- Department of Biological Sciences, Eck Institute for Global Health, University of Notre Dame, Notre Dame, IN, United States
Chikungunya virus (CHIKV) is an emerging mosquito-borne pathogen of significant public health importance. There are currently no prophylactic vaccines or therapeutics available to control CHIKV. One approach to arbovirus control that has been proposed is the replacement of transmission-competent mosquitoes with those that are refractory to virus infection. Several transgene effectors are being examined as potentially useful for this population replacement approach. We previously demonstrated the successful use of hammerhead ribozymes (hRzs) as an antiviral effector transgene to control CHIKV infection of, and transmission by, Aedes mosquitoes. In this report we examine a maxizyme approach to enhance the catalytic activity and prevent virus mutants from escaping these ribozymes. We designed a maxizyme containing minimized (monomer) versions of two hRzs we previously demonstrated to be the most effective in CHIKV suppression. Three versions of CHIKV maxizyme were designed: Active (Mz), inactive (ΔMz), and a connected CHIKV maxizyme (cMz). The maxizymes with their expression units (Ae-tRNA val promoter and its termination signal) were incorporated into lentivirus vectors with selection and visualization markers. Following transformation, selection, and single-cell sorting of Vero cells, clonal cell populations were infected with CHIKV at 0.05 and 0.5 MOI, and virus suppression was assessed using TCID50-IFA, RT-qPCR, and caspase-3 assays. Five transgenic mosquito lines expressing cMz were generated and transgene insertion sites were confirmed by splinkerette PCR. Our results demonstrate that Vero cell clones expressing Mz exhibited complete inhibition of CHIKV replication compared to their respective inactive control version or the two parent hRzs. Upon oral challenge of transgenic mosquitoes with CHIKV, three out of the five lines were completely refractory to CHIKV infection, and all five lines tested negative for salivary transmission. Altogether, this study demonstrates that maxizymes can provide a higher catalytic activity and viral suppression than hRzs.
Introduction
Chikungunya virus (CHIKV) is an enveloped, single-stranded, positive-sense RNA virus that belongs to the genus Alphavirus and family Togaviridae. CHIKV is transmitted to humans by Aedes aegypti and Ae. albopictus mosquitoes (Weaver and Lecuit, 2015; Higgs and Vanlandingham, 2018), causing chikungunya fever (CF), which is characterized by symptoms such as fever, myalgia, and debilitating joint pain that may last for months (Pialoux et al., 2007; Weaver and Lecuit, 2015; Halstead, 2018), and in some cases, can result in fatality (Cardona-Ospina et al., 2015). The disease can impact the economy in several spheres, significantly affecting the health system and national economies (Costa et al., 2023). Currently there are no effective prophylactic or therapeutic measures to control CF, although a putative vaccine is making its way through clinical trials (Schmidt and Schnierle, 2022; Schneider et al., 2023).
Since arboviruses such as CHIKV require mosquitoes to complete their transmission cycle (Franz et al., 2006), alternative approaches aimed at reducing or replacing naturally competent mosquitoes with virus refractory mosquitoes expressing various transgene effectors have been pursued to control arboviruses (Gantz et al., 2015; Aliota et al., 2016; Williams et al., 2020; Reid et al., 2021).
In our lab we have been exploring the potential of several types of antiviral ribozymes including hammerhead ribozyme (hRz) and maxizyme (Mz), among others (Nawtaisong et al., 2009; Carter et al., 2015; Mishra et al., 2016). Hammerhead ribozymes are small catalytic RNA molecules that can cleave target RNA in a sequence-specific manner. In contrast, maxizymes (Mz) are dual-catalytic RNA molecules capable of simultaneously cleaving multiple target sequences in an RNA molecule.
We previously identified two hRzs, #9 and #14, targeting the sub-genomic region of CHIKV that were effective at inhibiting CHIKV 181/25 replication both in vitro and in vivo (Mishra et al., 2016). However, these single hRzs target regions of 19 nt in size, making them potentially susceptible to escape variants.
In this study, we utilized a maxizyme approach to enhance the ribozyme activity and provide broad spectrum activity against escape variants (Haasnoot et al., 2007). Maxizymes (Mz) utilize minimized versions of two hRzs by combining them into a single catalytic unit (Kuwabara et al., 1998). A maxizyme consists of two minizymes (minimized hRz) that lack stem loop II of the hammerhead structure (Figure 1). While minizymes have lower catalytic activity compared to their parental hRzs, some have cleavage activity similar to or better than their parental hRzs when they are configured into a dimeric form such as maxizymes (Iyo et al., 2002, 2004; Kuwabara et al., 2002) (Figure 1).
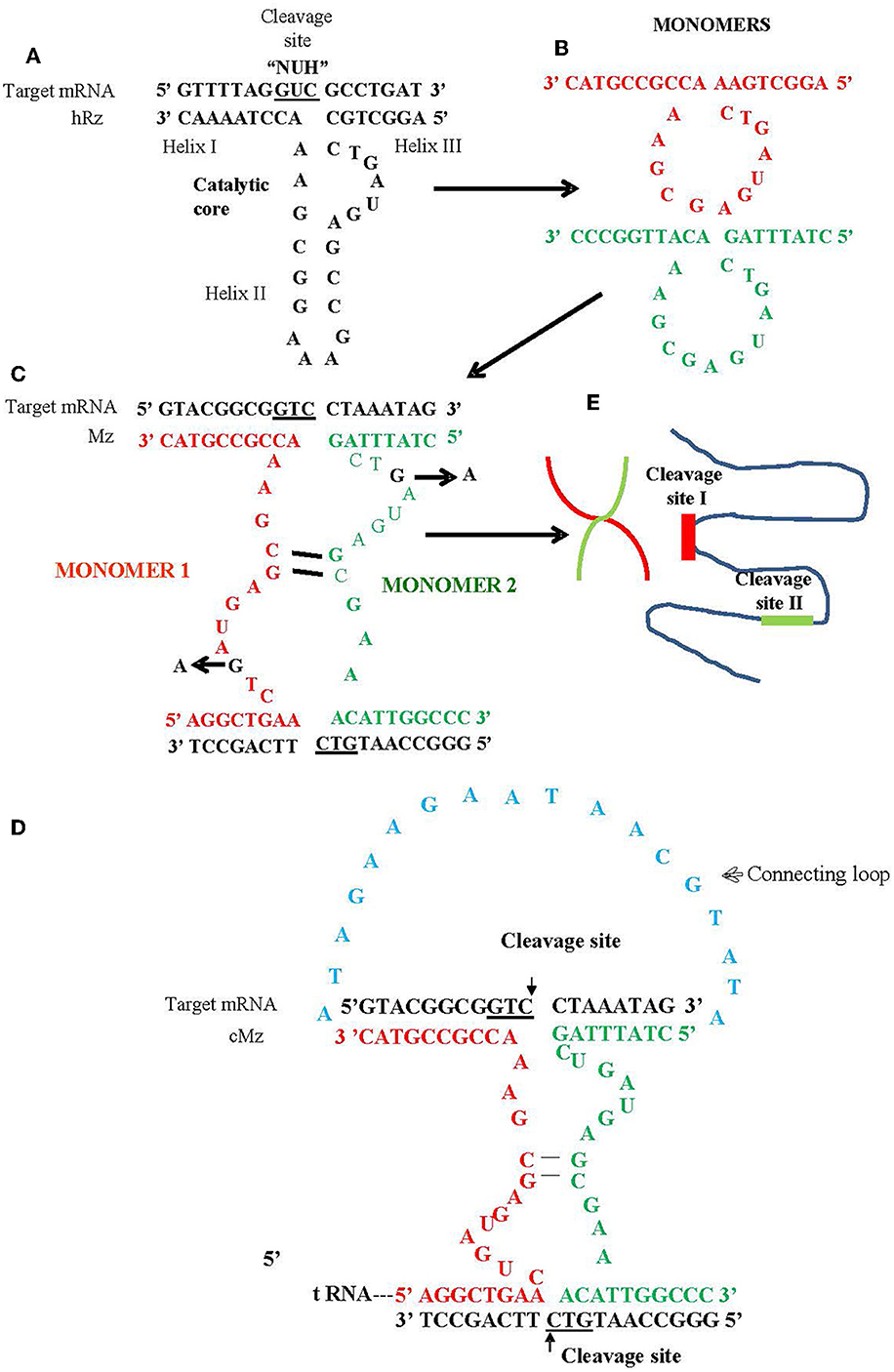
Figure 1. Schematic of hammerhead ribozyme (hRz) and maxizymes. (A) Structure of hRz. (B) Monomers derived from hRzs by deletion of helix II stem loop structure. (C) Monomers (Red or Green) undergoing bimolecular interaction leading to the formation of heterodimer/maxizyme, (guanine: G in Black) in the catalytic core for both monomers replaced by adenine (A) to generate inactive version of maxizyme (D) connected maxizyme with 13 nucleotide connecting chain (Blue) (E) Maxizyme binding to two different target sites on viral RNA.
Previous research has demonstrated that Mzs are catalytically more active compared to hRzs (Kuwabara et al., 1998, 2002; Hamada et al., 1999). The binding of one target site enhances unwinding of RNA secondary structures in the targeted RNA and serves as an alternative approach to the recruitment of RNA helicases, thereby cleaving less accessible sites in the target RNA molecule through binding two arms rather than one (Kuwabara et al., 2002).
Our maxizyme constructs demonstrated greater effectiveness against CHIKV infection than the hRzs they were derived from when expressed in both transformed cell cultures and transgenic mosquitoes. These results suggest that maxizyme can be an effective antiviral for arboviruses like CHIKV, DENV, and Zika as mosquito transgene effectors.
Materials and methods
Cells and viruses
African monkey kidney (Vero) cells (ATCC, USA) were maintained on Dulbecco's modified eagle medium (DMEM; Sigma Aldrich, USA) supplemented with 10% fetal bovine serum (FBS, Atlanta Biological, Flowery Branch, GA, USA) and non-essential amino acids [(1x), Gibco, USA)]. The CHIKV 181/25 strain is an attenuated vaccine strain (a gift from Dr. Scott Weaver, UTMB, Galveston) that was used for testing the effectiveness of our maxizymes in vitro and in vivo. We chose this strain for safety reasons since we do not have appropriate containment facilities for handling a virulent strain. Additionally, while the 181/25 strain is attenuated for human virulence, it does not exhibit significant reduction in mosquito infection.
Design and cloning strategy for CHIKV maxizyme
We designed two versions of a CHIKV maxizyme, active Mz and inactive ΔMz, by combining the most effective anti-CHIKV hammerhead ribozymes (hRzs) #9 and #14 (Mishra et al., 2016). Each maxizyme version consists of two monomers, I and II, with the following components: tRNAval promoter, partial target binding sites of hRz# 9 and #14, catalytic core (active: CTG, or inactive: CTA), and a termination signal (Figure 1 and Table 1).

Table 1. Sequence of maxizymes: each maxizyme sequence includes the Ae-tRNA val promoter (Black), monomer I (Red), monomer II (Green) and stop signal sequence (Purple).
The active Mz was cloned into an Aedes expression lentivirus plasmid, pLAeARH (Nawtaisong et al., 2009), in two steps. The first step amplified Mz monomer I from the pLAeRz#9ARH vector along with the promoter and termination signals using PCR primers Mz-I F and Mz-I R (Table 1) and cloned the amplified sequence into Bam HI and Not I sites of pLAeARH. In the second step, the Mz-II was amplified from the Mz-II template using the primers Mz-II F and Mz-II R (Table 2), and cloned into Pme I and Not I sites of the lentivirus plasmid pLAeMzIARH downstream of Mz-I to construct pLAeMzIAeMzIIARH.

Table 2. Primers/oligos used for the construction of anti-CHIKV maxizyme and connected maxizyme, and for confirmation of maxizyme expression.
The inactive ΔMz was made by introduction of a point mutation (CTG-CTA) to disrupt the catalytic activity of the maxizyme. Cloning of this inactive ΔMz involved generation of the ΔMz monomer I from the pLAeMzIARH vector with active monomer I through PCR amplification using Mz-I F and ΔMz R primers (Table 2). The amplified sequence was inserted at the restriction sites Bam HI and Not I pLAeARH, as described above. The remaining portion of inactive ΔMz was created by amplification from the template ΔMz monomer II using primers Mz-II F and ΔMz II R MI (Table 2) and cloned into Pme I and Not I sites of the lentivirus plasmid pLAeΔMzIARH. The final vector was pLAeΔMzIAeΔMzIIARH. Finally, a CMV-ds RED fluorescent marker was cloned into both the maxizyme plasmids, as previously described (Mishra et al., 2016).
Generation of clonal cell populations
Vero cells were seeded into 6 well plates and 24 hours (hrs) later the ribozyme expression plasmids were transfected using lipofectamine LTX and plus reagent (Invitrogen, USA), following the manufacturer's instructions. Forty-eight hrs later, the transfected cells were selected using 200 μg/ml of hygromycin B (Invitrogen, USA) and maintained for two passages before sorting into 96 well plates. Cell sorting and screening was performed as previously described (Mishra et al., 2016).
RT-PCR based detection of ribozyme expression
Total cellular RNA was TRIzol-extracted following the manufacturer's protocol (Invitrogen, USA). The concentration of extracted RNA was determined spectrophotometrically using a NanoDrop ND-1000 UV-Vis Spectrophotometer. A total of 5μg of RNA was treated with Turbo DNase I (Ambion, USA) and directly used for reverse transcriptase (RT) positive and negative reactions using the Superscript III one step RT-PCR kit (Invitrogen, USA). For the RT negative reaction, Taq DNA polymerase (Invitrogen) was used. A common Ae-tRNA val forward primer was used along with ribozyme-specific reverse primers (Table 2). The RT-PCR products were resolved on 2.0% agarose gels (Ethidium bromide concentration 10 mg/ml) at 105 V for 1 h. Similarly, RT-PCR was performed using twenty mosquitoes per reaction to check for the expression of maxizyme in transgenic mosquitoes.
For mosquito analysis, we collected mosquitoes and organized them into groups of 20. We manually homogenized these groups in 500 μL of Trizol (Invitrogen, USA) followed by centrifugation at 12, 000 g for 10 min at 4°C. After centrifugation, we processed the resulting supernatant for RNA extraction, following the manufacturer's instructions (Invitrogen, USA).
CHIKV infection of vero cells
Wild-type Vero cells and selected clonal Vero cells expressing effector molecules specific to CHIKV were plated at a density of 1 × 105 cells per well. After overnight incubation at 37 °C, the cells were washed once with serum-free DMEM and were challenged with CHIKV 181/25 at an MOI of 0.05 or 0.5 for 2 h. The infected cells were fed with fresh DMEM supplemented with 10% FBS. Two days post infection (dpi) supernatants were collected for TCID50, RT-qPCR, and caspase 3 assays.
TCID50-IFA analysis
CHIKV cell supernatants were collected at 2 dpi for assay. Briefly, 10-fold serial dilutions of virus supernatant were prepared in DMEM plus 10% FBS, and 100 μl of each dilution was aliquoted into 10 wells of a 96 well plate pre-seeded with 1 × 105 cells per well. After 3 dpi, the plates were fixed and stained with a primary antibody (1:100) specific to CHIKV capsid protein (Virostat, USA). Infected positive cells were recognized using a biotinylated secondary antibody (GE healthcare) and streptavidin detection system (Invitrogen). An inverted fluorescent microscope (Nikon, Japan) was used for observation of cytoplasmic fluorescence. Wells scored positive for the presence of green cytoplasmic fluorescence. The numbers of positive wells were counted and the virus titers calculated according to Karber's method (Kärber, 1931). The titer was expressed as log10TCID50/ml.
Caspase 3 assay
The caspase assay was performed using the Caspase-glo 3/7 kit (Promega, USA) according to the manufacturer protocol. Vero cells were plated in 96 well plates 24 h before infection. The cells were then infected with the clones exhibiting complete suppression at an MOI of 0.05. At 2dpi, the cells were incubated with Caspase-glo reagent for 1 h in the dark at room temperature. The caspase activity was measured by detecting the luminescence using LMAX-2 luminometer (Molecular Devices).
Quantitative real time PCR
Viral RNA was isolated from 2 dpi supernatants collected from clones exhibiting complete suppression at an MOI of 0.05 for CHIKV, using the viral RNA mini kit (Qiagen, Germany). All isolated nucleic acids were quantitated using a Nanodrop ND-1000 spectrophotometer (Thermofisher). Stock virus with known titer was used as a control to generate the standard curve. Complementary DNA synthesis was carried out using the Gene Amp RNA PCR MULV reverse transcriptase kit (Applied Biosystem) both for samples and standards. For CHIKV, the primer targeting the nsP2 region of the virus, nsP2 reverse: aaattcggcctgaaccttct, was utilized (Ho et al., 2010). One cycle of 30 minutes at 42°C and 5 min at 99°C was performed (Mishra et al., 2016). The absolute quantification was performed on the 7500 fast real-time PCR system (Applied Biosystem) using Power sybr green master mix (Applied Biosystem) at a particular thermocyclic condition of one cycle for 2 min at 50°C, one cycle for 10 min at 95°C, 40 cycles for 15 s at 95°C, and 1 min at 60°C. For quantification of CHIKV, the above mentioned primer nsP2 reverse along with nsP2 forward: ttctgggggtcagagaaaga was used (Ho et al., 2010). Beta-actin was used as an internal control for all RT-qPCR assays. The slope of the standard curve was −2.8 and the R2 value was 0.97. The absolute quantification of viral RNA copies/ml in the samples was performed by comparing them to the corresponding standards with known viral titer.
Construction of connected maxizyme-expressing transgenes in the piggyBac vector and mosquito injections
For the generation of transgenic mosquitoes expressing maxizymes, we adopted the connected maxizyme approach. In this approach, both monomers of the CHIKV-Mz were connected using a 13-nucleotide long linker sequence (atagaagaataacgtata) and expressed using a single Ae-tRNAval promoter (Figure 1). This was done to increase the efficiency of formation of a bi-molecular heterodimeric maxizyme structure (Hamada et al., 1999; Kuwabara et al., 2002) and reduce the chance of inactive homodimer formation. The cloning of the transgene into the piggyBac vector, pXL-BacII-3xP3-ECFP, involved PCR amplification of the Ae-tRNA val pol III promoter from the pLAeARz#9RH vector using the Ae-tRNA val F and R primers (Table 2) and inserting it into the pLAeARH plasmid Sal I and Not I sites. The complementary oligonucleotides of connected maxizyme (cMz-sense and cMz-antisense, Table 2) were annealed together and cloned into the Bam HI and Not I sites downstream of the promoter in pLAeARH, resulting in the plasmid pLAeAcMzRH. The connected Mz and expression units were PCR amplified from the pLAeAcMzRH vector using transgene F and R (Table 2) primers and subcloned into the Sac II and Bgl II sites of the piggyBac vector.
Generation of transgenic mosquitoes and identification of transgene integration sites using splinkertte PCR
Transgenic mosquitoes were generated by embryo injection of piggBac cMz expression vectors into mosquito embryos, essentially as previously described (Mishra et al., 2016). Higgs White Eye (HWE) mosquitoes were used in this study, facilitating detection of the 3×P3-ECFP eye-specific transgene marker gene and allowing transgenic mosquitoes to be detected from UV fluorescence in the eyes. Mosquitoes were reared in an Arthropod containment level-2 (ACL-2) facility at 28oC with 60–80% relative humidity. They were maintained on 10% sucrose solution and water and artificially fed with citrated sheep's blood (Colorado Serum Company, Denver, CO, USA) and 1 mM/10 ml of phagostimulant ATP. During infections, virus-infected cell culture medium was mixed with an equal volume of feeding solution.
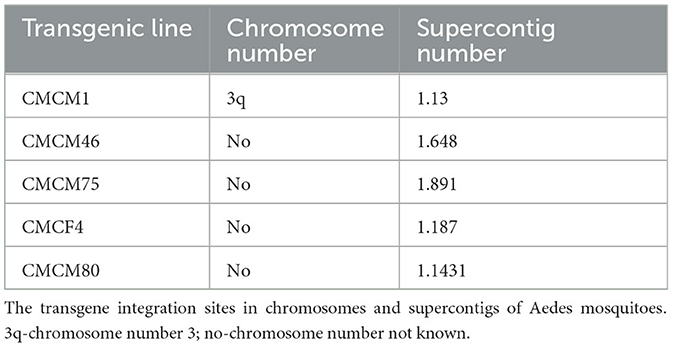
A total of five independent transgenic lines were established and a splinkerette protocol was adopted from Potter and Luo (2010) to assess the genomic location of each piggyBac-integrated transgene as previously described (Mishra et al., 2016). All five transgenic mosquito lines exhibited unique integration sites in Ae. aegypti genome (Table 3).
Analysis of CHIKV infection in cMz transgenic mosquitoes
Control and transgenic lines were fed infectious blood meals with a viral titer of 3 × 109 TCID50/mL. Both the infected controls and cMz transgenics were maintained for seven days on 10% sucrose solution prior to feeding infected bloodmeals. For each transgenic line, a total of 15 mosquitoes in small containers were allowed to feed for 2 h on 700 μL probing solution (50% FBS (164 mM) + NaCl (100 mM) + NaHCO3 (0.2 mM) + ATP (50 μg) + sucrose, pH 7.0) contained between two parafilm membranes, as described by Franz et al. (2006). Successful feeding was confirmed by observation of fully engorged mosquitoes in all groups. Mosquito homogenates were processed as previously described (Mishra et al., 2016).
Statistical analysis
The statistical analysis performed in this study is described in figure legends. All the statistical tests were carried out using GraphPad Prism version 9.3.0.3.
Results
Effect of maxizyme on CHIKV replication in vitro
To compare the effect of our maxizyme on CHIKV replication, we successfully constructed two lentivirus expression constructs with maxizyme: Mz, and ΔMz. Then, we transformed these maxizyme expression plasmids along with plasmids expressing the parental hRz #9 and # 14 and isolated several clonal cell populations for each. All these clonal populations were challenged with CHIKV at two different MOIs, and the effect was evaluated by the presence or absence of CPE. Our screening results revealed clones #2 and #15 of Mz, clones #5 and #27 of hRz#9, and clone #37 of hRz#14 were resistant to CHIKV CPE. However, all the clones from the inactive ΔMz had marked CPE (data not shown).
We employed two different MOIs in our study to more thoroughly assess the effectiveness of maxizyme against CHIKV replication. At a lower MOI (0.05), the Mz clones #M2 and #M15 showed eight logs of CHIKV suppression as compared to the negative controls: i.e., untransformed wild-type and ΔMz transformed Vero cells (Figure 2A). However, the hRzs #9/5, #9/37, and #14/27clones showed six and three logs of suppression relative to the negative controls (Figure 2A). We also determined the CHIKV viral RNA copies by RT-qPCR assay in the supernatant collected of the infected maxizyme clones. The results revealed that Mz clones M/2 and M/15 completely suppressed viral RNA production, unlike the negative controls (Figure 3A). Additionally, we performed a caspase-3 assay to measure the virus-induced apoptosis. The infected virus supernatant collected from the Mz clones was tested for caspase-3 activity and the supernatants of both Mz clones exhibited caspase-3 activity similar to the levels of uninfected Vero cells (Figure 3B). However, the supernatant of the negative control, ΔMz, showed 500 times more activity than uninfected controls and Mz clones (Figure 3B), reflecting no effect on CHIKV replication.
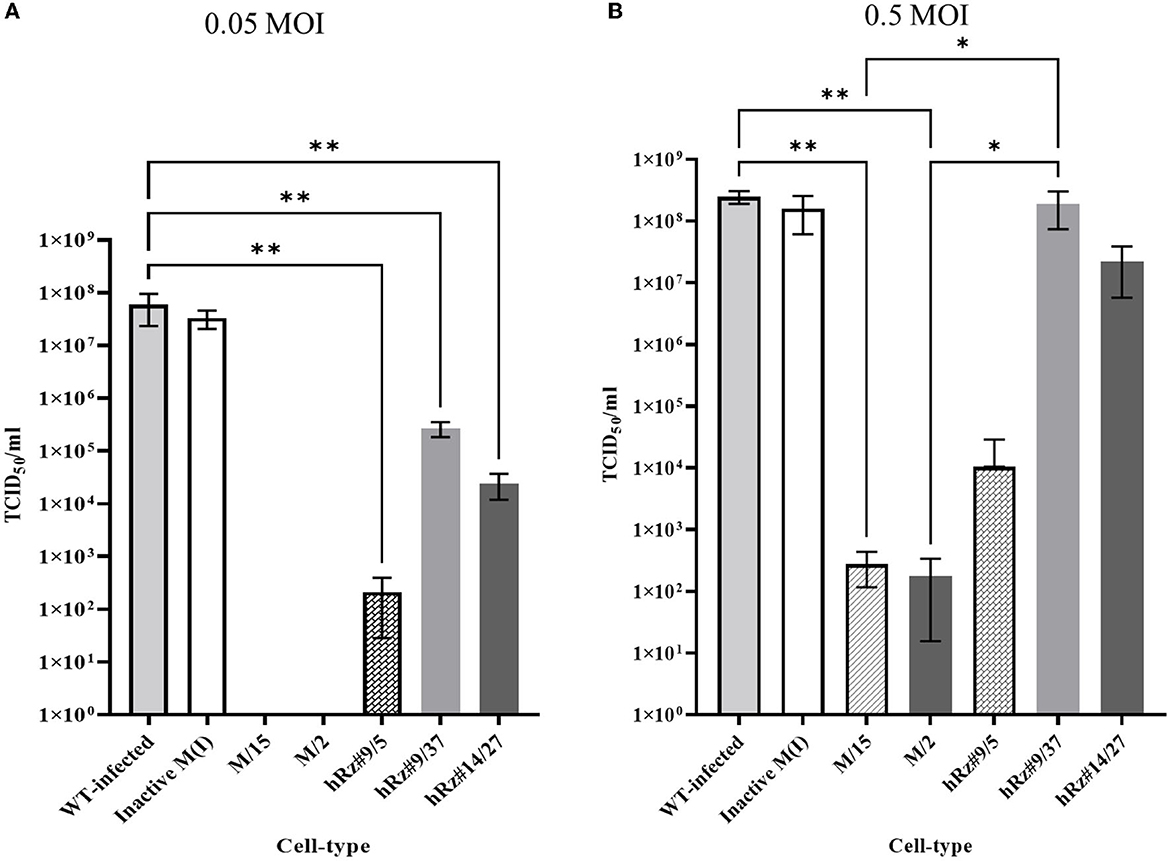
Figure 2. TCID50-IFA analysis of clonal populations expressing CHIKV specific maxizymes and hammerhead ribozymes with two different MOI. (A) Challenge MOI of 0.05 and (B) Challenge MOI of 0.5, cells were fixed and stained with anti-CHIKV capsid specific antibody 3 dpi. WT: Untransformed wild type Vero cells and M (I): inactive control for maxizyme. Each clone is represented as maxizyme or hRz number / clone number. Each bar represents an average CHIKV titer from three independent experiments. Error bars represent the standard deviation among the three independent replicates for each clone. Statistical analysis was performed using the Two-way ANOVA test and Tukey's multiple comparisons test (* p < 0.05,** p < 0.005).
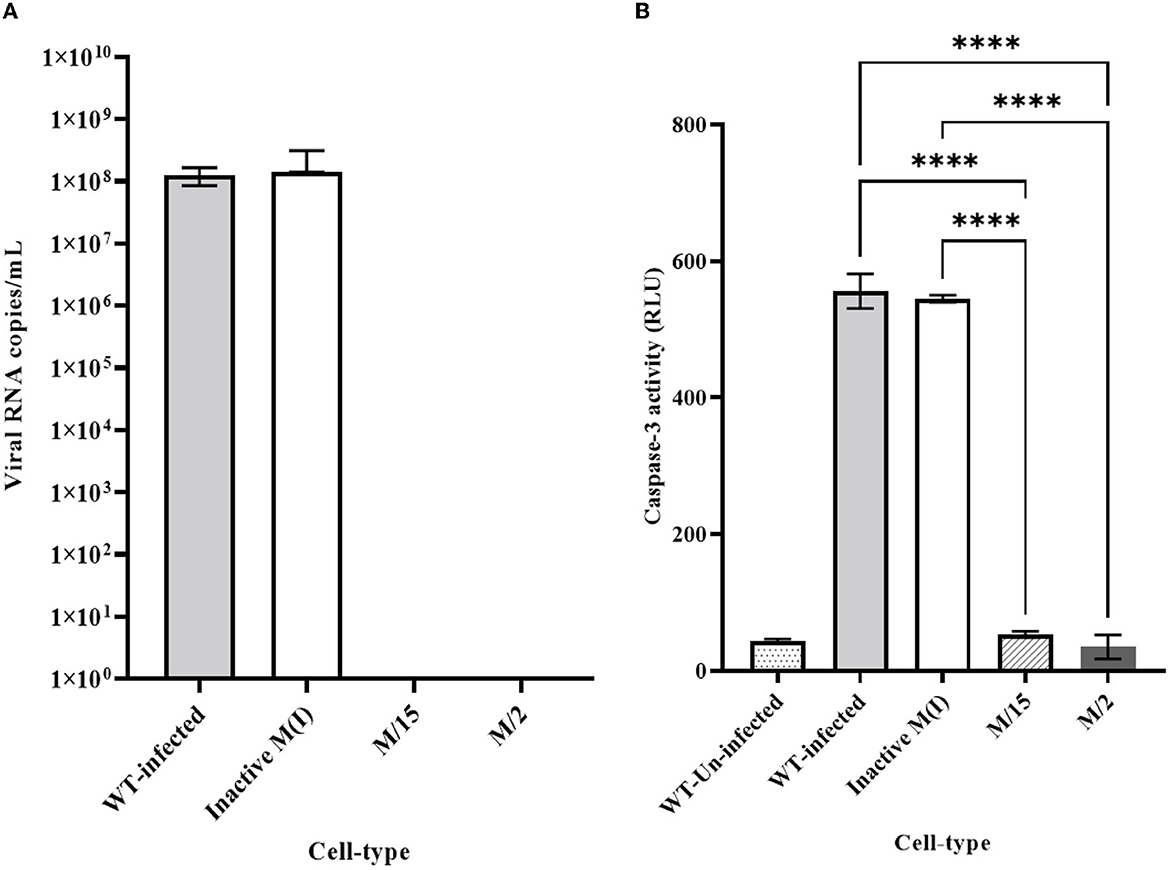
Figure 3. Quantitative RT-qPCR and caspase 3 analysis for clonal expressing antiviral ribozymes specific to CHIKV. (A) RT-qPCR performed on viral RNA isolated from the infected cell culture supernatants collected 2 dpi challenged at an MOI of 0.05. nsP2 specific primer were employed for quantification. (B) Caspase 3 analysis performed by infecting healthy control and transformed clonal cells using supernatant collected from 2dpi challenge experiment at an MOI of 0.05. WT designates untransformed Vero cells, either infected with virus as the infected control, or incubated with media as the Unifected control. M-maxizyme, M(I)- maxizyme Inactive, each clone is represented as maxizyme/clone number. Each bar is an average of three independent infection experiments. Bars denote the standard deviation among the three independent replicates done for each clone. Statistical analysis was performed using the Two-way ANOVA test and Tukey's multiple comparisons test (**** p < 0.0001).
Next, we compared the level of CHIKV suppression for the hRz and Mz clones at a higher MOI of 0.5. Our initial tests at a lower MOI of 0.05 demonstrated that the maxizyme was highly effective in suppressing CHIKV replication. However, it was important for us to understand how this maxizyme performed across a range of virus concentrations, as this reflects the diverse conditions found in the field. The results revealed the Mz clones # M5 and # M2 suppressed CHIKV replication by six logs as compared to the negative controls. However, the suppression level of hRz clones #9/5, #9/37, and #14/27 clones dropped to four, zero and half a log, respectively. The maxizyme expression in the clones was confirmed by RT-PCR (Figure 4). Overall, under in vitro conditions, the maxizyme was more effective in suppressing CHIKV replication than the parental hammerhead ribozymes.
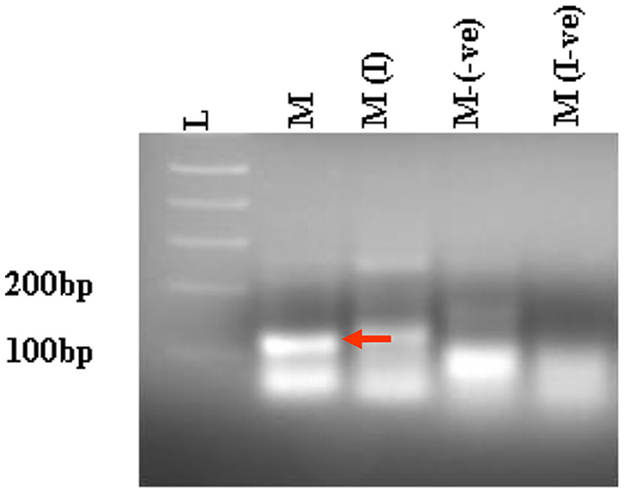
Figure 4. RT-PCR analysis of selected clonal cell populations expressing antiviral ribozyme constructs specific to CHIKV. The presence of the desired band specific to each CHIKV maxizyme is evident, where M: active maxizyme, M (I): inactive maxizymes, M(-ve) and M(I-ve) is RT-negative reaction. The presence of the correct band (110 bp size) is indicated by the red arrow. L- 1kb plus DNA ladder.
Effect of maxizyme on CHIKV replication in transgenic mosquitoes
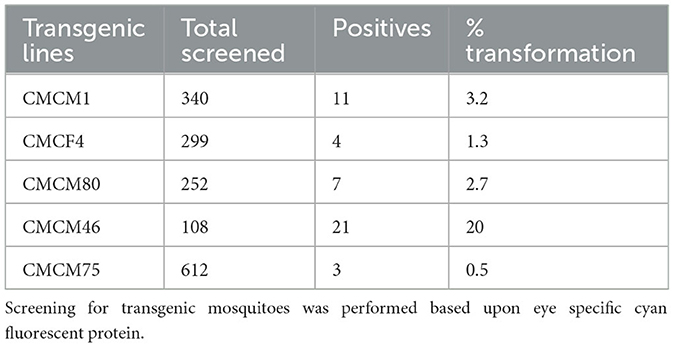
To test the effectiveness of Mz in controlling CHIKV transmission, we generated five transgenic lines of Ae. aegypti mosquitoes. For transgenesis, we used a connected maxizyme (cMz) approach for ease of integration into piggyBac vector (pXL-Bac-II-ECFP) and to increase the efficiency of formation of active heterodimeric structures. The pXL-Bac-II-ECFP-cMz expression and transposase helper plasmids were co-injected into Ae. aegypti embryos as previously described (Mishra et al., 2016), and the transformation efficiency ranged from 0.5 to 20% (Table 4). A percentage fluorescence of 57–96% was obtained from all the transgenic lines at G5 (data not shown). Generation 5 positive mosquitoes were then backcrossed to wild-type HWE mosquitoes as previously described (Mishra et al., 2016) to generate a heterozygous G6 transgenic mosquito population. cMz expression in these transgenic lines was confirmed by RT-PCR (Figure 5). Each of the integration sites of these five lines were identified in the mosquito genome at different super contigs (Table 3).
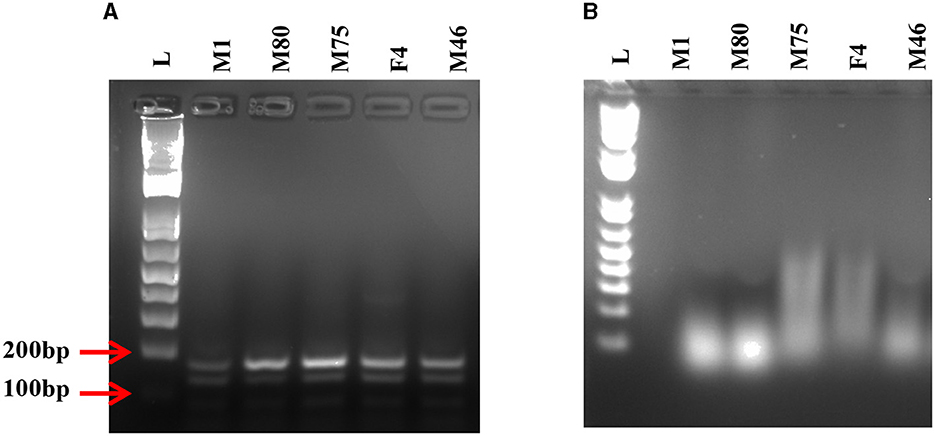
Figure 5. RT-PCR on transgenic mosquitoes. (A) Agarose gel displays the presence of desired 170 bp band confirming the expression of the connected maxizyme in each transgenic mosquito line. (B) RT negative reaction run using Taq polymerase shows the absence of the desired band. L- 1kb plus DNA ladder and M1, M80, M75, F4 and M46 are transgenic lines.
Challenge of transgenic mosquitoes with CHIKV
Heterozygous G6 transgenic lines were challenged with CHIKV by oral exposure to infectious CHIKV blood meal, as previously described (Mishra et al., 2016). Mosquitoes were collected at 7 days post-blood meal (7 dpbm) and individual homogenates were made of each mosquito in 200 μl of DMEM. Homogenates were filtered through a 0.2 μM membrane filter for further analysis. Mosquitoes from three of the lines, CMCM80, CMCM46, and CMCF4/F2, exhibited complete suppression and had a 0% infection rate. However, non-transgenic (wild-type) HWE mosquitoes had an infection rate of 81 % with an average infectious virus titre of 2 × 105 TCID50/ml (Figure 6). In contrast, the transgenic mosquito lines CMCM75 and CMCM1/F2 had an infection rate of 3.8 and 2 %, and infectious virus titer of 3.2 × 102 and 2.2 × 102 TCID50/ml, respectively (Figure 6).
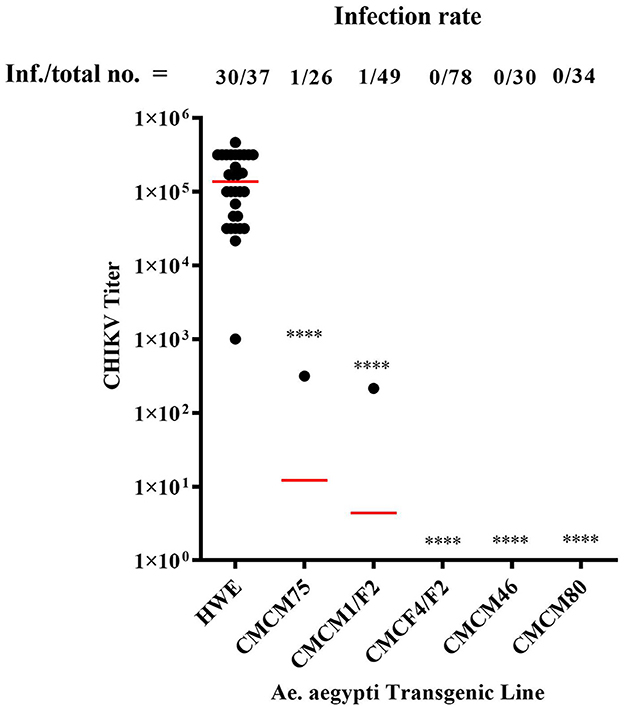
Figure 6. TCID50- IFA performed on CHIKV challenged transgenic G6 heterozygous mosquito population. TCID50-IFA was performed on homogenates of single transgenic mosquito fed an infectious blood meal compared to control HWE mosquitoes. Analysis was performed seven days post infectious blood meal. “Inf.” indicates the number of infected mosquitoes analyzed and “total no.” indicates the total number of mosquitoes analyzed. HWE-Higgs White Eye mosquitoes, CMCM75, CMCM1/F2, CMCF4/F2, CMCM46 and CMCM80 are connected maxizyme expressing transgenic mosquitoes. Statistical analysis was performed using the Two-way ANOVA test and Tukey's multiple comparisons test. All the transgenic lines had a statistical significance of ****p < 0.0001 as compared to the HWE line.
Next, we tested the transmission potential of these transgenic mosquitoes. As expected, the wild-type HWE mosquitoes effectively transmitted CHIKV to probe solution with an average titer of 2.3 × 104 TCID50/ml, along with an average whole-body titer of 5.9 × 105 TCID50/ml (Figure 7). However, no infectious virus was detected from either the probe solution or the whole-body homogenates of the five transgenic mosquito lines (Figure 7). Overall, these results indicate that the transgenic mosquito line expressing cMz completely inhibited CHIKV infection and transmission and did so from a heterozygous genetic background.
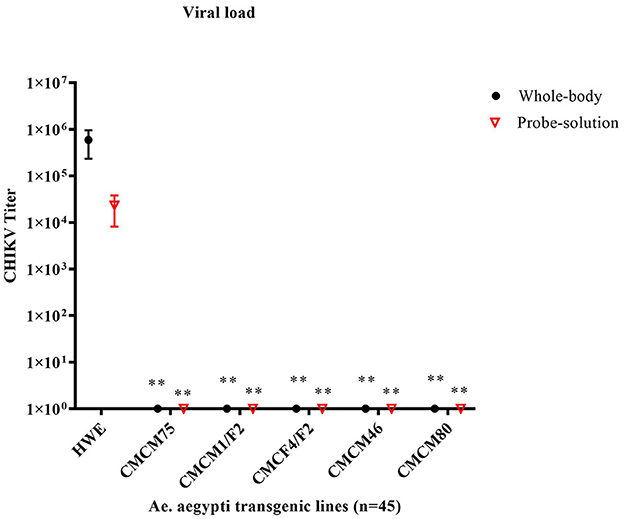
Figure 7. TCID50- IFA performed on the probe solution (saliva) and whole-body of CHIKV challenged transgenic G6 heterozygous mosquito population. Briefly, pooled probe solutions and whole-body homogenates were collected from the infected transgenic and infected wild-type mosquitoes at 7dpbm. The virus titer of these samples was determined by TCID50-IFA. Statistical analysis was performed using the Two-way ANOVA test and Tukey's multiple comparisons test. All the transgenic lines (whole body and probe solution) had a statistical significance of **p < 0.005 as compared to the HWE line.
Discussion
Anti-pathogen effectors have been investigated for several arboviruses such as CHIKV (Mishra et al., 2016), DENV-2 (Franz et al., 2006), DENV3 and CHIKV (Yen et al., 2018), and Zika (Buchman et al., 2019). The presumptive outcome of such investigations is the development of transgenic mosquitoes that could eventually be used for replacement of naturally arboviruses competent mosquitoes (Marshall et al., 2019). Thus, far, researchers have come close to releasing some of these transgenic approaches, but the caveat remaining is the difficulty of introgression into wild-type mosquitoes due to possible position effects on efficient expression, the presence of multiple strains of virus circulating in the target areas, and the size of the transgene. Wolbachia infection of mosquitoes has been successful at controlling arboviruses in various countries, however the stability and the effect of such use is not yet clear (Yen and Failloux, 2020). To overcome these limitations, we have developed a strategy of using maxizymes, which has the potential advantages of small size transgene, higher potency, effectiveness against escape variants, and possible adaptability to target multiple arboviruses simultaneously.
We first demonstrated the effectiveness of our maxizyme in inhibiting CHIKV replication by measuring virus production, viral RNA, and virus-induced apoptosis in Vero cells. To mimic the variability of virus doses in nature we tested the effectiveness of Mz under lower (0.05 MOI) and higher doses (0.5 MOI). The Mz was effective at inhibiting CHIKV replication at both doses of virus as compared to the parental hRzs. The observation two 2 logs of virus production at higher MOIs in Mz clones could be due to higher doses of the virus skewing the target-enzyme ratio, or it could be due to weaker expression of both the monomers (Figure 2). Nevertheless, the CHIKV suppression was more significant than the controls and parental hRzs (Mishra et al., 2016). Overall, Mz showed a higher potency and effectiveness in controlling CHIKV replication as compared to the parental counterparts in vitro.
We utilized a connected maxizyme in the transgenic mosquitoes owing to the advantages discussed in the results section. All five transgenic lines were highly refractory to CHIKV infection and prevented virus transmission, and this was true in a heterozygous state. This latter observation is important because if we envision release of these transgenes in a population control strategy, heterozygotes will predominate in the first and subsequent generations. Additionally, this approach is not susceptible to gene position effects, as demonstrated by cMz expression and CHIKV inhibition irrespective of the integration loci (Table 4).
Although these results are promising, our study has the following limitations. First, we have not tested the effectiveness of Mz against a virulent strain of CHIKV due to the unavailability of BSL-3 facility. While the 181/25 CHIKV strain is attenuated for human infection, and lacks the A226V mutation attributed to recent epidemic outbreaks of CHIKV, the maxizyme targets we chose are present in both the attenuated and virulent strains. Additionally, based upon our results in this and prior studies, infection of cell cultures and mosquitoes is not appreciably affected by the dual mutations in the E2 protein responsible for the attenuation in humans (Gorchakov et al., 2012). Second, the stability of the transgene and its effectiveness over generations in our transgenic mosquitoes was not addressed. This will take additional time and analyses, and we will pursue this in our future studies.
These results confirm that maxizymes can be potent inhibitors of CHIKV replication in mammalian cells or transmission in transgenic mosquitoes. Additionally, we could use this approach to design Mz against multiple arboviruses and possibly develop a universal transgenic mosquito resistant to several arboviruses (Carter et al., 2015).
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
PM: Conceptualization, Investigations, Validation, Resources, Data curation, Formal analysis, Methodology, Writing—original draft, Writing—review & editing. VB: Resources, Writing—original draft, Writing—review & editing. MF: Conceptualization, Funding acquisition, Resources, Supervision, Writing—review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by NIH/NIAID grant 1R01AI097554 to Principal Investigator MF.
Acknowledgments
The authors would like to thank Scott Weaver (University of Texas Medical Branch, Galveston, TX, USA), for providing the attenuated strain of CHIKV 181/25. We also acknowledge the invaluable instruction and assistance of Tresa Fraser in maintenance of cell cultures and in mosquito blood feeding and maintenance, Cheryl Kucharski in facilitating cell sorting and Pruksa Nawtaisong for providing the pLAeARH vector.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aliota, M. T., Walker, E. C., Uribe Yepes, A., Dario Velez, I., Christensen, B. M., Osorio, J. E., et al. (2016). The wMel strain of Wolbachia reduces transmission of chikungunya virus in Aedes aegypti. PLoS Negl. Trop. Dis. 10, e0004677. doi: 10.1371/journal.pntd.0004677
Buchman, A., Gamez, S., Li, M., Antoshechkin, I., Li, H-. H., Wang, H-. W., et al. (2019). Engineered resistance to Zika virus in transgenic Aedes aegypti expressing a polycistronic cluster of synthetic small RNAs. Proc. Nat. Acad. Sci. 116, 3656–61. doi: 10.1073/pnas.1810771116
Cardona-Ospina, J. A., Henao-San Martin, V., Paniz-Mondolfi, A. E., and Rodríguez-Morales, A. J. (2015). Mortality and fatality due to Chikungunya virus infection in Colombia. J. Clin. Virol. 70, 14–5. doi: 10.1016/j.jcv.2015.07.001
Carter, J. R., Taylor, S., Fraser, T. S., Kucharski, C. A., Dawson, J. L., Fraser, M. J., et al. (2015). Suppression of the arboviruses dengue and chikungunya using a dual-acting group-i intron coupled with conditional expression of the Bax C-terminal domain. PLoS ONE 10, e0139899. doi: 10.1371/journal.pone.0139899
Costa, L. B., Barreto, F. K. A., Barreto, M. C. A., Santos, T. H. P., Andrade, M. M. O., Farias, L. A. B. G., et al. (2023). Epidemiology and economic burden of chikungunya: a systematic literature review. Trop. Med. Inf. Dis. 8, 301. doi: 10.3390/tropicalmed8060301
Franz, A. W. E., Sanchez-Vargas, I., Adelman, Z. N., Blair, C. D., Beaty, B. J., James, A. A., et al. (2006). Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc. Nat. Acad. Sci. 103, 4198–203. doi: 10.1073/pnas.0600479103
Gantz, V. M., Jasinskiene, N., Tatarenkova, O., Fazekas, A., Macias, V. M., Bier, E., et al. (2015). Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Nat. Acad. Sci. 49, 112. doi: 10.1073/pnas.1521077112
Gorchakov, R., Wang, E., Leal, G., Forrester, N. L., Plante, K., Rossi, S. L., et al. (2012). Attenuation of chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J Virol. 86, 6084–96. doi: 10.1128/JVI.06449-11
Haasnoot, J., Westerhout, E. M., and Berkhout, B. R. N. A. (2007). interference against viruses: strike and counterstrike. Nat. Biotechnol. 25, 1435–43. doi: 10.1038/nbt1369
Hamada, M., Kuwabara, T., Warashina, M., Nakayama, A., and Taira, K. (1999). Specificity of novel allosterically trans - and cis -activated connected maxizymes that are designed to suppress BCR-ABL expression. FEBS Lett. 461, 77–85. doi: 10.1016/S0014-5793(99)01367-8
Higgs, S., and Vanlandingham, D. L. (2018). Chikungunya Virus and Zika Virus Transmission Cycles. Amsterdam: Elsevier, 15–68.
Ho, P. S., Ng, M. M. L., and Chu, J. J. H. (2010). Establishment of one-step SYBR green-based real time-PCR assay for rapid detection and quantification of chikungunya virus infection. Virol J. 7, 13. doi: 10.1186/1743-422X-7-13
Iyo, M., Kawasaki, H., Miyagishi, M., and Taira, K. (2004). Allosterically Controlled Ribozymes as Artificial Ribonucleases. Artif. Nucl. 12, 89–109. doi: 10.1007/978-3-642-18510-6_7
Iyo, M., Kawasaki, H., and Taira, K. (2002). Allosterically controllable maxizymes for molecular gene therapy. Curr. Opin. Mol. Ther. 4, 154–65. doi: 10.1016/B978-012476249-7/50013-8
Kärber, G. (1931). Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn-Schmiedebergs Archiv. Für Exp. Pathol. Und Pharmakol. 162, 480–3. doi: 10.1007/BF01863914
Kuwabara, T., Warashina, M., Orita, M., Koseki, S., Ohkawa, J., Taira, K., et al. (1998). Formation of a catalytically active dimer by tRNAVal -driven short ribozymes. Nat. Biotechnol. 16, 961–5. doi: 10.1038/nbt1098-961
Kuwabara, T., Warashina, M., and Taira, K. (2002). Cleavage of an inaccessible site by the maxizyme with two independent binding arms: an alternative approach to the recruitment of RNA helicases. J. Biochem. 132, 149–55. doi: 10.1093/oxfordjournals.jbchem.a003193
Marshall, J. M., Raban, R. R., Kandul, N. P., Edula, J. R., León, T. M., and Akbari, O. S. Winning the tug-of-war between effector gene design pathogen evolution in vector population replacement strategies. Front. Genet. (2019) 10:1072. doi: 10.3389/fgene.2019.01072.
Mishra, P., Furey, C., Balaraman, V., and Fraser, M. (2016). Antiviral hammerhead ribozymes are effective for developing transgenic suppression of chikungunya virus in Aedes aegypti mosquitoes. Viruses 8, 163. doi: 10.3390/v8060163
Nawtaisong, P., Keith, J., Fraser, T., Balaraman, V., Kolokoltsov, A., Davey, R. A., et al. (2009). Effective suppression of dengue fever virus in mosquito cell cultures using retroviral transduction of hammerhead ribozymes targeting the viral genome. Virol J. 6, 73. doi: 10.1186/1743-422X-6-73
Pialoux, G., Gaüzère, B-. A., Jauréguiberry, S., and Strobel, M. (2007). Chikungunya, an epidemic arbovirosis. Lancet Infect Dis. 7, 319–27. doi: 10.1016/S1473-3099(07)70107-X
Potter, C. J., and Luo, L. (2010). Splinkerette PCR for mapping transposable elements in drosophila. PLoS ONE 5, e10168. doi: 10.1371/journal.pone.0010168
Reid, W. R., Olson, K. E., and Franz, A. W. E. (2021). Current effector and gene-drive developments to engineer arbovirus-resistant Aedes aegypti (Diptera: Culicidae) for a sustainable population replacement strategy in the field. J. Med. Entomol. 58, 1987–96. doi: 10.1093/jme/tjab030
Schmidt, C., and Schnierle, B. S. (2022). Chikungunya vaccine candidates: current landscape and future prospects. Drug Design Dev. Ther. 16, 3663–73. doi: 10.2147/DDDT.S366112
Schneider, M., Narciso-Abraham, M., Hadl, S., McMahon, R., Toepfer, S., Fuchs, U., et al. (2023). Safety and immunogenicity of a single-shot live-attenuated chikungunya vaccine: a double-blind, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 401, 2138–47. doi: 10.1016/S0140-6736(23)00641-4
Weaver, S. C., and Lecuit, M. (2015). Chikungunya virus and the global spread of a mosquito-borne disease. New Eng. J. Med. 372, 1231–9. doi: 10.1056/NEJMra1406035
Williams, A., Franz, A., Reid, W., and Olson, K. (2020). Antiviral effectors and gene drive strategies for mosquito population suppression or replacement to mitigate arbovirus transmission by Aedes aegypti. Insects 11, 52. doi: 10.3390/insects11010052
Yen, P-. S., and Failloux, A-. B. (2020). A review: Wolbachia-based population replacement for mosquito control shares common points with genetically modified control approaches. Pathogens 9, 404. doi: 10.3390/pathogens9050404
Keywords: maxizyme (Mz), hammerhead ribozyme (hRz), chikungunya (CHIKV), connected maxizyme (cMz), mosquito transmission
Citation: Mishra P, Balaraman V and Fraser MJ Jr (2023) Maxizyme-mediated suppression of chikungunya virus replication and transmission in transgenic Aedes aegypti mosquitoes. Front. Microbiol. 14:1286519. doi: 10.3389/fmicb.2023.1286519
Received: 31 August 2023; Accepted: 14 November 2023;
Published: 22 December 2023.
Edited by:
Ke Liu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Pradip Barde, National Institute for Research in Tribal Health (ICMR), IndiaStephen Higgs, Kansas State University, United States
Copyright © 2023 Mishra, Balaraman and Fraser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Malcolm J. Fraser Jr., fraser.1@nd.edu
†Present addresses: Priya Mishra, Office of Research Affairs, Rush University Medical Center, Chicago, IL, United States
Velmurugan Balaraman, Department of Diagnostic Medicine and Pathobiology, Center of Excellence for Emerging and Zoonotic Animal Diseases, Kansas State University, Manhattan, KS, United States
‡These authors share senior authorship
 Priya Mishra
Priya Mishra Velmurugan Balaraman
Velmurugan Balaraman Malcolm J. Fraser Jr.
Malcolm J. Fraser Jr.