- Department of Functional Anatomy and Neuroscience, Graduate School of Medicine, Nagoya University, Nagoya, Japan
Peripheral nervous system (PNS) neurons survive and regenerate after nerve injury, whereas central nervous system (CNS) neurons lack the capacity to do so. The inability of the CNS to regenerate presumably results from a lack of intrinsic growth activity and a permissive environment. To achieve CNS regeneration, we can learn from successful nerve regeneration in the PNS. Neurons in the PNS elicit dynamic changes in gene expression in response to permissive environmental cues following nerve injury. To switch gene expression on and off in injured neurons, transcription factors and their networks should be carefully orchestrated according to the regeneration program. This is the so-called “intrinsic power of axonal growth.” There is an increasing repertoire of candidate transcription factors induced by nerve injury. Some of them potentiate the survival and axonal regeneration of damaged neurons in vivo; however, our knowledge of transcriptional events in injured neurons is still limited. How do these transcription factors communicate with each other? How does the transcriptional machinery regulate the wide variety of regeneration-associated genes (RAGs) in the properly coordinated manner? In this review, we describe our current understanding of the injury-inducible transcriptional factors that enhance the intrinsic growth capacity, and propose a potential role for specificity protein 1 (Sp1), which provides a platform to recruit injury-inducible transcription factors, in simultaneous gene regulation. Finally, we discuss an additional mechanism that is involved in epigenetic modifications in damaged neurons. A comprehensive understanding of the nuclear events in injured neurons will provide clues to clinical interventions for successful nerve regeneration.
Introduction
Unlike neurons of the peripheral nervous system (PNS), the central nervous system (CNS) neurons are unable to survive and regenerate after nerve injury (Filbin, 2003; Harel and Strittmatter, 2006). This serious problem has led to many studies being focused on how to drive axon elongation in the CNS. However, the molecular mechanisms underlying successful nerve regeneration in the PNS remains to be elucidated. It will be difficult to overcome the problem in the CNS without a basic understanding of the exact mechanisms underlying successful nerve regeneration. From this respect, an understanding of nerve regeneration in the PNS is of both basic and potential clinical interest.
There are two major determinants of successful nerve regeneration, intrinsic growth ability and an extrinsic permissive environment (Chen et al., 2007; Raivich and Makwana, 2007). Injured neurons switch the expression of regeneration-associated genes (RAGs) on and off according to the regeneration program (Raivich and Makwana, 2007; Sun and He, 2010). We refer to this process as “intrinsic growth ability.” The activation of intrinsic growth capacity depends on the existence of a local permissive environment, which consists of the molecular network organized by Schwann cells and macrophages at the injury site, and by astrocytes and microglia around the cell bodies (Figure 1; Leon et al., 2000; Moran and Graeber, 2004; Raivich, 2005). This environment produces neurotrophic factors such as nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), glial cell line-derived factor (GDNF), and neurotrophin-3 (NT-3) (Lindholm et al., 1987; Yan et al., 1992; Funakoshi et al., 1993; Li et al., 1994; Naveilhan et al., 1997); cytokines such as leukemia inhibitory factor (LIF), ciliary neurotrophic factor (CNTF), interleukin-6 (IL-6), and chemokines (Sendtner et al., 1990; Banner and Patterson, 1994; Curtis et al., 1994; Murphy et al., 1995; Cafferty et al., 2004; Cao et al., 2006); chemoattractants like chemokine (Namikawa et al., 2006; Yin et al., 2006; Gamo et al., 2008); neuropeptides like calcitonin gene-related peptide (CGRP), galanin, and vasoactive intestinal peptide (VIP) (Shadiack et al., 2001); cell adhesion molecules like CD44, integrin, and L1 (Nieke and Schachner, 1985; Jones et al., 1997; Kloss et al., 1999; Werner et al., 2000); and numerous extracellular matrix (ECM) molecules (Araki and Milbrandt, 2000; Murakami et al., 2006). On the contrary, in the non-permissive environment in the CNS, astrocytes form a physical barrier to growth and oligodendrocytes generate molecules that are highly inhibitory to axon growth (Silver and Miller, 2004; Benowitz and Yin, 2008; Shen et al., 2009; Lee et al., 2010). In the PNS, nerve injury signals from the permissive environment are transduced via cell surface receptors of the cell body and via retrograde axonal transport (Hanz et al., 2003; Cavalli et al., 2005; Yudin et al., 2008), and then activate intracellular cascades like the mitogen-activated protein kinase (MAPK)pathway and the phosphoinositide 3-kinase (PI3K) pathway in injured neurons (Kiryu et al., 1995a; Liu et al., 1997; Namikawa et al., 2000; Markus et al., 2002; Zhou and Snider, 2005; Park et al., 2008; Hammarlund et al., 2009). Ultimately, the signals are integrated into specific regulators of the nuclear transcription factors that induce the expression of large ensembles of distinct genes sequentially and under tight control. The repertoire of transcription factors that are induced by nerve injury is increasing. They include c-Jun, sox11, CREB, Smad1, ATF3, AKRD1, NFIL3, p53, STAT3, C/EBPβ, and several KLF family members (Herdegen et al., 1997; Tsujino et al., 2000; Schweizer et al., 2002; Nakagomi et al., 2003; Gao et al., 2004; Raivich et al., 2004; Kiryu-Seo et al., 2005; Nadeau et al., 2005; Qiu et al., 2005; Jankowski et al., 2006; Okuyama et al., 2007; Moore et al., 2009; Zou et al., 2009). Cox et al. have reported that CREB is locally synthesized in growth cone and retrogradely transported to the cell body (Cox et al., 2008). Axonally derived transcription factors may be involved in a specific response to the injury site.
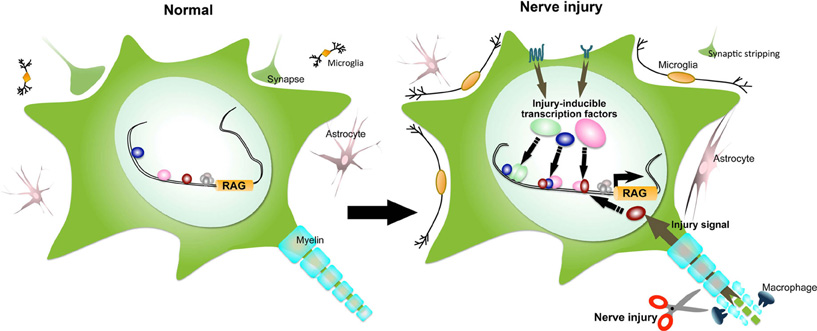
Figure 1. Peripheral nerve injury elicits a dynamic change in the transcription of regeneration-associated genes (RAGs) to enhance intrinsic growth capacity in neurons. The transcription factors involved in nerve regeneration are expressed at low-level in normal neurons. RAGs are expressed at low-level or switched off. Upon axotomy, microglial cells and subsequent astrocytes surround the cell body of injured neuron. Synaptic terminals are detached from the cell body. At the injury site, Schwann cells undergo dedifferentiation and proliferation, and Schwann cells and macrophages rapidly remove myelin debris. The local permissive environment provides the injury signals via cell surface receptors of the cell body and via retrograde axonal transport from growth cone. The signals induce the expression of numerous transcription factors, modify them at the post-translational level and lead to form various types of transcriptional complex, to promote the RAGs expression (See main text for details).
Recent large-scale screening approaches using a combination of microarrays with phosphoproteomics have identified 39 injury-inducible transcription factors (Michaelevski et al., 2010), while the comparative transcriptome microarray analysis of peripheral versus central nerve injured models of dorsal root ganglion (DRG) neurons found 30 candidate transcription factors (Stam et al., 2007). These injury-inducible transcription factors are presumed to control hundreds of transcriptional targets. It has been established that the 5′-flanking sequences of some RAGs such as an important regulator of growth cone motility, GAP43 (Kobayashi et al., 1994), intermediate filament, peripherin (Terao et al., 2000), and cytoskeletal protein, α-tubulin (Tetzlaff et al., 1988), have injury-responsive characteristics (Belecky-Adams et al., 1993; Gloster et al., 1994; Vanselow et al., 1994; Uveges et al., 2002). Recent genome-wide screening of gene promoters identified multiple potential target genes (MacGillavry et al., 2011). However, our knowledge of the targets is limited.
A principal goal in the field has been to identify the master regulators that coordinate the regenerative program, and to understand the global gene regulation processes that drive intrinsic growth ability. It is required to know how such a large number of transcription factors interplay with each other to regulate RAGs synchronously, and how global and dynamic changes in gene expressions are controlled. In this review, we describe the current understanding of the transcription factors associated with nerve regeneration and highlight our finding of the specificity protein 1 (Sp1)-dependent transcriptional machinery as a result of its interactions with injury-inducible transcription factors. This finding has raised several unanswered questions to be addressed in the future. Finally, we discuss the possible involvement of epigenetic regulation in nerve regeneration, including chromatin remodeling and DNA methylation.
Transcription Factors Involved in Nerve Regeneration
The physiological relevance of both injury-inducible transcription factors and their potential targets awaits in vivo study. Recent exciting work using knockout and transgenic mice is uncovering their relevance, as shown in Table 1. C/EBPβ knockout mice showed reduced expression of RAGs such as α-tubulin and GAP-43 after facial nerve injury (Nadeau et al., 2005). Deletion of the tumor suppressor p53 rescued the motor neuron death after hypoglossal nerve injury by preventing expression of the proapoptotic gene Noxa (Kiryu-Seo et al., 2005). However, Giovanni et al. demonstrated that the role of p53 is predominantly one of axonal regeneration rather than cell survival after facial nerve axotomy (Di Giovanni et al., 2006). It is possible that p53 has a dual role, because p53 is a multifunctional and stress-sensitive protein that initiates numerous transcriptional cascades in response to nerve injury. The absence of Krüppel-like factor-4 (KLF4) in retinal ganglion cells modestly enhanced axonal regeneration after optic nerve injury, suggesting that KLF4 is a negative regulator of intrinsic growth ability (Moore et al., 2009). Gao et al. have shown that the injection of the adenovirus containing constitutively active CREB into the DRG enhanced nerve regeneration, although these authors did not use a genetically modified mouse (Gao et al., 2004). All of these studies have provided valuable in vivo information about the gene regulation events that elevate intrinsic growth ability. Furthermore, the prominent effects of c-Jun, ATF3, and STAT3 on nerve regeneration have been particularly well examined. Here, we summarize the current understanding of the roles of these three injury-inducible transcription factors in nerve regeneration.
C-Jun
c-Jun is a major component of the activator protein-1 (AP-1) family of transcription factors and forms homo- and heterodimers together with Jun family proteins (c-Jun, JunB, and JunD) and Fos proteins (c-fos, FosB, Fra-1, and Fra-2) (Herdegen and Leah, 1998). Many Jun family proteins depend on the activation of N-terminal kinases (JNKs) and are involved in a large number of cellular functions. N-terminal phosphorylated c-Jun is rapidly accumulated in the nucleus where it promotes neuronal death (Herdegen et al., 1997). This phenomenon is observed during embryonic development as well as in a variety of nerve injury models (Herdegen et al., 1997; Raivich et al., 2004; Lindwall and Kanje, 2005). Thus, overexpression of c-Jun leads to cell death by inducing propapoptotic genes such as Bim, whereas c-Jun suppression or dominant-negative c-Jun expression prevents neuronal death (Ham et al., 1995; Whitfield et al., 2001; Palmada et al., 2002). Peripheral nerve injury induces, c-Jun expression, suggesting its involvement in nerve regeneration (Raivich et al., 2004). However, the situation is not so simple. Neuronal death is sometimes induced even by peripheral nerve axotomy. This phenomenon is more augmented in mice than in rats (Kiryu-Seo et al., 2005, 2006). Using this feature, Raivich et al. demonstrated that c-Jun seems to have a dual role like p53. They established mice with nestin-Cre-mediated neuronal deletion of c-Jun. The c-Jun-negative motor neurons blocked neuronal death after facial axotomy, although they still showed severe neuronal atrophy. Despite showing inhibition of neuronal death, the mice showed strongly diminished speed of target innervation regeneration after nerve injury. Furthermore, they showed reduced expression of several RAGs including cell adhesion molecules such as integrin α7β1 and CD44 (Jones et al., 1997; Werner et al., 2000) and neuropeptides such as galanin (Shadiack et al., 2001). This finding reinforces the notion that c-Jun has an important role in turning on the regeneration program after injury. This does not exclude the possibility that a lack of c-Jun affects non-neuronal cells in these mice in addition to neurons, because the nestin-Cre-mediated deletion of c-Jun also occurs in glial cells. c-Jun can drive Schwann cell dedifferentiation and is upregulated in non-neuronal cells like Schwann cells after axotomy. In line with this fact, mice with conditional inactivation of c-Jun in Schwann cells showed strikingly delayed myelin loss after sciatic nerve injury (Parkinson et al., 2008). Because rapid removal of myelin enables axon regeneration in the PNS (Vargas and Barres, 2007), these mice would be expected to have compromised functional recovery after nerve injury. Further study is required to elucidate these points.
ATF3
Activating transcription factor 3 (ATF3) is a stress-inducible transcription factor, which binds to the ATF/CRE site (Hai and Hartman, 2001). ATF3 is not normally found in neuronal cells but is highly expressed in response to nerve injury. Thus, ATF3 is known as an injury marker in the nervous system (Tsujino et al., 2000; Nakagomi et al., 2003; Ohba et al., 2003). The role of ATF3 diverges according to the cellular context and/or the components of the ATF3-containing complex. ATF3 forms homo- and heterodimers to function as both a repressor and activator of genes (Hai and Hartman, 2001).
ATF3 seems to be a major determinant of the intrinsic growth state of neurons. ATF3 expression is correlated with the survival and activation of intrinsic growth ability in CNS neurons (Ohba et al., 2003; Campbell et al., 2005; Zhang et al., 2011b). Overexpression of ATF3 in cultured superior cervical ganglion (SCG) neurons protects against cell death after NGF withdrawal, a popular apoptotic stimulus, while it enhances neurite outgrowth in both SCG neurons and DRG neurons (Nakagomi et al., 2003; Seijffers et al., 2006). Mice overexpressing ATF3 under the control of the neuron-specific Thy1.2 promoter revealed enhanced neurite elongation in vitro and in vivo after sciatic nerve injury (Seijffers et al., 2006). ATF3 is also up-regulated as an adaptive response in cultured demyelinated DRG neurons and increases the speed of axonal mitochondrial transport, probably to maintain axonal homeostasis (Kiryu-Seo et al., 2010). To achieve these roles, ATF3 has been believed to dimerize with c-Jun in a context-specific manner. The concurrent expression of c-Jun with ATF3 is frequently observed in damaged neurons after nerve injuries, including peripheral axotomy and Middle Cerebral Artery (MCA) occlusion (Nakagomi et al., 2003; Ohba et al., 2003). There is also in vitro evidence to show an interaction between ATF3 and c-Jun (Chen et al., 1996; Nakagomi te al., 2003). However, an interaction between ATF3 and c-Jun has not been reported in vivo to date. It is likely that some previously unidentified signaling pathway or associated proteins, which are activated by nerve injury, are involved in facilitating the dimerization of ATF3 and c-Jun.
Information about the target genes of ATF3 is still limited, although ATF3 can bind to an atypical ATF/CRE site in the Heat shock protein (Hsp) 27 promoter (Benn et al., 2002; Nakagomi et al., 2003). The aforementioned ATF3 transgenic mouse altered the expression of a set of RAGs in non-injured DRG neurons (Seijffers et al., 2006). They include c-Jun, Hsp27 which prevents neuronal death (Lewis et al., 1999), Small proline-rich repeat protein (SPRR) 1A which promotes axonal outgrowth (Bonilla et al., 2002), and CAP23 which is functionally related to GAP43 (Bomze et al., 2001). These genes might be candidate targets of ATF3. If a potential counterpart like c-Jun is activated at the same time, much greater induction of much more RAGs may be observed in non-injured DRG neurons.
To clarify the importance of ATF3 in injured neurons, ATF3 knockout mouse will be a useful tool (Hartman et al., 2004). To date, ATF3-deficient mouse lines have revealed no obvious abnormalities in neurite elongation or survival after nerve injury. This is probably because the mouse is not a complete null mutant, but still expresses the DNA-binding domain and leucine zipper domain of the protein, resulting in abundant expression of a small fragment protein and mRNA after nerve injury (Seijffers et al., 2007) and our observations.
STAT3
Signal transducer and activator of transcription 3 (STAT3) is activated in response to growth factors, cytokines, and hormones that are known to play protective role after nerve injury (Dziennis and Alkayed, 2008). Binding of these growth factors or cytokines to their cognate receptors, including the common receptor subunit gp130, activates Janus kinase (JAK), which phosphorylates tyrosine residues on the cytoplasmic portion of the receptor complex, and then phosphorylates STAT3. In general, axotomy increases the expression and phosphorylation of STAT3(Schwaiger et al., 2000; Qiu et al., 2005). To examine the role of STAT3 in vivo, Schweizer et al. used mice with neuron-specific ablation of STAT3 created by Cre recombinase expression under the control of the neurofilament light chain (NF-L) promoter (Schwaiger et al., 2002). The study indicated that STAT3 contributes to the survival of motor neurons after facial nerve lesions through activation of Reg2 and Bcl-XL (Gonzalez-Garcia et al., 1995; Livesey et al., 1997), which promote neuronal survival. Schweizer et al. did not investigate axonal regeneration using these mice. However, other studies implicated an effect of STAT3 on nerve regeneration using the peripheral conditioning injury paradigm and optic nerve injury model. The conditioning lesion paradigm is a widely accepted model in which elevated intrinsic growth ability caused by prior peripheral nerve axotomy facilitates subsequently lesioned dorsal column axons to regenerate over a short distance (Richardson and Issa, 1984; Neumann and Woolf, 1999). Qiu et al. demonstrated that STAT3 activation is necessary for increased growth ability of DRG neurons and improved axonal regeneration in the spinal cord after conditioning injury (Qiu et al., 2005). Furthermore, the deletion of suppressor of cytokine signaling 3 (SOCS3), which is a negative regulator of the JAK/STAT3 pathway, promotes axonal regeneration after optic nerve injury (Smith et al., 2009). Thus, a considerable body of evidence points to an involvement of STAT3 in nerve regeneration.
Apart from the well-understood job of STAT proteins in transmitting transcriptional signals from the cell surface to the nucleus, a new finding reveals that mitochondrial localization of STAT3 contributes to oxidative phosphorylation, leading to regulation of respiration (Gough et al., 2009). Furthermore, other studies claim that unphosphorylated STAT3 can be also imported into the nucleus independent of phosphorylation and, once there, regulate gene transcription by binding to other nuclear proteins (Liu et al., 2005; Yang et al., 2005). Although the physiological relevance of these newly discovered functions of STAT3 remains to be established, these findings expand our classical understanding of gene regulation by STAT3.
Cooperative Transcriptional Regulation by Injury-Inducible Transcription Factors
As mentioned above, numerous candidate injury-inducible transcription factors have been identified to date. How do those injury-inducible transcription factors communicate with each other and/or participate in the complex molecular machine? How do they access distinct RAGs with common or different injury-responsible DNA elements? We have assumed that one of the best ways to answer the questions is via the promoter analysis of a representative nerve RAG, which gives us clues to resolve these mysteries. From this point of view, damage-induced neuronal endopeptidase (DINE) is considered to be an ideal representative gene to provide further information about how injury-inducible transcription factors cooperate with each other during nerve regeneration.
Nerve Regeneration-Associated Gene DINE
Previous large-scale screening studies from our laboratory and others have identified numerous potential RAGs (Kiryu et al., 1995b; Tanabe et al., 1999; Bonilla et al., 2002; Costigan et al., 2002; Tanabe et al., 2003). The aim of our studies has been to understand the molecular basis underlying nerve regeneration. In addition, another important goal has been to find pan-nerve injury-inducible genes. These aims are based on the idea that there must be common injury-responsive transcriptional machinery in both the PNS and CNS, which triggers RAG expression and will be a potential key to enabling robust CNS regeneration to proceed in the future. In a search for RAGs, we identified DINE (Kiryu-Seo et al., 2000), which another group isolated independently as XCE later registered as endothelin converting enzyme-like 1 (ECEL-1); (Valdenaire et al., 1999). The most intriguing property of DINE is its extreme transcriptional response against various kinds of nerve injuries, both in the PNS and CNS, including motor, sensory and sympathetic nerve injuries, brain and spinal cord trauma, and cerebral ischemia (Kiryu-Seo et al., 2000; Boeshore et al., 2004), which led to its name (Kriyu-Seo et al., 2000). The enhanced expression of DINE is strikingly restricted to neuronal cells, and not seen in glial cells. DINE mRNA induction is always accompanied by the induction of ATF3, in various injury models (Nakagomi et al., 2003; Ohba et al., 2004). DINE mRNA is significantly induced by LIF treatment and NGF withdrawal both in vitro and in vivo, suggesting that c-Jun, ATF-3, and STAT3 are candidate transcription factors downstream of LIF-gp130 signaling and JNK signaling for transactivation of DINE gene (Kato et al., 2002; Kiryu-Seo and Kiyama, 2004).
At the protein level, DINE functions as a neuron-specific membrane-bound metalloprotease. It shares homology with neprilysin (NEP) and endothelin converting enzyme (ECE), which degrade or process neuropeptides such as amyloid β and endothelin, which play important roles in Alzheimer pathology and migration from the neural crest, respectively (Xu et al., 1994; Iwata et al., 2000). DINE is supposed to have a critical role in injured neurons, although its substrate remains unknown (Shirotani et al., 2001). DINE-deficient mice die immediately after birth due to respiratory failure (Schweizer et al., 1999; Nagata et al., 2010). This is caused by the failure of distal axonal arborization into muscle, resulting in poor formation of neuromuscular junctions (NMJ) (Nagata et al., 2010). Because DINE deficiency leads to abnormal axon behavior during development, DINE is expected to function during nerve regeneration as well (Nagata et al., 2006, 2010). Thus, DINE is considered to be a fascinating target gene with which to examine the transcriptional mechanisms involved in the response to nerve injury.
Sp1-Mediated Transcriptional Complex
Using the DINE promoter, we identified activation of the Sp1-mediated transcriptional machinery in response to nerve injury (Kiryu-Seo et al., 2008). Here, we describe this machinery and propose a potential explanation for how a cohort of nerve injury-induced transcription factors participates in the dynamic alteration of gene expression during nerve regeneration.
The 5′-flanking region of the DINE gene is thought to contain the injury-responsive element. DINE promoter activity was enhanced in response to LIF treatment and NGF withdrawal in cultured DRGs, mimicking the endogenous response of DINE mRNA (Kato et al., 2002 ). Co-expression of ATF3, c-Jun, and STAT3 increased DINE promoter activity by a small amount. This is because the efficient dimerization of c-Jun and ATF3 does not occur in this context. It is unclear what makes c-Jun and ATF3 dimerize, although the heterodimer is believed to promote axonal regeneration (Nakagomi et al., 2003 ). There may exist some unidentified signal that makes the heterodimer stable in injured neurons. Thus, we used a forced heterodimer of ATF3 and c-Jun in our experiment. The forced heterodimer up-regulated DINE promoter activity tremendously, with a further increase observed in the presence of STAT3, suggesting that these injury-inducible transcription factors contribute to the regulation of the DINE promoter. Unexpectedly, c-Jun, ATF3, and STAT3 did not directly bind to a specific element within the DINE promoter. Instead, Sp1 directly bound to the GC-rich region located in the region proximal to the transcription start site of the DINE promoter, and could function as a scaffolding protein to recruit c-Jun, ATF3, and STAT3 to elicit their functional synergy (Figure 2; Kiryu-Seo et al., 2008). Of the complex, ATF3, which makes subsequent dimerization with c-Jun, may be the most critical, because ATF3 is specifically expressed after nerve injury. The findings of an in vivo study using chromatin immunoprecipitation (ChIP) assay supported the existence of such a complex in injured hypoglossal neurons. We could not rule out the possibility that these transcription factors, like c-Jun, ATF3, and STAT3, regulate the DINE gene by binding to individual binding sites separately in an unidentified enhancer region, and that a combination of these proposed transcriptional mechanisms regulates the expression of DINE. However, consistent with our finding, it has been reported that gene promoters without the AP1 site or without the STAT3 binding site were activated by c-Jun or STAT3 through an interaction with Sp1 (Kardassis et al., 1999; Chen and Chang, 2000). Importantly, the mechanism by which Sp1 may provide c-Jun/ATF3/STAT3 with a platform would be practical and effective when increased gene expression is required in the event of a fatal emergency such as neuronal injury. Many neuronal genes contain the Sp1 binding site (Ross et al., 2002 ), suggesting that nerve injury-inducible transcription factors are capable of accessing many genes without specific binding sites.
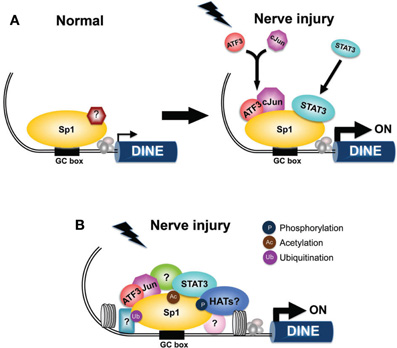
Figure 2. Possible mechanism of the Sp1-mediated transcriptional machinery to induce the regeneration-associated gene DINE in response to nerve injury. (A) Transcriptional regulation of DINE before and after nerve injury. In the normal conditions, Sp1 and the unknown transcriptional complex express DINE at the low-level. Following nerve injury, injury-inducible transcription factor ATF3 form a heterodimer with cJun and Sp1 provides a platform to recruit ATF3, c-Jun, and Stat3. (B) The activity of Sp1 can be regulated by post-translational modifications including phosphorylation, acetylation, ubiquitination. Additional transcription factors and chromatin modifiers may participate in the Sp1-mediated transcriptional complex shown in (A).
Multifunctional Transcription Factor Sp1
Sp1 is a general transcription factor that binds to GC-rich motifs with high affinity. Sp1 was thought to serve mainly as a constitutive activator of housekeeping genes in a classical view. Growing evidence suggests that Sp1 is a multifunctional transcription factor.
At the transcriptional level, Sp1 is not induced in injured neurons, but is expressed constitutively. The regulation of Sp1 is important at the protein level. The activity of Sp1 is regulated by post-translational modifications, as well as by interaction partners. Phosphorylation, acetylation, sumoylation, ubiquitination, and glycosylation are among the post-translational modification that can influence the transcriptional activity and stability of Sp1 (Hung et al., 2006; Tan and Khachigian, 2009). The accessibility of injury-inducible transcription factors may be determined by the modification of Sp1, suggesting the possibility that different modifications of Sp1 recruit different injury-inducible transcription factors. The activity of Sp1 is also regulated by interactions or interplay with other transcription factors such as p53, c-myc, Smad, AP-2, and E2F-1 (Wierstra, 2008). These additional transcription factors may participate in the Sp1/c-Jun/ATF3/ STAT3 complex to yield maximum effects in injured neurons (Figure 2).
The involvement of Sp1 is illuminated in the neurodegenerative disease Huntington disease (HD). Sp1 is induced and acetylated under the conditions of oxidative stress observed in HD neurons (Ryu et al., 2003b; Qiu et al., 2006). Sp1 induction confers a resistance to oxidative stress-induced cell death through transcriptional activation (Ryu et al., 2003a). In HD pathology, mutant huntingtin (Htt) interacts with Sp1 and core components of the transcriptional machinery, leading to disruption of transcriptional activation of neuronal genes such as the dopamine D2 receptor (Dunah et al., 2002; Freiman and Tjian, 2002; Zhai et al., 2005 ). Conversely, others have reported that a decrease in the Sp1 level has a beneficial effect on HD pathology (Qiu et al., 2006; Ravache et al., 2010 ). These results seem to be contradictory. The reason for this may be attributable to the diverging role of Sp1. Recently, Sleiman et al. demonstrated that a GC-rich DNA-binding drug mithramycin (MTM), which competes with Sp1 for its binding site, prevents neuronal death in the HD model of Drosophila as well as in cultured cortical neurons under conditions of oxidative stress. The protective role depends on the selectivity of MTM, meaning that MTM selectively binds to the GC-box of proapoptotic genes like myc but not that of anti-apoptotic genes like p21 (Sleiman et al., 2011). The mechanism of action remains unknown; however, this work suggests that Sp1 could be a target of therapeutic strategies.
Interestingly, Sp1 interacts with chromatin-modifying factors such as p300, histone acetylases (HATs) and histone deacetylases (HDACs) (Ryu et al., 2003a; Hung et al., 2006), while the Sp1 target sequence, the GC-box, can be a substrate for DNA methylation. It is therefore, an attractive possibility that Sp1 is a target for epigenetic regulation. Taking this possibility into consideration, our finding of the Sp1-mediated mechanism, which provides a platform the injury-inducible transcription factors, stretches the possibility of understanding global gene regulation during nerve regeneration.
Potential Involvement of Epigenetic Modification
Epigenetic modification of both histones and DNA is emerging as a fundamental mechanism by which neurons adapt their transcriptional responses to environmental cues (Jaenisch and Bird, 2003; Riccio, 2010). Histone modification including acetylation, methylation, phosphorylation, ubiquitination, and sumoylation modifies local chromatin structure or provides binding sites for non-DNA-binding chromatin proteins. The combination of such modifications can induce the formation of transiently closed or open chromatin domains. Open chromatin is associated with active gene expression, while closed chromatin is associated with DNA compaction and gene repression.
Chromatin-modifying enzymes are found in large multiprotein complexes that are recruited to gene promoters by the transcription factors that bind to specific DNA sequences and confer target specificity (Borrelli et al., 2008). In neurons, HATs and HDACs are the best-characterized chromatin-modifying enzymes. Multiple lines of evidence suggesting that epigenetic regulation is involved in neurodegenerative pathology (Juliandi et al., 2010; Ma et al., 2010), and that the use of HDAC inhibitors combats neurodegenerative conditions in cellular and disease models (Chuang et al., 2009), have been accumulated. In this context, it is conceivable that epigenetic regulation by chromatin modifiers influences nerve regeneration (Figure 3). Indeed, nerve injuries such as optic nerve injury and hypoglossal nerve injury enhance the expression of HDAC family proteins (Pelzel et al., 2010) and our unpublished data. Neuron-restrictive silencer factor (NRSF) is up-regulated after sciatic nerve injury and recruits HDACs to suppress the expression of Nav1.8 sodium channels, u-opioid receptor (MOP) and the voltage-gated potassium channel Kv4.3 in injured DRG neurons (Uchida et al., 2010a, b). Usually, NRSF represses the expression of neuronal genes in non-neuronal cells, although it can be an activator in some situations (Ballas et al., 2005). This suggests that the normal pattern of gene expression in neurons can be reversed in injured neurons to allow expression of non-neuronal genes as observed in the genes encoding ERK and Shc (Kiryu et al., 1995a; Tanabe et al., 1998). Another point is that HATs and HDACs participate in the activities of numerous transcription factors, such as p53, Sp1, Smad1, CREB NFκB, and STAT, which are also associated with nerve regeneration. Recently, Chen et al. and Jacob et al. reported the epigenetic regulation of Schwann cells, showing that HDACs are essential not only for chromatin condensation but also for the transcriptional activation events that regulate survival and myelination. HDACs have synergy with Sox10 and NFκB to activate myelin genes via a spatiotemporally controlled transcriptional cascade (Jessen and Mirsky, 2005; Chen et al., 2011; Jacob et al., 2011). It is implied that a similar mechanism works in Schwann cells as well as in neurons after nerve injury.
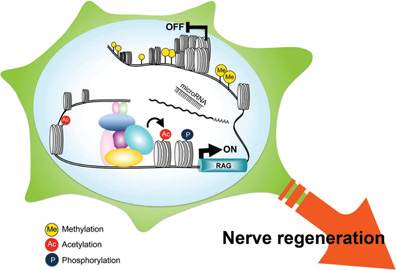
Figure 3. The possible involvement of epigenetic modification during nerve regeneration. In addition to the involvement of numerous transcriptional factors shown in Figure 1, the epigenetic modification of chromatin structure might affect the gene expression following nerve injury. Both histone modification including acetylation and phosphorylation and DNA modification such as methylation are associated with active gene expression as well as gene repression. Furthermore, microRNAs are implicated as potent silencers of gene expression.
DNA methylation at CpG dinucleotides in the genome is also a major epigenetic mechanism, and is associated with a condensed structure and transcriptional repression. DNA methylation status seems to be flexible rather than fixed to allow adaptation to environmental changes. Damaged cells are no longer able to maintain their prior level of DNA methylation (Ramchandani et al., 1999; Endres et al., 2000). Using the conditioning lesion model, which enables lesioned dorsal column axons to undergo lengthy regeneration in a segment of peripheral nerve transplanted into spinal cord (Hoffman, 2010), Iskandar et al. reported that the injury suppresses DNA methyltransferase (Dnmt) protein levels in spinal cord and that DNA methylation is decreased globally (Iskandar et al., 2010). The administration of folic acid, which is required for DNA methylation through the folate pathway, restored Dnmt protein expression as well as axonal regeneration. Although it is not apparent which cells are responsible for this effect, these findings imply that DNA methylation is a critical factor for gene regulation after nerve injury.
Recently, a new player has emerged as a key molecule in the epigenetic gene regulation of many biological processes. microRNAs (miRNAs) act as potent silencers of gene expression via translational repression and/or mRNA destabilization. Interestingly, some miRNAs are especially abundant in the nervous system, suggesting that they might be particularly important there. Exciting work has been reported in the fields of neurogenesis, plasticity, and neurodegeneration (Eacker et al., 2009; Siegel et al., 2011). Although the involvement of miRNAs in nerve regeneration is not clear, the altered expression of some miRNAs has been validated in DRG neurons after sciatic nerve injury (Zhang et al., 2011a; Zhou et al., 2011). A recent elegant study demonstrated that miRNA 206 in muscle promotes regeneration of neuromuscular synapses (Williams et al., 2009). Gaining further understanding of the roles of epigenetic modification and miRNAs in nerve regeneration is the next critical step and will advance our understanding of the gene regulation mechanisms operative in injured neurons.
Conclusions
The challenging goal is to reach a sufficient understanding of the global transcriptional machinery in injured neurons to realize robust CNS regeneration. Accumulating evidence supports the possibility that injury-inducible transcription factors like c-Jun, ATF3, and STAT3 are involved in the intrinsic regenerative growth ability in vivo. To achieve orchestrated expression of distinct RAGs, the transcriptional network and the multiprotein complex could function in damaged neurons in response to environmental cues. The Sp1 transcriptional machinery described in this review provides one explanation for how numerous RAGs are regulated simultaneously in injured neurons. Additionally, epigenetic regulation is also emerging as a crucial factor in controlling gene regulation during nerve regeneration. We are still far from a comprehensive understanding of how the interplay of transcriptional and epigenetic processes contributes to nerve regeneration. It will be challenging to define the exact mechanism underlying global transcriptional regulation in the nuclei of injured neurons.
As a biological tool that is currently available for studying nerve regeneration in vivo, knockout mice have proven to be very valuable (Zheng et al., 2006). However, for technical reasons, only a few knockout mice for the transcription factors described here have been studied using this method to date. Most transcription factors are critical in development and normal cellular function, so their knockout in mice would be lethal during development and/or the development of mice would be influenced by their ablation. The best approach is to perform gene deletions specifically in injured neurons using Cre/loxP technology. For this purpose, transgenic mice with Cre recombinase expressed only in injured neurons appear to be a suitable approach. Ongoing work using this approach will improve our understanding of how nuclear events activate the intrinsic ability to drive axonal growth and will provide clues to specific therapeutic strategies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Grants from The Ministry of Education, Culture, Sports, Science, and Technology (MEXT) and a Grant-in-Aid for Young Scientists (B). We would like to thank members of the laboratory for helpful discussions.
References
Araki, T., and Milbrandt, J. (2000). Ninjurin2, a novel homophilic adhesion molecule, is expressed in mature sensory and enteric neurons and promotes neurite outgrowth. J. Neurosci. 20, 187–195.
Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C., and Mandel, G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657.
Banner, L. R., and Patterson, P. H. (1994). Major changes in the expression of the mRNAs for cholinergic differentiation factor/leukemia inhibitory factor and its receptor after injury to adult peripheral nerves and ganglia. Proc. Natl. Acad. Sci. U.S.A. 91, 7109–7113.
Belecky-Adams, T., Wight, D. C., Kopchick, J. J., and Parysek, L. M. (1993). Intragenic sequences are required for cell type-specific and injury-induced expression of the rat peripherin gene. J. Neurosci. 13, 5056–5065.
Benn, S. C., Perrelet, D., Kato, A. C., Scholz, J., Decosterd, I., Mannion, R. J., Bakowska, J. C., and Woolf, C. J. (2002). Hsp27 upregulation and phosphorylation is required for injured sensory and motor neuron survival. Neuron 36, 45–56.
Benowitz, L., and Yin, Y. (2008). Rewiring the injured CNS: lessons from the optic nerve. Exp. Neurol. 209, 389–398.
Boeshore, K. L., Schreiber, R. C., Vaccariello, S. A., Sachs, H. H., Salazar, R., Lee, J., Ratan, R. R., Leahy, P., and Zigmond, R. E. (2004). Novel changes in gene expression following axotomy of a sympathetic ganglion: a microarray analysis. J. Neurobiol. 59, 216–235.
Bomze, H. M., Bulsara, K. R., Iskandar, B. J., Caroni, P., and Skene, J. H. (2001). Spinal axon regeneration evoked by replacing two growth cone proteins in adult neurons. Nat. Neurosci. 4, 38–43.
Bonilla, I. E., Tanabe, K., and Strittmatter, S. M. (2002). Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J. Neurosci. 22, 1303–1315.
Borrelli, E., Nestler, E. J., Allis, C. D., and Sassone-Corsi, P. (2008). Decoding the epigenetic language of neuronal plasticity. Neuron 60, 961–974.
Cafferty, W. B., Gardiner, N. J., Das, P., Qiu, J., McMahon, S. B., and Thompson, S. W. (2004). Conditioning injury-induced spinal axon regeneration fails in interleukin-6 knock-out mice. J. Neurosci. 24, 4432–4443.
Campbell, G., Hutchins, K., Winterbottom, J., Grenningloh, G., Lieberman, A. R., and Anderson, P. N. (2005). Upregulation of activating transcription factor 3 (ATF3) by intrinsic CNS neurons regenerating axons into peripheral nerve grafts. Exp. Neurol. 192, 340–347.
Cao, Z., Gao, Y., Bryson, J. B., Hou, J., Chaudhry, N., Siddiq, M., Martinez, J., Spencer, T., Carmel, J., Hart, R. B., and Filbin, M. T. (2006). The cytokine interleukin-6 is sufficient but not necessary to mimic the peripheral conditioning lesion effect on axonal growth. J. Neurosci. 26, 5565–5573.
Cavalli, V., Kujala, P., Klumperman, J., and Goldstein, L. S. (2005). Sunday Driver links axonal transport to damage signaling. J. Cell Biol. 168, 775–787.
Chen, B. K., and Chang, W. C. (2000). Functional interaction between c-Jun and promoter factor Sp1 in epidermal growth factor-induced gene expression of human 12(S)-lipoxygenase. Proc. Natl. Acad. Sci. U.S.A. 97, 10406–10411.
Chen, B. P., Wolfgang, C. D., and Hai, T. (1996). Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol. Cell. Biol. 16, 1157–1168.
Chen, Y., Wang, H., Yoon, S. O., Xu, X., Hottiger, M. O., Svaren, J., Nave, K. A., Kim, H. A., Olson, E. N., and Lu, Q. R. (2011). HDAC-mediated deacetylation of NF-kappaB is critical for Schwann cell myelination. Nat. Neurosci. 14, 437–441.
Chen, Z. L., Yu, W. M., and Strickland, S. (2007). Peripheral regeneration. Annu. Rev. Neurosci. 30, 209–233.
Chuang, D. M., Leng, Y., Marinova, Z., Kim, H. J., and Chiu, C. T. (2009). Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 32, 591–601.
Costigan, M., Befort, K., Karchewski, L., Griffin, R. S., D'Urso, D., Allchorne, A., Sitarski, J., Mannion, J. W., Pratt, R. E., and Woolf, C. J. (2002). Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 3, 16.
Cox, L. J., Hengst, U., Gurskaya, N. G., Lukyanov, K. A., and Jaffrey, S. R. (2008). Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat. Cell Biol. 10, 149–159.
Curtis, R., Scherer, S. S., Somogyi, R., Adryan, K. M., Ip, N. Y., Zhu, Y., Lindsay, R. M., and DiStefano, P. S. (1994). Retrograde axonal transport of LIF is increased by peripheral nerve injury: correlation with increased LIF expression in distal nerve. Neuron 12, 191–204.
Di Giovanni, S., Knights, C. D., Rao, M., Yakovlev, A., Beers, J., Catania, J., Avantaggiati, M. L., and Faden, A. I. (2006). The tumor suppressor protein p53 is required for neurite outgrowth and axon regeneration. EMBO J. 25, 4084–4096.
Dunah, A. W., Jeong, H., Griffin, A., Kim, Y. M., Standaert, D. G., Hersch, S. M., Mouradian, M. M., Young, A. B., Tanese, N., and Krainc, D. (2002). Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science 296, 2238–2243.
Dziennis, S., and Alkayed, N. J. (2008). Role of signal transducer and activator of transcription 3 in neuronal survival and regeneration. Rev. Neurosci. 19, 341–361.
Eacker, S. M., Dawson, T. M., and Dawson, V. L. (2009). Understanding microRNAs in neurodegeneration. Nat. Rev. Neurosci. 10, 837–841.
Endres, M., Meisel, A., Biniszkiewicz, D., Namura, S., Prass, K., Ruscher, K., Lipski, A., Jaenisch, R., Moskowitz, M. A., and Dirnagl, U. (2000). DNA methyltransferase contributes to delayed ischemic brain injury. J. Neurosci. 20, 3175–3181.
Filbin, M. T. (2003). Myelin-associated inhibitors of axonal regeneration in the adult mammalian CNS. Nat. Rev. Neurosci. 4, 703–713.
Freiman, R. N., and Tjian, R. (2002). Neurodegeneration. A glutamine-rich trail leads to transcription factors. Science 296, 2149–2150.
Funakoshi, H., Frisen, J., Barbany, G., Timmusk, T., Zachrisson, O., Verge, V. M., and Persson, H. (1993). Differential expression of mRNAs for neurotrophins and their receptors after axotomy of the sciatic nerve. J. Cell Biol. 123, 455–465.
Gamo, K., Kiryu-Seo, S., Konishi, H., Aoki, S., Matsushima, K., Wada, K., and Kiyama, H. (2008). G-protein-coupled receptor screen reveals a role for chemokine receptor CCR5 in suppressing microglial neurotoxicity. J. Neurosci. 28, 11980–11988.
Gao, Y., Deng, K., Hou, J., Bryson, J. B., Barco, A., Nikulina, E., Spencer, T., Mellado, W., Kandel, E. R., and Filbin, M. T. (2004). Activated CREB is sufficient to overcome inhibitors in myelin and promote spinal axon regeneration in vivo. Neuron 44, 609–621.
Gloster, A., Wu, W., Speelman, A., Weiss, S., Causing, C., Pozniak, C., Reynolds, B., Chang, E., Toma, J. G., and Miller, F. D. (1994). The T alpha 1 alpha-tubulin promoter specifies gene expression as a function of neuronal growth and regeneration in transgenic mice. J. Neurosci. 14, 7319–7330.
Gonzalez-Garcia, M., Garcia, I., Ding, L., O'Shea, S., Boise, L. H., Thompson, C. B., and Nunez, G. (1995). bcl-x is expressed in embryonic and postnatal neural tissues and functions to prevent neuronal cell death. Proc. Natl. Acad. Sci. U.S.A. 92, 4304–4308.
Gough, D. J., Corlett, A., Schlessinger, K., Wegrzyn, J., Larner, A. C., and Levy, D. E. (2009). Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324, 1713–1716.
Hai, T., and Hartman, M. G. (2001). The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273, 1–11.
Ham, J., Babij, C., Whitfield, J., Pfarr, C. M., Lallemand, D., Yaniv, M., and Rubin, L. L. (1995). A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron 14, 927–939.
Hammarlund, M., Nix, P., Hauth, L., Jorgensen, E. M., and Bastiani, M. (2009). Axon regeneration requires a conserved MAP kinase pathway. Science 323, 802–806.
Hanz, S., Perlson, E., Willis, D., Zheng, J. Q., Massarwa, R., Huerta, J. J., Koltzenburg, M., Kohler, M., van-Minnen, J., Twiss, J. L., and Fainzilber, M. (2003). Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron 40, 1095–1104.
Harel, N. Y., and Strittmatter, S. M. (2006). Can regenerating axons recapitulate developmental guidance during recovery from spinal cord injury? Nat. Rev. Neurosci. 7, 603–616.
Hartman, M. G., Lu, D., Kim, M. L., Kociba, G. J., Shukri, T., Buteau, J., Wang, X., Frankel, W. L., Guttridge, D., Prentki, M., Grey, S. T., Ron, D., and Hai, T. (2004). Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Mol. Cell. Biol. 24, 5721–5732.
Herdegen, T., and Leah, J. D. (1998). Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res. Brain Res. Rev. 28, 370–490.
Herdegen, T., Skene, P., and Bahr, M. (1997). The c-Jun transcription factor – bipotential mediator of neuronal death, survival and regeneration. Trends Neurosci. 20, 227–231.
Hoffman, P. N. (2010). A conditioning lesion induces changes in gene expression and axonal transport that enhance regeneration by increasing the intrinsic growth state of axons. Exp. Neurol. 223, 11–18.
Hung, J. J., Wang, Y. T., and Chang, W. C. (2006). Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol. Cell. Biol. 26, 1770–1785.
Iskandar, B. J., Rizk, E., Meier, B., Hariharan, N., Bottiglieri, T., Finnell, R. H., Jarrard, D. F., Banerjee, R. V., Skene, J. H., Nelson, A., Patel, N., Gherasim, C., Simon, K., Cook, T. D., and Hogan, K. J. (2010). Folate regulation of axonal regeneration in the rodent central nervous system through DNA methylation. J. Clin. Invest. 120, 1603–1616.
Iwata, N., Tsubuki, S., Takaki, Y., Watanabe, K., Sekiguchi, M., Hosoki, E., Kawashima-Morishima, M., Lee, H. J., Hama, E., Sekine-Aizawa, Y., and Saido, T. C. (2000). Identification of the major Abeta1-42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat. Med. 6, 143–150.
Jacob, C., Christen, C. N., Pereira, J. A., Somandin, C., Baggiolini, A., Lotscher, P., Ozcelik, M., Tricaud, N., Meijer, D., Yamaguchi, T., Matthias, P., and Suter, U. (2011). HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat. Neurosci. 14, 429–436.
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33(Suppl.), 245–254.
Jankowski, M. P., Cornuet, P. K., McIlwrath, S., Koerber, H. R., and Albers, K. M. (2006). SRY-box containing gene 11 (Sox11) transcription factor is required for neuron survival and neurite growth. Neuroscience 143, 501–514.
Jessen, K. R., and Mirsky, R. (2005). The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci. 6, 671–682.
Jones, L. L., Kreutzberg, G. W., and Raivich, G. (1997). Regulation of CD44 in the regenerating mouse facial motor nucleus. Eur. J. Neurosci. 9, 1854–1863.
Juliandi, B., Abematsu, M., and Nakashima, K. (2010). Chromatin remodeling in neural stem cell differentiation. Curr. Opin. Neurobiol. 20, 408–415.
Kardassis, D., Papakosta, P., Pardali, K., and Moustakas, A. (1999). c-Jun transactivates the promoter of the human p21(WAF1/Cip1) gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J. Biol. Chem. 274, 29572–29581.
Kato, R., Kiryu-Seo, S., and Kiyama, H. (2002). Damage-induced neuronal endopeptidase (DINE/ECEL) expression is regulated by leukemia inhibitory factor and deprivation of nerve growth factor in rat sensory ganglia after nerve injury. J. Neurosci. 22, 9410–9418.
Kiryu, S., Morita, N., Ohno, K., Maeno, H., and Kiyama, H. (1995a). Regulation of mRNA expression involved in Ras and PKA signal pathways during rat hypoglossal nerve regeneration. Brain Res. Mol. Brain Res. 29, 147–156.
Kiryu, S., Yao, G. L., Morita, N., Kato, H., and Kiyama, H. (1995b). Nerve injury enhances rat neuronal glutamate transporter expression: identification by differential display PCR. J. Neurosci. 15, 7872–7878.
Kiryu-Seo, S., Gamo, K., Tachibana, T., Tanaka, K., and Kiyama, H. (2006). Unique anti-apoptotic activity of EAAC1 in injured motor neurons. EMBO J. 25, 3411–3421.
Kiryu-Seo, S., Hirayama, T., Kato, R., and Kiyama, H. (2005). Noxa is a critical mediator of p53-dependent motor neuron death after nerve injury in adult mouse. J. Neurosci. 25, 1442–1447.
Kiryu-Seo, S., Kato, R., Ogawa, T., Nakagomi, S., Nagata, K., and Kiyama, H. (2008). Neuronal injury-inducible gene is synergistically regulated by ATF3, c-Jun, and STAT3 through the interaction with Sp1 in damaged neurons. J. Biol. Chem. 283, 6988–6996.
Kiryu-Seo, S., and Kiyama, H. (2004). DINE (damage induced neuronal endopeptidase). Protein Pept. Lett. 11, 451–460.
Kiryu-Seo, S., Ohno, N., Kidd, G. J., Komuro, H., and Trapp, B. D. (2010). Demyelination increases axonal stationary mitochondrial size and the speed of axonal mitochondrial transport. J. Neurosci. 30, 6658–6666.
Kiryu-Seo, S., Sasaki, M., Yokohama, H., Nakagomi, S., Hirayama, T., Aoki, S., Wada, K., and Kiyama, H. (2000). Damage-induced neuronal endopeptidase (DINE) is a unique metallopeptidase expressed in response to neuronal damage and activates superoxide scavengers. Proc. Natl. Acad. Sci. U.S.A. 97, 4345–4350.
Kloss, C. U., Werner, A., Klein, M. A., Shen, J., Menuz, K., Probst, J. C., Kreutzberg, G. W., and Raivich, G. (1999). Integrin family of cell adhesion molecules in the injured brain: regulation and cellular localization in the normal and regenerating mouse facial motor nucleus. J. Comp. Neurol. 411, 162–178.
Kobayashi, N., Kiyama, H., and Tohyama, M. (1994). GAP-43 (B50/F1) gene regulation by axonal injury of the hypoglossal nerve in the adult rat. Brain Res. Mol. Brain Res. 21, 9–18.
Lee, J. K., Geoffroy, C. G., Chan, A. F., Tolentino, K. E., Crawford, M. J., Leal, M. A., Kang, B., and Zheng, B. (2010). Assessing spinal axon regeneration and sprouting in Nogo-, MAG-, and OMgp-deficient mice. Neuron 66, 663–670.
Leon, S., Yin, Y., Nguyen, J., Irwin, N., and Benowitz, L. I. (2000). Lens injury stimulates axon regeneration in the mature rat optic nerve. J. Neurosci. 20, 4615–4626.
Lewis, S. E., Mannion, R. J., White, F. A., Coggeshall, R. E., Beggs, S., Costigan, M., Martin, J. L., Dillmann, W. H., and Woolf, C. J. (1999). A role for HSP27 in sensory neuron survival. J. Neurosci. 19, 8945–8953.
Li, L., Oppenheim, R. W., Lei, M., Houenou, L. J. (1994). Neurotrophic agents prevent motoneuron death following sciatic nerve section in the neonatal mouse. J. Neurobiol. 25, 759–766.
Lindholm, D., Heumann, R., Meyer, M., and Thoenen, H. (1987). Interleukin-1 regulates synthesis of nerve growth factor in non-neuronal cells of rat sciatic nerve. Nature 330, 658–659.
Lindwall, C., and Kanje, M. (2005). Retrograde axonal transport of JNK signaling molecules influence injury induced nuclear changes in p-c-Jun and ATF3 in adult rat sensory neurons. Mol. Cell. Neurosci. 29, 269–282.
Liu, L., McBride, K. M., and Reich, N. C. (2005). STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc. Natl. Acad. Sci. U.S.A. 102, 8150–8155.
Liu, Y. W., Chen, B. K., Chen, C. J., Arakawa, T., Yoshimoto, T., Yamamoto, S., and Chang, W. C. (1997). Epidermal growth factor enhances transcription of human arachidonate 12-lipoxygenase in A431 cells. Biochim. Biophys. Acta. 1344, 38–46.
Livesey, F. J., O'Brien, J. A., Li, M., Smith, A. G., Murphy, L. J., and Hunt, S. P. (1997). A Schwann cell mitogen accompanying regeneration of motor neurons. Nature 390, 614–618.
Ma, D. K., Marchetto, M. C., Guo, J. U., Ming, G. L., Gage, F. H., and Song, H. (2010). Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat. Neurosci. 13, 1338–1344.
MacGillavry, H. D., Cornelis, J., van der Kallen, L. R., Sassen, M. M., Verhaagen, J., Smit, A. B., and van Kesteren, R. E. (2011). Genome-wide gene expression and promoter binding analysis identifies NFIL3 as a repressor of C/EBP target genes in neuronal outgrowth. Mol. Cell. Neurosci. 46, 460–468.
Markus, A., Zhong, J., and Snider, W. D. (2002). Raf and akt mediate distinct aspects of sensory axon growth. Neuron 35, 65–76.
Michaelevski, I., Segal-Ruder, Y., Rozenbaum, M., Medzihradszky, K. F., Shalem, O., Coppola, G., Horn-Saban, S., Ben-Yaakov, K., Dagan, S. Y., Rishal, I., Geschwind, D. H., Pilpel, Y., Burlingame, A. L., and Fainzilber, M. (2010). Signaling to transcription networks in the neuronal retrograde injury response. Sci. Signal. 3, ra53.
Moore, D. L., Blackmore, M. G., Hu, Y., Kaestner, K. H., Bixby, J. L., Lemmon, V. P., and Goldberg, J. L. (2009). KLF family members regulate intrinsic axon regeneration ability. Science 326, 298–301.
Moran, L. B., and Graeber, M. B. (2004). The facial nerve axotomy model. Brain Res. Brain Res. Rev. 44, 154–178.
Murakami, K., Namikawa, K., Shimizu, T., Shirasawa, T., Yoshida, S., and Kiyama, H. (2006). Nerve injury induces the expression of EXT2, a glycosyltransferase required for heparan sulfate synthesis. Neuroscience 141, 1961–1969.
Murphy, P. G., Grondin, J., Altares, M., and Richardson, P. M. (1995). Induction of interleukin-6 in axotomized sensory neurons. J. Neurosci. 15, 5130–5138.
Nadeau, S., Hein, P., Fernandes, K. J., Peterson, A. C., and Miller, F. D. (2005). A transcriptional role for C/EBP beta in the neuronal response to axonal injury. Mol. Cell. Neurosci. 29, 525–535.
Nagata, K., Kiryu-Seo, S., and Kiyama, H. (2006). Localization and ontogeny of damage-induced neuronal endopeptidase mRNA-expressing neurons in the rat nervous system. Neuroscience 141, 299–310.
Nagata, K., Kiryu-Seo, S., Maeda, M., Yoshida, K., Morita, T., and Kiyama, H. (2010). Damage-induced neuronal endopeptidase is critical for presynaptic formation of neuromuscular junctions. J. Neurosci. 30, 6954–6962.
Nakagomi, S., Suzuki, Y., Namikawa, K., Kiryu-Seo, S., and Kiyama, H. (2003). Expression of the activating transcription factor 3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J. Neurosci. 23, 5187–5196.
Namikawa, K., Honma, M., Abe, K., Takeda, M., Mansur, K., Obata, T., Miwa, A., Okado, H., and Kiyama, H. (2000). Akt/protein kinase B prevents injury-induced motoneuron death and accelerates axonal regeneration. J. Neurosci. 20, 2875–2886.
Namikawa, K., Okamoto, T., Suzuki, A., Konishi, H., and Kiyama, H. (2006). Pancreatitis-associated protein-III is a novel macrophage chemoattractant implicated in nerve regeneration. J. Neurosci. 26, 7460–7467.
Naveilhan, P., ElShamy, W. M., and Ernfors, P. (1997). Differential regulation of mRNAs for GDNF and its receptors Ret and GDNFR alpha after sciatic nerve lesion in the mouse. Eur. J. Neurosci. 9, 1450–1460.
Neumann, S., and Woolf, C. J. (1999). Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron 23, 83–91.
Nieke, J., and Schachner, M. (1985). Expression of the neural cell adhesion molecules L1 and N-CAM and their common carbohydrate epitope L2/HNK-1 during development and after transection of the mouse sciatic nerve. Differentiation 30, 141–151.
Ohba, N., Kiryu-Seo, S., Maeda, M., Muraoka, M., Ishii, M., and Kiyama, H. (2004). Expression of damage-induced neuronal endopeptidase (DINE) mRNA in peri-infarct cortical and thalamic neurons following middle cerebral artery occlusion. J. Neurochem. 91, 956–964.
Ohba, N., Maeda, M., Nakagomi, S., Muraoka, M., and Kiyama, H. (2003). Biphasic expression of activating transcription factor-3 in neurons after cerebral infarction. Brain Res. Mol. Brain Res. 115, 147–156.
Okuyama, N., Kiryu-Seo, S., and Kiyama, H. (2007). Altered expression of Smad family members in injured motor neurons of rat. Brain Res. 1132, 36–41.
Palmada, M., Kanwal, S., Rutkoski, N. J., Gustafson-Brown, C., Johnson, R. S., Wisdom, R., and Carter, B. D. (2002). c-jun is essential for sympathetic neuronal death induced by NGF withdrawal but not by p75 activation. J. Cell Biol. 158, 453–461.
Park, K. K., Liu, K., Hu, Y., Smith, P. D., Wang, C., Cai, B., Xu, B., Connolly, L., Kramvis, I., Sahin, M., and He, Z. (2008). Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966.
Parkinson, D. B., Bhaskaran, A., Arthur-Farraj, P., Noon, L. A., Woodhoo, A., Lloyd, A. C., Feltri, M. L., Wrabetz, L., Behrens, A., Mirsky, R., and Jessen, K. R. (2008). c-Jun is a negative regulator of myelination. J. Cell Biol. 181, 625–637.
Pelzel, H. R., Schlamp, C. L., and Nickells, R. W. (2010). Histone H4 deacetylation plays a critical role in early gene silencing during neuronal apoptosis. BMC Neurosci. 11, 62.
Qiu, J., Cafferty, W. B., McMahon, S. B., and Thompson, S. W. (2005). Conditioning injury-induced spinal axon regeneration requires signal transducer and activator of transcription 3 activation. J. Neurosci. 25, 1645–1653.
Qiu, Z., Norflus, F., Singh, B., Swindell, M. K., Buzescu, R., Bejarano, M., Chopra, R., Zucker, B., Benn, C. L., DiRocco, D. P., Cha, J. H., Ferrante, R. J., and Hersch, S. M. (2006). Sp1 is up-regulated in cellular and transgenic models of Huntington disease, and its reduction is neuroprotective. J. Biol. Chem. 281, 16672–16680.
Raivich, G. (2005). Like cops on the beat: the active role of resting microglia. Trends Neurosci. 28, 571–573.
Raivich, G., Bohatschek, M., Da Costa, C., Iwata, O., Galiano, M., Hristova, M., Nateri, A. S., Makwana, M., Riera-Sans, L., Wolfer, D. P., Lipp, H. P., Aguzzi, A., Wagner, E. F., and Behrens, A. (2004). The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron 43, 57–67.
Raivich, G., and Makwana, M. (2007). The making of successful axonal regeneration: genes, molecules and signal transduction pathways. Brain Res. Rev. 53, 287–311.
Ramchandani, S., Bhattacharya, S. K., Cervoni, N., Szyf, M. (1999). DNA methylation is a reversible biological signal. Proc. Natl. Acad. Sci. U.S.A. 96, 6107–6112.
Ravache, M., Weber, C., Merienne, K., and Trottier, Y. (2010). Transcriptional activation of REST by Sp1 in Huntington's disease models. PLoS One 5, e14311. doi: 10.1371/journal.pone.0014311
Riccio, A. (2010). Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat. Neurosci. 13, 1330–1337.
Richardson, P. M., and Issa, V. M. (1984). Peripheral injury enhances central regeneration of primary sensory neurones. Nature 309, 791–793.
Ross, S., Tienhaara, A., Lee, M. S., Tsai, L. H., and Gill, G. (2002). GC box-binding transcription factors control the neuronal specific transcription of the cyclin-dependent kinase 5 regulator p35. J. Biol. Chem. 277, 4455–4464.
Ryu, H., Lee, J., Olofsson, B. A., Mwidau, A., Dedeoglu, A., Escudero, M., Flemington, E., Azizkhan-Clifford, J., Ferrante, R. J., and Ratan, R. R. (2003a). Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc. Natl. Acad. Sci. U.S.A. 100, 4281–4286.
Ryu, H., Lee, J., Zaman, K., Kubilis, J., Ferrante, R. J., Ross, B. D., Neve, R., and Ratan, R. R. (2003b). Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J. Neurosci. 23, 3597–3606.
Schwaiger, F. W., Hager, G., Schmitt, A. B., Horvat, A., Streif, R., Spitzer, C., Gamal, S., Breuer, S., Brook, G. A., Nacimiento, W., and Kreutzberg, G. W. (2000). Peripheral but not central axotomy induces changes in Janus kinases (JAK) and signal transducers and activators of transcription (STAT). Eur. J. Neurosci. 12, 1165–1176.
Schweizer, A., Valdenaire, O., Koster, A., Lang, Y., Schmitt, G., Lenz, B., Bluethmann, H., and Rohrer, J. (1999). Neonatal lethality in mice deficient in XCE, a novel member of the endothelin-converting enzyme and neutral endopeptidase family. J. Biol. Chem. 274, 20450–20456.
Schweizer, U., Gunnersen, J., Karch, C., Wiese, S., Holtmann, B., Takeda, K., Akira, S., and Sendtner, M. (2002). Conditional gene ablation of Stat3 reveals differential signaling requirements for survival of motoneurons during development and after nerve injury in the adult. J. Cell Biol. 156, 287–297.
Seijffers, R., Allchorne, A. J., and Woolf, C. J. (2006). The transcription factor ATF-3 promotes neurite outgrowth. Mol. Cell. Neurosci. 32, 143–154.
Seijffers, R., Mills, C. D., and Woolf, C. J. (2007). ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J. Neurosci. 27, 7911–7920.
Sendtner, M., Kreutzberg, G. W., and Thoenen, H. (1990). Ciliary neurotrophic factor prevents the degeneration of motor neurons after axotomy. Nature 345, 440–441.
Shadiack, A. M., Sun, Y., and Zigmond, R. E. (2001). Nerve growth factor antiserum induces axotomy-like changes in neuropeptide expression in intact sympathetic and sensory neurons. J. Neurosci. 21, 363–371.
Shen, Y., Tenney, A. P., Busch, S. A., Horn, K. P., Cuascut, F. X., Liu, K., He, Z., Silver, J., and Flanagan, J. G. (2009). PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science 326, 592–596.
Shirotani, K., Tsubuki, S., Iwata, N., Takaki, Y., Harigaya, W., Maruyama, K., Kiryu-Seo, S., Kiyama, H., Iwata, H., Tomita, T., Iwatsubo, T., and Saido, T. C. (2001). Neprilysin degrades both amyloid beta peptides 1-40 and 1-42 most rapidly and efficiently among thiorphan- and phosphoramidon-sensitive en-dopeptidases. J. Biol. Chem. 276, 21895–21901.
Siegel, G., Saba, R., and Schratt, G. (2011). MicroRNAs in neurons: manifold regulatory roles at the synapse. Curr. Opin. Genet. Dev. 21, 491–497
Silver, J., and Miller, J. H. (2004). Regeneration beyond the glial scar. Nat. Rev. Neurosci. 5, 146–156.
Sleiman, S. F., Langley, B. C., Basso, M., Berlin, J., Xia, L., Payappilly, J. B., Kharel, M. K., Guo, H., Marsh, J. L., Thompson, L. M., Mahishi, L., Ahuja, P., Maclellan, W. R., Geschwind, D. H., Coppola, G., Rohr, J., and Ratan, R. R. (2011). Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J. Neurosci. 31, 6858–6870.
Smith, P. D., Sun, F., Park, K. K., Cai, B., Wang, C., Kuwako, K., Martinez-Carrasco, I., Connolly, L., and He, Z. (2009). SOCS3 deletion promotes optic nerve regeneration in vivo. Neuron 64, 617–623.
Stam, F. J., MacGillavry, H. D., Armstrong, N. J., de Gunst, M. C., Zhang, Y., van Kesteren, R. E., Smit, A. B., and Verhaagen, J. (2007). Identification of candidate transcriptional modulators involved in successful regeneration after nerve injury. Eur. J. Neurosci. 25, 3629–3637.
Sun, F., and He, Z. (2010). Neuronal intrinsic barriers for axon regeneration in the adult CNS. Curr. Opin. Neurobiol. 20, 510–518.
Tan, N. Y., and Khachigian, L. M. (2009). Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell. Biol. 29, 2483–2488.
Tanabe, K., Bonilla, I., Winkles, J. A., and Strittmatter, S. M. (2003). Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J. Neurosci. 23, 9675–9686.
Tanabe, K., Kiryu-Seo, S., Nakamura, T., Mori, N., Tsujino, H., Ochi, T., and Kiyama, H. (1998). Alternative expression of Shc family members in nerve-injured motoneurons. Brain Res. Mol. Brain Res. 53, 291–296.
Tanabe, K., Nakagomi, S., Kiryu-Seo, S., Namikawa, K., Imai, Y., Ochi, T., Tohyama, M., and Kiyama, H. (1999). Expressed-sequence-tag approach to identify differentially expressed genes following peripheral nerve axotomy. Brain Res. Mol. Brain Res. 64, 34–40.
Terao, E., Janssens, S., van den Bosch de Aguilar, P., Portier, M., and Klosen, P. (2000). In vivo expression of the intermediate filament peripherin in rat motoneurons: modulation by inhibitory and stimulatory signals. Neuroscience 101, 679–688.
Tetzlaff, W., Bisby, M. A., and Kreutzberg, G. W. (1988). Changes in cytoskeletal proteins in the rat facial nucleus following axotomy. J. Neurosci. 8, 3181–3189.
Tsujino, H., Kondo, E., Fukuoka, T., Dai, Y., Tokunaga, A., Miki, K., Yonenobu, K., Ochi, T., and Noguchi, K. (2000). Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: a novel neuronal marker of nerve injury. Mol. Cell. Neurosci. 15, 170–182.
Uchida, H., Ma, L., and Ueda, H. (2010a). Epigenetic gene silencing underlies C-fiber dysfunctions in neuropathic pain. J. Neurosci. 30, 4806–4814.
Uchida, H., Sasaki, K., Ma, L., and Ueda, H. (2010b). Neuron-restrictive silencer factor causes epigenetic silencing of Kv4.3 gene after peripheral nerve injury. Neuroscience 166, 1–4.
Uveges, T. E., Shan, Y., Kramer, B. E., Wight, D. C., and Parysek, L. M. (2002). Intron 1 is required for cell type-specific, but not injury-responsive, peripherin gene expression. J. Neurosci. 22, 7959–7967.
Valdenaire, O., Richards, J. G., Faull, R. L., and Schweizer, A. (1999). XCE, a new member of the endothelin-converting enzyme and neutral endopeptidase family, is preferentially expressed in the CNS. Brain Res. Mol. Brain Res. 64, 211–221.
Vanselow, J., Grabczyk, E., Ping, J., Baetscher, M., Teng, S., and Fishman, M. C. (1994). GAP-43 transgenic mice: dispersed genomic sequences confer a GAP-43-like expression pattern during development and regeneration. J. Neurosci. 14, 499–510.
Vargas, M. E., and Barres, B. A. (2007). Why is Wallerian degeneration in the CNS so slow? Annu. Rev. Neurosci. 30, 153–179.
Werner, A., Willem, M., Jones, L. L., Kreutzberg, G. W., Mayer, U., and Raivich, G. (2000). Impaired axonal regeneration in alpha7 integrin-deficient mice. J. Neurosci. 20, 1822–1830.
Whitfield, J., Neame, S. J., Paquet, L., Bernard, O., and Ham, J. (2001). Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron 29, 629–643.
Wierstra, I. (2008). Sp1: emerging roles – beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 372, 1–13.
Williams, A. H., Valdez, G., Moresi, V., Qi, X., McAnally, J., Elliott, J. L., Bassel-Duby, R., Sanes, J. R., and Olson, E. N. (2009). MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549–1554.
Xu, D., Emoto, N., Giaid, A., Slaughter, C., Kaw, S., deWit, D., and Yanagisawa, M. (1994). ECE-1: a membrane-bound metalloprotease that catalyzes the proteolytic activation of big endothelin-1. Cell 78, 473–485.
Yan, Q., Elliott, J., and Snider, W. D. (1992). Brain-derived neurotrophic factor rescues spinal motor neurons from axotomy-induced cell death. Nature 360, 753–755.
Yang, J., Chatterjee-Kishore, M., Staugaitis, S. M., Nguyen, H., Schlessinger, K., Levy, D. E., and Stark, G. R. (2005). Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 65, 939–947.
Yin, Y., Henzl, M. T., Lorber, B., Nakazawa, T., Thomas, T. T., Jiang, F., Langer, R., and Benowitz, L. I. (2006). Oncomodulin is a macrophage-derived signal for axon regeneration in retinal ganglion cells. Nat. Neurosci. 9, 843–852.
Yudin, D., Hanz, S., Yoo, S., Iavnilovitch, E., Willis, D., Gradus, T., Vuppalanchi, D., Segal-Ruder, Y., Ben-Yaakov, K., Hieda, M., Yoneda, Y., Twiss, J. L., and Fainzilber, M. (2008). Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron 59, 241–252.
Zhai, W., Jeong, H., Cui, L., Krainc, D., and Tjian, R. (2005). In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell 123, 1241–1253.
Zhang, H. Y., Zheng, S. J., Zhao, J. H., Zhao, W., Zheng, L. F., Zhao, D., Li, J. M., Zhang, X. F., Chen, Z. B., and Yi, X. N. (2011a). MicroRNAs 144, 145, and 214 are down-regulated in primary neurons responding to sciatic nerve transection. Brain Res. 1383, 62–70.
Zhang, S. J., Buchthal, B., Lau, D., Hayer, S., Dick, O., Schwaninger, M., Veltkamp, R., Zou, M., Weiss, U., and Bading, H. (2011b). A signaling cascade of nuclear calcium-CREB-ATF3 activated by synaptic NMDA receptors defines a gene repression module that protects against extrasynaptic NMDA receptor-induced neuronal cell death and ischemic brain damage. J. Neurosci. 31, 4978–4990.
Zheng, B., Lee, J. K., and Xie, F. (2006). Genetic mouse models for studying inhibitors of spinal axon regeneration. Trends Neurosci. 29, 640–646.
Zhou, F. Q., and Snider, W. D. (2005). Cell biology. GSK-3beta and microtubule assembly in axons. Science 308, 211–214.
Zhou, S., Yu, B., Qian, T., Yao, D., Wang, Y., Ding, F., and Gu, X. (2011). Early changes of microRNAs expression in the dorsal root ganglia following rat sciatic nerve transection. Neurosci. Lett. 494, 89–93.
Keywords: ATF3, c-Jun, STAT3, Sp1, DINE, transcription, axotomy, nerve injury
Citation: Kiryu-Seo S and Kiyama H (2011) The nuclear events guiding successful nerve regeneration. Front. Mol. Neurosci. 4:53. doi: 10.3389/fnmol.2011.00053
Received: 30 June 2011; Accepted: 29 November 2011;
Published online: 12 December 2011.
Edited by:
Gennadij Raivich, University College London, UKCopyright: © 2011 Kiryu-Seo and Kiyama. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Sumiko Kiryu-Seo, Department of Functional Anatomy and Neuroscience, Graduate School of Medicine, Nagoya University, 65 Tsurumai-cho, Showa-ku, Nagoya 466-8550, Japan. e-mail:c2tpcnl1QG1lZC5uYWdveWEtdS5hYy5qcA==