- 1Laboratory of Food and Environmental Microbiology, Earth and Life Institute, Louvain-la-Neuve, Belgium
- 2Multi-Omics Laboratory, School of Pharmacy, Lebanese American University, Byblos, Lebanon
Horizontal gene transfer (HGT) is a major driving force in shaping bacterial communities. Key elements responsible for HGT are conjugation-like events and transmissible plasmids. Conjugative plasmids can promote their own transfer as well as that of co-resident plasmids. Bacillus cereus and relatives harbor a plethora of plasmids, including conjugative plasmids, which are at the heart of the group species differentiation and specification. Since the first report of a conjugation-like event between strains of B. cereus sensu lato (s.l.) 40 years ago, many have studied the potential of plasmid transfer across the group, especially for plasmids encoding major toxins. Over the years, more than 20 plasmids from B. cereus isolates have been reported as conjugative. However, with the increasing number of genomic data available, in silico analyses indicate that more plasmids from B. cereus s.l. genomes present self-transfer potential. B. cereus s.l. bacteria occupy diverse environmental niches, which were mimicked in laboratory conditions to study conjugation-related mechanisms. Laboratory mating conditions remain nonetheless simplistic compared to the complex interactions occurring in natural environments. Given the health, economic and ecological importance of strains of B. cereus s.l., it is of prime importance to consider the impact of conjugation within this bacterial group.
Introduction
Genetic diversity is key for long-term survival of various organisms, be it micro-ones like bacteria or fungi, all the way to plants and animals, in continuously changing ecosystems. For bacteria, although their reproduction is mostly asexual by binary fission, they have developed the capacity to diversify their genetic content and to acquire DNA material from relatively distant organisms, by horizontal gene transfer (HGT; Arnold et al., 2021). Defined as “the non-genealogical transmission of genetic material from one organism to another” (Goldenfeld and Woese, 2007), HGT has a strong impact on bacterial genomes by increasing the potential of acquiring advantageous genes, resulting in better adaptation and colonization of new ecological niches (Soucy et al., 2015; McInerney et al., 2017). Diverse mechanisms drive HGT among prokaryotes, and the three major mechanisms are natural transformation, transduction and conjugation (von Wintersdorff et al., 2016). While transformation consists in the capture of foreign DNA from the environment (Mell and Redfield, 2014) and transduction is mediated by bacteriophages (a.k.a. phages; Colavecchio et al., 2017), conjugation requires a physical contact between the cell partners prior to transfer of plasmid DNA (de la Cruz et al., 2010). Aside from the three major mechanisms of HGT, several non-canonical mechanisms of DNA transfer have been identified, including the so-called “gene-transfer agents,” transfer via membrane vesicles and intercellular nano-tubes (Arnold et al., 2021). Yet, these non-canonical modes of HGT are beyond the scope of this review.
The mechanism of conjugation – or conjugative transfer – occurs in a stepwise process and requires a secretion system, consisting of a type IV secretion system (T4SS), and a replication machinery to obtain two copies of the conjugative element at the end of the transfer event (for detailed reviews on conjugation mechanism, refer to Cabezón et al., 2015 and Virolle et al., 2020). During this process, a relaxase first recognizes the origin of transfer (oriT) of the conjugative element, nicks the oriT in a strand-specific manner and remains bound to the nicked strand. The resulting nucleoprotein complex is then recruited to the T4SS by a coupling protein (CP) and replication of the intact strand takes place in the donor cell, displacing the nicked strand for transfer and further replication in the recipient cell (de la Cruz et al., 2010). T4SS are usually made of three ATPases, including one CP and two motor ATPases, one peptidoglycan hydrolase and several membrane proteins inserted into the donor cell envelop and forming the core of the T4SS complex (for detailed description of T4SS, refer to Ilangovan et al., 2015 and Waksman, 2019). Studies have mainly focused on the conjugative systems of Gram-negative bacteria, but the global mechanism of conjugative transfer is conserved among Gram-positive bacteria, despite structural differences in the T4S machinery (for a review on Gram-positive conjugation systems, refer to Goessweiner-Mohr et al., 2013).
Conjugative plasmids are capable of self-transfer and encode all the genetic material necessary for a functional conjugative apparatus. Some plasmids, called mobilizable plasmids, lack the genes encoding for the T4SS, but usually encode an oriT and a relaxase. They can hijack the transfer apparatus of a co-resident conjugative plasmid to trigger their transfer (Fernández-López et al., 2014). Suh transfer process is called mobilization. Other plasmids remain however non-transmissible and are often called non-mobilizable, as they lack all the elements necessary for conjugation-like movements, i.e., an oriT, a relaxase and a T4SS (Smillie et al., 2010).
As suggested by its large diversity of plasmids and mobile genetic elements (MGEs), the B. cereus group is of prime interest when it comes to conjugation and conjugation-related plasmid transfer. The B. cereus group, or B. cereus sensu lato (s.l.), refers to a group of closely related bacterial species, yet displaying a large ecological diversity. B. cereus group members are ubiquitous sporulating Gram-positive bacteria, mainly studied because several members of the group are of health, environmental or commercial interests or concerns (Ramarao et al., 2015). Within this review, we look back at the earlier work as well as at the data collected on B. cereus conjugation and conjugative plasmids described so far. We also examine the proportion of potential conjugative plasmids among the B. cereus s.l. genome sequences available to date. Overall, we aim at providing a full picture on conjugative plasmids residing within the plasmid-rich group of B. cereus and their implications in both laboratory and environmental-like conditions, as known to date.
Bacillus cereus s.l., plasmid-borne toxins and horizontal gene transfer
Three main species form the core of the B. cereus group: the opportunistic pathogen B. cereus sensu stricto (s.s.), potentially associated with foodborne infections, the etiological agent of anthrax, B. anthracis, and the entomopathogen B. thuringiensis, well-known for its biopesticide application (Kolstø et al., 2009; Ehling-Schulz et al., 2019). In addition, the B. cereus group includes other species, like the rhizoid-growing B. mycoides and B. pseudomycoides (Flügge, 1886; Nakamura, 1998), the psychrotrophic B. weihenstephanensis (Lechner et al., 1998; Hoton et al., 2009) – recently reclassified as one species with B. mycoides (Liu et al., 2018) – and the thermotolerant and cytotoxic B. cytotoxicus (Guinebretière et al., 2013). Diverse classification methods have been proposed over the years (for a recent review, refer to Carroll et al., 2021) in an effort to resolve the ongoing taxonomic conflicts surrounding B. cereus s.l. due to (i) an insufficient differentiation at the genomic level and (ii) whether or not plasmids and MGEs should be included in the phylogenetic analyses. Members of B. cereus s.l. carry a wide range of plasmids, some of which are susceptible candidates for horizontal dissemination (Kolstø et al., 2009; Yuan et al., 2010). Although many studies presented evidence that MGEs and HGT can confound the phylogenetic process (Bazinet, 2017), there is no denying the importance of extra-chromosomal entities in defining a species’ lifestyle, hence their importance in the phylogenetic process (Liu et al., 2015; Patiño-Navarrete and Sanchis, 2017).
In fact, the three main members of the B. cereus group are discriminated based on specific toxins they secrete, whose genetic determinants are often encoded on large transmissible plasmids, or on specific plasmids they carry. For instance, B. anthracis is characterized by two large plasmids, pXO1 and pXO2, responsible for the production of the bipartite anthrax toxins and the anthrax capsule, respectively (Baillie and Read, 2001). Both pXO1 and pXO2 encode genes reminiscent of conjugation systems, but they show no ability for self-transfer, although they were proven as mobilizable (Grynberg et al., 2007). Other plasmids share the backbone of pXO1 and pXO2 and are therefore grouped into distinct plasmid families. pXO2-like plasmids are likely to depict self-transmissible capacities as some of their representatives (i.e., pAW63 from B. thuringiensis sv. kurstaki, pBT9727 from B. thuringiensis sv. konkukian and pXO2 from B. anthracis) harbor a tra (transfer) region with genes homologous to the main components of functional conjugative systems and T4SS. While both pAW63 and pBT9727 are effective conjugative plasmids, pXO2 has an interrupted virD4-like gene likely responsible for its inability to conjugate autonomously (Van der Auwera et al., 2005; Hu et al., 2009b; see Section “pAW63 and other pXO2-like plasmids”).
The pXO1-like plasmid family includes pCER270, a plasmid bearing the genetic determinants of the cereulide, a dodecadepsipeptide responsible for the emetic syndrome in B. cereus-mediated food infections (Hoton et al., 2005; Rasko et al., 2007). Other pXO1-like plasmids are pBMB67 from B. thuringiensis (Chao et al., 2007), pER272 and pBC10987 from B. cereus strains (Rasko et al., 2007; Mei et al., 2014) and pBCXO1 from B. cereus G9241 and B. cereus biovar anthracis (Hoffmaster et al., 2004; Brézillon et al., 2015). These plasmids share a high degree of synteny with pXO1 but lack or diverge in their pathogenicity genetic determinants. Contrary to the pXO2-like plasmids, none of the pXO1-like plasmids harbor self-transfer features, although pBMB67 displays CDS (Coding DNA Sequence) homologous to the VirB/D4 model for T4SS and was therefore described as a putative conjugative plasmid (Chao et al., 2007).
The main feature associated with B. thuringiensis is the production of entomopathogen delta-endotoxins (Cry toxins) in parasporal crystals, used worldwide in biopesticide preparations (Bravo et al., 2007; Palma et al., 2014). The genetic determinants of Cry toxins are plasmid-borne, encoded on cry plasmids, which are either transmissible upon mobilization, like pBtoxis from B. thuringiensis sv. israelensis (Berry et al., 2002; Hu et al., 2005), or conjugative themselves, like pHT73 from B. thuringiensis sv. kurstaki (Wilcks et al., 1998). Establishing a link between the presence of plasmids and the production of Cry toxins was at the base of the discovery of conjugation-like events between strains of B. thuringiensis (see Section “First hints of conjugation and premises of a new transfer tool”).
Besides toxin plasmids, B. cereus s.l. harbors a large pool of plasmids and other mobile elements. While plasmids typically promote intercellular genetic moves, other MGEs, like insertion sequences (IS) and transposable elements (Tn), are usually responsible for intracellular mobility, i.e., moving from one location to another within the same genome. Transposable elements are for instance more prevalent in toxin plasmids of B. thuringiensis, namely about 30% of the total “thuringiensis” plasmid pool, as they often flank cry genes (Fiedoruk et al., 2017; Fayad et al., 2019, 2021a). In natural environments, B. cereus s.l. isolates also present a flexible pool of extrachromosomal molecules, with HGT evidenced between closely related isolates (Hu et al., 2009a). HGT within the B. cereus group is not restricted to plasmids. Some chromosomally-encoded enterotoxin operons (hbl and cytK) of B. cereus were for example shown to have spread by HGT amongst different taxonomic clusters (Böhm et al., 2015). Although the cereulide genetic determinants (ces) are located on non-self-transmissible plasmids or in the chromosome, they are also prone to horizontal dissemination because they are typically flanked by MGEs, like Tnces in emetic strains of B. weihenstephanensis (Mei et al., 2014).
With their multiple combinations of plasmids and other MGEs, strains of B. cereus s.l. are provided with a strong potential for adaptability. Indeed, HGT is recognized as a major route for acquiring new potentially beneficial genetic material, enabling bacteria to adapt and migrate to new environmental niches (McInerney et al., 2017), and the B. cereus group makes no exception. However, acquiring foreign DNA can sometimes be harmful to bacteria thriving in some habitats. Therefore, bacteria developed protection systems preventing from such invasion, such as the antiviral CRISPR-Cas system (Barrangou and Marraffini, 2014). In staphylococci, the immune and protective function of the CRISPR-Cas system acts as barriers limiting the acquisition of MGEs by HGT (Marraffini and Sontheimer, 2008). Interestingly, CRISPR-Cas systems are less prevalent and mostly inactivated in B. cereus s.l. (Zheng et al., 2020). In addition, strains of B. cereus with functional CRISPR-Cas systems showed a restricted ecological niche, a poorer pool of MGEs and an impaired adaptation to harsh environments. On an evolutionary point of view, acquisition of MGEs and ecological adaptability were apparently favored by B. cereus s.l., rather than protective CRISPR-Cas systems (Zheng et al., 2020), further illustrating the importance of HGT within this specific bacterial group.
Historical accounts
Up until the discovery of conjugative transfer among the B. cereus group in the early 1980s, phage-mediated transduction was the main technique available for studying B. cereus s.l. at a molecular level (Thorne, 1978). Most early molecular studies focused on B. thuringiensis due to its entomocidal Cry toxins and their common application as biopesticide (for a review, see Palma et al., 2014). The historical milestones described in this section are illustrated on Figure 1.
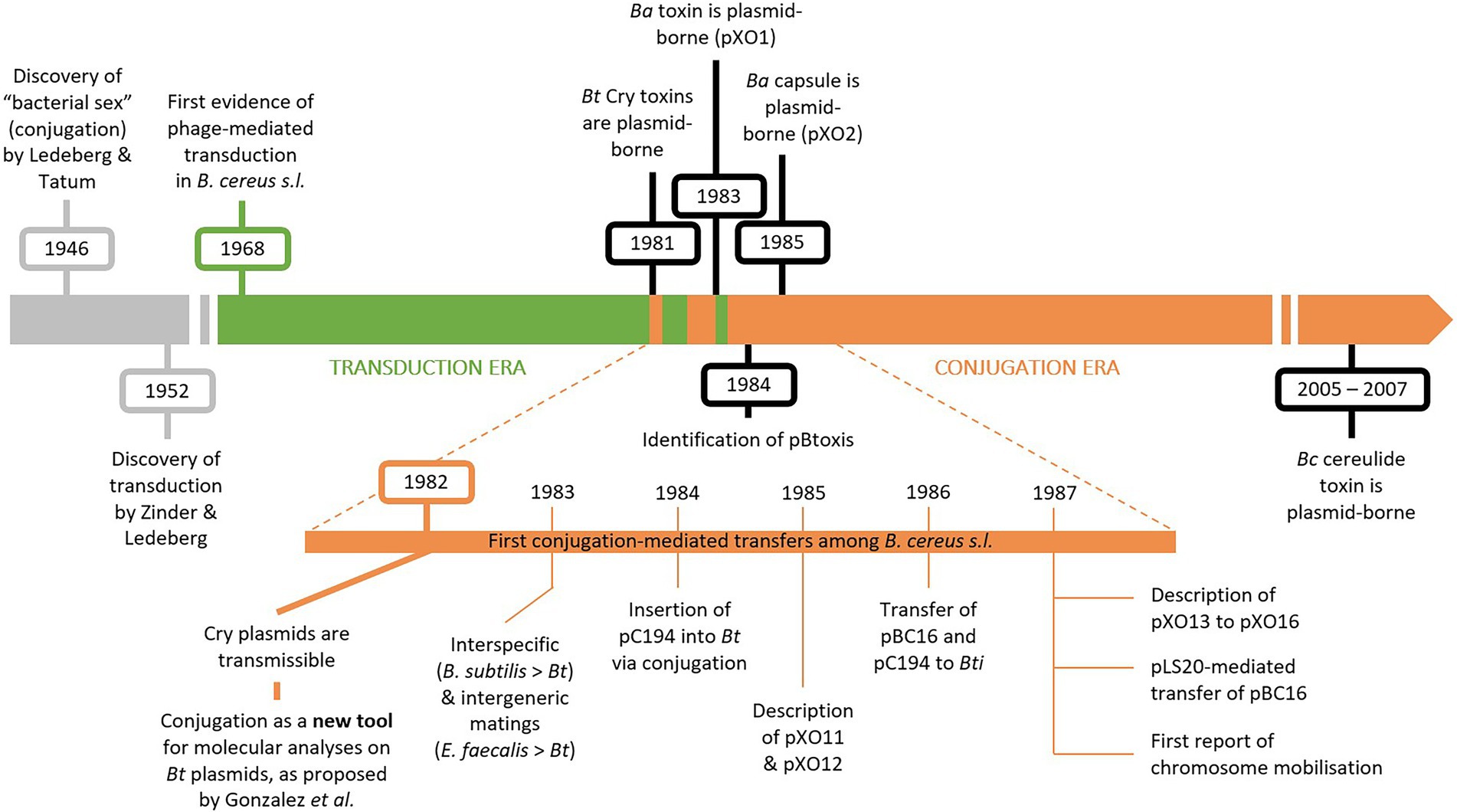
Figure 1. Timeline displaying milestones in Bacillus cereus s.l. molecular research. In the 1960’s and 1970’s, molecular research in B. cereus s.l. was mostly conducted using phage-mediated transduction (“transduction era” in green). Since the discovery of conjugative transfer in B. cereus s.l., plasmid-mediated conjugation became the prevalent technique (“conjugation era” in orange). Plasmid-borne toxins play a key role in B. cereus s.l. taxonomy and were mostly identified alongside conjugation (in black). The historical discovery of conjugation and transduction are depicted in grey. Ba, B. anthracis; Bt, B. thuringiensis; Bc, B. cereus; Bti, B. thuringiensis sv. israelensis.
Bacillus cereus s.l. in the pre-conjugation era
Transducing phages were mostly used for genetic experiments in B. cereus s.l. during the 60’s and 70’s, before the discovery of conjugative plasmids and their application as molecular tools (Figure. 1). The first evidence of phage-mediated transduction in B. cereus s.l. dates back to 1968, when a transducing phage, designated CP51, was isolated from a soil sample along with its host bacterium B. cereus strain 569 (Thorne, 1968). Phage CP51 was later shown to successfully mediate the transfer of the plasmidial vectors pBC16, conferring resistance to tetracycline (TetR), and pC194, conferring resistance to chloramphenicol (CmR), to specific strains of B. cereus, B. anthracis and B. thuringiensis (Ruhfel et al., 1984) and therefore became a tool for genetic manipulations in B. cereus s.l. Other CP-phages mediating generalized transduction in B. cereus s.l. were also described, such as CP53, stable at lower temperatures (Yelton and Thorne, 1970), CP54, displaying a broader host spectrum (Thorne, 1978) and CP54-Ber, isolated from CP-54 lysates and infecting B. thuringiensis sv. berliner strain 1715 (Lecadet et al., 1980). Many non CP-phages were also described for their potential in generalized transduction in the B. cereus group (for extensive reviews, refer to Gillis and Mahillon, 2014 or Sorokin, 2012).
Transducing phages of B. cereus s.l. are grouped into three categories, based on their size and host range (Sorokin, 2012). A restricted host spectrum characterizes smaller phages. Medium-size phages show rather poor transduction efficiency, although they include CP51, the most-predominantly used phage for transduction experiments in B. cereus s.l. (Thorne, 1978). Large phages exhibit broader host spectra and were good candidates for early genome mapping experiments. For instance, the genome of B. thuringiensis strain 4042B was mapped using the phages TP13 and TP18 (Barsomian et al., 1984). Given its insecticidal properties, B. thuringiensis was the main focus of most transduction-based studies in the B. cereus group (Sorokin, 2012).
First hints of conjugation and premises of a new transfer tool
Forty years ago, González and co-workers reported, for the first time, a conjugation-like process between strains of B. thuringiensis during growth in mixed cultures, while studying the genetic relationship between the presence of plasmids and the production of delta-endotoxin crystals. Notably, the plasmid-borne origin of B. thuringiensis delta-endotoxins was shown, along with the transmissible nature of the “cry plasmids” (González et al., 1981, 1982). The transfer of a cry plasmid was also observed between strains of B. thuringiensis and B. cereus s.s. This conjugation-like event consisted in the transfer of a cry plasmid accompanied by the transfer of a cryptic conjugative plasmid, namely a mobilization event (González et al., 1981, 1982). It is important to note that Cry toxins, and therefore cry plasmids, were, and still often are, considered a major feature distinguishing B. thuringiensis from other B. cereus s.l. strains. Thus, the transmissibility of cry plasmids reinforced the running hypothesis that both species were closely related (González et al., 1982) and further blurred the lines of species distinction among members of the B. cereus group.
One of the obstacles in the study of B. thuringiensis genetics has for long been the lack of effective transformation tools. The newly described conjugation-based system provided researchers with a potential new “transformation” system for studying B. thuringiensis plasmids (Figure 1; González and Carlton, 1982). The examples mentioned hereinafter rely on mobilization events, rather than on conjugative transfers per se. Conjugative plasmids are usually naturally cryptic and transfers were followed by assessing the transmissibility of detectable mobilizable plasmids, like cry plasmids or small plasmids encoding an antibiotic resistance marker.
The newly proposed transfer tool was used to transfer the cry genes of B. thuringiensis sv. berliner 1715, cloned firstly into B. subtilis, to an acrystalliferous strain of B. thuringiensis sv. kurstaki and an “israelensis” strain. The acrystalliferous strain could then produce toxin crystals and the “israelensis” strain could therefore produce two types of delta-endotoxins, broadening its range of susceptible target insects (Klier et al., 1983). Besides interspecies matings between B. subtilis and B. thuringiensis, pAMβ1 was successfully exchanged in intergeneric matings between Streptococcus (Enterococcus) faecalis and B. thuringiensis (Lereclus et al., 1983, 1985). The conjugation-mediated transfer tool also served to identify a large 75-MDa transmissible plasmid – later named pBtoxis – responsible for producing Cry toxins in the mosquitocidal B. thuringiensis sv. israelensis (González and Carlton, 1984). The CmR plasmid pC194 and the TetR plasmid pBC16 were also successfully introduced into strains of B. thuringiensis by mobilization (Fischer et al., 1984; Loprasert et al., 1986). Other interspecies matings were later performed between B. subtilis and B. cereus s.l., where the transfer of pBC16 was mediated by the B. subtilis conjugative plasmid pLS20 (Koehler and Thorne, 1987).
In parallel, the transfer features of the large plasmids pXO11, pXO12, pXO13, pXO14, pXO15 and pXO16 originating from different serovars of B. thuringiensis were investigated by following the mobilization of pBC16 (Battisti et al., 1985; Reddy et al., 1987). All six plasmids could successfully be transferred to B. cereus s.s. and B. anthracis, except for pXO15, which was only transferred to B. cereus s.s. Because pXO12 carries Cry toxin determinants, its conjugation-mediated transfer resulted in transcipients with “hybrid” phenotypes, as B. cereus s.s. and B. anthracis transcipients could produce Cry toxins besides their characteristic “cereus” and “anthracis” toxins (Battisti et al., 1985). On the contrary, no function other than conjugation could characterize the five other plasmids (Reddy et al., 1987). With their ability to transfer a large range of plasmids from their respective donor strains, plasmids pXO11 to pXO16 were useful exchange tools for analyzing genetic determinants on plasmids of B. anthracis, B. cereus and B. thuringiensis (Battisti et al., 1985). For instance, both pXO12 and pXO14 successfully triggered the transfer of the “anthracis” plasmids pXO1 and pXO2 to plasmid-cured B. anthracis and B. cereus, while pXO16 was only capable of transferring pXO2, and pXO13 could mobilize none (Battisti et al., 1985; Reddy et al., 1987). In the case of pXO12-mediated mobilization of pXO1 and pXO2, the specific mobilization process is called conduction. Conduction usually calls for recombination events either between homologous or site-specific sequences, or mediated by transposable elements on one of the plasmids involved (Andrup, 1998). Here, cointegrates of pXO12 and pXO1 (or pXO2) were observed, and the recombination events were most likely mediated by the pXO12-encoded transposon Tn4430 (Green et al., 1989). In opposition to conduction, pXO12 mediated the transfer of the small plasmid pBC16 by donation, a process whereby both plasmids remain physically distinct and do not associate (Green et al., 1989; Andrup, 1998).
Chromosomal fragments were also discovered to transfer laterally at low frequencies alongside the transfer of a conjugative plasmid, by a mobilization-like mechanism (Aronson and Beckman, 1987). Chromosome mobilization was first noticed between strains of B. thuringiensis and B. cereus s.s., invariably accompanied by the transfer of at least one plasmid from the donor strain. The transfer efficiency of chromosomal fragments was reduced compared to the transfer efficiency of plasmids. The efficiency also varied for different chromosomal markers, suggesting that the localization of the marker could impact the transfer efficacy (Aronson and Beckman, 1987). Then, a study focusing on plasmid and chromosome transfers between B. thuringiensis sv. israelensis and various B. thuringiensis serovars showed that the transfer frequencies varied depending on the recipient strain and on the transferred plasmid (Wiwat et al., 1990). While transfer frequencies of chromosomal fragments using broth mating technique were very low, it was suggested that filter mating might improve such frequencies (Wiwat et al., 1990). With current knowledge, the multiple mobilization events witnessed in B. thuringiensis sv. israelensis can most likely be attributable to pXO16. With that, researchers’ observations of chromosome transfer and suggestion about improving transfer frequencies by filter mating were later confirmed (Makart et al., 2017, 2018; Hinnekens et al., 2019; see Section “pXO16, the unique conjugative master”).
Circling back to toxin-carrying plasmids, establishing the connection between plasmids and cry genes in B. thuringiensis opened the possibility about the anthrax toxin genes being plasmid-borne (Mikesell et al., 1983). The genetic determinants of both the protective antigen, i.e., one of the three proteins forming the anthrax bipartite toxins, and the anthrax capsule were indeed demonstrated to be encoded on two distinct large plasmids, pXO1 (Vodkin and Leppla, 1983) and pXO2 (Green et al., 1985), respectively (Figure 1). Isolating and characterizing strains of B. anthracis carrying both plasmids led to the conclusion that both pXO1 and pXO2 are involved in the pathogenicity and the virulence of B. anthracis (Uchida et al., 1985, 1986).
Conjugative plasmids of Bacillus cereus s.l.
Over the years, several plasmids from strains of B. cereus s.l. were characterized as conjugative, based either on experimental transfer evidence or on strong bioinformatics predictions (Table 1). Although recent works keep focusing on the importance of plasmids in the B. cereus group, especially for B. thuringiensis and its Cry toxins (Bolotin et al., 2017; Gillis et al., 2018; Fayad et al., 2019, 2021a; Zheng et al., 2020), few studies have investigated conjugative transfer per se. Furthermore, reported transfer events often consist in mobilization events without incriminating the implicated conjugative plasmid (Fguira et al., 2014). Therefore, only 20 conjugative plasmids of B. cereus s.l. have been reported to date, but the conjugative capacities of eight of them lack experimental proofs (Table 1).
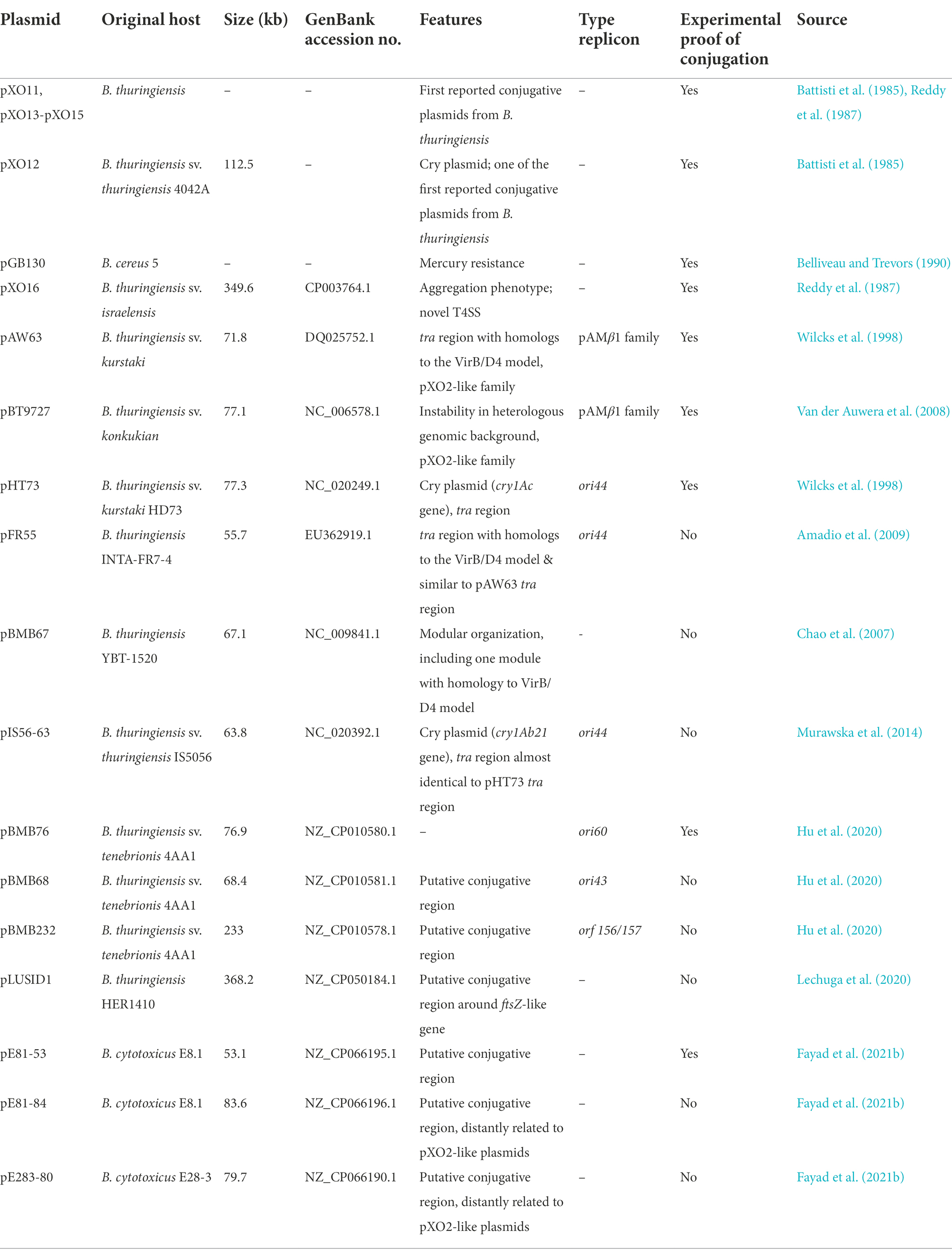
Table 1. Conjugative plasmids from B. cereus s.l. List of B. cereus s.l. plasmids reported as conjugative – or potentially conjugative – and their main characteristics.
The first plasmids described as conjugative, i.e., pXO11 to pXO15, were presented in Section “First hints of conjugation and premises of a new transfer tool.” In this section, the more recently reported B. cereus s.l. conjugative elements have been split into three groups for readily description: (i) the conjugative plasmids sharing the tra region of pAW63 and the pXO2-like plasmids, (ii) the unique conjugative system of pXO16, and (iii) other major conjugative plasmids, including pHT73 and plasmids sharing an ori44-like replicon.
pAW63 and other pXO2-like plasmids
The broad-host range plasmid pAW63
B. thuringiensis sv. kurstaki HD73, a common commercial biopesticide strain, was part of González and colleagues’ original study in early 1980s that discovered the transmissible nature of cry plasmids (González et al., 1981, 1982). This “kurstaki” strain was later shown to contain a second large plasmid, dubbed pAW63, aside from the cry plasmid pHT73 (Wilcks et al., 1998; see Section “Ori44-type replicon related plasmids”). This plasmid exhibited high transfer frequencies, reaching up to 100% efficiency between “kurstaki” strains in liquid mating, reminiscent of pXO16 transfer efficiency between strains of B. thuringiensis sv. israelensis (Wilcks et al., 1998; Van der Auwera et al., 2005; see next Section). pAW63 was classified as a broad host range plasmid, like the plasmid pIP501 initially isolated from Streptococcus (Evans and Macrina, 1983). It could indeed transfer to B. thuringiensis sv. israelensis, B. cereus, Bacillus licheniformis, B. subtilis and Bacillus (Lysinibacillus) sphaericus (Wilcks et al., 1998), as well as to more distantly-related species, such as Listeria innocua and E. faecalis (Van der Auwera et al., 2005). Studies also assessed pAW63 transfer efficiency in food matrices, especially dairy products and plant-based drinks (Van der Auwera and Mahillon, 2008; Modrie et al., 2010; see Section “Conjugation in foodstuffs”).
Studying pAW63 4.1-kb replicon highlighted a strong similarity between pAW63 largest replication protein, Rep63A, and the replication proteins RepR and RepE from pIP501 and pAMβ1, respectively (Wilcks et al., 1999). Both pIP501 and pAMβ1 belong to the pAMβ1-family of theta replicating plasmids, whose replication relies on the host DNA polymerase 1 and is single-stranded DNA-independent. As it shares these features, pAW63 was associated within the same plasmid family (Wilcks et al., 1999). The full sequence of pAW63 was published in 2005 and revealed a modular organization. One 42-kb module forms the tra region and encodes 15 CDSs putatively assigned to conjugative functions. Among these, three CDSs share homologies with the ATPases from the Vir secretion system, the CP VirD4, the motor ATPase VirB4 and the ATPase VirB11. Other CDSs share homologies with conjugation proteins from various Gram-positive T4SS (Van der Auwera et al., 2005). Interestingly, two group II-related introns interrupt two genes within the conjugative locus (Figure 2). On the one hand, ORFs 8 and 9 are split by the B.th.I1 intron and are predicted to encode the conjugative cell wall hydrolase. On the other hand, ORFs 14 and 15 are separated by the B.th.I2 intron but form one functional gene encoding the putative VirD4-like CP (Van der Auwera and Mahillon, 2008). Group II introns have also been found on the conjugative plasmid pRS01 from Lactococcus lactis (Belhocine et al., 2005) and on other plasmids from B. cereus s.l. (Tourasse et al., 2005). It is worth mentioning that most of the current knowledge on pAW63 tra region and associated conjugation mechanism was inferred from homologies with other T4SS and does not rely on experimental data.
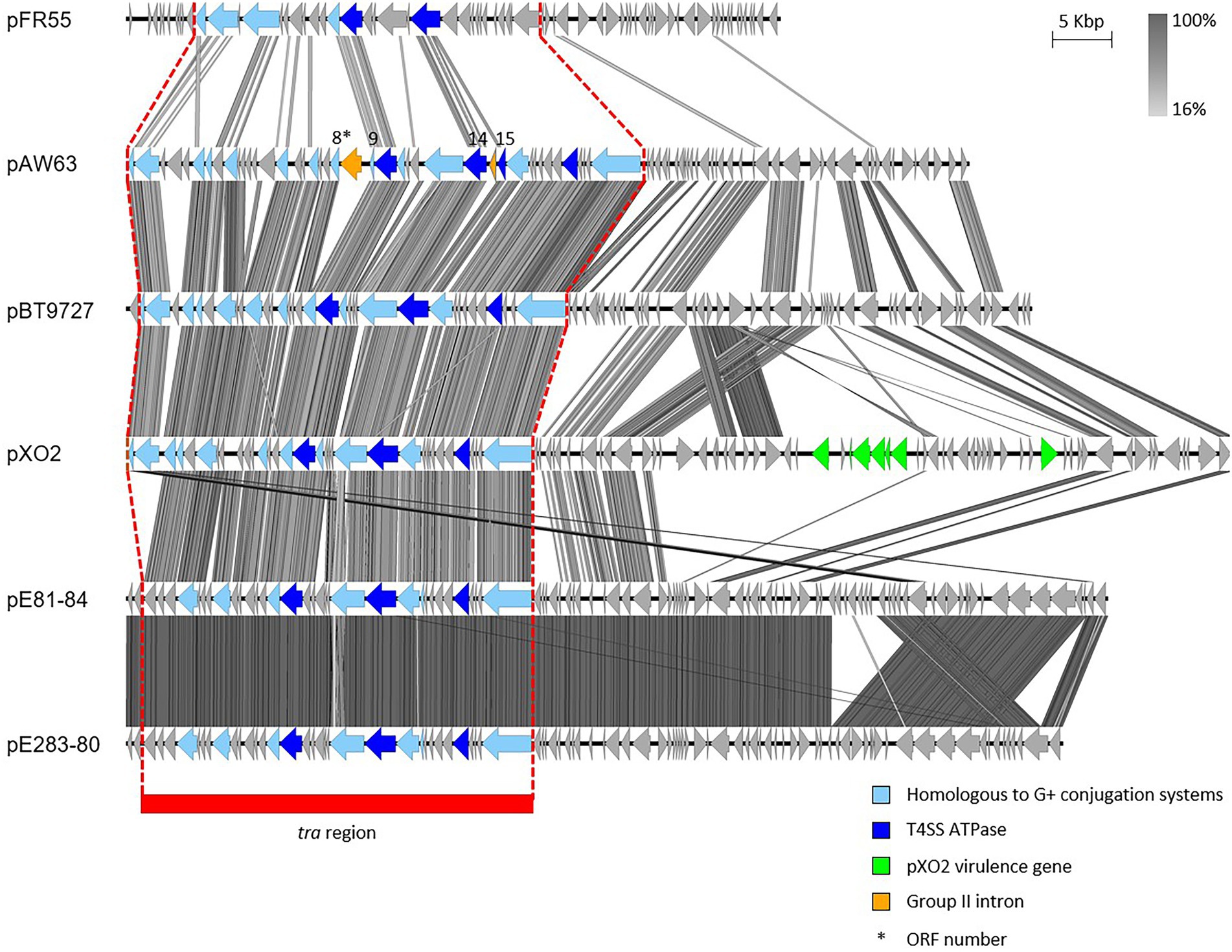
Figure 2. Sequence comparison of the (putative) conjugative plasmids of the pXO2-like family. The tra region of the plasmids pFR55, pAW63, pBT9727, pXO2, pE81-84 and pE283-80 share evident similarities. CDS depicted in light blue share homologies with other known conjugative systems; CDS depicted in dark blue are the putative ATPases; both CDS in orange in pAW63 sequence correspond to group II introns interrupting the genes encoded by the CDS 8–9 and 14–15; CDS colored in green encode Cap and Acp virulence factors of pXO2. The region highlighted in red corresponds to the tra region. The image was generated by Easyfig 2.0 software. Genbank accession numbers: pXO2: NC_007323.3; others: Table 1.
The family of pXO2-like plasmids
Analyzing pAW63 sequence indicated strong similarities with the “anthracis” plasmid pXO2 from B. anthracis and the pBT9727 plasmid from strain 97–27 of B. thuringiensis sv. konkukian (Van der Auwera et al., 2005). Indeed, among the 76 CDSs predicted on pAW63, 42 are common between the three plasmids, with 50 CDSs shared by pAW63 and pXO2 and 49 by pAW63 and pBT9727. Of specific interest is the high conservation of the region corresponding to the pAW63 transfer module in pXO2 and pBT9727 (Figure 2; Van der Auwera et al., 2005). Although pXO2 is mobilizable but non conjugative, pBT9727 was shown to promote its own transfer and to trigger the mobilization of small co-resident mobilizable plasmids (Van der Auwera et al., 2008). Based on comparative sequence analyses, all three plasmids were proposed to derive from a same ancestral conjugative plasmid, from which pAW63 and pBT9727 retained conjugation capacities, while pXO2 genetically drifted and lost the potential to conjugate (Van der Auwera et al., 2008). Most constituents of the tra region are intact in pXO2 compared to pAW63 and pBT9727. However, at least two putatively major elements for conjugation display punctual mutations in pXO2, including the virD4-like gene, presumably leading to frameshifts and disruption of the conjugative functions (Van der Auwera et al., 2005).
pXO2, pAW63 and pBT9727 compose the core of the pXO2-like plasmid family, but other conjugative – or putatively conjugative – plasmids also share some similarities. Three closely related plasmids from B. cytotoxicus were recently described as distantly related to the trio of pXO2-like plasmids (Fayad et al., 2021b). Two of them, pE81-84 and pE283-80, also harbor a region reminiscent of the tra region of pAW63 and kin, suggesting a conjugative potential for both plasmids. As illustrated on Figure 2, these two plasmids are highly similar, and a large portion of their sequence is very similar to that of pXO2. The putative conjugative region encodes three candidates for ATPase functions, including one putative CP, as well as candidates for transglycosylase and relaxase functions. Although they harbor all signature elements of a functional T4SS, experimental data are lacking to confirm the conjugative nature of pE81-84 and pE283-80.
pFR55, a plasmid isolated from B. thuringiensis INTA-FR7-4, presents a tra region with multiple CDSs sharing similarities to CDSs involved in pXO2-like plasmid conjugation (Figure 2; Amadio et al., 2009). pFR55 putative conjugative locus encodes a relaxase, a transglycosylase, a putative CP and an ATPase, all key elements of functional conjugative systems (Goessweiner-Mohr et al., 2013). Given these characteristics, pFR55 has a strong potential for conjugation, but experimental evidence of its self-transferability is lacking. Apart from its tra region, pFR55 does not share the common backbone of pXO2-like plasmids (Figure 2). For instance, its replication region belongs to the family of ori44-type replicon, like the well-studied cry plasmid pHT73 from B. thuringiensis sv. kurstaki HD73 (Amadio et al., 2009), and pFR55 forms therefore a separate group from pXO2-like plasmids.
Prevalence of pXO2-like plasmids
As pXO2-like transfer modules exhibit a significant degree of conservation amongst several described conjugative plasmids, the prevalence of such module was evaluated at a larger scale (Hu et al., 2009a, 2009b). pXO2-like plasmids/replicons were widespread in environmental isolates and had therefore a significant potential as conjugative plasmids. However, the proportion of strains demonstrating conjugative elements remained minor compared to the initial population of interest, especially from environmental samples. Indeed, in two independent studies, pXO2-like transfer modules were detected in only 5% of the B. cereus s.l. strains selected from both environmental samples and laboratory collections (Hu et al., 2009a, 2009b).
pXO16, the unique conjugative master
The conjugative plasmid pXO16, first identified by Reddy et al. in 1987, was thoroughly investigated by different research teams. Much attention was given to its phenotypic description, mainly for the aggregation phenotype induced by pXO16, as well as to the genetic components underlying its conjugation mechanism.
Aggregation-mediated conjugation
Aggregation-mediated conjugation mechanisms have been reported for various conjugative systems. The most studied example is the conjugative system of E. faecalis, which involves clumping of donor cells under the influence of a recipient-borne sex pheromone (Dunny et al., 1978). Cell aggregates also mediate plasmid transfer in conjugative systems of some lactic bacteria, such as L. lactis and Lactobacillus plantarum. In L. lactis, the aggregation-inducing factor is actually chromosomally encoded (Walsh and McKay, 1981; Van Der Lelie et al., 1991), while the aggregation-mediated conjugation system of L. plantarum is neither plasmid-encoded nor specific and facilitates the formation of mating pairs for various conjugative plasmids (Reniero et al., 1992). Macroscopic cell aggregates were also witnessed in broth matings of B. thuringiensis sv. israelensis and were attributed to pXO16 conjugation (Andrup et al., 1993; Jensen et al., 1995). pXO16-mediated aggregation was thoroughly studied and brought to light some key features of pXO16 conjugative system, e.g., the transmissibility of the aggregation phenotype alongside pXO16 transfer (Jensen et al., 1996), the mobilization of small co-resident plasmids (Andrup et al., 1996), and the importance of S-layer proteins in transfer efficiency (Wiwat et al., 1995).
The interest for pXO16-mediated aggregation got renewed when the aggregation-encoding genes were localized within a 25-kb agr region of pXO16 sequence (Makart et al., 2015). After over two decades of hypothesizing that aggregation was a prerequisite for pXO16 transfer, an aggregation-deficient mutant was constructed and exhibited reduced, yet effective, transfer in absence of the aggregation proteins (Makart et al., 2018). Thus, pXO16 aggregation facilitates its transfer but is not required. By studying B. thuringiensis sv. israelensis aggregation under atomic force microscopy (AFM), surface proteins encoded in the agr region were shown to mediate specific interactions between mating partners, resulting in strong adhesion forces (~2nN; Feuillie et al., 2018).
Kinetics of transfer and DNA capture properties
pXO16 transfer is characterized by high-speed, as transfer is completed in ca. 3.5–4 min, by the formation of mating pairs involving one donor and one recipient within large mating aggregates, and by the hypothetical low copy number of the plasmid (Andrup et al., 1998). Primary transfer of pXO16 can be detected within 10 min and peaks at 40–50 min after mating onset (Timmery et al., 2009). A 10-min recovery period is required in between two transfer events from a same donor, and a 40 min maturation time is necessary for newly formed transconjugants to turn into donors themselves (Andrup et al., 1998).
Back in 1987, pXO16 was first reported for triggering the transfer of the mobilizable plasmid pBC16 by a conjugation-like event between B. thuringiensis sv. israelensis and strains of B. cereus s.s. or B. anthracis (Reddy et al., 1987). Further work dived into pXO16 mobilization capacities and revealed that pXO16 can mobilize mobilizable – i.e. usually encoding an oriT and a relaxase, but no T4SS – as well as “non-mobilizable” – i.e. lacking all elements necessary for conjugation (oriT, relaxase and T4SS) – co-resident plasmids (Timmery et al., 2009). It can also trigger the capture of a plasmid from a recipient cell to the pXO16-harboring donor cell via a mechanism known as retro-mobilization (Timmery et al., 2009). More recently, studies reported that pXO16 could trigger the transfer of chromosomal loci between mating partners (Makart et al., 2017). Furthermore, chromosomal fragments of at least 791 kb could be transferred between strains of B. thuringiensis sv. israelensis and chromosomal markers were successfully transferred by pXO16 between strains of B. cytotoxicus (Makart et al., 2018; Koné et al., 2021). pXO16 transfer and capture capacities were also evaluated under different ecological and food-related conditions, such as the midgut of dipteran larvae or milk and rice pudding (Thomas et al., 2001; Van der Auwera et al., 2007; see Sections “Conjugation under ‘natural’ environmental conditions” and “Conjugation in foodstuffs”).
Conjugation apparatus, a novel type of T4SS
Despite detailed work on phenotypical features of pXO16 transfer, little was known about the genetics underlying its conjugation mechanism until pXO16 sequence was analyzed in 2015 (Makart et al., 2015). Analysis of the 434 CDSs, covering more than 85% of the 350-kb sequence, revealed very few relevant homologies with public databases. Among the few CDSs with a predicted functional annotation, none were related to any T4SS components identified so far, suggesting that pXO16 conjugative system is rather unique (Makart et al., 2015). Based on detailed bioinformatics studies and on experimental data, a 25-kb locus involved in conjugation was delimitated and referred to as the transfer of israelensis plasmid (tip) locus (Makart et al., 2018). Within the 16 CDSs forming the tip locus, genes encoding essential conjugative functions were identified, such as two ATPases and one cell wall hydrolase (Makart et al., 2018; Hinnekens et al., 2021). While these functions are widely recognized as essential in canonical T4SS, key elements, particularly the relaxase and the oriT, remain unidentified in pXO16 conjugative apparatus.
Transfer of pXO16 across the Bacillus cereus group
From first investigations, pXO16 host spectrum was thought to be rather restricted, with only few serovars of B. thuringiensis and few strains of B. cereus s.s. capable of receiving pXO16 (Jensen et al., 1996). These conclusions were based on the hypothesis that aggregation (Agr) was mandatory for the conjugation-mediated transfer of pXO16. Strains carrying pXO16, designated Agr+, could form macroscopic aggregates with some other strains (Agr− strains). Only those Agr− strains were considered as potential recipients for pXO16 (Table 2), while strains which could not form macroscopic aggregates with an Agr+ strain were ignored (Jensen et al., 1996). However, as indicated above, an aggregation-deficient variant of pXO16 was capable of conjugative transfer, albeit at dramatically reduced frequencies, indicating that the Agr phenotype is important but not mandatory for pXO16 transfer, as previously stated (Makart et al., 2018). Based on this new finding, previously overlooked non-aggregating strains of B. cereus s.l. were reported to successfully act as recipients for pXO16 transfer in liquid matings (Makart et al., 2018). Further work demonstrated that mimicking Agr by using solid mating conditions could compensate Agr deficiency and allow transfer to several B. cereus s.l. strains, including B. cytotoxicus, B. mycoides and strains appertaining to the same clade of B. anthracis but lacking the “anthracis” virulence genes (Hinnekens et al., 2019; Hinnekens and Mahillon, 2022; Table 2). Based on recent host spectrum data, assuming pXO16 transfer based on the Agr phenotype of the recipient strain is no longer relevant. Similarly, the Agr phenotype should not be confused with the wild-type or aggregation-deficient version of pXO16, as they, respectively, express strain-specific and plasmid-specific features.
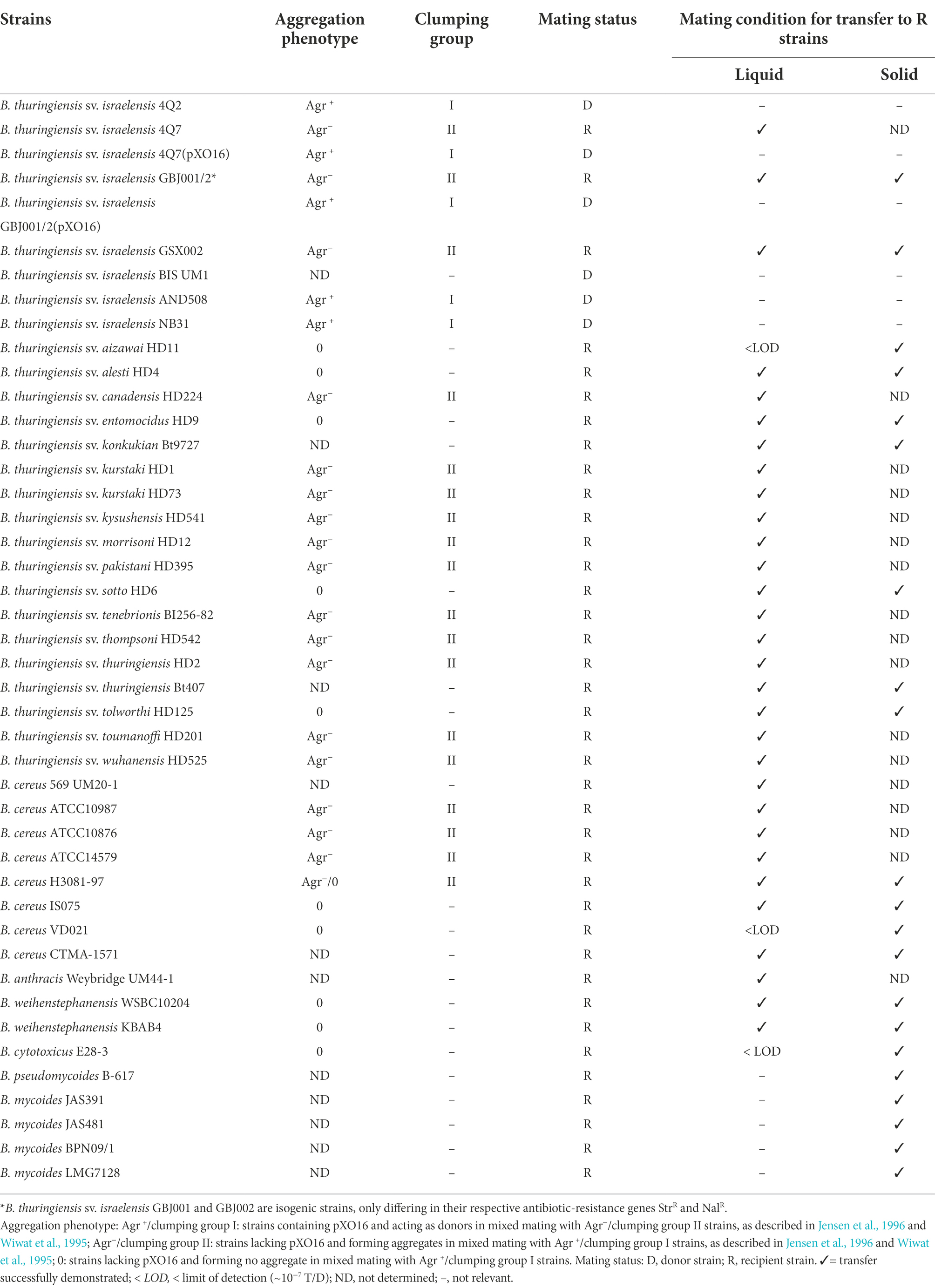
Table 2. pXO16 host spectrum. List of all the strains to which pXO16 transfer was successfully demonstrated under liquid or solid mating conditions, either by assessing the aggregation phenotype of the strain or by plating out transconjugants. This table also contains strains in which pXO16 was naturally found and that were used as donors in mating experiments. When appropriate, the aggregation phenotype/clumping status of the strain is mentioned. Original data can be found in the followings: Reddy et al. (1987); Andrup et al. (1993); Jensen et al. (1995, 1996); Makart et al. (2018); Hinnekens et al. (2019, 2021); Hinnekens and Mahillon (2022).
The highly efficient mobilization capacities of pXO16 could be of concern in terms of gene circulation, including antibiotic resistance genes, if these characteristics were to express in natural settings. However, pXO16 transferability within the B. cereus group was only detected under tightly controlled laboratory conditions, very distant from environmental conditions. For instance, the most efficient transfer set up tested to date consists in the highly artificial filter mating technique (Hinnekens et al., 2019). Also, pXO16 exerts a significant impact on its B. cereus s.l. hosts, apart from its natural host, suggesting a co-adaptation between pXO16 and its “israelensis” host (Hinnekens and Mahillon, 2022). Taken with the natural distribution of pXO16 apparently restricted to B. thuringiensis sv. israelensis (Gillis et al., 2018), the potential impact of pXO16-mediated conjugation within the B. cereus s.l. in natural environments seems rather restricted. Furthermore, to the best of our knowledge, pXO16 has never been transferred by conjugation outside of the B. cereus group, even under solid mating conditions, suggesting a limited impact of its incredible transfer features at a wider scale.
Ori44-type replicon related plasmids
The conjugative potential of the aforementioned pHT73 was first reported in early work on conjugation-like transfer within the B. cereus group (González et al., 1982; Wilcks et al., 1998). This cry plasmid encodes a cry1Ac gene and was successfully transferred to different members of the B. cereus group, including B. mycoides, under laboratory conditions (Hu et al., 2004). Despite successful transfer, pHT73 stability varied significantly between different recipient strains (Hu et al., 2004). Under environmental selective pressure, such instability could strongly impair the maintenance of the plasmid in those strains. Consequently, the highest stability rate was observed in “kurstaki” strains, indicating a biological adaptation of the plasmid to its original serotype (Hu et al., 2004). Such indications confirmed the positive correlation reported previously between the cry gene content of a specific strain and the strain serotype (Gaviria Rivera and Priest, 2003). pHT73 host spectrum was described as rather narrow at the time, reminiscent of the host limitations observed in early studies about pXO16 (Jensen et al., 1996). Transfer of pHT73 was later established to strains of B. anthracis (Yuan et al., 2010), and the compatibility of this cry plasmid with the anthrax plasmids pXO1 and pXO2 and the production of the Cry toxin by the “anthracis” recipient were also demonstrated. Such manipulations resulted in a laboratory strain of B. anthracis producing Cry toxin, similar to the strains of B. cereus s.s. and B. anthracis that could produce pXO12-borne Cry toxin upon conjugation-mediated acquisition of pXO12 (Battisti et al., 1985). Although the possibility of formation of such strains with hybrid phenotypes raised concerns about the spreading of insecticidal strains in the environment (Yuan et al., 2010), their actual occurrence in natural settings has so far never been reported, but remains to be further investigated.
Other potentially conjugative plasmids share the ori44-type replicon of pHT73, pFR55 (Amadio et al., 2009; see Section “pAW63 and other pXO2-like plasmids”) and pIS56-63 (Murawska et al., 2014). pIS56-63 sequence exhibits strong similarities with pHT73 as it carries a cry gene, multiple MGEs and conjugation-related genes, even though experimental data assessing its conjugative capacities are still missing (Figure 3). Another plasmid harbors a conjugative locus related to that of pHT73, the B. cytotoxicus plasmid pE81-53 (Figure 3), hosted by the same strain as the pXO2-like plasmid pE81-84 (Fayad et al., 2021b). Contrary to pE81-84, experimental evidence confirmed the conjugation potential of pE81-53 (Table 1). The conjugative loci of pHT73, pE81-84 and pIS56-63 encode three putative ATPases (Figure 3) and candidates for other conjugative functions, including homologues of TcpD and TcpE, two membrane proteins essential for the conjugative system of the clostridial plasmid pCW3 (Wisniewski et al., 2015).
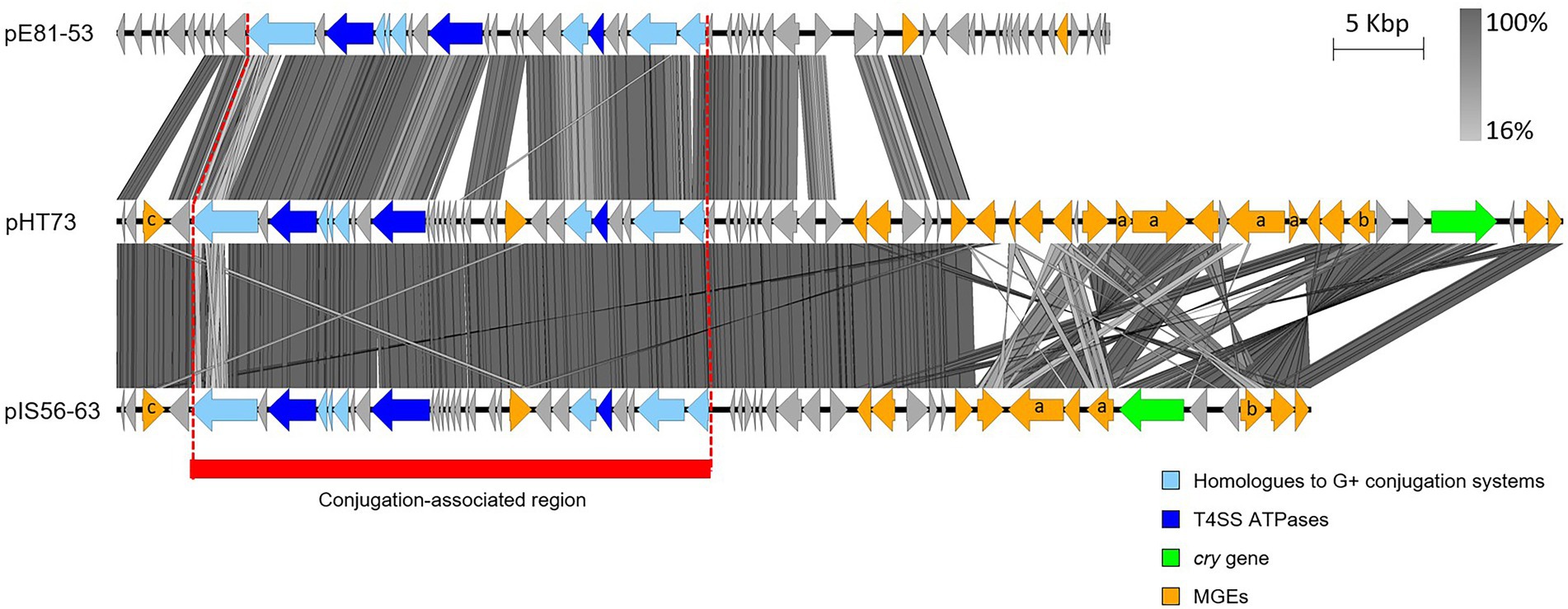
Figure 3. Sequence comparison of the (putative) conjugative plasmids similar to pHT73. The conjugation-associated region of the plasmids pHT73, pE81-53 and pIS56-63 share evident similarities. CDS depicted in light blue share homologies with other known conjugative systems; CDS depicted in dark blue are the putative ATPases; CDS in orange correspond to different types of MGEs (a = Tn/ b = group II intron / c = recombinase / no annotation = IS); CDS in green encode Cry toxins. The region highlighted in red corresponds to the region associated with conjugative functions. The image was generated by Easyfig 2.0 software. Genbank accession numbers: Table 1.
Bacillus cereus s.l.: A pool of conjugative candidates
As stated previously, the B. cereus s.l. group is rich with MGEs, including those with intercellular mobility, such as plasmids, or with intracellular mobility, such as IS, class II Tn and the group-specific B. cereus repeats (Bcr). These MGEs are considered as major contributors to the genomic plasticity seen in members of this group. Many studies have attributed the ecological specificity of each member of the group to lineage-specific plasmid acquisition throughout the species’ evolution (Méric et al., 2018). These plasmids differed in number and composition. For the former, a comparison of the average number of distinct plasmids per strain among B. cereus s.l. species showed that B. cereus s.s. carries an average of 2 plasmids per cell, whereas B. thuringiensis average plasmid content pikes at 6–7 plasmids per cell, followed closely by B. mycoides at 5–6 plasmids per cell (Fayad et al., 2019). B. thuringiensis plasmid numbers in fact ranged between 1 to 14 distinct plasmids per cell. B. thuringiensis sv. israelensis, for example, usually harbors nine extrachromosomal molecules, among which the toxin-encoding plasmid pBtoxis and the conjugative plasmid pXO16 (Gillis et al., 2018).
While the conjugative nature of some plasmids is classically uncovered through conjugation assays, nowadays, this nature could be highlighted by the in silico analysis of sequenced genomes, whose numbers are increasing day-by-day. Therefore, to get a better picture at the distribution of potentially conjugative plasmids within the B. cereus group, we compared the sequence of plasmids from sequenced B. cereus s.l. genomes with the sequence of known B. cereus s.l. conjugative plasmids presented in previous sections. In total, 365 complete B. cereus s.l. genomes were recovered from the NCBI genome assembly public database (https://www.ncbi.nlm.nih.gov/assembly; last accessed June 15, 2022) mainly from the “cereus” (115 genomes), “thuringiensis” (74 genomes) and “anthracis” (104 genomes) species. This amounted to 1,168 plasmids (Table 3). Among the 20 plasmids reported as conjugative in the B. cereus group (Table 1), no sequence nor information relative to the genes involved in conjugation are available for six of them, i.e., pXO11 to pXO15 and pGB130. Six other plasmids can be grouped into four classes based on the similarity of their putative transfer apparatus: pAW63 and pBT9727 form class 1, pFR55 forms class 2, pHT73 and pE81-53 form class 3 and pBMB76 forms class 4. A class 5 comprising the case of the conjugative plasmid pXO16 will also be discussed hereafter.
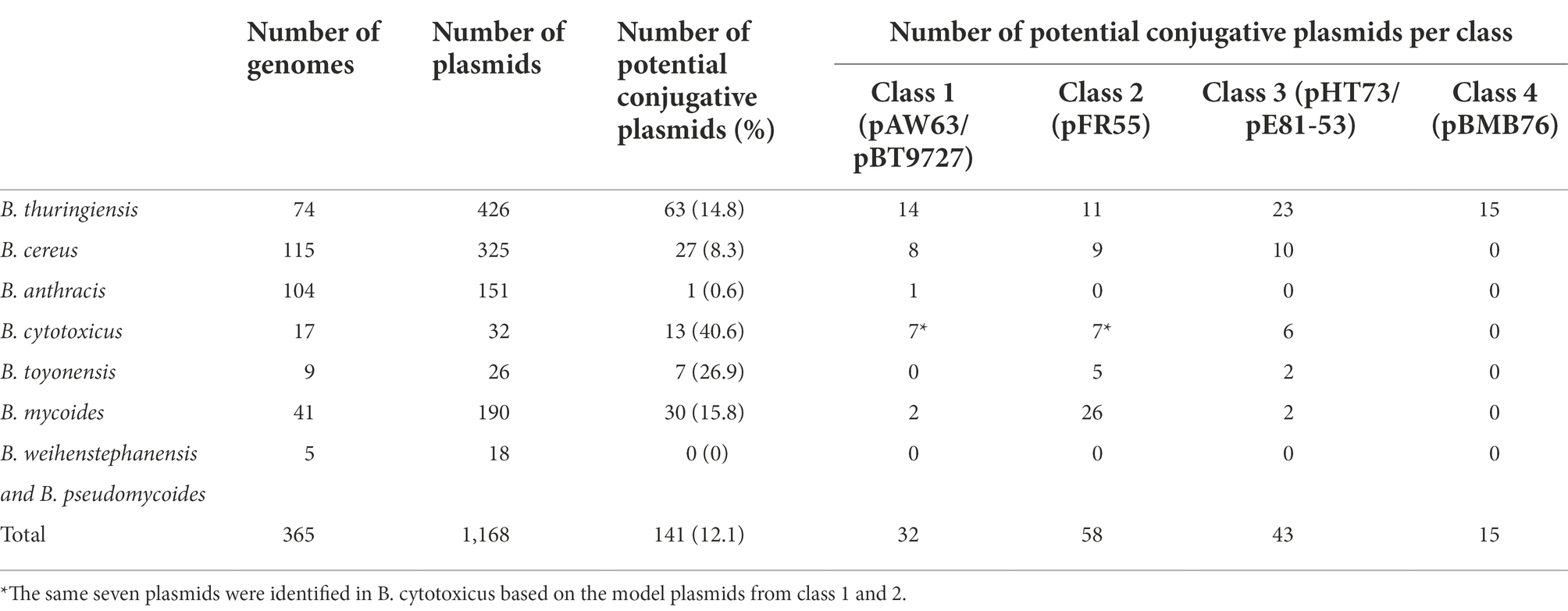
Table 3. Identification of potential conjugative plasmids within the B. cereus group. Plasmids retrieved from closed genomes of B. thuringiensis, B. cereus, B. anthracis, B. cytotoxicus, B. toyonensis, B. mycoides and B. weihenstephanensis-B. pseudomycoides were analyzed for their protein sequence similarities with the model conjugative plasmids of four distinct classes. The potential conjugative plasmids encode at least two ATPase-like and one peptidoglycan hydrolase-like proteins sharing ≥60% sequence coverage and ≥35% sequence identity with the respective conjugative elements of a model conjugative plasmid.
Based on proteins composition of model conjugative plasmids from each of the four classes, a tBLASTn approach was adopted to identify potential conjugative plasmids among the 1,168 analyzed via BLAST+ executables (Camacho et al., 2009; BLAST+ v2.12.0+). Results were then filtered based on the presence of hits with at least 60% sequence coverage and 35% sequence identity with two putative transfer ATPases and one putative conjugative peptidoglycan hydrolase. These three elements are often recognized as the basic components of conjugative T4SS (Goessweiner-Mohr et al., 2013; Schiwon et al., 2013). Plasmids previously described as conjugative (Table 1) were discarded from the results, as well as the model plasmids themselves. Overall, 12% (141 plasmids) were labelled as potentially conjugative, i.e., with self-transfer potential. Of these, 44.7% originated from B. thuringiensis, followed by 21.3% from B. mycoides and 19.1% from B. cereus. This was somewhat expected given that the highest number of analyzed plasmids (426, i.e., 36.47%) originated from B. thuringiensis.
A comparison based on the four classes of conjugative plasmids adopted in this mining process showed that 23% of putative conjugative plasmids were identified with the model plasmids from class 1, 41% from class 2, 30% from class 3 and 11% from class 4. The 3D graph shown in Figure 4 highlights the distribution of the 141 putative conjugative plasmids per model per species. Although the four mined classes were of conjugative plasmids originating from B. thuringiensis, this was not always reflected in their distribution per species. For instance, plasmids originally from B. mycoides formed 44.83% of the potential conjugative pFR55-like plasmids (class 2), vs. 18.97% from B. thuringiensis. Class 1, i.e., pAW63/pBT9727, both originally B. thuringiensis plasmids, were identified mostly in B. thuringiensis (43.75%) with B. cereus and B. cytotoxicus taking on second and third places with 25 and 21.88%, respectively. The distribution of these models across species raises the questions of the early speciation inside the B. cereus s.l. group, and whether some of these plasmids, or their likes, date back to the early days of B. cereus s.l. evolution. This is one of the reasons some studies suggest that these MGEs should be ignored as to not confound the phylogenetic inference process (Bazinet, 2017).
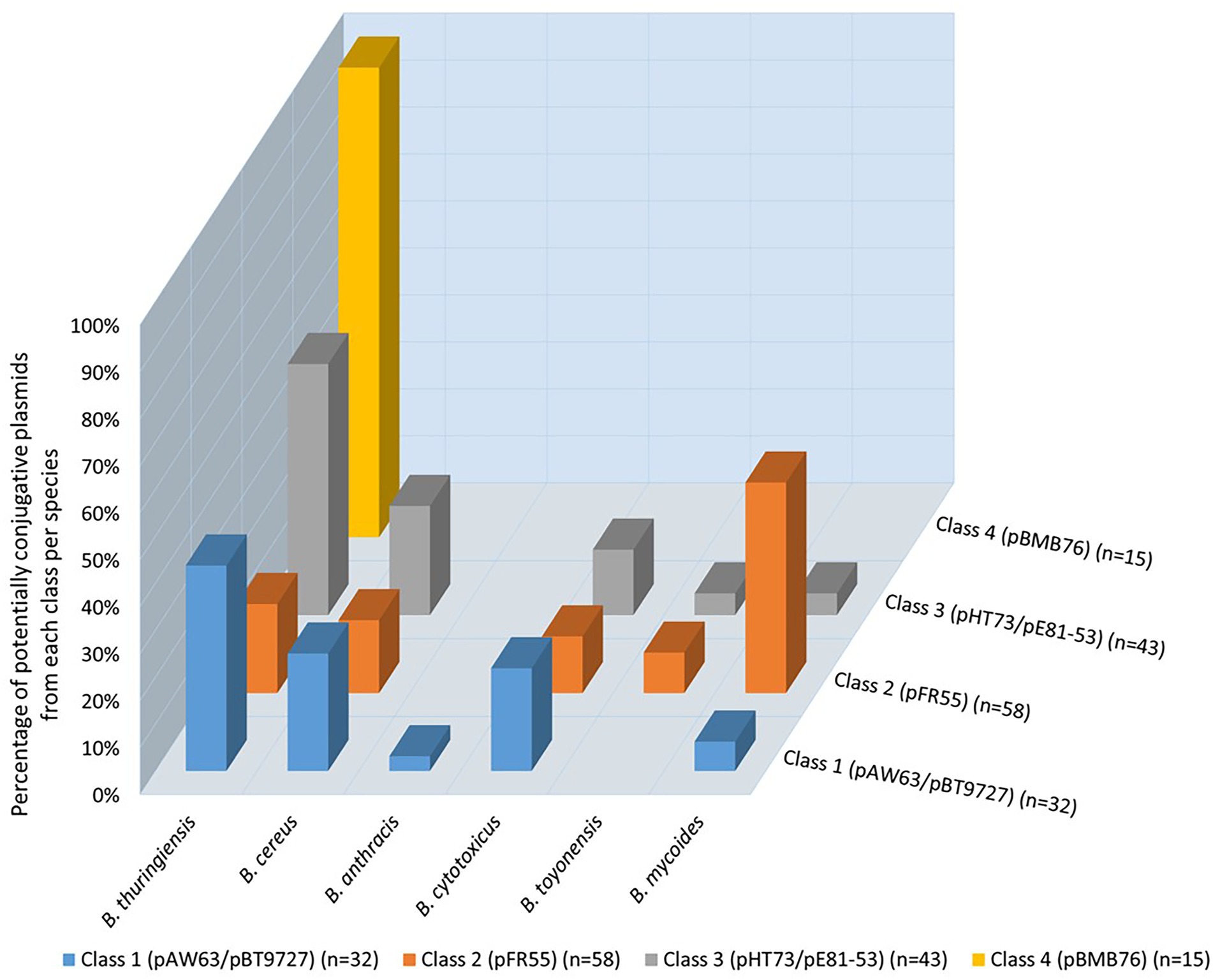
Figure 4. Distribution of potentially conjugative plasmids per species based on the four conjugative models (four classes). No hits were obtained with B. weihenstephanensis and B. pseudomycoides. Each bar shows the percentage of potentially conjugative plasmids identified based on the conjugative model of each of the four classes in the corresponding species. The total number of potentially conjugative plasmids identified is shown in parenthesis next to the corresponding class.
A fifth class included the conjugative model based on the putative transfer proteins of pXO16. However, when compared to the 1,168 analyzed plasmids, nine hits from strains of B. thuringiensis matched, five of which are confirmed as “israelensis” strains. The protein sequences from these nine plasmids share at least 99.8% of identity and 99% of coverage with the corresponding protein sequences from pXO16. The reported size of those plasmids is also almost identical to the size of pXO16. The data strongly indicate that the plasmids identified in the nine B. thuringiensis strains are actually copies of pXO16 itself. Based on this assumption, the nine plasmids were not counted into the putative conjugative plasmids and were considered copies of the conjugative model itself.
Conjugation into the wild and beyond
Conjugation under “natural” environmental conditions
Few years after the first description of conjugation-like events in B. thuringiensis, interest for plasmid transfer under environmentally relevant conditions started with cadavers of infected lepidopteran larvae as mating matrices (Jarrett and Stephenson, 1990). Transfer was assessed by following either the mobilization of plasmids conferring a specific trait, such as cry plasmids (Jarrett and Stephenson, 1990) or the mobilizable TetR plasmid pBC16 (Thomas et al., 2000), or by following the transfer of a conjugative plasmid itself, like pHT73 from B. thuringiensis sv. kurstaki (Vilas-Bôas et al., 1998; Yuan et al., 2007; Santos et al., 2010) or pXO16 from B. thuringiensis sv. israelensis (Thomas et al., 2001). Over the years, transfer was evaluated in multiple infected lepidopteran larvae, including Galleria mellonella and Bombyx mori (Jarrett and Stephenson, 1990; Santos et al., 2010), in dipteran larvae (Thomas et al., 2001) and from B. thuringiensis donor strains to B. thuringiensis or B. cereus s.s. recipient strains. As environment-like matrices are much more complex, results were sometimes diverging. For instance, a first study stated that pHT73 could only conjugate in dead larvae among growing vegetative bacteria, after spore germination post infection (Yuan et al., 2007). However, a second study nuanced the previous results by reporting efficient conjugative transfers in environments unsuitable for the multiplication of the recipient strain (Santos et al., 2010).
Preparations of B. thuringiensis are widely commercialized as biopesticide to fight against various insect pests. Upon application, numerous spores are released in the environment, especially in soil, where they can survive and multiply. Subsequent studies have focused on conjugal transmission in soils to evaluate the potential dissemination of plasmid-borne genes, e.g., cry genes. Significant conjugative transfer of the cry plasmid pHT73 was reported in non-amended sterile soil, indicating that genetic exchanges between strains of B. thuringiensis could take place in soil (Vilas-Bôas et al., 1998, 2000). However, no transfer could be witnessed when following pBC16 mobilization in non-sterile and non-manipulated soil (Thomas et al., 2000). Instead, rapid sporulation of the strains inoculated for conjugation was observed, which most probably prevented from plasmid exchange (Thomas et al., 2000). These results validated previous observations of transfer of the conjugative plasmid pFT30 between strains of B. cereus s.s. and B. subtilis in nutrient-amended soil, while no transfer could be detected in non-amended soil (Van Elsas et al., 1987). In this early study, the B. thuringiensis donor strain would also quickly germinate after introduction into a soil sample.
From another study ran two decades later, only one soil-borne isolate out of 75 exhibited mobilizing activities in triparental matings, i.e., carried a conjugative plasmid (Hu et al., 2009a). Such prevalence suggested that conjugation might be rare in soil, although possible, given results described above (Hu et al., 2009a). Moreover, mating conditions used in such studies might not be representative enough of the reality, despite the evident potential for plasmid exchange. Although survival and potential conjugation in soil microcosms is essential for evaluating the impact of B. thuringiensis-based pesticides, the question of the impact of conjugation on the genetic structure of local populations remains. In this context, recent work surveyed the transmissibility and occurrence of conjugative plasmids in confined soil plots (Hu et al., 2020). Within two years post application, the mobility in sprayed plots was 4–5 times higher compared to control plots. Collected data suggested that application of B. thuringiensis in specific confined habitats may result in this population supplanting the natural B. cereus population and therefore boosting mobility of genetic elements associated with entomocidal properties (Hu et al., 2020). The authors however conceded that the bacterial concentrations used in their study exceeded by far the concentrations applied on field for pest management.
Application of B. thuringiensis sv. israelensis-based products to waterways is essential for the fight against dipteran larvae (e.g., mosquitoes and blackflies). Similar to work in soil microcosms, the HGT potential in river water was assessed by using the conjugative plasmid pXO16 (Thomas et al., 2001). Besides significant transfer ratio, an increase in transfer rate at the onset of parental strain sporulation was revealed, suggesting that the optimal cell growth phase for conjugation corresponds to stationary or early sporulation phases (Thomas et al., 2001). Interestingly, another environment explored for the conjugative transfer of pXO16 was the intestine of gnotobiotic rats (Wilcks et al., 2008). In parallel to many studies focusing on conjugative transfer in natural environments between strains of B. thuringiensis, HGT between strains of B. anthracis was demonstrated within a rhizosphere model set up used to decipher B. anthracis survival in the rhizosphere of grass plants (Saile and Koehler, 2006). Although highly important in the case of B. cereus s.l., interest for transfer of genetic material in natural environments, and especially in the rhizosphere, goes beyond the B. cereus group (Normander et al., 1998; Sørensen and Jensen, 1998; Ronchel et al., 2000; Pearce et al., 2001).
Conjugation in foodstuffs
For food security and health management, bacterial contamination and gene transfer in food products are key issues. Studies have revealed the potential of genetic material exchange in microenvironments, including home cooking-related situations (Kruse and Sorum, 1994), and in common food products (Cocconcelli et al., 2003). Since some members of the group are opportunistic pathogens occasionally associated with food poisoning (Dierick et al., 2005; Naranjo et al., 2011; Delbrassinne et al., 2012), B. cereus s.l. behavior in food products has been documented, including its DNA exchange potential. Plasmid exchange was mostly illustrated by conjugation of the B. thuringiensis plasmids pAW63 and pXO16, as well as by mobilization of the small plasmids pUB110 and pC194. Studies focused on liquid and semi-liquid products, including cow milk, plant-based drinks and rice pudding (Van der Auwera et al., 2007; Timmery et al., 2009; Modrie et al., 2010). Overall, conjugative transfers usually occurred at least as efficiently in food preparations as in culture broth and mobilization of small plasmids was successfully reported. Kinetics studies revealed an impact of the different food matrices on both the potential for plasmid transfer and the timing of conjugation. Compared to frequencies obtained in typical culture broth, higher transfer frequencies were obtained in food matrices, resulting from an earlier onset of conjugation, an increased transfer frequency and an extended mating time (Modrie et al., 2010). Similarly, higher transfer frequencies of pHT73 were noticed in milk compared to LB medium (Yuan et al., 2010). Gene transfer can even occur in prepacked ready-to-eat food, and not solely in private or restaurant kitchens. Indeed, pAW63 showed hasty and more efficient conjugation properties in salt conditions mimicking those commonly found in food products like cheese and sausages (Beuls et al., 2012).
In a study looking at B. cereus strains used in probiotics, a large proportion of isolated strains were resistant to tetracycline and one of these strains harbored the genetic determinants of tetracycline resistance on an MGE, presenting a risk of antibiotic-resistance gene transfer (Zhu et al., 2016). From other strains of B. cereus s.l. isolated from both raw and ultra-heat-treated milk, around 20% of the non-B. anthracis strains carried pXO1-like and/or pXO2-like plasmids (Bartoszewicz and Marjańska, 2017). As described previously, the family of pXO2-like plasmids are well-known for their conjugative potential (i.e., pAW63 and pBT9727, see Section “pAW63 and other pXO2-like plasmids”), emphasizing the potential for conjugation in this case study. Based on work cited hereinabove, food products are proposed to favor exchange of genetic material (Bartoszewicz and Marjańska, 2017). By providing richer media, foodstuffs might indeed sustain bacterial proliferation, resulting in higher bacterial density, extended mating periods and more efficient plasmid transfer (Bartoszewicz and Marjańska, 2017).
Conjugation in confined environments
Bacteria are well-known for their adaptability to challenging environments and for their amazing genetic flexibility due to HGT. Strictly confined environments, such as the Antarctic Concordia Station (ACS) or the International Space Station (ISS), are especially rough due to space confinement, high levels of irradiation and/or microgravity. Microbial genetic exchange in such conditions raised scientists’ interest due to potential acquisition of new pathogenic traits. In spacecraft, modifications in microbiome could threaten astronaut health. Significant number of studies evaluated potential effects of real or simulated microgravity on bacterial characters, including growth, phenotype, virulence, metabolism and gene transfer (Senatore et al., 2018; Prasad et al., 2020; Bijlani et al., 2021; Mahnert et al., 2021). Most of these studies concluded in a strong influence of microgravity on bacterial communities, potentially leading to the evolution of space-specific properties that would not arise on Earth. Research groups also focused on the potential effects of microgravity on HGT. While an increase in genetic exchange between strains of Acinetobacter and Staphylococcus co-cultivated under simulated microgravity was reported, the root cause of this stimulation of genetic transfer was not investigated and transformation was proposed as the main underlying transfer mechanism (Urbaniak et al., 2021). Another research study examined strains of Staphylococcus and Enterococcus isolated from the ISS and the ACS for their conjugative properties. More than 80% of ISS strains and over 50% of ACS strains actually carried plasmids encoding signature transfer genes from well-characterized Gram-positive conjugative plasmids, like pIP501 (Schiwon et al., 2013).
Studying conjugation under space or confined conditions among the members of the B. cereus group was not left behind, particularly for strains of B. thuringiensis. The first study investigating plasmid-mediated conjugation under space flight conditions involved triparental matings of Gram-positive or Gram-negative bacteria and used B. thuringiensis as a Gram-positive model. Not only was it established that genetic exchange could occur by conjugation in space flight, but it was also suggested that conjugation and mobilization capacities were increased in the Gram-positive model (De Boever et al., 2007). However, such conclusions must be nuanced by the poor experimental controls and unexplained negative results in ground-based experiments. Furthermore, work published a couple of years later reported no statistically significant differences in conjugation and mobilization efficiency of B. thuringiensis in simulated microgravity – using three different simulation set-ups – compared to standard laboratory conditions (Beuls et al., 2012). Although no stimulation of genetic transfer was revealed, neither was inhibition. Further evidence of HGT in confined space habitats were gathered when one B. cereus s.l. strain, putatively harboring a conjugative plasmid, was identified among more than 40 Bacillus strains isolated from ISS and ACS and tested for conjugative abilities (Timmery et al., 2011). Strains were also tested for their capacity to receive foreign DNA by HGT, mimicking their potential recipient role in conjugative matings. Overall, two strains of B. cereus s.l. could acquire the conjugative plasmids pAW63 and pXO16, and four additional strains successfully acquired pXO16 (Timmery et al., 2011). Concerning pXO16 transfer, recent work demonstrated an extended host range of the plasmid via solid mating instead of liquid mating (Hinnekens et al., 2019). One should then consider the possibility that more ISS and ACS strains could act as pXO16 recipients when assessing transfer in solid matings. Altogether, collected data indicated that (i) most B. cereus s.l. strains analyzed for their plasmid profiles carried at least one plasmid and (ii) plasmid-mediated transfer, i.e., conjugation, could occur between strains of B. cereus s.l. originating from space environments and in space-mimicking conditions (De Boever et al., 2007; Beuls et al., 2009; Timmery et al., 2011). In light of astronaut health, such considerations are essential as modifications in “strain-plasmid” association could lead to harmless strains developing pathogenic characteristics (Timmery et al., 2011).
Concluding remarks and outlooks
Plasmids are key players in the lifestyle of B. cereus s.l., whether they encode major toxins, provide niche-specific traits, or promote exchange of genetic material. When it comes to conjugation, although a significant proportion of plasmids from B. cereus s.l. show potential for self-transferability, more elements come into play. Despite studies analyzing conjugal transfer in specific environments, like insect larvae, soil, foodstuff or confined environments, conjugation was mostly examined in laboratory environments, under highly artificial mating conditions. This applies to studies both in B. cereus s.l. and other bacterial groups. However, the natural environment for conjugation seems much more complex, likely occurring in dense bacterial communities in diverse natural niches (Shen et al., 2022). Furthermore, the different members of the B. cereus group tend to occupy different environmental niches, limiting their contact and interaction, and thereby limiting conjugation events. This is particularly true for B. anthracis which displays a specific and restricted natural lifestyle (Jensen et al., 2003). Moreover, all strains exhibiting a hybrid phenotype, e.g., strains of B. anthracis or B. cereus s.s. with B. thuringiensis cry plasmids, were so far obtained in laboratory settings and not isolated from natural environments (Battisti et al., 1985; Yuan et al., 2010).
In addition to the limited occurrence of “natural” conjugation per se, it is important to consider the relative stability of plasmids in different hosts. Broad-host spectrum plasmids have evolved to adapt to multi-host environments. However, in restricted niches, studies showed that in the hosts, plasmids have specifically adapted to alleviate their metabolic cost (Kottara et al., 2016). In other bacteria, maintaining such host-specific plasmids might be more costly, limiting the success of potential transfer overtime. Plasmid stability in a defined host relies on complex interactions between the plasmid itself, the host and the environment, which interact together to determine the balance between plasmid carriage burden and the profit from plasmid-borne genes (Hall et al., 2015; Kottara et al., 2018). More insight into the overall impact and risk of conjugation in the B. cereus group, and in other bacterial species, should be brought by further studies focusing on conjugation under environmental-relevant conditions, within complex bacterial communities.
Author contributions
PH, NF, and JM: conceptualization, methodology, validation, and writing – original draft preparation. PH and NF: Software, formal analysis, investigation, and data curation. JM: resources, Supervision, project administration, and funding. PH, NF, AG, and JM: writing – review and editing. All authors read and approved the final manuscript.
Funding
This work was supported by the National Foundation for Scientific Research (FNRS, Belgium – Research grants FNRS-CDR J.0144.20 to J.M. and FNRS 1.A682.19 to PH), by the International Office for Cooperation of the Université catholique de Louvain (UCLouvain; Bursaries to NF), and the Research Department of the Communauté française de Belgique (Concerted Research Action, ARC 17/22–084 grant to JM).
Acknowledgments
We really want to thank all our past and present collaborators on Bacillus cereus s.l. plasmids for their invaluable contributions. We are also particularly indebted to Lars Andrup, Gert B. Jensen and their colleagues for their original, seminal and inspiring works on the biology of B. cereus/B. thuringiensis plasmids.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amadio, A. F., Benintende, G. B., and Zandomeni, R. O. (2009). Complete sequence of three plasmids from Bacillus thuringiensis INTA-FR7-4 environmental isolate and comparison with related plasmids from the Bacillus cereus group. Plasmid 62, 172–182. doi: 10.1016/J.PLASMID.2009.07.005
Andrup, L. (1998). Conjugation in Gram-positive bacteria and kinetics of plasmid transfer. APMIS Suppl. 106, 47–55. doi: 10.1111/j.1600-0463.1998.tb05648.x
Andrup, L., Damgaard, J., and Wassermann, K. (1993). Mobilization of small plasmids in Bacillus thuringiensis subsp. israelensis is accompanied by specific aggregation. J. Bacteriol. 175, 6530–6536. doi: 10.1128/jb.175.20.6530-6536.1993
Andrup, L., Jørgensen, O., Wilcks, A., Smidt, L., and Jensen, G. B. (1996). Mobilization of “nonmobilizable” plasmids by the aggregation-mediated conjugation system of Bacillus thuringiensis. Plasmid 36, 75–85. doi: 10.1006/plas.1996.0035
Andrup, L., Smidt, L., Andersen, K., and Boe, L. (1998). Kinetics of conjugative transfer: a study of the plasmid pXO16 from Bacillus thuringiensis subsp. israelensis. Plasmid 40, 30–43. doi: 10.1006/plas.1998.1346
Arnold, B. J., Huang, I. T., and Hanage, W. P. (2021). Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 20, 206–218. doi: 10.1038/s41579-021-00650-4
Aronson, A. I., and Beckman, W. (1987). Transfer of chromosomal genes and plasmids in Bacillus thuringiensis. Appl. Environ. Microbiol. 53, 1525–1530. doi: 10.1128/aem.53.7.1525-1530.1987
Baillie, L., and Read, T. D. (2001). Bacillus anthracis, a bug with attitude! Curr. Opin. Microbiol. 4, 78–81. doi: 10.1016/S1369-5274(00)00168-5
Barrangou, R., and Marraffini, L. A. (2014). CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell 54, 234–244. doi: 10.1016/J.MOLCEL.2014.03.011
Barsomian, G. D., Robillard, N. J., and Thorne, C. B. (1984). Chromosomal mapping of Bacillus thuringiensis by transduction. J. Bacteriol. 157, 746–750. doi: 10.1128/JB.157.3.746-750.1984
Bartoszewicz, M., and Marjańska, P. S. (2017). Milk-originated Bacillus cereus sensu lato strains harbouring Bacillus anthracis-like plasmids are genetically and phenotypically diverse. Food Microbiol. 67, 23–30. doi: 10.1016/j.fm.2017.05.009
Battisti, L., Green, B., and Thorne, C. (1985). Mating system for transfer of plasmids among Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. J. Bacteriol. 162, 543–550. doi: 10.1128/JB.162.2.543-550.1985
Bazinet, A. L. (2017). Pan-genome and phylogeny of Bacillus cereus sensu lato. BMC Evol. Biol. 17, 1–16. doi: 10.1186/S12862-017-1020-1/TABLES/6
Belhocine, K., Yam, K. K., and Cousineau, B. (2005). Conjugative transfer of the Lactococcus lactis chromosomal sex factor promotes dissemination of the Ll.LtrB group II intron. J. Bacteriol. 187, 930–939. doi: 10.1128/JB.187.3.930-939.2005
Belliveau, B. H., and Trevors, J. T. (1990). Mercury resistance determined by a self-transmissible plasmid in Bacillus cereus 5. Biol. Met. 3, 188–196. doi: 10.1007/BF01140578
Berry, C., O’Neil, S., Ben-Dov, E., Jones, A. F., Murphy, L., Quail, M. A., et al. (2002). Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 68, 5082–5095. doi: 10.1128/AEM.68.10.5082-5095.2002
Beuls, E., Modrie, P., Deserranno, C., and Mahillon, J. (2012). High-salt stress conditions increase the pAW63 transfer frequency in Bacillus thuringiensis. Appl. Environ. Microbiol. 78, 7128–7131. doi: 10.1128/AEM.01105-12
Beuls, E., Van Houdt, R., Leys, N., Dijkstra, C., Larkin, O., and Mahillon, J. (2009). Bacillus thuringiensis conjugation in simulated microgravity. Astrobiology 9, 797–805. doi: 10.1089/ast.2009.0383
Bijlani, S., Stephens, E., Singh, N. K., Venkateswaran, K., and Wang, C. C. C. (2021). Advances in space microbiology. iScience 24:102395. doi: 10.1016/J.ISCI.2021.102395
Böhm, M.-E., Huptas, C., Krey, V. M., and Scherer, S. (2015). Massive horizontal gene transfer, strictly vertical inheritance and ancient duplications differentially shape the evolution of Bacillus cereus enterotoxin operons hbl, cytK and nhe. BMC Evol. Biol. 15:246. doi: 10.1186/s12862-015-0529-4
Bolotin, A., Gillis, A., Sanchis, V., Nielsen-LeRoux, C., Mahillon, J., Lereclus, D., et al. (2017). Comparative genomics of extrachromosomal elements in Bacillus thuringiensis subsp. israelensis. Res. Microbiol. 168, 331–344. doi: 10.1016/j.resmic.2016.10.008
Bravo, A., Gill, S. S., and Soberón, M. (2007). Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 49, 423–435. doi: 10.1016/j.toxicon.2006.11.022
Brézillon, C., Haustant, M., Dupke, S., Corre, J.-P., Lander, A., Franz, T., et al. (2015). Capsules, toxins and AtxA as virulence factors of emerging Bacillus cereus biovar anthracis. PLoS 9:e0003455. doi: 10.1371/journal.pntd.0003455
Cabezón, E., Ripoll-Rozada, J., Peña, A., de la Cruz, F., and Arechaga, I. (2015). Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. Cancer Res. 39, 81–95. doi: 10.1111/1574-6976.12085
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carroll, L. M., Cheng, R. A., Wiedmann, M., and Kovac, J. (2021). Keeping up with the Bacillus cereus group: taxonomy through the genomics era and beyond. Crit. Rev. Food Sci. Nutr. 62, 7677–7702. doi: 10.1080/10408398.2021.1916735
Chao, L., Qiyu, B., Fuping, S., Ming, S., Dafang, H., Guiming, L., et al. (2007). Complete nucleotide sequence of pBMB67, a 67-kb plasmid from Bacillus thuringiensis strain YBT-1520. Plasmid 57, 44–54. doi: 10.1016/J.PLASMID.2006.06.002
Cocconcelli, P. S., Cattivelli, D., and Gazzola, S. (2003). Gene transfer of vancomycin and tetracycline resistances among Enterococcus faecalis during cheese and sausage fermentations. Int. J. Food Microbiol. 88, 315–323. doi: 10.1016/S0168-1605(03)00194-6
Colavecchio, A., Cadieux, B., Lo, A., and Goodridge, L. D. (2017). Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the Enterobacteriaceae family – a review. Front. Microbiol. 8:1108. doi: 10.3389/fmicb.2017.01108
De Boever, P., Mergeay, M., Ilyin, V., Forget-Hanus, D., Van Der Auwera, G., and Mahillon, J. (2007). Conjugation-mediated plasmid exchange between bacteria grown under space flight conditions. Microgravity Sci. Technol. 19, 138–144. doi: 10.1007/BF02919469
de la Cruz, F., Frost, L. S., Meyer, R. J., and Zechner, E. L. (2010). Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34, 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
Delbrassinne, L., Andjelkovic, M., Dierick, K., Denayer, S., Mahillon, J., and Van Loco, J. (2012). Prevalence and levels of Bacillus cereus emetic toxin in rice dishes randomly collected from restaurants and comparison with the levels measured in a recent foodborne outbreak. Foodborne Pathog. Dis. 9, 809–814. doi: 10.1089/fpd.2012.1168
Dierick, K., Van Coillie, E., Swiecicka, I., Meyfroidt, G., Devlieger, H., Meulemans, A., et al. (2005). Fatal family outbreak of Bacillus cereus-associated food poisoning. J. Clin. Microbiol. 43:4277. doi: 10.1128/JCM.43.8.4277-4279.2005
Dunny, G. M., Brown, B. L., and Clewell, D. B. (1978). Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. U. S. A. 75, 3479–3483. doi: 10.1073/PNAS.75.7.3479
Ehling-Schulz, M., Koehler, T. M., and Lereclus, D. (2019). The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.GPP3-0032-2018
Evans, R. P., and Macrina, F. L. (1983). Streptococcal R plasmid pIP501: endonuclease site map, resistance determinant location, and construction of novel derivatives. J. Bacteriol. 154, 1347–1355. doi: 10.1128/JB.154.3.1347-1355.1983
Fayad, N., Kallassy Awad, M., and Mahillon, J. (2019). Diversity of Bacillus cereus sensu lato mobilome. BMC Genomics 20, 436–411. doi: 10.1186/s12864-019-5764-4
Fayad, N., Kambris, Z., El Chamy, L., Mahillon, J., and Awad, M. K. (2021a). A novel antidipteran Bacillus thuringiensis strain: unusual cry toxin genes in a highly dynamic plasmid environment. Appl. Environ. Microbiol. 87:e02294-20. doi: 10.1128/AEM.02294-20
Fayad, N., Koné, K. M., Gillis, A., and Mahillon, J. (2021b). Bacillus cytotoxicus genomics: chromosomal diversity and plasmidome versatility. Front. Microbiol. 12:789929. doi: 10.3389/fmicb.2021.789929
Fernández-López, C., Bravo, A., Ruiz-Cruz, S., Solano-Collado, V., Garsin, D. A., Lorenzo-Díaz, F., et al. (2014). Mobilizable rolling-circle replicating plasmids from Gram-positive bacteria: a low-cost conjugative transfer. Microbiol. Spectr. 2:8. doi: 10.1128/microbiolspec.PLAS-0008-2013
Feuillie, C., Valotteau, C., Makart, L., Gillis, A., Mahillon, J., and Dufrêne, Y. F. (2018). Bacterial sexuality at the nanoscale. Nano Lett. 18, 5821–5826. doi: 10.1021/acs.nanolett.8b02463
Fguira, I. B., Fourati, Z., Kamoun, F., Tounsi, S., and Jaoua, S. (2014). Isolation of the Bacillus thuringiensis plasmid carrying Bacthuricin F4 coding genes and evidence of its conjugative transfer. J. Infect. Dev. Ctries. 8, 727–732. doi: 10.3855/jidc.3552
Fiedoruk, K., Daniluk, T., Mahillon, J., Leszczynska, K., and Swiecicka, I. (2017). Genetic environment of cry1 genes indicates their common origin. Genome Biol. Evol. 9, 2265–2275. doi: 10.1093/GBE/EVX165
Fischer, H.-M., Lüthy, P., and Schweitzer, S. (1984). Introduction of plasmid pC194 into Bacillus thuringiensis by protoplast transformation and plasmid transfer. Arch. Microbiol. 139, 213–217. doi: 10.1007/BF00402002
Gaviria Rivera, A. M., and Priest, F. G. (2003). Pulsed field gel electrophoresis of chromosomal DNA reveals a clonal population structure to Bacillus thuringiensis that relates in general to crystal protein gene content. FEMS Microbiol. Lett. 223, 61–66. doi: 10.1016/S0378-1097(03)00347-1
Gillis, A., Fayad, N., Makart, L., Bolotin, A., Sorokin, A., Kallassy, M., et al. (2018). Role of plasmid plasticity and mobile genetic elements in the entomopathogen Bacillus thuringiensis serovar israelensis. FEMS Microbiol. Rev. 42, 829–856. doi: 10.1093/femsre/fuy034
Gillis, A., and Mahillon, J. (2014). Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future. Viruses 6, 2623–2672. doi: 10.3390/v6072623
Goessweiner-Mohr, N., Arends, K., Keller, W., and Grohmann, E. (2013). Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid 70, 289–302. doi: 10.1016/j.plasmid.2013.09.005
Goldenfeld, N., and Woese, C. (2007). Biology’s next revolution. Nature 445:369. doi: 10.1038/445369a
González, J. M., Brown, B. J., and Carlton, B. C. (1982). Transfer of Bacillus thuringiensis plasmids coding for delta-endotoxin among strains of B. thuringiensis and B. cereus. Proc. Natl. Acad. Sci. U. S. A. 79, 6951–6955. doi: 10.1073/PNAS.79.22.6951
González, J. M. J., and Carlton, B. C. (1982). “Plasmid transfer in Bacillus thuringiensis” in Genetic Exchange: A celebration and a New Generation. eds. E. U. N. Streips, S. H. Goodgal, W. R. Guild, and G. A. Wilson (New York: Dekker), 85–95.
González, J. M., and Carlton, B. C. (1984). A large transmissible plasmid is required for crystal toxin production in Bacillus thuringiensis variety israelensis. Plasmid 11, 28–38. doi: 10.1016/0147-619X(84)90004-0
González, J. M., Dulmage, H. T., and Carlton, B. C. (1981). Correlation between specific plasmids and delta-endotoxin production in Bacillus thuringiensis. Plasmid 5, 351–365. doi: 10.1016/0147-619X(81)90010-X
Green, B., Battisti, L., Koehler, T., Thorne, C., and Ivins, B. (1985). Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49, 291–297. doi: 10.1128/IAI.49.2.291-297.1985
Green, B. D., Battisti, L., and Thorne, C. B. (1989). Involvement of Tn4430 in transfer of Bacillus anthracis plasmids mediated by Bacillus thuringiensis plasmid pXO12. J. Bacteriol. 171, 104–113. doi: 10.1128/JB.171.1.104-113.1989
Grynberg, M., Li, Z., Szczurek, E., and Godzik, A. (2007). Putative type IV secretion genes in Bacillus anthracis. Trends Microbiol. 15, 191–195. doi: 10.1016/j.tim.2007.03.002
Guinebretière, M. H., Auger, S., Galleron, N., Contzen, M., de Sarrau, B., de Buyser, M. L., et al. (2013). Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int. J. Syst. Evol. Microbiol. 63, 31–40. doi: 10.1099/ijs.0.030627-0
Hall, J. P. J., Harrison, E., Lilley, A. K., Paterson, S., Spiers, A. J., and Brockhurst, M. A. (2015). Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ. Microbiol. 17, 5008–5022. doi: 10.1111/1462-2920.12901
Hinnekens, P., Koné, K. M., Fayad, N., Leprince, A., and Mahillon, J. (2019). pXO16, the large conjugative plasmid from Bacillus thuringiensis serovar israelensis displays an extended host spectrum. Plasmid 102, 46–50. doi: 10.1016/j.plasmid.2019.02.004
Hinnekens, P., Leprince, A., and Mahillon, J. (2021). TipB, a novel cell wall hydrolase, is required for efficient conjugative transfer of pXO16 from Bacillus thuringiensis sv. israelensis. Res. Microbiol. 172:103866. doi: 10.1016/j.resmic.2021.103866
Hinnekens, P., and Mahillon, J. (2022). Conjugation-mediated transfer of pXO16, a large plasmid from Bacillus thuringiensis sv. israelensis, across the Bacillus cereus group and its impact on host phenotype. Plasmid 122:102639. doi: 10.1016/J.PLASMID.2022.102639
Hoffmaster, A. R., Ravel, J., Rasko, D. A., Chapman, G. D., Chute, M. D., Marston, C. K., et al. (2004). Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. U. S. A. 101, 8449–8454. doi: 10.1073/pnas.0402414101
Hoton, F. M., Andrup, L., Swiecicka, I., and Mahillon, J. (2005). The cereulide genetic determinants of emetic Bacillus cereus are plasmid-borne. Microbiology 151, 2121–2124. doi: 10.1099/mic.0.28069-0
Hoton, F. M., Fornelos, N., N’Guessan, E., Hu, X., Swiecicka, I., Dierick, K., et al. (2009). Family portrait of Bacillus cereus and Bacillus weihenstephanensis cereulide-producing strains. Environ. Microbiol. Rep. 1, 177–183. doi: 10.1111/j.1758-2229.2009.00028.x
Hu, X., Hansen, B. M., Eilenberg, J., Hendriksen, N. B., Smidt, L., Yuan, Z., et al. (2004). Conjugative transfer, stability and expression of a plasmid encoding a cry1Ac gene in Bacillus cereus group strains. FEMS Microbiol. Lett. 231, 45–52. doi: 10.1016/S0378-1097(03)00925-X
Hu, X., Hansen, B. M., Yuan, Z., Johansen, J. E., Eilenberg, J., Hendriksen, N. B., et al. (2005). Transfer and expression of the mosquitocidal plasmid pBtoxis in Bacillus cereus group strains. FEMS Microbiol. Lett. 245, 239–247. doi: 10.1016/j.femsle.2005.03.012
Hu, X., Huang, D., Ogalo, J., Geng, P., Yuan, Z., Xiong, H., et al. (2020). Application of Bacillus thuringiensis strains with conjugal and mobilizing capability drives gene transmissibility within Bacillus cereus group populations in confined habitats. BMC Microbiol. 20, 363–312. doi: 10.1186/s12866-020-02047-4
Hu, X., Swiecicka, I., Timmery, S., and Mahillon, J. (2009a). Sympatric soil communities of Bacillus cereus sensu lato: population structure and potential plasmid dynamics of pXO1- and pXO2-like elements. FEMS Microbiol. Ecol. 70, 344–355. doi: 10.1111/j.1574-6941.2009.00771.x
Hu, X., Van Der Auwera, G., Timmery, S., Zhu, L., and Mahillon, J. (2009b). Distribution, diversity, and potential mobility of extrachromosomal elements related to the Bacillus anthracis pXO1 and pXO2 virulence plasmids. Appl. Environ. Microbiol. 75, 3016–3028. doi: 10.1128/AEM.02709-08
Ilangovan, A., Connery, S., and Waksman, G. (2015). Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol. 23, 301–310. doi: 10.1016/j.tim.2015.02.012
Jarrett, P., and Stephenson, M. (1990). Plasmid transfer between strains of Bacillus thuringiensis infecting Galleria mellonella and Spodoptera littoralis. Appl. Environ. Microbiol. 56, 1608–1614. doi: 10.1128/AEM.56.6.1608-1614.1990
Jensen, G. B., Andrup, L., Wilcks, A., Smidt, L., and Poulsen, O. M. (1996). The aggregation-mediated conjugation system of Bacillus thuringiensis subsp. israelensis: host range and kinetics of transfer. Curr. Microbiol. 33, 228–236. doi: 10.1007/s002849900105
Jensen, G. B., Hanssen, B. M., Eilenberg, G., and Mahillon, J. (2003). The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5, 631–640. doi: 10.1046/j.1462-2920.2003.00461.x
Jensen, G. B., Wilcks, A., Petersen, S. S., Damgaard, J., Baum, J. A., and Andrup, L. (1995). The genetic basis of the aggregation system in Bacillus thuringiensis subsp. israelensis is located on the large conjugative plasmid pXO16. J. Bacteriol. 177, 2914–2917. doi: 10.1128/jb.177.10.2914-2917.1995
Klier, A., Bourgouin, C., and Rapoport, G. (1983). Mating between Bacillus subtilis and Bacillus thuringiensis and transfer of cloned crystal genes. Mol. Gen. Genet. 191, 257–262. doi: 10.1007/BF00334823
Koehler, T. M., and Thorne, C. B. (1987). Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J. Bacteriol. 169, 5271–5278. doi: 10.1128/jb.169.11.5271-5278.1987
Kolstø, A.-B., Tourasse, N. J., and Økstad, O. A. (2009). What sets Bacillus anthracis apart from other Bacillus species? Annu. Rev. Microbiol. 63, 451–476. doi: 10.1146/annurev.micro.091208.073255
Koné, K. M., Hinnekens, P., Jovanovic, J., Rajkovic, A., and Mahillon, J. (2021). New insights into the potential cytotoxic role of Bacillus cytotoxicus Cytotoxin K-1. Toxins 13:698. doi: 10.3390/TOXINS13100698
Kottara, A., Hall, J. P. J., Harrison, E., and Brockhurst, M. A. (2016). Multi-host environments select for host-generalist conjugative plasmids. BMC Evol. Biol. 16, 1–4. doi: 10.1186/S12862-016-0642-Z/FIGURES/1
Kottara, A., Hall, J. P. J., Harrison, E., and Brockhurst, M. A. (2018). Variable plasmid fitness effects and mobile genetic element dynamics across Pseudomonas species. FEMS Microbiol. Ecol. 94:172. doi: 10.1093/femsec/fix172
Kruse, H., and Sorum, H. (1994). Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 60, 4015–4021. doi: 10.1128/AEM.60.11.4015-4021.1994
Lecadet, M. M., Blondel, M. O., and Ribier, J. (1980). Generalized transduction in Bacillus thuringiensis var. berliner 1715 using bacteriophage CP-54Ber. J. Gen. Microbiol. 121, 203–212. doi: 10.1099/00221287-121-1-203
Lechner, S., Mayr, R., Francis, K. P., Pruo, B. M., Kaplan, T., Wieoner-Gunkel, E., et al. (1998). Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int. J. Syst. Bacteriol. 48, 1373–1382. doi: 10.1099/00207713-48-4-1373
Lechuga, A., Lood, C., Salas, M., van Noort, V., Lavigne, R., and Redrejo-Rodríguez, M. (2020). Completed genomic sequence of Bacillus thuringiensis HER1410 reveals a cry-containing chromosome, two megaplasmids, and an integrative plasmidial prophage. G3 10, 2927–2939. doi: 10.1534/G3.120.401361
Lereclus, D., Lecadet, M. M., Klier, A., Ribier, J., Rapoport, G., and Dedonder, R. (1985). Recent aspects of genetic manipulation in Bacillus thuringiensis. Biochimie 67, 91–99. doi: 10.1016/S0300-9084(85)80234-0
Lereclus, D., Menou, G., and Lecadet, M.-M. (1983). Isolation of a DNA sequence related to several plasmids from Bacillus thuringiensis after a mating involving the Streptococcus faecalis plasmid pAMβ1. Mol. Gen. Genet. 191, 307–313. doi: 10.1007/BF00334831
Liu, Y., Lai, Q., Göker, M., Meier-Kolthoff, J. P., Wang, M., Sun, Y., et al. (2015). Genomic insights into the taxonomic status of the Bacillus cereus group. Sci. Rep. 5:14082. doi: 10.1038/srep14082
Liu, Y., Lai, Q., and Shao, Z. (2018). Genome analysis-based reclassification of Bacillus weihenstephanensis as a later heterotypic synonym of Bacillus mycoides. Int. J. Syst. Evol. Microbiol. 68, 106–112. doi: 10.1099/ijsem.0.002466
Loprasert, S., Pantuwatana, S., and Bhumiratana, A. (1986). Transfer of plasmids pBC16 and pC194 into Bacillus thuringiensis subsp. israelensis. J. Invertebr. Pathol. 48, 325–334. doi: 10.1016/0022-2011(86)90061-3
Mahnert, A., Verseux, C., Schwendner, P., Koskinen, K., Kumpitsch, C., Blohs, M., et al. (2021). Microbiome dynamics during the HI-SEAS IV mission, and implications for future crewed missions beyond Earth. Microbiome 9:27. doi: 10.1186/S40168-020-00959-X
Makart, L., Commans, F., Gillis, A., and Mahillon, J. (2017). Horizontal transfer of chromosomal markers mediated by the large conjugative plasmid pXO16 from Bacillus thuringiensis serovar israelensis. Plasmid 91, 76–81. doi: 10.1016/j.plasmid.2017.04.001
Makart, L., Gillis, A., Hinnekens, P., and Mahillon, J. (2018). A novel T4SS-mediated DNA transfer used by pXO16, a conjugative plasmid from Bacillus thuringiensis serovar israelensis. Environ. Microbiol. 20, 1550–1561. doi: 10.1111/1462-2920.14084
Makart, L., Gillis, A., and Mahillon, J. (2015). pXO16 from Bacillus thuringiensis serovar israelensis: almost 350 kb of terra incognita. Plasmid 80, 8–15. doi: 10.1016/j.plasmid.2015.03.002
Marraffini, L. A., and Sontheimer, E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845. doi: 10.1126/science.1165771
McInerney, J. O., McNally, A., and O’Connell, M. J. (2017). Why prokaryotes have pangenomes. Nat. Microbiol. 2, 1–5. doi: 10.1038/nmicrobiol.2017.40
Mei, X., Xu, K., Yang, L., Yuan, Z., Mahillon, J., and Hu, X. (2014). The genetic diversity of cereulide biosynthesis gene cluster indicates a composite transposon Tnces in emetic Bacillus weihenstephanensis. BMC Microbiol. 14, 1–11. doi: 10.1186/1471-2180-14-149
Mell, J. C., and Redfield, R. J. (2014). Natural competence and the evolution of DNA uptake specificity. J. Bacteriol. 196, 1471–1483. doi: 10.1128/JB.01293-13
Méric, G., Mageiros, L., Pascoe, B., Woodcock, D. J., Mourkas, E., Lamble, S., et al. (2018). Lineage-specific plasmid acquisition and the evolution of specialized pathogens in Bacillus thuringiensis and the Bacillus cereus group. Mol. Ecol. 27, 1524–1540. doi: 10.1111/MEC.14546
Mikesell, P., Ivins, B. E., Ristroph, J. D., and Dreier, T. M. (1983). Evidence for plasmid-mediated toxin production in Bacillus anthracis. Infect. Immun. 39, 371–376. doi: 10.1128/IAI.39.1.371-376.1983
Modrie, P., Beuls, E., and Mahillon, J. (2010). Differential transfer dynamics of pAW63 plasmid among members of the Bacillus cereus group in food microcosms. J. Appl. Microbiol. 108, 888–897. doi: 10.1111/J.1365-2672.2009.04488.X
Murawska, E., Fiedoruk, K., and Swiecicka, I. (2014). Modular genetic architecture of the toxigenic plasmid pIS56-63 harboring cry1Ab21 in Bacillus thuringiensis subsp. thuringiensis strain IS5056. Pol. J. Microbiol. 63, 147–156. doi: 10.33073/pjm-2014-020
Nakamura, L. K. (1998). Bacillus pseudomycoides sp. nov. Int. J. Syst. Bacteriol. 48, 1031–1035. doi: 10.1099/00207713-48-3-1031
Naranjo, M., Denayer, S., Botteldoorn, N., Delbrassinne, L., Veys, J., Waegenaere, J., et al. (2011). Sudden death of a young adult associated with Bacillus cereus food poisoning. J. Clin. Microbiol. 49, 4379–4381. doi: 10.1128/JCM.05129-11
Normander, B., Christensen, B. B., Molin, S., and Kroer, N. (1998). Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris). Appl. Environ. Microbiol. 64, 1902–1909. doi: 10.1128/AEM.64.5.1902-1909.1998
Palma, L., Muñoz, D., Berry, C., Murillo, J., Caballero, P., and Caballero, P. (2014). Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins (Basel) 6, 3296–3325. doi: 10.3390/toxins6123296
Patiño-Navarrete, R., and Sanchis, V. (2017). Evolutionary processes and environmental factors underlying the genetic diversity and lifestyles of Bacillus cereus group bacteria. Res. Microbiol. 168, 309–318. doi: 10.1016/J.RESMIC.2016.07.002
Pearce, D., Late Bazln, T. M., Lynch, J., Pearce, O., and Late Bazin, T. M. (2001). The impact of flow rate (simulated leaching) on plasmid transfer frequency between bacteria in a model rhizosphere system. J. Appl. Microbiol. 90, 953–961. doi: 10.1111/JAM.2001.90.6.953
Prasad, B., Richter, P., Vadakedath, N., Mancinelli, R., Krüger, M., Strauch, S. M., et al. (2020). Exploration of space to achieve scientific breakthroughs. Biotechnol. Adv. 43:107572. doi: 10.1016/J.BIOTECHADV.2020.107572
Ramarao, N., Lereclus, D., and Sorokin, A. (2015). The Bacillus cereus group. Mol. Med. Microbiol. 2–3, 1041–1078. doi: 10.1016/B978-0-12-397169-2.00059-7
Rasko, D. A., Rosovitz, M. J., Økstad, O. A., Fouts, D. E., Jiang, L., Cer, R. Z., et al. (2007). Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J. Bacteriol. 189, 52–64. doi: 10.1128/JB.01313-06
Reddy, A., Battisti, L., and Thorne, C. B. (1987). Identification of self-transmissible plasmids in four Bacillus thuringiensis subspecies. J. Bacteriol. 169, 5263–5270. doi: 10.1128/jb.169.11.5263-5270.1987
Reniero, R., Cocconcelli, P., Bottazzi, V., and Morelli, L. (1992). High frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J. Gen. Microbiol. 138, 763–768. doi: 10.1099/00221287-138-4-763
Ronchel, M. C., Ramos-Díaz, M. A., and Ramos, J. L. (2000). Retrotransfer of DNA in the rhizosphere. Environ. Microbiol. 2, 319–323. doi: 10.1046/J.1462-2920.2000.00109.X
Ruhfel, R. E., Robillard, N. J., and Thorne, C. B. (1984). Interspecies transduction of plasmids among Bacillus anthracis, B. cereus, and B. thuringiensis. J. Bacteriol. 157, 708–711. doi: 10.1128/JB.157.3.708-711.1984
Saile, E., and Koehler, T. M. (2006). Bacillus anthracis multiplication, persistence, and genetic exchange in the rhizosphere of grass plants. Appl. Environ. Microbiol. 72, 3168–3174. doi: 10.1128/AEM.72.5.3168-3174.2006
Santos, C. A., Vilas-Bôas, G. T., Lereclus, D., Suzuki, M. T., Angelo, E. A., and Arantes, O. M. N. (2010). Conjugal transfer between Bacillus thuringiensis and Bacillus cereus strains is not directly correlated with growth of recipient strains. J. Invertebr. Pathol. 105, 171–175. doi: 10.1016/J.JIP.2010.06.014
Schiwon, K., Arends, K., Rogowski, K. M., Fürch, S., Prescha, K., Sakinc, T., et al. (2013). Comparison of antibiotic resistance, biofilm formation and conjugative transfer of Staphylococcus and Enterococcus isolates from International Space Station and Antarctic Research Station Concordia. Microb. Ecol. 65, 638–651. doi: 10.1007/S00248-013-0193-4/FIGURES/1
Senatore, G., Mastroleo, F., Leys, N., and Mauriello, G. (2018). Effect of microgravity & space radiation on microbes. Future Microbiol. 13, 831–847. doi: 10.2217/fmb-2017-0251
Shen, Z., Tang, C. M., and Liu, G. Y. (2022). Towards a better understanding of antimicrobial resistance dissemination: what can be learnt from studying model conjugative plasmids? Mil. Med. Res. 9, 1–11. doi: 10.1186/S40779-021-00362-Z/FIGURES/2
Smillie, C., Pilar Garcillán-Barcia, M., Francia, M. V., Rocha, E. P. C., and De La Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/MMBR.00020-10
Sørensen, S. J., and Jensen, L. E. (1998). Transfer of plasmid RP4 in the spermosphere and rhizosphere of barley seedling. Antonie Van Leeuwenhoek 73, 69–77. doi: 10.1023/A:1000661115753
Sorokin, A. (2012). “Genetics and phages—from transduction and sequencing to recombineering” in Bacillus Thuringiensis Biotechnology. ed. E. Sansinenea (Dordrecht: Springer), 131–157.
Soucy, S. M., Huang, J., and Gogarten, J. P. (2015). Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472–482. doi: 10.1038/nrg3962
Thomas, D. J. I., Morgan, J. A. W., Whipps, J. M., and Saunders, J. R. (2000). Plasmid transfer between the Bacillus thuringiensis subspecies kurstaki and tenebrionis in laboratory culture and soil and in lepidopteran and coleopteran larvae. Appl. Environ. Microbiol. 66, 118–124. doi: 10.1128/AEM.66.1.118-124.2000
Thomas, D. J. I., Morgan, J. A. W., Whipps, J. M., and Saunders, J. R. (2001). Plasmid transfer between Bacillus thuringiensis subsp. israelensis strains in laboratory culture, river water, and dipteran larvae. Appl. Environ. Microbiol. 67, 330–338. doi: 10.1128/AEM.67.1.330-338.2001
Thorne, C. B. (1968). Transducing bacteriophage for Bacillus cereus. J. Virol. 2, 657–662. doi: 10.1128/JVI.2.7.657-662.1968
Thorne, C. B. (1978). Transduction in Bacillus thuringiensis. Appl. Environ. Microbiol. 35, 1109–1115. doi: 10.1128/AEM.35.6.1109-1115.1978
Timmery, S., Hu, X., and Mahillon, J. (2011). Characterization of bacilli isolated from the confined environments of the Antarctic Concordia station and the International Space Station. Astrobiology 11, 323–334. doi: 10.1089/ast.2010.0573
Timmery, S., Modrie, P., Minet, O., and Mahillon, J. (2009). Plasmid capture by the Bacillus thuringiensis conjugative plasmid pXO16. J. Bacteriol. 191, 2197–2205. doi: 10.1128/JB.01700-08
Tourasse, N. J., Stabell, F. B., Reiter, L., and Kolstø, A. B. (2005). Unusual group II introns in bacteria of the Bacillus cereus group. J. Bacteriol. 187, 5437–5451. doi: 10.1128/JB.187.15.5437-5451.2005
Uchida, I., Hashimoto, K., and Terakado, N. (1986). Virulence and immunogenicity in experimental animals of Bacillus anthracis strains harbouring or lacking 110 MDa and 60 MDa plasmids. J. Gen. Microbiol. 132, 557–559. doi: 10.1099/00221287-132-2-557
Uchida, I., Sekizaki, T., Hashimoto, K., and Terakado, N. (1985). Association of the encapsulation of Bacillus anthracis with a 60 megadalton plasmid. J. Gen. Microbiol. 131, 363–367. doi: 10.1099/00221287-131-2-363
Urbaniak, C., Grams, T., Mason, C. E., and Venkateswaran, K. (2021). Simulated microgravity promotes horizontal gene transfer of antimicrobial resistance genes between bacterial genera in the absence of antibiotic selective pressure. Life 11:960. doi: 10.3390/life11090960
Van der Auwera, G. A., Andrup, L., and Mahillon, J. (2005). Conjugative plasmid pAW63 brings new insights into the genesis of the Bacillus anthracis virulence plasmid pXO2 and of the Bacillus thuringiensis plasmid pBT9727. BMC Genomics 6, 1–14. doi: 10.1186/1471-2164-6-103
Van der Auwera, G., and Mahillon, J. (2008). Transcriptional analysis of the conjugative plasmid pAW63 from Bacillus thuringiensis. Plasmid 60, 190–199. doi: 10.1016/j.plasmid.2008.07.003
Van der Auwera, G. A., Timmery, S., Hoton, F., and Mahillon, J. (2007). Plasmid exchanges among members of the Bacillus cereus group in foodstuffs. Int. J. Food Microbiol. 113, 164–172. doi: 10.1016/j.ijfoodmicro.2006.06.030
Van der Auwera, G. A., Timmery, S., and Mahillon, J. (2008). Self-transfer and mobilisation capabilities of the pXO2-like plasmid pBT9727 from Bacillus thuringiensis subsp. konkukian 97-27. Plasmid 59, 134–138. doi: 10.1016/j.plasmid.2007.11.007
Van Der Lelie, D., Chavarri, F., Venema, G., and Gasson, M. J. (1991). Identification of a new genetic determinant for cell aggregation associated with lactose plasmid transfer in Lactococcus lactis. Appl. Environ. Microbiol. 57, 201–206. doi: 10.1128/AEM.57.1.201-206.1991
Van Elsas, J. D., Govaert, J. M., and Van Veen, J. A. (1987). Transfer of plasmid pFT30 between bacilli in soil as influenced by bacterial population dynamics and soil conditions. Soil Biol. Biochem. 19, 639–647. doi: 10.1016/0038-0717(87)90110-6
Vilas-Bôas, G. F., Vilas-Bôas, L. A., Lereclus, D., and Arantes, O. M. (1998). Bacillus thuringiensis conjugation under environmental conditions. FEMS Microbiol. Ecol. 25, 369–374. doi: 10.1111/j.1574-6941.1998.tb00488.x
Vilas-Bôas, L. A., Vilas-Bôas, G. F. L. T., Saridakis, H. O., Lemos, M. V. F., Lereclus, D., and Arantes, O. M. N. (2000). Survival and conjugation of Bacillus thuringiensis in a soil microcosm. FEMS Microbiol. Ecol. 31, 255–259. doi: 10.1111/J.1574-6941.2000.TB00691.X
Virolle, C., Goldlust, K., Djermoun, S., Bigot, S., and Lesterlin, C. (2020). Plasmid transfer by conjugation in gram-negative bacteria: from the cellular to the community level. Genes 11:1239. doi: 10.3390/genes11111239
Vodkin, M. H., and Leppla, S. H. (1983). Cloning of the protective antigen gene of Bacillus anthracis. Cells 34, 693–697. doi: 10.1016/0092-8674(83)90402-6
von Wintersdorff, C. J. H., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 1–10. doi: 10.3389/fmicb.2016.00173
Waksman, G. (2019). From conjugation to T4S systems in Gram-negative bacteria: a mechanistic biology perspective. EMBO Rep. 20:e47012. doi: 10.15252/embr.201847012
Walsh, P. M., and McKay, L. L. (1981). Recombinant plasmid associated cell aggregation and high-frequency conjugation of Streptococcus lactis ML3. J. Bacteriol. 146, 937–944. doi: 10.1128/JB.146.3.937-944.1981
Wilcks, A., Jayaswal, N., Lereclus, D., and Andrup, L. (1998). Characterization of plasmid pAW63, a second self-transmissible plasmid in Bacillus thuringiensis subsp. kurstaki HD73. Microbiology 144, 1263–1270. doi: 10.1099/00221287-144-5-1263
Wilcks, A., Smidt, L., Bahl, M. I., Hansen, B. M., Andrup, L., Hendriksen, N. B., et al. (2008). Germination and conjugation of Bacillus thuringiensis subsp. israelensis in the intestine of gnotobiotic rats. J. Appl. Microbiol. 104, 1252–1259. doi: 10.1111/j.1365-2672.2007.03657.x
Wilcks, A., Smidt, L., Økstad, O. A., Kolstø, A. B., Mahillon, J., and Andrup, L. (1999). Replication mechanism and sequence analysis of the replicon of pAW63, a conjugative plasmid from Bacillus thuringiensis. J. Bacteriol. 181, 3193–3200. doi: 10.1128/jb.181.10.3193-3200.1999
Wisniewski, J. A., Teng, W. L., Bannam, T. L., and Rood, J. I. (2015). Two novel membrane proteins, TcpD and TcpE, are essential for conjugative transfer of pCW3 in Clostridium perfringens. J. Bacteriol. 197, 774–781. doi: 10.1128/JB.02466-14
Wiwat, C., Panbangred, W., and Bhumiratana, A. (1990). Transfer of plasmids and chromosomal genes amongst subspecies of Bacillus thuringiensis. J. Ind. Microbiol. 6, 19–27. doi: 10.1007/BF01576173
Wiwat, C., Panbangred, W., Mongkolsuk, S., Pantuwatana, S., and Bhumiratana, A. (1995). Inhibition of a conjugation-like gene transfer process in Bacillus thuringiensis subsp. israelensis by the anti-s-layer protein antibody. Curr. Microbiol. 30, 69–75. doi: 10.1007/BF00294185
Yelton, D. B., and Thorne, C. B. (1970). Transduction in Bacillus cereus by each of two bacteriophages. J. Bacteriol. 102, 573–579. doi: 10.1128/JB.102.2.573-579.1970
Yuan, Y. M., Hu, X. M., Liu, H. Z., Hansen, B. M., Yan, J. P., and Yuan, Z. M. (2007). Kinetics of plasmid transfer among Bacillus cereus group strains within lepidopteran larvae. Arch. Microbiol. 187, 425–431. doi: 10.1007/s00203-006-0206-5
Yuan, Y., Zheng, D., Hu, X., Cai, Q., and Yuan, Z. (2010). Conjugative transfer of insecticidal plasmid pHT73 from Bacillus thuringiensis to B. anthracis and compatibility of this plasmid with pXO1 and pXO2. Appl. Environ. Microbiol. 76, 468–473. doi: 10.1128/AEM.01984-09
Zheng, Z., Zhang, Y., Liu, Z., Dong, Z., Xie, C., Bravo, A., et al. (2020). The CRISPR-Cas systems were selectively inactivated during evolution of Bacillus cereus group for adaptation to diverse environments. ISME J. 14, 1479–1493. doi: 10.1038/s41396-020-0623-5
Keywords: Bacillus anthracis, Bacillus cereus, conjugation, plasmid, T4SS, Bacillus thuringiensis, Bacillus mycoides, Bacillus cytotoxicus
Citation: Hinnekens P, Fayad N, Gillis A and Mahillon J (2022) Conjugation across Bacillus cereus and kin: A review. Front. Microbiol. 13:1034440. doi: 10.3389/fmicb.2022.1034440
Edited by:
Ravindra Soni, Indira Gandhi Krishi Vishva Vidyalaya, IndiaReviewed by:
Arun Kumar Rai, Sikkim University, IndiaTheresa M. Koehler, University of Texas Health Science Center at Houston, United States
Copyright © 2022 Hinnekens, Fayad, Gillis and Mahillon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacques Mahillon, amFjcXVlcy5tYWhpbGxvbkB1Y2xvdXZhaW4uYmU=
 Pauline Hinnekens
Pauline Hinnekens Nancy Fayad
Nancy Fayad Annika Gillis
Annika Gillis Jacques Mahillon
Jacques Mahillon