- 1Department of Cell Biology and Imaging, Institute of Zoology, Faculty of Biology and Earth Sciences, Jagiellonian University, Krakow, Poland
- 2Department of Medical Biotechnology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Krakow, Poland
- 3Department of Clinical Immunology and Transplantology, Polish-American Institute of Pediatrics, Medical College, Jagiellonian University, Krakow, Poland
The retina and the first optic neuropil (lamina) of Drosophila show circadian rhythms in various processes. To learn about the regulation of circadian rhythms in the retina and lamina and in two cell types, glial and the lamina L2 interneurons, we examined expression of the following clock genes; per, tim, clk, and cry and clock-controlled genes (ccgs); Atpα, nrv2, brp, Pdfr. We found that the expression of gene studied is specific for the retina and lamina. The rhythms of per and tim expression in the retina and glial cells are similar to that observed in the whole head and in clock neurons, while they differ in the lamina and L2 cells. In both the retina and lamina, CRY seems to be a repressor of clk expression. In L2 interneurons per expression is not cyclic indicating the other function of PER in those cells than in the circadian molecular clock. In contrast to per and tim, the pattern of clk and cry expression is similar in both the retina and lamina. The retina holds the autonomous oscillators but the expression of cry and ccgs, Atpα and nrv2, is also regulated by inputs from the pacemaker transmitted by PDF and ITP neuropeptides.
Introduction
The circadian clock is a self-sustaining generator of endogenous oscillations with a period of about a day that generates rhythmicity in biochemical and physiological processes of most organisms and in the behavior of animals. It maintains the homeostasis of organisms in time, anticipates daily changes of light in the environment and its function continues in constant darkness (DD). Under day/night or light/dark (LD) conditions the circadian clock is synchronized by light and the period of the rhythm is equal to 24 h. In DD the period becomes longer or shorter than 24 h.
In animals, circadian rhythms are mostly studied in terms of behavior, predominantly in locomotor activity, but they can also be observed in the activity and structure of cells, including neurons and glial cells (Pyza, 2010). In the visual system of flies, the morphology of cells, synaptic proteins and activity of ion pumps oscillate during the day and most of these rhythms are correlated with the species specific rhythm in locomotor activity. These cyclical changes also occur in DD, indicating that remodeling of the neuron structure and synaptic contacts in the day is regulated by the circadian clock. The changes in morphology of neurons and synapses are synchronized with animal activity and with daily changes of light intensity in the environment (Weber et al., 2009; Damulewicz et al., 2013; Górska-Andrzejak et al., 2005, 2013).
The visual system of the fruit fly Drosophila melanogaster provides an excellent model for studying circadian rhythms at the cellular level. It consists of the retina and three optic neuropils: lamina, medulla and lobula. The retina is composed of 700–800 single modules called ommatidia and each ommatidium comprises eight photoreceptor cells, R1–R8. Six of them, R1–R6 terminate in the first optic neuropil (lamina) while R7 and R8 pass the lamina and terminate in the medulla (Meinertzhagen and Hanson, 1993). The photic and visual information received by the retina photoreceptors are transmitted to the lamina by tetrad synapses formed between R1–R6 and the first order lamina interneurons, large monopolar cells L1 and L2 and two other cell types (Meinertzhagen and O’Neil, 1991). Like the retina, the lamina also has a modular structure and is composed of cylindrical units called cartridges. Each cartridge is surrounded by three glial cells and is composed of six photoreceptor terminals, five lamina monopolar cells, processes of amacrine cells and axons of neurons located in other visual neuropils and in the central brain (Meinertzhagen and Sorra, 2001). The lamina not only receive an efferent input from the retina through tetrad synapses but also sends feedback synapses back to the photoreceptor terminals R1–R6.
Circadian rhythms have been detected in both the retina and lamina of flies. In the retina, circadian oscillations have been found in the amplitude of the electroretinogram (ERG) and synthesis of photopigment (Chen et al., 1992). In the lamina, the number of tetrad and feedback synapses (Pyza and Meinertzhagen, 1993) and the size of monopolar cells L1 and L2 and glial cells (Pyza and Meinertzhagen, 1995, 1999; Pyza and Górska-Andrzejak, 2004) oscillate during the day and night.
The rhythms in the lamina are generated by circadian oscillators located in the brain, by the so-called central clock or pacemaker and by peripheral oscillators located in the retina photoreceptors and in some glial cells of the optic lobe (Damulewicz et al., 2013; Górska-Andrzejak et al., 2013). The central clock consists of about 150 clock cells, expressing clock genes, located in the proximal medulla, namely: ventral lateral neurons (LNvs) and dorsal lateral neurons (LNds) and in the dorsal protocerebrum, three groups of dorsal neurons (DNs1–3). The LNvs, except the so-called 5th small LNv, express the neuropeptide pigment-dispersing factor (PDF), while the 5th small LNv and one of the LNds express the ion transport peptide (ITP; Johard et al., 2009). The 5th small LNv projects to the lamina and this projection is immunoreactive to ITP, however, both peptides, PDF and ITP, are involved in regulating circadian rhythms in the lamina (Damulewicz and Pyza, 2011).
The circadian rhythms in the retina seem to be generated by circadian oscillators located in photoreceptors, as they expressed the clock genes period (per), timeless (tim; Siwicki et al., 1988; Zerr et al., 1990) and cryptochrome (cry; Yoshii et al., 2008). It has been suggested that the retina possesses an autonomous clock, independent from the pacemaker located in the brain (Siwicki et al., 1988; Cheng and Hardin, 1998). Moreover, in the retina photoreceptors per mRNA peaks later during the day than in the pacemaker and PER protein is degraded earlier in the day (Zerr et al., 1990). Nothing is known, however, about expression of clock genes in the lamina, a site of pronounced circadian rhythms in changes of synapse frequency, neurons and glial cell morphology and of protein level. It is also unknown how molecular clocks operate in the retina and in glial cells that express per and other clock genes.
To learn how the rhythms in the retina and lamina are regulated we examined the expression of clock genes and possible clock-controlled genes (ccgs) in the retina or lamina, isolated from the Drosophila head manually or by laser microdissection. We studied the expression of two core genes of the molecular clock, per and tim, as well as clock (clk), encoding the transcription factor CLK, that together with CYCLE (CYC), activates the transcription of per, tim and ccgs. We also examined the expression of the cry gene, a circadian photoreceptor in most of the clock neurons and a circadian transcriptional repressor of the molecular clock in the peripheral oscillators (Stanewsky et al., 1998; Collins et al., 2006). In addition to clock genes we examined the expression of the following ccgs; Atpα, nrv2 and brp. Atpα and nrv2 encode α and β subunits of the Na+/K+-ATPase, a major ion pump in all cells and brp encodes the presynaptic scaffolding protein Bruchpilot (BRP). ATPα and BRP show strong circadian rhythms in abundance in the lamina (Damulewicz et al., 2013; Górska-Andrzejak et al., 2013). The expression of these genes was examined at four time points in wild-type flies maintained in light/dark (LD12:12) or constant darkness (DD) conditions, in null mutants of per (except the per), cry (except the cry), Pdf and in a strain with decreased expression of itp in LD 12:12.
In addition, the expression of per, tim, Atpα, brp and Pdfr—encoding PDF receptors, was studied at four time points in LD 12:12 in two types of cells; the lamina L2 monopolar cells, the first order interneurons in the visual system, and glial cells, which show robust circadian rhythms in structural changes. In addition to checking the rhythms in the mRNAs of clock genes and ccgs in two different layers of the visual system and in two cell types, we also tested if those rhythms are affected by the lack of CRY or clock neurotransmitters PDF or ITP.
The expression of clock and ccgs have not been studied in the visual system—a site of circadian plasticity and in glial cells that participate in the regulation of circadian rhythms in the brain and in behavior (Jackson, 2011; Górska-Andrzejak, 2013).
Materials and Methods
Animals
The following strains of D. melanogaster were used: wild-type Canton S, cry01—a null mutant of the circadian photoreceptor gene cryptochrome (Dolezelova et al., 2007), Pdf 0—a null mutant of the clock neuropeptide Pigment dispersing factor gene, per01—null mutant of the clock gene period, cry-GAL4.39 expressing the yeast transcription factor GAL4 under control of cry promoter, 21D-GAL4/MKRS expressing GAL4 in L2 interneurons, repo-GAL4 expressing GAL4 under control of repo (reversed polarity gene) promoter, a specific marker of glial cells, UAS-dicer2; UAS-itp-RNAi/MKRS, expressing interfering RNA for itp (ITP gene) and dicer2 which catalyses the first step of RNA interference, under control of the UAS sequence, and UAS-gfp expressing Green Fluorescent Protein (GFP) under control of the UAS sequence. The following crosses were carried out: cry-GAL4 × UAS-dicer2; UAS-itp-RNAi/MKRS; 21D-GAL4 × UAS-gfp and repo-GAL4 × UAS-gfp. The first generation of cry > itpRNAi strain has the itp expression silenced only in two cells: in one of the LNds and in the 5th small ventral lateral neuron (s-LNv). 21D > gfp and repo > gfp strains were used to visualize L2 and glial cells, respectively, with GFP.
Flies were maintained on a standard cornmeal medium under a light/dark condition of LD12:12 (12 h of light and 12 h of darkness) or constant DD and at constant temperature 24°C.
Laser Microdissection
Males, 7-days old, were decapitated at the following five time points: ZT1, ZT4, ZT13, ZT16, ZT20 (where ZT0 means light-on, ZT12 means light-off). In constant darkness flies were collected at CT1, CT4, CT13, CT16, CT20 under dim red light illumination. Approximately 30 flies were used for each time point and experiments were repeated at least three times. Heads were fixed in 96% ethanol for 45 min, then they were washed in phosphate buffer saline (PBS, pH 7.4) twice and cryoprotected by incubation in 12.5% sucrose for 10 min and in 25% sucrose at 4°C overnight. Next they were embedded in Tissue-Tek, frozen in liquid nitrogen, and cryostat 16 μm sections were cut and collected on special RNase, DNase—free membrane slides (PEN).
Laminas were selected, marked and dissected using an UV laser microdissector Leica LMD7000 (power: 15; aperture: 2; speed: 6; specimen balance 20; pulse frequency: 5000; S1 Figure). Specimens were collected using gravity into caps of microcentrifuge tubes containing a lysis buffer (NucleoSpin RNA XS kit, Macherey Nagel). Thirty laminas were collected for each time point. The retinas were cut manually from the optic lobe.
Cell Sorting
Males of the repo > gfp and 21D > gfp strains were decapitated at four time points: ZT1, ZT4, ZT13, and ZT16. Brains were isolated on ice and digested in trypsin-EDTA (Sigma) for 1 h in 37°C. Cells were centrifuged for 10 min at 10,000 rpm, after which the cell pellet was resuspended in PBS and incubated with Hoechst dye for 15 min. Then the cells were centrifuged for 10 min at 10,000 rpm and resuspended in PBS. A MoFlo XDP cytofluorimeter with a sorter (Beckman Coulter) was used for cell sorting (laser: 488 nm, 633 and 375 nm). Cells were sorted depending on size and granulation, Hoechst fluorescence and GFP signal. At least 1000 cells were isolated from one sample. 10 flies were used for each time point and experiments were repeated three times.
RNA Isolation and Real Time PCR
Total RNA isolation was performed using a NucleoSpin RNA XS kit (Macherey Nagel) according to the manufacturer’s protocol. 500 ng of total RNA was used for reverse transcription and then 1:8 diluted cDNA was used for quantitative PCR. Gene expression was examined using TaqMan Gene Expression Assays with 6′-FAM as a labeling molecule (Applied Biosystems) and 7500 Fast Real-Time PCR System (Applied Biosystems). The following genes were examined: period (per, FBgn0003068, Assay ID: Dm01843683_g1), timeless (tim, FBgn0014396; Assay ID: Dm01814244_g1), clock (clk, FBgn0023076; Assay ID: Dm01795381_g1), cryptochrome (cry, FBgn0025680; Assay ID: Dm02149911_m1) nervana2 (nrv2, FBgn0015777; Assay ID: Dm01803292_g1), sodium pump α-subunit (Atpα, FBgn0002921; Assay ID: Dm02154298_m1), bruchpilot (brp, FBgn0259246; and Assay ID: Dm01794336_g1). Expression of the Pdfr (FBgn0260753) gene was examined using SybrGreen qPCR (Applied Biosystem; forward primer: 5′ACGGCCGTGTTACAAGCCG, reverse primer: 5′GGAGAGGCAGAGGCCCACGA). Each experiment was repeated at least three times.
Data were collected as raw CT values and analyzed using the 2−ΔΔ CT method. Ribosomal protein 32 (rpl32, FBgn0002626; Assay ID: Dm02151827_g1 or forward primer: 5′AGAAGCGCAAGGAGATTGTC and reverse primer: 5′ATGGTGCTGCTATCCCAATC) was used as a reference gene. Gene expression was normalized on an arbitrary scale with ZT1 as 1.0.
Statistical analysis was performed using initially the non-parametric ANOVA Kruskal-Wallis test and then Tukey’s test. Statistica 7.0 software was used for the analyses. GraphPad Prism was used to prepare Figures.
Results
Gene Expression in the Retina
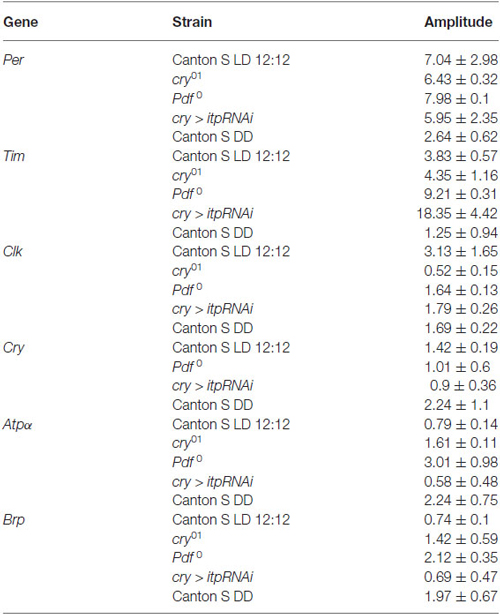
In the retina of wild-type flies maintained in LD 12:12, the expression of both core clock genes, per and tim, was cyclical with a peak reached at ZT16 but higher level of both gene mRNAs than in other time points was also detected at ZT13 (Figures 1A,B). In the cry01, Pdf 0 mutants and cry > itpRNAi strain, the daily expression of per and tim was maintained with the peak at ZT16, however, in contrast to Canton S the increase of mRNA levels at ZT13 was not observed. In all strain studied, except Canton S in LD 12:12 and Pdf 0, the amplitude of the per and tim mRNA rhythms was small (Table 1). The most striking changes in per and tim mRNA cycling were observed in the cry01 mutant and cry > itpRNAi strain, indicating that CRY and ITP are important for maintaining high levels of per and tim mRNAs during the night. In per01 mutant the expression of tim was arrhythmic (Figure 1B). In DD the pattern of per and tim gene expression in the retina of Canton S flies was similar to their expression in LD12:12 with a peak at CT16 but the amplitude of the rhythms was low (Figures 1A,B).
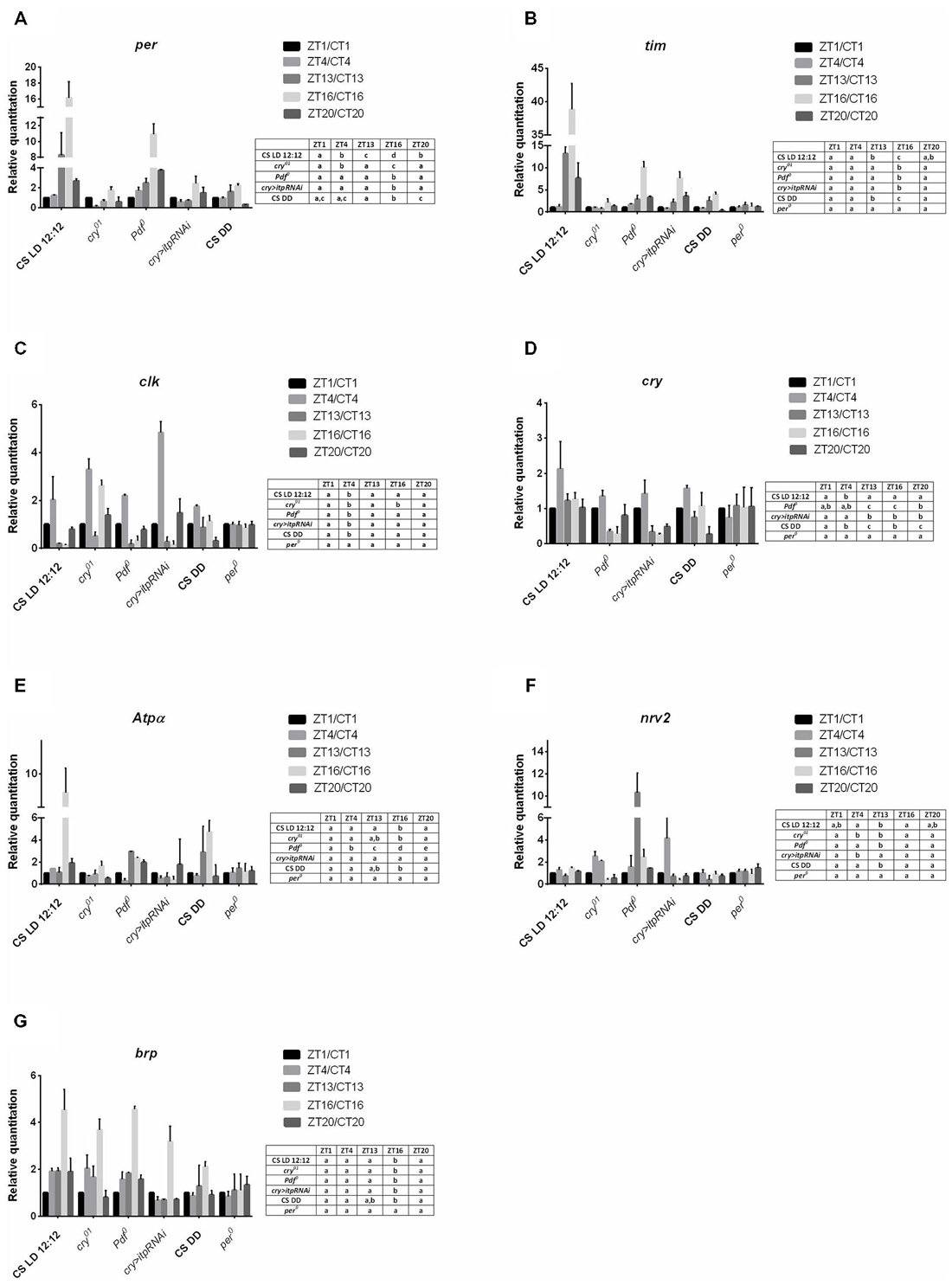
Figure 1. Gene expression in the retina of different strains of D. melanogaster. A—per gene; B—tim; C—clk, D—cry, E—Atpα, F—nrv2, G—brp. Tissues were collected at five time points: ZT1, ZT4, ZT13, ZT16, ZT20 in LD12:12 and CT1, CT4, CT13, CT16, CT20 in DD. Relative quantitation (RQ) determines the changes in steady-state mRNA level, data are normalized to ZT1 in LD 12:12 or CT1 in DD (value = 1.0). Gene expression was examined in LD 12:12 light conditions for Canton S, cry01, Pdf 0, cry>itp-RNAi, per01 and in constant darkness (DD) for Canton S. Statistical analyses were carried out to compare differences between time points for each genotype in the same condition. Statistically significant differences are indicated with small letters (A–D) in tables next to the figures. The same letter means no significant differences between those time points.
The expression of clk in the retina was also cyclic and the pattern was the same in Canton S, (in both LD 12:12 and DD conditions), Pdf 0 and cry > itpRNAi, reaching its peak at ZT4 (Figure 1C). However, this pattern was changed in the cry01 mutant. In addition to a peak of clk mRNA at ZT4, the second one was observed at ZT16. It suggests that CRY suppresses clk expression at ZT16. In per01 mutant the level of clk mRNA was the same at four time point studied.
In the retina, cry is regarded as a circadian photoreceptor and an element of the molecular circadian clock (Collins et al., 2006). In wild-type flies the cry expression was the highest at ZT4 and a similar level was observed at other time points (Figure 1D). In Pdf 0 and cry > itpRNAi strains, this peak was also observed, however, in Pdf 0 the second peak was at ZT20. The amplitude of the cry mRNA rhythm was similar in all the strains studied (Table 1). In per01 mutant cry expression was arrhythmic. In DD the rhythm was maintained with peaks at CT4 and CT16. The obtained results suggest that light and PDF, which is released from terminals of l-LNvs, affect cry expression in the retina.
The daily expression of two genes, Atpα and nrv2, encoding the α and β subunits of Na+/K+-ATPase, was detected in the retina but its pattern was specific for each gene (Figures 1E,F). The α subunit gene (Atpα) expression peaked at ZT16 while that of nrv2 showed two peaks at ZT4 and ZT16. However, at ZT1 and ZT20 the level of nrv2 mRNA was only slightly lower than at ZT4 and ZT16. In case of Atpα the pattern observed in Canton S was unchanged in the cry01 mutant (Figure 1E), but the amplitude of this rhythm was very low (Table 1). In contrast, the daily pattern of nrv2 mRNA was completely different in the cry01 mutant because the highest level of mRNA was at ZT4 and was still high at ZT13, when compared with other time points (Figure 1F). When both gene mRNAs were studied in the Pdf 0 and cry > itpRNAi strains their daily patterns were deemed to have changed. In the Pdf 0 mutant, the peak of AtpαmRNA was found at ZT13 and a high level of mRNA was also maintained throughout the rest of the night (Figure 1E). The amplitude of the AtpαmRNA rhythm in the cry01 and Pdf 0 flies was small (Table 1). In the cry > itpRNAi strain the rhythm of Atpα mRNA was abolished (Figure 1E). The mRNA of the nrv2 gene showed daily oscillations with a peak at ZT13 in the Pdf 0 mutants (Figure 1F). The pattern of this rhythm was different than that found in Canton S, and the cry01 and cry > itpRNAi strains. In cry > itpRNAi flies, the daily rhythm of nrv2 was also observed although the peak was determined at ZT4 and this pattern was different than that present in other strains (Figure 1F). In cry01, Pdf 0 and cry > itpRNAi, the amplitude of the nrv2 mRNA rhythm was larger than in Canton S (Table 1). In per01 mutant the expression of both Atpα and nrv2 genes was arrhythmic. In DD conditions nrv2 mRNA showed a trough at CT13 and Atpα had the similar pattern of expression as in LD 12:12 with a peak at CT16. The obtained results indicate that in the retina the expression of Atpα and nrv2 is cyclic and controlled by clock genes. Moreover CRY, PDF and ITP affect both rhythms.
The expression of brp gene in the retina of wild type flies was cyclic with the maximum found in the middle of the night at ZT16 and at CT16 in DD (Figure 1G). This pattern did not change in other experimental strains, except of per01 mutant. In this mutant the circadian rhythm of brp expression was abolished. The obtained results suggest that neither CRY nor neuropeptides PDF and ITP affect the circadian rhythm of brp expression in the retina.
Gene Expression in the Lamina
In wild type (Canton S) flies all the studied gene expression was detected in the lamina. All of them, except nrv2, also showed rhythmic mRNA changes in both LD 12:12 and DD. The expression of per and tim in the lamina was significantly higher during the night than during the day (Figures 2A,B) but the amplitude of per mRNA oscillations was high in comparing with that of tim (Table 2). In the cry01 mutant per and tim mRNA levels were the highest at ZT13 but the amplitude of the rhythm was high for per but low for tim. Moreover, in contrast to Canton S, the mRNA level of per was additionally high at ZT4 and low at ZT20 (Figure 2A). In the Pdf 0 mutant, which does not synthesize the clock neuropeptide PDF we observed a different pattern of per and tim expressions compared with wild-type flies. This strain showed maxima of per and tim mRNAs at ZT16 that were several times higher than at other time points (Figures 2A,B). After silencing itp using the cry-GAL4 > UAS-itpRNAi strain, the high level of per mRNA was found at ZT16 and ZT20 and of tim mRNA at ZT13, ZT16 and ZT20. The lack of PDF or reduced level of ITP strongly increased per and tim expressions during the night at ZT16. In both the Pdf 0 mutant and cry-GAL4 > UAS-itpRNAi strain, the daily pattern of per and tim expressions was similar in the lamina and retina. The expression of tim was arrhythmic in per01 mutant (Figure 2B). In DD conditions the expression of per and tim genes was similar as in LD 12:12 (Figures 2A,B) but the amplitude of oscillations was decreased (Table 2) in comparing with other strain studied.
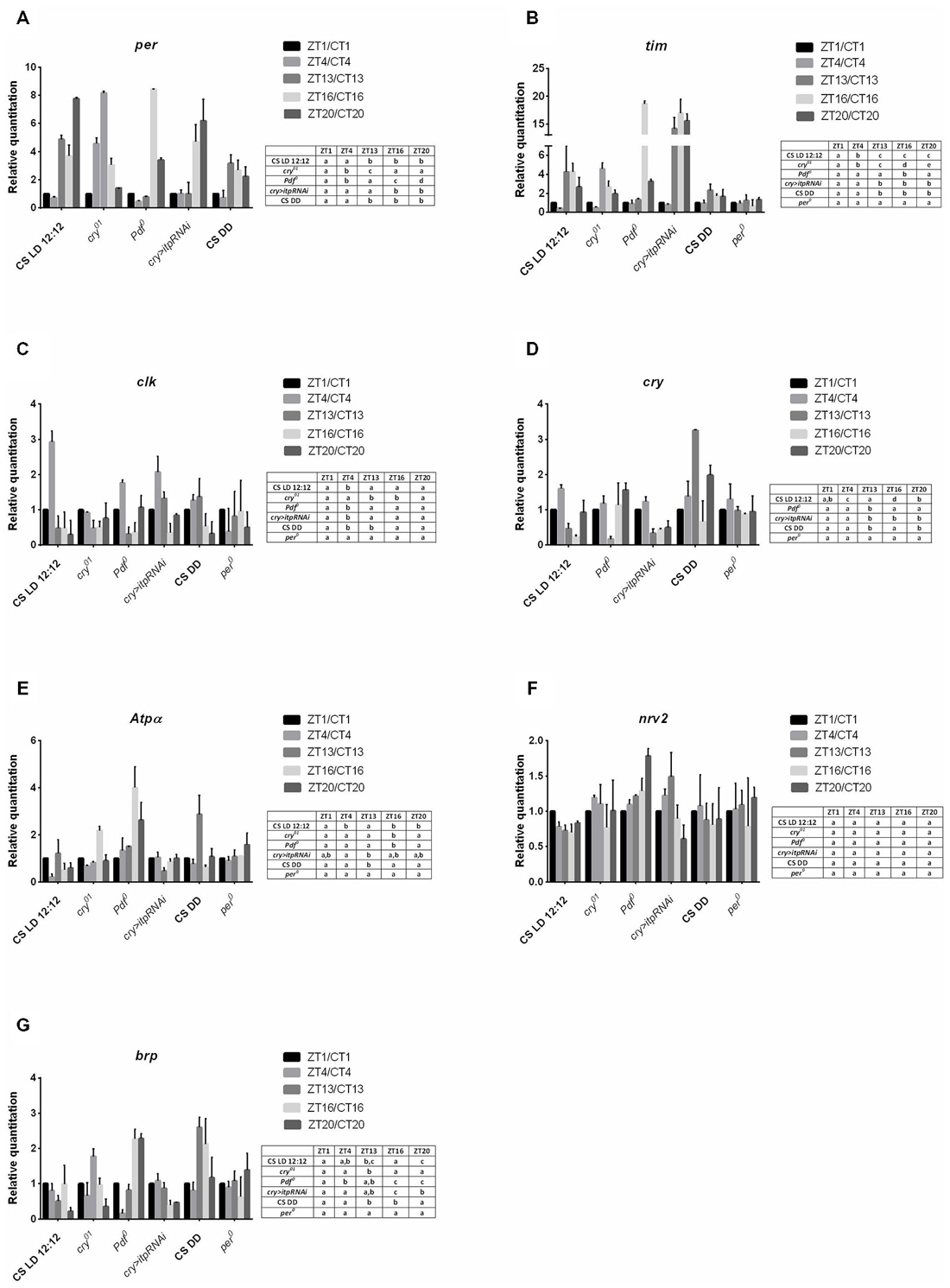
Figure 2. Gene expression in the lamina of different strains of D. melanogaster. A—per gene; B—tim; C—clk, D—cry, E—Atpα, F—nrv2, G—brp. Data are normalized to ZT1 or CT1 (value = 1.0). The following strains were examined in LD 12:12: Canton S, cry01, Pdf 0, cry > itp-RNAi, per01 and in DD: Canton S. Statistically significant differences are shown in tables next to the figures.
The clk expression was cyclic in the lamina of the wild-type, cry01, Pdf 0 and cry > itpRNAi flies. Moreover, the daily pattern of changes was similar in all strains studied, except the cry01 mutant. In Canton S, Pdf 0 and cry > itpRNAi, the mRNA level of this gene was the highest at ZT4 (Figure 2C) although the amplitude of the rhythms was lower in Pdf 0 and cry > itpRNAi than in Canton S (Table 2). In the cry01 mutant the clk expression pattern was completely changed. The higher level of mRNA was observed at ZT1, ZT4 and ZT20. The amplitude of this rhythm was low, since the mRNA level was only two times lower at ZT13 and ZT16 than at ZT1 (Table 2). The daily rhythm of clk expression was similar in the lamina and retina in all strains studied, except cry01. In per01 mutant the rhythm in changes of clk mRNA level was not detected. In DD mRNA level of clk was the highest at CT4 and CT13 and the amplitude of the circadian rhythm was low.
The mRNA level of cry also oscillates in the lamina in both LD 12:12 and DD. In wild-type flies the mRNA level of cry was the highest at ZT4, about nine times higher than in a trough at ZT16 (Figure 2D). In DD the expression pattern was changed with two peaks at CT13 and CT20 and similar level at CT1, CT4 and CT16. The amplitude of the rhythm was higher in DD than in LD 12:12. In the Pdf 0 mutant this pattern was different than in Canton S in LD 12:12 since the mRNA level was similar at the four time points studied but dropped significantly at ZT13. In turn in cry > itpRNAi flies the cry expression pattern was similar to that observed in wild-type flies with higher level of mRNA noted at ZT1 and ZT4. The daily pattern of cry expression was similar in the lamina and retina in Canton S and cry > itpRNAi flies but different in the Pdf 0 mutant. In per01 mutant the expression of cry was arrhythmic.
In addition to clock genes we also determined the expression of Atpα, nrv2 and brp genes in the lamina, since they are involved in rhythmic processes in this optic neuropil. Examination of the mRNA level of both Atpα and nrv2 genes in the lamina showed that Atpα mRNA oscillates during the day in Canton S, cry01, Pdf 0 and cry > itpRNAi flies (Figure 2E), but that the mRNA of nrv2 is constant (Figure 2F). In case of Atpα the expression displayed the daily rhythm with two, morning (ZT1) and evening (ZT13) peaks (Figure 2E) but in DD the peak at CT1 was not detected and the amplitude of the rhythm was higher than in LD 12:12 (Table 2). In cry01 and Pdf 0 mutants only one peak was observed at ZT16, while after silencing of itp in the clock neurons, the level of Atpα mRNA was the same during the day and night, except ZT13 where it significantly dropped when compared with ZT4. The amplitude of the rhythm was the highest in Pdf 0. In per01 mutant Atpα expression was arrhythmic.
The expression of the brp gene was rhythmic in the lamina. In wild-type flies brp mRNA was high at ZT1, ZT4 and ZT16 and in DD at CT13 and CT16 (Figure 2G). In cry01 the peak of brp mRNA was located at ZT13 and the amplitude of the rhythm was larger than in wild-type flies (Table 2). In the Pdf 0 mutant and after silencing itp, the pattern of brp expression was different than in Canton S. The highest level of brp mRNA in Pdf 0 was observed at the end of the night, at ZT16 and ZT20 while in the cry > itpRNAi strain the high level was during the day at ZT4. In per01 mutant brp mRNA level was similar at four time point studied.
Gene Expression in L2 Interneurons
In sorted cells labeled with GFP in 21D > gfp strain, the clock gene per was expressed but its mRNA level was constant during the day and night (Figure 3A). In turn tim expression was below the detection level during the day (ZT1, ZT4) but was higher during the night at ZT13 and ZT16 (Figure 3B). These results indicate the other functions of both clock genes in the lamina L2 monopolar cells rather than in photoreceptors and clock neurons. In turn the mRNA of the cry gene was not detected in L2 interneurons.
Figure 3. Gene expression in L2 interneurons labeled with GFP (21D > gfp strain). A—per gene; B—tim; C—Atpα, D—brp, E—Pdfr. Cells were isolated at four time points: ZT1, ZT4, ZT13, ZT16 in LD 12:12. RQ determines the changes in mRNA level and data are normalized to ZT1 (value = 1.0). Statistically significant differences are marked by different letters (A–C) above bars.
All non-clock genes, Atpα, brp and Pdfr—encoding PDF receptors, had cyclic expressions, although their mRNA daily patterns were specific for each gene (Figures 3C–E). The mRNA level of Atpα was the highest at ZT13 and the lowest at ZT16 (Figure 3C). The brp expression was rhythmic with the maximum detected at ZT4 (Figure 3D). The highest expression of Pdfr was observed at ZT1 and was more than two times higher than at other time points (Figure 3E). The mRNA of nrv2 was not detected in L2 cells.
Gene Expression in Glial Cells
We used the repo > gfp strain to obtain a purified population of glial cells from the brain of Drosophila by cell sorting. In these glial cells we detected oscillations of per and tim expression. The highest level of per mRNA was detected at ZT16 and the lowest at ZT4 (Figure 4A). This pattern was similar to that detected in the retina. The expression of tim peaked at ZT16 and was low during the day (ZT1, ZT4) and at the beginning of the night (ZT13; Figure 4B). The mRNA level of cry was very low, being almost at the limit of detection.
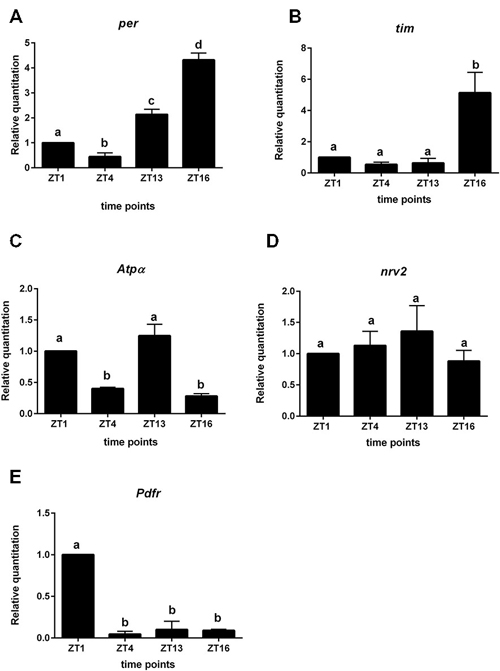
Figure 4. Gene expression in glial cells labeled with GFP (repo > gfp strain). A—per gene; B—tim; C—Atpα, D—nrv2, E—Pdfr. Cells were isolated at four time points: ZT1, ZT4, ZT13, ZT16 in LD 12:12. Data are normalized to ZT1 (value = 1.0). Statistically significant differences are marked by different letters (A–C) above bars.
The expression of Atpα was rhythmic in glial cells, with two peaks, at ZT1 and ZT13. This pattern was similar to that obtained for the lamina (Figure 4C). The nrv2 gene was expressed in glial cells but its expression was arrhythmic (Figure 4D). The mRNA of the Pdfr gene displayed the robust daily rhythm. The maximum of Pdfr mRNA was observed at ZT1 (Figure 4E), at the same time as in the L2 cells.
Discussion
Studying the expression of clock genes and potential ccgs, in two layers of nerve cells in the visual system and in two types of cells which show circadian rhythms in activity and structural plasticity (Chen et al., 1992; Pyza, 2010), we found that during the day the clock core genes, per and tim, and also clk and cry were cyclically expressed in all tissue and cells studied except for per in the L2 interneurons. In these cells the expression of per was arrhythmic, suggesting that it plays another non-clock function. Moreover, in the L2 cells, cry expression was not detected and tim expression was different to that of per and it was below the detection level during the day but present during the night. In our earlier studies, we found that the L2 dendritic tree morphology does not oscillate and its shape is different in the per01 mutant (Weber et al., 2009), suggesting that PER in these cells may regulate the morphology of dendrites but their circadian plasticity is probably controlled by the retina clock, the pacemaker and glial oscillators as for other rhythms in the lamina (Damulewicz et al., 2013; Górska-Andrzejak et al., 2013).
The cyclic expression of gene studied was also maintained in constant darkness and abolished in per01 mutant and this indicates that the expression of clock genes per, tim, clk, cry and ccgs atpα, brp is circadian in both the retina and lamina.
So far the expression of clock genes and other cycling transcripts have been studied only in whole head homogenates (Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Ceriani et al., 2002; Lin et al., 2002; Ueda et al., 2002) and in clock neurons, including LNs and DNs in the brain (Kula-Eversole et al., 2010; Nagoshi et al., 2010). However, the expression of clock genes in pacemaker cells or in the whole head does not explain how circadian rhythms in cellular processes and in structural plasticity, are regulated in different regions of the brain and in single neurons and glial cells. Our study is the first attempt to understand how circadian rhythms are regulated at the level of tissues and cells in the brain. Moreover, we successfully applied the laser microdissection method for the isolation of the lamina from the optic lobe of D. melanogaster, and also highlighted the fact that this method might be very valuable for other approaches as well.
In the fruit fly head homogenates, two peaks around ZT10 and ZT20 in the expression of per, tim and clk, cry have been detected (Ueda et al., 2002). In clock neurons the peak of per and tim mRNAs is at ZT16 (Kula-Eversole et al., 2010). In the present study, we found that the pattern of per and tim expression is similar in the retina and glial cells to that in the LNvs and maxima of per and tim expression are at ZT16 in LD 12:12 and CT16 in DD. In turn the expression of clk and cry peaks at ZT4/CT4. The rhythms of per and tim mRNAs in the retina were not affected by the lack of CRY, ITP and PDF, although their amplitudes were reduced. In contrast, the amplitude of the rhythm of clk was higher in cry01 and cry > itpRNAi than in Canton S flies. The lack of CRY and ITP increases per and tim expression at the end of the night and at the beginning of the day. It means that accumulation of CRY during the night in the photoreceptors, represses per and tim transcription at the end of the night and the beginning of the day, probably by inhibiting the clk expression at ZT16. These results show that CRY might be a repressor of the molecular clock in the retina, as it has already been reported for peripheral oscillators (Collins et al., 2006). It is also possible that in the cry01 mutant the molecular oscillators are not entrained and because of their desynchronisation the amplitude of oscillations is low. The expression of cry in the retina was only slightly changed by the lack of PDF and ITP by increasing the mRNA of cry during the night in the Pdf 0 mutant and after the silencing of itp. The obtained results confirm the finding of other authors suggesting that the retina possesses an autonomous clock (Cheng and Hardin, 1998).
In the lamina, the clock genes also have a cyclic expression but the daily patterns of their expression are different than in the retina. Moreover, the rhythms of per and tim expression are regulated by both PDF and ITP and in case of per, but not tim, they changed in the phase in the cry01 mutant.
The similar daily expression in the lamina and retina was found only in the case of clk and cry. The rhythm of cry expression in the lamina is affected by PDF but its mRNA level was very low in glial cells and not detected in the L2 cells. It suggests that the clock in the glial cell may not require CRY, but that expression of cry in the lamina may originate from the photoreceptor terminals in the lamina. In turn, the per and tim mRNA rhythms in the lamina may reflect the expression of per and tim in the epithelial glial cells that is modified by specific per and tim expression in neurons, for example in the L2 monopolar cells.
In contrast to clock genes, the daily rhythms of Atpα and nrv2 expression in the retina are controlled by the pacemaker, since in the Pdf 0 mutant and after silencing itp, the pattern of both rhythms was changed or the rhythm was disrupted as in the case of Atpα in cry > itpRNAi flies. The rhythm of the sodium pump α subunit confirms that the activity of this ion pump is cyclically regulated, not only in the lamina, but also in the retina by changes to the Atpα expression. In turn, the rhythmic expression of nrv2 only in the retina, suggests that this protein might be involved in other cyclic functions of Na+/K+-ATPase rather than in ion pumping. It has been reported that Na+/K+-ATPase participates in the formation of cell junctions and the β subunit forms bridges between cells (Paul et al., 2007; Vagin et al., 2012). In contrast, in the lamina, glial and L2 cells, only the expression of Atpα was rhythmic and the pattern of this rhythm was similar to the daily rhythm of the ATPα protein detected in the lamina glia (Damulewicz et al., 2013). However, the expression of Atpα was different in the L2 cells. The mRNA of Atpα and ATPα protein peak at ZT13 in neurons, while in the glia they peak twice at ZT1 and ZT13 (Damulewicz et al., 2013). In the lamina the expression of all genes studied was changed in cry01, Pdf 0 mutants and in cry > itpRNAi line, indicating that rhythms in the lamina are controlled by several oscillators, located in the retina, the pacemaker and in the lamina itself. Moreover, PDF and ITP neuropeptides are transmitters of circadian information from the pacemaker to the lamina. It confirms our earlier studies which showed that the circadian rhythms of two proteins in the lamina, the α subunit of Na+/K+-ATPase and BRP, are also regulated by these three types of oscillators (Damulewicz et al., 2013; Górska-Andrzejak et al., 2013). In addition, the oscillation of ATPα level is affected by the lack of both PDF and ITP.
Another ccg that was studied in the retina and lamina was brp. The presynaptic protein BRP, encoded by this gene, shows the circadian rhythm in abundance in the lamina, with two peaks, in the morning and in the evening (Górska-Andrzejak et al., 2013). Here, we found that brp expression oscillates in both the retina and lamina, however, the pattern is different than that found in the case of BRP protein. This can be explained by the fact that the protein was measured only in tetrad synapses in the lamina, while mRNA was examined in all lamina neurons. In the retina, brp mRNA was highest at ZT16 and this rhythm was unchanged in the cry01 mutant and by the lack of PDF and ITP. In the lamina, however, the daily pattern of brp expression was very different when CRY, PDF or ITP were eliminated or as in the case of ITP decreased.
In two selected cell types, namely: L2 interneurons and glial cells, we detected the robust rhythm of Pdfr mRNA with a peak at ZT1, suggesting that this gene is another ccg and that the rhythms in the lamina, at least in the L2, and in glial cells, are controlled by PDF and the cyclic expression of its receptors.
In the present study, we showed that clock genes are expressed differently in the retina and in the first optic neuropil (the lamina), of the brain optic lobe.
Moreover, we showed that cells which do not contain circadian clocks cyclically express genes and show circadian rhythms in cellular processes. In both cell types, L2 interneurons and glial cells, the α subunit of the sodium pump shows the cyclic expression. It suggests that many cellular processes in neurons and glial cells might be controlled over time by the cyclic activity of the sodium pump. In addition to cellular processes in neurons and glia, synaptic transmission is also controlled by the clock, since expression of brp and level of BRP (Górska-Andrzejak et al., 2013) as well as other genes and their proteins in synapses, oscillate during a 24 h period (Krzeptowski et al., 2014).
On the basis of our results and the findings of other authors (Gorostiza et al., 2014), we suggest that the pacemaker controls the circadian rhythms in the brain by cyclic remodeling of synaptic contacts, between the clock and target neurons. However, in distant brain tissues, as in the lamina, circadian rhythms are regulated by neuropeptides released from the pacemaker neurons and by the daily expression of PDF receptors in neurons and glial cells. Moreover, peripheral oscillators, as glial cells, locally shape the circadian rhythms of the structure of neurons and synapses in particular brain regions and cell types.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the following scientists for their kind donation of fly strains: Dr. E. Rosato (University of Leicester, Great Britain) for repo-GAL4, Dr. C. Helfrich-Förster (University of Würzburg, Germany) for Pdf 0 mutant and UAS-dicer2; UAS-itp-RNAi/MKRS, Dr. F. Rouyer (CNRS, France) for cry-GAL4.39 and Dr. T. Raabe (University of Würzburg, Germany) for 21D-GAL4/MKRS. This work was supported by the Polish Ministry of Science and High Education (grant no 56/7.PR/2014) and by the European FP7-People-2012-ITN INsecTIME project nr 316790 to EP. The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficiary of the structural funds from the European Union and the Polish Ministry of Science and Higher Education (grants No: POIG.02.01.00–12–064/08 and 02.02.00–00–014/08) and is a partner of the Leading National Research Center (KNOW) supported by the Ministry of Science and Higher Education. The study is exempt from the requirement of approval by the Animal Care and Use Committee of the Jagiellonian University.
References
Ceriani, M. F., Hogenesch, J. B., Yanovsky, M., Panda, S., Straume, M., and Kay, S. A. (2002). Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22, 9305–9319.
Chen, D. M., Christianson, J. S., Sapp, R. J., and Stark, W. S. (1992). Visual receptor cycle in normal and period mutant Drosophila: microspectrophotometry, electrophysiology and ultrastructural morphometry. Vis. Neurosci. 9, 125–135. doi: 10.1017/s0952523800009585
Cheng, Y., and Hardin, P. E. (1998). Drosophila photoreceptors contain an autonomous circadian oscillator that can function without period mRNA cycling. J. Neurosci. 18, 741–750.
Claridge-Chang, A., Wijnen, H., Naef, F., Boothroyd, C., Rajewsky, N., and Young, M. W. (2001). Circadian regulation of gene expression system in the Drosophila head. Neuron 32, 657–671. doi: 10.1016/s0896-6273(01)00515-3
Collins, B., Mazzoni, E. O., Stanewsky, R., and Blau, J. (2006). Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 16, 441–449. doi: 10.1016/j.cub.2006.01.034
Damulewicz, M., and Pyza, E. (2011). The clock input to the first optic neuropil of Drosophila melanogaster expressing neuronal circadian plasticity. PLoS One 6:e21258. doi: 10.1371/journal.pone.0021258
Damulewicz, M., Rosato, E., and Pyza, E. (2013). Circadian regulation of the Na+/K+-ATPase alpha subunit in the visual system is mediated by the pacemaker and by retina photoreceptors in Drosophila melanogaster. PLoS One 8:e73690. doi: 10.1371/journal.pone.0073690
Dolezelova, E., Dolezel, D., and Hall, J. C. (2007). Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics 177, 329–345. doi: 10.1534/genetics.107.076513
Gorostiza, E. A., Depetris-Chauvin, A., Frenkel, L., Pírez, N., and Ceriani, M. F. (2014). Circadian pacemaker neurons change synaptic contacts across the day. Curr. Biol. 24, 2161–2167. doi: 10.1016/j.cub.2014.07.063
Górska-Andrzejak, J. (2013). Glia-related circadian plasticity in the visual system of Diptera. Front. Physiol. 4:36. doi: 10.3389/fphys.2013.00036
Górska-Andrzejak, J., Keller, A., Raabe, T., Kilianek, L., and Pyza, E. (2005). Structural daily rhythms in GFP-labelled neurons in the visual system of Drosophila melanogaster. Photochem. Photobiol. Sci. 4, 721–726. doi: 10.1039/b417023g
Górska-Andrzejak, J., Makuch, R., Stefan, J., Görlich, A., Semik, D., and Pyza, E. (2013). Circadian expression of the presynaptic active zone protein bruchpilot in the lamina of Drosophila melanogaster. Dev. Neurobiol. 73, 14–26. doi: 10.1002/dneu.22032
Jackson, F. R. (2011). Glial cell modulation of circadian rhythms. Glia 59, 1341–1350. doi: 10.1002/glia.21097
Johard, H. A., Yoishii, T., Dircksen, H., Cusumano, P., Rouyer, F., Helfrich-Förster, C., et al. (2009). Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J. Comp. Neurol. 516, 59–73. doi: 10.1002/cne.22099
Krzeptowski, W., Górska-Andrzejak, J., Kijak, E., Görlich, A., Guzik, E., Moore, G., et al. (2014). External and circadian inputs modulate synaptic protein expression in the visual system of Drosophila melanogaster. Front. Physiol. 5:102. doi: 10.3389/fphys.2014.00102
Kula-Eversole, E., Nagoshi, E., Shang, Y., Rodriguez, J., Allada, R., and Rosbash, M. (2010). Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc. Natl. Acad. Sci. U S A 107, 13497–13502. doi: 10.1073/pnas.1002081107
Lin, Y., Han, M., Shimada, B., Wang, L., Gibler, T. M., Amarakone, A., et al. (2002). Influence of the period-dependent circadian clock on diurnal, circadian and aperiodic gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A 99, 9562–9567. doi: 10.1073/pnas.132269699
McDonald, M. J., and Rosbash, M. (2001). Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107, 567–578. doi: 10.1016/s0092-8674(01)00545-1
Meinertzhagen, I. A., and Hanson, T. E. (1993). “The development of the optic lobe,” in The Development of Drosophila Melanogaster, eds M. Bate and A. M. Arias (New York: Cold Spring Harbor Laboratory), 1363–1491.
Meinertzhagen, I. A., and O’Neil, S. D. (1991). Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J. Comp. Neurol. 305, 232–263. doi: 10.1002/cne.903050206
Meinertzhagen, I. A., and Sorra, K. E. (2001). Synaptic organization in the fly’s optic lamina: few cells, many synapses and divergent microcircuits. Prog. Brain Res. 131, 53–69. doi: 10.1016/s0079-6123(01)31007-5
Nagoshi, E., Sugino, K., Kula, E., Okazaki, E., Tachibana, T., Nelson, S., et al. (2010). Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat. Neurosci. 13, 60–68. doi: 10.1038/nn.2451
Paul, S. M., Palladino, M. J., and Beitel, G. J. (2007). A pump-independent function of the Na,K-ATPase is required for epithelial junction function and tracheal tube-size control. Development 134, 147–155. doi: 10.1242/dev.02710
Pyza, E. (2010). “Circadian rhythms in the fly’s visual system,” in Encyclopedia of the Eye (Vol. 1), ed A. D. Darlene (Oxford: Academic Press), 302–311.
Pyza, E., and Górska-Andrzejak, J. (2004). Involvement of glial cells in rhythmic size changes in neurons of the housefly’s visual system. J. Neurobiol. 59, 205–215. doi: 10.1002/neu.10307
Pyza, E., and Meinertzhagen, I. A. (1993). Daily and circadian rhythms of synaptic frequency in the first visual neuropile of the housefly’s (Musca domestica L.) optic lobe. Proc. Biol. Sci. 254, 97–105. doi: 10.1098/rspb.1993.0133
Pyza, E., and Meinertzhagen, I. A. (1995). Monopolar cell axons in the first optic neuropil of the housefly, Musca domesticaL., undergo daily fluctuations in diameter that have a circadian basis. J. Neurosci. 15, 407–418.
Pyza, E., and Meinertzhagen, I. A. (1999). Daily rhythmic changes of cell size and shape in the first optic neuropil in Drosophila melanogaster. J. Neurobiol. 40, 77–88. doi: 10.1002/(sici)1097-4695(199907)40:1<77::aid-neu7>3.0.co;2-0
Siwicki, K. K., Eastman, C., Petersen, G., Rosbash, M., and Hall, J. C. (1988). Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1, 141–150. doi: 10.1016/0896-6273(88)90198-5
Stanewsky, R., Kaneko, M., Emery, P., Beretta, B., Wager-Smith, K., Kay, S. A., et al. (1998). The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692. doi: 10.1016/s0092-8674(00)81638-4
Ueda, H. R., Matsumoto, A., Kawamura, M., Iino, M., Tanimura, T., and Hashimoto, S. (2002). Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J. Biol. Chem. 277, 14048–14052. doi: 10.1074/jbc.c100765200
Vagin, O., Dada, L. A., Tokhtaeva, E., and Sachs, G. (2012). The Na-K-ATPase α1β1 heterodimer as a cell adhesion molecule in epithelia. Am. J. Physiol. Cell Physiol. 302, C1271–C1281. doi: 10.1152/ajpcell.00456.2011
Weber, P., Kula-Eversole, E., and Pyza, E. (2009). Circadian control of dendrite morphology in the visual system of Drosophila melanogaster. PLoS One 4:e4290. doi: 10.1371/journal.pone.0004290
Yoshii, T., Todo, T., Wülbeck, C., Stanewsky, R., and Helfrich-Förster, C. (2008). Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J. Comp. Neurol. 508, 952–966. doi: 10.1002/cne.21702
Keywords: circadian rhythms, photoreceptors, visual interneurons, glial cells, BRP synaptic protein, sodium pump, PDF receptors
Citation: Damulewicz M, Loboda A, Bukowska-Strakova K, Jozkowicz A, Dulak J and Pyza E (2015) Clock and clock-controlled genes are differently expressed in the retina, lamina and in selected cells of the visual system of Drosophila melanogaster. Front. Cell. Neurosci. 9:353. doi: 10.3389/fncel.2015.00353
Received: 14 April 2015; Accepted: 24 August 2015;
Published: 15 September 2015.
Edited by:
Dieter Wicher, Max Planck Institute for Chemical Ecology, GermanyReviewed by:
Sebastian Kadener, The Hebrew University of Jerusalem, IsraelRodolfo Costa, University of Padova, Italy
Copyright © 2015 Damulewicz, Loboda, Bukowska-Strakova, Jozkowicz, Dulak and Pyza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution and reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elzbieta Pyza, Department of Cell Biology and Imaging, Institute of Zoology, Faculty of Biology and Earth Sciences, Jagiellonian University, Gronostajowa 9, 30-387 Krakow, Poland,ZWx6YmlldGEucHl6YUB1ai5lZHUucGw=
 Milena Damulewicz
Milena Damulewicz Agnieszka Loboda
Agnieszka Loboda Karolina Bukowska-Strakova
Karolina Bukowska-Strakova Alicja Jozkowicz2
Alicja Jozkowicz2 Jozef Dulak
Jozef Dulak Elzbieta Pyza
Elzbieta Pyza