- 1Key Laboratory of Agricultural Products Processing and Food Safety in Hunan Higher Education, Hunan Provincial Engineering Research Center for Fresh Wet Rice Noodles, Science and Technology Innovation Team for Efficient Agricultural Production and Deep Processing at General University in Hunan Province, Hunan Provincial Key Laboratory for Health Aquaculture and Product Processing in Dongting Lake Area, Hunan Provincial Key Laboratory for Molecular Immunity Technology of Aquatic Animal Diseases, State Key Laboratory of Developmental Biology of Freshwater Fish, College of Life and Environmental Sciences, Hunan University of Arts and Science, Changde, Hunan, China
- 2School of Life and Health Sciences, Hunan University of Science and Technology, Xiangtan, Hunan, China
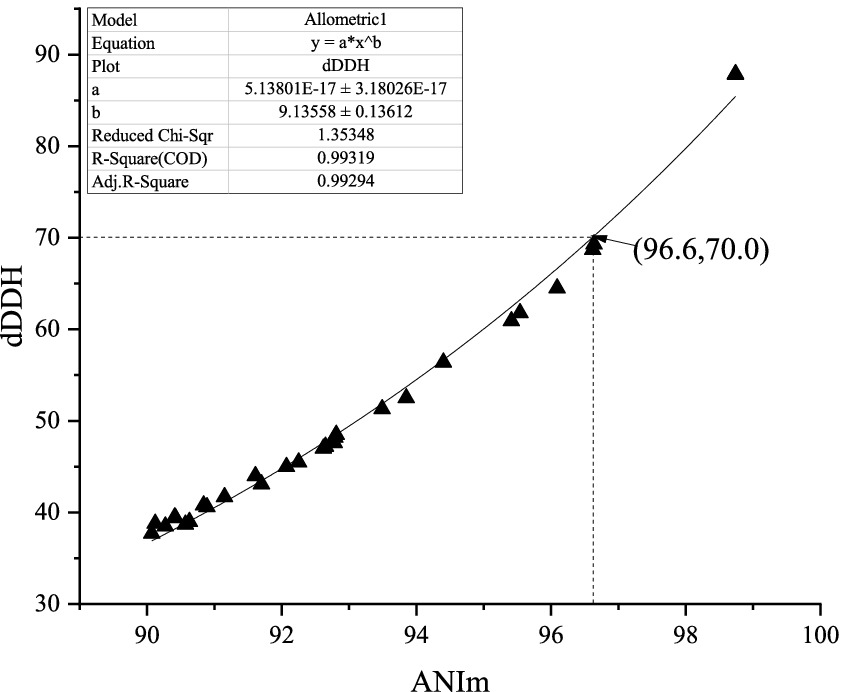
At present, it is widely believed that a 95–96% average nucleotide identity (ANI) value is equivalent to a 70% digital DNA–DNA hybridization (dDDH) value in the prokaryotic taxonomy. However, in the present study, comparative genome analysis of 29 pairs of Amycolatopsis type strains revealed that a 70% dDDH value did not correspond to a 95–96% ANI based on the MuMmer ultra-rapid aligning tool (ANIm) but approximately corresponded to a 96.6% ANIm value in the genus Amycolatopsis. Based on this corresponding relationship, phenotypic and chemotaxonomical characteristics, as well as phylogenetic analysis, an actinobacterial strain HUAS 11-8T isolated from the rhizosphere soil of Cynara scolymus, was subjected to a polyphasic taxonomic characterization. Based on EzBioCloud alignment, it was found that strain HUAS11-8T had the 16S rRNA gene similarities of 99.78% with A. rhizosphaerae JCM 32589T, 97.8% with A. dongchuanensis YIM 75904T, and < 97.8% sequence similarities to other Amycolatopsis species. Phylogenetic analysis of 16S rRNA gene sequences and whole-genome sequences revealed that strain HUAS 11-8T was closely related to A. rhizosphaerae JCM 32589T. ANIm and dDDH values between strains HUAS 11-8T and A. rhizosphaerae JCM 32589T were 96.3 and 68.5%, respectively, lower than the 96.6 and 70% thresholds recommended for the delineation of a novel Amycolatopsis species. Consequently, strain HUAS 11-8T should represent a novel Amycolatopsis species, for which the name Amycolatopsis cynarae sp. nov. (type strain HUAS 11-8T = MCCC 1K08337T = JCM 35980T) is proposed. Furthermore, based on comparative genomic analysis and rule 42 of the Prokaryotic Code, we propose that Amycolatopsis niigatensis is a later heterotypic synonym of Amycolatopsis echigonensis.
1 Introduction
The genus Amycolatopsis, belonging to the family Pseudonocardiaceae of the order Pseudonocardiales, was initially described using specific methods (Lechevalier et al., 1986) and then emended by Lee (2009) and Tang et al. (2010). At the time of writing, this genus included more than 80 species with validly published and correct names.1 The members of the genus Amycolatopsis are distributed in various environments such as peat swamp forest soil (Teo et al., 2020, 2021), deep-sea sediment (Zhang et al., 2016), arid soil (Tan et al., 2006; Zucchi et al., 2012; Busarakam et al., 2016), plant tissues (Wang et al., 2020; Tedsree et al., 2021), animals, and humans (Labeda et al., 2003; Huang et al., 2004). In the past 10 years, Amycolatopsis strains have gained widespread attention due to their potential in antibiotic production, bioremediation, bioconversion, and biodegradation processes (Dávila Costa and Amoroso, 2014; Adamek et al., 2018). Therefore, it is of great practical significance to search for Amycolatopsis strains, especially novel Amycolatopsis species.
Recently, in a survey on the diversity of actinobacteria from the rhizospheric soil of different plants, 100 s of strains were isolated using different media. Interestingly, the conclusions were contradictory if the taxonomic status of strain HUAS 11-8T, one of all those strains mentioned above, was described using the different classification criteria. In addition, in the course of analyzing the relatedness between average nucleotide identity based on the MuMmer ultra-rapid aligning tool (ANIm) and digital DNA–DNA hybridization (dDDH) in the genus Amycolatopsis, we found that Amycolatopsis niigatensis and Amycolatopsis echigonensis should be of the same genomic species. Thus, the main aims of the present study are to (1) elucidate the reasons for the abovementioned contradictory results, (2) evaluate the taxonomic status of strain HUAS 11-8T using a polyphasic taxonomic approach, and (3) clarify the taxonomic relation between A. niigatensis and A. echigonensis based on comparative genomic analysis.
2 Materials and methods
2.1 Genome data used to analyze the relationship between ANIm and dDDH values in the genus Amycolatopsis
A total of 29 genomes from type strains of Amycolatopsis species with validly published names were downloaded from the GenBank database. All genomes used in this study must meet the criteria of >95% completeness and < 5% contamination in order to obtain more reliable analysis results. The quality analysis and GenBank assembly of genomes are shown in Supplementary Table S1.
As Meier-Kolthoff and Göker (2019) proposed, when the ANI value between two genomes is more than 90%, ANIm can provide more credible results with respect to ANI based on the BLAST algorithm (ANIb). Thus, the ANIm value was selected for comparative analysis in this study. The ANIm and dDDH values were calculated by using the JSpeciesWS online service (Richter et al., 2016) and the Genome-to-Genome Distance Calculator with Formula 2 (Meier-Kolthoff et al., 2013), respectively. The coherence analysis between ANIm and dDDH values was performed using these methods (Hu et al., 2022).
2.2 Evaluation of the taxonomic status of strain HUAS 11-8T
2.2.1 Isolation and maintenance of strain HUAS 11-8T
Strain HUAS 11-8T was isolated from the rhizosphere soil of Cynara scolymus, which was collected in Changde city of Hunan Province, China (29.20201° N 111.98113° E), as described by Mo et al. (2017). The purified strain HUAS 11-8T was prepared for short-term preservation on Reasonerʼ2A (Reasoner and Geldreich, 1985) slopes at 4°C and suspended in a sterile 30% (w/v) glycerol solution for long-term conservation at −80°C. The type strain Amycolatopsis rhizosphaerae JCM 32589T was purchased from the Japan Collection of Microorganisms (JCM). Strains HUAS 11-8T and A. rhizosphaerae JCM 32589T were tested under the same conditions.
2.2.2 Genome sequencing and phylogenetic analysis
The genome sequencing of strain HUAS 11-8T was completed by using a Nanopore PromethION sequencing system at Wuhan Benagen Technology Co., Ltd. (Hubei, China). Genomic DNA extraction and PCR conditions of the 16S rRNA gene were carried out using the method described by Mo et al. (2023). The 16S rRNA gene of strain HUAS 11-8T was amplified using universal primers (27F and 1492R) (Lane, 1991). The 16S rRNA gene sequence of strain HUAS 11-8T was compared with the EzBioCloud database2 (Yoon et al., 2017). Closely related reference strains were downloaded and used for constructing phylogenetic trees using neighbor-joining (NJ) (Saitou and Nei, 1987), maximum-likelihood (ML) (Felsenstein, 1981), and maximum-parsimony (MP) (Kluge and Farris, 1969) with 1,000 bootstrap replications in MEGA 11 (Tamura et al., 2021). According to the result of 16S rRNA gene sequence analysis, the genome sequences of type strains that were closely related to strain HUAS 11-8T were selected for reconstructing the phylogenomic tree using the Type (Strain) Genome Server. The ANIm and dDDH values between the genomes of strain HUAS 11-8T and its relatives were calculated according to the aforementioned description. The gene prediction analysis and functional annotation of the genome of strain HUAS 11-8T were performed by the NCBI Prokaryotic Genome Annotation Pipeline v4.4 and Rapid Annotation using Subsystem Technology v.2.0 (RAST3) (Aziz et al., 2008). The secondary metabolism biosynthetic gene clusters of strain HUAS 11-8T and antibiotic resistance genes were analyzed using antiSMASH version 6.0.14 and the Comprehensive Antibiotic Resistance Database (CARD5), respectively (Blin et al., 2011; Alcock et al., 2020). Clustered regularly interspaced short palindromic repeat sequences (CRISPRs) of strain HUAS 11-8T were identified by CRISPR-case Finder6 (Makarova et al., 2019), and then the genomic islands were predicted by the Island Viewer 4 webserver (Claire et al., 2017).7
2.2.3 Morphological, cultural, and physio-biochemical characteristics
Spore features of HUAS 11-8T were observed by a light microscope (NE620, Ningbo Yongxin Optics) and scanning electron microscope (FEI-Quanta 450, America), using cultures grown on Reasonerʼ 2A after incubation for 21 days at 28°C. The cultural features of strains HUAS 11-8T and A. rhizosphaerae JCM 32589T were observed on various media, including Gauseʼs synthetic No. 1 medium (Atlas, 1993), Reasonerʼ 2A and ISP 2–7 media (Shirling and Gottlieb, 1966) for 21 days at 28°C. Color determinations, such as the aerial mycelium, the substrate mycelium, and diffusible soluble pigments, were delineated according to the methods of Shirling and Gottlieb (1966). Growth was carried out on tryptic soy broth (TSB) medium for 14 days at temperatures (4, 10, 15, 20, 22, 25, 27, 30, 35, 37, 40, 45, 50, and 55°C), pH (2.0–12.0, at intervals of 1.0 pH unit), and concentrations of NaCl (0–15%, w/v, at an interval of 1% w/v). The following tests, i.e., carbon and nitrogen source utilization, starch hydrolysis, gelatin liquefaction, nitrate reduction, and Tweens (20, 40, 60, and 80) degradation, were performed according to the methods described by Xu et al. (2007).
2.2.4 Chemotaxonomical characteristics
Biomass for chemotaxonomic analysis was collected by centrifugation after culturing for 5–7 days at 28°C in TSB in shake flasks. The cellular fatty acids of strains HUAS 11-8T and A. rhizosphaerae JCM 32589T were detected as described by Sherlock MIDI protocol (Sherlock Microbial Identification System, version 6.0B) (MIDI, 2005), which were carried out by the Marine Culture Collection of China (MCCC). The diaminopimelic acid isomers in the cell wall peptidoglycan were separated by thin-layer chromatography and analyzed using a solution of ninhydrin in acetone (Hasegawa et al., 1983). The whole-cell sugars were analyzed as described by Lechevalier and Lechevalier (1970). Polar lipids and menaquinones were extracted and analyzed using the method described by Ruan and Huang (2011).
3 Results and discussion
3.1 Relationship between ANIm and dDDH values in the genus Amycolatopsis
Over the past 50 years, the traditional DNA–DNA hybridization (DDH) technology has played a key role in the classification and identification of bacteria and archaea. However, there is a large amount of evidence that this technology has its limitations, such as being labor-intensive, error-prone, and difficult to generate cumulative databases. Thus, there has been an urgent need to look for an alternative standard (Stackebrandt et al., 2002; Gevers et al., 2005). At present, as alternative standards based on the genome level, ANI values of 95–96% and dDDH values of 70% have generally acted as a gold standard for bacterial species delineation (Richter and Rosselló-Móra, 2009; Auch et al., 2010). Nevertheless, during our search for novel actinobacteria capable of producing bioactive compounds, we found that strain HUAS 11-8T, one of all actinomycete strains isolated, should belong to a new Amycolatopsis species according to the 68.5% dDDH value between the strain and its relative, but this strain should belong to a known Amycolatopsis species according to the 96.3% ANIm value between the strain and its relative. What are the reasons for this contradictory phenomenon? To resolve this question, we first calculated the ANIm values between all validly published Amycolatopsis species whose genomes were available. Then, the dDDH values between all pairs of strains, whose ANIm values were ≥ 90%, were calculated for subsequent analysis. Finally, the dDDH and ANIm values of a total of 29 pairs of Amycolatopsis species (including synonyms) were randomly selected for correlation analysis (Supplementary Table S2). As shown in Figure 1, the dDDH value revealed an extremely high correlation (R2 = 0.99319) with the ANIm value based on an exponential regression model. However, a 70% dDDH value was not equivalent to a 95–96% ANIm value, but to an ANIm value of approximately 96.6%. Thus, the contradiction above can be well explained based on this corresponding relationship. At present, in addition to AINm, ANIb is also a mainstream ANI computing model. Then, how does dDDH correspond to ANIb in the genus Amycolatopsis? As shown in Supplementary Figure S1, a 70% dDDH value approximately corresponded to a 95.8% ANIb value, which is in the middle of 95–96%. Although the dDDH value also showed an extremely high correlation (>0.99) with the ANIb value based on an exponential regression model, this corresponding relationship did not well explain the contradictory phenomena above. Furthermore, the conclusions were completely consistent if all currently known Amycolatopsis species with validly published and correct names were delineated using a 96.6% ANIm value or 70% dDDH value (data not shown). Thus, we recommended that a 96.6% ANIm value could act as the threshold for delineating Amycolatopsis species.
3.2 Evaluation of the taxonomic status of strain HUAS 11-8T
3.2.1 Genome analysis
The genome sequence size of strain HUAS 11-8T is 7,474,574 bp with a DNA G + C content of 70.3%. A total of 7,267 genes (7,092 coding genes, 74 RNA genes, and 101 pseudogenes) and 7,193 CDSs (7,092 CDSs with protein and 101 CDSs without protein) were predicted. The analysis of the genome of strain HUAS 11-8T by the RAST Server revealed 316 subsystems that could be classified into 23 categories, and the subsystem coverage was 19%. The represented subsystem features identified were “Amino Acids and Derivatives” (378 CDSs), “Carbohydrates” (368 CDSs), “Fatty Acids, Lipids, and Isoprenoids” (214 CDSs), “Cofactors, Vitamins, Prosthetic Groups, Pigments” (213 CDSs), “Protein Metabolism” (173 CDSs), “DNA Metabolism” (121 CDSs), “Respiration” (109 CDSs), “Nucleosides and Nucleotides” (92 CDSs), “Metabolism of Aromatic Compounds” (91 CDSs), “Virulence, Disease and Defense” (63 CDSs), “Miscellaneous” (51 CDSs), “RNA Metabolism” (51 CDSs), “Stress Response” (44 CDSs), “Cell Wall and Capsule” (39 CDSs), “Membrane Transport” (35 CDSs), “Nitrogen Metabolism” (35 CDSs), “Phosphorus Metabolism” (32 CDSs), “Regulation and Cell signaling” (22 CDSs), “Sulfur Metabolism” (11 CDSs), “Iron acquisition and metabolism” (8 CDSs), “Dormancy and Sporulation” (7 CDSs), “Potassium metabolism” (6 CDSs), and “Secondary Metabolism” (3 CDSs). RAST revealed that strain HUAS 11-8T comprised lots of putative genes known to be associated with the abilities of dealing with harsh environmental conditions found in plant-associated environments, such as osmotic stress, oxidative stress, detoxification, stress response-no subcategory (SigmaB stress response regulation, dimethylarginine metabolism, and bacterial hemoglobins), and periplasmic stress. Three genes (Streptomyces venezuelae rox, vanR gene in vanO cluster, and Mycobacterium tuberculosis folC with mutation conferring resistance to para-aminosalicylic acid) of strain HUAS 11-8T related to antibiotic resistance were recognized by CRAD analysis, which might confer resistance to rifamycin antibiotic, glycopeptide antibiotic, and salicylic acid antibiotic. The potential secondary metabolite biosynthetic gene clusters in strain HUAS 11-8T were analyzed by antiSMASH, and 16 gene clusters were annotated. The three main biosynthetic gene clusters were aryl polyene, non-ribosomal peptide synthetase (NRPS), and type I polyketide synthase (PKS) [T1PKS]. The two T1PKS gene clusters had 96 and 8% similarities to macrotermycins and A54145, respectively. Meanwhile, nine CRISPR repeats of strain HUAS 11-8T were identified in the genome. In total, 10 genetic islands with a size range from 4,724 to 30,712 bp were identified in the genome of strain HUAS 11-8T.
3.2.2 Phylogenic analysis
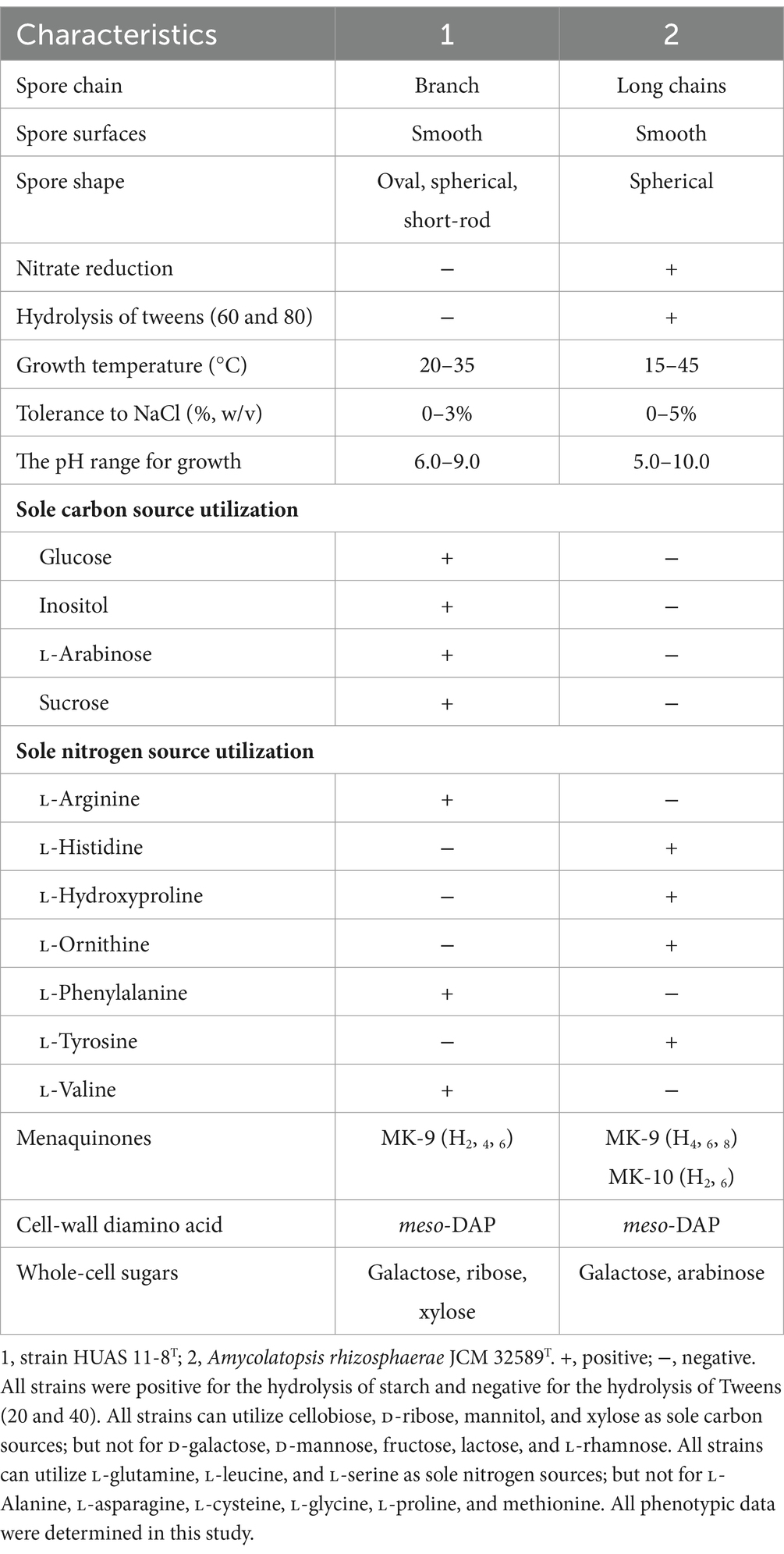
Based on EzBioCloud perform alignment, it was found that strain HUAS11-8T had 16S rRNA gene similarities of 99.78% with A. rhizosphaerae JCM 32589T, 97.8% with A. dongchuanensis YIM 75904T, and < 97.8% sequence similarities to other Amycolatopsis species. An ML phylogenetic tree based on 16S rRNA gene sequence demonstrated that strain HUAS 11-8T was most closely related to A. rhizosphaerae JCM 32589T (Figure 2). The relationship between HUAS 11-8T and A. rhizosphaerae JCM 32589T also appeared in the NJ and MP trees (Supplementary Figures S2, S3). This topological structure was further supported by the results of phylogenomic analysis (Figure 3). Nevertheless, the ANIm and dDDH values between strains HUAS 11-8T and A. rhizosphaerae JCM 32589T were 96.3 and 68.5%, respectively, lower than the 96.6 and 70% thresholds recommended for the delineation of a novel Amycolatopsis species. Furthermore, this result was further confirmed by phenotypic and chemotaxonomic differences between strains HUAS 11-8T and A. rhizosphaerae JCM 32589T (Table 1). For example, the spore chains of strain HUAS11-8T are branched, and the spores are oval, spherical, and short-rod. While A. rhizosphaerae JCM 32589T produces long spore chains and spherical spores. In addition, the dominant menaquinones are MK-9 (H4), MK-9 (H2), and MK-9 (H6). Galactose, ribose, and xylose were detected as the whole-cell reducing sugars in strain HUAS11-8T. The dominant menaquinone of strain A. rhizosphaerae JCM 32589T is MK-9 (H4, 6, 8) and MK-10 (H2, 6). Galactose and arabinose were detected as the whole-cell reducing sugars in strain A. rhizosphaerae JCM 32589T.
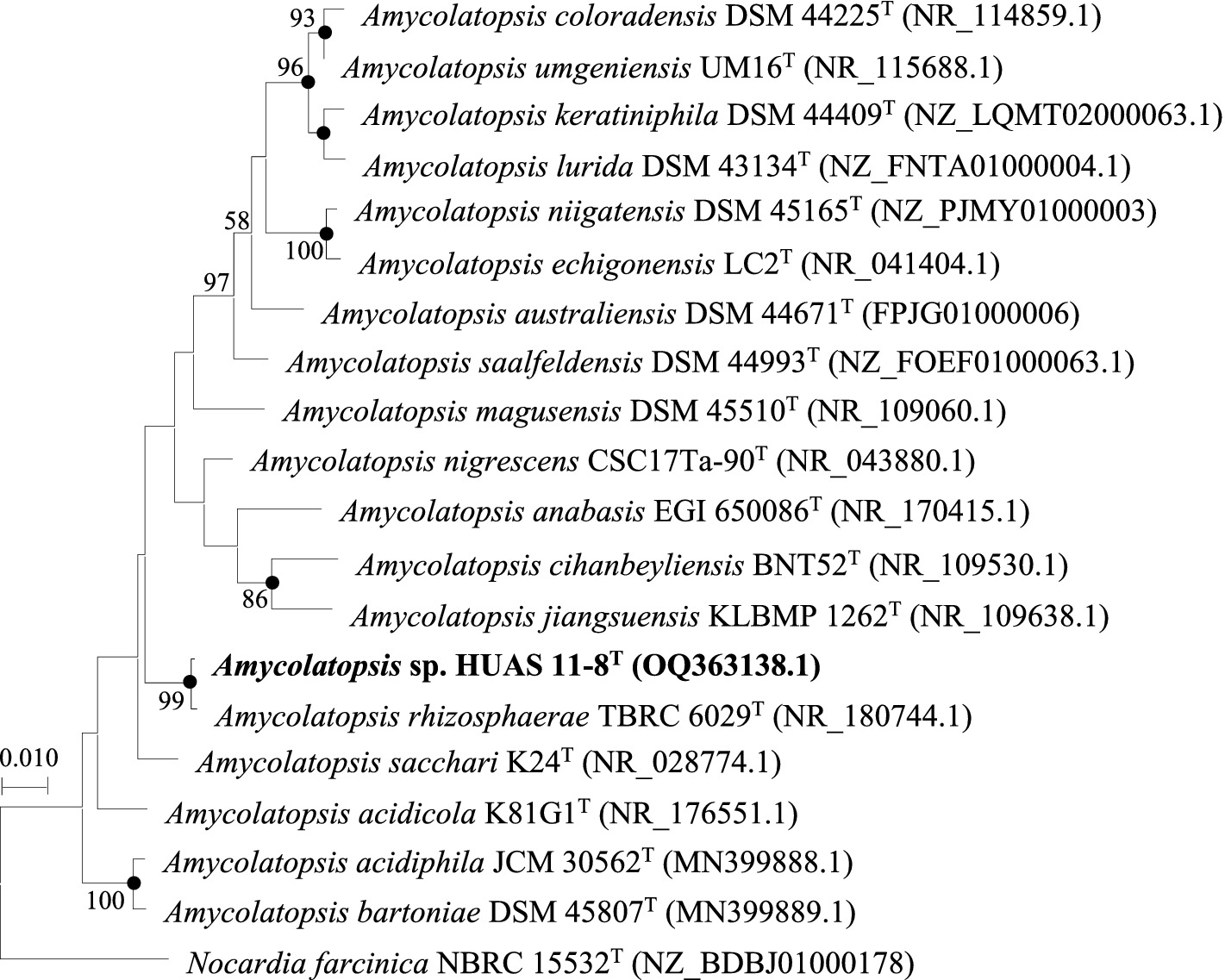
Figure 2. Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences showing the relationship between selected species of the genus Amycolatopsis. Nocardia farcinica NBRC 15532T was used as an outgroup. Bootstrap percentages over 50% derived from 1,000 replications are shown at the nodes. Dots indicate branches also recovered in the neighbor-joining and maximum-parsimony trees. Bar, 0.010 substitutions per site.
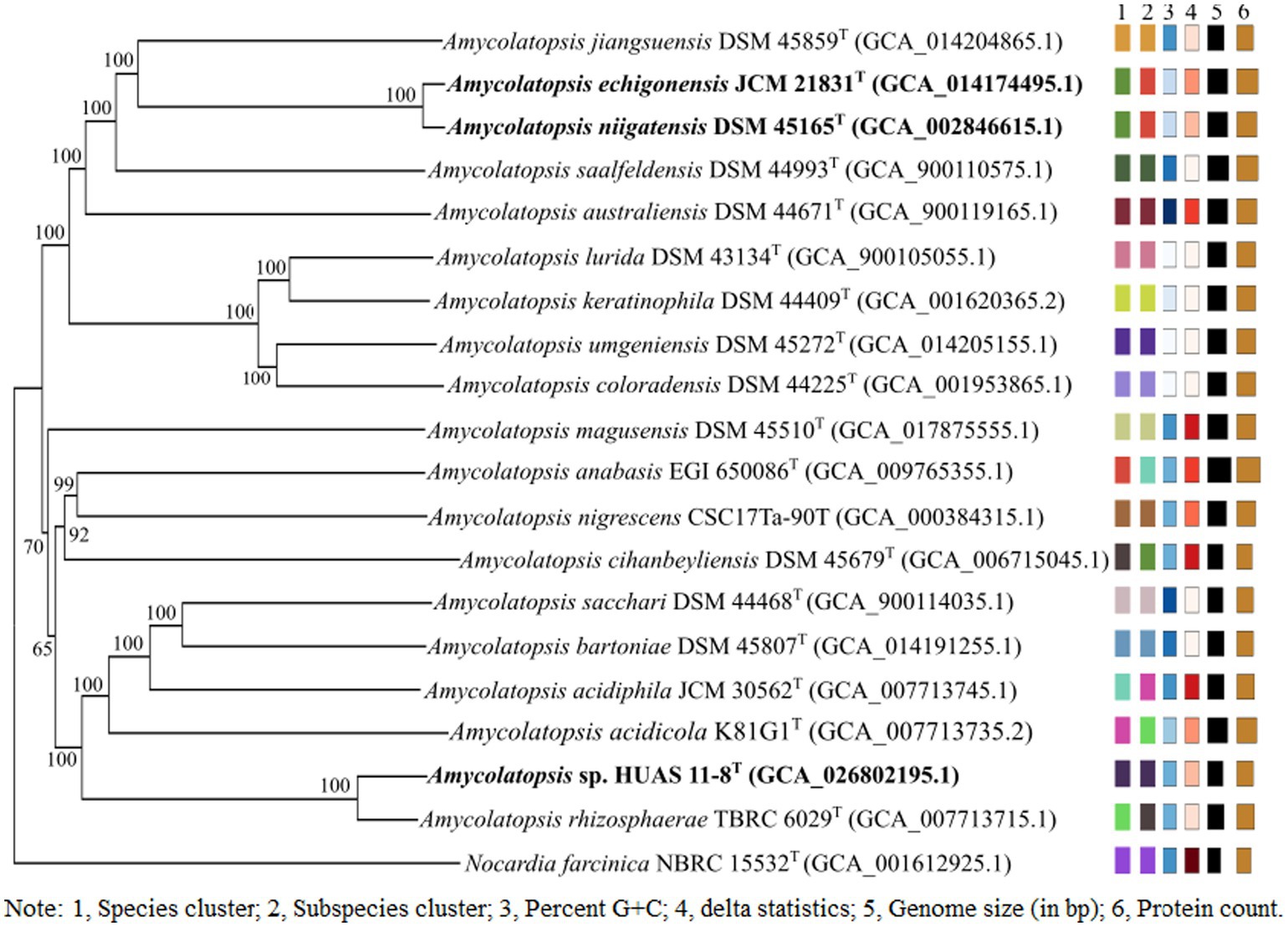
Figure 3. Phylogenetic tree based on whole-genome sequences of HUAS 11-8T and related reference strains. Tree inferred with FastME 2.1.6.1 (Vincent et al., 2015) from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of the GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values >60% from 100 replications, with an average branch support of 96.0%. The tree was rooted at the midpoint (Farris, 1972).
3.2.3 Morphological, physiological, and chemotaxonomic characteristics
Morphologically, strain HUAS 11-8T produced white substrate hyphae and aerial mycelia that differentiated into spore chains on Reasonerʼ2A medium after incubation for 21 days at 28°C. Spore chains were branched, and spore surfaces were smoothed (Figure 4). This strain was observed to grow well on Gauseʼs synthetic No. 1 medium, Reasonerʼ2A, and ISP 2–7 serial media (Supplementary Table S3). Growth was observed at 20–35°C (optimum, 30°C), at pH 6.0–9.0 (optimum, pH 7.0), and in the presence of 0–3% of NaCl (optimum, 0–1%). Detailed physiological and biochemical characteristics are provided in the species description. The dominant menaquinones were MK-9(H4) (75.2%), MK-9(H2) (20.6%), and MK-9(H6) (2.1%). The strain was found to contain meso-diaminopimelic acid as the cell wall amino acid. Galactose, ribose, and xylose were detected as whole-cell reducing sugars. The major fatty acids (≥10%) were iso-C16:0 (30.5%), C16:0 (10.8%), and C17:1 ω6c (10.0%). The detailed fatty acid composition is shown in Supplementary Table S4. The polar lipid pattern consisted of diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, and phosphatidylinositol (Supplementary Figure S4). All these morphological and chemotaxonomic data were consistent with the assignment of strain HUAS 11-8T to the genus Amycolatopsis.
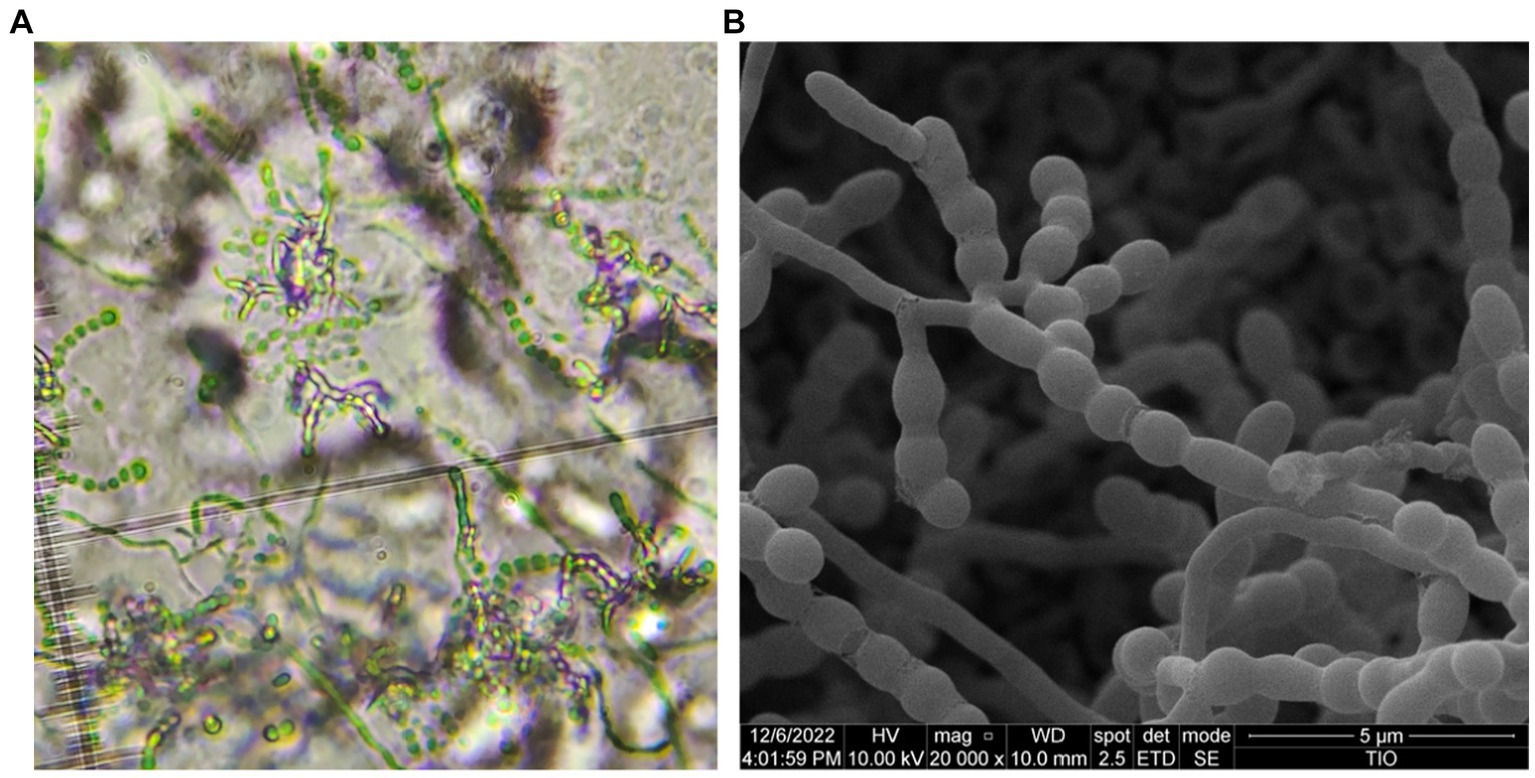
Figure 4. Optical microscope (A) and scanning electron microscope (B) images of strain HUAS 11-8T grown on Reasonerʼ2A after incubation for 21 days at 28°C.
In conclusion, based on phenotypic, chemotaxonomic, and genotypic data, strain HUAS 11-8T could be distinguished from A. rhizosphaerae JCM 32589T. Therefore, it is concluded that strain HUAS 11-8T represents a novel species of the genus Amycolatopsis, for which the name Amycolatopsis cynarae sp. nov. is proposed.
3.3 A proposal of Amycolatopsis niigatensis as a later heterotypic synonym of Amycolatopsis echigonensis
In order to determine the corresponding relationship between ANIm and dDDH values in the genus Amycolatopsis, we first analyzed the 16S rRNA gene sequence similarity between each validly published Amycolatopsis species and other validly published Amycolatopsis species. As a result, it was found that A. echigonensis JCM 21831T shared ≥98.7% sequence similarities to A. niigatensis DSM 45165T, A. halotolerans NRRL B-24428T, A. albidoflavus NRRL B-24149T, A. rubida NRRL B-24150T, A. circi S1.3T, A. nivea CFH S0261T, A. equina SE (8)3T, A. dendrobii DR6-1T, and A. hippodromi S3.6T, respectively (Supplementary Tables S5, S6). According to the proposal of Stackebrandt and Ebers (2006), a 16S rRNA gene sequence similarity threshold range of 98.7–99% is the point at which DNA–DNA reassociation experiments should be mandatory for testing the genomic uniqueness of a novel isolate. Thus, it is necessary to evaluate DNA–DNA relatedness between A. echigonensis JCM 21831T and its relatives. The result showed that the ANIm and dDDH values between A. echigonensis JCM 21831T and A. niigatensis DSM 45165T were 98.7 and 87.9%, respectively, much higher than the 96.6 and 70% thresholds recommended for the delineation of a novel Amycolatopsis species, suggesting that A. echigonensis JCM 21831T and A. niigatensis DSM 45165T belonged to the same genomic species. This result was further supported by the phylogenomic analysis (Figure 3). In addition, the ANIm/dDDH values between A. echigonensis JCM 21831T and type strains of other four species (A. rubida, A. circi, A. nivea, and A. dendrobii) were 92.5%/45.8, 92.2%/45.5, 92.6%/47.0, and 92.7%/47.2%, respectively, much less than the 96.6 and 70% thresholds recommended for the delineation of a novel Amycolatopsis species. Unfortunately, due to the unavailability of genomic data for type strains of A. halotolerans, A. albidoflavus, A. equina, and A. hippodrome, we could not evaluate DNA–DNA relatedness between A. echigonensis JCM 21831T and them. Whereas, there is evidence that a gyrB genetic distance of >0.02 or a recN genetic distance of >0.04 between two Amycolatopsis strains is proposed to provide a good indication that they belong to different species (Everest and Meyers, 2009; Everest et al., 2011). As shown in Supplementary Table S5, gyrB or recN genetic distances between A. echigonensis JCM 21831T and type strains of the above-mentioned four Amycolatopsis species were well over the thresholds recommended for assessing quickly whether an isolate is worthy of full taxonomic characterization.
Based on the analysis above and rule 42 of the Prokaryotic Code (Oren et al., 2022), we propose that Amycolatopsis niigatensis (Ding et al., 2007) (Approved Lists, 2007) (Euzeby, 2007) is a later synonym of Amycolatopsis echigonensis (Approved Lists, 2007).
4 Description
4.1 Description of Amycolatopsis cynarae sp. nov
Amycolatopsis cynarae [cy.na’rae. N.L. fem. n. Cynara, genus name of artichoke; N.L. gen. Fem. n. cynarae, of Cynara (Cynara scolymus L.)].
Aerobic, Gram-positive actinobacterium that forms white substrate hyphae and aerial mycelia that differentiate into branched spore chains consisting of smooth-surfaced oval, spherical, and short-rod spores on Reasonerʼ2A medium. Good growth is observed on all tested media. No diffusible pigment is produced on all tested media. Positive for the hydrolysis of starch, but negative for the hydrolysis of Tweens (20, 40, 60, and 80). Growth occurs at 20–35°C (optimum, 30°C), at pH 6.0–9.0 (optimum, pH 7.0), and in the presence of 0–3% of NaCl (optimum, 1%). Cellobiose, D-ribose, glucose, inositol, L-arabinose, mannitol, sucrose, and xylose can be utilized as sole carbon sources, but not for D-galactose, D-mannose, fructose, lactose, and L-rhamnose. The following substances, such as L-alanine, L-asparagine, L-cysteine, L-glycine, L-histidine, L-hydroxyproline, L-ornithine, L-proline, L-tyrosine, and methionine, can act as sole nitrogen sources, but not for L-arginine, L-glutamine, L-leucine, L-phenylalanine, L-serine, and L-valine. The cell wall diamino acid contains meso-diaminopimelic acid, and the whole-cell sugars contain galactose, ribose, and xylose. The main menaquinones are MK-9(H2), MK-9(H4), and MK-9(H6). The polar lipid profile contains diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, and phosphatidylinositol. The major fatty acids (>10%) were iso-C16:0, C16:0, and C17:1 ω6c.
The type strain, HUAS 11-8T (= JCM 35980T = MCCC 1K08337T), was isolated from the rhizosphere soil of Cynara scolymus collected in Changde city, Hunan Province, China. The DNA G + C content of the type strain genome is 70.3%. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence is OQ363138. The whole-genome shotgun sequence has been deposited at DDBJ/ENA/GenBank under the accession code CP113836.
4.2 Emended description of Amycolatopsis echigonensis
Later heterotypic synonym: Amycolatopsis niigatensis Ding et al. (2007) (Approved Lists 2007).
The description is as before, with the following modifications. The DNA G + C content of the type strain genome is 69.5%, its approximate size is 9.66 Mbp, and its GenBank accession number is NZ_JACJHR000000000.
The type strain is LC2T (=IAM 15387T = CCTCC AB206019T = DSM 45164T = JCM 21831T).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
AD: Conceptualization, Investigation, Writing–original draft. LF: Writing–review & editing, Resources, Methodology. PM: Writing–review & editing. YZ: Writing–original draft, Data curation, Formal analysis. TT: Writing–original draft, Investigation, Software. JG: Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Scientific Research Fund of Hunan Provincial Education Department, China (no. 22A0487), the Natural Science Foundation of Hunan Province, China (no. 2022JJ50249), and the project of Hunan Provincial Education Department (no. 23B0659).
Acknowledgments
The authors are grateful to Zhisheng Zhu, Xiupian Liu, and Li Gu (Marine Culture Collection of China, MCCC, Xiamen, China) for providing excellent technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1359021/full#supplementary-material
Footnotes
1. ^https://lpsn.dsmz.de/genus/Amycolatopsis
2. ^https://www.ezbiocloud.net/
4. ^https://antismash.secondarymetabolites.org
References
Adamek, M., Alanjary, M., Sales-Ortells, H., Goodfellow, M., Bull, A. T., Winkler, A., et al. (2018). Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genomics 19:426. doi: 10.1186/s12864-018-4809-4
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Auch, A. F., von Jan, M., Klenk, H. P., and Göker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2, 117–134. doi: 10.4056/sigs.531120
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., and Medema, M. H. (2011). Anti SMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Busarakam, K., Brown, R., Bull, A. T., Tan, G. Y. A., Zucchi, T. D., da Silva, L. J., et al. (2016). Classification of thermophilic actinobacteria isolated from arid desert soils, including the description of Amycolatopsis deserti sp. nov. Antonie Van Leeuwenhoek 109, 319–334. doi: 10.1007/s10482-015-0635-8
Claire, B., Laird, M. R., Williams, K. P., Lau, B. Y., Hoad, G., Winsor, G. L., et al. (2017). Island viewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Dávila Costa, J. S., and Amoroso, M. J. (2014). Current biotechnological applications of the genus Amycolatopsis. World J. Microbiol. Biotechnol. 30, 1919–1926. doi: 10.1007/s11274-014-1622-3
Ding, L. X., Hirose, T., and Yokota, A. (2007). Amycolatopsis echigonensis sp. nov. and Amycolatopsis niigatensis sp. nov., novel actinomycetes isolated from a filtration substrate. Int. J. Syst. Evol. Microbiol. 57, 1747–1751. doi: 10.1099/ijs.0.64791-0
Euzeby, J. P. (2007). Notification that new names and new combinations have appeared in volume 57, part 8 of the IJSEM. Int J Syst Evol Microbiol. 57, 2451–2452. doi: 10.1099/00207713-50-4-1419
Everest, G. J., Cook, A. E., Kirby, B. M., and Meyers, P. R. (2011). Evaluation of the use of recN sequence analysis in the phylogeny of the genus Amycolatopsis. Antonie Van Leeuwenhoek 100, 483–496. doi: 10.1007/s10482-011-9604-z
Everest, G. J., and Meyers, P. R. (2009). The use of gyrB sequence analysis in the phylogeny of the genus Amycolatopsis. Antonie Van Leeuwenhoek 95, 1–11. doi: 10.1007/s10482-008-9280-9
Farris, J. S. (1972). Estimating phylogenetic trees from distance matrices. Am. Nat. 106, 645–668. doi: 10.1086/282802
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Gevers, D., Cohan, F. M., Lawrence, J. G., Spratt, B. G., Coenye, T., Feil, E. J., et al. (2005). Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3, 733–739. doi: 10.1038/nrmicro1236
Hasegawa, T., Takizawa, M., and Tanida, S. (1983). A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol. 29, 319–322. doi: 10.2323/jgam.29.319
Hu, S. R., Li, K. Q., Zhang, Y. F., Wang, Y. F., Fu, L., Xiao, Y., et al. (2022). New insights into the threshold values of multi-locus sequence analysis, average nucleotide identity and digital DNA–DNA hybridization in delineating Streptomyces species. Front. Microbiol. 13:910277. doi: 10.3389/fmicb.2022.910277
Huang, Y., Pasciak, M., Liu, Z. H., Xie, Q., and Gamian, A. (2004). Amycolatopsis palatopharyngis sp. nov., a potentially pathogenic actinomycete isolated from a human clinical source. Int. J. Syst. Evol. Microbiol. 54, 359–363. doi: 10.1099/ijs.0.02685-0
Kluge, A. G., and Farris, F. S. (1969). Quantitative phyletics and the evolution of anurans. Syst. Zool. 18, 1–32. doi: 10.1093/sysbio/18.1.1
Labeda, D. P., Donahue, J. M., Williams, N. M., Sells, S. F., and Henton, M. M. (2003). Amycolatopsis kentuckyensis sp. nov., Amycolatopsis lexingtonensis sp. nov. and Amycolatopsis pretoriensis sp. nov., isolated from equine placentas. Int. J. Syst. Evol. Microbiol. 53, 1601–1605. doi: 10.1099/ijs.0.02691-0
Lane, D. J. (1991). “16S/23S rRNA sequencing” in Nucleic acid techniques in bacterial systematics. eds. E. Stackebrandt and M. Goodfellow (New York: Wiley), 115–175.
Lechevalier, M. P., and Lechevalier, H. (1970). Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Bacteriol. 20, 435–443. doi: 10.1099/00207713-20-4-435
Lechevalier, M. P., Prauser, H., Labeda, D. P., and Ruan, J. S. (1986). Two new genera of nocardioform actinomycetes: Amycolata gen. Nov. and Amycolatopsis gen. Nov. Int. J. Syst. Bacteriol. 36, 29–37. doi: 10.1099/00207713-36-1-29
Lee, S. D. (2009). Amycolatopsis ultiminotia sp. nov., isolated from rhizosphere soil, and emended description of the genus Amycolatopsis. Int. J. Syst. Evol. Microbiol. 59, 1401–1404. doi: 10.1099/ijs.0.006577-0
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2019). Evolutionary classification of CRISPR–Cassystems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83. doi: 10.1038/s41579-019-0299-x
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformat. 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
MIDI. Sherlock microbial identification system operating manual, version 6.0. Newark DE: MIDI Inc. (2005).
Mo, P., Li, K. Q., Zhou, J. H., Zhou, F. M., He, J., Zhou, W. S., et al. (2023). Nocardiopsis changdeensis sp. nov., an endophytic actinomycete isolated from the roots of Eucommia ulmoides Oliv. J. Antibiot. 76, 191–197. doi: 10.1038/s41429-023-00596-0
Mo, P., Yu, Y. Z., Zhao, J. R., and Gao, J. (2017). Streptomyces xiangtanensis sp. nov., isolated from a manganese-contaminated soil. Antonie Van Leeuwenhoek 110, 297–304. doi: 10.1007/s10482-016-0797-z
Oren, A., Arahal, D. R., Göker, M., Moore, E. R. B., Rossello-Mora, R., and Sutcliffe, I. C. (2022). International code of nomenclature of prokaryotes. Prokaryotic code (2022 revision). Int. J. Syst. Evol. Microbiol. 73:005585. doi: 10.1099/ijsem.0.005585
Reasoner, D. J., and Geldreich, E. E. (1985). A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49, 1–7. doi: 10.1128/aem.49.1.1-7.1985
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Glöckner, F. O., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Ruan, J. S., and Huang, Y. (2011). Rapid identification and systematics of Actinobacteria. Beijing: Science press. (in Chinese).
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Stackebrandt, E., and Ebers, J. (2006). Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33, 152–155.
Stackebrandt, E., Frederiksen, W., Garrity, G. M., Grimont, P. A. D., Kämpfer, P., Maiden, M. C., et al. (2002). Report of the ad hoc committee for the reevaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52, 1043–1047. doi: 10.1099/00207713-52-3-1043
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA 11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tan, G. Y. A., Robinson, S., Lacey, E., and Goodfellow, M. (2006). Amycolatopsis australiensis sp. nov., an actinomycete isolated from arid soils. Int. J. Syst. Evol. Microbiol. 56, 2297–2301. doi: 10.1099/ijs.0.64260-0
Tang, S. K., Wang, Y., Guan, T. W., Lee, J. C., Kim, C. J., and Li, W. J. (2010). Amycolatopsis halophila sp. nov., a halophilic actinomycete isolated from a salt lake. Int. J. Syst. Evol. Microbiol. 60, 1073–1078. doi: 10.1099/ijs.0.012427-0
Tedsree, N., Tanasupawat, S., Sritularak, B., Kuncharoen, N., and Likhitwitayawuid, K. (2021). Amycolatopsis dendrobii sp. nov. an endophytic actinomycete isolated from dendrobium heterocarpum lindl. Int. J. Syst. Evol. Microbiol. 71:004902. doi: 10.1099/ijsem.0.004902
Teo, W. F. A., Lipun, K., Srisuk, N., and Duangmal, K. (2021). Amycolatopsis acididurans sp. nov., isolated from peat swamp forest soil in Thailand. J. Antibiot. (Tokyo) 74, 199–205. doi: 10.1038/s41429-020-00382-2
Teo, W. F. A., Srisuk, N., and Duangmal, K. (2020). Amycolatopsis acidicola sp. nov., isolated from peat swamp forest soil. Int. J. Syst. Evol. Microbiol. 70, 1547–1554. doi: 10.1099/ijsem.0.003933
Vincent, L., Richard, D., and Olivier, G. (2015). Fast ME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evo. 32, 2798–2800. doi: 10.1093/molbev/msv150
Wang, H. F., Li, X. Y., Gao, R., Xie, Y. G., Xiao, M., Li, Q. L., et al. (2020). Amycolatopsis anabasis sp. nov., a novel endophytic actinobacterium isolated from roots of anabasis elatior. Int. J. Syst. Evol. Microbiol. 70, 3391–3398. doi: 10.1099/ijsem.0.004184
Xu, L. H., Li, W. J., Liu, Z. H., and Jiang, C. L. (2007). Actinomycete systematic principle, methods and practice. Beijing: Science press. (in Chinese).
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zhang, G. Y., Wang, L. N., Li, J. L., and Zhou, Y. (2016). Amycolatopsis albispora sp. nov., isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 66, 3860–3864. doi: 10.1099/ijsem.0.001277
Keywords: International Streptomyces Project, ANIm and dDDH, corresponding relationship, Amycolatopsis cynarae sp. nov., synonym, Amycolatopsis niigatensis, Amycolatopsis echigonensis
Citation: Deng A, Fu L, Mo P, Zheng Y, Tang T and Gao J (2024) New insights into the relationship between the average nucleotide identity and the digital DNA–DNA hybridization values in the genus Amycolatopsis and Amycolatopsis cynarae sp. nov., a novel actinobacterium from the rhizosphere soil of Cynara scolymus, and proposal of Amycolatopsis niigatensis as a synonym of Amycolatopsis echigonensis based on comparative genomic analysis. Front. Microbiol. 15:1359021. doi: 10.3389/fmicb.2024.1359021
Edited by:
Jesus L. Romalde, University of Santiago de Compostela, SpainReviewed by:
Geeta Chhetri, Korea Institute of Industrial Technology, Republic of KoreaGuohong Liu, Fujian Academy of Agricultural Sciences, China
Copyright © 2024 Deng, Fu, Mo, Zheng, Tang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Gao, xtgojian@hnust.edu.cn
†These authors have contributed equally to this work
 Aihua Deng1†
Aihua Deng1† Jian Gao
Jian Gao