- 1Department of Molecular and Cellular Biology, College of Science and Mathematics, Kennesaw State University, Kennesaw, GA, United States
- 2United States Department of Agriculture, Agricultural Research Service, Washington, DC, United States
- 3Department of Food, Bioprocessing and Nutrition Sciences, College of Agriculture and Life Sciences, North Carolina State University, Raleigh, NC, United States
Salmonella is one of the leading causes of foodborne illnesses worldwide. The rapid emergence of multidrug-resistant Salmonella strains has increased global concern for salmonellosis. Recent studies have shown that bacteriophages (phages) are novel and the most promising antibacterial agents for biocontrol in foods because phages specifically kill target bacteria without affecting other bacteria, do not alter organoleptic properties or nutritional quality of foods, and are safe and environmentally friendly. Due to the vast variation in Salmonella serotypes, large numbers of different and highly virulent Salmonella phages with broad host ranges are needed. This study isolated 14 Salmonella phages from turkey fecal and cecal samples. Six phages (Φ205, Φ206, Φ207, ΦEnt, ΦMont, and Φ13314) were selected for characterization. These phages were from all three families in the Caudovirales order. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) revealed that each phage had a unique structural protein profile. Each phage had a distinct host range. Φ207 and ΦEnt are both siphophages. They shared eight hosts, including seven different Salmonella serovars and one Shigella sonnei strain. These two phages showed different restriction banding patterns generated through EcoRI or HindIII digestion, but shared three bands from EcoRI digestion. ΦEnt displayed the broadest and very unusual host range infecting 11 Salmonella strains from nine serovars and three Shigella strains from two species, and thus was further characterized. The one-step growth curve revealed that ΦEnt had a short latent period (10 min) and relatively large burst size (100 PFU/infected cell). ΦEnt and its host showed better thermal stabilities in tryptic soy broth than in saline at 63 or 72°C. In the model food system (cucumber juice or beef broth), ΦEnt infection [regardless of the multiplicity of infections (MOIs) of 1, 10, and 100] resulted in more than 5-log10 reduction in Salmonella concentration within 4 or 5 h. Such high lytic activity combined with its remarkably broad and unusual host range and good thermal stability suggested that ΦEnt is a novel Salmonella phage with great potential to be used as an effective biocontrol agent against diverse Salmonella serovars in foods.
Introduction
Salmonella is one of the leading causes of foodborne illnesses worldwide. The disease caused by Salmonella is known as salmonellosis, which is a type of gastroenteritis characterized by acute onset of fever, abdominal pain, diarrhea, nausea, and vomiting (WHO, 2021). Some people may develop reactive arthritis which causes joint pain, eye irritation, and painful urination. It was estimated that each year Salmonella causes 93.8 million cases of salmonellosis and 155,000 deaths globally (Majowicz et al., 2010). In the United States, Salmonella causes about 1.35 million illnesses, 26,500 hospitalizations, 420 deaths, and $365 million in direct medical costs annually (Scallan et al., 2011; Sjölund-Karlsson et al., 2013; CDC, 2021a). Natural reservoirs of Salmonella are domestic and wild animals, such as turkeys, chickens, cows, pigs, fish, turtles, rodents, dogs, and cats. Salmonella is usually transmitted to humans through contaminated foods. Many Salmonella outbreaks occurred in multiple states of the United States, which were often linked to animal-derived foods such as turkey products (CDC, 2021b). An increasing number of salmonellosis outbreaks were linked to fruits and vegetables, such as cucumbers, melon, sprouts, and salad greens (CDC, 2016, 2021b,2021c). The most recent multistate outbreak of salmonellosis was linked to onions, resulting in 892 illnesses and 183 hospitalizations in 38 states and Puerto Rico (CDC, 2021d).
The genus Salmonella includes two species: S. enterica and S. bongori. S. enterica is the most pathogenic species consisting of six subspecies with more than 2,600 serovars (also referred to as serotypes) (Tindall et al., 2005; Andino and Hanning, 2015). Many S. enterica strains are multidrug resistant and have been frequently isolated from foods in many countries (Angelika et al., 2005; Zhao et al., 2017; Zhang et al., 2018; Nikiema et al., 2021). The rapid emergence of multidrug-resistant Salmonella strains has increased global concern about salmonellosis, due to an increase in the mortality rate of infected patients (Gong et al., 2019; Zhan et al., 2019).
Improving food safety is a public health priority. Effective control of Salmonella and other bacterial pathogens is essential for food safety. Many conventional methods, such as pasteurization, chemical treatment, and irradiation, have been used in the food industries to control Salmonella and other bacterial pathogens. However, these methods often indiscriminately kill beneficial bacteria in foods, can adversely alter organoleptic properties and nutritional quality of foods, and are not always environmentally friendly. This has led to many attempts to develop novel alternative methods to control Salmonella and other foodborne bacterial pathogens in foods. Recently, bacteriophages (phages) have gained renewed interest as antimicrobial agents to reduce or eliminate specific foodborne pathogens in food products (Gordillo Altamirano and Barr, 2019; Principi et al., 2019; Picozzi et al., 2021). Phages are viruses that specifically infect and kill the target bacteria without affecting other bacteria and other organisms, such as humans (Principi et al., 2019). Phages are safe for human consumption if they are in foods and safe for use to treat bacterial infections. To date, no reports have shown that phages have any significant side effects or risks of toxicity on mammalian cells (El-Shibiny and El-Sahhar, 2017; Picozzi et al., 2021). In addition, phages do not alter the color, taste, texture, and nutritional quality of foods. Moreover, phages are natural biocontrol agents and environmentally friendly. Furthermore, the process of isolation and selection of new phages is less expensive than the development process required for antibiotics (Golkar et al., 2014; Romero-Calle et al., 2019). All these attractive features make phages novel and promising antibacterial agents. Many studies showed that lytic phages can cause rapid bacterial lysis in various foods, and they can be used as safe, natural, and effective antibacterial agents to control foodborne bacterial pathogens such as Salmonella (Bao et al., 2015; Kawacka et al., 2020; Vikram et al., 2020; Esmael et al., 2021; Steffan et al., 2021). Significant progress in phage research for food safety has been made toward phage applications in foods. A number of phages and their derivatives have been approved by the US Food and Drug Administration (FDA) as natural food additives to control specific foodborne pathogens. Salmonella phage products, such as SalmoFresh (Intralytix, Inc., Baltimore, United States), have been used in meat products to control Salmonella. Due to the vast variation in Salmonella serovars, a wide variety and a large number of highly virulent Salmonella phages with a broad host range are in great need. The availability of a variety of Salmonella phages can also be useful to help combat the possible phage resistance. In this work, we isolated and characterized Salmonella phages obtained from turkeys, and evaluated the potential of the phage with the broadest and very unusual host range as an effective biocontrol agent against multiple Salmonella serovars in cucumber juice (CJ) and beef broth (BB) as model food systems.
Materials and Methods
Bacterial Strains and Culture Conditions
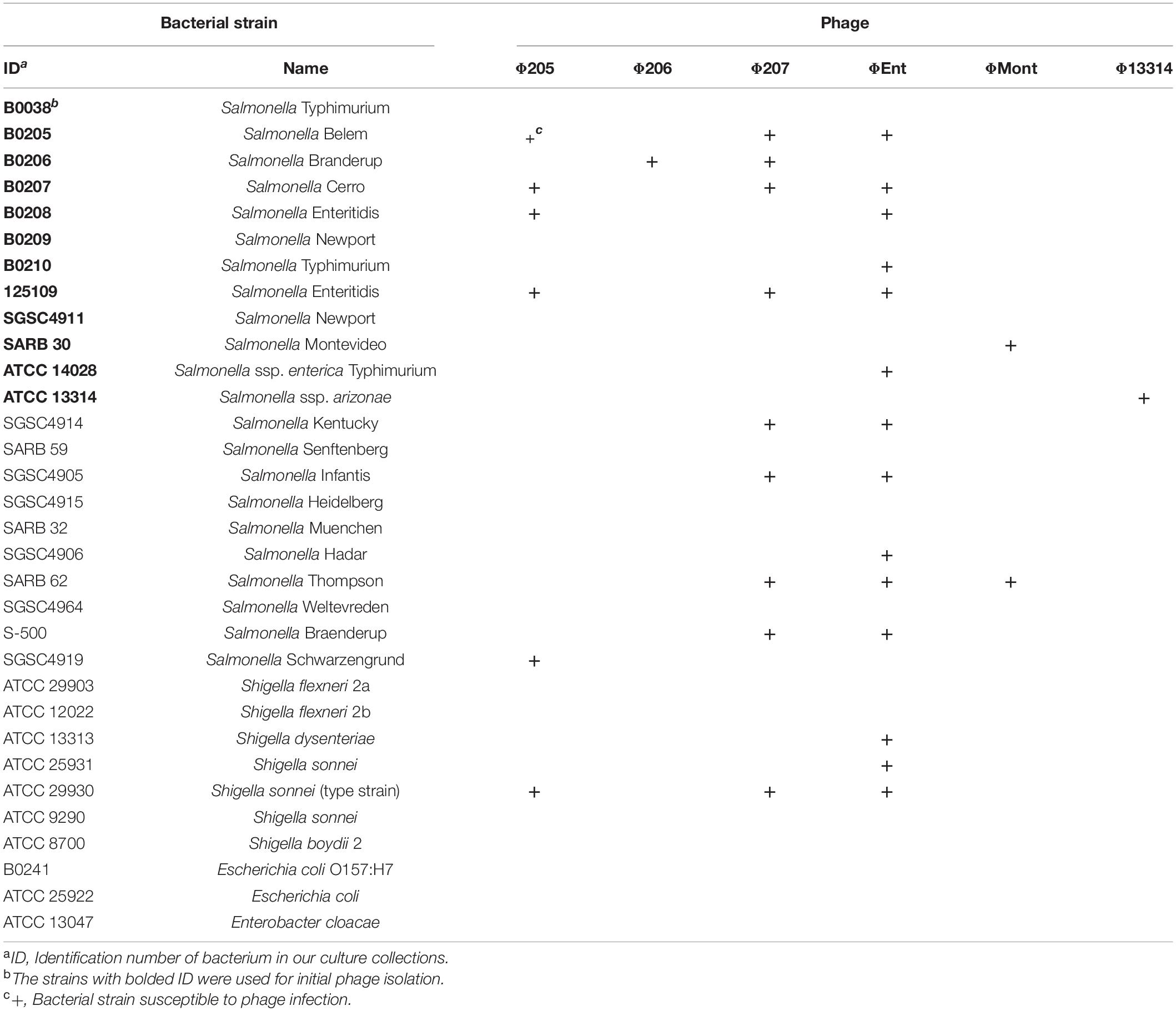
A total of 22 Salmonella enterica strains from various serovars and 10 other Gram-negative bacterial strains were used in this study (Tables 1, 2). Most of these strains were from our culture collections, whereas a few strains were purchased from the American Type Culture Collection (ATCC, Gaithersburg, MD, United States). All bacterial strains were maintained at –80°C in tryptic soy broth (TSB) supplemented with 20% glycerol (v/v). Working stocks were maintained on tryptic soy agar (TSA) at 4°C for 1 month. Fresh overnight cultures were prepared by inoculating 10 ml of TSB with an isolated colony from a TSA plate and incubated at 37°C statically for 12 h. Bacterial concentration was measured by the plate count method using TSA plates.
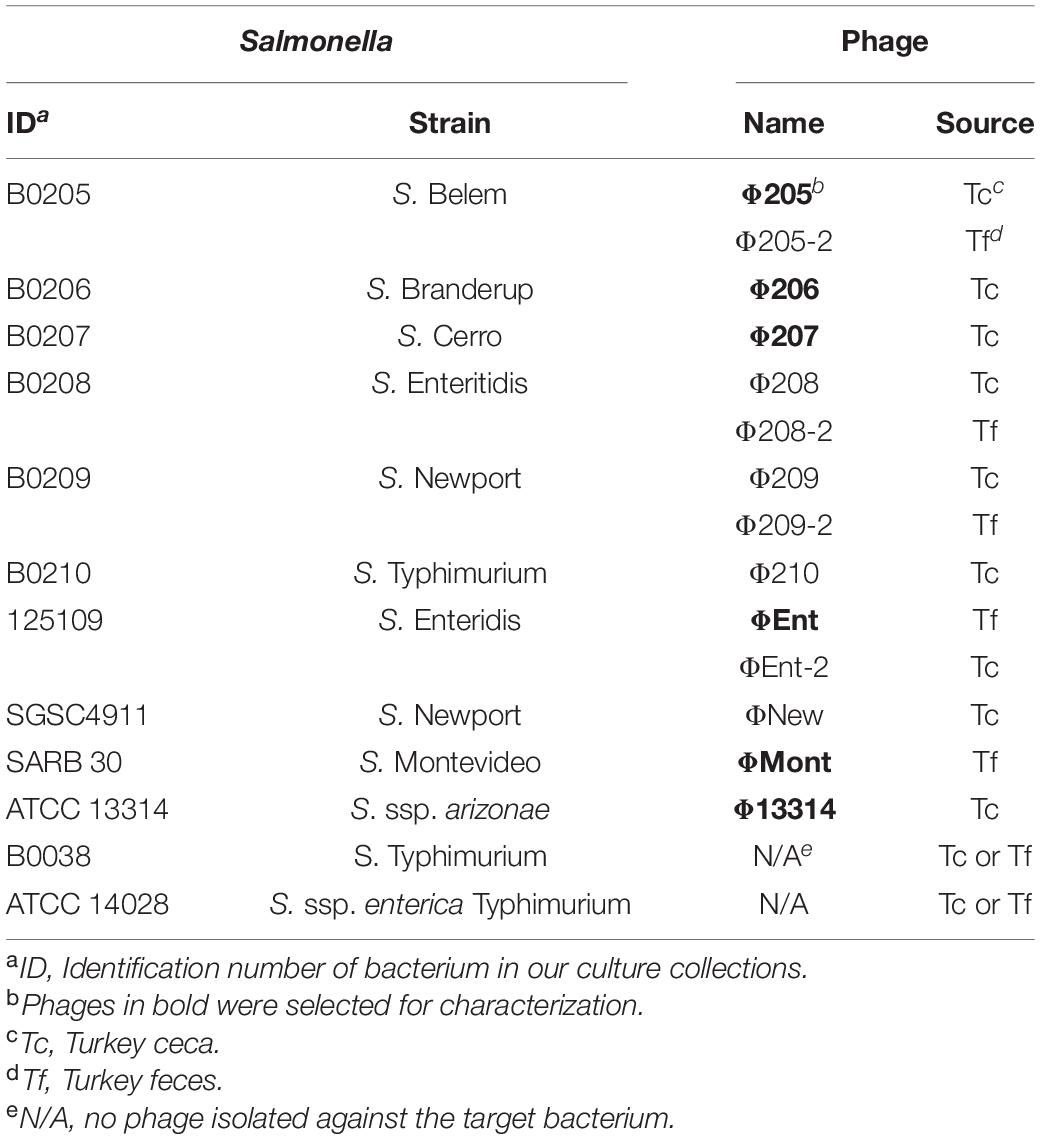
Table 1. Salmonella enterica serovars and strains used for phage isolation and phages isolated in this study.
Phage Propagation and Plaque Assay
Phages were propagated on their hosts at 37°C in TSB or on TSA unless otherwise specified. Phage titer was measured by double agar overlay plaque assay (Adams, 1959; Kropinski et al., 2009). Soft TSA (as top agar) was prepared with TSB supplemented with 0.5% agar. For plaque assay, 100 μl of overnight bacterial culture and 100 μl of phage sample were added into 3 ml of molten soft TSA maintained at 50°C. The resulting mixture was then poured onto a TSA plate. All plates were incubated overnight at 37°C before the examination.
Model Food Systems
Cucumber juice and beef broth were used as model food systems to represent vegetable and meat products in this study. To prepare CJ, fresh cucumbers (size 3A, 44–51 mm in diameter) were washed, cut into pieces, and blended at maximum speed for 1 min (Waring Co., Torrington, CT, United States). The resulting cucumber slurry was frozen overnight. After thawing, the slurry was centrifuged at 10,000 × g and 4°C for 30 min. The supernatant was filtered using a bottle-top filter with a pore size of 0.45 μm. The beef broth was purchased from a local grocery store and centrifuged at 10,000 × g and 4°C for 1 h. The resulting supernatant was filtered using a bottle-top filter (0.45 μm pore size). Both filtered CJ and BB were aliquoted and stored at –80°C until use.
Sample Collection and Processing
Salmonella phages were isolated from turkey ceca and feces. Whole ceca from 72 Hybrid tom turkeys were collected by the veterinarians of a turkey production company and were shipped overnight on ice to our lab at North Carolina State University (NCSU), Raleigh, NC, United States. Fecal materials from 10 turkeys were collected from a poultry field lab at NCSU. The cecal and fecal samples of the turkey were placed immediately in an ice box and transported to the laboratory, where they were temporarily stored in a refrigerator until processed. The contents from 72 ceca were collected and pooled together. The fecal materials from 10 turkeys were also pooled together. The pooled cecal content and pooled fecal material were separately diluted 10-fold with TSB and then centrifuged at 10,000 × g for 1 h. The resulting supernatants were filtered through Nalgene filtration units with 0.45 μm pore size. The two filtrates (one from ceca and one from feces) were stored at 4°C until used as potential phage sources for phage isolation.
Phage Enrichment and Detection
Twelve Salmonella strains (Table 1) were used as potential target hosts for phage isolation. The phages were enriched by inoculation of 10 ml of a filtrate with 500 μl of an overnight Salmonella culture (approximately 1 × 109 CFU/mL) and incubation at 37°C for 6 h. After incubation, 1 ml of each potentially enriched phage lysate was centrifuged at 16,110 × g (13,200 rpm, 5415D Benchtop Microcentrifuge, Eppendorf, United States) for 1 min. The supernatant was filtered through a syringe filter (0.45 μm pore size). The resulting filtrates were then screened for the presence of phage activity by the spot test method (Chopin et al., 1976). Briefly, 10 μl of a filtrate was spotted onto a double agar overlay plate with the top layer seeded with 100 μl of a fresh Salmonella culture. The spot test plates were left to dry on the bench for 20 min and then incubated overnight at 37°C. The plates were examined for phage activity indicated by the presence of a clear zone or plaques.
Phage Isolation, Purification, and Concentration
The plates containing a clear zone or plaques from spot tests were used for initial phage isolation. Briefly, the lysis zone or plaques from a spot test plate were picked with a truncated sterile pipette tip and transferred into a tube containing 1 ml of TSB and 100 μl of an overnight target Salmonella culture (the same strain used for enrichment). The tube was incubated at 37°C for about 6 h. The phage lysate was centrifuged at 16,110 × g for 1 min to precipitate bacterial cells. The supernatant was used for plaque assay. The plaque assay plates were incubated at 37°C for 6–12 h. A well-isolated plaque was picked for another round of plaque purification. Each isolated phage underwent multiple (3–5) rounds of plaque purification.
High-titer phage lysate was prepared through the propagation of purified phage on its host in TSB at 37°C for 12 h. The phage lysate was then centrifuged at 5,000 × g for 30 min. The supernatant was filtered through a bottle-top filter (0.45 μm pore size). The filtered high-titer phage stock (typically ca. in the range from 108 to 1010 PFU/ml) was stored at 4°C until use. Frozen phage stock was prepared with TSB supplemented with 20% glycerol (v/v) and stored at –80°C. A portion of high-titer phage stock was further purified and concentrated by using the methods described by Lu et al. (2003). Briefly, the filtered phage stock was treated with DNase I and RNase A, and then concentrated by polyethylene glycol precipitation. The phage pellet was resuspended in a small amount of Tris-EDTA (TE) buffer (pH 7.6). The resuspended phage was layered on a CsCl step density gradient (1.4, 1.5, and 1.7 g/ml) and ultracentrifuged at 600,000 × g for 6 h at 4°C. The phage bands were extracted and desalted through dialysis against 10 mM Tris buffer. The ultracentrifuge-purified high-titer (∼1010–1012 PFU/mL) phages were used for electron microscopy analysis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and phage DNA extraction.
Host Range
The host range of each selected phage was determined against a panel of 22 Salmonella strains and 10 other Gram-negative bacterial strains (Table 2) using spot tests as described previously. In these tests, a high-titer phage lysate (∼1010–1012 PFU/ml) was used. The clearing zone or plaques in the spotted area indicated that the phage infected the tested strain. Each test was done in duplicate. A positive spot test was confirmed by plaque assay.
Electron Microscopy
Each ultracentrifuge-purified high-titer phage was negatively stained with 2% (w/v) aqueous uranyl acetate (pH 4) on carbon-coated grids and examined with a transmission electron microscope (JEM 1200EX TEM, JEOL) at an accelerating voltage of 80 kV. Electron micrographs were taken at a magnification of × 50,000 in the Electron Microscopy Center, NCSU.
Phage Structural Proteins
The structural proteins of phages were analyzed using the method previously described by Lu et al. (2003) with some modifications. Briefly, an ultracentrifuge-purified phage was mixed with SDS-PAGE sample buffer and then heated in a boiling water bath for 10 min. The boiled sample was allowed to cool to room temperature before loading onto a NuPAGE precast gradient minigel (4–12% Bis-Tris, Invitrogen Corporation, Carlsbad, CA, United States). Electrophoresis was carried out at 75 V for 2 h. The gel was stained with SimplyBlue SafeStain (Invitrogen). Novex® Sharp Pre-stained Protein Standard (Invitrogen) was used to estimate the molecular weights of the separated phage proteins.
Phage DNA Extraction and Restriction Analysis
Genomic DNAs of the selected phages were extracted from highly concentrated phage lysates using the phenol-chloroform extraction method as described by Lu et al. (2003) and digested with restriction endonucleases (EcoRI and HindIII; New England BioLabs, Beverly, MA, United States) according to the manufacturer’s recommendations. The resulting DNA fragments were separated on the 1% agarose gel containing 0.001% SYBR Safe DNA gel stain (Invitrogen) by gel electrophoresis in Tris-borate-EDTA buffer at 70 V (constant voltage) for 2 h. The 1 kb DNA ladder (Promega, Madison, WI, United States) was used to estimate the size of the DNA fragments.
One-Step Growth Kinetics
The one-step growth curve of a selected phage was measured at the initial multiplicity of infection (MOI) of 0.01 in CJ. The infection tube containing phage (approximately 1 × 105 PFU/ml), host culture (1 × 107 PFU/ml), and CJ was incubated in a water bath at 37°C for 20 min (to allow for phage adsorption) and then centrifuged at 16,110 × g for 1 min. The supernatant (which contained un-adsorbed phage particles) was discarded. The remaining pellet was washed once with CJ and then re-suspended in CJ. The suspension was incubated in a water bath at 37°C. Samples were taken at 10-min intervals over a period of 100 min. Each sample was immediately diluted and subjected to plaque assay. All assays were carried out in duplicate. The experiment was independently repeated three times. The latent period was defined as the time interval between the end of the adsorption and the beginning of the first burst, as indicated by the initial rise in phage titer (Ellis and Delbruck, 1939; Adams, 1959). Burst size was calculated as the ratio of the final titer of liberated phage particles to the initial concentration of infected bacterial cells during the latent period (Adams, 1959).
Thermal Stabilities
The thermal stabilities of phage ΦEnt and host Salmonella Thompson were independently evaluated in saline (0.85% NaCl) and TSB at 37, 50, 63, and 72°C. The initial phage titer and host concentrations were approximately 106 PFU/ml and 106 CFU/ml, respectively. A set of 1.5-ml tubes containing 900 μl of saline or TSB were pre-heated in a water bath to reach the test temperature. Then 100 μl of phage or host solution was added to a pre-heated tube. After briefly vortexed, the tube was immediately placed back into the water bath and heated for the indicated time periods. A sample (100 μl) was taken from this tube and immediately transferred into a tube containing 900 μl of saline at room temperature. The original tube was discarded. A new preheated tube was used for another treatment at the same or different temperatures following the same procedure. The titer of the surviving phages and the concentration of the surviving cells were measured by plaque assay and plate count method, respectively. Each sample was plated in duplicate. All experiments were repeated at least three times.
Efficacy of Phage Infection Against Salmonella in Model Food Systems
The lytic activity of phage ΦEnt infection to control S. Thompson was evaluated at three different MOIs in CJ and BB as model food systems. The frozen CJ was thawed at 4°C and then equilibrated to room temperature before use. An overnight host culture was diluted with CJ to a concentration of ca. 105 CFU/ml. A 9.9 ml aliquot of the CJ-diluted bacterial culture was added to each of the four 15-ml tubes. To one of the three tubes, 100 μl of phage stock at a specific titer was added to achieve an initial MOI of 1, 10, or 100. To the fourth tube, 100 μl of saline was added. This tube without phage served as a control. After mixing briefly, the four tubes were incubated statically in a water bath at 37°C. Hourly samples were taken from each tube for a 5-h period. Each sample was then serially diluted and plated onto a TSA plate in duplicate. After incubation at 37°C overnight, each plate was examined to obtain a plate count for the calculation of cell concentration. The same procedure was followed when using BB as a model food system. Each experiment was independently conducted three times.
Statistical Analysis
All experiments were carried out in triplicate, and each sample was plated in duplicate. One-way analysis of variance (ANOVA) was performed using Statistica for Windows (StatSoft, Tulsa, OK, United States). Tukey’s Honest Significant Difference test was used to compare the mean values of data for significant difference (P ≤ 0.05).
Results and Discussion
Phage Isolation
A total of 14 Salmonella phages were isolated using 12 common Salmonella enterica serovars and/or subspecies as potential target hosts for phage enrichment and isolation (Table 1). Nine phages were isolated from turkey ceca (Tc), while the remaining five were isolated from turkey feces (Tf). Ten of the potential target hosts were susceptible to at least one isolated phage. No phage was isolated against two S. Typhimurium strains (B0038 and ATCC 14028). Six phages (Φ205, Φ206, Φ207, ΦEnt, ΦMont, and Φ13314) produced clear and relatively large plaques (1.3–3 mm in diameter), while the other eight phages produced clear but very small (<1 mm in diameter) or turbid plaques (data not shown). Turbid plaque on a host lawn resulted from incomplete lysis, which may indicate potential lysogeny as the induction of a prophage inside bacterial strains could lead to turbid plaques, which is often a characteristic of temperate phages (Campoy et al., 2006; Gallet et al., 2011; Thanki et al., 2019). Turbid clearing may also indicate partial resistance of the bacterial host to the phage, thus only a sub-population of cells was lysed (Bull et al., 2014). Phages that produce turbid plaques, therefore, may not be good candidates for use as biocontrol agents (Chan et al., 2013; Abedon et al., 2017). Thus, only the six phages that produce clear plaques with a diameter > 1 mm were selected for characterization.
Host Range
The host ranges of six selected phages were determined using spot tests against a panel of 22 Salmonella strains comprising diverse serovars that were commonly associated with the foodborne Salmonellosis and 10 other Gram-negative bacteria including seven Shigella strains, two Escherichia coli strains, and one Enterobacter cloacae strain (Table 2). The six phages displayed different host ranges, indicating that they are distinct from one another. Host range is largely dependent on the ability of phages to bind to either a single receptor (i.e., narrow host range) or multiple different receptors (i.e., broad host range; Fong et al., 2021). Φ206, ΦMont, and Φ13314 showed very narrow host ranges, infecting only one or two tested Salmonella strains. In contrast, Φ205, Φ207, and ΦEnt exhibited broader host ranges. Φ205 and Φ207 infect five and eight tested Salmonella strains, respectively. In addition, they both crossed the boundary of the genus and infected Shigella sonnei type strain (Table 2). ΦEnt displayed the broadest and very unusual host range, infecting 11 of the 22 (50%) tested Salmonella strains from nine different serovars (Belem, Cerro, Enteritidis, Typhimurium, Kentucky, Infantis, Hadar, Thompson, and Braenderup). Moreover, ΦEnt also broke the boundary of the genus and lysed three Shigella strains from two species (S. dysenteriae and S. sonnei). It is well-known that S. Enteritidis, Typhimurium, Kentucky, Infantis, Hadar, and Thompson belong to the most common and highly pathogenic Salmonella serovars (Anonymous, 2021), while Shigella dysenteriae and S. sonnei are important human pathogens causing bacterial dysentery and shigellosis (CDC, 2022). It is noted that ΦEnt and Φ207 (both siphophages) share seven Salmonella hosts from different serovars and one Shigella host (Table 2), which is not very common. The similarity in host ranges may indicate that these phages recognize similar receptors in the hosts (Kalatzis et al., 2016). Phages are generally species-specific or genus-specific (Hyman and Abedon, 2010; Cazares et al., 2021). Broad-host range Salmonella phages have been reported in the literature (Santos et al., 2011; Li, M. et al., 2020; Tao et al., 2021), but phages infecting bacteria from different genera are comparatively rare. It has been reported that phages SFP10 and KIT03 broke the boundary between Salmonella and Escherichia and infected both Salmonella enterica and E. coli O157:H7 (Park et al., 2012; Pham-Khanh et al., 2019). A few studies have reported the phages that can cross the genus boundary between Salmonella and Shigella. Hamdi et al. (2017) reported that phage SH7 was able to lyse strains of Salmonella paratyphi and Shigella dysenteriae (Hamdi et al., 2017). It is of interest to explore the genetic basis for ΦEnt and Φ207 to share similar host ranges and their ability to infect bacteria from two genera. Host range is critical in the selection of phage candidates for biocontrol agents and detection applications. To be considered as excellent candidates for use as biocontrol agents in foods, phages should be strictly lytic and possibly have a broad host range (Sharma et al., 2017; Fong et al., 2021; Picozzi et al., 2021). Since ΦEnt exhibited the broadest and unusual host range, formed clear plaques (possibly with high lytic activity), and had higher efficiency of plating (EOP) (data not shown), it was selected for further characterization of (with respect to) its growth kinetics, restriction pattern, and lytic profiles in model food systems and for determining its thermal stability.
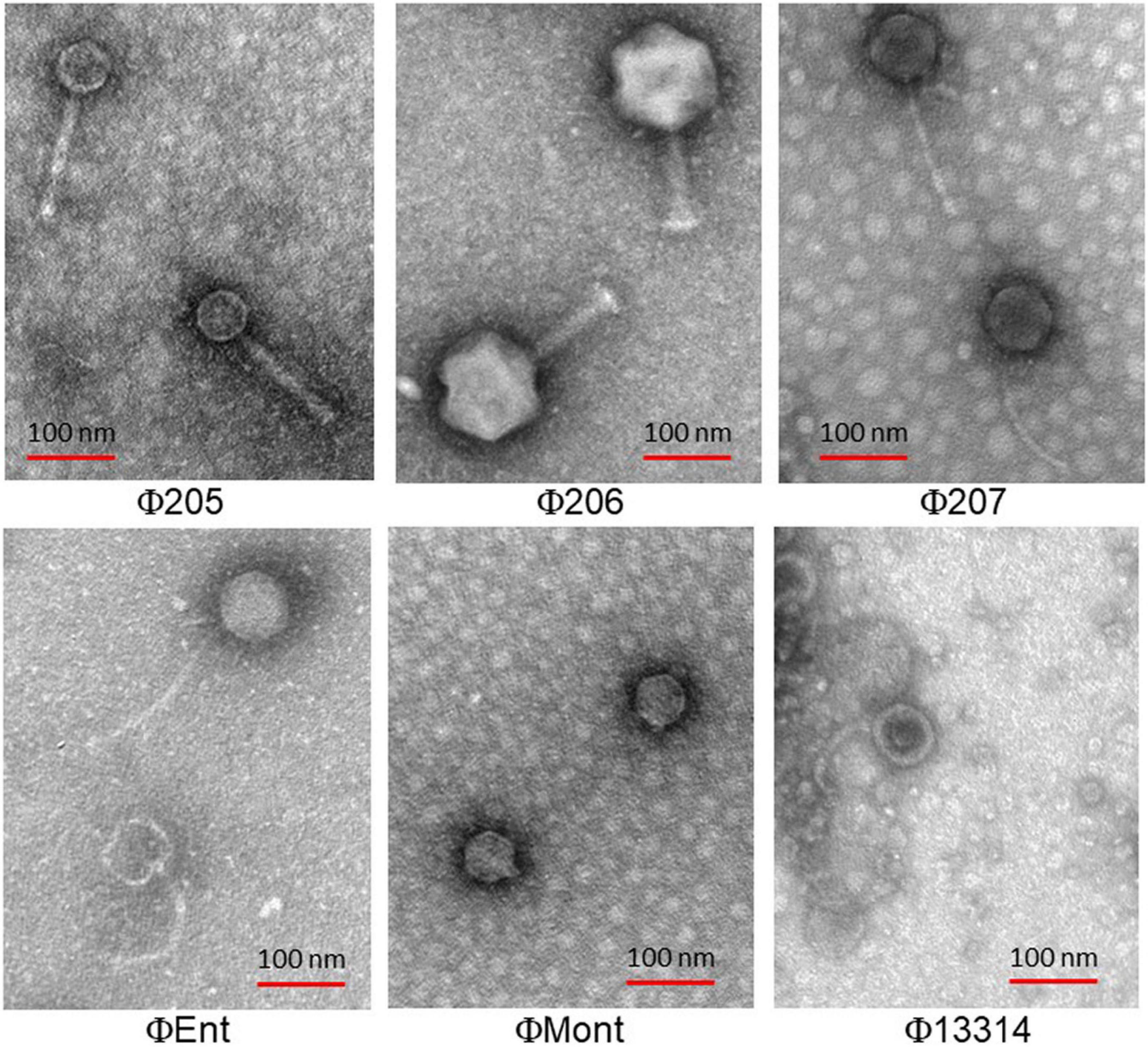
Electron Microscopy
The electron micrographs show that the six selected phages are all tailed phages with icosahedral heads and thus belong to the order Caudovirales (Figure 1). Three phages (Φ207, ΦEnt, and Φ13314) have long flexible, non-contractile tails and thus belong to the Siphoviridae family. Two phages (Φ205 and Φ206) have contractile tails and belong to the Myoviridae family. One phage (ΦMont) has a very short tail and belongs to the Podoviridae family.
Structural Protein Profiles
The SDS-PAGE analysis showed the structural protein profiles of the six Salmonella phages (Figure 2). Each phage showed at least one thick band representing the major structural proteins in the range of 30–60 kDa. However, each phage has a unique structural protein banding pattern, indicating that these phages are distinct from one another. Among the six phages, Φ13314 (a siphoviridae phage) showed the most bands (at least 12 bands). Φ207 and ΦEnt are also siphoviridae phages, but they showed much fewer bands than Φ13314. These two broadest host-range phages appeared to share two structural protein bands: one thick band (major structural protein) and a band between 50 and 60 kDa. Genetic analysis of the two phage genomes is needed to see if these two proteins are involved in phage attachment to the hosts, which might explain why these two phages shared seven Salmonella hosts. Φ205 and Φ206 (Myoviridae phages) showed 5–6 bands, while ΦMont (a Podoviridae phage) showed a few very faint bands besides a thick band. Lu et al. (2020) previously isolated eight phages and their Gram-negative bacterial hosts from a commercial cucumber fermentation. They demonstrated that the phages from the same phage family exhibited similar structural protein banding patterns. However, that was not the case with the six phages in this study.
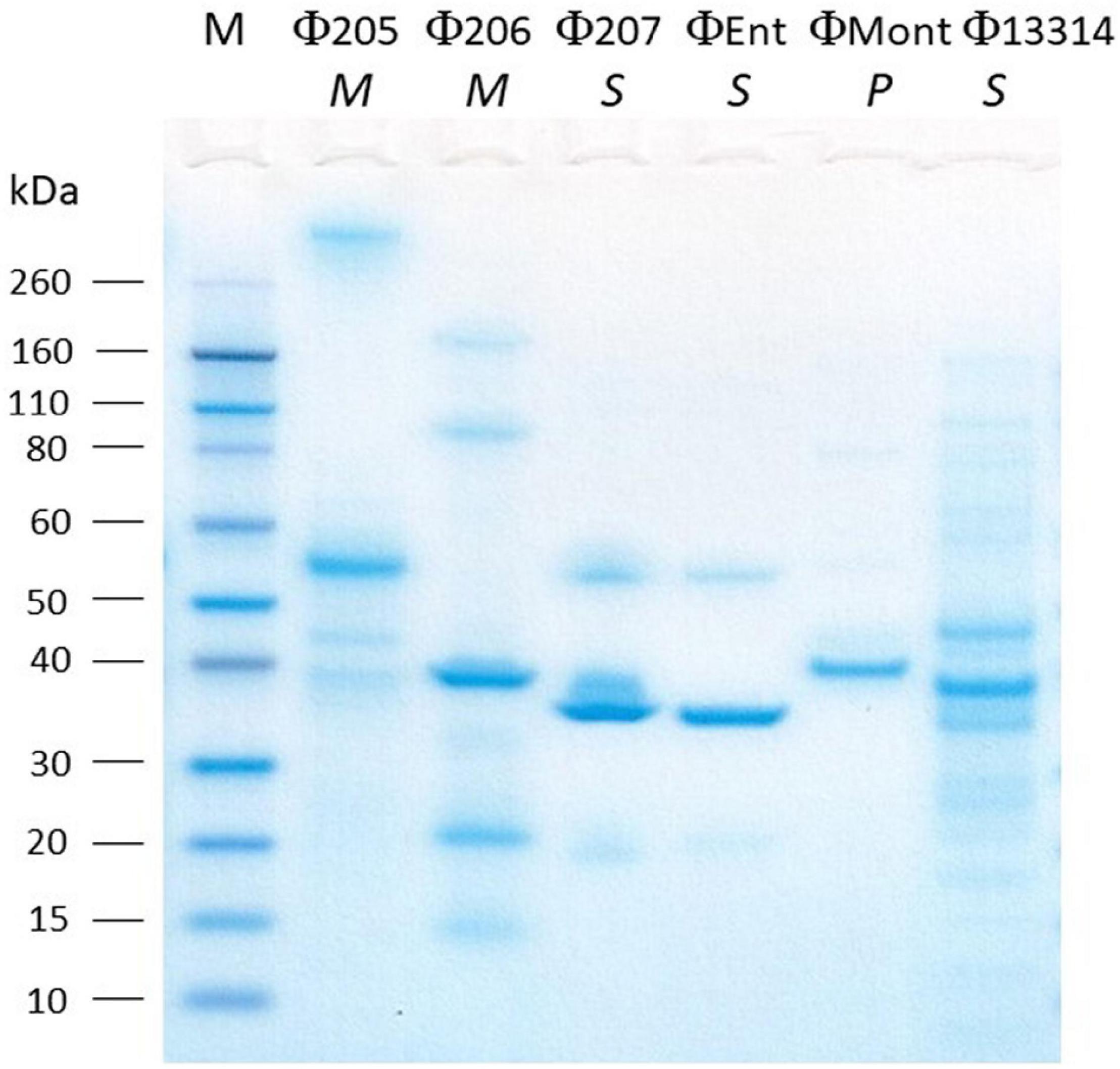
Figure 2. SDS-PAGE analysis of the structural proteins of six Salmonella phages isolated from turkeys. Lane M, molecular mass standard. The italicized letters (M, S, and P) below phage names stand for Myoviridae, Siphoviridae, and Podoviridae, respectively.
DNA Restriction Analysis of Phages Φ207 and ΦEnt Sharing Eight Hosts
The two broadest host-range phages, Φ207, and ΦEnt (both siphophages), share seven different Salmonella enterica serovars and one Shigella sonnei strain (Table 2), which is rare. It is of interest to see if their genomes may share some common features. The two phage genomes were digested with two restriction endonucleases, EcoRI and HindIII. Figure 3 showed that both phage genomes can be digested by EcoRI and HindIII and generate multiple bands on the resolved gel, confirming that the phage genomes are dsDNA. Φ207 and ΦEnt showed different restriction banding patterns from EcoRI digestion or from HindIII digestion, confirming the unique identities of these phages. Nevertheless, they shared three restriction bands (two bands with molecular weights above 10,000 bp and one band with molecular weight of approximately 3,200 bp) from EcoRI digestion, suggesting the conservation of some EcoRI-specific cutting sites. Although siphoviruses are known to exhibit remarkable genomic mosaicism (Santander et al., 2017), it is not surprising that phages from the same family share some components and thus exhibit some notable degree of similarity among siphophages (Fong et al., 2017). This may partially explain why Φ207 and ΦEnt (both siphophages) showed similar, but not identical, broad host ranges. Santander et al. (2017) reported that Salmonella phages fSE1C and fSE4C showed very similar banding patterns from EcoRI or HindIII digestion. They found that the two phage genomes shared 43% sequence similarity, with genes involved in structure, replication, host specificity, and DNA metabolism indicating remarkable conservation. Complete genome analyses of Φ207 and ΦEnt are needed to reveal the genetic basis for the two phages sharing seven Salmonella hosts and one Shigella host and for their relevance for practical application.
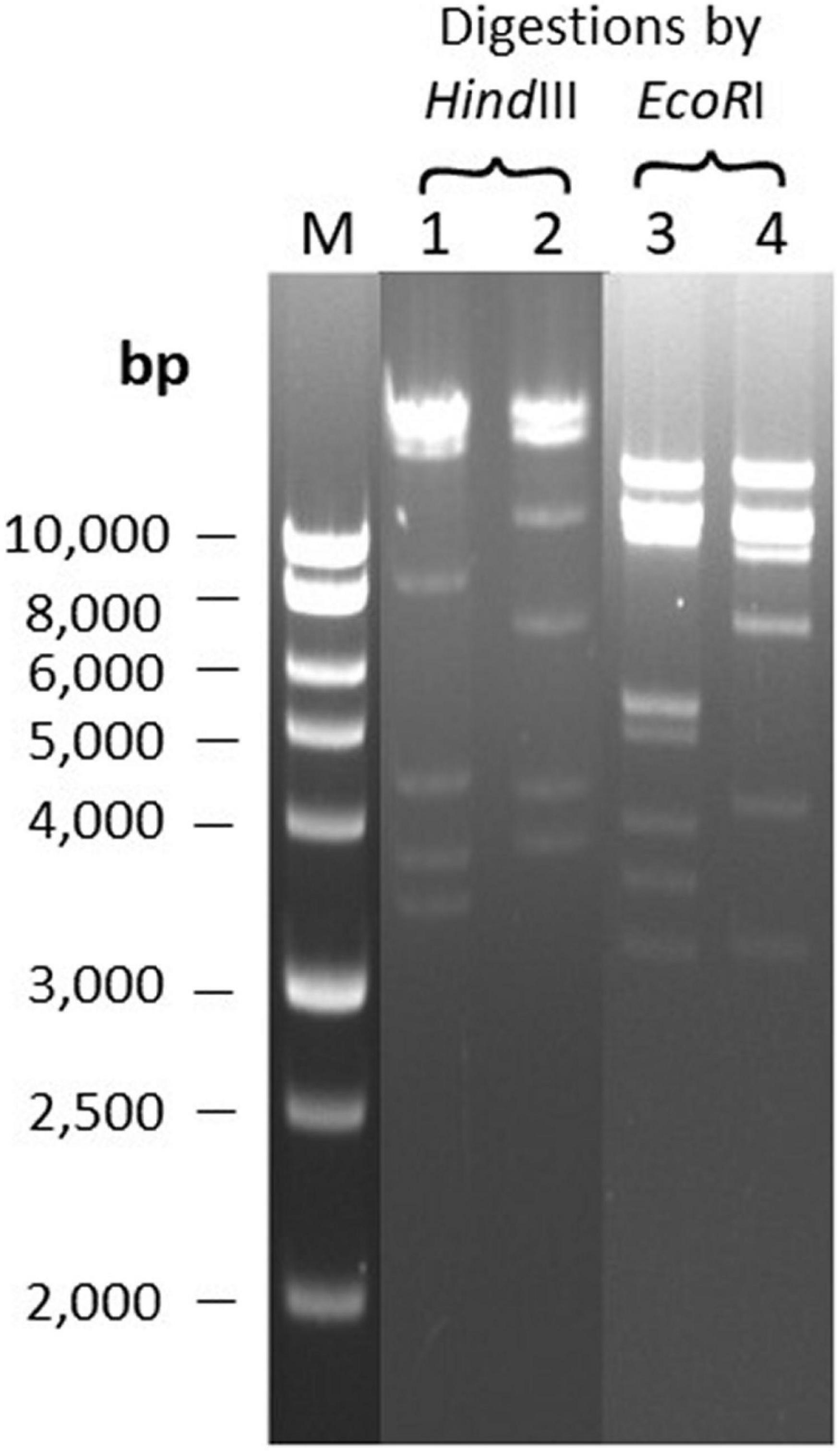
Figure 3. Restriction banding patterns of phage DNAs upon digestion by endonuclease EcoRI and HindIII. Lane M: 1 kb ladder; Lanes 1 and 3: digestion of Φ207 DNA; Lanes 2 and 4: digestion of ΦEnt DNA.
One-Step Growth Kinetics of Phage ΦEnt
One-step growth curve of phage ΦEnt was measured in CJ (as a model food system) at MOI of 0.01 against S. Thompson instead of S. Enteritidis (the target host used for phage isolation) because ΦEnt formed larger clear plaques on the lawn of S. Thompson, suggesting that ΦEnt has a higher efficacy against S. Thompson than the primary strain of isolation. Figure 4 shows that ΦEnt has a short latent period of 10 min, which is shorter (25–65 min) than that of many other reported Salmonella phages (Huang et al., 2018a; El-Dougdoug et al., 2019; Duc et al., 2020). A shorter latent period has been positively related to the lytic activity of the phage (Li, Z. et al., 2020). ΦEnt also exhibited a relatively large burst size (∼100 PFU/infected cell). For many Siphoviridae and Myoviridae phages, the typical burst size ranged es, the typical burst size raes, the typical burst size ra 50 and 100 PFU/infected cell (Park et al., 2012; Litt and Jaroni, 2017; Khalatbari-Limaki et al., 2020). A phage with both a short latent period (15 min or less) and a large burst size (> 50 PFU/cell) may have a selective advantage over competing phages, resulting in very high lytic activity (Park et al., 2012). Thus, such a phage is desirable for use in the food industry (Kalatzis et al., 2016).
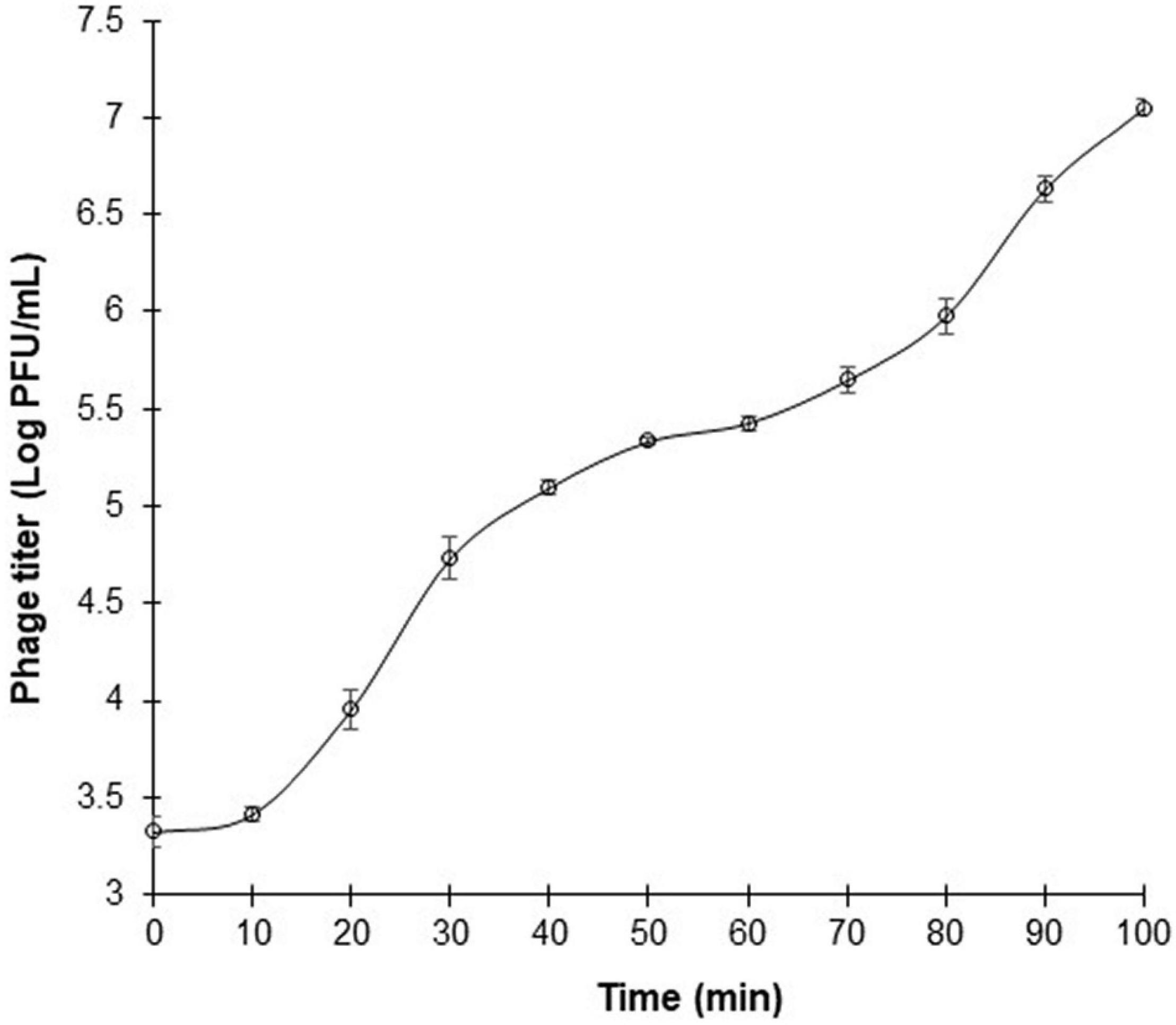
Figure 4. One-step growth curve of phage ΦEnt infecting Salmonella Thompson at MOI 0.01 in cucumber juice at 37°C. The latent period is approximately 10 min, and the burst size is about 100 phage particles per infected cell. Error bars indicate the standard error of the mean.
Thermal Stabilities of Phage ΦEnt and Host Salmonella Thompson
The stability of phages against adverse effects during food processing is important for phage application. Heat treatments, such as pasteurization, are often applied in the food industry. Therefore, it is necessary to evaluate the thermal stabilities of phages for their application under practical conditions. The thermal stabilities of phage ΦEnt and host S. Thompson were evaluated in saline (0.85% NaCl) at 37, 50, 63, and 72°C. It was found that both ΦEnt and the host were stable at 37°C (data not shown), but at higher temperatures, ΦEnt and the host exhibited different trends in thermal stability. During the first 30 s at 50, 63, and 72°C, phage titer declined rapidly, resulting in 1. 5-, 2. 5-, and 3-log10 reduction, respectively (Figure 5A). But after 30 s at these temperatures, the decline in ΦEnt titer was much slower. Exposure to 50, 63, and 72°C for 4 min resulted in 2-, 3. 3-, and 5-log10 reduction in ΦEnt titers, respectively. Significant numbers of the phage (104, 103, and 10 PFU/ml) survived the 4-min heating at 50, 63, and 72°C, respectively. In contrast, the host was stable at 50°C during the 4-min heating (Figure 5B). At 63°C, host concentration initially decreased slowly during the first 30 s, but rapidly thereafter. It only took 2 min at 63°C or 30 s at 72°C to completely inactivate all the host cells (6-log10 reduction). These results demonstrated that the host in saline was much more sensitive to 63 or 72°C than the phage. The thermal stabilities of ΦEnt and the host were further evaluated in the TSB medium. It was found that the phage titer and cell concentration remained unchanged at 37°C (data not shown) and 50°C (Figures 5C,D), indicating that both the phage and its host were very stable in TSB at these temperatures. As the temperature increased, their stabilities decreased, but at different rates. Exposure to 63 and 72°C for 4 min in TSB resulted in 2.6-log10 and 3.3-log10 unit reductions in ΦEnt titer, respectively. These reductions were much smaller than those (3.3-log10 at 63°C and 5-log10 at 72°C) observed in saline, demonstrating that ΦEnt had significantly (p < 0.05) better thermal stability in TSB than in saline. It should be noted that the exposure to 72°C for 15 s (the typical pasteurization condition used in food industries) caused less than 0.6-log10 unit reduction in phage titer (Figure 5C), indicating that ΦEnt retained the good stability required for use as biocontrol agent under commonly encountered pasteurization conditions. The host also showed better stability at 63 and 72°C in TSB (Figure 5D) than in saline (Figure 5B). However, compared with ΦEnt in TSB (Figure 5C), the host was significantly (p < 0.05) less stable at 63 and 72°C (Figure 5D). Exposure to 63°C for 4 min caused a 3.6-log10 reduction in host concentration in TSB (1-log10 more reduction compared to that for phage). At 72°C, the host was completely eliminated within 3 min, while many phage particles (5 × 102 PFU/ml) survived the 4-min heating.
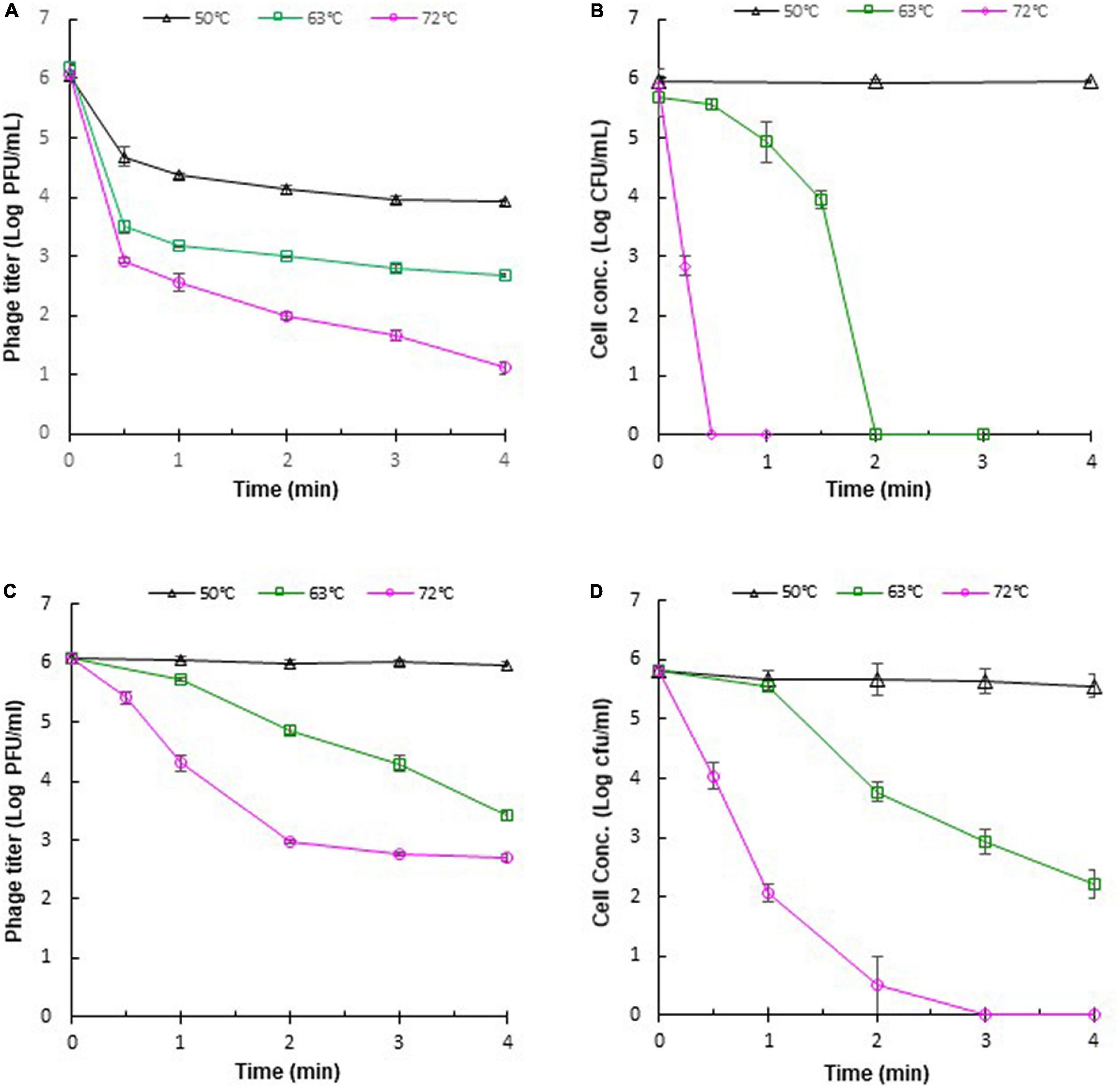
Figure 5. Thermal stabilities of Salmonella phage ΦEnt and its host Salmonella Thompson in saline (A,B, respectively) and in TSB (C,D, respectively). Error bars indicate the standard error of the mean.
The differences in the thermal stabilities of the phage or the host in TSB and saline could be attributed to the components other than salt (such as peptides and proteins) in TSB, which provided thermal protection to ΦEnt and the host. Several other studies also noted that the medium components of phage solution could affect the thermal stability of phages (Fabrizio et al., 1999; Sua’/rez and Reinheimer, 2002; Quiberoni et al., 2003; Briggiler Marco et al., 2009). It was reported that reconstituted non-fat dry skim milk is more protective than De Man, Rogosa and Sharpe (MRS) broth against thermal inactivation of some dairy phages (Daoust et al., 1965; Quiberoni et al., 2003). Whole milk provides better thermal protection than skim milk for Lactococcus lactis phages (Fabrizio et al., 1999). Tris-magnesium gelatin buffer was more effective in protecting L. lactis phages against heat exposures than M17 broth (Sua’/rez and Reinheimer, 2002). It can be expected that different foods provide different levels of thermal protection to phages or other organisms. The thermal stability of phages may depend on their DNA content (Yamagishi and Ozeki, 1972). It was reported that Listeria phage A511 with a denser head capsid content showed higher thermal stability than Listeria phage P100 (Ahmadi et al., 2017). Heating could cause irreversible damage to phage particles, such as irreversible unfolding and dissociation of structural proteins, damage to the portal complex, disintegration of phage into head and tail, and leakage of DNA (Ahmadi et al., 2017). Shang et al. (2021) visualized the heat-treated phages with a transmission electron microscope. The detrimental effects of heat on phage structure were evidenced by phage tail aggregation, detachment of phage head and tail, and empty capsid.
Efficacy of Phage ΦEnt Infection Against Salmonella in Model Food Systems
Many foodborne outbreaks of salmonellosis were linked to animal-based foods and vegetables. Thus, BB (simulating meats) and CJ (simulating vegetables) were used as model food systems in this study to evaluate the efficacy of phage ΦEnt infection against S. Thompson at three different MOIs (1, 10, and 100). Figure 6A shows changes in the host concentration in the control and infection tubes containing CJ during 6 h. During the first hour of incubation, the host concentrations in all tubes (control and infection tubes) were the same (with a slight decrease due to the medium change from TSB to CJ). But during the following 2 h (hours 2 and 3), the host concentration in the control tube and the infection tubes with initial an MOI of 1 or 10 increased rapidly at the same rate, while the host concentration in the infection tube with initial MOI of 100 remained unchanged. During the fourth hour of incubation, the host concentration in the control tube continued to increase rapidly and reached 2.4 × 107 CFU/mL. In contrast, the host concentration in all the three infection tubes dramatically decreased, resulting in ≥ 4-log10 unit reduction in host concentration (causing host concentration < 102 CFU/mL, Figure 5). At the end of the fifth hour, the final host concentrations in the control tube increased to 5.3 × 107 CFU/mL, while the final host concentration in all three infection tubes (regardless of MOI) was ≤ 102 CFU/mL. Such a large difference (more than 5-log10 units) in final host concentration between the control and the infection tubes indicated that ΦEnt infection is highly effective in inhibiting and even eliminating the Salmonella host in CJ. The results also indicated that given enough time (4 h) the phage infection with an initial MOI of 1 could achieve the same log reduction in host concentration as those with higher initial MOIs (10 and 100). Huang et al. (2018b) evaluated the efficacy of the phage infection by a Salmonella phage LPSE1 against S. Enteritidis ATCC 13076 on lettuce leaves and found that 5-h phage infection at an initial MOI of 1, 10, or 100 resulted in only 2-log10 reduction. Several other studies reported that Salmonella phage infection resulted in a 1.7-log10 reduction in host concentration in lettuce (Spricigo et al., 2013), 3-log10 reduction in Chinese cabbage (Bao et al., 2015), 1.37-log10 reduction in mustard, and 0.55-log10 reduction on broccoli (Pao et al., 2006). None of these studies achieved the same level (≥ 4-log10) of the reduction as achieved in CJ in our study. This is probably partially due to the fact that the solid food matrices limit mass transport/diffusion and reduce phage–host interaction (Whichard et al., 2003; Hudson et al., 2010). Figure 6A reveals that the increase in cell concentration occurred in all the three infection tubes at 5 h of incubation, indicating that phage resistance mutants emerged. Bacteria can become resistant to phages through different mechanisms, such as spontaneous mutations in bacterial cell surface receptors, restriction modification systems, and adaptive immunity via the CRISPR-Cas system (Labrie et al., 2010; Mangalea and Duerkop, 2020). It was observed that some surviving host cells formed much smaller and/or irregularly shaped colonies after 3 h of infection (data not shown), indicating a cytopathic effect from phage infection.
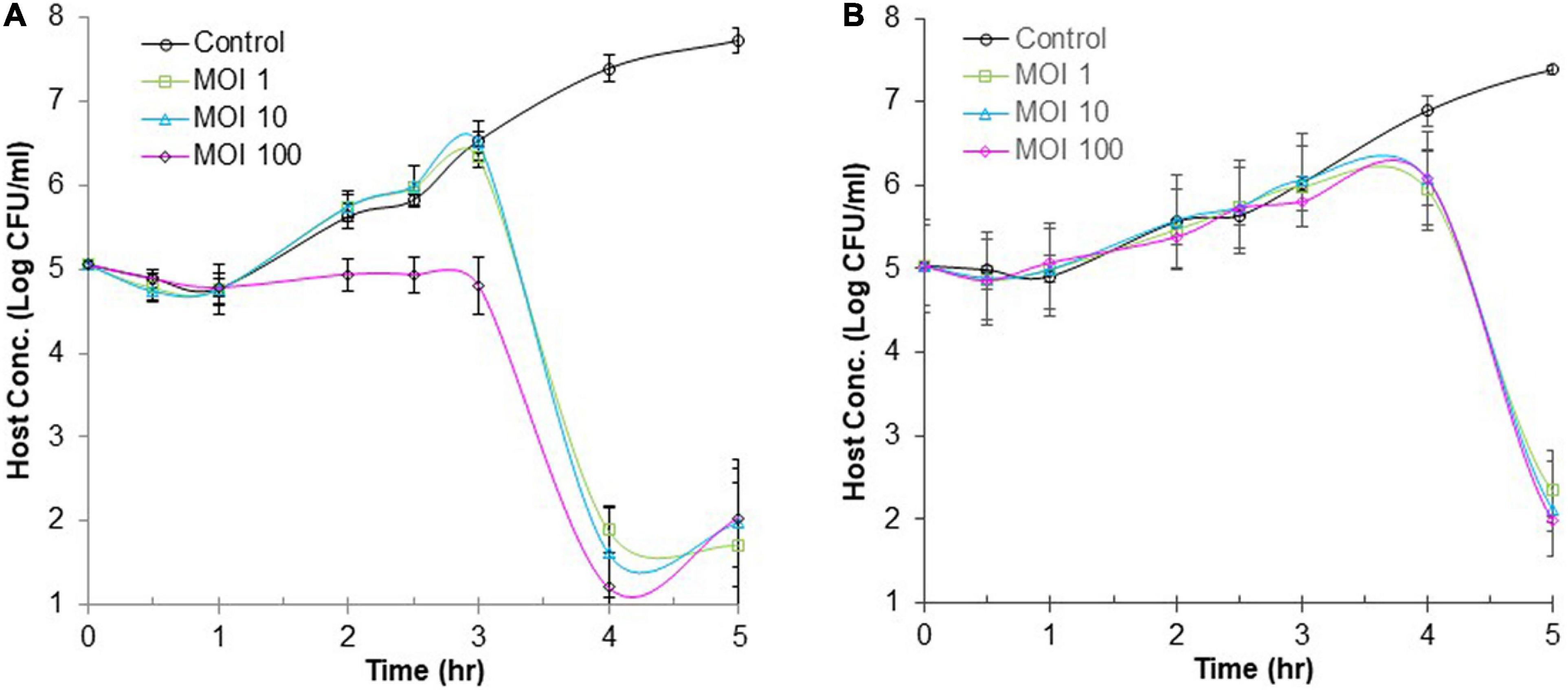
Figure 6. Efficacy of phage infection by ΦEnt against S. Thompson in cucumber juice (A) and beef broth (B). Each point represents the mean ± standard error based on three measurements.
Figure 6B shows changes in the host concentration in the control and infection tubes containing BB during 6 h. The host concentration in all tubes (regardless of MOI used) increased approximately by 1-log unit during the first 3.5 h of incubation. Thereafter, the host concentration in the control continued to increase and reached 3.7 × 107 CFU/mL at the end of the sixth hour. In contrast, the host concentration in the infection tubes (regardless of MOI) decreased rapidly after 3.5 h of treatment. At 6 h, the cell concentration in infection tubes was either below or slightly above 102 CFU/ml, indicating a 6-log10 reduction in host concentration compared to the controls. These results demonstrated that the 6-h ΦEnt infection at all MOIs tested is highly effective to suppress and even eliminate the host in BB. Huang et al. (2018b) assessed the anti-Salmonella efficacy of phage LPSE1 in pork sausage at 28°C and found that 6-h phage infection at MOIs of 1 and 100 only resulted in 2-log10 and 2.6-log10 reduction in host concentration, respectively, which were much lower than those achieved by ΦEnt in BB in our study. Such a big difference may be partially explained by the fact that BB as a liquid matrix provides better homogeneity and an opportunity for a phage to come in contact with its host. It is expected that higher MOI and longer incubation time may be needed to achieve the same log reduction in cell concentration if using solid food matrices. Shang et al. (2021) evaluated the ability of phage vB_SalP_TR2 to reduce Salmonella Albany contamination in pasteurized milk and chicken meat stored at 37°C for 6 h. They found that the phage was much more effective against S. Albany in pasteurized milk than in chicken meat (Shang et al., 2021).
The high efficacy of ΦEnt against S. Thompson observed in both model food systems is largely due to its short latent period (10 min) and large burst size (100 PFU/cell). The evaluation of the efficacies of ΦEnt against multiple other susceptible Salmonella serovars is underway. It is of great interest in evaluating the efficacy of ΦEnt against the three susceptible Shigella strains to further explore its biocontrol potential.
Conclusion
In conclusion, this study isolated and characterized the phages from turkeys against various Salmonella serovars that were commonly associated with foodborne Salmonellosis. Six selected phages were from all three families in the Caudovirales order. Each of them had a unique host range and a structural protein profile. The two broadest host range phages shared seven different Salmonella serovars and one Shigella sonnei strain. ΦEnt showed a remarkable and very unusual broad host range (infecting 11 Salmonella strains in nine serovars and three Shigella strains in two species, dysenteriae and sonnei), high lytic activity at all MOIs tested against Salmonella in two model food systems, and good thermal stability under pasteurization conditions. These desirable characteristics suggested that ΦEnt is a novel phage with great potential to be used as an effective biocontrol agent to control Salmonella in foods. Further studies are needed to assess the efficacy of ΦEnt against three susceptible Shigella strains to further explore its biocontrol potential. It would be also needed to evaluate ΦEnt and other isolated phages against multiple Salmonella serovars in various food matrices. Additionally, comparative genomic analysis of these phages, particularly the two broad host range phages, needs to be conducted to explore the genetic basis for phage–host interactions.
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
ZL contributed to the conceptualization, funding acquisition, experimental design, phage isolation and purification, all other experiments, data analysis, draft manuscript, and manuscript revision. JM, ST, HM, DI, SKo, RD, HT, RP, and AR contributed to the experiments. FB contributed to electron microscopy analysis and manuscript revision. SKa contributed to sample resources and manuscript revision. All authors contributed to the article and approved the submitted version.
Funding
This work was partially funded by the Department of Molecular and Cellular Biology and the College of Science and Mathematics at Kennesaw State University, United States.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jeffrey Niedermeyer for help with sample collection for this study.
References
Abedon, S. T., García, P., Mullany, P., and Aminov, R. (2017). Editorial: phage therapy: past, present and future. Front. Micro. 8:981. doi: 10.3389/fmicb.2017.00981
Ahmadi, H., Radford, D., Kropinski, A. M., Lim, L.-T., and Balamurugan, S. (2017). Thermal-stability and reconstitution ability of listeria phages P100 and A511. Front. Microbiol. 8:2375. doi: 10.3389/fmicb.2017.02375
Andino, A., and Hanning, I. (2015). Salmonella enterica.: survival, colonization, and virulence differences among serovars. Sci. World J. 2015:520179. doi: 10.1155/2015/520179
Angelika, M., Karin, P., Andreas, S., and Reiner, H. (2005). Molecular mechanisms of resistance in multidrug-resistant serovars of Salmonella enterica isolated from foods in Germany. J. Antimicrob Chemother. 56, 1025–1033. doi: 10.1093/jac/dki365
Anonymous (2021). Bacteriophage Products. SalmoFresh™. Available online at: http://www.intralytix.com/index.php?page=prod&id=3 (accessed December 1, 2021)
Bao, H., Zhang, P., Zhang, H., Zhou, Y., Zhang, L., and Wang, R. (2015). Bio-control of Salmonella enteritidis in foods using bacteriophages. Viruses 7, 4836–4853. doi: 10.3390/v7082847
Briggiler Marco, M., De Antoni, G. L., Reinheimer, J. A., and Quiberoni, A. (2009). Thermal, chemical, and photocatalytic inactivation of Lactobacillus plantarum bacteriophages. J. Food Prot. 72, 1012–1019. doi: 10.4315/0362-028x-72.5.1012
Bull, J. J., Vegge, C. S., Schmerer, M., Chaudhry, W. N., and Levin, B. R. (2014). Phenotypic resistance and the dynamics of bacterial escape from phage control. PLoS One 9:e94690. doi: 10.1371/journal.pone.0094690
Campoy, S., Hervàs, A., Busquets, N., Erill, I., Teixidó, L., and Barbé, J. (2006). Induction of the SOS response by bacteriophage lytic development in Salmonella Enterica. Virology 351, 360–367. doi: 10.1016/j.virol.2006.04.001
Cazares, D., Cazares, A., Figueroa, W., Guarneros, G., Edwards, R. A., and Vinuesa, P. (2021). A novel group of promiscuous podophages infecting diverse gammaproteobacteria from river communities exhibits dynamic intergenus host adaptation. mSystems 6:e00773-20. doi: 10.1128/mSystems.00773-20
CDC (2016). Multistate Outbreak Of Salmonella Poona infections Linked To Imported Cucumbers (Final Update).
CDC (2021a). Salmonella. Available online at: https://www.cdc.gov/salmonella/index.html (accessed December 1, 2021)
CDC (2021b). Reports of Selected Salmonella Outbreak Investigations. Available online at: https://www.cdc.gov/salmonella/outbreaks.html (accessed December 1, 2021)
CDC (2021c). Salmonella Outbreak Linked to BrightFarms Packaged Salad Greens. Available online at: https://www.cdc.gov/salmonella/typhimurium-07-21/index.html (accessed October 10, 2021)
CDC (2021d). Salmonella Outbreak Linked To Onions. Available online at: https://www.cdc.gov/salmonella/oranienburg-09-21/index.html (accessed December 1, 2021)
CDC (2022). Shigella – Shigellosis. Available online at: https://www.cdc.gov/shigella/index.html (accessed May 24, 2022)
Chan, B. K., Abedon, S. T., and Loc-carrillo, C. (2013). Phage cocktails and the future of phage therapy. Future Med. 8, 769–783. doi: 10.2217/fmb.13.47
Chopin, M. C., Chopin, A., and Roux, C. (1976). Definition of bacteriophage groups according to their lytic action on mesophilic lactic streptococci. Appl. Environ. Microbiol. 32, 741–746. doi: 10.1128/aem.32.6.741-746.1976
Daoust, D. R., El-Bisi, H. M., and Litsky, W. (1965). Thermal destruction kinetics of a lactic streptococcal bacteriophage. Appl. Microbiol. 13, 478–485. doi: 10.1128/am.13.3.478-485.1965
Duc, H. M., Son, H. M., Yi, H. P. S., Sato, J., Ngan, P. H., Masuda, Y., et al. (2020). Isolation, characterization and application of a polyvalent phage capable of controlling Salmonella and Escherichia coli O157:H7 in different food matrices. Food Res. Int. 131:108977. doi: 10.1016/j.foodres.2020.108977
El-Dougdoug, N. K., Cucic, S., Abdelhamid, A. G., Brovko, L., Kropinski, A. M., Griffiths, M. W., et al. (2019). Control of Salmonella newport on cherry tomato using a cocktail of lytic bacteriophages. Int. J. Food Microbiol. 293, 60–71. doi: 10.1016/j.ijfoodmicro.2019.01.003
El-Shibiny, A., and El-Sahhar, S. (2017). Bacteriophages: the possible solution to treat infections caused by pathogenic bacteria. Can. J. Microbiol. 63, 865–879. doi: 10.1139/cjm-2017-0030
Esmael, A., Azab, E., Gobouri, A., Nasr-Eldin, M., Moustafa, M., Mohamed, S., et al. (2021). Isolation and characterization of two lytic bacteriophages infecting a multi-drug resistant Salmonella typhimurium and their efficacy to combat salmonellosis in ready-to-use foods. Microorganisms 9:423. doi: 10.3390/microorganisms9020423
Fabrizio, S. V., Ledford, R. A., and Parada, J. L. (1999). Heat resistance of Lactococcus lactis bacteriophages. Microbiol. Aliments Nutr. 17, 191–198.
Fong, K., LaBossiere, B., Switt, A. I. M., Delaquis, P., Goodridge, L., Levesque, R. C., et al. (2017). Characterization of four novel bacteriophages isolated from British columbia for control of non-typhoidal Salmonella in vitro and on sprouting alfalfa seeds. Front. Microbiol. 8:2193. doi: 10.3389/fmicb.2017.02193
Fong, K., Wong, C. W. Y., Wang, S., and Delaquis, P. (2021). How broad is enough: the host range of bacteriophages and its impact on the agri-food sector. Phage 2, 83–91. doi: 10.1089/phage.2020.0036
Gallet, R., Kannoly, S., and Wang, I. N. (2011). Effects of bacteriophage traits on plaque formation. BMC Micro. 11:181. doi: 10.1186/1471-2180-11-181
Golkar, Z., Bagasra, O., and Pace, D. G. (2014). Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 8, 129–136. doi: 10.3855/jidc.3573
Gong, J., Zeng, X., Zhang, P., Zhang, D., Wang, C., and Lin, J. (2019). Characterization of the emerging multidrug-resistant Salmonella enterica serovar Indiana strains in China. Emerg. Microbes Infect. 8, 29–39. doi: 10.1080/22221751.2018.1558961
Gordillo Altamirano, F. L., and Barr, J. J. (2019). Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 32:e66-18. doi: 10.1128/CMR.00066-18
Hamdi, S., Rousseau, G. M., Labrie, S. J., Tremblay, D. M., Kourda, R. S., Karim, B. S., et al. (2017). Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci. Rep. 7:40349. doi: 10.1038/srep40349
Huang, C., Shi, J., Ma, W., Li, Z., Wang, J., Li, J., et al. (2018a). Isolation, characterization, and application of a novel specific Salmonella bacteriophage in different food matrices. Food Res. Int. 111, 631–641. doi: 10.1016/j.foodres.2018.05.071
Huang, C., Virk, S. M., Shi, J., Zhou, Y., Willias, S. P., Morsy, M. K., et al. (2018b). Isolation, characterization, and application of bacteriophage LPSE1 against Salmonella enterica in ready to eat (RTE) foods. Front. Microbiol. 9:1046. doi: 10.3389/fmicb.2018.01046
Hudson, J. A., Mcintyre, L., and Billington, C. (2010). “Application of bacteriophages to control pathogenic and spoilage bacteria in food processing and distribution,” in Bacteriophages in the Control of Food- and Waterborne Pathogens, eds P. M. Sabour and M. W. Griffiths (Washington, DC: ASM Press), 119–135. doi: 10.1128/9781555816629.ch7
Hyman, P., and Abedon, S. T. (2010). Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70, 217–248. doi: 10.1016/S0065-2164(10)70007-1
Kalatzis, P. G., Bastias, R., Kokkari, C., and Katharios, P. (2016). Isolation and characterization of two lytic bacteriophages, (St2 and (Grn1: phage therapy application for biological control of vibrio alginolyticus in aquaculture live feeds. PLoS One 11:e0151101. doi: 10.1371/journal.pone.0151101
Kawacka, I., Olejnik-Schmidt, A., Schmidt, M., and Sip, A. (2020). Effectiveness of phage-based inhibition of listeria monocytogenes in food products and food processing environments. Microorganisms 8:1764. doi: 10.3390/microorganisms8111764
Khalatbari-Limaki, S., Hosseinzadeh, S., Shekarforoush, S. S., and Berizi, E. (2020). The morphological and biological characteristics of a virulent PI phage isolated from slaughterhouse sewage in Shiraz, Iran. Iran J. Microbiol. 12, 616–624. doi: 10.18502/ijm.v12i6.5037
Kropinski, A. M., Mazzocco, A., Waddell, T. E., Lingohr, E., and Johnson, R. P. (2009). “Enumeration of bacteriophages by double agar overlay plaque assay,” in Bacteriophages. Methods in Molecular Biology, Vol. 501, eds M. R. Clokie and A. M. Kropinski (Humana Press), doi: 10.1007/978-1-60327-164-6_7
Labrie, S., Samson, J., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Li, M., Lin, H., Jing, Y., and Wang, J. (2020). Broad-host-range Salmonella bacteriophage STP4-a and its potential application evaluation in poultry industry. Poultry Sci. 99, 3643–3654. doi: 10.1016/j.psj.2020.03.051
Li, Z., Ma, W., Li, W., Ding, Y., Zhang, Y., Yang, Q., et al. (2020). A broad-spectrum phage controls multidrug-resistant Salmonella in liquid eggs. Food Res. Int. 132:109011. doi: 10.1016/j.foodres.2020.109011
Litt, P. K., and Jaroni, D. (2017). Isolation and physiomorphological characterization of Escherichia coli O157:H7-infecting bacteriophages recovered from beef cattle operations. Int. J. Microbiol. 2017:7013236. doi: 10.1155/2017/7013236
Lu, Z., Breidt, F., Fleming, H. P., Altermann, E., and Klaenhammer, T. R. (2003). Isolation and characterization of a Lactobacillus plantarum bacteriophage ΦJL-1 from a cucumber fermentation. Int. J. Food Microbiol. 84, 225–235. doi: 10.1016/S0168-1605(03)00111-9
Lu, Z., Pérez-Díaz, I. M., Hayes, J. S., and Breidt, F. (2020). Bacteriophages infecting gram-negative bacteria in a commercial cucumber fermentation. Front. Microbiol. 11:1306. doi: 10.3389/fmicb.2020.01306
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Mangalea, M. R., and Duerkop, B. A. (2020). Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immun. 88:e926-19. doi: 10.1128/IAI.00926-19
Nikiema, M. E. A., Pardos de la Gandara, M., Compaore, K. A. M., Soro, K. D., and Nikiema, P. A. (2021). Contamination of street food with multidrug-resistant Salmonella, in ouagadougou, burkina faso. PLoS One 16:e0253312. doi: 10.1371/journal.pone.0253312
Pao, S., Rolph, S. P., Westbrook, E. W., and Shen, H. (2006). Use of bacteriophages to control Salmonella in experimentally contaminated sprout seeds. J. Food Sci. 69, M127–M130. doi: 10.1111/j.1365-2621.2004.tb10720.x
Park, M., Lee, J. H., Shin, H., Kim, M., Choi, J., Kang, D. H., et al. (2012). Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl. Environ. Microbiol. 78, 58–69. doi: 10.1128/AEM.06231-11
Pham-Khanh, N. H., Sunahara, H., Yamadeya, H., Sakai, M., Nakayama, T., Yamamoto, H., et al. (2019). Isolation, characterisation and complete genome sequence of a tequatrovirus phage, Escherichia phage KIT03, which simultaneously infects Escherichia coli O157:H7 and Salmonella enterica. Curr. Microbiol. 76, 1130–1137. doi: 10.1007/s00284-019-01738-0
Picozzi, C., Garcia, P., and Vives, M. (2021). Editorial: bacteriophages to fight food-borne pathogens/phages struggling for food safety. Front. Microbiol. 12:741387. doi: 10.3389/fmicb.2021.741387
Principi, N., Silvestri, E., and Esposito, S. (2019). Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front. Pharmacol. 10:513. doi: 10.3389/fphar.2019.00513
Quiberoni, A., Guglielmotti, D. M., and Reinheimer, J. A. (2003). Inactivation of Lactobacillus delbrueckii bacteriophages by heat and biocides. Int. J. Food Microbiol. 84, 51–62. doi: 10.1016/s0168-1605(02)00394-x
Romero-Calle, D., Guimarães Benevides, R., Góes-Neto, A., and Billington, C. (2019). Bacteriophages as alternatives to antibiotics in clinical Care. Antibiotics 8:138. doi: 10.3390/antibiotics8030138
Santander, J., Vasquez, J. I., Segovia, C., Santos, L., Turra, G., Huber, K., et al. (2017). Complete genome sequence of the Salmonella enterica serovar enteritidis bacteriophages fSE1C and fSE4C isolated from food matrices. Stand. Genome Sci. 12:1. doi: 10.1186/s40793-016-0218-y
Santos, S. B., Kropinski, A. M., Ceyssens, P. J., Ackermann, H. W., Villegas, A., Lavigne, R., et al. (2011). Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: creation of a new phage genus. J. Virol. 85, 11265–11273. doi: 10.1128/JVI.01769-10
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the united states—major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.p11101
Shang, Y., Sun, Q., Chen, H., Wu, Q., Chen, M., Yang, S., et al. (2021). Isolation and characterization of a novel Salmonella phage vB_SalP_TR2. Front. Microbiol. 12:664810. doi: 10.3389/fmicb.2021.664810
Sharma, S., Chatterjee, S., Datta, S., Prasad, R., Dubey, D., Prasad, R. K., et al. (2017). Bacteriophages and its applications: an overview. Folia Microbiol. 62, 17–55. doi: 10.1007/s12223-016-0471-x
Sjölund-Karlsson, M., Howie, R. L., Blickenstaff, K., Boerlin, P., and Ball, T. (2013). Occurrence of β-lactamase genes among non-typhi Salmonella enterica isolated from humans, food animals, and retail meats in the United States and Canada. Microb. Drug Resist. 19, 191–197. doi: 10.1089/mdr.2012.0178
Spricigo, D. A., Bardina, C., Cortés, P., and Llagostera, M. (2013). Use of a bacteriophage cocktail to control Salmonella in food and the food industry. Int. J. Food Microbiol. 165, 169–174. doi: 10.1016/j.ijfoodmicro.2013.05.009
Steffan, S. M., Shakeri, G., Hammerl, J. A., Kehrenberg, C., Peh, E., Rohde, M., et al. (2021). isolation and characterization of group III Campylobacter jejuni–specific bacteriophages from Germany and their suitability for use in food production. Front. Microbiol. 12:761223. doi: 10.3389/fmicb.2021.761223
Sua’/rez, V. B., and Reinheimer, J. A. (2002). Effectiveness of thermal treatments and biocides in the inactivation of argentinian Lactococcus lactis phages. J. Food Prot. 65, 1756–1759. doi: 10.4315/0362-028x-65.11.1756
Tao, C., Yi, Z., Zhang, Y., Wang, Y., Zhu, H., Afayibo, D. J. A., et al. (2021). Characterization of a broad-host-range lytic phage SHWT1 against multidrug-resistant Salmonella and evaluation of its therapeutic efficacy in vitro and in vivo. Front. Vet. Sci. 8:683853. doi: 10.3389/fvets.2021.683853
Thanki, A. M., Brown, N., Millard, A. D., and Clokie, M. R. J. (2019). Genomic characterization of jumbo Salmonella phages that effectively target United Kingdom pig-associated Salmonella serotypes. Front. Microbiol. 10:1491. doi: 10.3389/fmicb.2019.01491
Tindall, B. J., Grimont, P. A., Garrity, G. M., and Euzeby, J. P. (2005). Nomenclature and taxonomy of the genus Salmonella. Int. J. Syst. Evol. Microbiol. 55, 521–524. doi: 10.1099/ijs.0.63580-0
Vikram, A., Tokman, J. I., Woolston, J., and Sulakvelidze, A. (2020). Phage biocontrol improves food safety by significantly reducing the level and prevalence of Escherichia coli O157:H7 in various foods. J. Food Prot. 83, 668–676. doi: 10.4315/0362-028X.JFP-19-433
Whichard, J. M., Sriranganathan, N., and Pierson, F. W. (2003). Suppression of Salmonella growth by wild-type and large-plaque variants of bacteriophage felix O1 in liquid culture and on chicken frankfurters. J. Food Prot. 66, 220–225. doi: 10.4315/0362-028x-66.2.220
WHO (2021). Salmonella (Non-Typhoidal). Available online at: https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) (accessed December 1, 2021)
Yamagishi, H., and Ozeki, H. (1972). Comparative study of thermal inactivation of phage 080 and lambda. Virology 48, 316–322. doi: 10.1016/0042-6822(72)90042-6
Zhan, Z., Xu, X., Gu, Z., Meng, J., Wufuer, X., Wang, M., et al. (2019). Molecular epidemiology and antimicrobial resistance of invasive non-typhoidal Salmonella in China, 2007–2016. Infect. Drug Resist. 12, 2885–2897. doi: 10.2147/IDR.S210961
Zhang, L., Fu, Y., Xiong, Z., Ma, Y., Wei, Y., Qu, X., et al. (2018). Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front. Microbiol. 9:2104. doi: 10.3389/fmicb.2018.02104
Keywords: Salmonella, bacteriophage, phage ΦEnt, biocontrol, food safety
Citation: Lu Z, Marchant J, Thompson S, Melgarejo H, Ignatova D, Kopić S, Damaj R, Trejo H, Paramo R, Reed A, Breidt F and Kathariou S (2022) Bacteriophages Isolated From Turkeys Infecting Diverse Salmonella Serovars. Front. Microbiol. 13:933751. doi: 10.3389/fmicb.2022.933751
Received: 01 May 2022; Accepted: 02 June 2022;
Published: 05 July 2022.
Edited by:
Paula Teixeira, Catholic University of Portugal, PortugalReviewed by:
Kitiya Vongkamjan, Kasetsart University, ThailandDomonkos Sváb, Veterinary Medical Research Institute, Hungary
Copyright © 2022 Lu, Marchant, Thompson, Melgarejo, Ignatova, Kopić, Damaj, Trejo, Paramo, Reed, Breidt and Kathariou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongjing Lu, zlu2@kennesaw.edu
 Zhongjing Lu
Zhongjing Lu John Marchant1
John Marchant1 Samantha Thompson
Samantha Thompson Henry Melgarejo
Henry Melgarejo Sandra Kopić
Sandra Kopić Fred Breidt
Fred Breidt Sophia Kathariou
Sophia Kathariou