- 1P. A. Hertsen Moscow Oncology Research Center, Branch of the National Medical Research Radiological Center, Ministry of Health of the Russian Federation, Moscow, Russia
- 2Faculty of Biology and Biotechnologies, Higher School of Economics, Moscow, Russia
- 3Darby Children’s Research Institute, Medical University of South Carolina, Charleston, SC, United States
- 4Department of Pediatrics, Medical University of South Carolina, Charleston, SC, United States
- 5Department of Pharmaceutical Sciences, University at Buffalo, Buffalo, NY, United States
- 6Hollings Cancer Center, Department of Biochemistry and Molecular Biology, Medical University of South Carolina, Charleston, SC, United States
- 7Department of Chemical Enzymology, M.V. Lomonosov Moscow State University, Moscow, Russia
- 8Department of Chemistry and Physical Sciences, Dyson College of Arts and Sciences, Pace University, Pleasantville, NY, United States
- 9Department of Neuroscience, Medical University of South Carolina, Charleston, SC, United States
- 10Department of Drug Discovery, Medical University of South Carolina, Charleston, SC, United States
The Keap1-Nrf2 signaling axis is a validated and promising target for cellular defense and survival pathways. This minireview discusses the potential off-target effects and their impact on future drug development originating from Keap1-targeting small molecules that function as displacement activators of the redox-sensitive transcription factor Nrf2. We argue that small-molecule displacement activators, similarly to electrophiles, will release both Nrf2 and other Keap1 client proteins from the ubiquitin ligase complex. This non-specificity is likely unavoidable and may result in off-target effects during Nrf2 activation by targeting Keap1. The small molecule displacement activators may also target Kelch domains in proteins other than Keap1, causing additional off-target effects unless designed to ensure specificity for the Kelch domain only in Keap1. A potentially promising and alternative therapeutic approach to overcome this non-specificity emerging from targeting Keap1 is to inhibit the Nrf2 repressor Bach1 for constitutive activation of the Nrf2 pathway and bypass the Keap1-Nrf2 complex.
Introduction
With the increase in the global population, the worldwide prevalence of neurodegenerative diseases is on the rise. They are among the leading causes of disability and death worldwide and will continue to grow in the coming decades due to increased life expectancy. Despite a considerable amount of basic and clinical research, most strategies to manage neurodegenerative diseases are palliative. A large body of evidence suggests that these diseases are multifactorial caused by genetic, environmental, and endogenous factors related to aging. An emerging target for neurodegenerative diseases that could modulate multiple etiological pathways involves drug-induced activation of a coordinated genetic program to maintain redox equilibrium through the expression of pro-survival proteins and cytoprotective genes (Ammal Kaidery et al., 2019; Brandes and Gray, 2020). A key transcription factor orchestrating this process is nuclear factor erythroid 2-related factor 2 (Nrf2), a member of the cap’n’collar family of basic leucine zipper transcription factors. By binding the antioxidant response elements (ARE) in promoter regions, Nrf2 regulates the transcription of over 250 genes, which together build a multifaceted network that integrates cellular activities including drug detoxification, immunomodulation, maintenance of both redox and protein homeostasis, and energy metabolism (Ammal Kaidery et al., 2019). The breadth of this endogenous response suggests that its activation might counterbalance many of the large numbers of etiological pathways implicated in neurodegenerative diseases. In recent years, therapies based on Nrf2 activation have been proposed to benefit neurodegenerative diseases (Johnson and Johnson, 2015; Gazaryan and Thomas, 2016; Cuadrado et al., 2019; Brandes and Gray, 2020). Currently, there are three known pathways for Nrf2 stabilization and activation: (a) the constitutively operating Kelch-like ECH associated protein 1 (Keap1)-dependent pathway where Nrf2 is negatively regulated by Keap1, which promotes the ubiquitination and subsequent proteasomal degradation; and (b) two recently described stress-induced pathways, where beta-transducin repeat-containing E3 ubiquitin-protein ligase (β-TrCP) and E3 ubiquitin-protein ligase synoviolin (Hrd1) negatively regulate Nrf2 through Keap1-independent mechanisms (Figure 1; Harder et al., 2015).
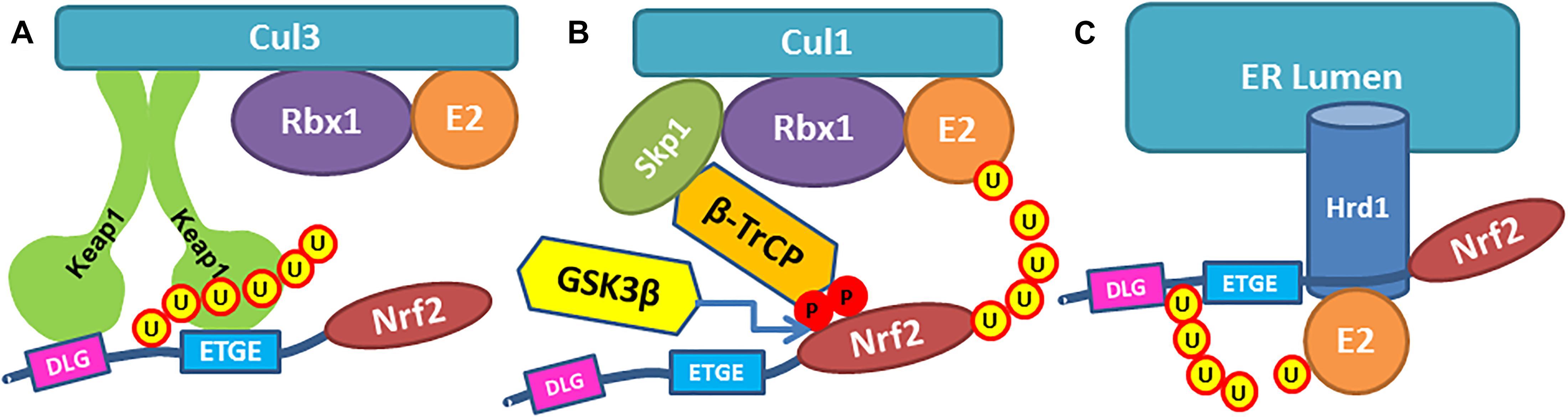
Figure 1. Three pathways of Nrf2 ubiquitinylation. (A) Nrf2 Neh2-domain is recognized by Keap1 dimer and binds Keap1 Kelch domains via ETGE and DLG binding motifs, so the lysine residues in Neh2 are ubiquitinylated by Cullin3 ligase. (B) β-TrCP in Cul1 ubiquitin ligase complex binds Neh6 domain in Nrf2 protein via DSAPGS and GSK3β- phosphorylated DSGIS binding motifs, so Nrf2 is getting ubiquitinylated. (C) Ubiquitin ligase Hrd1 ubiquitinylates Nrf2.
Keap1-Nrf2 Pathway
In the Keap1-dependent constitutive pathway, dimeric Keap1 (Ogura et al., 2010) recognizes the Neh2-domain in the Nrf2 protein. Keap1 binds to Nrf2 so that lysine residues in Neh2-domain are ubiquitinylated by Cullin3 (Cul3) ligase, and thus, Nrf2 is labeled for proteasomal degradation. Neh2 domain is stretched between two Kelch domains in the Keap1 dimer: Neh2 binds Kelch via DLG (weak) and ETGE (strong) binding motifs (Figures 1A, 2). As demonstrated recently1, the minimum recognition sequence in Neh2 domain includes 16–85 aa residues. Keap1 is a redox sensor protein (Baird and Yamamoto, 2020), and chemical modification of Keap1 Cys151 results in detachment of Cul3 ligase and stabilization of Nrf2 protein. Modification of the other redox-active cysteines in Keap1 induces conformational changes that break Keap1 dimeric structure and release Nrf2 protein (Baird and Yamamoto, 2020). A physiologically relevant Keap1 redox regulation with hydrogen peroxide is based on the formation of a disulfide bridge between Cys226/Cys613 (Fourquet et al., 2010) or any two of the Cys226/Cys613/Cys622/Cys624 residues (Figure 2), which likely results in Keap1 dimer destabilization and Nrf2 release (Suzuki et al., 2019). This constitutive pathway is characterized in detail and is commonly used to manipulate Nrf2 stability and activation.
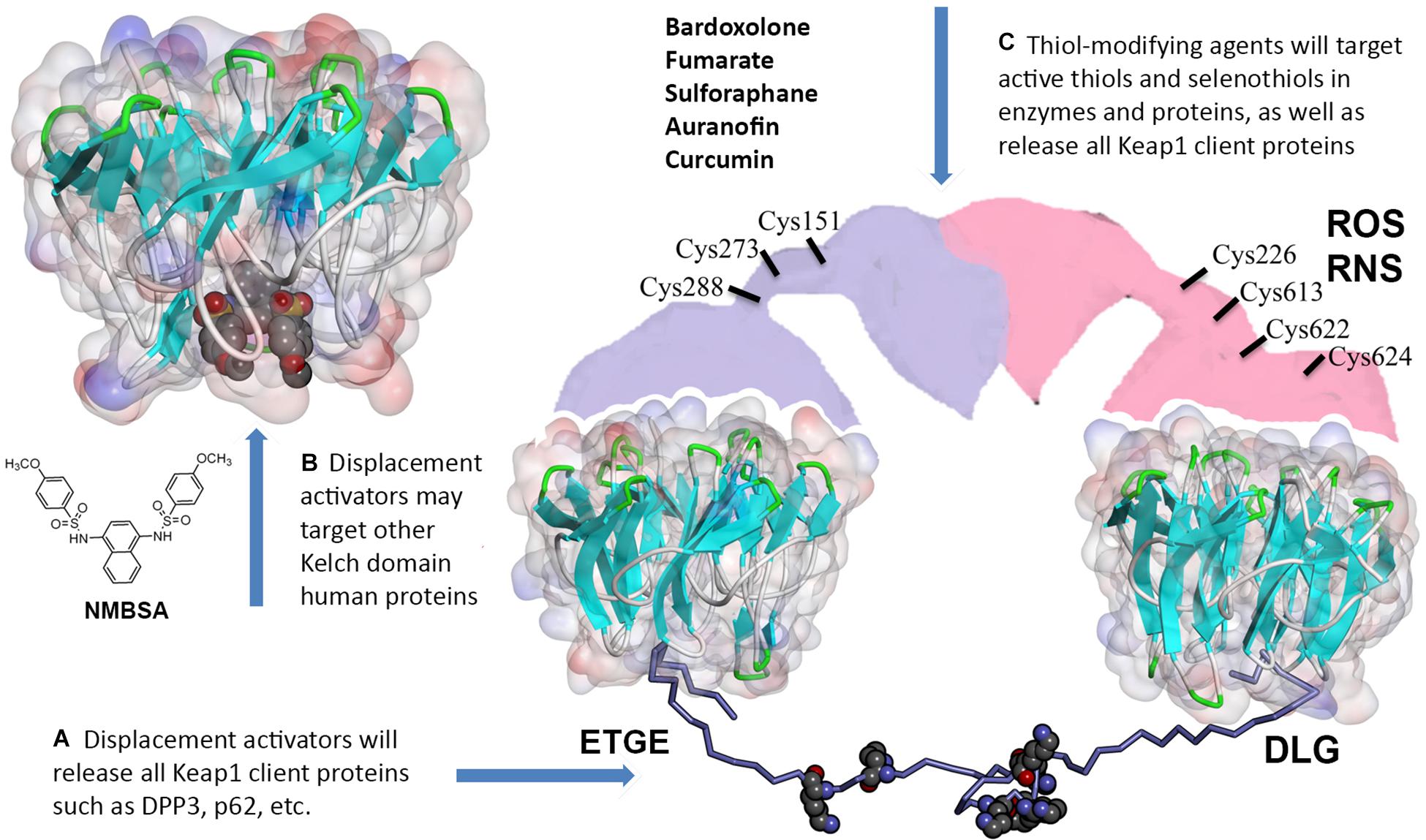
Figure 2. Off-target effects of Keap1 pharmacological targeting. (A) Thiol-modifying agents as well as displacement activators of Nrf2 (both peptides and small molecules) will release all other Keap1 client proteins. (B) Small molecule displacement activators may target Kelch domains in other proteins. (C) Thiol-modifying agents (bardoxolone and others) will inactivate redox active thiols and selenothiols in other proteins. The model for 16–85 aa Neh2 peptide (lysine residues shown with WdW radii) stretched between two Kelch domains was built using Kelch crystal structures with the corresponding Nrf2 binding motifs (4IFL.pdb and 2DYH.pdb) distanced at ca. 80 Å in accord with the Keap1 dimer conformation reported in Ogura et al. (2010). Full description of the modeling procedure is in https://www.x-mol.com/paper/1287827255333666816.
Nrf2 Neh6 Domain Phosphodegron
Nuclear factor erythroid 2-related factor 2 is more rapidly turned over in cells grown under homeostatic conditions than in those experiencing oxidative stress. The variable turnover of Nrf2 is accomplished through the use of at least two degrons and its redox-sensitive interaction with Keap1. McMahon et al. (2004) described a redox-insensitive degron (amino acids 329–379) in Nrf2’s Neh6- domain in 2004. They showed that if Nrf2’s half-life in Keap1 expressing cells was 10 min, in Keap1 null cells upon oxidative stress, Nrf2 was still unstable with a half-life of 40 min. The serine/threonine kinase, glycogen synthase kinase 3β (GSK3β), phosphorylates a group of serine residues in the Neh6-domain of mouse Nrf2 and promotes its degradation in a Keap1-independent manner (Rada et al., 2011; Figure 1B). There are two distinct motifs within Neh6-domain, namely DSAPGS and DSGIS, recognized by β-transducin repeat-containing protein (β-TrCP) in β-TrCP-Skp1-Cul1-Rbx1 E3/Roc1 ubiquitin ligase complex (Figure 1B). A phosphorylated DSGIS motif binds more tightly on β-TrCP than its non-phosphorylated version (Chowdhry et al., 2013). Thus, activation of GSK3β generates a phosphodegron resulting in destabilization of Nrf2. Consequently, GSK3β inhibition may result in Nrf2 stabilization and activation (in the unlikely scenario where there is no functional Keap1). Some of the reported GSK3β-inhibitors have strong pro-oxidant motifs in their structure and may likely function as direct stabilizers of Nrf2 working via the Keap1 pathway. For example, our unpublished data shows that GSK3β-inhibitor TDZD8 activates Neh2-luc reporter, which screens for direct Nrf2 stabilizers working via Keap1 (Smirnova et al., 2011). The contribution of GSK3β inhibition toward Nrf2 activation in the presence of functional Keap1 will be insignificant unless Keap1 is targeted simultaneously. This scenario also exists with TDZD8, and therefore it is not surprising that this GSK3β-inhibitor is protective in pretreatment paradigms in renal ischemia/reperfusion (Hu et al., 2016; Shen et al., 2017) or prolonged post-treatment after transplantation (Deng et al., 2020). Furthermore, GSK3β is implicated in numerous pathways, and its inhibition will unlikely be specific for Nrf2 activation. Moreover, again, in the presence of functionally active Keap1, merely inhibiting GSK3β will have no significant impact on Nrf2 activation.
Endoplasmic Reticulum Stress-Induced Activation of Nrf2 Pathway
A more recently described pathway leading to Nrf2 destabilization is based on endoplasmic reticulum (ER)-stress-induced 3-hydroxy-3-ethylglutaryl-CoA reductase degradation protein 1 (Hrd1) (Wu et al., 2014; Figure 1C). Ubiquitin ligase Hrd1 is the focus of prevailing research since it is the critical enzyme in the complex executing retro-translocation and poly-ubiquitinylation of misfolded luminal ER proteins (Wu X. et al., 2020). However, the Hrd1-Nrf2 link is very poorly studied. The recent work claiming octyl itaconate as Hrd1 inhibitor and, as such, an Nrf2 activator (Sun et al., 2019), completely ignored the fact that octyl itaconate is a classic Michael acceptor capable of alkylating Keap1 thiols, and Cys151 in particular as shown by Liu et al. (2018). Thus, the constitutive pathway is the best-studied and is the focus of numerous efforts for the therapeutic development of Nrf2 activators. The two stress-induced pathways, and the Hrd1-Nrf2 pathway, in particular, are far from being mechanistically well-understood. They are complementary to the constitutive pathway under the conditions of acute oxidative stress. Therefore, they have to be targeted together with the constitutive pathway to reach the full benefits of Nrf2 activation.
Nrf2 Stabilization via Targeting Keap1
Two principally different modes of Nrf2 stabilization in the constitutive pathway can be achieved: (a) irreversible, via Keap1 thiols modification followed by either dimer destabilization or Cul3 ubiquitin ligase detachment from Keap1, and (b) reversible, via competitive displacement of Nrf2 from the complex with Keap1 using Nrf2 peptides or small molecules specifically targeting Keap1 Kelch domain (Figure 2). The standard view is that the latter approach will be more beneficial because non-specific alkylation or oxidation of reactive thiols in proteins other than Keap1 can be avoided. However, one can expect off-target effects even from Nrf2 competitive displacement from Keap1, since (a) Keap1 has numerous client proteins, and (b) there are dozens of human BTB-Kelch proteins with Kelch domain structures very similar to the one in Keap1.
Identification of Keap1 Client Proteins
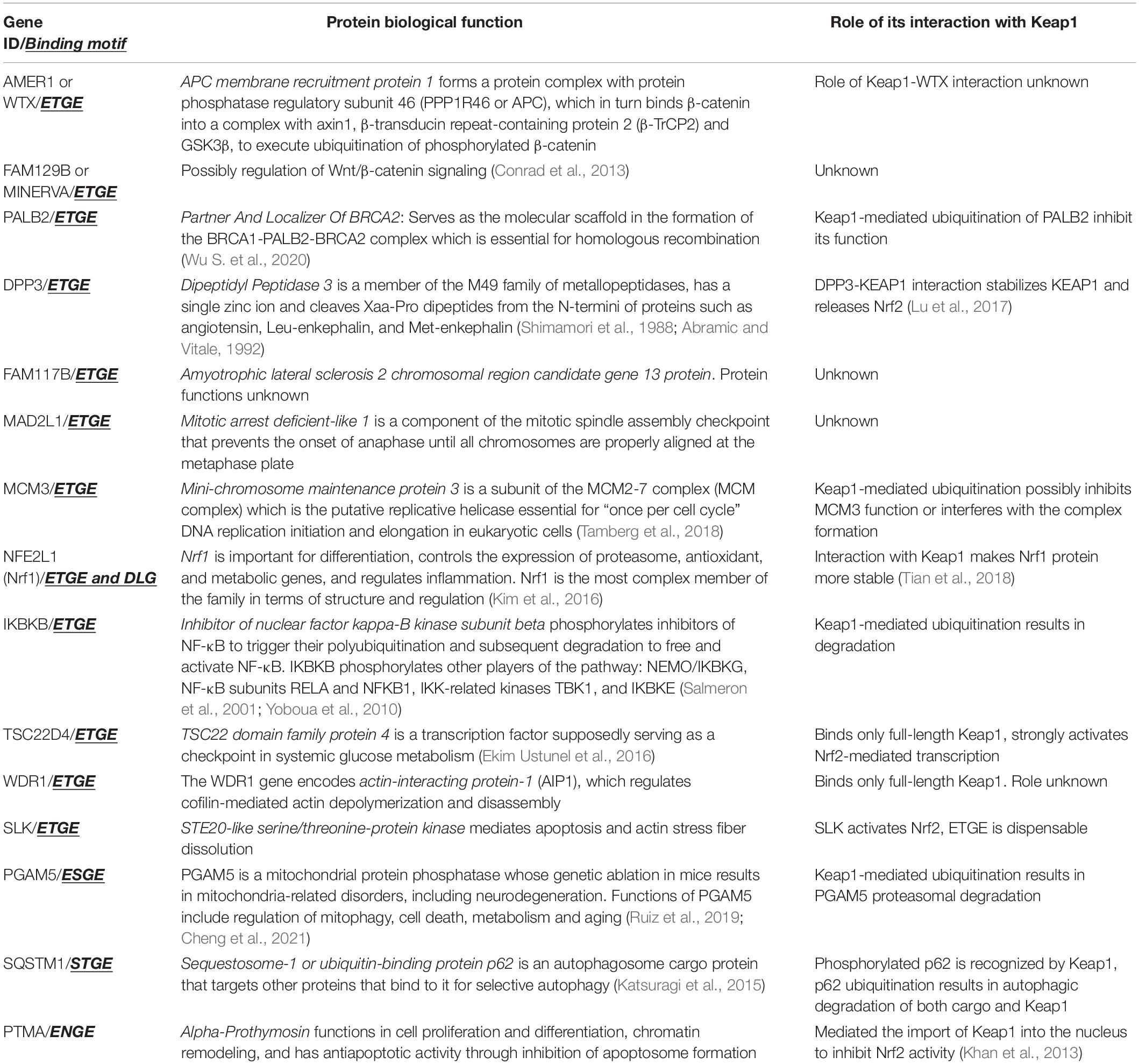
The strong binding peptide motif for the Keap1 Kelch domain has the sequence of ETGE or ESGE, and mutations in these sequences disrupts Nrf2 interaction with the Keap1 Kelch-domain and results in Nrf2 activation. A comprehensive proteomics study of Keap1 interaction network (Hast et al., 2013) identified all ETGE- or ESGE-containing proteins and determined whether they functionally control Nrf2. Despite the fact that 42 Keap1-interacting proteins have been reliably identified, only 17 of those contained an ETGE, ESGE, or both motifs. The current list of verified Keap1 interacting proteins (Hast et al., 2013; Kopacz et al., 2020; Zhang et al., 2020) are shown in Table 1.
As seen from Table 1, which lists only verified Keap1 interacting proteins via ETGE-like motifs, in many cases, the role played by such interaction is unclear. Moreover, very often, there is no ubiquitination of client proteins. Most intriguing is the recent work on Nrf1 binding to Keap1, which suggests that stretching of a peptide via ETGE and DLG between Kelch domains is not sufficient for ubiquitination and that there are specific requirements for the sequences inside the stretched peptide (Tian et al., 2018). In terms of the possibility of numerous side-effects of Keap1 targeting with either activator, the biggest concern is with the inhibitor of nuclear factor kappa-B kinase subunit beta (IKBKB), which in addition to phosphorylation of NF-κB pathway players (see in Table 1), also phosphorylates FOXO3, NCOA3, BCL10, IRS1, RIPK1, IRF5, to name just a few.
The list of Keap1 interactors covers a panoply of cellular processes, from DNA replication and licensing to cytoskeletal dynamics, transcription, and apoptosis. Predicting the plausible side effects is difficult given the absence of detailed information on Keap1 clients. However, based on the differences in the mechanism of Keap1 targeting by alkylating and displacement activators, some general predictions can be made. If we focus on thiol modifying Nrf2 activators targeting Keap1, they either destroy its dimeric structure or detach Keap1 from the ubiquitin ligase complex. In both cases, Keap1 will be no longer active with respect to ubiquitination of a client protein or its 2-point stretching between Kelch domains. However, this treatment should have no effect on the ability of individual Kelch domains to bind Keap1 client proteins via ETGE-like sequences. Contrary to this, high-affinity displacement activators targeting the Kelch domain will be capable of displacing all Keap1 interactors even if they are “hanging” onto one of the Kelch domains in the Keap1 dimer or are bound to the Keap1 monomer. So, one may expect more additional pathways to be triggered with displacement activators due to their tight binding to the Kelch domain in Keap1, which results in the release of all Keap1 dimer and monomer interactors. Displacement with peptides will be more selective since the affinity of Keap1 clients for Kelch is different, and only less tightly bound Keap1 client proteins will be displaced with a peptide (a typical dissociation constant for Nrf2-derived cell-permeable peptides is around 50 nM). So, peptide-targeted alkylating agents may turn out to be more specific than small-molecule displacement activators. However, there is an additional concern relevant to the small molecule displacement activator’s action, as we discuss below.
Kelch Domain Proteins Besides Keap1
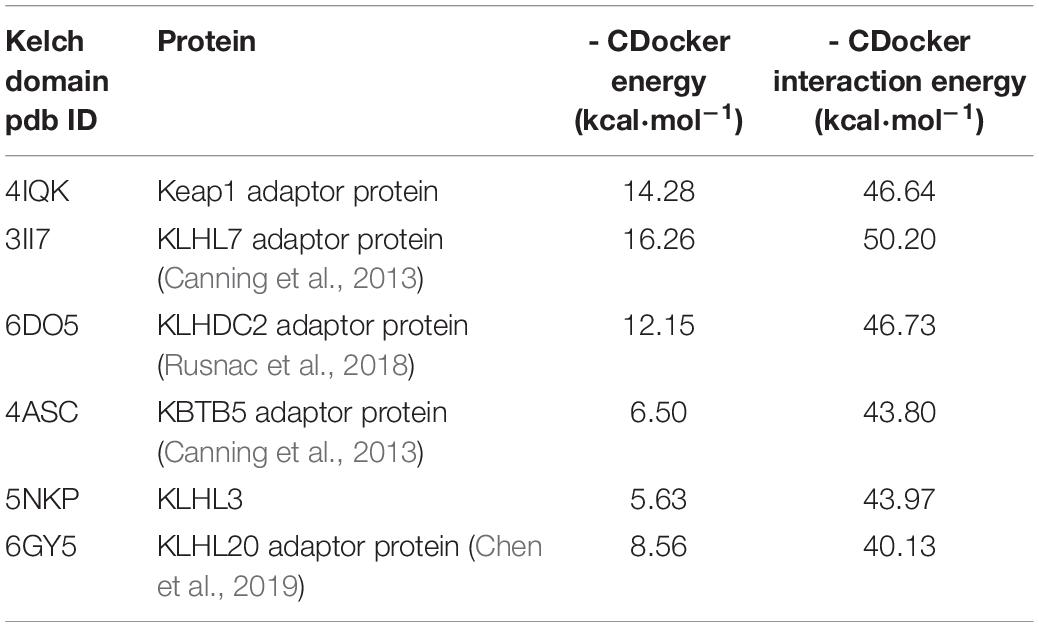
Small molecule displacement activators of Nrf2 are designed to target the Kelch domain in Keap1. However, Keap1 is only one among 42 Kelch-like gene family member (KLHLs) proteins in humans. Only a small portion of these proteins are currently being studied concerning their structure and functions. KLHLs are closely related to the pathogenesis of various human diseases, and specifically cancer. KLHL members associated with inherited forms of the human disease include KLHL3, KLHL7, KLHL9, KLHL12, and GAN (KLHL16), whereas KLHL6, KEAP1 (KLHL19), KLHL20, and ENC1 (KLHL37) are associated with cancer (Dhanoa et al., 2013). The KLHL family is conserved throughout evolution. According to KLHL family members’ phylogenetic analysis, it can be subdivided into three subgroups, with KLHL11 as the oldest member and KLHL9 as the youngest (Dhanoa et al., 2013). Increasing evidence shows that KLHLs exert essential biological functions through the ubiquitination of specific substrates (Shi et al., 2019). BTB-containing proteins are involved in various cellular functions such as control of the cytoskeletal organization, ion channel gating, transcription suppression, and protein targeting for ubiquitination. BTB-Kelch proteins form the largest subfamily of Cullin-RING E3 ligases, yet their substrate complexes are mapped and structurally characterized only for Keap1 and recently for some other adaptor proteins. A potential source of substrate diversity is possible through BTB cross-dimerization between different KLHL proteins, which could theoretically allow for differential substrate-binding depending on the spatial and temporal expression of KLHL proteins. Obviously, without running small molecule displacement activators against all other Kelch proteins structurally similar to the Keap1 Kelch domain, no one can guarantee the displacement activator is specific for Keap1.
We decided to pick available crystal structures of “6-blade propeller” Kelch domains and run docking for a well-known displacement activator N,N′-naphthalene-1,4-diylbis(4-methoxybenzenesulfonamide) or NMBSA, whose structure is shown in Figure 2. We found 15 structures of Kelch domains for nine proteins (KBTB5 – 4ASC.pdb; KLHDC2 – 6DO3, 6DO4, 6DO5; KLHL2 – 2XN4; KLHL3 -4CH9, 5NKP; KLHL7 – 3II7; KLHL12-6TTK, 6V7O; KLHL17 – 6HRL; KLHL20 – 5YQ4, 6GY5; NS1 protein – 5YY8, 6N3H); among those, 8 proteins, except NS1 protein (Zhang K. et al., 2018), were adaptor proteins for Cul3 ubiquitin ligase complexes. We took one structure per protein to perform NMBSA docking. To validate the docking procedure, we first docked NMBSA into the original crystal structure of Keap1 Kelch domain of (4IQK.pdb) to ensure that its docking position coincides with the actual position. Then the docking energies were taken as controls for comparison. This approach demonstrated reliable results when we used it previously to predict benfotiamine binding and its precursors to Keap1 Kelch [see docking protocol details in Tapias et al. (2018)]. As seen from Table 2, five out of the known eight crystal structures of Cul3 ubiquitin ligase adaptor proteins (or more than 60% of the available Kelch structures) showed reasonable docking energies, comparable to that for the original structure. Moreover, NMBSA docking into 3II7.pdb showed better energies than those for NMBSA docking into the original crystal structure 4IQK.pdb. Docking results do not prove that such complexes truly exist. However, their formation is permitted for these five Kelch domains, which belong to five other BTB-Kelch adaptor proteins for Cul3 ubiquitin ligase complexes. It is important to note that peptide sequences bound to various Kelch domains are not identical, and they are specific for a particular adaptor protein. Hence, using peptides with unique recognition sequences for each Kelch protein may be more specific than using small molecule displacement activators unless their specificity for Keap1 is experimentally demonstrated.
Counteracting Non-Specificity of Keap1 Targeting by De-Repression of Bach1
As we can see from the discussion on the molecular mechanisms of Nrf2 ubiquitination inhibition, there are principal concerns for using this approach to treat ongoing neurodegeneration. First, only one ubiquitination pathway is really well characterized at the molecular level – Keap1-mediated degradation of Nrf2 protein. Second, the existing thiol-modifying agents are potent inducers of the Nrf2-mediated transcriptional program and can be effectively used as preventive therapies. However, their use for existing and ongoing neurological conditions is problematic unless these agents are targeting Keap1 specifically, which is a big challenge. The excitement to use small molecule displacement activators to stabilize and activate Nrf2 by targeting the Kelch domain of Keap1 will likely diminish, as it becomes more and more evident that- (a) Keap1 has multiple client proteins, and they all will come to play upon Nrf2 displacement with high-affinity activators of this kind, and (b) there are dozens of Kelch-BTB protein adaptors for multiple ubiquitin ligase complexes having very similar structures of Kelch domains, and (c) working concentrations of displacement activators in the cells are high above their dissociation constant determined in fluorescent polarization Keap1 binding assay.
Acute and chronic oxidative stress generates intracellular ROS and RNS, and they, as we recently learned from Suzuki et al. (2019), attack Keap1 Cys226/Cys613/Cys622/Cys624 residues (Figure 2), resulting in Nrf2 activation. As we know from numerous publications on neurodegenerative diseases, Nrf2 is stabilized under these conditions. However, it fails to bring the antioxidant genetic program to the level needed to fight against the ongoing oxidative stress. The reason for that is continuous activation of the Nrf2-driven transcriptional program that has a feedback regulation – the program triggers the expression of Nrf2 transcriptional repressors. Therefore, the higher level of Nrf2 protein is compensated by the higher level of its repressors. It is suggested that stress-induced increases in Nrf2-dependent genes decline with aging. In an interesting study, Zhou and colleagues measured basal and inducible levels of Nrf2-regulated antioxidant genes in human bronchial epithelial cells from human subjects of young adults (21–29 years) and older (60–69 years) individuals and explored factors affecting their expression. The basal expression of Nrf2-regulated genes was higher in cells from the older individuals compared with cells from the young adult. Upon exposure to an Nrf2 activator, the expression of Nrf2 genes was increased in cells from both the young adults and the older individuals; however, the induction by Nrf2 activator in older adult cells was significantly less than those observed in young adult cells. They found that the basal expression of Bach 1, the heme-responsive transcriptional repressor of Nrf2, was higher in cells from older adults than from younger adults (Zhou et al., 2018). Bach1 is an Nrf2 target gene, and as such, provide feedback mechanism counteracting Nrf2 activation. Hence, under the conditions of chronic oxidative stress, an increase in Nrf2 protein stabilization is accompanied by a simultaneous increase in the expression of Bach1. Bach1 forms heterodimers with the small Maf proteins (MafF, MafG, and MafK) to bind to MARE (Maf recognition element), thus repressing the expression of Nrf2 target genes (Oyake et al., 1996; Igarashi and Sun, 2006). As a heme sensor, Bach1 binds heme through its multiple heme regulatory motifs (Ogawa et al., 2001), thereby losing its activity as a repressor. The DNA binding activity of Bach1 dramatically decreases upon binding to heme and porphyrin-like molecules resulting in Bach1’s cytoplasmic export and its subsequent ubiquitination and degradation by the proteasome resulting in activation of Nrf2 (Figure 3). A potential alternate pathway based on pharmacological de-repression of Bach1, and thus leading to constitutive activation of the Nrf2 pathway, may result in a better therapeutic outcome when compared to Keap1 targeting. Notably, several studies have demonstrated that Bach1 inhibition/deletion is beneficial in a wide range of disorders such as spinal cord injury (Yamada et al., 2008; Kanno et al., 2009), atherosclerosis (Watari et al., 2008), ischemia/reperfusion injury (Yu et al., 2020), Huntington’s disease (Casares et al., 2020), and experimental autoimmune encephalomyelitis (So et al., 2012). The protective role of Bach1 deletion against neuronal degeneration suggests that Bach1 may represent a promising target for drug development by activating the Nrf2 pathway. Future research should focus on additional experimental studies to obtain a global view of Nrf2-mediated gene regulation via Bach1 inhibition and develop selective and safe small molecule Bach1 inhibitors as therapeutic agents. Given that Nrf2 repressor Bach1 is an Nrf2 target gene (Zhang X. et al., 2018), overexpressed in chronic oxidative stress, there is a need for a combinatorial approach involving Bach1 inhibition and Nrf2 activation as a therapeutic strategy for neurodegenerative disorders.
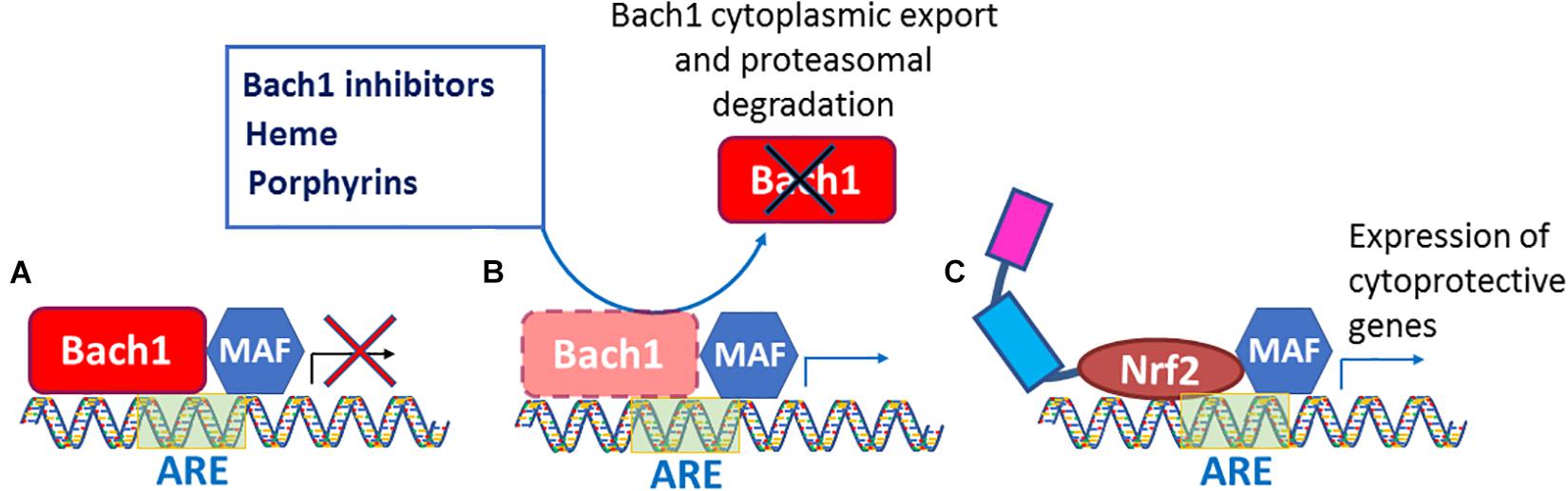
Figure 3. Bach1 inhibition leads to constitutive Nrf2 activation. (A) Bach1 binds to ARE site present on the promoter to repress Nrf2 target genes. (B) Bach1 inhibitors, heme, and porphyrins de-repress Bach1 resulting in its cytoplasmic export and proteasomal degradation. (C) The Bach1 de-repression allows Nrf2 to bind to ARE sites and leads to constitutive activation of Nrf2 pathway to overexpress cytoprotective genes.
Author Contributions
BT and IG conceived and wrote the manuscript. DH performed in silico modeling. DH, NA, MA, AP, and SS provided intellectual contribution and assisted in writing the manuscript. All authors approved the submitted version of the manuscript.
Funding
Financial support was provided by grants from National Institutes of Health (NS101967), Michael J. Fox Foundation for Parkinson’s Disease Research, Par for Parkinson, and Parkinson’s Foundation to BT. Modeling studies were supported by Russian Scientific Foundation grant 20-15-00207.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Abramic, M., and Vitale, L. (1992). Basic amino acids preferring broad specificity aminopeptidase from human erythrocytes. Biol. Chem. Hoppe Seyler. 373, 375–380. doi: 10.1515/bchm3.1992.373.2.375
Ammal Kaidery, N., Ahuja, M., and Thomas, B. (2019). Crosstalk between Nrf2 signaling and mitochondrial function in Parkinson’s disease. Mol. Cell Neurosci. 101:103413. doi: 10.1016/j.mcn.2019.103413
Baird, L., and Yamamoto, M. (2020). The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 40:e00099-20. doi: 10.1128/MCB.00099-20
Brandes, M. S., and Gray, N. E. (2020). NRF2 as a therapeutic target in neurodegenerative diseases. ASN Neuro. 12:1759091419899782. doi: 10.1177/1759091419899782
Canning, P., Cooper, C. D., Krojer, T., Murray, J. W., Pike, A. C., Chaikuad, A., et al. (2013). Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 288, 7803–7814. doi: 10.1074/jbc.M112.437996
Casares, L., Unciti-Broceta, J. D., Prados, M. E., Caprioglio, D., Mattoteia, D., Higgins, M., et al. (2020). Isomeric O-methyl cannabidiolquinones with dual BACH1/NRF2 activity. Redox Biol. 37:101689. doi: 10.1016/j.redox.2020.101689
Chen, Z., Picaud, S., Filippakopoulos, P., D’Angiolella, V., and Bullock, A. N. (2019). Structural Basis for Recruitment of DAPK1 to the KLHL20 E3 Ligase. Structure 27, 1395–1404.e4. doi: 10.1016/j.str.2019.06.005
Cheng, M., Lin, N., Dong, D., Ma, J., Su, J., and Sun, L. (2021). PGAM5: a crucial role in mitochondrial dynamics and programmed cell death. Eur. J. Cell Biol. 100:151144. doi: 10.1016/j.ejcb.2020.151144
Chowdhry, S., Zhang, Y., McMahon, M., Sutherland, C., Cuadrado, A., and Hayes, J. D. (2013). Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32, 3765–3781. doi: 10.1038/onc.2012.388
Conrad, W., Major, M. B., Cleary, M. A., Ferrer, M., Roberts, B., Marine, S., et al. (2013). FAM129B is a novel regulator of Wnt/beta-catenin signal transduction in melanoma cells. F1000Research 2:134. doi: 10.12688/f1000research.2-134.v2
Cuadrado, A., Rojo, A. I., Wells, G., Hayes, J. D., Cousin, S. P., Rumsey, W. L., et al. (2019). Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 18, 295–317. doi: 10.1038/s41573-018-0008-x
Deng, J., Wang, X., Zhou, Q., Xia, Y., Xiong, C., Shao, X., et al. (2020). Inhibition of glycogen synthase kinase 3beta alleviates chronic renal allograft dysfunction in rats. Transplantation doi: 10.1097/TP.0000000000003446
Dhanoa, B. S., Cogliati, T., Satish, A. G., Bruford, E. A., and Friedman, J. S. (2013). Update on the Kelch-like (KLHL) gene family. Hum. Genom. 7:13. doi: 10.1186/1479-7364-7-13
Ekim Ustunel, B., Friedrich, K., Maida, A., Wang, X., Krones-Herzig, A., Seibert, O., et al. (2016). Control of diabetic hyperglycaemia and insulin resistance through TSC22D4. Nat. Commun. 7:13267. doi: 10.1038/ncomms13267
Fourquet, S., Guerois, R., Biard, D., and Toledano, M. B. (2010). Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation. J. Biol. Chem. 285, 8463–8471. doi: 10.1074/jbc.M109.051714
Gazaryan, I. G., and Thomas, B. (2016). The status of Nrf2-based therapeutics: current perspectives and future prospects. Neural Regen. Res. 11, 1708–1711. doi: 10.4103/1673-5374.194706
Harder, B., Jiang, T., Wu, T., Tao, S., Rojo de la Vega, M., Tian, W., et al. (2015). Molecular mechanisms of Nrf2 regulation and how these influence chemical modulation for disease intervention. Biochem. Soc. Trans. 43, 680–686. doi: 10.1042/BST20150020
Hast, B. E., Goldfarb, D., Mulvaney, K. M., Hast, M. A., Siesser, P. F., Yan, F., et al. (2013). Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res. 73, 2199–2210. doi: 10.1158/0008-5472.CAN-12-4400
Hu, B., Wu, Y., Liu, J., Shen, X., Tong, F., Xu, G., et al. (2016). GSK-3beta inhibitor induces expression of Nrf2/TrxR2 signaling pathway to protect against renal ischemia/reperfusion injury in diabetic rats. Kidney Blood Press Res. 41, 937–946. doi: 10.1159/000452598
Igarashi, K., and Sun, J. (2006). The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid. Redox Signal. 8, 107–118. doi: 10.1089/ars.2006.8.107
Johnson, D. A., and Johnson, J. A. (2015). Nrf2–a therapeutic target for the treatment of neurodegenerative diseases. Free Radic. Biol. Med. 88(Pt B), 253–267. doi: 10.1016/j.freeradbiomed.2015.07.147
Kanno, H., Ozawa, H., Dohi, Y., Sekiguchi, A., Igarashi, K., and Itoi, E. (2009). Genetic ablation of transcription repressor Bach1 reduces neural tissue damage and improves locomotor function after spinal cord injury in mice. J. Neurotrauma 26, 31–39. doi: 10.1089/neu.2008.0667
Katsuragi, Y., Ichimura, Y., and Komatsu, M. (2015). p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 282, 4672–4678. doi: 10.1111/febs.13540
Khan, H., Cino, E. A., Brickenden, A., Fan, J., Yang, D., and Choy, W. Y. (2013). Fuzzy complex formation between the intrinsically disordered prothymosin alpha and the Kelch domain of Keap1 involved in the oxidative stress response. J. Mol. Biol. 425, 1011–1027. doi: 10.1016/j.jmb.2013.01.005
Kim, H. M., Han, J. W., and Chan, J. Y. (2016). Nuclear factor erythroid-2 Like 1 (NFE2L1): structure, function and regulation. Gene 584, 17–25. doi: 10.1016/j.gene.2016.03.002
Kopacz, A., Kloska, D., Forman, H. J., Jozkowicz, A., and Grochot-Przeczek, A. (2020). Beyond repression of Nrf2: an update on Keap1. Free Radic. Biol. Med. 157, 63–74. doi: 10.1016/j.freeradbiomed.2020.03.023
Liu, H., Feng, Y., Xu, M., Yang, J., Wang, Z., and Di, G. (2018). Four-octyl itaconate activates Keap1-Nrf2 signaling to protect neuronal cells from hydrogen peroxide. Cell Commun. Signal. 16:81. doi: 10.1186/s12964-018-0294-2
Lu, K., Alcivar, A. L., Ma, J., Foo, T. K., Zywea, S., Mahdi, A., et al. (2017). NRF2 induction supporting breast cancer cell survival is enabled by oxidative stress-induced DPP3-KEAP1 Interaction. Cancer Res. 77, 2881–2892. doi: 10.1158/0008-5472.CAN-16-2204
McMahon, M., Thomas, N., Itoh, K., Yamamoto, M., and Hayes, J. D. (2004). Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 279, 31556–31567. doi: 10.1074/jbc.M403061200
Ogawa, K., Sun, J., Taketani, S., Nakajima, O., Nishitani, C., Sassa, S., et al. (2001). Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20, 2835–2843. doi: 10.1093/emboj/20.11.2835
Ogura, T., Tong, K. I., Mio, K., Maruyama, Y., Kurokawa, H., Sato, C., et al. (2010). Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc. Natl. Acad. Sci. U.S.A. 107, 2842–2847. doi: 10.1073/pnas.0914036107
Oyake, T., Itoh, K., Motohashi, H., Hayashi, N., Hoshino, H., Nishizawa, M., et al. (1996). Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell Biol. 16, 6083–6095. doi: 10.1128/mcb.16.11.6083
Rada, P., Rojo, A. I., Chowdhry, S., McMahon, M., Hayes, J. D., and Cuadrado, A. S. C. F. (2011). /{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell Biol. 31, 1121–1133. doi: 10.1128/MCB.01204-10
Ruiz, K., Thaker, T. M., Agnew, C., Miller-Vedam, L., Trenker, R., Herrera, C., et al. (2019). Functional role of PGAM5 multimeric assemblies and their polymerization into filaments. Nat. Commun. 10:531. doi: 10.1038/s41467-019-08393-w
Rusnac, D. V., Lin, H. C., Canzani, D., Tien, K. X., Hinds, T. R., Tsue, A. F., et al. (2018). Recognition of the Diglycine C-End Degron by CRL2(KLHDC2) Ubiquitin Ligase. Mol. Cell 72, 813–822.e4. doi: 10.1016/j.molcel.2018.10.021
Salmeron, A., Janzen, J., Soneji, Y., Bump, N., Kamens, J., Allen, H., et al. (2001). Direct phosphorylation of NF-kappaB1 p105 by the IkappaB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 276, 22215–22222. doi: 10.1074/jbc.M101754200
Shen, X., Hu, B., Xu, G., Chen, F., Ma, R., Zhang, N., et al. (2017). Activation of Nrf2/HO-1 pathway by glycogen Synthase kinase-3beta inhibition attenuates renal ischemia/reperfusion injury in diabetic rats. Kidney Blood Press Res. 42, 369–378. doi: 10.1159/000477947
Shi, X., Xiang, S., Cao, J., Zhu, H., Yang, B., He, Q., et al. (2019). Kelch-like proteins: physiological functions and relationships with diseases. Pharmacol. Res. 148:104404. doi: 10.1016/j.phrs.2019.104404
Shimamori, Y., Watanabe, Y., and Fujimoto, Y. (1988). Human placental dipeptidyl aminopeptidase III: hydrolysis of enkephalins and its stimulation by cobaltous ion. Biochem. Med. Metab. Biol. 40, 305–310. doi: 10.1016/0885-4505(88)90133-8
Smirnova, N. A., Haskew-Layton, R. E., Basso, M., Hushpulian, D. M., Payappilly, J. B., Speer, R. E., et al. (2011). Development of Neh2-luciferase reporter and its application for high throughput screening and real-time monitoring of Nrf2 activators. Chem. Biol. 18, 752–765. doi: 10.1016/j.chembiol.2011.03.013
So, A. Y., Garcia-Flores, Y., Minisandram, A., Martin, A., Taganov, K., Boldin, M., et al. (2012). Regulation of APC development, immune response, and autoimmunity by Bach1/HO-1 pathway in mice. Blood 120, 2428–2437. doi: 10.1182/blood-2012-04-426247
Sun, X., Zhang, B., Pan, X., Huang, H., Xie, Z., Ma, Y., et al. (2019). Octyl itaconate inhibits osteoclastogenesis by suppressing Hrd1 and activating Nrf2 signaling. FASEB J. 33, 12929–12940. doi: 10.1096/fj.201900887RR
Suzuki, T., Muramatsu, A., Saito, R., Iso, T., Shibata, T., Kuwata, K., et al. (2019). Molecular mechanism of cellular oxidative stress sensing by Keap1. Cell Rep. 28, 746–758.e4. doi: 10.1016/j.celrep.2019.06.047
Tamberg, N., Tahk, S., Koit, S., Kristjuhan, K., Kasvandik, S., Kristjuhan, A., et al. (2018). Keap1-MCM3 interaction is a potential coordinator of molecular machineries of antioxidant response and genomic DNA replication in metazoa. Sci. Rep. 8:12136. doi: 10.1038/s41598-018-30562-y
Tapias, V., Jainuddin, S., Ahuja, M., Stack, C., Elipenahli, C., Vignisse, J., et al. (2018). Benfotiamine treatment activates the Nrf2/ARE pathway and is neuroprotective in a transgenic mouse model of tauopathy. Hum. Mol. Genet. 27, 2874–2892. doi: 10.1093/hmg/ddy201
Tian, W., Rojo de la Vega, M., Schmidlin, C. J., Ooi, A., and Zhang, D. D. (2018). Kelch-like ECH-associated protein 1 (KEAP1) differentially regulates nuclear factor erythroid-2-related factors 1 and 2 (NRF1 and NRF2). J. Biol. Chem. 293, 2029–2040. doi: 10.1074/jbc.RA117.000428
Watari, Y., Yamamoto, Y., Brydun, A., Ishida, T., Mito, S., Yoshizumi, M., et al. (2008). Ablation of the bach1 gene leads to the suppression of atherosclerosis in bach1 and apolipoprotein E double knockout mice. Hypertens. Res. 31, 783–792. doi: 10.1291/hypres.31.783
Wu, T., Zhao, F., Gao, B., Tan, C., Yagishita, N., Nakajima, T., et al. (2014). Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 28, 708–722. doi: 10.1101/gad.238246.114
Wu, X., Siggel, M., Ovchinnikov, S., Mi, W., Svetlov, V., Nudler, E., et al. (2020). Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 368:aaz2449. doi: 10.1126/science.aaz2449
Wu, S., Zhou, J., Zhang, K., Chen, H., Luo, M., Lu, Y., et al. (2020). Molecular mechanisms of PALB2 function and its role in breast cancer management. Front. Oncol. 10:301. doi: 10.3389/fonc.2020.00301
Yamada, K., Tanaka, N., Nakanishi, K., Kamei, N., Ishikawa, M., Mizuno, T., et al. (2008). Modulation of the secondary injury process after spinal cord injury in Bach1-deficient mice by heme oxygenase-1. J. Neurosurg. Spine 9, 611–620. doi: 10.3171/SPI.2008.10.08488
Yoboua, F., Martel, A., Duval, A., Mukawera, E., and Grandvaux, N. (2010). Respiratory syncytial virus-mediated NF-kappa B p65 phosphorylation at serine 536 is dependent on RIG-I, TRAF6, and IKK beta. J. Virol. 84, 7267–7277. doi: 10.1128/JVI.00142-10
Yu, S., Zhai, J., Yu, J., Yang, Q., and Yang, J. (2020). Downregulation of BACH1 protects AGAINST cerebral ischemia/reperfusion injury through the functions of HO-1 and NQO1. Neuroscience 436, 154–166. doi: 10.1016/j.neuroscience.2020.04.014
Zhang, K., Shang, G., Padavannil, A., Wang, J., Sakthivel, R., Chen, X., et al. (2018). Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression. Proc. Natl. Acad. Sci. U.S.A. 115, E12218–E12227. doi: 10.1073/pnas.1818012115
Zhang, X., Guo, J., Wei, X., Niu, C., Jia, M., Li, Q., et al. (2018). Bach1: function, regulation, and involvement in disease. Oxid. Med. Cell Longev. 2018:1347969. doi: 10.1155/2018/1347969
Zhang, Y., Shi, Z., Zhou, Y., Xiao, Q., Wang, H., and Peng, Y. (2020). Emerging Substrate Proteins of Kelch-like ECH associated protein 1 (Keap1) and potential challenges for the development of small-molecule inhibitors of the keap1-nuclear factor Erythroid 2-related factor 2 (Nrf2) protein-protein interaction. J. Med. Chem. 63, 7986–8002. doi: 10.1021/acs.jmedchem.9b01865
Keywords: Nrf2, Kelch domain, displacement activator, ubiquitylation pathways, BACH1
Citation: Hushpulian DM, Ammal Kaidery N, Ahuja M, Poloznikov AA, Sharma SM, Gazaryan IG and Thomas B (2021) Challenges and Limitations of Targeting the Keap1-Nrf2 Pathway for Neurotherapeutics: Bach1 De-Repression to the Rescue. Front. Aging Neurosci. 13:673205. doi: 10.3389/fnagi.2021.673205
Received: 27 February 2021; Accepted: 15 March 2021;
Published: 08 April 2021.
Edited by:
Bindu Diana Paul, Johns Hopkins University, United StatesReviewed by:
Zahoor A. Shah, University of Toledo, United StatesMin-Kyoo Shin, University Hospitals Cleveland Medical Center, United States
Copyright © 2021 Hushpulian, Ammal Kaidery, Ahuja, Poloznikov, Sharma, Gazaryan and Thomas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bobby Thomas, dGhvbWFzYm9AbXVzYy5lZHU=
 Dmitry M. Hushpulian1,2
Dmitry M. Hushpulian1,2 Sudarshana M. Sharma
Sudarshana M. Sharma Irina G. Gazaryan
Irina G. Gazaryan Bobby Thomas
Bobby Thomas