- 1Department of Animal Sciences, The Ohio State University, OARDC, Wooster, OH, United States
- 2School of Veterinary Medicine and Biomedical Sciences, University of Nebraska-Lincoln, Lincoln, NE, United States
- 3Los Alamos National Laboratory, Bioscience Division, Group B-10: Biosecurity and Public Health, Los Alamos, NM, United States
- 4Division of Medicinal Chemistry and Pharmacognosy, College of Pharmacy, The Ohio State University, Columbus, OH, United States
- 5College of Pharmacy, University of Findlay, OH, United States
Introduction: Control of Campylobacter from farm to fork is challenging due to the frequent emergence of antimicrobial-resistant isolates. Furthermore, poultry production systems are known reservoirs of Campylobacter. The twin-arginine translocation (Tat) pathway is a crucial bacterial secretion system that allows Campylobacter to colonize the host intestinal tract by using formate as the main source of energy. However, Tat pathway is also a major contributing factor for resistance to copper sulfate (CuSO4).
Methods: Since mammals and chickens do not have proteins or receptors that are homologous to bacterial Tat proteins, identification of small molecule (SM) inhibitors targeting the Tat system would allow the development of safe and effective control methods to mitigate Campylobacter in infected or colonized hosts in both pre-harvest and post-harvest. In this study, we screened 11 commercial libraries (n = 50,917 SM) for increased susceptibility to CuSO4 (1 mM) in C. jejuni 81–176, a human isolate which is widely studied.
Results: Furthermore, we evaluated 177 SM hits (2.5 μg/mL and above) that increased the susceptibility to CuSO4 for the inhibition of formate dehydrogenase (Fdh) activity, a Tat-dependent substrate. Eight Tat-dependent inhibitors (T1–T8) were selected for further studies. These selected eight Tat inhibitors cleared all tested Campylobacter strains (n = 12) at >10 ng/mL in the presence of 0.5 mM CuSO4in vitro. These selected SMs were non-toxic to colon epithelial (Caco-2) cells when treated with 50 μg/mL for 24 h and completely cleared intracellular C. jejuni cells when treated with 0.63 μg/mL of SM for 24 h in the presence of 0.5 mM of CuSO4. Furthermore, 3 and 5-week-old chicks treated with SM candidates for 5 days had significantly decreased cecal colonization (up to 1.2 log; p < 0.01) with minimal disruption of microbiota. In silico analyses predicted that T7 has better drug-like properties than T2 inhibitor and might target a key amino acid residue (glutamine 165), which is located in the hydrophobic core of TatC protein.
Discussion: Thus, we have identified novel SM inhibitors of the Tat pathway, which represent a potential strategy to control C. jejuni spread on farms.
Introduction
Campylobacter is a leading cause of bacterial foodborne gastroenteritis worldwide (Igwaran and Okoh, 2019) and a major public health problem. A recent estimate by the CDC indicates that Campylobacter is not only among the most common causes of domestically acquired foodborne illnesses in humans (over 845,024 cases per year) but also is among the leading causes of hospitalization (over 8,463 annually) in the United States (USDA ERS–Cost Estimates of Foodborne Illnesses, 2017; Igwaran and Okoh, 2019).
Poultry products represent a key source of human Campylobacter infections (Kittl et al., 2013; Skarp et al., 2016). C. jejuni and C. coli densely colonize the intestine of poultry (up to 109 bacteria per gram of ceca in chicken) and are the most common species encountered in human infections (Allos, 2001; Newell and Fearnley, 2003; Lee and Newell, 2006; Young et al., 2007; Facciolà et al., 2017; Connerton et al., 2018). Despite extensive intestinal colonization, Campylobacter infection produces little or no clinical diseases in poultry (Johnson et al., 2017). Prevalence studies conducted in Europe and United States have reported Campylobacter-positive flocks ranging up to 100% (Newell and Fearnley, 2003; Luangtongkum et al., 2006; Jorgensen et al., 2011; Sibanda et al., 2018; Koutsoumanis et al., 2020; Kuhn et al., 2020). Colonized chickens shed Campylobacter in their feces until slaughter (up to 109 CFU/g of cecum) (Newell and Fearnley, 2003; Luangtongkum et al., 2006; Jorgensen et al., 2011; Sibanda et al., 2018; Koutsoumanis et al., 2020; Kuhn et al., 2020), increasing the risk of horizontal transmission of Campylobacter to the whole flock and post-harvest contaminations of the meat products (over 50% in average and up to 100%) (Logue et al., 2003; Berrang et al., 2007; Young et al., 2007; Guerin et al., 2010; Hue et al., 2010; Kaakoush et al., 2015). The high prevalence of Campylobacter in poultry and detection of identical genotypes in both poultry and human isolates support poultry contamination as the predominant route of human infection (Allos, 2001; Newell and Fearnley, 2003; Lee and Newell, 2006; Young et al., 2007; Facciolà et al., 2017; Connerton et al., 2018).
On-farm Campylobacter control efforts have not been successful in mitigating human Campylobacter infections as evidenced by the continuous increase in human infections (Kittl et al., 2013; Skarp et al., 2016). Although improving biosecurity has beneficial effects on lowering the overall flock prevalence (Kaakoush et al., 2015), these measures have not resulted in consistent and predictable outcomes in controlling Campylobacter (Wagenaar et al., 2013; Kaakoush et al., 2015). In addition, stringent biosecurity measures are cost-prohibitive and hard to maintain and their effectiveness seems to vary with production systems and they do not apply to small farm poultry operations (e.g., backyard chickens) and pet chickens (Koutsoumanis et al., 2020). Currently, there are no effective competitive exclusion products, vaccines, bacteriocins, bacteriophages, or feed/water additives to exclude Campylobacter from chickens under production conditions (Lin, 2009; Richards et al., 2019; Vandeputte et al., 2019; Quintel et al., 2020; Hakeem and Lu, 2021). Furthermore, the widespread use of antibiotics in poultry production has been implicated in the emergence of highly resistant Campylobacter strains (Young et al., 2007; Agyare et al., 2018; Yang et al., 2019; Andrew Selaledi et al., 2020). Therefore, there is a critical need for novel anti-Campylobacter control strategies that can amend and/or replace on-going efforts and specifically target pathways and novel mechanisms to combat both Campylobacter spread and antibiotic resistance.
The transport of proteins from cytoplasm to extra-cytoplasmic locations is critical for bacterial survival, virulence, and stress resistance (Berks, 2015). Extra-cytoplasmic protein transport in bacteria most commonly occurs via two major export systems, namely, the general secretory (Sec) pathway and the twin-arginine translocation (Tat) pathway (Frain et al., 2019a,b). The Campylobacter Tat pathway is highly conserved between strains, and the inactivation of the Tat pathway induces a multitude of functional defects in Campylobacter including motility, outer membrane permeability, biofilm formation, antibiotic resistance, growth under oxygen-limiting conditions, iron acquisition, and copper homeostasis (Drozd et al., 2011; Kassem et al., 2011). Copper is crucial for the survival of pathogenic bacteria in the host and external environment; however, it also exhibits antimicrobial properties at high concentration (Hall et al., 2008; Stolle et al., 2016). As a defense mechanism, C. jejuni expresses two important proteins for copper homeostasis as follows: (1) CopA a copper transporting P-type ATPase responsible for transporting toxic Cu (I) from cytoplasm to periplasm, (2) CueO, a multicopper oxidase that converts toxic Cu (I) into the less toxic Cu (II) in periplasm and is dependent on the Tat system. Both of these copper homeostasis proteins are essential for copper resistance and avian host colonization (Hall et al., 2008; Stolle et al., 2016; Kassem et al., 2017; Gardner and Olson, 2018). In addition, the deletion of the tatC gene significantly reduced C. jejuni persistence in the intestinal tract of chickens and decreased fecal shedding (Rajashekara et al., 2009). Previous studies have demonstrated that Tat inhibitors are an effective control method to mitigate Pseudomonas aeruginosa (Vasil et al., 2012; Massai et al., 2019) and Escherichia coli (Panahandeh et al., 2008; Bageshwar et al., 2016; Blümmel et al., 2017). Furthermore, the use of SM has been shown to be effective strategy against multi-drug resistant pathogens (i.e., Salmonella, Escherichia coli, Campylobacter, Staphylococcus, Burkholderia, Pseudomonas, and Candida), where conventional antibiotics failed (Hong-Geller and Micheva-Viteva, 2013; Abouelhassan et al., 2014; Selin et al., 2015; Guo et al., 2016; Deblais et al., 2018, 2019; Helmy et al., 2018, 2020; Kathayat et al., 2018). Since chickens and mammals do not have Tat protein homologs, the inactivation of the Tat pathway using small molecule (SM) inhibitors is a promising approach to discover new antimicrobial agents with no toxicity to eukaryotes as an alternative to conventional Campylobacter control methods.
In this study, we used commercially available SM libraries (n = 50,917) to identify several, lead SM inhibitors that targeted the Tat-system and had anti-C. jejuni activity. Tat-dependent inhibitors were identified by (1) increased C. jejuni susceptibility to copper sulfate and (2) reduced C. jejuni Fdh activity. Eight potential Tat-dependent inhibitors (T1-T8) exhibited in vitro antimicrobial activity against C. jejuni. Two inhibitors (T2 and T7) were identified as promising lead compounds with good antimicrobial efficacy against C. jejuni in chickens and with minimal impact on the host microbiota. Furthermore, an in silico docking study identified that the most promising Tat inhibitor (T7) targets key amino acids in TatC.
Materials and methods
Bacterial strains
Campylobacter jejuni 81–176 was the primary model strain used for the selection of the potential Tat-dependent inhibitors by high-throughput screening. C. jejuni 81–176 is resistant to 0.5 and 1 mM CuSO4 (Supplementary Figure S1). C. jejuni 81–176 tatC knockout mutant (C. jejuni ΔtatC) was used to confirm the Tat-dependent inhibitory effect of the selected SM. C. jejuni ΔtatC is susceptible to 0.5 mM and higher of CuSO4 (Rajashekara et al., 2009). The specificity of the Tat-dependent inhibitors was also tested on 11 additional Campylobacter strains (C. coli ATCC33559 and 10 other C. jejuni strains isolated from chickens) and 7 commensal/beneficial bacteria (Supplementary Table S1). The 1 C. jejuni strains (Au-13, Au-20, Au-38, Au-39, Au-47, Au-50, Au-24, Au-32, Au-44, and Au-45) were selected based on their single nucleotide polymorphism (SNP)-type and percent of prevalence in chickens (Merchant-Patel et al., 2008; Kumar et al., 2016). Additional details about the bacterial strains are shown in Supplementary Table S1.
Eukaryotic models
Colonic epithelial cells (Caco-2) were used to evaluate the cytotoxicity and the ability of the selected eight SMs to clear intracellular C. jejuni. The Caco-2 cells were obtained from the American Type Culture Collection (ATCC HTB-37; Rockville, MD, United States). For in-vivo testing, 3 and 5-week-old White Cronish broiler chickens, obtained from Meyer’s hatchery (Polk, OH, USA), were used to validate the anti-C. jejuni efficacy (log reduction in cecal content) of the selected four SMs (T1, T2, T7, and T8) in poultry and their impact on the cecal microbiota. Broiler chickens were grown in accordance with The Ohio State University Animal Care and Use Program (accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International) and performed following the Institutional Animal Care and Use Committee. Chickens were fed a standard broiler diet (i.e., broiler starter phase from day 0 to 10; broiler growth phase from day 11 to 25; and broiler finisher phase from day 26 to 42; PNW extension #658) which was obtained from local feed mill at the Ohio Agricultural Research and Development Center (OARDC). Chickens and associated rooms were observed at least twice daily to assure that no shortage in feed or side effects occur due to the bacterial inoculation or treatments provided during the experiments.
SM libraries
A total of 50,917 SMs obtained from 11 libraries were screened in this study. SMs were suspended in 100% DMSO at concentration ranging from 2.5 μg/mL to 12.5 μg/mL between libraries and stored at −80°C (Supplementary Table S2). These libraries originated from The National Screening Laboratory for the Regional Center of Excellence in Biodefense and Emerging Infectious Disease (NSRB, United States)—The New England Regional Center of Excellence for Biodefense and Emerging Infectious Diseases (NERC, USA) collection. The NSRB and NERC libraries included FDA-approved bio-active SM, SM used by NIH in recent clinical trials, bio-active screens, and diversified SM synthesized for favorable physico-chemical properties (e.g., solubility, low/no toxicity and increased stability). Details about the origin and concentration of the libraries are shown in Supplementary Table S2.
High-throughput copper sulfate sensitivity assay
Our in silico study highlighted that the unique assembly and disassembly feature of Tat system is required for the translocation of several relatively large folded proteins, such as formate dehydrogenase (Fdh) and multi-copper oxidase (CueO) (Rajashekara et al., 2009). Since CueO function requires the transport of the Tat system, increased susceptibility to copper was used as an indicator during our in vitro screen for Tat-specific SM inhibitors. The identification of SM increasing sensitivity of C. jejuni 81–176 to copper sulfate (1 mM CuSO4, non-lethal dose) was performed using a high-throughput screening in 384 plate formats at NSRB facilities (Harvard Medical School, Cambridge, MA, USA). Columns 1 to 24 of assay plates were filled with 40 μL of MH broth +1 mM CuSO4 using a Matrix WellMate (Thermo Fisher, United States) automatic plate filler (Drozd, 2012). In total, 100 nL of SM were transferred to each well using The Institute of Chemistry and Cell Biology (ICCB, USA) Longwood Screen Facility Seiko D-TRAN XM3106-31 PN 4-axis cartesian robot (V&P Scientific), which was controlled by SRC-310A Controller/SPEL. Columns 1–23 of the assay plates were inoculated with 40 μL of C. jejuni 81–176 normalized at optical density (OD600) of 0.16 in fresh MH broth supplemented with 1 mM CuSO4. Wells in column 24 were filled with 40 μL of C. jejuni ΔtatC (susceptible to 1 mM CuSO4) normalized at 0.16 OD600 in the same conditions. The ΔtatC mutant is susceptible to 0.5 mM CuSO4; however, for screening purpose, we used the stringent 1 mM CuSO4. Plates were incubated at 42°C for 36 h under microaerophilic conditions (85% N2, 10% CO2, and 5% O2). Turbidimetric measurements (OD600) were recorded before and after incubation using a Biotek Synergy HT spectrophotometer. The positive and negative controls on each assay plate were used to calculate a Z’ value for that plate. If the Z’ value was >0, the threshold for defining a compound as positive was set at three standard deviations above the average positive control value. Z’ is calculated as described in the study mentioned in the reference (Zhang et al., 1999): Z’ = 1- (3σneg + 3σpos)/(μneg-μpos), where μ is the mean and σ is the standard deviation. The ΔtatC mutant is the positive control mimic, and wild type without drug is the negative control.
Furthermore, validation of the primary assay was performed in a 96-well plate format for the selected 177 SMs with high drug-like properties that significantly increased susceptibility of C. jejuni to CuSO4. In total, 100 μL of C. jejuni suspension normalized at 0.08 OD600 in fresh MH broth supplemented with 1 mM CuSO4 were transferred to each well of a 96-well plate and treated with 6.25 μg of SM. A parallel 96-well plate was prepared as described above but without CuSO4, to identify SM that did not inhibit the growth of C. jejuni in a Tat-specific manner. Plates were incubated at 42°C under microaerophilic conditions for 24 h. Turbidimetric measurements were recorded before and after incubation, to remove a potential increase in OD600 caused by the SM. Only SM inhibiting C. jejuni WT growth in the presence of CuSO4 without inhibiting C. jejuni WT growth in the absence of CuSO4 was selected for further analyses.
Counter screens using the screensaver SM database
Based on previous publicly available bioassay data, the ICCB-longwood/NSRB Screensaver database version v2010.10.29 and v2012.01.26 was used to eliminate SMs that were less likely to target the Tat pathway (Tolopko et al., 2010). Commercial and pharmaceutical based libraries with known antibacterial applications were cross-referenced with the selected SM. In addition, SMs that had positive effects on eukaryote-based screens were deprioritized because of the absence of eukaryotic homolog Tat system.
Counter-screen using medicinal chemistry software
A series of filters were established to select SM with the optimized drug-like properties. The criteria were based on physicochemical descriptors, potential liabilities (i.e., predicted toxicity), chemical structural diversity, and novelty. ChemDraw suite (CambridgeSoft, PerkinElmer, United States) was used to calculate molecular weight from the simplified molecular-input line-entry system (SMILES) notation. Molecular weights less than 200 Daltons (Da) and more than 550 Da were deprioritized due to the golden triangle measurements for drug-like characteristics (Johnson et al., 2009). ChemBioFinder (CambridgeSoft, PerkinElmer, United States) was used to identify structural redundancy between the SM and remove highly similar candidates. SciFinder® (Chemical Abstracts Service) was used to deprioritize SM, showing more than 90% similarity with previously investigated drugs.
Formate dehydrogenase inhibition activity assay
Formate is an essential source of energy for Campylobacter and plays a role in optimizing the adaptation of C. jejuni to the oxygen-limited gastrointestinal tract of the host (Kassem et al., 2017). Fdh is translocated by the Tat system in the periplasm, and thus, the inactivation of Tat system results in a reduction of the Fdh activity. Hence, this activity was used as an indicator during our in vitro screening procedures, to validate Tat-dependent SM inhibitors of C. jejuni. The Fdh inhibition activity assay was performed with the 177 SM with drug-like properties that increased the susceptibility of C. jejuni to CuSO4 (1 mM). C. jejuni 81–176 suspension normalized to 0.08 OD600 in fresh MH broth was incubated for 24 h in microaerophilic condition with 6.25 μg of SM. Subsequently, the treated cultures were suspended in an oxygen-restricted solution containing 25 mM sodium phosphate buffer (pH 7) with 1 mM benzyl-viologen and 10 mM sodium formate. The increase of OD578 was measured using a SpectraMax Plus 384 absorbance plate reader (Molecular Devices, USA), to monitor the reduction in benzyl viologen, an indicator of Fdh activity. N = three replicates per SM. C. jejuni 81–176 treated with 1% DMSO and C. jejuni ∆tatCmutant were used as controls. SMs that inhibited at least 30% Fdh activity (which is equivalent to the inhibitory effect of 1 mM of azide on FDH activity (Davies, 2017)) compared with the DMSO control were further down selected for analysis.
Activity spectrum of the selected 19 SMs against several Campylobacter jejuni, Campylobacter coli, and commensal/beneficial gut bacteria
In total, 19 SMs were selected using CuSO4 sensitivity and Fdh inhibition assay and further tested on 11 Campylobacter strains grown in MH broth supplemented with 0.5 mM CuSO4 and treated with 6.25 μg/mL of SM, as described above. A similar experiment was performed with seven commensal/beneficial gut bacteria to determine species specificity of these SMs. In brief, an overnight suspension was normalized at 0.05 OD600 using the appropriate medium and challenged with 6.25 μg/mL of SM. Details about the strains and their growing conditions are shown in Supplementary Table S1. Medium alone and 1% DMSO were used as controls (N = 3 replicates per SM).
Copper sulfate sensitivity dose–response assay in vitro using the selected 19 SM
A CuSO4 sensitivity dose–response assay was performed to determine the minimal concentration of SM that completely inhibited (MIC) or killed (MBC) C. jejuni 81–176 in the presence of CuSO4 (0.5 mM). In total, 19 SMs were two-fold serially diluted to obtain a final SM concentration ranging from 6.25 to 0.012 μg/mL. C. jejuni 81–176 was then treated with a determined concentration of SM, as described in the copper sensitivity assay. The lowest SM concentration that completely inhibited the growth without killing C. jejuni in the presence of 0.5 mM CuSO4 was considered the MIC (no increase in OD600 over time, but viable cells were recovered on agar plate after challenge). The lowest bactericidal SM concentration was considered the MBC (no increase in OD600 over time and no viable cells were recovered on agar plate after challenge) (N = 3 replicates per SM). A similar copper sulfate sensitivity dose–response assay was performed with the eight most potent SMs (T1-T8) against other Campylobacter strains (n = 11; Supplementary Table S1) using methodology described above. However, these SMs were two-fold serially diluted to obtain a final SM concentration ranging from 5 μg to 0.0625 μg for testing (N = two independent experiments with four technical replicate for each SM).
Cytotoxicity of the selected eight SM on Caco-2 colon epithelial cells
Cytotoxicity of the eight SMs (T1-T8) was tested on Caco-2 cells at 5, 25, and 50 μg/mL, as previously described (Kumar et al., 2016; Deblais et al., 2019). In brief, a 96-well plate seeded with approximately 1.4 × 105 Caco-2 cells/well in MEM medium was challenged with a final SM concentration of 5, 25, and 50 μg/mL. After 24 h of incubation at 37°C in a humidified 5% CO2 incubator, cytotoxic effects were determined using the PierceTM Lactate Dehydrogenase Cytotoxicity Assay Kit (Thermo Fisher Scientific). Cell death was measured based on the production of formazan (chromogenic dye), which can be read at OD570. Equal concentrations (5, 25, and 50 μg/mL) of kanamycin or chloramphenicol, 1% DMSO, and 10X lysis buffer were used as control. The cytotoxicity level was calculated according to the manufacturer’s instructions (N = two independent experiments with three technical replicate for each SM).
Copper sulfate sensitivity dose–response assay in infected Caco-2 colon epithelial cells
The intracellular reduction in C. jejuni in the presence of a given SM (T1-T8) and 0.5 mM CuSO4 were evaluated using Caco-2 cells, as previously described (Kumar et al., 2016; Deblais et al., 2019). In brief, cells were infected for 2 h using a multiplicity of infection (MOI) of 100. Infected cells were treated with SM at a final concentration ranging between 5 and 0.315 μg/mL and incubated at 37°C for 24 h in a humidified 5% CO2 incubator. Following incubation, cells were washed once with 1X PBS, lysed with 0.1% Triton-100X, serially 10-fold diluted in 1X PBS, and plated on MH agar plate. Plates were incubated in microaerophilic conditions at 42°C for 24 h, to determine the intracellular survival. Cells not infected and not treated and cells infected and treated with 1% DMSO were used as controls (N = two independent experiments with four technical replicate for each SM).
Effect of selected four SMs on Campylobacter jejuni persistence in chicken ceca
Four SMs (T1, T2, T7, and T8) were selected from the initial list of eight SMs based on their low MBC in Caco2 cells (<5 μg SM/ml), low cytotoxicity indexes (<20% toxicity at 5 μg SM /ml), and their minimal effect on commensal bacterial species (less than two species inhibited by the SM out of the seven strains tested; Table 1). The antimicrobial efficacy of our SM was tested on 5-week-old chickens, to assess the clearance of C. jejuni load right before slaughter age. Overall, 5-week-old Campylobacter-free, i.e., specific pathogen-free [SPF] chickens were inoculated orally with a mixture of five C. jejuni strains (105 CFU/chicken, Supplementary Table S3). Rectal swabs were collected 1 day post-inoculation (DPI) to confirm C. jejuni colonization (CFU/g of feces) in birds. From 2 DPI to 7 DPI, groups of 4–5 chickens were treated orally twice a day with one of the four SMs (T1, T2, T7, or T8; approximately 0.225 mg of SM per kg body weight). Details of the treatment groups and the inoculum are shown in Supplementary Table S3.
The antimicrobial efficacy of T1, T7, and T8 was also tested on 3-week-old chickens to assess the clearance of C. jejuni load in chicken right after inoculation. Overall, 3-week-old Campylobacter-free chickens were inoculated orally with a mixture of five C. jejuni strains (105 CFU/chicken, Supplementary Table S3). Rectal swabs were collected 1 DPI to confirm the intestinal colonization of the birds by C. jejuni (CFU/g of feces). From 2 DPI, chickens were treated orally twice a day for 5 days (from 2 DPI to 7 DPI) with one of the SMs (T1, T7, or T8; approximately 0.127 mg of SM per kg body weight; n = 5–6 chicken per group). Details of the treatment groups and the inoculum are shown in Supplementary Table S3.
For both experiments, C. jejuni colonized-chickens treated with 0.0001% DMSO and colonized chickens without additional carrier treatment (non-treated) were used as controls (n = 3–6 chicken per group). Details of the treatment groups and the inoculum are shown in Supplementary Table S3. Both 3 and 5-week-old chickens were euthanized at 7 DPI using carbon dioxide gas and ceca (cecal content and the pouch) and were aseptically collected. One of the ceca pairs was immediately stored at −80°C for microbiota studies. The other ceca were suspended in 1X PBS, homogenized, serially diluted, plated on MH media supplemented with Campylobacter Selective Supplement (CSS) agar plate, and incubated for 48 h at 42°C in microaerophilic conditions to determine C. jejuni load in the ceca (CFU/g of ceca).
DNA extraction and 16S MiSeq sequencing
All the samples collected during the chicken experiments (n = 58) were selected for microbiota analysis. Genomic DNA was extracted from 150 to 200 mg of cecal contents using the PureLink Microbiome DNA Purification Kit (Life Technologies, Invitrogen Corp.) and combined with RNAse treatment (10 units/h). After quality control with nanodrop, the 16S rRNA V4-V5 variable region was amplified, purified, and sequenced. Amplicon libraries were prepared by using Phusion® High-Fidelity PCR Kit (New England Biolabs Inc., Ipswich, MA, United States), as previously described (Kumar, 2015; Deblais et al., 2018, 2019; Kumar et al., 2018; Srivastava et al., 2020). PCR products were cleaned using AMPure XP PCR (Beckman Coulter Inc., Beverly MA, USA) and were sequenced using Illumina MiSeq 300-base paired-end kit at the Molecular and Cellular Imaging Center.1 Sequencing raw data files are publicly available at NCBI Bioproject #PRJNA1023035.
Bioinformatics analysis
Quality control of the raw reads was performed using FastQC (Babraham Bioinformatics, Cambridge, United States). Trimmomatic was used for trimming and removal of NexteraPE-PE adapter sequences (Bolger et al., 2014). Trimmed reads were processed using QIIME2 v. 2020.11 (Bolyen et al., 2019). The DADA2 plugin was used to process and check the quality of the reads (Callahan et al., 2016). A sequencing depth of 6,600 reads was used for the rarefaction. Taxonomic assignment was performed using QIIME2 and the latest SILVA reference database (version 138.1; 99% homology cut off) (Quast et al., 2013). The obtained reads were further filtered for eukaryotic, mitochondrial, and chloroplastic genetic signatures.
In silico docking study of the interactions between the selected compounds and the Tat system
Autodock 4.0 (BIOVIA discovery studio visualizer) (Morris et al., 2009) was used for docking with a homology model, which was generated using an online platform Phyere 2 (Kelley et al., 2015). Due to the unavailability of the 3D crystal structure of C. jejuni TatC, we turned to Aquifex aeolicus VF5, which possessed a TatC homolog. The sequence alignment and secondary structure of A. aeolicus VF5 closely resemble those of C. jejuni TatC (Ramasamy et al., 2013). Of particular significance, the 3D crystal structure of A. aeolicus VF5 is readily accessible in the Protein Data Bank (PDB ID: 4HTS). Consequently, we opted to utilize this structure for the generation of a homology model and Discovery Studio Visualizer along with Chimera to visualize protein–ligand (SM) interaction. Graphical User Interface: AutoDock Tool (ADT) was used for the preparation of pdbqt files for protein and ligand and grid box creation. AutoGrid was employed for the preparation of the grid map, and the grid size was set to 60X60X60 xyz points with grid spacing at 0.375 A. During docking, both protein and ligand were considered as rigid, and the outcomes of docking with 1.0 A in positional root-mean-square deviation (RMSD) were clustered together. The docking pose with most favorable parameters (i.e., lowest energy or binding affinity) was aligned with protein structure and was further analyzed using BIOVIA discovery studio visualizer.
Data analysis
A one-way ANOVA combined with a Tukey’s test was used to analyze the difference in the abundance of C. jejuni in ceca between treatments. A p-value ≤0.05 was considered statistically significant. Alpha diversity was analyzed using Shannon (richness and evenness) and Faith’s PD (phylodiversity) indices. The Kruskal–Wallis test was used to identify difference in alpha diversity. Based on Bray–Curtis distance matrices, permutational multivariate analysis of variances (PERMANOVA) was used to identify difference in beta diversity (unweighted and weighted uniFrac). An analysis of composition of microbes (ANCOM) was used to identify differences in the relative abundance between phylum and species levels (Mandal et al., 2015). A p-value ≤0.01 was considered statistically significant. A multivariate analysis was performed to identify potential correlations between the spectrum of activity and antimicrobial efficacy (MIC/MBC) of the selected SM and the microbiota data and the C. jejuni load in the chicken ceca. Table 2 shows summary of the cutoff selection criteria used in this study, to select the best Tat-dependent SM inhibitors (Ramasamy et al., 2013).
Table 2. Selection criteria used in this study to identify the most potent Tat-dependent SM inhibitors.
Results
The growth of Campylobacter jejuni 81–176 affected by 177 SMs only in the presence of copper sulfate
A total of 50,917 SMs divided into 11 libraries were screened against C. jejuni 81–176, to identify potential inhibitors of the Tat system (Supplementary Table S2). The hits were identified by screening in the presence of 1 mM CuSO4. Since CueO contributed to CuSO4 resistance and its function requires Tat system transport, increased susceptibility to copper was used as an indicator during high-throughput sequencing for Tat-specific SM inhibitors. The growth profile obtained for each SM tested was compared with the growth of the ΔtatC mutant (susceptible to 1 mM CuSO4; no OD600 increase over time) and C. jejuni 81–176 (resistant to 1 mM CuSO4; Supplementary Figure S1) in the presence of 1 mM CuSO4. Out of them, 679 SMs completely inhibited the growth of C. jejuni in the presence of 1 mM copper sulfate after comparison of the turbidimetric values obtained with the C. jejuni ΔtatC mutant. The number of SM identified per library was not proportional to the concentration of the libraries (ranging between 2.5 and 12.5 μg/mL). However, known bioactive collection (Biomol 4, MSDiscovery 1, Microsource 1, and MIH Clinical Collection 1 and 2) displayed higher hit rate (approximately 3.54%) than commercial libraries (Asinex, Chembridge 3, Chemdiv 4, Enamine 2, Lifechemicals 1, and Maybridge 5; approximately 1.20%), suggesting that each library might be composed of SM with distinct chemical structures (Supplementary Table S2). The average hit rate across all libraries was 1.33%, which is significantly higher than the optimal hit rate proposed by the NSRB screening guidelines (0.3% or approximately 1 hit/plate) (Drozd, 2012). Out of the 679 SMs identified, 177 SMs had no predicted bioactivity in eukaryotic cells (i.e., selected SM had no predicted interactions with known eukaryotic targets based on in silico counter screens) and followed the Lipinski rule of five based on in silica analyses (Lipinski et al., 2001; Lipinski, 2004). Among the 177 SMs, 66 SMs had a thiourea group, 46 SM had a benzimidazole group, 38 SM had an acylhydrazone group, and 11 SM had an oxadiazole group. These 177 SMs (107 SM from Asinex, 28 SM from Chembridge, 17 SM from Life chemicals, and 25 SM from Maybridge) were selected for the secondary screen upon resynthesis.
21 SMs increased the susceptibility of Campylobacter jejuni to CuSO4 and reduced FDH activity in vitro
Of the selected 177 SMs tested, only 33.3% of the SM (n = 59/177) reduced at least 30% growth of C. jejuni in the presence of 0.5 mM CuSO4 compared with the DMSO control (Figure 1A). Twenty-one SM completely inhibited C. jejuni growth, 19 SM reduced the growth of C. jejuni by 75 to 99%, and 19 SM reduced the growth of C. jejuni between 30 and 75% in the presence of 0.5 mM CuSO4 compared to the DMSO control. The Fdh inhibition activity assay showed that 56.5% of the SMs (n = 100/177) reduced C. jejuni Fdh activity by 30 to 100% compared with the DMSO control (Figure 1B). Overall, 21 SMs that sensitized C. jejuni to copper sulfate also displayed a significant reduction in FDH activity (>30%), suggesting that these SMs affect the growth of C. jejuni in a Tat-dependent manner. These 21 SMs were selected for further testing.
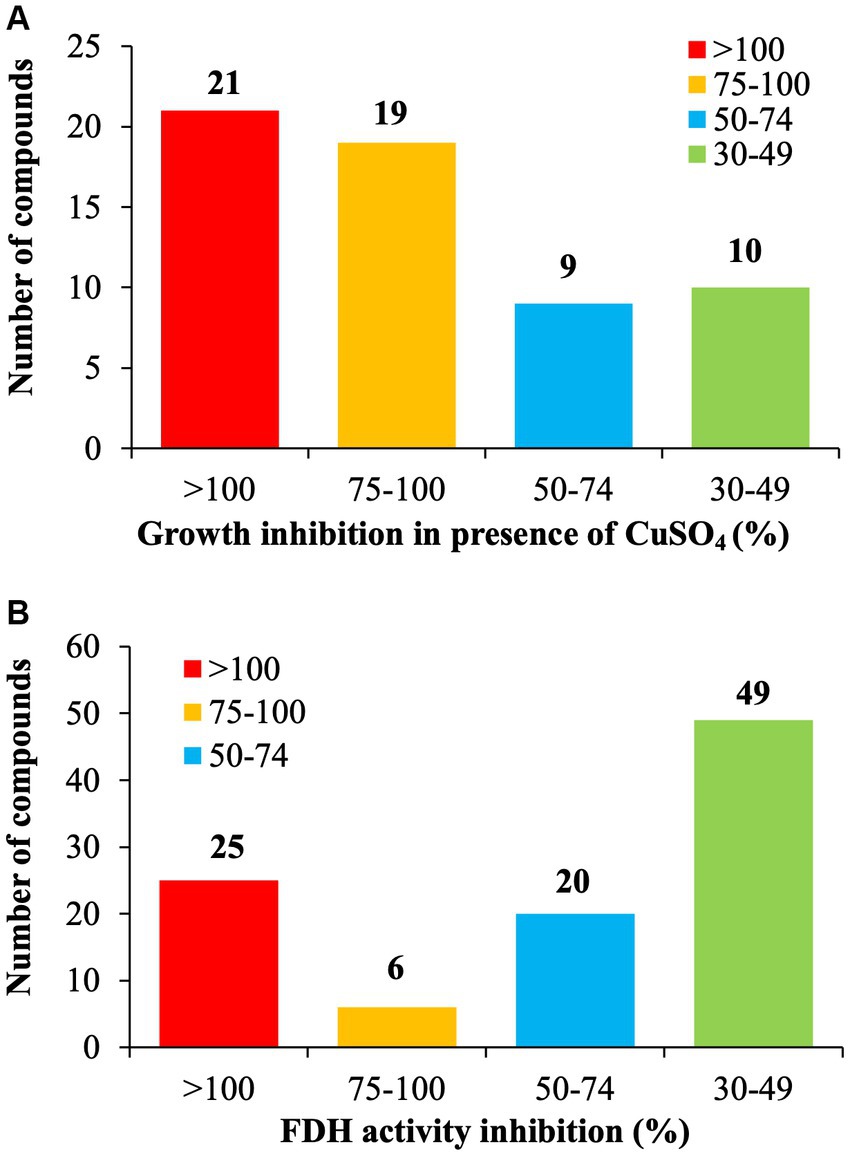
Figure 1. Identification of Tat dependent inhibitors in C. jejuni 81–176. (A) Copper sulfate sensitivity assay. C. jejuni 81–176 was challenged for 24 h with 6.25 μg/mL of SM plus 0.5 mM CuSO4 in microaerophilic condition. The growth inhibition was determined by measuring the optical density and being compared to the DMSO control. (B) Formate Dehydrogenase (FDH) inhibition activity assay. C. jejuni 81–176 was challenged for 24 h with 6.25 μg/mL of SM in microaerophilic condition. The FDH activity was determined by measuring the optical density and being compared to the DMSO control. N = 3 replicates per group.
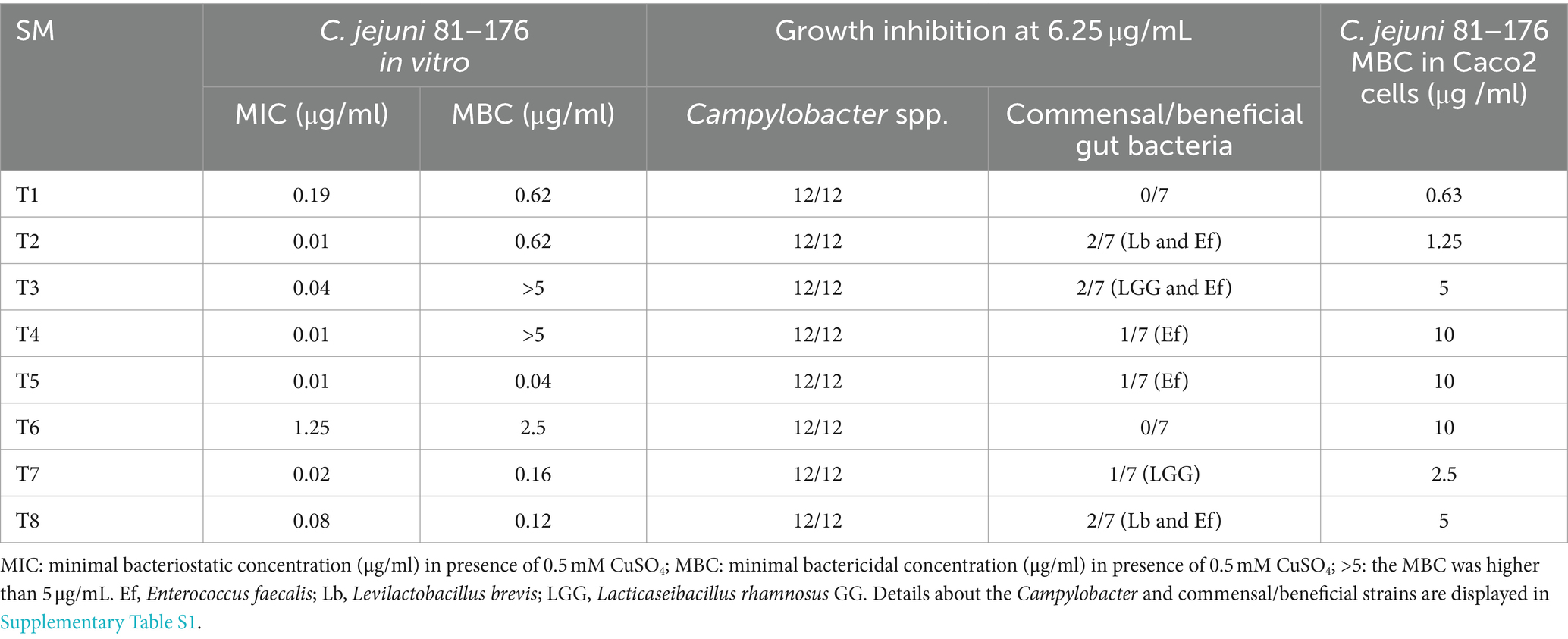
The inhibitory activity of the 19/21 SM (2 SM could not be resynthesized) was also tested on other Campylobacter strains (n = 11; Supplementary Table S1) and commensal/beneficial gut bacteria (n = 7) using a copper sensitivity assay at 0.5 mM CuSO4 with 6.25 μg of SM, as mentioned above. Overall, 19 SMs completely inhibited the growth of 12 Campylobacter strains in the presence of copper sulfate while having minimal impact on the growth of two commensal/beneficial gut bacteria in a similar growth condition (growth inhibition in the presence of copper sulfate lower than 50% compared with the DMSO control). The selected 19 SMs displayed a very similar spectrum of activity profiles. They completely inhibited the growth of all Campylobacter strains tested at 6.25 μg. Furthermore, the 19 SMs had no growth effect on Escherichia coli Nissle 1917, Streptococcus bovis, Bifidobacterium adolescentis, Bifidobacterium longum, and Bacteroides thetaiotaomicron at 6.25 μg in the presence of 0.5 mM CuSO4; however, they affected the growth of Lacticaseibacillus rhamnosus GG (n = 13 SM), Enterococcus faecalis (n = 16 SM), and Levilactobcillus brevis (n = 2 SM; Table 1).
Selection of eight most potent Tat-dependent inhibitors
A copper sulfate sensitivity dose–response assay was performed in vitro on C. jejuni 81–176 using the selected 19 Tat-dependent inhibitors (Figure 2). One SM had an MIC of 3.13 μg/mL; two SMs had MIC at 1.56 μg/mL; three SMs had MIC at 0.78 μg/mL; one SM had MIC at 0.39 μg/mL; two SMs had MIC at 0.19 μg/mL; two SMs had MIC at 0.098 μg/mL; two SMs had MIC at 0.049 μg/mL; three SMs had MIC at 0.024 μg/mL; and one SM had MIC at 0.012 μg/mL in the presence of 0.5 mM CuSO4. Interestingly, the number of strains affected by the compounds (i.e., spectrum of activity) was not correlated with their antimicrobial efficacy (e.g., MIC and MBC).
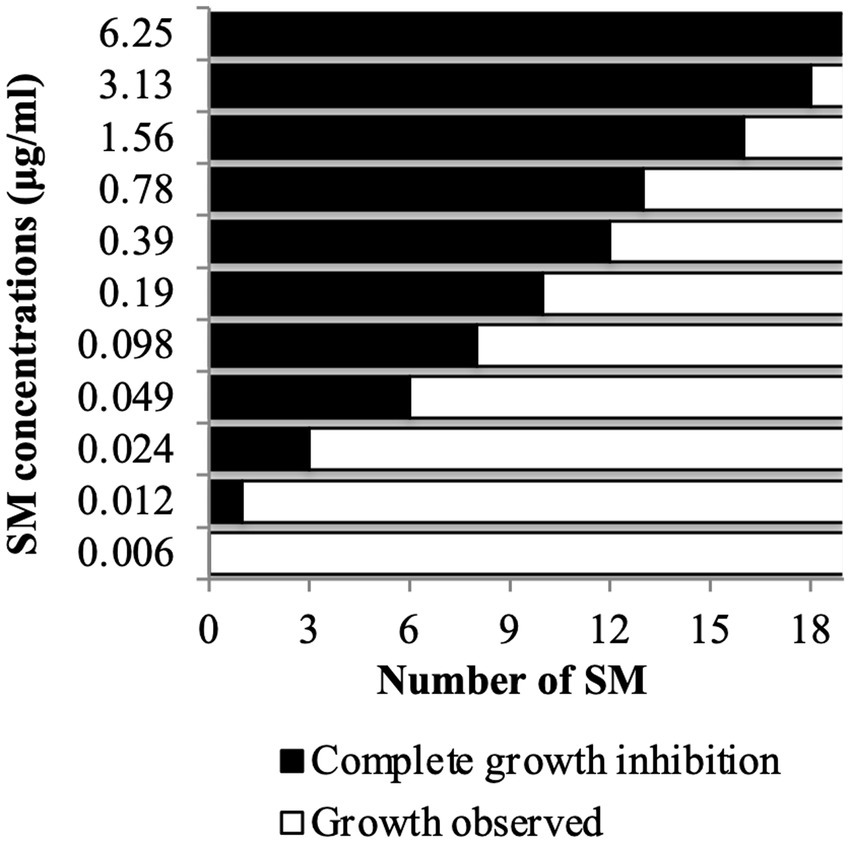
Figure 2. Copper sulfate sensitivity dose–response assay in vitro. C. jejuni 81–176 was challenged for 24 h with SM concentration ranging between 0.006 and 6.25 μg/mL in presence of 0.5 mM CuSO4. A total of 19 compounds were tested. N = four replicates per group.
Based on the antimicrobial activity (i.e., efficacy and spectrum of activity) and in silica data obtained, we selected eight SMs with high drug-like properties, little or no growth effect on commensal/beneficial gut bacteria, and high efficacy against several Campylobacter strains (Table 1; Supplementary Table S1). Overall, the antimicrobial efficacy of the selected eight SMs was similar across the 12 Campylobacter strains tested with MIC values of 0.01 μg/mL and higher, and MBC values of 0.04 μg/mL and higher (Supplementary Tables S4, S5, respectively). In addition, the three-dimensional analysis of the chemical structure of the eight selected SMs showed that T8 and T2, T6 and T1, and T7 and T3 displayed high structural similarities, while T4 and T5 had unique chemical structures (Figure 3). Furthermore, most of the SMs have sulfur and/or nitrogen-based functional groups (thiourea, imidazole uracil, sulfonamide, thiomorpholine dioxide, phenylurea, pyridine, piperidine, oxadiazole, and quinoline). However, no association was detected between the chemical structure of the SM and their antimicrobial properties.
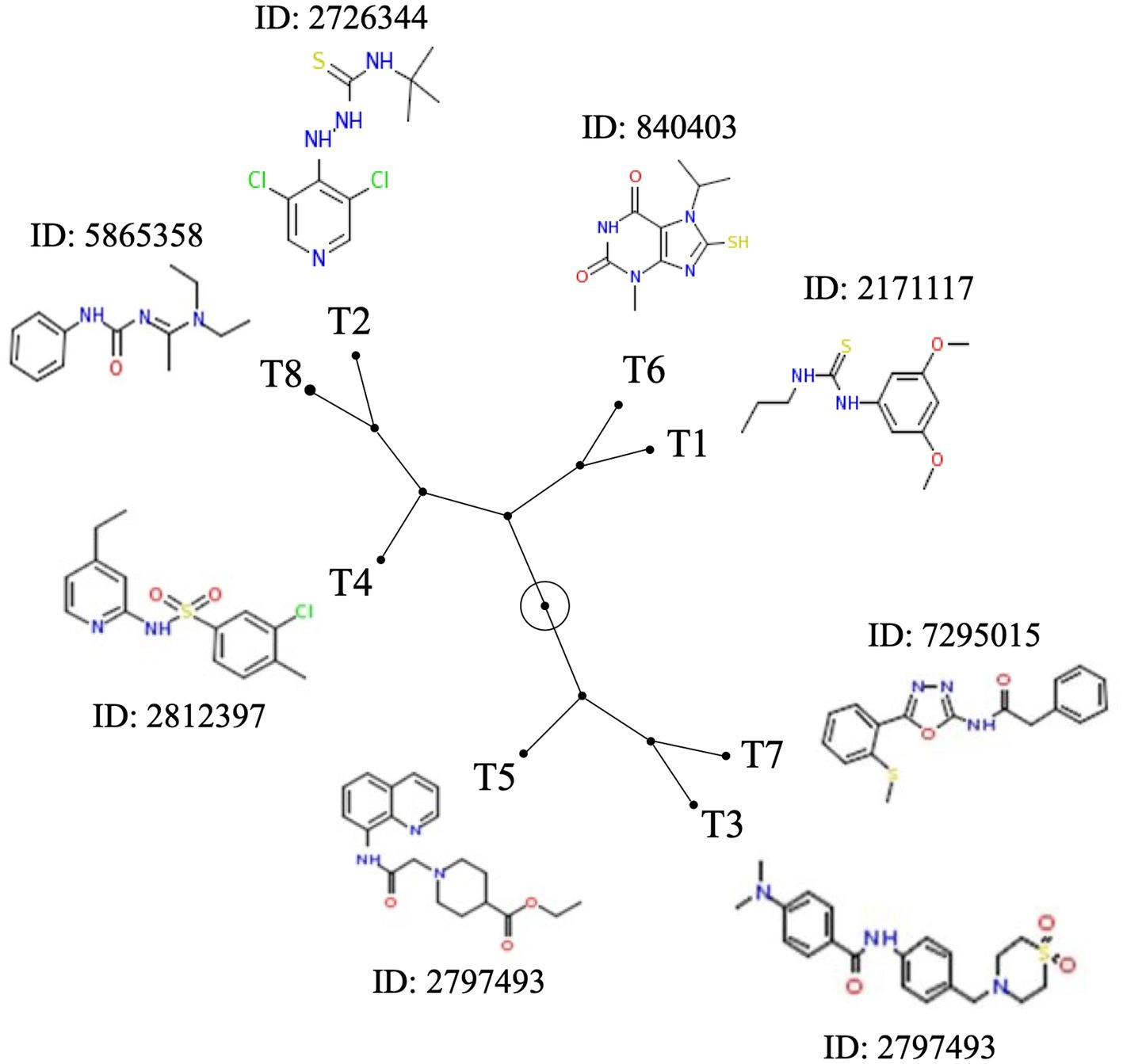
Figure 3. Chemical structure diversity of the eight most potent Tat-dependent inhibitors. The constellation tree was built based on the structure similarity score generated based on 3D Tanimoto scoring method in PubChem (https://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?p=clustering). The circled node represents the root of the tree. Each SM is associated with its chemical structure and its PubChem ID.
The eight Tat-dependent inhibitors reduced Campylobacter jejuni intracellular population In infected colon epithelial cells with low cytotoxicity level
All SMs completely cleared internalized C. jejuni 81–176 in Caco-2 cells after 24 h of treatments with a concentration of SM ranging from 0.63 μg/mL to 10 μg/mL (Table 1). Most of the SMs displayed low toxicity (at most 10%) to Caco-2 cells when treated with 50 μg/mL for 24 h (Figure 4). The toxicity values were comparable to the kanamycin and chloramphenicol-treated cells. Only T1 and T8 displayed toxicity level (36 and 33%, respectively) when treated with 25 μg/mL or 50 μg/mL of SM for 24 h; however, T1 and T8 cleared intracellular C. jejuni 81–176 at 0.63 μg/mL and 5 μg/mL, respectively, which represent concentrations up to 80-fold lower compared with ones used for the toxicity assay. Overall, T1, T2, T7, and T8 displayed the most promising antimicrobial properties in vitro were selected for further analyses.
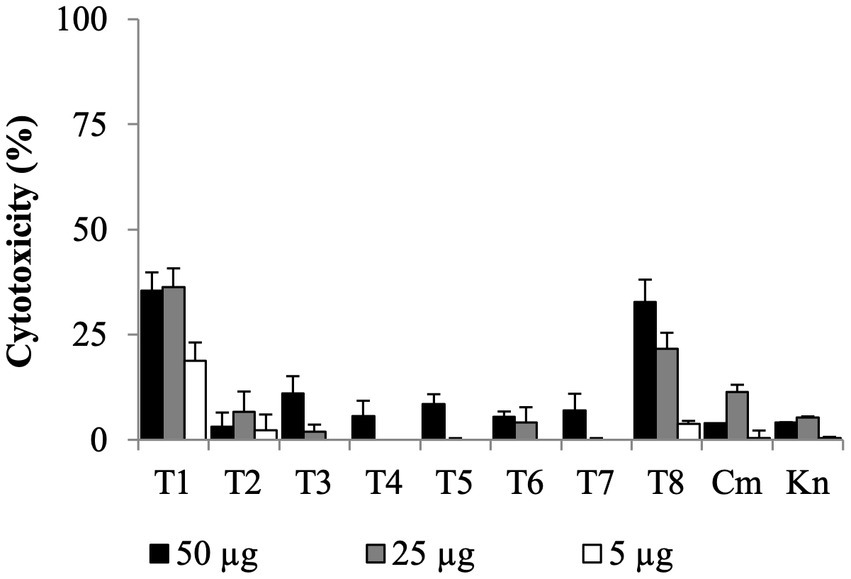
Figure 4. Dose-dependent cytotoxicity assay of the eight Tat-dependent inhibitors using colon epithelial (Caco-2) cells. Bar: standard deviation; Cm, chloramphenicol; Kn, kanamycin; LDH, lactate dehydrogenase; N = four replicates per group.
T2 and T7 treatments reduced Campylobacter jejuni load in colonized chicken ceca
As a proof of concept, the selected SMs (T1, T2, T7, and T8) were tested on 5-week-old chickens inoculated with a mixture of C. jejuni strains (Supplementary Table S1), to assess the clearance of C. jejuni in chicken right before the slaughter. Following 5 days of SM treatment (twice a day, approximately 0.225 mg of SM per kg body weight per treatment), colonized chickens treated with T2 displayed a significant reduction (1.2-log) in C. jejuni population per gram of cecum compared with the DMSO control group, which harbored approximately 5×107 CFU per gram of cecum (p < 0.01; Figure 5A). Colonized chickens treated with the T7 and T8 groups displayed a 0.5-log reduction in C. jejuni population in ceca compared with the DMSO control group (p > 0.05), while the T1 treatment did not reduce the abundance of C. jejuni in ceca compared with the DMSO group. No significant difference in body weight was recorded between treatment groups before or after 5 days of treatment (p > 0.05; Supplementary Figure S2A).
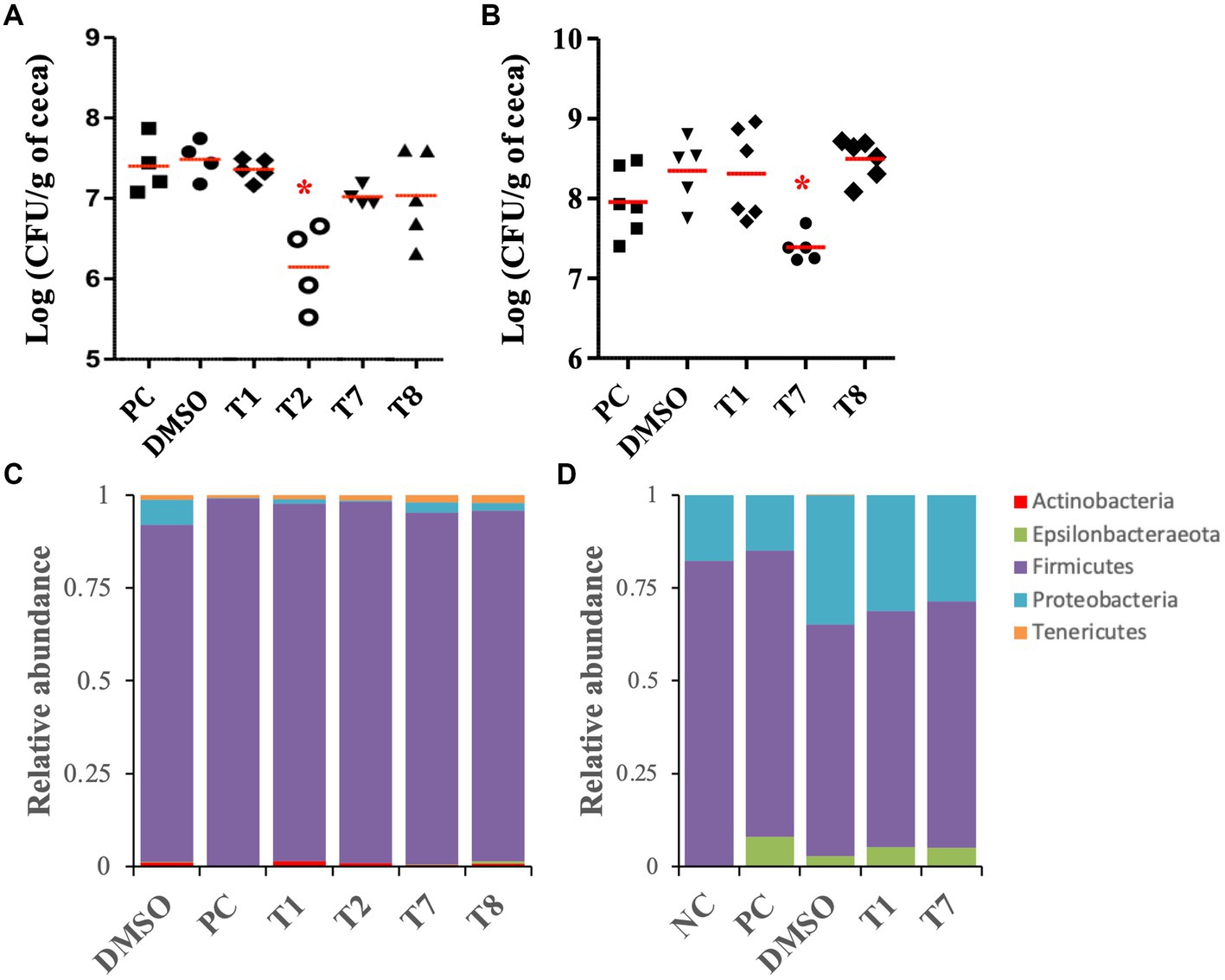
Figure 5. Effect of the four most potent Tat-dependent inhibitors on the persistence of C. jejuni in three- and five-week-old chicken ceca and its microbiota. (A) C. jejuni abundance in five-week-old chicken ceca after SM treatment (n = 5–4 chickens per group). The C. jejuni population in ceca was determined after 5 days of treatment with 0.127 mg/mL of SM. Each dot represents a chicken. Red bar represents the mean. *: significant reduction of the C. jejuni population in ceca compared to the DMSO control group (p < 0.01). (B) C. jejuni abundance in three-week-old chicken ceca after SM treatment. The C. jejuni population in ceca was determined after 5 days of treatment with 0.255 mg/mL of SM (n = six to five chickens per group). (C) Relative abundance at the phylum level in five-week-old chicken ceca after treatment. (D) Relative abundance at the phylum level in three-week-old chicken ceca after treatment. NC: not inoculated not treated chickens; PC: colonized not treated chickens; DMSO, T1, T2, T7, T8: colonized chickens treated with DMSO or one of the selected SM.
The antimicrobial activity of T1, T7, and T8 was also tested on 3-week-old chickens inoculated with a cocktail of C. jejuni strain (Supplementary Table S1), to assess the clearance of C. jejuni in young chicken right after inoculation. T2 could not be resynthesized in sufficient quantities and, thus, was not tested on this 3-week-old experiment. Following 5 days of SM treatment (approximately 0.127 mg of SM per kg body weight per treatment), 3-week-old chickens treated with T7 displayed a significant reduction (0.9-log) in C. jejuni population per gram of cecal contents compared with the DMSO-treated group, which harbored approximately 4 × 108 CFU per gram of cecal contents (p < 0.01; Figure 5B). Chickens treated with T1 and T8 harbored similar C. jejuni abundance in ceca compared with the DMSO control group. No significant difference in body weight was recorded between treatment groups before or after 5 days of treatment (p > 0.05; Supplementary Figure S2B).
The SM treatments had minimal impact on the chicken cecal microbiota
The impact of the SM treatments on the cecal microbiota of the 3 and 5-week-old chickens was studied using 16S sequencing. After processing and taxonomic assignment with the SILVA reference database, 682,777 sequences were obtained from the 58 samples studied. Sequencing depth varied between 6,648 and 15,749 reads per sample (mean = 9,753 reads per sample). Cecal samples were normalized to 6,600 sequences per sample for the data presented below.
For the 3-week-old chicken experiment, the analysis of the cecal alpha diversity, using Faith’s PD and Shannon diversity index, indicated no significant differences between the treatment groups. On the other hand, the DMSO treatment (DMSO, T1, T2, T7, and T8 groups) significantly increased the Shannon index value (approx. 6.3) compared with the colonized, non-treated groups (PC; approx. 5.5; p < 0.04). The beta diversity analysis demonstrated that the SM treatments (T1, T2, T7, and T8 groups) had minimal impact on the global microbiome composition compared with the DMSO group (Supplementary Figure S3A). Most of the variations were detected between the colonized chickens treated with DMSO (DMSO, T1 and T7 groups) and the chickens that were not treated with DMSO (NC and PC groups; p < 0.01). Additional details concerning the impact of the DMSO and C. jejuni colonization on the cecal microbiome at the genus and species levels are shown in Supplementary Figures S3A–C. Overall, the cecal microbiome was composed of Firmicutes (90–99%), followed by Proteobacteria (0.1–6.7%), Tenericutes (0.6–2.1%), Actinobacteria (0.06–1.4%), and Epsilonbacteraeota (0.02–0.1%, Figure 5C). No significant differences were detected at the phylum level between the SM-treated groups (T1, T2, T7, and T8) and the associated control group (DMSO). At the species level, T8 treatment significantly increased GCA-900066225 (11.6-fold), and T7 treatment significantly increased Ruminococcaceae UCG-014 (2.2-fold) compared with the DMSO group (p < 0.01).
For the 3-week-old chicken experiment, the analysis of the cecal alpha diversity, using Faith’s PD and Shannon diversity index, indicated no significant differences between the treatment groups of (Supplementary Figures S3D,E). Similarly, beta diversity analysis, using the weighted uniFrac, confirmed that the SM treatments (T1 and T7 groups; T8 was not included for microbiota study since it did not have positive impact on C. jejuni load in the cecum) had minimal impact on the global microbiome composition compared with the DMSO group (Supplementary Figure S3F). Most of the variations were detected between the colonized chickens treated with DMSO (DMSO, T1 and T7 groups) and the chickens not treated with DMSO (NC and PC groups; p < 0.01). Additional details concerning the impact of DMSO and inoculation of C. jejuni on the cecal microbiome at the genus and species levels are shown in Supplementary Figures S3D–F. Overall, the cecal microbiome was composed of Firmicutes (62–82%), followed by Proteobacteria (15–35%) and Epsilonbacteraeota (0–8%, Figure 5D). No significant differences were detected at the phylum level between the SM-treated groups (T1 and T7) and the associated control group (DMSO). At the species level, Caproiciproducens (6.5-fold and 4.4-fold, respectively) and Flavonifractor (1.7-fold) were significantly higher in T1 and T7 groups compared with the DMSO group. Eubacterium coprostanoligenes group (8-fold) and Neglecta timonensis (only detected in the T7 group at 0.8%) were significantly higher in the T7 group compared with the DMSO group (p < 0.01).
Docking studies predicted that T2 and T7 interact with the TatC protein of the Tat system
TatC subunit is the largest and most important part of Tat system, while Tat A and Tat B are much smaller units. The docking studies were conducted with all Tat subunits; however, TatA and B protein folding could not be predicted with high certainty due to smaller protein size. Therefore, we focused on TatC, which was modeled with a high degree of predictability and well-defined binding pockets, giving repeatable docking results. The in silico docking study demonstrated that both active compounds T2 and T7 bind in the same hydrophobic binding pocket associated with the key active residue Glu 165 (Figure 6). The calculated binding energy for T2 is −6.26Kcal/mol, while T7 has a higher binding energy of −8.0 Kcal/mol. The T2 thiourea nitrogen forms a hydrogen bond with Ile80, while the phenyl ring forms Pi-Pi stacking interaction with Phe87. Trp85 and Phe87 form Pi-sigma interaction with the T2 pyridine ring and tert-butyl group, respectively (Figures 6A,C). In addition, there are several hydrophobic interactions within carbon chains of T2 and Ile80, Ser 77, Ile162, Phe111, Ser107, Ile168, Met166, Gln83, and Ser17. The T7 functional groups (i.e., oxadiazole and phenyl groups) bind to several amino acids that are located inside the hydrophobic core of TatC (Figure 6B). More precisely, all aromatic rings of T7 have π-anion interactions with the key Glu165 (Figures 6B,D); the phenylacetamide group has π-alkyl interactions with Val169, π-π interactions with Phe84 and Trp85, and van der Waals interactions with Ile168 and Ser107; the methylthio-phenyl group has π-alkyl interactions with Ile80, Ile162, and Leu81, and van der Waals interactions with Phe111, Ser77, and Phe73. π- Σ interactions are also detected on the amide and methanethiol groups. Thus, anti-C. jejuni activity of both T2 and T7 may be attributed to their binding to hydrophobic pocket in TatC and the interaction with key residues Glu165 and Trp85 (Figure 6).
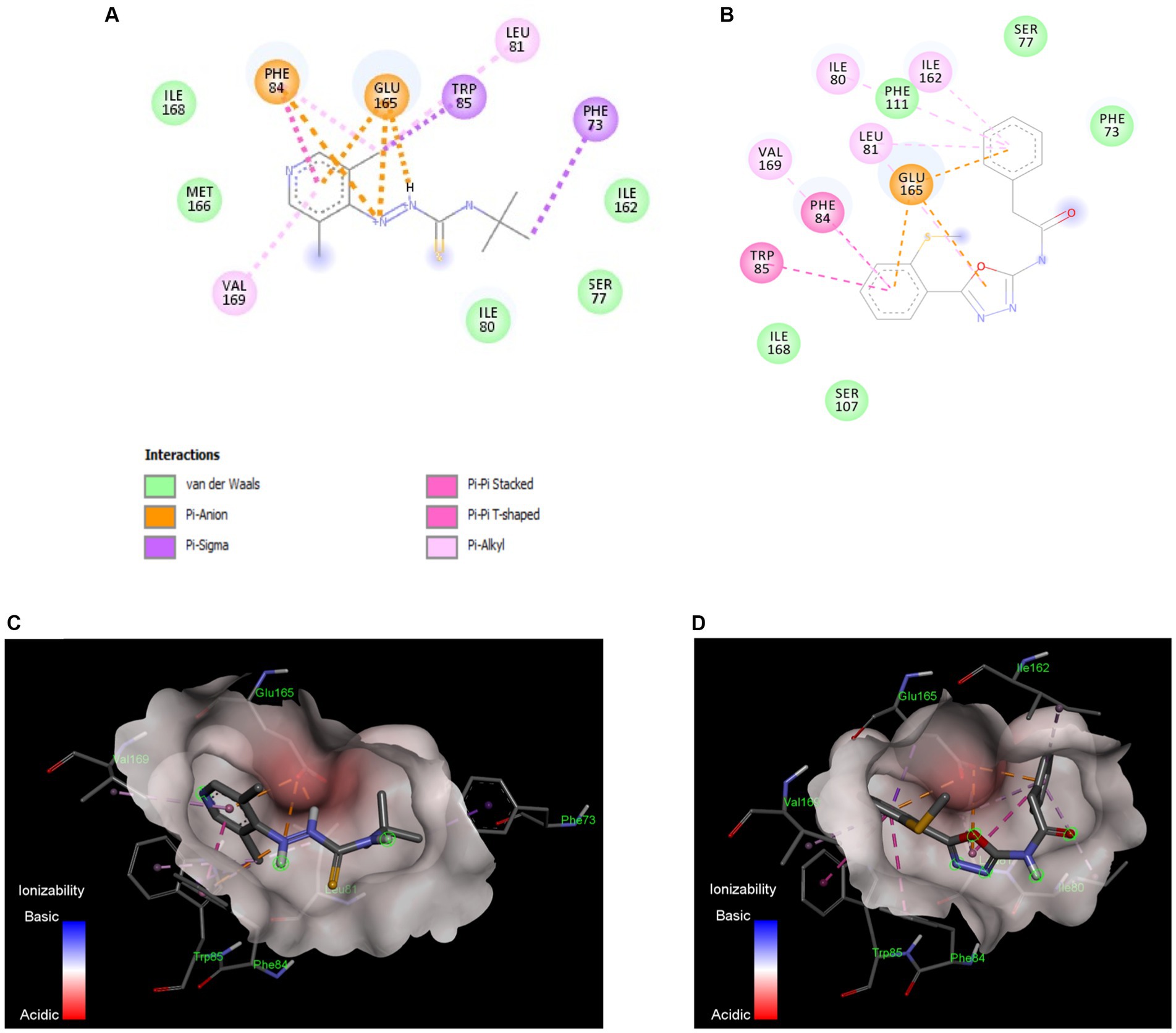
Figure 6. In silico docking model between the Tat inhibitors and the TatC system in C. jejuni. Binding interactions of the most active small molecule inhibitors with a homology model of TatC from Aquifex aeolicus, Compound T2 (A), Compound T7 (B). The compounds bind in the same pocket and the interaction with key residue Glu165 and Trp85 is responsible for Tat C inhibition. The ionizability model for T2 (C) and T7 (D) of residues in the binding pocket indicates the Pi-anion interactions with Glu165 and Pi-Pi interaction with Trp85. These docking models indicate that TatC inhibition may be responsible for the anti-C. jejuni activity of T2 and T7.
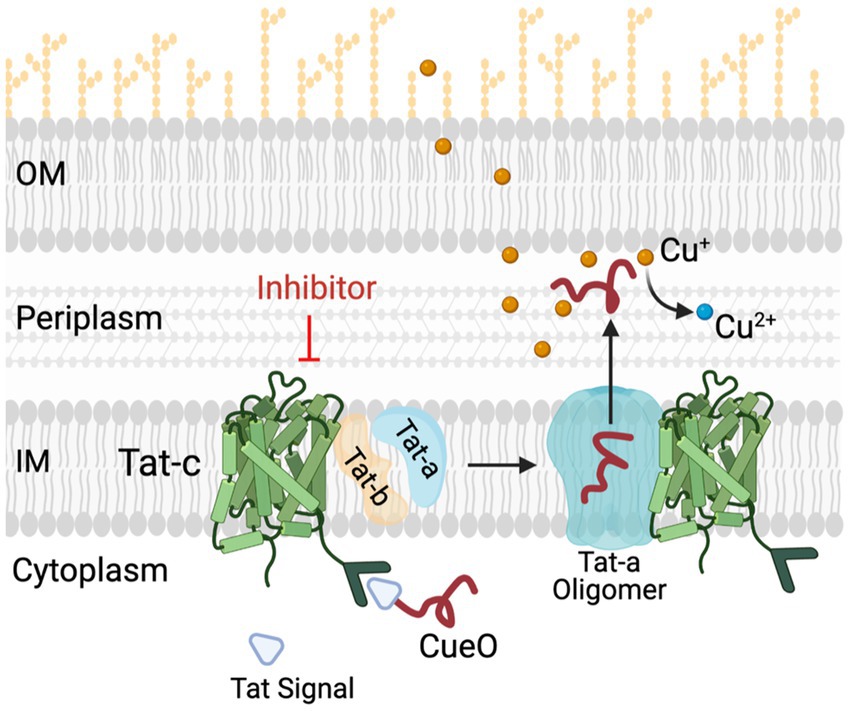
Figure 7. Role of Tat system in copper (Cu) homeostasis in C Jejuni. C jejuni employs a Tat complex (Tat-A, B, C) for the transport of proteins from cytoplasm to periplasm. TatC is the core transmembrane component of this complex located in inner membrane (IM), responsible for translocation of folded proteins such as multi-copper oxidase (CueO) and formate dehydrogenase (Fdh) from cytoplasm to periplasm. CueO is critical for oxidation of Cu+ which are highly toxic as compared to relatively non-toxic Cu2+ form. Thus, in the presence of TatC inhibitors the transportation of important cytoplasmic proteins such as CueO and Fdh is hampered. This results in increased sensitivity of C jejuni to copper. OM and IM, outer and inner membrane, respectively; CueO, copper oxidase; Cu, copper.
Discussion
The emergence of antibiotic-resistant isolates significantly reduces the antimicrobial efficacy of current control methods which are used to mitigate human campylobacteriosis (Kaakoush et al., 2015; Subbiah et al., 2016; Yang et al., 2019). Previous studies showed that the Tat system is highly conserved in Campylobacter spp. and is critical for the persistence of Campylobacter in the intestinal tract of poultry, a recurrent reservoir for Campylobacter (Rajashekara et al., 2009; Hitchcock et al., 2010; Hermans et al., 2011). Additionally, the absence of homologous proteins in chickens and mammals increases the likelihood that the Tat system-based control strategies would avoid damage or unwanted interaction with host cells (Kassem and Rajashekara, 2014). Previous studies demonstrated increased copper sulfate sensitivity and reduced Fdh activity in Campylobacter when the Tat system was non-functional; therefore, these two indicators were used to select the compounds that are likely to specifically inhibit the Tat system (hit compounds). A total of 50,917 SMs distributed among 11 “commercial” or “known bioactive” libraries provided by ICCB-Longwood version v2010.10.29 and v2012.01.26 were screened. A total of, eight SMs (T1-T8; 10 ng/mL and higher) sensitized several C. jejuni and C. coli isolated from human and poultry to sublethal dose of copper sulfate (0.5 mM) in vitro, and had minimal to absent toxicity in Caco-2 colon epithelial cells. These initial findings support our hypothesis that the Tat system is a promising target for the development of the anti-C. jejuni control method with minimal impact on eukaryotes. Further efforts will validate the molecular target of T2 and T7 to facilitate their uses against C. jejuni infections in humans. Distinct chemical backbones were observed among the eight SMs, suggesting that multiple chemical scaffolds can target the Tat system in C. jejuni or interact with chaperoning proteins. Most of the SMs were characterized by sulfur and/or nitrogen-containing functional groups (thiourea, imidazole uracil, sulfonamide, thiomorpholine dioxide, phenylurea, pyridine, piperidine, oxadiazole, and quinoline). Furthermore, SM with low molecular weights (< 400 Da) completely cleared intracellular C. jejuni between 0.63 μg/mL and 2.5 μg/mL, while SM with high molecular weights (> 400 Da) completely cleared intracellular C. jejuni at 5.0 μg/mL and higher. Therefore, the molecular weights could serve as a selection criterion of SM for testing them in a cell culture setting (Schuffenhauer et al., 2006).
This study identified two potential Tat inhibitors (T2 and T7) that significantly reduced the cecal C. jejuni load (up to 1.2-log) in colonized chickens after a 5-day treatment twice a day with 0.225 mg or 0.127 mg of SM per kg body weight per treatment, respectively. Furthermore, the SM treatments did not affect the chicken body weight gain or disturb the cecal microbiota. This preliminary in vivo data validated that the Tat system could represent a good target for the development of novel and safe control methods against C. jejuni. Future studies will focus on the development of T2 and T7 derivatives with improved antimicrobial efficacy, dosage titration, and testing these compounds in farm-like settings. Similarly, the addition of our SM to the feed will be tested, which is the preferred delivery method in commercial poultry operations. Since the solvent used to deliver the SM to the chickens had a significant impact on the cecal microbiota, future studies will focus on the development of water-soluble T2 and T7 derivatives, optimizing the dosage and bioavailability for these molecules in chicken tissues after treatment.
Differences in antimicrobial efficacy were observed with T7 and T8 between the 3 and 5-week-old chicken experiments. These differences might be due to SM dosage used and/or the microbial composition in chicken ceca. Interestingly, the abundance of the Eubacterium coprostanoligenes group was significantly increased in 3-week-old chickens treated with T7 compared with the DMSO-treated chickens. Eubacterium coprostanoligenes is an anaerobe genus involved in the reduction of cholesterol (Wei et al., 2020). However, it was also showed that depletion in the membrane cholesterol was associated with a reduction in C. jejuni cytolethal distending toxin-induced pathogenesis and, thus, attenuated the intoxication of host cells (Lin et al., 2011; Lai et al., 2013, 2015). Future studies will assess whether this bacterium has anti-C. jejuni properties. If true, the combination of our Tat inhibitor T7 with Eubacterium coprostanoligenes will also be investigated to mitigate C. jejuni in poultry. It was also observed that treating chickens with DMSO, using oral gavage, significantly affected the composition of the chicken ceca microbiota (Lactobacillus, Romboutsia, and Enterobacteriaceae). Enterobacteriaceae and Lactobacillaceae bacteria are major components of the initial intestinal microbiota and are essential for the installation and proliferation of aerobic sensitive bacteria over time (Lin and Zhang, 2017). Furthermore, certain Lactobacillus isolates have been shown to harbor potential antagonistic properties against Campylobacter in chicken (Dec et al., 2018). Therefore, the microbiome alterations caused by the DMSO treatment might have enhanced the antimicrobial efficacy of our selected compounds.
The in silico docking study demonstrated that our Tat inhibitors T2 and T7 bind with a key amino acid residue, Glu 165, are located in the hydrophobic core of TatC. Glu 165 is conserved as polar glutamine or glutamate across species of bacteria and plays an important role in TatC functioning. Glu 165 forms a hydrogen bonding network with Ser107 and Trp 85, which is important for generating electrochemical gradient for Tat energy function (Rollauer et al., 2012). Trp 85 forms an important interaction between Tat signal and W85G and suppresses Tat signaling and transportation (Ramasamy et al., 2013). It is also shown that TatC forms a glove- like shape, and Glu 165 sits in the concave surface, where it can interact with TatA (Figure 7). The ionized Glu 165 present in hydrophobic core of concave TatC is important for interactions with TatA. The mutation E165A severely compromises TatC functioning. Thus, our in silico data suggest that the binding of T2 and T7 in Glu-165 hydrophobic pocket of TatC might perturb the transport of CueO and Fdh. It is important to mention that the model displayed in this study to assess the binding affinity of our SM to TatC was conducted using the TatC crystal structure from Aquifex aeolicus. No crystal structure of other TatC proteins is available to date. Thereby, no predictive docking modeling of our SM with other TatC protein could have been conducted in this study to assess the specific binding of our SM to C. jejuni TatC. However, TatC is the most conserved protein of the Tat system across bacteria (i.e., E. coli, Aquiflex aeolicus, C. jejuni, Thermus thermophilus, and Staphylococcus aureus) (Lee et al., 2006; Ramasamy et al., 2013; Patel et al., 2014). Based on a NCBI Blastp analysis conducted on 01/10/2024, it was found that C. jejuni TatC displays at least 98.37 and 93.88% sequence similarity with other C. jejuni TatC sequences and C. coli TatC sequences (n = 100 sequenced tested per species), respectively. These observations support the antimicrobial efficacy of our SM against the C. jejuni and C. coli strains tested in this study. On the other hand, low sequence similarity was observed between C. jejuni TatC and the few TatC obtained from other non-thermophilic Campylobacter strains; Campylobacter fetus (similarity of 59.43% and above; n = 14), candidatus Campylobacter infans (similarity of 55.95%; n = 2), Campylobacter hyointestinalis (similarity of 61.76% and above; n = 12) and Campylobacter upsaliensis (similarity of 72.02% and above; n = 48). Future studies will investigate whether our best candidates could be also used against non-thermophilic Campylobacter in vivo or if better candidates can be identified from the initial libraries tested in this study. Furthermore, a similarity of 34.55 and 26.73% were observed between TatC obtained from C. jejuni and Escherichia coli Nissle 1917 and Bifidobacterium longum, respectively. To date, no TatC proteins were identified in L. brevis and rhamnosus L., Bifidobacterium adolescentis and lactis, and Enterococcus faecalis. These in silico results concord with the limited impact of our SM on commensal/beneficial bacteria.
In conclusion, data presented in this study represent a proof of concept that the Tat system represents a good target for the development of novel and safe control methods against C. jejuni. We have identified two Tat-dependent inhibitors (T2 and T7) with the potential to become effective control method to mitigate C. jejuni in poultry production systems. However, additional efforts are required to improve the antimicrobial efficacy of the Tat compounds before being used to control C. jejuni in large-scale poultry production systems.
Data availability statement
The datasets presented in this study can be found in the NCBI repository, accession number PRJNA1023035.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee (IUACUC) protocol n° 2010A00000149-R2-AM1. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LD: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. MD: Conceptualization, Methodology, Validation, Writing – review & editing. AK: Conceptualization, Methodology, Validation, Writing – review & editing. JA: Methodology, Writing – review & editing. JF: Conceptualization, Investigation, Methodology, Supervision, Writing – review & editing. RK: Formal analysis, Methodology, Validation, Visualization, Writing – review & editing. GR: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing. YH: Investigation, Methodology, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The research in GR laboratory is supported by National Institute for Food and Agriculture (NIFA) Grant # 2013–67018-21240, United States Department of Agriculture; the Poultry CRC, established and supported under the Australian Government’s Cooperative Research Centers Program; and the Ohio Agricultural Research and Development Center (OARDC).
Acknowledgments
The authors thank ICCB-L for assistance with high throughput screening. The authors also thank Rosario A. Candelero for the technical support. The authors thank Saranga Wijeratne, Molecular and Cellular Imaging Center, Ohio Agricultural Research and Development Center (http://oardc.osu.edu/mcic/), The Ohio State University for providing assistance with bioinformatics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1342573/full#supplementary-material
Footnotes
References
Abouelhassan, Y., Garrison, A. T., Burch, G. M., Wong, W., Norwood, V. M., and Huigens, R. W. (2014). Discovery of quinoline small molecules with potent dispersal activity against methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis biofilms using a scaffold hopping strategy. Bioorg. Med. Chem. Lett. 24, 5076–5080. doi: 10.1016/j.bmcl.2014.09.009
Agyare, C., Boamah, V. E., Zumbi, C. N., and Osei, F. B. (2018). “Antibiotic use in poultry production and its effects on bacterial resistance” in Antimicrobial resistance–a global threat. ed. Y. Kumar (London: IntechOpen)
Allos, B. M. (2001). Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 32, 1201–1206. doi: 10.1086/319760
Andrew Selaledi, L., Mohammed Hassan, Z., Manyelo, T. G., and Mabelebele, M. (2020). The current status of the alternative use to antibiotics in poultry production: an African perspective. Antibiot. Basel Switz. 9:594. doi: 10.3390/antibiotics9090594
Bageshwar, U. K., VerPlank, L., Baker, D., Dong, W., Hamsanathan, S., Whitaker, N., et al. (2016). High throughput screen for Escherichia coli twin arginine translocation (tat) inhibitors. PLoS One 11:e0149659. doi: 10.1371/journal.pone.0149659
Berks, B. C. (2015). The twin-arginine protein translocation pathway. Annu. Rev. Biochem. 84, 843–864. doi: 10.1146/annurev-biochem-060614-034251
Berrang, M. E., Bailey, J. S., Altekruse, S. F., Patel, B., Shaw, W. K., Meinersmann, R. J., et al. (2007). Prevalence and numbers of campylobacter on broiler carcasses collected at rehang and postchill in 20 U.S. processing plants. J. Food Prot. 70, 1556–1560. doi: 10.4315/0362-028x-70.7.1556
Blümmel, A.-S., Drepper, F., Knapp, B., Eimer, E., Warscheid, B., Müller, M., et al. (2017). Structural features of the TatC membrane protein that determine docking and insertion of a twin-arginine signal peptide. J. Biol. Chem. 292, 21320–21329. doi: 10.1074/jbc.M117.812560
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Connerton, P. L., Richards, P. J., Lafontaine, G. M., O’Kane, P. M., Ghaffar, N., Cummings, N. J., et al. (2018). The effect of the timing of exposure to Campylobacter jejuni on the gut microbiome and inflammatory responses of broiler chickens. Microbiome 6:88. doi: 10.1186/s40168-018-0477-5
Davies, J. A. (2017). Characterisation of the reversible formate dehydrogenases of Shewanella. University of East Anglia. School of Biological Sciences. Available at: https://ueaeprints.uea.ac.uk/id/eprint/66856/ (Accessed September 28, 2023).
Deblais, L., Helmy, Y. A., Kathayat, D., Huang, H., Miller, S. A., and Rajashekara, G. (2018). Novel imidazole and Methoxybenzylamine growth inhibitors affecting salmonella cell envelope integrity and its persistence in chickens. Sci. Rep. 8:13381. doi: 10.1038/s41598-018-31249-0
Deblais, L., Helmy, Y. A., Kumar, A., Antwi, J., Kathayat, D., Acuna, U. M., et al. (2019). Novel narrow spectrum benzyl thiophene sulfonamide derivatives to control campylobacter. J. Antibiot. (Tokyo) 72, 555–565. doi: 10.1038/s41429-019-0168-x
Dec, M., Nowaczek, A., Urban-Chmiel, R., Stępień-Pyśniak, D., and Wernicki, A. (2018). Probiotic potential of lactobacillus isolates of chicken origin with anti-campylobacter activity. J. Vet. Med. Sci. 80, 1195–1203. doi: 10.1292/jvms.18-0092
Drozd, M. R. (2012). Campylobacter jejuni survival strategies and counter-attack: an investigation of campylobacter phosphate mediated biofilms and the design of a high-throughput small-molecule screen for TAT inhibition. The Ohio State University. Available at: https://etd.ohiolink.edu/pg_10?0::NO:10:P10_ETD_SUBID:76976 (Accessed April 30, 2018).
Drozd, M., Gangaiah, D., Liu, Z., and Rajashekara, G. (2011). Contribution of TAT system translocated PhoX to Campylobacter jejuni phosphate metabolism and resilience to environmental stresses. PLoS One 6:e26336. doi: 10.1371/journal.pone.0026336
Facciolà, A., Riso, R., Avventuroso, E., Visalli, G., Delia, S. A., and Laganà, P. (2017). Campylobacter: from microbiology to prevention. J. Prev. Med. Hyg. 58, E79–E92.
Frain, K. M., Robinson, C., and van Dijl, J. M. (2019a). Transport of folded proteins by the tat system. Protein J. 38, 377–388. doi: 10.1007/s10930-019-09859-y
Frain, K. M., van Dijl, J. M., and Robinson, C. (2019b). The twin-arginine pathway for protein secretion. EcoSal Plus 8:2018. doi: 10.1128/ecosalplus.ESP-0040-2018
Gardner, S. P., and Olson, J. W. (2018). Interaction of copper toxicity and oxidative stress in Campylobacter jejuni. J. Bacteriol. 200, e00208–e00218. doi: 10.1128/JB.00208-18
Guerin, M. T., Sir, C., Sargeant, J. M., Waddell, L., O’Connor, A. M., Wills, R. W., et al. (2010). The change in prevalence of campylobacter on chicken carcasses during processing: a systematic review. Poult. Sci. 89, 1070–1084. doi: 10.3382/ps.2009-00213
Guo, Q., Wei, Y., Xia, B., Jin, Y., Liu, C., Pan, X., et al. (2016). Identification of a small molecule that simultaneously suppresses virulence and antibiotic resistance of Pseudomonas aeruginosa. Sci. Rep. 6:srep19141. doi: 10.1038/srep19141
Hakeem, M. J., and Lu, X. (2021). Survival and control of campylobacter in poultry production environment. Front. Cell. Infect. Microbiol. 10:5049. doi: 10.3389/fcimb.2020.615049
Hall, S. J., Hitchcock, A., Butler, C. S., and Kelly, D. J. (2008). A multicopper oxidase (Cj1516) and a CopA homologue (Cj1161) are major components of the copper homeostasis system of Campylobacter jejuni. J. Bacteriol. 190, 8075–8085. doi: 10.1128/JB.00821-08
Helmy, Y. A., Deblais, L., Kassem, I. I., Kathayat, D., and Rajashekara, G. (2018). Novel small molecule modulators of quorum sensing in avian pathogenic Escherichia coli (APEC). Virulence 9, 1640–1657. doi: 10.1080/21505594.2018.1528844
Helmy, Y. A., Kathayat, D., Ghanem, M., Jung, K., Closs, G., Deblais, L., et al. (2020). Identification and characterization of novel small molecule inhibitors to control Mycoplasma gallisepticum infection in chickens. Vet. Microbiol. 247:108799. doi: 10.1016/j.vetmic.2020.108799
Hermans, D., Van Deun, K., Martel, A., Van Immerseel, F., Messens, W., Heyndrickx, M., et al. (2011). Colonization factors of Campylobacter jejuni in the chicken gut. Vet. Res. 42:82. doi: 10.1186/1297-9716-42-82
Hitchcock, A., Hall, S. J., Myers, J. D., Mulholland, F., Jones, M. A., and Kelly, D. J. (2010). Roles of the twin-arginine translocase and associated chaperones in the biogenesis of the electron transport chains of the human pathogen Campylobacter jejuni. Microbiol. Read. Engl. 156, 2994–3010. doi: 10.1099/mic.0.042788-0
Hong-Geller, E., and Micheva-Viteva, S. (2013). Small Molecule Screens to Identify Inhibitors of Infectious Disease.
Hue, O., Le Bouquin, S., Laisney, M.-J., Allain, V., Lalande, F., Petetin, I., et al. (2010). Prevalence of and risk factors for campylobacter spp. contamination of broiler chicken carcasses at the slaughterhouse. Food Microbiol. 27, 992–999. doi: 10.1016/j.fm.2010.06.004
Igwaran, A., and Okoh, A. I. (2019). Human campylobacteriosis: A public health concern of global importance. Heliyon 5:e02814. doi: 10.1016/j.heliyon.2019.e02814
Johnson, T. W., Dress, K. R., and Edwards, M. (2009). Using the Golden triangle to optimize clearance and oral absorption. Bioorg. Med. Chem. Lett. 19, 5560–5564. doi: 10.1016/j.bmcl.2009.08.045
Johnson, T. J., Shank, J. M., and Johnson, J. G. (2017). Current and potential treatments for reducing campylobacter colonization in animal hosts and disease in humans. Front. Microbiol. 8:487. doi: 10.3389/fmicb.2017.00487
Jorgensen, F., Ellis-Iversen, J., Rushton, S., Bull, S. A., Harris, S. A., Bryan, S. J., et al. (2011). Influence of season and geography on Campylobacter jejuni and C. coli subtypes in housed broiler flocks reared in Great Britain. Appl. Environ. Microbiol. 77, 3741–3748. doi: 10.1128/AEM.02444-10
Kaakoush, N. O., Castaño-Rodríguez, N., Mitchell, H. M., and Man, S. M. (2015). Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 28, 687–720. doi: 10.1128/CMR.00006-15
Kassem, I. I., Candelero-Rueda, R. A., Esseili, K. A., and Rajashekara, G. (2017). Formate simultaneously reduces oxidase activity and enhances respiration in Campylobacter jejuni. Sci. Rep. 7:40117. doi: 10.1038/srep40117
Kassem, I. I., and Rajashekara, G. (2014). Formate dehydrogenase localization and activity are dependent on an intact twin arginine translocation system (tat) in Campylobacter jejuni 81-176. Foodborne Pathog. Dis. 11, 917–919. doi: 10.1089/fpd.2014.1797
Kassem, I. I., Zhang, Q., and Rajashekara, G. (2011). The twin-arginine translocation system: contributions to the pathobiology of Campylobacter jejuni. Future Microbiol. 6, 1315–1327. doi: 10.2217/fmb.11.107
Kathayat, D., Helmy, Y. A., Deblais, L., and Rajashekara, G. (2018). Novel small molecules affecting cell membrane as potential therapeutics for avian pathogenic Escherichia coli. Sci. Rep. 8:15329. doi: 10.1038/s41598-018-33587-5
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kittl, S., Korczak, B. M., Niederer, L., Baumgartner, A., Buettner, S., Overesch, G., et al. (2013). Comparison of genotypes and antibiotic resistances of Campylobacter jejuni and Campylobacter coli on chicken retail meat and at slaughter. Appl. Environ. Microbiol. 79, 3875–3878. doi: 10.1128/AEM.00493-13
Koutsoumanis, K., Allende, A., Alvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., Davies, R., et al. (2020). Update and review of control options for campylobacter in broilers at primary production. EFSA J. 18:e06090. doi: 10.2903/j.efsa.2020.6090
Kuhn, K. G., Nygård, K. M., Guzman-Herrador, B., Sunde, L. S., Rimhanen-Finne, R., Trönnberg, L., et al. (2020). Campylobacter infections expected to increase due to climate change in northern Europe. Sci. Rep. 10:13874. doi: 10.1038/s41598-020-70593-y
Kumar, A. (2015). Understanding the gut transcriptome responses to lactobacillus probiotics and investigating the impact of nutrition and rotavirus infection on the infant gut microbiome. The Ohio State University. Available at: https://etd.ohiolink.edu/pg_10?0::NO:10:P10_ETD_SUBID:109268 (Accessed December 26, 2017).
Kumar, A., Drozd, M., Pina-Mimbela, R., Xu, X., Helmy, Y. A., Antwi, J., et al. (2016). Novel anti-campylobacter compounds identified using high throughput screening of a pre-selected enriched small molecules library. Front. Microbiol. 7:405. doi: 10.3389/fmicb.2016.00405
Kumar, A., Vlasova, A. N., Deblais, L., Huang, H.-C., Wijeratne, A., Kandasamy, S., et al. (2018). Impact of nutrition and rotavirus infection on the infant gut microbiota in a humanized pig model. BMC Gastroenterol. 18:93. doi: 10.1186/s12876-018-0810-2
Lai, C.-H., Lai, C.-K., Lin, Y.-J., Hung, C.-L., Chu, C.-H., Feng, C.-L., et al. (2013). Characterization of putative cholesterol recognition/interaction amino acid consensus-like motif of Campylobacter jejuni Cytolethal distending toxin C. PLoS One 8:e66202. doi: 10.1371/journal.pone.0066202
Lai, C.-K., Su, J.-C., Lin, Y.-H., Chang, C.-S., Feng, C.-L., Lin, H.-J., et al. (2015). Involvement of cholesterol in Campylobacter jejuni cytolethal distending toxin-induced pathogenesis. Future Microbiol. 10, 489–501. doi: 10.2217/fmb.14.119
Lee, M. D., and Newell, D. G. (2006). Campylobacter in poultry: filling an ecological niche. Avian Dis. 50, 1–9. doi: 10.1637/7474-111605R.1
Lee, P. A., Tullman-Ercek, D., and Georgiou, G. (2006). The bacterial twin-arginine translocation pathway. Ann. Rev. Microbiol. 60, 373–395. doi: 10.1146/annurev.micro.60.080805.142212
Lin, J. (2009). Novel approaches for campylobacter control in poultry. Foodborne Pathog. Dis. 6, 755–765. doi: 10.1089/fpd.2008.0247
Lin, C.-D., Lai, C.-K., Lin, Y.-H., Hsieh, J.-T., Sing, Y.-T., Chang, Y.-C., et al. (2011). Cholesterol depletion reduces entry of Campylobacter jejuni Cytolethal distending toxin and attenuates intoxication of host cells. Infect. Immun. 79, 3563–3575. doi: 10.1128/IAI.05175-11
Lin, L., and Zhang, J. (2017). Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 18:2. doi: 10.1186/s12865-016-0187-3
Lipinski, C. A. (2004). Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 1, 337–341. doi: 10.1016/j.ddtec.2004.11.007
Lipinski, C. A., Lombardo, F., Dominy, B. W., and Feeney, P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26. doi: 10.1016/S0169-409X(00)00129-0
Logue, C. M., Sherwood, J. S., Elijah, L. M., Olah, P. A., and Dockter, M. R. (2003). The incidence of campylobacter spp. on processed Turkey from processing plants in the midwestern United States*. J. Appl. Microbiol. 95, 234–241. doi: 10.1046/j.1365-2672.2003.01969.x
Luangtongkum, T., Morishita, T. Y., Ison, A. J., Huang, S., McDermott, P. F., and Zhang, Q. (2006). Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of campylobacter spp. in poultry. Appl. Environ. Microbiol. 72, 3600–3607. doi: 10.1128/AEM.72.5.3600-3607.2006
Mandal, S., Treuren, W. V., White, R. A., Eggesbø, M., Knight, R., and Peddada, S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. doi: 10.3402/mehd.v26.27663
Massai, F., Saleeb, M., Doruk, T., Elofsson, M., and Forsberg, Å. (2019). Development, optimization, and validation of a high throughput screening assay for identification of tat and type II secretion inhibitors of Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 9:250. doi: 10.3389/fcimb.2019.00250
Merchant-Patel, S., Blackall, P. J., Templeton, J., Price, E. P., Miflin, J. K., Huygens, F., et al. (2008). Characterisation of chicken Campylobacter jejuni isolates using resolution optimised single nucleotide polymorphisms and binary gene markers. Int. J. Food Microbiol. 128, 304–308. doi: 10.1016/j.ijfoodmicro.2008.09.002
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi: 10.1002/jcc.21256
Newell, D. G., and Fearnley, C. (2003). Sources of campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69, 4343–4351. doi: 10.1128/AEM.69.8.4343-4351.2003
Panahandeh, S., Maurer, C., Moser, M., DeLisa, M. P., and Müller, M. (2008). Following the path of a twin-arginine precursor along the TatABC translocase of Escherichia coli. J. Biol. Chem. 283, 33267–33275. doi: 10.1074/jbc.M804225200
Patel, R., Smith, S. M., and Robinson, C. (2014). Protein transport by the bacterial tat pathway. Biochim. Biophys. Acta 1843, 1620–1628. doi: 10.1016/j.bbamcr.2014.02.013
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Quintel, B. K., Prongay, K., Lewis, A. D., Raué, H.-P., Hendrickson, S., Rhoades, N. S., et al. (2020). Vaccine-mediated protection against campylobacter-associated enteric disease. Sci. Adv. 6:eaba4511. doi: 10.1126/sciadv.aba4511
Rajashekara, G., Drozd, M., Gangaiah, D., Jeon, B., Liu, Z., and Zhang, Q. (2009). Functional characterization of the twin-arginine translocation system in Campylobacter jejuni. Foodborne Pathog. Dis. 6, 935–945. doi: 10.1089/fpd.2009.0298
Ramasamy, S., Abrol, R., Suloway, C. J. M., and Clemons, W. M. (2013). The glove-like structure of the conserved membrane protein TatC provides insight into signal sequence recognition in twin-arginine translocation. Structure 21, 777–788. doi: 10.1016/j.str.2013.03.004
Richards, P. J., Connerton, P. L., and Connerton, I. F. (2019). Phage biocontrol of Campylobacter jejuni in chickens does not produce collateral effects on the gut microbiota. Front. Microbiol. 10:476. doi: 10.3389/fmicb.2019.00476
Rollauer, S. E., Tarry, M. J., Graham, J. E., Jääskeläinen, M., Jäger, F., Johnson, S., et al. (2012). Structure of the TatC core of the twin-arginine protein transport system. Nature 492, 210–214. doi: 10.1038/nature11683
Schuffenhauer, A., Brown, N., Selzer, P., Ertl, P., and Jacoby, E. (2006). Relationships between molecular complexity, biological activity, and structural diversity. J. Chem. Inf. Model. 46, 525–535. doi: 10.1021/ci0503558
Selin, C., Stietz, M. S., Blanchard, J. E., Gehrke, S. S., Bernard, S., Hall, D. G., et al. (2015). A pipeline for screening small molecules with growth inhibitory activity against Burkholderia cenocepacia. PLoS One 10:e0128587. doi: 10.1371/journal.pone.0128587
Sibanda, N., McKenna, A., Richmond, A., Ricke, S. C., Callaway, T., Stratakos, A. C., et al. (2018). A review of the effect of management practices on campylobacter prevalence in poultry farms. Front. Microbiol. 9:2002. doi: 10.3389/fmicb.2018.02002
Skarp, C. P. A., Hänninen, M.-L., and Rautelin, H. I. K. (2016). Campylobacteriosis: the role of poultry meat. Clin. Microbiol. Infect. 22, 103–109. doi: 10.1016/j.cmi.2015.11.019
Srivastava, V., Deblais, L., Huang, H.-C., Miyazaki, A., Kandasamy, S., Langel, S. N., et al. (2020). Reduced rotavirus vaccine efficacy in protein malnourished human-faecal-microbiota-transplanted gnotobiotic pig model is in part attributed to the gut microbiota. Benef. Microbes 11, 733–751. doi: 10.3920/BM2019.0139
Stolle, P., Hou, B., and Brüser, T. (2016). The tat substrate CueO is transported in an incomplete folding state*. J. Biol. Chem. 291, 13520–13528. doi: 10.1074/jbc.M116.729103
Subbiah, M., Mitchell, S. M., and Call, D. R. (2016). Not all antibiotic use practices in food-animal agriculture afford the same risk. J. Environ. Qual. 45, 618–629. doi: 10.2134/jeq2015.06.0297
Tolopko, A. N., Sullivan, J. P., Erickson, S. D., Wrobel, D., Chiang, S. L., Rudnicki, K., et al. (2010). Screensaver: an open source lab information management system (LIMS) for high throughput screening facilities. BMC Bioinformatics 11:260. doi: 10.1186/1471-2105-11-260
USDA ERS–Cost Estimates of Foodborne Illnesses. (2017). Available at: https://www.ers.usda.gov/data-products/cost-estimates-of-foodborne-illnesses.aspx#.VDW27r4mUfy (Accessed September 28, 2017).
Vandeputte, J., Martel, A., Canessa, S., Van Rysselberghe, N., De Zutter, L., Heyndrickx, M., et al. (2019). Reducing Campylobacter jejuni colonization in broiler chickens by in-feed supplementation with hyperimmune egg yolk antibodies. Sci. Rep. 9:8931. doi: 10.1038/s41598-019-45380-z
Vasil, M. L., Tomaras, A. P., and Pritchard, A. E. (2012). Identification and evaluation of twin-arginine translocase inhibitors. Antimicrob. Agents Chemother. 56, 6223–6234. doi: 10.1128/AAC.01575-12
Wagenaar, J. A., French, N. P., and Havelaar, A. H. (2013). Preventing campylobacter at the source: why is it so difficult? Clin. Infect. Dis. 57, 1600–1606. doi: 10.1093/cid/cit555
Wei, Z.-Y., Rao, J.-H., Tang, M.-T., Zhao, G.-A., Li, Q.-C., Wu, L.-M., et al. (2020). Characterization of dynamic age-dependent changes and driver microbes in primate gut microbiota during host’s development and healthy aging via captive crab-eating macaque model. bioRxiv 2020:015305. doi: 10.1101/2020.03.30.015305
Yang, Y., Feye, K. M., Shi, Z., Pavlidis, H. O., Kogut, M., Ashworth, A, J., et al. (2019). A historical review on antibiotic resistance of foodborne campylobacter. Front. Microbiol. 10:1509. doi: 10.3389/fmicb.2019.01509
Young, K. T., Davis, L. M., and Dirita, V. J. (2007). Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5, 665–679. doi: 10.1038/nrmicro1718
Keywords: Campylobacter jejuni , poultry production system, twin arginine translocase, small molecule inhibitor, microbiome
Citation: Deblais L, Drozd M, Kumar A, Antwi J, Fuchs J, Khupse R, Helmy YA and Rajashekara G (2024) Identification of novel small molecule inhibitors of twin arginine translocation (Tat) pathway and their effect on the control of Campylobacter jejuni in chickens. Front. Microbiol. 15:1342573. doi: 10.3389/fmicb.2024.1342573
Edited by:
Stuart A. Thompson, Augusta University, United StatesReviewed by:
Fang Liu, University of New South Wales, AustraliaMarja-Liisa Hänninen, University of Helsinki, Finland
Copyright © 2024 Deblais, Drozd, Kumar, Antwi, Fuchs, Khupse, Helmy and Rajashekara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gireesh Rajashekara, rajashekara.2@osu.edu
†These authors have contributed equally to this work
 Loïc Deblais
Loïc Deblais Mary Drozd
Mary Drozd Anand Kumar
Anand Kumar Janet Antwi4
Janet Antwi4 James Fuchs
James Fuchs Rahul Khupse
Rahul Khupse Gireesh Rajashekara
Gireesh Rajashekara