- 1Department of Food Hygiene, Faculty of Veterinary Medicine, Shahid Chamran University of Ahvaz, Ahvaz, Iran
- 2Department of Microbiology, Faculty of Agriculture and Forestry, University of Helsinki, Helsinki, Finland
In order to develop strategies for preventing biofilm formation in the dairy industry, a deeper understanding of the interaction between different species during biofilm formation is necessary. Bacterial strains of the P. fluorescens group are known as the most important biofilm-formers on the surface of dairy processing equipment that may attract and/or shelter other spoilage or pathogenic bacteria. The present study used different strains of the P. fluorescens group as background microbiota of milk, and evaluated their interaction with Staphylococcus aureus, Bacillus cereus, Escherichia coli O157:H7, and Salmonella Typhimurium during dual-species biofilm formation on stainless steel surfaces. Two separate scenarios for dual-species biofilms were considered: concurrent inoculation of Pseudomonas and pathogen (CI), and delayed inoculation of pathogen to the pre-formed Pseudomonas biofilm (DI). The gram-positive pathogens used in this study did not form dual-species biofilms with P. fluorescens strains unless they were simultaneously inoculated with Pseudomonas strains. E. coli O157:H7 was able to form dual-species biofilms with all seven P. fluorescens group strains, both in concurrent (CI) and delayed (DI) inoculation. However, the percentage of contribution varied depending on the P. fluorescens strains and the inoculation scenario. S. Typhimurium contributed to biofilm formation with all seven P. fluorescens group strains under the CI scenario, with varying degrees of contribution. However, under the DI scenario, S. Typhimurium did not contribute to the biofilm formed by three of the seven P. fluorescens group strains. Overall, these are the first results to illustrate that the strains within the P. fluorescens group have significant differences in the formation of mono-or dual-species biofilms with pathogenic bacteria. Furthermore, the possibility of forming dual-species biofilms with pathogens depends on whether the pathogens form the biofilm simultaneously with the P. fluorescens group strains or whether these strains have already formed a biofilm.
Introduction
More than 50 validly named species and a large number of unclassified isolates make up Pseudomonas fluorescens complex group, one of the most diverse groups within the Pseudomonas genus (Mulet et al., 2010; Gomila et al., 2015; Garrido-Sanz et al., 2016). There are nine subgroups within this complex group, including P. fluorescens, P. jessenii, P. fragi, P. gessardii, P. corrugata, P. chlororaphis, P. mandelii, P. koreensis and P. protegens (Garrido-Sanz et al., 2016). Bacterial strains of the P. fluorescens group are among the most important spoilage bacteria in milk and dairy products, and have been frequently isolated from fresh milk and cheese (Wang and Jayarao, 2001; De Jonghe et al., 2011; Martin et al., 2011; Carrascosa et al., 2015). Their ability to grow at low temperatures allows them to outgrow other bacteria in cold raw milk (Fricker et al., 2011; von Neubeck et al., 2015; Meng et al., 2017). Furthermore, P. fluorescens group strains have also been reported to cause repeat and sporadic post-pasteurization contamination, which results in shorter shelf life for pasteurized milk (Reichler et al., 2018).
P. fluorescens group bacterial strains are important in the dairy industry not only because of their heat-resistant enzymes, but also because of their ability to adhere to and from biofilms on the surface of milk tanks and other dairy processing equipment (Shpigel et al., 2015; Stoeckel et al., 2016). They are therefore able to withstand harsh conditions such as cleaning-in-place (CIP) processes, allowing them to remain within the dairy processing plant for extended periods of time (Cherif-Antar et al., 2016). Aside from contaminating subsequent batches of milk passing the biofilm region, Pseudomonas biofilms may also attract and/or provide shelter for other (spoilage or pathogenic) bacteria.
Biofilm formation by spoilage and pathogenic bacteria is a serious concern in the dairy industry, and hence has received much attention. A variety of different spoilage and pathogenic bacteria are known to attach to the internal stainless steel surfaces of the raw milk storage tanks and pipelines, where they can grow and form mono-or multi-species biofilms (Austin and Bergeron, 1995; Sharma and Anand, 2002; Latorre et al., 2010; Marchand et al., 2012; Anand et al., 2014; Cherif-Antar et al., 2016). Although biofilms in the dairy industry are more likely to be formed by spoilage bacteria (due to the higher population), pathogenic bacteria may also participate in the biofilm formation process, which may result in dual-or multi-species biofilms (Shi and Zhu, 2009; van Houdt and Michiels, 2010; Schirmer et al., 2013; Makovcova et al., 2017). In such cases, the cooperative interspecies interactions within dual-or multi-species biofilms are likely increase their resistance to adverse conditions compared to single-species biofilms. On the contrary, interspecific competition may occur and cause antagonistic effects (Yang et al., 2011; Elias and Banin, 2012; Rendueles and Ghigo, 2012; Liu et al., 2016).
To our knowledge, there is no information on the interaction between P. fluorescens group strains and bacterial pathogens from different genera during biofilm formation. Hence, in the present study, we used different strains of the P. fluorescens complex group as background microbiota of milk and evaluated their interaction with Staphylococcus aureus, Bacillus cereus, Escherichia coli O157:H7, and Salmonella Typhimurium during dual-species biofilm formation on stainless steel surfaces. We considered two separate scenarios for dual-species biofilms; (i) concurrent inoculation of Pseudomonas and the pathogen, and (ii) delayed inoculation of the pathogen to the pre-formed Pseudomonas biofilm.
Materials and methods
Bacterial strains and culture conditions
This study used P. fluorescens ATCC 13525, six other P. fluorescens group bacterial strains previously isolated from cold raw milk: P. fluorescens 5a, P. fluorescens 21c, P. fluorescens 68a, P. veronii 25d, P. cedrina 69a, and P. simiae 77a (Zarei et al., 2020). S. aureus (ATCC 25923), B. cereus (PTCC 1154), E. coli O157:H7 (ATCC 43895), and S. Typhimurium (ATCC 14028) were also used in this study. Stock cultures of bacterial strains were stored at-70°C in Tryptic Soy Broth (TSB; Merck, Germany) supplemented with 25% (v/v) sterile glycerol (Merck, Germany). Bacterial strains were first activated by two successive transfers in TSB at 30°C for 48 h. To prepare the inoculum, 0.1 ml of activated culture was added to 10 ml of ultra-high temperature (UHT) milk (3.79% protein and 1.5% fat), and incubated at 30°C for 48 h. Commercial UHT milk from the same batch was used for all experiments throughout the study.
Biofilm formation on stainless steel surfaces
Single-species biofilm
To evaluate the ability of individual Pseudomonas strains and the pathogenic bacteria to produce a biofilm on stainless steel surfaces, overnight cultures of each strain were diluted to the final concentration of 106 CFU/ml in UHT milk. The inoculated UHT milk (2 ml) was added to 12-well plates containing 1 × 1 cm stainless steel coupons (AISI 304, 2B, Norsk Stål AS, Norway), and incubated at 7°C for 48 h before quantifying the biofilm.
Dual-species biofilms
Each of the seven Pseudomonas strains was tested in dual-culture with each individual pathogen. The individual strain cultures were added to UHT milk and combined to contain approximately 106 CFU/ml of each strain.
To investigate dual-species biofilms, two scenarios were considered. In the first scenario, an individual strain of Pseudomonas and the pathogen were inoculated concurrently (106 CFU/ml of each strain) in UHT milk samples, and the formation of dual-species biofilms was evaluated. To achieve this, (hereinafter referred to as concurrent inoculation; CI), the combined dual-cultures in UHT milk (2 ml) were added to 12-well plates containing 1 × 1 cm stainless steel coupons, and incubated at 7°C for 48 h before quantifying the biofilm.
In the second scenario, the possibility of the pathogen being added to the pre-formed Pseudomonas biofilms was evaluated. For this scenario, (hereinafter referred to as delayed inoculation; DI), the UHT milk inoculated with 106 CFU/ml of each individual Pseudomonas strain was added to 12-well plates containing stainless steel coupons, and incubated at 7°C. After 48 h of incubation, the wells were drained and replaced with 2 ml of UHT milk containing 106 CFU/ml of each individual pathogen. The plates were then incubated for another 48 h before quantifying the biofilm.
Biofilm quantification
At the end of the incubation period, the wells were drained and the plates were gently washed three times by adding sterile dH2O (2 ml) to the coupon wells, followed by swirling of the plates and pipetting to remove non-attached bacteria. Biofilm cells were scraped into 1 ml of physiological saline solution using a cell scraper, and resuspended by vigorous pipetting for 15 s. Serial decimal dilutions of the cells were plated onto Tryptic Soy Agar (TSA) plates, and colonies were counted after 36 h of incubation at 30°C (Zarei et al., 2020).
Bacterial contribution to biofilm communities
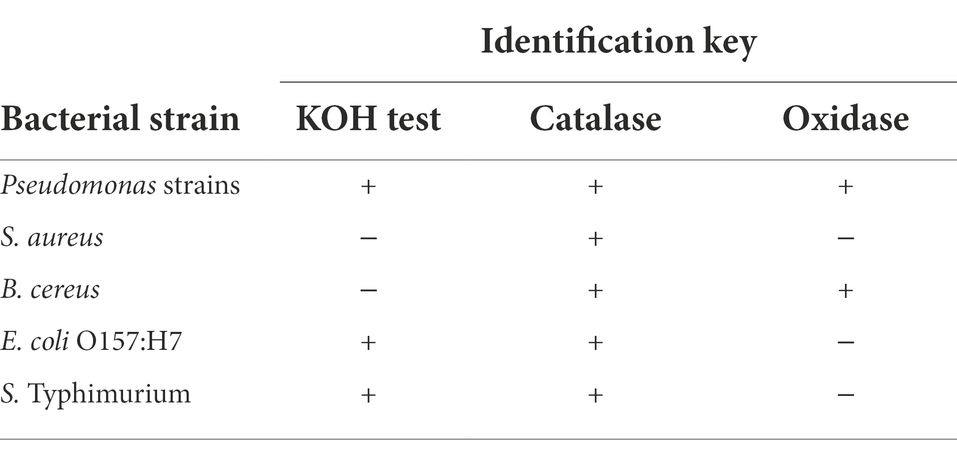
To determine the percent contribution of bacterial strains to dual-species biofilms, all colonies within a zone of the TSA plates were selected and identified using KOH test, oxidase, and catalase tests, as shown in Table 1. The size of the zones for colony selection was adjusted to have approximately 30 colonies within the zone of TSA plates, with a total of 30–300 colonies (Heir et al., 2018).
Statistical analysis
All experiments were replicated at least three times on different days. Results were analyzed using One-Way ANOVA (SPSS 20, SPSS Inc., Chicago, IL). The significance levels are expressed at a 95% confidence level (p ≤ 0.05) throughout.
Results
Single-species biofilms
Biofilm-forming ability of the selected strains of the P. fluorescens group and the pathogenic bacteria on stainless steel surfaces was evaluated in UHT milk. In general, after 48 h incubation at 7°C, the number of biofilm cells of the Pseudomonas strains was higher than the pathogens (p < 0.05). As shown in Figure 1, comparing Pseudomonas strains revealed that the highest number of biofilm cells on the stainless steel surfaces were from P. simiae 77a (5.89 ± 0.51 log CFU/cm2), and the lowest number was from P. cedrina 69a (3.66 ± 0.47 CFU/cm2). Among the pathogens, the highest and lowest number of biofilm cells were from S. aureus (2.87 ± 0.43 CFU/cm2) and E. coli O157:H7 (2.08 ± 0.31 CFU/cm2), respectively.
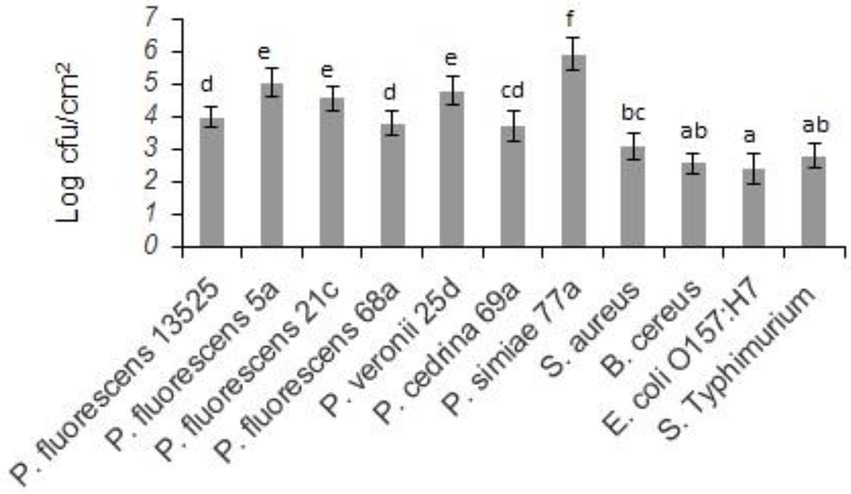
Figure 1. Biofilm cell counts of the selected P. fluorescens group strains and the pathogenic bacteria in mono-species biofilms on stainless steel surfaces after 48 h of incubation in UHT milk at 7°C. Different letters indicate significant differences between the strains.
Dual-species biofilms
Pseudomonas fluorescens group strains and Staphylococcus aureus
Evaluating the formation of dual-species biofilm by different P. fluorescens group strains and S. aureus showed differences between P. fluorescens group strains within each scenario, and also between scenarios. In the CI scenario, S. aureus was found in six of the seven biofilms, with varying degrees of contribution. As shown in Figure 2, the highest contributions were observed with P. simiae 77a (24.3%) and P. veronii 25d (20.4%), while the lowest was observed with P. fluorescens 5a (3.6%). In this scenario, S. aureus did not contribute to the biofilm formed by P. fluorescens 21c. Additionally, there were significantly more dual-species biofilm cells with P. fluorescens ATCC 13525, P. fluorescens 68a, P. veronii 25d, and P. simiae 77a than in their single-species biofilms (p < 0.05; Figure 3). In contrast, S. aureus did not contribute to the structure of any biofilms in the DI scenario. Biofilms formed in pure cultures of P. fluorescens group strains did not differ significantly from those formed in the DI scenario in terms of the number of biofilm cells (p > 0.05; Figure 3).
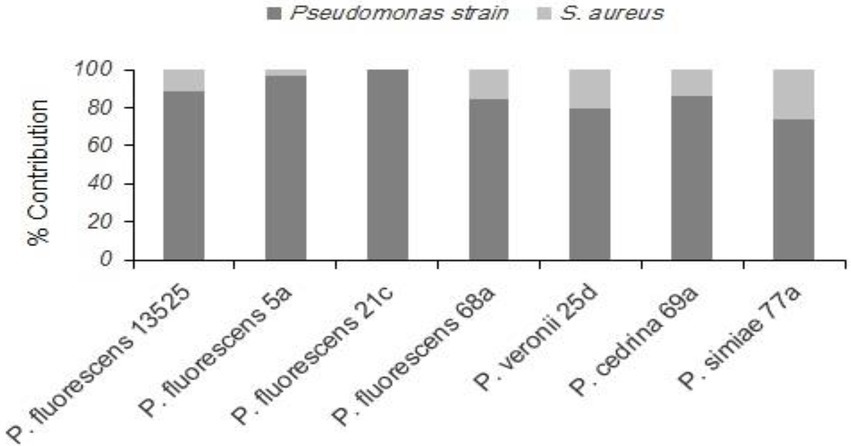
Figure 2. Contributions of different P. fluorescens group strains and S. aureus to dual-species biofilms on stainless steel surfaces under the CI scenario.
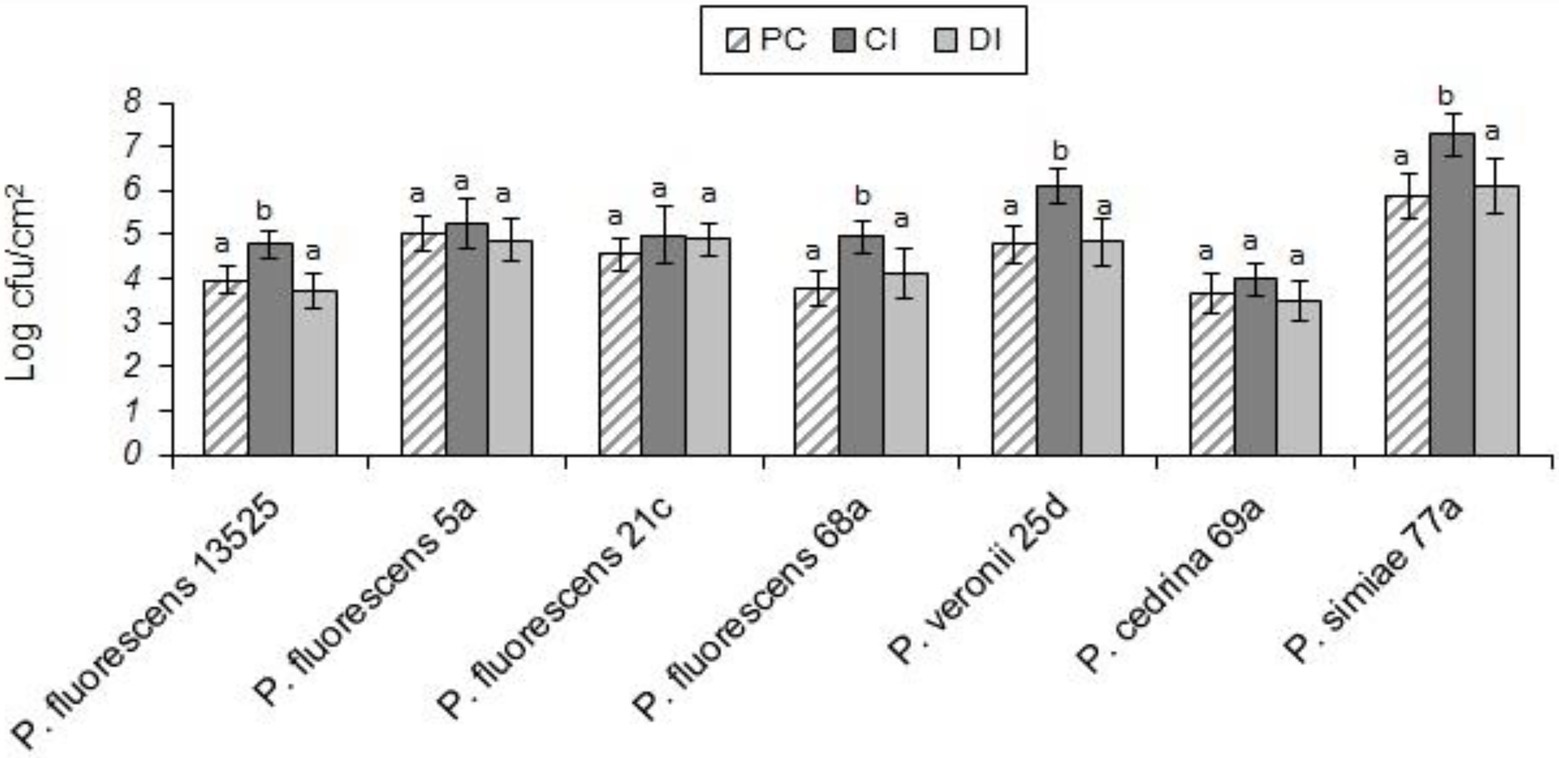
Figure 3. Total number of biofilm cells in mono- and dual-species biofilms of the seven P. fluorescens group strains with S. aureus on stainless steel surfaces. PC: pure culture biofilms; CI: dual-species biofilms under the CI scenario; DI: dual-species biofilms under the DI scenario. For each P. fluorescens strain, different letters indicate significant differences in the total number of biofilm cells.
Pseudomonas fluorescens group strains and Bacillus cereus
As with S. aureus, B. cereus did not contribute to the biofilm formation in the DI scenario, and the total number of biofilm cells did not differ significantly from those formed in pure cultures of P. fluorescens group strains. In contrast, B. cereus contributed to five out of seven biofilms in the CI scenario. As shown in Figure 4, the highest contributions were observed with P. simiae 77a (19.4%) and P. veronii 25d (14.3%), while the lowest was observed with P. fluorescens 5a (6.9%); B. cereus did not contribute to the biofilm formed by P. fluorescens 21c and P. cedrina 69a. The total number of biofilm cells in dual-species biofilms with B. cereus and P. fluorescens ATCC 13525, P. fluorescens 5a, P. fluorescens 68a, P. veronii 25d and P. simiae 77a were significantly higher than those in the pure cultures of P. fluorescens group strains (p < 0.05; Figure 5).
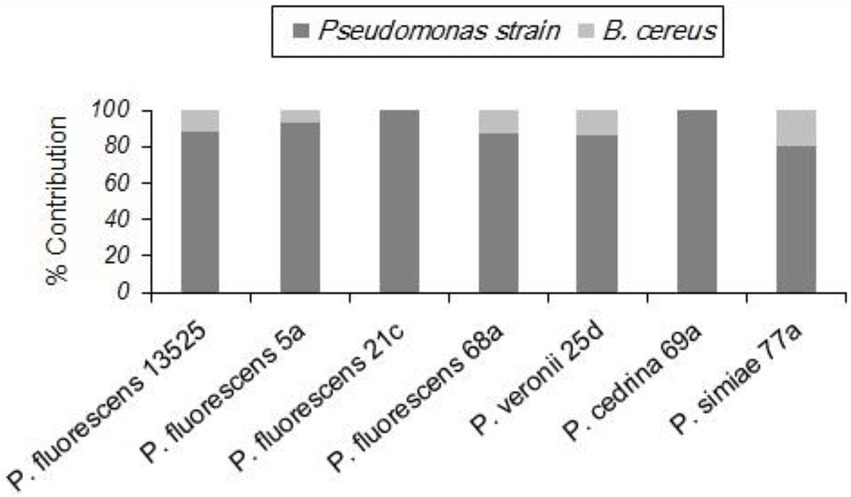
Figure 4. Contributions of different P. fluorescens group strains and B. cereus to dual-species biofilms on stainless steel surfaces in CI scenario.
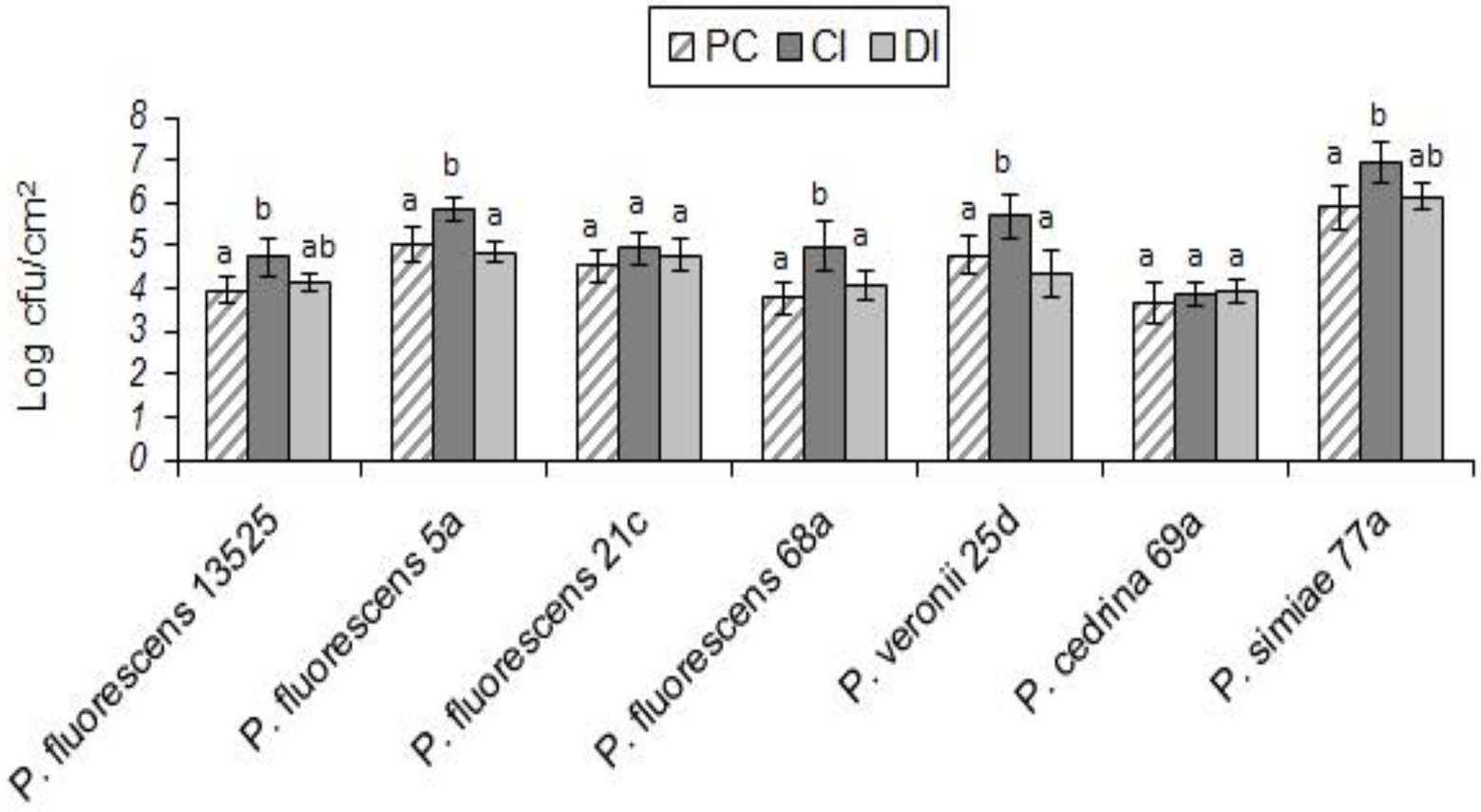
Figure 5. Total number of biofilm cells in mono- and dual-species biofilms of the seven P. fluorescens group strains with B. cereus on stainless steel surfaces. PC: pure culture biofilms; CI: dual-species biofilms under the CI scenario; DI: dual-species biofilms under the DI scenario. For each P. fluorescens strain, different letters indicate significant differences in the total number of biofilm cells.
Pseudomonas fluorescens group strains and Escherichia coli O157:H7
Unlike the two gram-positive bacteria that did not contribute to the biofilm structure in the DI scenario, E. coli O157:H7 contributed to biofilm formation with all seven P. fluorescens group strains in both CI and DI scenarios, to varying degrees. As shown in Figure 6, the highest percent contribution was found with P. cedrina 69a (27.8 and 16.7% in the CI and DI scenarios, respectively), and the lowest was found with P. fluorescens 5a (2.1 and 4.7% in the CI and DI scenarios, respectively).
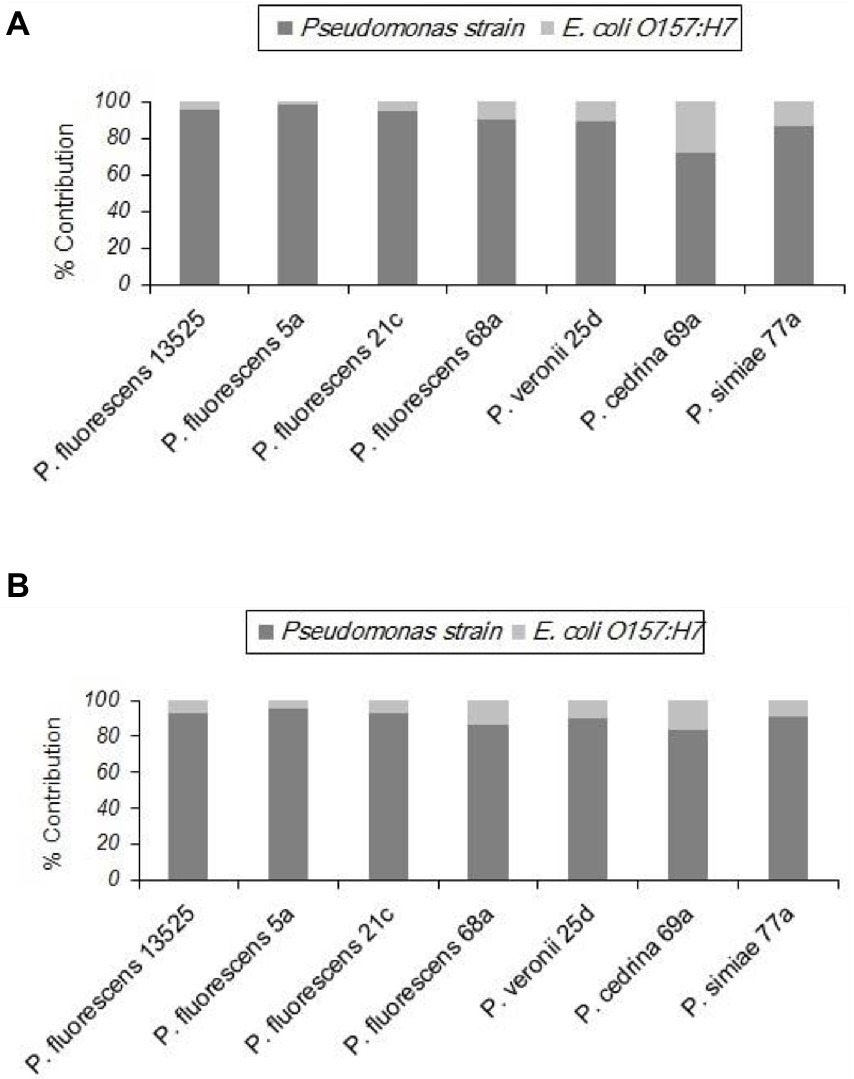
Figure 6. Contributions of different P. fluorescens group strains and E. coli O157:H7 to dual-species biofilms on stainless steel surfaces under CI (A) and DI (B) scenarios.
In both scenarios, the total number of biofilm cells in dual-species biofilms with E. coli O157:H7 and P. fluorescens 68a, P. cedrina 69a and P. simiae 77a were significantly higher than those in pure cultures of P. fluorescens group strains (Figure 7; p < 0.05). For P. veronii 25d, the total number of biofilm cells in dual-species biofilms was significantly higher than in the pure culture (p < 0.05), but only in the CI scenario. No significant differences were observed between CI and DI scenarios in terms of the number of biofilm cells in dual-species biofilms of the seven P. fluorescens group strains with E. coli O157:H7 (p > 0.05; Figure 7).
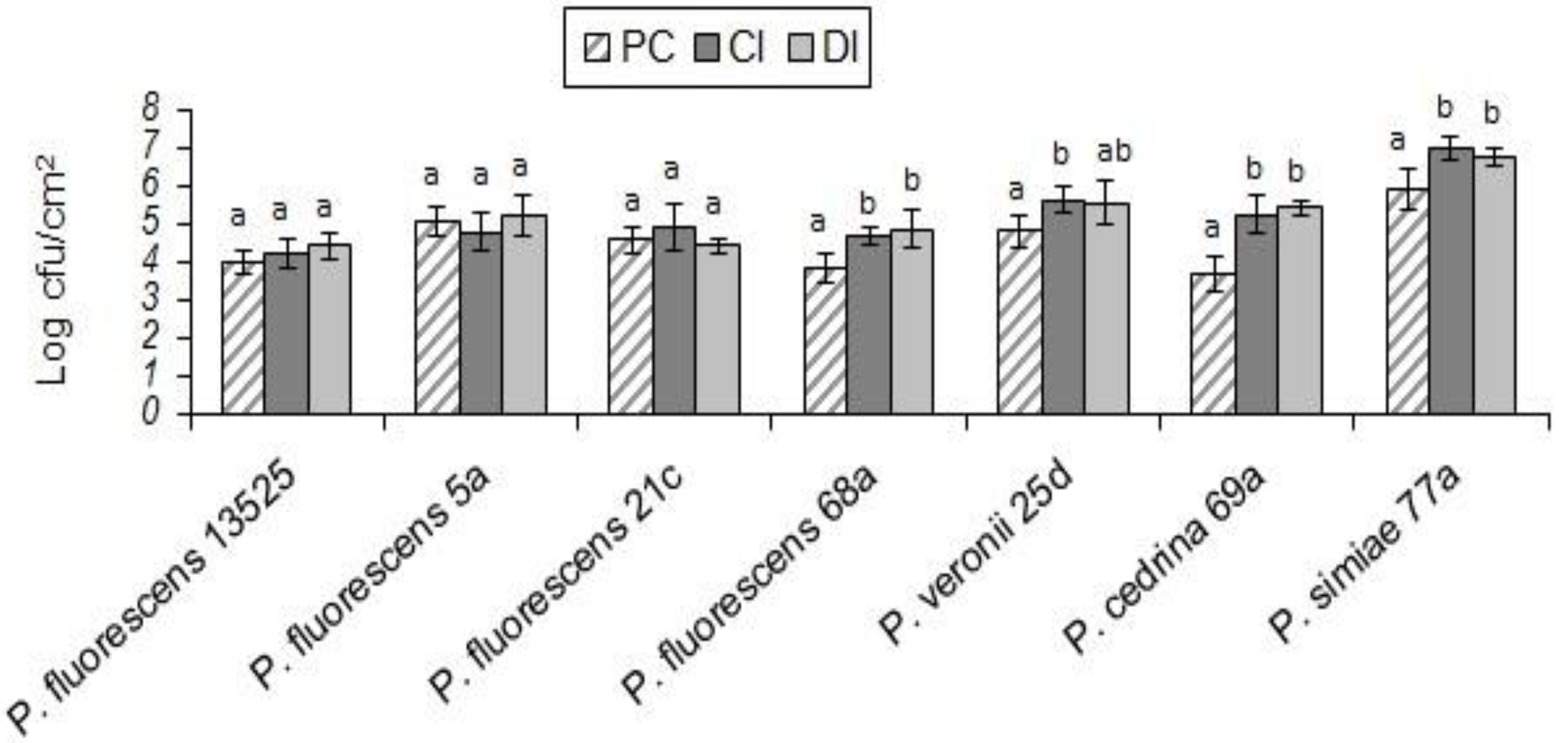
Figure 7. Total number of biofilm cells in mono-and dual-species biofilms of the seven P. fluorescens group strains with E. coli O157:H7 on stainless steel surfaces. PC: pure culture biofilms; CI: dual-species biofilms under the CI scenario; DI: dual-species biofilms under the DI scenario. For each P. fluorescens strain, different letters indicate significant differences in the total number of biofilm cells.
Pseudomonas fluorescens group strains and Salmonella Typhimurium
With S. Typhimurium, the situation was somewhat different from that of the other Gram-negative bacteria tested (E. coli O157:H7). S. Typhimurium contributed to biofilm formation with all seven P. fluorescens group strains in the CI scenario, to varying degrees. As shown in Figure 8A, the highest contribution was found with P. fluorescens 68a (25.2%), and the lowest was with P. fluorescens 5a (4.6%). However, in the DI scenario, S. Typhimurium did not contribute to the biofilm formed by P. fluorescens 5a, P. fluorescens 21c, or P. veronii 25d. It did contribute to dual-species biofilms with other P. fluorescens group strains, but to a lesser extent than in the CI scenario (Figure 8B).
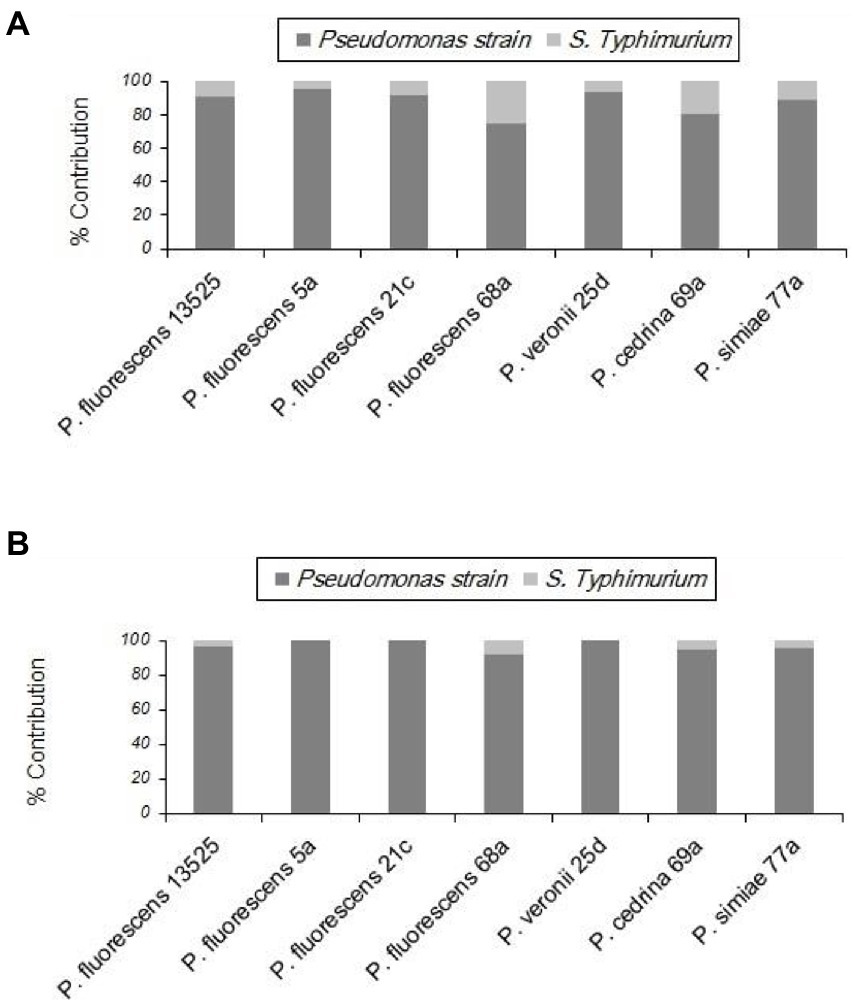
Figure 8. Contributions of different P. fluorescens strain groups and S. Typhimurium to dual-species biofilms on stainless steel surfaces under CI (A) and DI (B) scenarios.
As shown in Figure 9, in both scenarios the total numbers of biofilm cells in dual-species biofilms of P. fluorescens 5a, P. fluorescens 21c and P. veronii 25d were not significantly different from those in pure cultures of P. fluorescens group strains (p > 0.05). In contrast, the total number of biofilm cells in dual-species biofilms with S. Typhimurium and P. fluorescens 68a and P. cedrina 69a in the CI scenario were significantly higher than those in the DI scenario, and in pure cultures (p < 0.05). Furthermore, the total number of biofilm cells in dual-species biofilms with S. Typhimurium and P. fluorescens 13,525 and P. simiae 77a in the CI scenario were significantly higher than that in the DI scenario, and in the pure culture, respectively (p < 0.05).
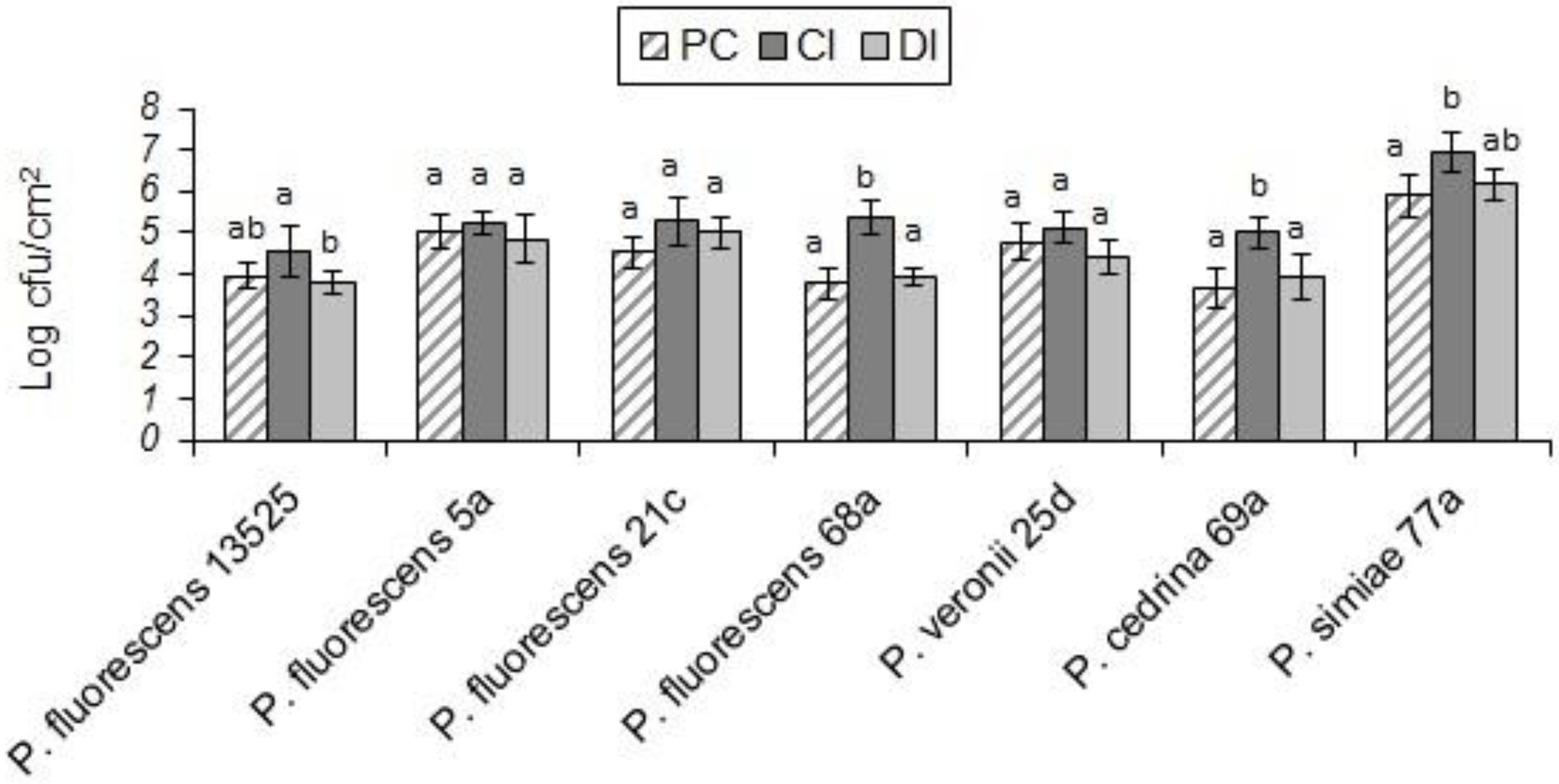
Figure 9. Total number of biofilm cells in mono- and dual-species biofilms of the seven P. fluorescens group strains with S. typhimurium on stainless steel surfaces. PC: pure culture biofilms; CI: dual-species biofilms under the CI scenario; DI: dual-species biofilms under the DI scenario. For each P. fluorescens strain, different letters indicate significant differences in the total number of biofilm cells.
Discussion
P. fluorescens group strains are among the bacteria most frequently isolated from surfaces in the food industry, and are characterized as quick and thick biofilm producers on various surfaces (Simoes et al., 2008; Mann and Wozniak, 2012; Marchand et al., 2012; Puga et al., 2016; Zarei et al., 2020). Results of the present study re-confirmed the high ability of these bacteria to produce biofilm on stainless steel surfaces. In general, at 7°C these bacteria produced more biofilm than the pathogenic strains tested, however the P. fluorescens strains differed significantly in their ability to produce biofilms on stainless steel surfaces in UHT milk. Previous research has highlighted strain diversity of the P. fluorescens group isolated from dairy products and raw milk in terms of blue pigment production and lipoproteolytic activity (Chierici et al., 2016; Longhi et al., 2022). Other differences in the proteolytic activity of P. fluorescence group strains have also been reported previously (Zarei et al., 2020).
The presence of spoilage and pathogenic bacteria in milk and dairy products is a worldwide problem that not only leads to shelf-life reduction and alteration of organoleptic properties, but is also related to many disease outbreaks. The coexistence and interactions between foodborne pathogens and resident background microbiota are likely to occur on the surface of milk tanks and other dairy processing equipment between sessile cells, particularly in biofilms. It has been well documented that most naturally occurring biofilms are composed of multiple bacterial species (Yannarell et al., 2019). Hence, in terms of pathogen densities in dual-or multi-species biofilms, interactions with background microbiota strains can have neutral, positive, or antagonistic effects (Langsrud et al., 2016; Møretrø and Langsrud, 2017; Heir et al., 2018). These interactions can protect bacteria from environmental stresses, and can also influence the growth and survival of the individual members of these microbial consortia (Røder et al., 2015; Sanchez-Vizuete et al., 2015; Møretrø and Langsrud, 2017; Papaioannou et al., 2018).
In the present study, the interactions between P. fluorescens group bacterial strains and S. aureus, B. cereus, E. coli O157:H7, and S. Typhimurium during dual-species biofilm formation on stainless steel surfaces in UHT milk were evaluated. Two scenarios (concurrent inoculation and delayed inoculation of pathogens) were examined to better understand the possibility of dual-species biofilm formation. Given that all experiments in this study were performed at 7°C, the predominance of the Pseudomonas population in biofilms in both scenarios was expected.
In dual-species biofilms with S. aureus, this pathogen was found in six of the seven biofilms, with varying degrees of contributions in the CI scenario, which revealed differences between various strains within the P. fluorescens group. Moreover, our findings indicated that although S. aureus could co-produce biofilms with P. fluorescens group bacterial strains, it could not be incorporated into the pre-formed biofilms. Almost the same results were observed for another gram-positive bacterium used in this study – B. cereus. This pathogen was found in five of the seven biofilms, also with varying degrees of contributions in the CI scenario, while it did not contribute to biofilm formation in the DI scenario. The metabolites secreted around the biofilm matrix of P. fluorescens group strains may have prevented these two gram-positive pathogens from entering the pre-formed biofilms. Insufficient places to attach to the surface can be another explanation for the lack of contribution of these two gram-positive bacteria to the biofilm formation in the DI scenario (Balaure and Grumezescu, 2020). Evidently, biofilm formation in the CI scenario was not hampered by this challenge, leading to dual-species biofilms. Simoes et al. (2008) demonstrated that dual-species biofilms of P. fluorescens and B. cereus were significantly more metabolically active than P. fluorescens mono-species biofilms. Furthermore, Davies and Marques (2011) found that the fatty acid cis-2-decenoic acid produced by Pseudomonas group strains yielded dispersion of biofilms formed by S. aureus.
In contrast to the two gram-positive bacteria, E. coli O157:H7 and S. Typhimurium formed dual-species biofilms with all seven strains of the P. fluorescens group in the CI scenario, and with seven (E. coli O157:H7) and four (S. Typhimurium) strains in the DI scenario. Nevertheless, for both pathogens, significant differences were observed between P. fluorescens group strains in both scenarios, indicating strong and weak competitor strains within the P. fluorescens group. It has been previously reported that the biofilm formation of E. coli O157:H7 increased in the presence of background microbiota of meat (Dourou et al., 2011). This might explain why E. coli O157:H7 had a higher percent contribution to biofilm formation in the CI scenario than the DI scenario.
Overall, this study provides the first demonstration of differences between strains of the P. fluorescens group in terms of the formation of mono-or dual-species biofilms with pathogenic bacteria. Moreover, the ability to produce dual-species biofilms with pathogenic bacteria depends on whether the pathogens form the biofilm simultaneously with the P. fluorescens group strains or whether P. fluorescens group strains have already formed a biofilm. Such differences between strains and the inoculation time makes comparison of the results difficult, but also highlights the complexity of bacterial interactions involving the strains of the P. fluorescens group that requires further understanding for improved control of biofilms in the dairy industry. This study also contributes to the understanding of the role of different strains of the P. fluorescens group in the ability of four important foodborne pathogens to establish, survive and persist in dairy processing premises.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
MZ designed the research. AY and SR performed the experiments. MZ and PS performed data analysis. MZ and AY prepared the figures. MZ wrote the manuscript, with contributions from AY and PS. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the research grant provided by Shahid Chamran University of Ahvaz (SCU.VF99.245) and open access funded by Helsinki University Library.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anand, S., Singh, D., Avadhanula, M., and Marka, S. (2014). Development and control of bacterial biofilms on dairy processing membranes. Compr. Rev. Food Sci. 13, 18–33. doi: 10.1111/1541-4337.12048
Austin, J. W., and Bergeron, G. (1995). Development of bacterial biofilms in dairy processing lines. J. Dairy Res. 62, 509–519. doi: 10.1017/s0022029900031204
Balaure, P. C., and Grumezescu, A. M. (2020). Recent advances in surface nanoengineering for biofilm prevention and control. Part I: molecular basis of biofilm recalcitrance. Passive anti-biofouling nanocoatings. Nano 10:1230. doi: 10.3390/nano10061230
Carrascosa, J. M., Cuevas, R., Gonzalez, R., Azcorra, A., and Garcia, D. (2015). Quantifying the economic and cultural biases of social media through trending topics. PLoS One 10:e0134407. doi: 10.1371/journal.pone.0134407
Cherif-Antar, A., Moussa-Boudjemaa, B., Didouh, N., Medjahdi, K., Mayo, B., and Florez, A. B. (2016). Diversity and biofilm-forming capability of bacteria recovered from stainless steel pipes of a milk-processing dairy plant. Dairy Sci. Technol. 96, 27–38. doi: 10.1007/s13594-015-0235-4
Chierici, M., Picozzi, C., La Spina, M. G., Orsi, C., Vigentini, I., Zambrini, V., et al. (2016). Strain diversity of Pseudomonas fluorescens group with potential blue pigment phenotype isolated from dairy products. J. Food Prot. 79, 1430–1435. doi: 10.4315/0362-028X.JFP-15-589
Davies, D. G., and Marques, C. N. (2011). A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403. doi: 10.1128/JB.01214-08
de Jonghe, V., Coorevits, A., van Hoorde, K., Messens, W., van Landschoot, A., de Vos, P., et al. (2011). Influence of storage conditions on the growth of Pseudomonas species in refrigerated raw milk. Appl. Environ. Microbiol. 77, 460–470. doi: 10.1128/AEM.00521-10
Dourou, D., Beauchamp, C. S., Yoon, Y., Geornaras, I., Belk, K. E., Smith, G. C., et al. (2011). Attachment and biofilm formation by Escherichia coli O157:H7 at different temperatures, on various food-contact surfaces encountered in beef processing. Int. J. Food Microbiol. 149, 262–268. doi: 10.1016/j.ijfoodmicro.2011.07.004
Elias, S., and Banin, E. (2012). Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36, 990–1004. doi: 10.1111/j.1574-6976.2012.00325
Fricker, M., Skånseng, B., Rudi, K., Stessl, B., and Ehling-Schulz, M. (2011). Shift from farm to dairy tank milk microbiota revealed by a polyphasic approach is independent from geographical origin. Int. J. Food Microbiol. 145, S24–S30. doi: 10.1016/j.ijfoodmicro.2010.08.025
Garrido-Sanz, D., Meier-Kolthoff, J. P., Göker, M., Martín, M., and Rivilla, R. (2016). Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS One 11:e0153733. doi: 10.1371/journal.pone.0153733
Gomila, M., Peña, A., Mulet, M., Lalucat, J., and García-Valdés, E. (2015). Phylogenomics and systematics in pseudomonas. Front. Microbiol. 6:214. doi: 10.3389/fmicb.2015.00214
Heir, E., Møretrø, T., Simensen, A., and Langsrud, S. (2018). Listeria monocytogenes strains show large variations in competitive growth in mixed culture biofilms and suspensions with bacteria from food processing environments. Int. J. Food Microbiol. 275, 46–55. doi: 10.1016/j.ijfoodmicro.2018.03.026
Langsrud, S., Moen, B., Møretrø, T., Loype, M., and Heir, E. (2016). Microbial dynamics in mixed culture biofilms of bacteria surviving sanitation of conveyor belts in salmon processing plants. J. Appl. Microbiol. 120, 366–378. doi: 10.1111/jam.13013
Latorre, A. A., van Kessel, J. S., Karns, J. S., Zurakowski, M. J., and Pradhan, A. K. (2010). Biofilm in milking equipment on a dairy farm as a potential source of bulk tank milk contamination with listeria monocytogenes. J. Dairy Sci. 93, 2792–2802. doi: 10.3168/jds.2009-2717
Liu, W., Røder, H. L., Madsen, L. S., Bjarnsholt, T., Sørensen, S. J., and Burmølle, M. (2016). Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front. Microbiol. 7:1366. doi: 10.3389/fmicb.2016.01366
Longhi, R., Correia, S., Bruzaroski, S., Poli-Frederico, R., Fagnani, R., and Santana, E. (2022). Pseudomonas fluorescens and Pseudomonas putida from refrigerated raw milk: genetic diversity and lipoproteolytic activity. J. Dairy Res. 89, 86–89. doi: 10.1017/S0022029922000048
Makovcova, J., Babak, V., Kulich, P., Masek, J., Slany, M., and Cincarova, L. (2017). Dynamics of mono-and dual-species biofilm formation and interactions between Staphylococcus aureus and Gram-negative bacteria. Microb. Biotechnol. 10, 819–832. doi: 10.1111/1751-7915.12705
Mann, E. E., and Wozniak, D. J. (2012). Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 36, 893–916. doi: 10.1111/j.1574-6976.2011.00322.x
Marchand, S., De Block, J., De Jonghe, V., Coorevits, A., Heyndrickx, M., and Herman, L. (2012). Biofilm formation in milk production and processing environments; influence on milk quality and safety. Compr. Rev. Food Sci. Food Saf. 11, 133–147. doi: 10.1111/j.1541-4337.2011.00183.x
Martin, N. H., Murphy, S. C., Ralyea, R. D., Wiedmann, M., and Boor, K. J. (2011). When cheese gets the blues: Pseudomonas fluorescens as the causative agent of cheese spoilage. J. Dairy Sci. 94, 3176–3183. doi: 10.3168/jds.2011-4312
Meng, L., Zhang, Y., Liu, H., Zhao, S., Wang, J., and Zheng, N. (2017). Characterization of Pseudomonas spp. and associated proteolytic properties in raw milk stored at low temperatures. Front. Microbiol. 8, 1–7. doi: 10.3389/fmicb.2017.02158
Møretrø, T., and Langsrud, S. (2017). Residential bacteria on surfaces in the food industry and their implications for food safety and quality. Compr. Rev. Food Sci. Food Saf. 16, 1022–1041. doi: 10.1111/1541-4337.12283
Mulet, M., Lalucat, J., and García-Valdés, E. (2010). DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 12, 1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x
Papaioannou, E., Giaouris, E. D., Berillis, P., and Boziaris, I. S. (2018). Dynamics of biofilm formation by Listeria monocytogenes on stainless steel under mono-species and mixed culture simulated fish processing conditions and chemical disinfection challenges. Int. J. Food Microbiol. 267, 9–19. doi: 10.1016/j.ijfoodmicro.2017.12.020
Puga, C. H., Orgaz, B., and San Jose, C. (2016). Listeria monocytogenes impact on mature or old Pseudomonas fluorescens biofilms during growth at 4 and 20°C. Front. Microbiol. 7:134. doi: 10.3389/fmicb.2016.00134
Reichler, S. J., Trmčić, A. H., Martin, N. H., Boor, K. J., and Wiedmann, M. (2018). Pseudomonas fluorescens group bacterial strains are responsible for repeat and sporadic post pasteurization contamination and reduced fluid milk shelf life. J. Dairy Sci. 101, 7780–7800. doi: 10.3168/jds.2018-14438
Rendueles, O., and Ghigo, J. M. (2012). Multi-species biofilms: how to avoid unfriendly neighbors. FEMS Microbiol. Rev. 36, 972–989. doi: 10.1111/j.1574-6976.2012.00328.x
Røder, H. L., Raghupathi, P. K., Herschend, J., Brejnrød, A., Knøchel, S., Sørensen, S. J., et al. (2015). Interspecies interactions result in enhanced biofilm formation by co-cultures of bacteria isolated from a food processing environment. Food Microbiol. 51, 18–24. doi: 10.1016/j.fm.2015.04.008
Sanchez-Vizuete, P., Orgaz, B., Aymerich, S., Le Coq, D., and Briandet, R. (2015). Pathogens protection against the action of disinfectants in multispecies biofilms. Front. Microbiol. 6:12. doi: 10.3389/fmicb.2015.00705
Schirmer, B. C. T., Heir, E., Møretrø, T., Skaar, I., and Langsrud, S. (2013). Microbial background flora in small-scale cheese production facilities does not inhibit growth and surface attachment of Listeria monocytogenes. J. Dairy Sci. 96, 6161–6171. doi: 10.3168/jds.2012-6395
Sharma, M., and Anand, S. K. (2002). Characterization of constitutive microflora of biofilms in dairy processing lines. Food Microbiol. 19, 627–636. doi: 10.1006/fmic.2002.0472
Shi, X., and Zhu, X. (2009). Biofilm formation and food safety in food industries. Trends Food Sci. Technol. 20, 407–413. doi: 10.1016/j.tifs.2009.01.054
Shpigel, N. Y., Pasternak, Z., Factor, G., and Gottlieb, Y. (2015). Diversity of bacterial biofilm communities on sprinklers from dairy farm cooling systems in Israel. PLoS One 10:e0139111. doi: 10.1371/journal.pone.0139111
Simoes, M., Simoes, L. C., and Vieira, M. J. (2008). Physiology and behavior of Pseudomonas fluorescens single and dual strain biofilms under diverse hydrodynamics stresses. Intl. J. Food Microbiol. 128, 309–316. doi: 10.1016/j.ijfoodmicro.2008.09.003
Stoeckel, M., Lidolt, M., Achberger, V., Glück, C., Krewinkel, M., Stressler, T., et al. (2016). Growth of Pseudomonas weihenstephanensis, Pseudomonas proteolytica and Pseudomonas spp. in raw milk: impact of residual heat-stable enzyme activity on stability of UHT milk during shelf-life. Int. Dairy J. 59, 20–28. doi: 10.1016/j.idairyj.2016.02.045
van Houdt, R., and Michiels, C. W. (2010). Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 109, 1117–1131. doi: 10.1111/j.1365-2672.2010.04756.x
von Neubeck, M., Baur, C., Krewinkel, M., Stoeckel, M., Kranz, B., Stressler, T., et al. (2015). Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int. J. Food Microbiol. 211, 57–65. doi: 10.1016/j.ijfoodmicro.2015.07.001
Wang, L., and Jayarao, B. M. (2001). Phenotypic and genotypic characterization of Pseudomonas fluorescens isolated from bulk tank milk. J. Dairy Sci. 84, 1421–1429. doi: 10.3168/jds.S0022-0302(01)70174-9
Yang, L., Liu, Y., Wu, H., Høiby, N., Molin, S., and Song, Z. (2011). Current understanding of multi-species biofilms. Int. J. Oral Sci. 3, 74–81. doi: 10.4248/IJOS11027
Yannarell, S. M., Grandchamp, G. M., Chen, S., Daniels, K. E., Shank, E. A., and Elizabeth, A. (2019). A dual-species biofilm with emergent mechanical and protective properties. J. Bacteriol. Res. 201:e00670-18. doi: 10.1128/JB.00670-18
Zarei, M., Yousefvand, A., Maktabi, S., Pourmahdi Borujeni, M., and Mohammadpour, H. (2020). Identification, phylogenetic characterisation and proteolytic activity quantification of high biofilm-forming Pseudomonas fluorescens group bacterial strains isolated from cold raw milk. Int. Dairy J. 109:104787. doi: 10.1016/j.idairyj.2020.104787
Keywords: Biofilm, Pseudomonas fluorescens, Staphylococcus aureus, Bacillus cereus, Escherichia coli O157:H7, Salmonella Typhimurium
Citation: Zarei M, Rahimi S, Saris PEJ and Yousefvand A (2022) Pseudomonas fluorescens group bacterial strains interact differently with pathogens during dual-species biofilm formation on stainless steel surfaces in milk. Front. Microbiol. 13:1053239. doi: 10.3389/fmicb.2022.1053239
Edited by:
Agapi Doulgeraki, Hellenic Agricultural Organization, GreeceReviewed by:
Steve Flint, Massey University, New ZealandQingli Dong, University of Shanghai for Science and Technology, China
Copyright © 2022 Zarei, Rahimi, Saris and Yousefvand. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mehdi Zarei, zarei@scu.ac.ir; Amin Yousefvand, amin.yousefvand@helsinki.fi
 Mehdi Zarei
Mehdi Zarei Saeid Rahimi1
Saeid Rahimi1 Per Erik Joakim Saris
Per Erik Joakim Saris Amin Yousefvand
Amin Yousefvand