- 1Department of Bacteriology, Mycology, and Immunology, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt
- 2U.S. National Poultry Research Center, Agricultural Research Service, United States Department of Agriculture, Athens, GA, United States
- 3Department of Hygiene and Zoonoses, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt
Aim: Campylobacter is the leading bacterial pathogen that causes foodborne illnesses worldwide. Pasture farming is regarded as an important source of agricultural production for small farming communities. Consumer preference for pasture-raised animal products has increased; however, there is a paucity of information on the microbiological quality of pasture-raised poultry products. The purpose of this study was to explore genetic relatedness of thermophilic Campylobacter isolates, to assess antibiotic resistance phenotypically and genotypically, and to screen the presence of virulence determinants of Campylobacter isolates from pasture-raised poultry farms from southeastern United States.
Methods: Ninety-seven Campylobacter isolates previously identified by Q7 BAX® System Real-Time PCR were genotyped by multilocus sequence typing (MLST). Campylobacter isolates were then evaluated for their phenotypic antimicrobial susceptibility against nine antimicrobial agents using Sensititre plates. Additionally, Campylobacter isolates were tested for the presence of antimicrobial resistance-associated elements. Furthermore, Campylobacter isolates were screened for the presence of 13 genes encoding putative virulence factors by PCR. These included genes involved in motility (flaA and flhA), adhesion and colonization (cadF, docC, racR, and virB11), toxin production (cdtA, cdtB, cdtC, wlaN, and ceuE) and invasion (ciaB and iamA).
Results: Among 97 Campylobacter isolates, Campylobacter jejuni (n = 79) and Campylobacter coli (n = 18) were identified. By MLST, C. jejuni isolates were assigned to seven clonal complexes. Among them, ST-353, ST-607 and ST-21 were the most common STs recognized. All C. coli (n = 18) isolates were included in CC-828. Interestingly, eight STs identified were not belonging any previous identified clonal complex. Campylobacter isolates displayed a high resistance rate against tetracycline (81.4%), while a low rate of resistance was observed against macrolides (azithromycin and erythromycin), quinolones and fluoroquinolones (nalidixic acid and ciprofloxacin), aminoglycosides (gentamicin), ketolide (telithromycin), amphenicol (florfenicol) and lincomycin (clindamycin). Thirteen isolates (13.54%) were pan-susceptible to all tested antibiotics, while nine isolates were multi-antimicrobial resistant (MAR; resist to three or more antimicrobial classes). Interestingly, there were no isolates resistant to all antimicrobial classes. Thr86Ile mutation was identified in all quinolones resistant strains. Erythromycin encoding gene (ermB) was identified in 75% of erythromycin resistant isolates. The A2075 mutation was detected in one erythromycin resistant strain, while A2074 could not be identified. The tetO gene was identified in 93.7% of tetracycline resistant isolates and six tetracycline susceptible isolates. In conclusion, the results of this study revealed that Campylobacter isolates from pasture-raised poultry farms showed the ST relatedness to Campylobacter isolates commonly associated with humans, indicating pasture-raised broiler flocks, similar to conventionally-reared broiler flocks, as a potential vector for antibiotic-resistant and pathogenic strains of thermophilic Campylobacter to humans.
Introduction
Thermophilic Campylobacter spp., particularly Campylobacter jejuni and Campylobacter coli, have been established as leading causes of food-borne illnesses worldwide (Vetchapitak and Misawa, 2019; European Food Safety Authority, 2021; Sher et al., 2021). The U.S. Centers for Disease Control and Prevention (Centers for Disease Control and Prevention, 2022) estimated that 1.5 million U.S. residents are infected with Campylobacter each year. Most patients have acute, self-limiting gastroenteritis, but some may have severe and long-lasting illnesses, which require antibiotic treatment, particularly in immunocompromised patients (Ma et al., 2014). Additionally, the infection by Campylobacter may be associated with a number of complications such as polyarthralgia, Guillain-Barre syndrome (GBS), Miller Fisher syndrome and even death (Kaakoush et al., 2015).
Campylobacteriosis is transmitted by eating raw or undercooked poultry meat (Centers for Disease Control and Prevention, 2022). Campylobacter contaminates poultry meats prior to or during processing representing a potential health threat to consumers (Suzuki and Yamamoto, 2009). Campylobacter contamination in poultry farms could occur via feed, water, soil, contact animals, biosecurity threats, and vehicles (Ghareeb et al., 2013).
The survival and pathogenicity of Campylobacter species are all influenced by several virulence factors (Casabonne et al., 2016). Bacterial motility, adherence to the intestinal epithelial walls, colonization and cytotoxin production are the main bacterial virulence factors. Several genes related to Campylobacter virulence factors have recently been identified including adhesion and colonization (flaA, flhA, cadF, and racR), invasion-associated markers (ciaB, iam, and virB11), and ganglioside mimicry (wlaN) (Bolton, 2015).
There is a growing antibiotic resistance crisis in clinical medicine since antibiotics were historically used in food animal production either for treatment or for growth promotion, which led to human exposure and infection through a variety of pathways, including meat and poultry products (Price et al., 2007). Moreover, a significant portion of the antibiotics provided are not absorbed by the animals and are excreted in the urine and feces. In Campylobacter infections, antibiotic therapy is commonly required for immunocompromised patients and those with severe campylobacteriosis (Kaakoush et al., 2015). Generally, Campylobacter infections are treated with macrolides (erythromycin, clarithromycin, and azithromycin), although fluoroquinolones (ciprofloxacin) are the most effective drugs to treat diarrhea (Aarestrup et al., 2008). Additional alternative drugs for treatment are tetracycline, doxycycline, and chloramphenicol (Skirrow and Blaser, 2000).
Pastured poultry farms in the USA are considered an important source of animal production that may provide an important opportunity to strengthen rural communities (Conner et al., 2008). Consumer preference of free-range and pasture-raised animal products such as meat, milk, and eggs has grown (Stampa et al., 2020). Because there is a paucity of information on the quality of pasture-raised chickens, many customers feel that these products are of superior quality in contrast to conventionally-farmed chickens, due to their more natural growing conditions (Yeung and Morris, 2001). There is insufficient research on genotyping, presence of virulence determinants, and antibiotic resistance of Campylobacter isolates from pasture-raised poultry farms; therefore, the purpose of this study was to explore genetic relatedness of thermophilic Campylobacter isolated from pasture-raised poultry farms and the following processing operations of broiler carcasses, and to assess antibiotic resistance phenotypes and genotypes as well as to screen the presence of virulence determinants in the retrieved isolates.
Materials and methods
The farm description, sample collection and processing, and Campylobacter isolation methods were previously described (Rothrock et al., 2016). Briefly, the samples were collected from feces, pasture soil, cecal content at processing, whole carcass rinsates and final whole carcass products. All samples were collected in the field and were brought back to the laboratories in a cooler packed in ice. The total amount of fecal and soil samples was at least 25 grams per sample. For homogenization, three grams (feces, cecal and soil samples) were diluted 1:3 in 10 mM phosphate buffered saline (PBS) in sterile filtered stomacher bags (Seward Laboratories, Inc., Bohemia, NY, United States). For rinsates, 100 mL of 10 mM PBS were added to each carcass within the storage bag, and the bags were vigorously shaken for 1 minute. The rinsates were collected into the sterile filtered stomacher bags (Seward Laboratories, Inc.). All samples were homogenized for 1 minute with a Stomacher® 400 Blender (Seward Laboratories, Inc.), and these homogenates were used for the downstream Campylobacter isolation. A volume of 100 μL from the above homogenized suspension was plated onto Campy-Cefex agar (prepared in the laboratory; Stern et al., 1992). The plates were incubated at 42 ± 1°C for 36 to 48 h in a microaerobic condition (85% N2, 10% CO2 and 5% O2) (Hiett et al., 2008; Yeh et al., 2013). Presumptive Campylobacter colonies were selected and enumerated on Brucella agar supplemented with 10% lyzed horse blood for isolation (prepared in the laboratory; Stern et al., 1992). The plates were incubated as described above. Speciation of Campylobacter was carried out using a Q7 BAX Real-Time PCR system according to the manufacturer’s instructions as described previously (Yeh et al., 2022). An end-point multiplex PCR assays were also performed. The 16S rRNA primers specific to Campylobacter in the PCR assays generated amplicons both in C. jejuni and C. coli samples, verifying the isolates as Campylobacter (Linton et al., 1997). The PCR assays with hipO primers amplified a 323-bp product in the C. jejuni samples, but not in the C. coli samples, verifying the isolates as C. jejuni (Caner et al., 2008). The PCR with primers from the ask gene generated about a 550-bp gene fragment that identified the samples of C. coli (Linton et al., 1997). Campylobacter isolates were frozen at −80°C in Luria-Bertani broth with 20% glycerol until downstream analyses were performed.
Bacterial cultures and genomic DNA isolation
Campylobacter jejuni (n = 79) and C. coli (n = 18) isolates from our stock in the U.S. National Poultry Research Center, Agricultural Research Service, U.S. Department of Agriculture, Athens, GA, United States were used in this study. Bacterial cultures were revived in Mueller-Hinton agar plates at 42°C for 48 h under the microaerobic condition as described as above.
DNA was extracted from pure bacterial cultures of 79 C. jejuni and 18 C. coli using the DNeasy Blood & Tissue Kit (Qiagen Inc., Germantown, MD, United States) in accordance with the manufacturer’s instructions. DNA concentrations were measured spectrophotometrically using a DeNovix DS-11 FX spectrophotometer (DeNovix Inc., Wilmington, DE, United States).
Multilocus sequence typing of Campylobacter isolates
Amplification of seven housekeeping genes was performed according to the procedures described by Dingle et al. (2001) using the primer sets given in the Campylobacter MLST website.1 All PCR products were purified with a DNA Clean & Concentrator™-5 kit (Zymo Research, Irvine, CA, United States). The purified PCR products were sent to the core facilities at the USDA ARS Genomics and Bioinformatics Research Unit (Stoneville, MS, United States) for DNA sequencing with an ABI 3730xl Genetic Analyzer (Thermo Fisher Scientific, Foster City, CA, United States) using a BigDye terminator v.3.1 Chemistry. Allelic profile, sequence type (ST) and clonal complex (CC) were assigned to the isolates using the allelic profile query function in the MLST database. Minimum spanning tree (MST) of MLST allelic differences was generated using BioNumerics (version 7.6; Applied Maths, Austin, TX, United States).
Antimicrobial susceptibility test
Antimicrobial susceptibility of C. jejuni and C. coli isolates was determined using a Sensititre™ system (Thermo Fisher Scientific) according to the manufacturer’s instructions described previously (Yeh et al., 2022). Sensititire™ Campylobacter CAMPY AST plates were used in this study (Thermo Fisher Scientific). The results were read photometrically using Sensititre™ Vizion™ Digital MIC Viewing System (Thermo Fisher Scientific) in associated with the SWIN software (version 3.3). Quality control was performed using C. jejuni, ATCC 33560. The breakpoints for Campylobacter resistance were interpreted according to the guidelines from Clinical and Laboratory Standards Institute M45, 3rd Edition (Clinical and Laboratory Standards Institute (CLSI), 2015) as follows: azithromycin, ≥8 μg/mL; erythromycin, ≥32 μg/mL; gentamicin, ≥8 μg/mL; tetracycline, ≥16 μg/mL; ciprofloxacin, ≥4 μg/mL; florfenicol, ≥16 μg/mL; nalidixic acid, ≥32 μg/mL; and clindamycin, ≥8 μg/mL.
Molecular detection of antibiotic resistance-associated genes
Resistance-associated genes of tetracycline, quinolones and macrolides in resistant isolates were determined. For tetracycline, the presence of the tetO gene was determined as described previously by Gibreel et al. (2004). Primers DMT 1 and DMT 2 (Table 1) were used to amplify a 559-bp product of the tetO gene in Campylobacter genomes. The mismatch amplification mutation assay (MAMA-PCR) was used to detect point mutations at Thr-86-Ile in QRDR of the gyrA gene (Zirnstein et al., 1999) and Ala-2074-Cys and Ala-2075-Gly in 23S rRNA gene (Alonso et al., 2005) for quinolone- and erythromycin-resistant isolates, respectively. Also, the ermB gene was used for screening the erythromycin resistant isolation according to the protocol described by Zhou et al. (2016). Primer sequences for PCR amplification are listed in Table 1.
Detection of virulence-associated genes
Campylobacter isolates were screened for the presence of some virulence determinants by PCR, including the genes responsible for motility (flaA and flhA), adhesion and colonization (cadF, docA, racR, and virB11), cytotoxin production (cdtA, cdtB, cdtC, ceuE, and wlaN) and invasion-associated markers (iam and ciaB). Primer sequences and protocol for PCR amplification of the above virulence factors are listed in Table 1.
Statistical analysis
To determine if the differences in the frequencies of isolate recovery was significant among the examined sources as well as frequencies of virulence genes among the examined isolates, these frequencies were used as inputs to create contingency tables and the significance was determined by Chi-square (X2) test, with a cutoff level for p-value equal to 0.05. The results of resistance phenotypes and frequencies of virulence genes were converted into binary data (0/1), where the presence of a virulence gene received scores of 1, whereas susceptibility to antimicrobials and the absence of a virulence gene received scores of 0. To determine the association of resistance phenotypes and virulence genes to sequence types (STs) among the examined Campylobacter, a heatmap with hierarchical clustering based on the binary data (0/1) of antimicrobial resistance and virulence genes was created using the package “pheatmap” in R software (version 217 3.4.2).
Results
Genetic diversity of Campylobacter isolates using MLST
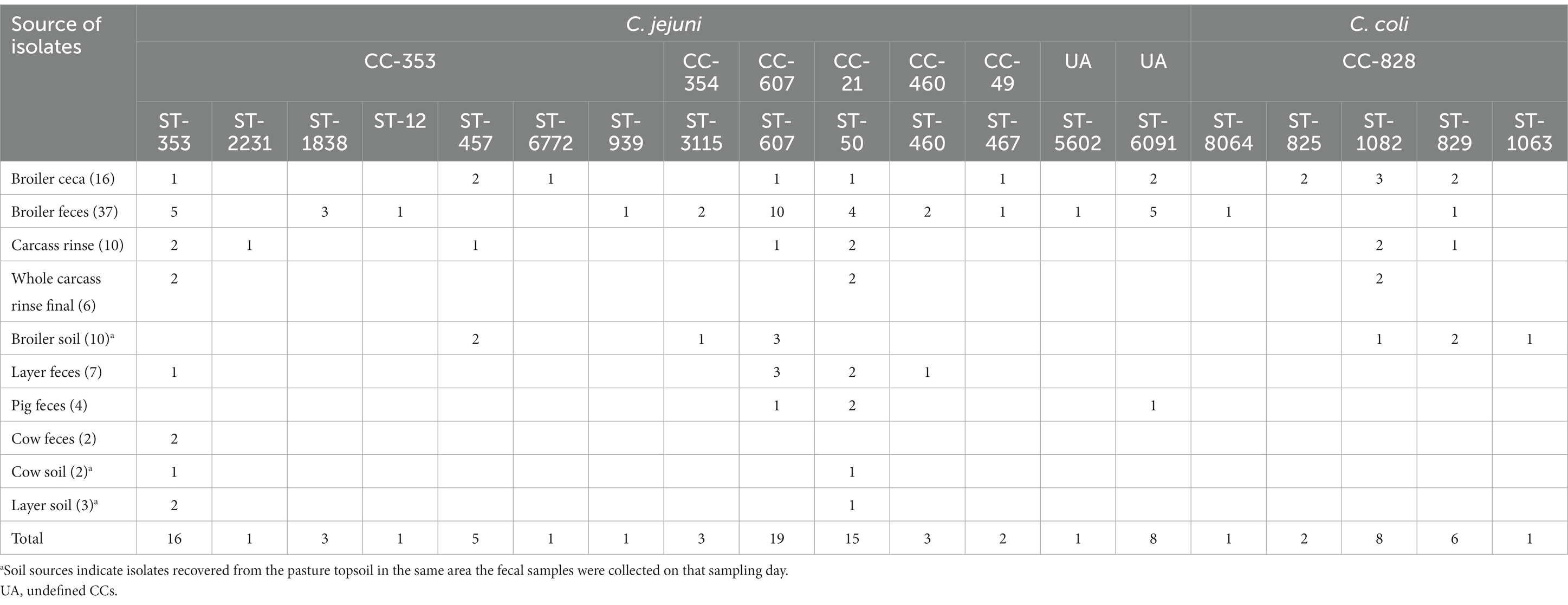
MLST analysis showed high genetic diversity among both C. jejuni and C. coli isolates (Figure 1). A total of 19 different STs were identified: 14 for C. jejuni and five for C. coli (Table 2). The STs found in C. jejuni included ST-607 (n = 19), ST-353 (n = 16), ST-50 (n = 15), ST-6091 (n = 8), ST-457 (n = 5), ST-460 (n = 3), ST-1838 (n = 3), ST-3115 (n = 3), ST-467 (n = 2), ST-12 (n = 1), ST-939 (n = 1), ST-2231 (n = 1), ST-5602 (n = 1) and ST-6772 (n = 1). C. jejuni isolates from broiler feces showed the most diversity, including 11 STs, followed by seven STs found in broiler cecae. Further, 12 C. jejuni STs could be assigned to six previously described CCs (CC21, CC607, CC353, CC49, CC354, and CC460), whereas two (ST-5602 and ST-6091) belonged to undefined CCs. The STs found in C. coli were assigned to a single previously described CC828 included ST-8064 (n = 1), ST-829 (n = 6), ST-825 (n = 2), ST-1082 (n = 8) and ST-1063 (n = 1).
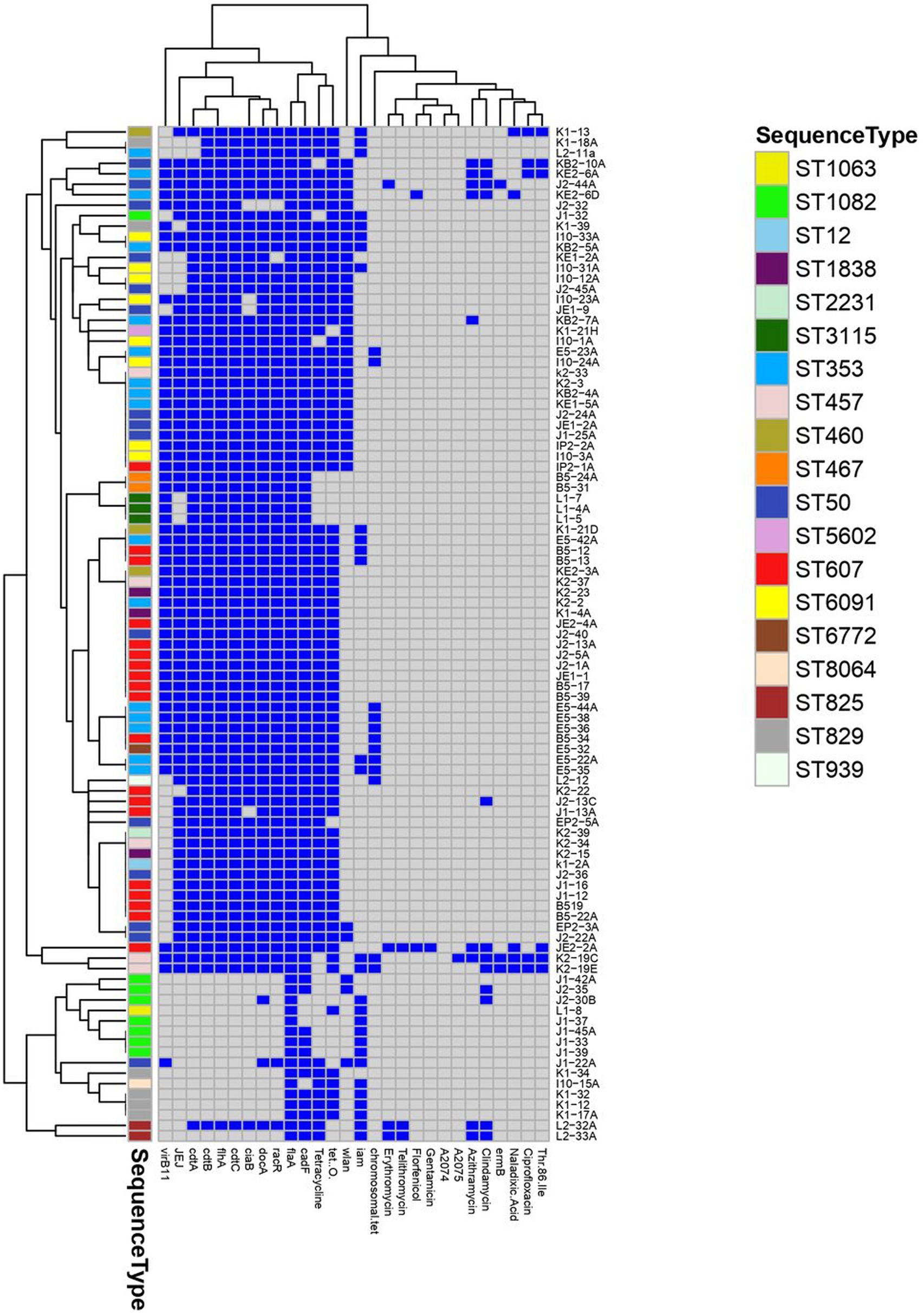
Figure 1. A heatmap supported by a dendrogram showing the distribution of antimicrobial resistance phenotypes, resistance and virulence genes among the examined Campylobacter assigned to various multilocus sequence types (ST). Dark blue squares indicate the presence of virulence and resistance genes and phenotypic resistance; gray squares indicate absent genes and phenotypic susceptibility.
Thermophilic isolates used in this study were originating from nine pastured poultry and livestock raised flocks, including broiler feces, broiler soil, broiler ceca, whole carcass rinse, pig feces, layer feces, layer soil, final whole carcass rinse, cow feces, and cow soil. By studying the frequency distribution of the recovered C. jejuni and C. coli from different sources, a significant (p < 0.05) associations of isolate recovery to the examined sources was obvious (Supplementary Table S1).
Concerning the distribution of C. jejuni and C. coli STs according to the source of samples, 7STs were detected belonging to CC-353 which was the most frequent clonal complex identified including ST-353 from broiler ceca, broiler feces, whole carcass rinse, layer feces, cow feces, cow soil and layer soil, ST-2231 from carcass rinse, ST-1838 from broiler feces (3), ST-12 from broiler feces (1), ST-457 was detected from carcass rinse and broiler soil, ST-6772 was detected from broiler ceca (1) and ST-939 was detected from broiler feces. Regarding CC 607, only ST-607 from broiler ceca, broiler feces, carcass rinse, broiler soil, layer feces and pig feces was identified. From CC-21 only ST- 50 was detected from broiler ceca, broiler feces, carcass rinse, whole carcass rinse, broiler soil, layer feces and pig feces. In addition to CC 460, ST- 460 was identified from broiler feces and layer feces and CC49 from which ST-467 was detected from broiler ceca and broiler feces. Furthermore, two STs not assigned to any clonal complex were also identified including ST-5602 from broiler feces and ST-6091 was detected from broiler ceca, broiler feces and pig feces. Regarding C. coli only CC-828 were detected and 5 STs were identified including ST- 8064 (broiler feces), ST- 825 (broiler ceca), ST-1082 (broiler ceca, carcass rinse, whole carcass rinse, broiler soil), ST-829 (broiler ceca, broiler feces, carcass rinse, broiler soil) and ST-1063 from broiler soil (Table 2).
Antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli isolates
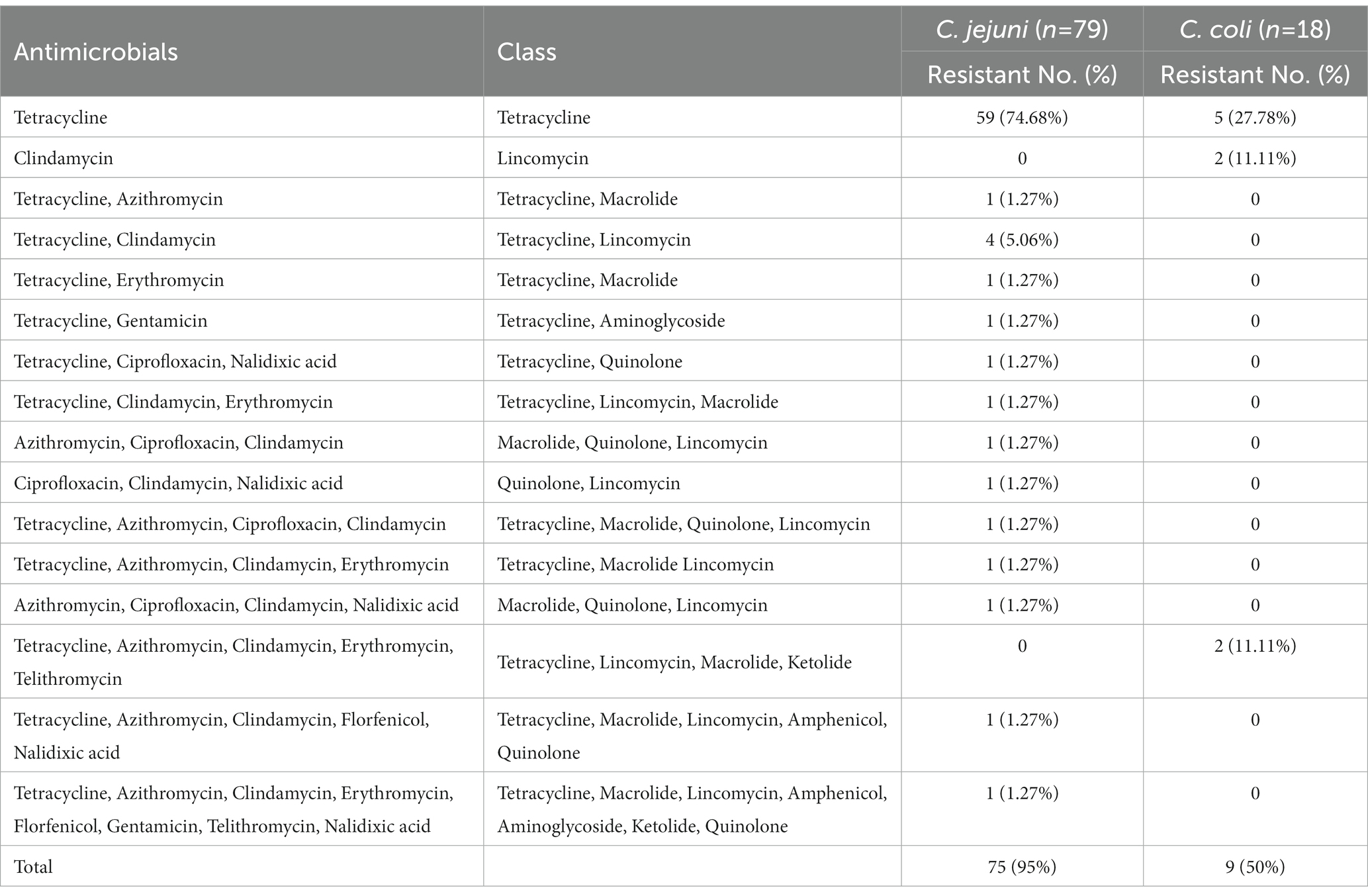
Frequency of antibiotic resistance of the C. jejuni and C. coli isolates to various antibiotics is presented in Table 3 and Figure 1. In total, 75 (95%) of the C. jejuni isolates were resistant to various numbers of antibiotics tested. Fifty-nine isolates (75%) were resistant to tetracycline alone, four isolates (5%) were resistant to two antibiotics (azithromycin and tetracycline), and another four (5%) were pan susceptible to all nine antibiotics tested. Twelve isolates were resistant to at least three antibiotics, and therefore considered multi-drug resistant (MDR), with one isolate showing resistance to eight antibiotics (azithromycin, clindamycin, erythromycin, florfenicol, gentamicin, telithromycin, tetracycline, and nalidixic acid). For C. coli, five (28%) and two (11%) isolates were resistant to tetracycline and clindamycin, respectively. However, two (11%) C. coli isolates were resistant to five antibiotics (tetracycline, azithromycin, clindamycin, erythromycin, and telithromycin). All C. coli isolates were sensitive to the quinolone class antibiotics (nalidixic acid and ciprofloxacin).
Detection of antimicrobial resistance mechanisms
The tetO gene that is responsible for tetracycline-resistant was detected in 80 isolates (82.5%) including 71 (89.9%) for C. jejuni and 9 (50%) for C. coli. Interestingly, the tetO gene was not detected in five phenotypically resistant isolates, but was detected from six phenotypically sensitive strains. The point mutation in gyrA responsible for quinolone resistance of C. jejuni (n = 5) and C. coli (n = 1) isolates was detected using MAMA-PCR. All phenotypically resistant isolates had a point mutation in the gyrA. For erythromycin-resistant isolates, ermB was detected in three isolates. The mutated A2075 was found in one isolate, while the A2074 mutation could not be identified.
Distribution of virulence genes
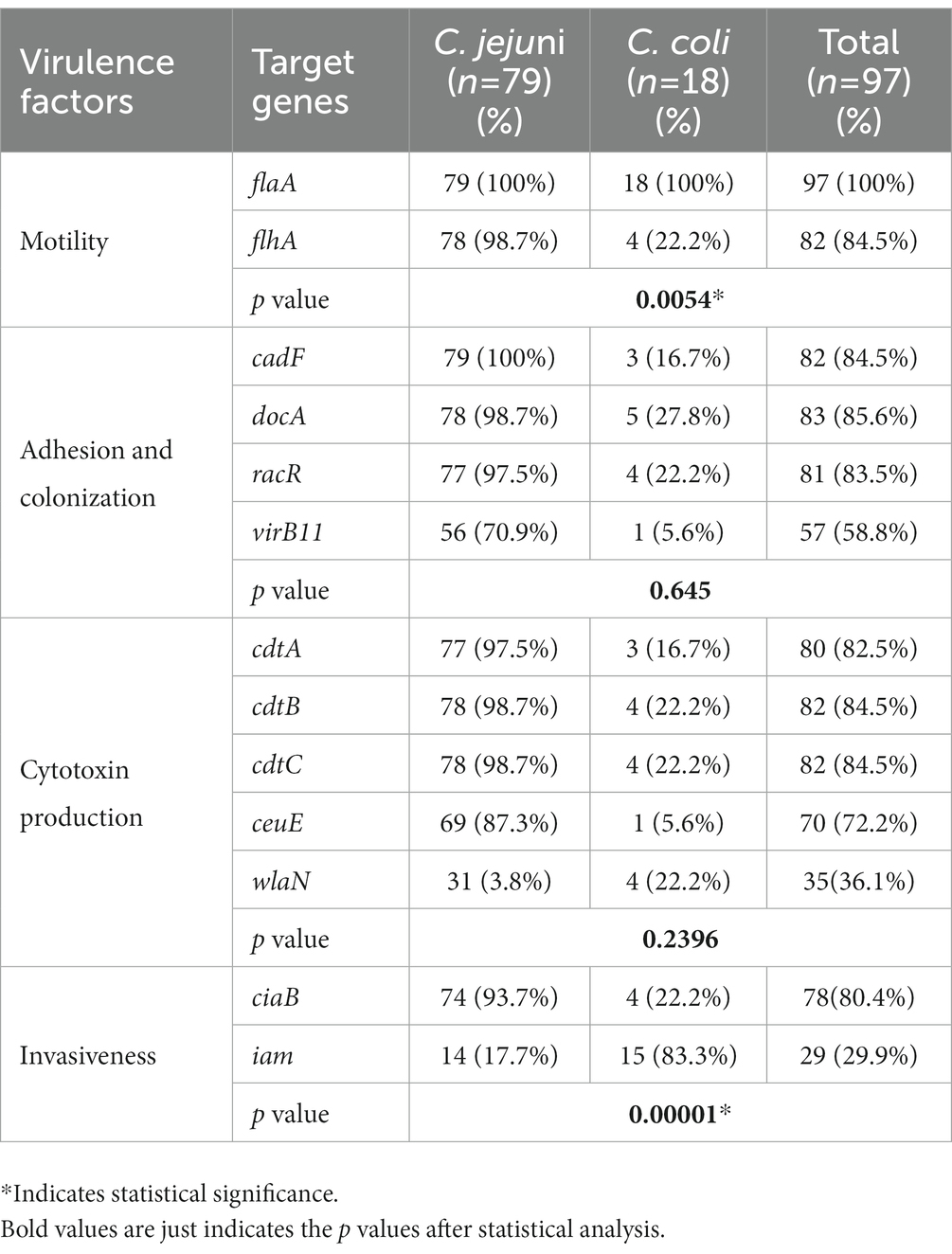
Analysis of virulence gene distribution among C. jejuni and C. coli isolates revealed that all 97 Campylobacter isolates harbored the virulence genes tested (Table 4). All isolates contained the flaA gene (100%) and the other genes were detected in a high prevalence rate, including flhA (84.5%; 82/97), cadF (84.5%; 82/97), docA (85.6%, 83/97), ciaB (80.4%, 79/97), racR (83.5%, 81/97), cdtC (84.5%; 82/97), cdtB (84.5%, 82/97), cdtA (82.5%, 80/97), ceuE (72.2, 70/97), and VirB11 (58.8%, 57/97). On the other hand, the wlaN gene was detected in only 35 isolates (36.1%) and iam gene was found in only 29 isolates (29.9%). In addition, the frequency of genes encoding adhesion and colonization factors in C. jejuni was significantly higher than that in C. coli (Table 4).
Discussion
Within the poultry industry, concerns have been expressed over the microbiological safety of pasture-raised poultry products despite consumer confidence in these types of production. The continuous exposure of the flocks to the pasture environment increases the possibility of contact with other sources of Campylobacter such as wild birds, insects, etc. (Berg, 2001). Due to the growing preference of this type of meat product, the question of whether the welfare benefits for this type of production is aligned with appropriate food safety should be explored. As a result, the current study was carried out to explore the genetic relatedness, virulence, and antimicrobial susceptibility of thermophilic Campylobacter by characterizing 97 isolates from pasture-raised poultry farms and the following processing operations.
Campylobacter sequence-based genotyping techniques yield data that is consistent across host sources, reproducible, and suitable for population genetic study (Dingle et al., 2001). Multi-locus sequence typing (MLST) identifies clonal complexes and links Campylobacter species to specific animal sources (Dingle et al., 2002; Colles et al., 2008). In this study, Campylobacter genotypes identified by MLST were diverse based on the number of samples taken from each flock. These results are in an agreement with the results reported by Colles et al. (2010) who found a great diversity in Campylobacter genotypes isolated from free-range broiler flocks. However, Bull et al. (2006) and Lindmark et al. (2006) reported a lower ST diversity of up to three STs within housed flocks. These discrepant findings highlight the importance of collecting large numbers of samples from a flock in order to identify the full range of variability within a flock. The most common clonal complexes CC607, CC21 and CC353 were predominant among C. jejuni strains in our study. These CCs were reported also as the most common CCs identified from human samples in various geographic regions (Dingle et al., 2001; de Haan et al., 2010; Smid et al., 2013). On the ST level, ST-353 and ST-50 were reported also as the most widely distributed STs among human and broiler C. jejuni isolates (Harvala et al., 2016; Elhadidy et al., 2018). These results highlight the importance of poultry sources for human campylobacteriosis.
The presence of thirteen virulence genes was investigated by PCR to confirm the pathogenic potential of these isolates. Significant differences in the occurrence of virulence genes were observed, C. jejuni isolates had a higher virulence potential than C. coli isolates. These results are in an agreement with those reported by Casabonne et al. (2016) and Wieczorek et al. (2013) from conventionally raised broiler flocks. Our results showed that the flaA gene was detected in all strains, and the flhA gene was found in most of the isolates examined. Similar findings are also reported by Rossler et al. (2020) that flaA and flhA genes were detected in all their isolates collection. Mobility of Campylobacter, involving the coordination of many genes (such as flaA and flhA), is important for passage through the stomach and gut (Gilbreath et al., 2011). The presence of flaA and flhA genes in a high proportion of the isolates examined suggests that motility and virulence mechanism are synchronized during Campylobacter pathogenesis (Wieczorek et al., 2015; Zhang et al., 2016; Frazão et al., 2017; Rossler et al., 2020).
The Campylobacter adhesion to fibronectin F (cadF) gene, encoding an adhesin and fibronectin-binding protein that involves in the process of invasion and influences the microfilament organization in host cells (Zhang et al., 2016), was also detected in most of our isolates (Table 4). Similar observations have been reported that the high frequency of the cadF gene in Campylobacter species was detected from poultry productions and processing operations (Rozynek et al., 2005; Frazão et al., 2017; Rossler et al., 2020). Additionally, Ziprin et al. (1999) demonstrated that Campylobacter cadF-negative strains are not able to colonize in chicken gastrointestinal tract. Therefore, the cadF gene product may play a similar function in human pathogenesis and causing disease.
The Guillain-Barré syndrome associated gene (wlaN) and its gene product have ganglioside-like structures and is responsible for specific lipooligosaccharides (LOS) synthesis (Hermans et al., 2011). This LOS synthesis is thought to be involved in the development of Guillain-Barré and Miller-Fischer syndromes after C. jejuni infection (Gilbert et al., 2000; Linton et al., 2000). The presence of this gene in Campylobacter may increase the risk for suffering post-neurological conditions. Our findings indicate its presence in 36.1% of the total thermophilic Campylobacter isolates, which is in line with many previous studies in conventional poultry management systems (Talukder et al., 2008; Koolman et al., 2015; Wieczorek et al., 2018).
Campylobacter toxins are important virulence marker determinants. One of the toxin groups is the cytolethal distending cytotoxins, which are encoded by the cdt genes and form polycistronic cdt operons. The gene products include CdtA, CdtB, and CdtC cytotoxins, which are toxic to host enterocytes (Carvalho et al., 2013). These cytotoxins play important roles in development of diarrhea by interfering with the proliferation and differentiation of intestinal crypt cells (Scuron et al., 2016). The three subunits are required for the full activity of the toxins (Lapierre et al., 2016). CdtB displays enzymatic Dnase activity resulting in cell-cycle arrest and cell death, while CdtA and CdtC are responsible for the translocation of CdtB across the target cell membrane (Lara-Tejero and Galan, 2001). In this study, these toxin genes were detected in majority of C. jejuni isolates (98.7–97.5%).
Campylobacter survival in the digestive tract is highly dependent on the ciaB gene. This gene can secret a CiaB protein that is responsible for the invasion and colonization of this microorganism in chicken intestines (Hermans et al., 2011). Among Campylobacter isolates, it was found in a frequency of 80.4%. Similarly, in conventionally reared broilers, the ciaB gene was detected in a similar prevalence by Raeisi et al. (2017) and Wieczorek et al. (2018). Because the ciaB gene is important in the early stages of colonization, their removal may causes bacterial failure to survive the stress of passage through the gut followed by colonization failure (Ziprin et al., 2001). Additionally, regulatory protein R (racR) gene and its gene product regulate temperature during growth and colonization of this microorganism in the hosts. The prevalence of racR in our study is 83.5%, which is similar to that reported in conventional broiler flocks by Datta et al. (2003) and Talukder et al. (2008); however, Hanning et al. (2012) demonstrated a lower racR prevalence rate (34%) in a pasture-raised broiler flock study.
The enterochelin binding lipoprotein encoded by siderophore transport (ceuE), which has an important role in virulence and regulation of the siderophore transport system (Hermans et al., 2011), was detected in 72.2% of the Campylobacter isolates.
The use of antibiotics, either overuse or abuse, in food animals contributes to the establishment of antimicrobial resistance (AMR) in commensal and zoonotic enteric bacteria (Varga et al., 2009; van Boeckel et al., 2015). To prevent the spread of AMR Campylobacter through the food chain, it is critical to continuously monitor its antimicrobial resistance and resistance mechanisms. In this study, five (5.2%) Campylobacter isolates were resistant to quinolones and fluroquinolones [nalidixic acid (n = 4) and ciprofloxacin (n = 1)]. The lower resistance of quinolones and fluoroquinolone-resistant isolates in this study may be related to that the U.S. Food and Drug Administration (FDA) banned the use of fluoroquinolones in poultry production in the United States in 2005 (Griggs et al., 2005). However, other studies argued that the FDA’s restriction on fluoroquinolones in chicken production may not be enough to mitigate the resistant Campylobacter in poultry products, because fluoroquinolone-resistant Campylobacter was found in persistent pollutants of poultry products even after discontinuous on-farm fluoroquinolone use (Price et al., 2007). Monitoring the prevalence of resistant strains in chicken flocks, production facilities, consumer poultry products, and human diseases is therefore crucial in order to accurately evaluate the effectiveness of this policy. The low frequency of resistance to quinolones and fluoroquinolones in this study may also related to the fact that antibiotics were not utilized by any farms during the duration of this study. Luangtongkum et al. (2006) reported a significant difference between antimicrobial resistance rates of <2% vs. 46–67% in organic and conventional raised poultry farms, respectively.
The Campylobacter isolates that displayed resistant to quinolones and fluroquinolones [ciprofloxacin (MIC = 8 μg/mL) and nalidixic acid (MIC = 64 μg/mL), respectively] were further examined for the presence of the most common mutation site. A point mutation at position 86 leading to threonine replacement by isoleucine was detected in the QRDR of the gyrA gene from our isolates. A MAMA-PCR was used to determine the presence of this type of mutation (Zirnstein et al., 1999; Payot et al., 2004). In this protocol, a conserved forward primer, CampyMAMAgyrA1, and a reverse mutation detection primer, CampyMAMAgyrA5 were used to generate a 265-bp PCR product, indicating the presence of the Thr-86-Ile (ACA to ATA) mutation in the C. jejuni gyrA gene. This method was used as an alternative to nucleotide sequencing because it is not accessible in ordinary microbiology laboratories. Our results revealed that this mutation was found among all phenotypically resistant isolates. In contrast, this mutation was found to be absent in some quinolone resistance isolates, leading researchers to speculate that it could be linked to alternative resistance mechanisms (Bolton et al., 2013; Elhadidy et al., 2018; Yeh et al., 2022).
Emergence of resistance to erythromycin by Campylobacter isolates has been reported (Deng et al., 2015; Liu et al., 2019; Jehanne et al., 2021). Resistance of Campylobacter to this macrolide is chromosomally mediated, most commonly due to a shift in the target site on the 23S rRNA subunit. These mutations have been identified at locations of 2074 and 2075 (Vacher et al., 2003). The transitory mutation A2075G is the most prevalent among erythromycin-resistant Campylobacter isolates, while the A2074C mutation is less identified among the resistant strains (Vacher et al., 2003). In erythromycin resistant isolates in this study (MIC >64 μg/mL), A2075G was detected in one isolate, while A2074G could not be identified from any isolate. Additionally, ermB was found in three out of nine erythromycin-resistant isolates, while Elhadidy et al. (2020) could not identify ermB gene from any erythromycin-resistant Campylobacter isolates. These findings on the molecular basis of macrolide resistance in Campylobacter revealed the importance of additional resistance mechanisms in Campylobacter encoding erythromycin resistance such as CmeABC is a multi-drug efflux pump system broadly distributed in Campylobacter, representing an important mechanism for antibiotic resistance (Lin et al., 2002).
Interestingly, one isolate in this study was gentamicin resistant (MIC>32 μg/mL). This result is congruent with that of Luangtongkum et al. (2006), who reported that none of the Campylobacter species isolated from conventionally or organically raised broilers were gentamicin resistant. Similarly, Giacomelli et al. (2014) and Elhadidy et al. (2018) could not identify gentamicin resistant isolates among Campylobacter isolates from poultry in Italy and Belgium, respectively. On the other hand, Saenz et al. (2000) found a 25% prevalent rate of gentamicin resistant isolates from broilers in Spain. The common low prevalence of gentamicin resistance may contribute to few usages of this antibiotic during the poultry production (Saenz et al., 2000; Roth et al., 2019).
Our results showed the high prevalence of tetracycline resistance among the isolates (MIC >64 μg/mL) (Table 3). These findings are also reported by other researchers from Kenya, Finland, Iraq, Poland, and USA (Luangtongkum et al., 2006; Nguyen et al., 2016; Pohjola et al., 2016; Wieczorek et al., 2018; Shakir et al., 2021) where C. jejuni and C. coli were isolated from small scale and backyard chicken flocks. In addition, Bailey et al. (2019) found that tetracycline resistances in organic farms were more common than the conventional farms. The reports have demonstrated that the plasmid-encoded tet (O) gene is responsible for tetracycline resistance in Campylobacter (Gibreel et al., 2004; Wozniak-Biel et al., 2018; Elhadidy et al., 2019), and this gene can be horizontally transferred between C. jejuni and C. coli isolates in the intestines of food animals and humans (Kim et al., 2010). Interestingly, the presence of phenotypic tetracycline resistant isolates that did not harbored tet (O) gene may be related to the genetic inactivated of efflux pumps (Jeon et al., 2011). The high rates of resistance reported for tetracycline could be attributed to the overuse during the poultry production (Giacomelli et al., 2014).
In conclusion, MLST analysis showed high genetic diversity among both C. jejuni and C. coli isolates. The identified STs were reported also as the most common STs identified from human samples in various geographic regions. These results highlight the importance of poultry sources for human campylobacteriosis. Additionally, Campylobacter isolated from pasture-raised poultry flocks from this study were generally consistent with Campylobacter previously isolated from conventionally reared broiler flocks in regard to ST prevalence and diversity, antibiotic resistance patters, and virulence. Thus, in terms of public health risk of campylobacteriosis, these results indicate that pasture-raised poultry products appear to be equivalent conventionally reared products, but still represents a potential zoonotic source of Campylobacter that requires further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
AA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. H-YY: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft, Writing – review & editing. HR: Data curation, Investigation, Methodology, Writing – review & editing. MR: Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. AA, an Egyptian Junior Scientist, was supported by the U.S. – Egypt Science and Technology Joint Fund, Cairo, Egypt. This study was supported by the USDA Agricultural Research Service CRIS Project No. 6040-32000-071-00D.
Acknowledgments
We thank Susan Q. Brooks and Manju Amin of Poultry Microbiological Safety and Processing Research Unit, U.S. National Poultry Research Center, Agricultural Research Service, U.S. Department of Agriculture, Athens, GA, United States for the technical supports. AA, an Egyptian Junior Scientist, was supported by the U.S. – Egypt Science and Technology Joint Fund, Cairo, Egypt. This study was supported by the USDA Agricultural Research Service CRIS Project No. 6040-32000-071-00D. Mention of trade names or commercial products in this paper is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture, which is an equal opportunity provider and employer.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1271551/full#supplementary-material
Footnotes
References
Aarestrup, F. M., McDermott, P. F., and Wegener, H. C. (2008). “Transmission of antibiotic resistance from food animals to humans” in Campylobacter. eds. I. Nachamkin, C. M. Szymanski, and M. J. Blaser (Washington, D. C.: ASM Press), 645–665.
Alonso, R., Mateo, E., Churruca, E., Martinez, I., Girbau, C., and Fernández-Astorga, A. (2005). MAMA-PCR assay for the detection of point mutations associated with high-level erythromycin resistance in Campylobacter jejuni and Campylobacter coli strains. J. Microbiol. Methods 63, 99–103. doi: 10.1016/j.mimet.2005.03.013
Bacon, D. J., Alm, R. A., Burr, D. H., Hu, L., Kopecko, D. J., Ewing, C. P., et al. (2000). Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68, 4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000
Bailey, M. A., Taylor, R. M., Brar, J. S., Corkran, S. C., Velásquez, C., Novoa Rama, E., et al. (2019). Prevalence and antimicrobial resistance of Campylobacter from antibiotic-free broilers during organic and conventional processing. Poult. Sci. 98, 1447–1454. doi: 10.3382/ps/pey486
Bang, D. D., Moller Nielsen, E., Scheutz, F., Pedersen, K., Handberg, K., and Madsen, M. (2003). PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J. Appl. Microbiol. 94, 1003–1014. doi: 10.1046/j.1365-2672.2003.01926.x
Berg, C. (2001). Health and welfare in organic poultry production. Acta Vet. Scand. 43, 1–9. doi: 10.1186/1751-0147-43-S1-S37
Bolton, D. J. (2015). Campylobacter virulence and survival factors. Food Microbiol. 48, 99–108. doi: 10.1016/j.fm.2014.11.017
Bolton, D., Patriarchi, A., Fox, Á., and Fanning, S. (2013). A study of the molecular basis of quinolone and macrolide resistance in a selection of Campylobacter isolates from intensive poultry flocks. Food Control 30, 222–226. doi: 10.1016/j.foodcont.2012.06.044
Bull, S. A., Allen, V. M., Domingue, G., Jørgensen, F., Frost, J. A., Ure, R., et al. (2006). Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl. Environ. Microbiol. 72, 645–652. doi: 10.1128/AEM.72.1.645-652.2006
Caner, V., Cokal, Y., Cetin, C., Sen, A., and Karagenc, N. (2008). The detection of hipO gene by real-time PCR in thermophilic Campylobacter spp. with very weak and negative reaction of hippurate hydrolysis. Antonie Van Leeuwenhoek. 94, 527–532. doi: 10.1007/s10482-008-9269-4
Carvalho, A. F., da Silva, D. M., Azevedo, S. S., Piatti, R. M., Genovez, M. E., and Scarcelli, E. (2013). Detection of CDT toxin genes in Campylobacter spp. strains isolated from broiler carcasses and vegetables in São Paulo, Brazil. Braz. J. Microbiol. 44, 693–699. doi: 10.1590/s1517-83822013000300005
Carvalho, A. C. T., Ruiz-Palacios, G. M., Ramos-Cervantes, P., Cervantes, L.-E., Jiang, X., and Oickering, L. K. (2001). Molecular characterization of invasive and noninvasive Campylobacter jejuni and Campylobacter coli isolates. J. Clin. Microbiol. 39, 1353–1359. doi: 10.1128/jcm.39.4.1353-1359.2001
Casabonne, C., Gonzalez, A., Aquili, V., Subils, T., and Balague, C. (2016). Prevalence of seven virulence genes of Campylobacter jejuni isolated from patients with diarrhea in Rosario, Argentina. Int. J. Inf. Secur. 3:e37727. doi: 10.17795/iji-37727
Centers for Disease Control and Prevention. (2022). Campylobacter. Available at: https://www.cdc.gov/campylobacter/faq.html (Accessed October 1, 2023).
Clinical and Laboratory Standards Institute (CLSI) (2015) in M45: methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. eds. J. A. Hindler and S. S. Richter. 3rd ed
Colles, F. M., Jones, T. A., McCarthy, N. D., Sheppard, S. K., Cody, A. J., Dingle, K. E., et al. (2008). Campylobacter infection of broiler chickens in a free-range environment. Environ. Microbiol. 10, 2042–2050. doi: 10.1111/j.1462-2920.2008.01623.x
Colles, F. M., McCarthy, N. D., Sheppard, S. K., Layton, R., and Maiden, M. C. J. (2010). Comparison of Campylobacter populations isolated from a free-range broiler flock before and after slaughter. Int. J. Food Microbiol. 137, 259–264. doi: 10.1016/j.ijfoodmicro.2009.12.021
Conner, D. S., Campbell-Arvai, V., and Hamm, M. W. (2008). Consumer preferences for pasture-raised animal products: results from Michigan. J. Food Distrib. Res. 39, 12–25. doi: 10.22004/ag.econ.55972
Datta, S., Niwa, H., and Itoh, K. (2003). Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 52, 345–348. doi: 10.1099/jmm.0.05056-0
de Haan, C. P., Kivisto, R., Hakkinen, M., Rautelin, H., and Hanninen, M. L. (2010). Decreasing trend of overlapping multilocus sequence types between human and chicken Campylobacter jejuni isolates over a decade in Finland. Appl. Environ. Microbiol. 76, 5228–5236. doi: 10.1128/AEM.00581-10
Deng, F., Shen, J., Zhang, M., Wu, C., Zhang, Q., and Wang, Y. (2015). Constitutive and inducible expression of the rRNA methylase gene erm (B) in Campylobacter. Antimicrob. Agents Chemother. 59, 6661–6664. doi: 10.1128/AAC.01103-15
Dingle, K. E., Colles, F. M., Ure, R., Wagenaar, J. A., Duim, B., Bolton, F. J., et al. (2002). Molecular characterization of Campylobacter jejune clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8, 949–955. doi: 10.3201/eid0809.02-0122
Dingle, K. E., Van Den Braak, N., Colles, F. M., Price, L. J., Woodward, D. L., Rodgers, F. G., et al. (2001). Sequence typing confirms that Campylobacter jejuni strains associated with Guillain-Barre and Miller-fisher syndromes are of diverse genetic lineage, serotype, and flagella type. J. Clin. Microbiol. 39, 3346–3349. doi: 10.1128/JCM.39.3.3346-3349.2001
Elhadidy, M., Ali, M. M., El-Shibiny, A., Miller, W. G., Elkhatib, W. F., Botteldoorn, N., et al. (2020). Antimicrobial resistance patterns and molecular resistance markers of Campylobacter jejuni isolates from human diarrheal cases. PLoS One 15:e0227833. doi: 10.1371/journal.pone.0227833
Elhadidy, M., Miller, W. G., Arguello, H., Álvarez-Ordóñez, A., Dierick, K., and Botteldoorn, N. (2019). Molecular epidemiology and antimicrobial resistance mechanisms of Campylobacter coli from diarrhoeal patients and broiler carcasses in Belgium. Transbound. Emerg. Dis. 66, 463–475. doi: 10.1111/tbed.13046
Elhadidy, M., Miller, W. G., Arguello, H., Álvarez-Ordóñez, A., Duarte, A., Dierick, K., et al. (2018). Genetic basis and clonal population structure of antibiotic resistance in Campylobacter jejuni isolated from broiler carcasses in Belgium. Front. Microbiol. 9:1014. doi: 10.3389/fmicb.2018.01014
European Food Safety Authority (2021). The European Union one health 2019 zoonoses report. EFSA J. 19:e06406. doi: 10.2903/j.efsa.2021.6406
Frazão, M. R., Medeiros, M., Duque, S., and Falcão, J. P. (2017). Pathogenic potential and genotypic diversity of Campylobacter jejuni: a neglected food-borne pathogen in Brazil. J. Med. Microbiol. 66, 350–359. doi: 10.1099/jmm.0.000424
Ghareeb, K., Awad, W. A., Mohnl, M., Schatzmayr, G., and Boehm, J. (2013). Control strategies for Campylobacter infection in poultry production. Worlds Poult. Sci. J. 69, 57–76. doi: 10.1017/S0043933913000068
Giacomelli, M., Salata, C., Martini, M., Montesissa, C., and Piccirillo, A. (2014). Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli from poultry in Italy. Microb. Drug Resist. 20, 181–188. doi: 10.1089/mdr.2013.0110
Gibreel, A., Tracz, D. M., Nonaka, L., Ngo, T. M., Connell, S. R., and Taylor, D. E. (2004). Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet (O)-mediated tetracycline resistance. Antimicrob. Agents Chemother. 48, 3442–3450. doi: 10.1128/AAC.48.9.3442-3450.2004
Gilbert, M., Brisson, J.-R., Karwaski, M.-F., Michniewicz, J., Cunningham, A.-M., Wu, Y., et al. (2000). Biosynthesis of ganglioside mimics in Campylobacter jejuni OH 4384: identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-MHz 1H and 13C NMR analysis. J. Biol. Chem. 275, 3896–3906. doi: 10.1074/jbc.275.6.3896
Gilbreath, J. J., Cody, W. L., Merrell, D. S., and Hendrixson, D. R. (2011). Change is good: variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter. Microbiol. Mol. Biol. Rev. 75, 84–132. doi: 10.1128/MMBR.00035-10
Griggs, D. J., Johnson, M. M., Frost, J. A., Humphrey, T., Jorgensen, F., and Piddock, L. J. V. (2005). Incidence and mechanism of ciprofloxacin resistance in Campylobacter spp. isolated from commercial chicken flocks in the United Kingdom before, during and after fluoroquinolone treatment. Antimicrob. Agents Chemother. 49, 699–707. doi: 10.1128/AAC.49.2.699-707.2005
Hanning, I., Biswas, D., Herrera, P., Roesler, M., and Ricke, S. C. (2012). Prevalence and characterization of Campylobacter jejuni isolated from pasture flock poultry. J. Food Sci. 75, M496–M502. doi: 10.1111/j.1750-3841.2010.01747.x
Harvala, H., Rosendal, T., Lahti, E., Engvall, E. O., Brytting, M., Wallensten, A., et al. (2016). Epidemiology of Campylobacter jejuni infections in Sweden, November 2011–October 2012: is the severity of infection associated with C. jejuni sequence type? Infect. Ecol. Epidemiol. 6:31079. doi: 10.3402/iee.v6.31079
Hermans, D., Van Deun, K., Martel, A., Immerseel, F., Messens, W., Heyndrickx, M., et al. (2011). Colonization factors of Campylobacter jejuni in the chicken gut. Vet. Res. 42:82. doi: 10.1186/1297-9716-42-82
Hickey, T. E., McVeigh, A. L., Scott, D. A., Michieluth, R. E., Bixby, A., Carroll, S. A., et al. (2000). Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect. Immun. 68, 6535–6541. doi: 10.1128/iai.68.12.6535-6541.2000
Hiett, K. L., Stintzi, A., Andacht, T. M., Kuntz, R. L., and Seal, B. S. (2008). Genomic differences between Campylobacter jejuni isolates identify surface membrane and flagellar function gene products potentially important for colonizing the chicken intestine. Funct. Integr. Genomics 8, 407–420. doi: 10.1007/s10142-008-0087-6
Jehanne, Q., Bénéjat, L., Ducournau, A., Domingues-Martins, C., Cousinou, T., Bessède, E., et al. (2021). Emergence of erythromycin resistance methyltransferases in Campylobacter coli strains in France. Antimicrob. Agents Chemother. 65:e0112421. doi: 10.1128/AAC.01124-21
Jeon, B., Wang, Y., Hao, H., Barton, Y. W., and Zhang, Q. (2011). Contribution of Cme G to antibiotic and oxidative stress resistance in Campylobacter jejuni. J. Antimicrob. Chemother. 66, 79–85. doi: 10.1093/jac/dkq418
Kaakoush, N. O., Castaño-Rodríguez, N., Mitchell, H. M., and Man, S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28, 687–720. doi: 10.1128/CMR.00006-15
Kim, J. M., Hong, J., Bae, W., Koo, H. C., Kim, S. H., and Park, Y. H. (2010). Prevalence, antibiograms, and transferable tet (O) plasmid of Campylobacter jejuni and Campylobacter coli isolated from raw chicken, pork, and human clinical cases in Korea. J. Food Prot. 73, 1430–1437. doi: 10.4315/0362-028X-73.8.1430
Konkel, M. E., Garvis, S. G., Tipton, S. L., Anderson, Jr., D. E., and Cieplak, Jr., W. (1997). Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol. Microbiol. 24, 953–963. doi: 10.1046/j.1365-2958.1997.4031771.x
Koolman, L., Whyte, P., Burgess, C., and Bolton, D. (2015). Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 12, 424–432. doi: 10.1089/fpd.2014.1883
Lapierre, L., Gatica, M. A., Riquelme, V., Vergara, C., Yañez, J. M., San Martin, B., et al. (2016). Characterization of antimicrobial susceptibility and its association with virulence genes related to adherence, invasion, and cytotoxicity in Campylobacter jejuni and Campylobacter coli isolates from animals, meat, and humans. Microb. Drug Resist. 22, 432–444. doi: 10.1089/mdr.2015.0055
Lara-Tejero, M., and Galan, J. E. (2001). CdtA, CdtB, and CdtC form a tripartite complex that is required for cytolethal distending toxin activity. Infect. Immun. 69, 4358–4365. doi: 10.1128/IAI.69.7.4358-4365.2001
Lin, J., Michel, L. O., and Zhang, Q. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46, 2124–2131. doi: 10.1128/aac.46.7.2124-2131.2002
Lindmark, H., Diedrich, I. C., Andersson, L., Lindqvist, R., and Engvall, E. O. (2006). Distribution of Campylobacter genotypes on broilers during slaughter. J. Food Prot. 69, 2902–2907. doi: 10.4315/0362-028x-69.12.2902
Linton, D., Gilbert, M., Hitchen, P. G., Dell, A., Morris, H. R., Wakarchuk, A. A., et al. (2000). Phase variation of a beta-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 37, 501–514. doi: 10.1046/j.1365-2958.2000.02020.x
Linton, D., Lawson, A. J., Owen, R. J., and Stanley, J. (1997). PCR detection, Identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35, 2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997
Liu, D., Liu, W., Lv, Z., Xia, J., Li, X., Hao, Y., et al. (2019). Emerging erm (B)-mediated macrolide resistance associated with novel multidrug resistance genomic islands in Campylobacter. Antimicrob. Agents Chemother. 63, e00153–e00119. doi: 10.1128/AAC.00153-19
Luangtongkum, T., Morishita, T. Y., Ison, A. J., Huang, S., McDermott, P. F., and Zhang, Q. (2006). Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl. Environ. Microbiol. 72, 3600–3607. doi: 10.1128/AEM.72.5.3600-3607.2006
Ma, L., Wang, Y., Shen, J., Zhang, Q., and Wu, C. (2014). Tracking Campylobacter contamination along a broiler chicken production chain from the farm level to retail in China. Int. J. Food Microbiol. 181, 77–84. doi: 10.1016/j.ijfoodmicro.2014.04.023
Müller, J., Schulze, F., Muller, W., and Hanel, I. (2006). PCR detection of virulence-associated genes in Campylobacter jejuni strains with differential ability to invade Caco-2 cells and to colonize the chick gut. Vet. Microbiol. 113, 123–129. doi: 10.1016/j.vetmic.2005.10.029
Nguyen, T. N., Hotzel, H., Njeru, J., Mwituria, J., El-Adawy, H., Tomaso, H., et al. (2016). Antimicrobial resistance of Campylobacter isolates from small scale and backyard chicken in Kenya. Gut Pathog. 8:39. doi: 10.1186/s13099-016-0121-5
Payot, S., Avrain, L., Magras, C., Praud, K., Cloeckaert, A., and Chaslus-Dancla, E. (2004). Relative contribution of target gene mutation and efflux to fluoroquinolone and erythromycin resistance, in French poultry and pig isolates of Campylobacter coli. Int. J. Antimicrob. Agents 23, 468–472. doi: 10.1016/j.ijantimicag.2003.12.008
Pohjola, L., Nykäsenoja, S., Kivistö, R., Soveri, T., Huovilainen, A., Hänninen, M. L., et al. (2016). Zoonotic public health hazards in backyard chickens. Zoonoses Public Health 63, 420–430. doi: 10.1111/zph.12247
Price, L. B., Lackey, L. G., Vailes, R., and Silbergeld, E. (2007). The persistence of fluoroquinolone-resistant Campylobacter in poultry production. Environ. Health Perspect. 115, 1035–1039. doi: 10.1289/ehp.10050
Raeisi, M., Khoshbakht, R., Ghaemi, E. A., Bayani, M., Hashemi, M., Seyedghasemi, N. S., et al. (2017). Antimicrobial resistance and virulence-associated genes of Campylobacter spp. isolated from raw milk, fish, poultry, and red meat. Microb. Drug Resist. 23, 925–933. doi: 10.1089/mdr.2016.0183
Rossler, E., Olivero, C., Soto, L. P., Frizzo, L. S., Zimmermann, J., Rosmini, M. R., et al. (2020). Prevalence, genotypic diversity and detection of virulence genes in thermotolerant Campylobacter at different stages of the poultry meat supply chain. Int. J. Food Microbiol. 326:108641. doi: 10.1016/j.ijfoodmicro.2020.108641
Roth, N., Käsbohrer, A., Mayrhofer, S., Zitz, U., Hofacre, C., and Domig, K. J. (2019). The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: a global overview. Poult. Sci. 98, 1791–1804. doi: 10.3382/ps/pey539
Rothrock, M. J., Hiett, K. L., Guard, J. Y., and Jackson, C. R. (2016). Antibiotic resistance patterns of major zoonotic pathogens from all-natural, antibiotic-free, pasture-raised broiler flocks in the southeastern United States. J. Environ. Qual. 45, 593–603. doi: 10.2134/jeq2015.07.0366
Rozynek, E., Dzierzanowska-Fangrat, K., Jozwiak, P., Popowski, J., Korsak, D., and Dzierzanowska, D. (2005). Prevalence of potential virulence markers in polish Campylobacter jejuni and Campylobacter coli isolates obtained from hospitalized children and from chicken carcasses. J. Med. Microbiol. 54, 615–619. doi: 10.1099/jmm.0.45988-0
Saenz, Y., Zarazaga, M., Lantero, M., Gastanares, M. J., Baquero, F., and Torres, C. (2000). Antimicrobial resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997-1998. Antimicrob. Agents Chemother. 44, 267–271. doi: 10.1128/AAC.44.2.267-271.2000
Scuron, M. D., Boesze-Battaglia, K., Dlakićcy, M., and Shenker, B. J. (2016). The cytolethal distending toxin contributes to microbial virulence and disease pathogenesis by acting as a tri-perditious toxin. Front. Cell. Infect. Microbiol. 6:168. doi: 10.3389/fcimb.2016.00168
Shakir, Z. M., Alhatami, A. O., Ismail Khudhair, Y., and Muhsen Abdulwahab, H. (2021). Antibiotic resistance profile and multiple antibiotic resistance index of Campylobacter species isolated from poultry. Arch. Razi Inst. 76, 1677–1686. doi: 10.22092/ari.2021.356400.1837
Sher, A. A., Ashraf, M. A., Mustafa, B. E., and Raza, M. M. (2021). Epidemiological trends of foodborne Campylobacter outbreaks in the United States of America, 1998–2016. Food Microbiol. 97:103751. doi: 10.1016/j.fm.2021.103751
Skirrow, M. B., and Blaser, M. J. (2000). “Clinical aspects of Campylobacter infection” in Campylobacter. eds. I. Nachamkin and M. J. Blaser. 2nd ed (Washington, D. C.: ASM Press), 69–88.
Smid, J. H., Mughini Gras, L., de Boer, A. G., French, N. P., Havelaar, A. H., Wagenaar, J. A., et al. (2013). Practicalities of using non-local or non-recent multilocus sequence typing data for source attribution in space and time of human campylobacteriosis. PLoS One 8:e55029. doi: 10.1371/journal.pone.0055029
Stampa, E., Schipmann-Schwarze, C., and Hamm, U. (2020). Consumer perceptions, preferences, and behavior regarding pasture-raised livestock products: a review. Food Qual. Prefer. 82:103872. doi: 10.1016/j.foodqual.2020.103872
Stern, N. J., Wojton, B., and Kwiatek, K. A. (1992). Differential-selective medium and dry ice-generated atmosphere for recovery of Campylobacter jejuni. J. Food Prot. 55, 514–517. doi: 10.4315/0362-028X-55.7.514
Suzuki, H., and Yamamoto, S. (2009). Campylobacter contamination in retail poultry meats and by-products in the world: a literature survey. J. Vet. Med. Sci. 71, 255–261. doi: 10.1292/jvms.71.255
Talukder, K., Aslam, M., Islam, Z., Azmin, I., and Duttad, D. (2008). Prevalence of virulence genes and cytolethal distending toxin production in Campylobacter jejuni isolates from diarrheal patients in Bangladesh. J. Clin. Microbiol. 46, 1485–1488. doi: 10.1128/JCM.01912-07
Vacher, S., Me Nard, A., Bernard, E., and Me Graud, F. (2003). PCR restriction fragment length polymorphism analysis for detection of point mutations associated with macrolide resistance in Campylobacter spp. Antimicrob. Agents Chemother. 47, 1125–1128. doi: 10.1128/AAC.47.3.1125-1128.2003
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U. S. A. 112, 5649–5654. doi: 10.1073/pnas.1503141112
Varga, C., Rajić, A., McFall, M. E., Reid-Smith, R. J., and McEwen, S. A. (2009). Associations among antimicrobial use and antimicrobial resistance of Salmonella spp. isolates from 60 Alberta finishing swine farms. Foodborne Pathog. Dis. 6, 23–31. doi: 10.1089/fpd.2008.0118
Vetchapitak, T., and Misawa, N. (2019). Current status of Campylobacter food poisoning in Japan. Food Saf. (Tokyo) 7, 61–73. doi: 10.14252/foodsafetyfscj.D-19-00001
Wieczorek, K., Denis, E., Lynch, O., and Osek, J. (2013). Molecular characterization and antibiotic resistance profiling of Campylobacter isolated from cattle in polish slaughterhouses. Food Microbiol. 34, 130–136. doi: 10.1016/j.fm.2012.12.003
Wieczorek, K., Denis, E., and Osek, J. (2015). Comparative analysis of antimicrobial resistance and genetic diversity of Campylobacter from broilers slaughtered in Poland. Int. J. Food Microbiol. 210, 24–32. doi: 10.1016/j.ijfoodmicro.2015.06.006
Wieczorek, K., Wołkowicz, T., and Osek, J. (2018). Antimicrobial resistance and virulence-associated traits of Campylobacter jejuni isolated from poultry food chain and humans with diarrhea. Front. Microbiol. 9:1508. doi: 10.3389/fmicb.2018.01508
Wozniak-Biel, A., Bugla-Płoskonska, G., Kielsznia, A., Korzekwa, K., Tobiasz, A., Korzeniowska-Kowal, A., et al. (2018). High prevalence of resistance to fluoroquinolones and tetracycline Campylobacter spp. isolated from poultry in Poland. Microb. Drug Resist. 24, 314–322. doi: 10.1089/mdr.2016.0249
Yeh, H., Cox, N. A., Hinton, A. Jr., Berrang, M. E., Plumblee-Lawrence, J. R., and Thompson, T. M. (2022). Prevalence and characterization of quinolone resistance in Campylobacter spp. isolates in chicken livers from retail stores in Georgia, USA. J. Food Prot. 85, 406–413. doi: 10.4315/JFP-21-357
Yeh, H., Hiett, K. L., Line, J. E., Oakley, B. B., and Seal, B. S. (2013). Construction, expression, purification and antigenicity of recombinant Campylobacter jejuni flagellar proteins. Microbiol. Res. 168, 192–198. doi: 10.1016/j.micres.2012.11.010
Yeung, R. M. W., and Morris, J. (2001). Consumer perception of food risk in chicken meat. Nutr. Food Sci. 31, 270–279. doi: 10.1108/00346650110409092
Zhang, T., Luo, Q., Chen, Y., Li, T., Wen, G., Zhang, R., et al. (2016). Molecular epidemiology, virulence determinants and antimicrobial resistance of Campylobacter spreading in retail chicken meat in central China. Gut Pathog. 8:48. doi: 10.1186/s13099-016-0132-2
Zhou, J., Zhang, M., Yang, W., Fang, Y., Wang, G., and Hou, F. (2016). A seventeen-year observation of the antimicrobial susceptibility of clinical Campylobacter jejuni and the molecular mechanisms of erythromycin-resistant isolates in Beijing, China. Int. J. Infect. Dis. 42, 28–33. doi: 10.1016/j.ijid.2015.11.005
Ziprin, R. L., Young, C. R., Byrd, J. A., Stanker, L. H., Hume, M. E., Gray, S. A., et al. (2001). Role of Campylobacter jejuni potential virulence genes in cecal colonization. Avian Dis. 45, 549–557. doi: 10.2307/1592894
Ziprin, R. L., Young, C. R., Stanker, L. H., Hume, M. E., and Konkel, M. E. (1999). The absence of coecal colonization of chicks by a mutant of Campylobacter jejuni not expressing bacterial fibronectin-binding protein. Avian Dis. 43, 586–589. doi: 10.2307/1592660
Zirnstein, G., Li, Y., Swaminathan, B., and Angulo, F. (1999). Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J. Clin. Microbiol. 37, 3276–3280. doi: 10.1128/JCM.37.10.3276-3280.1999
Keywords: Campylobacter, pasture-raised poultry, MLST, antimicrobial resistance, virulence
Citation: Awad A, Yeh H-Y, Ramadan H and Rothrock MJ (2023) Genotypic characterization, antimicrobial susceptibility and virulence determinants of Campylobacter jejuni and Campylobacter coli isolated from pastured poultry farms. Front. Microbiol. 14:1271551. doi: 10.3389/fmicb.2023.1271551
Edited by:
Beatrix Stessl, University of Veterinary Medicine Vienna, AustriaReviewed by:
Hosny El-Adawy, Friedrich Loeffler Institut, GermanyIgori Balta, University of Life Sciences "King Mihai I", Romania
Copyright © 2023 Awad, Yeh, Ramadan and Rothrock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hung-Yueh Yeh, hungyueh.yeh@usda.gov
 Amal Awad
Amal Awad Hung-Yueh Yeh
Hung-Yueh Yeh Hazem Ramadan
Hazem Ramadan Michael J. Rothrock2
Michael J. Rothrock2