- College of Veterinary Medicine and Research Institute for Veterinary Science, Seoul National University, Seoul, Republic of Korea
Campylobacter is a major zoonotic pathogen that causes gastrointestinal and, rarely, immune diseases in humans. The antimicrobial-resistance gene cfr(C) carried by Campylobacter and is a cfr-like gene that targets bacterial 23S rRNA through A2503 methylation. cfr(C) confers cross-resistance to five antimicrobial classes (PhLOPSA), including lincosamide, streptogramin A, and pleuromutilin, which are classified as critically important antimicrobials to human by the World Health Organization. To elucidate the genetic variation and horizontal transfer mechanism of cfr(C), we analyzed the genetic background and horizontal transfer unit of Campylobacter-derived cfr(C) through comparative genomic analysis. We identified nine cfr(C)-positive C. coli strains of 157 strains isolated from swine sources. Three novel cfr(C) gene single nucleotide polymorphism (SNP) sites (19delA, 674C > A, and 890 T > C) were identified from nine cfr(C)-positive strains. Among six identified cfr(C) SNP variant types (SNP-I to -VI), five types of randomly inserted cfr(C)-cassettes on chromosome and one type of plasmid-like element were identified, their gene cassette composition differing depending on the cfr(C) variants. Three of six cfr(C) cassette types contained aminoglycoside-streptothricin resistance cluster “aphA3-sat4-aadE.” The cfr(C) gene cassette with pcp gene (GC-1, GC-4, and GC-5) formed a pcp-mediated circular intermediate “pcp-hp-cfr(C)-aphA3,” which has not been previously reported. Other two cfr(C) cassette-types with ISChh1 formed circular intermediate “ISChh1-aphA3-cfr(C)-lnu (G)-pnp-ant1-hp-ATPase” and “ISChh1-aphA3-cfr(C)-hp.” In conjugation assay, the pcp-mediated circular intermediate was naturally transferred to the plasmid of recipient C. coli wild-type strain from swine source, and comparative genomic analysis revealed that cfr(C) encoded in pcp-mediated circular intermediate was inserted into the plasmid of recipient by homologous recombination with pcp and aphA3. This study revealed that novel multidrug resistance gene cfr(C) carried by C. coli from swine sources can be highly genetically diverse and transferable. Moreover, we suggest that the transferability of chromosomal cfr(C) may contribute to the global spread of multidrug resistance against clinically important antimicrobials.
1 Introduction
Campylobacter species, particularly C. jejuni and C. coli, are leading zoonotic foodborne pathogen that can cause gastrointestinal infections (such as diarrhea) and immune diseases (such as Guillain-Barré syndrome) in humans (Silva et al., 2011). Unlike in humans, Campylobacter causes mostly asymptomatic infections in food-animals, including poultry, cattle, and pigs, and is considered a commensal bacterium (Burnham and Hendrixson, 2018). According to the World Health Organization (WHO), the major route of human campylobacteriosis is via the food-chain of animal products (such as meat and milk) which are contaminated with Campylobacter, present in animal feces, during the slaughter process (Jaakkonen et al., 2020).
Since 2013, the US Centers for Disease Control and Prevention has classified drug-resistant Campylobacter as a serious antibiotic resistance threat (CDC, 2013). In recent years, Campylobacter has become increasingly resistant to clinically important antimicrobials and has developed multiple mechanisms of antimicrobial resistance. These include mutations in target genes, such as 23S rRNA mutations that confer resistance to macrolides and, gyrA mutations associated with fluoroquinolone resistance; multidrug efflux pump CmeABC/RE-CmeABC extruding structurally diverse compounds and antimicrobials; and horizontally acquired antimicrobial resistance genes, such as tet(O), erm(B), fexA, and fosXCC (Aarestrup et al., 2008). Among the two major species of Campylobacter, C. jejuni and C. coli, which are mainly responsible for human infection, C. coli is known to be more resistant to antimicrobials (Liu et al., 2019b).
The cfr gene, which encodes 23S rRNA methyltransferase, confers resistance to five antimicrobial classes, namely phenicols, lincosamides, oxazolidionones, pleuromutilins, and streptogramin A, known as PhLOPSA phenotype (Schwarz et al., 2000; Long et al., 2006). WHO classified lincosamides, pleuromutilins, and streptogramin A as “critically important antimicrobials: highly important” and warned that resistance to these antimicrobial agents would make it difficult to treat bacterial infection (WHO, 2019). Since the first report of cfr gene in Staphylococcus sciuri in 2006, five types of the cfr gene family have been reported: cfr, cfr(B), cfr(C), cfr(D), and cfr(E) (Schwarz et al., 2000; Deshpande et al., 2015; Hansen and Vester, 2015; Candela et al., 2017; Tang et al., 2017; Sassi et al., 2019; Stojković et al., 2019). Tang et al. (2017) first identified plasmid-mediated multi-drug resistance gene cfr(C) in C. coli isolates of feedlot cattle origin (Tang et al., 2017). Since the first report of cfr(C) in Campylobacter, three additional studies reported the presence of cfr(C) in Campylobacter species (Zhao et al., 2019; Liu et al., 2019a; Tang et al., 2020). However, the horizontal transferability of Campylobacter-derived cfr(C) has been reported in only one study (Tang et al., 2017), and the transfer mechanism of cfr(C) is still unclear and requires further research.
An understanding of genetic variation of cfr(C) and their transmission mechanisms can help control the propagation of antimicrobial multi-resistance, a serious problem when it is transferred to humans. Thus, in this study we analyzed genetic background and horizontal transfer unit of cfr(C) in C. coli strains through comparative genomic analysis to elucidate the genetic variation and horizontal transfer mechanism of the Campylobacter-derived cfr(C) gene from swine sources.
2 Materials and methods
2.1 Identification of cfr(C)-positive Campylobacter coli strains
A total of 157 swine-derived C. coli strains were used to screen cfr(C) presence. Of these, 130 C. coli strains were isolated from six swine farms in South Korea in our previous study (Guk et al., 2021). In addition, 15 C. coli strains were collected from 30 swine fecal samples from one slaughterhouse in South Korea in November 2017 and 12 C. coli strains from 30 fecal samples from one swine farm in South Korea in April 2018. Confirmation of cfr(C) was conducted using polymerase chain reaction (PCR). The primer used and annealing temperature are presented in Supplementary Table 1.
2.2 Antimicrobial susceptibility testing
Minimum inhibitory concentration (MIC) tests were conducted using Sensititre CAMPY2 plates (ThermoFisher Scientific, USA) following the manufacturer’s instructions for the following eight antimicrobial agents: azithromycin, ciprofloxacin, erythromycin, gentamicin, tetracycline, florfenicol, nalidixic acid, and clindamycin. The MIC value of each antimicrobial agent was interpreted according to the criteria of the National Antimicrobial Resistance Monitoring System for enteric bacteria.1 C. jejuni ATCC 33560 was used as the quality control strain for the MIC test.
2.3 Whole genome sequencing (WGS)
Total genomic DNA was extracted using NucleoSpin Microbial DNA kit (Macherey-Nagel, Germany) following the manufacturer’s instructions. Total genomic DNA was sequenced via a combination of NextSeq® 500 technology (Illumina, Inc., USA) and MinION platforms (Oxford Nanopore Technologies, UK). Raw short-read and long-read data were assembled using hybrid-assembly strategy in Unicycler (v0.5.0) (Wick et al., 2017) and annotated using prokka (v1.14.5) (Seemann, 2014).
2.4 Identification of novel cfr(C) variants via single nucleotide polymorphism (SNP) analysis
To identify cfr(C) variant types, the nucleotide sequences of cfr(C) of nine isolated C. coli strains were aligned with that of the reference strain, C. coli Tx40 (National Center for Biotechnology Information [NCBI] accession number: NG_060579). Thereafter, we conducted cfr(C) gene SNP-based comparative genomic analysis by comparing the cfr(C) sequences of nine strains with that of 28 C. coli strains, the WGS of which were deposited in the NCBI database. In addition, SNP-based phylogenetic analysis was performed for cfr(C) of nine cfr(C)-positive strains together with that of other strains published in NCBI database. Phylogenetic tree was constructed with MEGAX software by using UPGMA method (Kumar et al., 2018). Bootstrap values were calculated with 1,000 replications. The accession numbers of nucleotide sequences are listed in Supplementary Table 2.
2.5 Functional confirmation of cfr(C) variants through cloning experiments
To determine the role of cfr(C) in conferring florfenicol resistance, a 1,425 bp DNA fragment was amplified from eight C. coli strains (CC021, 027, 032, 041, 042, 046, 135, and 159) through PCR. This fragment included the coding sequence of cfr(C) (1,140 bp) as well as 63 bp upstream and 163 bp downstream sequences of the gene. For C. coli strain CC024, a 1,327 bp DNA fragment was amplified through PCR. This fragment included the coding sequence of cfr(C) as well as the 122 bp upstream and 163 bp downstream sequences of the gene. The amplicon was ligated with an E. coli/C. jejuni shuttle plasmid pUC19 (Yanisch-Perron et al., 1985) to construct plasmid pUC19-cfr(C). The pUC19-cfr(C) plasmid was then transformed into E. coli DH5α competent cells by heat shock transformation. The E. coli transformants were selected from Luria-Bertani agar (Sigma-Aldrich, USA) plates containing 50 mg/L ampicillin and 50 mg/L kanamycin. The confirmation for carrying plasmid pUC19-cfr(C) was performed thorough PCR using the cfr(C)-specific primers. MIC tests were also conducted for E. coli transformants to evaluate the antimicrobial resistance. Used primer and annealing temperature used in the experiments are listed in Supplementary Table 2.
2.6 Analysis of cfr(C)-carrying gene cassettes and circular intermediates
The cfr(C)-carrying gene cassettes of nine C. coli strains were aligned against the corresponding region of reference strain C. coli Tx40 ([NCBI] accession number: NG_060579) and C. coli SHP40 ([NCBI] accession number: MF037584). Additionally, the inserted positions of cfr(C)-carrying gene cassette in nine C. coli strains were identified by alignment and comparison with C. coli strain ATCC33559. The comparative genomic visualization was performed using “Easyfig (v2.2.3)” (Sullivan et al., 2011). The circular intermediate form of cfr(C)-carrying gene cassette was investigated by using inverse PCR and Sanger sequencing. Amplicon sequences were aligned and compared with sequences submitted in the NCBI GenBank database using the Basic Local Alignment Search Tool program (BLAST).2 The primers and annealing temperature used in the experiments are listed in Supplementary Table 2.
2.7 Conjugation assay
To evaluate the horizontal transferability of cfr(C)-carrying gene cassette, we conducted conjugation assay following the previously described methods with slight modification (Davis et al., 2008). The conjugation assay was performed with a gentamicin-resistant wild-type C. coli as the recipient and eight cfr(C)-positive C. coli strains as the donors: CC021, CC027, CC032, CC041, CC042, CC046, CC135, and CC159. Among nine cfr(C)-positive strains included in this study, CC024 was excluded from the conjugation assay with the gentamicin-resistant recipient strain, since it carried resistance to gentamicin. To select the transconjugants, Mueller-Hinton agar (Sigma Aldrich) plates containing 1 mg/L gentamicin and 4 mg/L florfenicol were used. The presence of cfr(C) gene and circular intermediate form in transconjugants were confirmed using PCR. The antimicrobial resistance was evaluated for transconjugants though MIC tests.
3 Results
3.1 Novel cfr(C) SNP variants
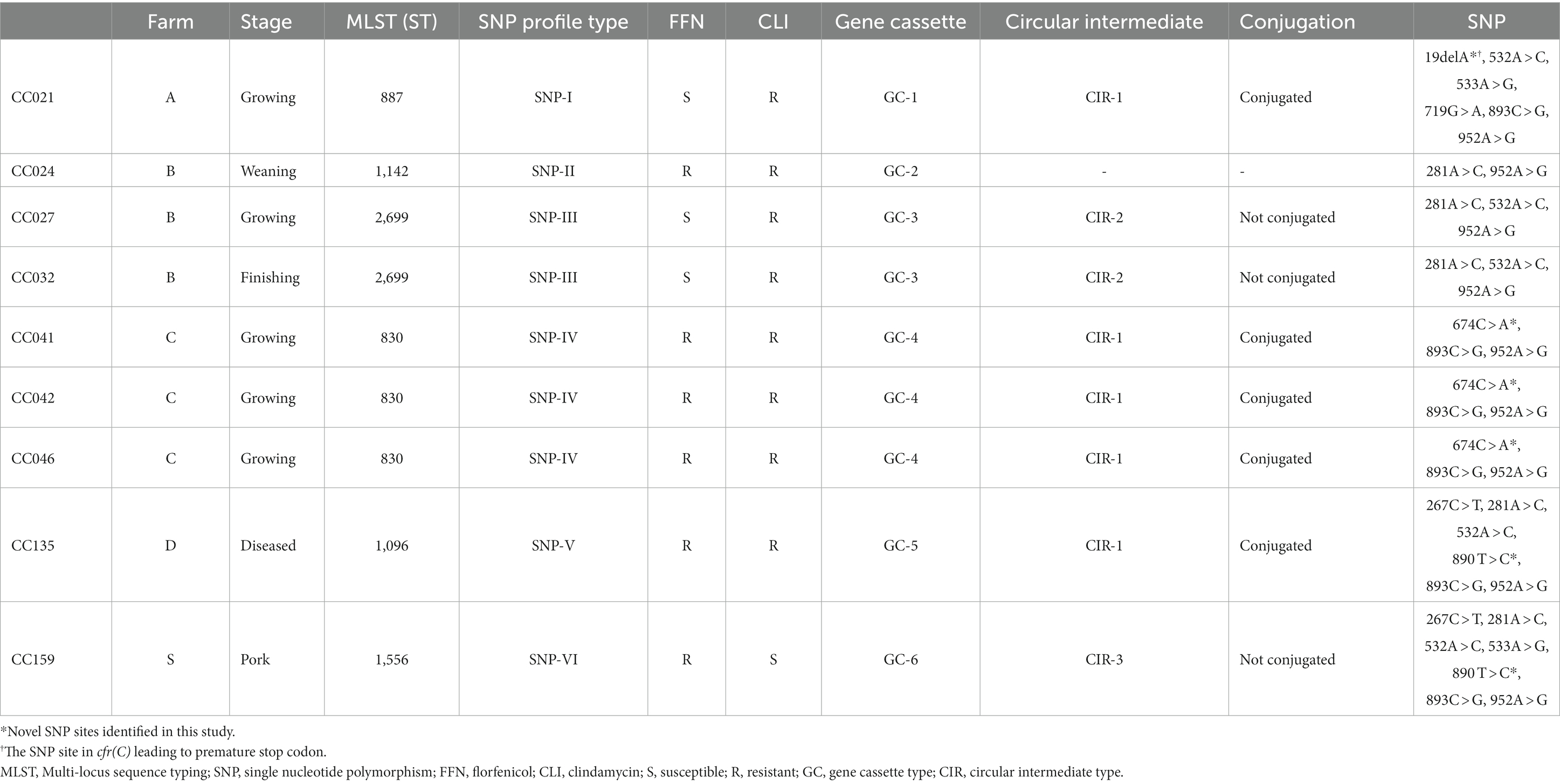
Among the 157 swine-derived C. coli strains, nine cfr(C)-positive strains were identified (Table 1). A total of six SNP variant types were identified in SNP-based comparative genomic analysis: SNP-I (CC021), SNP-II (CC024), SNP-III (CC027 and CC032), SNP-IV (CC041, CC042, and CC046), SNP-V (CC135), and SNP-VI (CC159) (Figure 1 and Supplementary Figure 1). SNP-I carried six SNPs, including a nonsense SNP (19delA, M7fs), which caused frameshift mutation of cfr(C) in strain CC021. SNP-II carried two SNPs, 281A > C (E94A) and 952A > G (I318V), and this type was truncated at the 3′ end (1,101 bp). SNP-III carried three SNPs, 281A > C, 532A > C (K178Q), and 952A > G. The SNP-IV type carried three SNPs, including the novel SNP 674C > A (T225K), which was not identified in previously reported strains uploaded in the NCBI database. The SNP-V type carried six SNPs, including the novel SNP 890T > C (sSNP), which was not identified in previously reported strains uploaded in the NCBI database. The SNP-VI type carried seven SNPs, including 533A > G (K178R) and the novel SNP 890T > C.
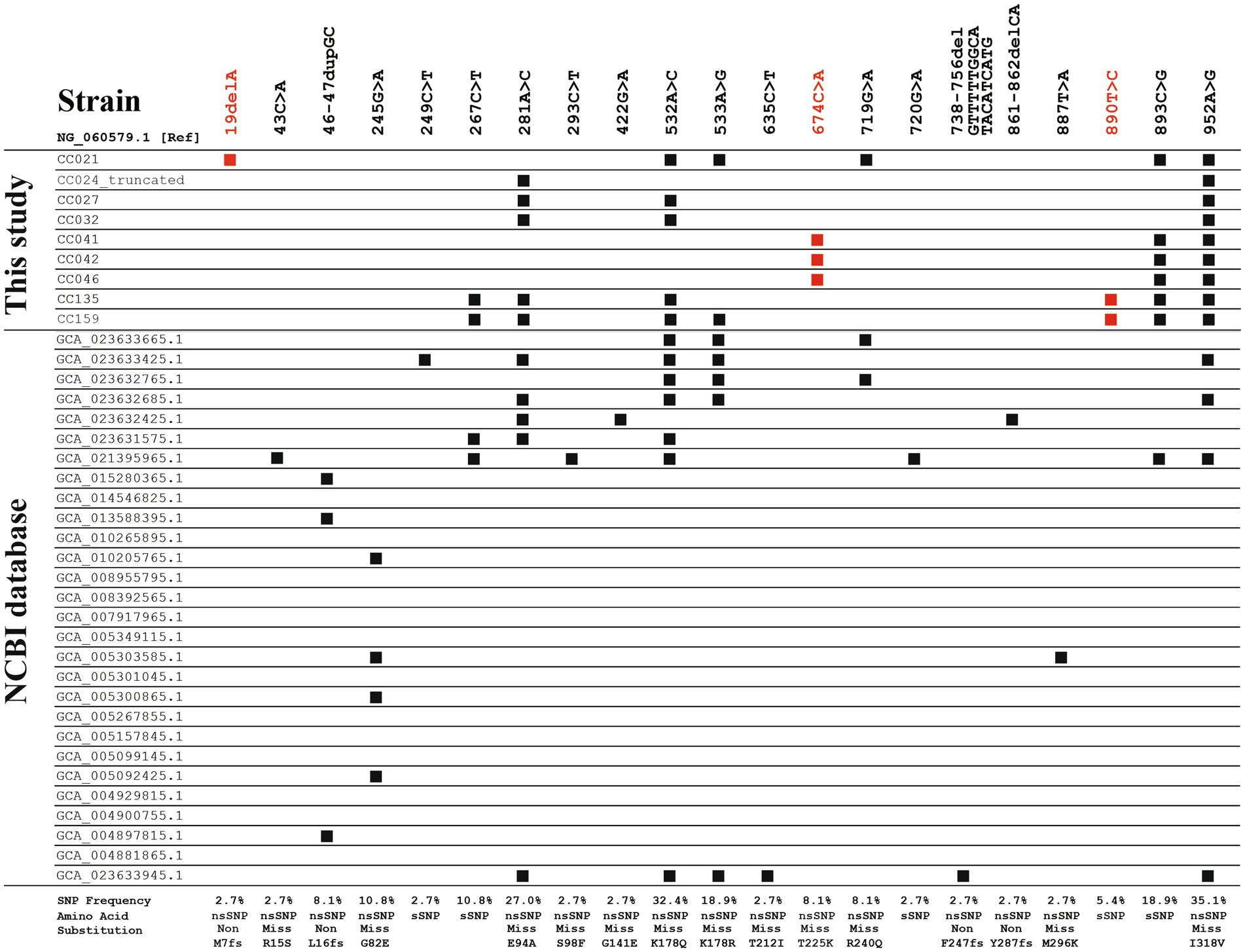
Figure 1. Single nucleotide polymorphic (SNP) sites of cfr(C) in 37 strains, including nine strains identified in this study and 28 strains obtained from NCBI database. Nucleotide sequence of cfr(C) (1,140 bp) of C. coli Tx40 strain (NG_060579.1) was used as reference. SNP sites shown in red font represent the novel SNP sites identified in this study. nsSNP, non-synonymous SNP; sSNP, synonymous SNP; Non, non-sense nsSNP; Miss, Miss-sense nsSNP; fs, frameshift mutation.
In the SNP-based phylogenetic tree, the cfr(C) nucleotide sequences of the nine strains were genetically similar to those of swine- and human-derived C. coli strains isolated from China rather than that of cattle, chicken, and human-derived C. coli strains isolated from USA (Supplementary Figure 2). In addition, the cfr(C) genes reported from the USA were genetically homogeneous regardless of region and year of isolation, whereas the cfr(C) genes reported from China showed high genetic variations.
3.2 Antimicrobial resistance of nine cfr(C)-positive Campylobacter coli strains and transformants
Two SNP types showed susceptibility to florfenicol: SNP-I (CC021) and SNP-III (CC027 and 032), with MIC values of 2 and 4 mg/L, respectively (Supplementary Table 3). The other types showed resistance to florfenicol (MIC: 8 mg/L). Resistance to clindamycin was identified in three SNP types: SNP-II, -III, and -V. Regardless of the SNP variants, strains CC024, CC027, CC032 and CC135 exhibited resistance to the macrolide class, namely azithromycin (MIC: ≥64 mg/L) and erythromycin (MIC: ≥64 mg/L). Except for the CC024 strain, all other types showed susceptibility to gentamicin (MIC: ≤4 mg/L). All strains except for CC159 strain showed resistance to the quinolone class, including ciprofloxacin (MIC: ≥1 mg/L) and nalidixic acid (MIC: ≥32 mg/L).
All six SNP types of cfr(C) were transformed into E. coli DH5α competent cells. The transformant of SNP-I type (CC021) exhibited susceptibility to florfenicol (MIC: 2 mg/L). The other SNP types (SNPs II–VI) exhibited resistance to florfenicol (MICs: 4–8 mg/L). For the clindamycin-resistant phenotype, MICs increased at least two-fold compared to that of E. coli DH5α competent cells (increase to ≥32 mg/L from 16 mg/L).
3.3 Gene-cassette carrying cfr(C)
A total of six cfr(C)-carrying gene cassette types (GC-1–6) were identified, differing based on SNP-types (Figure 2 and Table 1). Except for the gene cassette of strain CC024, all five gene cassettes carrying cfr(C) were identified to be encoded on the chromosome. Of the 6 GC types, three types (GC-1, −4, and −5) were flanked by the pcp gene in the upstream of cfr(C). Two types (GC-3 and -6) were bracketed by two copies of the IS607 family member ISChh1 upstream and downstream of cfr(C). GC-5 was bracketed by one ISChh1 upstream of cfr(C) and downstream of pcp. In all six GC types, aphA3 was encoded downstream of cfr(C). The aminoglycoside-streptothricin resistance cluster “aphA3-sat4-ΔaadE” was identified from 3 GC types (GC-1, -2, and -4). The GC-1 type (CC021) was “Δpcp-hp-cfr(C)-aphA3-sat4-ΔaadE-pcp,” in which the ppo gene was deleted compared to corresponding region of the reference C. coli SHP40 strain. The GC-4 type (CC041, 042, and 046) was “Δpcp-hp-hp-cfr(C)-aphA3-sat4-ΔaadE,” with pcp deleted upstream of ΔaadE. The GC-2 type (CC024) was “hp-Δcfr(C)-aphA3-sat4-ΔaadE-hp,” with pcp and hp. deleted in the gene cassette compared to the corresponding region of the GC-1 type. The GC-5 type (CC135) was “Δpcp-hp-cfr(C)-aphA3-ISChh1.” The GC-6 type (CC159) was “ISChh1-cfr(C)-aphA3-ISChh1.” The gene cassette of the SNP-III type (GC-3) consisted of “ΔISChh1-ATPase-hp-ant1-pnp (deoD)-lnu (G)-cfr(C)-aphA3-ISChh1” and contained the insert ATPase-hp-ant1-pnp-lnu(G) compared with the corresponding region of GC-6.
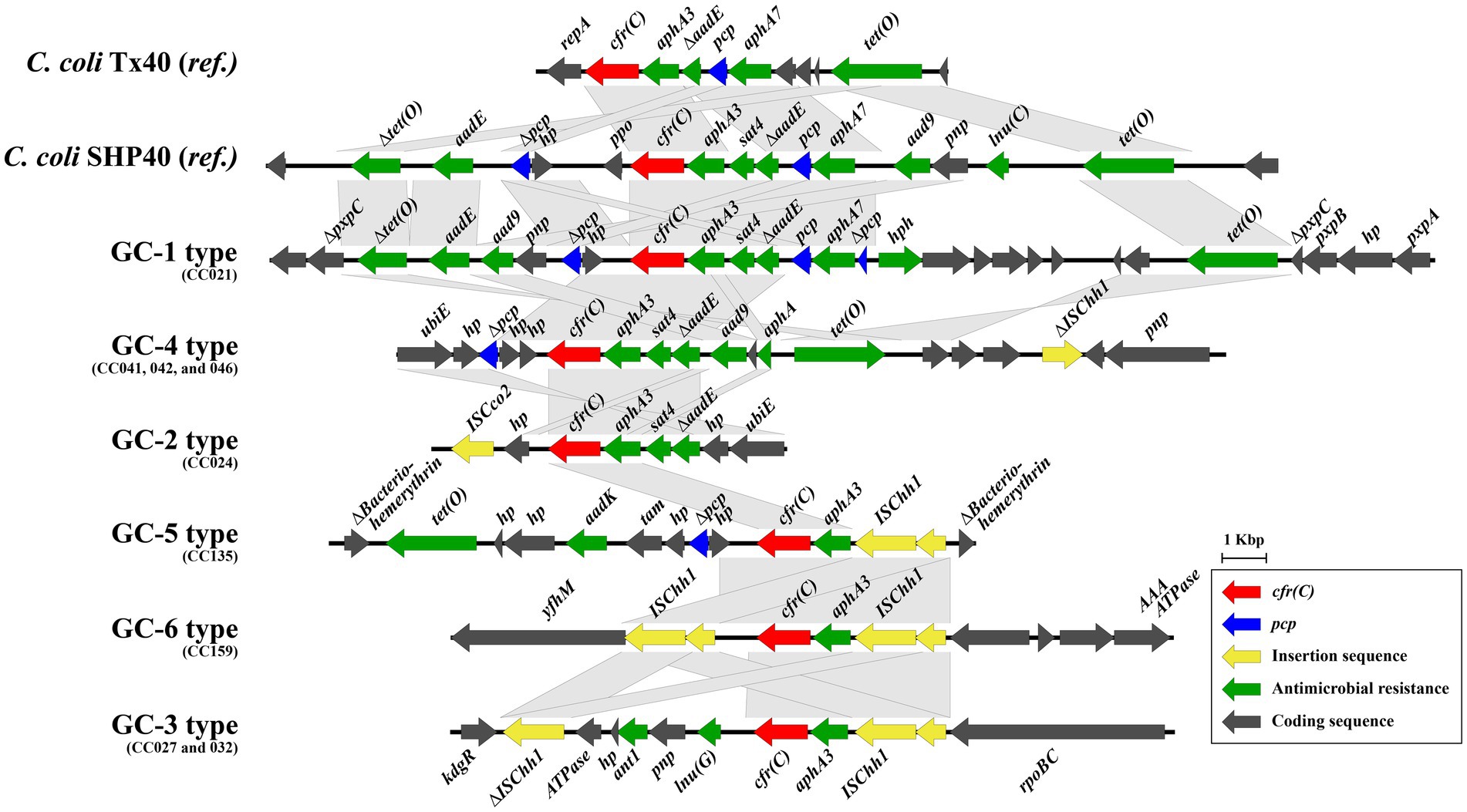
Figure 2. Six types of gene cassettes carrying cfr(C) identified in comparative genomic analysis of nine strains. The nucleotide sequences of cfr(C)-carrying gene cassettes were aligned and compared by using Easyfig (v2.2.3).
3.4 Circular intermediate form of cfr(C)-carrying gene cassettes and horizontal transferability
Of the six GC types identified in this study, five formed circular intermediates of the cfr(C)-cassette. In total, three types of circular intermediates (CIR-1, CIR-2, and CIR-3) were identified (Figure 3A). The CIR-1 type, which was 3,678-bp long and consisted of “pcp-hp-cfr(C)-aphA3,” was identified from three GC types, GC-1, -4, and -5. The CIR-2 type, which was 7,833-bp long and consisted of “ΔISChh1-aphA3-cfr(C)-lnu(G)-pnp-ant1-hp-ATPase,” was identified from GC-3. CIR-3, which was 4,854-bp long and consisted of ‘“ISChh1-aphA3-cfr(C)-hp,” was identified from GC-6. The circular intermediate form of GC-2 was not identified.
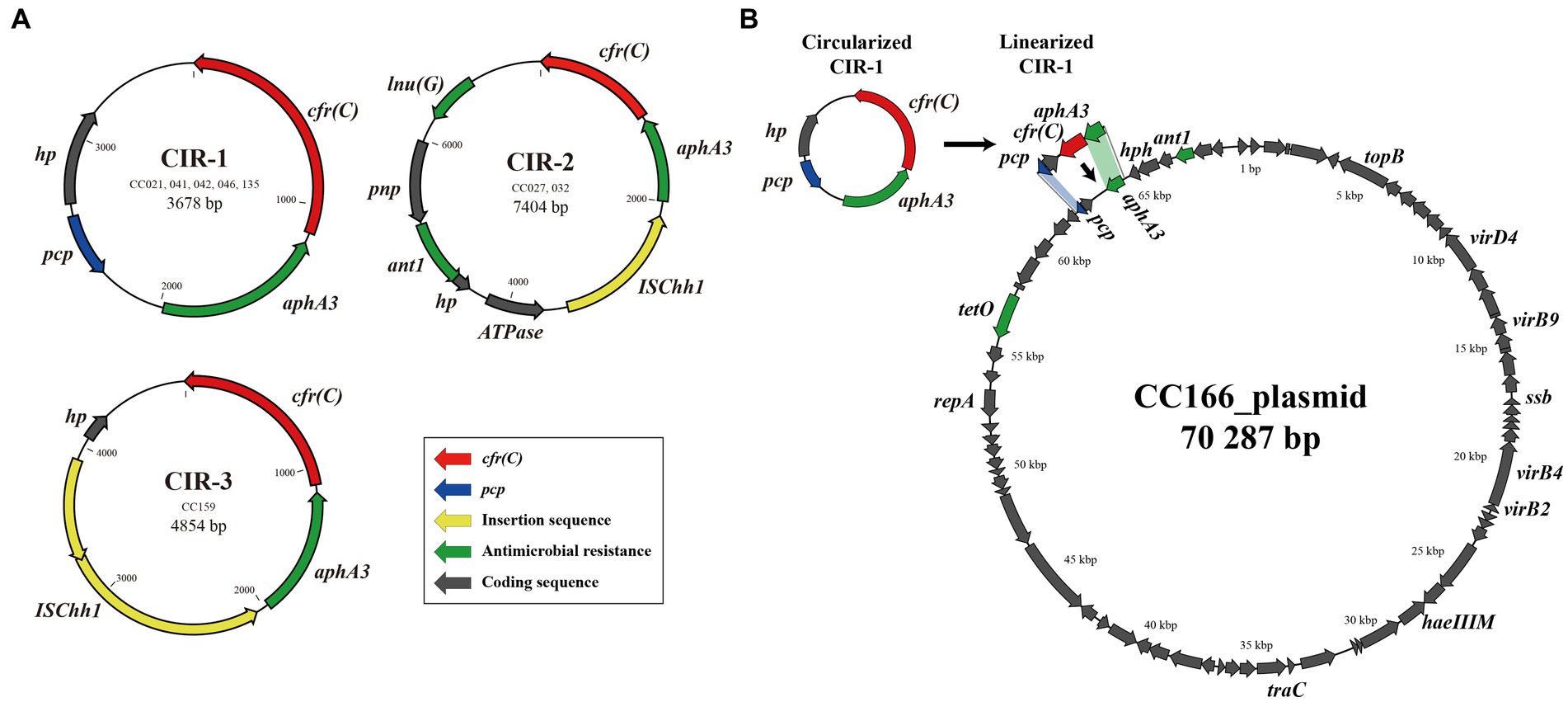
Figure 3. cfr(C)-carrying circular intermediates identified by inverse PCR and amplicon sequencing. Three types of circular intermediates were identified from cfr(C)-carrying C. coli, except for strain CC024. (A) Three types of circular intermediates formed by eight C. coli strains, (B) the genetic environment of recipient strain C. coli CC166 plasmid and insertion site of CIR-1.
In the conjugation assay, CIR-1 was confirmed to be horizontally transferred to recipient strain. The horizontal transferability of other two types, CIR-2 and CIR-3, was not identified. The transconjugants of CIR-1 exhibited acquired resistance to florfenicol (MIC: 8 mg/L) and clindamycin (MIC: 8 mg/L). In WGS analysis, the “pcp-hp-aphA3” region encoded on the plasmid of wild-type strain CC166 was replaced into “pcp-hp-cfr(C)-aphA3” after conjugation with a CIR-1 type strain (Figure 3B). As a result, the size of the plasmid increased from 70,287 to 71,195 bp.
3.5 Favored insertion site of cfr(C)-carrying gene cassette
The genomic island of the GC-1 type was composed of ΔtetO-aadE-aad9-pnp-Δpcp-hp-cfr(C)-aphA3-satA-ΔaadE-Δpcp-hph-hp (2)-traC-hp (6)-tetO, which was inserted into the pxpC gene of the pxpABC complex at positions 249,516–268,882 bp (19,367 bp), based on the location of the dnaA gene that encodes the chromosomal replication protein (Figure 4 and Supplementary Figure 3). GC-4, which was part of a genomic island composed of ΔISChh1-hp (3)-tetO-aphA-hp-aad9-hp-sat4-aphA3-cfr(C)-hp (2)-Δpcp-hp-ubiE (14,424 bp), was inserted at positions 1,184,369–1,198,793 bp encoding lptD-pnp-hp upstream and hp (2)-sbnD downstream. For GC-5, the cfr(C)-carrying genomic island (11,787 bp), which was composed of ISChh1-aphA3-cfr(C)-hp (2)-tam-aadK-hp (2)-tetO, was inserted at positions 165,574–177,361 bp on the gene encoding bacteriohemerythrin. As a result of this insertion, both sides of the GC-5 genomic island’s insertion site encoded a truncated bacteriohemerythrin gene. The GC-6 genomic island consisted of ISChh1-aphA3-cfr(C)-ISChh1 and was found to be inserted at positions 227,744–234,515 bp, flanked by “purE-dapB-AAA ATPase-hp (3)” and “yfhM-mtgA-hp” on each side. The GC-3 type was composed of ΔISChh1-ATPase-hp-ant1-pnp-lnu(G)-cfr(C)-aphA3-ISChh1, inserted at positions 1,261,026–1,270,356 bp and flanked by “uxaA-dapA-kdgR” and “rpoBC-rplL” on each side. Lastly, GC-2 of strain CC024 was identified within a circular genetic element with a size of 119,851 bp. The genomic island containing GC-2 was inserted into the traG gene.
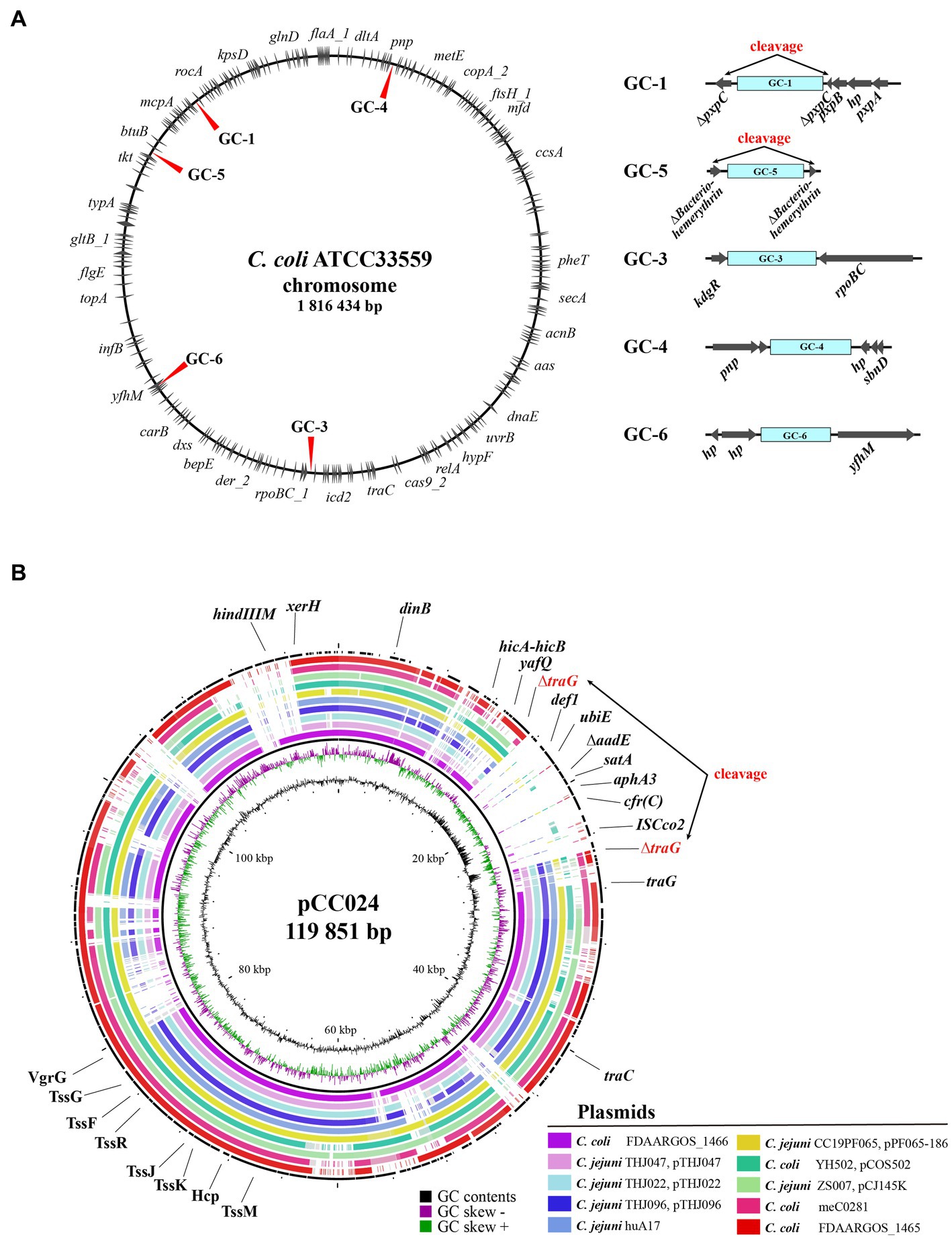
Figure 4. Insertion sites of cfr(C)-carrying gene cassette in wild type cfr(C)-carrying C. coli. (A) Chromosomal insertion sites of each cfr(C)-carrying gene cassette types based on nucleotide sequence of C. coli strain ATCC33559. (B) Comparative genomic analysis of cfr(C)-carrying plasmid-like genetic element of CC024 strain by using BRIG. For comparison, nucleotide sequences of 10 plasmids of Campylobacter species with high genetic homology to the plasmid of CC024 were obtained from NCBI database by using BLAST.
4 Discussion
The rRNA methyltransferase N, encoded by cfr gene, causes m8-modification of A2503 of 23S rRNA, which is known to confer resistance to two antimicrobial agents, florfenicol and clindamycin (Long et al., 2006). Even though cfr(C) is known to induce antimicrobial resistance to critically important antimicrobials to humans, its transfer mechanism is still unclear and requires further research. In this study, nine novel cfr(C)-positive C. coli strains were isolated from swine sources (swine farms and slaughterhouses). To elucidate the horizontal transfer mechanism of the Campylobacter-derived cfr(C) gene, we analyzed the genetic background and horizontal transfer unit of cfr(C) in C. coli strains through comparative genomic analysis.
Compared to the previously reported sequences of cfr(C), two strains (CC021 and CC024) carried incomplete amino acid coding sequences of the cfr(C) gene. Insertion or deletion of one or two adenines at the polyadenine site, such as 19delA (amino acid: M7fs) identified in CC021, generates premature stop codons by downstream frameshifting, leading to premature chain termination (Park et al., 2000). Tang et al. (2020) reported loss of resistance to florfenicol and clindamycin of cfr(C) due to other nucleotide deletion mutations in the C. coli JP10 strain, in which a frameshift mutation occurred due to nucleotide deletion at position 738–756 of cfr(C). The cfr(C) amino acid sequence of CC024 (366 amino acids) exhibited 95.51% identity to that of C. coli Tx40 (379 amino acids). Notably, the original CC024 strain, as well as its transformant strain, showed resistance to florfenicol. According to a study of cfr protein structure and function, the function of the cfr protein analog was ensured by a highly conserved amino acid sequence around the C-terminus (G.DIdAACGQL) (Atkinson et al., 2013). In this study, although the amino acid sequence of the cfr(C) gene was truncated in the CC024 strain, the conserved C-terminal region of cfr(C) was preserved, which may ensure the function of cfr(C).
Remarkably, the transformants of CC027 and CC032 with three missense non-synonymous (ns) SNPs (281A > C, 532A > C, and 952A > G) showed weak resistance to florfenicol (4–8 mg/L). These three missense nsSNPs were also identified in the florfenicol resistant cfr(C)-positive strains, CC135 and CC159. The amino acid substitutions (E94A, K178Q, and I318V), due to three SNP sites, have also been identified in the florfenicol and clindamycin-resistant C. coli JZ_1_79 and SH96 strains reported by Tang et al. (2020). The cfr(C)-encoded rRNA methyltransferase has functional amino acid sites including three substrate entrance sites and eight substrate modulation sites (Atkinson et al., 2013). Considering that the mutation sites of CC027 and CC032 do not overlap with the mutation sites of the previous report, it can be inferred that the three SNPs of these two strains are not associated with the phenotype for florfenicol resistance. Therefore, in relation to decreased resistance to phenicols found in strains CC027 and CC032, further research on the genetic background and expression of cfr(C) is needed.
Except for CC159, all eight cfr(C)-positive C. coli strains showed resistance to clindamycin, although the resistance level was different between strains depending on the carriage of 23S rRNA A2075G substitution: four strains with 23S rRNA A2075G substitutions showed high-level clindamycin resistance, whereas strains without the 23S rRNA A2075G substitution showed low-level resistance. The 23S rRNA A2075G substitution is known to be the most common mechanism underlying high-level resistance to the macrolide and lincosamide classes of antimicrobial agents in Campylobacter (Perez-Boto et al., 2010). Considering that the resistance of bacteria to lincosamide is generally mediated by the combination of three mechanisms (ribosomal target methylation or mutation, efflux of antimicrobials, and drug inactivation) (Leclercq, 2002), strains with simultaneous cfr(C)-induced A2503 methylation and A2075G substitution could exhibit high resistance to the lincosamide class. Our result suggests that cfr(C)-induced A2503 methylation may lead to low-level resistance to clindamycin, which could be enhanced by the acquisition of A2075G substitution.
cfr(C) is encoded on transferable plasmids (Tang et al., 2017;Liu et al., 2019a; Tang et al., 2020) or the chromosome of cfr(C)-positive C. coli strains (Liu et al., 2019a; Tang et al., 2020). In this study, the cfr(C) gene was encoded on the bacterial chromosome in the form of gene cassettes in eight cfr(C)-positive C. coli strains, except the gene cassette of strain CC024, and a total of six different gene cassettes were identified. Consistent with previous studies, all six cfr(C) gene cassettes carried the aminoglycoside resistance gene, aphA3, which was encoded upstream of cfr(C) in the same direction (Tang et al., 2017; Zhao et al., 2019; Liu et al., 2019a; Tang et al., 2020). Three cassette types, namely GC-1 (CC021), GC-2 (CC041, 042, and 046), and GC-5 (CC024), carried the aminoglycoside-streptothricin resistance gene cluster “aphA3-sat4-aadE” in the upstream of the cfr(C)-cassette gene. The “aphA3-sat4-aadE” cluster was also reported to be encoded in the upstream of the cfr(C)-cassette in the C. coli SHP40 strain, where the cfr(C) gene was first reported on the chromosome (Liu et al., 2019a). In contrast, the aminoglycoside resistance gene cluster “aphA3-aadE” without “sat4” was encoded in the upstream of the cfr(C)-cassette in the plasmids of 2 C. coli strains, Tx40 and N61925F (Tang et al., 2017; Zhao et al., 2019). The aphA3 gene is an aminoglycoside-resistance determinant, encoding aminoglycoside 3′-phosphotransferase, and the sat4 gene is a streptothricin-resistance determinant, encoding streptothricin N-acetyltransferase (Lysnyansky and Borovok, 2021b). The “aphA3-sat4-aadE” cluster is one of the highly transmissible and disseminated clusters, mainly found in the plasmids of gram-positive (Staphylococcus and Enterococcus) and gram-negative bacteria (Campylobacter) (Boerlin et al., 2001; Lysnyansky and Borovok, 2021a; Guirado et al., 2022). The aphA3-sat4-aadE cluster is transmitted with various antimicrobial-resistance genes, such as ermB, optrA, and tet (O) (Werner et al., 2001, 2003; Palmieri et al., 2012). Although the aphA3-sat4-aadE cluster has been reported to be mediated by mobile genetic elements, such as Tn5405 (Werner et al., 2001), recent studies have also shown that in Campylobacter spp., this resistance gene cluster may be mediated by a process of homologous recombination (Qin et al., 2012). Homologous recombination may enable horizontal transfer and chromosomal integration of gene clusters without mobile genetic elements (de Vries and Wackernagel, 2002). In the case of GC-4, another resistance gene cluster, “ant1-pnp-lnu(G),” was encoded in the upstream of the cfr(C)-cassette. The lincosamide resistance gene, lnu (E), is encoded in the upstream of the ISEnfa5-flanked cfr gene cassette in the Streptococcus suis RN4220 strain (Zhao et al., 2014).
The random insertion of a gene cassette into cfr(C) has the potential to significantly affect the fitness and survival of Campylobacter, as the insertion of the cfr(C) gene cluster may interfere with crucial cellular processes of the bacterium. The insertion sites of GC-1 and -5, harboring genomic islands, were different from those of the potD gene reported by Tang et al. (2020). GC-1 was inserted into the pxpC gene, which encodes 5-oxoprolinase. 5-Oxoproline, a metabolite from the breakdown of glutamine, has been reported to inhibit bacterial growth when it accumulates without proper disposal (Niehaus et al., 2017). GC-5 was inserted into the bacteriohemerythrin-encoding gene. Hemerythrin helps maintain bacterial homeostasis through intracellular oxygen- and redox-sensing functions. Cells cannot rapidly adapt to environmental changes without hemerythrin (Kitanishi, 2022). Our results suggest that the cleavage of these two genes, caused by the insertion of cfr(C)-carrying GCs, could have detrimental effects on the survivability of C. coli strains. The GC-3, -4, and -6 types were inserted between genes without gene cleavage. The insertion site of GC-3 downstream of the rpoBC gene has been previously reported (Tang et al., 2020). However, the other two types of insertion sites, pnp to sbnD and ATPase-encoding gene to yfhM, are novel positions. In contrast, GC-2 (CC024) was inserted into a circular genetic element without a self-replication module, such as the replication initiation protein gene repA. This genetic element of CC024 was similar in genetic composition to the previously reported plasmids of C. jejuni and C. coli. In addition, differences in GC contents between insertion sites and other coding regions indicated that the cfr(C)-carrying cluster was not included in ancestral genetic elements. TraG, truncated by GC-2, plays major roles in conjugation, and is associated with the invasion of C. jejuni into epithelial cells (Poly et al., 2005). Thus, truncation of the traG gene could attenuate the pathogenicity and survivability of Campylobacter.
Of the six types of gene cassette, GC-1, -4, and -5 formed circular intermediates and were horizontally transferred to recipient C. coli strains. The transferred circular intermediate form was 3,678 bp in length and consisted of the four coding sequences “pcp-hp-cfr(C)-aphA3.” Although the present study found differences in the length and composition of cfr(C)-positive strains, the pcp gene-mediated circular intermediate “pcp-hp-ppo-cfr(C)-aphA3-sat4-aadE” has also been reported in the C. coli SHP40 strain (Liu et al., 2019a). Similar to our study, the cfr(C)-cassette was also encoded in the bacterial chromosome of the C. coli SHP40 strain. The pcp gene encodes an enzyme that is conserved in a variety of organisms, from prokaryotes (archaea and bacteria) to eukaryotes (Barrett et al., 2012). The pcp gene is found to be inserted at multiple sites in the bacterial genome, including chromosomes and plasmids, and this multiple insertion of pcp could mediate the intermolecular transfer of gene clusters by forming the circular intermediate: the circular intermediates released from chromosome/plasmids could be integrated into other chromosome/plasmids containing a homologous sequence, such as the pcp gene (Liu et al., 2019a). These characteristics of the pcp gene can facilitate the transmission of cfr(C), which forms a cluster with pcp. Similarly, tetO-mediated circular intermediates of ermB, escaping from original plasmids/chromosome, could be integrated into other bacterial genomes by targeting the multiple inserted tetO homologous sequence (Liu et al., 2019b). In particular, the CC041, CC042, and CC046 strains that form GC-4 have been reported as hyper-aerotolerant strains, along with ST830, which is related to clinical isolates, according to our previous study (Guk et al., 2021). Since these strains not only transfer cfr(C) to other strains but also have a higher probability for transmission to humans than other C. coli strains, hyper-aerotolerant cfr(C)-positive C. coli potentially carry higher risk than other cfr(C)-positive C. coli strains.
In GC-3, 4, 5, and 6, cfr(C) was flanked by the IS607 family member ISChh1. The IS607 family is one of the major insertion sequences (ISs) that mediate transfer of antimicrobial resistance in Campylobacter spp. (Chen et al., 2018). For example, IS607 has been reported to mediate the transfer of the optrA gene, which induces resistance to oxazolidinones and phenicols, as well as of fexA, a florfenicol–chloramphenicol resistance gene (Tang et al., 2021). The IS607 family encoded in the upstream of the cfr(C)-gene cassette was also identified in the plasmids of the JZ_1_79, SH89, and JZ_1_74 strains reported by Tang et al. (2020). Among the three cfr(C)-cassette types flanked by IS607, GC-3 and GC-6 were bracketed by two copies of ISChh1 in the same orientation at both sides of the gene cassette and formed the ISChh1-mediated circular intermediates “ISChh1-aphA3-cfr(C)-lnu(G)-pnp-antl1-hp-ATPase” and “ISChh1-aphA3-cfr(C)-hp,” respectively. Even though the circular intermediates of GC-3 and -6, namely CIR-2 and -3, were not horizontally transferred in our study, two copies of IS elements in the same orientation can form a circular intermediate and can be transferred using the mechanism “copy-out-paste-in,” since the IS607 family is a highly active IS element (He et al., 2019). Tang et al. (2020) confirmed the existence of IS607 only upstream of the cfr(C)-cassette but could not confirm the formation of circular intermediates by the IS607 family. GC-4 and -5, bracketed by the ISChh1 segment and pcp, formed a circular intermediate mediated by pcp regardless of the position of the ISChh1 segment. These results suggest that two copies of IS607 in the same orientation play an important role in the formation of a circular intermediate form of gene cassettes.
The CIR-1s of five strains (CC021, 041, 042, 046, and 135) were horizontally transferred from the chromosomes into the plasmid of the recipient strain CC166 in the conjugation assay. For all five strains, the pcp and aphA3 genes were present at the insertion sites, suggesting that insertion of CIR-1 may have been mediated by homologous recombination with pcp and aphA3. The conjugation of gene cassettes targeting homologous gene regions has been reported in several studies. Qin et al. (2012) suggested that the aminoglycoside resistance gene island of C. coli strain SX81 may be acquired from the C. coli strain RM2228 and mediated by homologous recombination of the cadF and CCO1582 genes (Qin et al., 2012). Taken together with our results, this suggests that the use of pcp, which is a commonly encoded gene in Campylobacter, as a vector in the horizontal transfer of cfr(C)-carrying gene cassettes may be a possible reason for the global success of cfr(C) propagation.
This is the first study to elucidate the horizontal transferability of cfr(C) and random insertion site of the chromosome-encoded cfr(C) cassette in Campylobacter spp. The results reveal novel cfr(C) variants along with their associated genetic environments in C. coli isolates and indicate the flexibility of C. coli in acquiring new antimicrobial resistance genes. The transferability of chromosomal cfr(C) to humans may be attributed to the global spread of multidrug resistance against clinically important antimicrobials. Thus, enhanced surveillance is needed to monitor the emergence and spread of cfr(C) in Campylobacter from swine sources as well as other pathogens.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1000181.
Author contributions
J-UA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. SL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Visualization, Writing – original draft, Writing – review & editing. J-HG: Conceptualization, Formal analysis, Methodology, Writing – original draft. JW: Conceptualization, Formal analysis, Methodology, Writing – original draft. HS: Writing – review & editing. SC: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Research Foundation of Korea (NRF-2021R1A2C2005907).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1274245/full#supplementary-material
Abbreviations
BLAST, Basic Local Alignment Search Tool; IS, insertion sequence; MIC, minimum inhibitory concentration; NCBI, National Center for Biotechnology Information; Ns, non-synonymous; PCR, polymerase chain reaction; SNP, single nucleotide polymorphism; WGS, whole genome sequencing; WHO, World Health Organization.
Footnotes
References
Aarestrup, F. M., Mcdermott, P. F., and Wegener, H. C. (2008). “Transmission of antibiotic resistance from food animals to humans” in Campylobacter. eds. I. Nachamkin, C. M. Szymanski, and M. J. Blaser, vol. 3 (Washington, DC: ASM Press), 645–665.
Atkinson, G. C., Hansen, L. H., Tenson, T., Rasmussen, A., Kirpekar, F., and Vester, B. (2013). Distinction between the Cfr methyltransferase conferring antibiotic resistance and the housekeeping RlmN methyltransferase. Antimicrob. Agents Chemother. 57, 4019–4026. doi: 10.1128/AAC.00448-13
Barrett, A.J., Woessner, J.F., and Rawlings, N.D. (2012). Handbook of proteolytic enzymes. London: Elsevier.
Boerlin, P., Burnens, A. P., Frey, J., Kuhnert, P., and Nicolet, J. (2001). Molecular epidemiology and genetic linkage of macrolide and aminoglycoside resistance in Staphylococcus intermedius of canine origin. Vet. Microbiol. 79, 155–169. doi: 10.1016/S0378-1135(00)00347-3
Burnham, P. M., and Hendrixson, D. R. (2018). Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol. 16, 551–565. doi: 10.1038/s41579-018-0037-9
Candela, T., Marvaud, J.-C., Nguyen, T. K., and Lambert, T. (2017). A cfr-like gene cfr (C) conferring linezolid resistance is common in Clostridium difficile. Int. J. Antimicrob. Agents 50, 496–500. doi: 10.1016/j.ijantimicag.2017.03.013
CDC (2013). Antibiotic resistance threats in the United States, 2013. Available at: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
Chen, W., Mandali, S., Hancock, S. P., Kumar, P., Collazo, M., Cascio, D., et al. (2018). Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition. Elife 7:e39611. doi: 10.7554/eLife.39611
Davis, L., Young, K., and Dirita, V. (2008). Genetic manipulation of Campylobacter jejuni. Curr. Protoc. Microbiol. 10, Unit 8A 2 1–8A 2 17. doi: 10.1002/9780471729259.mc08a02s10
de Vries, J., and Wackernagel, W. (2002). Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc. Natl. Acad. Sci. U. S. A. 99, 2094–2099. doi: 10.1073/pnas.042263399
Deshpande, L. M., Ashcraft, D. S., Kahn, H. P., Pankey, G., Jones, R. N., Farrell, D. J., et al. (2015). Detection of a new cfr-like gene, cfr (B), in Enterococcus faecium isolates recovered from human specimens in the United States as part of the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 59, 6256–6261. doi: 10.1128/AAC.01473-15
Guirado, P., Miro, E., Iglesias-Torrens, Y., Navarro, F., Campoy, S., Alioto, T. S., et al. (2022). A new variant of the aad E-sat 4-aph A-3 gene cluster found in a conjugative plasmid from a MDR Campylobacter jejuni isolate. Antibiotics 11:466. doi: 10.3390/antibiotics11040466
Guk, J. H., Song, H., Yi, S., An, J. U., Lee, S., Kim, W. H., et al. (2021). Hyper-Aerotolerant Campylobacter coli from swine may pose a potential threat to public health based on its quinolone resistance, virulence potential, and genetic relatedness. Front. Microbiol. 12:703993. doi: 10.3389/fmicb.2021.703993
Hansen, L. H., and Vester, B. (2015). A cfr-like gene from Clostridium difficile confers multiple antibiotic resistance by the same mechanism as the cfr gene. Antimicrob. Agents Chemother. 59, 5841–5843. doi: 10.1128/AAC.01274-15
He, Y. Z., Li, X. P., Miao, Y. Y., Lin, J., Sun, R. Y., Wang, X. P., et al. (2019). The ISApl1 (2) dimer circular intermediate participates in mcr-1 transposition. Front. Microbiol. 10:15. doi: 10.3389/fmicb.2019.00015
Jaakkonen, A., Kivisto, R., Aarnio, M., Kalekivi, J., and Hakkinen, M. (2020). Persistent contamination of raw milk by Campylobacter jejuni ST-883. PLoS One 15:e0231810. doi: 10.1371/journal.pone.0231810
Kitanishi, K. (2022). Bacterial hemerythrin domain-containing oxygen and redox sensors: versatile roles for oxygen and redox signaling. Front. Mol. Biosci. 9:967059. doi: 10.3389/fmolb.2022.967059
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Leclercq, R. (2002). Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34, 482–492. doi: 10.1086/324626
Liu, D., Li, X., Liu, W., Yao, H., Liu, Z., Fessler, A. T., et al. (2019a). Characterization of multiresistance gene cfr(C) variants in Campylobacter from China. J. Antimicrob. Chemother. 74, 2166–2170. doi: 10.1093/jac/dkz197
Liu, D., Liu, W., Lv, Z., Xia, J., Li, X., Hao, Y., et al. (2019b). Emerging erm (B)-mediated macrolide resistance associated with novel multidrug resistance Genomic Islands in Campylobacter. Antimicrob. Agents Chemother. 63:e00153-19. doi: 10.1128/AAC.00153-19
Long, K. S., Poehlsgaard, J., Kehrenberg, C., Schwarz, S., and Vester, B. (2006). The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505. doi: 10.1128/AAC.00131-06
Lysnyansky, I., and Borovok, I. (2021a). A GC-rich prophage-like genomic region of Mycoplasma bovirhinis HAZ141_2 carries a gene cluster encoding resistance to kanamycin and neomycin. Antimicrob. Agents Chemother. 65:e01010-20. doi: 10.1128/AAC.01010-20
Lysnyansky, I., and Borovok, I. (2021b). The aad E*-sat 4-aph A-3 gene cluster of Mycoplasma bovirhinis HAZ141_2 undergoes genomic rearrangements influencing the primary promoter sequence. Antibiotics 10:1335. doi: 10.3390/antibiotics10111335
Niehaus, T. D., Elbadawi-Sidhu, M., De Crecy-Lagard, V., Fiehn, O., and Hanson, A. D. (2017). Discovery of a widespread prokaryotic 5-oxoprolinase that was hiding in plain sight. J. Biol. Chem. 292, 16360–16367. doi: 10.1074/jbc.M117.805028
Palmieri, C., Mingoia, M., Massidda, O., Giovanetti, E., and Varaldo, P. E. (2012). Streptococcus pneumoniae transposon Tn1545/Tn6003 changes to Tn6002 due to spontaneous excision in circular form of the erm (B)- and aph A3-containing macrolide-aminoglycoside-streptothricin (MAS) element. Antimicrob. Agents Chemother. 56, 5994–5997. doi: 10.1128/AAC.01487-12
Park, B. K., Jang, J. S., and Park, K. (2000). Deletion of one adenine base within the polyadenine tract of transforming growth factor-beta receptor type II in human MDA-MB-231 breast cancer cell line. Int. J. Oncol. 17, 473–478. doi: 10.3892/ijo.17.3.473
Perez-Boto, D., Lopez-Portoles, J. A., Simon, C., Valdezate, S., and Echeita, M. A. (2010). Study of the molecular mechanisms involved in high-level macrolide resistance of Spanish Campylobacter jejuni and Campylobacter coli strains. J. Antimicrob. Chemother. 65, 2083–2088. doi: 10.1093/jac/dkq268
Poly, F., Threadgill, D., and Stintzi, A. (2005). Genomic diversity in Campylobacter jejuni: identification of C. jejuni 81-176-specific genes. J. Clin. Microbiol. 43, 2330–2338. doi: 10.1128/JCM.43.5.2330-2338.2005
Qin, S., Wang, Y., Zhang, Q., Chen, X., Shen, Z., Deng, F., et al. (2012). Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob. Agents Chemother. 56, 5332–5339. doi: 10.1128/AAC.00809-12
Sassi, M., Guérin, F., Zouari, A., Beyrouthy, R., Auzou, M., Fines-Guyon, M., et al. (2019). Emergence of optr A-mediated linezolid resistance in enterococci from France, 2006–16. J. Antimicrob. Chemother. 74, 1469–1472. doi: 10.1093/jac/dkz097
Schwarz, S., Werckenthin, C., and Kehrenberg, C. (2000). Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44, 2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Silva, J., Leite, D., Fernandes, M., Mena, C., Gibbs, P. A., and Teixeira, P. (2011). Campylobacter spp. as a foodborne pathogen: a review. Front. Microbiol. 2:200. doi: 10.3389/fmicb.2011.00200
Stojković, V., Ulate, M. F., Hidalgo-Villeda, F., Aguilar, E., Monge-Cascante, C., Pizarro-Guajardo, M., et al. (2019). cfr (B), cfr (C), and a new cfr-like gene, cfr (E), in Clostridium difficile strains recovered across Latin America. Antimicrob. Agents Chemother. 64:e01074-19. doi: 10.1128/AAC.01074-19
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tang, Y., Dai, L., Sahin, O., Wu, Z., Liu, M., and Zhang, Q. (2017). Emergence of a plasmid-borne multidrug resistance gene cfr(C) in foodborne pathogen Campylobacter. J. Antimicrob. Chemother. 72, 1581–1588. doi: 10.1093/jac/dkx023
Tang, Y. Z., Lai, Y., Wang, X. Y., Lei, C. W., Li, C., Kong, L. H., et al. (2021). Novel insertion sequence ISChh1-like mediating acquisition of optr a gene in foodborne pathogen Campylobacter coli of swine origin. Vet. Microbiol. 252:108934. doi: 10.1016/j.vetmic.2020.108934
Tang, Y., Lai, Y., Yang, X., Cao, X., Hu, Y., Wang, X., et al. (2020). Genetic environments and related transposable elements of novel cfr(C) variants in Campylobacter coli isolates of swine origin. Vet. Microbiol. 247:108792. doi: 10.1016/j.vetmic.2020.108792
Werner, G., Hildebrandt, B., and Witte, W. (2001). Aminoglycoside-streptothricin resistance gene cluster aad E-sat 4-aph A-3 disseminated among multiresistant isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 45, 3267–3269. doi: 10.1128/AAC.45.11.3267-3269.2001
Werner, G., Hildebrandt, B., and Witte, W. (2003). Linkage of erm (B) and aad E-sat 4-aph A-3 in multiple-resistant Enterococcus faecium isolates of different ecological origins. Microb. Drug Resist. 9, 9–16. doi: 10.1089/107662903322541847
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Yanisch-Perron, C., Vieira, J., and Messing, J. (1985). Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33, 103–119. doi: 10.1016/0378-1119(85)90120-9
Zhao, S., Mukherjee, S., Hsu, C. H., Young, S., Li, C., Tate, H., et al. (2019). Genomic analysis of emerging florfenicol-resistant Campylobacter coli isolated from the cecal contents of cattle in the United States. mSphere 4:e00367-19. doi: 10.1128/mSphere.00367-19
Zhao, Q., Wendlandt, S., Li, H., Li, J., Wu, C., Shen, J., et al. (2014). Identification of the novel lincosamide resistance gene lnu (E) truncated by ISEnfa5-cfr-ISEnfa5 insertion in Streptococcus suis: de novo synthesis and confirmation of functional activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 1785–1788. doi: 10.1128/AAC.02007-13
Keywords: Campylobacter coli , chromosome-encoded, circular intermediate, horizontal transfer, whole genome sequencing
Citation: An J-U, Lee S, Guk J-H, Woo J, Song H and Cho S (2023) Circular intermediate-mediated horizontal transfer of the chromosome-encoded cfr(C) gene in multi-drug resistant Campylobacter coli from swine sources. Front. Microbiol. 14:1274245. doi: 10.3389/fmicb.2023.1274245
Edited by:
Xunde Li, University of California, Davis, United StatesReviewed by:
Ben Pascoe, University of Oxford, United KingdomMónica Oleastro, National Health Institute Doutor Ricardo Jorge (INSA), Portugal
Copyright © 2023 An, Lee, Guk, Woo, Song and Cho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seongbeom Cho, chose@snu.ac.kr
 Jae-Uk An
Jae-Uk An Soomin Lee
Soomin Lee Jae-Ho Guk
Jae-Ho Guk Jungha Woo
Jungha Woo Hyokeun Song
Hyokeun Song Seongbeom Cho
Seongbeom Cho