- 1School of Animal Sciences, College of Veterinary Medicine, Virginia Polytechnic Institute and State University, Blacksburg, VA, United States
- 2Department of Population Health, University of Georgia, Athens, GA, United States
- 3Department of Biomedical Sciences and Pathobiology, College of Veterinary Medicine, Virginia Polytechnic Institute and State University, Blacksburg, VA, United States
- 4Department of Microbiology and Molecular Genetics, University of California, Irvine, Irvine, CA, United States
- 5Department of Microbiology, University of Georgia, Athens, GA, United States
Baby chicks administered a fecal transplant from adult chickens are resistant to Salmonella colonization by competitive exclusion. A two-pronged approach was used to investigate the mechanism of this process. First, Salmonella response to an exclusive (Salmonella competitive exclusion product, Aviguard®) or permissive microbial community (chicken cecal contents from colonized birds containing 7.85 Log10Salmonella genomes/gram) was assessed ex vivo using a S. typhimurium reporter strain with fluorescent YFP and CFP gene fusions to rrn and hilA operon, respectively. Second, cecal transcriptome analysis was used to assess the cecal communities’ response to Salmonella in chickens with low (≤5.85 Log10 genomes/g) or high (≥6.00 Log10 genomes/g) Salmonella colonization. The ex vivo experiment revealed a reduction in Salmonella growth and hilA expression following co-culture with the exclusive community. The exclusive community also repressed Salmonella’s SPI-1 virulence genes and LPS modification, while the anti-virulence/inflammatory gene avrA was upregulated. Salmonella transcriptome analysis revealed significant metabolic disparities in Salmonella grown with the two different communities. Propanediol utilization and vitamin B12 synthesis were central to Salmonella metabolism co-cultured with either community, and mutations in propanediol and vitamin B12 metabolism altered Salmonella growth in the exclusive community. There were significant differences in the cecal community’s stress response to Salmonella colonization. Cecal community transcripts indicated that antimicrobials were central to the type of stress response detected in the low Salmonella abundance community, suggesting antagonism involved in Salmonella exclusion. This study indicates complex community interactions that modulate Salmonella metabolism and pathogenic behavior and reduce growth through antagonism may be key to exclusion.
Introduction
Day-of-hatch chicks are susceptible to clinical disease in response to Salmonella exposure (Gast and Beard, 1989); however, chicks challenged at 2 days of age are easily colonized but do not exhibit clinical disease symptoms (Cheng et al., 2015). Young chicks’ intestinal microbiota contains low community diversity, with Enterobacteriaceae as a dominant bacterial group, indicating that the community may have a large niche for Gram-negative enterics (Lu et al., 2003; Ballou et al., 2016; Zhou et al., 2021). Salmonella colonization and Enterobacteriaceae abundance diminish with age as community diversity increases (Pedroso et al., 2021). Firmicutes and Actinomycetota become the abundant Gram-positive bacteria, while the Bacteroidetes, are the dominant Gram-negatives in the chicken intestinal microbiome as it matures (Lu et al., 2003; Xiao et al., 2017; Bhogoju et al., 2018; Khan et al., 2020). In the mature chicken, small intestinal and cecal intestinal compartments contain distinctly different communities (Lu et al., 2003; Choi et al., 2014; Xiao et al., 2017; Khan et al., 2020). This community segregation, however, does not occur until birds are approximately 3 weeks old (Lu et al., 2003), and at this time the abundance of intestinal Salmonella begins to decline (Cheng et al., 2015).
Day-of-hatch chicks exposed to a mature intestinal microbiota rapidly develop high community diversity (Pedroso et al., 2016) and are resistant to Salmonella infection and colonization (Nurmi and Rantala, 1973). One of the earliest examples of fecal transplants was done in chickens, where cecal bacteria from mature hens were shown to block Salmonella colonization in chicks (Nurmi and Rantala, 1973). This phenomenon was later termed competitive exclusion, even though it was not known whether competition (Fujikawa, 2016) was at the heart of the exclusion mechanism. Fecal transplants with the intestinal microbiota from adult chickens have also been shown to dramatically reduce Salmonella prevalence in broiler chickens in the field (Wierup et al., 1988; Hirn et al., 1992). Nurmi suggested that competitive exclusion may be attributed to: (1) competition for limiting nutrients; (2) competition for attachment sites on the mucosa; or (3) the production of antibacterial substances, including volatile fatty acids (VFAs) or bacteriocins (Nurmi et al., 1992). Another possibility is that competitive exclusion (4) modulates the virulence of the enteropathogens, reducing their ability to modify the intestinal environment or create a tissue reservoir to enable persistence (Watkins and Miller, 1983).
A great deal of culture-based and molecular studies have focused on identifying potential intestinal community members involved in pathogen exclusion. Most molecular studies have used a 16S census approach to characterize the intestinal microbiota (Stenkamp-Strahm et al., 2018; Pedroso et al., 2021; Valeris-Chacin et al., 2022; Falardeau et al., 2023). Candidate taxons have been identified that positively or negatively correlate with Salmonella abundance (Azcarate-Peril et al., 2018; Ding et al., 2021; Pedroso et al., 2021; Pottenger et al., 2023). However, the identified intestinal species have not been repeatedly encountered across studies, suggesting a multifactorial role (Pedroso et al., 2021). The key to pathogen exclusion likely involves a diverse microbial community exhibiting complex metabolic and inhibitory effects (Lone et al., 2013; Stanley et al., 2014; Zhang et al., 2015; Chopyk et al., 2016; Pedroso et al., 2021).
With the success of undefined, diverse intestinal communities in competitive exclusion, efforts led to the development of undefined intestinal communities free of any avian pathogens (Schneitz et al., 1991). Aviguard® is a competitive exclusion product derived from the cecal contents of adult hens that has been shown to be effective at reducing Salmonella and other enteropathogens in the intestine of chickens (Hofacre et al., 1998, 2000; Weschka et al., 2021). This competitive exclusion product has also been shown to promote intestinal development in young chicks (Lee et al., 2023a). Undefined intestinal communities used as competitive exclusion products have been limited in their distribution or application by regulatory agencies like the U.S. Food and Drug Administration (Lee et al., 2023b). This has turned emphasis toward the development of single or multiple microbial species formulations capable of excluding an animal pathogen (Khan et al., 2020). These microbes have either been isolated from the intestine (Pascual et al., 1999; Kubasova et al., 2019), chosen based on similarities with intestinal species (Nair et al., 2021), or based on microbial properties of competition or antagonism so as to be detrimental to the pathogen in question (Latorre et al., 2016).
While the chicken small intestine is rich in nutrients, Salmonella and other Proteobacteria are a minor population within the bacterial community in this environment (Lu et al., 2003; Wang et al., 2022). In chickens, Salmonella’s niche appears to be the cecum (Snoeyenbos et al., 1982). While this compartment, in comparison to the small intestine, is scarce in free sugars (Józefiak et al., 2004), Salmonella abundance can reach 107 cells per gram of cecal content (Cheng et al., 2015), despite the fact that the cecum expels its content daily. In general, Proteobacteria are not capable of degrading the fibers and mucin polysaccharides that are plentiful in the cecal compartment. They are therefore largely dependent on Clostridia, which are primary degraders capable of breaking down complex polysaccharides into free mono- and disaccharides that Salmonella and other scavengers can then metabolize. This scavenger activity would be able to occur through cooperation, in that members of the microbiota could provide nutrition for other members. However, the pathogen is in competition with other intestinal species for even these resources. One possible adaptation for this competition would be for Salmonella to metabolize fermentation end products produced by the primary degraders through respiration (Shelton et al., 2022). Their metabolism of these waste products would also help maintain an environment favorable to continued metabolism and growth for the primary degraders, illustrating another example of cooperation (Benoit et al., 2020). In ecological terms, intestinal member species can interact with Salmonella favorably or unfavorably through cooperation, competition, or antagonism (Hammarlund et al., 2021; Rogers et al., 2021). Some intestinal member species can only compete with Salmonella for the same limited resources by producing metabolites that suppress growth or kill their competitor through antagonism (Sibinelli-Sousa et al., 2022). Competitive exclusion therefore works by failing to support pathogen growth either through competition or antagonism, which has been the guiding principle of many groups in developing probiotics (Zhao et al., 2006; Maltby et al., 2013). Competition may only be effective under conditions where primary degraders and other member species stop feeding Salmonella nutrients that only it can metabolize, forcing it to compete for other resources equally metabolized by other intestinal community members.
Because the mechanism of competitive exclusion is currently unknown and is likely multifactorial, in this study, an ex vivo transcriptomic approach was used to reveal Salmonella physiology and behavior in response to permissive or exclusive communities. The ex vivo approach would allow direct analysis of Salmonella transcriptional response even if their growth was inhibited. In addition, chickens were experimentally infected with Salmonella, and its abundance was quantified in order to allow in vivo analysis of the cecal microbial transcriptomes to identify community metabolism that favors or excludes Salmonella. These experiments revealed significant metabolic differences in Salmonella’s response to the intestinal community and vice versa; however, failure to metabolize liberated sugars or microbial metabolites did not fully account for the mechanism of exclusion by the Salmonella exclusive community. The chicken intestinal community was found to reduce pathogen behavior (attenuation) and metabolism (cooperation/competition) and produce antimicrobials that limit Salmonella growth (antagonism), suggesting that the mechanism of competitive exclusion was multifactorial.
Materials and methods
Experimental approach
The mechanism underlying competitive exclusion involves one of the principles of population biology: cooperation, competition, antagonism, or attenuation. Table 1 describes the research approach to identifying the role of each in the competitive exclusion of Salmonella. One methodological challenge, addressed by this research approach, was detecting Salmonella growth rate and transcriptional signal in low abundance in the cecal microbiota, whose abundance may exceed Salmonella’s by 1,000-fold (Cheng et al., 2015). Therefore, a non-culture-based approach was used to determine cecal community metabolism correlating with Salmonella abundance in vivo in chickens (Figure 1A) and Salmonella gene expression and growth ex vivo, in response to communities that “permit” or “exclude” Salmonella (Figure 1B). The permissive community used in the ex vivo experiments was acquired from the in vivo chicken experiment and consisted of cecal contents from 35-day-old birds with high Salmonella abundance (7.86 Log10 Salmonella genomes/g cecal content) (Pedroso et al., 2021). The exclusive community used in the ex vivo experiments was the commercially competitive exclusion product, Aviguard® (Lallemand Animal Nutrition; Montreal, Canada), which reduces Salmonella colonization in chickens by at least 5.0 Log10 CFU (Lee et al., 2023b).
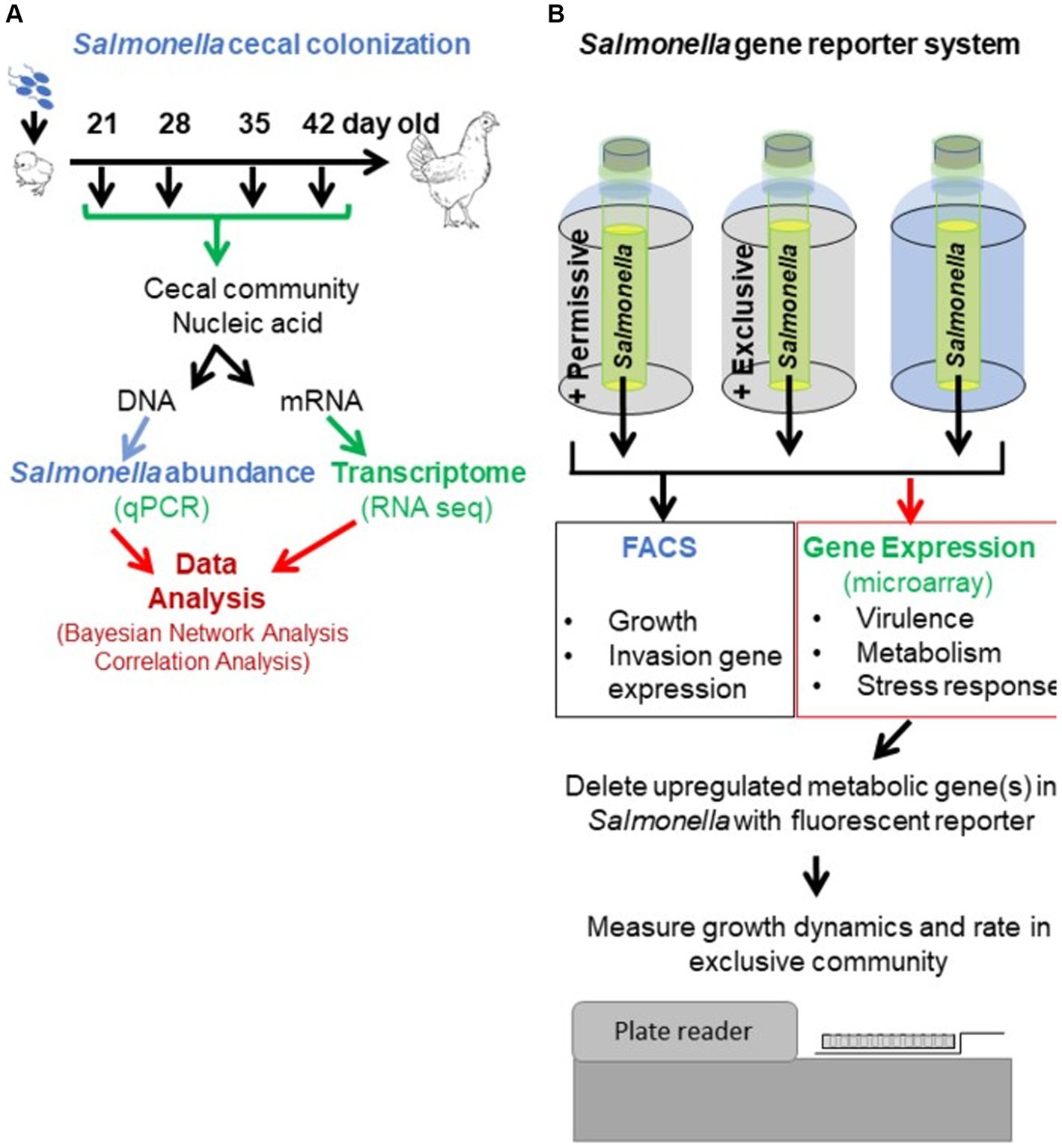
Figure 1. Experimental approach to revealing the mechanism of action of competitive exclusion. A two-prong approach involved determining cecal community metabolism relative to Salmonella abundance in vivo (A) and Salmonella expression in response to a permissive or exclusive community ex vivo (B). In the in vivo study, 2-day-old, specific pathogen-free white leghorn chickens were orally administered Salmonella Typhimurium SL1344 (1 × 105 CFU). At different ages, chickens were sacrificed, the ceca were aseptically collected from each bird, and nucleic acids were extracted. Salmonella invA qPCR was used to determine Salmonella abundance in the cecal community samples. The cecal community RNA was sequenced and annotated using the MG-RAST pipeline. The ex vivo study focused on Salmonella response when grown microaerophilically in a simulated cecal medium, Ex Vivo Cecal Contents (EVCC), containing a permissive or exclusive community. A fluorescent S. typhimurium SL1344 rrn promoter::yfp, hilA operon::cfp reporter strain was placed within dialysis tubing, physically separate from the permissive or exclusive community, enabling the collection of Salmonella cells for analysis. Jellyfish green fluorescent protein variants YFP and CFP served as reporters for Salmonella growth and SPI-1 invasion gene reporter, respectively. Fluorescence was measured using fluorescence-activated cell sorting. The cecal community from a 35-day-old chicken with high Salmonella abundance served as the permissive community, while the exclusive community consisted of the cecal community comprising the competitive exclusion product Aviguard®. Salmonella grown in EVCC alone served as a control. Select Salmonella metabolic genes, upregulated in either or both communities, were deleted in a S. typhimurium SL1344 fluorescent reporter strain, and mutants were compared to the wild-type reporter strain for growth in the exclusive community.
The ex vivo system was developed to monitor Salmonella growth, virulence (SPI-1 expression), and gene expression (microarray) in response to the permissive and exclusive communities. The jellyfish green fluorescent protein variants, yellow fluorescent protein (yfp) and cyan fluorescent protein (cfp), were fused to rrn growth-dependent promoter and hilA operon (SPI-1 cell invasion locus), respectively, in Salmonella. Fluorescence associated with the YFP and CFP reporters was used to monitor Salmonella growth and SPI-1 virulence gene expression in co-culture with cecal communities ex vivo. The Salmonella reporter strain was grown in dialysis tubing in a simulated cecal medium, Ex Vivo Cecal Contents (EVCC), submerged in permissive or exclusive communities to enable collection of Salmonella cells for study even if there was poor growth. Initially, the fluorescent reporters were used to empirically determine the earliest time point at which the exclusive community had the most significant impact on Salmonella growth or virulence expression relative to the permissive community. Six-hour co-culture of the reporter strain with the communities was determined based on YFP brightness measured by flow cytometry. Cells were then harvested for gene expression (microarray) comparisons, and genes within metabolic pathways that were differentially expressed in permissive vs. exclusive communities were deleted in Salmonella using pGLOW, a fluorescent reporter visible in anaerobic conditions. Salmonella mutants’ growth dynamics when co-cultured with the exclusive community were monitored continuously over 48 h using a fluorescence plate reader.
Table 1 also describes expected outcomes based on the roles of cooperation, competition, antagonism, or attenuation in the mechanism of Salmonella competitive exclusion with respect to the in vivo and ex vivo community transcriptomics and Salmonella response. The detailed methodology for each research approach is described in detail below (in vivo: “The cecal communities’ transcriptome relative to Salmonella abundance”; ex vivo: “Assessing Salmonella growth rate and SPI-1 T3SS expression in an ex vivo system with permissive or exclusive communities,” “Microarray analysis of S. Typhimurium YC 1104 reporter strain growth in permissive or exclusive microbial communities,” and “Contribution of metabolic gene(s) to Salmonella Typhimurium growth in an exclusive community”).
Construction of Salmonella reporter strains and mutants
λ Red (Datsenko and Wanner, 2000) was used to construct rrn and iag (SPI-1) reporters, tagged with jellyfish green fluorescent protein variants: yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP), respectively (Heim and Tsien, 1996; Miyashiro and Goulian, 2007). The growth-dependent rrn promoter was cloned upstream of a “promoter-less” yfp in tandem with the chloramphenicol resistance marker cat (Supplementary Tables S1, S2). The rrnB P1 promoter was engineered into the forward primer targeting 5′ yfp with its ribosome binding site (RBS) (Supplementary Table S2). The reverse primer was initially designed with a 3′ overlap with yfp and the rrnBT12 transcriptional terminator, previously cloned downstream of yfp in plasmid pMG32 to create pMG32T1T2 (Supplementary Tables S1, S2). These primers, using pMG32T1T2 as template, were used to amplify yfp, now with rrnB P1 promoter and T1T2 transcriptional terminators. The resulting amplicon was cloned into pCR-XL-TOPO and subsequently subcloned into Π-dependent suicide vector pGP704 (Miller and Mekalanos, 1988) to create pCY01 (Supplementary Figure S1). The rrnBT1T1 was replaced with λT0 transcriptional terminator (pCY02), to which cat was introduced 3′ from this terminator by cloning amplicons with engineered restriction enzyme sites for directional cloning to generate the final plasmid pCY03 (Supplementary Tables S1, S2). PCR primers were designed to target a P22 insertion site, an intergenic site between thrW and STM0324, and contained overlap with the rrn promoter and cat sequences. These primers were used to amplify the reporter construct using pCY03 as template (Supplementary Figure S1; Supplementary Table S2). λ Red was used to introduce Salmonella growth reporter into a P22 integration site with the P22att-rrnB P1-yfb-cat amplicon (Datsenko and Wanner, 2000).
The hilA operon reporter using cfp was constructed as follows. Tetracycline resistance gene tetA was first introduced into the intergenic region between iagB and sptP, just 5′ of the transcriptional terminator for the hilA operon, using λ Red (Datsenko and Wanner, 2000), with primers described in Supplementary Table S2 and pKD3 to generate the amplicon used to transform S. typhimurium SL1344. iagB is the last gene in the hilA operon. The tetA was later replaced with a “promoter-less” cfp following transformation with the cfp amplicon generated with iagB, sptB, and cfp primers (Supplementary Table S2) and selection for fusaric acid resistance. Fusaric-resistant colonies were screened for tetracycline sensitivity (Bochner et al., 1980). P22 was used to move rrnB P1-yfp-cat into Salmonella strain with hilA operon-cfp reporter to construct the final S. typhimurium strain YC1104 with yfp growth and cfp, hilA operon reporters (Provence and Curtiss Iii, 1994).
λ Red was also used to create single and double mutations in metabolic pathways in Salmonella (Supplementary Tables S1, S2). P22 generalized transduction was used to introduce λred-generated knockouts with intact antibiotic resistance gene cassettes into S. typhimurium SL1344 or SL1344 mutants before removing the antibiotic resistance marker with pCP20. YFP was replaced with pGLOW as a reporter of Salmonella growth (evocatal GmbH, Monheim am Rhein, Germany). This reporter was chosen because fluorescence is not oxygen-dependent (Drepper et al., 2007), which allows monitoring of Salmonella growth continuously under strictly anaerobic conditions. Electroporation was used to introduce plasmids into Salmonella strains (Dower et al., 1988).
Measuring YFP and CFP expression using fluorescence-activated cell sorting
Flow cytometry was conducted with a CyAn ADP Analyzer (Beckman Coulter; Brea, CA) using the software Summit (ver. 4.3, Beckman Coulter). The 468 nm excitation and FL1 530/30 BP filter were used for YFP detection. The 409 nm excitation and FL6 450/50 BP filter were used for detecting CFP. For each sample, a total of 20,000 events were collected. In some instances, samples were read multiple times to detect variations between sample collections. The flow cytometer reported fluorescence as median intensity values of arbitrary units. The cytometer flow rate was adjusted to keep the average event rate between 1,000 and 3,000 events per second to avoid coincidental detection of bacterial cells. Data were analyzed using FlowJo software (ver. 9.4, TreeStar, Inc.). Particles that did not fall into the expected size and shape of a bacterial cell were not included in the analyses. Positive (YFP, CFP; Escherichia coli with pMG32 or pMG34, respectively) and negative (S. typhimurium SL1344, wild-type strain) controls were included with every run.
Assessing Salmonella growth and SPI-1 T3SS expression in an ex vivo system with permissive or exclusive communities
The ex vivo study entailed the growth of Salmonella reporter strain YC1104 in the presence of a permissive or exclusive community. The S. typhimurium reporter strain has rrn-yfp and hilA operon-cfp chromosomal promoter fusions for monitoring growth and SPI-1 expression, respectively. This approach allowed for growth of Salmonella to cell densities and volumes to later harvest sufficient Salmonella mRNA for microarray analysis. The reporter strain is a derivative of S. typhimurium SL1344, the same wild-type strain used in the in vivo study (see below).
A medium was developed to mimic the luminal cecal nutritional environment, based on published literature, and contained porcine gastric mucin, uric acid, six amino acids (arginine, cysteine, isoleucine, lysine, methionine, and threonine), and phytone peptone. All amino acids, except cysteine, were at concentrations previously reported to be found in the chicken cecum (Babinszky et al., 2006). Uric acid was added at a concentration found in the cecal contents of white leghorn chickens (Beck and Chang, 1980). A redox indicator, resazurin, and a reducing agent, cysteine, were also added at concentrations to enhance anaerobiosis (Demain and Solomon, 1986). This ex vivo chicken cecal (EVCC) medium formulation is described in Supplementary Table S3. Stock solutions were made for resazurin (1,000×), hemin (1,000×), uric acid (100×, pH 8.8–9.0), and amino acid supplement (10×, pH 7). The pH of the basal medium was adjusted to 6.1 ± 0.1 prior to autoclaving. After the addition of uric acid and amino acid supplements, the final pH of the EVCC medium was approximately 6.5.
All cultures, media preparation, and dilutions were performed in an anaerobic glove box. Media and diluent were sparged with gas to make conditions microaerophilic. Autoclaved, 30-ml-capacity serum bottles (Fisher Scientific) with rubber stoppers served as culture vessels. About 20 mL of EVCC medium (Supplementary Table S3) was added to each vessel in an anaerobic cabinet, to which a Salmonella permissive (7.86 Log10 Salmonella genomes/g cecal contents) or exclusive community was added (Supplementary Figure S2). One culture vessel was left uninoculated (EVCC alone) (Supplementary Figure S2). The lyophilized Aviguard® product was rehydrated in sterile saline according to the manufacturer’s recommendation. This product is generally administered to chickens in drinking water and has been shown to reduce Salmonella colonization by at least 5 Log10 in chickens (Lee et al., 2023b). Direct bacterial cell counts were determined microscopically at 1,000× magnification using a Petroff-Hausser counting chamber. The cell density of the starting material was estimated for both communities at 1011 cells/ml. The rehydrated Aviguard viability was determined by a LIVE/DEAD BacLight stain (Molecular Probes; Grand Island, NY). The viability of the commercial lots varied between 30 and 75%. Both communities were diluted 100-fold in sterile saline before adding 2 mL of this cell suspension to EVCC medium to generate a final cell density of 108 cells/ml. Spectrum™ (Fisher Scientific) cellulose ester membrane with 100,000 Daltons exclusion limit and 20 mm width served as the internal culture vessel for the Salmonella reporter. Approximately 80 mm of dialysis tubing was cut, rehydrated in sterile H2O, and clamped at one end with a dialysis clip, pre-treated with 70% ethanol. The clamped end was placed in a serum bottle first, and the open dialysis end was pulled over the mouth of the bottle. The Salmonella reporter strain was prepared as follows. Strain YC1104 was grown anaerobically on MacConkey agar at 39°C for 48 h. A cell suspension was made from growth on the MacConkey agar plate in sterile saline to 0.2 OD (λ 600 nm) (~108 CFU/mL). The cell suspension was diluted 100-fold in sterile saline. Subsequently, 1 mL of the 100-fold dilution was used to inoculate 5 mL of EVCC medium (105 CFU/mL final cell density), which was transferred to the dialysis tubing. A rubber stopper and metal crimp were used to seal the vessel. A sterile syringe needle was used to sparge the vessel with a gas mix of 6% CO2, 6% O2, 85% N2, and 3% H2 or sample the dialysis tubing as depicted (Supplementary Figure S2). The oxygen concentration was chosen based on jellyfish green fluorescent proteins and its variants’ requirements for oxygen to fluoresce (Heim et al., 1994) and the likely minimal oxygen tension necessary for supporting the microaerophile Campylobacter jejuni (Pennie et al., 1984), an abundant (8 Log10 CFU/g) bacterial species in the chicken cecum (Hue et al., 2011). The top was sealed with tape. Culture vessels were placed in an anaerobic jar, flushed with the same gas mix described previously, and incubated at 39°C to simulate the chicken’s internal body temperature. This and subsequent samplings (see below) were all done within an anaerobe glove box. The dialysis tubing’s exclusion limit allowed free exchange of metabolites, peptides, and proteins between Salmonella and the external microbial community. Due to the nature of the cellulose dialysis material and because the cecal anaerobic community may secrete cellulases, the integrity of the membrane during experimentation was determined earlier by incubating tubing containing just dextran blue with a cecal community grown anaerobically in EVCC for 24 h at 39°C. There was no visible leakage of the dextran blue into the external culture, confirming the integrity of the dialysis membrane. Culture vessels were incubated at 39°C. The Salmonella reporter strain was periodically sampled over time (3, 6, 9, 12, and 24 h) using a sterile needle and syringe to sample the dialysis tubing, and fluorescence was monitored by fluorescence-activated cell sorting (FACS) to empirically identify the best time point, for harvesting Salmonella, where growth or SPI-1 expression was significantly reduced in the exclusive community, compared to the permissive community and the uninoculated, EVCC control. 0.2 mL aliquots were added to cryotubes containing 40 μL of 60% sterile glycerol (12% final concentration) and stored at −80°C for later FACS analysis.
Microarray analysis of Salmonella typhimurium YC 1104 strain growth in permissive or exclusive microbial communities
Microarray analysis was used to assess Salmonella’s transcriptome response to microbial communities in the ex vivo system, as previously described, with the following modifications. Salmonella strain YC1104 was grown anaerobically on MacConkey agar at 39°C for 48 h. A cell suspension was made in 5-ml sterile saline from the MacConkey plate, adjusted to a cell density of 0.2 OD (λ 600 nm) (~108 CFU/mL). This cell suspension was used to inoculate 5 mL of EVCC medium (106 CFU/mL) and incubate anaerobically at 39°C for 12 h. Five milliliter of EVCC was inoculated with a 4-fold dilution of the 12 h culture to 107 CFU/mL and transferred to dialysis tubing. The higher cell density per dialysis volume was needed to provide a sufficient amount of Salmonella RNA for microarray analysis. This experiment was done in triplicate, where Salmonella was grown and harvested after 6-h incubation in EVCC alone or in EVCC with the permissive or exclusive community (Supplementary Figure S2). Salmonella grown in culture vessels with EVCC alone served as the comparison control in the microarray analysis. Total RNA was harvested from Salmonella grown in the dialysis tubing, and microarray analysis was performed as previously described (Cheng et al., 2015). Briefly, the samples were treated with 0.1 mL volume of 95% ethanol and 5% acidic phenol (pH 4.3), and total RNA was extracted using MasterPure Complete DNA and RNA purification kits (Epicentra; Madison, WI) and High Pure RNA isolation kit (Roche; Indianapolis, IN). The samples were subsequently treated two times with the Turbo DNA-free kit (Ambion; Austin, TX). Salmonella-specific hilA PCR (Cheng et al., 2015) was used to confirm that RNA was free of contaminating DNA, and the quality and integrity of the RNA were evaluated by gel electrophoresis (Sambrook et al., 1989). Genomic Salmonella DNA served as a labeling and hybridization reference control. Hybridization and fluorescent labeling of RNA were performed as described by Fink et al. (2007). Microarray chips were scanned with a ScanArray 5,000 laser scanner (GSI Lumonics; Watertown, MA). The data were analyzed using QuantArray v.2.01 software, and background intensity was subtracted from spot boundary signal intensities to derive Cy3 and Cy5 median signal intensities. Differential gene expression and statistical significance were determined with the Significance Analysis of Microarrays software package (Stanford University).1 The data were analyzed using paired Student’s t-test, and statistical significance from multiple comparisons was determined by one-way analysis of variance followed by Bonferroni posttest, with statistically significant p-values set at <0.001.
Contribution of metabolic gene(s) to Salmonella Typhimurium growth in an exclusive community
Mutant or wild-type strains, with the pGLOW reporter, were grown in EVCC + Aviguard. Oxyrase (Sigma-Aldrich; St. Louis, MO) was added to the medium (1:20 final dilution) and overlaid with mineral oil to create low oxygen conditions. Salmonella starting cell density was 105 CFU/mL. The lyophilized Aviguard was reconstituted as recommended by the manufacturer (Lallemand Animal Nutrition) in sterile saline with Oxyrase and used to inoculate EVCC cecal medium (1:20 dilution) (7.99 Log10 viable cells/mL). The viability of the rehydrated Aviguard was determined microscopically using LIVE/DEAD BacLight stain (Molecular Probes). Sterile 96-well, black-walled polystyrene microtiter plates with a clear bottom (Fisher Scientific) were used to monitor Salmonella fluorescence with a BioTek Synergy HT 96-well fluorescence microtiter plate reader at 37°C. A filter was added to the BioTek Synergy HT fluorescent plate reader (BioTek; Winooski, VT) to record pGLOW fluorescence based on its excitation frequency (Drepper et al., 2007). Fluorescence was recorded every 30 min. Strains were run in triplicate.
The cecal communities’ transcriptome relative to Salmonella abundance
The in vivo study involved oral administration of Salmonella Typhimurium SL1344 (1 × 105 CFU) to 2-day-old, specific pathogen-free (certified Salmonella-free), white leghorn chickens (Charles River Laboratories; Wilmington, MA; n = 100), dispersed into five HEPA-filtered isolator units with wire mesh floors to reduce re-exposure due to coprophagy. To ensure birds were Salmonella-free, day-of-hatch chicks and their isolator environment were also tested for Salmonella by enrichment (Pedroso et al., 2021). Chicks and their environment were culture-negative for Salmonella. The birds were fed a non-medicated, commercial starter feed ad libitum. The bird density per isolator unit was reflective of commercial standards; culling birds was necessary periodically to maintain this stocking density. No probiotic(s), competitive exclusion product, or cocktail of intestinal bacterial species was administered to chickens in this study. One bird from each of five isolator units (n = 5) was collected at 21, 28, 35, and 42 days of age, euthanized, and the ceca was aseptically obtained. One bird in isolator 5 died after day 35, and therefore, there were only four subjects for day 42. Salmonella invA qPCR was used to enumerate Salmonella present in cecal DNA as previously described (Pedroso et al., 2021). Nucleic acid was extracted from cecal bacteria using the Mo Bio Soil DNA extraction kit, vortexing samples with glass beads at maximum speed for 40 min (Lu et al., 2003). Lysate was treated with 0.5% SDS and proteinase K (0.1 mg/mL) at 37°C for 30 min before phenol–chloroform–isoamyl alcohol (25:24:1) extraction. Sample nucleic acid was portioned equally, one being stored at −80°C for RNA purification (see below) and the other treated with DNAse-free RNase for Salmonella qPCR.
RNAse-free DNAse was added to nucleic acid (Pedroso et al., 2021) for each sample (n = 19) and incubated at 37°C. The quality and quantity of RNA were assessed by agarose gel electrophoresis. PCR, using universal 16S rRNA primers (Supplementary Table S2; Suzuki et al., 2000), assessed and confirmed that the RNA was free of DNA. Nucleic acid preps were repeatedly treated with RNAse-free DNAse until no PCR amplicon was detected to confirm RNA purity. Ribosomal RNA was removed using the MICROBExpress™ Additional rRNA removal was performed until samples were free of the detectable 23S and 16S rRNA bands (2.9 and 1.5 Kb, respectively) on an agarose gel. Finally, the Bacterial mRNA Enrichment Kit (Thermo Fisher Scientific; Waltham, MA) was used to further purify and concentrate cecal community mRNA, as described by the manufacturer’s instructions. mRNA was submitted to the Georgia Genomics and Bioinformatics Core for sequencing using RNA-seq (Illumina; San Diego, CA). The resulting sequence reads were uploaded to the MG-RAST server for annotation (Keegan et al., 2016).
Carbohydrate and volatile fatty acid analysis of the chicken cecum
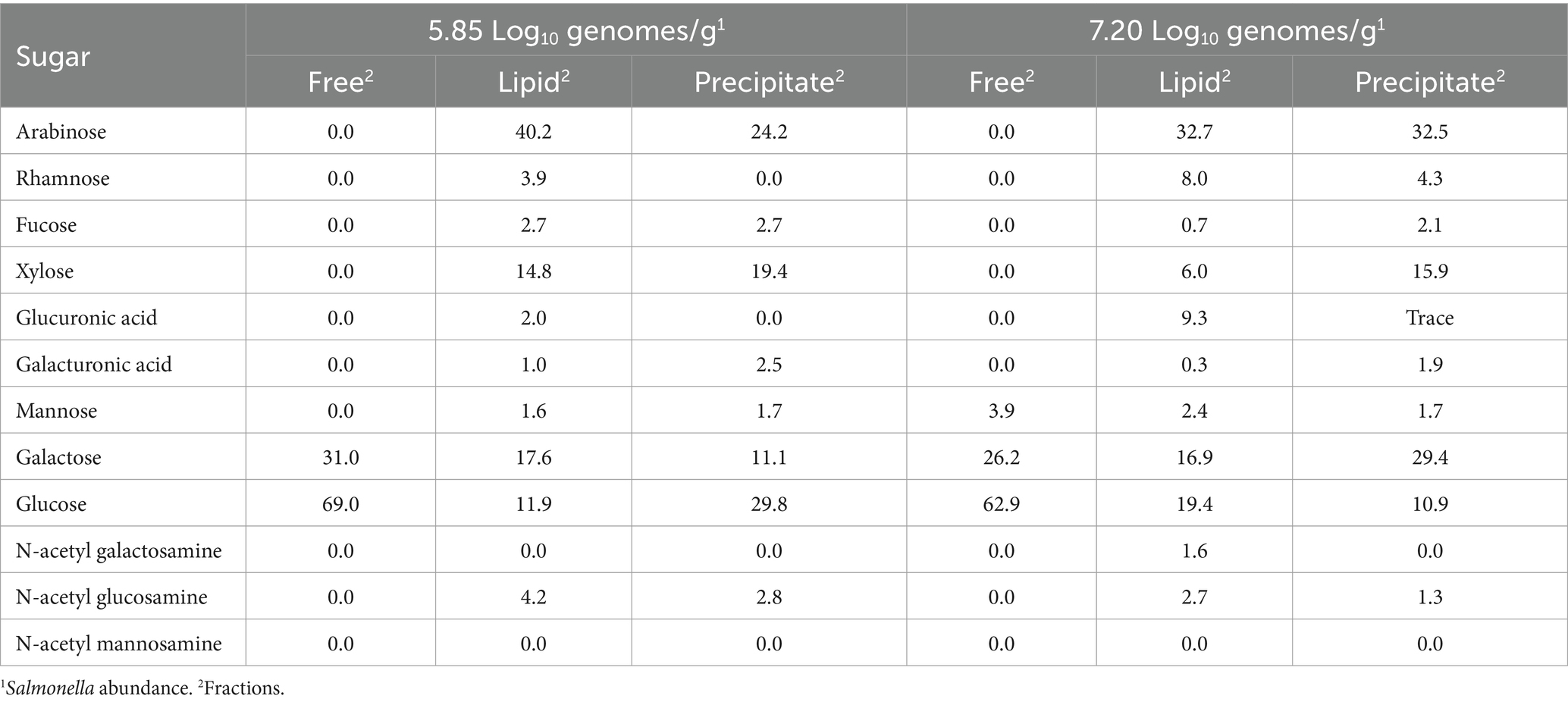
In order to reveal carbohydrates present in the cecal glycome of a permissive community, two samples with high (7.20 Log10 Salmonella genomes/g) vs. low Salmonella abundance (5.85 Log10 Salmonella genomes/g) were submitted to the University of Georgia Complex Carbohydrate Center for glycome analysis. Glycosyl composition analysis was performed by combined gas chromatography–mass spectrometry (GC–MS) of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides produced by acidic methanolysis as previously described (Santander et al., 2013). GC–MS analysis of the resulting TMS methyl glycosides was performed on an Agilent 7890A GC interfaced to a 5975C MSD (Agilent; Santa Clara, CA), using a Supelco Equity-1 fused silica capillary column (30 mm × 0.25 mm, internal diameter).
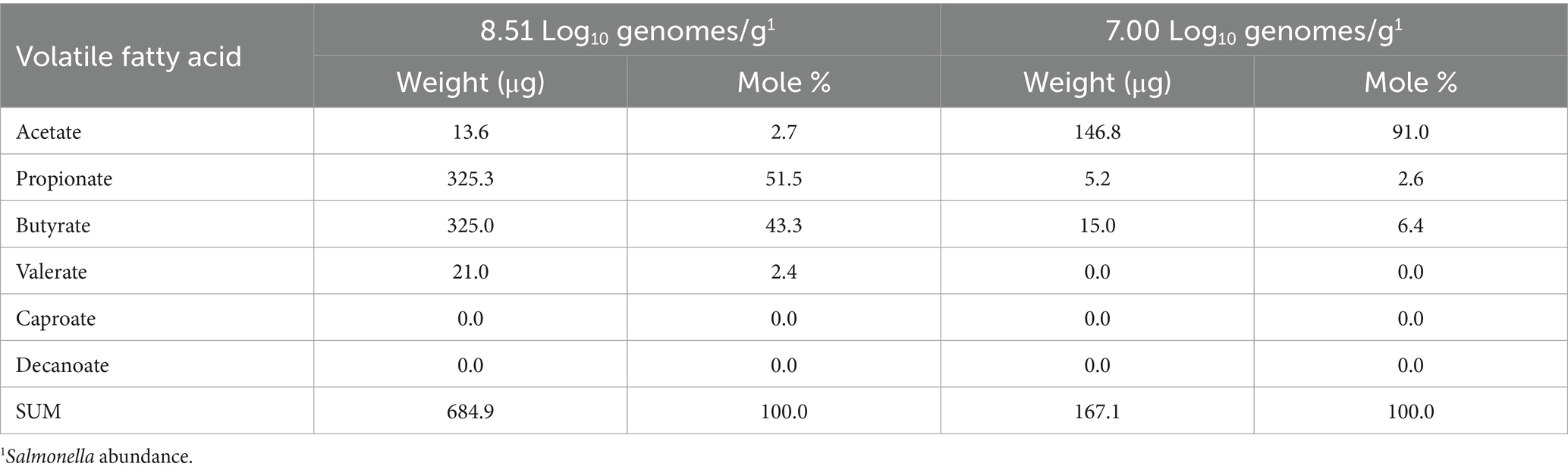
Because some short-chain fatty acids can attenuate Salmonella virulence, inhibit, or promote pathogen growth (Kwon and Ricke, 1999; Zhang S. et al., 2020; Zhang Z.J. et al., 2020), cecal contents were obtained from two birds with high Salmonella abundance (7.00 Log10 Salmonella genomes/g cecal contents, 8.51 Log10 Salmonella genomes/g cecal contents). VFA analysis was performed by combined gas chromatography–mass spectrometry (GC–MS) of the butyl ester derivatives produced from the samples. The cecal contents were lyophilized and subsequently suspended in hexane, to which octadecane was added as an internal standard. Butyl esters were prepared by adding BF3-1-butanol to the sample and heating it at 100°C for 2 h in a sealed tube. The reaction was subsequently quenched with 1–2 mL H2O. The aqueous layer was discarded, and the organic phase was washed once with an equal volume of H2O. The organic layer was then used for GC–MS analysis, performed using HP 6890 GC (Agilent) interfaced to a 5975b MSD (Agilent) using an All Tech EC-1 fused silica capillary column (30 mm × 0.25 mm) (Fisher Scientific). A mixture containing known concentrations of VFAs and octadecane was treated under the same conditions and served as standards for quantification.
Network and statistical analyses of chicken cecal meta-transcriptomes for cecal communities with high or low Salmonella abundance
Chicken cecum transcriptome read counts were normalized as a percentage of total reads for the MG-RAST categories KO metabolism, COG metabolism, virulence, and stress response. Enzymes were mapped and counted in the raw data set for sequence reads through Python version 3.7 (Ekmekci et al., 2016). These counted reads were merged in R through scripting, and then calculated as the percentage of these reads against total reads in Python to create a data set with normalized read counts of all enzymes in all samples. The read count data were edited, transposed, combined with Salmonella abundance, associated with individual samples, and filtered according to abundance from highest to lowest. The mean value of Salmonella abundance was calculated, and the value above the mean (5.85 Log10 Salmonella genomes/g cecal contents) was considered high abundance; a value equal to or below the mean was considered low abundance.
For analysis of the fermentation meta-transcriptome, the data set was first limited to 45 enzymes associated with fermentation in the KO data set, listed in Supplementary Table S5. This data set was expanded to 164 enzyme transcripts to include the enzymes listed in Supplementary Table S6. In the analysis of the microbiome’s stress response, data were pulled from stress response and virulence data sets and categorized into groups based on function ascribed to the enzyme, for example, DnaK and heat shock, and consolidated into the individual categories. Two hundred thirty-four enzymes extracted from these data sets included those associated with the following categories: heat shock; carbon starvation; extra-cytoplasmic/envelop stress response; regulation; translation and protein export; oxidative stress; osmotic shock; acid tolerance; iron metabolism; antimicrobials; miscellaneous; and polyketide synthesis (Supplementary Table S7).
Bayesian network analysis was performed in R with the bnlearn package (Friedman et al., 2013; Scutari and Denis, 2021). Bayesian network analysis is a probabilistic model in which a graph structure represents a qualitative dependency relationship among random variables and a conditional probability expresses a quantitative link between individual variables. This method is comparable to approaches used to infer gene regulatory networks based on microarray or RNA-seq data and represents a directed edge connecting two genes used to determine a biochemical process such as a reaction, transformation, interaction, activation, or inhibition. The arc or arrows represent the probability of connectivity. A graph structure showing dependency relationships between nodes, obtained from a collection of conditional probabilities, defines the model (Ebana and Furukawa, 2019). The conditional probability with the state of the node close to the arrow propagates through the arrow one after the other. The probability of each node is used to generate the graph structure. The effective final network produced automatically from the data indicates the impact and the association link between the data. The strength of probabilistic links, reflected by the arc of a connection, was measured and presented.
Several different network models were used, including the score-based learning algorithm—Hill-Climbing (HC) (Friedman et al., 2013); constraint-based structure learning algorithms—Max–Min Parents and Children (MMPC) (Lagani and Tsamardinos, 2010); hybrid structure learning algorithms—Max–Min Hill-Climbing (MMHC) (Song et al., 2022), Hybrid Parents and Children (H2PC) (Anonymous, 2021), and General 2-Phase Restricted Maximization (RSMAX2) (Yu et al., 2019) to identify a common network structure for the various enzymes or categories described. Among all these implemented algorithms, HC performed best and produced the highest number of connections with various data sets. Therefore, for model fitting, a network structure was inferred using the Hill-Climbing approach, and the quality of the fit was evaluated using the Bayesian information criterion (BIC). The HC algorithm trains the network in a score-based “greedy search” (Scutari et al., 2019). This algorithm ranks network architectures based on the increase/decrease in BIC score induced by the removal of the arc. The network score was used to build arcs connecting two nodes if there were direct relationships between them. The arc strength value >0.5 was considered reliable. Illogical arcs were “blacklisted” and excluded from analysis to improve model fitting and avoid circular structure in the structure learning process (Ebana and Furukawa, 2019).
A more detailed analysis was performed at the individual enzyme level for those enzymes associated with carbohydrate metabolism, fermentation/respiration, antimicrobials, and stress response (Supplemental Cecal Transcriptome Data Set; n = 598). Pearson and Spearman-rank correlations were determined for enzyme transcript abundance compared to Salmonella abundance. Significance was given to Pearson or Spearman r or ρ values >0.6999 or < −0.6999 and p-values <0.05.
The identity of enzyme transcripts, with network connections or statistically significant correlation with Salmonella abundance, was determined by BLAST search at the nucleotide level (Altschul et al., 1990). Species or genus were assigned to the enzyme transcript if there was ≥99% coverage and 98–100% sequence match. Some matches based on these criteria identified Clostridia yet to be classified, which were assigned strain name or descriptor of the matching deposited sequence or genome.
Results
Salmonella growth and expression of virulence genes were reduced in the presence of an exclusive community
A fluorescence-reporter system was developed to monitor the Salmonella response to permissive and exclusive communities ex vivo. The growth-dependent promoter for rRNA operon, rrn (Bartlett and Gourse, 1994), was fused to the jellyfish fluorescent protein variant YFP and inserted into the P22 prophage integration site within the Salmonella chromosome, while CFP was inserted downstream of iag, the last gene in SPI-1 hilA operon, to monitor expression of the type 3 secretion system (T3SS) cell invasion genes. Both reporter insertions were expected to be neutral, as one was placed within the intragenic site for P22 prophage integration and the other involved insertion behind the last gene of the hilA operon iag, leaving all genes within this operon intact. The rrn-yfp promoter fusion reported was used to monitor Salmonella growth. Fluorescence varied depending on growth conditions, with intensive signal observed from cells grown in complex medium (LB vs. minimal medium with glycerol as a carbon/energy source) or at higher growth temperatures in LB broth (37°C vs. 42°C) (Figure 2). Salmonella reporter strain’s growth dynamics and growth rate were the same as the wild type under these ex vivo conditions with regard to doubling time (Dt) and growth rate (μ) at 30°C vs. 37°C (Supplementary Figure S3; Parish, 1985). When Salmonella was grown in EVCC medium, its growth was reduced after 6 h in the exclusive community compared to the permissive community (Figure 3). For Salmonella grown in the EVCC alone (control), the intensity of YFP fluorescence diminished over time until there was no detectable fluorescence after 24 h. When grown with either cecal community, Salmonella YFP fluorescence decreased to undetectable levels after 9 h. Expression of Salmonella T3SS invasion hilA operon increased over 6 h in the EVCC control, then decreased to undetectable levels by 24 h. Two peaks were observed in the FACS analysis: a major peak that overlapped with the non-fluorescent Salmonella negative control and a minor CFP fluorescent peak, indicating that the majority of cells were not expressing hilA (Figure 4). The proportion of Salmonella expressing CFP was diminished when grown in the presence of either cecal community after 3 h, but after 6 h of growth in the exclusive community, CFP expression was at its lowest compared to the other two conditions (Figure 4). The data indicate that after 6 h with the exclusive community, Salmonella growth was reduced and T3SS invasion expression was repressed. This time point was therefore selected to examine the Salmonella transcriptome response to growth in the communities.
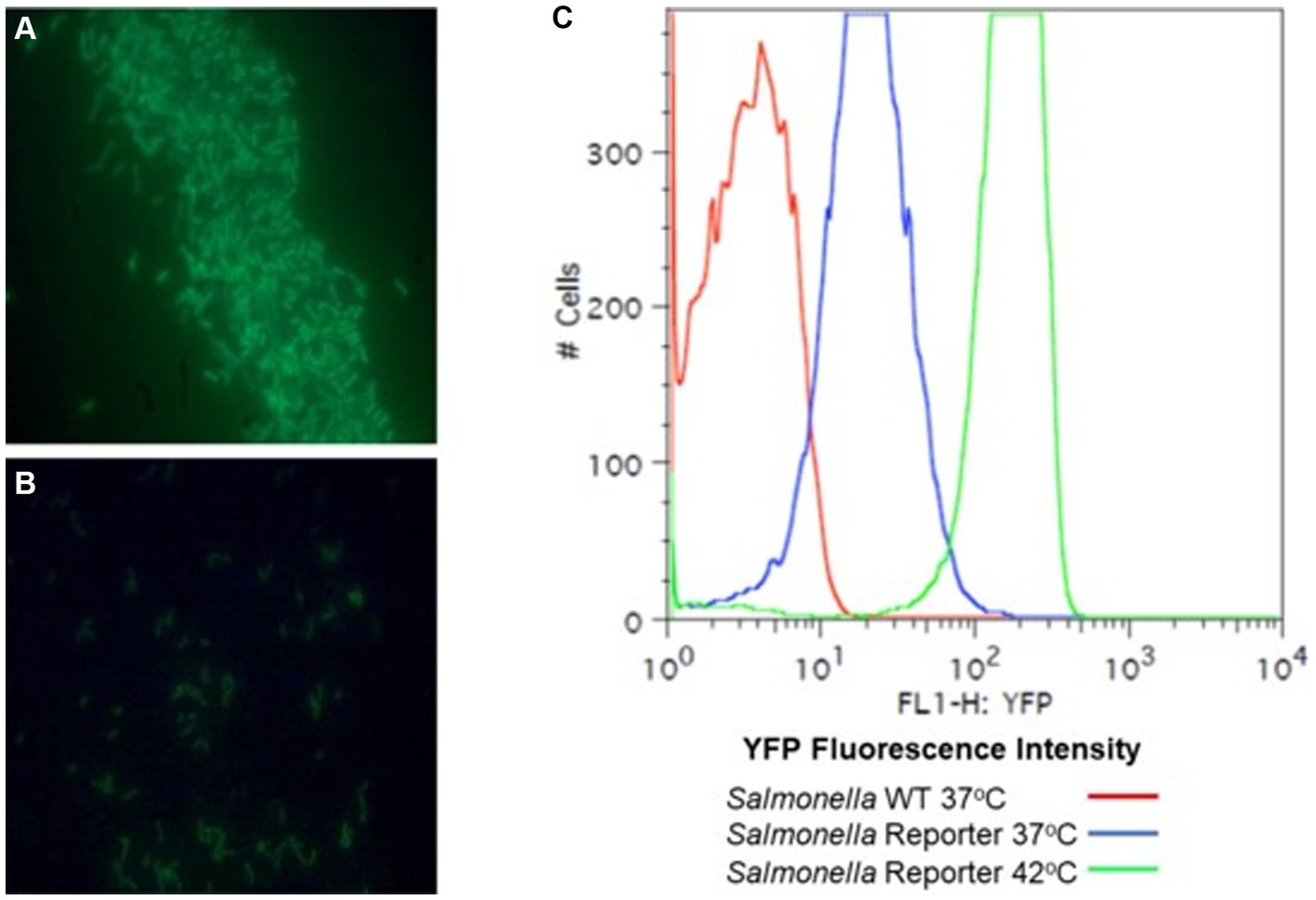
Figure 2. A rrn promoter jellyfish yellow fluorescent protein (YFP) gene fusion as a reporter for Salmonella growth. Lambda red was used to construct rrn tagged with YFP. The rrn promoter was cloned upstream of a “promoter-less” yfp and in tandem with cat. PCR primers were designed to overlap with the target insertion site, an intergenic site between thrW and STM0324. The λ Red system (Datsenko and Wanner, 2000) was used to introduce this reporter into S. enterica Typhimurium SL1344. S. typhimurium SL1344 strain YC1104 with the rrn-yfp reporter was grown aerobically to OD 1.0, λ 600 nm, in LB broth (A) or M9 minimal medium with 0.4% glycerol as a carbon source (B) with aeration and observed with a fluorescence microscope. (C) Fluorescence-activated cell sorting (FACS) analysis of the Salmonella rrn-yfp reporter strain grown aerobically, in LB broth to mid-exponential phase (OD 0.5, λ 600 nm) at 37°C or 42°C to demonstrate fluorescence intensity with a high growth rate.
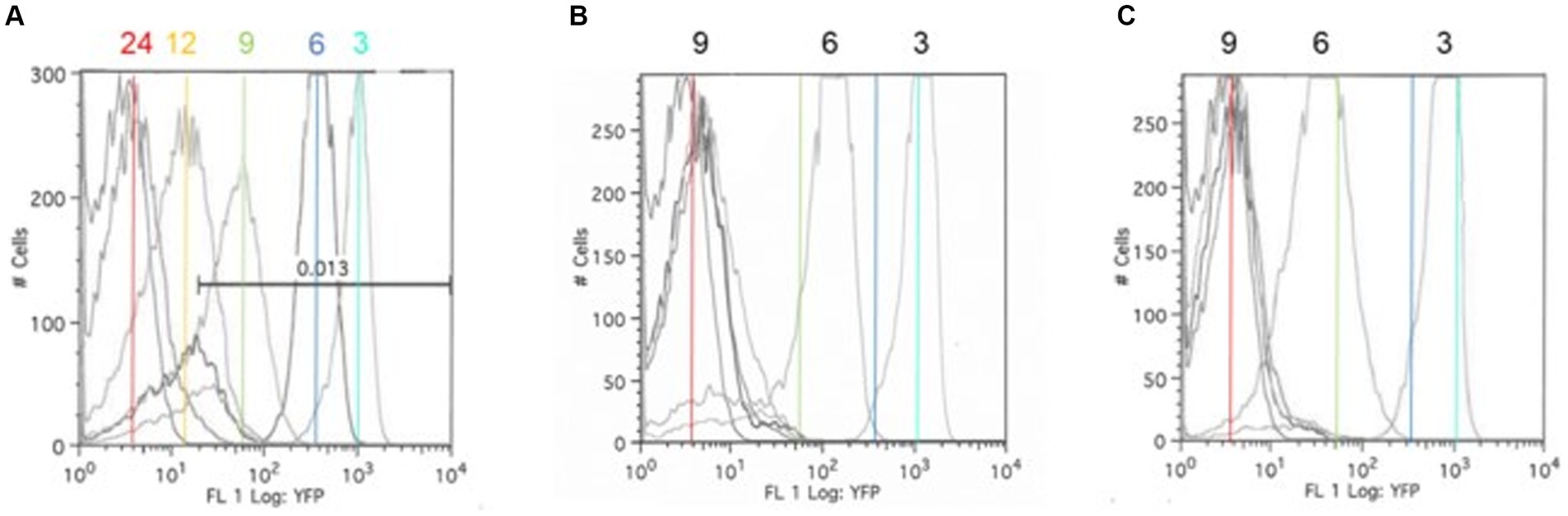
Figure 3. The Salmonella ex vivo growth in permissive (B) or exclusive (C) communities. FACS analysis was used to measure the rrn-yfp promoter activity in S. typhimurium SL1344 grown in simulated cecal medium (EVCC) (A) within a permissive or exclusive community. Salmonella YFP expression was monitored by FACS analysis at 3, 6, 9, 12, and 24 h using the parental strain as a negative fluorescence control. Cyan (3 h), blue (6 h), green (9 h), and red (24 h) lines mark the peak fluorescence levels of the reporter strain grown alone in EVCC (A) in order to demonstrate changes in intensity when grown in a permissive (B) or exclusive (C) community.
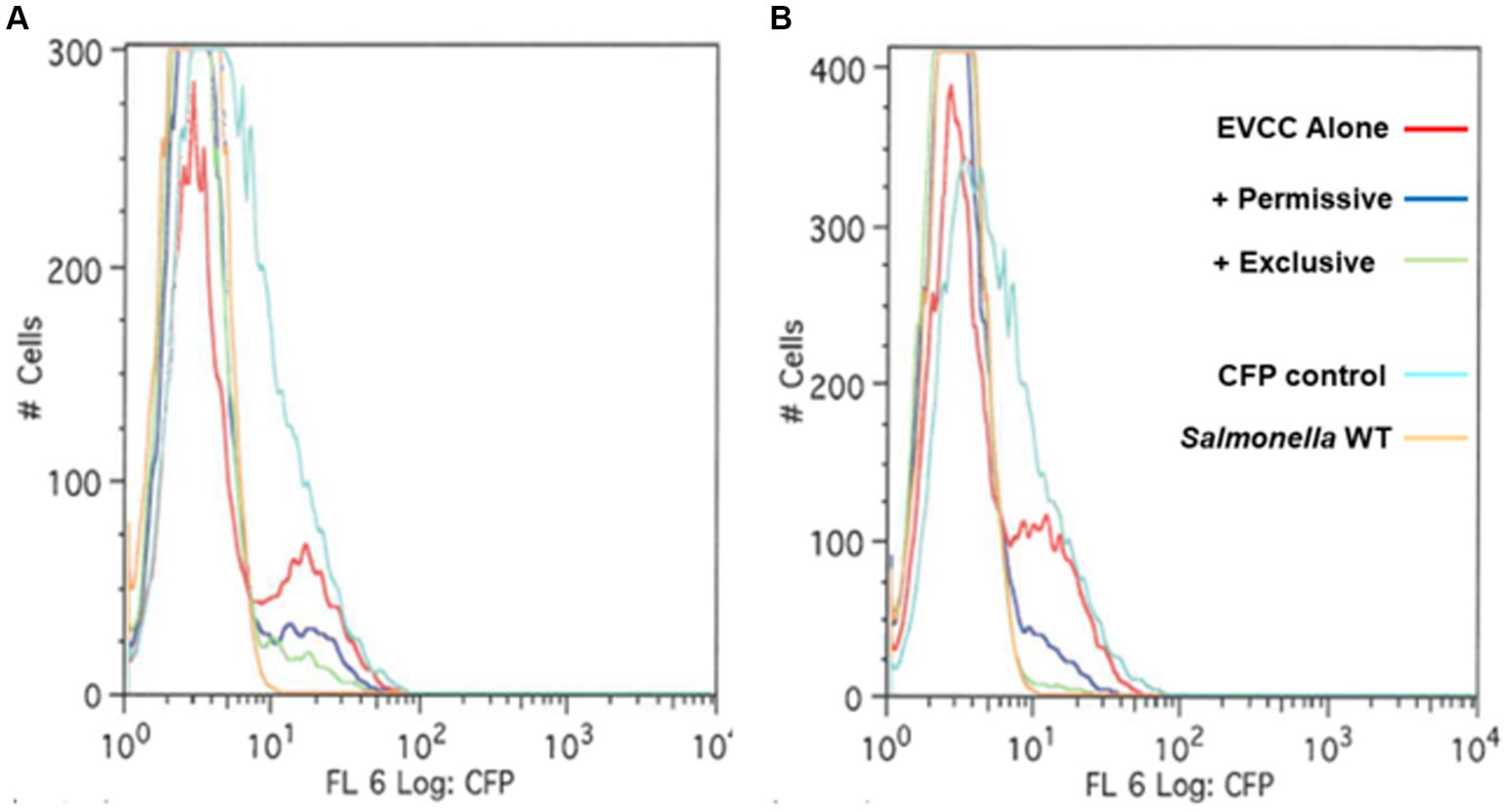
Figure 4. A hilA operon-cfp promoter fusion using jellyfish cyan fluorescent protein (CFP) as a reporter for Salmonella pathogenicity. A “promoter-less” cfp was placed between iag and the transcriptional terminator for the hilA operon using λ Red recombineering (Datsenko and Wanner, 2000). The Salmonella reporter strain was grown in EVCC medium alone or EVCC medium inoculated with the permissive or exclusive cecal community at (A) 3 h or (B) 6-h incubation. FACS analysis was used to measure the hilA operon-cfp promoter fusion in S. typhimurium SL1344 grown in EVCC medium alone (red line) or with a permissive (blue line) or exclusive (green line) community. The S. typhimurium SL1344 parental strain (orange line) served as a negative fluorescence control in FACS analysis. The CFP plasmid vector, pMG34, was used as a fluorescence-positive control (cyan line) (Miyashiro and Goulian, 2007). While a fluorescent population of cells were detected for Salmonella grown in EVCC alone or in a permissive community, an exclusive community repressed the Salmonella hilA locus, which is a global regulator of the pathogenicity island 1 (SPI-1) associated type 3 secretion system (T3SS).
Microarray transcriptome analysis of Salmonella revealed that the exclusive community had a profound effect on the expression of pathogenic behavior, significantly affecting the expression of 52 virulence genes (15.9%), compared to 7 that were altered when Salmonella was grown in the presence of the permissive community (Figure 5). The major differences in virulence gene expression in response to these communities were associated with LPS synthesis and Salmonella pathogenicity island 1 (Figures 6, 7). Most of the important virulence genes in SPI-1 as well as its ancillary T3SS effectors were repressed when Salmonella was grown with the exclusive community, compared to growth in the permissive community or the EVCC control (p < 0.001) (Figure 6). AvrA, an anti-inflammatory/anti-virulence factor (Jones et al., 2008), was elevated in Salmonella grown with the exclusive community. The only fimbrial operon expressed under these experimental conditions was the type I fimbrial operon, an important adhesin essential to the cell invasion process (Ernst et al., 1990). While the major fimbrial subunit FimA was expressed under all conditions, there was significantly less expression of export apparatus (fimC,D) and fimbrial adhesin (fimF) (p < 0.001). The polymyxin resistance operon pmr, responsible for modifying the Salmonella LPS (Gunn, 2008), was also repressed by the exclusive community. These results indicate that attenuation is likely involved in the mechanism of competitive exclusion.
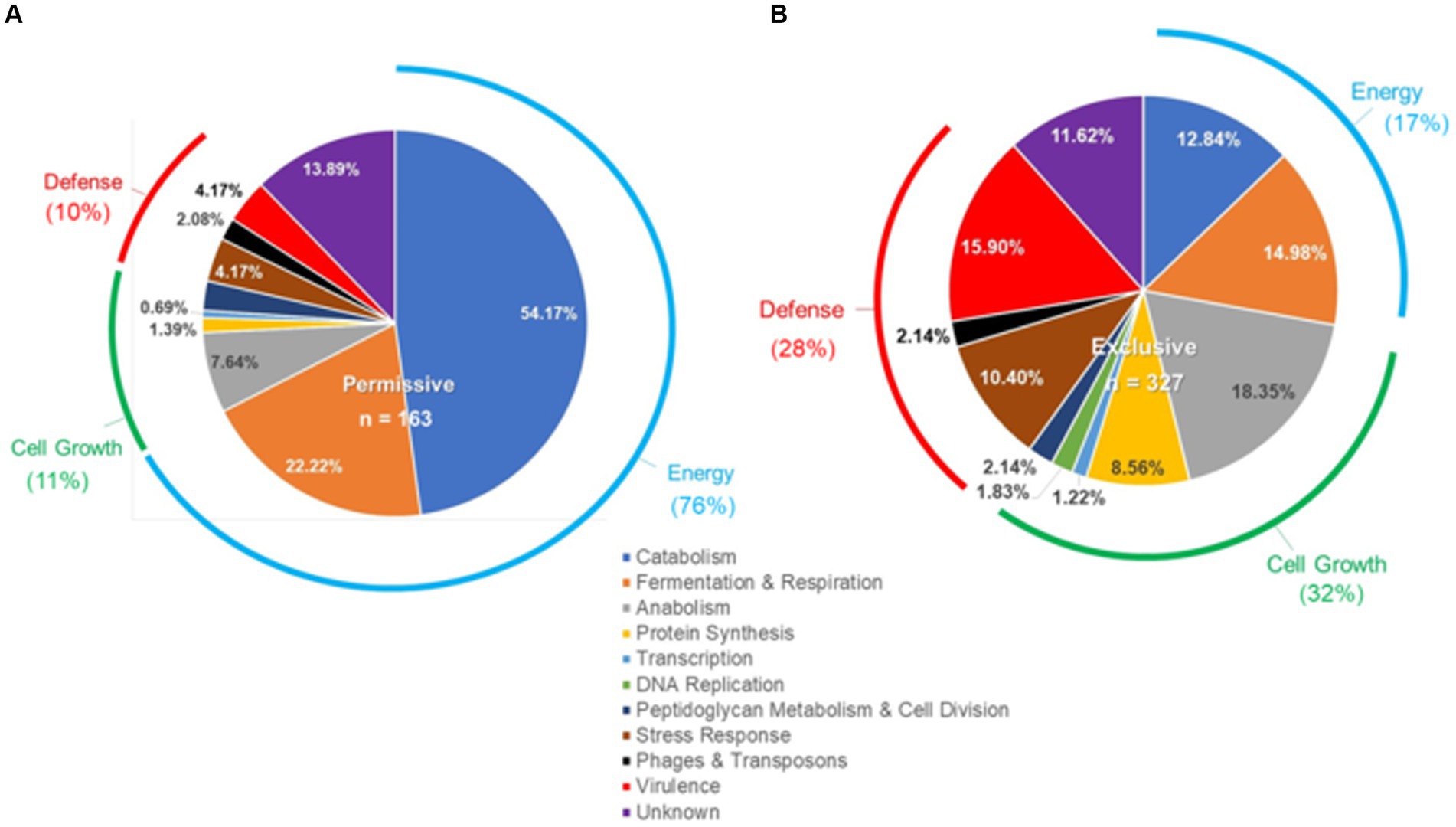
Figure 5. Differences in the Salmonella response to cecal permissive (A) and exclusive (B) communities ex vivo. The Salmonella SL1344 rrn-yfp, hilA operon-cfp reporter strain was grown in EVCC alone or with a permissive or exclusive community. Total RNA was extracted from Salmonella after 6-h growth. Genes reported with elevated expression and significant differences in expression (p < 0.0001) for growth in the permissive or exclusive community versus the negative community control were categorized based on function.
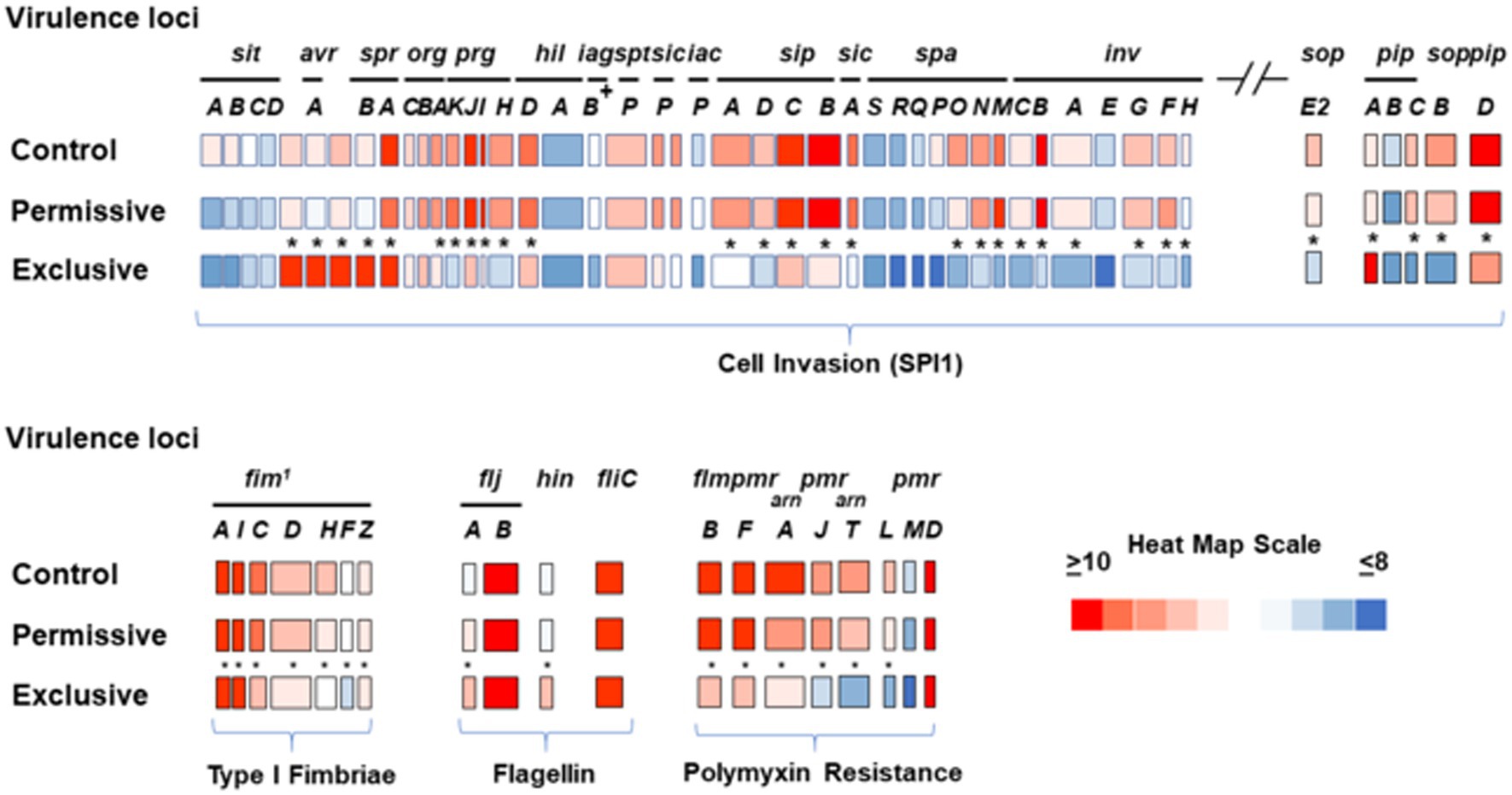
Figure 6. Transcriptional analysis of Salmonella virulence expression after growth in permissive or exclusive communities. FACS analysis was used to measure the hilA operon-cfp promoter fusion expression in S. typhimurium SL1344 grown in EVCC or with permissive or exclusive communities. Because Cfp expression was diminished in the Salmonella reporter strain after 6 h of growth in the exclusive community, the reporter strain was harvested after 6 h for microarray analysis. Significant transcription differences (*p < 0.0001) were observed in the SPI-1 locus, its ancillary T3SS effectors sopE2, pipA-D, and sopB, the type I fimbriae operon, flagellin, and polymyxin resistance expression. A heat map scale depicts the magnitude of microarray signal.
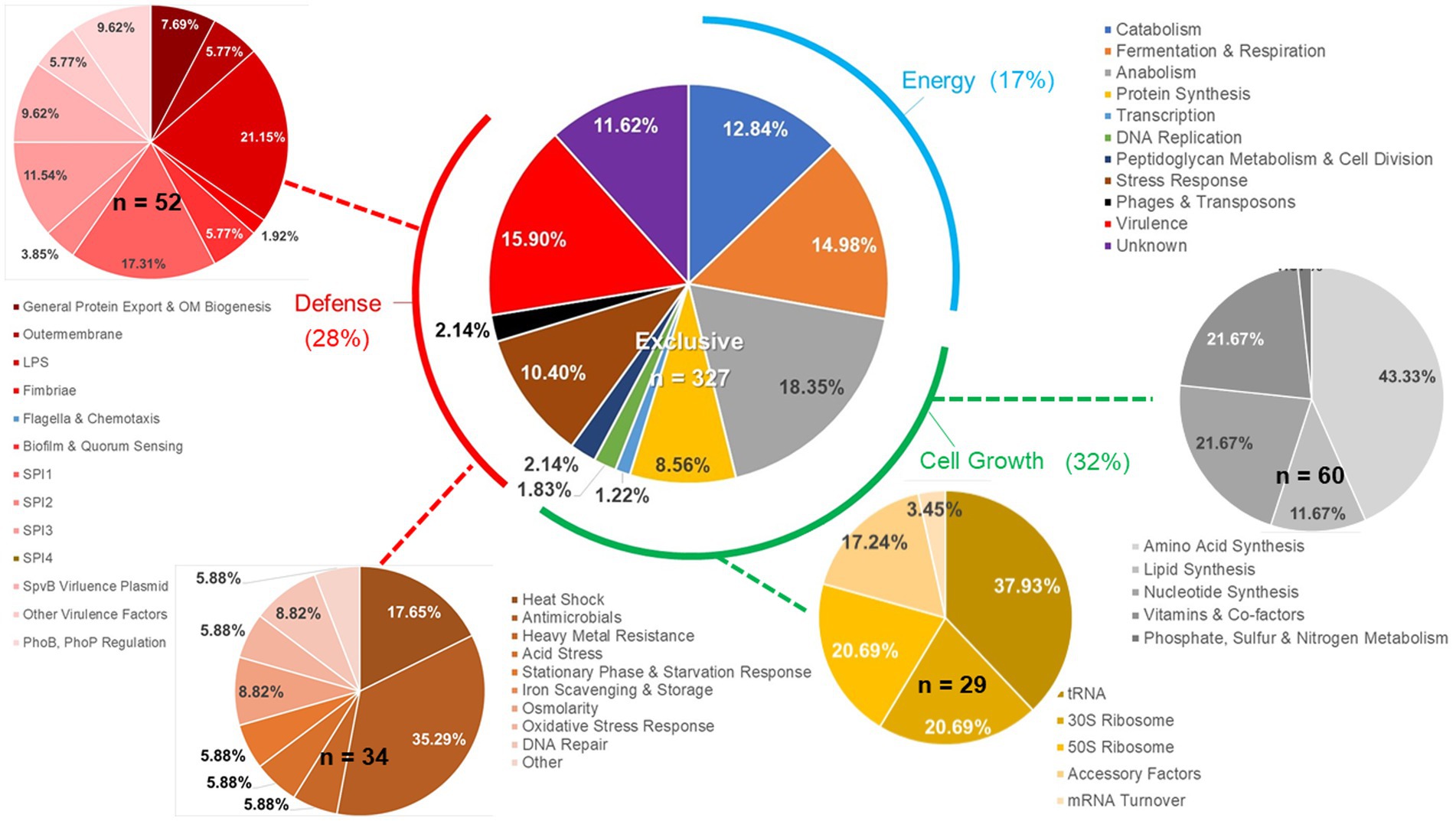
Figure 7. Details of ex vivo Salmonella transcriptional response to an exclusive community. Salmonella SL1344 rrn-yfp, hilA operon-cfp reporter strain was grown in co-culture in EVCC with an exclusive community or media alone. Total RNA was extracted from Salmonella after 6-h growth. Genes reported with significant differences in expression (p < 0.0001) versus the medium control were categorized based on function.
Salmonella catabolic and anabolic response to permissive and exclusive communities ex vivo
Individual gene transcript levels for salmonellae grown with either permissive or exclusive communities were compared to those of Salmonella grown in the EVCC medium alone. Of the 6,069 genes analyzed, 161 were significantly elevated (p < 0.0001) in Salmonella grown with either community (Figure 8). Thirty-six percent of these transcripts were associated with energy production, and most of these genes (87.5%; n = 48) were responsible for propanediol metabolism, the end product of fucose and rhamnose fermentation (Obradors et al., 1988). However, there was low to no expression of transcripts associated with propionate metabolism (prpB-E), the end product of propanediol metabolism (Supplemental: Salmonella Coculture Data Set). In addition, genes associated with vitamin B12 synthesis (cbi operon), a key co-factor involved in both propanediol and ethanolamine metabolism pathways (Jeter, 1990; Brinsmade et al., 2005), were also elevated in Salmonella grown with either microbial community, compared to the EVCC control (Table 2). In the presence of the exclusive community, Salmonella did not express vitamin B12 transporters (btuBCD) compared to EVCC medium alone or with the permissive community. Moreover, ethanolamine utilization transcripts were only expressed in Salmonella grown in EVCC alone (Table 2).
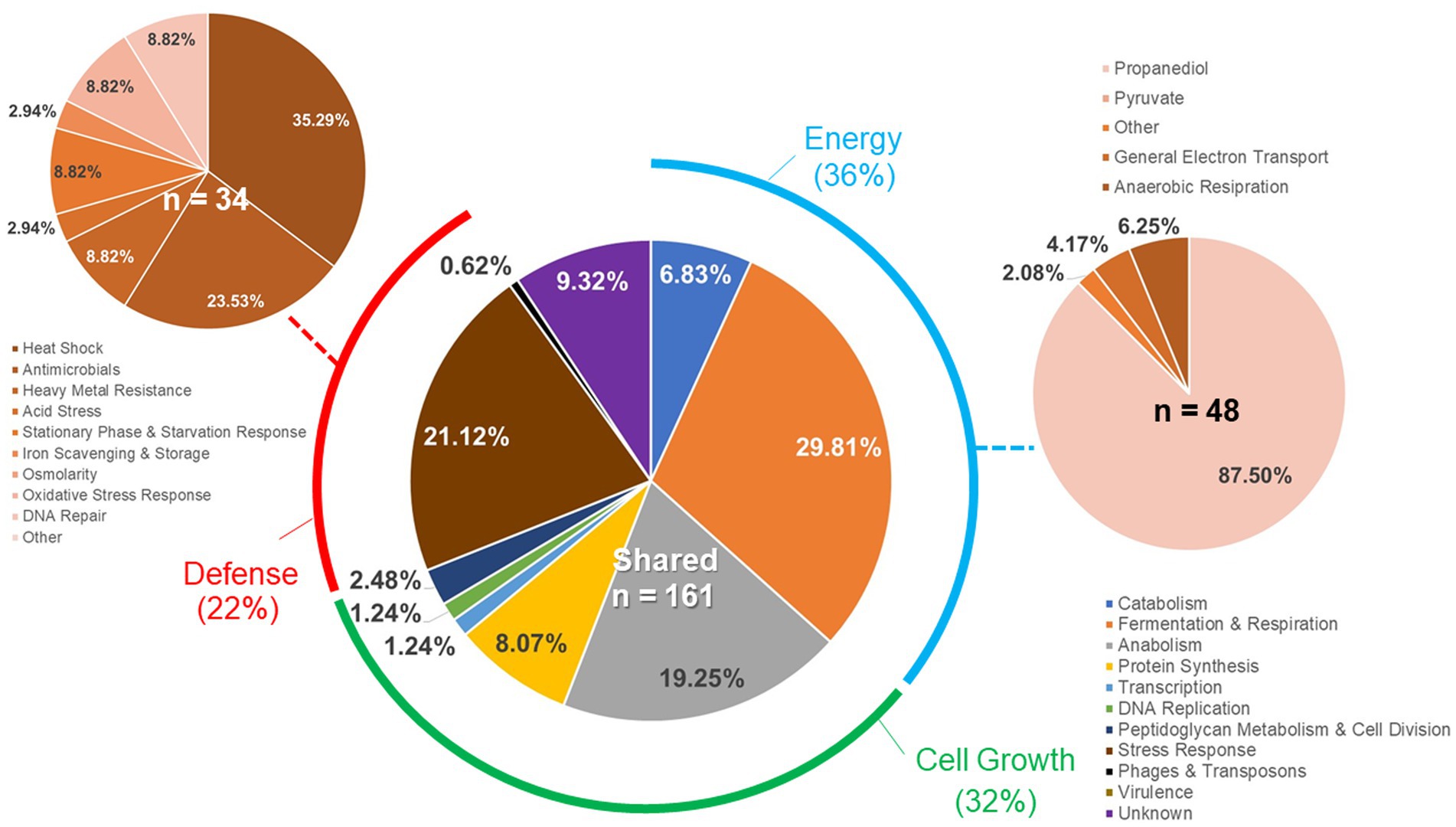
Figure 8. Differences in the Salmonella response to cecal permissive or exclusive communities ex vivo. The Salmonella SL1344 rrn-yfp, hilA operon-cfp reporter strain was grown in EVCC alone or with a permissive or exclusive community. Total RNA was extracted from Salmonella after 6-h growth. Genes reported with significant differences in expression (p < 0.0001) for both communities versus the negative community control were categorized based on function.
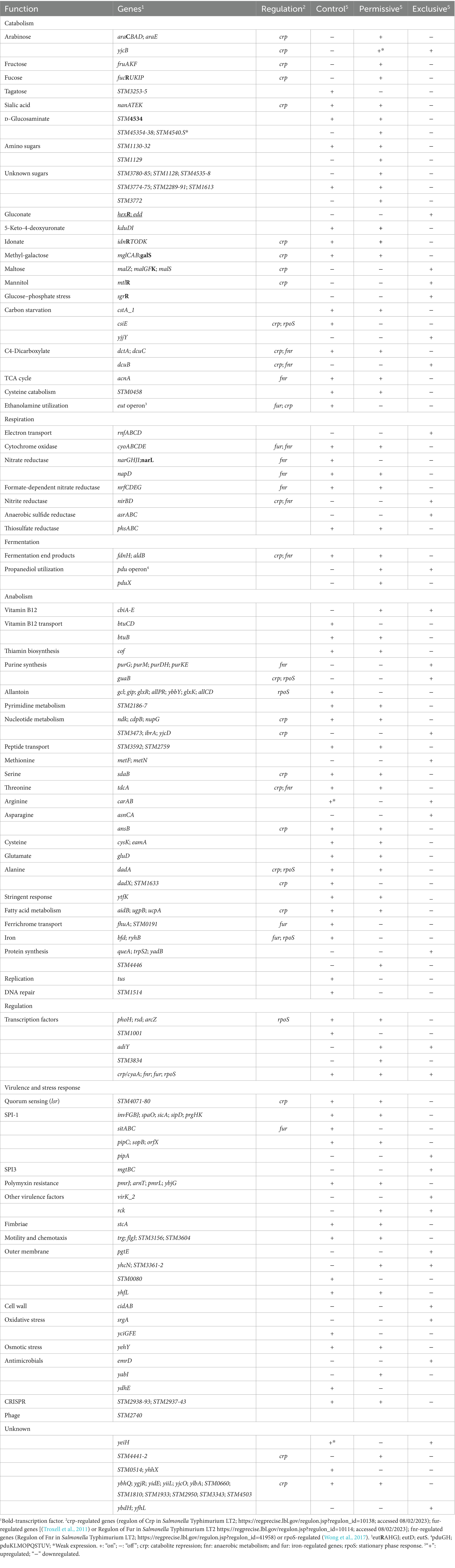
Table 2. Salmonella transcriptome response when co-cultured with cecal communities with high Salmonella abundance (permissive), with the competitive exclusion product (exclusive), or in EVCC medium (control).
There were significant differences in the Salmonella global transcriptome response to the microbial communities. Over 75% of 163 elevated gene transcripts from Salmonella grown with the permissive community, compared to the control, were dedicated to energy metabolism, whereas this was only the case for 17% of 327 elevated gene transcripts for Salmonella grown with the exclusive community (Figure 5). Salmonella grown with the permissive community directed fewer resources toward anabolism, as a percentage of upregulated genes (8%) than those grown with the exclusive community, where 18% of the upregulated genes were devoted to anabolism (Figure 5). Salmonella metabolic genes associated with the catabolism of arabinose, fructose, fucose, D-glucosaminate, and other amino-sugars were exclusively upregulated in Salmonella grown with the permissive community, while maltose was the only sugar utilization pathway expressed by Salmonella in co-culture with the exclusive community (Table 2). Several genes annotated as sugar transporters and associated enzymes, whose substrates have yet to be identified, were also expressed in Salmonella grown with the permissive community. In addition, Salmonella grown in EVCC medium alone or with the permissive community also upregulated genes responsible for sialic acid metabolism (nanATEK) and transport of methyl-galactose (mglCAB, galS). Catabolite repression may be central to regulating Salmonella gene expression in either microbial community, as several of these and other catabolic genes tied to energy generation (i.e., respiration) and a few genes with a role in the anabolic pathways responsible for nucleotide and amino acid metabolism possessed the signature nucleotide sequence recognized by the catabolite repressor protein Crp (Gunasekera et al., 1992). Transcripts for crp and the adenylate cyclase cyaA were produced in Salmonella under all growth conditions. Ex vivo, Salmonella produced enzymes under all conditions for metabolizing glucose (ptsG, glk, pfkAB, fdaB, tpiA, pgk, gpmA, eno, pykF), galactose (galMKTE), mannose (manA, manXYZ), glucuronate/galacturonate (kdgT, uxaA, uxaC), N-acetyl-galactosamine (gatY), and N-acetyl-glucosamine (nag operon) (Supplemental: Salmonella Coculture Data Set). There was greater expression for several of these genes (ptsG, glk, fdaB, manXYZ) in Salmonella grown with the permissive cecal community versus the exclusive community.
As propanediol is an end product of fucose and rhamnose fermentation, the focus of the Salmonella transcriptome analysis was shifted toward metabolism of microbial metabolites, including volatile fatty acids. Several genes tied to acetate (ackA, pta, poxB), D-lactate (dld), and hydrogen (hydrogenase-1, hydrogenase-2, and hydrogenase-3) metabolism were expressed under all conditions (Supplemental: Salmonella Coculture Data Set). However, neither acetate nor lactate was likely to be serving as a resource provided by its microbial community members, as the acetate permeases ActP and YaaH and lactate permease LldP were not elevated in Salmonella under any growth conditions. Differential expression was observed for formate (fdnH) and aldehyde (aldB) dehydrogenases in Salmonella grown in the presence of the microbial communities. However, the highest transcript levels of these genes were detected in Salmonella grown in the absence of either community.
Respiration may be central to Salmonella growth in that NAD appeared to be regenerated via electron transport, and NADH dehydrogenase genes (nuo operon) were strongly expressed under all growth conditions. In addition, cytochrome d oxidase genes (cydAB) were expressed under all growth conditions, whereas there was a significant decrease in cytochrome o oxidase cyo operon transcripts in Salmonella grown with the exclusive community (Table 2). While aerobic respiration appeared to be involved in Salmonella metabolism, several enzymes involved in anaerobic respiration were also differentially transcribed by Salmonella grown in EVCC alone (nitrate reductase: narGHIJ, narL), with the permissive community (nitrate reductases: napD; nrfCDEG; thiosulfate reductase: phsABC) or the exclusive community (nitrite reductase: nirBD; anaerobic sulfide reductase: asrABC).
Finally, there were substantial differences in the upregulation of anabolic enzyme transcripts between Salmonella grown with either community compared to EVCC medium alone (7% vs. 20%, respectively; Figure 5). In Salmonella grown in the presence of the exclusive community, a total of 65% of these anabolic enzymes were dedicated to amino acid and nucleotide synthesis. Salmonella grown with the exclusive community-expressed enzymes needed to synthesize nucleotides (STM3473; ibrA; yicD), especially purines (purG; purM; purDH; purKE; guaB) (Table 2). The peptide transporters STM3592 and STM2759 were expressed by Salmonella with the permissive community, as well as several enzymes associated with amino acid metabolism for serine (sdaB), threonine (tdcA), asparagine (asparaginase ansB), cysteine (cysK, eamA), glutamate (gluD), and D-alanine (dadA). There were significantly fewer transcripts found in the expression of enzymes involved in catabolism of serine/threonine (sdaA, sdaBC; tdc operon) in Salmonella grown with the exclusive community (Supplemental: Salmonella Coculture Data Set). These catabolic enzymes are subject to catabolite repression by Crp (Table 2). Methionine (metF, metN), arginine (carAB), and asparagine (asnCA) anabolism enzyme transcripts were upregulated in Salmonella grown with the exclusive community.
Cecal communities in chickens with high Salmonella abundance have a volatile fatty acid metabolic network
A Bayesian network analysis was used to analyze the cecal transcriptome pulled from MG-RAST-generated metabolism data sets. Guided by the findings of the ex vivo transcriptome, analysis of the cecal transcriptome was focused on enzyme transcripts involved in fermentation. A Hill-Climbing network identified 36 enzymes with 20 total connections (total enzymes, n = 45), of which there were seven single connections and five multiple connections with three or more enzymes (Figure 9) for the cecal communities with high Salmonella abundance (>5.85 Log10 Salmonella genomes/g cecal contents). Twelve enzymes associated with propanediol fermentation were identified in this network analysis in cecal communities with high Salmonella abundance. Propionate kinase PduW was a major node connected to enzymes involved in propanediol and acetate metabolism. The network connections for the high Salmonella abundance group mirrored their enzymatic positions within their respective metabolic pathways for some enzyme transcripts. Ten enzyme connections involving 18 enzymes were identified from the low Salmonella abundance cecal transcriptome. Here, propanediol-associated enzymes: methylglyoxal synthase, glycerol dehydratase, propionyl-CoA carboxylase, propionyl-CoA synthase, and propionaldehyde dehydrogenase were identified. Fourteen enzymes were present in both groups and included: methylglyoxal synthase; glycerol dehydratase; propionyl-CoA carboxylase; propionyl-CoA transferase; glutaconate-CoA transferase; hydroxybutyryl-CoA dehydratase; hydroxybutyryl-CoA dehydrogenase; pyruvate oxidase; acetyl-CoA synthase; acetyl-CoA hydrolase; ethanolamine ammonia-lyase; 4-aminobutyrate aminotransferase; cobalamin (vitamin B12); and cobalamin biosynthesis; although some of these were in different enzyme connections between these two groups.
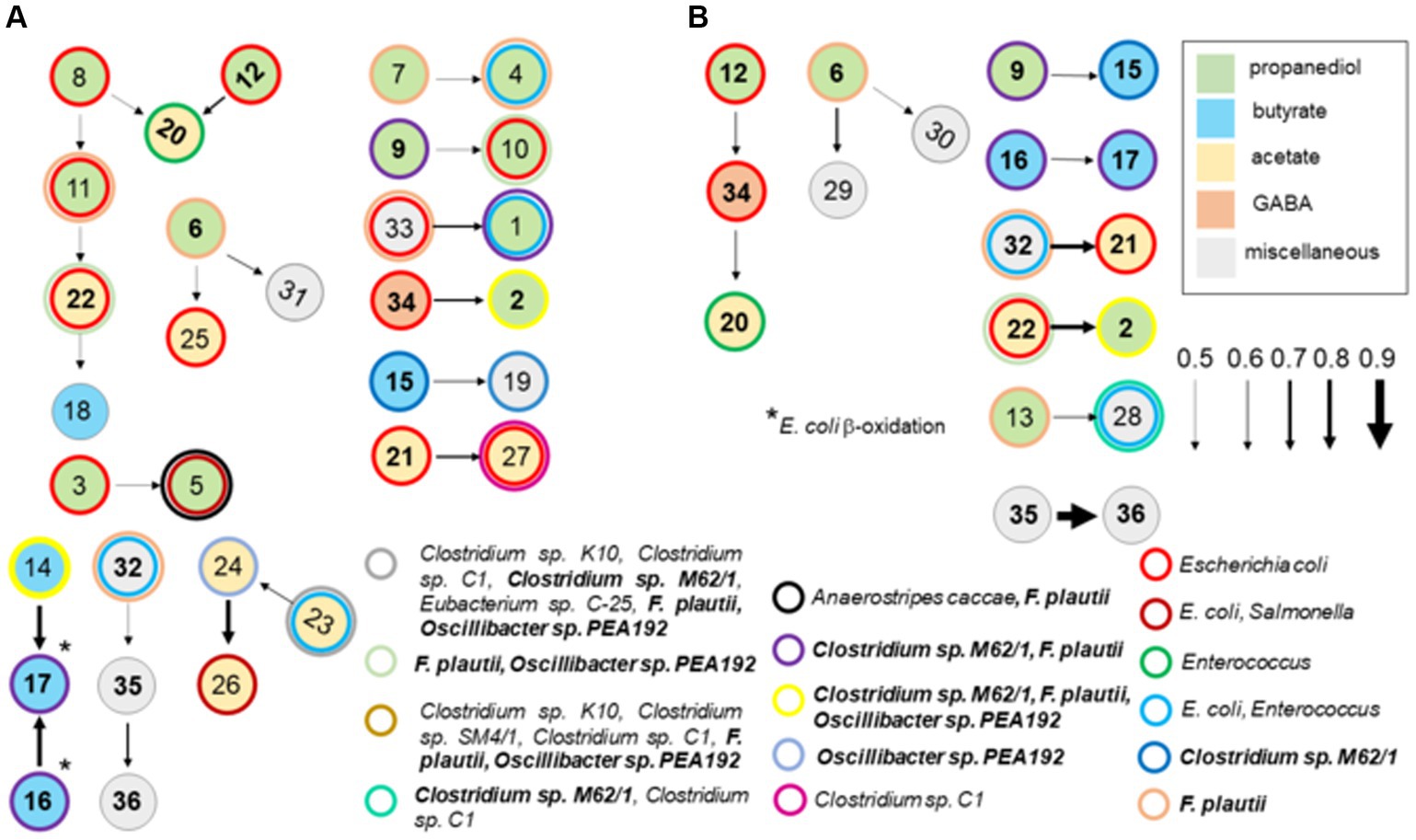
Figure 9. A Bayesian network analysis of cecal transcriptome, focused on fermentation, from chickens with high (>5.85 Log10 Salmonella genomes/g cecal contents) (A) or low (B) Salmonella abundance. The network depicted was identified using the score-based learning algorithm Hill-Climbing. The data were obtained from KO metabolism data sets in MG-RAST. Connections were identified among the 45 enzyme transcripts by network analysis, of which 36 were identified as single or multiple connections, producing 20 and 10 connections from the transcriptomes of cecal communities with high or low Salmonella abundance, respectively. Arrows indicate the direction and strength of the connection. Enzymes are color-coded based on their associated propanediol (green), butyrate (blue), or acetate (yellow) fermentation pathways. Enzymes identified in these networks were: methyl-malonyl-CoA decarboxylase (1); methylglyoxal synthase (2); lactaldehyde dehydrogenase (3); hydroxyacylglutathione hydrolase (4); glycerol dehydrogenase (5); glycerol dehydratase (6); propanediol dehydratase (7); propionate kinase (8); propionyl-CoA carboxylase (9); propionate-CoA transferase (10); methyl-malonyl-CoA mutase (11); propionyl-CoA synthase (12); propionaldehyde dehydrogenase (13); butyryl-CoA dehydrogenase (14); glutaconate-CoA transferase (15); hydroxybutyryl-CoA dehydratase (16); hydroxybutyryl-CoA dehydrogenase (17); thiolase (18); ferredoxin hydrogenase (19); pyruvate oxidase (20); acetyl-CoA synthase (21); acetyl-CoA hydrolase (22); acetate kinase (23); pyruvate ferredoxin oxidoreductase (24); acetyl-CoA synthetase (25); aldehyde dehydrogenase (26); phosphotransacetylase (27); lactate dehydrogenase (28); phosphoketolase (29); glutamate synthase (30); methylaspartate ammonia-lyase (31); ethanolamine ammonia-lyase (32); ethanolamine utilization protein (33); 4-aminobutyrate aminotransferase (34); cobalamin (vitamin B12) (35); and cobalamin biosynthesis (36). The following enzymes were not identified in any network analysis: lactaldehyde reductase, butyrate kinase, pyruvate dehydrogenase, citramalate synthase, serine dehydratase, alanine dehydrogenase, formate hydrogenlyase, formate C-acetyltransferase, and glutamate mutase. Numbers in bold are enzymes shared between cecal communities with high and low Salmonella abundance. Circles with thick-colored borders are for enzymatic transcripts with 98–100% nucleotide identity and 99–100% coverage by BLAST scores for intestinal species including Escherichia coli, Enterococcus faecium, and Flavonifractor plautii.
As these enzymatic pathways are associated with carbohydrate or amino acid fermentation, network analysis was expanded to include metabolic enzyme transcripts (164 total enzymes) with polysaccharide degradation and fucose/rhamnose catabolism responsible for producing the fermentation end product propanediol. Forty-four were identified in single or multiple connections for the two groups, producing 26 and 17 connections from the cecal transcriptomes for the high and low Salmonella abundance groups, respectively (Supplementary Figure S4). Propanediol and acetate metabolism remain dominant metabolic pathways in this new network analysis, especially for the high Salmonella abundance group, pulling in additional enzymes mostly associated with glucose metabolism and the TCA cycle. In fact, the same 30 fermentation enzymes from the first network analysis were identified in the expanded network analysis, except there were fewer single-enzyme connections for both Salmonella abundance groups. As expected, the expanded network analysis identified similar connections in the high Salmonella abundance group compared to the original 45-gene analysis (Supplementary Figure S3; Figure 9). Three of 10 enzyme transcripts involved with fermentation (n = 25), propionyl-CoA carboxylase, butyryl-CoA dehydrogenase, and pyruvate dehydrogenase, also had a significantly positive correlation with low Salmonella abundance (<6.00 Log10 Salmonella genomes/g cecal contents) (Table 3), while propionyl-CoA synthase and pyruvate oxidase had a significantly negative correlation with Salmonella abundance at Salmonella levels <6.00 Log10 Salmonella genomes/g of cecal contents. Other propionate enzyme transcripts (propionaldehyde dehydrogenase and propionate kinase) exhibited a negative correlation (p < 0.05) related to the age of the bird (not shown).
Most cecal transcripts identified as propionate kinase, methyl-malonyl CoA mutase, propionyl-CoA synthase, acetyl-CoA hydrolase, lactate dehydrogenase, or acetyl-CoA synthetase had 98–100% identity to Escherichia coli at the nucleotide level (Supplementary Table S8). The identities of other genera within the Enterobacteriaceae were all <98% nucleotide identities. However, some of these same transcripts had 98–100% identity, at the nucleotide level, to Enterococcus faecium, Flavonifractor plautii, or Clostridia. Some butyryl-CoA dehydrogenase and propionyl-CoA carboxylase enzyme transcripts had >98% nucleotide identity with Clostridia yet to be given a genus or species designation. However, there were many enzyme transcripts with little to no homology to any nucleotide sequences in BLAST but contained pfam domains characteristic of these enzymes. Pyruvate oxidase transcripts were primarily from Enterococcus or Lactobacillus species.
Analysis of the volatile fatty acid profile of the cecal contents of two birds with high Salmonella abundance showed variability. One sample had high acetate levels relative to the other VFAs, whereas another had high propionate and butyrate concentrations (Table 4). Therefore, in this small sample set, the volatile fatty acid profile and abundance did not correlate with the community transcriptome associated with fermentation.
Cecal community transcripts that correlate with Salmonella abundance in vivo
Carbohydrate analysis revealed that arabinose and glucose were the most abundant sugars present in the cecal lipid fraction (Table 5), with varying levels of rhamnose, fucose, xylose, glucuronic acid, galacturonic acid, galactose, N-acetyl galactosamine, and N-acetyl glucosamine in the lipid or precipitate fractions of both groups. However, mannose was present in the cecal contents of birds with high Salmonella abundance. The cecal community transcriptome contained many enzyme transcripts associated with liberating the sugars from complex carbohydrates, including transcripts annotated as: α-L-arabinofuranosidase, arabinogalactan endo-β-1,4-galactanase, β-fructofuranosidase, β-fructosidase, α-fucosidase, β-N-acetylhexosaminidase, β-hexosaminidase, α,β-galactosidases, α,β-glucosidases, α-mannosidase, α-glucuronidase, β-xylosidase, glycosidases, endoglucanase, cellulase, chitinase, and xylanases. Sialidase was present as a rare transcript in the cecal transcriptome. The transcriptome also contained various enzyme transcripts for transport and channeling of these sugars into central catabolic pathways. Twenty-four enzyme transcripts associated with carbohydrate utilization, out of 70 analyzed, had a significantly positive or negative correlation with Salmonella abundance as determined by Pearson or Spearman correlation coefficients (p < 0.05), and 5 and 12 of these enzyme transcripts correlated with high or low Salmonella abundance, respectively (Table 3). The enzymes α-galactosidase, L-fuculokinase, and rhamnulokinase exhibited a significant (p < 0.05) positive correlation with high Salmonella abundance (7.00–9.22 Log10 Salmonella genomes/g cecal contents), while arabinogalactan endo-4,4-β-galactosidase, ribose/xylose/arabinose/galactoside ABC-type transport, ABC-type maltose transport system, β-hexosaminidase, and dehydro-3-deoxyphosphogluconate aldolase were positively correlated with Salmonella low abundance (<6.00 Log10 Salmonella genomes/g cecal contents; p < 0.05). There was also a significant positive correlation with fucose isomerase and Salmonella abundance (2.11–9.22 Log10 Salmonella genomes/g, cecal content). Most fuculokinase (fucK) transcripts were identified as E. coli transcripts, while L-fucose isomerase (fucI) transcripts had >98% sequence identity with Anaerotruncus colihominis or Clostridium sp. M62/1 (Supplementary Table S8).
Cytochrome bd type quinol oxidase and cytochrome o ubiquinol oxidase had a significant positive and negative correlation, respectively, with Salmonella abundance. Nickel-dependent hydrogenase had a positive correlation with Salmonella abundance from 2.11 to 9.22 Log10 Salmonella genomes/g cecal contents (p < 0.05) (Table 3). The majority of the enzyme transcripts associated with respiration were from E. coli, with a few identified as Salmonella enterica (hybD, narI) (Supplementary Table S8).
Finally, a detailed analysis of the cecal community transcriptome focused on amino acid metabolism and Salmonella abundance identified 43 enzyme transcripts associated with amino acid/nitrogen transport or metabolism and Salmonella abundance (p < 0.05 by Pearson or Spearman) (Table 3). These transcripts were responsible for arginine, aspartate, glutamate, glutamine, glycine, histidine, phenylalanine, threonine, tryptophan, and tyrosine metabolism. Five (ABC-type polar amino acid transport system; periplasmic component/domain involved in amino acid transport; isopropylmalate/homocitrate/citramalate synthases; acetylglutamate kinase; and glutamate 5-kinase) and 10 enzyme transcripts (glutamate dehydrogenase; 5-enolpyruvylshikimate-3-phosphate synthase; allophanate hydrolase; aspartate-semialdehyde dehydrogenase; glutaminase; imidazolonepropionase; arginine/lysine/ornithine decarboxylases; threonine efflux protein; phosphoribosylanthranilate isomerase; and uncharacterized protein involve in cysteine biosynthesis) from cecal transcriptomes with low Salmonella levels had positive or negative correlations, respectively, with Salmonella abundance (p < 0.05), while seven enzyme transcripts (transglutaminase-like enzyme; tryptophanase; anthranilate phosphoribosyltransferase; anthranilate/para-aminobenzoate synthase; di- and tripeptidases; glutamine synthetase; and histidine ammonia-lyase) had a significantly positive correlation with high Salmonella abundance. Enzyme transcripts that focused primarily on glutamate metabolism (glt, gadB, gadC, gabT, and gabD) were from E. coli, Enterococcus, Lactobacillus, Clostridia, and unknown organisms (Supplementary Table S8).
Salmonella exclusion by the intestinal community ex vivo was not due to scavenger metabolism and competition
Single and double mutations were examined in Salmonella that affected vitamin B12 synthesis (Δcbi) or transport (ΔbtuC); propanediol (Δpdu) and ethanolamine utilization (Δeut); propionate metabolism (Δprp), fucose (ΔfucI) metabolism; anaerobic respiration (Δnar or Δnir), or glucose/galactose (Δmgl) transport (Figure 10). A fluorescent reporter that functions under anaerobic conditions was used to monitor Salmonella growth over time. The Salmonella wild-type strain had a longer growth lag in co-culture with the exclusive community, and its growth plateaued sooner compared to growth in EVCC medium alone. At 18.5- to 26-h incubation, the exponential phase growth rate, as determined from the slope of the line, was the same for the Salmonella wild-type and most metabolic mutants when grown with the exclusive community (m = 4.27 + 0.04; range 4.04–4.57). However, the Salmonella Δpdu single and Δcbi, ΔbtuC double mutants had slower (3.84) or faster (4.76) growth rates, respectively, compared to the wild type (4.23; Student’s t-test p < 0.05), when grown with the exclusive community. Biphasic growth was observed over the 48-h incubation, but Salmonella growth was significantly retarded in co-culture with the exclusive community during the last 8 h (m = 4.27 vs. 1.35). The biphasic growth exhibited by Salmonella wild type in EVCC medium alone exhibited a slower growth rate in the first phase (m = 2.74) compared to the second phase (m = 3.14). These results using mutants deficient in scavenger metabolism, anaerobic respiration, and multiple carbohydrate utilization indicate that Salmonella’s metabolic versatility augments its ability to compete for nutrients with an exclusive community.
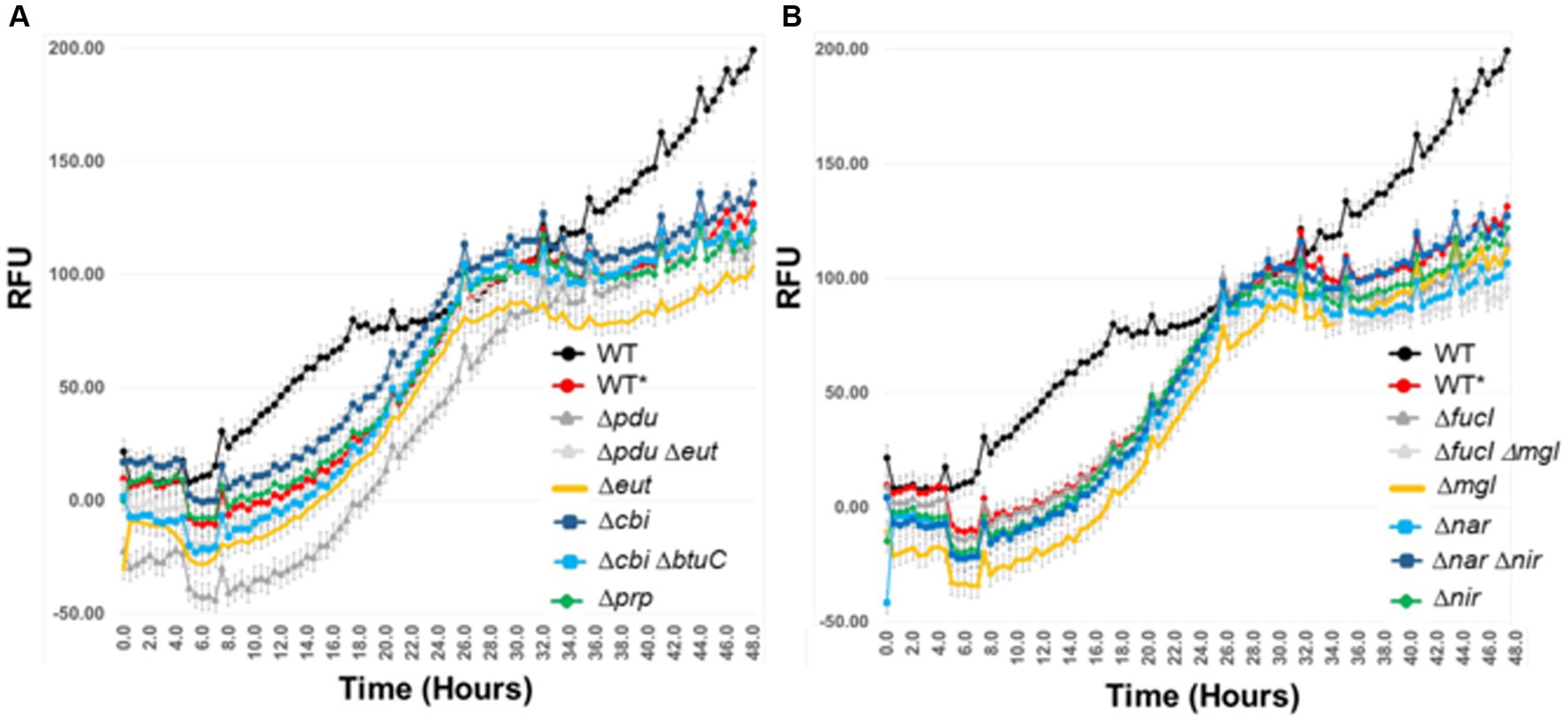
Figure 10. The effect of mutations in one or more metabolic pathways involved in Salmonella catabolism in an ex vivo intestinal environment containing an exclusive community. λ Red was used to create single and double mutations in Salmonella SL1344. pGLOW, a fluorescent reporter that fluoresces under aerobic and anaerobic conditions, was used to monitor Salmonella growth over 48 h. Mutant and wild-type (WT*) S. typhimurium SL1344 strains were grown in EVCC with exclusive community; WT was grown in EVCC alone as a control. Oxyrase (Sigma-Aldrich; St. Louis, MO) was added to the medium and overlaid with mineral oil to create and maintain low oxygen conditions. The selected mutations affect the catabolism of: (A) microbial metabolites (eut, pdu, prp), vitamin B12 synthesis (cbi), vitamin B12 uptake (btuC), or (B) sugar utilization (fucI, mgl) or anaerobic respiration (nar, nir).
The role of antagonism in the mechanism of competitive exclusion
Salmonella ex vivo transcripts in response to both communities were elevated for 34 of the total 161 genes associated with the stress response (Figure 8), compared to their growth in EVCC alone. Approximately 70% of the response was associated with heat shock, acid tolerance, and antimicrobials. There was also a significant difference in the Salmonella stress response in the exclusive community, where transcription of 32 genes was increased compared to the permissive community. One-third of these genes were associated with response to antimicrobials (Figure 7). In a network analysis of the cecal community transcriptomes’ stress response, a pattern emerged between cecal communities with high versus low Salmonella abundance (Figure 11). Oxidative stress appeared to be a major stress response in both cecal communities, but none of these or other stress response transcripts were shared, indicating that the stressors experienced in the communities were different. This contrasts with the results from the fermentation network analysis, where 30 enzyme transcripts were shared between Salmonella abundance groups and network analyses. For the high Salmonella abundance group, the organic hydroperoxide resistance protein (ohr) was the central node linked to various enzymes involved in carbon starvation, cold stress, envelope stress, osmotic stress, and acid tolerance. It also connected with glutamate-1-semialdehyde aminotransferase, an enzyme with a role in antibiotic synthesis. Yersiniabactin non-ribosomal peptide synthase was the major node in the stress response network for the cecal community with low Salmonella abundance (Figure 11). Forty-one of the enzyme transcripts from the two cecal communities were identified as E. coli or Enterococcus transcripts (>98% nucleotide identity with >99% coverage by BLAST) (Figure 11; Supplementary Table S8). There was a significantly negative correlation between transcript and Salmonella abundance for 72.5% of the enzyme transcripts associated with cecal transcriptomes’ stress response (Table 3). Several of these transcripts were associated with polyketide synthesis and annotated as: polyketide synthase module; thioesterase domains of type I polyketide synthases; Yersiniabactin non-ribosomal peptide synthase, non-ribosomal peptide/polyketide synthase, synthetase, and salicyl-AMP ligase; mycobactin salicyl-AMP ligase; oligoketide cyclase/lipid transport protein; and predicted thioesterase involved in non-ribosomal peptide biosynthesis. For yersiniabactin non-ribosomal peptide synthase, a few transcripts displayed a significant nucleotide identity with E. coli and Klebsiella pneumoniae. These sequence matches were identified as polyketide synthases irp responsible for producing the iron-scavenging yersiniabactin siderophore.
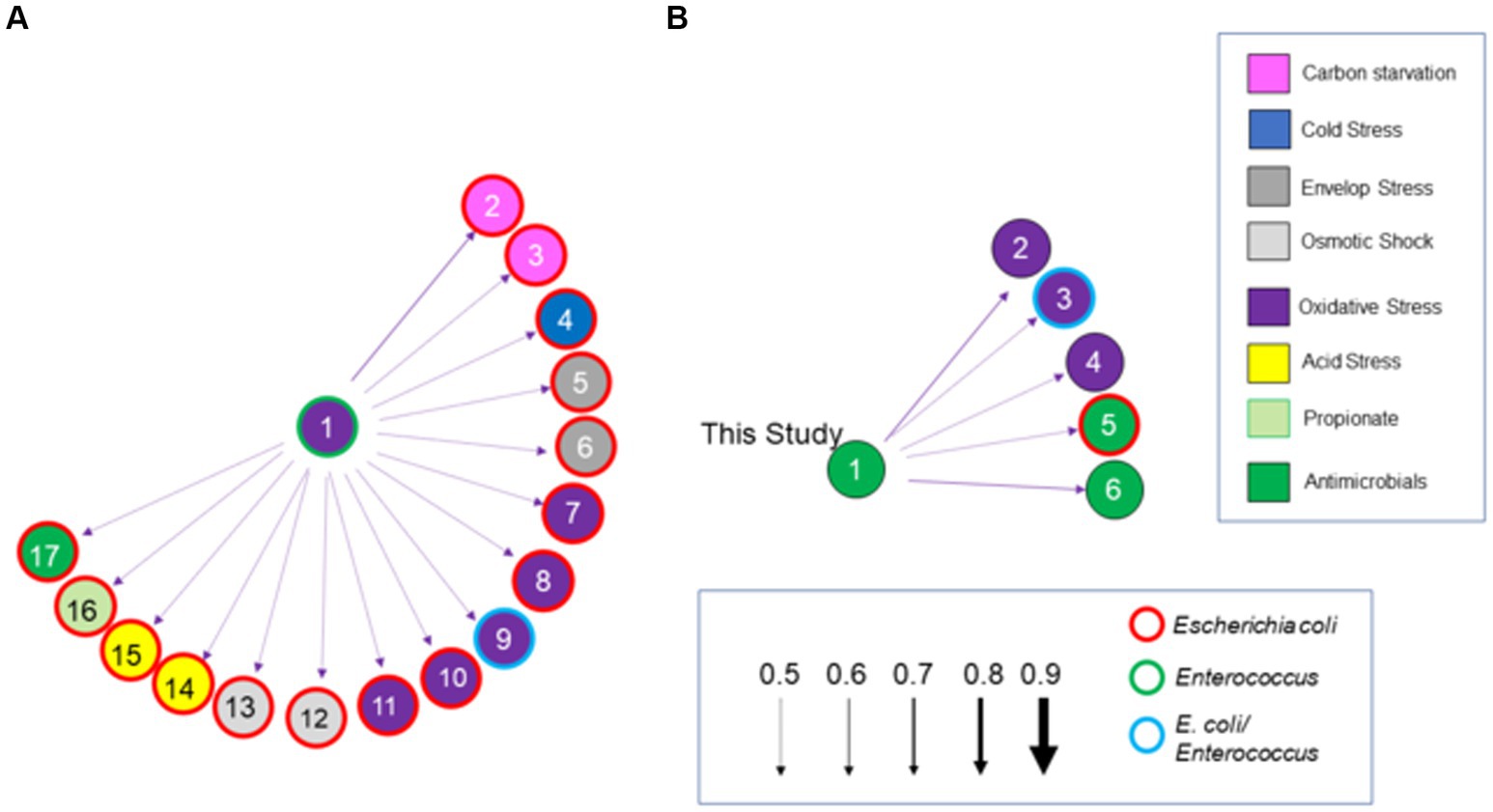
Figure 11. A Bayesian network analysis of the cecal transcriptome, focused on stress response, from chickens with high (>5.85 Log10 Salmonella genomes/g) (A) or low (B) Salmonella abundance. The network depicted was constructed using the score-based learning algorithm Hill-Climbing (arrows with solid line). Arrows indicate the direction and strength of the connection. Enzymes are color-coded based on the stress response. Enzymes or genes identified in the high Salmonella abundance community (A) are: (1) organic hydroperoxide resistance protein (ohr); (2) universal stress protein E (uspE); (3) yciT; (4) cspI; (5) pspC; (6) rseA; (7) arcA; (8) glutaredoxin; (9) nrd; (10) yncG; (11) fmrR; (12) betI; (13) yehZ; (14) ybaT; (15) hdeA; (16) propanediol diffusion facilitator; (17) glutamate-1-semialdehyde aminotransferase. Enzymes or genes identified in the low Salmonella abundance community (B) are: (1) yersiniabactin non-ribosomal peptide synthase; (2) putative bacterial hemoglobin; (3) gshF; (4) hemW; (5) tehB; (6) thioesterase domains of type I polyketides. Circles with thick red, green, or blue borders are for enzymatic transcripts with 98–100% nucleotide identity, by BLAST scores, to Escherichia coli, Enterococcus spp. or both, respectively.
Several of the antimicrobial transcripts with a positive correlation to Salmonella abundance were also associated with antimicrobial resistance, such as colicin E2 resistance creA; rRNA small subunit methyltransferase I (rsmI); spectinomycin 9-O-adenylyltransferase [ant (9)]; and translation elongation factor lepA. The creA transcript was from E. coli (≥98% nucleotide identity; ≥99 coverage by BLAST). The aminoglycoside resistance gene ant (9) was commonly associated with a variety of diverse bacterial species with near-identical nucleotide sequences, including spectinomycin-resistant Bacteroides, Campylobacter jejuni, Lactobacillus crispatus, Clostridioides difficile, and Clostridium sp. C1. Additionally, the following antimicrobial resistance genes were identified in the chicken cecal transcriptome: aadA1, spw, erm(B), and ermG. The latter two antibiotic resistance genes were from many bacterial species, including those commonly inhabiting the gastrointestinal tract (Supplementary Table S8). The lepA and rsmI enzyme transcripts had identity to sequences in E. coli, Enterococcus sp., various Clostridia, and unknown bacteria.
These results indicate that many of the members of the intestinal bacterial community were expressing resistance to antimicrobial compounds such as bacteriocins and antibiotics; however, there were no exogenous antimicrobials administered to the birds. The polyketide pathways suggest that some members of the microbiota may have been secreting antimicrobials that modulated the bacterial community, indicating the role of antagonism in competitive exclusion.
Discussion
An exclusive bacterial community attenuates the expression of virulence
These experiments illustrated that the chicken intestinal microbiota can significantly modulate Salmonella’s pathogenic behavior, indicating that attenuation may be involved in competitive exclusion. This suppression may be due to microbial metabolites like butyrate (Zhang Z.J. et al., 2020) or indole (Kohli et al., 2018) being shown to repress the SPI-1 T3SS cell invasion locus. Butyrate was not likely the main repressor, as this pathway was expressed in the cecal transcriptome of chickens with low and high Salmonella abundance, and there were differences in butyrate levels in chickens with high Salmonella abundance. Tryptophanase, the enzyme that liberates indole from tryptophan, was also present in the cecal transcriptome and directly proportional to Salmonella levels in the cecum, suggesting indole as a likely modulator of Salmonella virulence by the intestinal community. However, repression of SPI-1 T3SS by the exclusive community may be indirect, as this community also slowed Salmonella growth ex vivo, and growth rate is important in regulating the cell invasion of T3SS (Lee and Falkow, 1990). In addition, the exclusive community turned on avrA, a gene within the SPI-1 locus that lowers inflammation (Jones et al., 2008) and restores intestinal barrier integrity (Liao et al., 2008). It is the combination of SPI-1 T3SS repression and avrA activation that is responsible for Salmonella’s reduced virulence in the presence of an exclusive community. Others have shown repression of an enteropathogen’s virulence by the intestinal community or its member species (Le Bihan et al., 2015; Girinathan et al., 2021; Saenz et al., 2023).
In avian and mammalian species, juveniles are more likely to exhibit disease symptoms than adults when infected with Salmonella (Barrow and Methner, 2013). Day-of-hatch chicks may succumb to Salmonella infection after challenge (Bythwood et al., 2019), while 2-day-old birds will not. Instead, they shed high levels of Salmonella for 2–3 weeks before there is a precipitous drop in Salmonella shedding at approximately 3 weeks of age (Cheng et al., 2015; Pedroso et al., 2021). This suggests that the hatchling’s intestinal community is not effective in reducing the expression of Salmonella pathogenic behavior, but the microbiota rapidly gains this ability or at least counters inflammation induced by the pathogen (Yu et al., 2023). The exclusive community repressed LPS modification by the pmr locus in Salmonella, which is associated with resistance to cationic antimicrobial peptides (Matamouros and Miller, 2015). The host’s intestinal surface expresses β-defensins, a group of cationic antimicrobial peptides toxic to Salmonella (Milona et al., 2007). The exclusive community repressed pmr and therefore would make Salmonella susceptible to these and other defensins. While this early community can reduce the pathogen’s virulence early in the chick’s life, it’s not sufficient to exclude the pathogen from the intestinal luminal environment. The mechanism of competitive exclusion must involve more than suppression of the pathogen’s virulence in its mode of action.
Competition did not overcome Salmonella’s metabolic versatility
Salmonella appeared to spend less energy toward anabolism when grown in the permissive community compared to the exclusive community. There was a substantial difference in transcript abundance for amino acid metabolism and peptide transport, nucleotide biosynthesis, and vitamin B12 uptake between Salmonella grown in these two communities. There were also differences in cecal community transcript abundance in the comparison of enzymes involved in amino acid metabolism. These findings suggest substantial differences in the availability of free amino acids, nucleotides, and vitamins within permissive and exclusive communities. Low availability of these nutrients in an exclusive community should result in Salmonella’s slow growth because of the energy cost of de novo synthesis (Ushijima and Seto, 1991; Ha et al., 1994; Yang et al., 2016, 2017).
However, the diverse sugars available for Salmonella utilization were reflected in the glycome analysis of the cecal contents of birds with high Salmonella abundance. In addition, the cecal community transcriptome displayed a diversity of glycosyl hydrolases that would liberate these sugars from complex carbohydrates and glycoproteins. Carbohydrate metabolism appeared to be central to Salmonella growth, as revealed by the array of Salmonella enzyme transcripts associated with catabolism and the significant number of differentially expressed genes subject to catabolite repression. This concurs with the finding that mutations in crp and cya attenuate Salmonella (Curtiss and Kelly, 1987) and other intestinal pathogens (Peighambari et al., 2002; Sun et al., 2010; Zhou et al., 2020). The ex vivo transcriptome revealed that the exclusive community appeared to contain fewer sugars but more fermentation end products, such as propanediol, for Salmonella growth. Network connections and cecal community enzyme transcript abundance relative to Salmonella levels indicated that propanediol may be linked to Salmonella growth through community cooperation. Transcriptional analysis of Salmonella grown anaerobically ex vivo in the exclusive community showed abundant respiration transcripts, suggesting that it was involved in growth on a poor energy source such as 1,2-propanediol. Propanediol utilization has previously been shown to be a contributor to early Salmonella colonization in mice and chickens; however, these studies were done with animals lacking a mature intestinal microbiota (Harvey et al., 2011; Faber et al., 2017). These results suggest that cooperation may be involved in promoting Salmonella growth in cecal communities.
Several metabolic genes were identified that were differentially regulated in the cecal communities with high or low Salmonella abundance. Single and multiple deletions were introduced into these genes to assess their contribution to Salmonella growth in the exclusive community. The genes pdu, cbi, and vitamin B12 receptors are all necessary for propanediol metabolism, and they were transcribed during the ex vivo growth of Salmonella with the cecal communities. Only the Salmonella Δpdu single mutation significantly reduced Salmonella growth rates compared to wild type. There were no significant growth defects between the other mutants and wild type in the presence of the exclusive community. This finding suggests that, when presented with a diverse array of nutrients, Salmonella is metabolically versatile and adapts to the changing availability of nutrients in the intestine. Its metabolic versatility is reflected in the different metabolic genes shown to be involved in the colonization of different animal and plant species (Kwan et al., 2018). Therefore, neither cooperation nor competition appears to be the sole mechanism behind competitive exclusion.
Cecal communities may exhibit antagonism via the production of antimicrobials
The network analysis of the cecal stress response revealed that a polyketide synthase was the central node linked to similar enzymes in the cecal community containing low Salmonella abundance. With a few exceptions, these enzyme transcripts could not be assigned to known intestinal species in the NCBI database but exhibited similarity to as yet to be identified clostridial species. Most polyketide synthase modules responsible for antibiotic synthesis have been found in aerobic soil microbes because much research has focused on these organisms (Behnken and Hertweck, 2012a). However, recent studies have identified potential antimicrobial polyketides in Clostridia (Seedorf et al., 2008; Behnken and Hertweck, 2012b; Li et al., 2020), and polyketide synthases have been identified within the human intestinal microbiome (Fritz et al., 2018). Clostridial polyketide synthase may also be involved in siderophore synthesis (Singh et al., 2017). There was a negative correlation between transcript abundance for many of these enzymes and Salmonella levels in the cecum, as well as other enzyme transcripts associated with antibiotic and colicin resistance. This finding suggests that the exclusive community was likely producing antimicrobial substances. In addition, transcripts annotated as resistance genes with homology to many intestinal species, including Salmonella and E. coli, were detected, indicating the expression of antibiotic resistance. The chickens used in this study were reared without any antibiotics; therefore, the in vivo detection of antimicrobial resistance gene expression was not due to iatrogenic usage. Alternatively, the contact-dependent antibacterial type 6 secretion system (T6SS) (Alteri et al., 2013) might be at play in Salmonella exclusion. The ex vivo experiment would have precluded the detection of this mechanism. However, E. coli T6SS transcripts were present in the cecal transcriptomes, demonstrating the strength of a combined ex vivo/in vivo approach to unraveling the mechanism of competitive exclusion. While there was a negative correlation between the cecal T6SS transcript and Salmonella abundance, this is not the likely mechanism, as this correlation was only observed in chickens with high Salmonella colonization.
Early work demonstrated inhibition of Salmonella growth by cecal microbiota in vivo (Bohnhoff et al., 1964a; Royal and Mutimer, 1972), and pretreatment of mice with streptomycin reduced the inhibitory effect of the cecal microbiota on Salmonella colonization (Bohnhoff et al., 1964b). Others have looked at the ability of intestinal species to compete with bacterial pathogens for limiting resources (Barrow et al., 1987; Ushijima and Seto, 1991) or adherence to mucosal surfaces, indicating that competition was accepted as the mechanism of competitive exclusion (Gusils et al., 2003; Collado et al., 2005). However, the inhibitory effect on Salmonella growth was hypothesized to be related to volatile fatty acid (VFA) production and the potentiating effect of pH, suggesting that the role of inhibition was also considered (Bohnhoff et al., 1964b; Royal and Mutimer, 1972). This led to bio-prospecting for intestinal bacteria species for their production of inhibitory VFAs (Hinton et al., 1991). Antimicrobial production has also been used to select candidate probiotics; however, most have failed to reduce Salmonella colonization (Pan et al., 2008; Grant et al., 2018; Lu et al., 2023). There has also been considerable focus on microbes that produce bacteriocins (Grant et al., 2018; Angelopoulou et al., 2019; Khan et al., 2020). As an example, Wooley et al. demonstrated the inhibitory effects of a microcin-producing avian E. coli isolate on Salmonella colonization in chickens (Wooley et al., 1999). More recently, Bacillus spp. have been shown to be quite effective at excluding or controlling some intestinal pathogens in chickens (Grant et al., 2018). The genus Bacillus contains many member species containing antimicrobial polyketide synthases (pks) (Olishevska et al., 2019). B. subtilis pks has been shown to produce substances that inhibit Salmonella growth in co-culture (Podnar et al., 2022). It has been long established that prophylactic use of antibiotics, even at sub-therapeutic levels, can adversely affect Salmonella colonization in several animal species, including chickens (Sieburth, 1957; Evangelisti et al., 1975; Smith and Tucker, 1975). The logical next step would be to screen, isolate, and characterize presumptive antimicrobials from exclusive communities to demonstrate their effects on colonization.
Conclusion
The mode of action of competitive exclusion in reducing Salmonella in chickens appears to involve a combination of competition, attenuation, and antagonism by member species in the cecal community. Studies have demonstrated the importance of microbial community diversity in pathogen exclusion (recently reviewed in Pedroso et al., 2021). There have been many studies seeking to identify one or several intestinal species that inversely correlate with Salmonella abundance in the chicken intestine in an effort to describe a defined competitive exclusion formulation. Except for the Bacillus commercial products, no single species has been shown to directly affect Salmonella abundance in chickens across multiple studies. Even the competitive exclusion product Aviguard® exhibits considerable variability in community composition while retaining product efficacy for reducing Salmonella levels in chickens (Lee et al., 2023b). This diversity may reflect the multitude of intestinal species required to collectively affect Salmonella’s metabolic versatility and compete for the available resources in the intestine, reduce pathogenic behavior, or produce antimicrobials that inhibit Salmonella. Overlap of single functions may occur between different members of the collective community, rendering individual species dispensable. Alternatively, all three functions may be required for pathogen exclusion within the collective community, with a diversity of members necessary to achieve pathogen control.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by the University of Georgia Animal Care and Use and Procedures Committee. The study was conducted in accordance with the USDA and institutional requirements.
Author contributions
JM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Writing – original draft, Writing – review & editing. YC: Conceptualization, Formal analysis, Investigation, Methodology, Writing – review & editing. AP: Investigation, Methodology, Writing – review & editing. KT: Formal analysis, Investigation, Methodology, Writing – review & editing. SA: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. TK: Investigation, Methodology, Writing – review & editing. GM: Formal analysis, Supervision, Writing – review & editing. SK: Data curation, Formal analysis, Writing – review & editing. SP: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. MM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Writing – review & editing. JE-S: Methodology, Writing – review & editing. ML: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the United States Department of Agriculture (2005–01378, 2009–03561, and VA-160130). JE-S was supported by NIH grant R35-GM130399. Carbohydrate analysis was supported by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, U.S. Department of Energy grant (DE-SC0015662) to Parastoo Azadi at the University of Georgia Complex Carbohydrate Research Center.
Acknowledgments
We wish to thank Beatrice Kolwaski’s assistance with measuring the growth rate of the Salmonella reporter strain YC1104.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1342887/full#supplementary-material
Footnotes
References
Alteri, C. J., Himpsl, S. D., Pickens, S. R., Lindner, J. R., Zora, J. S., Miller, J. E., et al. (2013). Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 9:e1003608. doi: 10.1371/journal.ppat.1003608
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Angelopoulou, A., Warda, A. K., Hill, C., and Ross, R. P. (2019). Non-antibiotic microbial solutions for bovine mastitis - live biotherapeutics, bacteriophage, and phage lysins. Crit. Rev. Microbiol. 45, 564–580. doi: 10.1080/1040841X.2019.1648381
Anonymous (2021). Salmonella outbreak linked to wild songbirds. Center for Disease Control and Prevention. Available at: https://www.cdc.gov/salmonella/typhimurium-04-21/index.html (Accessed April 29, 2021).
Azcarate-Peril, M. A., Butz, N., Cadenas, M. B., Koci, M., Ballou, A., Mendoza, M., et al. (2018). An attenuated Salmonella enterica Serovar typhimurium strain and Galacto-oligosaccharides accelerate clearance of Salmonella infections in poultry through modifications to the gut microbiome. Appl. Environ. Microbiol. 84:e02526-17. doi: 10.1128/AEM.02526-17
Babinszky, I. J., Tossenberger, K. N., and Halas, V. (2006). Determination of amino acid distibility with different methods in birds (a comparative study). Slovak J. Anim. Sci. 39, 74–78.
Ballou, A. L., Ali, R. A., Mendoza, M. A., Ellis, J. C., Hassan, H. M., Croom, W. J., et al. (2016). Development of the Chick microbiome: how early exposure influences future microbial diversity. Front. Vet. Sci. 3:2. doi: 10.3389/fvets.2016.00002
Barrow, P. A., and Methner, U. (2013). Salmonella in domestic animals. Wallingford, United Kingdom: CABI.
Barrow, P. A., Tucker, J. F., and Simpson, J. M. (1987). Inhibition of colonization of the chicken alimentary tract with Salmonella Typhimurium gram-negative facultatively anaerobic bacteria. Epidemiol. Infect. 98, 311–322. doi: 10.1017/S0950268800062063
Bartlett, M. S., and Gourse, R. L. (1994). Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J. Bacteriol. 176, 5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994
Beck, J. R., and Chang, T. S. (1980). Measurement of uric acid levels in chicken cecal contents. Poult. Sci. 59, 1193–1196. doi: 10.3382/ps.0591193
Behnken, S., and Hertweck, C. (2012a). Anaerobic bacteria as producers of antibiotics. Appl. Microbiol. Biotechnol. 96, 61–67. doi: 10.1007/s00253-012-4285-8
Behnken, S., and Hertweck, C. (2012b). Cryptic polyketide synthase genes in non-pathogenic Clostridium SPP. PLoS One 7:e29609. doi: 10.1371/journal.pone.0029609
Benoit, S. L., Maier, R. J., Sawers, R. G., and Greening, C. (2020). Molecular hydrogen metabolism: a widespread trait of pathogenic Bacteria and Protists. Microbiol. Mol. Biol. Rev. 84:e00092-19. doi: 10.1128/MMBR.00092-19
Bhogoju, S., Nahashon, S., Wang, X., Darris, C., and Kilonzo-Nthenge, A. (2018). A comparative analysis of microbial profile of Guinea fowl and chicken using metagenomic approach. PLoS One 13:e0191029. doi: 10.1371/journal.pone.0191029
Bochner, B. R., Huang, H. C., Schieven, G. L., and Ames, B. N. (1980). Positive selection for loss of tetracycline resistance. J. Bacteriol. 143, 926–933. doi: 10.1128/jb.143.2.926-933.1980
Bohnhoff, M., Miller, C. P., and Martin, W. R. (1964a). Resistance of the mouse's intestinal tract to experimental Salmonella infection. I. Factors which interfere with the initiation of infection by oral inoculation. J. Exp. Med. 120, 805–816. doi: 10.1084/jem.120.5.805
Bohnhoff, M., Miller, C. P., and Martin, W. R. (1964b). Resistance of the mouse's intestinal tract to experimental Salmonella infection. II. Factors responsible for its loss following streptomycin treatment. J. Exp. Med. 120, 817–828. doi: 10.1084/jem.120.5.817
Brinsmade, S. R., Paldon, T., and Escalante-Semerena, J. C. (2005). Minimal functions and physiological conditions required for growth of salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 187, 8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005
Bythwood, T. N., Soni, V., Lyons, K., Hurley-Bacon, A., Lee, M. D., Hofacre, C., et al. (2019). Antimicrobial resistant Salmonella enterica typhimurium colonizing chickens: the impact of plasmids, genotype, bacterial communities, and antibiotic administration on resistance. Front. Sustain. Food Syst. 3:20. doi: 10.3389/fsufs.2019.00020
Cheng, Y., Pedroso, A. A., Porwollik, S., Mcclelland, M., Lee, M. D., Kwan, T., et al. (2015). rpoS-regulated core genes involved in the competitive fitness of Salmonella enterica Serovar Kentucky in the intestines of chickens. Appl. Environ. Microbiol. 81, 502–514. doi: 10.1128/AEM.03219-14
Choi, J. H., Kim, G. B., and Cha, C. J. (2014). Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult. Sci. 93, 1942–1950. doi: 10.3382/ps.2014-03974
Chopyk, J., Moore, R. M., Dispirito, Z., Stromberg, Z. R., Lewis, G. L., Renter, D. G., et al. (2016). Presence of pathogenic Escherichia coli is correlated with bacterial community diversity and composition on pre-harvest cattle hides. Microbiome 4, 1–11. doi: 10.1186/s40168-016-0155-4
Collado, M. C., Gueimonde, M., Hernández, M., Sanz, Y., and Salminen, S. (2005). Adhesion of selected Bifidobacterium strains to human intestinal mucus and the role of adhesion in Enteropathogen exclusion. J. Food Prot. 68, 2672–2678. doi: 10.4315/0362-028X-68.12.2672
Curtiss, R., and Kelly, S. M. (1987). Salmonella Typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect. Immun. 55, 3035–3043. doi: 10.1128/iai.55.12.3035-3043.1987
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645. doi: 10.1073/pnas.120163297
Demain, A. L., and Solomon, N. A. (1986). Manual of industrial microbiology and biotechnology. Washington DC, United States: American Society for Microbiology.
Ding, J., Zhou, H., Luo, L., Xiao, L., Yang, K., Yang, L., et al. (2021). Heritable gut microbiome associated with Salmonella enterica Serovar Pullorum infection in chickens. mSystems 6:e01192-20. doi: 10.1128/mSystems.01192-20
Dower, W. J., Miller, J. F., and Ragsdale, C. W. (1988). High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16, 6127–6145. doi: 10.1093/nar/16.13.6127
Drepper, T., Eggert, T., Circolone, F., Heck, A., Krauss, U., Guterl, J. K., et al. (2007). Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25, 443–445. doi: 10.1038/nbt1293
Ebana, Y., and Furukawa, T. (2019). Networking analysis on superior vena cava arrhythmogenicity in atrial fibrillation. Int J Cardiol Heart Vasc 22, 150–153. doi: 10.1016/j.ijcha.2019.01.007
Ekmekci, B., Mcanany, C. E., and Mura, C. (2016). An introduction to programming for bioscientists: a Python-based primer. PLoS Comput. Biol. 12:e1004867. doi: 10.1371/journal.pcbi.1004867
Ernst, R. K., Dombroski, D. M., and Merrick, J. M. (1990). Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella Typhimurium. Infect. Immun. 58, 2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990
Evangelisti, D., English, A., Girard, A., Lynch, J., and Solomons, I. (1975). Influence of subtherapeutic levels of oxytetracycline on Salmonella Typhimurium in swine, calves, and chickens. Antimicrob. Agents Chemother. 8, 664–672. doi: 10.1128/AAC.8.6.664
Faber, F., Thiennimitr, P., Spiga, L., Byndloss, M. X., Litvak, Y., Lawhon, S., et al. (2017). Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog. 13:e1006129. doi: 10.1371/journal.ppat.1006129
Falardeau, J., Yildiz, E., Yan, Y., Castellarin, S. D., and Wang, S. (2023). Microbiome and physicochemical features associated with differential Listeria monocytogenes growth in soft, surface-ripened cheeses. Appl. Environ. Microbiol. 89:e0200422. doi: 10.1128/aem.02004-22
Fink, R. C., Evans, M. R., Porwollik, S., Vazquez-Torres, A., Jones-Carson, J., Troxell, B., et al. (2007). FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar typhimurium (ATCC 14028s). J. Bacteriol. 189, 2262–2273. doi: 10.1128/JB.00726-06
Friedman, N., Goldszmidt, M., and Wyner, A. (2013). Data analysis with Bayesian networks: A bootstrap approach. arXiv preprint arXiv:1301.6695.
Fritz, S., Rajaonison, A., Chabrol, O., Raoult, D., Rolain, J. M., and Merhej, V. (2018). Full-length title: NRPPUR database search and in vitro analysis identify an NRPS-PKS biosynthetic gene cluster with a potential antibiotic effect. BMC Bioinform. 19:463. doi: 10.1186/s12859-018-2479-5
Fujikawa, H. (2016). Prediction of competitive microbial growth. Biocontrol Sci. 21, 215–223. doi: 10.4265/bio.21.215
Gast, R. K., and Beard, C. W. (1989). Age-related changes in the persistence and pathogenicity of Salmonella Typhimurium in chicks. Poult. Sci. 68, 1454–1460. doi: 10.3382/ps.0681454
Girinathan, B. P., Dibenedetto, N., Worley, J. N., Peltier, J., Arrieta-Ortiz, M. L., Immanuel, S. R. C., et al. (2021). In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe 29, 1693–1708.e1697. doi: 10.1016/j.chom.2021.09.007
Grant, A., Gay, C. G., and Lillehoj, H. S. (2018). Bacillus spp. as direct-fed microbial antibiotic alternatives to enhance growth, immunity, and gut health in poultry. Avian Pathol. 47, 339–351. doi: 10.1080/03079457.2018.1464117
Gunasekera, A., Ebright, Y. W., and Ebright, R. H. (1992). DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J. Biol. Chem. 267, 14713–14720. doi: 10.1016/S0021-9258(18)42099-6
Gunn, J. S. (2008). The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16, 284–290. doi: 10.1016/j.tim.2008.03.007
Gusils, C., Oppezzo, O., Pizarro, R., and González, S. (2003). Adhesion of probiotic lactobacilli to chick intestinal mucus. Can. J. Microbiol. 49, 472–478. doi: 10.1139/w03-055
Ha, S. D., Ricke, S. C., Nisbet, D. J., Corrier, D. E., and Deloach, J. R. (1994). Serine utilization as a potential competition mechanism between Salmonella and a chicken Cecal bacterium. J. Food Prot. 57, 1074–1079. doi: 10.4315/0362-028X-57.12.1074
Hammarlund, S. P., Gedeon, T., Carlson, R. P., and Harcombe, W. R. (2021). Limitation by a shared mutualist promotes coexistence of multiple competing partners. Nat. Commun. 12:619. doi: 10.1038/s41467-021-20922-0
Harvey, P. C., Watson, M., Hulme, S., Jones, M. A., Lovell, M., Berchieri, A. Jr., et al. (2011). Salmonella enterica serovar typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect. Immun. 79, 4105–4121. doi: 10.1128/IAI.01390-10
Heim, R., Prasher, D. C., and Tsien, R. Y. (1994). Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 91, 12501–12504. doi: 10.1073/pnas.91.26.12501
Heim, R., and Tsien, R. Y. (1996). Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6, 178–182. doi: 10.1016/S0960-9822(02)00450-5
Hinton, A., Spates, G. E., Corrier, D. E., Hume, M. E., Deloach, J. R., and Scanlan, C. M. (1991). In vitro inhibition of the growth of Salmonella Typhimurium and Escherichia coli 0157:H7 by Bacteria isolated from the Cecal contents of adult chickens. J. Food Prot. 54, 496–501. doi: 10.4315/0362-028X-54.7.496
Hirn, J., Nurmi, E., Johansson, T., and Nuotio, L. (1992). Long-term experience with competitive exclusion and salmonellas in Finland. Int. J. Food Microbiol. 15, 281–285. doi: 10.1016/0168-1605(92)90059-C
Hofacre, C. L., Froyman, R., Gautrias, B., George, B., Goodwin, M. A., and Brown, J. (1998). Use of Aviguard and other intestinal bioproducts in experimental Clostridium perfringens-associated necrotizing enteritis in broiler chickens. Avian Dis. 42, 579–584. doi: 10.2307/1592685
Hofacre, C. L., Primm, N. D., Vance, K., Goodwin, M. A., and Brown, J. (2000). Comparison of a lyophilized chicken-origin competitive exclusion culture, a lyophilized probiotic, and fresh Turkey Cecal material against Salmonella colonization. J. Appl. Poult. Res. 9, 195–203. doi: 10.1093/japr/9.2.195
Hue, O., Allain, V., Laisney, M. J., Le Bouquin, S., Lalande, F., Petetin, I., et al. (2011). Campylobacter contamination of broiler caeca and carcasses at the slaughterhouse and correlation with Salmonella contamination. Food Microbiol. 28, 862–868. doi: 10.1016/j.fm.2010.11.003
Jeter, R. M. (1990). Cobalamin-dependent 1,2-propanediol utilization by Salmonella Typhimurium. J. Gen. Microbiol. 136, 887–896. doi: 10.1099/00221287-136-5-887
Jones, R. M., Wu, H., Wentworth, C., Luo, L., Collier-Hyams, L., and Neish, A. S. (2008). Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe 3, 233–244. doi: 10.1016/j.chom.2008.02.016
Józefiak, D., Rutkowski, A., and Martin, S. (2004). Carbohydrate fermentation in the avian ceca: a review. Anim. Feed Sci. Technol. 113, 1–15. doi: 10.1016/j.anifeedsci.2003.09.007
Keegan, K. P., Glass, E. M., and Meyer, F. (2016). MG-RAST, a metagenomics Service for Analysis of microbial community structure and function. Methods Mol. Biol. 1399, 207–233. doi: 10.1007/978-1-4939-3369-3_13
Khan, S., Moore, R. J., Stanley, D., and Chousalkar, K. K. (2020). The gut microbiota of laying hens and its manipulation with prebiotics and probiotics to enhance gut health and food safety. Appl. Environ. Microbiol. 86:e00600-20. doi: 10.1128/AEM.00600-20
Kohli, N., Crisp, Z., Riordan, R., Li, M., Alaniz, R. C., and Jayaraman, A. (2018). The microbiota metabolite indole inhibits Salmonella virulence: involvement of the PhoPQ two-component system. PLoS One 13:e0190613. doi: 10.1371/journal.pone.0190613
Kubasova, T., Kollarcikova, M., Crhanova, M., Karasova, D., Cejkova, D., Sebkova, A., et al. (2019). Gut anaerobes capable of chicken Caecum colonisation. Microorganisms 7:597. doi: 10.3390/microorganisms7120597
Kwan, G., Plagenz, B., Cowles, K., Pisithkul, T., Amador-Noguez, D., and Barak, J. D. (2018). Few differences in metabolic network use found between Salmonella enterica colonization of plants and Typhoidal mice. Front. Microbiol. 9:695. doi: 10.3389/fmicb.2018.00695
Kwon, Y., and Ricke, S. (1999). Salmonella Typhimurium poultry isolate growth response to propionic acid and sodium propionate under aerobic and anaerobic conditions. Int. Biodeterior. Biodegradation 43, 161–165. doi: 10.1016/S0964-8305(99)00045-1
Lagani, V., and Tsamardinos, I. (2010). Structure-based variable selection for survival data. Bioinformatics 26, 1887–1894. doi: 10.1093/bioinformatics/btq261
Latorre, J. D., Hernandez-Velasco, X., Wolfenden, R. E., Vicente, J. L., Wolfenden, A. D., Menconi, A., et al. (2016). Evaluation and selection of Bacillus species based on enzyme production, antimicrobial activity, and biofilm synthesis as direct-fed microbial candidates for poultry. Front. Vet. Sci. 3:95. doi: 10.3389/fvets.2016.00095
Le Bihan, G., Jubelin, G., Garneau, P., Bernalier-Donadille, A., Martin, C., Beaudry, F., et al. (2015). Transcriptome analysis of Escherichia coli O157:H7 grown in vitro in the sterile-filtrated cecal content of human gut microbiota associated rats reveals an adaptive expression of metabolic and virulence genes. Microbes Infect. 17, 23–33. doi: 10.1016/j.micinf.2014.09.008
Lee, C. A., and Falkow, S. (1990). The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. U. S. A. 87, 4304–4308. doi: 10.1073/pnas.87.11.4304
Lee, M. D., Pedroso, A. A., Lumpkins, B., Cho, Y., and Maurer, J. J. (2023a). Pioneer colonizers: Bacteria that alter the chicken intestinal morphology and development of the microbiota. Front. Physiol. 14:1139321. doi: 10.3389/fphys.2023.1139321
Lee, M. D., Pedroso, A. A., and Maurer, J. J. (2023b). Bacterial composition of a competitive exclusion product and its correlation with product efficacy at reducing Salmonella in poultry. Front. Physiol. 13:1043383. doi: 10.3389/fphys.2022.1043383
Li, J. S., Barber, C. C., Herman, N. A., Cai, W., Zafrir, E., Du, Y., et al. (2020). Investigation of secondary metabolism in the industrial butanol hyper-producer Clostridium saccharoperbutylacetonicum N1-4. J. Ind. Microbiol. Biotechnol. 47, 319–328. doi: 10.1007/s10295-020-02266-8
Liao, A. P., Petrof, E. O., Kuppireddi, S., Zhao, Y., Xia, Y., Claud, E. C., et al. (2008). Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PLoS One 3:e2369. doi: 10.1371/journal.pone.0002369
Lone, A. G., Selinger, L. B., Uwiera, R. R., Xu, Y., and Inglis, G. D. (2013). Campylobacter jejuni colonization is associated with a dysbiosis in the cecal microbiota of mice in the absence of prominent inflammation. PLoS One 8:e75325. doi: 10.1371/journal.pone.0075325
Lu, J., Idris, U., Harmon, B., Hofacre, C., Maurer, J. J., and Lee, M. D. (2003). Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69, 6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003
Lu, H., Yang, P., Zhong, M., Bilal, M., Xu, H., Zhang, Q., et al. (2023). Isolation of a potential probiotic strain Bacillus amyloliquefaciensLPB-18 and identification of antimicrobial compounds responsible for inhibition of food-borne pathogens. Food Sci. Nutr. 11, 2186–2196. doi: 10.1002/fsn3.3094
Maltby, R., Leatham-Jensen, M. P., Gibson, T., Cohen, P. S., and Conway, T. (2013). Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One 8:e53957. doi: 10.1371/journal.pone.0053957
Matamouros, S., and Miller, S. I. (2015). S. typhimurium strategies to resist killing by cationic antimicrobial peptides. Biochim. Biophys. Acta 1848, 3021–3025. doi: 10.1016/j.bbamem.2015.01.013
Miller, V. L., and Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988
Milona, P., Townes, C. L., Bevan, R. M., and Hall, J. (2007). The chicken host peptides, gallinacins 4, 7, and 9 have antimicrobial activity against Salmonella serovars. Biochem. Biophys. Res. Commun. 356, 169–174. doi: 10.1016/j.bbrc.2007.02.098
Miyashiro, T., and Goulian, M. (2007). Stimulus-dependent differential regulation in the Escherichia coli PhoQ PhoP system. Proc. Natl. Acad. Sci. USA 104, 16305–16310. doi: 10.1073/pnas.0700025104
Nair, D. V. T., Johnson, T. J., Noll, S. L., and Kollanoor Johny, A. (2021). Effect of supplementation of a dairy-originated probiotic bacterium, Propionibacterium freudenreichii subsp. freudenreichii, on the cecal microbiome of turkeys challenged with multidrug-resistant Salmonella Heidelberg. Poult. Sci. 100, 283–295. doi: 10.1016/j.psj.2020.09.091
Nurmi, E., Nuotio, L., and Schneitz, C. (1992). The competitive exclusion concept: development and future. Int. J. Food Microbiol. 15, 237–240. doi: 10.1016/0168-1605(92)90054-7
Nurmi, E., and Rantala, M. (1973). New aspects of Salmonella infection in broiler production. Nature 241, 210–211. doi: 10.1038/241210a0
Obradors, N., Badía, J., Baldomà, L., and Aguilar, J. (1988). Anaerobic metabolism of the L-rhamnose fermentation product 1,2-propanediol in Salmonella Typhimurium. J. Bacteriol. 170, 2159–2162. doi: 10.1128/jb.170.5.2159-2162.1988
Olishevska, S., Nickzad, A., and Déziel, E. (2019). Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl. Microbiol. Biotechnol. 103, 1189–1215. doi: 10.1007/s00253-018-9541-0
Pan, X., Wu, T., Zhang, L., Song, Z., Tang, H., and Zhao, Z. (2008). In vitro evaluation on adherence and antimicrobial properties of a candidate probiotic Clostridium butyricum CB2 for farmed fish. J. Appl. Microbiol. 105, 1623–1629. doi: 10.1111/j.1365-2672.2008.03885.x
Parish, J. H. (1985) in Biochemistry of bacterial growth. eds. J. Mandelstam, K. McQuillen, and I. Dawes. 3rd ed (Oxford: Blackwell Scientific Publications)
Pascual, M., Hugas, M., Badiola, J. I., Monfort, J. M., and Garriga, M. (1999). Lactobacillus salivarius CTC2197 prevents Salmonella enteritidis colonization in chickens. Appl. Environ. Microbiol. 65, 4981–4986. doi: 10.1128/AEM.65.11.4981-4986.1999
Pedroso, A. A., Batal, A. B., and Lee, M. D. (2016). Effect of in ovo administration of an adult-derived microbiota on establishment of the intestinal microbiome in chickens. Am. J. Vet. Res. 77, 514–526. doi: 10.2460/ajvr.77.5.514
Pedroso, A. A., Lee, M. D., and Maurer, J. J. (2021). Strength lies in diversity: how community diversity limits Salmonella abundance in the chicken intestine. Front. Microbiol. 12:694215. doi: 10.3389/fmicb.2021.694215
Peighambari, S. M., Hunter, D. B., Shewen, P. E., and Gyles, C. L. (2002). Safety, immunogenicity, and efficacy of two Escherichia coli cya crp mutants as vaccines for broilers. Avian Dis. 46, 287–297. doi: 10.1637/0005-2086(2002)046[0287:SIAEOT]2.0.CO;2
Pennie, R. A., Zunino, J. N., Rose, C. E. Jr., and Guerrant, R. L. (1984). Economical, simple method for production of the gaseous environment required for cultivation of Campylobacter jejuni. J. Clin. Microbiol. 20, 320–322. doi: 10.1128/jcm.20.3.320-322.1984
Podnar, E., Erega, A., Danevčič, T., Kovačec, E., Lories, B., Steenackers, H., et al. (2022). Nutrient availability and biofilm polysaccharide shape the Bacillaene-dependent antagonism of Bacillus subtilis against Salmonella Typhimurium. Microbiol. Spectr. 10:e0183622. doi: 10.1128/spectrum.01836-22
Pottenger, S., Watts, A., Wedley, A., Jopson, S., Darby, A. C., and Wigley, P. (2023). Timing and delivery route effects of cecal microbiome transplants on Salmonella Typhimurium infections in chickens: potential for in-hatchery delivery of microbial interventions. Anim. Microbiome 5:11. doi: 10.1186/s42523-023-00232-0
Provence, D. L., And Curtiss Iii, R. (1994). Gene transfer in gram-negative bacteria. Washington, DC: ASM Press.
Rogers, A. W. L., Tsolis, R. M., and Bäumler, A. J. (2021). Salmonella versus the microbiome. Microbiol. Mol. Biol. Rev. 85:e00027-19. doi: 10.1128/MMBR.00027-19
Royal, W. A., and Mutimer, M. D. (1972). Inhibition of Salmonella Typhimurium by fowl Caecal cultures. Res. Vet. Sci. 13, 184–185. doi: 10.1016/S0034-5288(18)34069-4
Saenz, C., Fang, Q., Gnanasekaran, T., Trammell, S. A. J., Buijink, J. A., Pisano, P., et al. (2023). Clostridium scindens secretome suppresses virulence gene expression of Clostridioides difficile in a bile acid-independent manner. Microbiol. Spectr. 11:e0393322. doi: 10.1128/spectrum.03933-22
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Santander, J., Martin, T., Loh, A., Pohlenz, C., Gatlin, D. M., and Curtiss, R. (2013). Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology 159, 1471–1486. doi: 10.1099/mic.0.066639-0
Schneitz, C., Nuotio, L., Kiiskinen, T., and Nurmi, E. (1991). Pilot-scale testing of the competitive exclusion method in chickens. Br. Poult. Sci. 32, 881–884. doi: 10.1080/00071669108417414
Scutari, M., and Denis, J.-B. (2021). Bayesian networks: with examples in R. Boca Raton, Florida, United States: CRC Press.
Scutari, M., Vitolo, C., and Tucker, A. (2019). Learning Bayesian networks from big data with greedy search: computational complexity and efficient implementation. Stat. Comput. 29, 1095–1108. doi: 10.1007/s11222-019-09857-1
Seedorf, H., Fricke, W. F., Veith, B., Brüggemann, H., Liesegang, H., Strittmatter, A., et al. (2008). The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. U. S. A. 105, 2128–2133. doi: 10.1073/pnas.0711093105
Shelton, C. D., Yoo, W., Shealy, N. G., Torres, T. P., Zieba, J. K., Calcutt, M. W., et al. (2022). Salmonella enterica serovar Typhimurium uses anaerobic respiration to overcome propionate-mediated colonization resistance. Cell Rep. 38:110180. doi: 10.1016/j.celrep.2021.110180
Sibinelli-Sousa, S., De Araújo-Silva, A. L., Hespanhol, J. T., and Bayer-Santos, E. (2022). Revisiting the steps of Salmonella gut infection with a focus on antagonistic interbacterial interactions. FEBS J. 289, 4192–4211. doi: 10.1111/febs.16211
Sieburth, J. M. (1957). The effect of Furazolidone on the cultural and serological response of Salmonella typhi-murium infected chickens. Avian Dis. 1, 180–194. doi: 10.2307/1587730
Singh, M., Chaudhary, S., and Sareen, D. (2017). Non-ribosomal peptide synthetases: identifying the cryptic gene clusters and decoding the natural product. J. Biosci. 42, 175–187. doi: 10.1007/s12038-017-9663-z
Smith, H. W., and Tucker, J. (1975). The effect of antibiotic therapy on the faecal excretion of Salmonella Typhimurium by experimentally infected chickens. Epidemiol. Infect. 75, 275–292. doi: 10.1017/S0022172400047306
Snoeyenbos, G. H., Soerjadi, A. S., and Weinack, O. M. (1982). Gastrointestinal colonization by salmonellae and pathogenic Escherichia coli in monoxenic and holoxenic chicks and poults. Avian Dis. 26, 566–575. doi: 10.2307/1589903
Song, W., Qiu, L., Qing, J., Zhi, W., Zha, Z., Hu, X., et al. (2022). Using Bayesian network model with MMHC algorithm to detect risk factors for stroke. Math. Biosci. Eng. 19, 13660–13674. doi: 10.3934/mbe.2022637
Stanley, D., Wu, S. B., Rodgers, N., Swick, R. A., and Moore, R. J. (2014). Differential responses of cecal microbiota to fishmeal, Eimeria and Clostridium perfringens in a necrotic enteritis challenge model in chickens. PLoS One 9:e104739. doi: 10.1371/journal.pone.0104739
Stenkamp-Strahm, C., Mcconnel, C., Magzamen, S., Abdo, Z., and Reynolds, S. (2018). Associations between Escherichia coli O157 shedding and the faecal microbiota of dairy cows. J. Appl. Microbiol. 124, 881–898. doi: 10.1111/jam.13679
Sun, W., Roland, K. L., Kuang, X., Branger, C. G., and Curtiss, R. (2010). Yersinia pestis with regulated delayed attenuation as a vaccine candidate to induce protective immunity against plague. Infect. Immun. 78, 1304–1313. doi: 10.1128/IAI.01122-09
Suzuki, M. T., Taylor, L. T., and Delong, E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66, 4605–4614. doi: 10.1128/AEM.66.11.4605-4614.2000
Troxell, B., Fink, R. C., Porwollik, S., Mcclelland, M., and Hassan, H. M. (2011). The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol. 11:236. doi: 10.1186/1471-2180-11-236
Ushijima, T., and Seto, A. (1991). Selected faecal bacteria and nutrients essential for antagonism of Salmonella Typhimurium in anaerobic continuous flow cultures. J. Med. Microbiol. 35, 111–117. doi: 10.1099/00222615-35-2-111
Valeris-Chacin, R., Weber, B., Johnson, T. J., Pieters, M., and Singer, R. S. (2022). Longitudinal changes in Campylobacter and the litter microbiome throughout the broiler production cycle. Appl. Environ. Microbiol. 88:e0066722. doi: 10.1128/aem.00667-22
Wang, M., Hu, J., Yu, H., Li, W., He, G., Dong, J., et al. (2022). Lactobacillus fermentum 1.2133 display probiotic potential in vitro and protect against Salmonella pullorum in chicken of infection. Lett. Appl. Microbiol. 76:ovac041. doi: 10.1093/lambio/ovac041
Watkins, B. A., and Miller, B. F. (1983). Competitive gut exclusion of avian pathogens by Lactobacillus acidophilus in gnotobiotic chicks. Poult. Sci. 62, 1772–1779. doi: 10.3382/ps.0621772
Weschka, D., Mousavi, S., Biesemeier, N., Bereswill, S., and Heimesaat, M. M. (2021). Survey of pathogen-lowering and Immuno-modulatory effects upon treatment of Campylobacter coli-infected secondary abiotic IL-10(−/−) mice with the probiotic formulation Aviguard®. Microorganisms 9:1127. doi: 10.3390/microorganisms9061127
Wierup, M., Wold-Troell, M., Nurmi, E., and Häkkinen, M. (1988). Epidemiological evaluation of the Salmonella-controlling effect of a Nationwide use of a competitive exclusion culture in poultry. Poult. Sci. 67, 1026–1033. doi: 10.3382/ps.0671026
Wong, G. T., Bonocora, R. P., Schep, A. N., Beeler, S. M., Lee Fong, A. J., Shull, L. M., et al. (2017). Genome-wide transcriptional response to varying RpoS levels in Escherichia coli K-12. J. Bacteriol. 199:e00755-16. doi: 10.1128/JB.00755-16
Wooley, R. E., Gibbs, P. S., and Shotts, E. B. Jr. (1999). Inhibition of Salmonella Typhimurium in the chicken intestinal tract by a transformed avirulent avian Escherichia coli. Avian Dis. 43, 245–250. doi: 10.2307/1592614
Xiao, Y., Xiang, Y., Zhou, W., Chen, J., Li, K., and Yang, H. (2017). Microbial community mapping in intestinal tract of broiler chicken. Poult. Sci. 96, 1387–1393. doi: 10.3382/ps/pew372
Yang, H. J., Bogomolnaya, L. M., Elfenbein, J. R., Endicott-Yazdani, T., Reynolds, M. M., Porwollik, S., et al. (2016). Novel two-step hierarchical screening of mutant pools reveals mutants under selection in chicks. Infect. Immun. 84, 1226–1238. doi: 10.1128/IAI.01525-15
Yang, H. J., Bogomolnaya, L., Mcclelland, M., and Andrews-Polymenis, H. (2017). De novo pyrimidine synthesis is necessary for intestinal colonization of Salmonella Typhimurium in chicks. PLoS One 12:e0183751. doi: 10.1371/journal.pone.0189541
Yu, H., Campbell, M. T., Zhang, Q., Walia, H., and Morota, G. (2019). Genomic Bayesian confirmatory factor analysis and Bayesian network to characterize a wide Spectrum of Rice phenotypes. G3 9, 1975–1986. doi: 10.1534/g3.119.400154
Yu, Z., Chen, J., Liu, Y., Meng, Q., Liu, H., Yao, Q., et al. (2023). The role of potential probiotic strains Lactobacillus reuteri in various intestinal diseases: new roles for an old player. Front. Microbiol. 14:1095555. doi: 10.3389/fmicb.2023.1095555
Zhang, S., Dogan, B., Guo, C., Herlekar, D., Stewart, K., Scherl, E. J., et al. (2020). Short chain fatty acids modulate the growth and virulence of Pathosymbiont Escherichia coli and host response. Antibiotics 9:462. doi: 10.3390/antibiotics9080462
Zhang, L., Dong, D., Jiang, C., Li, Z., Wang, X., and Peng, Y. (2015). Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe 34, 1–7. doi: 10.1016/j.anaerobe.2015.03.008
Zhang, Z. J., Pedicord, V. A., Peng, T., and Hang, H. C. (2020). Site-specific acylation of a bacterial virulence regulator attenuates infection. Nat. Chem. Biol. 16, 95–103. doi: 10.1038/s41589-019-0392-5
Zhao, T., Podtburg, T. C., Zhao, P., Schmidt, B. E., Baker, D. A., Cords, B., et al. (2006). Control of Listeria spp. by competitive-exclusion bacteria in floor drains of a poultry processing plant. Appl. Environ. Microbiol. 72, 3314–3320. doi: 10.1128/AEM.72.5.3314-3320.2006
Zhou, P., Han, X., Ye, X., Zheng, F., Yan, T., Xie, Q., et al. (2020). Phenotype, virulence and immunogenicity of Edwardsiella piscicida cyclic AMP receptor protein (Crp) mutants in catfish host. Microorganisms 8:517. doi: 10.3390/microorganisms8040517
Keywords: pathogen, exclusion, Salmonella, antimicrobials, competition, attenuation
Citation: Maurer JJ, Cheng Y, Pedroso A, Thompson KK, Akter S, Kwan T, Morota G, Kinstler S, Porwollik S, McClelland M, Escalante-Semerena JC and Lee MD (2024) Peeling back the many layers of competitive exclusion. Front. Microbiol. 15:1342887. doi: 10.3389/fmicb.2024.1342887
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
Na Zhang, Harbin University of Commerce, ChinaSundus Javed, COMSATS University, Islamabad Campus, Pakistan
Copyright © 2024 Maurer, Cheng, Pedroso, Thompson, Akter, Kwan, Morota, Kinstler, Porwollik, McClelland, Escalante-Semerena and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margie D. Lee, mlee2@vt.edu
 John J. Maurer
John J. Maurer Ying Cheng2
Ying Cheng2 Shamima Akter
Shamima Akter Tiffany Kwan
Tiffany Kwan Gota Morota
Gota Morota Sydney Kinstler
Sydney Kinstler Michael McClelland
Michael McClelland Margie D. Lee
Margie D. Lee