- 1 Department of Evolutionary Neuroethology, Max Planck Institute for Chemical Ecology, Jena, Germany
- 2 Institute of Physiology, University of Hohenheim, Stuttgart, Germany
As odor information plays a vital role in the life of moths, their olfactory sense has evolved into a highly specific and sensitive apparatus relevant to reproduction and survival. The key players in the detection of odorants are olfactory receptor (OR) proteins. Here we identify four OR-encoding genes differentially expressed in the antennae of males and females of the sphingid moth Manduca sexta. Two male-specific receptors (the previously reported MsexOR-1 and the newly identified MsexOR-4) show great resemblance to other male moth pheromone ORs. The putative pheromone receptors are co-expressed with the co-receptor involved in general odorant signal transduction, the DmelOr83b homolog MsexOR-2. One female-specific receptor (MsexOR-5) displays similarities to BmorOR-19, a receptor in Bombyx mori tuned to the detection of the plant odor linalool.
Introduction
Volatile cues elicit complex behaviors crucial to fitness in many insect species. Among these cues sex pheromones have attracted considerable interest, both due to their involvement in eliciting a clear and quantifiable behavior, and their potential application in pest control (reviewed in Witzgall et al., 2010). The identification of a number of pheromones has led to extensive efforts in analyzing the olfactory sense in a variety of moth species (Vogt and Blomquist, 2003). Besides pheromones, a multitude of other behaviorally active compounds have been identified, including both green leaf volatiles and flower odors (Renwick and Chew, 1994; Daly et al., 2001). The different types of volatile molecules are detected by olfactory receptor neurons (ORN) in the moth antenna. The odor specificity of the neuron is determined by the expression of a specific olfactory receptor (OR) protein (Hallem et al., 2004; Sakurai et al., 2004; Nakagawa et al., 2005; Grosse-Wilde et al., 2007; Anderson et al., 2009). In all higher insects investigated so far, the odor-specific OR is co-expressed with a ubiquitous OR-like protein first identified in Drosophila melanogaster, and there named DmelOR83b (Vosshall et al., 1999, 2000). Binding of a ligand by the odor-specific OR leads to activation of OR83b via both an ionotropic and a metabotropic pathway (Sato et al., 2008; Wicher et al., 2008), with OR83b acting as an ion channel (Wicher et al., 2008). In Lepidoptera the detection of pheromones is mediated by highly specific odorant receptors tuned to the respective pheromone components (Sakurai et al., 2004; Nakagawa et al., 2005; Grosse-Wilde et al., 2006). The genes encoding these receptors form a comparatively conserved subgroup in the family of OR-encoding genes (Krieger et al., 2004, 2005). The Manduca sexta pheromone is composed of two major constituents: E10, Z12 hexadecadienal (bombykal) and E10, E12, Z14 hexadecatrienal (EEZ). In addition a number of other minor components have been shown to have behavioral effects (Tumlinson et al., 1989, 1994). On the male antenna 70% of the sensilla contain a pair of ORNs, where one is tuned to bombykal and the other to EEZ (Kaissling et al., 1989; Kalinova et al., 2001).
The life history of the moth sexes has provided differentiated selection pressures on the olfactory sense. The dominance of sex pheromone-specific ORNs on the male antenna (Schneider, 1969; Lee and Strausfeld, 1990), and the corresponding volume increase of the glomeruli receiving information from these neurons is well documented (Boeckh and Boeckh, 1979; Matsumoto and Hildebrand, 1981; Christensen and Hildebrand, 1987; Hansson et al., 1991). The female generally lacks ORNs tuned to her own pheromone, but exceptions are known (Anton and Hansson, 1994; Kalinova et al., 2001). The female antenna might instead possess ORNs tuned to odors specifically involved in detection of suitable oviposition sites and of male aphrodisiac pheromones, neurons that may not be present in the male antenna (Hansson et al., 1989; Shields and Hildebrand, 2000). Therefore it is reasonable to expect some degree of sexual dimorphism in the presence of different physiological types of ORNs in the moth antenna.
The identification of the first sex pheromone bombykol by Butenandt et al. (1959) was one reason for the silkworm Bombyx mori to become a model species. Bombyx mori is today the only moth species for which nearly complete genomic data is publicly available (The International Silkworm Genome Consortium, 2008). Another model species of importance is the tobacco hornworm M. sexta, but the genome of this species is still not characterized. In contrast to the domesticated B. mori, data on the olfactory sense of M. sexta is valuable in an ecological context; in fact, its interaction with its food source and hosts is a topic of extensive research. Besides the behavioral and ecological data available, both the morphology and the physiology of the olfactory system have been scrutinized in great detail. At the molecular level, many genes and proteins involved in olfactory signal detection have been identified and characterized (Gyorgyi et al., 1988; Rybczynski et al., 1989; Vogt et al., 1991; Feng and Prestwich, 1997; Robertson et al., 1999; Rogers et al., 2001; Nardi et al., 2003). However, with the OR proteins the key players determining the specificity of ORNs have mostly eluded identification. So far, only three ORs have been identified in M. sexta: one putative male-specific sex pheromone receptor (MsexOR-1), the DmelOR83b homolog MsexOR-2, and a putative general odorant receptor (MsexOR-3) (Patch et al., 2009). Based on the number of ORs known in B. mori (Tanaka et al., 2009) and the finding that the number of OR genes roughly correlates with the number of glomeruli in the antennal lobe (de Bruyne and Baker, 2008), which is 63(±1) in M. sexta (Rospars and Hildebrand, 1992, 2000; Huetteroth and Schachtner, 2005), the majority of OR-coding genes are thus still unknown.
We decided to use the possible differences found between the sexes to identify putative male- and female-specific OR-coding genes in M. sexta. In the present study, we report the use of subtractive antennal cDNA libraries between male and female M. sexta. We identified four putative ORs; two male-specific members of the pheromone OR subclade, including the previously reported MsexOR-1, and two female-specific OR subtypes.
Materials and Methods
Animal Rearing
Animals were taken from a culture maintained at the Max Planck Institute for Chemical Ecology, Jena, Germany. In short, adults were allowed to mate inside a flight cage at room temperature, providing a Nicotiana attenuata plant for oviposition and sugar solution inside artificial flowers for nutrition. Eggs were collected from the plants three times a week and transferred to small boxes with artificial diet (46 g agar, 144 g wheat germ, 140 g corn meal, 76 g soz flour, 75 g casein, 24 g salt, 36 g sugar, 5 g cholesterol, 12 g ascorbic acid, 6 g sorbic acid, 3 g methyl paraben, 9 ml linseed oil, 60 ml 37% formalin, 30 mg nicotinic acid, 15 mg riboflavin, 7 mg thiamine, 7 mg pyridoxine, 7 mg folic acid and 0.6 mg biotin per 1.8 ml water). Eggs and emerging larvae were kept inside a climate controlled chamber at 27°C and 70% humidity. Hatched larvae were kept inside boxes on metal lattices covered with artificial diet. Wandering fifth instar larvae were transferred to small plastic boxes filled with paper to allow pupation. Pupae were kept inside climate chambers until 1 week before likely emergence and then either transferred to paper bags inside a climate controlled chamber (to keep experimental animals separate from each other but still provide enough room for emergence) or an open box inside the flight cage (for animals used in breeding).
Total RNA Extraction
Adult M. sexta were sorted by sex; the antennae of two animals per sex were cut off close to the scapulum and immediately immersed in 2 ml of Trizol reagent (Sigma-Aldrich, St. Louis, MO, USA). The antennae were then homogenized in the Trizol using a mortar and pestle. The homogenate was transferred into Eppendorf cups, with 1 ml per cup, and incubated for 10 min at room temperature. Afterwards, 120 μl of 1-bromo-3-chloro-propane was added to each tube. After vortexing and 10 min of incubation, the preparations were centrifuged. The watery phase was added to 0.7 volumes of isopropanol and kept at −86°C overnight. The next day, preparations were centrifuged, the supernatant removed and the pellet washed once with 80% ethanol. After the next centrifugation, the ethanol was removed and the pellets left to dry. Finally, pellets were diluted with 20 μl of RNA Storage Solution (Ambion, Austin, TX, USA) and RNA content and quality measured photometrically. We prepared three samples per sex.
Construction and Sequencing of Subtractive cDNA Libraries
Total RNA (10 μg) from the antennae of each sex were sent to Evrogen (Moscow, Russia) for construction of subtractive cDNA libraries using suppression subtractive hybridization with mirror orientation selection (MOS). Picking and sequencing of single clones was performed by Eurofins MWG Biotech (Ebersberg, Germany).
Sequence Analysis
Sequences were stripped of vector sequences using Seqclean and subsequently assembled using the Seqman assembler (DNAStar, Madison, WI, USA) with standard settings. Contigs were identified using the tblastx algorithm with the NCBI nr database and a database containing published insect odorant receptor genes. For sequences without clear results in tblastx, the largest identified ORF was further analyzed using InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/).
Cloning of Full Length or Genes
We used a normalized cDNA of M. sexta antennae provided by Heiko Vogel (Department of Entomology, MPI for Chemical Ecology, Jena, Germany) to extend identified cDNA fragments coding for putative ORs to full length. The cDNA was normalized using the SMART directional library kit (Clontech/Takara, Mountain View, CA, USA) together with the TRIMMER direct normalization kit (Evrogen). Using sequence-specific primers together with primers directed against the TRIMMER adapters and the normalized cDNA as template, we performed touchdown PCR to extend identified fragments in 3′- as well as 5′-direction. All PCRs were performed using Advantage Taq 2 (Clontech). Gel purified PCR products were sequenced. The sequences were assembled using Seqman and contigs analyzed by hand. The identified full-length cDNAs were then amplified in new PCR reactions using the same conditions but primers directed against 5′- and 3′-ends of the largest predicted open reading frames. Products were gel purified and subcloned using the pGEM-T easy kit (Promega, WI, USA). Inserts of the resultant plasmids were sequenced in full to verify their identity.
MsexOR-2 was PCR-amplified from antennal cDNA using gene-specific primers and subcloned into the pCRII vector (dual promoter TA cloning kit, Invitrogen, Carlsbad, CA, USA).
RT-PCR
New total RNA was extracted as described above and reverse transcribed into cDNA using the Superscript III-kit (Invitrogen) as per the manufacturers instructions. Touchdown PCR was performed using the gene-specific primers used for fragment extension with the new cDNA as template. PCR products were used in agarose gel electrophoresis to analyze for successful amplification. The fragment amplified by primers directed against MsexOR-5 was 354 bases in size. The same reaction with genomic DNA led to the amplification of a ca. 250 bases larger fragment, indicating the presence of an intron. This allowed us to test for genomic contamination of our cDNA templates.
Synthesis of Probes for in situ Hybridization
Biotin- or DIG-labeled antisense riboprobes were generated using the SP6/T7 RNA transcription system (Roche Diagnostics, Risch, Switzerland) and linearized recombinant plasmids following recommended protocols. Probes were subsequently shortened to ca. 600 bp length using a carbonate buffer (80 mM NaHCO3, 120 mM Na2CO3, pH 10.2) following the protocol of Angerer and Angerer (1992).
Cryosections
Antennae of adult moths were embedded in Tissue-Tek O.C.T. compound (Sakura Finetek Europe, Zoeterwoude, The Netherlands) and frozen at −20°C. The embedded specimens were mounted on an object holder and sectioned using a HM560 cryostat (Thermo Scientific, Waltham, MA, USA). 12 μm thick cryosections of antennae were thaw mounted on slides coated with Vectabond (Vector Laboratories, Burlingame, CA, USA) and air dried at room temperature for at least 30 min. Subsequently, slides were frozen at −86°C.
In situ Hybridization
Slides were treated with 4% paraformaldehyde in 0.1 M NaCO3, pH 9.5 for 30 min at 4°C. Subsequently, slides were incubated in PBS (phosphate-buffered saline = 0.85% NaCl, 1.4 mM KH2PO4, 8 mM Na2HPO4, pH 7.1) for 1 min, followed by 0.2 M HCl for 10 min and PBS with 1% Triton X-100 for 2 min, then two 30-s washes in PBS. Finally, slides were rinsed for 10 min in 50% formamide, 5× SSC (1× SSC = 0.15 M NaCl, 0.015 M Na-citrate, pH 7.0) and drained. For hybridization tissue sections were covered with 100 μl hybridization solution (50% formamide, 2× SSC, 10% dextran sulphate, 20 mg/ml yeast t-RNA, 0.2 mg/ml herring sperm DNA) containing riboprobes. A coverslip was placed onto the specimens and slides were incubated in a humid box (50% formamide) at 55°C overnight.
Post hybridization, slides were washed twice in 0.1× SSC at 60°C for 30 min, rinsed with TBS (100 mM Tris, pH 7.5, 150 mM NaCl) and incubated in TBS with 0.03% Triton X-100 and 1% blocking reagent (Roche Diagnostics) for 30 min at room temperature. Afterwards, slides were incubated with anti-DIG antibodies (linked to alkaline phosphatase) and anti-biotin-SA (linked to horseradish peroxidase) in the same buffer for 60 min at 37°C in a humid box. After washing three times for 5 min with TBST (TBS with 0.05% Tween 20), slides were incubated with HNPP/FastRedTR (Roche Diagnostics, for the detection of anti-DIG-AP-conjugates) for 30 min at 37°C, washed again three times for 5 min in TBST-buffer and then treated for 10 min with Fluorophore Tyramide Working-solution (part of TSA kit, Perkin Elmer, Waltham, MA, USA). After three additional TBST washes (5 min each), slides were rinsed with dH2O and sealed with coverslips in 10% glycerol. Signals were visualized using a LSM 510 Meta confocal microscope (Zeiss, Jena, Germany).
Results
Antennal cDNA libraries of M. sexta enriched in sex-specific transcripts were created as described. For analysis, 192 (male library) and 480 (female library) clones were picked and sequenced. Resulting sequences were assembled and annotated (data not shown). The majority of reads in both libraries corresponded to pheromone binding protein (PBP, male) and general odorant-binding protein (GOBP, female) coding genes. Besides binding proteins, a small number of genes encoding enzymes, including members of the cytochrome oxidase family, were found. Additionally, in each library five clones were identified that assembled into two putative OR-coding gene fragments per sex. One of the gene fragments present in the male library encoded the previously reported putative pheromone receptor MsexOR-1 (Patch et al., 2009), with the encoded protein being 100% identical. The other three identified genes encoded novel OR subtypes.
Since the subtraction procedure is an enrichment of tissue-specific transcripts (Diatchenko et al., 1996), non-specific transcripts will still be present in low amounts in each library. Therefore, the sex-specific expression of the identified gene fragments had to be verified. We performed RT-PCR with primers specific for the putative OR-encoding genes, using cDNA from either male of female antennae as template (Figure 1). Both MsexOR-1 and MsexOR-4 could only be amplified from male, but not from female antennal cDNA. In contrast, MsexOR-5 and MsexOR-6 could only be detected in female antennal cDNA. This verified that the identified putative ORs were indeed differentially expressed in the antennae of either male (MsexOR-1 and MsexOR-4) or female (MsexOR-5 and MsexOR-6) individuals.
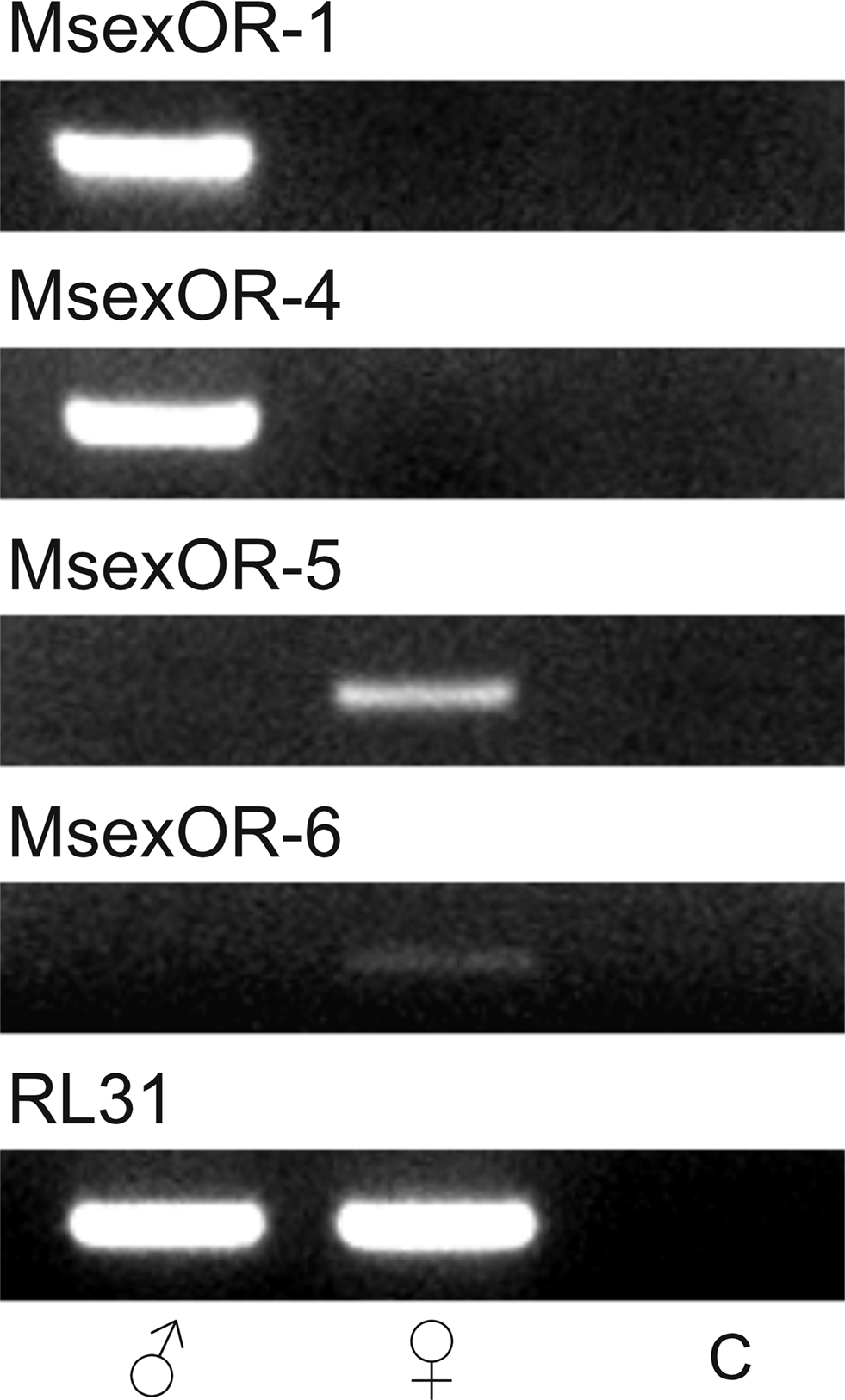
Figure 1. Tissue-specific expression of identified OR-coding genes. Reverse transcriptase (RT)-PCR was performed using gene-specific primers for each receptor type and cDNAs of male (left) and female (middle) M. sexta antennae (control using water instead of cDNA, right). PCR products were visualized after gel electrophoresis using ethidium bromide staining. All bands were of the expected size, based on primer design. MsexOR-4 and MsexOR-1 showed male-specific expression, while MsexOR-5 and MsexOR-6 could only be detected in cDNA of female antennae.
The gene fragments were extended in both 5′- and 3′-prime direction by RACE-PCR using gene-specific primers in conjunction with a SMART-amplified antennal cDNA and SMART-adapter-specific primers. The gene-specific primers were selected to yield amplified sequences with at least 150 bases overlap to the already identified fragments to facilitate assembly of all fragments. The assembled sequences were used to design gene-specific primers to allow extension to full length using RACE-PCR. The PCR products were TA-cloned and sequenced in total to verify the initial sequence. While this procedure led to the identification of putative full-length transcripts of MsexOR-1, MsexOR-4 and MsexOR-5, similar identification of a transcript including a full-length ORF did not succeed for MsexOR-6.
Multiple sequence alignment of the predicted protein sequences encoded by the identified genes with known Lepidopteran OR proteins was performed using the MUSCLE program (Figure 2, neighbor-joining dendrogram) (Edgar, 2004). The predicted proteins showed clear similarity to specific ORs of other species. The male-specific MsexOR-1 and MsexOR-4 belong to the subfamily of previously identified male-specific pheromone ORs. As previously reported (Patch et al., 2009) the closest relative of MsexOR-1 is the B. mori receptor BmOR-3, which mediates the male response to bombykal (Nakagawa et al., 2005; Grosse-Wilde et al., 2006), the minor pheromone component of B. mori and the main pheromone component of M. sexta. The closest relative of MsexOR-4 is HR13, the Heliothis virescens pheromone receptor detecting the main pheromone component Z11-hexadecenal (Grosse-Wilde et al., 2007). The closest relative of the female-specific putative receptor MsexOR-5 is BmorOR-19, a linalool-detecting, likewise female-specific receptor of B. mori (Anderson et al., 2009).
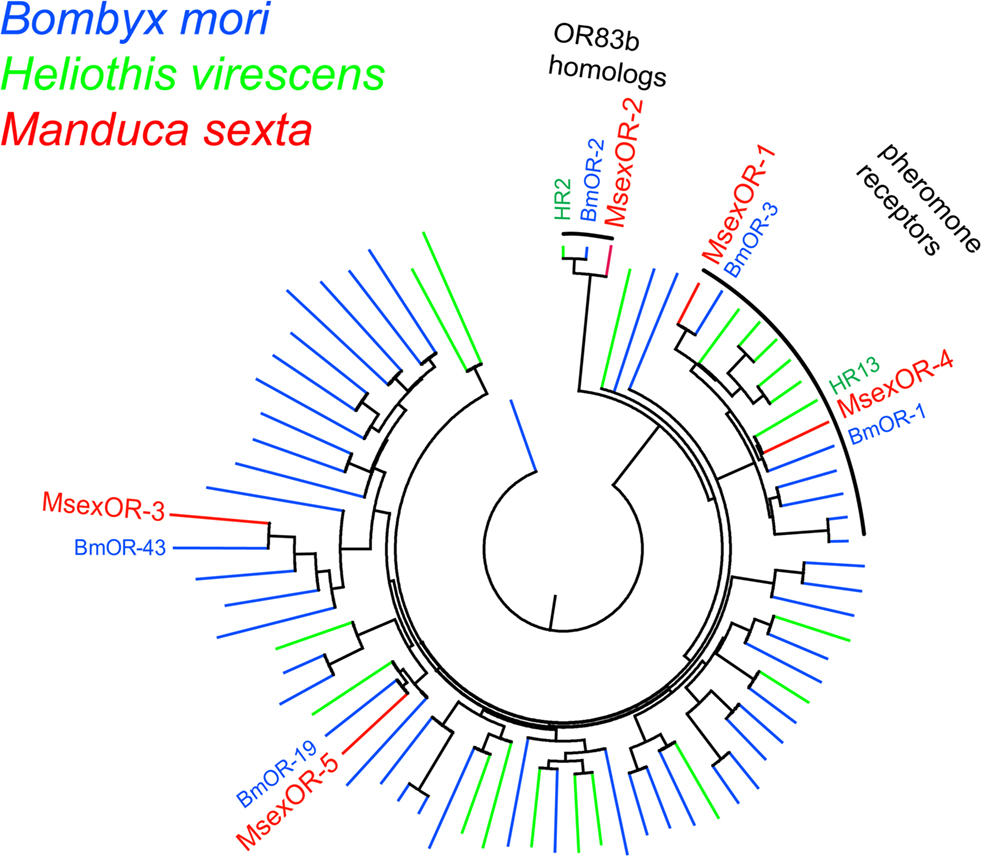
Figure 2. Sequence relatedness of M. sexta ORs to other Lepidopteran ORs. Neighbor-joining tree based on a MUSCLE multiple sequence alignment of MsexOR-1 to MsexOR-5 (red) predicted amino acid sequences with protein sequences of B. mori (blue) and H. virescens (green) ORs. MsexOR-1 and MsexOR-4 belong to the subgroup of male-specific pheromone receptors. In addition, the group of highly conserved R2 (OR83b) receptor types (Krieger et al., 2003) is indicated. The closest homolog of MsexOR-5 is the female-specific receptor BmorOR-19; MsexOR-3 (Patch et al., 2009) is most related to BmorOR-43.
Using the full-length constructs as basis, DIG- or biotin-labeled antisense RNA-probes directed against the respective mRNAs were generated. To visualize cells expressing the putative OR-encoding genes, we performed fluorescence in situ hybridization (ISH) using these probes on longitudinal and cross sections of M. sexta antennae.
Probes directed against MsexOR-1 or MsexOR-4 led to labeling of ORN cell bodies located laterally in the male antenna, close to the trichoid sensilla (Figure 3A), while no cells were labeled in the female antenna (data not shown). Double-ISH with differently labeled probes directed against both male-specific OR-coding genes revealed expression of the two genes in neighboring cells (longitudinal section and cross section, Figures 3A,B) closely associated with the trichoid sensilla that house the dendrites of pheromone- sensitive ORNs. Weak, axonal-like structures were stained in rare cases (Figure 3B), most likely due the fluorescent residue leaking into the axon. MsexOR-5 derived probes led to similar labeling of cells associated with trichoid sensilla in the female antenna (Figure 4), with no labeling in the male antenna (data not shown).
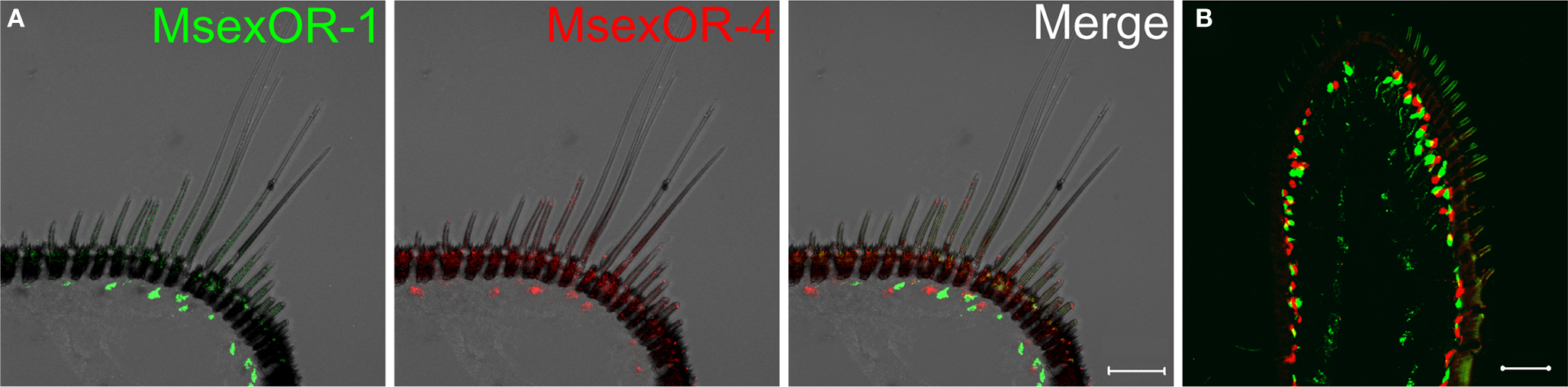
Figure 3. MsexOR-1 and MsexOR-4 expression in the male antenna. (A) Double-in situ hybridization (ISH) using gene-specific probes was performed on longitudinal cryosections of male M. sexta antennae. Probes were directed against MsexOR-1 (biotin-labeled probe, visualized in green, left) and MsexOR-4 (DIG-labeled, red, middle). Both probes label cells close to the long, pheromone-sensitive sensilla trichodea. Labels do not overlap (Merge, right); in many but not all instances MsexOR-1 and MsexOR-4 positive cells are neighbors. (Scale bar = 50 μm). (B) Labeling on a cross section. Note the close proximity of cells expressing the two receptor types to each other and to sensilla trichodea. (Scale bar = 50 μm).
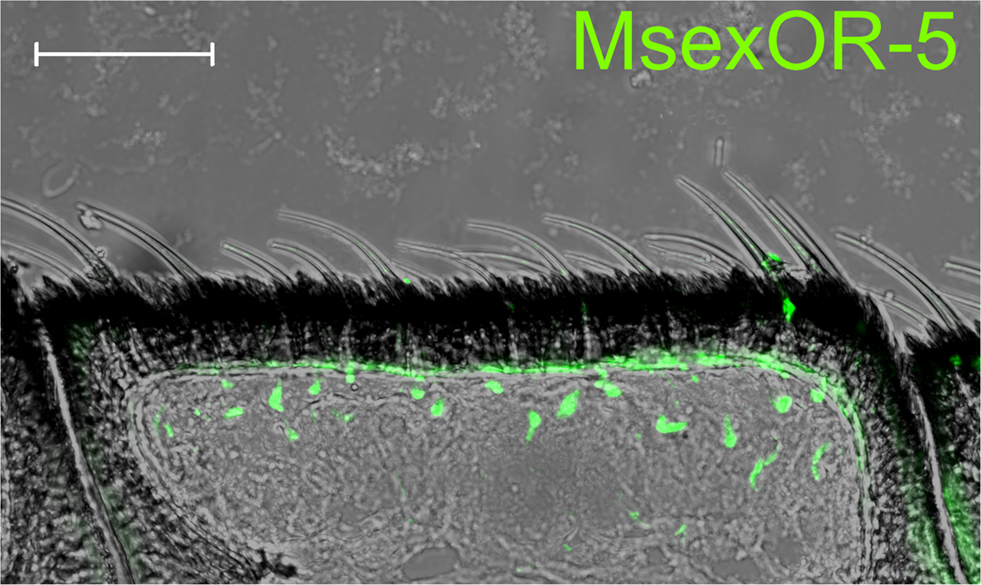
Figure 4. MsexOR-5 expression in the female antenna. ISH using a biotin-labeled probe directed against MsexOR-5 on a longitudinal cryosection of the female M. sexta antenna. Labeled cells are associated with sensillar hairs. (Scale bar = 50 μm).
To confirm the identity of the cells expressing MsexOR-4 as ORN, double-ISH with probes directed against this receptor and the olfactory co-receptor MsexOR-2 were performed (Figure 5). The probes labeled distinct cell bodies in the male antenna in close proximity of the pheromone-sensitive sensilla trichodea. Most MsexOR-4 expressing cells clearly co-expressed MsexOR-2 (Figure 5, left labeled cell). Close scrutiny of seemingly non-co-expressing cells in confocal imaging revealed a weak but distinct red staining, indicating a weak but clear expression of MsexOR-2.
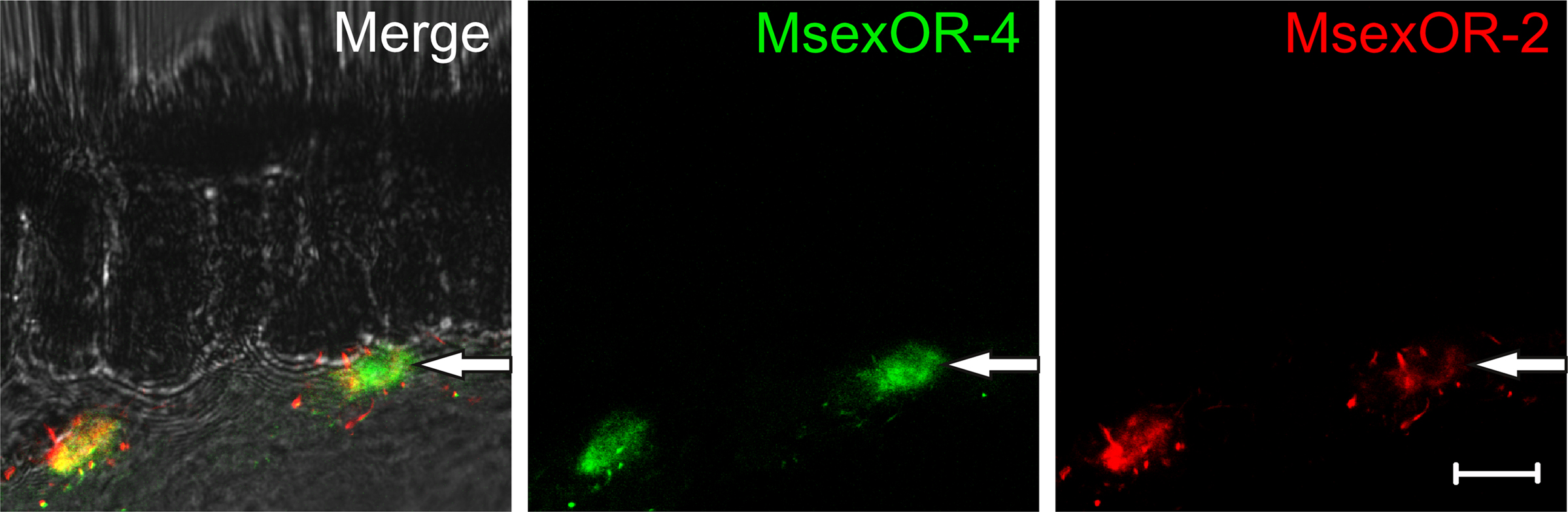
Figure 5. Expression of MsexOR-2 and MsexOR-4 in the male antenna. Double-ISH using probes directed against MsexOR-2 (DIG, red) and MsexOR-4 (Biotin, green) on a cross section of the male antenna. The pictures represent a single optical slice of a confocal stack. In close proximity to sensilla trichodea, probes label the same cells. While in some cases a clear double labeling is not apparent (arrow), close scrutiny reveals a weak red staining. (Scale bar = 10 μm).
Discussion
Using subtracted cDNA libraries we were able to identify four putative OR-coding genes of the tobacco hornworm M. sexta. The male-specific OR type MsexOR-4 belongs to the subgroup of pheromone receptor coding genes (Krieger et al., 2005) also containing the previously reported MsexOR-1 (Patch et al., 2009); the two OR types are expressed in distinct cells, which are in many cases neighbors. In the male M. sexta antenna three different pheromone-detecting ORNs (A–C) are found housed in mixed pairs in sensilla trichodea, with one type detecting the main pheromone component bombykal and the other two subtypes responding to the minor pheromone components E10, E12, E14–16:Al and E10, E12, Z14–16:Al, respectively (Kaissling et al., 1989; Kalinova et al., 2001). The gene expression pattern of the two OR types MsexOR-1 and MsexOR-4 closely matches the distribution of pheromone-specific ORNs, providing substantial support to the notion that the two ORs detect two components of the pheromone blend. Until functional analysis of the encoded OR proteins has been performed it remains unclear if one of the two ORs is responsible for detection of the main pheromone component bombykal. However, as MsexOR-1 is closely related to the bombykal-detecting receptor BmorOR-3 of B. mori (Nakagawa et al., 2005; Grosse-Wilde et al., 2006) this OR may detect this compound. The second potentially pheromone-specific OR, MsexOR-4 was found to be colocalized with MsexOR-1 under most sensilla. Taking into consideration that 70% of the pheromone-specific sensilla on the male antenna houses a pair of ORNs tuned to bombykal and EEZ respectively, MsexOR-4 is very likely tuned to the detection of EEZ. We were unable to identify additional OR types involved in pheromone component detection, which might be due to overall low expression levels of the respective genes. It is e.g., known that ORNs detecting the EEE isomer of the pheromone blend are very rare in the antenna, ca. 15% of the subpopulation of pheromone-sensitive trichoid sensilla contain a neuron primarily tuned to EEZ (Kaissling et al., 1989; Kalinova et al., 2001). The glomerulus innervated by these neurons is also considerably smaller than the glomeruli receiving input from bombykal- and EEZ-detecting neurons (Hansson et al., 1991, 2003; Heinbockel et al., 2004). Colocalization of the expression of MsexOR-2 with the expression of the putative pheromone receptor coding MsexOR-4 indicates a role for the co-receptor MsexOR-2 in pheromone signal transduction in M. sexta, similar to the role of its homolog OR83b in general odorant detection in Drosophila melanogaster.
The closest related sequence to the female-specific MsexOR-5 in another Lepidopteran species is BmOR-19, a likewise female-specific OR of B. mori involved in the detection of the floral scent linalool. While linalool is known to elicit antennal responses in both sexes in M. sexta, it is known also to elicit responses in a specific class of ORNs situated below female trichoid sensilla, in turn leading to activation of the lateral large female glomerulus (latLFG). Specifically, the latLFG is tuned to (+) linalool, showing a high preference even in comparison to (−) linalool (Reisenman et al., 2004). It can thus be speculated that MsexOR-5 is involved in this female-specific physiological response.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Christopher Koenig for assistance with in situ hybridization and Sylke Dietel for Manduca rearing. This study was supported by the Max Planck Society.
References
Anderson, A. R., Wanner, K. W., Trowell, S. C., Warr, C. G., Jaquin-Joly, E., Zagatti, P., Robertson, H., and Newcomb, R. D. (2009). Molecular basis of female-specific odorant responses in Bombyx mori. Insect Biochem. Mol. Biol. 39, 189–197.
Angerer, L. M., and Angerer, R. C. (1992) “In situ hybridization to cellular RNA with radiolabeled RNA probes,” in In Situ Hybridization, ed. D. G Wilkinson (Oxford: IRL Press), 17.
Anton, S., and Hansson, B. S. (1994). Central processing of sex pheromone, host odour, and oviposition deterrent information by interneurons in the antennal lobe of female Spodoptera littoralis (Lepidoptera: Noctuidae). J. Comp. Neurol. 350, 199–214.
Boeckh, J., and Boeckh, V. (1979). Threshold and odor specificity of pheromone-sensitive neurons in the deutocerebrum of Antheraea pernyi and A. polyphemus (Saturnidae). J. Comp. Physiol. A 132, 235–242.
Butenandt, V. A., Beckmann, R., Stamm, D. and, Hecker, E. (1959). über den Sexuallockstoff des Seidenspinners Bombyx mori. Reindarstellung und Konstitution Z. Naturforschung 14, 283–284.
Christensen, T. A., and Hildebrand, J. G. (1987). Male-specific, sex pheromone-selective projection neurons in the antennal lobes of the moth Manduca sexta. J. Comp. Physiol. A 160, 553–569.
Daly, K. C., Durtschi, M. L., and Smith, B. H. (2001). Olfactory-based discrimination learning in the moth, Manduca sexta. J. Insect Physiol. 47, 375–384.
de Bruyne, M., and Baker, T. C. (2008). Odor detection in insects: volatile codes. J. Chem. Ecol. 34, 882–897.
Diatchenko, L., Lau, Y. F., Campbell, A. P., Chenchik, A., Moqadam, F., Huang, B., Lukyanov, S., Lukyanov, K., Gurskaya, N., Sverdlov, E. D., and Siebert, P. D. (1996). Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. U.S.A. 93, 6025–6030.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Feng, L., and Prestwich, G. D. (1997). Expression and characterization of a Lepidopteran general odorant binding protein. Insect Biochem. Mol. Biol. 27, 405–412.
Grosse-Wilde, E., Gohl, T., Bouché, E., Breer, H., and Krieger, J. (2007). Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 25, 2364–2373.
Grosse-Wilde, E., Svatos, A., and Krieger, J. (2006). A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem. Senses 31, 547–555.
Gyorgyi, T. K., Roby-Shemkovitz, A. J., and Lerner, M. R. (1988). Characterization and cDNA cloning of the pheromone-binding protein from the tobacco hornworm, Manduca sexta: a tissue-specific developmentally regulated protein. Proc. Natl. Acad. Sci. U.S.A. 85, 9851–9855.
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979.
Hansson, B. S., Carlsson, M. A., and Kalinovà, B. (2003). Olfactory activation patterns in the antennal lobe of the sphinx moth, Manduca sexta. J. Comp. Physiol. A 189, 301–308.
Hansson, B. S., Christensen, T. A., and Hildebrand, J. G. (1991). Functionally distinct subdivisions of the macroglomerular complex in the antennal lobe of the male sphinx moth Manduca sexta. J. Comp. Neurol. 312, 264–278.
Hansson, B. S., van der Pers, J. N. C., and Loefquist, J. (1989). Comparison of male and female olfactory cell response to pheromone compounds and plant volatiles in the turnip moth, Agrotis segetum. Physiol. Entomol. 14, 147–155.
Heinbockel, T., Christensen, T. A., and Hildebrand, J. G. (2004). Representation of binary pheromone blends by glomerulus-specific olfactory projection neurons. J. Comp. Physiol. A 190, 1023–1037.
Huetteroth, W., and Schachtner, J. (2005). Standard three-dimensional glomeruli of the Manduca sexta antennal lobe: a tool to study both developmental and adult neuronal plasticity. Cell Tissue Res. 319, 513–524.
Kaissling, K.-E., Hildebrand, J. G., and Tumlinson, J. H. (1989). Pheromone receptor cells in the male moth Manduca sexta. Arch. Insect Biochem. Physiol. 10, 273–279.
Kalinova, B., Hoskovec, M., Liblikas, I., Unelius, C. R., and Hansson, B. S. (2001). Detection of sex pheromone components in Manduca sexta (L.). Chem. Senses 26, 1175–1186.
Krieger, J., Große-Wilde, E., Gohl, T., and Breer, H. (2005). Candidate pheromone receptors of the silkmoth Bombyx mori. Eur. J. Neurol. 21, 2167–2176.
Krieger, J., Grosse-Wilde, E., Gohl, T., Dewer, Y. M., Raming, K., and Breer, H. (2004). Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc. Natl. Acad. Sci. U.S.A. 101, 11845–11850.
Krieger, J., Klink, O., Mohl, C., Raming, K., and Breer, H. (2003). A candidate olfactory receptor subtype highly conserved across different insect orders. J. Comp. Physiol. A 189, 519–526.
Lee, J. K., and Strausfeld, N. J. (1990). Structure, distribution and number of surface sensilla and their receptor cells on the olfactory appendage of the male moth Manduca sexta. J. Neurocytol. 19, 519–538.
Matsumoto, S. G., and Hildebrand, J. G. (1981). Olfactory mechanisms in the moth Manduca sexta: response characteristics and morphology of central neurons in the antennal lobes. Proc. R. Soc. Lond. B 213, 249–277.
Nakagawa, T., Sakurai, T., Nishioka, T., and Touhara, K. (2005). Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–1642.
Nardi, J., Miller, L., Walden, K. O., Rovelstad, S., Wang, L., Frye, J., Ramsdell, K., Deem, L., and Robertson, H. (2003). Expression patterns of odorant-binding proteins in antennae of the moth Manduca sexta. Cell Tissue Res. 313, 321–333.
Patch, H. M., Velarde, R. A., Walden, K. K. O., and Robertson, H. M. (2009). A candidate pheromone receptor and two odorant receptors of the hawkmoth Manduca sexta. Chem. Senses 34, 305–316.
Reisenman, C. E., Christensen, T. A., Francke, W., and Hildebrand, J. G. (2004). Enantioselectivity of projection neurons innervating identified olfactory glomeruli. J. Neurosci. 24, 2602–2611.
Renwick, J. A. A., and Chew, F. S. (1994). Oviposition Behavior in Lepidoptera. Annu. Rev. Entomol. 39, 377–400.
Robertson, H. M., Martos, R., Sears, C. R., Todres, E. Z., Walden, K. K. O., and Nardi, J. B. (1999). Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol. Biol.8, 501–518.
Rogers, M. E., Krieger, J., and Vogt, R. G. (2001). Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J. Neurobiol. 49, 47–61.
Rospars, J. P., and Hildebrand, J. G. (1992). Anatomical identification of glomeruli in the antennal lobes of the male sphinx moth Manduca sexta. Cell Tissue Res. 270, 205–227.
Rospars, J. P., and Hildebrand, J. G. (2000). Sexually dimorphic and isomorphic glomeruli in the antennal lobes of the sphinx moth Manduca sexta. Chem. Senses 25, 119–129.
Rybczynski, R., Reagan, J., and Lerner, M. (1989). A pheromone-degrading aldehyde oxidase in the antennae of the moth Manduca sexta. J. Neurosci. 9, 1341–1353.
Sakurai, T., Nakagawa, T., Mitsuno, H., Mori, H., Endo, Y., Tanoue, S., Yasukochi, Y., Touhara, K., and Nishioka, T. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 101, 16653–16658.
Sato, K., Pellegrino, M., Nakagawa, T., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006.
Schneider, D. (1969). Insect olfaction: deciphering system for chemical messages. Science 163, 1031–1037.
Shields, V. D., and Hildebrand, J. G. (2000). Responses of a population of antennal olfactory receptor cells in the female moth Manduca sexta to plant- associated volatile organic compounds. J. Comp. Physiol. A 186, 1135–1151.
Tanaka, K., Uda, Y., Ono, Y., Nakagawa, T., Suwa, M., Yamaoka, R., and Touhara, K. (2009). Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr. Biol. 19, 881–890.
The International Silkworm Genome Consortium. (2008). The genome of a Lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38, 1036–1045.
Tumlinson, J. H., Brennan, M. M., Doolittle, R. E., Mitchell, E. R., Brabham, A., Mazomenos, B. E., Baumhover, A. H., and Jackson, D. M. (1989). Identification of a pheromone blend attractive to Manduca sexta (L.) males in a wind tunnel. Arch. Insect Biochem. Physiol. 10, 255–271.
Tumlinson, J. H., Mitchell, E. R., Doolittle, R. E., and Jackson, D. M. (1994). Field tests of synthetic Manduca sexta sex pheromone. J. Chem. Ecol. 20, 579–591.
Vogt, R., and Blomquist, G. ed. (2003). Insect Pheromone Biochemistry and Molecular Biology. Maryland Heights, MO: Elsevier.
Vogt, R., Rybczynski, R., and Lerner, M. (1991). Molecular cloning and sequencing of general odorant-binding proteins GOBP1 and GOBP2 from the tobacco hawk moth Manduca sexta: comparisons with other insect OBPs and their signal peptides. J. Neurosci. 11, 2972–2984.
Vosshall, L. B., Amrein, H., Morozov, P. S., Rzhetsky, A., and Axel, R. (1999). A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96, 725–736.
Vosshall, L. B., Wong, A. M., and Axel, R. (2000). An olfactory sensory map in the fly brain. Cell 102, 147–159.
Wicher, D., Schäfer, R., Bauernfeind, R., Stensmyr, M. C., Heller, R., Heinemann, S. H., and Hansson, B. S. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011.
Keywords: olfaction, Manduca sexta, Lepidoptera, odorant receptor, pheromone receptor
Citation: Große-Wilde E, Stieber R, Forstner M, Krieger J, Wicher D and Hansson BS (2010) Sex-specific odorant receptors of the tobacco hornworm Manduca sexta. Front. Cell. Neurosci. 4:22. doi: 10.3389/fncel.2010.00022
Received: 25 March 2010;
Paper pending published: 17 May 2010;
Accepted: 03 June 2010;
Published online: 03 August 2010
Edited by:
Randolf Menzel, Freie Universiät Berlin, GermanyReviewed by:
John Hildebrand, University of Arizona, USAKlemens F. Störtkuhl, Ruhr-Universität Bochum, Germany
Copyright: © 2010 Große-Wilde, Stieber, Forstner, Krieger, Wicher and Hansson. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Ewald Große-Wilde, Department of Evolutionary Neuroethology, Max Planck Institute for Chemical Ecology, Hans-Knöll-Strasse 8, 07745 Jena, Germany. e-mail:Z3Jvc3NlLXdpbGRlQGljZS5tcGcuZGU=





