- 1Department of Neurosciences, Mental Health and Sensory Organs – Sant'Andrea Hospital, Sapienza University of Rome, Rome, Italy
- 2Department of Neurosciences, Ohio State University, Columbus, OH, USA
- 3Department of Psychiatry and Behavioral Neurobiology, University of Alabama at Birmingham, Birmingham, AL, USA
- 4Section of Psychiatry, Department of Neuroscience, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health University of Genova, Genova, Italy
- 5Department of Cell and Developmental Biology, Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel
Introduction
Major affective disorders are one of the foremost causes of morbidity worldwide; such disabling conditions are also frequently associated with suicidal behavior (Innamorati et al., 2011; Gonda et al., 2012; Serafini et al., 2012). Although many psychopharmacological agents are currently available, in particular for the treatment of major depressive disorder (MDD) (Serafini et al., 2013), our knowledge concerning the molecular and cellular mechanisms underlying this complex condition is still limited. Indeed, even minor alterations in the expression of genes regulating neural and structural plasticity may be crucial to understanding the pathogenesis of major affective disorders (Dwivedi et al., 2009a,b; Serafini et al., 2011, 2012).
MiRNAs are gene expression regulators critically affecting brain development that have been investigated as potential biomarkers for the diagnosis, management, treatment, and progression of neuropsychiatric disorders (Machado-Vieira et al., 2010; Saugstad, 2010; Dwivedi, 2011). Several facets of miRNA expression alterations are currently under investigation to gain insight into the pathology of neuronal disorders (Hansen et al., 2007; Lopez et al., 2013): miRNA expression alterations in pathophysiological models of disease (Ziu et al., 2011; Brandenburger et al., 2012); miRNA expression alterations in the blood of patients, most of which represent further downstream or compensatory effects (Schipper et al., 2007; Gallego et al., 2012); and miRNAs and their effectors acting as targets for the action of psychoactive drugs such as antidepressants and mood stabilizers (Zhou et al., 2009; Baudry et al., 2010; Oved et al., 2012).
Several miRNAs have emerged as potential mediators of depressive pathophysiology. The existence of polymorphisms in pre-miR-30e (Xu et al., 2010) and pre-miR-182 (Saus et al., 2010) has been associated with an increased risk of major depression. Depressive behavioral responses have been induced by miR-16 up-regulation in the raphe nuclei and hippocampus, with the latter associated with subsequent down-regulation of BNDF (Bai et al., 2012). MiR-16 down-regulation within the locus coeruleus was also induced in depressive mouse models (Launay et al., 2011). Smalheiser et al. (2012) investigated the expression of miRNAs in the prefrontal cortex (specifically in Brodmann Area 9) of 18 antidepressant-free depressed suicide victims, and 17 well-matched non-psychiatric controls, whose information was collected using the psychological autopsy method. In this study, global miRNA expression was significantly down-regulated by 17%, and 24 miRNAs were down-regulated by at least 30%. The authors also found significant down-regulation in an extensive inter-connected network of 21 miRNAs involved in cellular growth and differentiation. Notably, both the global decrease of miRNA expression, as well as its decreased variability, are consistent with hypo-activation of the frontal cortex in depressed subjects. Interestingly, the noted miRNAs have been suggested to be down-regulated in the frontal cortex of rats treated with corticosterone, and therefore, might be crucial in regulating stress-mediated miRNA expression in depressed subjects (Dwivedi et al., unpublished data). Hence, alterations in miRNA expression may be a fundamental event underlying gene network reorganization associated with major depression.
Nevertheless, a comprehensive understanding of miRNA networks dysregulated in major depression and induced by antidepressant medications as a function of brain region is currently unknown (Mouillet-Richard et al., 2012). It is also generally poorly understood how miRNA regulation affects cellular signaling networks in these biological processes. Here we provide an overview and critical review of the published work, particularly examining the role of miR-185 in major depression and suicidal behavior.
The Challenges of miRNA:mRNA Target Prediction in Modeling Pathology
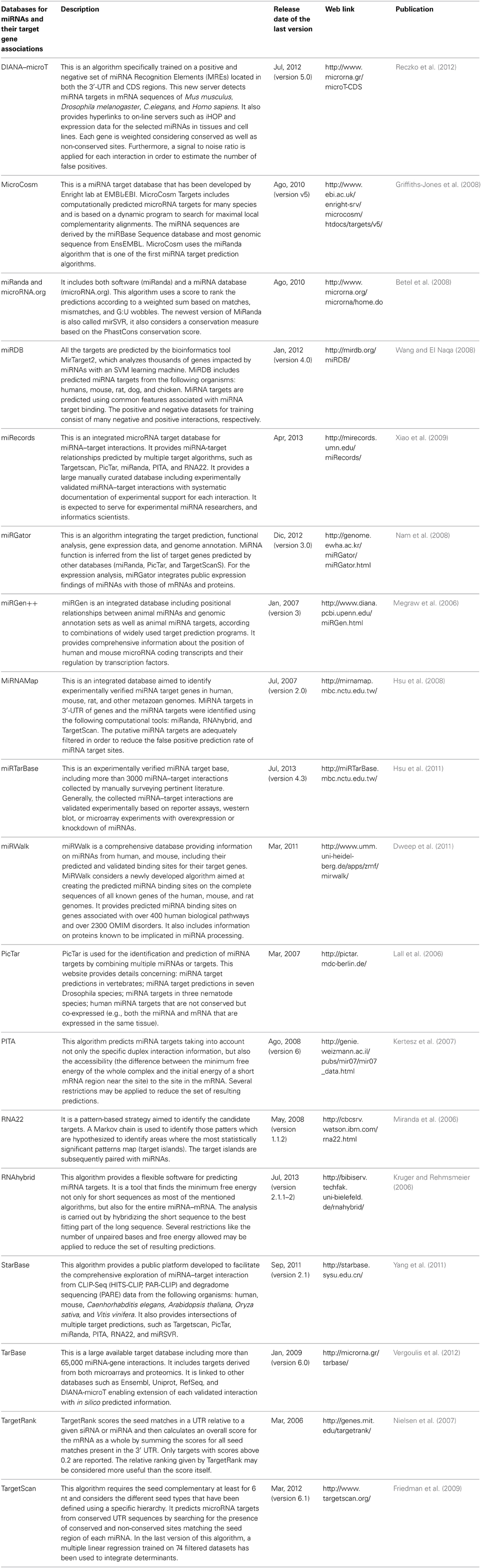
Expression profiling and RNA sequencing of miRNAs have increased our understanding of which miRNAs are present in specific tissues, and how they may change under pathological conditions (Oved et al., 2012). However, once identified, linking a miRNA to its mRNA targets can be a challenging task, and the authenticity of functional miRNA:mRNA target pairs should be validated. A very small fraction of software-predicted miRNA targets are validated in vivo. As suggested by Kuppers et al. (2011), only a subset of predicted targets are consistently reduced in phenotypes that overexpress some miRNAs. Validation could be provided by showing a direct interaction between the miRNA and mRNA target. Further, miRNA silencing, overexpression, and luciferase reporter based assays are commonly used as supporting evidence for a functional interaction (Kuhn et al., 2008).
Many databases for miRNA target prediction have been created using an array of interrogation approaches. Considering varying degrees of sequence similarity, conservation, site accessibility, and variation in the targeted regions of the mRNA, the numerous databases can provide a surprising level of divergent results. This divergence speaks to the difficulty that scientists have had in developing a clear and concise set of rules underlying miRNA:mRNA interaction: clearly target prediction software should only be viewed as a tool to guide bench science.
Table 1 Provides a summary of current miRNA target prediction software that may be used to guide in silico investigations into miRNA and their putative targets. The analytical paradigms for each database are describes as well as advantages of each mode of inquiry.
MiR-185: A Role in Major Affective Disorders and Suicidal Behavior?
Preliminary studies have suggested the importance of miR-185 and miR-491-3p in the pathogenesis of major depression and suicidal behavior. MiR-185 is expressed in several brain regions such as the hippocampus and cortex, predominantly in synapses (Lugli et al., 2008; Xu et al., 2013). Earls et al. (2012) reported that miR-185 regulates cognitive and psychiatric symptoms of patients with the 22q11 deletion syndrome. Recently, Xu et al. (2013) suggested that miR185 controls the expression of Golgi-apparatus related genes including a new inhibitor of neuronal maturation. In particular, a reduction of miR-185 altered dendritic and spine development resulting in structural alterations of the hippocampus.
With respect to MDD and suicidality, miR-185 was shown to be upregulated in patients who completed suicide (Maussion et al., 2012). These increases in expression were correlated with reduced TrkB-T1, a truncated TrkB transcript whose downregulation has been associated with suicide (Ernst et al., 2009). The downregulation of TrkB-T1was associated with suicidal behavior in a sample of 38 suicide completers (60.5% having been previously diagnosed with MDD). Of note, five putative binding sites for miR-185 were found in the 3′ UTR of TrkB-T1 (using an in silico investigation). Array findings were confirmed with RT-PCR investigation and three of the five potential binding sites for miR-185 in the TrkB-T1 3′ UTR were demonstrated to be functional by luciferase assay. The authors did not find any confounding effect of age, pH, PMI, or suicide method. Through Pearson correlation and subsequent in vitro functional analyses (using silencing or exogenous expression of miR-185), TrkB-T1 levels and hsa-miR-185 levels were reported to be inversely correlated.
A few notes of caution should be mentioned with regard to the Maussion et al. (2012) study. The authors acknowledge that the underlying mechanism of increased miR-185 expression remains unclear. The study used HEK293 cells that yielded TrkB-T1 expression levels that were 10-fold greater than neuronal cell lines. Furthermore, RNA binding proteins, such as ELAVL1 or PABPC1, may be expressed in HEK293 cells (Drury et al., 2010) and potentially bind TrkB mRNA (Jain et al., 2011). Therefore, despite disproportionate increases in TrkB-T1 expression, the functional effect of hsa-miR-185 on TrkB-T1 observed in HEK293 cells might have been attenuated by the expression of these genes and their binding activity (George and Tenenbaum, 2006). Further, the study is limited by the small sample size and the negative findings in other brain regions, such as the cerebellum. Indeed, presumably only a limited part of the total variability in miRNAs that might regulate TrkB-T1 has been identified.
Of note, the subjects of the Maussion et al. (2012) study were not assessed for microduplications in the 22q11.2 region. This is of potential interest because the miR-185 locus maps to the 22q11.2 region, which has been associated with mood disorders such as depression and anxiety (Jolin et al., 2012; Weisfeld-Adams et al., 2012; Tang et al., 2013). Deletions of this region have also been consistently associated with schizophrenia (Karayiorgou et al., 2010) whereas duplications have been found in patients with autism (Lo-Castro et al., 2009). Alterations in the 22q11.2 region are also associated with morphological alterations in dendritic spines at glutamatergic synapses (Mukai et al., 2008), and abnormal maturation of miRNAs (Stark et al., 2008). Fénelon et al. (2013) have suggested that mice with a 22q11.2 microdeletion show significant alterations in high-frequency synaptic transmission, short- and long-term plasticity, and dendritic spine stability. The authors reported that variation in synaptic plasticity occurs by subtle changes in neuronal density and a reduction in inhibitory neuron. All of these alterations in neuronal function could play critical roles in depressive pathophysiology.
Understanding the Limitations of Studies Examining the Role of miRNAs in Major Affective Disorders
Since the first detection in Caenorhabditis elegans in 1993 (Lee et al., 1993), small interfering RNAs have raised great interest among neurobiologists for their potential role in neuropathological regulation. In line with this notion, large-scale analyses on post-mortem brains, as well as investigations in animal models of depression, have evaluated the impact of psychoactive medications on global miRNA expression. Transcriptome studies are now commonly used as a starting point to investigate the association between dysregulated miRNAs and major affective disorders. However, there are a number of conflicting studies with regard to the magnitude and direction of biologically-relevant miRNA expression changes in psychiatric disorders (Perkins et al., 2007; Beveridge et al., 2010). This could be due to tissue-specific variations in expression levels as well as heterogeneity in quantification and normalization procedures (Belzeaux et al., 2012). Furthermore, some studies on miRNAs and depression were conducted in peripheral blood despite uncertainties regarding how closely changes in peripheral miRNA expression reflect modifications in the central nervous system (e.g., Bocchio-Chiavetto et al., 2013). It is also worth noting that, “control” RNAs commonly used to normalize miRNA data (U6, U44, and U48) are very sensitive to post-mortem decay (Sadikovic et al., 2011) and thus, should be carefully matched among groups to prevent the emergence of artifactual shifts in miRNA expression. Finally, neuronal shrinkage, loss of glial cells, or loss of dendritic spines may also contribute to changes in miRNA levels. Clearly, changes in tissue composition or cellular compartments should be carefully taken into account when examining the available studies.
Conclusion
Our understanding of the molecular mechanisms underlying major affective disorders may be significantly enriched by the knowledge of miRNAs' mechanisms of action. MiRNA targets are critically involved in stress-related disorders, neuroplasticity, and neurodevelopmental disorders (Rogaev, 2005). Given that miRNAs have been hypothesized to modulate ~50% of protein-coding genes and hundreds of mRNAs (Krol et al., 2010), a new level of complexity regarding gene expression has emerged. The entire miRNA context (both mRNA networks and their cellular environments) should be critically investigated when interpreting the effects of changes in miRNA levels. Much remains to be examined in order to translate these investigations into novel therapeutics for the treatment of psychiatric conditions.
Acknowledgments
Part of the funding was provided by R01MH082802, R01MH 101890 to Dr. Dwivedi.
References
Bai, M., Zhu, X., Zhang, Y., Zhang, S., Zhang, L., Xue, L., et al. (2012). Abnormal hippocampal bdnf and mir-16 expression is associated with depression-like behaviors induced by stress during early life. PLoS ONE 7:e46921. doi: 10.1371/journal.pone.0046921
Baudry, A., Mouillet-Richard, S., Schneider, B., Launay, J. M., and Kellermann, O. (2010). miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science 329, 1537–1541. doi: 10.1126/science.1193692
Belzeaux, R., Bergon, A., Jeanjean, V., Loriod, B., Formisano-Tréziny, C., Verrier, L., et al. (2012). Responder and non-responder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl. Psychiatry 2, e185. doi: 10.1038/tp.2012.112
Betel, D., Wilson, M., Gabow, A., Marks, D. S., and Sander, C. (2008). The microRNA. org resource. targets and expression. Nucl. Acids Res. 36, D149–D153. doi: 10.1093/nar/gkm995
Beveridge, N. J., Gardiner, E., Carroll, A. P., Tooney, P. A., and Cairns, M. J. (2010). Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry 15, 1176–1189. doi: 10.1038/mp.2009.84
Bocchio-Chiavetto, L., Maffioletti, E., Bettinsoli, P., Giovannini, C., Bignotti, S., Tardito, D., et al. (2013). Blood microRNA changes in depressed patients during antidepressant treatment. Eur. Neuropsychopharmacol. 23, 602–611. doi: 10.1016/j.euroneuro.2012.06.013
Brandenburger, T., Castoldi, M., Brendel, M., Grievink, H., Schlösser, L., Werdehausen, R., et al. (2012). Expression of spinal cord microRNAs in a rat model of chronic neuropathic pain. Neurosci. Lett. 506, 281–286. doi: 10.1016/j.neulet.2011.11.023
Drury, G. L., Di Marco, S., Dormoy-Raclet, V., Desbarats, J., and Gallouzi, I. E. (2010). FasL expression in activated T lymphocytes involves HuR-mediated stabilization. J. Biol. Chem. 285, 31130–31138. doi: 10.1074/jbc.M110.137919
Dweep, H., Sticht, C., Pandey, P., and Gretz, N. (2011). miRWalk–database: prediction of possible miRNA binding sites by walking the genes of 3 genomes. J. Biomed. Inform. 44, 839–837. doi: 10.1016/j.jbi.2011.05.002
Dwivedi, Y. (2011). Evidence demonstrating role of microRNAs in the etiopathology of major depression. J Chem Neuroanat 42, 142–156. doi: 10.1016/j.jchemneu.2011.04.002
Dwivedi, Y., Rizavi, H., Zhang, H., Mondal, A. C., Roberts, R. C., Conley, R. R., et al. (2009a). Neurotrophin receptor activation and expression in human postmortem brain: effect of suicide. Biol. Psychiatry 65, 319–328. doi: 10.1016/j.biopsych.2008.08.035
Dwivedi, Y., Rizavi, H. S., Zhang, H., Roberts, R. C., Conley, R. R., and Pandey, G. N. (2009b). Aberrant extracellular signal-regulated kinase (ERK)1/2 signaling in suicide brain: role of ERK kinase 1 (MEK1). Int. J. Neuropsychopharmacol. 12, 1337–1354. doi: 10.1017/S1461145709990575
Earls, L. R., Fricke, R. G., Yu, J., Berry, R. B., Baldwin, L. T., and Zakharenko, S. S. (2012). Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J. Neurosci. 32, 14132–14144. doi: 10.1523/JNEUROSCI.1312-12.2012
Ernst, C., Deleva, V., Deng, X., Sequeira, A., Pomarenski, A., and Klempan, T. (2009). Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Arch. Gen. Psychiatry 66, 22–32. doi: 10.1001/archpsyc.66.1.22
Fénelon, K., Xu, B., Lai, C. S., Mukai, J., Markx, S., Stark, K. L., et al. (2013). The pattern of cortical dysfunction in a mouse model of a schizophrenia-related microdeletion. J. Neurosci. 33, 14825–14839. doi: 10.1523/JNEUROSCI.1611-13.2013
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Gallego, J. A., Gordon, M. L., Claycomb, K., Bhatt, M., Lencz, T., and Malhotra, A. K. (2012). In vivo microRNA detection and quantitation in cerebrospinal fluid. J. Mol. Neurosci. 47, 243–248. doi: 10.1007/s12031-012-9731-7
George, A. D., and Tenenbaum, S. A. (2006). MicroRNA modulation of RNA-binding protein regulatory elements. RNA Biol. 3, 57–59. doi: 10.4161/rna.3.2.3250
Gonda, X., Pompili, M., Serafini, G., Montebovi, F., Campi, S., Dome, P., et al. (2012). Suicidal behavior in bipolar disorder: epidemiology, characteristics and major risk factors. J. Affect. Disord. 143, 16–26. doi: 10.1016/j.jad.2012.04.041
Griffiths-Jones, S., Saini, H. K., van Dongen, S., and Enright, A. J. (2008). miRBase: tools for microRNA genomics. Nucl. Acids Res. 36, D154–D158. doi: 10.1093/nar/gkm952
Hansen, T., Olsen, L., Lindow, M., Jakobsen, K. D., Ullum, H., Jonsson, E., et al. (2007). Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE 2:e873. doi: 10.1371/journal.pone.0000873
Hsu, S. D., Chu, C. H., Tsou, A. P., Chen, S. J., Chen, H. C., Hsu, P. W., et al. (2008). miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucl. Acids Res. 36, D165–D169. doi: 10.1093/nar/gkm1012
Hsu, S. D., Lin, F. M., Wu, W. Y., Liang, C., Huang, W. C., and Chan, W. L. (2011). miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucl. Acids Res. 39, D163–169. doi: 10.1093/nar/gkq1107
Innamorati, M., Pompili, M., Gonda, X., Amore, M., Serafini, G., Niolu, C., et al. (2011). Psychometric properties of the gotland scale for depression in Italian psychiatric inpatients and its utility in the prediction of suicide risk. J. Affect. Disord. 132, 99–103. doi: 10.1016/j.jad.2011.02.003
Jain, R., Devine, T., George, A. D., Chittur, S. V., Baroni, T. E., Penalva, L. O., et al. (2011). RIP-chip analysis: RNA-binding protein immunoprecipitation-microarray (chip) profiling. Methods Mol. Biol. 703, 247–263. doi: 10.1007/978-1-59745-248-9_17
Jolin, E. M., Weller, R. A., and Weller, E. B. (2012). Occurrence of affective disorders compared to other psychiatric disorders in children and adolescents with 22q11.2 deletion syndrome. J. Affect. Disord. 136, 222–228. doi: 10.1016/j.jad.2010.11.025
Karayiorgou, M., Simon, T. J., and Gogos, J. A. (2010). 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat. Rev. Neurosci. 11, 402–416. doi: 10.1038/nrn2841
Kertesz, M., Iovino, N., Unnerstall, U., Gaul, U., and Segal, E. (2007). The role of site accessibility in microRNA target recognition. Nat. Genet. 39, 1278–1284. doi: 10.1038/ng2135
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA bio-genesis, function and decay. Nat. Rev. Genet. 11, 597–610. doi: 10.1038/nrg2843
Kruger, J., and Rehmsmeier, M. (2006). RNAhybrid: microRNA target prediction easy, fast and flexible. Nucl. Acids Res. 34, W451–W454. doi: 10.1093/nar/gkl243
Kuhn, D. E., Martin, M. M., Feldman, D. S., Terry, A. V. Jr., Nuovo, G. J., and Elton, T. S. (2008). Experimental validation of miRNA targets. Methods 44, 47–54. doi: 10.1016/j.ymeth.2007.09.005
Kuppers, D. A., Hwang, H. C., Jackson, A. L., Linsley, P. S., Clurman, B. E., and Fero, M. L. (2011). Effect of Xpcl1 activation and p27(Kip1) loss on gene expression in murine lymphoma. PLoS ONE 6:e14758. doi: 10.1371/journal.pone.0014758
Lall, S., Grün, D., Krek, A., Chen, K., Wang, Y. L. L., Dewey, C. N. N., et al. (2006). A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 16, 460–471. doi: 10.1016/j.cub.2006.01.050
Launay, J. M., Mouillet-Richard, S., Baudry, A., Pietri, M., and Kellermann, O. (2011). Raphe-mediated signals control the hippocampal response to SRI antidepressants via miR-16. Transl. Psychiatry 1, e56. doi: 10.1038/tp.2011.54
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
Lo-Castro, A., Galasso, C., Cerminara, C., El-Malhany, N., Benedetti, S., Nardone, A. M., et al. (2009). Association of sindromi mental retardation and autismwith 22q11.2 duplication. Neuropediatrics 40, 137–140. doi: 10.1055/s-0029-1237724
Lopez, J. P., Fiori, L. M., Gross, J. A., Labonte, B., Yerko, V., Mechawar, N., et al. (2013). Regulatory role of miRNAs in polyamine gene expression in the prefrontal cortex of depressed suicide completers. Int. J. Neuropsychopharmacol. 12, 1–10. doi: 10.1017/S1461145713000941
Lugli, G., Torvik, V. I., Larson, J., and Smalheiser, N. R. (2008). Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J. Neurochem. 106, 650–661. doi: 10.1111/j.1471-4159.2008.05413.x
Machado-Vieira, R., Salvadore, G., DiazGranados, N., Ibrahim, L., Latov, D., Wheeler-Castillo, C., et al. (2010). New therapeutic targets for mood disorders. Sci. World. J. 10, 713–726. doi: 10.1100/tsw.2010.65
Maussion, G., Yang, J., Yerko, V., Barker, P., Mechawar, N., Ernst, C., et al. (2012). Regulation of a truncated form of tropomyosin-related kinase B (TrkB) by Hsa-miR-185* in frontal cortex of suicide completers. PLoS ONE 7:e39301. doi: 10.1371/journal.pone.0039301
Megraw, M., Sethupathy, P., Corda, B., and Hatzigeorgiou, A. G. (2006). miRGen: a database for the study of animal microRNA genomic organization and function. Nucl. Acids Res. 35, D149–D155. doi: 10.1093/nar/gkl904
Miranda, K. C., Huynh, T., Tay, Y., Ang, Y. S. S., Tam, W. L. L., Thomson, A. M., et al. (2006). A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217. doi: 10.1016/j.cell.2006.07.031
Mouillet-Richard, S., Baudry, A., Launay, J. M., and Kellermann, O. (2012). MicroRNAs and depression. Neurobiol. Dis. 46, 272–278. doi: 10.1016/j.nbd.2011.12.035
Mukai, J., Dhilla, A., Drew, L. J., Stark, K. L., Cao, L., MacDermott, A. B., et al. (2008). Palmitoylationdependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat. Neurosci. 11, 1302–1310. doi: 10.1038/nn.2204
Nam, S., Kim, B., Shin, S., and Lee, S. (2008). miRGator: an integrated system for functional annotation of microRNAs. Nucl. Acids Res. 36, 159–164. doi: 10.1093/nar/gkm829
Nielsen, C. B., Shomron, N., Sandberg, R., Hornstein, E., Kitzman, J., and Burge, C. B. (2007). Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 13, 1894–1910. doi: 10.1261/rna.768207
Oved, K., Morag, A., Pasmanik-Chor, M., Oron-Karni, V., Shomron, N., Rehavi, M., et al. (2012). Genome-wide miRNA expression profiling of human lymphoblastoid cell lines identifies tentative SSRI antidepressant response biomarkers. Pharmacogenomics 13, 1129–1139. doi: 10.2217/pgs.12.93
Perkins, D. O., Jeffries, C. D., Jarskog, L. F., Thomson, J. M., Woods, K., Newman, M. A., et al. (2007). microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 8, R27. doi: 10.1186/gb-2007-8-2-r27
Reczko, M., Maragkakis, M., Alexiou, P., Grosse, I., and Hatzigeorgiou, A. G. (2012). Functional microRNA targets in protein coding sequences. Bioinformatics 28, 771–776. doi: 10.1093/bioinformatics/bts043
Rogaev, E. I. (2005). Small RNAs in human brain development and disorders. Biochemistry (Mosc) 70, 1404–1407. doi: 10.1007/s10541-005-0276-z
Sadikovic, B., Pearson, R., Meng, L., and Beaudet, A. (2011). “Identification of miRNAs and target mRNAs with degulated expression in schizophrenia and bipolar brains,” in 12th International Congress of Human Genetics, (Montreal), 586T, 219.
Saugstad, J. A. (2010). MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J. Cereb. Blood Flow. Metab. 30, 1564–1576. doi: 10.1038/jcbfm.2010.101
Saus, E., Soria, V., Escaramis, G., Vivarelli, F., Crespo, J. M., Kagerbauer, B., et al. (2010). Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum. Mol. Genet. 19, 4017–4025. doi: 10.1093/hmg/ddq316
Schipper, H. M., Maes, O. C., Chertkow, H. M., and Wang, E. (2007). MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul. Syst. Bio. 1, 263–274.
Serafini, G., Pompili, M., Innamorati, M., Dwivedi, Y., Brahmachari, G., and Girardi, P. (2013). Pharmacological properties of glutamatergic drugs targeting NMDA receptors and their application in major depression. Curr. Pharm. Des. 19, 1898–1922. doi: 10.2174/13816128113199990293
Serafini, G., Pompili, M., Innamorati, M., Giordano, G., Montebovi, F., Sher, L., et al. (2012). The role of microRNAs in synaptic plasticity, major affective disorders and suicidal behavior. Neurosci. Res. 73, 179–190. doi: 10.1016/j.neures.2012.04.001
Serafini, G., Pompili, M., Innamorati, M., Giordano, G., Tatarelli, R., Lester, D., et al. (2011). Glycosides, depression and suicidal behaviour: the role of glycoside-linked proteins. Molecules 16, 2688–2713. doi: 10.3390/molecules16032688
Smalheiser, N. R., Lugli, G., Rizavi, H. S., Torvik, V. I., Turecki, G., and Dwivedi, Y. (2012). MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS ONE 7:e33201. doi: 10.1371/journal.pone.0033201
Stark, K. L., Xu, B., Bagchi, A., Lai, W. S., Liu, H., Hsu, R., et al. (2008). Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 40, 751–760. doi: 10.1038/ng.138
Tang, S. X., Yi, J. J., Calkins, M. E., Whinna, D. A., Kohler, C. G., Souders, M. C., et al. (2013). Psychiatric disorders in 22q11.2 deletion syndrome are prevalent but undertreated. Psychol. Med. 9, 1–11. doi: 10.1017/S0033291713001669
Vergoulis, T. I., Vlachos, P., Alexiou, G., Georgakilas, M., Maragkakis, M., Reczko, S., et al. (2012). Tarbase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucl. Acids Res. 40, D222–D229. doi: 10.1093/nar/gkr1161
Wang, X., and El Naqa, I. M. (2008). Prediction of both conserved and non-conserved microRNA targets in animals. Bioinformatics 24, 325–332. doi: 10.1093/bioinformatics/btm595
Weisfeld-Adams, J. D., Edelmann, L., Gadi, I. K., and Mehta, L. (2012). Phenotypic heterogeneity in a family with a small atypical microduplication of chromosome 22q11.2 involving TBX1. Eur. J. Med. Genet. 55, 732–736. doi: 10.1016/j.ejmg.2012.08.011
Xiao, F., Zuo, Z., Cai, G., Kang, S., Gao, X., and Li, T. (2009). miRecords: an integrated resource for microRNA-target interactions. Nucl. Acids Res. 37, D105–D110. doi: 10.1093/nar/gkn851
Xu, B., Hsu, P. K., Stark, K. L., Karayiorgou, M., and Gogos, J. A. (2013). Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell 152, 262–275. doi: 10.1016/j.cell.2012.11.052
Xu, Y., Liu, H., Li, F., Sun, N., Ren, Y., Liu, Z., et al. (2010). A polymorphism in the microRNA-30e precursor associated with major depressive disorder risk and P300 waveform. J. Affect. Disord. 127, 332–336. doi: 10.1016/j.jad.2010.05.019
Yang, J. H., Li, J. H., Shao, P., Zhou, H., Chen, Y. Q., and Qu, L. H. (2011). starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucl. Acids Res. 39, D202–D209. doi: 10.1093/nar/gkq1056
Zhou, R., Yuan, P., Wang, Y., Hunsberger, J. G., Elkahloun, A., Wei, Y., et al. (2009). Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology 34, 1395–1405. doi: 10.1038/npp.2008.131
Keywords: microRNAs, synaptic plasticity, major affective disorders, suicidal behavior, gene expression
Citation: Serafini G, Pompili M, Hansen KF, Obrietan K, Dwivedi Y, Amore M, Shomron N and Girardi P (2013) MicroRNAs: fundamental regulators of gene expression in major affective disorders and suicidal behavior?. Front. Cell. Neurosci. 7:208. doi: 10.3389/fncel.2013.00208
Received: 03 September 2013; Accepted: 21 October 2013;
Published online: 15 November 2013.
Edited by:
Tommaso Pizzorusso, CNR, ItalyReviewed by:
Joseph A. Gogos, Columbia University, USACopyright © 2013 Serafini, Pompili, Hansen, Obrietan, Dwivedi, Amore, Shomron and Girardi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:Z2lhbmx1Y2Euc2VyYWZpbmlAdW5pcm9tYTEuaXQ=
 Gianluca Serafini
Gianluca Serafini Maurizio Pompili1
Maurizio Pompili1 Noam Shomron
Noam Shomron