- 1Institute of Horticulture, Henan Academy of Agricultural Sciences, Graduate T&R Base of Zhengzhou University, Zhengzhou, Henan, China
- 2School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
- 3National Water Research Center, Shubra El Kheima, Egypt
- 4Henan Natural Products Biotechnology Co., Ltd., Zhengzhou, China
In recent years, bacterial-based biocontrol agents (BCA) have become a new trend for the control of fungal diseases such as fusarium wilt that seriously threaten the yield and quality of cucumber, which are transmitted through infested soil and water. This study was set out with the aim of figuring the mechanism of the isolated rhizobacterial strain Paenibacillus polymyxa PJH16 in preventing Fusarium oxysporum f. sp. cucumerinum (Foc). Biocontrol and growth-promoting experiments revealed that bacterial strain causes effective inhibition of the fungal disease through a significant growth-promoting ability of plants, and had activities of β-1,3-glucanase, cellulase, amylase and protease. It could produce siderophore and indole-3-acetic acid, too. Using the high-throughput sequencing tool PacBio Sequel II system and the database annotation, the bacterial strain was identified as P. polymyxa PJH16 and contained genes encoding for presence of biofilm formation, antimicrobial peptides, siderophores and hydrolyases. From comparing data between the whole genome of P. polymyxa PJH16 with four closely related P. polymyxa strains, findings revealed markedly the subtle differences in their genome sequences and proposed new antifungal substances present in P. polymyxa PJH16. Therefore, P. polymyxa PJH16 could be utilized in bioengineering a microbial formulation for application as biocontrol agent and bio-stimulant, in the future.
1 Introduction
Cucumber is an important economic crop. China is the main producer of cucumber, and its planting area and scale have ranked first in the world for many years. One cucumber disease that poses a serious threat to yield and quality is cucumber fusarium wilt caused by the fungus Fusarium oxysporum f. sp. cucumerinum which belongs to Ascomycota (Gao et al., 2014). This fungus is a soil-borne pathogen that can infect plants at any stage of growth. It begins invading the wounds and root hair growth points, then colonizes the vascular bundles resulting in blocked absorption of water and nutrients for plants, and ultimately leads to plant wilting and death. Due to the disease damaging effect and difficulty to control, it has limited greatly the development of cucumber global industry (Zhou et al., 2017).
To combat the phytopathogen, chemical fungicides are usually used which may cause environmental and health concerns, as well as the problem of pathogen resistance. Biological control is a powerful option for treating bacterial and fungal infections. Therefore, microbial biocontrol agents (BCA) have been developed over the past few decades such as employing biocontrol bacteria to manage plant diseases, decrease the reliance on chemical pesticides and promote sustainable agriculture (Maciag et al., 2023). At present, many bacteria from different genera have been identified as biocontrol agents, including Bacillus, Paenibacillus, Agrobacterium, Bradyrhizobium, Acinetobacter, Azospirillum, Azotobacter, Pseudomonas, Rhizobium, and Streptomyces (Ayaz et al., 2021). Since Paenibacillus polymyxa belongs to the genus Paenibacillus of the family Bacillaceae. It is a Gram-positive bacteria widely distributed in nature, non-pathogenic to plants, their cell wall does not contain endotoxins, and they are harmless to humans and animals. Accordingly, it is an ideal bacteria to use as a biocontrol agent. Applying BCA to crops allows them to biologically suppress plant diseases through variety of modes of action (Köhl et al., 2019). BCA may directly interact with the pathogen through antibiosis or hyperparasitism. Hyperparasites infiltrate and destroy bacterial and fungal pathogen. Without directly engaging in antagonistic interactions with the pathogen, they can either improve or produce resistance against phytopathogen (Conrath et al., 2015). From that point, part of plant defense systems is to enhance resistance by producing reactive oxygen species, phytoalexins, phenolic compounds, pathogenesis-related proteins, and physical barriers including modifying cell walls and cuticles (Wiesel et al., 2014). Another direct form of action is the production of secondary metabolites that have antagonistic effects on a range of phytopathogen (Ghorbanpour et al., 2018), and are commonly including ribosomal peptides (RPs), non-ribosomal peptides (NRPs), antibiotics, polyketides (PKs), and volatile organic compounds (VOCs) (Farzand et al., 2019a). Surfactin, iturin, mixirin, and fengycin are examples of lipopeptides with antifungal properties. Besides, the formation of biofilms reduces the number of pathogens. Extracellular enzymes like chitosanase, protease, glucanase, and cellulase are produced by Bacillus species, which are known to adhere to mycelial cell walls and deform hyphae (Farzand et al., 2019b).
In recent years, the rapid development of sequencing technology has becomeincreasingly more efficient, providing a major key to in-depth study of the function and mechanism of bacteria in disease control. Sequencing efforts help to understand the gene function or are used to design gene engineering experiments. Additionally, annotation in the same species can act as a template for reading maps, and can be aligned and compared to study molecular evolution (Giani et al., 2020). There are few studies on the complete genome sequence of this bacterial strain and other on its ability to regulate phytopathogens through the processes of antagonistic activity (production of antibiotics), colonization (biofilm formation), and induced resistance (Luo et al., 2018).
This study focused on a bacterial strain named Paenibacillus polymyxa PJH16 isolated from the rhizosphere soil of healthy cucumber plants, and investigates its inhibitory effect on the fungus disease Fusarium oxysporum f. sp. cucumerinum in vitro and in vivo. To elucidate its mechanisms in promoting cucumber plant growth and controlling fusarium wilt, 16S rRNA gene sequence phylogenetic tree and average nucleotide identity (ANI) analysis are investigated.
2 Materials and methods
2.1 Isolation of bacteria and screening of soil antagonistic bacteria
Through the application of dilution plate methodology, we successfully extracted the bacterial strain P. polymyxa PJH16 from the soil within the cucumber cultivation region of Beiwang Village, Luolong District, Luoyang City, Henan Province, China—a region known for being a high incidence area of cucumber fusarium wilt. The bacterial strain P. polymyxa PJH16 was nurtured on Luria Bertani medium (LB) plates. Simultaneously, the Fusarium oxysporum f. sp. cucumerinum FJH36 (Foc FJH36) fungi were isolated and conserved at the Institute of Horticulture, Henan Academy of Agricultural Sciences (Yang et al., 2023).
As for Antagonistic bacterial screening, the plate confrontation culture method was used with Foc FJH36 fungi as indicator fungi. A puncher with a diameter of 5 mm was used to punch the pathogenic fungi cake at the edge of the Foc plate into the center of the Potato Dextrose Agar (PDA) plate, and the bacteria were inoculated at a distance of 25 mm from the bacteria cake, which was located in the same line as the midpoint of the bacteria cake. The control was a PDA plate inoculated with Foc fungi in the absence of bacteria. Triplicate plates were inoculated at 30°C for 7 days. The fungal growth inhibition was assessed in fungi co-cultured with bacteria in comparison with control plates (Xu et al., 2020).
2.2 Observation of antagonistic mycelium
The mycelium of Foc FJH36 in the antagonistic plate was cut, and the volume of the sample was not more than 3 mm3. After the sample being gently rinsed with phosphate buffer saline (PBS) (2.7 mM KCl, 2.0 mM KH2PO4, 137 mM NaCl, 10 mM Na2HPO4, pH 7.4), it was quickly placed into the electron microscope fixative at room temperature for 2 h, then transferred to 4°C preservation. The fixed sample was rinsed three times for 15 min each time with 0.1 M phosphate buffer PB (2.7 mM KCl, 2.0 mM KH2PO4, 10 mM Na2HPO4, pH 7.4). A 0.1 M phosphate buffer PB (pH 7.4) was used to prepare 1% osmic acid, and the samples were fixed at room temperature in the dark for 2 h. Rinse three times for 15 min each with 0.1 M phosphate buffer PB (pH 7.4). The tissue was subjected to gradient dehydration in alcohol concentrations of 30, 50, 70, 80, 90, 95, 100% for 15 min each time, followed by 15 min soak in isoamyl alcohol. For drying, the samples were placed in a Quorum K850 critical point dryer. To examine and take photographs, the samples were tightly adhered to the double-sided adhesive of the conductive carbon film and placed on the sample table of the Hitachi MC1000 ion sputtering instrument for gold spraying for 30 s, and Hitachi SU8100 scanning electron microscope (Hitachi Higher Technologies Corp., Tokyo, Japan) was employed.
2.3 Biocontrol effect of PJH16 on cucumber fusarium wilt
To evaluate the biocontrol effect of bacteria on fungal disease, the pot experiment was carried out. The first step in this process was to prepare suspensions of Foc FJH36 fungi and bacterial strain PJH16, and germinating the seeds of cucumber as the following:
After growing Foc FJH36 on a PDA plate for 5 days at 30°C, ten fungal cakes were taken from the edge of the pathogen using a 5 mm puncher and cultured in a 100 mL Potato Dextrose Broth (PDB). The cultures were shaken for 3 days at 30°C, 180 rpm, and then the filtered cultures using 4 layers of sterile gauze were diluted with sterile water to a volume of 1 × 106 spores/mL pathogen spore suspension. For the bacterial strain PJH16, it was cultured in LB medium at 37°C, 180 rpm for 24 h, centrifuged at 4000 xg for 5 min, and diluted to 1 × 108 CFU/mL with LB medium.
For seed germination, cucumber seeds of the Bojie 616 variety, characterized by their robust, full, and uniform size, were carefully chosen. Initially, the cucumber seeds underwent a series of steps: they were immersed in 75% ethanol (v/v) for 30 s, rinsed five times with sterile water, and soaked in 55°C warm water for 10 min. Subsequently, the seeds were soaked in distilled water for 4 h, and gauze moistened with sterile water was utilized. Following thorough rinsing, the seeds were neatly arranged on the gauze and placed in a constant temperature incubator at 28°C. White cucumber seeds were then sown in a seedling plug (5 × 8) with sterile matrix for a duration of 10 days. Once the seeds had developed into seedlings, two-leaf seedlings of uniform size were selected and transplanted into pots with a diameter and height of 10 cm, along with 400 g of sterile matrix (Figure 1A).
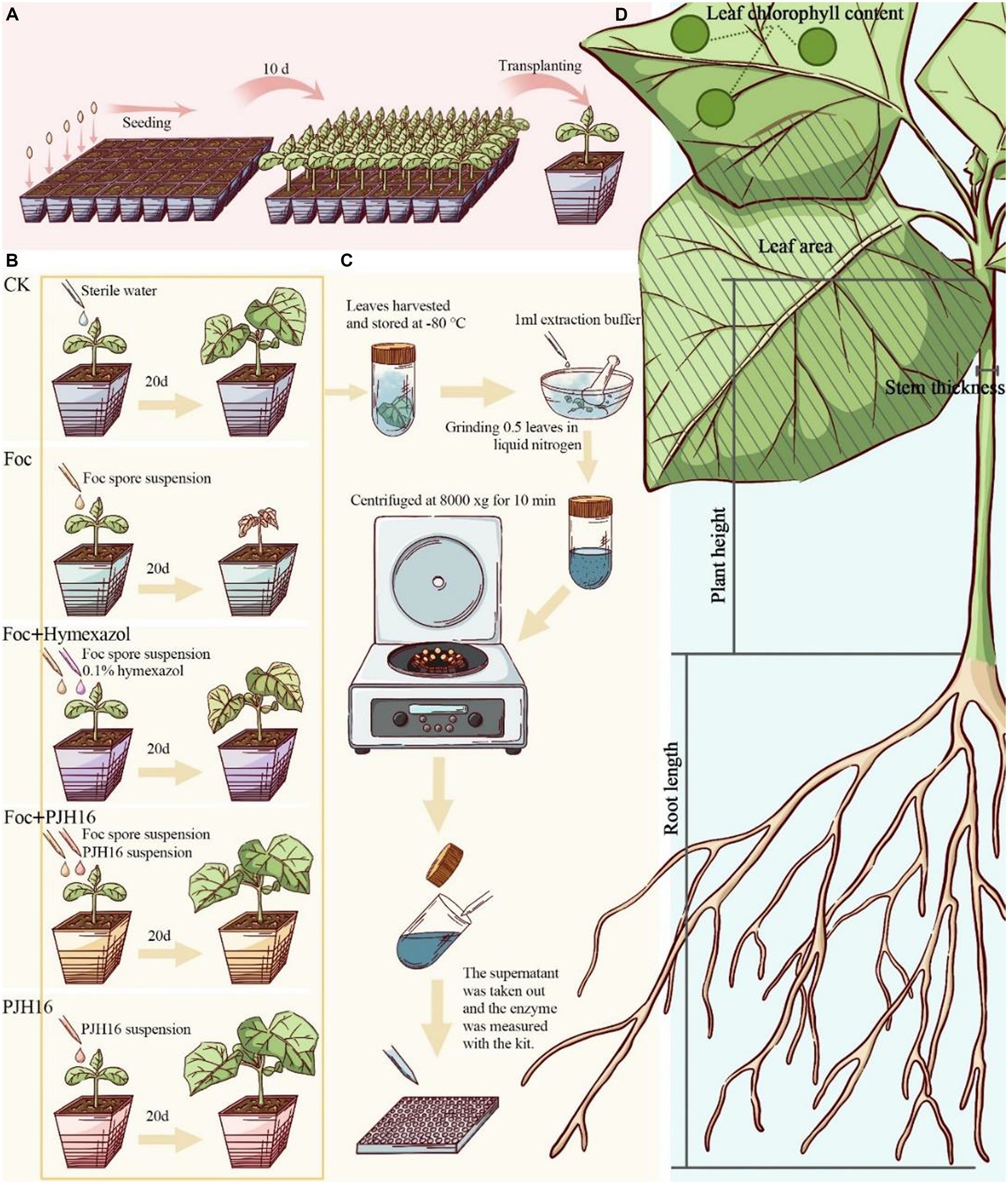
Figure 1. Experimental flow for this study. (A) Plug seedling, and transferred to the pot after 10 days; (B) procedure for the pot experiment; (C) after 20 days of inoculation, the activity of defense enzymes in cucumber leaves was determined; (D) the data of cucumber seedlings were measured 40 days after inoculation.
Subsequent to the initial phase, this study incorporated four treatments and a control for the seedlings. Watering was performed once every two days initially, followed by the continued use of only sterile distilled water throughout the growth period. The treatments were implemented as outlined in Figure 1B.
For the experimental setup, five treatments with distinct procedures were employed, each with five replicates, and five pots for each replicate, totaling 125 pots. The greenhouse conditions maintained an ambient temperature ranging from 14 to 25°C, with a photoperiodic lighting schedule of 16 h of light followed by 8 h of darkness.
We measured plant height, stem diameter, leaf area, chlorophyll content, shoot fresh weight, root fresh weight, shoot dry weight and root dry weight on the 20th and 40th days during the whole growth period (Yang et al., 2023; Figure 1D).
Finally, in order to come into view of the biocontrol effect of PJH16 on cucumber fusarium wilt, several formulas were calculated such as: Strong Seedling index, Disease severity, Disease Index (DI) and Disease Control efficacy (Chen H. et al., 2010; Chen F. et al., 2010). For the strong seedling index, it was calculated with the following formula: Strong seedling index = (stem diameter/plant height + root dry weight/shoot dry weight) × whole plant dry weight. As for estimation of the disease severity and disease indices of plants, they were recorded at 40 days. Disease severity was measured by the assessment of disease symptoms of leaves, that divided into 5 levels: level 0, leaves without symptoms; level 1 grade, and the leaf wilting was lower than 1/4 of cucumber seedlings; level 2, 1/4–1/2 of cucumber seedling leaves wilting; level 3, more than 1/2 leaves of cucumber seedlings wilted; level 4, the whole cucumber plant withered and died. The disease index was calculated using DI = [[(0 × N0) + (1 × N1) + (2 × N2) + (3 × N3) + (4 × N4)]/T × 4] × 100, where N is the number of cucumber seedlings scored for each disease, and T is the total number of cucumber seedlings. Incidence = [N1 + N2 + N3 + N4]/T × 100%. The disease control efficacy was figured through the next calculation: Control efficacy = (control DI-treatment DI)/control DI × 100%.
2.4 Determination of defense enzyme activity in cucumber leaves
As shown in Figure 1C, on the 20th day after inoculation, plant leaves were collected for determination of enzyme activity. Polyphenol oxidase (PPO), superoxide dismutase (SOD), catalase (CAT), phenylalanine ammonia-lyase (PAL) and lipoxidase (LOX) detection kits were used to determine the enzyme activity according to the manufacturer ‘s (Solarbio, Beijing, China) guidelines. The absorbance was measured by a microplate reader (Tecan Spark, Tecan, Switzerland).
2.5 Detection of plant growth-promoting bacteria and enzymatic activity
In order to evaluate the plant growth-promoting and biocontrol characteristics of strain PJH16, we referred to the previous research methods (Yang et al., 2023) to detect the β-glucanase activity, amylase activity, siderophore production ability, cellulase activity, protease activity and IAA production ability of strain PJH16.
Single colony of PJH16 strain was inoculated in β-glucan agar medium (0.5 g yeast extract, 1.0 g peptone, 0.05 g glucose, 0.5 g NaCl, 0.01 g Congo red and 18.0 g agar/L, pH 7.0), and cultured in an incubator at 30°C for 48 h. A clear area around the flora was observed, indicating that the strain had β-glucanase activity.
Single colony of PJH16 strain were inoculated in starch agar medium (10.0 g soluble starch, 10.0 g tryptone, 5.0 g glucose, 5.0 g NaCl, 5.0 g beef extract and 18.0 g agar/L, pH 7.0), and cultured in an incubator at 30°C for 48 h. Lugol ‘s iodine solution (1% iodine in 2% potassium iodide w/v) was added to the starch agar medium to make it evenly spread on a flat plate. After standing for 1 min, a colorless halo around the colony was observed, indicating that the strain had amylase activity.
Single colony of PJH16 strain was inoculated in Chrome Azurol S blue agar medium (10.0 mL 20% sucrose solution, 30.0 mL 10% acid hydrolyzed casein, 1.0 mL 1.0 mmol/L CaCl2, 5.0 mL 0.1 mol/L phosphate buffered saline (pH 6.8), 50.0 mL CAS staining solution and 18.0 g agar/L, pH 7.0), and cultured in an incubator at 30°C for 48 h. The surrounding of the colony changed from blue to orange, indicating that the strain could produce siderophores.
Single colony of PJH16 were inoculated in carboxymethyl cellulose agar medium (5.0 g CMC-Na, 0.1 g MgSO4·7H2O, 0.25 g K2HPO4, and 18.0 g agar/L, pH 5.5) and cultured at 30°C for 48 h. After staining with 1% (m/v) Congo red stain, the medium was washed with sterile distilled water. If there is a transparent halo around the colony, it indicates that the strain can produce cellulase.
The single colony of PJH16 strain was inoculated in skim milk agar medium (0.1 g CaCl2, 5.0 g NaCl, 10.0 g peptone and 18.0 g agar/L, pH 7.0), and cultured in an incubator at 30°C for 48 h. If a transparent area was present around the colony, it indicated that the strain had protease activity.
Single colonies of PJH16 were inoculated in L-tryptophan nutrient broth (3.0 g beef extract, 10.0 g peptone, 5.0 g NaCl, 0.5 g L-tryptophan/L, pH 7.2) and cultured in an incubator at 30°C for 48 h. After centrifugation at 14,000 xg for 10 min, 1 mL supernatant was mixed with 2 mL Salkowski dye, and then placed in dark at room temperature for 30 min. The color changed from yellow to orange, indicating the production of IAA.
2.6 DNA extraction, genome sequencing, and assembly of strain PJH16
In order to obtain the genomic information of strain PJH16, the single colony of strain PJH16 was first inoculated in LB medium and cultured for 18 h at 37°C and 150 rpm. Subsequently, according to the manufacturer’s instructions, genomic DNA was extracted using the Mini BEST bacterial genomic DNA extraction kit Ver.3.0. In order to construct an insertion sequencing library, about 10 kb DNA insertion fragments were prepared and sequenced using the PacBio Sequel II system (Pacific Biosciences, Menlo Park, CA, United States) of Frasergen (Wuhan, Hubei, China). The sequencing reads were assembled from scratch using HGAP4 (Chin et al., 2013) and Canu (v.1.6) (Koren et al., 2017) software, and the coverage depth of the genome was analyzed by comparison tools. Finally, the complete genome sequence of the assembled strain PJH16 was stored in NCBI GenBank, and the accession number was CP137742. In addition, a circular map of the genome of strain PJH16 was constructed by using Circos (Krzywinski et al., 2009).
2.7 Genome annotation of strain PJH16
The annotation of the PJH16 strain genome was conducted using Glimmer (v3.02) (Delcher et al., 2007). Identification of tRNA and rRNA genes was performed using tRNAscan-SE (v2.0) (Lowe and Eddy, 1997) and RNAmmer (v1.2) (Lagesen et al., 2007), respectively. BLASTx analysis utilized the NCBI non-redundant protein database (NR), SwissProt, Orthologous Group (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO). The blastp command of diamond (v2.0.9.147) software was employed to align the protein sequences of predicted genes with the NR microbial library (20210213 download version), using an E value of 1e−5, and the hit with the highest score was chosen as the final annotation.
The same diamond blastp command was applied to align the predicted gene protein sequences with the SwissProt (v2021_03) library, COG2020 library, and the full library of KOBAS-v3.0 (Xie et al., 2011), each with an E value of 1e−5. The hits with the highest scores were selected as the final annotation results. For GO annotation, SwissProt annotation results were mapped by ID Mapping (20210616), and go-basic.obo (v2021-09-01) was used for hierarchical annotations.
Specific functional annotation modules were constructed by integrating various function-related databases and annotation methods. These modules included drug resistance genes (CARD), pathogenic virulence factors (VFDB), and membrane transporter classification (TCDB). The predicted gene protein sequences were aligned to the CARD-v3.1.4 library, the SetA library of pathogenic virulence factors (VFDB, Last updated: Sep 24, 2021), and the TCDB library using diamond blastp with an E value of 1e−5. The hits with the highest scores were selected as the final annotation results based on the predicted gene amino acid sequence file.
2.8 Analysis and identification of strain PJH16
The 16S rRNA gene sequence tree of 114 strains (Supplementary Table S1) based on strain PJH16 and 20 strains of P. terrae, 15 strains of P. polymyxa, 23 strains of P. xylanexedens, 14 strains of P. thiaminolyticus, 11 strains of B. subtilis, 12 strains of B. mycoides and 19 strains of B. cereus was constructed by MEGA 7.0 using Neighbor Joining method (Kumar et al., 2016). The average nucleotide identity (ANI) of 10 strains of P. polymyxa and 3 strains of B. mycoides with PJH16 was calculated using the ANI calculator (Yoon et al., 2017).
2.9 Analysis of secondary metabolic genes in CAZymes of strain PJH16
During the genome analysis of strain PJH16, we employed dbCAN2 (Zheng et al., 2023) and HMMER (v3.1b2) (Finn et al., 2011) to align the predicted protein sequences with the Carbohydrate Active Enzyme (CAZy) database (Lombard et al., 2014), with a set E value threshold of 1e−15. Additionally, utilizing antiSMASH version 7.0.0, we conducted a comparative analysis of the protein-coding genes in the genome of strain PJH16 against the CAZy database to pinpoint gene clusters associated with the synthesis of secondary metabolites (Blin et al., 2019). Secondary metabolites are generally controlled by multiple genes, and their coding genes are usually clustered in the genome. This gene cluster is the secondary metabolite synthesis gene cluster (BGC).
2.10 The collinearity analysis of strain PJH16
We used Mauve (v2.3.1) (Darling et al., 2004) to globally align the chromosomes of P. polymyxa PJH16, P. polymyxa HY96-2, P. polymyxa SQR-21, P. polymyxa CF05 and P. polymyxa DSM 36. In order to visualize the alignment results, we used TBtools (v1.0697) (Chen C. et al., 2020) to draw the collinearity diagram of PJH16 and four P. polymyxa strains.
2.11 Statistical analysis
We used SPSS v21.0 for statistical analysis by one-way analysis of variance (ANOVA). The mean was compared with Duncan’s multi-range test with a probability of p ≤ 0.05.
3 Results
3.1 Antagonistic characteristics of PJH16 strain
On the purpose of figuring out the antagonistic activity against phytopathogen of the genus Fusarium, it was carried out the antagonistic bacterial screening for PJH16 strain isolated from soil of cucumber planting area with high incidence of cucumber fusarium wilt. It has been illustrated that the inhibition rate of PJH16 was 88.36%, with antagonistic band width was 11 mm (Figures 2A,B). At the same time, it was verified by scanning electron microscopy and observed the morphology of the mycelium of the antagonistic Fusarium oxysporum showed abnormal morphology, such as folding (Figure 2D), twisting (Figure 2E) and shrinkage (Figure 2F), comparing with the mycelium of the control treatment that was smooth and no abnormal changes (Figure 2C). Thus, the Paenibacillus polymyxa PJH16 strain presented anatogonistic behavior when paired with the genus Fusarium oxysporum f. sp. cucumerinum Foc FJH36, affecting the development pattern.
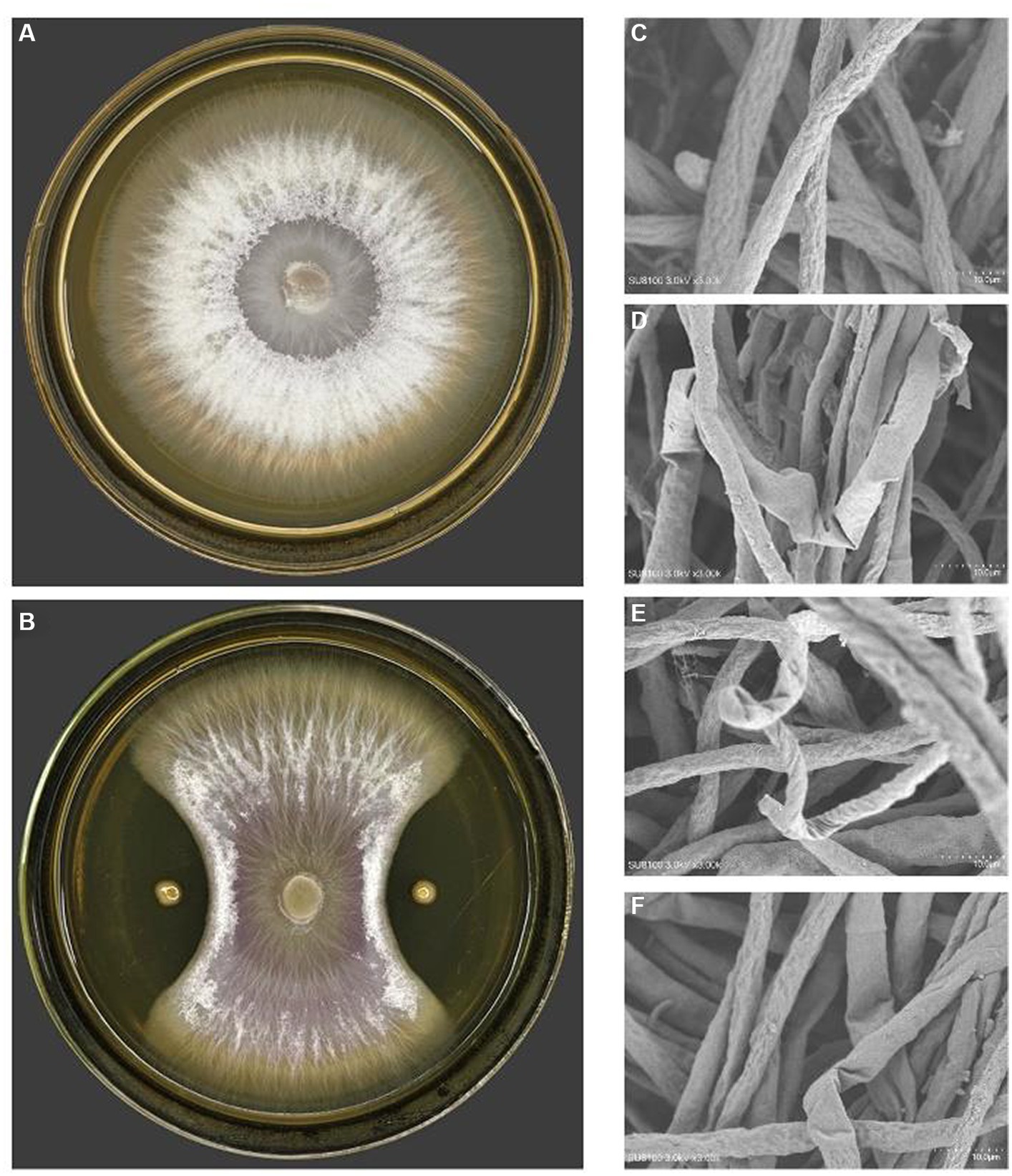
Figure 2. The antagonistic effect of PJH16 on Foc FJH36. (A) The colony morphology of Foc FJH36 without PJH16 strain on PDA plate; (B) the colony morphology of Foc FJH36 containing PJH16 strain on PDA plate; (C) Foc FJH36 normal development of mycelial morphology; (D) PJH16 strain caused the mycelium of Foc FJH36 to fold; the (E) PJH16 strain caused the mycelium of Foc FJH36 to distort; (F) PJH16 strain caused the mycelium of Foc FJH36 to shrink.
3.2 Anti-fusarium wilt activity of PJH16 strain in vivo
The experiment of phytopathogenic effect in vivo under controlled condition is a well-established approach in highlighting the anti-fusarium wilt activity. For such a goal, four treatments for cucumber seedlings were compared with control to assess the most effective anti-disease treatment. The first treatment was to evaluate the effect of disease alone on the seedlings; the second to estimate the fungicidal effect, the third to determine whether the bacterial strain can reduce the disease effect; and the fourth one with only the isolated bacterial strain to evaluate its influence. Afterwards, the severity of disease, disease incidence, disease index and control efficacy were determined for cucumber seedlings.
It is noteworthy that the severity of fusarium wilt in cucumber seedlings treated with PJH16 decreased 20 days after Foc inoculation. Furthermore, at 40 days, the disease incidence, disease index, and control efficacy of the three treatments indicated that the PJH16 strain significantly alleviated the wilt symptoms caused by Foc, with a reduction of 98.08% in disease incidence and 92.89% in disease index. Notably, PJH16 exhibited a control efficacy against Fusarium oxysporum higher than that of the chemical fungicide Hymexazol (92.71, 88.83%) (Table 1).
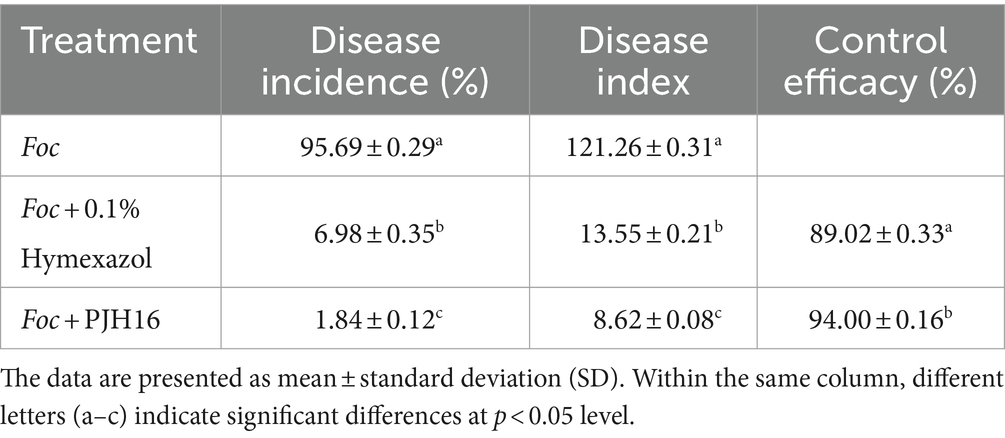
Table 1. The disease incidence, disease index, and control efficacy of hymexazol and Paenibacillus polymyxa PJH16 against cucumber wilt disease.
3.3 Effects of PJH16 strain on defense-related enzyme activities in cucumber seedlings
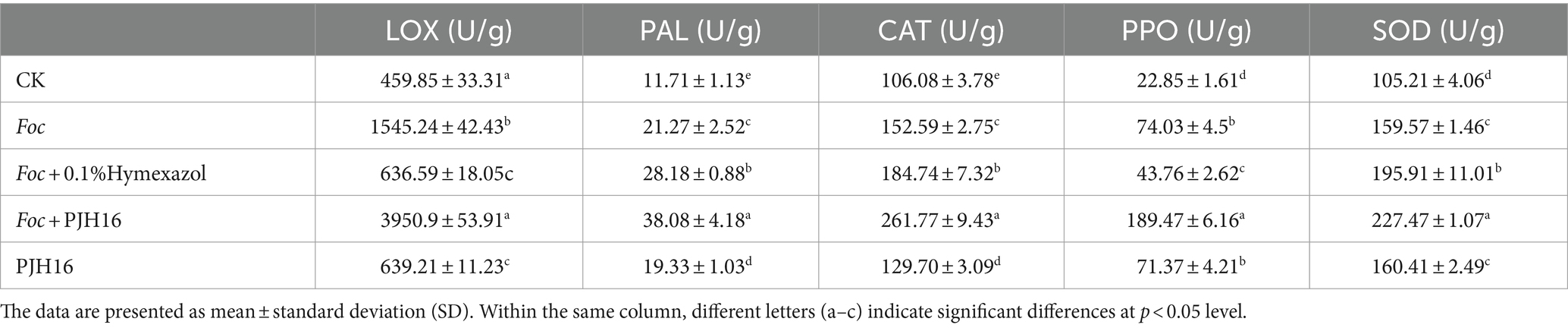
At 20th day, the enzyme activities of LOX, PAL, CAT, PPO and SOD in cucumber seedlings treated with four treatments were significantly higher than those in CK (cucumber seedlings irrigated with sterile distilled water). Furthermore, all enzyme activities of cucumber seedlings inoculated with Foc after inoculation with PJH16 strain (treatment 4) were significantly higher than those of cucumber seedlings inoculated with Foc alone (treatment 2), with an increase of 155.68, 79.03, 71.55, 155.94 and 42.55%, respectively. However, the activities of PAL, CAT and SOD were significantly increased after inoculation with Foc and Hymexazol, but LOX and PPO were significantly decreased (Table 2).
3.4 Cucumber seedling growth promoting activity of PJH16 strain
At 20 and 40 days, the chlorophyll content, plant height, stem diameter, root length, leaf area, shoot fresh weight and dry weight, and root fresh weight and dry weight of seedlings treated with only PJH16 strain (treatment 5) were remarkably increased (Figure 3; Supplementary Table S2). At 20 days, all indexes of treatment 5 were significantly higher than those of the control treatment. Among them compared with the control, not only the true leaf area increased the most, but also the area of the first true leaf and the second true leaf treated with treatment 5 increased by 129.63 and 503.40%, respectively (Figures 3B,C,E). As well, the fresh weight of the aboveground part increased by 88.99% (Figure 3F). Moreover, by the 40th day, the most substantial increase occurred in the average leaf area of the fourth true leaf in treatment 5, reaching 90.75 cm2, while the leaf area of the fourth true leaf in the control treatment remained below 5 cm2 (Figures 3D,E). Consequently, the seedling index of the PJH16 bacterial treatment (0.06) exhibited a significant increase compared to the control (0.05) at the 20-day mark (Figure 3J). By the 40th day, the seedling index for the PJH16 treatment (0.11) remained notably higher than that of the control (0.07) (Figure 3J). This outcome underscores the positive impact of the isolated bacterial strain PJH16 on enhancing plant growth.
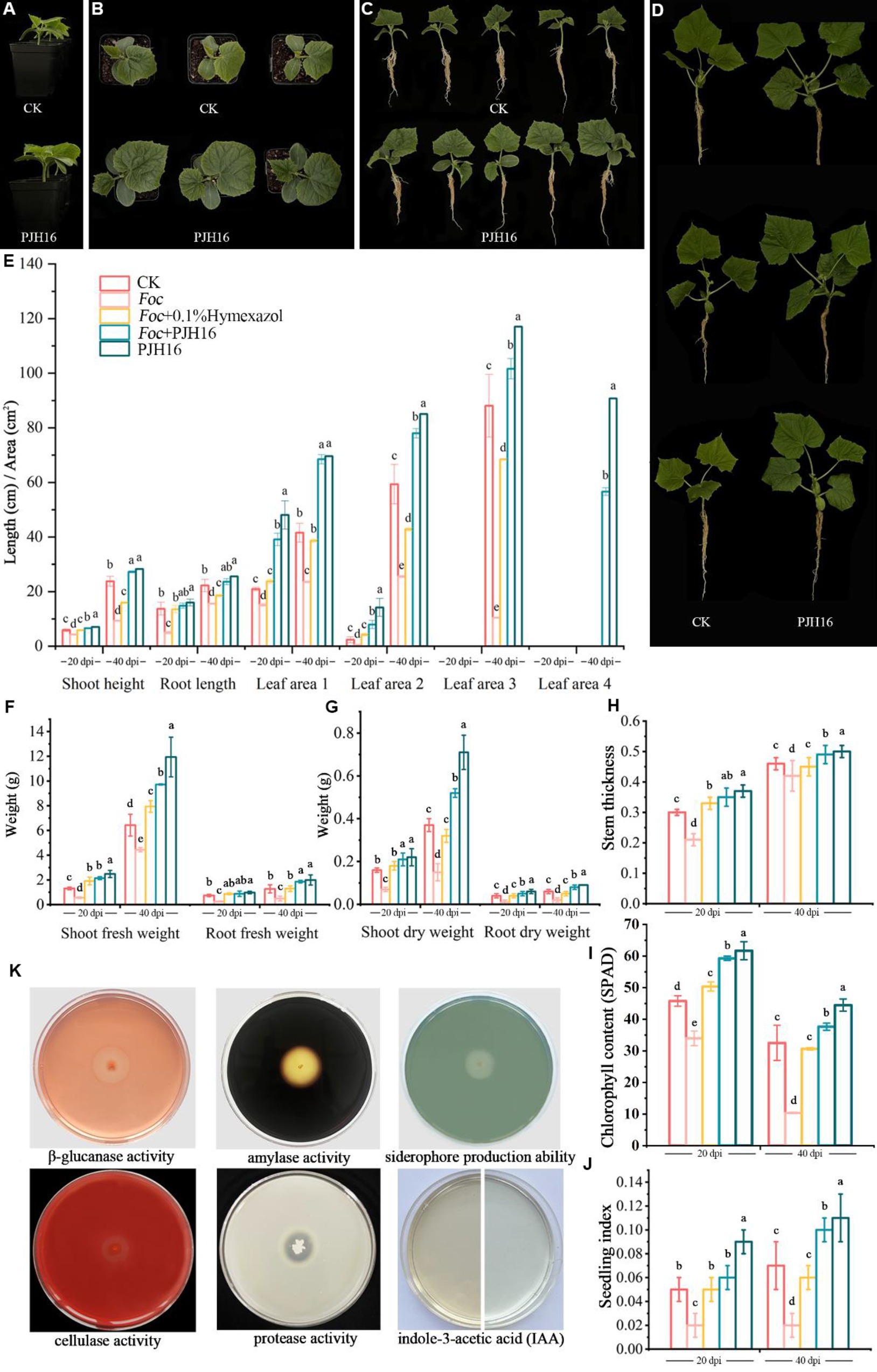
Figure 3. The growth-promoting effect of PJH16 strain on cucumber seedlings and its antifungal and growth-promoting properties in vitro were detected. (A–C) Control treatment (CK) and PJH16-inoculated cucumber seedlings at 20 days post-inoculation (dpi); (D) Control treatment (CK) and cucumber seedlings inoculated with PJH16 at 40 dpi; (E) Evaluation of plant height, root length, and leaf area of cucumber seedlings inoculated at 20 and 40 dpi; (F) Measurement of shoot fresh weight and root fresh weight for cucumber seedlings inoculated at 20 and 40 dpi; (G) Assessment of shoot dry weight and root dry weight for cucumber seedlings inoculated at 20 and 40 dpi; (H) Determination of stem thickness for cucumber seedlings inoculated at 20 and 40 dpi across five treatments; (I) Quantification of chlorophyll content in cucumber seedlings inoculated at 20 and 40 dpi; (J) Calculation of the robust seedling index for cucumber seedlings inoculated with five treatments at 20 and 40 dpi; The first row of (K) from left to right showcases the detection of β-glucanase activity, amylase activity, and siderophore production ability of the PJH16 strain; the second row from left to right displays the detection of cellulase activity, protease activity, and indole-3-acetic acid (IAA) production ability of the PJH16 strain. Within the same column, different letters (a–e) indicate significant differences at p < 0.05 level.
3.5 Detection of antifungal and growth-promoting properties in vitro
The in vitro antifungal and growth-promoting properties of PJH16 showed that PJH16 had the activities of β-1,3-glucanase, cellulase, amylase and protease, and could act on the cell wall of fungi by secreting these enzymes to antagonize pathogens. In addition, in terms of promoting plant growth, we found that PJH16 can produce indole acetic acid and siderophores, which is beneficial to plant growth and development (Figure 3K).
3.6 Whole genome sequencing and assembly of PJH16 strain
In this research endeavor, we employed a dual approach utilizing PacBio RSII to construct third-generation sequencing library. This strategy facilitated the efficient and accurate completion of whole-genome sequencing for the strains, allowing for a comprehensive exploration of their disease resistance mechanisms. The DNA sequencing of the strain yielded a total of 24,869 High Fidelity reads, characterized by an average length of 10,450.15 bp and an N50 of 10,764 bp. The genome showcased an average coverage depth of 45.18X (Figure 4A), and the assembled genome sequence has been meticulously cataloged in GenBank with the accession number CP137742. Comprising a circular chromosome spanning 5,746,138 bp, the strain’s genome exhibited a GC content of 45.81% (Figure 4D). Through analysis, a total of 5,187 protein-coding genes were predicted, encompassing 86.36% of the genome. The remaining 208 genes comprised tRNA (111 genes), rRNA (42 genes), and other non-coding RNAs (55 genes) (Figure 4B). Annotation of the protein sequences from the predicted genes against various databases (E value = 1e−5) unveiled matches for 5,058 (99.12%), 4,088 (76.44%), 2,536 (54.85%), and 3,518 (75.39%) protein-coding genes in NR, SwissProt, COG, KEGG, and GO databases, respectively (Figure 4D; Supplementary Table S3).
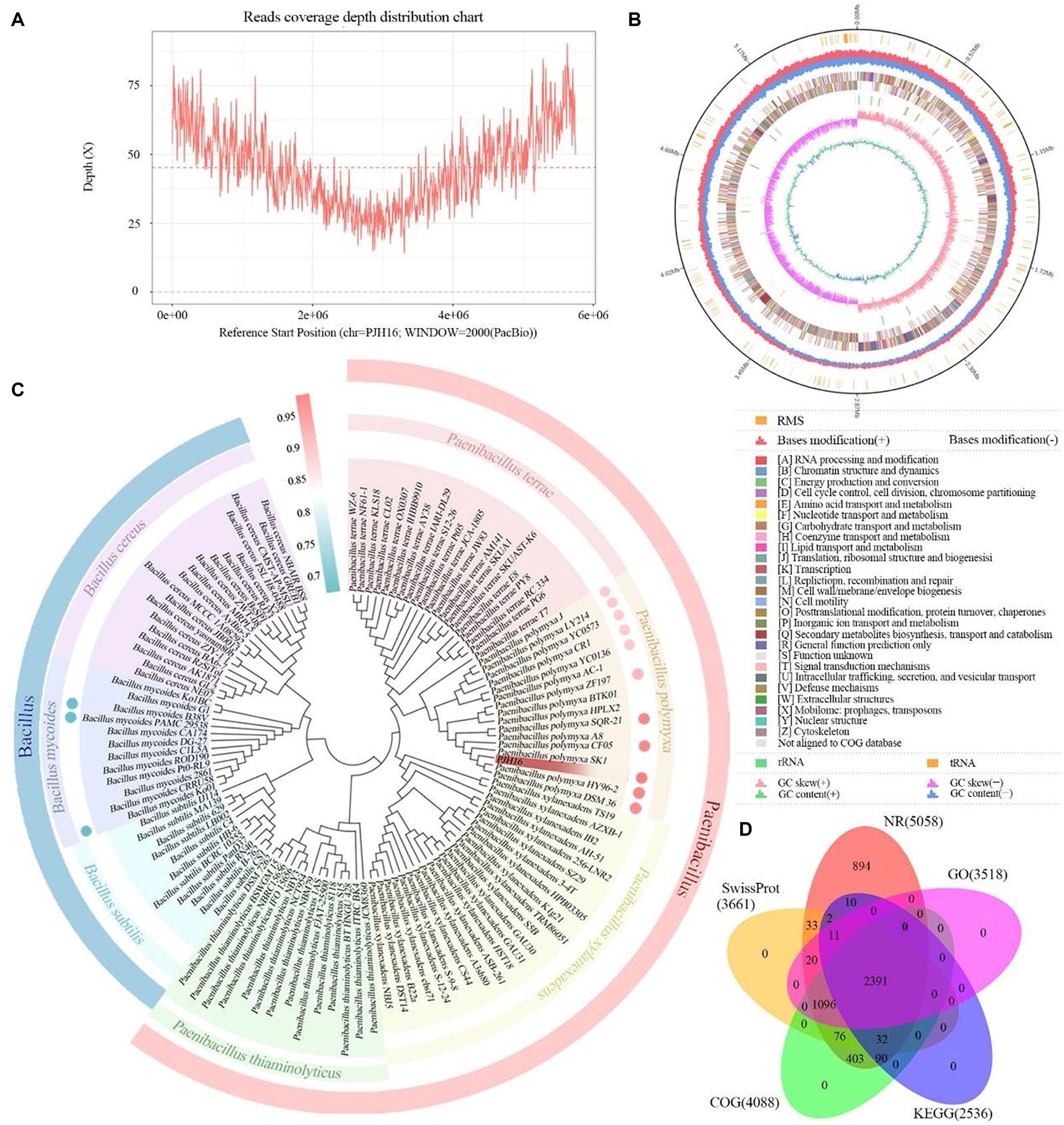
Figure 4. The basic information of whole genome sequencing, assembly, identification and functional annotation of P. polymyxa PJH16. (A) Visualization of genome coverage depth distribution. (B) Comprehensive genomic map of P. polymyxa PJH16. The concentric circles represent, from the innermost to the outermost: genome size (depicted by a black line); distribution of restriction modification systems on the forward (depicted in red) and reverse (depicted in blue) strands; COG classification of protein-coding genes, spanning both the forward and reverse strands; distribution of tRNA (depicted in brown) and rRNA (depicted in green); GC skewness; GC content. (C) Taxonomic identification of P. polymyxa PJH16. The concentric circles represent: Phylogenetic relationships of P. polymyxa PJH16 with 114 strains across 7 genera of Bacillus and Paenibacillus; Average Nucleotide Identity (ANI) analysis of P. polymyxa PJH16 compared to 10 strains of P. polymyxa and 3 strains of B. mycoides. (D) Comprehensive analysis and statistical representation of shared and unique annotations in the foundational database.
In the NR database, 53.5% of the sequences exhibited the highest similarity to species in the Paenibacillus group, followed by 41% of the sequences showing significant similarity to P. polymyxa. Additionally, 1.86% of the sequences were most similar to those of P. polymyxa SQR-21 (Supplementary Figure S1A). When compared with the SwissProt database, 49.17% of the sequences displayed the highest similarity to Bacillus subtilis (Supplementary Figure S1B).
3.7 Species identification and phylogenetic analysis of PJH16 strain
Based on the results of the phylogenetic tree, we found that the strain PJH16 clustered with 15 P. polymyxa strains, indicating that it had a close relationship with the P. polymyxa strain (Figure 4C). Based on ANI analysis, the gene correlation between PJH16 and 10 P. polymyxa strains with known whole genome sequences was analyzed. It was found that the ANI value between PJH16 and P. polymyxa HY96-2, P. polymyxa DSM36, P. polymyxa SQR-21 and P. polymyxa CF05 was >98%, which was higher than the 96% threshold (Figure 4C; Supplementary Table S4). The strain PJH16 had the highest ANI value (98.63%) with P. polymyxa SQR-21, and the strain PJH16 had greater homology with P. polymyxa SQR-21.
3.8 Genome function annotation of Paenibacillus polymyxa PJH16
Basic database annotations include NR (Supplementary Figure S1A), SwissProt (Supplementary Figure S1B), KEGG (Supplementary Figure S2A), GO (Supplementary Figure S2B), COG/KOG (Supplementary Figure S2C). Here, we use appropriate annotation tools to annotate the protein sequence of the predicted gene to the corresponding database. The whole genome of P. polymyxa PJH16 was annotated in the basic database to contain a large number of genes related to siderophore biosynthesis such as feuA, feuB, feuC, fepC, and fhuC (Supplementary Table S7).
3.9 Specific function database annotation
In our investigation, a comprehensive analysis of the P. polymyxa PJH16 genome revealed a total of 499 antibiotic resistance genes, encompassing various categories such as antibiotic synthesis genes, antibiotic resistance genes, and others, as identified through a meticulous comparison and analysis against the CARD database (Supplementary Table S6). Furthermore, a thorough annotation against the VFDB database uncovered 988 genes, featuring notable elements like motA and motB, associated with flagellar movement, luxS, a key quorum sensing regulatory gene, pgaC, involved in extracellular polysaccharide synthesis, as well as fimB and fimE genes contributing to Type 1 fimbriae formation (Supplementary Table S8).
A total of 1,390 genes in P. polymyxa PJH16 genome were annotated to the membrane transporter (TCDB) database, of which 532 genes were annotated to primary active transporters, 317 genes were annotated to electrochemical potential-driven transporters, and 274 genes were annotated to incompletely characterized transport systems. A total of 94 genes were annotated to channel/pores, 89 genes were annotated to group translocators, 72 genes were annotated to accessory factors involved in iransport, and 12 genes were annotated to transmembrane electron carriers (Figure 5D).
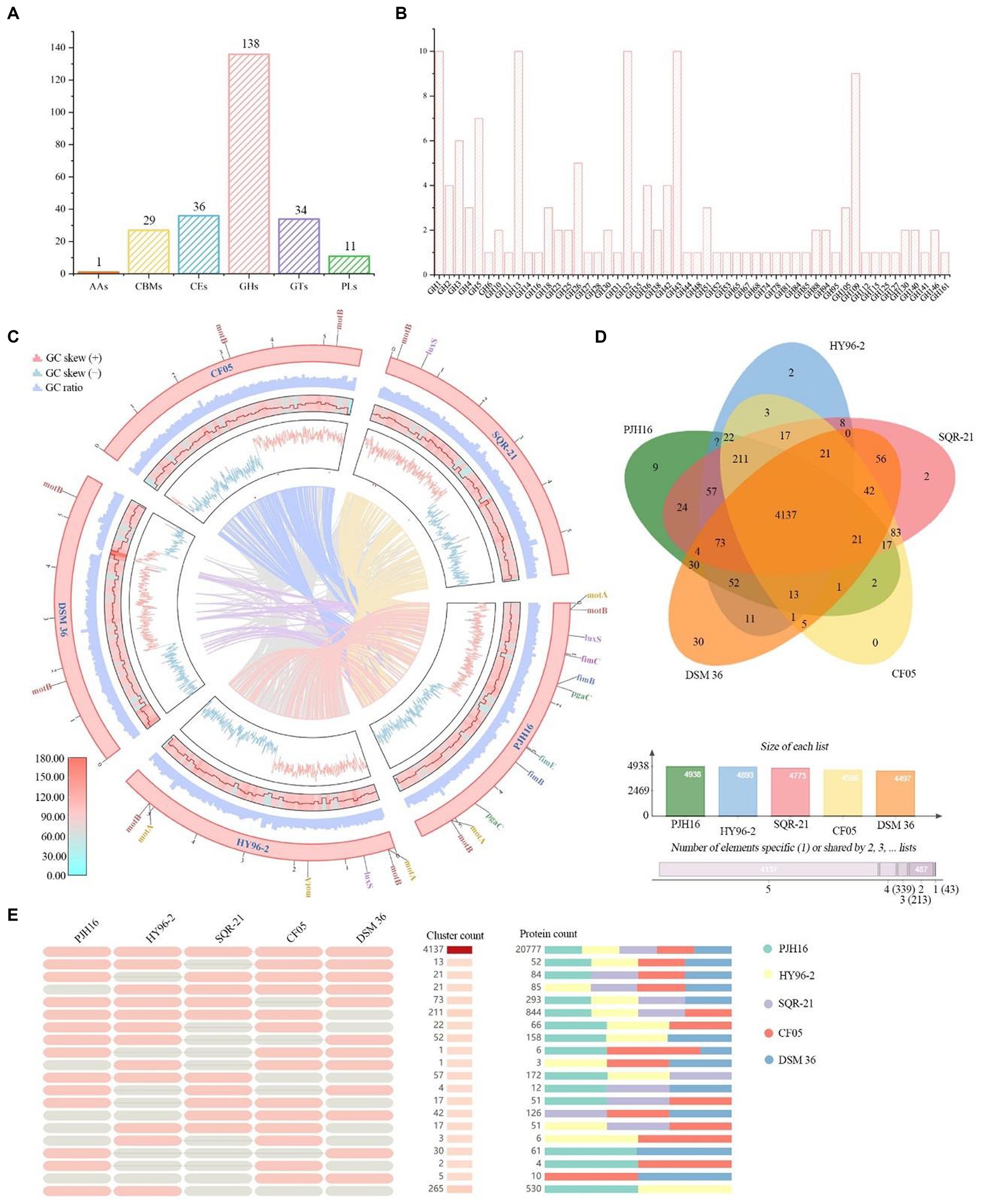
Figure 5. CAZymes analysis of P. polymyxa PJH16 genome and genome comparison of P. polymyxa PJH16 and four other P. polymyxa strains. (A) Distribution of gene counts across carbohydrate-active enzyme (CAZy) families in the P. polymyxa PJH16 genome. (B) Functional characterization of the glycoside hydrolase family based on the CAZy database. (C) Collinearity analysis of P. polymyxa PJH16 with three other P. polymyxa strains. The innermost circle depicts the comparison of P. polymyxa PJH16 with three other P. polymyxa strains, followed by the nratio, GC skews, gene density, and GC ratio of the genomes of the five P. polymyxa strains. Additionally, it illustrates five genes associated with biofilm formation in the P. polymyxa strains’ genomes. (D) Venn diagram showcasing the number of unique and shared gene clusters between P. polymyxa PJH16 and four other P. polymyxa strains. (E) Presence or absence of orthologous gene clusters in different species, where pink denotes presence and gray indicates absence.
3.10 CAZymes analysis of Paenibacillus polymyxa PJH16 genome
A comprehensive analysis revealed a total of 224 genes identified as potential Carbohydrate-Active Enzymes (CAZymes), encompassing 136 Glycoside Hydrolases (GHs), 34 Glycosyl Transferases (GTs), 36 Carbohydrate Esterases (CEs), 27 Carbohydrate-Binding Modules (CBMs), 11 Polysaccharide Lyases (PLs), and 1 Auxiliary Activity (AAs) (Figure 5A). These enzymes play a pivotal role in facilitating the binding of enzymes to their substrates. Notably, 18 genes (orf00422, orf00562, orf01040, orf01052, and orf01565, etc.) were concurrently classified as GHs and CBMs, while one gene (orf00227) was categorized as CBMs and PLs simultaneously. Another gene (orf04406) was concomitantly classified as CEs and PLs. Within the GH family, antifungal CAZymes were identified, including chitinase (GH18), cellulase (GH5), endoglucanase (GH5, GH51), β-glucanase (GH1), starch debranching enzyme (GH13_31, GH13_28, GH13_29), and L-fructosyltransferase (GH32), all of which have the potential to impede the growth of plant pathogens (Figure 5B). The GH43 family exhibited a diverse range of enzymes such as α-L-arabinofuranosidase, β-D-xylosidase, α-L-arabinosidase, and β-D-galactosidase involved in the debranching and degradation of hemicellulose and pectin polymers. Additionally, the GH3 enzyme, predominantly comprised of stereochemically retained β-glucosidase, and the β-N-acetylglucosaminidase subfamily were identified. These enzymes, widely distributed in bacteria, fungi, and plants, serve multiple functions such as cellulose biomass degradation, plant and bacterial cell wall remodeling, energy metabolism, and defense against pathogens. The genomic distribution of CAZymes in strain PJH16 underscores its potential to counteract fungal pathogens.
3.11 Secondary metabolic potential of Paenibacillus polymyxa PJH16
Within the genomic landscape of P. polymyxa PJH16, a total of 19 gene clusters associated with the biosynthesis of secondary metabolites were discerned (Table 3). These putative gene clusters are diverse, encompassing 9 clusters dedicated to the encoding of NRPS (nonribosomal peptide synthetase), 3 clusters for transAT-PKS (trans-acyltransferase polyketide synthase), 4 clusters affiliated with lanthipeptide-class-i, 3 clusters for cyclic-lactone-autoinducer, 1 cluster each for RRE-containing, proteusin, lassopeptide, T1PKS, betalactone, and ranthipeptide. Notably, Cluster 4 and Cluster 6 have the capability to encode both NRPS and transAT-PKS, while Cluster 9 encodes both NRPS-like and cyclic-lactone-autoinducer. Cluster 12 can encode both T1PKS and NRPS. Cluster 15 encodes transAT-PKS, NRPS, T3PKS and PKS-like. Among the nine clusters encoding NRPS, four clusters had 100% similarity to the known fusaricidin B, tridecaptin and polymyxin synthetic clusters, respectively, and the clusters encoding T1PKS and NRPS had 5% similarity to the laterocidine synthetic cluster. The cluster that encodes transAT-PKS, NRPS, T3PKS and PKS-like has 35% similarity with aurantinin B/aurantinin C/aurantinin D synthetic cluster. The four clusters encoding lanthipeptide-class-i are 100% similar to known synthetic clusters of paenibacillin, paenilan, S-layer glycan, and paenicidin A, respectively. The cluster similar to the paenibacillin synthetic cluster also encodes cyclic-lactone-autoinducer. Within the antiSMASH database, nine distinct biosynthetic gene clusters were identified, exhibiting dissimilarity to known clusters. These novel gene clusters appear to be potential candidates for the biosynthesis of secondary metabolites, necessitating further isolation and characterization. Notably, two of these clusters are implicated in transAT-PKS and NRPS biosynthesis, while one cluster is associated with the synthesis of NRPS-like and cyclic-lactone-autoinducer. The remaining clusters encompass a variety of functionalities, including RRE-containing, proteusin, Lassopeptide, NRPS, cyclic-lactone-autoinducer, and ranthipeptide. It’s noteworthy that the 19 gene clusters responsible for secondary metabolite biosynthesis in the genome of P. polymyxa PJH16 were also observed in the genomes of the other four P. polymyxa strains. However, the specific composition of gene clusters in P. polymyxa PJH16 differs from each of the three other strains (Supplementary Table S9).
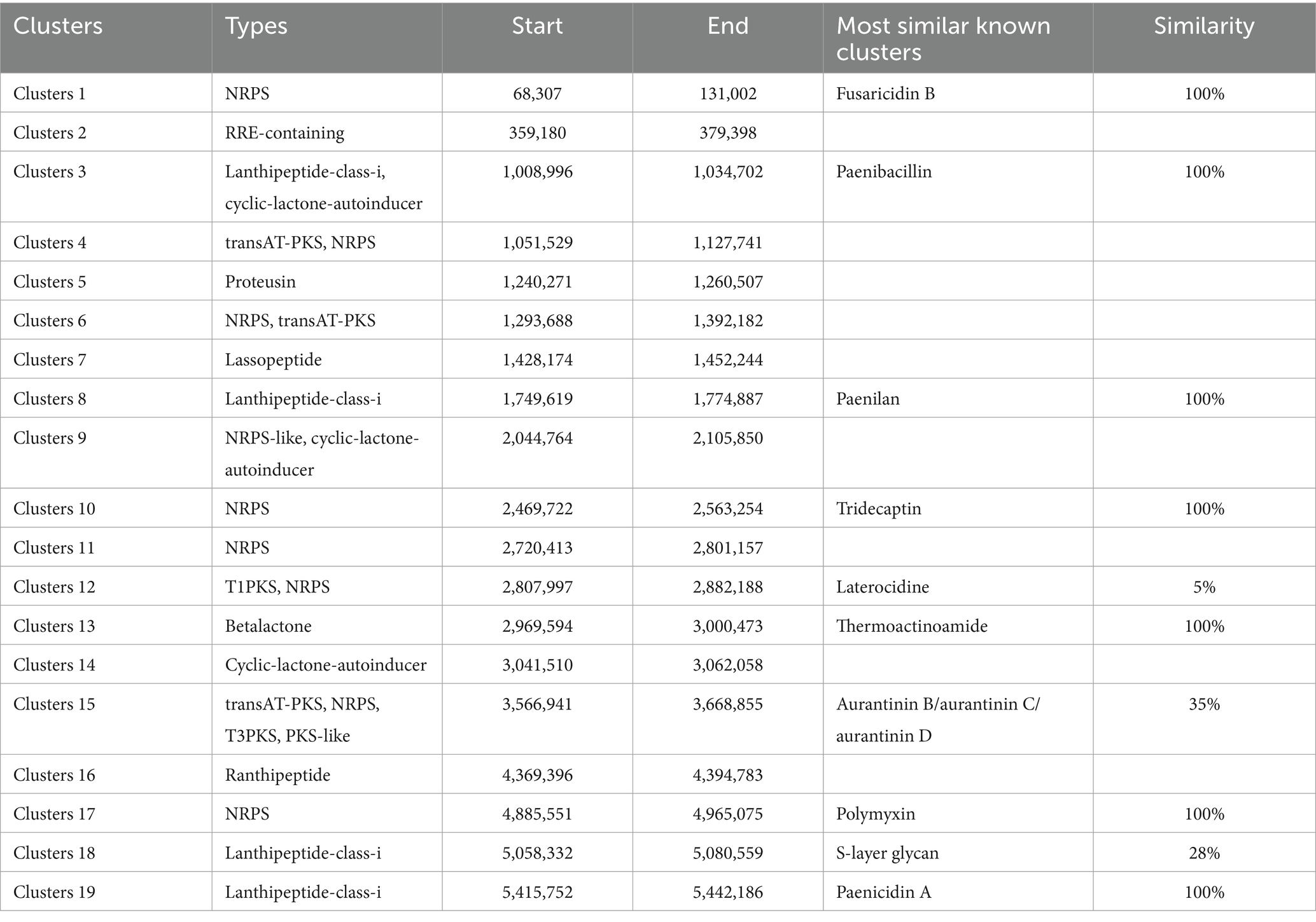
Table 3. The hypothetical gene cluster lists encoding secondary metabolites predicted by antiSMASH extracted from genomes of P. polymyxa PJH16.
3.12 Comparison of genomes
In order to obtain the collinearity analysis between the P. polymyxa PJH16 genome and the other four P. polymyxa strains, the whole genome sequences (P. polymyxa HY96-2, P. polymyxa SQR-21, P. polymyxa CF05 and P. polymyxa DSM 36) in the NCBI sequence library were analyzed for collinearity (Figure 5C). The results showed that P. polymyxa PJH16 had the best collinearity with P. polymyxa HY96-2 and P. polymyxa SQR-21, and the genome structure arrangement was basically similar without gene mutation, which proved the close relationship between the two strains and supported our phylogenetic results. There was strong chances Horizontal gene transfer (HGT) between P. polymyxa PJH16 strain and P. polymyxa CF05 and P. polymyxa DSM 36, respectively (Figure 5C). At the same time, we also annotated the genes related to biofilm formation in these five strains. We found that the genes related to biofilm formation were the most annotated in the genome of P. polymyxa PJH16.
We also performed pan-genome analysis to compare P. polymyxa PJH16 with these four P. polymyxa strains. As shown in the figure, the number of homologous genes of P. polymyxa PJH16, P. polymyxa HY96-2, P. polymyxa SQR-21, P. polymyxa CF05 and P. polymyxa DSM 36 were 4,938, 4,893, 4,773, 4,596, and 4,497, respectively. The number of single copy genes was 205, 261, 225, 71, and 549, respectively (Table 4). The number of homologous genes shared by the four strains was 4,137, and there were 9 unique gene clusters of P. polymyxa PJH16. There were 2 unique gene clusters in P. polymyxa HY96-2. There are 2 unique gene clusters in P. polymyxa SQR-21; there are 30 specific gene clusters in P. polymyxa DSM 36; P. polymyxa CF05 had no specific gene cluster (Figures 5D,E).
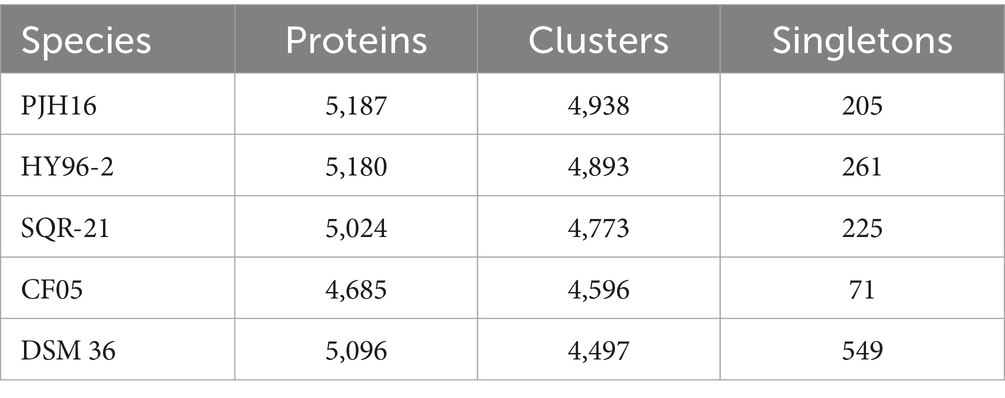
Table 4. The number of proteins, clusters, and singletons in P. polymyxa PJH16 and four other P. polymyxa strains.
4 Discussion
In our study, it was examined the effect of bacterial strain PJH16 on the disease Fusarium oxysporum f. sp. cucumerinum both in vitro and in vivo, and followed by investigating the mechanism of illness biological control by using the molecular identification of the bacterial strain PJH16 to provide information for efficient Fusarium wilt disease management in cucumber.
This research began with the isolation of P. polymyxa PJH16 strain from the rhizosphere soil of healthy cucumber that was verified by double culture experiments to check the effective inhibition for the Fusarium oxysporum f. sp. cucumerinum, and its antagonistic band width that reached 11 mm. The obtained results showed that the genome comprises 110450.15 bp circular chromosome, and its GC content is 45.81%.
When defining a species, the phylogenetic theory considers the evolutionary and genealogical relationships between organisms. Thus, the phylogenetic tree analysis based on 16S rRNA gene sequence achieved that the strain PJH16 had a close relationship with P. polymyxa genus and the P. polymyxa HY96-2 was the closest, followed by P. polymyxa DSM 36 strain. According to ANI analysis, PJH16 was the most similar to P. polymyxa HY96-2, followed by P. polymyxa SQR-21, P. polymyxa DSM 36 and P. polymyxa CF05. This analysis indicated that PJH16 strain belongs to P. polymyxa genera, additionally is closely related to P. polymyxa HY96-2, P. polymyxa SQR-21, P. polymyxa DSM 36 and P. polymyxa CF05. In such a case, horizontal gene transfer (HGT) plays a vital role in the structure of bacterial genomes and provides the raw material for natural selection in diversifying bacterial lineages, especially in the evolution of specific lineages, species and subspecies (Nowell et al., 2014). Through the global alignment of P. polymyxa PJH16, P. polymyxa SQR-21, P. polymyxa CF05 and P. polymyxa DSM 36 chromosomes by Mauve and TBtools visualization, we found that the genome of P. polymyxa PJH16 was highly homologous to P. polymyxa HY96-2 and P. polymyxa SQR-21. At the same time, collinearity analysis showed that there may be horizontal gene transfer between P. polymyxa PJH16 and the other two strains. In addition, genome comparative analysis showed that P. polymyxa PJH16 had the largest number of encoded proteins and gene clusters. Such results indicate that P. polymyxa PJH16 has a certain specificity, and its biological control mechanism may differ from that of other P. polymyxa.
Many bacteria belonging to P. polymyxa have the ability to promote plant growth and inhibit plant pathogens. They have potential plant growth promoting ability and known as Plant Growth Promoting Rhizobacteria (PGPR) (Timmusk and Wagner, 1999) and biological control agents (BCAs) (Lee et al., 2020). Despite this strain is widely distributed naturally, has a wide rage of industrial, agricultural, and environmental applications, and is generally regarded as an environmentally safe bacterium (Timmusk and Wagner, 1999), there have been few reports on the analysis of P. polymyxa gene sequence to date. Therefore, it was important to understand the biological control mechanism of P. polymyxa PJH16 at the molecular level. The predicted mechanisms are biofilm formation, the production of antifungal compounds, resistance induction in plants, and plant growth promotion.
Biofilm formation helps bacteria establish self-protection mechanisms, affects the bacterial response to nutrients in the environment and antibiotic resistance, and promotes root colonization (Ongena and Jacques, 2008). For instance, the isolated P. polymyxa B1 and B2 strains can colonize in the intercellular space after invading the roots of Arabidopsis thaliana, and then form a biological protective film on its surface, which can effectively prevent pathogenic fungus from invading Arabidopsis thaliana (Timmusk et al., 2005). With a deeper understanding of genetic identification, the key genes affecting biofilm formation included flagellar movement-related genes, quorum sensing genes, and extracellular polysaccharide production-related genes. The motA and motB genes related to the formation and movement of flagella (Platzer et al., 1997), were found in the genome of PJH16. In the context of biofilm formation, the quorum sensing (QS) system plays a pivotal role in orchestrating gene expression and coordinating cellular activities. This regulatory mechanism is recognized as a significant factor in governing biofilm development across various bacterial species (Miller and Bassler, 2001; Raafat et al., 2019). Notably, our genomic analysis revealed the presence of the luxS gene in the PJH16 strain. LuxS is a pivotal regulatory gene within the quorum sensing system, specifically mediated by autoinducer 2 (AI-2) (Ma et al., 2017). Another gene involved in biofilm formation in PJH16 is pgaC gene, which is involved in the synthesis of polymer poly β-1,6-N-acetyl-D-glucosamine (PNAG) (Chen et al., 2014). The type 1 fimbriae regulated by fimB and fimE genes can help P. polymyxa to bio-attach along the roots in the soil, positively impacting plant growth and health. From the above results, it can be speculated that the PJH16 strain can hinder the attachment and invasion of fungal pathogens by forming a tightly bound biofilm, and seize the nutrition and growth space required by fungi. In addition, extracellular polysaccharides and antifungal substances can interfere with the key processes of fungal cell wall synthesis, nutrient uptake and metabolism, thereby inhibiting the reproduction and spread of fungal pathogens (Figure 6A). However, whether P. polymyxa PJH16 can produce biofilm still needs to be verified by relevant experiments.
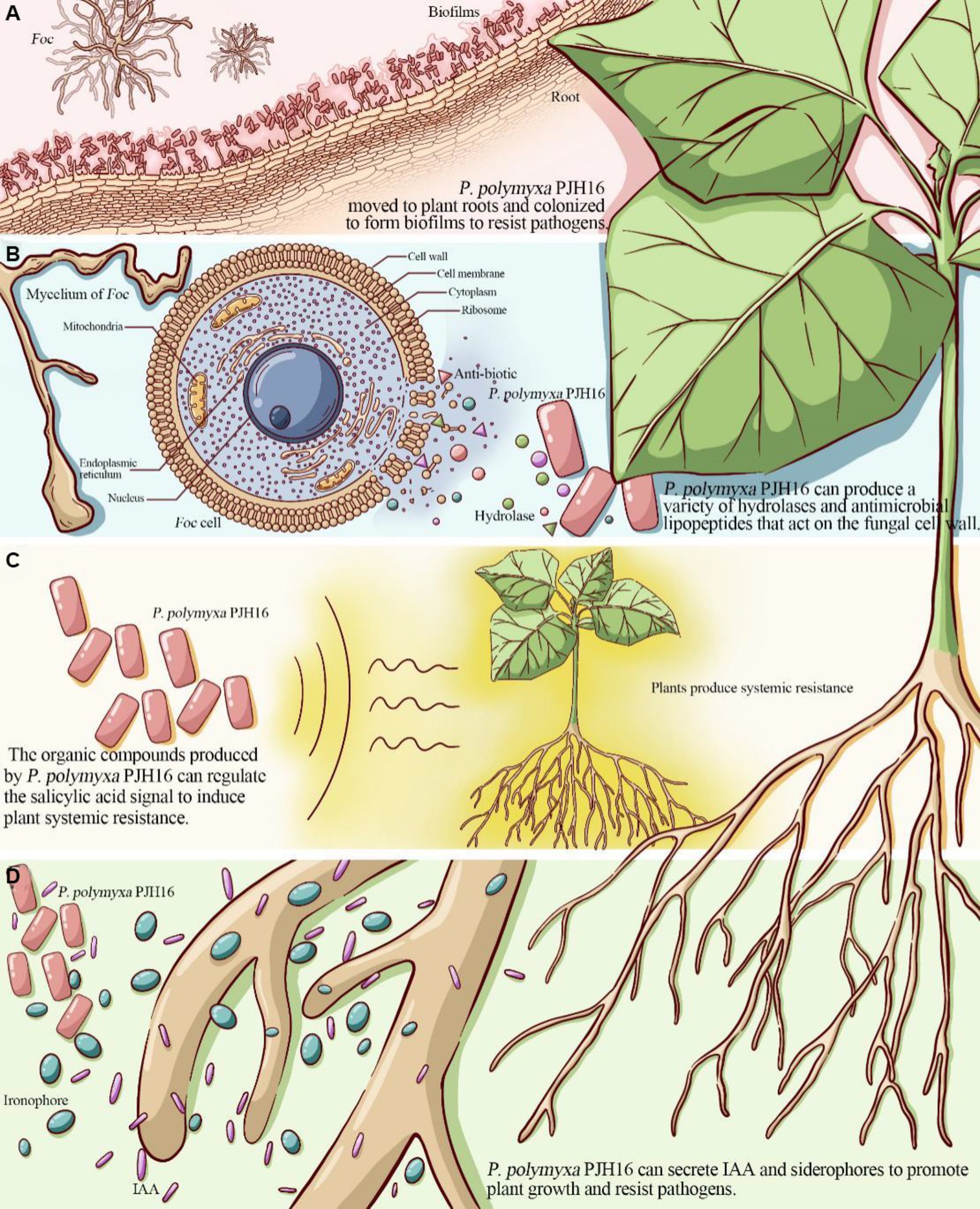
Figure 6. The proposed mechanism of P. polymyxa PJH16 antagonizing Foc. (A) P. polymyxa PJH16 moved to plant roots and colonized to form biofilms to resist pathogens. (B) P. polymyxa PJH16 can produce a variety of hydrolases and antimicrobial lipopeptides that act on the fungal cell wall. (C) The organic compounds produced by P. polymyxa PJH16 can regulate the salicylic acid signal to induce plant systemic resistance. (D) P. polymyxa PJH16 can secrete IAA and siderophores to promote plant growth and resist pathogens.
One of the biological control mechanisms is to secrete antagonistic substances for inhibiting fungal disease (XueLi et al., 2008). According to their material types, they can be divided into hydrolases (Beatty and Jensen, 2002) such as protease, β-1,3-glucanase, cellulase, xylanase and chitinase (Kwon et al., 2016), polypeptide antibiotics (Raza et al., 2008) such as polymyxin, fusaricidin and colistin (Naghmouchi et al., 2012), and plant hormones (Niu et al., 2013) such as cytokinin, auxin and indole phenol (Mei et al., 2014; Park et al., 2014). Previously, it was shown that P. polymyxa can inhibit spore germination and mycelial growth of Fusarium by producing glucanase, chitinase, cellulase and protease related to the destruction of eukaryotic cell wall (Nielsen and Sorensen, 1997; Huang et al., 2020). By employing scanning electron microscopy approach to observe the mycelium of Foc in the dual culture antagonism experiment, the P. polymyxa PJH16 strain caused deformation of the mycelium, as twisting, folding and shrinkage. Simultaneously, in vitro antifungal properties test showed that P. polymyxa PJH16 could secrete protease, amylase, β-glucanase and cellulase. These findings pointed out that there is a role of P. polymyxa PJH16 strain in inhibiting the growth and development of F. oxysporum by producing substances such as hydrolases and antibiotics. Furthermore, the genome of P. polymyxa PJH16 has the synthetic gene clusters of fusaricidin and paenibacillin, which have inhibitory effects on fungi. Nine gene clusters in the genome of P. polymyxa PJH16 have not been found to have homologous proteins in the database. Among them, ranthipeptides are free radical non-α-sulfur-containing peptides, which are a new class of natural products and belong to the ribosome synthesis and post-translational modification peptide (RiPP) superfamily. Studies have shown that ranthipeptides may be involved in quorum sensing and control of cell metabolism (Chen Y. L. et al., 2020). Similar to cyclic peptides, cyclic-lactone-autoinducer is a class of compounds that play a role in intercellular communication in bacteria. By binding to specific receptor proteins, it triggers signal transduction pathways within bacteria and regulates bacterial growth, development, biofilm formation, biological attachment, and toxin production (Mull et al., 2018). Whereas, P. polymyxa SQR-21 and P. polymyxa CF05 lacked the synthetic gene cluster of paenibacillin. Taken together, the differences in antibiotic synthesis gene clusters between P. polymyxa PJH16 and other P. polymyxa may lead to differences in their ability to control plant diseases (Figure 6B). However, this is only based on the prediction of the genome of P. polymyxa PJH16, and does not specifically detect whether the strain can produce related compounds. Therefore, subsequent experiments are needed to verify its metabolites.
The results of the existing research on the mechanism of biocontrol bacteria confirmed that some biocontrol bacteria have induced systemic resistance (ISR) in plants under the ‘plant-pathogen-growth-promoting bacteria’ interaction mode, and can be used as non-pathogenic microorganisms to induce host plants to produce a series of systemic resistance, thereby assisting host crops to defend against pathogenic microorganisms (Qiao et al., 2017; Wang et al., 2017). ISR regulated by jasmonic acid and ethylene signaling pathways is one of the important mechanisms for the prevention and control of plant pathogen infection by plant growth promoting rhizobacteria (Pieterse et al., 2014). Antagonistic bacteria P. polymyxa W3 suspension and its filtrate can induce systemic resistance of tomato leaves to Botrytis cinerea. The activities of PAL, peroxidase POD, PPO and SOD in leaves were significantly enhanced after P. polymyxa W3 and its filtrate treatment (Tong et al., 2004). The current study found that inoculation of P. polymyxa PJH16 strain significantly increased the activities of LOX, PAL, CAT, PPO and SOD in cucumber seedlings, indicates that the strain can help cucumber plants resist Fusarium oxysporum infection through ISR. In addition, many previous studies have shown that fusaricidin can protect plants against pathogens through ISR. Fusaricidin produced by P. polymyxa WLY78 strain can induce systemic resistance to fusarium wilt of cucumber through salicylic acid (SA) signal, and significantly inhibit the production of disease (Li and Chen, 2019). Hence, we speculated that the isolated strain P. polymyxa PJH16 can regulate salicylic acid signals by producing fusaricidin and other compounds to make cucumbers induce systemic resistance. Such findings need to be further explored (Figure 6C).
As an important PGPR resource, P. polymyxa can produce auxin analogs, ethylene, indole acetic acid, cytokinins and siderophores to directly promote plant growth. P. polymyxa SQR-21 is not only an effective biocontrol agent against watermelon fusarium wilt, but also a plant growth-promoting rhizobacteria. It can promote the growth and development of watermelon plants by inducing the expression of several proteins involved in growth, photosynthesis and other metabolic and physiological activities (Yaoyao et al., 2017). It has been proved that the fermentation broth and bacterial suspension of P. polymyxa CF05 strain have growth promoting effect on tomato seedlings. The fresh weight of tomato seedlings treated with P. polymyxa CF05 fermentation broth increased by 272.0%, and the dry weight increased by 266.7% (Guo et al., 2014). Likewise, the growth-promoting test of cucumber seedlings verified that P. polymyxa PJH16 affected growth-promotion and could secrete indoleacetic acid and siderophore. It is well known that indole-3-acetic acid (IAA) is an essential plant hormone for plant growth. The competition of siderophores for iron is an important feature of many bacterial biocontrol agents against plant pathogenic fungi. Iron is one of the key elements necessary for plant growth and development, and is essential for normal metabolism and biochemical reactions of plants. Along with the same effect, an appropriate amount of iron helps to enhance the stress resistance of plants, consequently, the plants can better cope with environmental stress, such as salt stress, drought and cold. However, iron in soil is not always easily absorbed by plants. The siderophores secreted by bacteria can effectively convert insoluble iron in soil into soluble forms, making plants more easily absorbed, thus helping to prevent leaf yellowing and wilting caused by iron deficiency. Many biocontrol bacteria have the ability to produce siderophores, such as P. polymyxa (Padda et al., 2017), P. riograndensis (Beneduzi et al., 2010), P. sinopodophylli (Chen et al., 2016) and P. panacihumi (Kim et al., 2018). At the same time, the genome analysis of PJH16 showed that it contained a large number of genes related to siderophore biosynthesis, such as feuA, feuB, feuC, fepC, and fhuC. A broadly similar point has been confirmed that PJH16 can promote plant growth by secreting indole acetic acid and siderophores, which in turn helps plants resist pathogens (Figure 6D).
5 Conclusion
This study supported the idea that the P. polymyxa PJH16 strain can be an eco-biopesticide agent demonstrating multifunctional positive features. The phenotypic, phylogenetic, and genomic analyses have elucidated the mechanism of P. polymyxa PJH16 antagonizing Fusarium cucumber disease (Figure 6). Several factors could explain this mechanism. Firstly, P. polymyxa PJH16 moved to plant roots and colonized to form biofilms to resist pathogens and protect plants. Secondly, P. polymyxa PJH16 can produce a variety of hydrolases and antimicrobial lipopeptides that act on the fungal cell wall, such as glucanase, proteinase and cellulase; therefore, it can directly inhibit the growth of pathogenic fungi. Thirdly, the organic compounds produced by P. polymyxa PJH16 can regulate the salicylic acid signal to induce plant systemic resistance. Finally, P. polymyxa PJH16, as an important PGPR, can secrete IAA and siderophores to promote plant growth and resist pathogens. However, the two conclusions of the formation of biofilm and the production of antifungal compounds based on genome-wide analysis have not been experimentally verified, and related experiments will be carried out in the future. Although this strain has not been used in different environmental conditions, plants and pathogenic fungi to detect its broad-spectrum antifungal activity, it can be used as an ideal strain for biological control and plant growth promotion. We will also carry out relevant broad-spectrum experiments to verify whether it can be widely used in different regions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
FY: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. HJ: Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. KM: Funding acquisition, Writing – review & editing. AH: Writing – review & editing. XW: Supervision, Writing – review & editing. SL: Resources, Validation, Writing – review & editing. GC: Formal analysis, Project administration, Writing – review & editing. LY: Validation, Visualization, Writing – review & editing. BT: Resources, Supervision, Writing – review & editing. XS: Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Technology System of Bulk Vegetable Industry in Henan Province (HARS-22-07-G2), Major Science and Technology Special Projects in Henan Province (201111110500), National Characteristic Vegetable Industry Technology System (CARS-24-G-15) and Key R & D projects in Henan Province (221111110100).
Conflict of interest
Author LY was employed by Henan Natural Products Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1359263/full#supplementary-material
References
Ayaz, M., Ali, Q., Farzand, A., Khan, A. R., Ling, H., and Gao, X. (2021). Nematicidal volatiles from Bacillus atrophaeus GBSC56 promote growth and stimulate induced systemic resistance in tomato against Meloidogyne incognita. Int. J. Mol. Sci. 22:5049. doi: 10.3390/ijms22095049
Beatty, P. H., and Jensen, S. E. (2002). Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can. J. Microbiol. 48, 159–169. doi: 10.1139/w02-002
Beneduzi, A., Costa, P. B., Parma, M., Melo, I. S., Bodanese-Zanettini, M. H., and Passaglia, L. M. P. (2010). Paenibacillus riograndensis sp nov., a nitrogen-fixing species isolated from the rhizosphere of Triticum aestivum. Int. J. Syst. Evol. Microbiol. 60, 128–133. doi: 10.1099/ijs.0.011973-0
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, K. M., Chiang, M. K., Wang, M. L., Ho, H. C., Lu, M. C., and Lai, Y. C. (2014). The role of pgaC in Klebsiella pneumoniae virulence and biofilm formation. Microb. Pathog. 77, 89–99. doi: 10.1016/j.micpath.2014.11.005
Chen, H., Lin, J., Liao, F., and Lin, B. (2010). Antibacterial activity and mechanism of Paenibacillus Polymyxa CP7 against Peronophythora litchi. Acta Horticult. Sinica 37, 1047–1056. doi: 10.16420/j.issn.0513-353x.2010.07.003
Chen, F., Wang, M., Zheng, Y., Luo, J., Yang, X., and Wang, X. (2010). Quantitative changes of plant defense enzymes and phytohormone in biocontrol of cucumber fusarium wilt by Bacillus subtilis B579. World J. Microbiol. Biotechnol. 26, 675–684. doi: 10.1007/s11274-009-0222-0
Chen, C. Q., Xin, K. Y., Li, M. H., Li, X., Cheng, J. L., Zhang, L., et al. (2016). Paenibacillus sinopodophylli sp nov., a siderophore-producing endophytic bacterium isolated from roots of Sinopodophyllum hexandrum (Royle) Ying. Int. J. Syst. Evol. Microbiol. 66, 4993–4999. doi: 10.1099/ijsem.0.001458
Chen, Y. L., Yang, Y. P., Ji, X. J., Zhao, R., Li, G. Q., Gu, Y., et al. (2020). The SCIFF-derived Ranthipeptides participate in quorum sensing in Solventogenic Clostridia. Biotechnol. J. 15:8. doi: 10.1002/biot.202000136
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Conrath, U., Beckers, G. J., Langenbach, C. J., and Jaskiewicz, M. R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. doi: 10.1146/annurev-phyto-080614-120132
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Farzand, A., Moosa, A., Zubair, M., Khan, A. R., Hanif, A., Tahir, H. A. S., et al. (2019a). Marker assisted detection and LC-MS analysis of antimicrobial compounds in different Bacillus strains and their antifungal effect on Sclerotinia sclerotiorum. Biol. Control 133, 91–102. doi: 10.1016/j.biocontrol.2019.03.014
Farzand, A., Moosa, A., Zubair, M., Khan, A. R., Massawe, V. C., Tahir, H. A. S., et al. (2019b). Suppression of Sclerotinia sclerotiorum by the induction of systemic resistance and regulation of antioxidant pathways in tomato using Fengycin produced by Bacillus amyloliquefaciens FZB42. Biomol. Ther. 9:17. doi: 10.3390/biom9100613
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Gao, X., Wang, J., Ma, L., and Yun, X. (2014). Research advances on the mechanism of pathogenesis and allelopathy of fusarium oxysporium. Microbiol. China 41, 2143–2148. doi: 10.13344/j.microbiol.china.140406
Ghorbanpour, M., Omidvari, M., Abbaszadeh-Dahaji, P., Omidvar, R., and Kariman, K. (2018). Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control 117, 147–157. doi: 10.1016/j.biocontrol.2017.11.006
Giani, A. M., Gallo, G. R., Gianfranceschi, L., and Formenti, G. (2020). Long walk to genomics: history and current approaches to genome sequencing and assembly. Comput. Struct. Biotechnol. J. 18, 9–19. doi: 10.1016/j.csbj.2019.11.002
Guo, F., Xie, Z., Lu, P., Guo, Y., Zhang, L., and Wang, Y. (2014). Identification of a Novelpaenibacillus polymyxa strain and its biocontrol and plant growth-promoting effects. Chin. J. Biol. Control 30, 489–496. doi: 10.16409/j.cnki.2095-039x.2014.04.011
Huang, Y., Xie, X., Shi, Y., Chai, A., Li, L., and Li, B. (2020). Biocontrol effect of Paenibacillus polymyxa strain ZF197 against base stem rot of Chinese cabbage. Acta Horticult. Sinica 47, 1059–1071. doi: 10.16420/j.issn.0513-353x.2019-0915
Kim, C. G., Kang, J. P., Huo, Y., Chokkalingam, M., Kim, Y. J., Kim, D. H., et al. (2018). Paenibacillus panacihumi sp nov., a potential plant growth-promoting bacterium isolated from ginseng-cultivated soil. Arch. Microbiol. 200, 1049–1055. doi: 10.1007/s00203-018-1517-z
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kwon, Y. S., Lee, D. Y., Rakwal, R., Baek, S. B., Lee, J. H., Kwak, Y. S., et al. (2016). Proteomic analyses of the interaction between the plant-growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics 16, 122–135. doi: 10.1002/pmic.201500196
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lee, Y., Kim, Y. S., Balaraju, K., Seo, Y. S., Park, J., Ryu, C. M., et al. (2020). Molecular changes associated with spontaneous phenotypic variation of Paenibacillus polymyxa, a commonly used biocontrol agent, and temperature-dependent control of variation. Sci. Rep. 10:16586. doi: 10.1038/s41598-020-73716-7
Li, Y. L., and Chen, S. S. (2019). Fusaricidin produced by Paenibacillus polymyxa WLY78 induces systemic resistance against fusarium wilt of cucumber. Int. J. Mol. Sci. 20:19. doi: 10.3390/ijms20205240
Lombard, V., Ramulu, H. G., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Luo, Y. C., Cheng, Y. J., Yi, J. C., Zhang, Z. J., Luo, Q., Zhang, D. J., et al. (2018). Complete genome sequence of industrial biocontrol strain Paenibacillus polymyxa HY96-2 and further analysis of its biocontrol mechanism. Front. Microbiol. 9:14. doi: 10.3389/fmicb.2018.01520
Ma, Y. P., Hao, L., Ke, H., Liang, Z. L., Ma, J. Y., Liu, Z. X., et al. (2017). LuxS/AI-2 in Streptococcus agalactiae reveals a key role in acid tolerance and virulence. Res. Vet. Sci. 115, 501–507. doi: 10.1016/j.rvsc.2017.07.032
Maciag, T., Koziel, E., Rusin, P., Otulak-Koziel, K., Jafra, S., and Czajkowski, R. (2023). Microbial consortia for plant protection against diseases: more than the sum of its parts. Int. J. Mol. Sci. 24:20. doi: 10.3390/ijms241512227
Mei, L., Liang, Y., Zhang, L., Wang, Y., and Guo, Y. (2014). Induced systemic resistance and growth promotion in tomato by an indole-3-acetic acid-producing strain of Paenibacillus polymyxa. Ann. Appl. Biol. 165, 270–279. doi: 10.1111/aab.12135
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Ann. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Mull, R. W., Harrington, A., Sanchez, L. A., and Tal-Gan, Y. (2018). Cyclic peptides that govern signal transduction pathways: from prokaryotes to multi-cellular organisms. Curr. Top. Med. Chem. 18, 625–644. doi: 10.2174/1568026618666180518090705
Naghmouchi, K., Hammami, R., Fliss, I., Teather, R., Baah, J., and Drider, D. (2012). Colistin a and colistin B among inhibitory substances of Paenibacillus polymyxa JB05-01-1. Arch. Microbiol. 194, 363–370. doi: 10.1007/s00203-011-0764-z
Nielsen, P., and Sorensen, J. (1997). Multi-target and medium-independent fungal antagonism by hydrolytic enzymes in Paenibacillus polymyxa and Bacillus pumilus strains from barley rhizosphere. FEMS Microbiol. Ecol. 22, 183–192. doi: 10.1016/s0168-6496(96)00089-x
Niu, B., Vater, J., Rueckert, C., Blom, J., Lehmann, M., Ru, J. J., et al. (2013). Polymyxin P is the active principle in suppressing phytopathogenic Erwinia spp. by the biocontrol rhizobacterium Paenibacillus polymyxa M-1. BMC Microbiol. 13:13. doi: 10.1186/1471-2180-13-137
Nowell, R. W., Green, S., Laue, B. E., and Sharp, P. M. (2014). The extent of genome flux and its role in the differentiation of bacterial lineages. Genome Biol. Evol. 6, 1514–1529. doi: 10.1093/gbe/evu123
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Padda, K. P., Puri, A., Zeng, Q. W., Chanway, C. P., and Wu, X. Q. (2017). Effect of GFP-tagging on nitrogen fixation and plant growth promotion of an endophytic diazotrophic strain of Paenibacillus polymyxa. Botany 95, 933–942. doi: 10.1139/cjb-2017-0056
Park, S. J., Park, S. H., and Ghim, S. Y. (2014). The effects of Paenibacillus polymyxa E681 on antifungal and crack remediation of cement paste. Curr. Microbiol. 69, 412–416. doi: 10.1007/s00284-014-0604-x
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., and Bakker, P. (2014). Induced systemic resistance by beneficial microbes. Ann. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Platzer, J., Sterr, W., Hausmann, M., and Schmitt, R. (1997). Three genes of a motility operon and their role in flagellar rotary speed variation in Rhizobium meliloti. J. Bacteriol. 179, 6391–6399. doi: 10.1128/jb.179.20.6391-6399.1997
Qiao, J., Zhang, X., Liang, X., Liu, Y., and Liu, Y. (2017). Plant system resistance triggered by root-colonizing Bacillus subtilis PTS-394 and its control effect on tomato gray Mold. Chin. J. Biol. Control 33, 219–225. doi: 10.16409/j.cnki.2095-039x.2017.02.012
Raafat, M. M., Ali-Tammam, M., and Ali, A. E. (2019). Quorum quenching activity of Bacillus cereus isolate 30b confers antipathogenic effects in Pseudomonas aeruginosa. Infect. Drug Resist. 12, 1583–1596. doi: 10.2147/idr.S182889
Raza, W., Yang, W., and Shen, Q. R. (2008). Paenibacillus polymyxa: antibiotics, hydrolytic enzymes and hazard assessment. J. Plant Pathol. 90, 419–430. doi: 10.4454/JPP.V90I3.683
Timmusk, S., Grantcharova, N., and Wagner, E. G. H. (2005). Paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 71, 7292–7300. doi: 10.1128/aem.71.11.7292-7300.2005
Timmusk, S., and Wagner, E. G. H. (1999). The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. Mol. Plant-Microbe Interact. 12, 951–959. doi: 10.1094/mpmi.1999.12.11.951
Tong, W., Guo, G., Xu, J., Ji, Z., and Chen, X. (2004). Induced resistance to gray Mould in tomato plant by antagonistic Bacteria. Chin. J. Biol. Control 20, 187–189. doi: 10.16409/j.cnki.2095-039x.2004.03.010
Wang, L., Jiang, P., Gan, Y., Chen, Z., and Liu, Y. (2017). Increase of defense enzyme activity and up-expression of defense gene in tomato induced by Bacillus amyloliquefaciens B1619. Chin. J. Biol. Control 33, 234–240. doi: 10.16409/j.cnki.2095-039x.2017.02.014
Wiesel, L., Newton, A. C., Elliott, I., Booty, D., Gilroy, E. M., Birch, P. R., et al. (2014). Molecular effects of resistance elicitors from biological origin and their potential for crop protection. Front. Plant Sci. 5:655. doi: 10.3389/fpls.2014.00655
Xie, C., Mao, X. Z., Huang, J. J., Ding, Y., Wu, J. M., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, W., Zhang, L. Y., Goodwin, P. H., Xia, M. C., Zhang, J., Wang, Q., et al. (2020). Isolation, identification, and complete genome assembly of an endophytic Bacillus velezensis YB-130, potential biocontrol agent against fusarium graminearum. Front. Microbiol. 11:12. doi: 10.3389/fmicb.2020.598285
XueLi, C., GuangHua, W., Jian, J. I. N., and BaoLin, L. U. (2008). Biocontrol effect of PaenibaciUus polymyxa BRF-1 and Bacillus subtilis BRF-2 on fusarium wilt disease of cucumber and tomato. Chin. J. Eco-Agric. 16, 446–450. doi: 10.3724/sp.J.1011.2008.00446
Yang, F., Jiang, H. Y., Ma, K., Wang, X., Liang, S., Cai, Y. X., et al. (2023). Genome sequencing and analysis of Bacillus velezensis VJH504 reveal biocontrol mechanism against cucumber fusarium wilt. Front. Microbiol. 14:18. doi: 10.3389/fmicb.2023.1279695
Yaoyao, E., Yuan, J., Yang, F., Wang, L., Ma, J., Li, J., et al. (2017). PGPR strain Paenibacillus polymyxa SQR-21 potentially benefits watermelon growth by re-shaping root protein expression. AMB Express 7:104. doi: 10.1186/s13568-017-0403-4
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Anton. Leeuw. Int. J. Gen. Mol. Microbiol. 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zheng, J. F., Ge, Q. W., Yan, Y. C., Zhang, X. P., Huang, L., and Yin, Y. B. (2023). dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 51, W115–W121. doi: 10.1093/nar/gkad328
Keywords: Fusarium oxysporum f.sp. cucumerinum, Paenibacillus polymyxa, biocontrol agent, genome sequencing and assembly, biocontrol mechanism
Citation: Yang F, Jiang H, Ma K, Hegazy A, Wang X, Liang S, Chang G, Yu L, Tian B and Shi X (2024) Genomic and phenotypic analyses reveal Paenibacillus polymyxa PJH16 is a potential biocontrol agent against cucumber fusarium wilt. Front. Microbiol. 15:1359263. doi: 10.3389/fmicb.2024.1359263
Edited by:
Tofazzal Islam, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Tahsin Islam Sakif, West Virginia University, United StatesTanja Berić, University of Belgrade, Serbia
Kunal Singh, Institute of Himalayan Bioresource Technology (CSIR), India
Copyright © 2024 Yang, Jiang, Ma, Hegazy, Wang, Liang, Chang, Yu, Tian and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqin Yu, yu1980011@163.com; Baoming Tian, tianbm@zzu.edu.cn; Xuanjie Shi, shixuanjie@hnagri.org.cn
†These authors have contributed equally to this work
 Fan Yang
Fan Yang Huayan Jiang1,2†
Huayan Jiang1,2† Abeer Hegazy
Abeer Hegazy