- State Environmental Protection Key Laboratory for Wetland Conservation and Vegetation Restoration & Jilin Provincial Key Laboratory of Ecological Restoration and Ecosystem Management & Key Laboratory of Vegetation Ecology of Ministry of Education, School of Environment, Northeast Normal University, Changchun, China
Antibiotic resistance genes (ARGs) are a new environmental contaminant that poses a major hazard to humans and the environment. This research discusses the methods and drawbacks of two ARG removal approaches, constructed wetlands (CWs) and photocatalysis. CWs primarily rely on the synergistic effects of substrate adsorption, plant uptake, and microbial processes to remove ARGs. The removal of ARGs can be influenced by wetland plants, substrate type, wetland type, and hydraulic conditions. The absolute abundance of ARGs in effluent decreased, but their relative abundance increased. Photocatalysis deactivates ARGs predominantly through reactive oxygen species, with removal effectiveness determined by catalyst type, radiation type, and radiation intensity. The drawback is that it exposes intracellular resistance genes, perhaps increasing the risk of ARG spread. To address the current shortcomings, this paper proposes the feasibility of combining a constructed wetland with photocatalysis technology, which provides a novel strategy for ARG removal.
1. Introduction
Antibiotics have accumulated massively in the environment as a result of their widespread use in recent years. Antibiotics not only cause chemical pollution, but they may also induce the production of resistance genes (ARGs) and resistant bacteria (ARB) in the environment, hastening resistance spread and diffusion. As a result, the evolution and variation of bacterial resistance, as well as the spread of ARGs, have received increased attention in the field of environmental research. ARGs have been found in abundance in a variety of environmental media, including surface water, groundwater (Jiang et al., 2013), sediment, soil (Wang et al., 2019), and air detection. Antibiotic resistance has emerged as a serious global environmental health issue (Ying et al., 2017; Li et al., 2020). This antibiotic resistance can be spread between microorganisms via a horizontal gene transfer (HGT) mechanism (Berendonk et al., 2015). ARGs are classified as extracellular ARGs (eARGs) and intracellular ARGs (iARGs), both of which are transmitted via HGT.
ARGs enter the environment via a variety of routes, including municipal water, sewer runoff, livestock wastewater, landfill leachate, and hospital wastewater (Ezeuko et al., 2021; Koch et al., 2021). Antibiotics leave significant levels of ARGs in humans and animals, which are eventually discharged into wastewater treatment plants via fecal wastewater runoff. The wastewater discharged from wastewater treatment plants, as well as the ARGs present in biosolids, enter the soil and aquatic environments and are absorbed in a cycle by plants, animals, and so on. ARGs have been found in a range of water environments, and some investigations have revealed that they are present in tap water (Bergeron et al., 2015). ARG removal solutions that are cost-effective are urgently needed, and ARGs pollution must be addressed.
Some of the current tactics for removing ARGs from wastewater include disinfection procedures, membrane treatment technologies, advanced oxidation technologies, and constructed wetlands (CWs). Disinfection techniques, such as chlorine disinfection, which increases antibiotic resistance and the genera of bacteria that can carry antibiotic resistance, are ineffective in eliminating ARGs and also encourage the transmission and spread of ARGs (Cheng et al., 2021). ARGs are physically removed by membrane treatment technologies; nevertheless, when membrane filtration is used, ARGs accumulate in membrane fouling and sewage sludge, which can re-enter the environment. Advanced oxidation technologies and CWs, to the contrary hand, have relatively good ARG removal. ARGs can be effectively eliminated by photocatalytic advanced oxidation based on hydroxyl radicals, and a TiO2/UV treatment can reduce 5.8 log of mecA and 4.7 log of ampC (Guo et al., 2017). Furthermore, CWs is an efficient and sustainable wastewater treatment technology that efficiently removes organic matter, bacteria, antibiotics, pharmaceuticals and personal care products (PPCPs) from wastewater (Hartl et al., 2021), and has great potential in ARGs removal (Chen et al., 2016a; Huang et al., 2017). Thus, this study summarizes recent research and uses of CWs and photocatalysis in the removal of ARGs. The basic mechanisms, affecting factors, and limitations of CWs and photocatalysis for ARG removal are summarized. The feasibility of using CWs in conjunction with photocatalysis to remove ARGs is considered. Some novel approaches to removing ARGs from aquatic environments are proposed.
2. CWs for the removal of ARGs
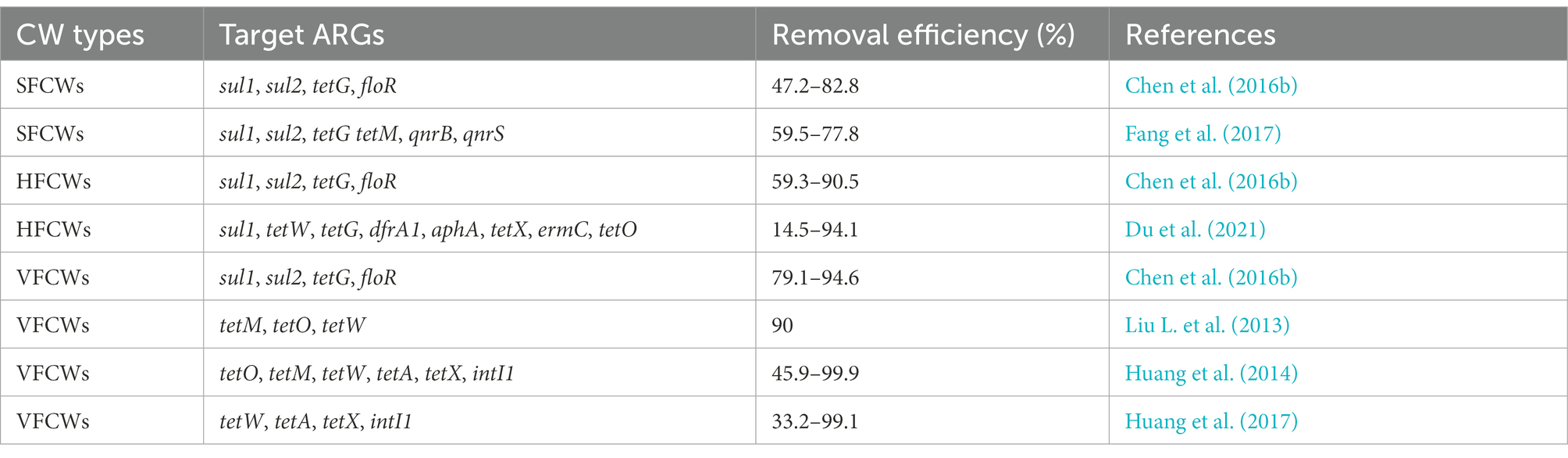
CWs are an ecosystem made up of water, microbial communities, plants, and substrate (Chen et al., 2019). It uses a synergistic process of physical, chemical, and biological processes to remove contaminants. Its advantages over conventional wastewater treatment technologies include low cost, simplicity of use, and good maintenance (Faulwetter et al., 2009). CWs are currently being utilized to treat domestic wastewater (Adrados et al., 2014), agricultural wastewater (Liu J. et al., 2013), and landfill leachate (Sundberg et al., 2007). CWs are effective in removing both new pollutants as well as conventional contaminants like nitrogen and phosphorus. CWs remove ARGs mainly by substrate sorption, plant uptake, and microbial removal (Figure. 1). The type of wetlands, the plants used, the type of substrate, and other factors all have an impact on how successfully CWs remove ARGs. ARGs can currently be removed from aquatic habitats using CWs, albeit the bulk of these techniques are still in the experimental stage and the precise process is unknown, needing additional research.
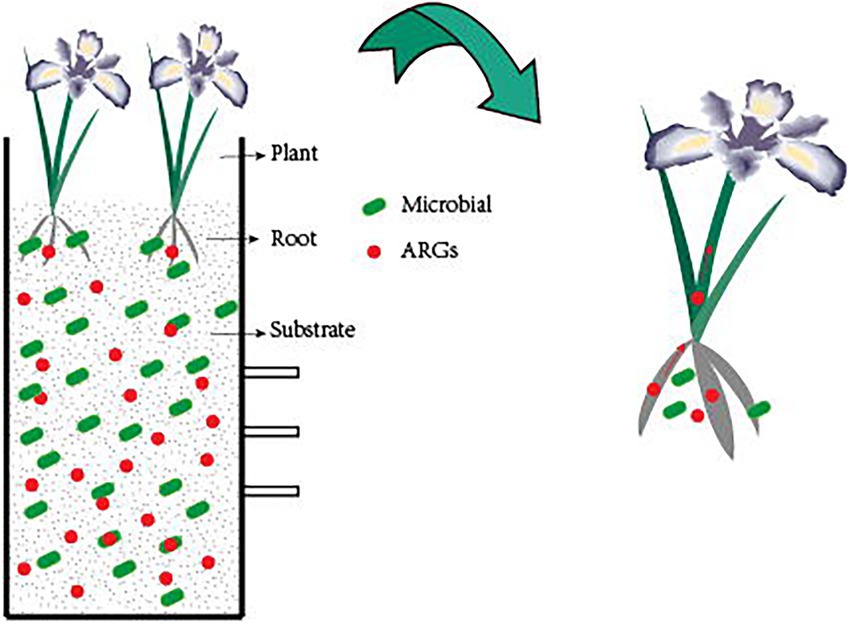
Figure 1. Mechanisms for the removal of ARGs in CWs. The substrate can absorb a high amount of ARGs, while the plant can also absorb ARGs. As the plant grows, ARGs migrate from the roots to the stems and leaves, and the plant root system’s biofilm structure removes ARGs by mechanisms of filtration, adsorption, absorption, and transformation of ARGs, and bacteria within the CWs can also remove ARGs.
2.1. Removal efficiency of ARGs for different CWs types
Surface flow constructed wetlands (SFCWs) and subsurface flow CWs are the two types of CWs. Subsurface flow CWs are classified as either horizontal subsurface flow constructed wetlands (HFCWs) or vertical subsurface flow constructed wetlands (VSFWs) based on the direction of the water flow (VFCWs). The removal effectiveness of ARGs varied dramatically amongst CW types (Table 1).
CWs have the ability to remove most common ARGs, with removal rates ranging from 14.5% to 100%. Furthermore, the ARG removal efficiency of several CWs was much higher than that of conventional wastewater treatment plants (Xu et al., 2015). Various types of CWs have their own targeted ARG species that are efficiently removed. For example, VFCWs significantly reduced the concentration of tetracycline resistance genes in livestock effluent (Liu L. et al., 2013), with the absolute abundance of tetM, tetW, and tetO being reduced by 90%. HFCWs was also effective in removing ARGs, especially sulfonamide ARGs. The abundance of sul1, sul2, tetM, tetO, tetQ, tetW, and intI1 was reduced by 1–3 orders of magnitude by HFCWs (Chen and Zhang, 2013a). HFCWs was more effective in eliminating sul1-carrying bacteria than some typical wastewater treatment facilities (Czekalski et al., 2012), indicating that it can be used as a supplement to standard wastewater treatment plants, particularly to reduce the amount of sulfonamide ARGs. HFCWs and VFCW are more effective at removing contaminants than SFCWs, with VFCWs having the highest removal efficacy, ranging from 33.2 to 99.9%. The differences in removal effectiveness may be connected to the adsorptive filtration, biochemical processes, and redox conditions found in each wetland. Furthermore, the direction of water flow in VFCWs induced disparities in removal results, with upflow VFCWs having a higher relative abundance of tetracycline ARGs and intI1 than downflow VFCWs (Chen et al., 2019).
2.2. ARGs uptake by plants in CWs and biofilm degradation in the root system
ARGs are primarily removed by plants in CWs via two pathways: absorption and root biofilm breakdown. Bacteria can reproduce in plant tissues and expand their populations during plant growth via hydraulic transport and active plant absorption. This allows plants to effectively reduce the abundance of microorganisms in the feed water, thus facilitating the removal of ARGs (Vacca et al., 2005). There were significant differences in the distribution of ARGs among different tissues of the plant, with ARG abundance higher in plant leaves than stems (Guo et al., 2021). The total abundance of ARGs observed in plants, however, was much lower than that found in the substrate.
The huge root systems of wetland plants can form a special biofilm structure together with the filler surface to remove ARGs through the processes of filtration, adsorption, absorption, and transformation of ARGs. Tetracycline ARGs, especially tetW, can rapidly migrate to the biofilm surface and are therefore more easily removed by CWs (Cheng et al., 2013). Furthermore, it has been demonstrated that plants can indirectly participate in the removal of ARGs, primarily by filtering solid particles and delivering oxygen to the microbial community, enhancing the role of inter-rhizosphere bacteria, or providing a medium for biofilm development, which improves the removal capacity of microorganisms and reduces the accumulation of ARGs, resulting in a reduction in ARGs abundance (Anderson et al., 2013; Fang et al., 2017). The type and amount of inter-root secretions vary among plants, thus affecting the inter-root microorganisms, resulting in different removal efficiencies of ARGs by different plants, with Thalia dealbata Fraser being more effective than Iris tectorum Maxim in removing ARGs (Chen et al., 2016b). Reed is a major aquatic plant for reducing ARGs contamination, with a removal effectiveness of more than 90% for ARGs such as sul1, sul2, ermB, qnrS, and blaTEM-1 (Avila et al., 2021). However, biological processes in CWs not only degrade ARGs as described above, but also lead to the transfer and increase of ARGs (Ghosh and LaPara, 2007; Diehl and LaPara, 2010; Guo et al., 2014; Yang et al., 2014), thus the role in ARGs removal is complex and needs to be studied in more depth.
2.3. Substrate effect on ARGs removal
In CWs, the matrix serves as an essential vehicle for physicochemical processes (Chen et al., 2019). The particle size distribution, surface charge, porosity, and pH of the matrix all have an impact on ARGs removal (He et al., 2021). Substrate adsorption and microbial degradation on the substrate surface are two important pathways for ARG removal. The inadequate elimination of macrolide resistance genes by CWs may be explained by substrate adsorption (Chen et al., 2019). Bacteria, particularly gut microorganisms, are easily absorbed by substrates (Huang et al., 2017). Furthermore, because ARGs are generally carried by gut bacteria, the high removal effectiveness of ARGs by CWs is most likely due to the substrate’s high adsorption efficiency on intestinal microorganisms (Huang et al., 2014). ARGs from tenericutes, cyanobacteria, and acidobacteria were more likely to be lost (Su et al., 2019). In addition, small pore size substrates enable bacterial filtration and precipitation and have a great ability to remove microorganisms from water. Gravel, zeolite, oyster shell, medicinal stone, ceramic, and tuff are the most typical substrates used in CWs for ARGs removal (Table 2). Zeolites have a microporous structure and silica hydroxyl groups, which provide surface area for chemisorption and microbial attachment, while silica hydroxyl groups are catalytically active for various chemical reactions, and their average pore size (4.32 nm) is smaller than that of volcanic rocks (10.78 nm; Liu L. et al., 2013), making zeolites more efficient for the removal of ARGs (Gorra et al., 2007). Ceramics have a porous morphology with a greater specific surface area, but they have a macroporous structure (Chen et al., 2016a), hence they are less effective at removing ARGs than zeolites. Tuff has a more porous structure and a bigger surface area, allowing for more bacterial adsorption and stronger biofilm development (Abou-Kandil et al., 2021), and hence has a better ability to remove ARGs. Both oyster shell and medical stone have an ordered lamellar structure, but oyster shell has considerable agglomeration that medical stone does not, and hence oyster shell is more efficient at removing ARGs than medical stone.
2.4. CWs remove the shortcomings of ARGs
The mechanism of ARG removal by CWs is complex, and the mechanism of migration and removal of ARGs in CWs is not well understood. While CWs are effective in removing ARGs, they also have the risk of enriching them. Despite a decrease in absolute abundance, the relative abundance of resistance genes in CWs effluent increased (Yi et al., 2017). ARGs are generally transmitted via vertical and horizontal gene transfer, i.e., genetic transfer between parents and transmission between microbes. Once ARGs produce resistance in pathogenic microbes or spread across pathogenic microorganisms, CWs can evolve into a large reservoir of ARGs, with potentially disastrous ecological and health effects. According to research, reducing the overall number of microorganisms in wastewater can successfully limit ARGs transmission (Song et al., 2018). Therefore, research on the removal of ARGs by CWs still faces a great challenge.
3. Photocatalytic removal of ARGs
Photocatalytic oxidation is a method of removing pollutants from water or the atmosphere that involves a sequence of reactions between a catalyst and oxygen in solution that produce powerful oxidizing ·OH under the action of solar radiation. The photocatalyst is central to photocatalytic technology, and there are many different types of photocatalysts, including TiO2, ZnO, and WO3, which are now the most researched photocatalysts. Because of its non-toxicity, low cost, and great photocatalytic efficiency, TiO2 is the most often used photocatalyst.
In comparison to the activated sludge method, membrane separation method, and chemical oxidation method, the photocatalytic oxidation method has the advantages of low energy consumption, rapid reaction, simple operation, and no secondary pollution, and it has become a popular research direction in recent years. Advanced photocatalytic oxidation processes have the potential to remove microbial contaminants, such as photocatalytic titanium dioxide, which can generate reactive oxygen species (ROS, e.g., OH), which kill microorganisms by oxidative damage to cell membranes, RNA, DNA, proteins, and lipids (Umar et al., 2019). TiO2 multiphase photocatalytic oxidation offers various advantages as a “green” water disinfection technology, including better efficiency in eliminating ARB from wastewater than standard disinfection processes and the absence of disinfection by-products.
3.1. Mechanism of photocatalytic removal of ARGs
Photocatalytic creation of holes and electrons under UVA irradiation causes redox processes in which oxidants first target microbial cell walls, membranes, and enzymes, and later interior components such as RNA and DNA (Koivunen and Heinonen-Tanski, 2005). Although bacteria have self-defense systems to defend themselves from ROS damage, excessive ROS can cause oxidative stress and assault membrane lipids, eventually leading to DNA damage (Li et al., 2011). ARG abundance is decreasing owing to photocatalyst light exposure, which causes DNA damage in bacterial cells (Figure 2). Furthermore, photocatalytic treatment breaks down long DNA strands into shorter nucleotides, allowing the deoxyribose phosphate backbone to be broken (Hirakawa et al., 2009). Free ARGs-containing deoxyribonucleic acids are due to a lack of protective bacterial cell walls, which can also be rapidly removed by photocatalysis. TiO2 can produce hydroxyl radicals, which are thought to be the most active oxidants for destroying ARGs (Pham and Lee, 2014). Therefore, the majority of current research employs TiO2 or modified TiO2 as a photocatalyst.
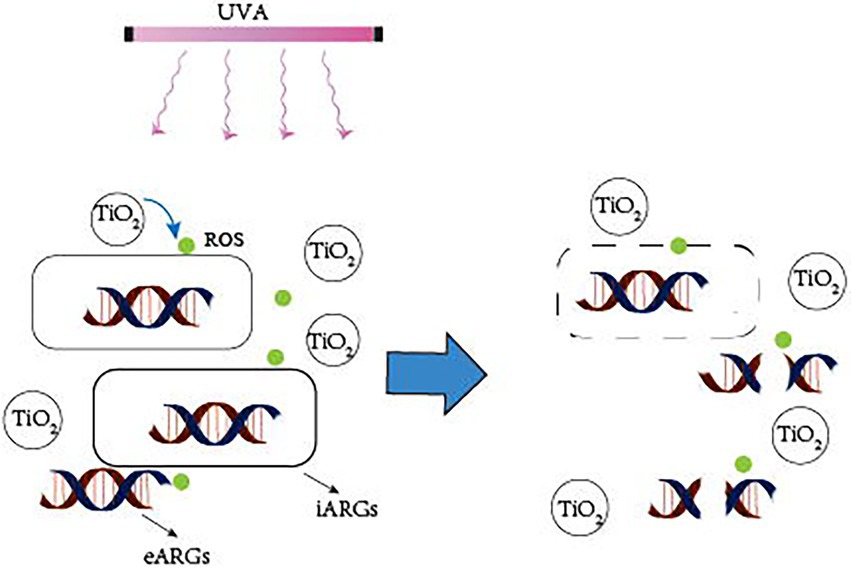
Figure 2. Mechanism of photocatalytic removal of ARGs. UVA irradiation causes the catalyst to create ROS, which attack the bacterial cell wall, breaking it down and converting iARGs to eARGs. Simultaneously, ROS attack eARGs, breaking the DNA strand, and inactivating ARGs.
3.2. Factors influencing photocatalytic removal of ARGs
Among the large number of photocatalytic materials, TiO2 and graphitic carbon nitride (g-C3N4) are two materials that have been studied more. The effects of different catalyst types, radiation types, and radiation intensities on the removal of ARGs varied significantly (Table 3). Radiation intensity is significant in photocatalysis efficiency (Yu et al., 2020), and the higher the radiation intensity, the more effective the removal of ARGs. UV removal of ARGs is more effective than solar radiation, and the decrease of ARGs by TiO2 under UV irradiation is 4–5 log, whereas it is only 0.5 log under sunlight irradiation, which may be related to TiO2’s low activation effectiveness under sunshine. To address TiO2’s low activation efficiency in solar radiation, the synthesis and application of semiconductor-reinforced TiO2 composites have emerged as a hot research area. TiO2-rGO composites are found to be much more effective than TiO2 at removing ARGs. The removal effect of different ARGs under the same treatment conditions is different, most likely because their sensitivity to oxidation radicals varies depending on their composition. Because reactive oxygen species react more quickly with guanine bases in DNA, ARGs with a high GC% content, such as sulfonamide ARGs, are degraded to a greater extent (Ren et al., 2018). In addition, the amount of photocatalyst used also affects the removal of ARGs, and the size of the composite also influences the treatment efficacy, as smaller composites are more likely to cross cell membranes and promote rupture (Guo and Tian, 2019). Hence, the development of non-toxic nanocomposites should be explored in the development of new photocatalysts for the removal of ARGs. Inorganic ions and organic debris in wastewater can both limit photocatalytic activity by adsorbing on the TiO2 surface and blocking active sites, and some anions can function as cavity scavengers, slowing the rate of chemical breakdown or disinfection on the TiO2 surface. In addition, organic matter in wastewater can also reduce the disinfection rate through a variety of mechanisms, including ROS scavenging, competition for active sites on the TiO2 surface, and direct UV absorption.
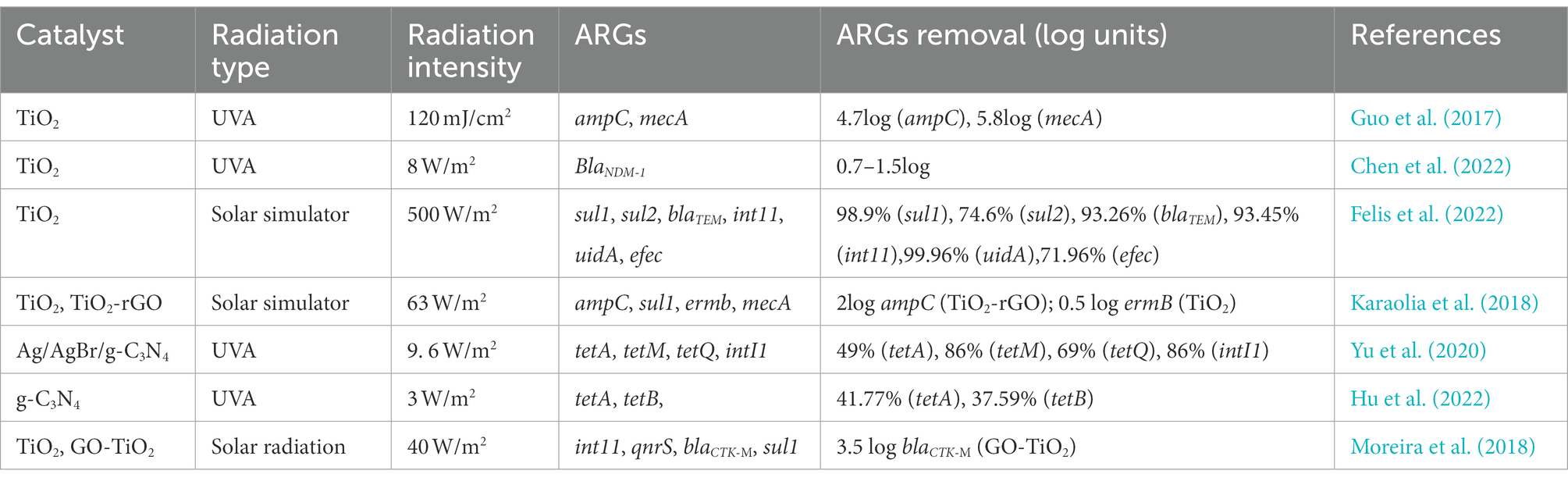
Table 3. Effect of different catalysts, radiation type and radiation intensity on the removal effectivness of ARGs.
3.3. Effect of nanomaterials on the diffusion of ARGs
The effect of nanomaterials on the HGT of ARGs in the environment has aroused the interest of many researchers. Related research has revealed that gene level transfer is a significant factor in the spread of ARGs (Cheng et al., 2021). ARGs can move from one bacterium to another when combined with mobile genetic elements like plasmids, integrons, transposons, and so on, allowing bacteria to acquire ARGs. Most nanomaterials can contribute to the diffusion of ARGs in pure bacterial systems. Nano-TiO2 significantly improved the splice transfer between RP4 plasmids in E. coli (Qiu et al., 2015), and with slight inhibition of bacterial growth, it could increase the splice transfer efficiency by 56-fold. ARB activity was reduced by graphene oxide (GO). However, at the level of ARGs, all GO greatly boosted transfer efficiency (Guo and Zhang, 2017). These findings suggest that the excellent adsorption characteristics of insoluble nanoparticles boost the binding of ARGs and bacteria, which can considerably contribute to the transfer efficiency of ARGs. The environmental conditions in actual water and wastewater treatment are complex, as are the technologies used. The environmental conditions in actual water and wastewater treatment are complicated, with a generally mixed flora of microorganisms. The impact of nanomaterials on the spread of ARGs in the actual world is more complicated. The effluent from a secondary wastewater treatment plant was treated using polyvinylidene fluoride ultrafiltration membranes enhanced with nano-TiO2 (Ren et al., 2018). The results showed that nano-TiO2 on this membrane was successful in eliminating 98% of ARGs when exposed to UV light, thus effectively controlling the HGT of ARGs. To remove resistant bacteria and genes from municipal wastewater, TiO2-rGO material was utilized (Karaolia et al., 2018). These results can be attributed to the ability of the reactive oxygen species generated by photoexcitation of nanomaterials to oxidatively damage DNA, thus enabling the control of the proliferation of ARGs in the real environment.
3.4. Photocatalysis’ shortcomings in ARG removal
Photocatalysis is an excellent method for removing ARGs by directly destroying cellular deoxyribonucleic acid; however, the continuous use of high-intensity UV light during wastewater treatment is difficult (Chen and Zhang, 2013b; Lee et al., 2017; Mauter et al., 2018). Moreover, after UV damage to the cell wall, iARGs flow out and are converted into eARGs, which can survive in the environment and bind to other bacteria via transformation, transduction, and other mechanisms, resulting in the spread of ARGs in the environment. In addition, genes fragmented by oxidation can integrate with other pathogens in the wastewater (De Vries and Wackernagel, 2002; Nielsen et al., 2007), thus requiring additional treatment. Bacteria with ruptured cell membranes can cause cell lysis and an increase in eARGs levels, resulting in secondary water contamination. The removal of ARGs released by ARBs is critical. Furthermore, injured bacteria treated with photocatalysis will recover, and in addition to bacterial regrowth, ARGs transfer may rise if pathogens are not completely inactivated. The release and transfer of ARGs may contribute to the subsequent development of resistant bacteria in the aquatic environment unless the duration of treatment is managed (Dunlop et al., 2015).
4. Prospect
CWs can efficiently remove ARGs, and the removal efficiency of VFCWs and HFCWs is greater than that of SFCWs. VFCWs effectively remove tetracycline ARGs, whereas HFCWs effectively remove sulfonamide ARGs. Furthermore, photocatalysis successfully removes ARGs, and the higher the intensity of the irradiation, the better the efficacy of ARGs removal. For the elimination of ARGs, nanocomposites outperformed TiO2. Combining the advantages of CWs and photocatalysis in the removal of ARGs, a combination of CWs and photocatalysis could be proposed to remove ARGs. With remarkable success, the combination of photocatalysis and CWs has been studied for the treatment of high-salt chromium-containing wastewater (Li et al., 2020) and municipal wastewater. More importantly, in the combination system of CWs and photocatalysis, CWs could further eliminate the eARGs. Under UVA irradiation, the catalyst generates ROS, and the ROS break the cell wall and allow intracellular DNA to flow out, converting the difficult-to-remove iARGs into eARGs. While the ROS also attacks the eARGs, breaking the DNA strands and inactivating the ARGs. eARGs and iARGs are removed further in the artificial wetland by substrate adsorption, plant uptake, and microbial action. Furthermore, the combination would be increased the removal efficiency of COD, BOD5, and Cr (VI) with no negative effect on plant production indicators (Li et al., 2019). In addition, photocatalysis can extend the life of the wetland because it eliminates clogging with refractory substances, reduces total phosphorus concentrations and reduces wetland loading.
Therefore, CWs paired with photocatalytic wastewater treatment technology provide the advantages of steady water quality, minimal investment, and low running costs. When CWs are combined with photocatalytic technology, they may remove both iARGs and eARGs, and the treatment is complete, lowering the risk of ARGs spreading. Moverover, CWs in combination with photocatalysis could produce a flexible and operable wastewater treatment system. This method combines ecological treatment technology with photocatalysis technology to produce a new technology that is superior to traditional wastewater treatment methods to remove resistance genes.
Author contributions
PC: data curation and analysis, writing—original draft. XY: worked on the technical details, supervised the findings of the work and helped in the development of manuscript. JZ: aided in interpreting the results and worked on the manuscript. YW: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (42222102).
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abou-Kandil, A., Shibli, A., Azaizeh, H., Wolff, D., Wick, A., and Jadoun, J. (2021). Fate and removal of bacteria and antibiotic resistance genes in horizontal subsurface constructed wetlands: effect of mixed vegetation and substrate type. Sci. Total Environ. 759:144193. doi: 10.1016/j.scitotenv.2020.144193
Adrados, B., Sánchez, O., Arias, C., Becares, E., Garrido, L., Mas, J., et al. (2014). Microbial communities from different types of natural wastewater treatment systems: vertical and horizontal flow constructed wetlands and biofilters. Water Res. 55, 304–312. doi: 10.1016/j.watres.2014.02.011
Anderson, J. C., Carlson, J. C., Low, J. E., Challis, J. K., Wong, C. S., Knapp, C. W., et al. (2013). Performance of a constructed wetland in Grand Marais, Manitoba, Canada: removal of nutrients, pharmaceuticals, and antibiotic resistance genes from municipal wastewater. Chem. Cent. J. 7, 54–69. doi: 10.1186/1752-153X-7-54
Avila, C., Garcia-Galan, M. J., Borrego, C. M., Rodriguez-Mozaz, S., Garcia, J., Barcelo, D., et al. (2021). New insights on the combined removal of antibiotics and ARGs in urban wastewater through the use of two configurations of vertical subsurface flow constructed wetlands. Sci. Total Environ. 755:142554. doi: 10.1016/j.scitotenv.2020.142554
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317. doi: 10.1038/nrmicro3439
Bergeron, S., Boopathy, R., Nathaniel, R., Corbin, A., and LaFleur, G. (2015). Presence of antibiotic resistant bacteria and antibiotic resistance genes in raw source water and treated drinking water. Int. Biodeterior. Biodegrad. 102, 370–374. doi: 10.1016/j.ibiod.2015.04.017
Chen, X., Han, W. X., Patel, M., Wang, Q., Li, Q. L., Zhao, S., et al. (2022). Inactivation of a pathogenic NDM-1-positive Escherichia coli strain and the resistance gene blaNDM-1 by TiO2/UVA photocatalysis. Sci. Total Environ. 846:157369. doi: 10.1016/j.scitotenv.2022.157369
Chen, J., Wei, X. D., Liu, Y. S., Ying, G. G., Liu, S. S., He, L. Y., et al. (2016a). Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: optimization of wetland substrates and hydraulic loading. Sci. Total Environ. 565, 240–248. doi: 10.1016/j.scitotenv.2016.04.176
Chen, J., Ying, G. G., Wei, X. D., Liu, Y. S., Liu, S. S., Hu, L. X., et al. (2016b). Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: effect of flow configuration and plant species. Sci. Total Environ. 571, 974–982. doi: 10.1016/j.scitotenv.2016.07.085
Chen, H., and Zhang, M. (2013a). Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in eastern China. Environ. Int. 55, 9–14. doi: 10.1016/j.envint.2013.01.019
Chen, H., and Zhang, M. M. (2013b). Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou, China. Environ. Sci. Technol. 47, 8157–8163. doi: 10.1021/es401091y
Chen, M., Zhu, M. X., Zhu, Y., Wang, D. B., Li, Z. W., Zeng, G. M., et al. (2019). Collision of emerging and traditional methods for antibiotics removal: taking constructed wetlands and nanotechnology as an example. NanoImpact 15:100175. doi: 10.1016/j.impact.2019.100175
Cheng, W. X., Chen, H., Su, C., and Yan, S. H. (2013). Abundance and persistence of antibiotic resistance genes in livestock farms: a comprehensive investigation in eastern China. Environ. Int. 61, 1–7. doi: 10.1016/j.envint.2013.08.023
Cheng, X. X., Xu, J. N., Smith, G., and Zhang, Y. Y. (2021). Metagenomic insights into dissemination of antibiotic resistance across bacterial genera in wastewater treatment. Chemosphere 271:129563. doi: 10.1016/j.chemosphere.2021.129563
Czekalski, N., Berthold, T., Caucci, S., Egli, A., and Bürgmann, H. (2012). Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front. Microbiol. 3, 1–18. doi: 10.3389/fmicb.2012.00106
De Vries, J., and Wackernagel, W. (2002). Integration of foreign DNA during natural transformation of Acinetobacter sp by homology facilitated illegitimate recombination. Proc. Natl. Acad. Sci. U. S. A. 99, 2094–2099. doi: 10.1073/pnas.042263399
Diehl, D. L., and LaPara, T. M. (2010). Effect of temperature on the fate of genes encoding tetracycline resistance and the integrase of class 1 integrons within anaerobic and aerobic digesters treating municipal wastewater solids. Environ. Sci. Technol. 44, 9128–9133. doi: 10.1021/es102765a
Du, J. P., Xu, T., Guo, X. P., and Yin, D. Q. (2021). Characteristics and removal of antibiotics and antibiotic resistance genes in a constructed wetland from a drinking water source in the Yangtze River Delta. Sci. Total Environ. 813:152540. doi: 10.1016/j.scitotenv.2021.152540
Dunlop, P. S. M., Ciavola, M., Rizzo, L., McDowell, D. A., and Byrne, J. A. (2015). Effect of photocatalysis on the transfer of antibiotic resistance genes in urban wastewater. Catal. Today 240, 55–60. doi: 10.1016/j.cattod.2014.03.049
Ezeuko, A. S., Ojemaye, M. O., Okoh, O. O., and Okoh, A. I. (2021). Potentials of metallic nanoparticles for the removal of antibiotic resistant bacteria and antibiotic resistance genes from wastewater: a critical review. J. Water Proc. Eng. 41:102041. doi: 10.1016/j.jwpe.2021.102041
Fang, H. S., Zhang, Q., Nie, X. P., Chen, B. W., Xiao, Y. D., Zhou, Q. B., et al. (2017). Occurrence and elimination of antibiotic resistance genes in a long-term operation integrated surface flow constructed wetland. Chemosphere 173, 99–106. doi: 10.1016/j.chemosphere.2017.01.027
Faulwetter, J. L., Gagnon, V., Sundberg, C., Chazarenc, F., Burr, M. D., Brisson, J., et al. (2009). Microbial processes influencing performance of treatment wetlands: a review. Ecol. Eng. 35, 987–1004. doi: 10.1016/j.ecoleng.2008.12.030
Felis, E., Buta-Hubeny, M., Zieliński, W., Hubeny, J., Harnisz, M., Bajkacz, S., et al. (2022). Solar-light driven photodegradation of antimicrobials, their transformation by-products and antibiotic resistance determinants in treated wastewater. Sci. Total Environ. 836:155447. doi: 10.1016/j.scitotenv.2022.155447
Ghosh, S., and LaPara, T. M. (2007). The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 1, 191–203. doi: 10.1038/ismej.2007.31
Gorra, R., Coci, M., Ambrosoli, R., and Laanbroek, H. J. (2007). Effects of substratum on the diversity and stability of ammonia-oxidizing communities in a constructed wetland used for wastewater treatment. J. Appl. Microbiol. 103, 1442–1452. doi: 10.1111/j.1365-2672.2007.03357.x
Guo, X. P., Li, J., Yang, F., Yang, J., and Yin, D. Q. (2014). Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci. Total Environ. 493, 626–631. doi: 10.1016/j.scitotenv.2014.06.035
Guo, Y. J., Qiu, T. L., Gao, M., Sun, Y. M., Cheng, S. T., Gao, H. Z., et al. (2021). Diversity and abundance of antibiotic resistance genes in rhizosphere soil and endophytes of leafy vegetables: Focusing on the effect of the vegetable species. J. Hazard. Mater. 415:125595. doi: 10.1016/j.jhazmat.2021.125595
Guo, M. T., and Tian, X. B. (2019). Impacts on antibiotic-resistant bacteria and their horizontal gene transfer by graphene-based TiO2&ag composite photocatalysts under solar irradiation. J. Hazard. Mater. 380:120877. doi: 10.1016/j.jhazmat.2019.120877
Guo, C. S., Wang, K., Hou, S., Wan, L., Lv, J. P., Zhang, Y., et al. (2017). H2O2 and/or TiO2 photocatalysis under UV irradiation for the removal of antibiotic resistant bacteria and their antibiotic resistance genes. J. Hazard. Mater. 323, 710–718. doi: 10.1016/j.jhazmat.2016.10.041
Guo, M. T., and Zhang, G. S. (2017). Graphene oxide in the water environment could affect tetracycline-antibiotic resistance. Chemosphere 183, 197–203. doi: 10.1016/j.chemosphere.2017.04.145
Hartl, M., Garcia-Galan, M. J., Matamoros, V., Fernandez-Gatell, M., Rousseau, D. P. L., Du Laing, G., et al. (2021). Constructed wetlands operated as bioelectrochemical systems for the removal of organic micropollutants. Chemosphere 271:129593. doi: 10.1016/j.chemosphere.2021.129593
He, Y., Zhang, L., Jiang, L., Wagner, T., Sutton, N. B., Ji, R., et al. (2021). Improving removal of antibiotics in constructed wetland treatment systems based on key design and operational parameters: a review. J. Hazard. Mater. 407:124386. doi: 10.1016/j.jhazmat.2020.124386
Hirakawa, K., Mori, M., Yoshida, M., Oikawa, S., and Kawanishi, S. (2009). Photo-irradiated titanium dioxide catalyzes site specific DNA damage via generation of hydrogen peroxide. Free Radic. Res. 38, 439–447. doi: 10.1080/1071576042000206487
Hu, X. M., Chen, X. Y., Tang, Y., Xu, Z. G., Zeng, Y. L., Wang, Y. H., et al. (2022). Effects of g-C3N4 on bacterial community and tetracycline resistance genes in two typical sediments in tetracycline pollution remediation. Front. Microbiol. 13:964401. doi: 10.3389/fmicb.2022.964401
Huang, X., Liu, C. X., Li, K., Su, J. Q., Zhu, G. F., and Liu, L. (2014). Performance of vertical up-flow constructed wetlands on swine wastewater containing tetracyclines and tet genes. Water Res. 70, 109–117. doi: 10.1016/j.watres.2014.11.048
Huang, X., Zheng, J. L., Liu, C. X., Liu, L., Liu, Y. H., and Fan, H. Y. (2017). Removal of antibiotics and resistance genes from swine wastewater using vertical flow constructed wetlands: effect of hydraulic flow direction and substrate type. Chem. Eng. J. 308, 692–699. doi: 10.1016/j.cej.2016.09.110
Jiang, L., Hu, X. L., Xu, T., Zhang, H. C., Sheng, D., and Yin, D. Q. (2013). Prevalence of antibiotic resistance genes and their relationship with antibiotics in the Huangpu River and the drinking water sources, Shanghai, China. Sci. Total Environ., 458–460. doi: 10.1016/j.scitotenv.2013.04.038
Karaolia, P., Michael-Kordatou, I., Hapeshi, E., Drosou, C., Bertakis, Y., Christofilos, D., et al. (2018). Removal of antibiotics, antibiotic resistant bacteria and their associated genes by grapheme based TiO2 composite photocatalysts under solar radiation in urban wastewaters. Appl. Catal. B-Environ. 224, 810–824. doi: 10.1016/j.apcatb.2017.11.020
Koch, N., Islam, N. F., Sonowal, S., Prasad, R., and Sarma, H. (2021). Environmental antibiotics and resistance genes as emerging contaminants: methods of detection and bioremediation. Curr. Res. Microb. Sci. 2:100027. doi: 10.1016/j.crmicr.2021.100027
Koivunen, J., and Heinonen-Tanski, H. (2005). Inactivation of enteric microorganisms with chemical disinfectants, UV irradiation and combined chemical/UV treatments. Water Res. 39, 1519–1526. doi: 10.1016/j.watres.2005.01.021
Lee, J., Jeon, J. H., Shin, J., Jang, H. M., Kim, S., Song, M. S., et al. (2017). Quantitative and qualitative changes in antibiotic resistance genes after passing through treatment processes in municipal wastewater treatment plants. Sci. Total Environ. 605-606, 906–914. doi: 10.1016/j.scitotenv.2017.06.250
Li, D. D., Chen, F. Z., and Han, J. Q. (2019). A study of the treatment of high-salt chromium-containing wastewater by the photocatalysis-constructed wetland combination method. Water Sci. Technol. 80, 1956–1966. doi: 10.2166/wst.2020.017
Li, M., Noriega-Trevino, M. E., Nino-Martinez, N., Marambio-Jones, C., Wang, J., Damoiseaux, R., et al. (2011). Synergistic bactericidal activity of ag-TiO2 nanoparticles in both light and dark conditions. Environ. Sci. Technol. 45, 8989–8995. doi: 10.1021/es201675m
Li, P., Wu, Y. F., He, Y. L., Zhang, B., Huang, Y. S., Yuan, Q. Y., et al. (2020). Occurrence and fate of antibiotic residues and antibiotic resistance genes in a reservoir with ecological purification facilities for drinking water sources. Sci. Total Environ. 707:135276. doi: 10.1016/j.scitotenv.2019.135276
Liu, L., Liu, C. X., Zheng, J. Y., Huang, X., Wang, Z., Liu, Y. H., et al. (2013). Elimination of veterinary antibiotics and antibiotic resistance genes from swine wastewater in the vertical flow constructed wetlands. Chemosphere 91, 1088–1093. doi: 10.1016/j.chemosphere.2013.01.007
Liu, J., Lu, Z., Zhang, J., Xing, M., and Yang, J. (2013). Phylogenetic characterization of microbial communities in a full-scale vermifilter treating rural domestic sewage. Ecol. Eng. 61, 100–109. doi: 10.1016/j.ecoleng.2013.09.015
Mauter, M. S., Zucker, I., Perreault, F., Werber, J. R., Kim, J. H., and Elimelech, M. (2018). The role of nanotechnology in tackling global water challenges. Nat. Sustain. 1, 166–175. doi: 10.1038/s41893-018-0046-8
Moreira, N. F. F., Narciso-da-Rocha, C., Polo-López, M. I., Pastrana-Martínez, L. M., Faria, J. L., Manaia, C. M., et al. (2018). Solar treatment (H2O2, TiO2-P25 and GO-TiO2 photocatalysis, photo-Fenton) of organic micropollutants, human pathogen indicators, antibiotic resistant bacteria and related genes in urban wastewater. Water Res. 135, 195–206. doi: 10.1016/j.watres.2018.01.064
Nielsen, K. M., Johnsen, P. J., Bensasson, D., and Daffonchio, D. (2007). Release and persistence of extracellular DNA in the environment. Environ. Biosaf. Res. 6, 37–53. doi: 10.1051/ebr:2007031
Pham, T. D., and Lee, B. K. (2014). Cu doped TiO2/GF for photocatalytic disinfection of Escherichia coli in bioaerosols under visible light irradiation: application and mechanism. Appl. Surf. Sci. 296, 15–23. doi: 10.1016/j.apsusc.2014.01.006
Qiu, Z. G., Shen, Z. Q., Qian, D., Jin, M., Yang, D., Wang, J. F., et al. (2015). Effects of nano-TiO2 on antibiotic resistance transfer mediated by RP4 plasmid. Nanotoxicology 9, 895–904. doi: 10.3109/17435390.2014.991429
Ren, S. J., Boo, C., Guo, N., Elimelech, M., and Wang, Y. K. (2018). Photocatalytic reactive ultrafiltration membrane for removal of antibiotic resistant bacteria and antibiotic resistance genes from wastewater effluent. Environ. Sci. Technol. 52, 8666–8673. doi: 10.1021/acs.est.8b01888
Song, H. L., Zhang, S., Guo, J. H., Yang, Y. L., Zhang, L. M., Li, H., et al. (2018). Vertical up-flow constructed wetlands exhibited efficient antibiotic removal but induced antibiotic resistance genes in effluent. Chemosphere 203, 434–441. doi: 10.1016/j.chemosphere.2018.04.006
Su, Y. L., Wang, J. X., Xia, H. P., and Xie, B. (2019). Comparative network analysis revealing the mechanisms of antibiotic resistance genes removal by leachate recirculation under different hydraulic loadings. Sci. Total Environ. 649, 318–326. doi: 10.1016/j.scitotenv.2018.08.361
Sundberg, C., Tonderski, K., and Lindgren, P. E. (2007). Potential nitrification and de-nitrification and the corresponding composition of the bacterial communities in a compact constructed wetland treating landfill leachates. Water Sci. Technol. 56, 159–166. doi: 10.2166/wst.2007.524
Umar, M., Roddick, F., and Fan, L. H. (2019). Moving from the traditional paradigm of pathogen inactivation to controlling antibiotic resistance in water-role of ultraviolet irradiation. Sci. Total Environ. 662, 923–939. doi: 10.1016/j.scitotenv.2019.01.289
Vacca, G., Wand, H., Nikolausz, M., Kuschk, P., and Kastner, M. (2005). Effect of plants and filter materials on bacteria removal in pilot-scale constructed wetlands. Water Res. 39, 1361–1373. doi: 10.1016/j.watres.2005.01.005
Wang, L. J., Zhao, X., Wang, J. H., Wang, J., Zhu, L. S., and Ge, W. L. (2019). Macrolide-and quinolone-resistant bacteria and resistance genes as indicators of antibiotic resistance gene contamination in farmland soil with manure application. Ecol. Indic. 106:105456. doi: 10.1016/j.ecolind.2019.105456
Xu, J., Xu, Y., Wang, H. M., Guo, C. S., Qiu, H. Y., He, Y., et al. (2015). Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 119, 1379–1385. doi: 10.1016/j.chemosphere.2014.02.040
Yang, Y., Li, B., Zou, S. C., Fang, H. P., and Zhang, T. (2014). Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 62, 97–106. doi: 10.1016/j.watres.2014.05.019
Yi, X. Z., Tran, N. H., Yin, T. R., He, Y. L., and Gin, K. Y. H. (2017). Removal of selected PPCPs, EDCs, and antibiotic resistance genes in landfill leachate by a full-scale constructed wetlands system. Water Res. 121, 46–60. doi: 10.1016/j.watres.2017.05.008
Ying, G. G., He, L. Y., Ying, A. J., Zhang, Q. Q., Liu, Y. S., Zhao, J. L., et al. (2017). China must reduce its antibiotic use. Environ. Sci. Technol. 51, 1072–1073. doi: 10.1021/acs.est.6b06424
Keywords: antibiotic resistance genes, constructed wetlands, photocatalysis, removal mechanism, combination
Citation: Chen P, Yu X, Zhang J and Wang Y (2023) New and traditional methods for antibiotic resistance genes removal: Constructed wetland technology and photocatalysis technology. Front. Microbiol. 13:1110793. doi: 10.3389/fmicb.2022.1110793
Edited by:
Huai Li, Northeast Institute of Geography and Agroecology (CAS), ChinaReviewed by:
Feng Li, Institute of Subtropical Agriculture (CAS), ChinaWeiqi Wang, Fujian Normal University, China
Copyright © 2023 Chen, Yu, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofei Yu,  yuxf888@nenu.edu.cn
yuxf888@nenu.edu.cn
 Pingping Chen
Pingping Chen Xiaofei Yu
Xiaofei Yu Jingyao Zhang
Jingyao Zhang Yiqi Wang
Yiqi Wang