- 1National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, South China Agricultural University, Guangzhou, China
- 2Laboratory of Veterinary Pharmacology, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 3Guangdong Provincial Center for Disease Control and Prevention, Guangzhou, China
- 4Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
Salmonella enterica can lead to intestinal diarrhea, and the emergence and spread of cephalosporin-resistant Salmonella have brought great challenges to clinical treatment. Therefore, this study investigated the prevalence and transmission of blaCTX-M genes among S. Typhimurium from diarrhoeal outpatients in Guangdong, China, from 2010 to 2017. A total of 221 blaCTX-M-positive isolates were recovered from 1,263 S. Typhimurium isolates from the facal samples of diarrhoea patients in 45 general hospitals from 11 cities. The most popular CTX-M gene was blaCTX-M-55 (39.6%, 72/182) in the CTX-M-1 group, followed by blaCTX-M-14 (22.5%, 41/182) and blaCTX-M-65 (19.2%, 35/182) in the CTX-M-9 group. The isolates that carried blaCTX-M-9G had significantly higher resistance rates to multiple antibacterials compared with blaCTX-M-1G (p < 0.01). Meanwhile, PFGE analysis not only showed the clonal transmission of blaCTX-M-55/14/65-positve isolates of diarrhoeal outpatients’ origins from different hospitals in Guangdong province, but also the characteristic of blaCTX-M-55/14/65-positve isolates’ bacterial persistence. Multilocus sequence typing (MLST) analysis indicated that these S. Typhimurium isolates possessed ST34 and ST19. Furthermore, genomic Beast phylogenomic analysis provided the evidence of a close relationship of blaCTX-M-positive S. Typhimurium isolates between the outpatients and pork. Most blaCTX-M-55/14/65 genes were transmitted by non-typeable or IncI1/IncFII/IncHI2 plasmids with the size of ranging from ~80 to ~280 kb. Moreover, whole-genome sequencing (WGS) analysis further revealed that blaCTX-M-55/14/65 coexisted with other 25 types of ARGs, of which 11 ARGs were highly prevalent with the detection rates >50%, and it first reported the emergence of blaTEM-141 in S. Typhimurium. This study underscores the importance of surveillance for blaCTX-M-positive microbes in diarrhea patients.
Introduction
Salmonella enterica is a zoonotic pathogen of substantial concern to human and animal health (Yin and Zhou, 2018). What’s more, it is a leading cause of morbidity and mortality in people worldwide, with approximately 90 million cases of gastroenteritis and 150,000 associated deaths (Xu et al., 2021). So far, more than 2,610 Salmonella serovars have been identified, while salmonellosis is caused mainly by S. enterica serovars Typhimurium, Enteritidis and Dublin (Shi, 2015; Mohammed et al., 2017). Nontyphoidal S. Typhimurium is a dominant factor of human gastroenteritis, and improper handling and digestion of inadequately looked food primarily result in the infection. Invasive complications, including meningitis, sepsis and bacteraemia, are very common in infants, the elderly and immunocompromised patients. The disease of S. Typhimurium is usually related to contaminated foods, such as pork and fruits, unpasteurized milk and dairy products, and undercooked eggs (Wegener et al., 2003).
In these potentially life-threatening S. Typhimurium cases, the antibiotics of choice are fluoroquinolones and extended-spectrum cephalosporins (Diard and Hardt, 2017). Third-generation cephalosporins (3GCs) are used across the world to threat infections caused by Salmonella, and subsequently the emergence of resistance attracts particular attention (Whichard et al., 2007). Multidrug-resistant (MDR) Salmonella spp. potentially arising for the selective pressure from sustained antimicrobial exposure are more likely to be the causative agents of invasive disease (Okoro et al., 2015). Moreover, the ESBL-producing strains of Salmonella have been reported in many regions in China, including Beijing, Shanghai, Guangdong, and Shandong (Cao C. et al., 2021). Worse, ESBL-producing S. Typhimurium have increasingly been detected from food animals, even environmental water and human patients (Fu et al., 2020; Ma et al., 2020). Hence, the number of ESBL-Salmon has increased worldwide.
TEM, SHV, and CTX-M were the most prevalent ESBL types. It has commonly been found that ESBL-CTX-M is located on plasmids and considered as the most prevalent type of ESBLs in many European countries (Paterson and Bonomo, 2005). At the same time, there is tremendous diversity of blaCTX-M genotypes isolated from food animals and human populations. Usually, among the reported bacteria with blaCTX-M-55-positive or blaCTX-M-14-positive or blaCTX-M-65-positive, most are isolated from food and animal sources (Xiang et al., 2015; Zhang et al., 2015; Nadimpalli et al., 2019). A practice was selected for antibiotic resistant S. enterica that can spread to human through contaminated foods. However, this practice is not currently monitored or regulated in Guangdong Province.
Therefore, in this study, ESBL-producing S. Typhimurium isolates, mainly from diarrheal patients, isolated from Guangdong province, and collected at the Guangdong Provincial CDC during the period of 2010–2017, were investigated to gain insight into their public health impacts.
Materials and Methods
Bacterial Isolates, Detection of ESBL/pAmpC Genes, and Antimicrobial Susceptibility Testing
A total of 1263 S. Typhimuriums were recovered from facal samples of diarrhoea patients in 45 general hospitals from 11 cities of Guangdong province between 2010 and 2017. These isolates were collected by the Guangdong Provincial Center for Disease Control and Prevention (CDC) in a clinic-based Salmonella infection surveillance of outpatients with diarrhea, as described previously (Zhang et al., 2013). All 1263 S. Typhimurium isolates were incubated on MacConkey agar plates, containing 4 mg/L cefotaxime. The cefotaxime-resistant S. Typhimurium isolates were subjected to screening for CTX-M, CTX-M-1G, CTX-M-9G, CMY-2G, SHV, and DHA genes (Supplementary Table S1; Liu et al., 2007), and blaCTX-M-1G/9G-positive isolates were further subjected to determine the subtypes of ESBL-encoding genes, as previously reported (Zhao and Hu, 2013). The DNA sequences and deduced amino acid sequences were compared with the reported sequences from GenBank. Antimicrobial susceptibility testing was performed on all the CTX-M-producing isolates by the agar dilution method, except for colistin with the broth dilution method. The following antimicrobials were tested: cefotaxime, ceftriaxone, ceftazidime, ceftiofur, meropenem, ciprofloxacin, nalidixic acid, sulfamethoxazole/trimethoprim, gentamicin, amikacin, florfenicol, fosfomycin, azithromycin, doxycycline, olaquindox, tigecycline, and colistin. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI, 2018: M100-S25), and veterinary CLSI (VET01-A4/VET01-S2) guidelines (Humphries et al., 2019), and the resistance breakpoints for colistin were interpreted based on EUCAST (>2 mg/L) criteria, respectively. Escherichia coli ATCC25922 was used as the quality control strain.
Molecular Typing
The genetic relatedness of blaCTX-M-positive S. Typhimurium isolates was analyzed by PFGE with the XbaI digestion of genomic DNA (Palhares et al., 2014). PFGE patterns were analyzed using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) with the Dice similarity coefficient, and a cut-off value of 85% of the similarity values was chosen to indicate identical or similar PFGE types.
WGS and Phylogenetic Analysis
Based on the results of PFGE types and resistance profiles analysis, representative blaCTX-M-positive S. Typhimurium isolates (n = 57) were selected and their genomic DNA were subjected to 250-bp paired-end whole-genome sequencing (WGS), which at a depth of 100X, using the Illumina MiSeq system (Illumina, San Diego, CA, United States), using default parameters, followed by assembling the 150 bp paired-end Illumina reads using SPAdes v3.6.2 (Humphries et al., 2019). Multi locus sequence typing (MLST), antibiotic resistance genes (ARGs), and plasmid replicon types were analyzed using the CGE server.1 Phylogenetic tree for CTX-M-producing isolates was structured on the basis of the core genome using Harvest version 1.1.2 (Treangen et al., 2014), and the corresponding characteristics of each isolate were visualized using online tool iTOL version 4 (Letunic and Bork, 2019). The population structure of each phylogenetic tree was defined using hierBAPS v6.0 (Cheng et al., 2013).
Conjugation Assay, Gene Location, and Plasmids Analysis
To test the transferability of blaCTX-M genes, conjugation experiment was carried out by the liquid mating-out assay, with the streptomycin-resistant E. coli C600 as the recipient. Transconjugants were selected on MacConkey agar plates that were supplemented with cefotaxime (2 mg/L) and streptomycin (1,500 mg/L). Antimicrobial susceptibility testing was conducted on transconjugants and the blaCTX-M gene was confirmed by PCR, as described above. PCR-based replicon typing was performed for transconjugants, as previously described (Bankevich et al., 2012). To determine the location of blaCTX-M, plasmids from the selected transconjugants were linearized using S1 nuclease and subjected to PFGE, followed by Southern blot hybridization using a digoxigenin-labeled probe specific for blaCTX-M-1G/9G, as previously described (Liu et al., 2007).
Data Availability
All genome assemblies of the 57 blaCTX-M-positive strains were deposited in GenBank and are registered under BioProject accession number PRJNA797940 and PRJNA629650.
Results
Prevalence of CTX-M Genes
A total of 221 (17.5%) isolates displayed resistance to cefotaxime among the 1,263 S. Typhimurium isolates collected in 45 hospitals across 11 cities from Guangdong, China. Of which, 82.4% (182/221) carried one or two blaCTX-M variants. In addition, 20.8% (46) isolates contained blaCMY-2G gene and 16 (7.2%) isolates harbored blaDHA gene, and no blaSHV gene was detected among these isolates. A total of nine blaCTX-M variants (blaCTX-M-55, blaCTX-M-14, blaCTX-M-65, blaCTX-M-64, blaCTX-M-130, blaCTX-M-27, blaCTX-M-15, blaCTX-M-104, and blaCTX-M-123) were detected in 182 blaCTX-M-producing isolates, and the most predominant was blaCTX-M-55 (39.6%, 72/182), followed by blaCTX-M-14 (22.5%, 41/182) and blaCTX-M-65 (19.2%, 35/182; Figure 1B). Furthermore, one isolate harbored both blaCTX-M-55 and blaCTX-M-14.
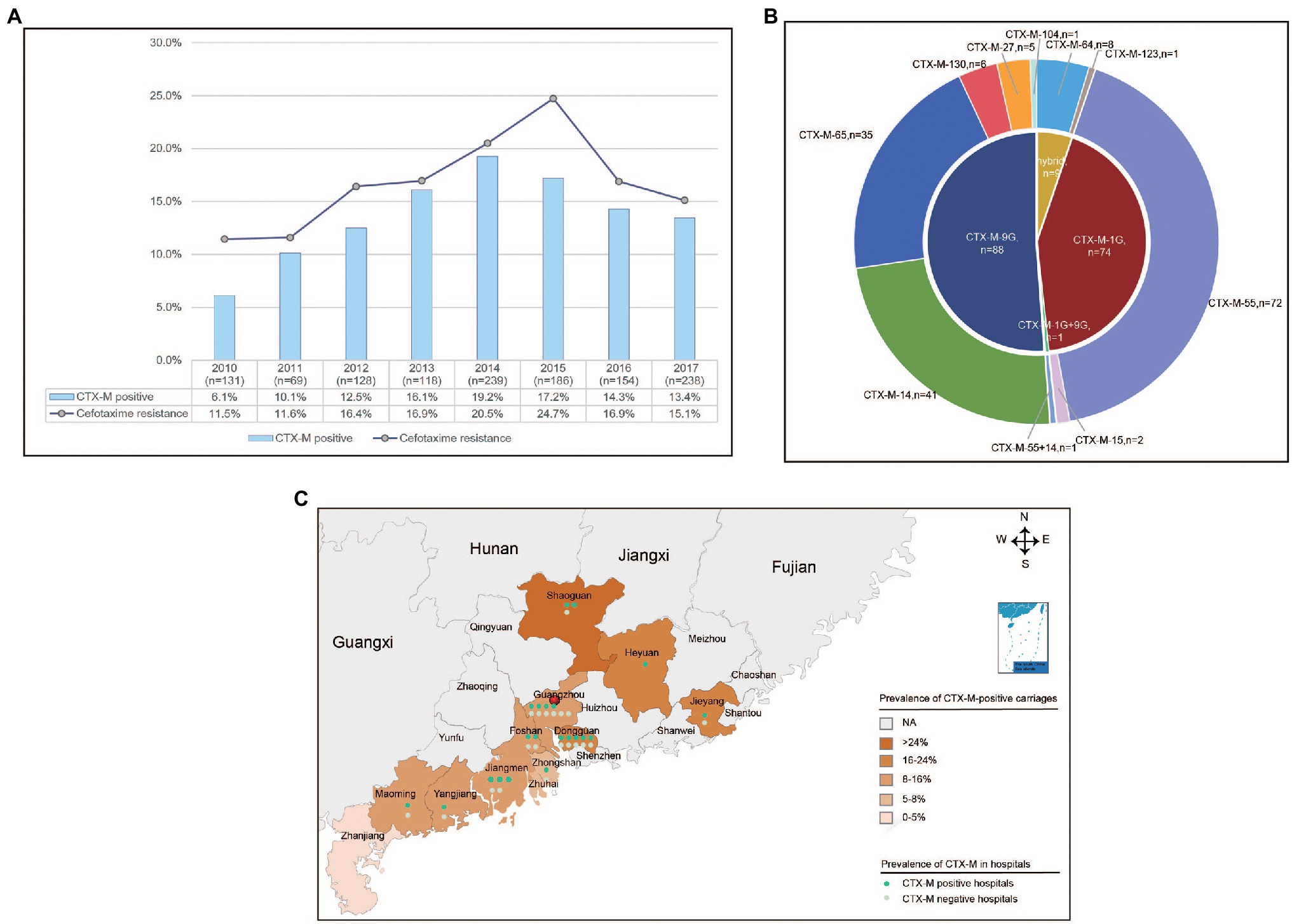
Figure 1. The prevalence of blaCTX-M-positive Salmonella Typhimurium isolates from diarrhoeal patients in 11 hospitals in Guangdong Province, China from 2010 to 2017. (A) The rate of resistance to cefotaxime and detection rate of blaCTX-M-positive S. Typhimurium isolates in Guangdong Province from 2010 to 2017. (B) The subtypes and numbers of variants in CTX-M. (C) The detection rate of blaCTX-M-positive S. Typhimurium isolates in 11 cities in Guangdong Province.
The percentages of cefotaxime-resistant isolates and blaCTX-M-positive isolates had been shifted significantly from 11.5% and 6.1% in 2010 to 24.7% and 17.2% in 2015, but decreased to 15.1% and 13.4% in 2017, respectively (Figure 1A). The blaCTX-M-positive isolates were identified in 35 hospitals among 11 cities. Of which, Shaoguan had the highest detection power of 25.0%. The mean positive prevalence of blaCTX-M carriages was 13.5% among the 12 cities (Figure 1C). Among the patients who were found to be positive for blaCTX-M-positive S. Typhimurium isolates, the median age of patients with blaCTX-M-producing isolates was 1 year (range 0–90 years), and 90% of patients were <5 years of age. In addition, 70.3% patients were male (Table 1).

Table 1. blaCTX-M-Positive S. Typhimurium isolates collected from patient demographics characteristics, Guangdong, 2010–2017 (N = 182).
Antimicrobial Susceptibility
Antimicrobial susceptibility was tested among the 182 blaCTX-M-positive S. Typhimurium isolates, and most of the isolates showed resistance to sulfamethoxazole/trimethoprim (81.3%), and florfenicol (70.9%), followed by gentamicin (48.4%) and ciprofloxacin (31.3%). Relatively low resistance rates were observed for colistin (14.8%), fosfomycin (14.3%), and amikacin (1.7%). All the 182 isolates were susceptible to meropenem. Of note, the isolates that carried blaCTX-M-9G had significantly higher resistance rates to nine antibacterials compared with blaCTX-M-1G (p < 0.01), including florfenicol, amikacin, gentamicin, ciprofloxacin, nalidixic acid, polymyxin, fosfomycin, azithromycin, and sulfamethoxazole/trimethoprim (Figure 2A). The same scenario was also observed in blaCTX-M-55, blaCTX-M-14, and blaCTX-M-65 positive isolates. However, the isolates that carried blaCTX-M-1G, including blaCTX-M-55, had remarkably higher rates of resistance to ceftazidime compared with blaCTX-M-9G including blaCTX-M-14 and blaCTX-M-65 (p < 0.001).
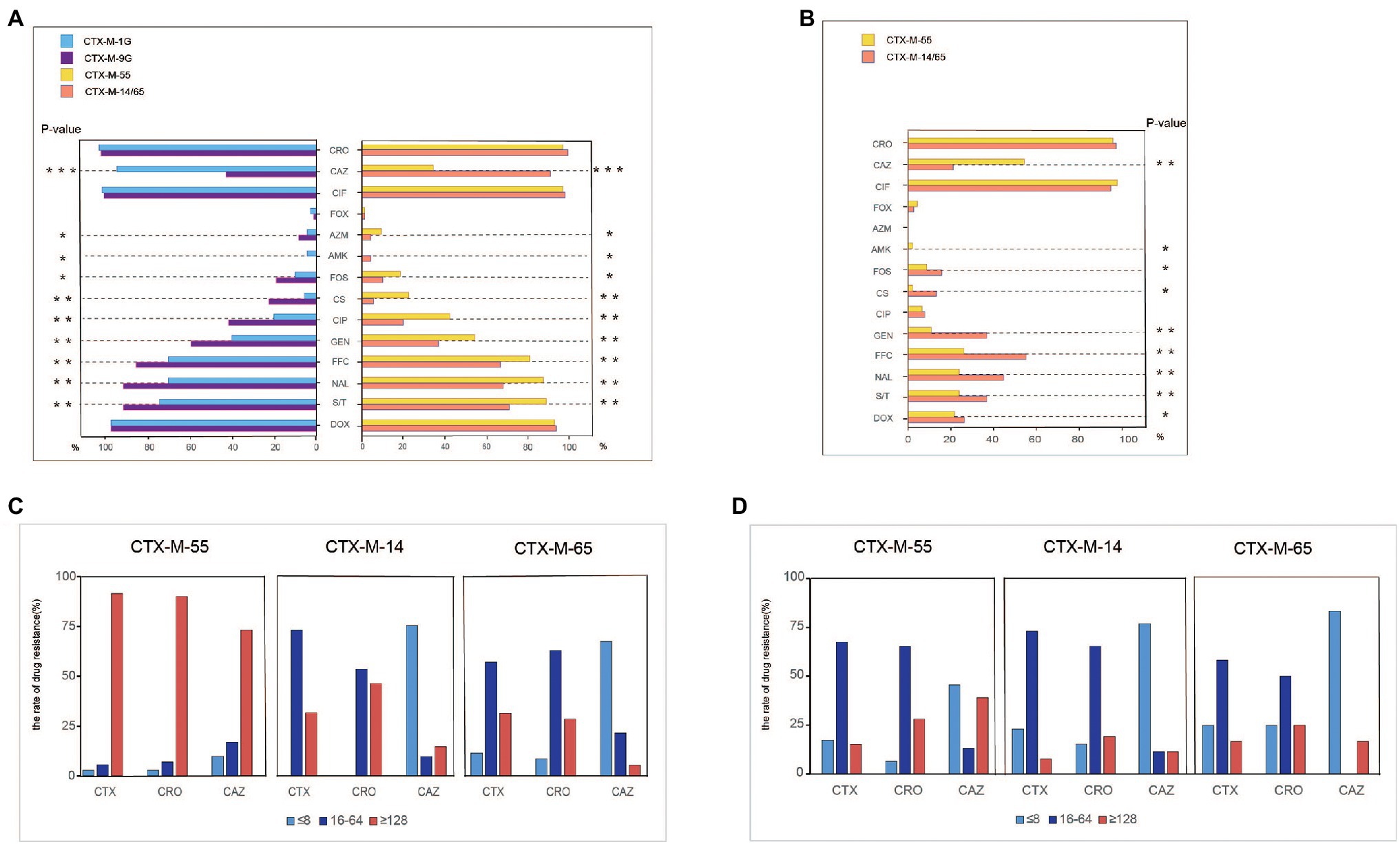
Figure 2. Antimicrobial-resistant phenotypes analysis of blaCTX-M-positive S. Typhimurium isolates and transconjugants isolates. (A) Antimicrobial-resistant phenotypes analysis of blaCTX-M-1G/9G/55/14/65-positive S. Typhimurium isolates. (B) Antimicrobial-resistant phenotypes analysis of blaCTX-M-1G/9G/55/14/65-positive S. Typhimurium’ transconjugants. (C) Anslysis of resistant to CTX, CRO, and CAZ in blaCTX-M-55/14/65-positive S. Typhimurium isolates. (D) Anslysis of resistant to CTX, CRO, and CAZ in blaCTX-M-55/14/65 S. Typhimurium’ transconjugants. CTX, cefotaxime; CRO, ceftriaxone; CAZ, ceftazidime; CIF, ceftiofur; FOX, cefoxitin; AZM, azithromycin; AMK, amikacin; FOS, fosfomycin; CS, polymyxin; CIP, ciprofloxacin; GEN, gentamicin; FFC, florfenicol; NAL, nalidixic acid; S/T, sulfamethoxazole/trimethoprium; and DOX, doxycycline. *means statistically different (p < 0.05), **means the difference is more significant (p < 0.01), ***means the difference is particularly significant (p < 0.001).
Furthermore, to determine the association between the dominant blaCTX-M genes and the 3GCs susceptibility, MICs of cefotaxime, ceftriaxone and ceftazidime were grouped into three levels, namely low resistance level (≤8 mg/ml), medium resistance level (16–64 mg/ml), and high resistance level (≥128 mg/ml; Figure 2C). The majority of blaCTX-M-55, blaCTX-M-14, and blaCTX-M-65-positive S. Typhimurium isolates showed moderate and high levels resistance to cefotaxime and ceftriaxone. However, the proportion of high levels of resistance to cefotaxime and ceftriaxone in blaCTX-M-55-positive S. Typhimurium isolates was higher than that of blaCTX-M-14/65-positive S. Typhimurium. It was obvious that most blaCTX-M-55-positive isolates are resistant to ceftazidime at high levels. In contrast, most isolates blaCTX-M-14-positive or blaCTX-M-65-positive were presented low-level resistant to ceftazidime.
Molecular Typing
The genetic relatedness of blaCTX-M-55-positive, blaCTX-M-14-positive and blaCTX-M-65-positive S. Typhimurium isolates were analyzed by PFGE, respectively. PFGE was successfully performed in 71 blaCTX-M-55-positive isolates and distributed into 26 pulsotypes. The 22 isolates in Type III were obtained in nine hospitals across four cities during 2014–2016. Similarly, the 17 isolates in Type VII were originated in six hospitals from four cities during 2010–2015 (Supplementary Figures S1A, S2). The clonal transmission of blaCTX-M-55-positive strains was observed at different hospitals in the same city between 2014 and 2016. A total of 21 different pulsotypes were detected among 41 blaCTX-M-14-positve isolates, and Type I was predominant (n = 9, 21.95%; Supplementary Figures S1B, S2). The clonal transmission of blaCTX-M-14-positive strains was observed at the same hospitals in the same city in 2012. Most importantly, all 35 blaCTX-M-65-positve isolates were distributed into 15 pulsotypes, and the most predominant Type VIII contained 19 isolates (54.3%) and was originated from nine hospitals in seven cities during 2013–2017 (Supplementary Figures S1C, S4). The spread of blaCTX-M-65-positve isolates’ clones from Guangzhou and Jieyang was observed.
According to PFGE typing and resistance phenotype, 57 (27 blaCTX-M-55, 14 blaCTX-M-14, 15 blaCTX-M-65, and 1 blaCTX-M-55/14) S. Typhimurium isolates were selected for WGS. In silico MLST analysis revealed that these isolates belong to ST34 (n = 40) and ST19 (n = 17; Figure 3). Among them, most ST34 (3.8%, 15/40) and ST19 (4.1%, 7/17) belong to cluster 1 from Guangzhou and cluster 2/3/4 from Dongguan, respectively.
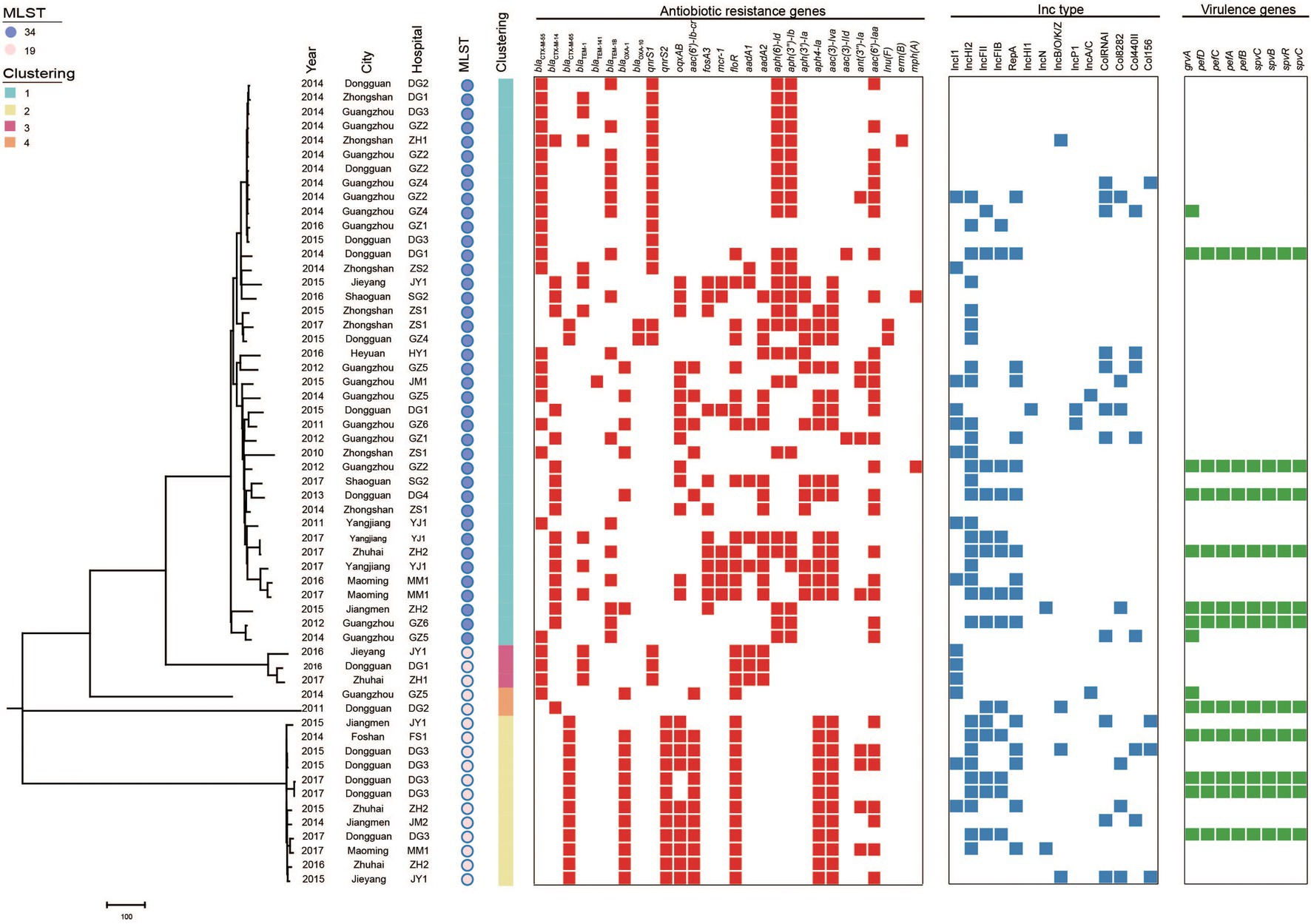
Figure 3. Phylogenetic analysis of blaCTX-M-positive S. Typhimurium isolates in this study (n = 57). Bayesian evolutionary tree was constructed using core-genome SNPs. Each isolate is labeled with the city of isolation year, hospital, ST and cluster. The red-filled squares indicate the possession of the indicated antimicrobial resistance genes (ARGs), the blue-filled squares indicate plasmid (Inc type), and the green-filled squares indicate virulence genes.
Phylogenetic Analysis of blaCTX-M-55/14/65- Positive Salmonella Typhimurium
The population structure was further analyzed by constructing phylogenetic trees based on the core genomes of the 57 blaCTX-M-positive isolates. Bayesian analysis displayed that all isolates were classified into four different lineages. The major Lineage I belong to ST34 and Lineage II-IV belong to ST19.
To explore the genetic relationships of blaCTX-M-55/14/65-positive S. Typhimurium isolates in this study and other resources in China, 84 blaCTX-M-positive S. Typhimurium isolates (including 36 blaCTX-M-55, 33 blaCTX-M-14, and 15 blaCTX-M-65) were selected from GenBank. A maximum likelihood phylogenetic tree was constructed on the basis of 32,165 core genome single nucleotide polymorphisms (cgSNPs) among 138 isolates (Figure 4). These 138 isolates were mainly distributed in Guangdong province (n = 134) and other provinces (n = 4), such as Shanghai, Hebei, Jiangxi, and Zhejiang. Notably, these 138 isolates were primarily ST34 and ST19 members and originated from diverse sample types, including humans (patients, synviol fluid and blood culture), food (beef, chicken, feces, pork) and the environments (stool). It should be noted that four blaCTX-M-14-positive ST34 S. Typhimurium isolates from patient samples in three cities, Guangdong (own isolate’ number: 17E74), shared only 64 SNPs with a blaCTX-M-14-positive ST34 S. Typhimurium isolate from a blood culture sample in Jiangxi (accession number SAMN10914546). In addition, a blaCTX-M-55-positive ST34 S. Typhimurium isolate from a patient in Dongguan, Guangdong, in this study (own isolate’ number: L-S2816) shared only six SNPs with a blaCTX-M-55-positive ST34 S. Typhimurium isolates from pork in Shenzhen, Guangdong (accession number SAMN16986615). Finally, one blaCTX-M-65-positive ST19 S. Typhimurium isolate from a patient sample in Zhongshan, Guangdong, in this study (own isolate’ number: 17E594) shared only 28 SNPs with a blaCTX-M-65-positive ST19 S. Typhimurium isolate from pork sample in Shenzhen, Guangdong (accession number SAMN16986937). These data may demonstrate that blaCTX-M-positive S. Typhimurium isolates from human were likely to be closely related to food and environment in China, and the environment and food chain may play an important role in the transmission of blaCTX-M-positive S. Typhimurium isolates.
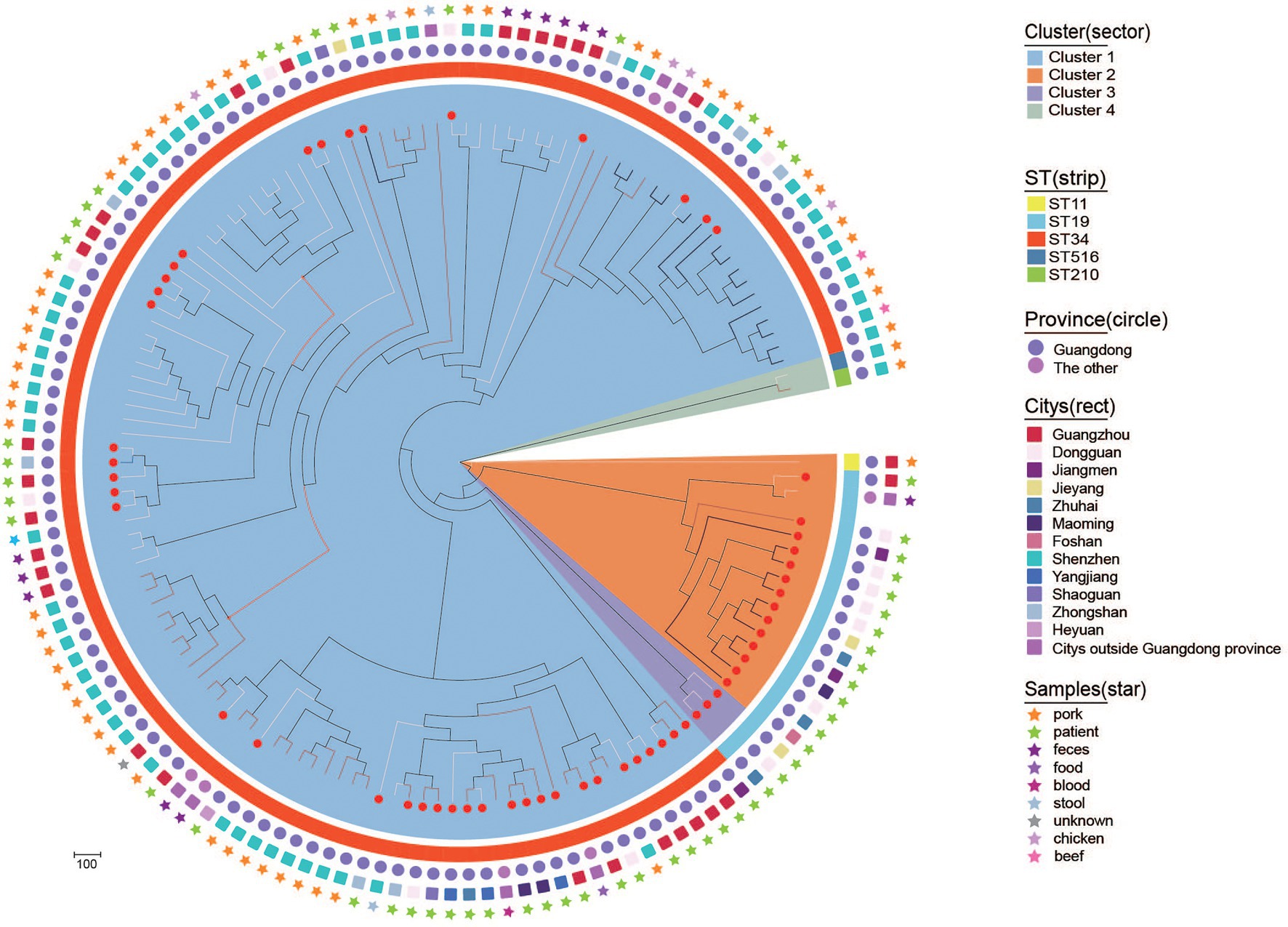
Figure 4. Phylogenetic structures of the blaCTX-M-positive S. Typhimurium isolates from this study and the GenBank database. The Bayesian evolutionary tree shows the relationships among the 141 blaCTX-M-55/14/65-positive S. Typhimurium isolates. Sample source, city, province and ST from which isolates obtained are indicated as star, rect, circle, and strip. blaCTX-M-55-positive, blaCTX-M-14-positive, and blaCTX-M-65-positive S. Typhimurium are indicated with a white, black, and red line, respectively. The red dots on the branches represent the bacteria that belong to this study.
Plasmid Analysis
Conjugation experimental results proved that 84 blaCTX-M-positive plasmids were successfully transferred to E. coil C600 recipient strains among 148 blaCTX-M-55/14/65 positive S. Typhimurium isolates. The transconjugants blaCTX-M-55/14/65-positive mainly showed moderate levels resistance to cefotaxime and ceftriaxone and low levels resistance to ceftazidime. Notably, the proportion of blaCTX-M-55-positive transconjugants with high levels of resistance to ceftazidime was higher than that of blaCTX-M-14/65-positive transconjugants (Figure 2B). Meanwhile, the blaCTX-M-55-positive transconjugants had significantly higher resistance rates to ceftazidime compared with blaCTX-M-14/65-positive transconjugants (p < 0.001; Figure 2B).
In addition to cephalosporins, partial transconjugants displayed resistance to florfenicol (n = 33), doxycline (n = 29), sulfamethoxazole/trimethoprim (n = 28), gentamicin (n = 25), fosfomycin (n = 17), and azithromycin (n = 17). Obviously, the transconjugants that carried blaCTX-M-9G had significantly higher resistance rate to seven antibiotics compared with blaCTX-M-1G (p < 0.01), including amikacin, fosfomycin, gentamicin, polymyxin, florfenicol, sulfamethoxazole/trimethoprim, nalidixic, acid and doxycycline (Figure 2D).
Through conjugation assay and gene location methods, replicon analysis was performed on the blaCTX-M-55/14/65-positive transconjugants, mainly IncI1(n = 49), followed by IncHI2(n = 23) and IncFII (n = 8; Supplementary Figure S7). Based on PFGE profiles, one isolate from each clonal lineage was selected for S1-PFGE and hybridization. For the blaCTX-M-55/14/65-positive isolates (n = 12, 8, and 6, respectively), S1-PFGE and hybridization analyses confirmed that blaCTX-M-55-positive genes (n = 10) from 12 isolates were mainly located on ~76.8 kb plasmids, blaCTX-M-14-positive genes (n = 4) from eight isolates were mainly located on 54.7–80 kb plasmids, and blaCTX-M-65-positive genes (n = 4) were from six isolates mainly located on 216.9–244.4 kb plasmids (Supplementary Figures S5, S6). Primers connecting contigs containing the backbone of different plasmids and blaCTX-M genes were used. The results illustrated that IncI1 (65.3%, 32/49), IncFII (50.0%, 4/8) and IncHI2 (30.4%, 7/23) plasmids may be major vectors for the wide dissemination of blaCTX-M-55, blaCTX-M-14, and blaCTX-M-65 genes in S. Typhimurium isolates. In addition, WGS analysis revealed that sequenced strains also carry other plasmids, such as IncFIB-type, IncHI1-type, IncN-type and other different kinds of plasmids.
Resistance Profiles
WGS analysis demonstrated that 57 blaCTX-M-producing isolates possessed 47 distinct ARGs. Several clinically important ARGs were identified to co-carry with blaCTX-M, including mcr-1, fosA3, oqxAB, qnrS1, qnrS2, aac-(6′)-Ib-cr, and floR, with a prevalence rate from 12.3% to 52.6%. Moreover, blaTEM-141 (n = 1) was first detected in S. Typhimurium isolates.
Notably, some ARGs were co-existence with a specific blaCTX-M variant. For example, mcr-1 and fosA3 were unique to blaCTX-M-14-positive isolates. qnrS2, aac(6′)-Ib-cr, and blaOXA-1 were primarily found in blaCTX-M-65-positive isolates. In contrast, qnrS1 and blaTEM-1B were largely present in blaCTX-M-55-positive isolates. Additionally, both oqxAB and floR mostly presented in blaCTX-M-65- and blaCTX-M-14-positive isolates.
Discussion
In this study, the detection rate of blaCTX-M-positive S. Typhimurium from diarrhoeal outpatients increased from 2010 to 2015 in Guangdong Province, China. It was speculated that blaCTX-M-positive S. Typhimurium outbreaks are linked to the consumption of food animal or raw meat, particularly pork. Firstly, previous studies showed that the swine is one of the major reservoirs for Salmonella (Wang et al., 2007; Jackson et al., 2013). Secondly, the data from China’s National Nutrition Survey also displayed that the total pork intake of Chinese residents increased by 73% from 1992 to 2012 (He et al., 2015). It’s worth noting that the overall percentage of cephalosporin use had an upward trend from 2012 to 2017 in hospitals (Branch, 2020), which can give us some hints that the transmission of blaCTX-M may be relevant to the selective pressure of cephalosporin antibiotics. Then, it was obvious that the detection rate of cefotaxime-resistant S. Typhimurium in Guangdong province steadily decreased from 2016 to 2017. Meanwhile, according to CHINET bacterial resistance monitoring, the detection rate of cefotaxime-resistant Enterobacter decreased gradually from 2015 to 2017. Therefore, the long-term monitoring of cephalosporin usage and the prevalence of the blaCTX-M-positive Salmonella are necessary for public health.
In this study, nine blaCTX-M variants were detected in 182 blaCTX-M-producing isolates, and the most predominant was blaCTX-M-55, followed by blaCTX-M-14 and blaCTX-M-65, which is consistent with previous studies (Zhang et al., 2018). Currently, blaCTX-M-55-positive Salmonella has been reported as the dominant genotype in other countries, including Germany, Cambodia, Korea and Vietnam, and was frequently detected in food animals, especially in poultry and pork (Nguyen et al., 2016; Kim et al., 2017; Lay et al., 2021; Pietsch et al., 2021); blaCTX-M-14-positive Enterobacteriaceae has been reported as the dominant genotype in some countries, including China, South Korea, Japan, and Spain, and was frequently detected in food, especially in retail chicken meat and pork (Bai et al., 2016; Shigemura et al., 2020; Wang et al., 2021); blaCTX-M-65-positive Salmonella has been found in China, the United States, and Germany, and was commonly found in food animal sources, especially in chicken (Brown et al., 2018; Martínez-Puchol et al., 2021; Pietsch et al., 2021).
The blaCTX-M-65-positive S. Typhimurium isolates with the same profile were found in the three hospitals (GZ5, JY1, and DG3), which indicated that the clonal dissemination of blaCTX-M-65-positive S. Typhimurium occurred in hospital. Furthermore, a few blaCTX-M-55/14/65-positive S. Typhimurium isolates from 2010 to 2017 showed 100% homology by PFGE analysis, which suggested that the possible long-term outbreaks were caused by clonal transfer of blaCTX-M-55/14/65-positive S. Typhimurium strains within the hospital. WGS demonstrated that these S. Typhimurium isolates belonged to ST34 and ST19. In fact, it has been shown that ST34 and ST19 are common S. Typhimurium STs responsible for infections worldwide, especially in China (Woh et al., 2021). It has been proved previously that the ST34 S. Typhimurium isolates with the highest percentage of MDR are mainly recovered from diarrhea patients (Biswas et al., 2019; Jiu et al., 2020; Luo et al., 2020; Sun et al., 2020). Furthermore, ST19 has been found mostly in human clinical Salmonella isolates, but also in animals and the environment, and successful in South African and China. ST19 was only occasionally found in United States and Mexico and coexists with quinolone resistance genes qnrS (Kariuki and Onsare, 2015; Gómez-Baltazar et al., 2019; Monte et al., 2020; Yao et al., 2020; Xiaoting et al., 2021).
Our genomic Beast tree analysis provided evidence for the closer relationship among blaCTX-M-positive strains from the outpatients in this study and pork. Pig has been singled out as the most likely reservoir for the amplification and spread of Enterobacteriaceae that are resistant to ESBL and other antibiotics (Nordmann and Poirel, 2016), The same major blaCTX-M, as presented in this study, was also detected in isolates from a pig farm in China (Zhang et al., 2019). Therefore, our study provides strong genome epidemiology-based evidence that the consumption of pork is the likely contamination source of blaCTX-M-positive S. Typhimurium.
In the current study, most blaCTX-M-55/14/65 genes identified were carried by IncI1, IncHI2, and IncFII plasmids, which indicated that plasmids belonged to blaCTX-M-55/14/65-positive isolates and were diverse. Among them, IncI1 has become one of the most common plasmid families in contemporary Enterobacteriaceae from both human and animal sources. In clinical epidemiology, IncI1 ranks first as the confirmed vehicle of the transmission of extended spectrum beta-lactamase and AmpC genes in isolates from food-producing animals (Carattoli et al., 2021). The second, HI2, followed by FII plasmid, was found to be associated with the transfer of the mcr-1 and ESBL encoding genes all over the world, especially in European and African countries. The coexistence of mcr-1 and ESBL encoding genes in HI2 plasmids was less reported in China in recent years (Biswas et al., 2019; Wang et al., 2020). Worryingly, as the vector of drug resistance gene, FII plasmid not only carries mcr-1, but also is one of the common carriers of NDM gene (Wu et al., 2019).
WGS analysis further revealed that blaCTX-M-55/14/65 coexisted with other 25 types of ARGs, of which 11 ARGs were highly prevalent with detection rates >50%. Of note, mcr-1, conferring resistance to the last-resort antibiotic colistin, was detected in seven blaCTX-M-positive S. Typhimurium isolates. To begin with, the coexistence of mcr-1 and blaCTX-M-55 was first reported in the literature from colistin-resistant clinical source E. coli isolates in Ecuador in 2016 (Ortega-Paredes et al., 2016). Next, the coexistence of mcr-1 and ESBL encoding genes (including blaCTX-M-55/14) has been found in Tunisian from chicken, in China from food animal (including pigs, cattle and chickens) and in France from human E. coli (Birgy et al., 2018; Hassen et al., 2020; Shafiq et al., 2021). The coexistence of mcr-1 and blaCTX-M in Salmonella isolates was mostly reported from food animal sources and found in Asian countries, including China, Cambodia and Laos (Ma et al., 2017; Lay et al., 2021). Last but not least, the coexistence of mcr-1, blaNDM-5, and blaCTX-M-55 in Klebsiella pneumoniae ST485 Clinical Isolates appeared in China (Cao X. et al., 2021), which further alerted us to the dangers of multidrug-resistant strains.
Conclusion
In summary, our study investigated the epidemiology of S. Typhimurium in Guangdong province, China. which could supplement important local epidemiological data. Among them, ST34 S. Typhimurium dominated the cefotaxime-resistant strains and the major resistance mechanism of cefotaxime-resistant Salmonella produced the CTX-M-type ESBLs, in which blaCTX-M-55 was most prevalent. Obviously, the prevalence of blaCTX-M-positive S. Typhimurium carried multiple resistance genes, which indicated the potential risk of Salmonella infections. In the current study, blaCTX-M-55/65-positive S. Typhimurium isolates were found from different outpatients with community acquired diarrhoea at same hospital, which suggested the nosocomial cloning transmission. This study underscored the importance of surveillance for blaCTX-M-positive microbes in patients and indicated a high likelihood for the spread of cephalosporin resistance from pig chain to humans.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
QJ wrote the first draft of the manuscript. QJ and L-xF contributed to conception and design of the study. B-xK, D-sW, DW, M-gW, R-yS, J-eL, and ZS performed the statistical analysis. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was jointly supported by the International Cooperation and Exchange of the National Natural Science Foundation of China (grant no. 31520103918), the Program for Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China IRT_17R39, the National Natural Science Fund of China (grant no. 31802244), and the National Science and Technology Major Project (2018ZX10714002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.865254/full#supplementary-material
Supplementary Figure S1 | Genetic relatedness, year, city and hospital of the blaCTX-M-positive Salmonella Typhimurium isolates in Guangdong from 2010 to 2017. (A,B) Genetic relatedness, year, city, hospital and pulsotype of the blaCTX-M-55-positive S. Typhimurium isolates (C) genetic relatedness, year, city, hospital and pulsotype of the blaCTX-M-14-positive S. Typhimurium isolates (D) genetic relatedness, year, city, hospital and pulsotype of the blaCTX-M-65-positive S. Typhimurium isolates.
Supplementary Figure S2 | Genetic relatedness, year, city and hospital of the blaCTX-M-55-positive S. Typhimurium isolates in Guangdong from 2010 to 2017.
Supplementary Figure S3 | Genetic relatedness, year, city and hospital of the blaCTX-M-14-positive S. Typhimurium isolates in Guangdong from 2010 to 2017.
Supplementary Figure S4 | Genetic relatedness, year, city and hospital of the blaCTX-M-65-positive S. Typhimurium isolates in Guangdong from 2010 to 2017.
Supplementary Figure S5 | S1 endonuclease pulsed-field gel electrophoresis analysis of plasmids from the blaCTX-M-1G-positive S. Typhimurium isolates.
Supplementary Figure S6 | S1 endonuclease pulsed-field gel electrophoresis analysis of plasmids from the blaCTX-M-9G-positive S. Typhimurium isolates.
Supplementary Figure S7 | PBRT types that blaCTX-M-positive S. Typhimurium isolates (A) all PBRT types that blaCTX-M-positive S. Typhimurium isolates (B) PBRT types that blaCTX-M-55-positive S. Typhimurium isolates. (C) PBRT types that blaCTX-M-14-positive S. Typhimurium isolates. (D) PBRT types that blaCTX-M-65-positive S. Typhimurium isolates.
Footnotes
References
Bai, L., Zhao, J., Gan, X., Wang, J., Zhang, X., Cui, S., et al. (2016). Emergence and diversity of Salmonella enterica serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime from patients and food-producing animals in China. Antimicrob. Agents Chemother. 60, 3365–3371. doi: 10.1128/aac.02849-15
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Birgy, A., Madhi, F., Hogan, J., Doit, C., Gaschignard, J., Caseris, M., et al. (2018). CTX-M-55-, MCR-1-, and FosA-producing multidrug-resistant Escherichia coli infection in a child in France. Antimicrob. Agents Chemother. 62, 00127–18. doi: 10.1128/aac.00127-18
Biswas, S., Li, Y., Elbediwi, M., and Yue, M. (2019). Emergence and dissemination of mcr-carrying clinically relevant Salmonella Typhimurium monophasic clone ST34. Microorganisms 7:298. doi: 10.3390/microorganisms7090298
Brown, A. C., Chen, J. C., Watkins, L. K. F., Campbell, D., Folster, J. P., Tate, H., et al. (2018). CTX-M-65 extended-spectrum β-lactamase-producing Salmonella enterica serotype infantis, United States(1). Emerg. Infect. Dis. 24, 2284–2291. doi: 10.3201/eid2412.180500
Cao, C., Niu, Q., Chen, J., Xu, X., Sheng, H., Cui, S., et al. (2021). Epidemiology and characterization of CTX-M-55-type extended-spectrum β-lactamase-producing Salmonella enterica serovar enteritidis isolated from patients in Shanghai, China. Microorganisms 9:260. doi: 10.3390/microorganisms9020260
Cao, X., Zhong, Q., Guo, Y., Hang, Y., Chen, Y., Fang, X., et al. (2021). Emergence of the coexistence of mcr-1, bla (NDM-5), and bla (CTX-M-55) in Klebsiella pneumoniae ST485 clinical isolates in China. Infect. Drug Resist. 14, 3449–3458. doi: 10.2147/idr.s311808
Carattoli, A., Villa, L., Fortini, D., and García-Fernández, A. (2021). Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid 118:102392. doi: 10.1016/j.plasmid.2018.12.001
Cheng, L., Connor, T. R., Siren, J., Aanensen, D. M., and Corander, J. (2013). Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 30, 1224–1228. doi: 10.1093/molbev/mst028
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Eighth Edition: M100.
Diard, M., and Hardt, W. D. (2017). Basic processes in Salmonella-host interactions: within-host evolution and the transmission of the virulent genotype. Microbiol. Spectr. 5:5. doi: 10.1128/microbiolspec.MTBP-0012-2016
Fu, Y., Xu, X., Zhang, L., Xiong, Z., Ma, Y., Wei, Y., et al. (2020). Fourth generation cephalosporin resistance among Salmonella enterica serovar enteritidis isolates in Shanghai, China conferred by bla (CTX-M-55) harboring plasmids. Front. Microbiol. 11:910. doi: 10.3389/fmicb.2020.00910
Gómez-Baltazar, A., Vázquez-Garcidueñas, M. S., Larsen, J., Kuk-Soberanis, M. E., and Vázquez-Marrufo, G. (2019). Comparative stress response to food preservation conditions of ST19 and ST213 genotypes of Salmonella enterica serotype Typhimurium. Food Microbiol. 82, 303–315. doi: 10.1016/j.fm.2019.03.010
Hassen, B., Abbassi, M. S., Ruiz-Ripa, L., Mama, O. M., Hassen, A., Torres, C., et al. (2020). High prevalence of mcr-1 encoding colistin resistance and first identification of bla(CTX-M-55) in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int. J. Food Microbiol. 318:108478. doi: 10.1016/j.ijfoodmicro.2019.108478
He, Y., Yang, X., Xia, J., Zhao, L., and Yang, Y. (2015). Consumption of meat and dairy products in China: a review. Proc. Nutr. Soc. 75, 385–391. doi: 10.1017/S0029665116000641
Humphries, R. M., Abbott, A. N., and Hindler, J. A. (2019). Understanding and addressing CLSI breakpoint revisions: a primer for clinical laboratories. J. Clin. Microbiol. 57, 00203–19. doi: 10.1128/JCM.00203-19
Jackson, B. R., Griffin, P. M., Cole, D., Walsh, K. A., and Chai, S. J. (2013). Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerg. Infect. Dis. 19, 1239–1244. doi: 10.3201/eid1908.121511
Jiu, Y., Meng, X., Hong, X., Huang, Q., Wang, C., Chen, Z., et al. (2020). Prevalence and characterization of Salmonella in three typical commercial pig abattoirs in Wuhan, China. Foodborne Pathog. Dis. 17, 620–627. doi: 10.1089/fpd.2019.2737
Kariuki, S., and Onsare, R. S. (2015). Epidemiology and genomics of invasive nontyphoidal Salmonella infections in Kenya. Clin. Infect. Dis. 61(Suppl. 4), S317–S324. doi: 10.1093/cid/civ711
Kim, J. S., Kim, S., Park, J., Shin, E., Yun, Y. S., Lee, D. Y., et al. (2017). Plasmid-mediated transfer of CTX-M-55 extended-spectrum beta-lactamase among different strains of Salmonella and Shigella spp. in the Republic of Korea. Diagn. Microbiol. Infect. Dis. 89, 86–88. doi: 10.1016/j.diagmicrobio.2017.03.014
Lay, K. K., Jeamsripong, S., Sunn, K. P., Angkititrakul, S., Prathan, R., Srisanga, S., et al. (2021). Colistin resistance and ESBL production in Salmonella and Escherichia coli from pigs and pork in the Thailand, Cambodia, Lao PDR, and Myanmar border area. Antibiotics 10:657. doi: 10.3390/antibiotics10060657
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Liu, J. H., Wei, S.-Y., Ma, J.-Y., Zeng, Z.-L., Lü, D.-H., Yang, G.-X., et al. (2007). Detection and characterisation of CTX-M and CMY-2 β-lactamases among Escherichia coli isolates from farm animals in Guangdong Province of China. Int. J. Antimicrob. Agents 29, 576–581. doi: 10.1016/j.ijantimicag.2006.12.015
Luo, Q., Wan, F., Yu, X., Zheng, B., Chen, Y., Gong, C., et al. (2020). MDR Salmonella enterica serovar Typhimurium ST34 carrying mcr-1 isolated from cases of bloodstream and intestinal infection in children in China. J. Antimicrob. Chemother. 75, 92–95. doi: 10.1093/jac/dkz415
Ma, S., Lei, C., Kong, L., Jiang, W., Liu, B., Men, S., et al. (2017). Prevalence, antimicrobial resistance, and relatedness of Salmonella isolated from chickens and pigs on farms, abattoirs, and markets in Sichuan Province, China. Foodborne Pathog. Dis. 14, 667–677. doi: 10.1089/fpd.2016.2264
Ma, Y., Xu, X., Gao, Y., Zhan, Z., Xu, C., Qu, X., et al. (2020). Antimicrobial resistance and molecular characterization of Salmonella enterica serovar corvallis isolated from human patients and animal source foods in China. Int. J. Food Microbiol. 335:108859. doi: 10.1016/j.ijfoodmicro.2020.108859
Martínez-Puchol, S., Riveros, M., Ruidias, K., Granda, A., Ruiz-Roldán, L., Zapata-Cachay, C., et al. (2021). Dissemination of a multidrug resistant CTX-M-65 producer Salmonella enterica serovar infantis clone between marketed chicken meat and children. Int. J. Food Microbiol. 344:109109. doi: 10.1016/j.ijfoodmicro.2021.109109
Mohammed, M., Le Hello, S., Leekitcharoenphon, P., and Hendriksen, R. (2017). The invasome of Salmonella Dublin as revealed by whole genome sequencing. BMC Infect. Dis. 17:544. doi: 10.1186/s12879-017-2628-x
Monte, D. F. M., Sellera, F. P., Lopes, R., Keelara, S., Landgraf, M., Greene, S., et al. (2020). Class 1 integron-borne cassettes harboring blaCARB-2 gene in multidrug-resistant and virulent Salmonella Typhimurium ST19 strains recovered from clinical human stool samples, United States. PLoS One 15:e0240978. doi: 10.1371/journal.pone.0240978
Nadimpalli, M., Fabre, L., Yith, V., Sem, N., Gouali, M., Delarocque-Astagneau, E., et al. (2019). CTX-M-55-type ESBL-producing Salmonella enterica are emerging among retail meats in Phnom Penh, Cambodia. J. Antimicrob. Chemother. 74, 342–348. doi: 10.1093/jac/dky451
Nguyen, D. T., Kanki, M., Nguyen, P. D., Le, H. T., Ngo, P. T., Tran, D. N., et al. (2016). Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 236, 115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017
Nordmann, P., and Poirel, L. (2016). Plasmid-mediated colistin resistance: an additional antibiotic resistance menace. Clin. Microbiol. Infect. 22, 398–400. doi: 10.1016/j.cmi.2016.03.009
Okoro, C. K., Barquist, L., Connor, T. R., Harris, S. R., Clare, S., Stevens, M. P., et al. (2015). Signatures of adaptation in human invasive Salmonella Typhimurium ST313 populations from sub-Saharan Africa. PLoS Negl. Trop. Dis. 9:e0003611. doi: 10.1371/journal.pntd.0003611
Ortega-Paredes, D., Barba, P., and Zurita, J. (2016). Colistin-resistant Escherichia coli clinical isolate harbouring the mcr-1 gene in Ecuador. Epidemiol. Infect. 144, 2967–2970. doi: 10.1017/s0950268816001369
Palhares, J. C., Kich, J. D., Bessa, M. C., Biesus, L. L., Berno, L. G., and Triques, N. J. (2014). Salmonella and antimicrobial resistance in an animal-based agriculture river system. Sci. Total Environ. 472, 654–661. doi: 10.1016/j.scitotenv.2013.11.052
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/CMR.18.4.657-686.2005
Pietsch, M., Simon, S., Meinen, A., Trost, E., Banerji, S., Pfeifer, Y., et al. (2021). Third generation cephalosporin resistance in clinical non-typhoidal Salmonella enterica in Germany and emergence of bla (CTX-M)-harbouring pESI plasmids. Microb. Genom. 7:000698. doi: 10.1099/mgen.0.000698
Shafiq, M., Huang, J., Shah, J. M., Wang, X., Rahman, S. U., Ali, I., et al. (2021). Characterization and virulence factors distribution of bla(CTX-M) and mcr-1carrying Escherichia coli isolates from bovine mastitis. J. Appl. Microbiol. 131, 634–646. doi: 10.1111/jam.14994
Shi, C. (2015). Molecular methods for serovar determination of Salmonella. Crit. Rev. Microbiol. 41, 309–325. doi: 10.3109/1040841x.2013.837862
Shigemura, H., Sakatsume, E., Sekizuka, T., Yokoyama, H., Hamada, K., Etoh, Y., et al. (2020). Food workers as a reservoir of extended-spectrum-cephalosporin-resistant Salmonella strains in Japan. Appl. Environ. Microbiol. 86, 00072–20. doi: 10.1128/aem.00072-20
Sun, R. Y., Ke, B. X., Fang, L. X., Guo, W. Y., Li, X. P., Yu, Y., et al. (2020). Global clonal spread of mcr-3-carrying MDR ST34 Salmonella enterica serotype Typhimurium and monophasic 1,4,[5],12:i:- variants from clinical isolates. J. Antimicrob. Chemother. 75, 1756–1765. doi: 10.1093/jac/dkaa115
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Wang, S., Duan, H., Zhang, W., and Li, J. W. (2007). Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol. Med. Microbiol. 51, 8–13. doi: 10.1111/j.1574-695X.2007.00305.x
Wang, J., Jiang, Y., Ji, R. Y., Wang, Z. Y., Lu, M. J., Wu, H., et al. (2021). Colistin- and tigecycline-resistant CTX-M-14-producing Salmonella enterica serovar Kentucky ST198 from retail chicken meat, China. Int. J. Antimicrob. Agents 59:106504. doi: 10.1016/j.ijantimicag.2021.106504
Wang, Z., Xu, H., Tang, Y., Li, Q., and Jiao, X. (2020). A multidrug-resistant monophasic Salmonella Typhimurium co-harboring mcr-1, fosA3, Bla (CTX-M-14) in a transferable IncHI2 plasmid from a healthy catering worker in China. Infect. Drug Resist. 13, 3569–3574. doi: 10.2147/idr.s272272
Wegener, H. C., Hald, T., Lo Fo Wong, D., Madsen, M., Korsgaard, H., and Bager, F. (2003). Salmonella control programs in Denmark. Emerg. Infect. Dis. 9, 774–780. doi: 10.3201/eid0907.030024
Whichard, J. M., Gay, K., Stevenson, J. E., Joyce, K. J., Cooper, K. L., Omondi, M., et al. (2007). Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg. Infect. Dis. 13, 1681–1688. doi: 10.3201/eid1311.061438
Woh, P. Y., Yeung, M. P. S., Goggins, W. B. 3rd, Lo, N., Wong, K. T., Chow, V., et al. (2021). Genomic epidemiology of multidrug-resistant nontyphoidal Salmonella in young children hospitalized for gastroenteritis. Microbiol. Spectr. 9:e0024821. doi: 10.1128/Spectrum.00248-21
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., and Zong, Z. (2019). NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32, 00115–18. doi: 10.1128/cmr.00115-18
Xiang, D. R., Li, J. J., Sheng, Z. K., Yu, H. Y., Deng, M., Bi, S., et al. (2015). Complete sequence of a novel IncR-F33:A-:B- plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence type 11 strain in China. Antimicrob. Agents Chemother. 60, 1343–1348. doi: 10.1128/aac.01488-15
Xiaoting, W., Chengcheng, N., Chunhui, J., Yan, L., Jing, L., Qingling, M., et al. (2021). Antimicrobial resistance profiling and molecular typing of ruminant-borne isolates of Clostridium perfringens from Xinjiang, China. J. Glob. Antimicrob. Resist. 27, 41–45. doi: 10.1016/j.jgar.2021.08.003
Xu, Z., Wang, M., Wang, C., Zhou, C., Liang, J., Gu, G., et al. (2021). The emergence of extended-spectrum β-lactamase (ESBL)-producing Salmonella London isolates from human patients, retail meats and chickens in southern China and the evaluation of the potential risk factors of Salmonella London. Food Control 128:108187. doi: 10.1016/j.foodcont.2021.108187
Yao, Z., Jiayin, W., Xinyi, Z., Ling, C., Mingyuan, H., Simin, M., et al. (2020). Identification of group B Streptococcus serotypes and genotypes in late pregnant women and neonates that are associated with neonatal early-onset infection in a South China population. Front. Pediatr. 8:265. doi: 10.3389/fped.2020.00265
Yin, Y., and Zhou, D. (2018). Organoid and enteroid modeling of Salmonella infection. Front. Cell. Infect. Microbiol. 8:102. doi: 10.3389/fcimb.2018.00102
Zhang, C. Z., Ding, X. M., Lin, X. L., Sun, R. Y., Lu, Y. W., Cai, R. M., et al. (2019). The emergence of chromosomally located Bla (CTX-M-55) in Salmonella from foodborne animals in China. Front. Microbiol. 10:1268. doi: 10.3389/fmicb.2019.01268
Zhang, L., Fu, Y., Xiong, Z., Ma, Y., Wei, Y., Qu, X., et al. (2018). Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front. Microbiol. 9:2104. doi: 10.3389/fmicb.2018.02104
Zhang, L., Lü, X., and Zong, Z. (2013). The emergence of blaCTX-M-15-carrying Escherichia coli of ST131 and new sequence types in Western China. Ann. Clin. Microbiol. Antimicrob. 12:35. doi: 10.1186/1476-0711-12-35
Zhang, W. H., Ren, S. Q., Gu, X. X., Li, W., Yang, L., Zeng, Z. L., et al. (2015). High frequency of virulence genes among Escherichia coli with the blaCTX-M genotype from diarrheic piglets in China. Vet. Microbiol. 180, 260–267. doi: 10.1016/j.vetmic.2015.08.017
Keywords: Salmonella Typhimurium, blaCTX-M, diarrhoeal outpatients, Guangdong, bacterial persistence
Citation: Jiang Q, Ke B-x, Wu D-s, Wang D, Fang L-x, Sun R-y, Wang M-g, Lei J-e, Shao Z and Liao X-p (2022) Epidemiology of blaCTX-M-Positive Salmonella Typhimurium From Diarrhoeal Outpatients in Guangdong, China, 2010–2017. Front. Microbiol. 13:865254. doi: 10.3389/fmicb.2022.865254
Edited by:
Guo-bao Tian, Sun Yat-sen University, ChinaReviewed by:
Hong-Ning Wang, Sichuan University, ChinaManal Mohammed, University of Westminster, United Kingdom
Copyright © 2022 Jiang, Ke, Wu, Wang, Fang, Sun, Wang, Lei, Shao and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-ping Liao, xpliao@scau.edu.cn
 Qi Jiang
Qi Jiang Bi-xia Ke3
Bi-xia Ke3 Liang-xing Fang
Liang-xing Fang Ruan-yang Sun
Ruan-yang Sun Jing-er Lei
Jing-er Lei Xiao-ping Liao
Xiao-ping Liao