EtBr and were visualized under UV light.
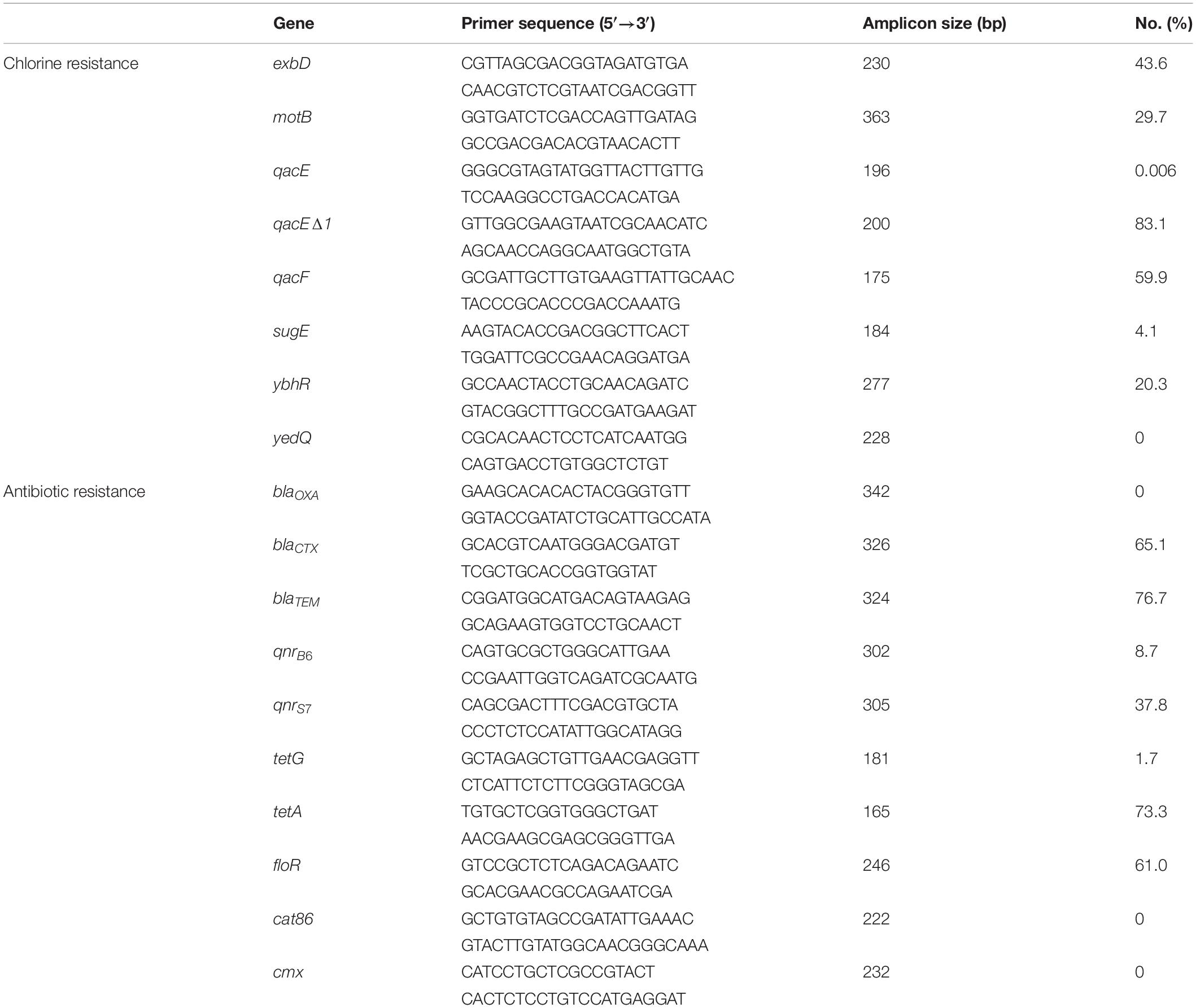
Table 1. Frequency of chlorine tolerance and antibiotic resistance genes determined by PCR among Salmonella isolates (n = 172).
Statistical Analysis
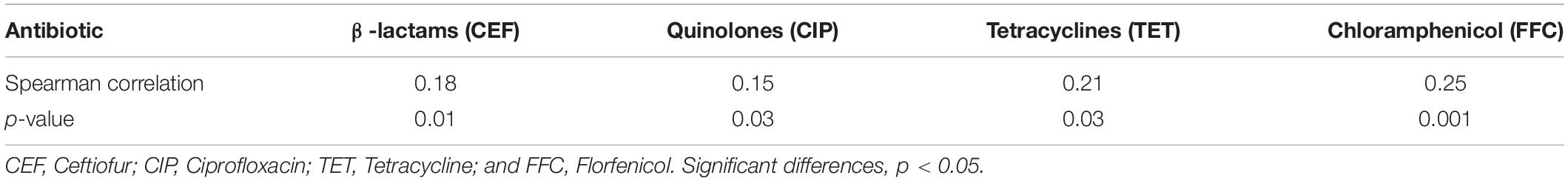
Cross-resistance between chlorine and antibiotics was assessed by comparing antibiotic MIC data with OD600 values growth in the presence of NaClO (128 mg/L) using the non-parametric Spearman correlation test in SPSS Statistics 20 software (IBM, Chicago, IL, United States). The Spearman coefficients ranged from –1 to + 1 indicating no association (r = 0) to monotonic relationships (r = –1 or + 1) (Schober et al., 2018). Significant differences were set at p < 0.05.
Results
Diversity of Salmonella Serotypes
We identified numerous Salmonella serovars from the wide range of samples we collected. These included samples from 10 farms (1 serotype), 102 slaughter houses (8 serotypes) and 60 retail markets (7 serotypes). A total of 10 serotypes were identified in 172 isolates and included S. Enteritidis, S. Kentucky, S. Typhimurium, S. Indiana, S. Agona, S. Stanley, S. Thompson, S. Derby, S. Mbandaka, and S. Montevideo. The most frequent serovars in the poultry supply chain were S. Enteritidis (77/172, 44.8%), S. Kentucky (44/172, 25.6%) and S. Typhimurium (18/172, 10.5%) and these accounted for > 80% of the total serovars isolated (Table 2).
NaClO Tolerance Evaluation
Minimum Inhibitory Concentrations and OD600 Determination
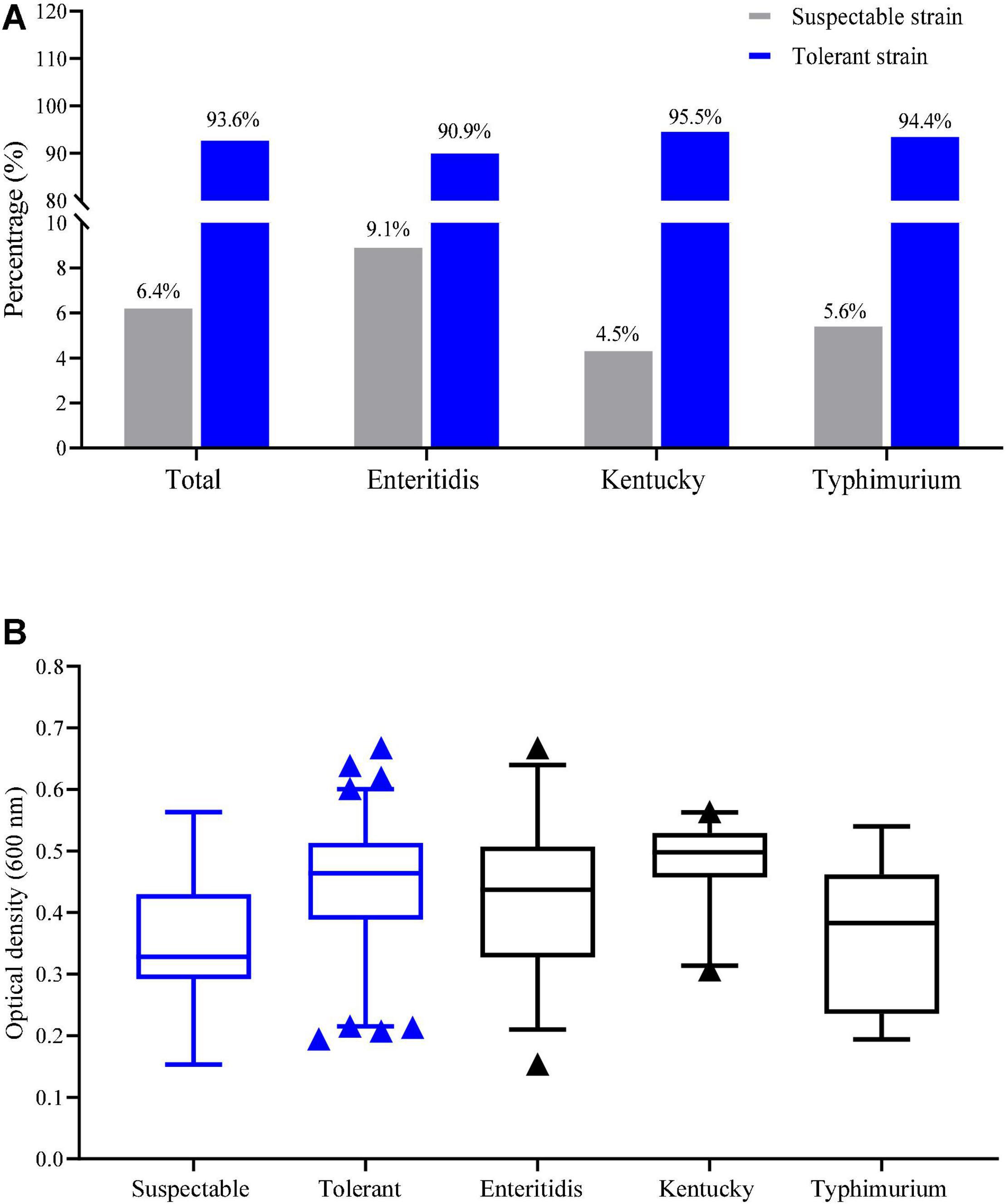
Tolerance to NaClO at MIC > 256 mg/L was detected in 93.6% of our Salmonella isolates (161/172). Tolerance based on the MIC values in the serovars S. Enteritidis, S. Kentucky and S. Typhimurium isolates were 90.9, 95.5, and 94.4%, respectively (Figure 1A). The average of OD600 values for tolerance (MIC > 256 mg/L) and susceptibility were 0.44 ± 0.10 and 0.33 ± 0.09, respectively and this difference was significant (p < 0.05). The average of OD600 values for growth in the presence of chlorine for S. Enteritidis, S. Kentucky and S. Typhimurium isolates were 0.42 ± 0.11, 0.48 ± 0.06, and 0.37 ± 0.11, respectively (Figure 1B). These data indicated that S. Kentucky displayed the highest chlorine tolerance compared with other two serotypes (p < 0.05).
Inactivation Kinetics
We more closely examined the mechanisms for choline tolerance using two defined laboratory strains that were susceptible (S. Enteritidis CVCC 1806) and tolerant (S. Enteritidis S2002-13). We found that growth inhibition for these strains significantly (p < 0.05) differed when cultured for 120 min in the presence of NaClO resulting in 0.83 ± 0.04 (S. Enteritidis CVCC 1806) and 0.46 ± 0.02 (S. Enteritidis S2002-13) Log CFU/mL, respectively. The data were fitted to the log-linear model and the D values of NaClO required to inactivate 1 Log unit for S. Enteritidis CVCC 1806 and S. Enteritidis S2002-13 were 175, and 249 min, respectively (Figure 2). These data indicated that S. Enteritidis S2002-13 displayed reduced susceptibility to NaClO.
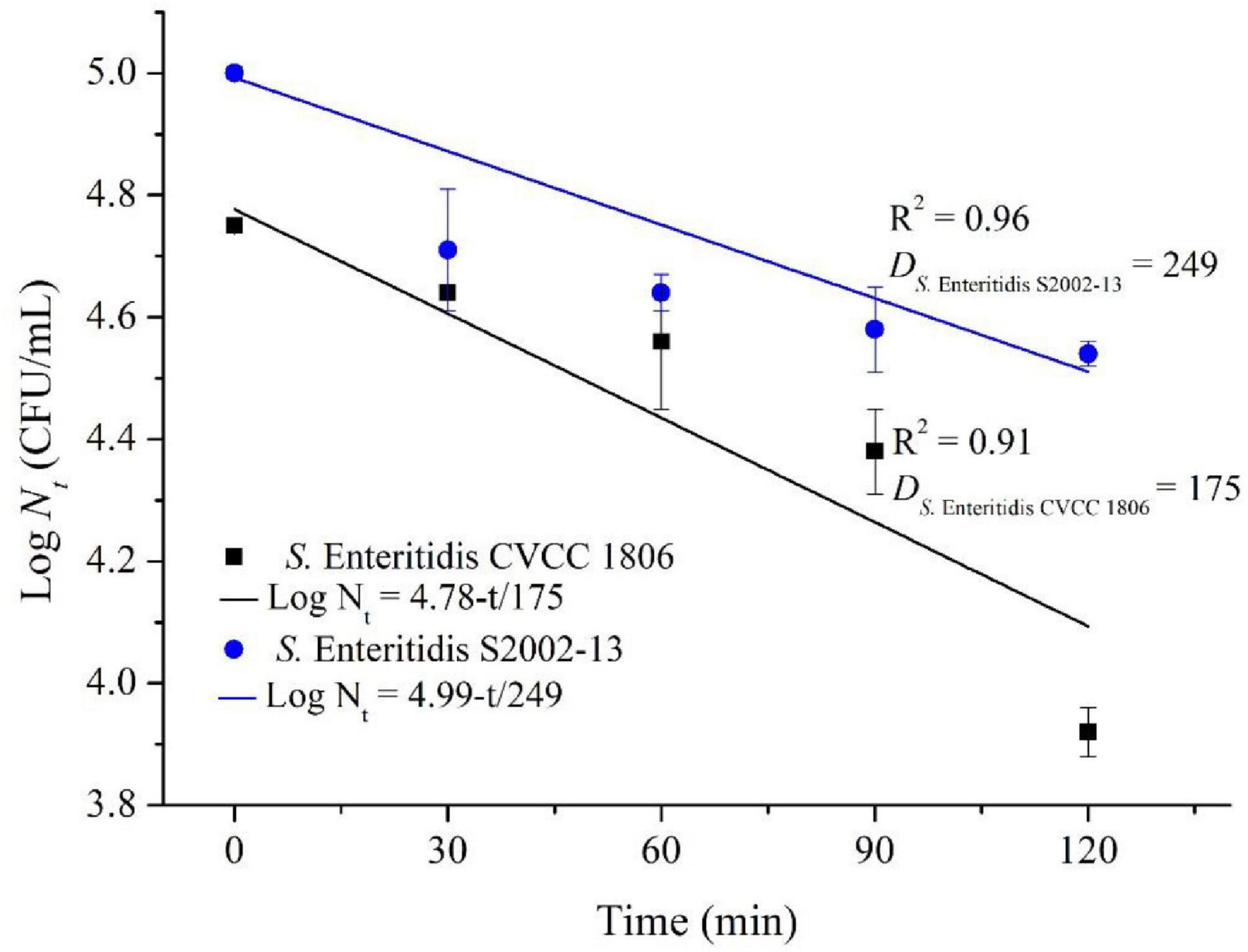
Figure 2. Inactivation kinetics of S. Enteritidis CVCC 1806 and S. Enteritidis S2002-13 exposed to concentration of NaClO at 100 mg/L for 0, 30, 60, 90, and 120 min.
Membrane Damage
We further assessed membrane damage to the bacteria using the membrane-impermeable green fluorescent nucleic acid dye SYTO9 and chlorine-treated cells had higher levels of staining (Figures 3A,D). Exposure to NaClO for 10 min resulted in staining of 11.6% of the susceptible S. Enteritidis CVCC 1806 and 6.9% of tolerant S. Enteritidis S2002-13 cells (Figures 3B,E). Exposure for 30 min resulted in 68% of S. Enteritidis CVCC 1806 as positive for PI staining compared with 19.2% of S. Enteritidis S2002-13 (Figures 3C,F). These data clearly showed that S. Enteritidis S2002-13 was more tolerant to NaClO.
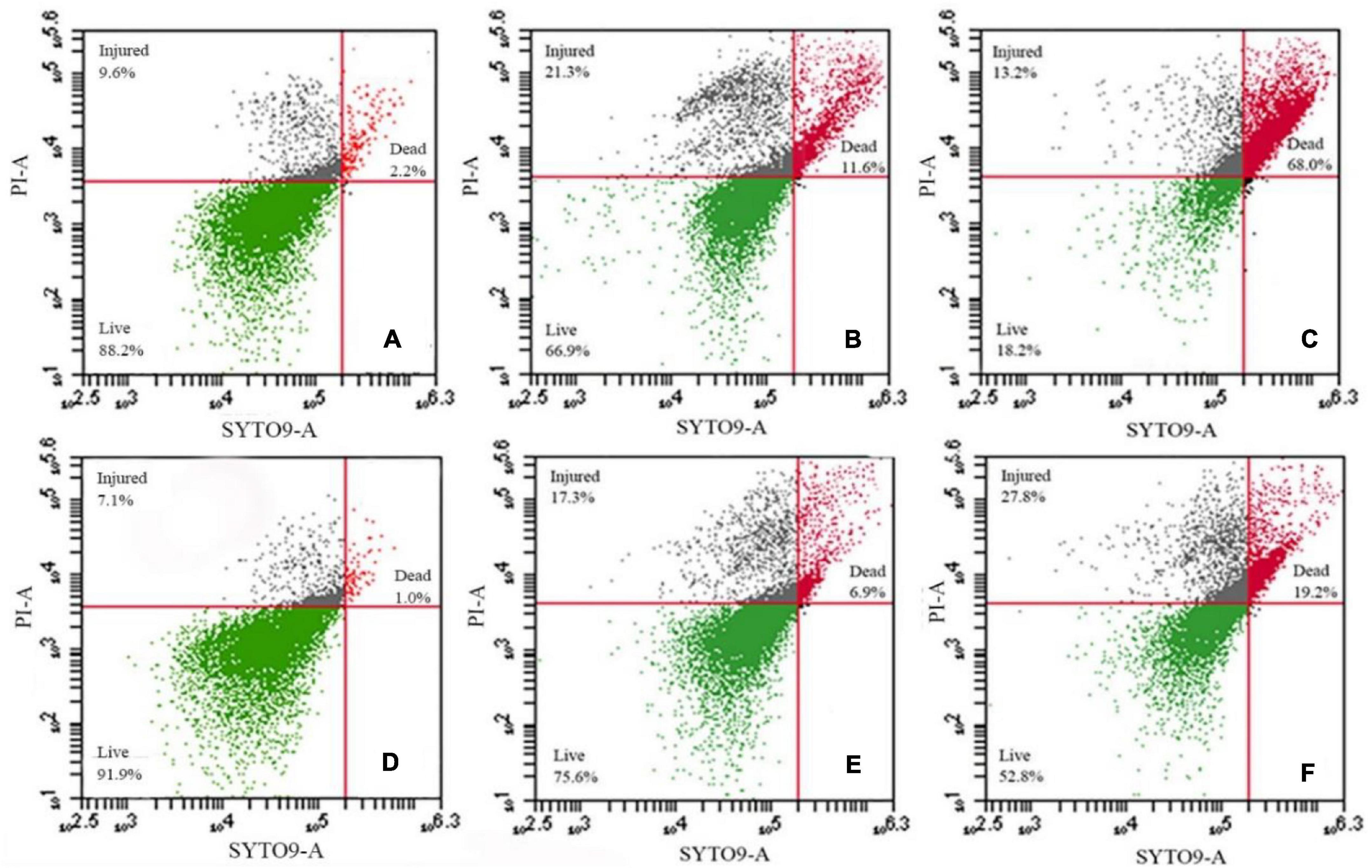
Figure 3. S. Enteritidis CVCC 1806 (A–C) and S. Enteritidis S2002-13 (D–F) stained with SYTO9 plus PI after treatment with 100 mg/L NaClO for 0, 10 and 30 min.
Ultrastructural analyses indicated the presence of severe cell ruptures for S. Enteritidis CVCC 1806 following NaClO treatment and cytoplasmic contents were displaced (Figures 4A–D). In contrast, structural damage was less common in S. Enteritidis S2002-13 and most cells showed no internal rearrangements of internal cellular content (Figures 4E–H). These data clearly demonstrated showed that S. Enteritidis S2002-13 was more tolerant to NaClO than S. Enteritidis CVCC 1806.
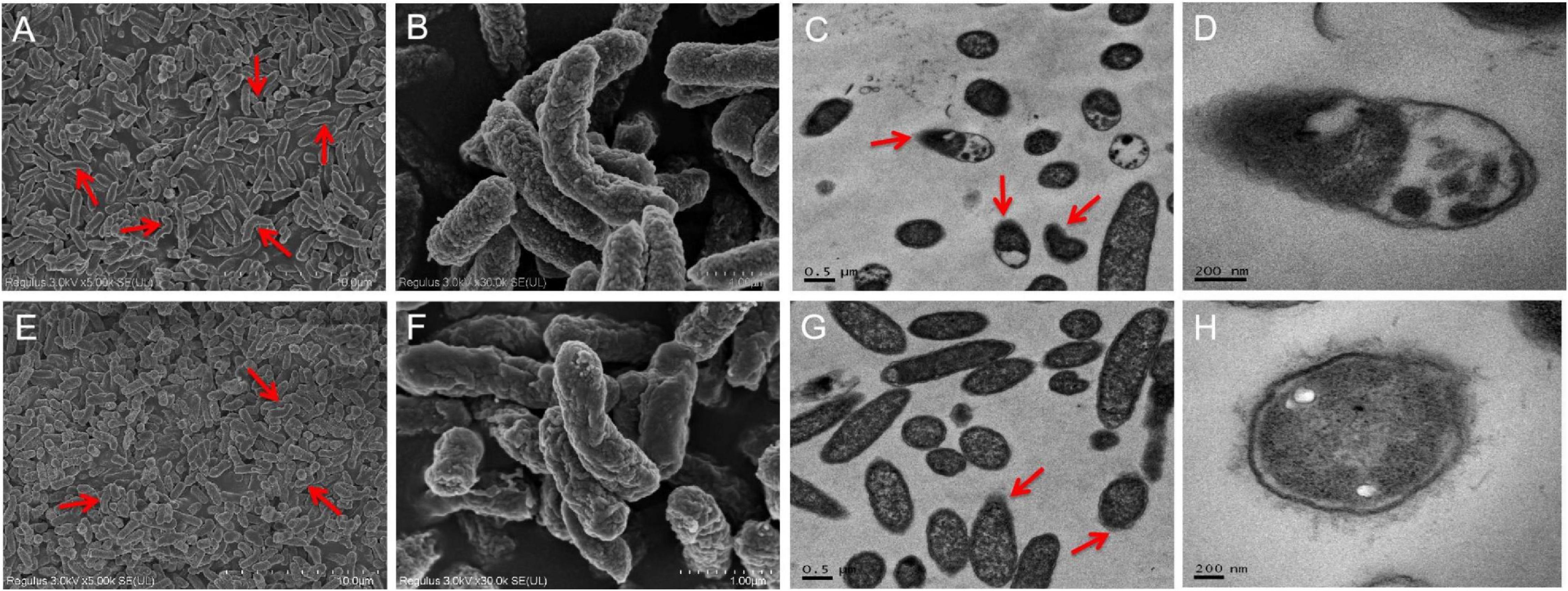
Figure 4. SEM and TEM photomicrographs of S. Enteritidis CVCC 1806 (A–D) and S. Enteritidis S2002-13 (E–H) after treatment with 100 mg/L NaClO for 30 min. The red arrows indicate regions of bacterial cell damage.
Antibiotic Resistance Phenotypes
We also assessed our population for antibiotic resistance and 155 of our Salmonella isolates (90.1%) were resistant to at least one and 117 (68%) isolates were resistant to at least 3 antibiotics. These results were similar to a previous study of Salmonella isolates from poultry in China (Tang et al., 2020). A majority of the Salmonella isolates were resistant to AMC (71.5%), AMP (70.9%), and TET (71.5%) and the least prevalent resistance phenotype was found for MEM (0.58%). Additionally, we found no evidence for differences in antibiotic resistance with sampling location. However, resistance did vary by serovar and S. Kentucky displayed the highest prevalence of multidrug resistance (93.2%) than the other two most common serotypes (Table 3).
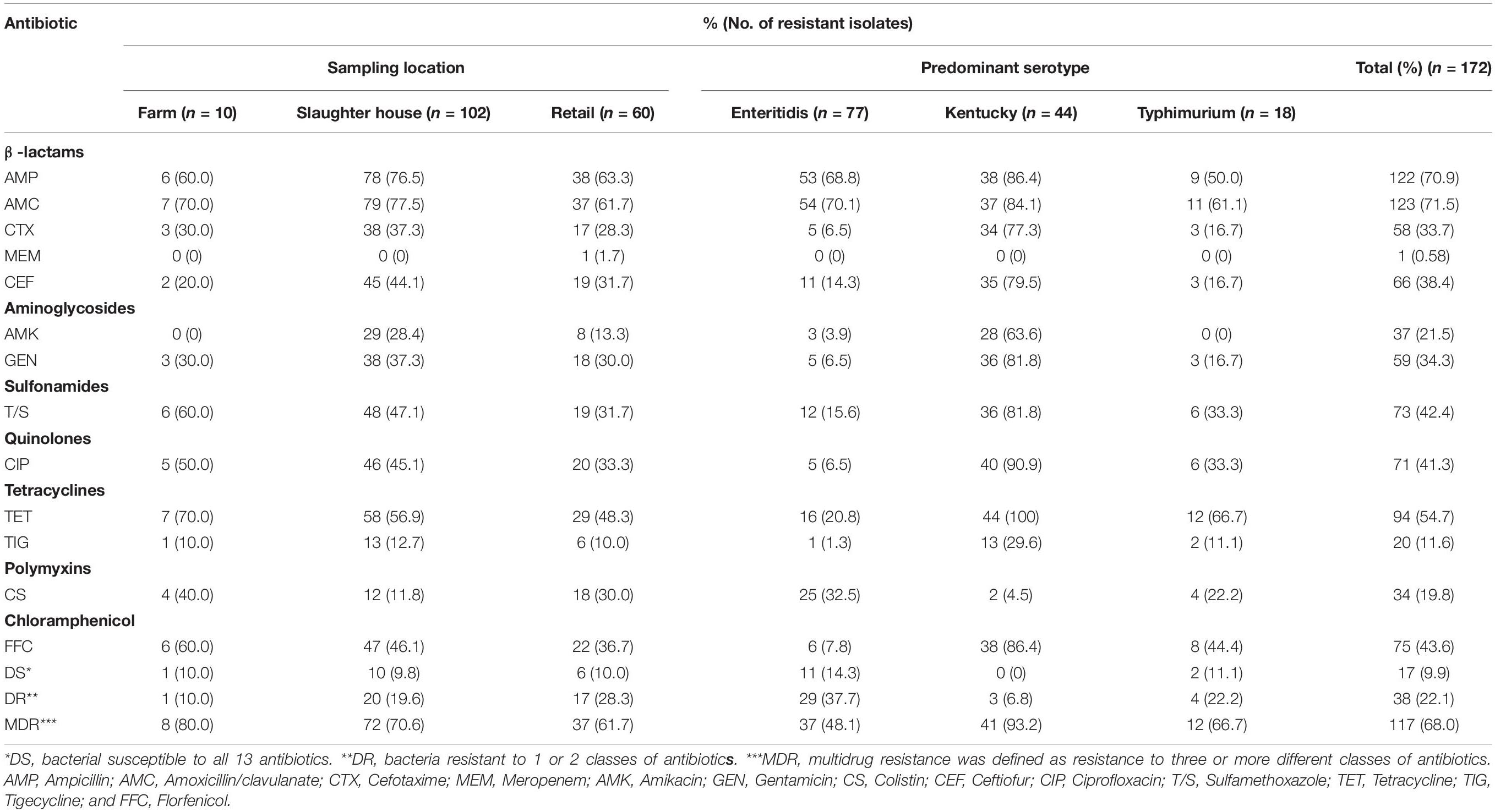
Table 3. Frequency (% of total isolates) of resistance to antibiotic agents among Salmonella isolates from poultry supply chains.
Determination of Chlorine and Antibiotic Resistance Genes
Our Salmonella isolates were also screened for the presence of genes enabling chlorine resistance and qacEΔ1 gene was the most prevalent (83.1%; n = 143) and qacF (59.9%; n = 103), exbD (43.6%; n = 75), and motB (29.7%; n = 51) were also highly represented. We found a positive correlation between the presence of chlorine resistance genes and higher MIC for chlorine that were associated with possession of qacEΔ1 (r = 0.66, p < 0.0001) and to lesser extents qacF (r = 0.36, p < 0.0001), exbD (r = 0.26, p < 0.0001), and motB (r = 0.19, p < 0.01). We also found that the antibiotic resistance genes blaTEM (76.7%, n = 132), tetA (73.3%, n = 126), blaCTX (65.1%, n = 112), and floR (61%, n = 105) were highly represented in our isolates from the different nodes of the poultry supply chain (Table 1).
Association Between Chlorine and Antibiotic Resistance
We compared our Salmonella isolates based on the antibiotic MIC values against OD600 values for NaClO (128 mg/L) using non-parametric Spearman correlation tests to determine whether chlorine and antibiotic resistance were correlated. We found significant positive relationships between chlorine and ceftiofur, tetracycline, ciprofloxacin and florfenicol resistance and was most likely related to the over-expression of drug efflux pumps (Table 4). Salmonella isolates with resistance to chlorine and antibiotics were analyzed for their content of resistance genes. In our group of 172 isolates, chlorine tolerant strains were more likely to also be antibiotic resistant (p < 0.05) (Figure 5A). This was especially for the qac genes that displayed a relatedness with correlation coefficients in the range of 0.10–0.44 (Figure 5B).
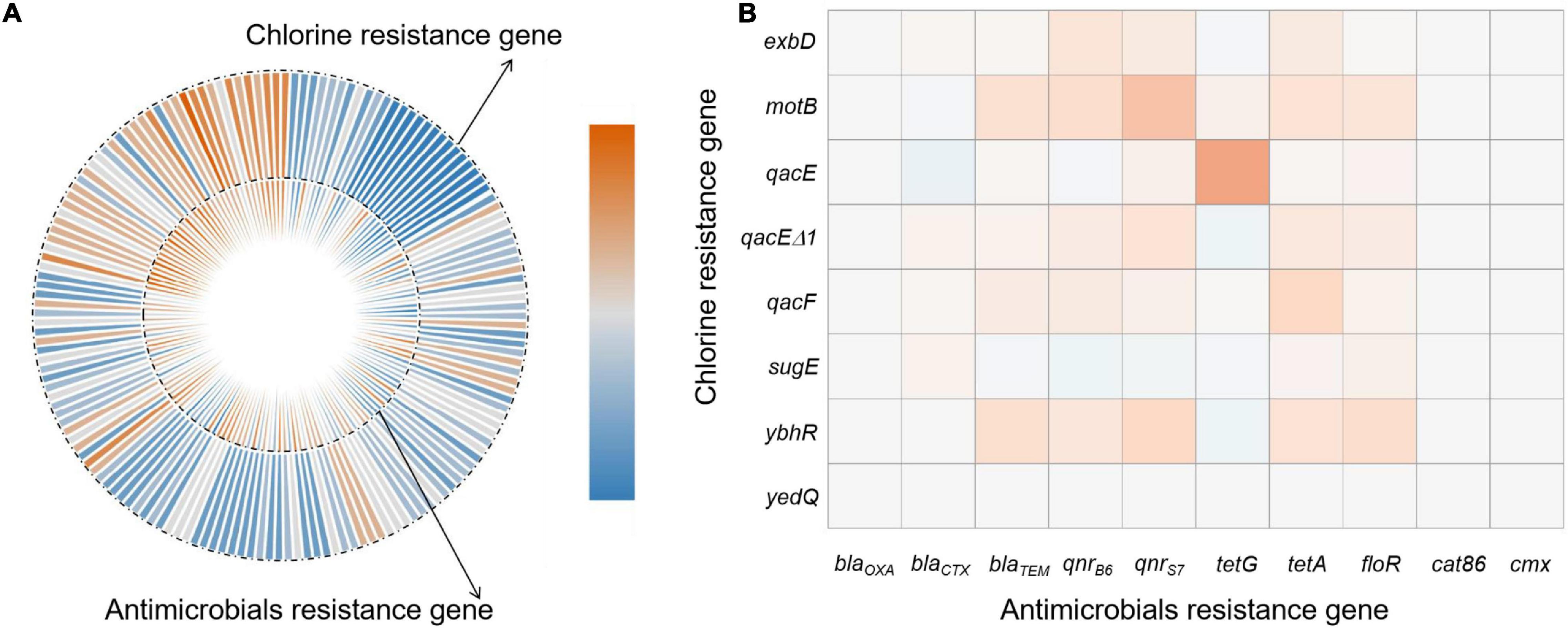
Figure 5. Correlations of bacterial NaClO and antibiotic l resistance genes based on (A) correlation coefficient and (B) frequency of resistance gene detection.
Discussion
Chlorine Tolerance
There is no widely accepted scientific definition of chlorine tolerance in bacteria or for quantification of the chlorine tolerance phenotype. Previous studies have utilized MIC, inhibition zone, logarithmic removal rate and membrane damage methods for determining bacterial chlorine tolerance (Jia et al., 2020; Luo et al., 2020). The MIC method is suitable for comparison of chlorine tolerance among isolates, but it cannot reflect the inactivation effect of typical MIC value (Garcia and Pelaz, 2008). Growth inhibition can be measured using 96 well plates and chlorine potency can be judged from growth inhibition quantified using absorbance at 600 nm (Kolarević et al., 2016). In our study, we combined MIC and OD600 methods to evaluate chlorine tolerance. Therefore, the disadvantage that for the MIC method that it does not reflect inactivation effects at the same MIC values was overcome.
The inhibition zone method is suitable for comparison of chlorine tolerance of a large number of isolates but becomes problematic with bacteria with differing growth rates and test results are often inaccurate (Khan et al., 2016; Luo et al., 2020). A previous study had isolated 87 bacterial strains in 22 genera from drinking water and chlorine tolerance could be differentiated at the species and genus levels (Khan et al., 2016). Logarithmic inactivation rates are suitable for comparison of chlorine tolerance between studies but disinfection concentration and time have not been standardized between studies (Zeng et al., 2020). Additionally, the antimicrobial activity of disinfectants is routinely tested by determination of survival curves and this procedure is labor-intensive and time consuming (Puangseree et al., 2021).
Culture-independent flow cytometry combined with fluorescent dyes has been used to investigate the effects of chlorine disinfection on bacterial physiological properties including membrane integrity and potential and respiratory activity (Song et al., 2019). Chlorine inactivates microorganisms by reacting and damaging cellular components including cell walls, membranes and nucleic acids. Chlorine-susceptible and chlorine-tolerant bacteria possess differences in the relative percentages of fatty acids in cell walls and membranes. Chlorine tolerance has been explained by replacement of 95.7% of membranes lipids as saturated long-chain fatty acids that act to limit chlorine diffusion (Chen et al., 2012).
Mechanisms of Chlorine Tolerance in Bacteria
Chlorine disinfection induces the expression of many functional gene families including responses to oxidative stress, DNA repair, energy metabolism, membrane damage and efflux pumps (Hou et al., 2019; Luo et al., 2020). The SOS response (a conserved response to DNA damage) triggered by oxidative stress renders bacteria chlorine resistant (Tong et al., 2021). Nutrient limitations result in quiescence of growth and metabolism that may subsequently lead to bacteria in the viable but non-culturable (VBNC) state for chlorine stress adaptations (Zhu et al., 2022). E. coli, Enterococcus and Salmonella can enter into a VBNC state after disinfection, and Salmonella was more resistant to chlorine in reclaimed water (Li et al., 2013). In addition, membrane permeability is a primary barrier to uptake of foreign or extracellular DNA and NaClO exposure alters the expression of cell membrane-related genes and especially those regulating cell membrane permeability as has been demonstrated in Pseudomonas spp. (Tong et al., 2021). The upregulation of efflux genes also contributes to the persistence of chlorine-treated cells (Hou et al., 2019). In our current study, qacEΔ1 was tightly linked to elevated MIC for NaClO. Previous studies had reported that the qacEΔ1 gene was the most frequently present in E. coli (69.70%), followed by K. pneumoniae (50.00%) and Salmonella (39.62%), respectively. The qacEΔ1 gene is common in enteric bacteria and encodes an efflux pump conferring resistance to chlorine disinfectants via an electrochemical proton gradient (Wu et al., 2015). The gene is also frequently associated with mobile genetic elements such as class I integrons resulting in co-selection of antibiotic resistance genes (Hu et al., 2018).
Cross-Resistance Between Chlorine Disinfectants and Antibiotics
Cross-resistance between chlorine and antibiotics has become a particular concern due to the possible contribution to the persistence of antibiotic resistance despite antibiotic withdrawn (Liao et al., 2020). In this study, exposure to chlorine resulted in cross-resistance to ceftiofur, tetracycline, ciprofloxacin, and florfenicol. Expression of multidrug efflux pumps is a major mechanism mediating such cross-resistance (Hou et al., 2019; Jin et al., 2020; Puangseree et al., 2021). The over-expression of the drug efflux pump MexEF-OprN in Pseudomonas aeruginosa following chlorine exposure promoted resistance to diverse antibiotics including tetracyclines, fluoroquinolones, β-lactams, chloramphenicol, macrolides, novobiocin, trimethoprim, and sulfonamides (Lister et al., 2009). This directly linked increased antibiotic resistance to chlorine exposure. Similarly, pretreatment of S. Enteritidis with chlorine, sodium nitrite, sodium benzoate or acetic acid induced the over-expression of the marRAB operon, a global antibiotic resistance regulator involved in the production of AcrAB efflux pumps that extrude antibiotics (Potenski et al., 2003). The development of VBNC bacteria via chlorination increases their resistance to antibiotics (Lin et al., 2017). Chlorine-tolerant injured bacteria that are physiologically competent cells present higher plasmid transformation frequencies than the corresponding untreated bacteria. Since the transferable plasmids released from killed sensitive antibiotic resistant bacteria have a consistent resistance to degradation through disinfection, the chlorination process can promote the horizontal transfer of the released plasmid into chlorine-injured bacteria through natural transformation. This leads to the enrichment of antibiotic resistance genes in viable bacteria (Jin et al., 2020). Therefore, using chlorine disinfectant could provide selection pressure for strains that acquire antibiotic resistance.
Conclusion
Salmonella spp. from poultry supply chains and possessed tolerance to NaClO at MIC > 256 mg/L in > 90% of the 172 isolates. These tolerant strains displayed higher D values and a more integrated cellular shape. The presence of NaClO tolerance genes and the MIC for NaClO were positively correlated especially for qacEΔ1 (r = 0.66, p < 0.0001). Significant positive relationships existed between NaClO and the antibiotics ceftiofur, tetracycline, ciprofloxacin, and florfenicol indicating that chlorine-tolerant bacteria were more likely to also be antibiotic resistant. The generated data could provide parts of the input data for microbial risk assessment of Salmonella with different chlorine tolerance profile to capture the variability in the strains of interest while decreasing the uncertainty in some model input parameters.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
WW and HY: writing—review and editing. XX and LB: investigation and writing—original draft preparation. SW and LL: data curation. XQ and JZ: software. YX, BT, and YL: resources. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This research was supported by the Walmart Foundation (UA2020-152, UA2021-247) and Ministry of Agriculture and Rural Affairs (14215033)
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jiele Ma and Mengjia Qiu for their help in the experiments.
Footnotes
References
Ahn, J., Balasubramaniam, V., and Yousef, A. (2007). Inactivation kinetics of selected aerobic and anaerobic bacterial spores by pressure-assisted thermal processing. Int. J. Food Microbiol. 113, 321–329. doi: 10.1016/j.ijfoodmicro.2006.08.012
Andoh, L., Dalsgaard, A., Obiri-Danso, K., Newman, M., Barco, L., and Olsen, J. (2016). Prevalence and antimicrobial resistance of Salmonella serovars isolated from poultry in Ghana. Epidemiol. Infect. 144, 3288–3299. doi: 10.1017/S0950268816001126
Barros, C. P., Pires, R. P. S., Guimarães, J. T., Abud, Y. K. D., Almada, C. N., Pimentel, T. C., et al. (2021). Ohmic heating as a method of obtaining paraprobiotics: impacts on cell structure and viability by flow cytometry. Food Res. Int. 140:110061. doi: 10.1016/j.foodres.2020.110061
Bernardez, L., and de Andrade Lima, L. (2015). Improved method for enumerating sulfate-reducing bacteria using optical density. MethodsX 2, 249–255. doi: 10.1016/j.mex.2015.04.006
Bhojani, G., Binod Kumar, S., Kumar Saha, N., and Haldar, S. (2018). Membrane biofouling by chlorine resistant Bacillus spp.: effect of feedwater chlorination on bacteria and membrane biofouling. Biofouling 34, 426–439. doi: 10.1080/08927014.2018.1461213
Carey, D. E., and McNamara, P. J. (2015). The impact of triclosan on the spread of antibiotic resistance in the environment. Front. Microbiol. 5:780. doi: 10.3389/fmicb.2014.00780
Chen, H., Wilson, J., Ercanbrack, C., Smith, H., Gan, Q., and Fan, C. (2021). Genome-wide screening of oxidizing agent resistance genes in Escherichia coli. Antioxidants 10:861. doi: 10.3390/antiox10060861
Chen, Y. Q., Chao, C., Zhang, X. J., Zheng, Q., and Liu, Y. Y. (2012). Inactivation of resistant Mycobacteria mucogenicum in water: chlorine resistance and mechanism analysis. Biomed. Environ. Sci. 25, 230–237. doi: 10.3967/0895-3988.2012.02.016
Elnekave, E., Hong, S., Mather, A. E., Boxrud, D., Taylor, A. J., Lappi, V., et al. (2018). Salmonella enterica serotype 4,[5], 12: i:-in swine in the United States Midwest: an emerging multidrug-resistant clade. Clin. Infect. Dis. 66, 877–885. doi: 10.1093/cid/cix909
Garcia, M., and Pelaz, C. (2008). Effectiveness of disinfectants used in cooling towers against Legionella pneumophila. Chemotherapy 54, 107–116. doi: 10.1159/000118662
Grimont, P. A., and Weill, F. X. (2007). Antigenic formulae of the Salmonella serovars, 9th Edn. (Paris: Institut Pasteur), 1–166.
Hou, A., Yang, D., Miao, J., Shi, D., Yin, J., Yang, Z., et al. (2019). Chlorine injury enhances antibiotic resistance in Pseudomonas aeruginosa through over expression of drug efflux pumps. Water Res. 156, 366–371. doi: 10.1016/j.watres.2019.03.035
Hu, Y., Jiang, L., Zhang, T., Jin, L., Han, Q., Zhang, D., et al. (2018). Occurrence and removal of sulfonamide antibiotics and antibiotic resistance genes in conventional and advanced drinking water treatment processes. J. Hazard. Mater. 360, 364–372. doi: 10.1016/j.jhazmat.2018.08.012
Hu, Z., Peng, Z., Zhang, X., Li, Z., Jia, C., Li, X., et al. (2021). Prevalence and molecular characterization of antimicrobial-resistant Escherichia coli in pig farms, slaughterhouses, and terminal markets in henan province of China. Foodborne Pathog. Dis. 18, 733–743. doi: 10.1089/fpd.2021.0011
Jia, S., Jia, R., Zhang, K., Sun, S., Lu, N., Wang, M., et al. (2020). Disinfection characteristics of Pseudomonas peli, a chlorine-resistant bacterium isolated from a water supply network. Environ. Res. 185:109417. doi: 10.1016/j.envres.2020.109417
Jin, M., Liu, L., Wang, D., Yang, D., Liu, W., Yin, J., et al. (2020). Chlorine disinfection promotes the exchange of antibiotic resistance genes across bacterial genera by natural transformation. ISME J. 14, 1847–1856. doi: 10.1038/s41396-020-0656-9
Jun, W., Guo, Y. C., and Ning, L. (2013). Prevalence and risk assessment of Campylobacter jejuni in chicken in China. Biomed. Environ. Sci. 26, 243–248. doi: 10.3967/0895-3988.2013.04.002
Karumathil, D. P., Yin, H. B., Kollanoor-Johny, A., and Venkitanarayanan, K. (2014). Effect of chlorine exposure on the survival and antibiotic gene expression of multidrug resistant Acinetobacter baumannii in water. Int. J. Environ. Res. Public Health 11, 1844–1854. doi: 10.3390/ijerph110201844
Khan, S., Beattie, T. K., and Knapp, C. W. (2016). Relationship between antibiotic-and disinfectant-resistance profiles in bacteria harvested from tap water. Chemosphere 152, 132–141. doi: 10.1016/j.chemosphere.2016.02.086
Kolarević, S., Milovanović, D., Avdović, M., Oalde, M., Kostić, J., Sunjog, K., et al. (2016). Optimisation of the microdilution method for detection of minimum inhibitory concentration values in selected bacteria. Bot. Serb. 40, 29–36. doi: 10.5281/zenodo.48751
Langsrud, S., Sidhu, M. S., Heir, E., and Holck, A. L. (2003). Bacterial disinfectant resistance-a challenge for the food industry. Int. Biodeter. Biodegr. 51, 283–290. doi: 10.1016/S0964-8305(03)00039-8
Lee, N., Park, S., Kang, I., and Ha, S. (2014). The evaluation of combined chemical and physical treatments on the reduction of resident microorganisms and Salmonella Typhimurium attached to chicken skin. Poultry Sci. 93, 208–215. doi: 10.3382/ps.2013-03536
Li, D., Zeng, S., Gu, A. Z., He, M., and Shi, H. (2013). Inactivation, reactivation and regrowth of indigenous bacteria in reclaimed water after chlorine disinfection of a municipal wastewater treatment plant. J. Environ. Sci. 25, 1319–1325. doi: 10.1016/S1001-0742(12)60176-4
Liao, X., Ma, Y., Daliri, E. B., Koseki, S., Wei, S., Liu, D., et al. (2020). Interplay of antibiotic resistance and food-associated stress tolerance in foodborne pathogens. Trends Food Sci. Technol. 95, 97–106. doi: 10.1016/j.tifs.2019.11.006
Lin, H., Ye, C., Chen, S., Zhang, S., and Yu, X. (2017). Viable but non-culturable E. coli induced by low level chlorination have higher persistence to antibiotics than their culturable counterparts. Environ. Pollut. 230, 242–249. doi: 10.1016/j.envpol.2017.06.047
Lister, P. D., Wolter, D. J., and Hanson, N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610. doi: 10.1128/CMR.00040-09
Luo, L., Wu, Y., Yu, T., Wang, Y., Chen, G. Q., Tong, X., et al. (2020). Evaluating method and potential risks of chlorine-resistant bacteria (CRB): a review. Water Res. 188:116474. doi: 10.1016/j.watres.2020.116474
Mechesso, A. F., Moon, D. C., Kim, S. J., Song, H. J., Kang, H. Y., Na, S. H., et al. (2020). Nationwide surveillance on serotype distribution and antimicrobial resistance profiles of non-typhoidal Salmonella serovars isolated from food-producing animals in South Korea. Int. J. Food Microbiol. 335:108893. doi: 10.1016/j.ijfoodmicro.2020.108893
Potenski, C. J., Gandhi, M., and Matthews, K. R. (2003). Exposure of Salmonella enteritidis to chlorine or food preservatives increases susceptibility to antibiotics. FEMS Microbiol. Lett. 220, 181–186. doi: 10.1016/S0378-1097(03)00099-5
Puangseree, J., Jeamsripong, S., Prathan, R., Pungpian, C., and Chuanchuen, R. (2021). Resistance to widely-used disinfectants and heavy metals and cross resistance to antibiotics in Escherichia coli isolated from pigs, pork and pig carcass. Food Control 124:107892. doi: 10.1016/j.foodcont.2021.107892
Roy, P. K., and Ghosh, M. (2017). Chlorine resistant bacteria isolated from drinking water treatment plants in West Bengal. Desal. Water Treat. 79, 103–107. doi: 10.5004/dwt.2017.20697
Schober, P., Boer, C., and Schwarte, L. A. (2018). Correlation coefficients: appropriate use and interpretation. Anesth. Analg. 126, 1763–1768. doi: 10.1213/ANE.0000000000002864
Slotved, H. C., Dalby, T., Harboe, Z. B., Valentiner-Branth, P., Casadevante, V. F. D., Espenhain, L., et al. (2016). The incidence of invasive pneumococcal serotype 3 disease in the Danish population is not reduced by PCV-13 vaccination. Heliyon 2:00198. doi: 10.1016/j.heliyon.2016.e00198
Song, Y., Mao, G., Gao, G., Bartlam, M., and Wang, Y. (2019). Resistance and resilience of representative low nucleic acid-content bacteria to free chlorine exposure. J. Hazard. Mater. 365, 270–279. doi: 10.1016/j.jhazmat.2018.10.080
Tang, B., Chang, J., Zhang, L., Liu, L., Xia, X., Hassan, B. H., et al. (2020). Carriage of distinct mcr-1-harboring plasmids by unusual serotypes of Salmonella. Adv. Biosyst. 4:1900219. doi: 10.1002/adbi.201900219
Templeton, M. R., Oddy, F., Leung, W. K., and Rogers, M. (2013). Chlorine and UV disinfection of ampicillin-resistant and trimethoprim-resistant Escherichia coli. J. Environ. Eng. 8, 231–237. doi: 10.1139/L09-040
Tong, C., Hu, H., Chen, G., Li, Z., Li, A., and Zhang, J. (2021). Chlorine disinfectants promote microbial resistance in Pseudomonas sp. Environ. Res. 199, 111296–111296. doi: 10.1016/j.envres.2021.111296
Tyagi, A. K., and Malik, A. (2012). Morphostructural damage in food-spoiling bacteria due to the lemon grass oil and its vapour: SEM, TEM, and AFM investigations. Evid Based Complement. Alternat. Med. 2012:692625. doi: 10.1155/2012/692625
Wang, J., Sui, M., Yuan, B., Li, H., and Lu, H. (2019). Inactivation of two mycobacteria by free chlorine: effectiveness, influencing factors, and mechanisms. Sci. Total Environ. 648, 271–284. doi: 10.1016/j.scitotenv.2018.07.451
Wu, G., Yang, Q., Long, M., Guo, L., Li, B., Meng, Y., et al. (2015). Evaluation of agar dilution and broth microdilution methods to determine the disinfectant susceptibility. J. Antibiot. 68, 661–665. doi: 10.1038/ja.2015.51
Xiao, X., Wang, W., Zhang, J., Liao, M., Yang, H., Fang, W., et al. (2019). Modeling the reduction and cross-contamination of Salmonella in poultry chilling process in China. Microorganisms 7:448. doi: 10.3390/microorganisms7100448
Yang, X., Guo, P., Li, M., Li, H., Hu, Z., Liu, X., et al. (2020a). Optimization of culture conditions for amoxicillin degrading bacteria screened from pig manure. Int. J. Environ. Res. Public Health 17:1973. doi: 10.3390/ijerph17061973
Yang, X., Huang, J., Zhang, Y., Liu, S., Chen, L., Xiao, C., et al. (2020b). Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 713:136385. doi: 10.1016/j.scitotenv.2019.136385
Zeng, F., Cao, S., Jin, W., Zhou, X., Ding, W., Tu, R., et al. (2020). Inactivation of chlorine-resistant bacterial spores in drinking water using UV irradiation, UV/Hydrogen peroxide and UV/Peroxymonosulfate: efficiency and mechanism. J. Clean. Prod. 243:118666. doi: 10.1016/j.jclepro.2019.118666
Zeng, X., Lv, S., Qu, C., Lan, L., Tan, D., Li, X., et al. (2021). Serotypes, antibiotic resistance, and molecular characterization of non-typhoidal Salmonella isolated from diarrheic patients in Guangxi Zhuang Autonomous Region, China, 2014–2017. Food Control 120:107478. doi: 10.1016/j.foodcont.2020.107478
Keywords: poultry, bacterial resistance, sodium hypochlorite (NaClO), qacEΔ1, efflux pump
Citation: Xiao X, Bai L, Wang S, Liu L, Qu X, Zhang J, Xiao Y, Tang B, Li Y, Yang H and Wang W (2022) Chlorine Tolerance and Cross-Resistance to Antibiotics in Poultry-Associated Salmonella Isolates in China. Front. Microbiol. 12:833743. doi: 10.3389/fmicb.2021.833743
Received: 12 December 2021; Accepted: 28 December 2021;
Published: 04 February 2022.
Edited by:
Qingli Dong, University of Shanghai for Science and Technology, ChinaReviewed by:
Ivanhoe Leung, The University of Melbourne, AustraliaLikou Zou, Sichuan Agricultural University, China
Copyright © 2022 Xiao, Bai, Wang, Liu, Qu, Zhang, Xiao, Tang, Li, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Yang, yanghua@zaas.ac.cn; Wen Wang, ww_hi1018@163.com
 Xingning Xiao
Xingning Xiao Li Bai
Li Bai Sheng Wang
Sheng Wang Lisha Liu2
Lisha Liu2 Jianmin Zhang
Jianmin Zhang Yingping Xiao
Yingping Xiao Biao Tang
Biao Tang Yanbin Li
Yanbin Li Wen Wang
Wen Wang