- 1Laboratory of Microbiology, Department of Disease Control, Faculty of Veterinary Medicine, Hokkaido University, Sapporo, Hokkaido, Japan
- 2Faculty of Veterinary Medicine, College of Agriculture, Can Tho University, Can Tho, Vietnam
- 3BioApplications, Inc., Pohang, Gyeongsangbuk, Republic of Korea
- 4Nansei Livestock Hygiene Service Center, Matsuzaka, Mie, Japan
- 5One Health Research Center, Hokkaido University, Sapporo, Hokkaido, Japan
- 6International Collaboration Unit, International Institute for Zoonosis Control, Hokkaido University, Sapporo, Hokkaido, Japan
- 7Institute for Vaccine Research and Development (HU-IVReD), Hokkaido University, Sapporo, Hokkaido, Japan
- 8Celltrix Co., Ltd., Seongnam, Gyeonggi, Republic of Korea
Background: It is essential to consider a practical antibody test to successfully implement marker vaccines and validate vaccination efficacy against classical swine fever virus (CSFV). The test should include a serological antibody assay, combined with a tool for differentiating infected from vaccinated animals (DIVA). The immunochromatographic test strip (ICS) has been exclusively designed for detecting CSFV E2 antibodies while lacking in detecting Erns antibodies, which can be employed and satisfy DIVA strategy. This study developed a novel ICS for detecting CSFV E2/Erns dual-antibody. The effectiveness of ICS in evaluating the DIVA capability of two novel chimeric pestivirus vaccine candidates was assessed.
Methods: Recombinant E2 or Erns protein was transiently expressed in the plant benthamiana using Agrobacterium tumefaciens. ICS was subsequently assembled, and goat anti-rabbit IgG and recombinant CSFV E2 or Erns protein were plated onto the nitrocellulose membrane as control and test lines, respectively. The sensitivity and specificity of ICS were evaluated using sera with different neutralizing antibody titers or positive for antibodies against CSFV and other pestiviruses. The coincidence rates for detecting E2 and Erns antibodies between ICS and commercial enzyme-linked immunosorbent assay (ELISA) kits were also computed. ICS performance for DIVA capability was evaluated using sera from pigs vaccinated with conventional vaccine or chimeric vaccine candidates.
Results: E2 and Erns proteins were successfully expressed in N. benthamiana-produced recombinant proteins. ICS demonstrated high sensitivity in identifying CSFV E2 and Erns antibodies, even at the low neutralizing antibody titers. No cross-reactivity with antibodies from other pestiviruses was confirmed using ICS. There were high agreement rates of 93.0 and 96.5% between ICS and two commercial ELISA kits for E2 antibody testing. ICS also achieved strong coincidence rates of 92.9 and 89.3% with two ELISA kits for Erns antibody detection. ICS confirmed the absence of CSFV Erns-specific antibodies in sera from pigs vaccinated with chimeric vaccine candidates.
Conclusion: E2 and Erns proteins derived from the plant showed great potential and can be used to engineer a CSFV E2/Erns dual-antibody ICS. The ICS was also highly sensitive and specific for detecting CSFV E2 and Erns antibodies. Significantly, ICS can fulfill the DIVA concept by incorporating chimeric vaccine candidates.
1 Introduction
Classical swine fever (CSF) is a highly infectious and fatal disease that has caused enormous mortality in domestic pigs and wild boars (Blome et al., 2017; Ganges et al., 2020). The disease is caused by CSF virus (CSFV), a positive-sense, enveloped single-stranded RNA virus belonging to the Pestivirus genus within the Flaviviridae family (Postel et al., 2021). The complete genome of CSFV is ~12.3 kb long, with a single large open reading frame encoding four structural proteins (C, Erns, E1, and E2) and eight nonstructural proteins (Npro, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) flanked by two untranslated regions (UTRs), 5′-UTR and 3′-UTR (Meyers and Thiel, 1996). After CSFV infection, an antibody response is elicited against envelope glycoproteins (E2 and Erns) and a nonstructural protein (NS3; König et al., 1995; Rau et al., 2006; Panyasing et al., 2018). Notably, envelope glycoprotein E2 is a prominent, protective antigen that can elicit neutralizing antibodies and is the primary target antigen of molecular and serological diagnostic assays used to assess vaccination effectiveness against CSFV (van Rijn et al., 1999; van Rijn, 2007). Furthermore, Erns can also induce the neutralizing antibody production, and the antibody response to Erns can be used as a marker to diagnose CSFV infection in pigs (König et al., 1995; Moormann et al., 2000; Lin et al., 2004).
In CSF-endemic countries, strategies involving biological safety precautions and routine vaccination using traditional live attenuated vaccines (LAVs) and subunit vaccines have been implemented to control and eradicate CSFV (Postel et al., 2018; Coronado et al., 2021). Nevertheless, failure of vaccination programs is an essential factor that impedes CSF control due to immunosuppression in pig herds caused by persistent infection with moderately or low virulent CSFV strains and the interference between vaccine types and maternally derived antibodies (MDA) in piglets (Coronado et al., 2021). These challenges contribute to reduced vaccine efficacy, resulting in sporadic outbreaks and CSF reemergence in vaccinated herds and leading to CSF-endemic disease (Pérez et al., 2012; Muñoz-González et al., 2015; Coronado et al., 2019). Hence, with the enforcement of improved surveillance systems and the advancement of efficient vaccines, developing efficient antibody detection systems is crucial for enhanced CSFV control. Considering this scenario, a suitable serological antibody assessment through extensive screening is needed to monitor the vaccination immunity of pigs against CSFV in practice (Blome et al., 2006; Schroeder et al., 2012).
Currently, the evaluation of antibody responses against CSFV is performed by serum neutralization test (SNT) and the enzyme-linked immunosorbent assay (ELISA) designed to detect antibody responses specific to the E2 or Erns glycoprotein (Postel et al., 2018; Wang et al., 2020). Notably, detecting Erns antibodies is a reliable diagnostic test for differentiating infected from vaccinated animals (DIVA) using marker vaccines that induce a protective immune response specific to CSFV E2 while showing an absence of specific antibodies to CSFV Erns (Langedijk et al., 2001; Lin et al., 2005; Meyer et al., 2017). Given this context, traditional immunoassays, such as SNT or ELISA, can validate DIVA properties and vaccination efficacy of several marker vaccine candidates that have been recently generated, including chimeric pestiviruses CP_E2alf (Reimann et al., 2004) and Flc-LOM-BErns (Lim et al., 2019) or two novel chimeric viruses vGPE−/PAPeV Erns and vGPE−/PhoPeV Erns (Huynh et al., 2023) and E2 subunit vaccines (Gong et al., 2019; Park et al., 2021). However, these assays must be conducted in a laboratory setting and involve complex experimental procedures, significant time consumption, and substantial equipment and maintenance costs despite their high sensitivity and accuracy (Postel et al., 2018; Ganges et al., 2020; Wang et al., 2020). Therefore, a reliable and rapid DIVA assay should be considered to successfully implement marker vaccines and validate vaccination efficacy.
An immunochromatographic strip test (ICS) is the most rapid and reliable point-of-care diagnostic tool for prompt policy regarding effective disease control strategies (Manessis et al., 2022). ICS provides benefits compared to traditional immunoassays, such as affordability, rapidness, user-friendliness, and high specificity and sensitivity (Wang et al., 2020; Manessis et al., 2022). Previously, an ICS for CSFV antibody detection was developed based on the chimeric protein CnC2, generated from a specific region encoding Erns (amino acids 109–160; assigned as Cn) and E2 (amino acids 1–176; assigned as C2) antigen epitopes of CSFV by the Escherichia coli expression system (Li et al., 2012). However, protein expression by the E. coli system can lead to protein misfolding, which may reduce antibody detection efficacy (Bai et al., 2022). Several ICS studies have been performed to improve CSFV E2 antibody detection by enhancing the protein expression system, such as the baculovirus expression system (Bai et al., 2022) or transgenic rice endosperm (Xu et al., 2022). However, ICS for CSFV E2 antibody detection can only be used to monitor vaccination efficacy and early detection of antibody responses, whereas the DIVA test for detecting Erns antibody is lacking.
Plant-based production systems are promising platforms for recombinant protein production due to their numerous advantages over traditional expression systems (Qiu et al., 2014; Ma et al., 2015). Among these plant systems, Nicotiana benthamiana, a close relative of tobacco, has gained widespread attention for its exceptional suitability for recombinant protein expression. N. benthamiana offers several advantages for recombinant protein production (Ward et al., 2020; Hager et al., 2022; Kallolimath et al., 2023) and is fast-growing, easily cultivated, and amenable to genetic manipulation, making it an attractive host for protein expression studies. Furthermore, N. benthamiana also possesses a robust protein folding and posttranslational modification machinery, allowing for the production of complex, biologically active proteins. A key advantage of N. benthamiana is its ability to produce glycosylated proteins with mammalian-like glycosylation patterns which is particularly advantageous for producing therapeutic proteins because proper glycosylation is often critical for protein stability, functionality, and immunogenicity (Strasser, 2023). Therefore, N. benthamiana-based expression system highlight their advantages, applications, and prospects.
In a previous study, two chimeric pestiviruses, namely vGPE−/PAPeV Erns and vGPE−/PhoPeV Erns, were generated based on the backbone of a CSFV LAV strain combined with the glycoprotein Erns distantly related to CSFV (Huynh et al., 2023). These two chimeric viruses were proposed as promising marker vaccine candidates, suggesting a feasible approach for establishing an enhanced serological negative marker in DIVA vaccine development for CSF. However, DIVA capability was still not assessed for these chimeric vaccine candidates.
Thus, in this study, a novel ICS was developed to rapidly and reliably detect CSFV E2 and Erns antibodies. Recombinant E2 and Erns proteins were transiently expressed in N. benthamiana using Agrobacterium tumefaciens. Subsequently, ICS was constructed to detect CSFV E2 and Erns antibodies. In addition, ICS performance was compared to commercial ELISA kits for CSFV E2 or Erns antibody detection. Furthermore, a panel of serum samples derived from pigs vaccinated with chimeric vaccine candidates was utilized, incorporating ICS to evaluate DIVA capability.
2 Materials and methods
2.1 Cloning of CSFV E2 and Erns genes
The full-length E2 (derived from CSFV Alfort A19 strain) and Erns (derived from LOM strain) encoding sequences were obtained from the National Center for Biotechnology Information (NCBI) database (GenBank accession numbers AAB50409.1 and ABD83960, respectively) and optimized for expression in N. benthamiana1 before gene synthesis. For CSFV E2 expression, genes encoding the signal peptide of BiP (immunoglobulin heavy-chain binding protein) with a length of 271 nucleotides (nt) -the endoplasmic reticulum (ER) chaperone, full-length CSFV E2 sequence (1,023 nt), linker (CAGATTACGACATTCCAACAACTGATGCA), 6 histidines, cellulose binding domain (CBD; 454 nt) derived from xynA of Clostridium stercorarium, and ER retention signal (His-Asp-Glu-Leu) were fused sequentially and then cloned into the pCAMBIA1300 vector (Park et al., 2019) harboring the 35S promoter and heat shock protein terminator. The expected size of the fusion protein was approximately 58 kDa. For Erns expression, genes encoding the signal peptide of chaperone binding protein (251 nt), full-length CSFV Erns (681 nt), linker (GGTGGGGGAGGCAGT), 10 histidines, and ER retention signal (His-Asp-Glu-Leu) were fused sequentially and cloned into a pTEX vector harboring the MacT promoter and RD29B terminator. The expected size of the fusion protein was nearly 27.7 kDa. The nucleotide sequence was verified by sequencing (Bioneer, Daejeon, Republic of Korea).
2.2 Transient expression of recombinant CSFV E2 and Erns
The plasmid for expressing recombinant CSFV E2 or Erns was transformed into the A. tumefaciens GV3101 strain (Lifeasible, Shirley, NY, United States) by electroporation. Transformed A. tumefaciens were grown for 16 h in a 5 mL yeast extract peptone (YEP) liquid medium supplemented with 50 mg/L kanamycin and 25 mg/L rifampicin. Next, 1 mL of cultured bacteria was inoculated into 1 L of fresh YEP medium and cultured for a further 16 h at 28°C. Bacteria were pelleted by centrifugation at 7,341 × g for 5 min at 4°C and resuspended at the desired concentration (determined by measuring OD600) in a solution consisting of 10 mM 2-(N-morpholino) ethane sulfonic acid (MES) (Duksan, Seoul, Republic of Korea), 10 mM magnesium chloride (Sigma-Aldrich, St. Louis, MO, United States), and 100 μM acetosyringone (Sigma-Aldrich) at pH 5.6. Agroinfiltration was performed under a vacuum. After 4 days, the leaves were harvested for protein purification.
2.3 Purification of CSFV E2 and Erns protein
In the purification of E2 protein, the leaves were crushed and incubated for 30 min in an extraction buffer 1 (EB 1) consisting of 50 mM sodium phosphate (pH 8.0), 300 mM sodium chloride (NaCl), 10 mM imidazole, 0.5% (v/v) Triton X-100, 50 mM glycine, 100 mM sodium sulfite, 50 mM ascorbic acid, and 1.5% polyvinylpolypyrrolidone. With the purification of Erns protein, an extraction buffer 2 (EB 2) consisting of 50 mM Tris-Cl (pH 8.5), 300 mM NaCl, 50 mM imidazole, 0.3% (v/v) Triton X-100, 100 mM glycine, 100 mM sodium sulfite, 1 mM phenyl methyl sulfonyl fluoride, and 1.5% polyvinylpolypyrrolidone was used. After centrifugation, the supernatant from EB 1 or EB 2 was mixed with His60 Ni Superflow Resin (Takara, Kusatsu, Japan) for 1 h at 4°C, and the bound proteins were eluted with 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, and 250 mM imidazole for E2 protein or 300 mM imidazole for Erns protein purification. The eluates were adjusted with 50 mM Tris-Cl (pH 7.2) and 150 mM NaCl. The purified proteins were treated with β-mercaptoethanol, assessed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by Western blotting with an anti-His antibody (BioLegend, San Diego, CA, United States), anti-LOM vaccinated serum, and anti-E2 subunit vaccinated serum.
2.4 Manufacture of the prototype immunochromatographic test strip
2.4.1 Colloidal gold solution preparation
The colloidal gold solution (in-house) was prepared by adding 90 mL of distilled water into a triangular flask and heating it to 100°C while stirring on a hot stirrer plate (DAIHAN Scientific, Wonju, Republic of Korea). Once the temperature reached 100°C, 1% (w/v) gold chloride trihydrate (10 mL) was added and dissolved. After the gold chloride trihydrate solution was completely dissolved, 1 mL of 1% sodium citrate tribasic dehydrate was added and dissolved for 10 min at room temperature until the color of the reaction solution changed from yellow to the final red color. The resulting colloidal gold solution was diluted with distilled water, and the absorbance was measured using a UV-1650PC spectrophotometer (Shimazu, Tokyo, Japan).
2.4.2 Conjugation of the CSFV E2 and Erns and pretreatment of the conjugate pad
For the conjugation of recombinant proteins to the colloidal gold, 50 mL of colloidal gold solution was adjusted to pH 7.2 using 0.1 M potassium carbonate. Next, the recombinant E2 or Erns solution with a concentration of 50 μg/mL in phosphate-buffered saline (PBS) was slowly added dropwise, one drop at a time, into the colloidal gold solution, and the mixture was stirred for 30 min on a stirrer. Then, 10% (w/v) bovine serum albumin (BSA; 5 mL) was added and stirred for an additional 30 min. The resulting conjugation solution was centrifuged to separate the upper layer for 30 min at 10,000 rpm, and the pellet was suspended in a solution containing 1% (w/v) BSA in PBS. Subsequently, to pretreat the conjugate pad with glass fiber material, a solution containing PBS, 3% sucrose, 0.1% sodium azide, 1% (w/v) BSA, recombinant E2 or Erns conjugate (1.1 μg/kit), and rabbit IgG (in-house) conjugate (0.88 μg/kit), was evenly absorbed into the glass fiber material of the conjugate pad. The conjugate pad was dried for >4 h in an environment with a relative humidity of <20%.
2.4.3 Fixation of the control and test lines and kit assembly
The control and test lines were fixed using nitrocellulose membranes (Adventec, Irvine, CA, United States). The control line was coated with 1 mg/mL goat anti-rabbit IgG (Arista Biologicals, Allentown, PA, United States), and the test line was coated with each recombinant protein (E2 or Erns) at 1 mg/mL. The membranes were dispensed using a dispenser, Matrix 1600 (Kinematic Automation, Sonora, CA, United States), at a rate of 1 μL/cm and dried for >4 h in an environment with a relative humidity of <20%. In the next step, the kit was assembled by attaching the membrane coated with the antigen onto the adhesive card, followed by the conjugate pad and sample pad overlaid to overlap at the bottom. The assembled kit was cut to a width of 4 mm and inserted into a plastic device.
2.5 Swine serum samples
2.5.1 Samples for evaluating the sensitivity and specificity of immunochromatographic test strip
ICS sensitivity was assessed using 23 serum samples with varying SNT titers obtained from a previous study (Oh et al., 2021), sourced from two distinct groups: LOM-vaccinated pigs (n = 10) and nonvaccinated pigs with MDA (n = 13). Furthermore, ICS specificity was evaluated using swine serum samples (n = 5) that were positive for antibodies against CSFV, bovine viral diarrhea virus (BVDV), or border disease virus (BDV) from a previous study (Sakoda et al., 2012). Additionally, CSFV antibody-negative serum samples (n = 50) from naïve pigs were included in the specificity analysis. Furthermore, the serum samples (n = 9) were obtained from pigs infected with African swine fever virus (ASFV), porcine circovirus 2 (PCV2), and porcine reproductive and respiratory syndrome virus (PRRSV) that were confirmed positive using commercial ELISA kits: ID Screen® African Swine Fever Indirect (Innovative Diagnostics, Grabels, France), PCV2 Antibody Test Kit (BioChek, Scarborough, ME, United States), and IDEXX PRRS X3 Ab Test (IDEXX, Westbrook, ME, United States), respectively.
2.5.2 Sera for determining the coincidence rates between the immunochromatographic test strip and commercial enzyme-linked immunosorbent assay kits
Six CSFV-negative piglets were purchased from a CSFV-negative farm located in Jeju Island, Republic of Korea, and confirmed by screening with an ELISA assay for E2 antibody detection. Piglets were injected intramuscularly twice on the neck at 40 days of age [0 day postvaccination (dpv)] and 60 days (20 dpv) after birth using the CSFV E2 subunit vaccine (BioApplications, Pohang, Republic of Korea), which was developed and evaluated in previous studies (Park et al., 2020, 2021). Sera were collected at 40 days of age (before vaccination), 60 days (20 days after the first vaccination), and 80 days (40 days after the first vaccination). Eighteen serum samples were used to compare the agreement rate between ICS and ELISA kits.
Recently, the LOM strain initially employed as a vaccine strain, was introduced to Jeju Island, where is known for its no-vaccination policy against CSFV. Unfortunately, the CSFV has started spreading from sows to piglets and has undergone mutations during its transmission. Consequently, the Government is promoting a CSFV marker vaccine policy to combat and eliminate CSF (Oh et al., 2021). To gather clinical sera for analysis, 57 serum samples were randomly obtained from CSFV-positive and-negative farms, subsequently determined to be positive and-negative for E2 and Erns antibodies by ELISA kits, respectively. These samples were used to evaluate the coincidence rates of ICS with ELISA kits for detecting Erns and E2 antibodies.
The definition and way of calculation of total coincidence rates and positive coincidence rates are as followed (Pan et al., 2019): total coincidence rates = 100% × [(both positive of ICS and ELISA kit + both negative of ICS and ELISA kit)/total samples]; compared to ELISA kit, the positive coincidence rates of ICS = 100% × [both positive of ICS and ELISA kit/(both positive of ICS and ELISA kit + ICS positive but ELISA kit negative)].
The performance of ICS was compared with commercial ELISA kits in terms of relative sensitivity and specificity, as followed (Zhong et al., 2021): relative sensitivity = 100% × [(the number of true positive samples)/(the number of true positive samples + the number of false negative samples)]; relative specificity = 100% × [(the number of true negative samples)/(the number of true negative samples + the number of false positive samples)].
2.5.3 Evaluation of the immunochromatographic test strip using serum samples from pigs vaccinated with chimeric virus and commercial live attenuated-vaccine
Serum samples from pigs vaccinated with novel chimeric vaccine candidates in a previous study (Huynh et al., 2023) were used to evaluate DIVA properties using a novel ICS. In detail, 52 serum samples from 26 pigs vaccinated with each chimeric virus candidate (vGPE−/PAPeV Erns or vGPE−/PhoPeV Erns) or vGPE− derived from a conventional LAV were evaluated at 0 and 21 dpv. Additionally, 45 serum samples from 15 pigs were collected at 0, 7 dpv and 14 days postchallenge (dpc) in challenge studies that were also assessed. In these trials, each chimeric virus candidate or vGPE− was independently administered to pigs 7 days before exposure to the moderately virulent CSFV vALD-A76 strain. Subsequently, continuous monitoring was conducted for 14 days. Furthermore, the vaccination efficacy of piglets with MDA (n = 6) immunized with a commercial conventional CSFV GPE− LAV strain was also investigated, involving the collection of 24 serum samples at different time intervals (0, 45, and 90 dpv) and before slaughtering.
2.6 Enzyme-linked immunosorbent assay
The assay was performed using the PrioCHECK CSFV Erns Antibody ELISA Kit (Thermo Fisher Scientific, Waltham, MA, United States), CSFV Antibody Test Kit (IDEXX, Westbrook, ME, United States), CSFV Antibody B-ELISA (Bionote, Gyenggi-do, Republic of Korea), and Pigtype CSFV Erns Antibody (Indical Bioscience, Leipzig, Germany). The assay procedures were performed according to the manufacturers’ protocol. The absorbance of the solution was subsequently read at 450 nm on a Muliskan ELISA reader (Thermo Fisher Scientific).
2.7 Serum neutralization test
The test was performed using a luciferase-based assay, as described previously (Tetsuo et al., 2020). The determination of neutralizing antibody titers in serum samples derived from pigs vaccinated with each chimeric virus candidate, vGPE−, or commercial conventional CSFV GPE− LAV strain was accomplished through a luciferase assay employing the Nano-Glo HiBiT lytic detection system (Promega, Madison, WI, USA) and POWERSCAN®4 (Agilent Technologies International Japan Ltd., Tokyo, Japan).
The neutralizing antibodies against CSFV in serum samples from a previous study (Oh et al., 2021) were tested using a neutralizing peroxidase-linked assay described previously (Park et al., 2020). In brief, equal volumes of serially diluted serum and 200 TCID50 of the CSFV LOM LAV strain were mixed well and incubated at 37°C in 5% CO2 for 1 h. The mixture and 100 μL cloned porcine kidney cell suspension were incubated in 96-well plates at 37°C in 5% CO2 for 72 h. Cells were fixed with 80% acetone and dried at 37°C, followed by staining with commercial anti-LOM antibody as the primary antibody for immunoperoxidase staining (Park et al., 2020). The SNT titers represent the reciprocal of the highest dilution that achieved 50% neutralization.
3 Results
3.1 Expression and purification of recombinant CSFV E2 and Erns proteins
The sequences encoding CSFV E2 and Erns proteins were cloned into the A. tumefaciens GV3101 strain, and either the E2 or Erns recombinant proteins were expressed in N. benthamiana plants. The harvested leaves were crushed and subjected to His60 Ni Superflow Resin for protein purification. The purified proteins were then assessed by Western blotting and SDS-PAGE. As illustrated in Figure 1, the glycosylated forms of recombinant E2 and Erns proteins were confirmed by anti-His antibody. The recombinant E2 protein exhibited a visible band of ~69 kDa, whereas the recombinant Erns protein appeared at ~45 kDa under denaturing conditions (Figure 1). Moreover, verification of the aforementioned findings was achieved by anti-LOM vaccinated serum (demonstrating positivity for E2 and Erns) and anti-E2 subunit vaccinated serum (displaying positivity exclusively for E2) for the purpose of validating protein expression via Western blot analysis (Supplementary Figure S1). Although the bands appeared to be diffuse in the analysis of Erns protein, the deglycosylation with glycosidase PNGase F resulted in a single intense band, and the molecular weights of Erns were approximately 28 kDa, which corresponded to its calculated molecular weights (Supplementary Figure S1D). Moreover, the intense band exceeded the anticipated molecular weights of recombinant E2 (58 kDa) or Erns (28 kDa) fusion protein, suggesting that two proteins were extensively glycosylated, particularly with N-glycosylation contributing to sizes larger than anticipated that were observed when expressed in plants (Park et al., 2019, 2020). Taken together, the different glycosylation levels of each protein indicate that these two proteins are properly glycosylated and can serve as suitable antigens for further experiments.
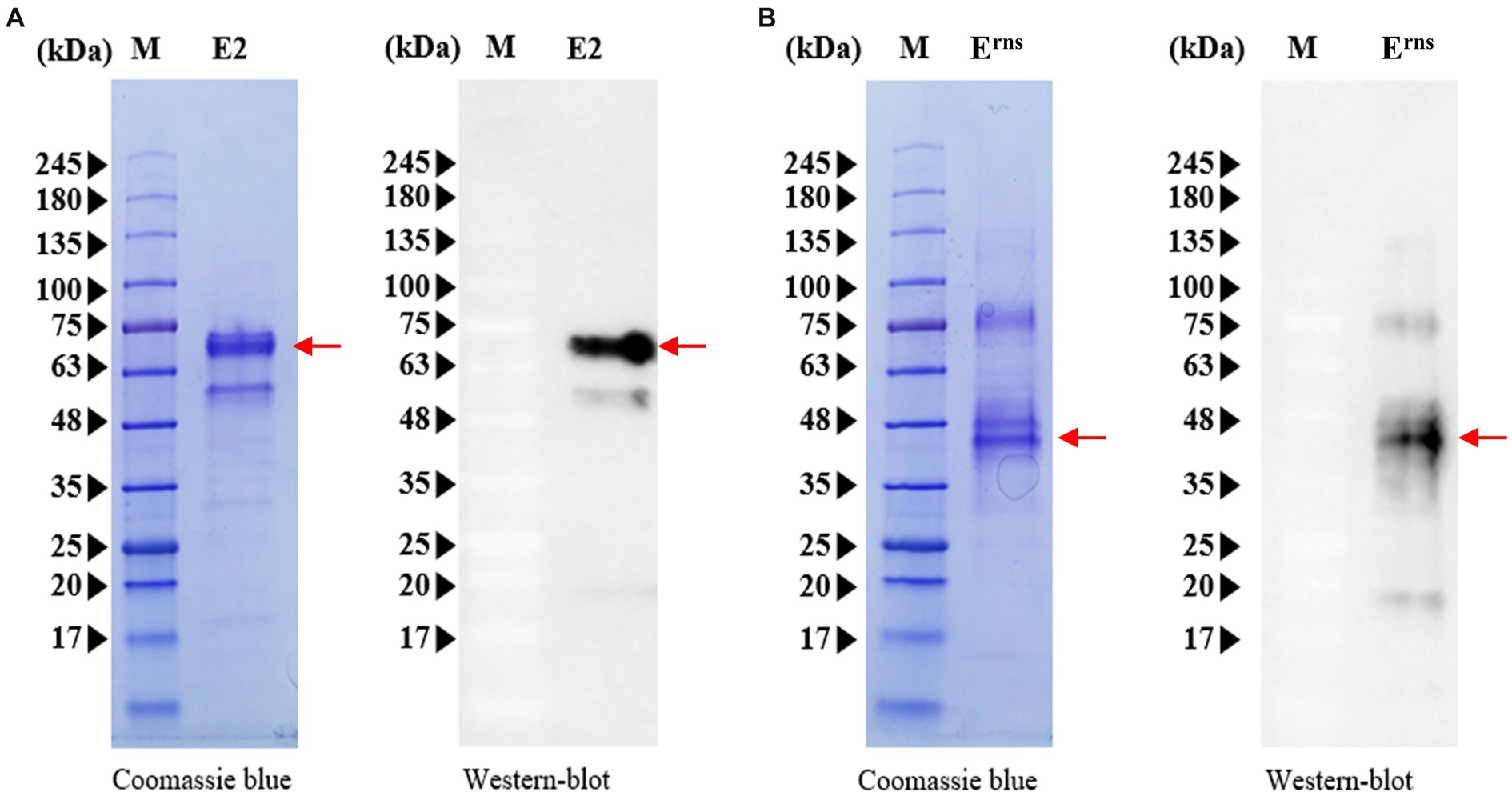
Figure 1. Expression and characterization of CSFV E2 and Erns proteins. (A) Purified E2 protein was subjected to Western blotting (right panels) using an anti-His monoclonal antibody, and gels were stained with Coomassie brilliant blue (left panels) after SDS-PAGE analysis. (B) Western blot analysis of purified Erns protein (right panels) using an anti-His monoclonal antibody and SDS-PAGE analysis (left panels). M, protein size marker. Red arrows represent the expression of E2 or Erns protein.
3.2 Conjugation of recombinant proteins with the gold nanoparticles and implementation of the working principle of the developed CSFV E2 and Erns detection kit
The optimal protein concentration for creating the E2 or Erns conjugate was 1.25 O.D., using a spectrophotometer at a wavelength of 540 nm and a particle size of 40 nm. The ICS assembly is illustrated in Figure 2. The assembled ICS comprised a membrane coated with the antigen onto the adhesive card, followed by a conjugate pad and sample pad overlaid to overlap at the bottom. Nitrocellulose membranes were used to fix the control line (C) and test line (T) membranes in the ICS. Specifically, the control line was plated with a second antibody (goat anti-rabbit IgG). Meanwhile, the test line was plated with recombinant E2 and Erns proteins corresponding to numbers 1 and 2 in the ICS (Figure 2A). In accordance with usage principles, serum samples from pigs were loaded on the application pad and diffused with gold-rabbit IgG and gold-E2 or Erns on the conjugate pad. For the control line, gold-rabbit IgG is bound to anti-rabbit IgG antibodies and accumulated. The anti-E2 or Erns antibody in the serum sample is bound and diffused with gold-E2 or Erns on the conjugate pad, and the complex bound to the E2 or Erns protein on the test line. Consequently, a red color appeared on the test line for the positive samples due to the accumulated complex. In contrast, the test line will not display red color for the negative samples that did not have anti-E2 or Erns antibodies (Figures 2B,C).
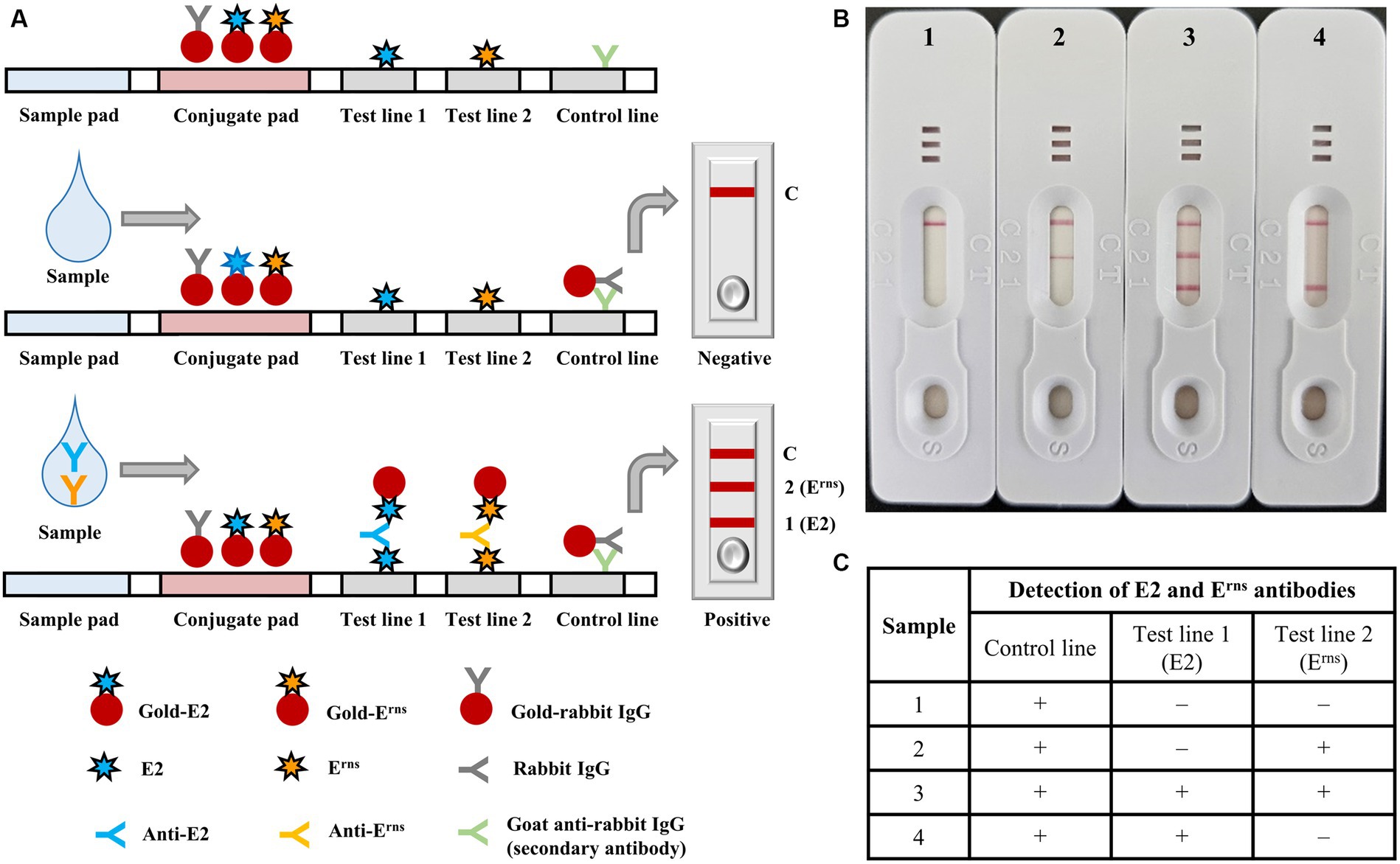
Figure 2. Diagrammatic illustration and implementation example of the immunochromatographic test strip. (A) Structural composition of the immunochromatographic test strip, including sample and conjugate pads, test lines 1 and 2, and control line; negative and positive results were also indicated. (B) The implementation of representative serum samples using the immunochromatographic test strip 1 (negative control), 2 (positive with Erns), 3 (positive with both E2 and Erns), and 4 (positive with E2), in which the serum samples were diluted with dilution buffer at a ratio of 1:1. (C) Representative results of each sample were summarized in the table.
3.3 Specificity and sensitivity of the immunochromatographic test strip
The sensitivity of ICS was evaluated using various serum samples positive for CSFV with different SNT titers. Additionally, the specificity of ICS was assessed using sera from CSFV-and pestivirus-inoculated pigs, and sera from naïve pigs. Twenty-three serum samples ranging from the lowest SNT titer of 4 to the highest SNT titer of 2,048 were subjected to ICS testing. The results in Table 1 demonstrated that ICS effectively detected antibodies against CSFV E2 and Erns in all samples. The E2 and Erns antibodies were detected at the lowest SNT titers of 16 and 4, respectively. These findings demonstrated the high sensitivity of the ICS for detecting E2 and Erns antibodies, with particular sensitivity for CSFV Erns antibody detection. Furthermore, the specificity of ICS for CSFV E2 and Erns antibody detection was assessed. Table 2 revealed that two CSFV-positive serum samples were also confirmed for detecting both E2 and Erns antibodies using ICS. In contrast, other pestivirus-positive serum samples (n = 3), including BVDV strains Nose (BVDV genotype 1a) and KZ-91-NCP (BVDV genotype 2a), BDV, and CSFV-negative sera from naïve pigs (n = 50) tested negative for E2 and Erns antibodies. Furthermore, additional serum samples (n = 9), consisting of ASFV (n = 3), PCV2 (n = 3), and PRRSV (n = 3) were also confirmed as negative. Data suggested that the ICS exhibited no cross-reactivity with antibodies from other swine pestiviruses and related swine viral diseases.
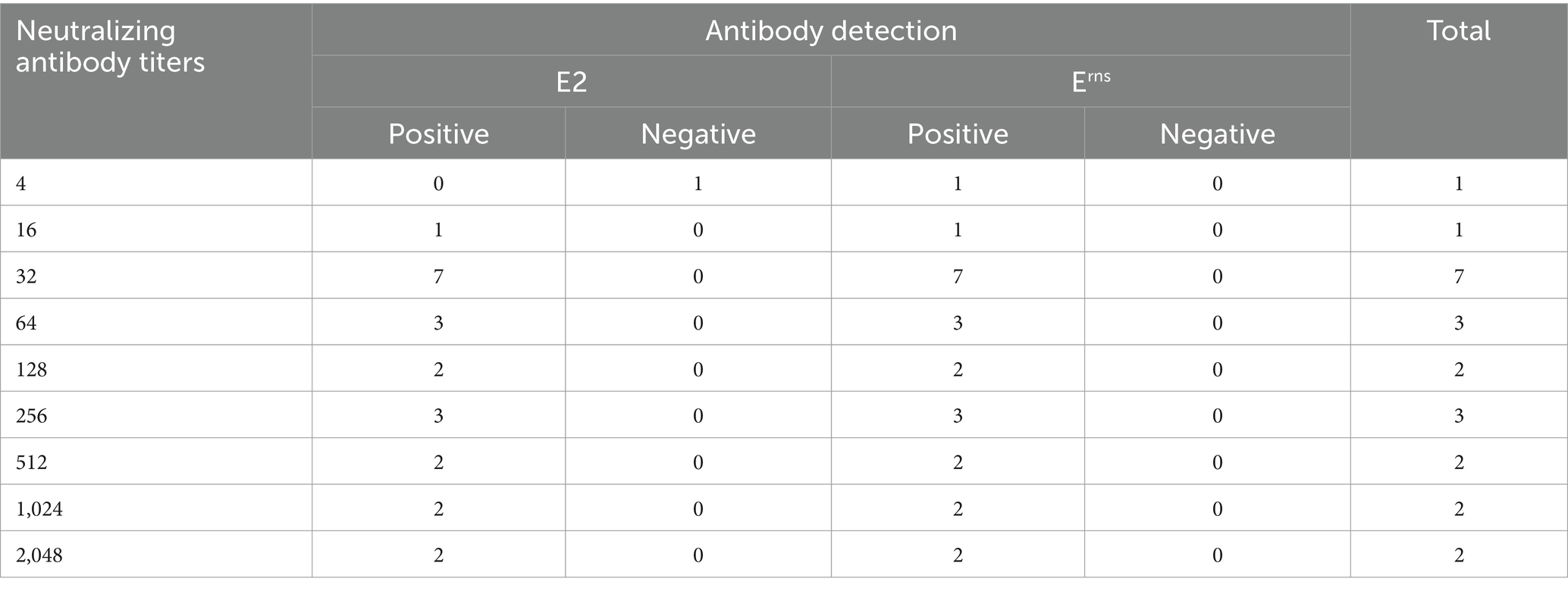
Table 1. Sensitivity of immunochromatographic test strip for detecting E2 and Erns antibodies using various swine serum samples with different neutralizing antibody titers.
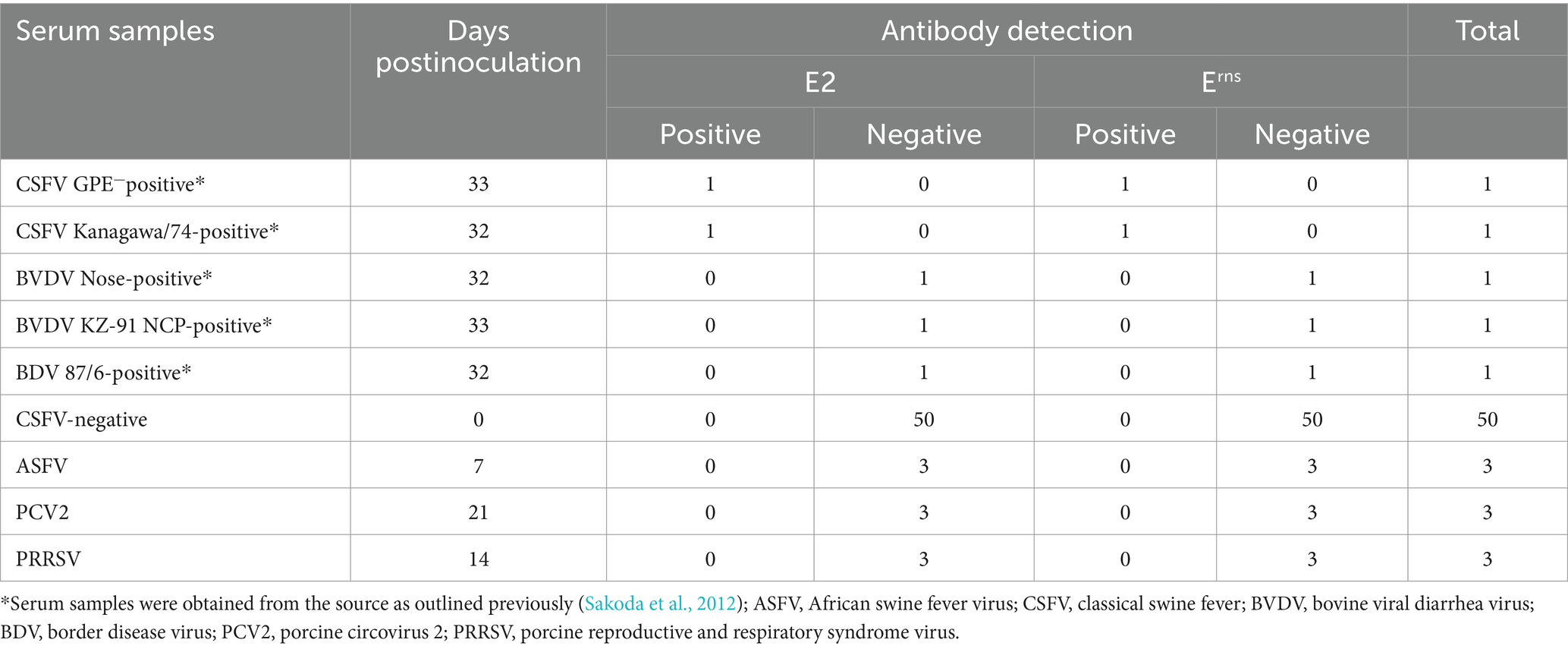
Table 2. Specificity of the immunochromatographic test strip for detecting E2 and Erns antibodies in reference swine serum samples.
3.4 CSFV E2 and Erns antibodies detection using immunochromatographic test strip and commercial enzyme-linked immunosorbent assay kits
The correlation between ICS and ELISA kits was then examined. Fifty-seven serum samples were randomly obtained from CSFV-positive farms positive for E2 antibody using ELISA kits (Bionote and IDEXX ELISA kits) and CSFV-negative farms that were tested to be negative for Erns antibody using two commercial ELISA kits (Indical Bioscience and Thermo Fisher Scientific ELISA kits). However, one serum sample was in poor condition during analysis and was positive for E2 antibody but could not be pinpointed as the band for Erns antibody detection. Therefore, the total number of E2 and Erns antibody analysis samples was 57 and 56, respectively. The results in Table 3 indicated that E2 positive proportions of the ICS corresponding to the Bionote and IDEXX ELISA kits were 100% (34 of 34) and 94.1% (32 of 34), respectively. The sensitivity and specificity of ICS corresponding to the Bionote ELISA kits were 89.5% (34 of 38) and 100% (19 of 19), respectively. In addition, the high sensitivity and specificity of ICS corresponding to the IDEXX ELISA kits were 100% (32 of 32) and 92.0% (23 of 25), respectively. A high agreement rate of ICS was determined to be 93.0% (53 of 57) and 96.5% (55 of 57), corresponding to the Bionote and IDEXX ELISA kits, respectively. In Table 4, the Erns positive percentages of ICS corresponding to the Indical Bioscience and Thermo Fisher Scientific ELISA kits were 86.2% (25 of 29) and 79.3% (23 of 29), respectively. ICS sensitivity for Erns antibody detection corresponding to the ELISA kits was 100% for the Indical Bioscience ELISA kits (25 of 25) and the Thermo Fisher Scientific ELISA kits (23 of 23). ICS showed high specificity for Erns detection at 87.1%, corresponding to the Indical Bioscience ELISA kits (27 of 31) and 81.8%, conforming to the Thermo Fisher Scientific ELISA kits (27 of 33). The agreement rates of ICS with the Indical Bioscience and Thermo Fisher Scientific ELISA kits were 92.9% (52 of 56) and 89.3% (50 of 56), respectively.
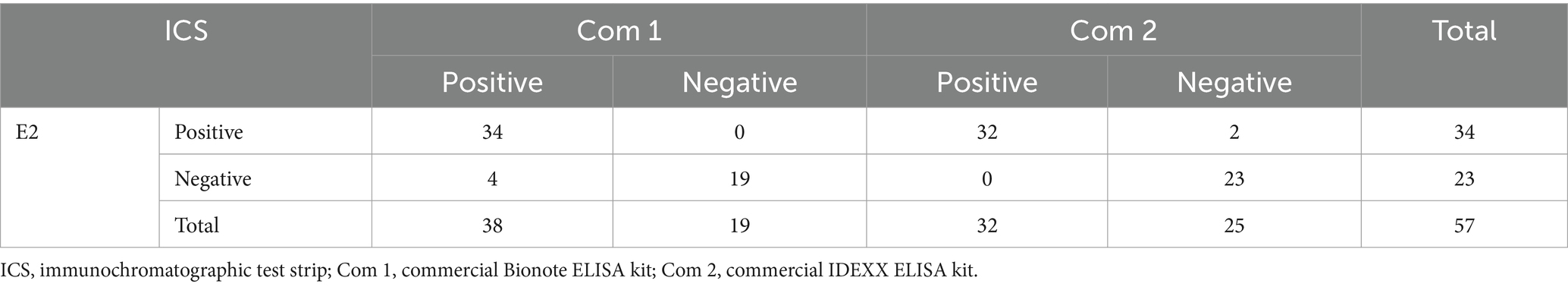
Table 3. Correlation between the immunochromatographic test strip and the two commercial ELISA kits for detecting CSFV E2 antibody.
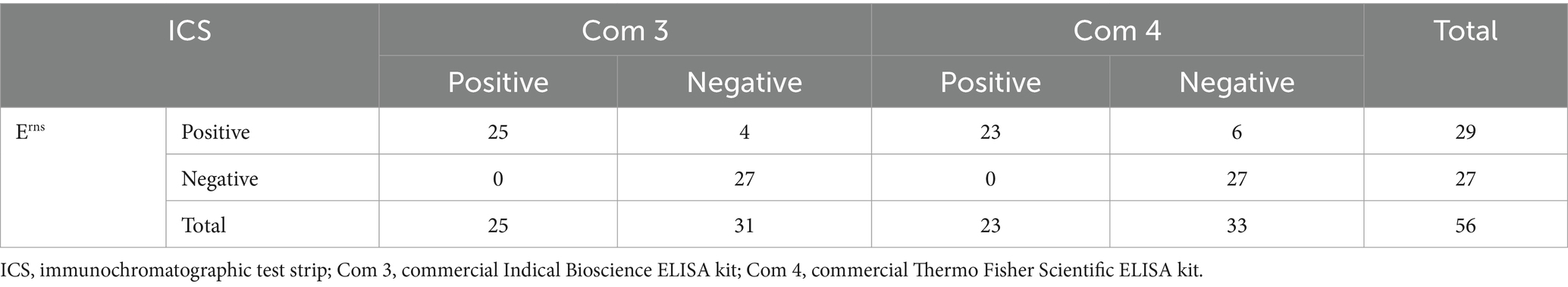
Table 4. Correlation between the immunochromatographic test strip and the two commercial ELISA kits for detecting CSFV Erns antibody.
ICS also strongly agreed with commercial ELISA kits in detecting E2 and Erns antibody responses to the subunit CSFV vaccine. This was observed in serum samples collected from pigs immunized twice with the CSFV E2 subunit vaccine at 40 and 60 days of age. As presented in Supplementary Table S1, all swine sera tested negative for the Erns antibody using commercial Bionote and IDEXX ELISA kits and ICS. The E2 antibody detection results using ICS aligned with those obtained from the commercial Bionote and IDEXX ELISA kits using serum samples collected at 40 dpv. This confirmation suggested that the ICS could detect the E2 antibody at a late stage in subunit-vaccinated pigs (Supplementary Table S1). Taken together, ICS was coincident at a high rate with the commercial ELISA kits used for detecting E2 and Erns antibodies, which would emerge as a superior choice.
3.5 Diagnostic capability of the immunochromatographic test strip in differentiating infected from vaccinated animals using sera of pigs vaccinated with the chimeric vaccine candidates vGPE−/PAPeV Erns and vGPE−/PhoPeV Erns
The swine serum samples obtained from two independent experiments from a previous study (Huynh et al., 2023) were used to assess the applicability of ICS for DIVA. In detail, serum samples from 26 pigs in the optimal infectious dose experiments were tested at 0 and 21 dpv using the ICS (Table 5). As shown in Table 5, all serum samples were negative for E2 and Erns antibodies at 0 dpv, consistent with SNT results. In addition, CSFV E2 neutralizing antibody was detected in the serum samples at 21 dpv of pigs immunized with vGPE−/PAPeV Erns (6 of 9) or vGPE−/PhoPeV Erns (4 of 4). At the same time, there were undetectable against Erns antibody in all pigs vaccinated with each chimeric virus. Conversely, the Erns antibody was detected in almost all serum samples from pigs vaccinated with conventional vGPE− strain, whereas only three of nine swine serum samples showed a positive result for the E2 antibody. Likewise, serum samples taken at different stages postvaccination from pigs vaccinated with commercial live attenuated GPE− vaccine were used to evaluate the performance of ICS. Most serum samples exhibiting high SNT titers showed a high positive rate for either E2 or Erns antibody or both, whereas samples with low SNT titers or a vaccination period <45 days were negative using ICS. Here, the anti-CSFV E2 antibody was detected in 18 of 24 serum samples, whereas 14 of 24 tested sera were positive for Erns antibody, primarily observed between 90 dpv and before slaughtering (Supplementary Table S2). Furthermore, antibody responses to E2 and Erns were assessed using serum samples collected at 0 and 7 dpv, and 14 dpc (or 21 dpv) in challenge studies, in which each chimeric virus candidate (vGPE−/PAPeV Erns or vGPE−/PhoPeV Erns) or a conventional LAV vGPE− was independently administered 7 days before challenge with the moderately virulent CSFV vALD-A76 strain and continuously monitored for 14 dpc. As presented in Table 6, the results showed that the E2 and Erns antibodies were undetected in all serum samples at 0 and 7 dpv. However, an increasing tendency of the E2 antibody was confirmed with positive results in several serum samples from pigs vaccinated with each chimeric virus at 14 dpc, whereas the Erns antibody was negative for all serum samples, as expected. It is crucial to notice that the Erns antibody was detected only in all pigs immunized with conventional LAV and the control groups (unvaccinated pigs), whereas it was negative for those vaccinated with each chimeric virus mentioned above at 14 dpc. The results of E2 antibody detection in sera from unvaccinated pigs were negative in all serum samples at 14 dpc. In contrast, the E2 antibody against CSFV vGPE− required a longer time to reach the detection threshold on the ICS than the SNT test.
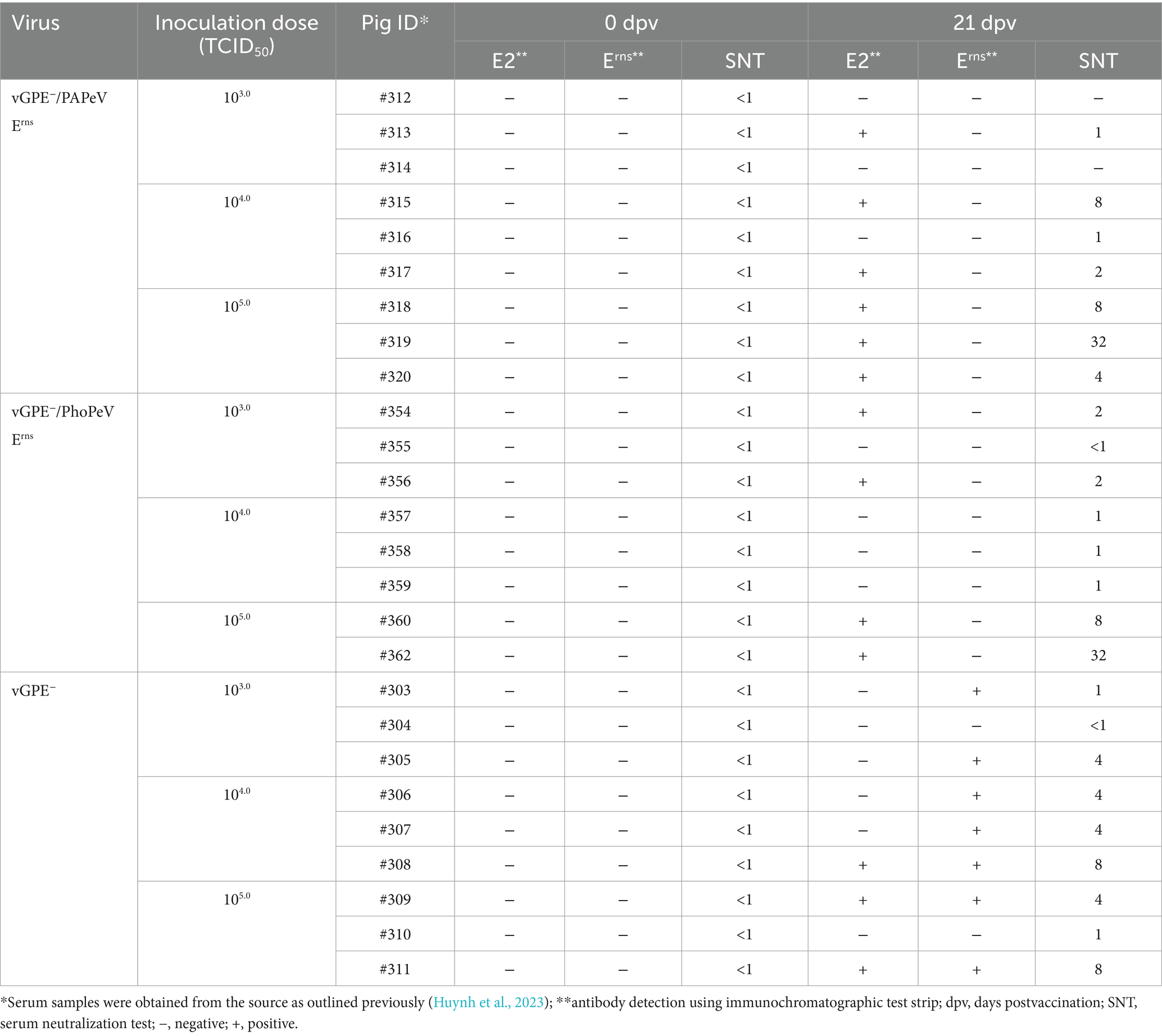
Table 5. Detection of E2 and Erns antibody responses from serum samples in pigs inoculated with vGPE−, vGPE−/PAPeV Erns, or vGPE−/PhoPeV Erns at different doses of TCID50 using the immunochromatographic test strip and serum neutralization test.
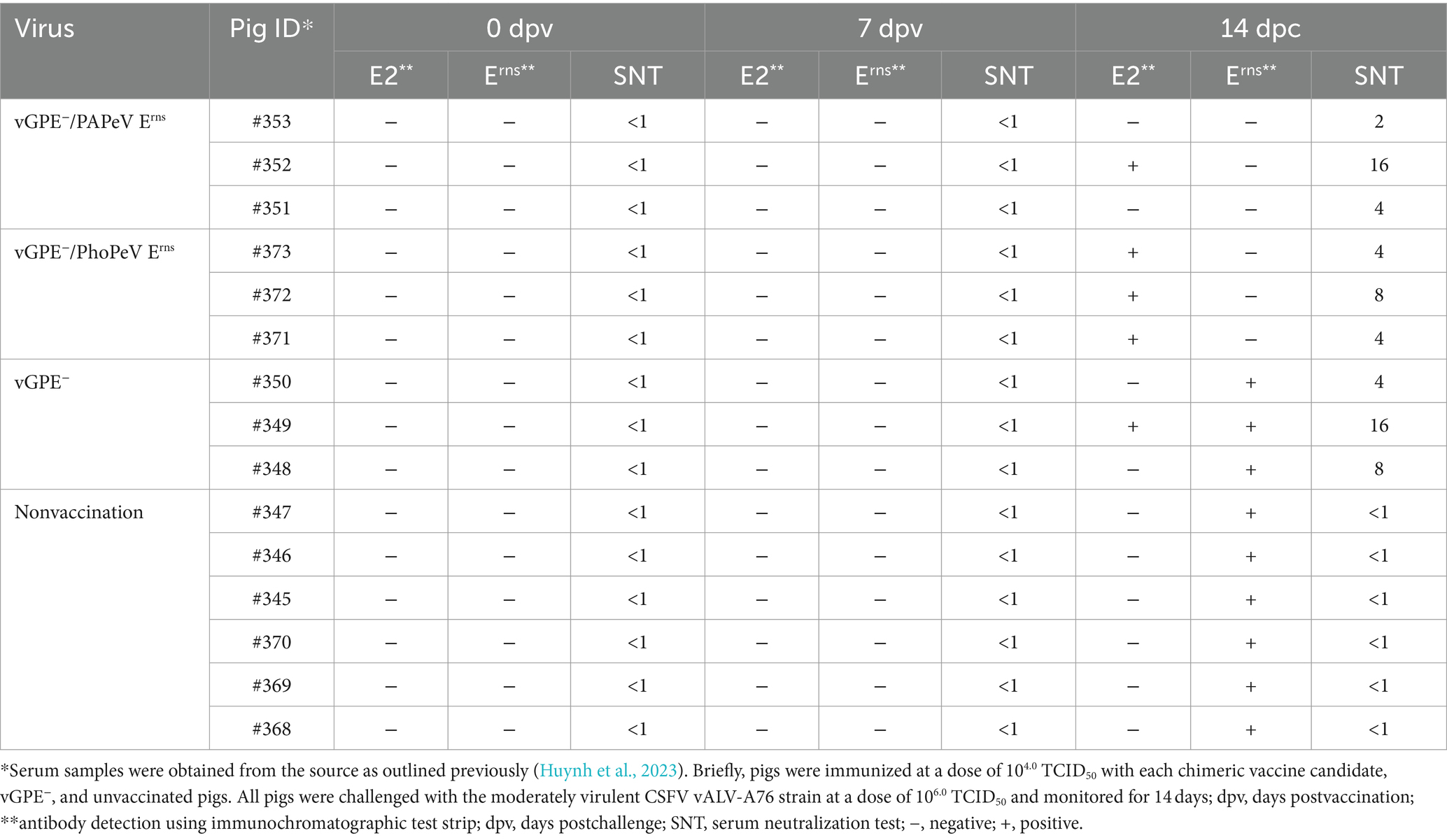
Table 6. Detection of E2 and Erns antibody responses from serum samples in pigs vaccinated with vGPE−, vGPE−/PAPeV Erns, or vGPE−/PhoPeV Erns in the challenge studies using the immunochromatographic test strip and serum neutralization test.
4 Discussion
Vaccination programs employing conventional CSFV LAVs have been demonstrated to protect pigs against CSFV infection and clinical signs in early-stage vaccination (Blome et al., 2006; Graham et al., 2012). Nevertheless, these conventional vaccines cannot resolve the challenges associated with the DIVA concept and the interference of MDAs. Various CSFV marker vaccines have been developed to address this challenge, such as subunit vaccines or chimeric pestiviruses designed to fulfill the DIVA strategy for controlling and eradicating CSF (Reimann et al., 2004; Gong et al., 2019; Lim et al., 2019; Park et al., 2021; Huynh et al., 2023). With the improvement of vaccine candidates, DIVA diagnostic tools using ELISA kits for detecting CSFV Erns antibody or monitoring the induction of neutralizing antibodies using some commercial ELISA kits for E2 antibody detection have been developed and employed as adjunctive diagnostic assessments together with the E2 subunit vaccine and chimeric pestiviruses (Meyer et al., 2017; Wang et al., 2020). However, ELISA procedures are time-consuming and require skilled personnel, which might not be suitable for rapid field testing if E2 and Erns antibody detection needs to be evaluated simultaneously with a large sample number in the field. Colloidal gold ICS have been established and are a valuable diagnostic tool for monitoring antibodies against CSFV in vaccinated pigs (Li et al., 2012; Bai et al., 2022; Xu et al., 2022). Compared to alternative antibody detection methodologies such as SNT or ELISA, ICS is notably user-friendly, rapid, and easily transportable (Li et al., 2012; Wang et al., 2020). Thus far, ICS development has preferably focused on detecting antibodies against the CSFV E2 protein, a powerful tool for the CSFV antibody assessment (Bai et al., 2022; Xu et al., 2022). Meanwhile, ICS development for CSFV Erns antibody detection for the DIVA diagnostic approach still has no specific innovation (Wang et al., 2020). Furthermore, considering the benefits of transient protein expression in plant-derived systems (Yamamoto et al., 2018; Laughlin et al., 2019; Park et al., 2019), a novel ICS has been developed for the simultaneous detection of dual CSFV E2 and Erns antibodies. This study established ICS based on colloidal gold recombinant E2 and Erns proteins derived from N. benthamiana plants to rapidly evaluate CSFV antibodies and integrate DIVA diagnostic features.
In protein manipulation, previous studies have indicated that plant-derived transient expression enhances high protein expression and yield, immunogenicity in animal experiments, and high binding affinity for antibody detection absent in certain conventional expression systems (Sohn et al., 2018; Yamamoto et al., 2018; Xu et al., 2022). Compared to the transgenic approach for CSFV E2 protein expression using baculovirus (Wei et al., 2021; Bai et al., 2022), mammalian cells (Ji et al., 2018; Chen et al., 2023), or the E. coli system (Li et al., 2012; Yi et al., 2022), the CSFV E2-expressing transgenic plant-derived system was preferable in terms of a large amount and stable expression of the target protein and inexpensiveness, as in recent studies (Sohn et al., 2018; Laughlin et al., 2019; Park et al., 2019, 2020; Xu et al., 2022). This approach facilitated the purification of recombinant E2 from N. benthamiana extracts. Although the E2 protein is typically dimeric form, earlier investigations (Park et al., 2019) have confirmed a multimeric form of E2 protein expressed in plant tissue through glycosylation. A few systems have been established for recombinant Erns protein expression in E. coli (Aebischer et al., 2013; Yi et al., 2022), baculovirus (Wei et al., 2021), or yeast (Huang et al., 2006; Luo et al., 2015), but there are several concerns regarding inappropriate posttranslational modifications, particularly the absence or low proportion of glycosylation in Erns or E2 protein that may influence the binding affinity of antibodies (Gavrilov et al., 2011). The most important feature of CSFV E2 and Erns is their heavy glycosylation, particularly Erns, which contains seven putative N-linked glycosylation sites. N-glycosylation results in a complex mixture of neutral and monosialylated multiantennary N-glycans, accounting for nearly half of the molecular mass of mature glycoproteins (Branza-Nichita et al., 2004; Gavrilov et al., 2011). Interestingly, in terms of N-glycosylation, a previous study (Strasser, 2023) reported that N. benthamiana plant-based expression platforms exhibit remarkable versatility in that they can effectively accommodate transient and stable engineering of the N-glycan processing pathways without inducing cell death or generating adverse growth phenotypes associated with biomass production, protein expression or yield, features lacking in conventional expression systems. In this study, protein expression using a plant-derived system was employed as a first report for Erns protein expression in which the yielded Erns protein had a molecular mass of ~45 kDa, comparable to the mature Erns protein in previous studies (Thiel et al., 1991; Bhattacharya et al., 2019). Although it was speculated that E2 and Erns proteins would be highly glycosylated in the plant expression system, the purified proteins were successfully conjugated with colloidal gold in this study to develop the ICS, thereby enhancing their availability.
This study collected various sera to assess the sensitivity and specificity of ICS and its correlation with commercial ELISA kits. As mentioned, ICS showed high sensitivity or specificity for detecting CSFV E2 and Ems antibodies under different vaccination or infection backgrounds, particularly Erns detection, even at low SNT titers (Tables 1, 5), which would be difficult to confirm through ELISA kits. The performance of ICS exhibited high coincidence rates with ELISA kits detecting E2 and Ems antibodies. However, the preference for detecting E2 antibodies was evident, particularly later when the E2 subunit vaccine was employed in this study (Supplementary Table S1). Despite ICS exhibiting sensitivity with SNT titers of 16 with serum samples from pigs vaccinated with the modified live LOM strain (Table 1), it surprisingly demonstrated high sensitivity in detecting E2 antibodies in serum samples from pigs vaccinated with commercial conventional CSFV GPE− LAV or chimeric vaccines, even at lower SNT titers of 16 (Tables 5, 6; Supplementary Table S2). Although this study confirmed high sensitivity and specificity across a diverse range of sera, the sample size was small. Therefore, future studies should employ larger sample sizes to assess the reproducibility of ICS. Given the impact of storage temperature, time, and manufacturing batches on the reliability of detection, further research is essential to evaluate the repeatability and stability of ICS. In addition, because the dependence of CSFV-specific antibody responses on several factors, such as CSFV infection with different genotypes, the induction of E2-or Erns-specific antibody (Popescu et al., 2019), and the failure of vaccination or the delay of antibody responses against CSFV induced by PRRSV (Chen et al., 2019; Li et al., 2020), a comprehensive approach involving a combination of various methods and continuous monitoring is essential to accurately determine the antibody response against CSFV.
ICS was used to evaluate the DIVA profiles of two chimeric pestiviruses, generated based on an infectious cDNA clone derived from the GPE− LAV strain. These chimeric viruses possess the Erns glycoprotein, which is phylogenetically distant from CSFV, and are potential DIVA vaccine candidates (Huynh et al., 2023). Serum samples obtained from optimal vaccination dose experiments and vaccine-efficient studies were reused to evaluate the DIVA profiles of these chimeric vaccine candidates. In the optimal vaccination dose experiment, successful detection of anti-CSFV Erns antibodies in serum samples from conventionally vaccinated pigs was confirmed, whereas all chimeric-vaccinated pigs were negative, indicating the effectiveness of this ICS in detecting specific CSFV Erns antibodies. Meanwhile, the presence of an E2-specific antibody indicated that antibody responses depended on the individual and the time point postvaccination (Supplementary Table S2). Significantly, the DIVA capability of chimeric vaccine candidates was validated in vaccine efficacy studies focusing on CSFV Erns detection, by which ICS successfully distinguished chimeric-vaccinated animals from infected and conventionally vaccinated animals. The outcomes obtained provide valuable insights into the potential of this ICS to address the limitations of previous ICS that lack DIVA diagnostic capabilities with Erns antibody detection (Li et al., 2012; Bai et al., 2022; Xu et al., 2022). Although CSFV Erns is used as a negative marker to evaluate the feasibility of ICS for DIVA in this study, future studies should concentrate on developing a serological antibody assay capable of detecting antibodies against recombinant PAPeV Erns and PhoPeV Erns. This development will facilitate the implementation of marker vaccine candidates vGPE−/PAPeV Erns and vGPE−/PhoPeV Erns to control and eradicate CSF in the future. In addition, DIVA application with Erns antibody detection was not feasible to evaluate CSF-free status individually in animals, a limitation of the sera used in this study. Instead, detection of Erns CSFV-specific antibodies was suggested for implementation at the herd level, as validated in previous studies (Schroeder et al., 2012; Pannhorst et al., 2015). Therefore, further investigations are necessary to comprehensively validate ICS specificity and sensitivity across larger populations under various field conditions. Because serum samples derived from CSFV genotype 1 were mainly used in this study, evaluating its performance against diverse strains, genotypes, and its potential cross-reactivity with antibodies from other swine pathogens in further studies will enhance its reliability and applicability in practical DIVA strategies.
Until now, the gold standard for determining antibodies against CSFV is SNT. However, existing commercial ELISA kits evaluate E2 or Erns separately, which incurs additional costs and labor and is more time-consuming. Although ICS development for antibody detection has been established, previous efforts have primarily focused on detecting CSFV E2 protein antibodies. This study is a pioneering effort to develop an ICS capable of detecting E2 and Erns antibodies simultaneously. This ICS addresses the prior limitation of being unable to differentiate infected from vaccinated animals from previous studies (Bai et al., 2022; Xu et al., 2022). Extensive studies and long-term monitoring are also essential to evaluate the DIVA potential of subunit or chimeric vaccines that contribute significantly to CSF surveillance and control.
5 Conclusion
E2 and Erns and antigens derived from the plant showed great potential and can be used to engineer a CSFV E2/Erns dual-antibody ICS, which is suitable for application in the field to monitor CSFV antibodies in vaccinated pig herds. ICS not only demonstrated high correlation with commercial ELISA kits but also exhibited sensitivity and specificity for detecting E2 and Erns antibodies. Thus, ICS is a rapid diagnostic tool for detecting antibodies against CSFV. The most vital point of ICS developed in this study is that it can satisfy the DIVA concept using two chimeric vaccine candidates, vGPE−/PAPeV Erns and vGPE−/PhoPeV Erns. Future work should be conducted to evaluate the stability of ICS and verify its repeatability.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by the Institutional Animal Care and Use Committee of the Faculty of Veterinary Medicine, Hokkaido University (approval no. 18-0038, approved on March 26, 2018, and approval no. 23-0029, approved on March 23, 2023) and performed according to the guidelines of this committee. The genetic modification experiments were performed according to the Cartagena Protocol on Biosafety under the Convention on Biological Diversity and approved by the Institutional Committee on the Safety of Genetic Modification Experiments of Hokkaido University and the Ministry of Education, Culture, Sports, Science and Technology (approval no. 2020-009 and 5-Monkashin-Dai-479, approved on June 8, 2021 and August 31, 2023, respectively). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. E-JS: Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing. YP: Conceptualization, Data curation, Investigation, Methodology, Writing – review & editing. JK: Conceptualization, Data curation, Investigation, Methodology, Writing – review & editing. TS: Data curation, Investigation, Writing – review & editing. TH: Data curation, Investigation, Writing – review & editing. NI: Data curation, Investigation, Writing – review & editing. S-HH: Data curation, Investigation, Writing – review & editing. H-NL: Data curation, Investigation, Writing – review & editing. YS: Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was partly conducted within the research project “Regulatory Research Projects for Food Safety, Animal Health and Plant Protection (JPJ008617.20319390),” funded by the Ministry of Agriculture, Forestry and Fisheries of Japan. The Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (MEXT KAKENHI grant no. JP22H02504) also partly funded this work. Assistance was also provided through the WISE Grant-in-Aid for Graduate Students, Program for One Health Frontier of the Graduate School of Excellence, Hokkaido University (grant no. PH36210001).
Acknowledgments
We thank the officials and farmers of the field farm vaccination project conducted in Jeju Island and Pohang City, Republic of Korea.
Conflict of interest
E-JS, YP, and JK were employed by BioApplications Inc. S-HH and H-NL were employed by Celltrix Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1383976/full#supplementary-material
Footnotes
References
Aebischer, A., Müller, M., and Hofmann, M. A. (2013). Two newly developed Erns-based ELISAs allow the differentiation of classical swine fever virus-infected from marker-vaccinated animals and the discrimination of pestivirus antibodies. Vet. Microbiol. 161, 274–285. doi: 10.1016/j.vetmic.2012.07.046
Bai, Y., Jia, R., Wei, Q., Wang, L., Sun, Y., Li, Y., et al. (2022). Development and application of a high-sensitivity immunochromatographic test strip for detecting classical swine fever virus antibodies. Transbound. Emerg. Dis. 69, e788–e798. doi: 10.1111/tbed.14367
Bhattacharya, S., Saini, M., Bisht, D., Rana, M., Bachan, R., Gogoi, S. M., et al. (2019). Lentiviral-mediated delivery of classical swine fever virus Erns gene into porcine kidney-15 cells for production of recombinant ELISA diagnostic antigen. Mol. Biol. Rep. 46, 3865–3876. doi: 10.1007/s11033-019-04829-0
Blome, S., Meindl-Böhmer, A., Loeffen, W., Thuer, B., and Moennig, V. (2006). Assessment of classical swine fever diagnostics and vaccine performance. Rev. Sci. Tech. 25, 1025–1038. doi: 10.20506/rst.25.3.1715
Blome, S., Moß, C., Reimann, I., König, P., and Beer, M. (2017). Classical swine fever vaccines—state-of-the-art. Vet. Microbiol. 206, 10–20. doi: 10.1016/j.vetmic.2017.01.001
Branza-Nichita, N., Lazar, C., Dwek, R. A., and Zitzmann, N. (2004). Role of N-glycan trimming in the folding and secretion of the pestivirus protein E(rns). Biochem. Biophys. Res. Commun. 319, 655–662. doi: 10.1016/j.bbrc.2004.05.039
Chen, W.-T., Liu, H.-M., Chang, C.-Y., Deng, M.-C., Huang, Y.-L., Chang, Y.-C., et al. (2023). Cross-reactivities and cross-neutralization of different envelope glycoproteins E2 antibodies against different genotypes of classical swine fever virus. Front. Vet. Sci. 10:1169766. doi: 10.3389/fvets.2023.1169766
Chen, D., Liu, X., Xu, S., Chen, D., Zhou, L., Ge, X., et al. (2019). TNF-α induced by porcine reproductive and respiratory syndrome virus inhibits the replication of classical swine fever virus C-strain. Vet. Microbiol. 234, 25–33. doi: 10.1016/j.vetmic.2019.05.007
Coronado, L., Perera, C. L., Rios, L., Frías, M. T., and Pérez, L. J. (2021). A critical review about different vaccines against classical swine fever virus and their repercussions in endemic regions. Vaccines 9:154. doi: 10.3390/vaccines9020154
Coronado, L., Rios, L., Frías, M. T., Amarán, L., Naranjo, P., Percedo, M. I., et al. (2019). Positive selection pressure on E2 protein of classical swine fever virus drives variations in virulence, pathogenesis and antigenicity: implication for epidemiological surveillance in endemic areas. Transbound. Emerg. Dis. 66, 2362–2382. doi: 10.1111/tbed.13293
Ganges, L., Crooke, H. R., Bohórquez, J. A., Postel, A., Sakoda, Y., Becher, P., et al. (2020). Classical swine fever virus: the past, present and future. Virus Res. 289:198151. doi: 10.1016/j.virusres.2020.198151
Gavrilov, B. K., Rogers, K., Fernandez-Sainz, I. J., Holinka, L. G., Borca, M. V., and Risatti, G. R. (2011). Effects of glycosylation on antigenicity and immunogenicity of classical swine fever virus envelope proteins. Virology 420, 135–145. doi: 10.1016/j.virol.2011.08.025
Gong, W., Li, J., Wang, Z., Sun, J., Mi, S., Xu, J., et al. (2019). Commercial E2 subunit vaccine provides full protection to pigs against lethal challenge with 4 strains of classical swine fever virus genotype 2. Vet. Microbiol. 237:108403. doi: 10.1016/j.vetmic.2019.108403
Graham, S. P., Everett, H. E., Haines, F. J., Johns, H. L., Sosan, O. A., Salguero, F. J., et al. (2012). Challenge of pigs with classical swine fever viruses after C-strain vaccination reveals remarkably rapid protection and insights into early immunity. PLoS One 7:e29310. doi: 10.1371/journal.pone.0029310
Hager, K. J., Pérez Marc, G., Gobeil, P., Diaz, R. S., Heizer, G., Llapur, C., et al. (2022). Efficacy and safety of a recombinant plant-based adjuvanted Covid-19 vaccine. N. Engl. J. Med. 386, 2084–2096. doi: 10.1056/NEJMoa2201300
Huang, C., Chien, M.-S., Hu, C.-M., Chen, C.-W., and Hsieh, P.-C. (2006). Secreted expression of the classical swine fever virus glycoprotein E(rns) in yeast and application to a sandwich blocking ELISA. J. Virol. Methods 132, 40–47. doi: 10.1016/j.jviromet.2005.08.020
Huynh, L. T., Isoda, N., Hew, L. Y., Ogino, S., Mimura, Y., Kobayashi, M., et al. (2023). Generation and efficacy of two chimeric viruses derived from GPE− vaccine strain as classical swine fever vaccine candidates. Viruses 15:1587. doi: 10.3390/v15071587
Ji, S., Luo, Y., Zhang, T., Shao, L., Meng, X.-Y., Wang, Y., et al. (2018). An improved indirect ELISA for specific detection of antibodies against classical swine fever virus based on structurally designed E2 protein expressed in suspension mammalian cells. Arch. Virol. 163, 1831–1839. doi: 10.1007/s00705-018-3809-7
Kallolimath, S., Palt, R., Föderl-Höbenreich, E., Sun, L., Chen, Q., Pruckner, F., et al. (2023). Glyco engineered pentameric SARS-CoV-2 IgMs show superior activities compared to IgG1 orthologues. Front. Immunol. 14:1147960. doi: 10.3389/fimmu.2023.1147960
König, M., Lengsfeld, T., Pauly, T., Stark, R., and Thiel, H. J. (1995). Classical swine fever virus: independent induction of protective immunity by two structural glycoproteins. J. Virol. 69, 6479–6486. doi: 10.1128/JVI.69.10.6479-6486.1995
Langedijk, J. P. M., Middel, W. G. J., Meloen, R. H., Kramps, J. A., and de Smit, J. A. (2001). Enzyme-linked immunosorbent assay using a virus type-specific peptide based on a subdomain of envelope protein Erns for serologic diagnosis of pestivirus infections in swine. J. Clin. Microbiol. 39, 906–912. doi: 10.1128/JCM.39.3.906-912.2001
Laughlin, R. C., Madera, R., Peres, Y., Berquist, B. R., Wang, L., Buist, S., et al. (2019). Plant-made E2 glycoprotein single-dose vaccine protects pigs against classical swine fever. Plant Biotechnol. J. 17, 410–420. doi: 10.1111/pbi.12986
Li, Y.-C., Chiou, M.-T., and Lin, C.-N. (2020). Serodynamic analysis of the piglets born from sows vaccinated with modified live vaccine or E2 subunit vaccine for classical swine fever. Pathogens 9:427. doi: 10.3390/pathogens9060427
Li, X., Wang, L., Shi, X., Zhao, D., Yang, J., Yang, S., et al. (2012). Development of an immunochromatographic strip for rapid detection of antibodies against classical swine fever virus. J. Virol. Methods 180, 32–37. doi: 10.1016/j.jviromet.2011.12.006
Lim, S., Choe, S., Kim, K.-S., Jeoung, H.-Y., Cha, R. M., Park, G.-S., et al. (2019). Assessment of the efficacy of an attenuated live marker classical swine fever vaccine (Flc-LOM-BErns) in pregnant sows. Vaccine 37, 3598–3604. doi: 10.1016/j.vaccine.2019.04.076
Lin, M., Trottier, E., and Pasick, J. (2005). Antibody responses of pigs to defined Erns fragments after infection with classical swine fever virus. Clin. Diagn. Lab. Immunol. 12, 180–186. doi: 10.1128/CDLI.12.1.180-186.2005
Lin, M., Trottier, E., Pasick, J., and Sabara, M. (2004). Identification of antigenic regions of the Erns protein for pig antibodies elicited during classical swine fever virus infection. J. Biochem. 136, 795–804. doi: 10.1093/jb/mvh189
Luo, Y., Li, L., Austermann-Busch, S., Dong, M., Xu, J., Shao, L., et al. (2015). Enhanced expression of the Erns protein of classical swine fever virus in yeast and its application in an indirect enzyme-linked immunosorbent assay for antibody differentiation of infected from vaccinated animals. J. Virol. Methods 222, 22–27. doi: 10.1016/j.jviromet.2015.05.006
Ma, J. K.-C., Drossard, J., Lewis, D., Altmann, F., Boyle, J., Christou, P., et al. (2015). Regulatory approval and a first-in-human phase I clinical trial of a monoclonal antibody produced in transgenic tobacco plants. Plant Biotechnol. J. 13, 1106–1120. doi: 10.1111/pbi.12416
Manessis, G., Gelasakis, A. I., and Bossis, I. (2022). Point-of-care diagnostics for farm animal diseases: from biosensors to integrated lab-on-chip devices. Biosensors 12:455. doi: 10.3390/bios12070455
Meyer, D., Fritsche, S., Luo, Y., Engemann, C., Blome, S., Beyerbach, M., et al. (2017). The double-antigen ELISA concept for early detection of Erns-specific classical swine fever virus antibodies and application as an accompanying test for differentiation of infected from marker vaccinated animals. Transbound. Emerg. Dis. 64, 2013–2022. doi: 10.1111/tbed.12611
Meyers, G., and Thiel, H.-J. (1996). “Molecular characterization of pestiviruses” in Advances in virus research. eds. K. Maramorosch, F. A. Murphy, and A. J. Shatkin (Cambridge: Academic Press), 53–118.
Moormann, R. J. M., Bouma, A., Kramps, J. A., Terpstra, C., and De Smit, H. J. (2000). Development of a classical swine fever subunit marker vaccine and companion diagnostic test. Vet. Microbiol. 73, 209–219. doi: 10.1016/S0378-1135(00)00146-2
Muñoz-González, S., Perez-Simó, M., Muñoz, M., Bohorquez, J. A., Rosell, R., Summerfield, A., et al. (2015). Efficacy of a live attenuated vaccine in classical swine fever virus postnatally persistently infected pigs. Vet. Res. 46:78. doi: 10.1186/s13567-015-0209-9
Oh, Y., Park, Y., Choi, B.-H., Park, S., Gu, S., Park, J., et al. (2021). Field application of a new CSF vaccine based on plant-produced recombinant E2 marker proteins on pigs in areas with two different control strategies. Vaccines 9:537. doi: 10.3390/vaccines9060537
Pan, Q., Wu, W., Liao, S., Wang, S., Zhao, C., Li, C., et al. (2019). Comparison of the detection performance of two different one-step-combined test strips with fluorescent microspheres or colored microspheres as tracers for influenza A and B viruses. Virol. J. 16:91. doi: 10.1186/s12985-019-1190-0
Pannhorst, K., Fröhlich, A., Staubach, C., Meyer, D., Blome, S., and Becher, P. (2015). Evaluation of an Erns-based enzyme-linked immunosorbent assay to distinguish classical swine fever virus-infected pigs from pigs vaccinated with CP7_E2alf. J. Vet. Diagn. Invest. 27, 449–460. doi: 10.1177/1040638715592446
Panyasing, Y., Thanawongnuwech, R., Ji, J., Giménez-Lirola, L., and Zimmerman, J. (2018). Detection of classical swine fever virus (CSFV) E2 and Erns antibody (IgG, IgA) in oral fluid specimens from inoculated (ALD strain) or vaccinated (LOM strain) pigs. Vet. Microbiol. 224, 70–77. doi: 10.1016/j.vetmic.2018.08.024
Park, Y., An, D.-J., Choe, S., Lee, Y., Park, M., Park, S., et al. (2019). Development of recombinant protein-based vaccine against classical swine fever virus in pigs using transgenic Nicotiana benthamiana. Front. Plant Sci. 10:624. doi: 10.3389/fpls.2019.00624
Park, Y., Lee, S., Kang, H., Park, M., Min, K., Kim, N. H., et al. (2020). A classical swine fever virus E2 fusion protein produced in plants elicits a neutralizing humoral immune response in mice and pigs. Biotechnol. Lett. 42, 1247–1261. doi: 10.1007/s10529-020-02892-3
Park, Y., Oh, Y., Wang, M., Ganges, L., Bohórquez, J. A., Park, S., et al. (2021). A novel E2 glycoprotein subunit marker vaccine produced in plant is able to prevent classical swine fever virus vertical transmission after double vaccination. Vaccines 9:418. doi: 10.3390/vaccines9050418
Pérez, L. J., Díaz de Arce, H., Perera, C. L., Rosell, R., Frías, M. T., Percedo, M. I., et al. (2012). Positive selection pressure on the B/C domains of the E2-gene of classical swine fever virus in endemic areas under C-strain vaccination. Infect. Genet. Evol. 12, 1405–1412. doi: 10.1016/j.meegid.2012.04.030
Popescu, L. N., Panyasing, Y., Giménez-Lirola, L., Zimmerman, J., and Rowland, R. R. R. (2019). E2 and Erns isotype-specific antibody responses in serum and oral fluid after infection with classical swine fever virus (CSFV). Vet. Microbiol. 235, 265–269. doi: 10.1016/j.vetmic.2019.07.007
Postel, A., Austermann-Busch, S., Petrov, A., Moennig, V., and Becher, P. (2018). Epidemiology, diagnosis and control of classical swine fever: recent developments and future challenges. Transbound. Emerg. Dis. 65, 248–261. doi: 10.1111/tbed.12676
Postel, A., Smith, D. B., and Becher, P. (2021). Proposed update to the taxonomy of Pestiviruses: eight additional species within the genus Pestivirus, family Flaviviridae. Viruses 13:1542. doi: 10.3390/v13081542
Qiu, X., Wong, G., Audet, J., Bello, A., Fernando, L., Alimonti, J. B., et al. (2014). Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53. doi: 10.1038/nature13777
Rau, H., Revets, H., Balmelli, C., McCullough, K. C., and Summerfield, A. (2006). Immunological properties of recombinant classical swine fever virus NS3 protein in vitro and in vivo. Vet. Res. 37, 155–168. doi: 10.1051/vetres:2005049
Reimann, I., Depner, K., Trapp, S., and Beer, M. (2004). An avirulent chimeric Pestivirus with altered cell tropism protects pigs against lethal infection with classical swine fever virus. Virology 322, 143–157. doi: 10.1016/j.virol.2004.01.028
Sakoda, Y., Wakamoto, H., Tamura, T., Nomura, T., Naito, M., Aoki, H., et al. (2012). Development and evaluation of indirect enzyme-linked immunosorbent assay for a screening test to detect antibodies against classical swine fever virus. Jpn. J. Vet. Res. 60, 85–94. doi: 10.14943/jjvr.60.2-3.85
Schroeder, S., von Rosen, T., Blome, S., Loeffen, W., Haegeman, A., Koenen, F., et al. (2012). Evaluation of classical swine fever virus antibody detection assays with an emphasis on the differentiation of infected from vaccinated animals. Rev. Sci. Tech. 31, 997–1010. doi: 10.20506/rst.31.3.2173
Sohn, E.-J., Lee, Y., Park, N., Park, M., Kim, N. H., Park, S., et al. (2018). Development of plant-produced E2 protein for use as a green vaccine against classical swine fever virus. J. Plant Biol. 61, 241–252. doi: 10.1007/s12374-018-0133-4
Strasser, R. (2023). Plant glycoengineering for designing next-generation vaccines and therapeutic proteins. Biotechnol. Adv. 67:108197. doi: 10.1016/j.biotechadv.2023.108197
Tetsuo, M., Matsuno, K., Tamura, T., Fukuhara, T., Kim, T., Okamatsu, M., et al. (2020). Development of a high-throughput serum neutralization test using recombinant pestiviruses possessing a small reporter tag. Pathogens 9:188. doi: 10.3390/pathogens9030188
Thiel, H. J., Stark, R., Weiland, E., Rümenapf, T., and Meyers, G. (1991). Hog cholera virus: molecular composition of virions from a pestivirus. J. Virol. 65, 4705–4712. doi: 10.1128/JVI.65.9.4705-4712.1991
van Rijn, P. A. (2007). A common neutralizing epitope on envelope glycoprotein E2 of different pestiviruses: implications for improvement of vaccines and diagnostics for classical swine fever (CSF)? Vet. Microbiol. 125, 150–156. doi: 10.1016/j.vetmic.2007.05.001
van Rijn, P. A., van Gennip, H. G. P., and Moormann, R. J. M. (1999). An experimental marker vaccine and accompanying serological diagnostic test both based on envelope glycoprotein E2 of classical swine fever virus (CSFV). Vaccine 17, 433–440. doi: 10.1016/S0264-410X(98)00215-1
Wang, L., Madera, R., Li, Y., McVey, D. S., Drolet, B. S., and Shi, J. (2020). Recent advances in the diagnosis of classical swine fever and future perspectives. Pathogens 9:658. doi: 10.3390/pathogens9080658
Ward, B. J., Makarkov, A., Séguin, A., Pillet, S., Trépanier, S., Dhaliwall, J., et al. (2020). Efficacy, immunogenicity, and safety of a plant-derived, quadrivalent, virus-like particle influenza vaccine in adults (18-64 years) and older adults (≥65 years): two multicentre, randomised phase 3 trials. Lancet 396, 1491–1503. doi: 10.1016/S0140-6736(20)32014-6
Wei, Q., Bai, Y., Song, Y., Liu, Y., Yu, W., Sun, Y., et al. (2021). Generation and immunogenicity analysis of recombinant classical swine fever virus glycoprotein E2 and Erns expressed in baculovirus expression system. Virol. J. 18:44. doi: 10.1186/s12985-021-01507-1
Xu, Q., Sun, Y., Yang, J., Ma, F., Wang, Y., Zhang, S., et al. (2022). An improved immunochromatographic strip based on plant-derived E2 for detection of antibodies against classical swine fever virus. Microbiol. Spectr. 10:e0105022. doi: 10.1128/spectrum.01050-22
Yamamoto, T., Hoshikawa, K., Ezura, K., Okazawa, R., Fujita, S., Takaoka, M., et al. (2018). Improvement of the transient expression system for production of recombinant proteins in plants. Sci. Rep. 8:4755. doi: 10.1038/s41598-018-23024-y
Yi, W., Zhu, H., Wu, Y., Li, Q., Lou, W., Zhao, H., et al. (2022). The recombinant Erns and truncated E2-based indirect enzyme-linked immunosorbent assays to distinguishably test specific antibodies against classical swine fever virus and bovine viral diarrhea virus. Virol. J. 19:121. doi: 10.1186/s12985-022-01851-w
Keywords: classical swine fever, immunochromatographic test strip, E2, Erns, antibody detection
Citation: Huynh LT, Sohn E-J, Park Y, Kim J, Shimoda T, Hiono T, Isoda N, Hong S-H, Lee H-N and Sakoda Y (2024) Development of a dual immunochromatographic test strip to detect E2 and Erns antibodies against classical swine fever. Front. Microbiol. 15:1383976. doi: 10.3389/fmicb.2024.1383976
Edited by:
Jianchao Wei, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Bin Zhou, Nanjing Agricultural University, ChinaFei Gao, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Huynh, Sohn, Park, Kim, Shimoda, Hiono, Isoda, Hong, Lee and Sakoda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eun-Ju Sohn, ejsohn@bioapp.co.kr; Yoshihiro Sakoda, sakoda@vetmed.hokudai.ac.jp
 Loc Tan Huynh
Loc Tan Huynh Eun-Ju Sohn3*
Eun-Ju Sohn3* Takahiro Hiono
Takahiro Hiono Norikazu Isoda
Norikazu Isoda Yoshihiro Sakoda
Yoshihiro Sakoda