- 1Department of Food and Nutrition, Sookmyung Women’s University, Seoul, Republic of Korea
- 2Risk Analysis Research Center, Sookmyung Women’s University, Seoul, Republic of Korea
In this study, genetic variations and characteristics of Listeria monocytogenes isolates from enoki mushrooms (23), smoked ducks (7), and processed ground meat products (30) were examined with respect to hemolysis, virulence genes, growth patterns, and heat resistance. The isolates that showed the highest pathogenicity and the lowest pathogenicity were analyzed to obtain the whole-genome sequence, and the sequences were further analyzed to identify genetic variations in virulence, low-temperature growth-related, and heat resistance-related factors. All isolates had β-hemolysis and virulence genes (actA, hlyA, inlA, inlB, and plcB). At low temperatures, isolates with high growth (L. monocytogenes strains SMFM 201803 SD 1-1, SMFM 201803 SD 4-2, and SMFM 201804 SD 5-3) and low growth (L. monocytogenes strains SMFM 2019-FV43, SMFM 2019-FV42, and SMFM 2020-BT30) were selected. Among them, L. monocytogenes SMFM 201804 SD 5-3 showed the highest resistance at 60°C and 70°C. The strains SMFM 201804 SD 5-3 (high-risk) and SMFM 2019-FV43 (low-risk) harbored 45 virulence genes; 41 single nucleotide variants (SNVs) were identified between these two isolates. A comparison of 26 genes related to low-temperature growth revealed 18 SNVs between these two isolates; a comparison of the 21 genes related to heat resistance revealed 16 SNVs. These results indicate that the differences in the pathogenicity of L. monocytogenes SMFM 201804 SD 5-3 and L. monocytogenes SMFM 2019-FV43 are associated with the SNVs identified in virulence genes, low-temperature growth-related genes, and heat resistance-related genes.
1. Introduction
Listeria monocytogenes contaminates various foods, such as meat, fresh agricultural products, and smoked salmon; thus, foodborne illnesses caused by L. monocytogenes are frequent, throughout the year (Sheng et al., 2018). In Korea, recalls of processed meat products due to L. monocytogenes contamination occur continuously (Lee and Yoon, 2021). L. monocytogenes is a gram-positive facultative anaerobe that causes listeriosis in humans (Chen and Knabel, 2007; Yang and Moon, 2021). The pathogen may also cause severe infectious diseases in pregnant women and the immunocompromised (Shin et al., 2007). The fatality rate of individuals infected with L. monocytogenes is 20–30%; this pathogen may cause infection even when present in low numbers compared to other pathogenic bacteria (Lee, 2010). The minimum infectious dose varies among patients and foods, but it is generally estimated 102–103 CFU/g (Park et al., 2014). L. monocytogenes can grow under anaerobic conditions, and thus, they can grow under vacuum or nitrogen-filled packaging (Duffy et al., 1994). It can grow even at refrigerated temperatures and survive under conditions of high-salt (≥10%), extreme acidity, and carbon-source depletion (Liu et al., 2006). Also, L. monocytogenes is one of the most heat-resistant pathogens and poses a notable risk to food safety, particularly when mild heat is treated in food processing and preparation (Pöntinen et al., 2017). These survival and viability characteristics of L. monocytogenes raise concerns regarding foodborne illnesses caused by the consumption of foods contaminated with this pathogen.
The advancement of next-generation sequencing (NGS) techniques has allowed the cost- and time-effective sequencing of DNA for microbial genome research. NGS techniques are used in various fields, including verification of the functionality of probiotics and investigation of the causes of foodborne outbreaks. They have also been used to establish a database of genomic information for foodborne pathogenic bacteria and to analyze the safety of microorganisms (Kwon et al., 2019; Cao et al., 2021). Whole-genome sequencing (WGS)-based methods have contributed to the identification of outbreak sources in several listeriosis outbreaks (Lüth et al., 2018). According to Hilliard et al. (2018), WGS analysis is a valuable methodology for classifying L. monocytogenes isolates and identifying virulence islands that may influence infectivity. WGS is increasingly used in the United States to facilitate detecting, investigating, and controlling foodborne bacterial outbreaks (Brown et al., 2019). In Ireland, WGS and phenotypic assays were used to explain the virulence of L. monocytogenes isolates (Stratakos et al., 2020). WGS provides an opportunity to determine strain characteristics typically obtained through resource-intensive traditional methodologies such as species identification, serotyping, virulence, and antimicrobial resistance profiling, all of which can be consolidated into a single WGS workflow (Ribot et al., 2019). The objective of this study was to identify phenotypes and genetic variations among L. monocytogenes isolates using WGS.
2. Materials and methods
2.1. Isolation of Listeria monocytogenes
The L. monocytogenes isolates from enoki mushrooms and other isolates (7 isolates from smoked ducks and 30 isolates from processed ground meat products) previously studied by Park et al. (2021) were used in the present study.
A total of 127 enoki mushrooms were collected from supermarkets in Korea between August 2019 and March 2020. The samples were transported in a cooler to our laboratory. Briefly, 25 g of each sample was placed in sample bags (3 M™, St. Paul, MN, USA) containing 225 mL of Listeria Enrichment Broth (Becton Dickinson and Company). The sample bags were hand-shaken for 1 min and incubated at 30°C for 48 h to enrich L. monocytogenes.
One-milliliter aliquots of the enriched cultures were spread-plated on Chrom agar (CHROM™ agar Listeria; Paris, France), and the plates were incubated at 37°C for 24 h. The isolated colonies were subjected to real-time PCR analysis to investigate the presence of the iap gene for the confirmatory test of L. monocytogenes [Food and Drug Administration (FDA), 2018] with a 264-bp DNA fragment, using suitable primers (F: 5′-TGG GAT TGC GGT AAC AGC AT-3′ R: 5′-TA TCA ACA CCA GCG CCA CT-3′) (Kim, 2019). A single colony was suspended in 50 μL of 0.25% sodium dodecyl sulfate (Biosesang, Gyeonggi-do, Korea)–0.05 N NaOH solution (Daejung, Gyeonggi-do, Korea) and 100 μL of dH2O. The suspensions were heated at 99°C for 15 min, left at room temperature (25°C) for 3 min, and centrifuged at 5,989 × g at 4°C for 3 min; the supernatants were used as DNA templates. Real-time PCR amplification was performed using a Rotor-Gene SYBR® Green PCR Kit (Qiagen, Hilden, Germany). Briefly, 1 μL DNA template was mixed with 2.5 μL of forward primer, 2.5 μL of reverse primer, 12.5 μL of SYBR green master mix, and 6.5 μL of dH2O. For iap amplification, an initial denaturation step at 95°C for 90 s and 35 cycles of denaturation at 95°C for 5 s, annealing at 56°C for 10 s, and extension at 72°C for 5 s were conducted. Finally, a 5-min extension was performed at 72°C. Amplified PCR products were loaded onto a 1.5% agarose gel and visualized under UV light.
2.2. Analysis of hemolytic property
Isolated colonies of L. monocytogenes were inoculated in 10 mL tryptic soy broth with 0.6% yeast extract (TSBYE; Becton Dickinson and Company) and incubated at 30°C overnight. Then, 100-μL aliquots of the cultures were transferred to fresh 10 mL TSBYE and incubated at 30°C for another 24 h. The sub-cultures were streaked onto Columbia agar with 5% sheep blood (bioMérieux, Mercy 1′Etoile, France) and incubated at 30°C for 48 h. Hemolytic properties (α-, β-, or γ-hemolysis) were determined by observing the zones formed around the bacterial colonies. β-hemolysis (complete hemolysis) was confirmed if a clear zone around the bacteria was observed.
2.3. Detection of virulence genes
Isolated colonies of L. monocytogenes isolates were suspended in 50 μL of 0.25% sodium dodecyl sulfate–0.05 N NaOH solution. One hundred microliters of sterile dH2O was added to the suspension, and the mixtures were incubated at 99°C for 15 min. Aliquots (2 μL) of the mixtures were mixed with the components of the Phire Hot Start II DNA Polymerase Kit (Thermo Fisher Scientific, Waltham, MA, US), Taq DNA polymerase mix (20 mM Tris–HCl; pH 7.4 at 25°C, 0.1 mM EDTA, 1 mM DTT, 100 mM KCl, 200 μg/mL BSA, and 50% glycerol), 1X reaction buffer (1.5 mM MgCl2), 200 μM deoxynucleoside triphosphates (dNTPs), and 0.5 μM of each of the virulence gene primers. Five virulence genes (actA, hlyA, inlA, inlB, and plcB) were detected with PCR using the primers listed in Table 1. PCR was performed on a Rotor-Gene Q thermal cycler (Qiagen) at the following conditions: initial denaturation at 98°C for 30 s, followed by 35 cycles of 98°C for 5 s, 60°C for 5 s, and 72°C for 10 s, with a final extension step at 72°C for 1 min. Amplified PCR products were loaded onto a 1.5% agarose gel and visualized under UV light.
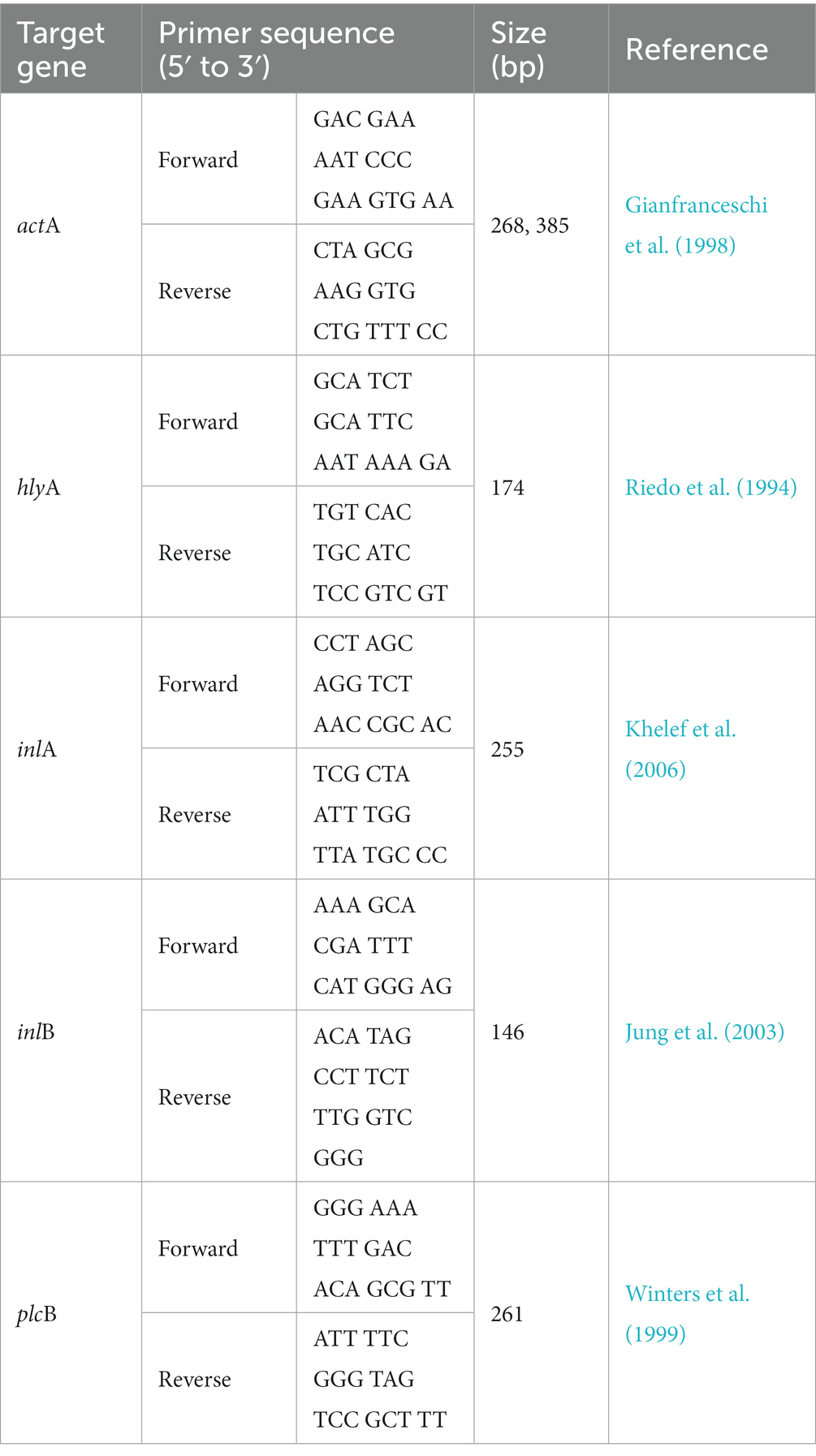
Table 1. Information of primers used for the detection of virulence genes (actA, hlyA, inlA, inlB, and plcB) in Listeria monocytogenes.
2.4. Comparing the growth patterns of Listeria monocytogenes isolates cultured at low temperature
As described in the section “2.2. Analysis of hemolytic property,” L. monocytogenes isolates were cultured and then sub-cultured in TSBYE at 30°C overnight. The sub-cultures were centrifuged at 1,912 × g and 4°C for 15 min. Cell pellets were washed twice with phosphate-buffered saline (PBS), and the suspensions were diluted with PBS and inoculated at 5 Log CFU/mL. The aliquots (0.2 mL) of the inocula of each L. monocytogenes isolates were inoculated into tubes containing 20 mL of TSBYE and incubated at 4°C for 288 h. During incubation, the cultures of L. monocytogenes were diluted with buffered peptone water (BPW; Bacto, Becton, Dickinson, Sparks, MD, USA), and 0.1-mL aliquots of the diluents were plated on tryptic soy agar with 0.6% yeast extract (TSAYE; Becton Dickinson and Company). The plates were incubated at 30°C for 48 h, and the colonies on the plates were manually counted. The six for each strain were repeated for each time point.
2.5. Comparing the growth patterns of Listeria monocytogenes isolates after vacuum treatment
Aliquots (0.1 mL) of the inocula were placed on a 10-g beef rump. The inoculated samples were vacuum-packed and stored at 4°C for 20 days. The samples were analyzed every 5 days. The samples were aseptically removed from the vacuum bags and placed in filter bags (3 M™) containing 20 mL BPW. The samples were then pummeled for 1 min in a pummeler (BagMixer; Interscience, St. Nom, France). Aliquots (1 mL) of the homogenates were diluted with BPW, and 0.1 mL of the diluents was spread-plated on Palcam agar (Becton Dickinson and Company). The plates were incubated at 30°C for 48 h, and the colonies on the plates were manually counted. The six for each strain were repeated for each time point.
2.6. Heat resistance
Aliquots (0.1 mL) of the inoculum were inoculated into tubes containing 9.9 mL of TSBYE; the tubes were preheated at 60°C and 70°C in a water bath. After 0, 3, 5, 8, and 10 min at 60°C and 0, 0.17, 3, 5, 8, and 10 min at 70°C, one-milliliter aliquots of the samples were retrieved and diluted with BPW, and 0.1-mL aliquots of the diluents were spread-plated on TSAYE, respectively. The plates were incubated at 30°C for 48 h, and the colonies on the plates were manually counted. The cell counts were then used to calculate D-values (decimal reduction time) as follows:
Where T is temperature, t is time, Log N0 is the initial number of bacteria, and Log Nt is the number of bacteria remaining at time. The six for each strain were repeated for each time point.
2.7. Whole-genome sequencing
2.7.1. DNA extraction and library preparation
After sub-culturing as described in the section “2.2. Analysis of hemolytic property” 3-mL aliquots of the sub-cultures were precipitated at 5,264 × g for 30 s. The cell pellets were used for extraction of DNA with the DNeasy Blood and Tissue Kit (Qiagen), according to the manufacturer’s protocol. Briefly, 5 μg of each extracted DNA sample was used to construct a library using the SMRTbell™ Template Prep Kit 1.0 (PN 100-259-100) (Pacific Biosciences, Menlo Park, CA, USA) following the manufacturer’s instructions. Fragments smaller than 20 kb of the SMRTbell template were removed using the BluePippin size selection system (Sage Science, Beverly, MA, USA) to construct large-insert libraries. The constructed library was validated using the 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) for quality control.
2.7.2. Sequencing and de novo assembly
The SMRTbell libraries were annealed to sequencing primers, and DNA polymerase was bound to the complex using the DNA/Polymerase Binding Kit P6 (Pacific Biosciences). This polymerase-SMRTbell-adaptor complex was loaded into SMRT cells (Pacific Biosciences) and sequenced using the PacBio RS II sequencing platform (Pacific Biosciences) which produce continuous long read (CLR). Long contigs were generated via de novo assembly (DNA LinkInc., Seoul, South Korea), and gene annotation and prediction were performed to analyze their genetic properties.
2.7.3. Analysis of genetic factors and genetic variations
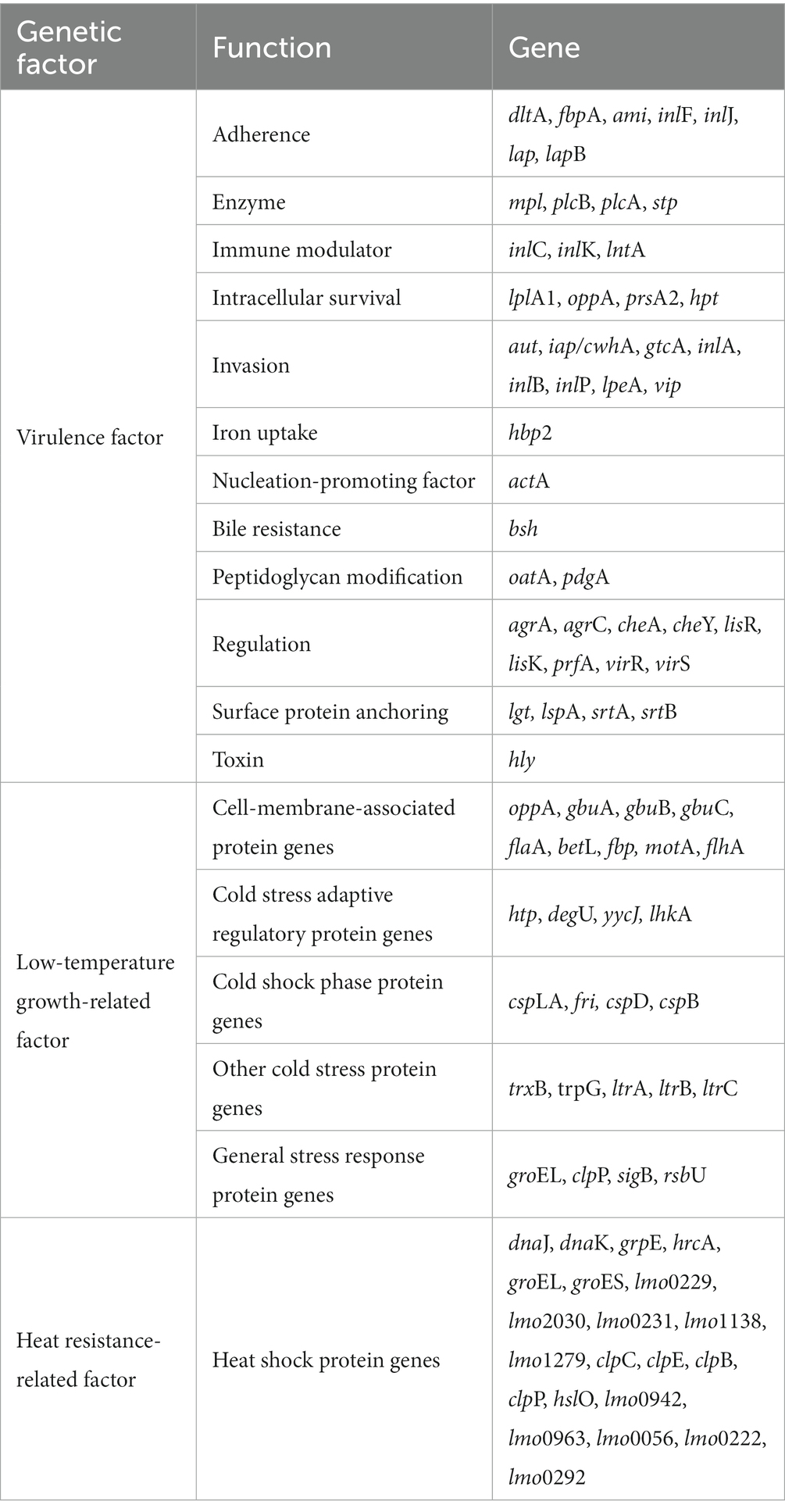
Genetic characteristics of two high-risk and low-risk L. monocytogenes isolates were analyzed for 45 virulence factors, 26 low-temperature growth-related factors, and 21 heat resistance-related factors using CLC Genomics Workbench ver. 12.0 (Qiagen) (Table 2). The sequences of these factors were obtained from the NCBI GenBank database. The presence of genetic factors in the two L. monocytogenes isolates was determined using a Basic Local Alignment Search Tool (BLAST), and genetic variations were assessed by comparing the sequences of each gene.
2.8. Statistical analysis
Statistical analysis was performed using SAS® version 9.4 (SAS Institute Inc., Cary, NC, USA). Significant differences among strains were determined using analysis of covariance and a general linear model at α = 0.05.
3. Results and discussion
3.1. Isolation and identification of Listeria monocytogenes
In this study, 23 L. monocytogenes isolates from 127 enoki mushrooms (Table 3), and 37 L. monocytogenes isolates (7 isolates from smoked ducks and 30 isolates from processed ground meat products) previously isolated (Table 4) were obtained. The iap gene for the confirmatory test of L. monocytogenes was detected in all 23 L. monocytogenes isolates from 127 enoki mushrooms (data not shown).
Table 3. Information regarding Listeria monocytogenes isolates from enoki mushrooms purchased from a market in Korea.
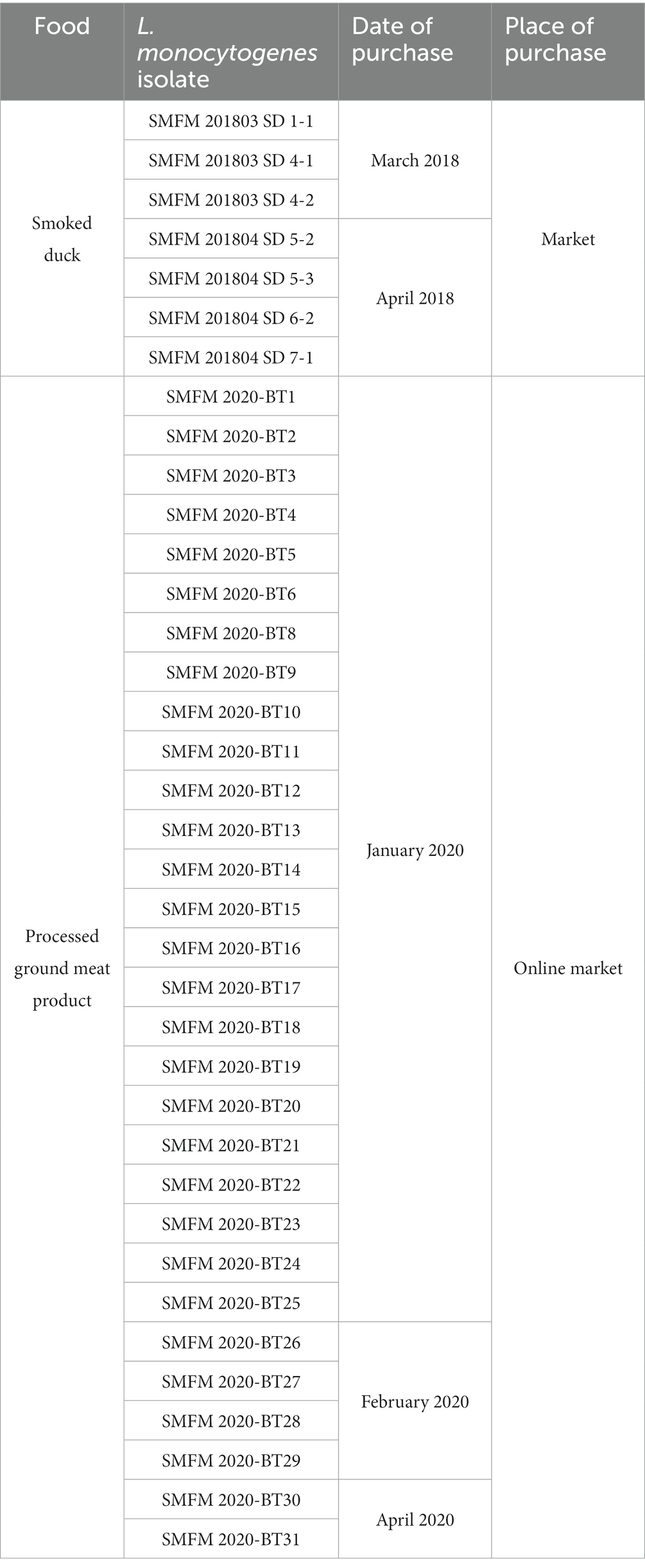
Table 4. Information regarding Listeria monocytogenes isolates from smoked ducks and processed ground meat products.
3.2. Hemolytic property
Listeria spp. which causes β-hemolysis is mostly pathogenic, and most of the isolated L. monocytogenes cause β-hemolysis (Farber and Peterkin, 1991). Examinations showed that all L. monocytogenes isolates caused β-hemolysis with narrow zone (Figure 1).

Figure 1. β-hemolytic property of Listeria monocytogenes SMFM 201803 SD 4-2, Listeria monocytogenes SMFM 201804 SD 5-2, and Listeria monocytogenes SMFM 201804 SD 6-2.
3.3. Virulence genes
Five virulence genes (actA, hlyA, inlA, inlB, and plcB) were detected in all L. monocytogenes isolates (Figure 2). In another study, most L. monocytogenes isolates harbored these virulence genes, and polymorphisms in the actA gene have been found between carcass and human isolates (Oh et al., 2018). ActA in L. monocytogenes encodes the surface protein actA, which is responsible for actin-based motility and cell-to-cell spread (Vàzquez-Boland et al., 2001). Rezaei et al. (2019) showed that the presence of hlyA and iap in L. monocytogenes is effective in increasing its aggressiveness and pathogenicity. Cellular invasion requires the presence of bacterial surface proteins internalin A (inlA) and/or inlB and their interaction with the cellular receptors E-cadherin and/or Met, respectively (Kühbacher et al., 2018). Virulence-associated genes plcA and plcB encode phosphatidylinositol phospholipase C (PI-PLC) and phosphatidyl-choline phospholipase (PC-PLC), respectively. Both phospholipases are related to effective escape from the phagocytic vacuole into the cytoplasm (Gründling et al., 2003; Hadjilouka et al., 2016). These results indicate that the isolates examined in this study are pathogenic to humans.
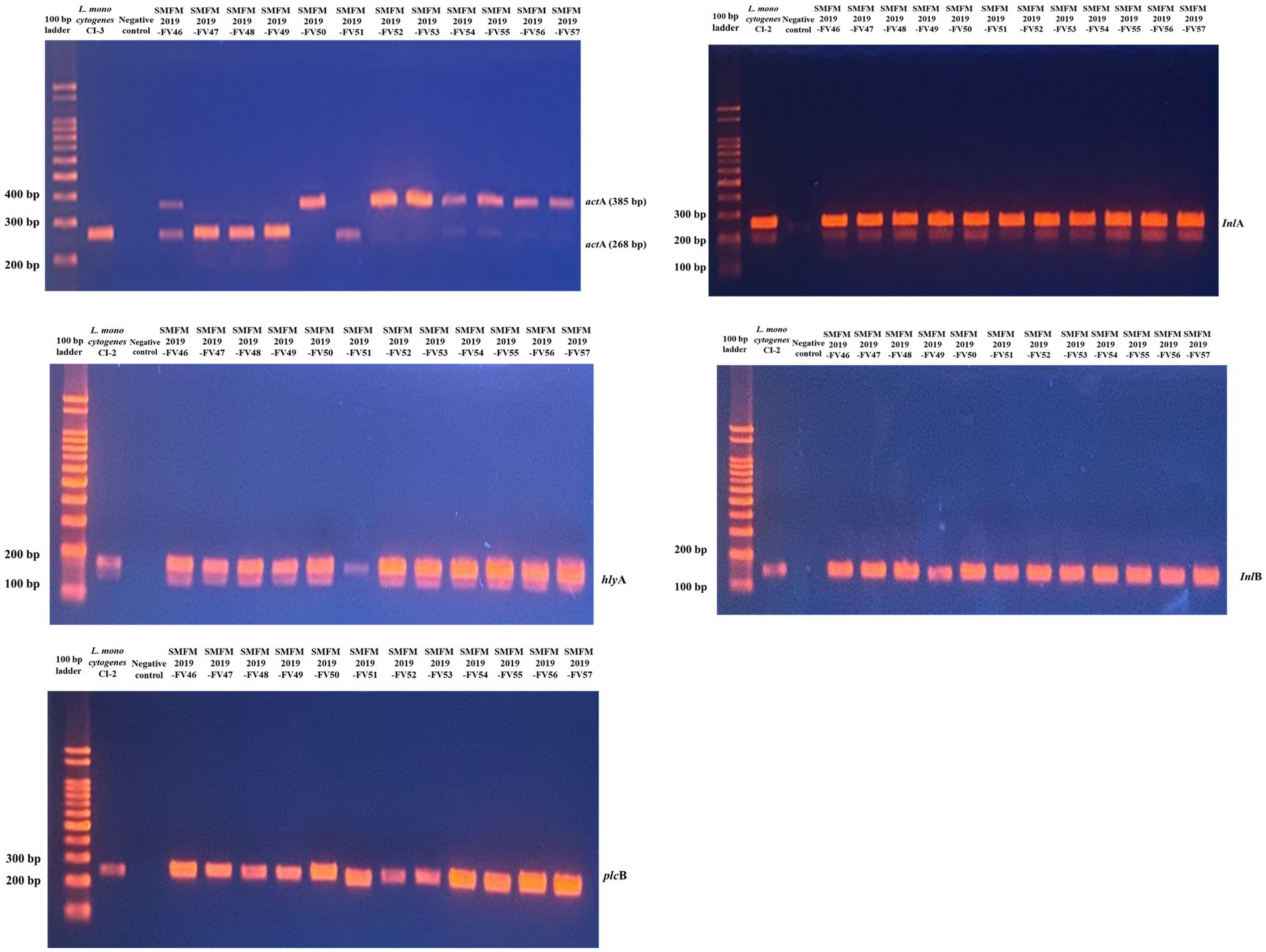
Figure 2. Gel image of PCR results for 12 Listeria monocytogenes isolates targeting actA, hlyA, inlA, inlB, and plcB genes.
3.4. Growth of Listeria monocytogenes isolates at 4°C
According to Kim et al. (1995), L. monocytogenes isolates from humans do not grow at 4°C; however, Seeliger and Jones (1986) reported that most L. monocytogenes could grow at 4°C. In this study, all the L. monocytogenes isolates showed growth at 4°C (Figure 3). Considering growth rates based on statistical difference over all time points (0, 48, 96, 144, 192, 240, and 288 h), high and low growth group were divided. L. monocytogenes SMFM 201803 SD 1-1 was in high growth group at all time points (48, 96, 144, 192, 240, and 288 h), and L. monocytogenes SMFM 201803 SD 4-2 was in high growth group at five time points (48, 144, 192, 240, and 288 h). L. monocytogenes SMFM 201804 SD 5-3 was in the high growth group at four points (48, 144, 240, and 288 h). L. monocytogenes SMFM 2019-FV43 was in the low growth group at all time points (48, 96, 144, 192, 240, and 288 h). L. monocytogenes SMFM 2020-BT30 was in low growth group at four time points (144, 192, 240, and 288 h), and L. monocytogenes SMFM 2019-FV42 was in low growth group at three time points (48, 96, and 240 h). Three (L. monocytogenes SMFM 201803 SD 1-1, L. monocytogenes SMFM 201803 SD 4-2, and L. monocytogenes SMFM 201804 SD 5-3) with high growth and three (L. monocytogenes SMFM 2019-FV42, L. monocytogenes SMFM 2019-FV43, and L. monocytogenes SMFM 2020-BT30) with low growth at 4°C were selected (Figure 3). The average cell counts (288 h – 0 h) of the high and low growth group were 5.3 and 4.6 Log CFU/mL, respectively. The three high-growth isolates were from smoked ducks; two low-growth isolates were from enoki mushrooms, while one low-growth isolate was from to the processed ground meat product.
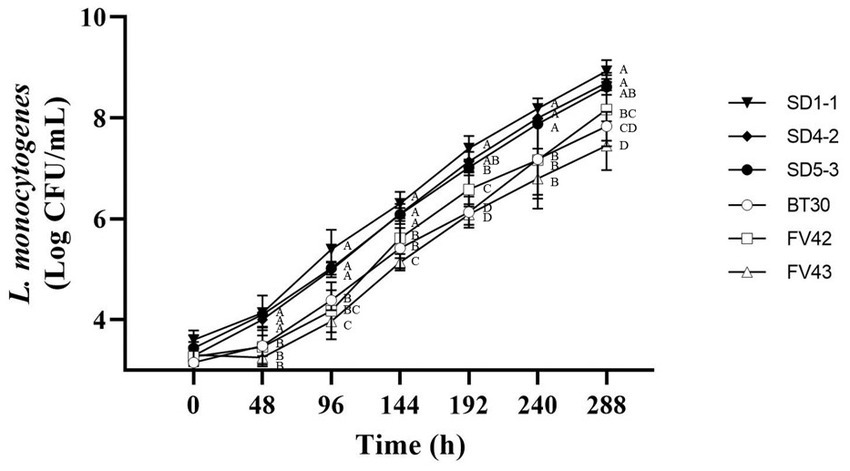
Figure 3. Cell counts of Listeria monocytogenes isolates in tryptic soy broth with 0.6% yeast extract during storage at 4°C. Error bars in the figure mean standard error. A–D means in the figure with different letters are significantly different (p < 0.05).
3.5. Growth of Listeria monocytogenes isolates in vacuum
Isolates with high and low growth (three each) at 4°C were further analyzed to compare their growth under vacuum conditions at 4°C. Duffy et al. (1994) showed that L. monocytogenes can grow in vacuum-packed foods stored in a refrigerator. However, in our study, the L. monocytogenes isolates showed high mortality rates (Figure 4). Isolates with high growth at 4°C (L. monocytogenes SMFM 201803 SD 1-1, L. monocytogenes SMFM 201803 SD 4-2, and L. monocytogenes SMFM 201804 SD 5-3) had better survival under vacuum conditions than those with low growth (L. monocytogenes SMFM 2019-FV42, L. monocytogenes SMFM 2019-FV43, and L. monocytogenes SMFM 2020-BT30) (Figure 4). These results showed that high-growth L. monocytogenes isolates in low temperature survive long in vacuum conditions.
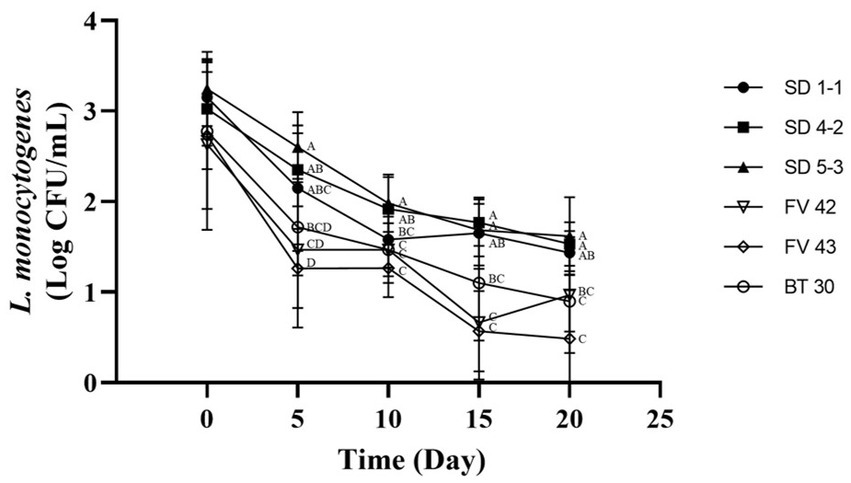
Figure 4. Cell counts of Listeria monocytogenes isolates inoculated in beef during storage under vacuum at 4°C. Error bars in the figure mean standard error. A–D means in the figure with different letters are significantly different (p < 0.05).
3.6. Heat resistance
The D60 and D70 values of the L. monocytogenes isolates, which describe heat resistance at 60°C and 70°C, ranged 2.37–3.55 min and 1.83–2.20 min, respectively (Table 5). According to Tangwatcharin et al. (2019), D-values of L. monocytogenes in inoculated restructured goat steaks ranged from 7.27 min at 60°C to 0.46 min at 70°C. In our study, at 60°C, L. monocytogenes SMFM 201804 SD 5-3 (3.55 min) had higher D60 value than the others followed by L. monocytogenes SMFM 2019-FV42 (3.17 min), and the D70 value (2.20 min) of L. monocytogenes SMFM 201804 SD 5-3 was also higher than that of the other isolates (Table 5). This indicated that L. monocytogenes SMFM 201804 SD 5-3 had the highest heat resistance.
There was association between cold and heat resistance. The growth of L. monocytogenes SMFM 201804 SD 5-3 at 4°C was significantly higher than that of L. monocytogenes SMFM 2020-BT30 and L. monocytogenes SMFM 2019-FV43 (p < 0.05, Figure 3). The D60 and D70 of L. monocytogenes SMFM 201804 SD 5-3 were significantly higher than those of L. monocytogenes SMFM 2019-FV43 (p < 0.05, Table 5). These results show a correlation between L. monocytogenes SMFM 201804 SD 5-3 (high cold- and heat-resistance) and L. monocytogenes SMFM 2019-FV43 (low cold- and heat-resistance) for growth at low temperature and survival in heat resistance. In the case of L. monocytogenes SMFM 201803 SD 1-1, the growth of the isolate was significantly higher than L. monocytogenes SMFM 2020-BT30, L. monocytogenes SMFM 2019-FV42, and L. monocytogenes SMFM 2019-FV43 at 4°C (p < 0.05, Figure 3), and there was no significant difference in D60. However, D70 of L. monocytogenes SMFM 201803 SD 1-1 was higher than that of L. monocytogenes SMFM 2019-FV42 and L. monocytogenes SMFM 2020-BT30 (p < 0.05, Table 5). L. monocytogenes SMFM 201803 SD 4-2 grew significantly higher than L. monocytogenes SMFM 2020-BT30, L. monocytogenes SMFM 2019-FV42, and L. monocytogenes SMFM 2019-FV43 at 4°C (p < 0.05, Figure 3), however, there was no significant difference in both D60 and D70.
3.7. Selection of high- and low-risk Listeria monocytogenes isolates
Analyses for hemolytic property, virulence genes, growth patterns, and the heat resistance test showed that L. monocytogenes SMFM 201804 SD 5-3 had the highest risk, while L. monocytogenes SMFM 2019-FV43 had the lowest risk. Thus, L. monocytogenes SMFM 201804 SD 5-3 and L. monocytogenes SMFM 2019-FV43 were defined as a high- and low-risk isolates, respectively. L. monocytogenes SMFM 201804 SD 5-3, isolated from smoked duck, showed relatively high growth at 4°C, the highest heat resistance, and relatively slow reduction in vacuum condition. L. monocytogenes SMFM 2019-FV43, isolated from the enoki mushrooms, had the lowest growth at 4°C, fast reduction under vacuum conditions, and low heat resistance compared with L. monocytogenes SMFM 201804 SD 5-3. For tracing the causes of these different characteristics between high- and low-risk L. monocytogenes, WGS was performed.
3.8. De novo assembly results
The de novo assembly results showed that L. monocytogenes SMFM 2019-FV43 and L. monocytogenes SMFM 201804 SD 5-3 had three contigs. The genome sizes of contigs 1, 2, and 3 for L. monocytogenes SMFM 2019-FV43 were 3,071,014, 60,439, and 6,273 bp, respectively, with GC contents of 37.92, 36.69, and 50.39%, respectively. The genome sizes of contigs 1, 2, and 3 for L. monocytogenes SMFM 201804 SD 5-3 were 3,038,302, 57,472, and 6,011 bp, with GC contents of 38.05, 36.04, and 50.39%, respectively.
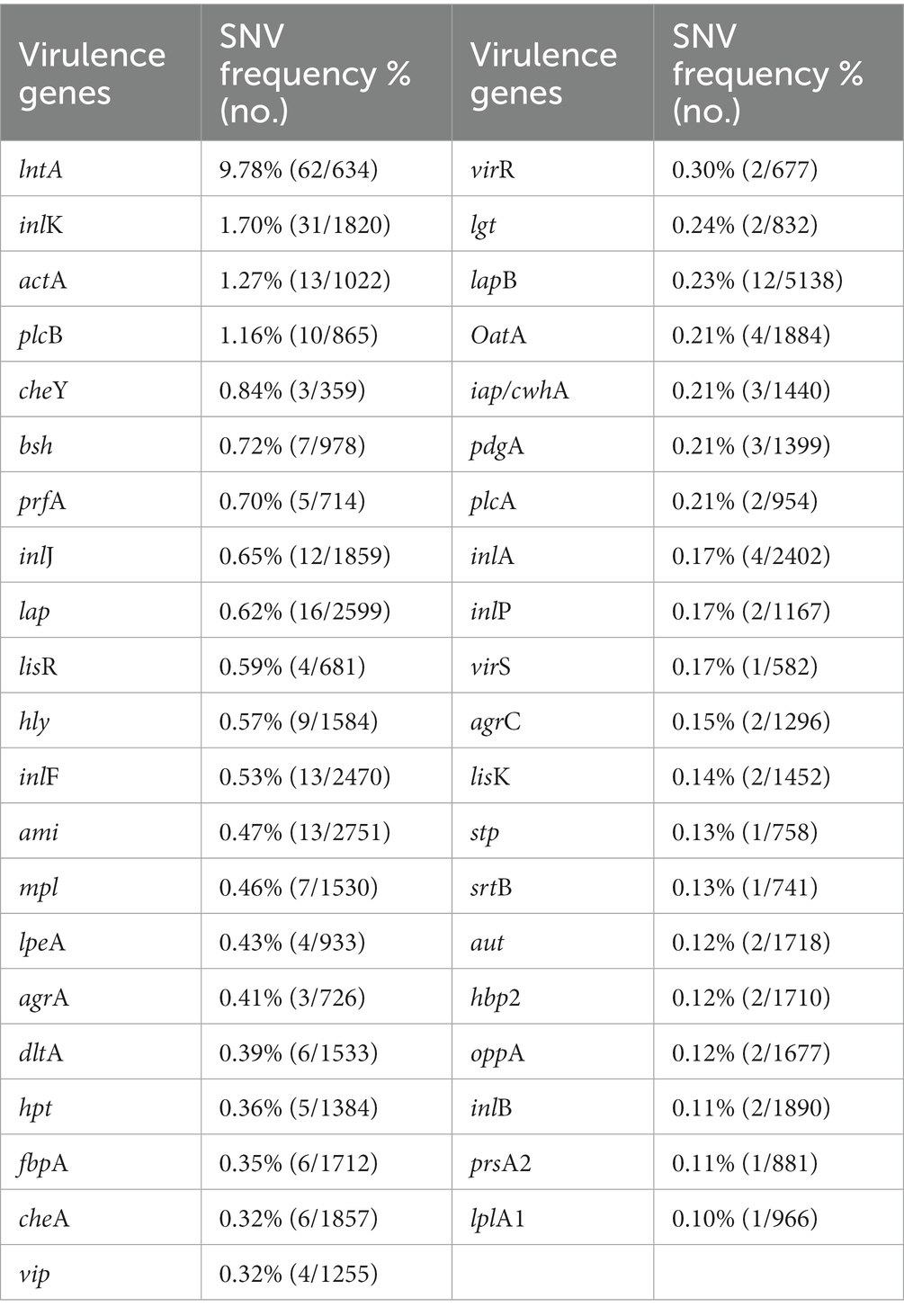
3.9. Genetic variations for virulence factors
Single nucleotide variants (SNVs) are variations reflecting differences between a single base [Ministry of Food and Drug Safety (MFDS), 2019]. The gene sequences of the virulence factors of L. monocytogenes SMFM 201804 SD 5-3 and L. monocytogenes SMFM 2019-FV43 were mapped to identify genetic variations. We identified 45 virulence genes in the two isolates. For these 45 virulence genes, 41 SNVs were found between the isolates (Table 6). The SNVs were found in 91% (41/45) of virulence genes. Among the genes, the four pathogenic genes (lntA, plcB, inlK, and actA), in which the frequency of SNVs was high, were focused (Figure 5). lntA in L. monocytogenes encodes a secretory protein that controls the expression of IFN-stimulated genes. This allows the bacterium to govern both the induction and repression of the host cell immune response to optimize conditions for specific stages of infection or colonization (Camejo et al., 2011). plcB encodes PC-PLC, which is involved in effective escape from the phagocytic vacuole to the cytoplasm (Gründling et al., 2003; Hadjilouka et al., 2016). Dortet et al. (2011) confirmed inlK as a gene highly activated during infection and that it may play a role in the infection process. actA is responsible for actin-based motility and the spread of L. monocytogenes to neighboring cells (Vàzquez-Boland et al., 2001).
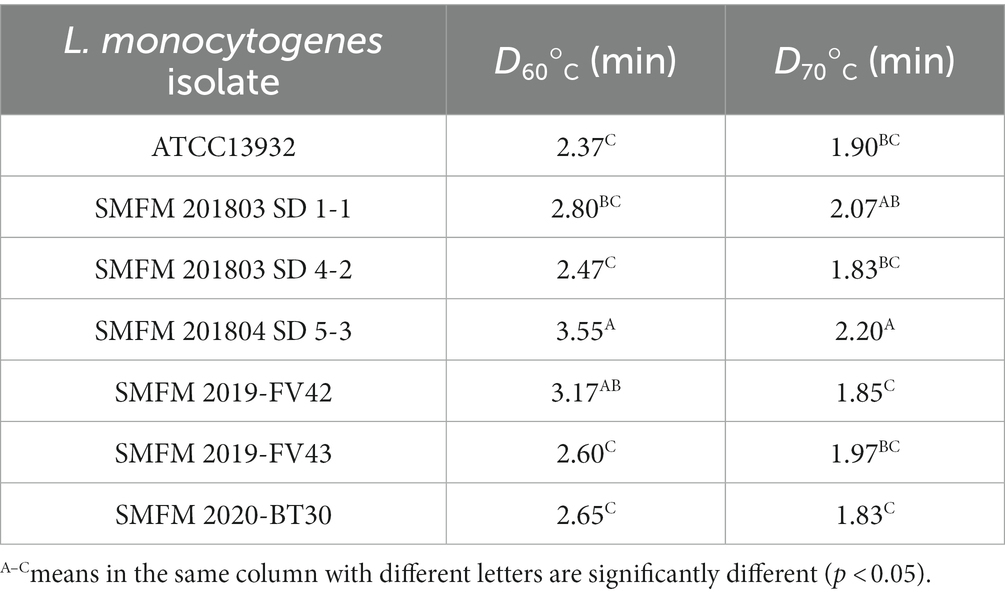
Table 6. Single nucleotide variants in virulence genes of Listeria monocytogenes SMFM 2019-FV43 and Listeria monocytogenes SMFM 201804 SD 5-3.

Figure 5. Single nucleotide variants in virulence factors (lntA, plcB, inlK, and actA) of Listeria monocytogenes SMFM 2019-FV43 and Listeria monocytogenes SMFM 201804 SD 5-3. contig.1.cir: Listeria monocytogenes SMFM 2019-FV43 contig 1; blue line: nucleotide sequence boundary. All dissimilar nucleotides are indicated in red.
Therefore, these variations of virulence genes may affect the high risk of L. monocytogenes SMFM 201804 SD 5-3. In particular, pathogenic genes (lntA, plcB, inlK, and actA) of the strain could improve attachment and invasion of host cells and affect the host cell immune response.
3.10. Genetic variations related to growth at 4°C
We identified genes related to growth at 4°C for L. monocytogenes SMFM 2019-FV43 and L. monocytogenes SMFM 201804 SD 5-3 and also found genetic variations in these genes. In all, 26 genes were identified in the two isolates. Among these 26 genes, 18 SNVs were identified (Table 7). The SNVs were found in 69% (18/26) of cold resistance genes. Among the genes, the four pathogenic genes (motA, ltrC, betL, and gbuB), in which the frequency of SNVs was high, were focused (Figure 6). Reportedly, gbu (gbuA, gbuB, and gbuC) and betL encode proteins involved in the betaine uptake system, and L. monocytogenes accumulates betaine when grown at low temperature, which functions as a cryoprotectant and osmoprotectant (Ko et al., 1994; Wemekamp-Kamphuis, 2003). Single and multiple deletions of these genes significantly reduce the viability of L. monocytogenes when exposed to low temperatures (Ko and Smith, 1999; Wemekamp-Kamphuis, 2003). ltrC is a stress-response gene essential for the growth of L. monocytogenes at cold temperatures (e.g., 4°C) (Zheng and Kathariou, 1995; Chan et al., 2007); flhA and motA also play a role in the cold tolerance of L. monocytogenes (Mattila et al., 2011). In this study, L. monocytogenes SMFM 201804 SD 5-3 had a high growth rate at 4°C than L. monocytogenes SMFM 2019-FV43 did, with a difference of 1.1 Log CFU/mL after storage for 288 h (Figure 3). Thus, it is reasonable to consider that L. monocytogenes SMFM 201804 SD 5-3 can sustain and grow at low temperatures, compared with L. monocytogenes SMFM 2019-FV43, given the genetic variations gbuB, betL, motA, and ltrC (which are related to cold stress) in the former.
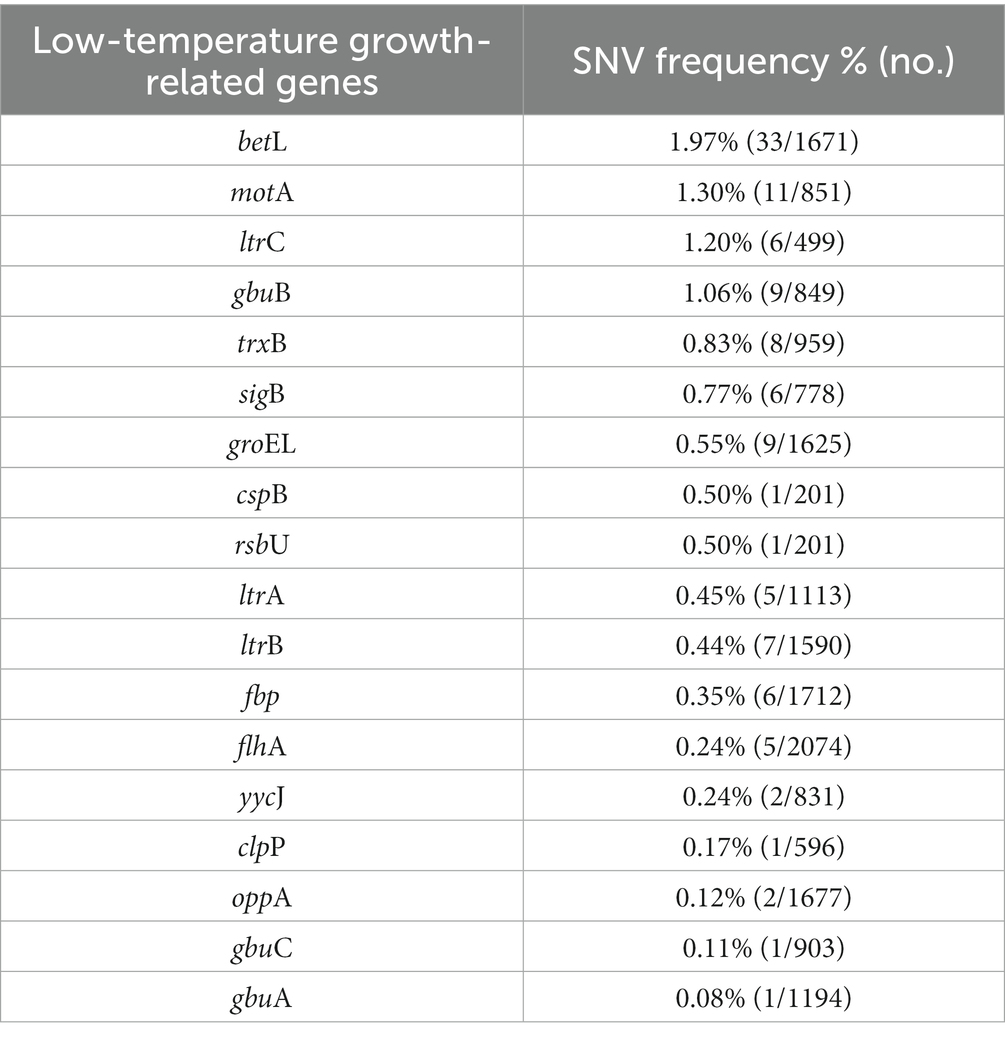
Table 7. Single nucleotide variants in low-temperature growth-related genes of Listeria monocytogenes SMFM 2019-FV43 and Listeria monocytogenes SMFM 201804 SD 5-3.

Figure 6. Single nucleotide variants in low-temperature growth related factors of Listeria monocytogenes SMFM 2019-FV43 and Listeria monocytogenes SMFM 201804 SD 5-3 (motA, ltrC, betL, and gbuB). contig.1.cir: Listeria monocytogenes SMFM 2019-FV43 contig 1; blue line: nucleotide sequence boundary. All dissimilar nucleotides are indicated in red.
3.11. Genetic variations related to heat resistance
We analyzed heat resistance-related genes and genetic variations in L. monocytogenes SMFM 2019-FV43 and L. monocytogenes SMFM 201804 SD 5-3. We identified 21 genes related to heat resistance. For these 21 genes, 16 SNVs were identified (Table 8). The SNVs were found in 76% (16/21) of heat resistance-related genes. The ctsR involved in regulating stress and heat shock, may coordinate the expression of stress genes (clpC, clpP, and clpE) in L. monocytogenes under stress conditions and in infected hosts. Stress-induced clpC, clpP, and clpE proteins are crucial for virulence (Nair et al., 2000). Regulation of gene expression in response to environmental stress is essential for bacterial survival (Akbar et al., 1997). Stress proteins play an important role in virulence, and therefore, variations in the heat resistance-related genes may be the plausible reasons underlying the heat resistance of L. monocytogenes SMFM 201804 SD 5-3.
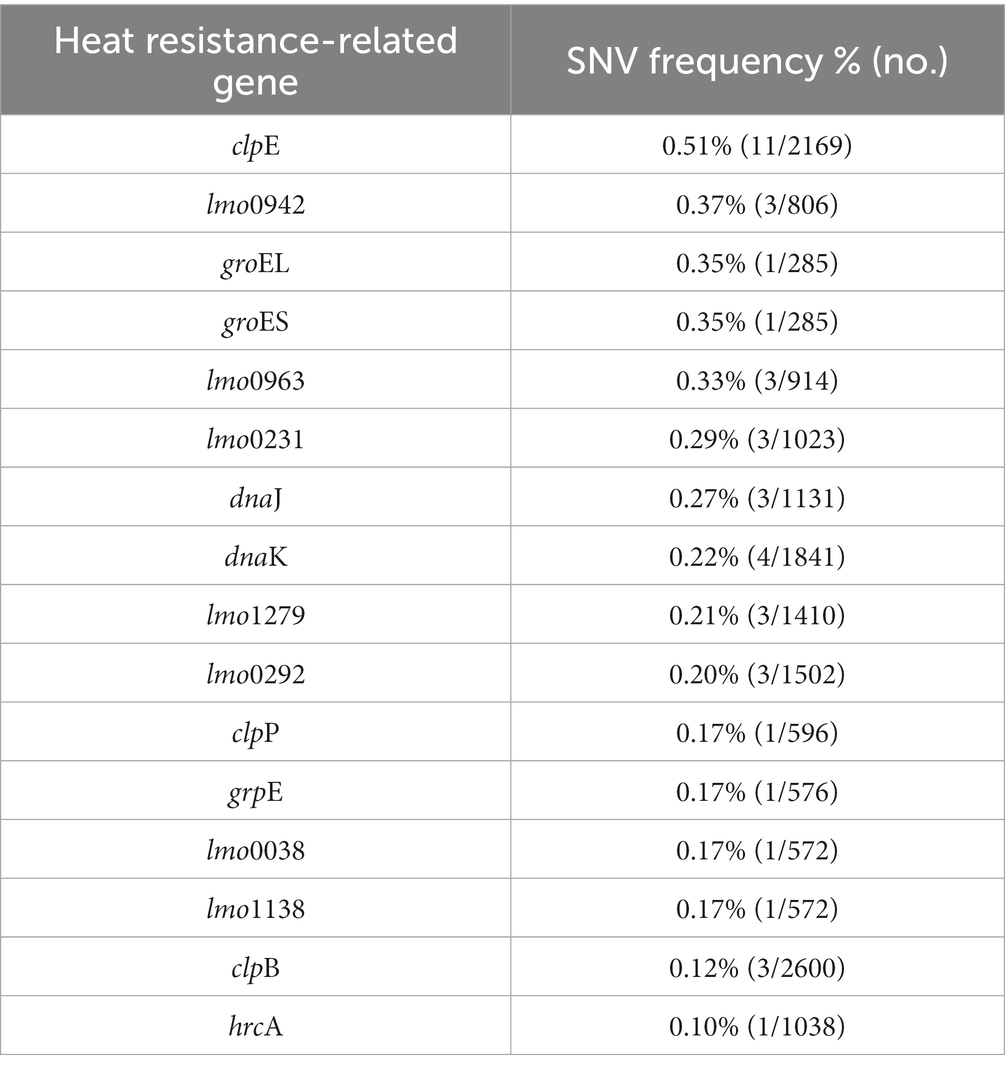
Table 8. Single nucleotide variants in heat resistance-related genes of Listeria monocytogenes SMFM 2019-FV43 and Listeria monocytogenes SMFM 201804 SD 5-3.
4. Conclusion
Listeria monocytogenes isolates have different risks, with different survival responses and pathogenicity under stressful conditions. These differences are caused by SNVs in virulence genes, low-temperature-related genes, and heat resistance-related genes. Our results support the position that L. monocytogenes SMFM 201804 SD 5-3 is a high-risk isolate whereas L. monocytogenes SMFM 2019-FV43 is a low-risk one.
Data availability statement
The datasets presented in this study can be found in the NCBI database. The Bioproject IDs are PRJNA958457 and PRJNA958458.
Author contributions
JR, YC, and YY made significant contributions to the manuscript and agree to its publication. YY and YC conceived and designed the study. JR performed laboratory experiments. All authors analyzed the data, draft the manuscript, and approved the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akbar, S., Kang, C. M., Gaidenko, T. A., and Price, C. W. (1997). Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24, 567–578. doi: 10.1046/j.1365-2958.1997.3631732.x
Brown, E., Dessai, U., McGarry, S., and Gerner-Smidt, P. (2019). Use of whole-genome sequencing for food safety and public health in the United States. Foodborne Pathog. Dis. 16, 441–450. doi: 10.1089/fpd.2019.2662
Camejo, A., Carvalho, F., Reis, O., Leitão, E., Sousa, S., and Cabanes, D. (2011). The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2, 379–394. doi: 10.4161/viru.2.5.17703
Cao, H., Yan, Y., Wang, L., Dong, L., Pang, X., Tang, S., et al. (2021). High-throughput sequencing reveals bacterial diversity in raw milk production environment and production chain in Tangshan city of China. Food Sci. Anim. Resour. 41, 452–467. doi: 10.5851/kosfa.2021.e10
Chan, Y. C., Raengpradub, S., Boor, K. J., and Wiedmann, M. (2007). Microarray-based characterization of the Listeria monocytogenes cold regulon in log-and stationary-phase cells. Appl. Environ. Microbiol. 73, 6484–6498. doi: 10.1128/AEM.00897-07
Chen, Y., and Knabel, S. J. (2007). Multiplex PCR for simultaneous detection of bacteria of the genus Listeria, Listeria monocytogenes, and major serotypes and epidemic clones of L. monocytogenes. Appl. Environ. Microbiol. 73, 6299–6304. doi: 10.1128/AEM.00961-07
Dortet, L., Mostowy, S., Louaka, A. S., Gouin, E., Nahori, M. A., Wiemer, E. A., et al. (2011). Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 7:e1002168. doi: 10.1371/journal.ppat.1002168
Duffy, L. L., Vanderlinde, P. B., and Grau, F. H. (1994). Growth of Listeria monocytogenes on vacuum-packed cooked meats: effects of pH, aw, nitrite and ascorbate. Int. J. Food Microbiol. 23, 377–390. doi: 10.1016/0168-1605(94)90164-3
Farber, J. M., and Peterkin, P. I. (1991). Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55, 476–511. doi: 10.1128/mr.55.3.476-511.1991
Food and Drug Administration (FDA). (2018) BAM protocol: simultaneous confirmation of Listeria species and L. monocytogenes isolates by real-time PCR. Available at: https://www.fda.gov/food/laboratory-methods-food/bam-protocol-simultaneous-confirmation-listeria-species-and-l-monocytogenes-isolates-real-time-pcr (Accessed April 2, 2023).
Gianfranceschi, M., Fransiosa, G., Gattuso, A., and Aureli, P. (1998). Detection of two phospholipases C by means of plate tests for the rapid identification of pathogenic Listeria monocytogenes. Archiv. fuer Lebensmittelhygiene (Germany) 49, 54–57.
Gründling, A., Gonzalez, M. D., and Higgins, D. E. (2003). Requirement of the Listeria monocytogenes broad-range phospholipase PC-PLC during infection of human epithelial cells. J. Bacteriol. 185, 6295–6307. doi: 10.1128/JB.185.21.6295-6307.2003
Hadjilouka, A., Molfeta, C., Panagiotopoulou, O., Paramithiotis, S., Mataragas, M., and Drosinos, E. H. (2016). Expression of Listeria monocytogenes key virulence genes during growth in liquid medium, on rocket and melon at 4, 10 and 30°C. Food Microbiol. 55, 7–15. doi: 10.1016/j.fm.2015.11.008
Hilliard, A., Leong, D., O’Callaghan, A., Culligan, E. P., Morgan, C. A., DeLappe, N., et al. (2018). Genomic characterization of Listeria monocytogenes isolates associated with clinical listeriosis and the food production environment in Ireland. Genes 9:171. doi: 10.3390/genes9030171
Jung, Y. S., Frank, J. F., Brackett, R. E., and Chen, J. (2003). Polymerase chain reaction detection of Listeria monocytogenes on frankfurters using oligonucleotide primers targeting the genes encoding internalin AB. J. Food Prot. 66, 237–241. doi: 10.4315/0362-028x-66.2.237
Khelef, N., Lecuit, M., Bierne, H., and Cossart, P. (2006). Species specificity of the Listeria monocytogenes InlB protein. Cell. Microbiol. 8, 457–470. doi: 10.1111/j.1462-5822.2005.00634.x
Kim, W. R. (2019) Development of Listeria monocytogenes detection technology in mushroom based on real-time PCR through development of enrichment medium and primer. Thesis for the Master’s degree. Sookmyung Women’s University.
Kim, Y. M., Park, U. Y., Mok, J. S., and Chang, D. S. (1995). Physiological characteristics of Listeria monocytogenes YM-7. Korean J. Fish Aquat. Sci. 28, 443–450.
Ko, R., and Smith, L. T. (1999). Identification of an ATP-driven, osmoregulated glycine betaine transport system in Listeria monocytogenes. Appl. Environ. Microbiol. 65, 4040–4048. doi: 10.1128/aem.65.9.4040-4048.1999
Ko, R., Smith, L. T., and Smith, G. M. (1994). Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J. Bacteriol. 176, 426–431. doi: 10.1128/jb.176.2.426-431.1994
Kühbacher, A., Novy, K., Quereda, J. J., Sachse, M., Moya-Nilges, M., Wollscheid, B., et al. (2018). Listeriolysin O-dependent host surfaceome remodeling modulates Listeria monocytogenes invasion. Pathog. Dis. 76, 1–12. doi: 10.1093/femspd/fty082
Kwon, J. G., Kim, S. K., and Lee, J. H. (2019). Recent next-generation sequencing and bioinformatic analysis methods for food microbiome research. Food Sci. 52, 220–228. doi: 10.23093/FSI.2019.52.3.220
Lee, J. Y. (2010) Molecular typing and antimicrobial susceptibility of Listeria monocytogenes in meats. Thesis for the degree of Philosophiae Doctor. Konkuk University. Seoul.
Lee, H., and Yoon, Y. (2021). Etiological agents implicated in foodborne illness world wide. Food Sci. Anim. Resour. 41, 1–7. doi: 10.5851/kosfa.2020.e75
Liu, D., Lawrence, M. L., Gorski, L., Mandrell, R. E., Ainsworth, A. J., and Austin, F. W. (2006). Listeria monocytogenes serotype 4b strains belonging to lineages I and III possess distinct molecular features. J. Clin. Microbiol. 44, 214–217. doi: 10.1128/JCM.44.1.214-217.2006
Lüth, S., Kleta, S., and Al Dahouk, S. (2018). Whole genome sequencing as a typing tool for foodborne pathogens like Listeria monocytogenes–the way towards global harmonisation and data exchange. Trends Food Sci. Technol. 73, 67–75. doi: 10.1016/j.tifs.2018.01.008
Mattila, M., Lindstrom, M., Somervuo, P., Markkula, A., and Korkeala, H. (2011). Role of flhA and motA in growth of Listeria monocytogenes at low temperatures. Int. J. Food Microbiol. 148, 177–183. doi: 10.1016/j.ijfoodmicro.2011.05.022
Ministry of Food and Drug Safety (MFDS) (2019) Guideline of next generation sequencing (NGS). Available at: http://www.kmdia.or.kr/board/board_10V.asp?mode=VIEW&bid=notice&bidsrl=2&bidssrl=5&intseq=10141 (Accessed May 18, 2023).
Nair, S., Derré, I., Msadek, T., Gaillot, O., and Berche, P. (2000). CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35, 800–811. doi: 10.1046/j.1365-2958.2000.01752.x
Oh, H., Kim, S., Lee, S., Lee, H., Ha, J., Lee, J., et al. (2018). Prevalence, serotype diversity, genotype and antibiotic resistance of Listeria monocytogenes isolated from carcasses and human in Korea. Food Sci. Anim. Resour. 38, 851–865. doi: 10.5851/kosfa.2018.e5
Park, M. S., Cho, J. I., Lee, S. H., and Bahk, G. J. (2014). The analysis for minimum infective dose of foodborne disease pathogens by meta-analysis. J. Food Hyg. Saf. 29, 305–311. doi: 10.13103/JFHS.2014.29.4.305
Park, E., Ha, J., Oh, H., Kim, S., Choi, Y., Lee, Y., et al. (2021). High prevalence of Listeria monocytogenes in smoked duck: antibiotic and heat resistance, virulence, and genetics of the isolates. Food Sci. Anim. Resour. 41, 324–334. doi: 10.5851/kosfa.2021.e2
Pöntinen, A., Aalto-Araneda, M., Lindström, M., and Korkeala, H. (2017). Heat resistance mediated by pLM58 plasmid-borne ClpL in Listeria monocytogenes. mSphere 2, e00364–e00317. doi: 10.1128/mSphere.00364-17
Rezaei, M., Kazemipour, N., Vand Yousefi, J., Rokhbakhsh Zamin, F., and Irajian, G. (2019). Determination of dominant serovars and molecular analysis of hly and iap genes related to Listeria monocytogenes strains isolated from spontaneous human abortions in Tehran. Iran. J. Microbiol. 13, 102–113. doi: 10.30699/ijmm.13.2.102
Ribot, E. M., Freeman, M., Hise, K. B., and Gerner-Smidt, P. (2019). PulseNet: entering the age of next-generation sequencing. Foodborne Pathog. Dis. 16, 451–456. doi: 10.1089/fpd.2019.2634
Riedo, F. X., Pinner, R. W., Tosca, M. L., Cartter, M. L., Graves, L. M., Reeves, M. W., et al. (1994). A point-source foodborne listeriosis outbreak: documented incubation period and possible mild illness. J. Infect. Dis. 170, 693–696. doi: 10.1093/infdis/170.3.693
Seeliger, H., and Jones, D. (1986). “Listeria” in Bergey's Manual of Systematic Bacteriology. ed. H. Sneath (Baltimore, MD: Williams and Wilkins), 1235–1245.
Sheng, J., Tao, T., Zhu, X., Bie, X., Lv, F., Zhao, H., et al. (2018). A multiplex PCR detection method for milk based on novel primers specific for Listeria monocytogenes 1/2a serotype. Food Control 86, 183–190. doi: 10.1016/j.foodcont.2017.11.028
Shin, D. H., Song, D. E., and Kim, K. R. (2007). Placental findings of Listeria monocytogenes infection in twin pregnancy. A Case Report. Korean J. Pathol. 41, 119–122. Available at: https://www.jpatholtm.org/journal/view.php?number=2550
Stratakos, A. C., Ijaz, U. Z., Ward, P., Linton, M., Kelly, C., Pinkerton, L., et al. (2020). In vitro and in vivo characterisation of Listeria monocytogenes outbreak isolates. Food Control 107:106784. doi: 10.1016/j.foodcont.2019.106784
Tangwatcharin, P., Sorapukdee, S., and Kongsrirat, K. (2019). Sous-vided restructured goat steaks: process optimized by thermal inactivation of Listeria monocytogenes and their quality characteristics. Food Sci. Anim. Resour. 39, 863–876. doi: 10.5851/kosfa.2019.e64
Vàzquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Dominguez-Bernal, G., Goebe, W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640. doi: 10.1128/CMR.14.3.584-640.2001
Wemekamp-Kamphuis, H. H. (2003) Survival strategies of Listeria monocytogenes: Roles of regulators and transporters. Wageningen: Wageningen University and Research.
Winters, D. K., Johnson, M. G., and Maloney, T. P. (1999). Rapid detection of Listeria monocytogenes by a PCR assay specific for an amino peptidase. Mol. Cell. Probes 13, 127–131. doi: 10.1006/mcpr.1999.0224
Yang, J. M., and Moon, G. S. (2021). Partial characterization of an anti-Listerial bacteriocin from Enterococcus faecium CJNU 2524. Food Sci. Anim. Resour. 41, 164–171. doi: 10.5851/kosfa.2020.e98
Zheng, W., and Kathariou, S. (1995). Differentiation of epidemic-associated strains of Listeria monocytogenes by restriction fragment length polymorphism in a gene region essential for growth at low temperatures (4 degrees C). Appl. Environ. Microbiol. 61, 4310–4314. doi: 10.1128/aem.61.12.4310-4314.1995
Keywords: enoki mushroom, smoked duck, processed ground meat product, next-generation sequencing, genetic variation, Listeria monocytogenes
Citation: Ryu J, Choi Y and Yoon Y (2023) Comparison of genetic variations between high- and low-risk Listeria monocytogenes isolates using whole-genome de novo sequencing. Front. Microbiol. 14:1163841. doi: 10.3389/fmicb.2023.1163841
Reviewed by:
Arthur William Pightling, United States Food and Drug Administration, United StatesAmit Mathews, Canadian Food Inspection Agency, Canada
Copyright © 2023 Ryu, Choi and Yoon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yohan Yoon, yyoon@sm.ac.kr
†These authors have contributed equally to this work
 Jihye Ryu
Jihye Ryu Yukyung Choi
Yukyung Choi Yohan Yoon
Yohan Yoon