- 1Key Laboratory of Protection and Utilization of Biological Resources in Tarim Basin of Xinjiang, Production & Construction Corps, Alar City, China
- 2School of Life Science and Technology, Tarim University, Alar, China
By the end of 2021, the pear yield in Xinjiang reached 1,795,900 tons, accounting for 1/9 of the country. Pear black spot, caused by Alternaria gaisen disease, has had a significant impact on the pear industry. A. gaisen can infect nearly all pear plants, resulting in black spots on the fruit that negatively affect both yield and quality. This study focused on the TRM76323 strain of Streptomyces, which was isolated from the soil of Tamarix chinensis in Xinjiang Province. Through a multiphase classification and identification method, the genetic classification status of the antagonistic strains was determined. The study also identified the antibacterial active components of streptochlorin using modern isolation and purification techniques. The antagonistic activity of Streptomyces against Alternaria was analyzed through in vitro and in vivo experiments. This research not only expanded the resource bank of antagonistic microorganisms in extreme environments in Xinjiang, but also identified active components that could contribute to the development of new drug lead compounds. Additionally, this study presents a novel approach for the prevention and control of pear black spot disease.
Introduction
Pear is an important fruit tree across the globe. However, Alternaria gaisen disease, known as pear disease, seriously causes a substantial loss in the pear industry. Alternaria species are a group of fungi that can cause plant diseases. These fungi, which usually exist in plant residues and living plants, are very harmful to crops. Specifically, they can cause pre- and post-harvest diseases in wheat, corn, celery, pepper, and other vegetables and fruits, including Aksu apples, red dates, and Korla fragrant pears, resulting in considerable crop losses (Chauhan et al., 2009). Currently, the main method for controlling A. gaisen involves the application of chemical agents; physical and chemical methods are not effective in controlling the spread of A. gaisen disease. Therefore, the prevention and control of microbial-derived biological agents is receiving increasing attention (Dale et al., 2017).
Actinobacteria produce large amounts of bioactive secondary metabolites (Jose et al., 2021). Streptomyces was first proposed in the early 1940s (Waksman and Henrici, 1943). So far, the genus Streptomyces includes a large number of well-described new species and has the ability to produce strong antibacterial properties (Brown et al., 1976; Ser et al., 2016). They have a wide variety of activities and are commonly used. In addition to their bactericidal, bacteriostatic, anti-infective, and other effects, their properties also enable them to be used as insecticides, anti-tumor agents, herbicides, and immunosuppressants. They are extremely important for various industries, including medicine, health and agriculture while also being applicable for environmental protection (Waksman et al., 1944; Ser et al., 2016). Phuakjaiphaeo et al. (2016) identified antifungal compounds from endophytic Streptomyces CEN26 against Alternaria. Sharma and Manhas (2022) validated the biological control potential of Streptomyces M4 against Alternaria leaf spot disease. Separated Streptomyces lydicus M01 regulates soil microbial community and alleviates foliar disease caused by Alternaria on cucumbers (Wang et al., 2020). Xinjiang, China, contains diverse landforms, such as deserts (e.g., Gobi), lakes, basins, grasslands, mountains, rivers, and swamps. Because of its special geographic location, Xinjiang is exposed to various environmental stresses, including drought, high temperatures, and salinity, which may induce the production of specific antibiotics by microbes.
In this study, a novel Streptomyces strain (TRM 76323T) was isolated from the soil, after which its broad-spectrum antifungal activities were revealed. The morphological, physiological, and biochemical characteristics of TRM 76323T were analyzed. Moreover, streptochlorin was isolated and purified from the fermentation broth of this strain. The effects of a crude extract on A. gaisen mycelial growth, morphology, and cell ultrastructure were examined. In addition, the utility of TRM 76323T as a biocontrol agent was investigated in an in vitro experiment. The objective of this study was to identify and charammcterize important actinobacterial resources for controlling fungal phytopathogens. The findings of this study may be useful for protecting agriculturally important plants from diseases.
Materials and methods
Isolation of strain TRM 76323T
Strain TRM 76323T was isolated from a Salix rhizosphere soil sample collected along the Ahe Highway in Xinjiang (36.888 E, 82.553 N; at an altitude of 1,664.8 m). The strain was isolated on glycerol arginine medium (2.00 g arginine, 12.00 g glycerol, 0.50 g MgSO4, 1.00 g K2HPO4, and 17.00 g agar), as previously described (Sun et al., 2020). The pH of the medium was adjusted to 7.5 using Gause's medium. Strain TRM 76323T was grown for 1 week at 30°C.
Analysis of morphological, cultural, physiological, and biochemical characteristics
To determine the best medium, strain TRM 76323T was cultured on various International Streptomyces Project (ISP) media, including ISP1, ISP2, ISP3, ISP4, ISP5, ISP6, and ISP7 (Xing et al., 2022). Additionally, it was cultured on Gause's medium, potato dextrose agar medium, and nutrient agar medium. The pH of all media was adjusted to 7.0. The strain was grown and maintained on an ISP4 synthetic medium. The JSM-6360 scanning electron microscope (JEOL, Ltd., Tokyo, Japan) was used to examine the cell morphological characteristics of the spores and mycelia on ISP4 medium in plates that were incubated at 28°C for 1 week. Carbon-source utilization tests were conducted following the method described by Law et al. (2017), using the basal medium recommended by Pridham (Gottlieb, 1948). The growth ability of the strain TRM 76323T was assessed at different temperatures ranging from 4 to 50°C (4, 12, 15, 20, 25, 28, 30, 37, 40, 45, and 50°C) and pH levels ranging from 4 to 11 (pH 4, 5, 6, 7, 8, 9, 10, and 11). The study aimed to determine the tolerance of the system to different concentrations of NaCl (0, 5, 10, 15, 20, and 25%; w/v). Additionally, the production of peroxidase, urease, esterase, and catalase was analyzed following the methods described by Gerhardt et al. (1994). The study also investigated the use of a sole carbon source (0.5%; w/v), cellulose decomposition, starch hydrolysis, liquefaction of gelatin, milk peptization and solidification, nitrate reduction, and H2S production.
Chemotaxonomic features
The types of amino acids in cell wall hydrolysates and the whole-cell sugar contents were determined (Tindall et al., 2007). Polar lipids were identified via two-dimensional thin-layer chromatography involving 10% ethanolic molybdophosphoric acid, as described by Minnikin et al. (1984).
16S rRNA gene and multi-locus sequence analyses
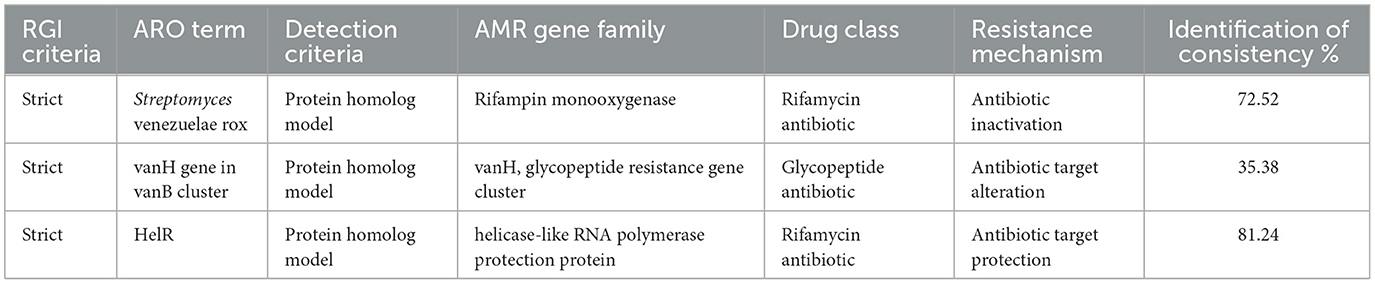
Strain TRM 76323T was identified through the analysis of its morphological, physiological, and biochemical characteristics. The molecular identification of the strain was performed by amplifying the 16S rRNA gene sequence using the prokaryotic universal primers 27F (5′-AGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′; Qi et al., 2019). The following housekeeping genes used in a previous Streptomyces multi-locus sequence analysis were selected: atpD (ATP synthase, beta subunit), gyrB (DNA gyrase B subunit), recA (recombinase A), rpoB (RNA polymerase, beta subunit), and trpB (tryptophan synthase, beta subunit). The average nucleotide identity (ANI) value was calculated as described by Lee et al. (2016). The digital DNA–DNA hybridization value was calculated according to formula 1 described by Meier-Kolthoff et al. (2013), which is available on the Genome-to-Genome Distance Calculator website. The strain TRM76323T draft genome sequence was analyzed using the Illumina platform. ABYSS software was used to assemble the sequencing data (Alanjary et al., 2019). The antiSMASH program (version 6.0) was used to detect biosynthetic gene clusters predicted to produce secondary metabolites (Blin et al., 2021). The Comprehensive Antimicrobial Research Database (CARD) was used to predict resistance genes (https://card.mcmaster.ca/analyze/rgi).
Fermentation and purification of TRM 76323T
Strain TRM 76323T from the ISP4 culture medium was inoculated into 1 L shake flasks for a 4-day fermentation at 28°C. The resulting seed culture was, then, transferred to a fermentation tank containing 50 L millet culture medium (15 g/L millet, 5 g/L glucose, 4 g/L peptone, and 3 g/L sodium chloride, pH 7). The fermentation at 28°C was carried out for 7 days. After completion, the samples were subjected to spray drying and further treated three times with 100% methanol to obtain extracts. For the TLC dot plate experiment, a solution consisting of dichloromethane:ethyl acetate (2:1) was used. Each 250 ml bottle was washed with 3 L of eluent, and the sample was, then, centrifuged and dried. The sample components were separated and purified through chromatography using a Sephadex LH-20 column. Finally, the obtained results were compared with published Microspectral data (http://www.nmrdata.com/; Lin et al., 2016).
Analysis of active components in the fermentation broth
The proteins in the fermentation broth were precipitated using 75% ethanol to determine their activity as ingredients. The crude extract was mixed with pre-cooled ethanol (final concentration of 75%). The resulting solution was, then, incubated at 4°C for 4 h and centrifuged at 10,000 rpm at 4°C for 20 min. The collected crude substance was resuspended in 20 mM Tris buffer (pH 6.8) before being used in subsequent experiments. To identify the active ingredients of TRM 76323T, freeze-dried bacteria of TRM76323T were treated with dichloromethane, ethyl acetate, methanol, and water to obtain extracts (Zou et al., 2021). A 100 μl aliquot of an A. gaisen suspension was spread over a PDA medium. The perforated confrontation method was employed to analyze the inhibitory effects of the fermentation broth on A. gaisen. Holes were created in the solid medium using a sterile blue gun head, and these holes were filled with 100 μl of the fermentation broth. The medium was, then, incubated at 28°C. The inhibitory effects were assessed using the dug plate confrontation method, with the fermentation broth being tested in triplicate. Alternaria gaisen was activated and cultured on a PDA medium for 48 h. The PDA medium served as the control for culturing A. gaisen, and the compound was inoculated with a concentration of 5 μl at 1 mg/ml using the filter paper method. One plate and three treatment groups were cultivated for 7 days for SEM analysis (Zou et al., 2021).
The effects of streptochlorin (2 mg/ml) on A. gaisen were examined according to the filter paper method. A 5 μl aliquot of streptochlorin or chloroform (control) was added to individual pieces of filter paper. The experiment was repeated three times. To further explore how the TRM 76323T fermentation broth was used to treat fragrant pears and branches that inhibited the growth and development of A. gaisen, in vitro experiments were conducted using fragment pears and branches that were washed three times with 75% ethanol and 5% sodium hypochlorite (Zhu et al., 2022). The lost inoculation method and sterile water treatment were used. Appropriate blank control groups were included. Samples were inoculated only with A. gaisen and then sprayed with the 100 μl fermentation broth after 24 h. Samples were placed in sterile bags and then incubated at 30°C for 7 days.
Results
Description of TRM 76323T
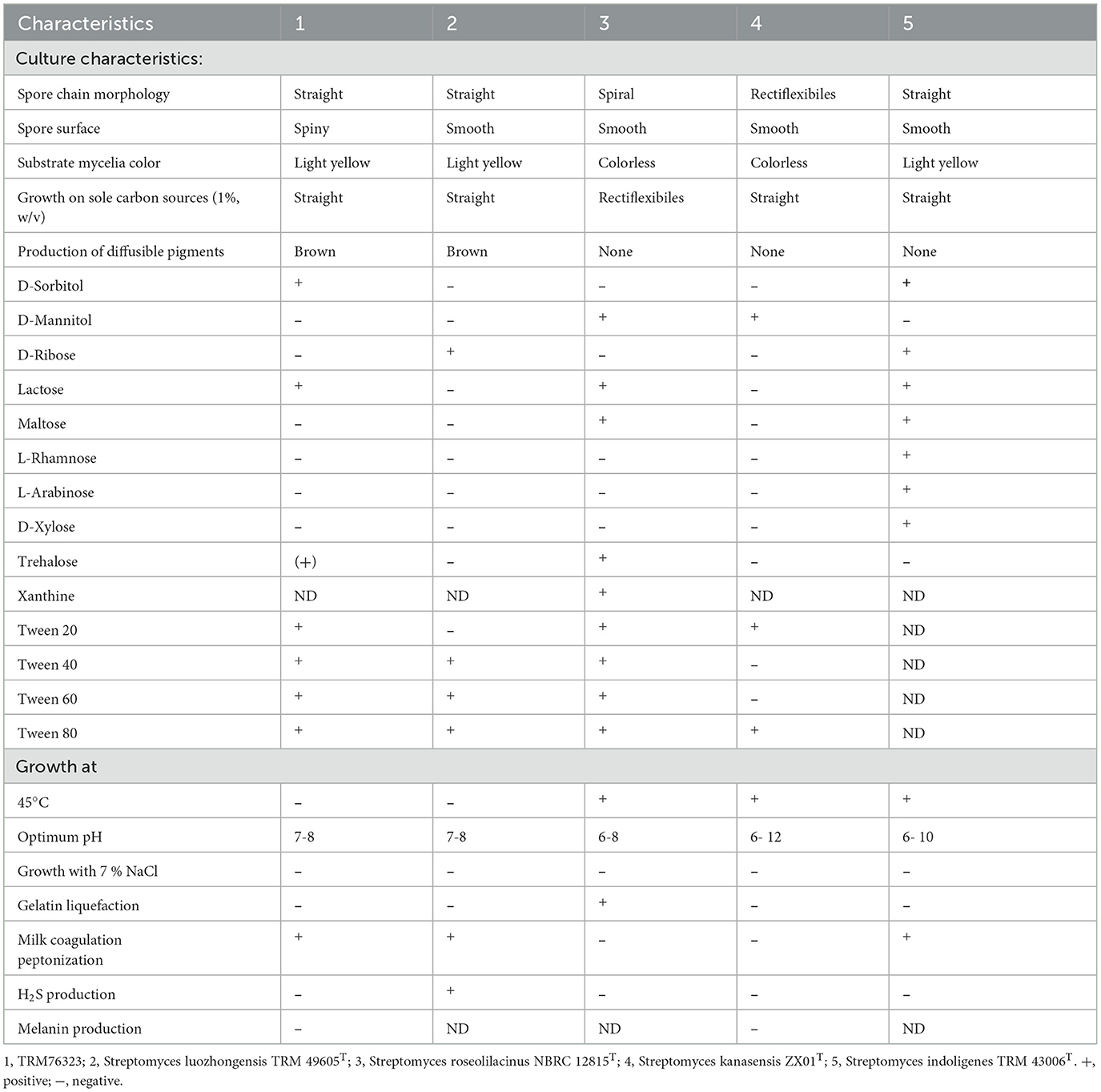
Strain TRM 76323T exhibited good growth on ISP4, GS, ISP1, and ISP5 while showing relatively poor growth on ISP6 and NA. It was unable to grow on ISP3. The morphology of the TRM76323T strain cultured in ISP 4 medium is shown in Figure 1B. Optimal growth was observed at a temperature of 28°C and a pH of 7.0, with suitable growth occurring within the temperature range of 16–37°C and pH range of 5.0–9.0. Although the cells were capable of growing in NaCl concentrations up to 5% (w/v), they exhibited the best growth in the absence of NaCl. The physiological characteristics of strain TRM 76323T are presented in Table 1. Examination of a 7-day-old TRM 76323T culture using an electron microscope revealed the presence of branched aerial hyphae and several long, straight, rod-shaped spores (Figure 1C). Positive results were obtained for nitrate reduction, catalase production, milk peptization, starch hydrolysis, cellulose hydrolysis, and urease production, while negative results were obtained for gelatin hydrolysis, oxidase production, melanin production, and H2S production.
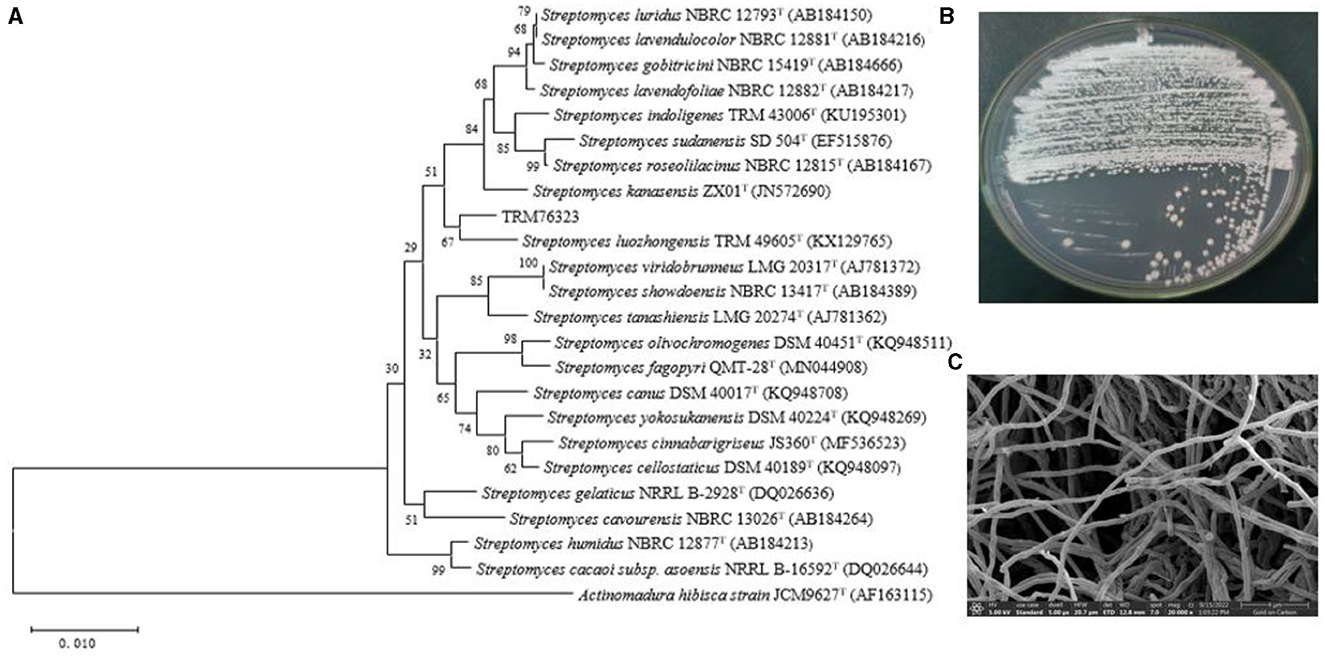
Figure 1. Morphological and evolutionary relationship of TRM76323. (A) Neighbor-joining unrooted tree based on 16S rRNA gene sequences for strain TRM 76323T and related taxa. Numbers at nodes are the bootstrap values (%) based on 1,000 resampled data sets. Bar, 0.0010 substitutions per nucleotide. (B) Strain morphology of TRM76323 strain cultured in ISP 4 medium. (C) Scanning electron micrograph of the aerial mycelium of the strain after 7 days at 28°C. Bar, 5 μm.
Chemotaxonomic features
The detected polar lipids were phospholipids, phosphatidylinositol, phosphatidylinositol mannoside, and phosphatidylinositol dimannoside (Supplementary Figure 1). Moreover, L,L-DAP was identified as the diagnostic cell wall diamino acid (Supplementary Figure 2A), and mannose (Man) was the main whole-cell sugar (Supplementary Figure 2B).
Genomic and phylogenetic analyses
The draft genome sequence for the strain TRM 76323T consisted of 7,338,911 bp and comprised 127 contigs, 5 rRNAs, 103 tRNAs, and 6,330 coding sequences. The G+C content was 72.1%. Based on the comparison with strain TRM 49605, the ANI value and dDDH value for TRM 76323T were 86.09 and 30.60%, respectively. A phylogenetic analysis of 16S rRNA sequences was performed using the neighbor-joining method (Figure 1A). In addition, the atpD, gyrB, recA, rpoB, and trpB gene sequences were concatenated for further analysis using the neighbor-joining (Supplementary Figure 3), maximum-likelihood (Supplementary Figure 4), and maximum-parsimony (Supplementary Figure 5) algorithms. Housekeeping genes were utilized for the multi-locus sequence analysis, which identified TRM76323T as a new species. A genome-wide collinearity analysis of TRM 76323T and Streptomyces luozhongensis TRM 49605 was conducted using Geneious Prime. Although the collinearity was relatively high, the strains were determined to be different species (Figure 2). Thirty gene clusters were detected in TRM 76323T, including 12 terpene, 7 NRPS, 1 T2PKS, 1 T3PKS, 1 phosphonate, and 3 T1PKS gene clusters. Among these, six gene clusters showed more than 50% similarity to known gene clusters (Supplementary Table 1). The resistance genes predicted in TRM 76323T using the Comprehensive Antimicrobial Research Database (CARD) belonged to the rifampicin monooxygenase and glycopeptide resistance gene clusters and helicase-like RNA polymerase protection protein gene families, with sequence similarities of 75.52, 35.38, and 81.24%, respectively (Table 2).
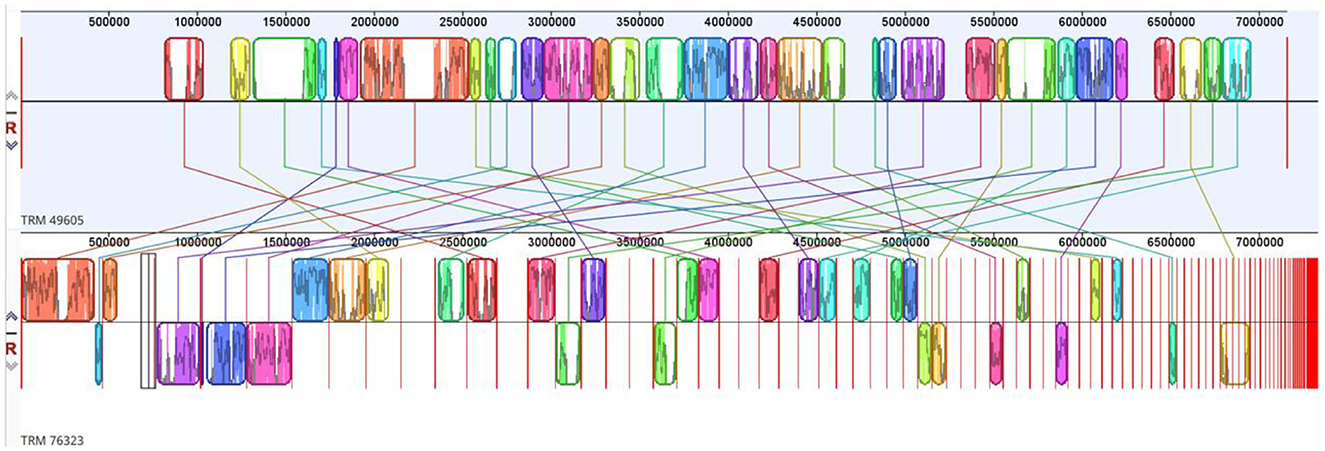
Figure 2. Genome-wide collinearity between TRM 76323T and Streptomyces luozhongensis TRM 49605T as determined using Geneious Prime.
Identification of bioactive compounds
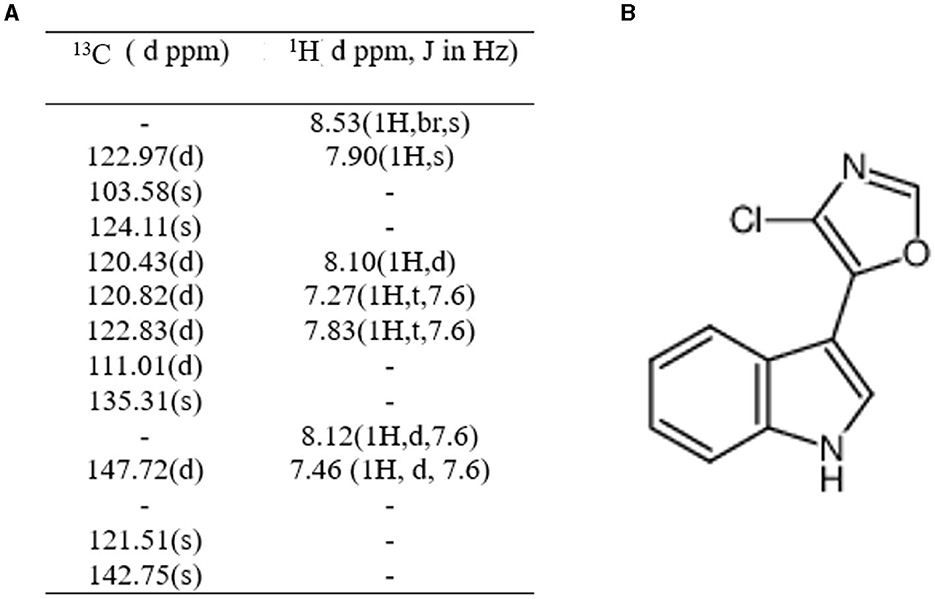
One compound, streptochlorin, which exhibits tyrosinase inhibitory activity, was isolated from the methanol extract of the TRM 76323T bacterial cake. Purified samples containing 12 mg of streptochlorin were obtained from the bacterial cake derived from a 50 L fermentation broth. The NMR data obtained were consistent with those obtained for the antibiotics, and their C-H data were also consistent (Supplementary Figures 6A, B). The antibiotics were identified as anti-nematode compounds based on the Combined Chemical Dictionary. Furthermore, the NMR analysis of these compounds and streptochlorin yielded very similar results (Figure 3).
Activity screening and validation
The TRM 76323T active components were mainly in the ethyl acetate extract (Supplementary Figure 7). The spray drying of the fermentation broth did not alter activities, implying that the components can tolerate exposure to high temperatures. The TRM 76323T extracts altered the morphological characteristics of A. gaisen mycelia collected from the edge of the invasion zone. More specifically, the SEM indicated that the control mycelia had a smooth surface and were normal and full (Figure 4A). In contrast, the mycelia treated with TRM 76323T extracts were rough, with an irregular shape and a collapsed cell wall (Figure 4B). The activity of streptochlorin was tested at a concentration of 2 mg/ml; the bacteriostatic effect resulted in an inhibition of ~1.5 ± 0.22 cm (Figure 5A). The minimum bacteriostatic concentration determined using the MIC method was 512 μg/ml (i.e., good bacteriostatic activity; Figure 5B). The effects of the TRM 76323T fermentation broth on fragrant pear fruits infected with A. gaisen were investigated. The fragrant pear fruit inoculated with A. gaisen had black spot disease symptoms and the incubation was for 7 days. Branches infected with A. gaisen also showed symptoms of black spot disease over an incubation period of 7 days. However, treating the affected fragrant pear fruit and branches with the TRM 76323T fertilization broth inhibited the severity of the black spot disease symptoms over the incubation period of 7 days (Figures 6A, B).
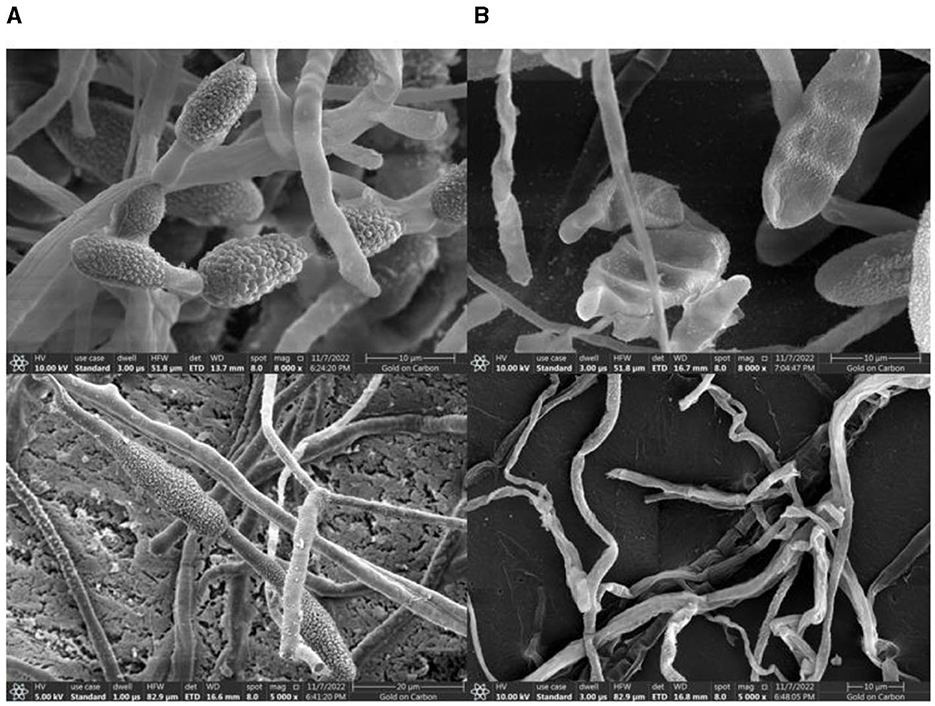
Figure 4. Effect of streptochlorin under SEM. (A) Alternaria gaisen under normal conditions. (B) Alternaria gaisen treated with the streptochlorin.
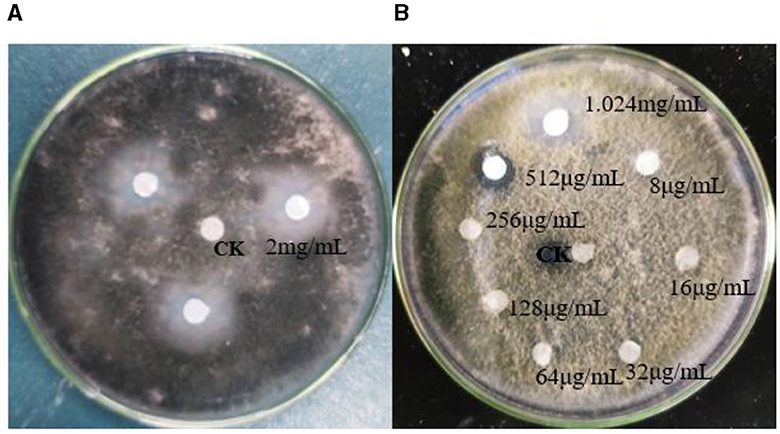
Figure 5. Effects of streptochlorin. (A) Effects of streptochlorin on Alternaria gaisen. (B) Antibacterial activity of streptochlorin at different concentrations under MIC (minimum inhibitory concentration).
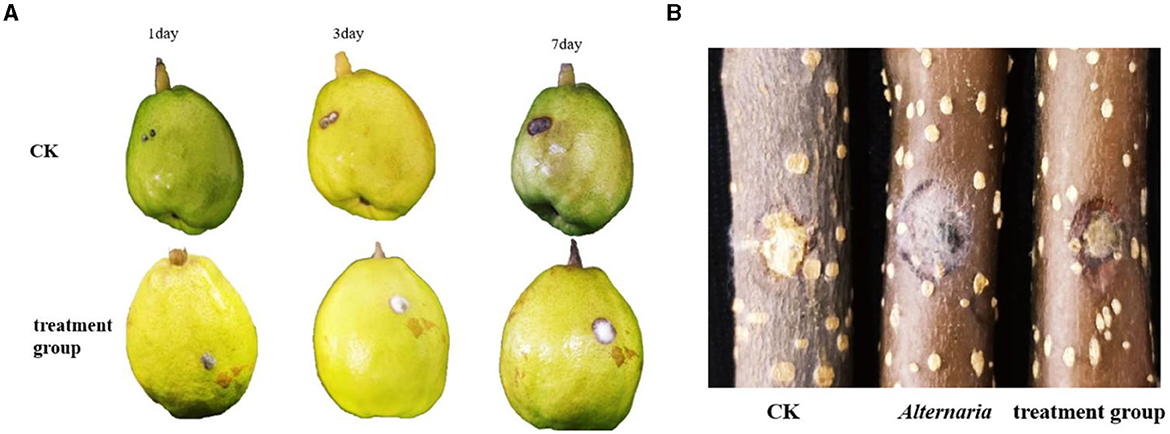
Figure 6. Control effect of fermentation liquid on Alternaria gaisen. (A) The inhibitory effect of streptomycin on Streptomyces on fruits. (B) Effect of streptochlorin on Streptomyces on tree stem. The inhibitory effect of bacteria.
Discussion
Pear black spot, caused by A. gaisen, is a destructive disease that negatively impacts pear tree yield. The use of Streptomyces-based biological control strategy presents a viable alternative to chemical fungicides as previous research has shown promising results (Song et al., 2020; Yuan et al., 2021). However, there is a scarcity of biological control strains with antagonistic activity against pear black spot disease. Hence, this study focuses on isolating strain TRM76323T from Tamarix and evaluating its ability to inhibit the growth of pear black spot through activity tracking. Strain TRM76323T produces streptochlorin, which effectively inhibits the hyphal growth of A. gaisen and induces hyphal malformation, aligning with previous studies. Overall, these findings highlight the potential of strain TRM76323T as a promising biological control resource for combating pear black spot disease. The survival of biocontrol strains in plants is crucial for their agricultural application. Microorganism-produced compounds can negatively impact fungal cells' growth and morphology, potentially leading to cell death (Bowman and Free, 2006). Notably, the culture filtrate of strain TRM76323T exhibits antifungal activity against A. gaisen, indicating the presence of antibacterial compounds. Numerous studies have demonstrated the contribution of microorganism secondary metabolites to plant resistance (Piasecka et al., 2015). In conclusion, these findings suggest that TRM76323T can induce antifungal defense in pears primarily by regulating the biosynthesis pathway of secondary metabolites.
Conclusion
In this study, on the basis of polyphasic classification and identification, physiological and biochemical indicators, chemical indicators, and genome sequence comparisons, TRM 76323T was identified as a new species. Its inhibitory activities against A. gaisen were verified, and its active components were identified. Microscopic examinations and in vivo experiments confirmed the inhibitory effects of TRM 76323T on A. gaisen at the micro- and macro-levels. Furthermore, streptochlorin was purified, with the subsequent analysis, revealing that its minimum inhibitory concentration was 512 μg/ml. In summary, our results showed not only the biosynthetic potential of strain TRM76323T for the biopharmaceutical field but also its distinguishable phenotypic and genotypic properties, confirming that it represents a novel species of the genus Streptomyces for which the name Streptomyces sp. nov. is proposed. The findings of this study form the basis of future research conducted to increase the utility of strain TRM 76323T for controlling agriculturally important plant diseases.
Description of Streptomyces sp.nov.
Strain TRM76323 can grow well on ISP1, ISP4, ISP5, and Gause's medium and can produce abundant spores after 7 days of cultivation. However, the growth on ISP6 and NA media is relatively poor, and it cannot grow on ISP3 medium. The optimal pH is 7.0, the optimal growth temperature is 28°C, and the strain can grow at a concentration of 5% (w/v) NaCl, with the best growth at a salt concentration of 0%. Strain TRM76323 can use D-sorbitol, lactose, and trehalose as the only carbon sources. Nitrate reduction, catalase, milk coagulation and peptonization, starch hydrolysis, cellulose hydrolysis, and urease production all obtained positive results, but gelatin hydrolysis, oxidase production, and melanin production all obtained negative results. TRM76323 was able to grow on media supplemented with Tween 20, Tween 40, Tween 60, and Tween 80, producing hydrolysis loops, indicating that the strain has lipase activity. The polar lipids of strain TRM76323 are PI (phosphatidylinositol), PIM (phosphatidylinositol mannoside), PIDM (phosphatidylinositol diamandoside), and PLS (phospholipid). The amino acid component of the cell wall is Diaminopimelic acid; TRM76323 whole-cell hydrolytic sugar component contains mannose, TRM76323, and S. The DNA–DNA hybridization (DDH) result of Streptomyces luozhongensis TRM49605T is 30.60%, which is far below from the threshold defined species by 70%, and the ANI value is 86.09%, which is lower than the threshold describing prokaryote species by 95%, indicating that TRM76323 and S. luozhongensis TRM49605T are different species. Strain TRM76323 was identified as a new species of Streptomyces through polyphasic classification. This study is activity-oriented, with the goal of preventing and controlling pear black spot disease. Using modern separation and purification techniques, the active component of strain TRM76323 was determined to be located in the ethyl acetate phase using the plate confrontation method. According to the HPLC method, the active product was separated, and the active product was identified as streptochlorin, so the minimum inhibitory concentration was 512 μg/ml. This study analyzes the antagonistic effect of the new species of Streptomyces sp. TRM76323 on pear black spot disease, which can more effectively carry out the prevention and control of pear black spot disease, reduce economic losses, and promote sustainable development of agriculture to lay the foundation for the healthy development of the pear industry and ensure the economic stability of the pear region. As a new means of disease control, microbial lead compound is characterized by safety, economy, and no resistance to pests and diseases. It provides a new vision and technical basis for sustainable agricultural development strategy and provides a theory for the formulation of pathogen control measures.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: 菌株保藏于 CCTCC M2023092.
Author contributions
Y-hC: Writing – original draft, Validation. YW: Validation, Investigation, Data curation, Writing – original draft. QL: Conceptualization, Supervision, Writing – review & editing. ZX: Supervision, Writing – review & editing. JW: Conceptualization, Writing – review & editing. X-XL: Funding acquisition, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Tarim University President gene project Preparation and application of phosphonic acid producing biocontrol agents (TDZKZD202202), supported by Bintuan Science and Technology Program (2021BB007), and the Xinjiang Uygur Autonomous Region graduate scientific research innovation project (XJ2022G240).
Acknowledgments
The authors would like to thank Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1273842/full#supplementary-material
References
Alanjary, M., Steinke, K., and Ziemert, N. (2019). AutoMLST: an automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 47, W276–W282. doi: 10.1093/nar/gkz282
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Bowman, S. M., and Free, S. J. (2006). The structure and synthesis of the fungal cell wall. Bioessays 28, 799–808. doi: 10.1002/bies.20441
Brown, A. G., Butterworth, D., Cole, M., Hanscomb, G., Hood, J. D., Reading, C., et al. (1976). Naturally-occurring β-lactamase inhibitors with antibacterial activity. J. Antibiot. 29, 668–669. doi: 10.7164/antibiotics.29.668
Chauhan, J. S., Badoni, A., Indrakumar Singh, N., and Ali, S. (2009). Effect of alternaria on some members of family brassicaceae of garhwal himalaya. N. Y. Sci. J. 6, 1200–1554. Available online at: http://www.sciencepub.net/newyork/
Dale, J., James, A., Paul, J. Y., Khanna, H., Smith, M., Peraza-Echeverria, S., et al. (2017). Transgenic cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 8:1496. doi: 10.1038/s41467-017-01670-6
Gerhardt, P., Murray, R. G. E., Wood, W. A., Smibert, R. M., and Krieg, N. R. (1994). Methods for General and Molecular Bacteriology. Washington, DC: American Society for Microbiology.
Gottlieb, T. G. P. D. (1948). The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J. Bacteriol. 56, 107–144. doi: 10.1128/jb.56.1.107-114.1948
Jose, P. A., Maharshi, A., and Jha, B. (2021). Actinobacteria in natural products research: progress and prospects. Microbiol. Res. 246:126708. doi: 10.1016/j.micres.2021.126708
Law, J. W.-F., Ser, H.-L., Duangjai, A., Saokaew, S., Bukhari, S. I., Khan, T. M., et al. (2017). Streptomyces colonosanans sp. nov., A novel actinobacterium isolated from Malaysia mangrove soil exhibiting antioxidative activity and cytotoxic potential against human colon cancer cell lines. Front. Microbiol. 8:877. doi: 10.3389/fmicb.2017.00877
Lee, I., Ouk Kim, Y., Park, S.-C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst.Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Lin, L., Shan, H., Li, J. D., and Xiao, J. Y. (2016). Efficient preparation of streptochlorin from marine Streptomyces sp. SYYLWHS-1-4 by combination of response surface methodology and highspeed counter-current chromatography. Molecules 21:693. doi: 10.3390/molecules21060693
Meier-Kolthoff, P. J., Alexander, F. A., Klenk, H. P., and Goker, M. (2013). Genome sequence based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Minnikin, D. E., O'Donnell, A. G., Goodfellow, M., Alderson, G., and Athalye, M. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2:233. doi: 10.1016/0167-7012(84)90018-6
Phuakjaiphaeo, C., Chang, C. I., Ruangwong, O., and Kunasakdakul, K. (2016). Isolation and identification of an antifungal compound from endophytic Streptomyces sp. CEN26 active against Alternaria brassicicola. Lett. Appl. Microbiol. 63, 38–44. doi: 10.1111/lam.12582
Piasecka, A., Jedrzejczak-Rey, N., and Bednarek, P. (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206, 948–964. doi: 10.1111/nph.13325
Qi, D., Zou, L., Zhou, D., Chen, Y., Gao, Z., Feng, R., et al. (2019). Taxonomy and broad spectrum antifungal activity of Streptomyces sp. SCA3-4 isolatedfrom rhizosphere soil of Opuntia stricta. Front. Microbiol. 10:1390. doi: 10.3389/fmicb.2019.01390
Ser, H. L., Law, J. W. F., Chaiyakunapruk, N., Jacob, S. A., Palanisamy, U. D., Chan, K.-G., et al. (2016). Fermentation conditions that affect clavulanic acid production in Streptomyces clavuligerus: a systematic review. Front. Microbiol. 7:522. doi: 10.3389/fmicb.2016.00522
Sharma, M., and Manhas, R. K. (2022). Biocontrol potential of Streptomyces sp. M4 and salvianolic acid B produced by it against Alternaria black leaf spot. Microb. Pathog. 173(Pt A):105869. doi: 10.1016/j.micpath.2022.105869
Song, X., Han, M., He, F., Wang, S., Li, C., Wu, G., et al. (2020). Antifungal mechanism of dipicolinic acid and its efficacy for the biocontrol of pear Valsa canker. Front. Microbiol. 11:958. doi: 10.3389/fmicb.2020.00958
Sun, B., Yuan, L., Xia, Z., Wan, C., and Zhang, L. (2020). Streptomyces albicerus sp. nov., a novel actinomycete isolated from the sediments of the Tailan River in Xinjiang, China. Arch. Microbiol. 202, 1639–1646. doi: 10.1007/s00203-020-01871-6
Tindall, B. J., Sikorski, J., Smibert, R. A., and Krieg, N. R. (2007). “Phenotypic characterization and the principles of comparative systematics,” in Methods for General and Molecular Microbiology, eds C. A. Reddy, T. J. Beveridge, J. A. Breznak, G. Marzluf, T. M. Schmidt, and L. R. Snyder (Washington, DC: ASM Press), 330–393. doi: 10.1128/9781555817497.ch15
Waksman, S. A., Bugie, E., and Schatz, A. (1944). Isolation of antibiotic substances from soil micro-organisms, with special reference to Streptothricin and Streptomycin. Proc. Staff Meet. Mayo Clin. 19, 537–548.
Waksman, S. A., and Henrici, A. T. (1943). The nomenclature and classification of the activomycetes. J. Bacteriol. 46, 337–341. doi: 10.1128/jb.46.4.337-341.1943
Wang, M., Xue, J., Ma, J., Feng, X., Ying, H., and Xu, H. (2020). Streptomyces lydicus M01 regulates soil microbial community and alleviates foliar disease caused by Alternaria alternata on cucumbers. Front. Microbiol. 11:942. doi: 10.3389/fmicb.2020.00942
Xing, L., Xia, Y., Zhang, Q., Xia, Z., and Wan, C. (2022). Streptomyces griseicoloratus sp. Nov., isolated from soil in cotton fields in Xinjiang, China. Arch. Microbiol. 204:254. doi: 10.1007/s00203-022-02818-9
Yuan, H., Shi, B., Huang, T., Zhou, Z., Wang, L., Hou, H., et al. (2021). Biological control of pear Valsa canker caused by Valsa pyriusing Penicillium citrinum. Horticulturae 7:198. doi: 10.3390/horticulturae7070198
Zhu, H., Ahmad, T., Zheng, Y., Moosa, A., Chengrong, N., and Liu, Y. (2022). First report of leaf blight disease of Livistona chinensis caused by Alternaria alternata in Pakistan. Plant Dis. doi: 10.1094/PDIS-08-21-1851-PDN. [Epub ahead of print].
Keywords: Alternaria gaisen, streptochlorin, antibacterial mechanism, antagonistic, Streptomyces
Citation: Chen Y-h, Wu YZ, Liu Q, Xia Z, Wang J and Luo X-X (2023) Streptomyces tamarix sp. nov.: antagonism against Alternaria gaisen producing streptochlorin, isolated from Tamarix root soil. Front. Microbiol. 14:1273842. doi: 10.3389/fmicb.2023.1273842
Received: 10 August 2023; Accepted: 05 October 2023;
Published: 21 November 2023.
Edited by:
Ajay Kumar, Amity University, IndiaReviewed by:
Yashavantha Rao H. C., Indian Institute of Science (IISc), IndiaTaswar Ahsan, University of Central Punjab, Pakistan
Xiufang Hu, Zhejiang Sci-Tech University, China
Copyright © 2023 Chen, Wu, Liu, Xia, Wang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Xia Luo, xxluo415@163.com
 Yi-huang Chen
Yi-huang Chen Yi Zheng Wu1,2
Yi Zheng Wu1,2 Xiao-Xia Luo
Xiao-Xia Luo