- 1State Key Laboratory of Cognitive Neuroscience and Learning and IDG/McGovern Institute for Brain Research, Beijing Normal University, Beijing, China
- 2Center for Cognition and Brain Disorders, Hangzhou Normal University, Hangzhou, China
- 3Zhejiang Key Laboratory for Research in Assessment of Cognitive Impairments, Hangzhou, China
- 4McConnell Brain Imaging Center, Montreal Neurological Institute, Montreal, QC, Canada
A Corrigendum on
GRETNA: a graph theoretical network analysis toolbox for imaging connectomics
by Wang, J., Wang, X., Xia, M., Liao, X., Evans, A., and He, Y. (2015). Front. Hum. Neurosci. 9:386. doi: 10.3389/fnhum.2015.00386
Here, we would like to correct the two points as follows.
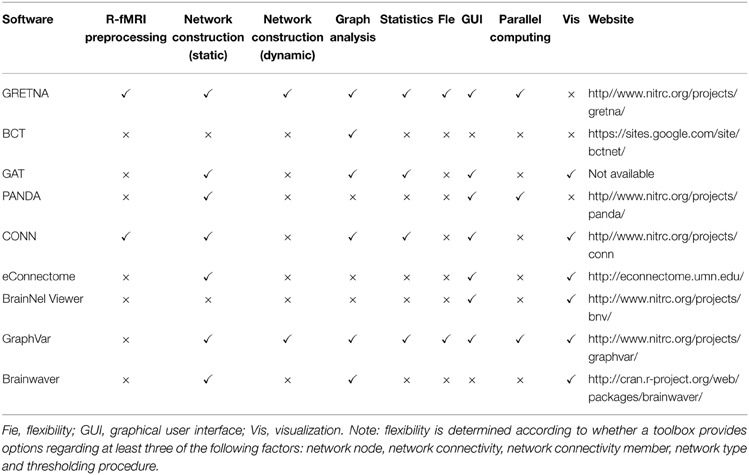
1) The updated Table 1:
2) Discussion: “Specifically, compared with the recent developed GraphVar (Kruschwitz et al., 2015), GRETNA has distinct features in parallel computing, capability to preprocess R-fMRI data.”
We would like to further clarify the description as
“Specifically, compared with the recent developed GraphVar (Kruschwitz et al., 2015), GRETNA has distinct features in parallel computing. The GraphVar (beta v0.611) can assign several jobs to different CPUs by calling Matlab's parallel computing toolbox. The GRETNA can assign parallel tasks by calling the PSOM toolbox (Bellec et al., 2012), which helps GRETNA to record and manage the data generated during fMRI preprocessing or graph-based network analyses and to restart the pipeline from the failure steps.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bellec, P., Lavoie-Courchesne, S., Dickinson, P., Lerch, J. P., Zijdenbos, A. P., and Evans, A. C. (2012). The pipeline system for Octave and Matlab (PSOM): a lightweight scripting framework and execution engine for scientific workflows. Front. Neuroinform. 6:7. doi: 10.3389/fninf.2012.00007
Keywords: network, graph theory, connectome, resting fMRI, small-world, hub
Citation: Wang J, Wang X, Xia M, Liao X, Evans A and He Y (2015) Corrigendum: GRETNA: a graph theoretical network analysis toolbox for imaging connectomics. Front. Hum. Neurosci. 9:458. doi: 10.3389/fnhum.2015.00458
Received: 20 July 2015; Accepted: 03 August 2015;
Published: 19 August 2015.
Edited by:
Wei Gao, Cedars-Sinai Medical Center, USAReviewed by:
Qingbao Yu, The Mind Research Network, USACopyright © 2015 Wang, Wang, Xia, Liao, Evans and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong He,eW9uZy5oZUBibnUuZWR1LmNu
 Jinhui Wang
Jinhui Wang Xindi Wang
Xindi Wang Mingrui Xia
Mingrui Xia Xuhong Liao
Xuhong Liao Alan Evans
Alan Evans Yong He
Yong He