- 1KOKORO-Biology Group, Laboratories for Integrated Biology, Graduate School of Frontier Biosciences, Osaka University, 1-3 Yamadaoka, Suita, Osaka 565-0871, Japan
- 2Institute of Experimental Animal Research, Gunma University Graduate School of Medicine, 3-39-22 Showa-machi, Maebashi, Gunma 371-8511, Japan
- 3Department of Ultrastructural Research, National Institute of Neuroscience, Kodaira, Tokyo 187-8551, Japan
- 4CREST, Osaka University, 1-3 Yamadaoka, Suita, Osaka 565-0871, Japan
The generation of complex neural circuits depends on the correct wiring of neurons with diverse individual characteristics. To understand the complexity of the nervous system, the molecular mechanisms for specifying the identity and diversity of individual neurons must be elucidated. The clustered protocadherins (Pcdh) in mammals consist of approximately 50 Pcdh genes (Pcdh-α, Pcdh-β, and Pcdh-γ) that encode cadherin-family cell surface adhesion proteins. Individual neurons express a random combination of Pcdh-α and Pcdh-γ, whereas the expression patterns for the Pcdh-β genes, 22 one-exon genes in mouse, are not fully understood. Here we show that the Pcdh-β genes are expressed in a 3′-polyadenylated form in mouse brain. In situ hybridization using a pan-Pcdh-β probe against a conserved Pcdh-β sequence showed widespread labeling in the brain, with prominent signals in the olfactory bulb, hippocampus, and cerebellum. In situ hybridization with specific probes for individual Pcdh-β genes showed their expression to be scattered in Purkinje cells from P10 to P150. The scattered expression patterns were confirmed by performing a newly developed single-cell 3′-RACE analysis of Purkinje cells, which clearly demonstrated that the Pcdh-β genes are expressed monoallelically and combinatorially in individual Purkinje cells. Scattered expression patterns of individual Pcdh-β genes were also observed in pyramidal neurons in the hippocampus and cerebral cortex, neurons in the trigeminal and dorsal root ganglion, GABAergic interneurons, and cholinergic neurons. Our results extend previous observations of diversity at the single-neuron level generated by Pcdh expression and suggest that the Pcdh-β cluster genes contribute to specifying the identity and diversity of individual neurons.
Introduction
The central and peripheral nervous systems contain an enormous number of neurons, that have diverse characteristics and unique identities that enable them to generate complex neural circuits. Therefore, elucidating the molecular mechanisms that specify the unique characteristics of individual neurons is important for understanding the overall complexity of the nervous system.
Recent studies reported that two families of cell-cell recognition molecules, Dscam1 in insects and clustered protocadherin (Pcdh) in vertebrates, are promising candidates for specifying the unique characteristics of individual neurons (Zipursky and Sanes, 2010; Yagi, 2012). Dscam1 proteins are single-pass transmembrane domain proteins of the immunoglobulin superfamily that play important roles in neural circuit formation (Hattori et al., 2008; Schmucker and Chen, 2009). The fly Dscam1 gene encodes 19,008 different ectodomain isoforms, which are expressed in a biased stochastic fashion in individual neurons, thereby conferring a unique molecular identity on each Drosophila neuron (Neves et al., 2004). In addition, Hattori et al. (2007) showed that Dscam1 diversity is essential for generating complex neural circuits.
Approximately 100 of the diverse cadherin-superfamily genes are highly expressed in vertebrate brain (Takeichi, 2007; Yagi, 2008), and about 50 of these are encoded by the Pcdh gene clusters, Pcdh-α, Pcdh-β, and Pcdh-γ (Kohmura et al., 1998; Wu and Maniatis, 1999; Sugino et al., 2000; Hirayama and Yagi, 2006; Morishita and Yagi, 2007). Cadherin-superfamily proteins play key roles in the morphogenesis and function of the brain, including the formation and maintenance of the neuroepithelium, neurite extension, migration of neuronal cells, synaptogenesis, and synaptic plasticity (Takeichi, 2007; Suzuki and Takeichi, 2008). Previous studies revealed that the clustered Pcdhs play important roles in neuronal survival, axonal projection, synaptic connectivity, and several brain functions, including learning and memory (Wang et al., 2002b; Fukuda et al., 2008; Hasegawa et al., 2008; Katori et al., 2009). Since the clustered Pcdhs show unique gene regulations and form heteromultimeric Pcdh protein tetramers with homophilic binding specificity, the clustered Pcdhs are considered to be candidates for generating complex neural circuitry by providing molecular codes that determine neuronal individuality in the vertebrate central and peripheral nervous systems (Schreiner and Weiner, 2010; Zipursky and Sanes, 2010; Yagi, 2012).
The Pcdh-α, Pcdh-β, and Pcdh-γ gene clusters are tandemly localted on the mouse 18 chromosome. All isoforms of Pcdh-α, Pcdh-β, and Pcdh-γ genes possess their own promoters and variable exons in each gene cluster in the same direction (Wu et al., 2001). The Pcdh-α or Pcdh-γ variable exons are spliced onto each Pcdh-α or Pcdh-γ constant exons, respectively, that encode the common 3′ regions. In contrast, the Pcdh-β gene cluster does not contain constant exons and is instead comprised of unspliced single exon genes.
Extensive investigation of the gene regulation mechanisms in the clustered Pcdhs has identified some unique features. In situ hybridization analysis showed that the Pcdh-α and Pcdh-γ genes are differentially regulated at the level of individual neurons (Kohmura et al., 1998; Esumi et al., 2005; Frank et al., 2005), and single-cell analysis of Purkinje cells using multiplex RT-PCRs established that one or two variable exons in the Pcdh-α and Pcdh-γ clusters are stochastically expressed from each allele in individual Purkinje cells (Esumi et al., 2005; Kaneko et al., 2006). The expression of the clustered Pcdh genes is governed by the activation of individual promoters from among multiple promoters in the cluster (Tasic et al., 2002; Wang et al., 2002a). Furthermore, the discovery of a long-range regulatory element in the Pcdh-α (Ribich et al., 2006; Kehayova et al., 2011) and Pcdh-β (Yokota et al., 2011) gene clusters provided further support for a model in which promoter choice among multiple promoters drives unique expression patterns in single neurons.
A number of transcription factors or DNA-binding proteins have been implicated in the regulation of clustered Pcdh gene transcription: that is, CTCF (Golan-Mashiach et al., 2012; Hirayama et al., 2012; Monahan et al., 2012), NRSF/REST (Tan et al., 2010; Kehayova et al., 2011), SOX4 (Castillo et al., 2012), Mecp2 (Chahrour et al., 2008; Miyake et al., 2011), Egr-1 (Schippert et al., 2009), and Nipbl (Kawauchi et al., 2009). In addition, Pcdh promoter CpG methylation is inversely correlated with Pcdh expression (Tasic et al., 2002; Kawaguchi et al., 2008; Dallosso et al., 2009, 2012; Kaneko et al., 2009). These results are consistent with the idea that the Pcdh cluster genes and their sophisticated gene regulation mechanisms could be involved in establishing the unique molecular identities of individual neurons in the brain (Chess, 2005; Zipursky and Sanes, 2010; Yagi, 2012). Although the regulatory mechanisms for generating Pcdh diversity at the single-neuron level have been addressed to some extent, whether this diversity contributes to the functional complexity of the mammalian nervous system remains to be determined.
To date, little information has been published on the expression of the Pcdh-β genes, which consist of 22 single exon genes located on mouse chromosome 18 (Vanhalst et al., 2001), although the postsynaptic localization of one Pcdh-β protein, Pcdh-β16, was reported for mouse and primate retina (Junghans et al., 2008; Puller and Haverkamp, 2011). Here, we show that the expression patterns of the Pcdh-β genes are similar to those of the Pcdh-α and Pcdh-γ genes. 3′-RACE analysis of single Purkinje cells showed that Pcdh-β isoforms derived from monoallelic chromosomes were combinatorially expressed in individual Purkinje cells. Furthermore, in situ hybridization analysis showed, for the first time, scattered expression patterns at the level of individual neurons in the trigeminal ganglion, dorsal root ganglion, GABAergic interneurons, and cholinergic neurons. Our results extend previous findings on the single-neuron diversity generated by Pcdh expression and suggest that Pcdh-β cluster genes may contribute to specifying the identity and diversity of individual neurons.
Materials and Methods
Animals
C57BL/6 (B6) mice were purchased from Charles River Japan. F1 hybrid offspring were obtained by intercrossing mice of the laboratory strain B6 with the Japanese wild mouse strain JF1, obtained from the National Institute for Genetics (Mishima, Shizuoka, Japan). All procedures undertaken in this study were approved by our institute's Animal Care and Use Committee and conform to Japanese guidelines.
RT-PCR
Total RNA was prepared from P21 B6 mouse cerebellum using Trizol reagent (Invitrogen) and reverse-transcribed by Superscript III reverse transcriptase (Invitrogen), using random primers, according to the manufacturer's protocol. The PCR of each Pcdh-β isoform was performed using cDNA as the template, 2 μl of 10× Ex-taq PCR buffer, 2 μl of 2.5 mM each dNTP mix, 0.4 μl of RT-PCR primer set, 0.1 μl of Ex-taq HS polymerase (Takara, Japan), and water to a final volume of 20 μl. The PCR conditions were 3 min at 94°C and then 35 cycles of 30 s at 94°C, 30 s at 60°C (for Pcdh-β1) or 58°C (for other Pcdh-β genes), and 2 min at 72°C. The primer sets and sequences are shown in Table S1.
In situ Hybridization
In situ hybridization was performed as described previously (Esumi et al., 2005; Kaneko et al., 2006; Watakabe et al., 2007; Katori et al., 2009; Noguchi et al., 2009; Watakabe et al., 2010; Yokota et al., 2011) on 10-μm-thick frozen sagittal and coronal sections prepared from the B6 mouse using Pcdh-β highly conserved and isoform-specific cRNA probes. The mouse ages and regions examined are indicated in the figure legends. A probe for a highly conserved region of the Pcdh-β2–β22 genes, pan-Pcdh-β probe, was designed based on the sequence in mouse β12 (nucleotides 1422–2152 according to GeneBank™ accession number NM_053137). The Pcdh-β isoform-specific probes were designed based on mouse β3 (nucleotides 237–1364; GeneBank™ accession number NM_053128), β15 (nucleotides 161–1362; GeneBank™ accession number NM_053140), β16 (nucleotides 277–1293; GeneBank™ accession number NM_053141), β19 (nucleotides 227–1147; GeneBank™ accession number NM_053144), and β22 (nucleotides 213–1146; GeneBank™ accession number NM_053147). The similarity of each Pcdh-β isoform-specific probe to other Pcdh-α, Pcdh-β, and Pcdh-γ genes and to other mouse genes was low enough (less than 65%) to detect specific Pcdh-β transcripts of interest. These specific isoform sequences were amplified using the primers shown in Supplemental Table S1 (for in situ hybridization probe) and KOD-plus polymerase (Toyobo, Japan). The PCR product was cloned into the pCRII vector (Invitrogen), and the sequence was confirmed by DNA sequencing. The resultant plasmids were used as templates to synthesize digoxigenin (DIG)-11-UTP-labeled (Roche) cRNA probes.
To obtain tissue samples, mice were deeply anesthetized with diethyl ether, then the brain was removed, embedded in O.C.T. compound (Sakura), and quickly frozen in isopentane cooled with dry ice. Sections (10-μm thick) were cut on a cryostat (Leica CM3050), thaw-mounted on slides (Matsunami), and air-dried. The sections were fixed in 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB, pH 7.3) for 10 min, washed three times in PBS, pH 7.4, acetylated for 10 min in 0.25% acetic anhydride in 0.1 M triethanolamine-HCl, pH 8.0, and washed three more times with PBS. Prehybridization was performed with hybridization buffer [50% formamide, 5 × SSC (20 × SSC is 3 M NaCl, 0.3 M sodium citrate, pH 7.0), 5× Denhardt's, 250 μg/ml yeast tRNA, 500 μg/ml salmon sperm DNA, and 0.2% RNasin RNase inhibitor (Promega)] for 30 min. The DIG-labeled cRNA probes were denatured for 5 min at 82°C and chilled on ice. The sections were covered with hybridization buffer containing 1 μg/ml of the cRNA probes, added drop wise, and a coverslip was added. The slides were incubated for 12 hrs at 72°C in a humidified chamber (50% formaldehyde, 5 × SSC), washed three times with 0.2 × SSC at 72°C, washed three times with TBS, pH 7.5 (100 mM Tris-HCl, pH 7.5, 100 mM NaCl), and rinsed with TNT (0.05% Tween 20 in TBS, pH 7.5). To detect the hybridized probes, the sections were blocked with 1× blocking solution (Roche) in TNT for 30 min, and then incubated with alkaline phosphatase (AP)-conjugated anti-DIG antibody (1:1000 dilution, Roche) in the blocking solution for 1 h. The sections were rinsed three times with TBS, pH 7.5, and the enzymatic activity was visualized with 0.2 mM 5-bromo-4-chloro-3-indolyl-phosphate, 0.2 mM nitro blue tetrazolium (NBT/BCIP) in 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 20 mM MgCl2, in the dark, until the signal reached a satisfactory intensity.
We also carried out double fluorescence in situ hybridization. Fluorescein (FITC)-labeled cRNA probes against glutamate decarboxylase 1 (GAD67), choline O-acetyltransferase (ChAT), and vesicular acetylcholine transporter (VAChT) were synthesized with a FITC-UTP RNA labeling Kit (Roche). The probes were designed based on mouse GAD67 (nucleotides 1073 ~ 2012; GeneBank™ accession number NM_008077), rat ChAT (nucleotides 243 ~ 745; GeneBank™ accession number NM_001170593), and rat VAChT (Slc18a3) (nucleotides 1201 ~ 2194; GeneBank™ accession number NM_031663). The DIG-labeled Pcdh-β probe and a FITC-labeled GAD67, ChAT, or VAChT cRNA probe were hybridized to sections, which were washed as above.
To detect the FITC-labeled probes, the sections were incubated with an anti-FITC antibody conjugated with horseradish peroxidase (Jackson Immuno Research Laboratory, 1:4000 in the blocking buffer) for 2–5 hrs at room temperature. After being washed in TNT three times for 15 min, the sections were treated with 1:50 diluted TSA-Plus [dinitrophenol (DNP)] reagents for 5 min, according to the manufacturer's instructions (Perkin-Elmer, Wellesley, MA), which converted the FITC signals to DNP signals. After being washed in TNT three times for 10 min, the sections were incubated overnight at 4°C with an anti-DNP antibody conjugated with Alexa488 (1/500, Molecular Probes) in 1% blocking buffer. At this point, an anti-DIG antibody conjugated with AP (1:1000, Roche Diagnostics) was included in the incubation, to detect the DIG-labeled probes. The AP activity was visualized with NBT/BCIP (see above) or with HNPP fluorescence.
To detect the DIG-labeled probes with HNPP fluorescence, the sections were washed three times in TNT, once in TS 8.0 (0.1 M Tris-HCl, pH 8.0, 0.1 M NaCl, 50 mM MgCl2), and the AP activity was detected using an HNPP fluorescence detection set (Roche Diagnostics), according to the manufacturer's instructions. The incubation for this substrate was carried out for 30 min and stopped by washing in TS 8.0. The sections were then counterstained with 4′,6-diamidino-2-phenylindope, dihydrochloride (DAPI, Invitrogen, USA) diluted in TS 8.0–100 ng/ml for 3 min. After a brief wash in TS 8.0, the sections were mounted with CC/Mount (Diagnostic Biosystems, Pleasanton, CA) mounting medium.
Split Single-Cell 3′-RACE of Purkinje Cells
A split single-cell 3′-RACE method for single Purkinje cells was developed to analyze the one-exon Pcdh-β3, β9, β10, β15, β19, and β22 genes, and was based on our previously published method for single-cell RT-PCR (Esumi et al., 2006). The primer sets and sequences are shown in Table S1 (for split single-cell 3′-RACE). In brief, Purkinje cells were prepared from the cerebellum of P21 mice, from the F1 litter of a B6 female × JF1 male. The tissue was dissected and digested with 90 units of papain (Worthington) at 37°C for 30 min in 10 ml of dissociation solution [0.002% DL-cysteine HCl (Sigma), 0.05% DNase I (Sigma), 0.1% bovine serum albumin (Sigma), and 0.05% glucose (Nacalai Tesque)]. The digested tissue was spun for 8 min at 300× g, and the pellet was resuspended in Dulbecco's modified Eagle's medium (Sigma). To remove debris, the cells were filtered through a 100-μm cell strainer (Falcon). Single Purkinje cells were picked up by glass capillary and placed in thin, 200-μl PCR tubes, with 6 μl of RNase-free water. Complementary DNA was synthesized from the single-cell samples after adding 12.5 pmol/reaction of adaptor-attached oligo dT primer and 2.5 pmol/reaction of Pcp-2 RT primer in a total volume of 10 μl, using six units of Thermoscript reverse transcriptase (Invitrogen), according to the manufacturer's protocol, at 55°C for 60 min. The reaction was stopped by heating at 85°C for 5 min.
The cDNAs derived from a single Purkinje cell were split into three PCR tubes as 3.3-μl aliquots and used as a template for the first round of a multiplex PCR, which was performed using the 3.3 μl of cDNA, 2.5 μl of 10× Ex-taq PCR buffer, 2 μl of 2.5 mM each dNTP mix, 1.5 μl of the first PCR primer set, 0.1 μl of Ex-taq HS polymerase (Takara), and 15.6 μl of water. The PCR conditions were an initial 3 min at 95°C and then 28 cycles of 20 s at 95°C, 20 s at 58°C, and 5 min at 68°C. The first PCR products were split into 0.5-μl aliquots, which were used as the template in the second round of nested PCR with 2 μl of 10× Ex-taq PCR buffer, 1.6 μl of 2.5 mM each dNTP mix, 0.4 μl of the second PCR primer set, 0.1 μl of Ex-taq HS polymerase (Takara), and 15.4 μl of water. The PCR conditions for the Pcdh-β genes were an initial 3 min at 96°C, 40 cycles of 30 s at 96°C, 15 s at 56°C (for β3, β9, and β15) or 58°C (for β10, β19, and β22), and 1 min at 72°C. The PCR conditions for the β-actin and Pcp-2 were described in previous reports (Esumi et al., 2005, 2006; Kaneko et al., 2006). The primer sets and sequences are shown in Table S1.
The secondary PCR products were divided into two tubes. One PCR-amplified product was analyzed by agarose gel electrophoresis: before loading, the sample was mixed with electrophoresis loading buffer (Takara) and heated to 72°C for 5 min, to sharpen the electrophoretic bands. The other half of the PCR sample was purified by polyethylene glycol precipitation and sequenced by direct sequencing, using a BigDye™ DNA sequencing kit (version 3.1) (ABI). The data were analyzed on an ABI Prism 3100 or an ABI Prism 3730 Genetic Analyzer. Direct sequencing permitted us to identify the allele from which each purified PCR product was derived. The discrimination of monoallelic expression from biallelic expression was accomplished using SeqScape software (ABI). The sequencing results from each 3′-RACE PCR product were confirmed by performing direct sequencing twice, using two distinct pairs of forward and reverse primers.
Results
Expression of the Pcdh-β Transcripts in Mouse Brain
To examine the expression pattern of each Pcdh-β gene in the brain, we first carried out an RT-PCR analysis of all 22 Pcdh-β gene family exons, using cDNA from the P21 mouse cerebellum (Figure 1A). This analysis clearly showed that all 22 Pcdh-β gene family exons were transcribed in the cerebellum. Next, we identified the 3′ ends of some of the Pcdh-β transcripts by 3′-RACE analysis using RNA from P21 mouse cerebellum (Table S2). No spliced product of Pcdh-β with constant region exons of Pcdh-α or Pcdh-γ was detected by first-round RT-PCR analysis (data not shown). These data re-confirmed that the Pcdh-β genes are transcribed as 3′ polyadenylated mRNAs (Vanhalst et al., 2001).
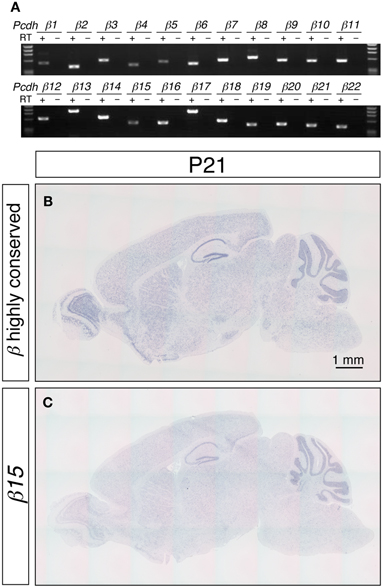
Figure 1. Expression of the Pcdh-β transcripts in the mouse brain. (A) RT-PCR analysis of P21 cerebellum using isoform-specific primers demonstrated that all 22 of the Pcdh-β transcripts were expressed. In situ hybridization analysis with a Pcdh-β highly conserved probe (B) and with a probe specific for β15 (C). Sagittal sections of P21 mouse brain are shown. The distributions of the signals were almost identical, but the intensity of the β15-specific probe at the macroscopic level was weaker than that obtained with the Pcdh-β highly conserved probe, suggesting that the expression of the Pcdh-β transcripts overlapped at the macroscopic level.
The distribution of the Pcdh-β transcripts in the P21 mouse brain was examined by in situ hybridization (Figures 1B,C). Because the DNA sequence that corresponds to extracellular cadherin domain 4 (EC4)-EC5-EC6 of Pcdh-β12 is highly conserved from β2 to β22 (more than 84% similarity), we used this sequence as a pan-Pcdh-β probe (Pcdh-β highly conserved probe, hereafter) to detect all 21 isoforms of Pcdh-β. We excluded the β1 gene from this study because of its low similarity to β2–β22. The Pcdh-β highly conserved probe hybridized widely in the brain, and prominent signals were observed in the olfactory bulb, hippocampus, and cerebellum. No signal was observed with the sense probe (data not shown). These results are consistent with findings obtained for rat brain (Sago et al., 1995).
To examine the distribution of each member of the Pcdh-β subfamily genes, we performed an in situ hybridization analysis using probes that detected specific Pcdh-β isoforms. Sequences showing low similarity (<65%) with other Pcdh-β gene members were used as probes. Although the Pcdh-β genes are very similar to each other, the similarity of β3, β15, β16, β19, and β22 is relatively low, and could be reliably discriminated. When examined at low magnification, the signals for β15 (Figure 1C) and the other 4 Pcdh-β genes (data not shown) hybridized in a pattern that was identical to that obtained using the Pcdh-β highly conserved probe, but with lower signal intensity. These results suggested that the Pcdh-β isoforms, at least β3, β15, β16, β19, and β22, have macroscopically overlapping expression patterns in the mouse brain.
Monoallelic and Combinatorial Gene Regulation of Pcdh-β Genes in Single Purkinje Cells
To investigate the gene regulation of the Pcdh-β cluster, we examined the expression patterns of the Pcdh-β transcripts in cerebellar Purkinje cells, since the regulation of the Pcdh-α, and Pcdh-γ clusters was extensively studied in these cells (Esumi et al., 2005; Kaneko et al., 2006). In the P21 cerebellum, the Pcdh-β highly conserved probe intensely stained most Purkinje cells (Figure 2A). In contrast, the specific probes for β3, β15, β16, β19, and β22 stained only a subset of the Purkinje cells (Figures 2B–F), showing that the Pcdh-β isoforms, at least β3, β15, β16, β19, and β22, were expressed in a scattered pattern among the Purkinje cells. Next, we analyzed the age-dependence of the expression patterns of the Pcdh-β genes in Purkinje cells. From P10 to P150, all the Purkinje cells were intensely stained by the Pcdh-β highly conserved probe (Figures 2A,G,I,K,M), whereas the specific probe for β15 stained only a subset of Purkinje cells (Figures 2C,H,J,L,N). Likewise, the specific probes for β3, β16, β19, and β22 stained only subsets of the Purkinje cells, resulting in scattered patterns such as those seen at P21 (data not shown). Therefore, the expression of the β3, β15, β16, β19, and β22 isoforms, and probably all of the Pcdh-β isoforms, was scattered among Purkinje cells from P10 to P150. On the other hand, in granule cells stained signals for both Pcdh-β highly conserved and specific β15 probes were reduced in P90 and P150.
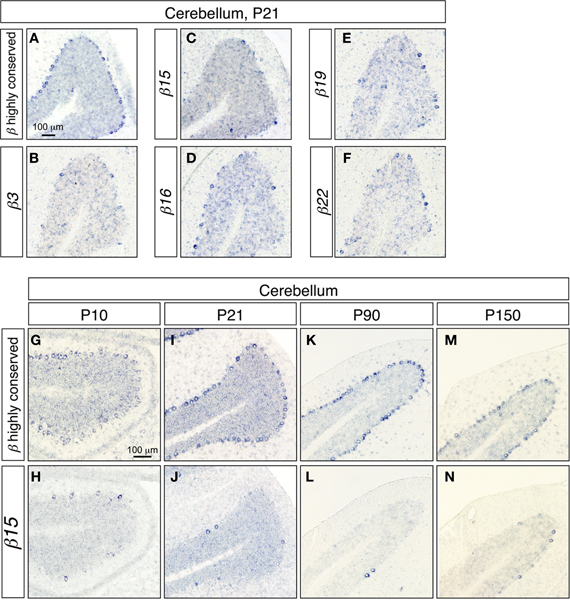
Figure 2. In situ hybridization analysis of Pcdh-β in the mouse cerebellum. (A–F) Coronal sections. (A) In situ hybridization with the Pcdh-β highly conserved probe; (B–F) in situ hybridization using probes specific for β3 (B), β15 (C), β16 (D), β19 (E), and β22 (F). (G–M) Comparison of staining with the Pcdh-β highly conserved probe (G,I,K,M) and the β15-specific probe (H,J,L,N) at P10–P150.
The next step was to confirm the differential expression revealed by the in situ hybridization, and to further investigate whether the gene regulation of Pcdh-β transcripts in single neurons is monoallelic and combinatorial. For this purpose, we modified our split single-cell RT-PCR method. Since the Pcdh-β genes consist of only one exon (Vanhalst et al., 2001), conventional RT-PCR against Pcdh-β exon could amplify not only the Pcdh-β transcripts but also the genomic DNA, leading us to erroneous conclusions. Fortunately, the Pcdh-β transcripts are polyadenylated (Table S2), so we adapted our method to 3′-RACE. First, we searched for a region of strong similarity shared by several of the Pcdh-β genes to use in the first multiplex primer annealing and found that base pairs 2289–2307 of β15 are highly conserved in β3, β9, β10, β15, β19, and β22. We next sequenced the region proximal to the functional polyadenylation signal of these six Pcdh-β genes in the JF1 mouse strain, and detected polymorphisms against the B6 strain (Figure S1). These data ultimately led us to choose β3, β9, β10, β15, β19, and β22 as the targets for this analysis.
We tested our split single-cell 3′-RACE method using Purkinje cells of P21 F1 mice from a B6 × JF1 cross. After optimizing the experimental conditions, we reproducibly obtained the predicted fragments for β3, β9, β10, β15, β19, β22, β-actin (positive control for 3′-RACE amplification), and Pcp-2 (a marker for Purkinje cells). Only single Purkinje cell-derived cDNA samples that showed intense expressions of Pcp-2 and β-actin were used for the expression analysis of the individual Pcdh-β isoforms in subsequent experiments.
Next, we analyzed the expression patterns of the β3, β9, β10, β15, β19, and β22 transcripts in single Purkinje cells using the split single-cell 3′-RACE method (Figure 3). Of the 28 single Purkinje cells analyzed, all were positive for the PCR products of Pcp-2 and β-actin, confirming that they were mature Purkinje cells, and their mRNAs were successfully reverse-transcribed. None of the control samples (#1–8, #1–16, #1–24, #1–32, or #1–40), which did not contain Purkinje cells, yielded PCR products, confirming the reliability of this method.
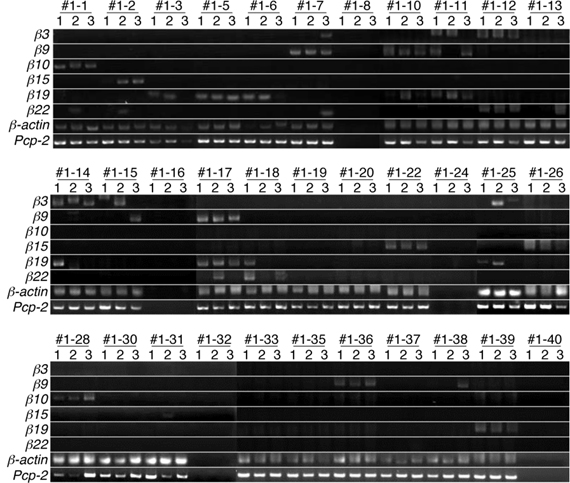
Figure 3. Split single-cell 3′-RACE analysis of Pcdh-β in Purkinje cells. Purkinje cells were from the P21 F1 progeny of a cross between the B6 and JF1 strains. Individual Purkinje cells are indicated as #1–1 to #1–39. Samples #1–8, #1–16, #1–24, #1–32, and #1–40 were negative controls that contained no cells. The RNAs in each Purkinje cell were reverse transcribed using an adapter-attached oligo dT primer and a Pcp-2-specific primer set. Each RT sample was divided into three separate tubes and subjected to 3′-RACE (PCR). The three independent PCR products are referred to as 1, 2, and 3. The results for electrophoresis of the second-round 3′-RACE (PCR)-products are shown.
When the cells were subjected to the Pcdh-β gene-specific second-round PCR, 21 cells were positive for some of the Pcdh-β genes. Of these, 15 showed specific amplification of one of the six Pcdh-β genes investigated in all three (3/3) sample tubes (for example, β10 in Cell #1–1, β15 in Cell #1–2, and β19 in Cell #1–5). In addition to the 3/3 PCR amplifications, five Purkinje cells showed specific PCR amplifications in 2/3 tubes (for example, β19 in Cells #1–3, β3 in Cell #1–15, and β22 in Cell #1–18), and specific PCR amplification was seen in 1/3 tubes for two Purkinje cells (β22 in Cell #1–13 and β9 in Cell #1–38). Obtaining only 2/3 and 1/3 positive tubes may suggest that these cells contained a low amount of the corresponding mRNAs. The remaining seven cells were negative for any of the Pcdh-β genes, probably indicating that other Pcdh-β genes not included in this analysis were expressed by these cells. Finally, more than one Pcdh-β isoform was amplified in 3/3 tubes of three Purkinje cells (β9 and β19 in Cell #1–10, β3 and β22 in Cell #1–12, and β9 and β19 in Cell #1–17). These results strongly suggested that the Pcdh-β genes are expressed in Purkinje cells in a differential and combinatorial manner.
Next, each PCR product was directly sequenced to distinguish among three possible patterns of expression: monoallelic expression of the maternal allele, monoallelic expression of the paternal allele, and biallelic expression. In the case of β-actin, all the PCR products showed a biallelic expression pattern. This was consistent with our previous analysis (Esumi et al., 2005; Kaneko et al., 2006), which was obtained by split single-cell RT-PCR. Thus, these findings confirmed the reliability of the present method. In contrast to β-actin, a monoallelic expression pattern was found for the Pcdh-β genes in 17 of the 18 3/3 3′-RACE-positive products (94%) (Figure 4) as follows: β10 in Cell #1–1, β15 in Cell #1–2, β19 in Cell #1–5, β9 in Cell #1–7, β19 in Cell #1–5, β9 and β19 in Cell #1–10, β19 in Cell #1–11, β3 and β22 in Cell #1–12, β3 in Cell #1–14, β9 and β19 in Cell #1–17, β15 in Cell #1–22, β15 in Cell #1–26, β10 in Cell #1–28, β19 in Cell #1–39. Biallelic expression was observed only for β9 in Cell #1–36. Statistically, if this assay amplified only a single cDNA molecule, even though biallelic products were present in the PCR tube, an artifactual 3/3 monoallelic expression pattern would theoretically be observed for 25% of the cells (Rhoades et al., 2000). Clearly, at 94% vs. 25%, there was a highly significant difference, which was upheld by a χ–square analysis between monoallelic and biallelic expression (P < 10−4). Of the 17 monoallelically expressed genes, seven were derived from the B6 allele and 10 from the JF1 allele. Thus, no allelic distortion from the paternal or maternal allele was found in this assay. Taken together, these results clearly showed that the Pcdh-β cluster genes are regulated monoallelically and combinatorially in individual Purkinje cells. This gene regulation would provide the molecular basis for the random expression of the Pcdh-β cluster genes, similar to the Pcdh-α and Pcdh-γ clusters.
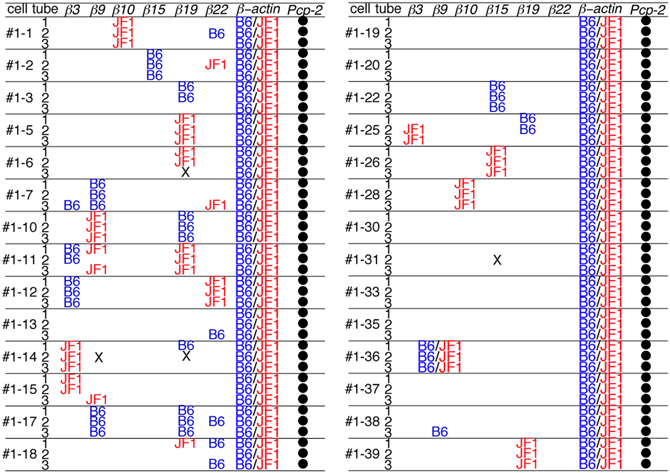
Figure 4. Distribution of the B6 and JF1 alleles as the source of each Pcdh-β gene in individual Purkinje cells. To determine whether the amplified 3′-RACE (PCR) products were from the B6 or JF1 allele, we performed direct sequencing of the products in each of the three tubes from 28 Purkinje cells. The samples that contained an insufficient quantity of product for sequencing are indicated with an X. The amplified Pcp-2 products were not sequenced and are indicated with a closed circle.
Diversity of the 3′-Terminus of the Pcdh-β Transcripts within Single Purkinje Cells
During the expressional analysis of the Pcdh-β transcripts using single-cell 3′-RACE, we simultaneously analyzed whether the 3′-terminus of each Pcdh-β transcript in single neurons was homogeneous or heterogeneous (Figure 5). In most individual Purkinje cells, the 3′-terminus of each Pcdh-β transcript was homogeneous, although we observed 3′-terminal heterogeneity in two cells: Pcdh-β22 in Cell #2–10 (Figures 5A,C) and Pcdh-β15 in Cell #2–21 (Figures 5B,D). The β22 transcripts in Cell #2–10 showed three types of 3′-termini, while the β15 transcripts in Cell #2–21 showed two types of 3′-termini. These data indicate that the polyadenylation of each Pcdh-β gene within a single neuron is not always homogeneous, suggesting the existence of transcriptional diversity for each Pcdh-β gene.
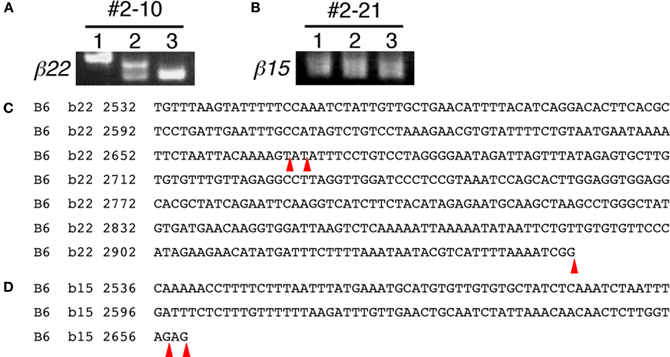
Figure 5. 3′-terminal diversity of Pcdh-β transcripts within single Purkinje cells. Single-cell 3′-RACE analysis of β22 in Cell #2–10 (Figures 5A,C) and β15 in Cell #2–21 (Figures 5B,D). The position of polyadenylation is indicated by red arrowheads. The β22 transcripts in Cell #2–10 showed three types of 3′-termini. The β15 transcripts in Cell #2–21 showed two types of 3′-termini.
Scattered Expression of Pcdh-β Transcripts in the Central and Peripheral Nervous Systems
To extend our knowledge of the neuron types that showed random expression of the Pcdh-β cluster genes, which might correspond to the specification of individual neuron identity, we examined the expression of Pcdh-β genes in a wide range of neurons using in situ hybridization. In the P21 hippocampus, intense signals from the Pcdh-β highly conserved probe were observed in all the pyramidal cell–layer neurons in CA1 and CA3 (Figures 6A,D,G). In contrast, the specific probes for β15 and β22 stained only subsets of these neurons (Figures 6B,E,H, and C,F,I, respectively). The other specific probes (for β3, β16, and β19) also stained only subsets of these neurons (data not shown). Furthermore, in the P21 cerebral cortex, most neurons showed intense staining for the Pcdh-β highly conserved probe (see Figures 6J and K for staining in the P21 motor cortex and Figures 6N and O for staining in the P21 visual cortex). In contrast, the specific probe for β22 stained only a subset of neurons in each of these regions (Figures 6L,M,P,Q), as did the other specific probes (data not shown). These results clearly demonstrated that these Pcdh-β gene isoforms were expressed only in subsets of neurons in the cerebral cortex and of pyramidal neurons in the hippocampal CA1 and CA3 areas, indicating that these Pcdh-β gene isoforms are expressed randomly and differentially in these neurons.
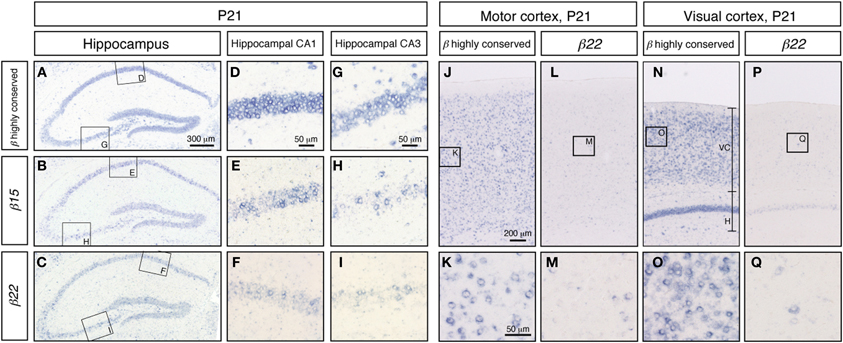
Figure 6. In situ hybridization analysis of Pcdh-β in mouse hippocampus and cerebral cortex. Pcdh-β gene expression in P21 mouse hippocampus at low (A,B,C) and high (D–I) magnification, showing the CA1 and CA3 fields. (A,D,G) β2 to β22 expression using the Pcdh-β highly conserved probe. (B,E,H) β15 expression. (C,F,I) β22 expression. Examination of the cerebral motor cortex (J–M), and cerebral visual cortex (N–Q) with specific probes for β22 (L,M,P,Q), and the Pcdh-β highly conserved probe (J,K,N,O). Most neurons were stained by the Pcdh-β highly conserved probe, but the specific probes stained only subsets of these neurons, suggesting that each member of the Pcdh-β subfamily is expressed differentially in subpopulations of these neurons.
We also examined the expression of the Pcdh-β transcripts in the trigeminal and dorsal root ganglia. As in the cerebral cortex and the hippocampus, these peripheral nervous system neurons showed prominent signals from the Pcdh-β highly conserved probe (Figures 7A,D), while distinct subsets were labeled with the β15 and β22 probes (Figures 7B,C,E,F) as well as the β3, β16, and β19 probes (data not shown). These results indicated that each Pcdh-β gene isoform is expressed randomly and differentially in the peripheral nervous system (at least in the trigeminal and dorsal root ganglia).
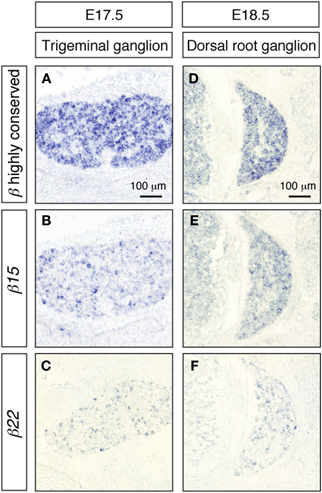
Figure 7. In situ hybridization analysis of Pcdh-β genes in mouse trigeminal and dorsal root ganglia. β2 to β22 expression using the Pcdh-β highly conserved probe on embryonic mouse trigeminal ganglion (A) and dorsal root ganglion (D). (B,E) Specific expression of β15; (C,F) specific expression of β22. Most neurons were stained by the Pcdh-β highly conserved probe, but the specific probes stained only subsets of these neurons, suggesting that each member of the Pcdh-β subfamily is expressed differentially in subpopulations of these neurons.
To examine whether the Pcdh-β cluster genes showed random-scattered expression in GABAergic and cholinergic neurons, we performed double in situ hybridization analysis of the Pcdh-β genes with marker genes for GABAergic and cholinergic neurons. In the hippocampus, GABAergic interneurons constitute an inhibitory network that regulates excitatory neurons (Kullmann, 2011). The GABAergic interneurons were labeled with a GAD67 probe (Figures 8D–F) and showed strong labeling with the Pcdh-β highly conserved probe (Figures 8A,G). Distinct subsets of GABAergic interneurons were labeled with the β16 and β22 probes (Figures 8B,H,C,I). In the brain stem facial nucleus, cholinergic neurons, which were labeled with ChAT + VAChT probes (Figures 8L,M) (Arvidsson et al., 1997), showed prominent labeling by the Pcdh-β highly conserved probe (Figures 8J,N), and distinct subsets were labeled with the β16 probe (Figures 8K,O). Taken together, these results revealed that each member of the Pcdh-β cluster genes is expressed in a scattered pattern in a wide range of neuron types in the central and peripheral nervous systems in mouse.
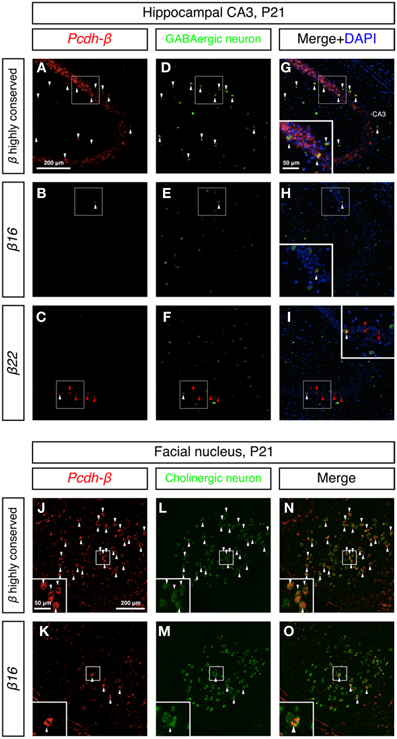
Figure 8. Differential expression of Pcdh-β genes in GABAergic interneurons and cholinergic neurons. (A–I) Double in situ hybridization analysis of Pcdh-β genes (red: HNPP fluorescence) and GAD67 (green) in sagittal sections of P21 mouse hippocampus. (J–O) Double in situ hybridization analysis of Pcdh-β genes (red: false color of NBT/BCIP) and cholinergic neuron markers (ChAT + VAChT, green) in coronal sections of P21 mouse brain stem facial nucleus. Insets: High-magnification photomicrograph of the outlined area. Arrowheads indicate neurons expressing both Pcdh-β and GAD67 or ChAT + VAChT mRNAs (white or black), and β22 mRNA alone (red), respectively. Most neurons were stained by the Pcdh-β highly conserved probe, but the specific probes stained only subsets of these neurons, suggesting that each member of the Pcdh-β subfamily is expressed differentially in subpopulations of GABAergic interneurons in the hippocampus and cholinergic neurons in the facial nucleus.
Discussion
In this study, we extended previous analyses on how Pcdh genes contribute to single-neuron diversity, by examining the expression patterns of the Pcdh-β genes using in situ hybridization analysis and the newly developed single-cell 3′-RACE analysis. Our results revealed that the Pcdh-β isoforms showed random-like scattered expression in both the central and the peripheral nervous systems, and that the Pcdh-β genes were regulated monoallelically and combinatorially in single Purkinje cells. We argue that the Pcdh-α, Pcdh-β, and Pcdh-γ gene clusters confer unique molecular identities on individual neurons that could be responsible for appropriate neuronal circuit formation.
The newly developed split single-cell 3′-RACE method revealed that a variety of 3′-ends of Pcdh-β mRNAs could be expressed by a single Purkinje cell. There are numerous examples of genes with multiple polyadenylation signals, and the 3′ ends of their transcripts sometimes vary with the developmental stage or state of the tissue in which they are expressed (Edwalds-Gilbert et al., 1997). However, it is unclear if the variation in the 3′-ends of mRNAs is constant or changeable within a single cell during a single time-frame. The present study revealed that some of the Pcdh-β genes have more than two functional polyadenylation signals, and that the polyadenylation site can vary in a single cell. For example, in Cell #2–10, three different variations of the 3′ end of β22 were detected. Two of the three transcripts utilized the same polyadenylation signal, but the polyadenylation was added on a different nucleotide, while the other transcript utilized a different polyadenylation signal (Figure 5). This finding shows that even within a single cell the polyadenylation site can be variable, and suggests that the polyadenylation of the 3′ ends of Pcdh-β transcripts may be tightly or stochastically regulated.
A previous study detected the expression of several members of the Pcdh-β genes in mouse and human brain as a single exonic and 3′-polyadenylated form (Vanhalst et al., 2001). Furthermore, several studies have reported the expression of Pcdh-β transcripts in the central nervous system including rat brain, rat hippocampus, and mouse retina (Sago et al., 1995; Schippert et al., 2009; McGowan et al., 2011). Our results confirm these findings and expand them to show that some, if not all, of the 22 Pcdh-β genes are expressed in the central and peripheral nervous system, including the cerebellum, hippocampus, cerebral cortex, trigeminal ganglion, dorsal root ganglion, and brain stem.
Focusing on the individual Pcdh-β genes, our findings of random-like and combinatorial expression of the individual Pcdh-β genes extensively broaden the potential for Pcdh-driven single-neuron diversity. The random-like scattered expression of individual Pcdh-α and Pcdh-γ genes has been previously observed in olfactory bulb periglomerular neurons, hippocampal neurons, Purkinje cells, dorsal root ganglia neurons, and choroid plexus epithelial cells (Kohmura et al., 1998; Esumi et al., 2005; Frank et al., 2005; Kaneko et al., 2006; Prasad and Weiner, 2011; Lobas et al., 2012). In this study, we observed that individual Pcdh-β genes are expressed with a similar scattered pattern in cerebellar Purkinje cells, hippocampus, cerebral cortex, trigeminal ganglia, dorsal root ganglia, GABAergic interneurons, and cholinergic neurons. This pattern persisted during development, in that the random-like scattered expression of the Pcdh-β genes in cerebellar Purkinje cells was observed from P10 to P150. Furthermore, our data clearly showed that the individual Pcdh-β genes are regulated monoallelically and combinatorially in cerebellar Purkinje cells, similar to the Pcdh-α and Pcdh-γ genes.
Our present results suggest that the individual Pcdh-β genes are differentially expressed among the various neuron types in the central and peripheral nervous systems from the juvenile to adult stages, and imply that the Pcdh-β gene cluster is regulated by similar mechanisms to the Pcdh-α and Pcdh-γ genes. The similar genomic organization, involving multiple promoters with a conserved sequence element (CGCT), long-range enhancer element, and multiple CTCF-binding sites, supports this hypothesis (Wu et al., 2001; Ribich et al., 2006; Handoko et al., 2011; Yokota et al., 2011). To date, we have not found any rules governing the spatial distribution of the Pcdh-β transcripts; however it remains to be elucidated whether or not the pattern is truly random. One intriguing possibility is that the Pcdh-β expressional repertoires in individual neurons determine their target status during neural circuit formation; i.e., pre- and post-synaptic neurons might express the same sets of Pcdh-β genes. The present finding that Pcdh-β genes show a scattered expression pattern on possible pre- and post-synaptic neurons in the hippocampal inhibitory interneuron network (Figures 6D–I and 8B,C,E,F,H,I) supports this idea.
The present study showed that the expression of the Pcdh-β gene cluster is monoallelic and combinatorial in single Purkinje cells. This strongly suggests that individual Purkinje cells express a variable repertoire of some, if not all, of the 22 Pcdh-β genes. In this study, we investigated six of the 22 Pcdh-β genes and showed that at least two types of Pcdh-β per allele can be expressed by a single Purkinje cell (see β9 and β19 in Cell #1–17, and β3 and β9 in Cell #2–22). The variety of Pcdh-β species expressed by a single Purkinje cell can be extrapolated from the present results. In the experiments shown in Figures 3 and 4, among the 28 Purkinje cells analyzed, in 18 of them, one or more clear bands representing the PCR product were obtained from all three split tubes (β10 in Cell #1–1, β15 in Cell #1–2, β19 in Cell #1–5, β9 in Cell #1–7, β9 and β19 in Cell #1–10, β19 in Cell #1–11, β3 and β22 in Cell #1–12, β3 in Cell #1–14, β9 and β19 in Cell #1–17, β15 in Cell #1–22, β15 in Cell #1–26, β10 in Cell #1–28, β9 from biallelic expression in Cell #1–36, β19 in Cell #1–39). Therefore, 0.64 (18/28) Pcdh-β genes were expressed per Purkinje cell. Assuming that there is no distortion of expression frequencies between isoforms, and given that there are 22 genes in the Pcdh-β cluster, we can estimate that 2.3 Pcdh-β genes (0.64 multiplied by 22/6) should be expressed per individual Purkinje cell. Thus, at least two Pcdh-β isoforms would be expressed by any given Purkinje cell. If these assumptions prove correct, the Pcdh-β transcripts could, in theory, produce more than 462 (22 × 21) possible combinations (one Pcdh-β isoform from each allele) in single Purkinje cells.
The combinatorial co-expression of the mouse Pcdh cluster genes, including the Pcdh-α, Pcdh-β, and Pcdh-γ genes, could provide diversity sufficient to represent more than 20 million unique Purkinje cells, among which Pcdh-α transcripts could produce 132 (12 × 11) possible combinations (one Pcdh-α isoform from each allele), Pcdh-β transcripts could produce 462 (22 × 21) possible combinations (one Pcdh-β isoform from each allele), and Pcdh-γ transcripts could produce 342 (19 × 18) possible combinations (one Pcdh-γ isoform from each allele). Interestingly, rats have about 500 thousand cerebellar Purkinje cells, and humans have 14 million (Ito, 1984). Thus, the possible combinations of Pcdh cluster gene expressions would be sufficient to distinguish each cell.
The hypothesis that the Pcdh cluster genes play roles in providing cell-surface diversity is supported by reports that showed synaptic and surface localization of the Pcdh-α, Pcdh-β, and Pcdh-γ proteins and their formation of multimeric complexes (Kohmura et al., 1998; Wang et al., 2002b; Murata et al., 2004; Junghans et al., 2008; Chen et al., 2009; Fernandez-Monreal et al., 2009; Han et al., 2010; Puller and Haverkamp, 2011). Schreiner and Weiner (2010) showed that seven Pcdh-γ members exhibit isoform-specific homophilic binding, and that heteromultimeric cis-tetramers function as a homophilic binding unit. Although whether heteromultimeric cis-tetramers include Pcdh-β proteins is unclear, it is an intriguing possibility that heteromultimeric cis-tetramers with Pcdh-β proteins extend the homophilic binding specificity and play important roles in neural circuit formation.
This scenario could be supported by the following two findings. First, Pcdh-β proteins associate with Pcdh-γ proteins (Han et al., 2010). Second, the Cys-(X)5-Cys (C-X5-C) motif is important for the formation and cell-surface expression of covalently bound cis-tetramers, and the C-X5-C motif in the EC1 domain is completely conserved among all clustered Pcdh proteins in vertebrates (Schreiner and Weiner, 2010; Yagi, 2012). Our group plans to examine the formation of heteromultimeric cis-tetramers that include Pcdh-β proteins and their binding activities.
The importance of highly diverse transmembrane molecules in the formation of neuronal circuits has been shown in Drosophila, where Dscam1 molecules are responsible for the correct patterning of neuronal circuits (Hattori et al., 2007, 2008; Schmucker and Chen, 2009). We therefore speculate that the requirement for single-neuron diversity in neural circuit formation is conserved from insects to vertebrates, and that the Pcdh cluster is responsible for this role in the vertebrate nervous system (Zipursky and Sanes, 2010; Yagi, 2012). Future studies aimed at testing this hypothesis could shed light on the fundamental mechanisms underlying vertebrate neural circuit formation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. T. Shiroishi and The National Institute of Genetics for providing the JF1 strain, and members of the laboratory of Takeshi Yagi for suggestions and discussions during the course of this work. We appreciate Mses. C. Choji, K. Shimazu, H. Masuda, Y. Hidaka, A. Kinjo, E. Naruto, M. Katagiri, and A. Morita for their technical support, and Ms. H. Morita for her excellent secretarial assistance. This work was supported by Grant-in-Aid for Young Scientists (B) (JSPS) (No. 19770147, 22700326), and the Life Science Foundation of Japan (Ryosuke Kaneko), and Grant-in-Aid for Scientific Research (S) (JSPS) (No. 19100006), Innovative Areas “Mesoscopic Neurocircuitry” (No. 23115513) and (Comprehensive Brain Science Network) from the Ministry of Education, Science, Sports, and Culture of Japan (MEXT), JST-CREST, and the Uehara Memorial Foundation, and the Takeda Foundation (Takeshi Yagi).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Molecular_Neuroscience/10.3389/fnmol.2012.00090/abstract
FIGURE S1. Partial sequence, of a region close to the 3-polyadenylated site, of some of the Pcdh-β genes analyzed in this study. We found 27 polymorphisms (indicated by a yellow background) that enabled us to distinguish whether the amplified product was from the B6 or JF1 chromosome. The putative polyadenylation signal sequences are shown in red-underlined letters.
Table S1. List of primers.
Table S2. The possible poly adenylation signals of some of the Pcdh-β transcripts.
References
Arvidsson, U., Riedl, M., Elde, R., and Meister, B. (1997). Vesicular acetylcholine transporter (VAChT) protein: a novel and unique marker for cholinergic neurons in the central and peripheral nervous systems. J. Comp. Neurol. 378, 454–467.
Castillo, S. D., Matheu, A., Mariani, N., Carretero, J., Lopez-Rios, F., Lovell-Badge, R., and Sanchez-Cespedes, M. (2012). Novel transcriptional targets of the SRY-HMG box transcription factor SOX4 link its expression to the development of small cell lung cancer. Cancer Res. 72, 176–186.
Chahrour, M., Jung, S. Y., Shaw, C., Zhou, X., Wong, S. T., Qin, J., and Zoghbi, H. Y. (2008). MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229.
Chen, J., Lu, Y., Meng, S., Han, M. H., Lin, C., and Wang, X. (2009). alpha- and gamma-Protocadherins negatively regulate PYK2. J. Biol. Chem. 284, 2880–2890.
Dallosso, A. R., Hancock, A. L., Szemes, M., Moorwood, K., Chilukamarri, L., Tsai, H. H., Sarkar, A., Barasch, J., Vuononvirta, R., Jones, C., Pritchard-Jones, K., Royer-Pokora, B., Lee, S. B., Owen, C., Malik, S., Feng, Y., Frank, M., Ward, A., Brown, K. W., and Malik, K. (2009). Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms' tumor. PLoS Genet. 5, e1000745.
Dallosso, A. R., Oster, B., Greenhough, A., Thorsen, K., Curry, T. J., Owen, C., Hancock, A. L., Szemes, M., Paraskeva, C., Frank, M., Andersen, C. L., and Malik, K. (2012). Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene. doi: 10.1038/onc.2011.609. [Online].
Edwalds-Gilbert, G., Veraldi, K. L., and Milcarek, C. (1997). Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 25, 2547–2561.
Esumi, S., Kakazu, N., Taguchi, Y., Hirayama, T., Sasaki, A., Hirabayashi, T., Koide, T., Kitsukawa, T., Hamada, S., and Yagi, T. (2005). Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat. Genet. 37, 171–176.
Esumi, S., Kaneko, R., Kawamura, Y., and Yagi, T. (2006). Split single-cell RT-PCR analysis of Purkinje cells. Nat. Protoc. 1, 2143–2151.
Fernandez-Monreal, M., Kang, S., and Phillips, G. R. (2009). Gamma-protocadherin homophilic interaction and intracellular trafficking is controlled by the cytoplasmic domain in neurons. Mol. Cell. Neurosci. 40, 344–353.
Frank, M., Ebert, M., Shan, W., Phillips, G. R., Arndt, K., Colman, D. R., and Kemler, R. (2005). Differential expression of individual gamma-protocadherins during mouse brain development. Mol. Cell. Neurosci. 29, 603–616.
Fukuda, E., Hamada, S., Hasegawa, S., Katori, S., Sanbo, M., Miyakawa, T., Yamamoto, T., Yamamoto, H., Hirabayashi, T., and Yagi, T. (2008). Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur. J. Neurosci. 28, 1362–1376.
Golan-Mashiach, M., Grunspan, M., Emmanuel, R., Gibbs-Bar, L., Dikstein, R., and Shapiro, E. (2012). Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res. 40, 3378–3391.
Han, M. H., Lin, C., Meng, S., and Wang, X. (2010). Proteomics analysis reveals overlapping functions of clustered protocadherins. Mol. Cell. Proteomics 9, 71–83.
Handoko, L., Xu, H., Li, G., Ngan, C. Y., Chew, E., Schnapp, M., Lee, C. W., Ye, C., Ping, J. L., Mulawadi, F., Wong, E., Sheng, J., Zhang, Y., Poh, T., Chan, C. S., Kunarso, G., Shahab, A., Bourque, G., Cacheux-Rataboul, V., Sung, W. K., Ruan, Y., and Wei, C. L. (2011). CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638.
Hasegawa, S., Hamada, S., Kumode, Y., Esumi, S., Katori, S., Fukuda, E., Uchiyama, Y., Hirabayashi, T., Mombaerts, P., and Yagi, T. (2008). The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol. Cell Neurosci. 38, 66–79.
Hattori, D., Demir, E., Kim, H. W., Viragh, E., Zipursky, S. L., and Dickson, B. J. (2007). Dscam diversity is essential for neuronal wiring and self-recognition. Nature 449, 223–227.
Hattori, D., Millard, S. S., Wojtowicz, W. M., and Zipursky, S. L. (2008). Dscam-mediated cell recognition regulates neural circuit formation. Annu. Rev. Cell Dev. Biol. 24, 597–620.
Hirayama, T., Tarusawa, E., Yoshimura, Y., Galjart, N., and Yagi, T. (2012). CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. doi: 10.1016/j.celrep.2012.06.014. [Online].
Hirayama, T., and Yagi, T. (2006). The role and expression of the protocadherin-alpha clusters in the CNS. Curr. Opin. Neurobiol. 16, 336–342.
Junghans, D., Heidenreich, M., Hack, I., Taylor, V., Frotscher, M., and Kemler, R. (2008). Postsynaptic and differential localization to neuronal subtypes of protocadherin beta16 in the mammalian central nervous system. Eur. J. Neurosci. 27, 559–571.
Kaneko, R., Kato, H., Kawamura, Y., Esumi, S., Hirayama, T., Hirabayashi, T., and Yagi, T. (2006). Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J. Biol. Chem. 281, 30551–30560.
Kaneko, R., Kawaguchi, M., Toyama, T., Taguchi, Y., and Yagi, T. (2009). Expression levels of Protocadherin-alpha transcripts are decreased by nonsense-mediated mRNA decay with frameshift mutations and by high DNA methylation in their promoter regions. Gene 430, 86–94.
Katori, S., Hamada, S., Noguchi, Y., Fukuda, E., Yamamoto, T., Yamamoto, H., Hasegawa, S., and Yagi, T. (2009). Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J. Neurosci. 29, 9137–9147.
Kawaguchi, M., Toyama, T., Kaneko, R., Hirayama, T., Kawamura, Y., and Yagi, T. (2008). Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-alpha gene cluster. J. Biol. Chem. 283, 12064–12075.
Kawauchi, S., Calof, A. L., Santos, R., Lopez-Burks, M. E., Young, C. M., Hoang, M. P., Chua, A., Lao, T., Lechner, M. S., Daniel, J. A., Nussenzweig, A., Kitzes, L., Yokomori, K., Hallgrimsson, B., and Lander, A. D. (2009). Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 5:e1000650. doi: 10.1371/journal.pgen.1000650
Kehayova, P., Monahan, K., Chen, W., and Maniatis, T. (2011). Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. U.S.A. 108, 17195–17200.
Kohmura, N., Senzaki, K., Hamada, S., Kai, N., Yasuda, R., Watanabe, M., Ishii, H., Yasuda, M., Mishina, M., and Yagi, T. (1998). Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 20, 1137–1151.
Kullmann, D. M. (2011). Interneuron networks in the hippocampus. Curr. Opin. Neurobiol. 21, 709–716.
Lobas, M. A., Helsper, L., Vernon, C. G., Schreiner, D., Zhang, Y., Holtzman, M. J., Thedens, D. R., and Weiner, J. A. (2012). Molecular heterogeneity in the choroid plexus epithelium: the 22-member gamma-protocadherin family is differentially expressed, apically localized, and implicated in CSF regulation. J. Neurochem. 120, 913–927.
McGowan, P. O., Suderman, M., Sasaki, A., Huang, T. C., Hallett, M., Meaney, M. J., and Szyf, M. (2011). Broad epigenetic signature of maternal care in the brain of adult rats. PLoS ONE 6:e14739. doi: 10.1371/journal.pone.0014739
Miyake, K., Hirasawa, T., Soutome, M., Itoh, M., Goto, Y., Endoh, K., Takahashi, K., Kudo, S., Nakagawa, T., Yokoi, S., Taira, T., Inazawa, J., and Kubota, T. (2011). The protocadherins, PCDHB1 and PCDH7, are regulated by MeCP2 in neuronal cells and brain tissues: implication for pathogenesis of Rett syndrome. BMC Neurosci. 12, 81.
Monahan, K., Rudnick, N. D., Kehayova, P. D., Pauli, F., Newberry, K. M., Myers, R. M., and Maniatis, T. (2012). Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc. Natl. Acad. Sci. U.S.A. 109, 9125–9130.
Morishita, H., and Yagi, T. (2007). Protocadherin family: diversity, structure, and function. Curr. Opin. Cell Biol. 19, 584–592.
Murata, Y., Hamada, S., Morishita, H., Mutoh, T., and Yagi, T. (2004). Interaction with protocadherin-gamma regulates the cell surface expression of protocadherin-alpha. J. Biol. Chem. 279, 49508–49516.
Neves, G., Zucker, J., Daly, M., and Chess, A. (2004). Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat. Genet. 36, 240–246.
Noguchi, Y., Hirabayashi, T., Katori, S., Kawamura, Y., Sanbo, M., Hirabayashi, M., Kiyonari, H., Nakao, K., Uchimura, A., and Yagi, T. (2009). Total expression and dual gene-regulatory mechanisms maintained in deletions and duplications of the Pcdha cluster. J. Biol. Chem. 284, 32002–32014.
Prasad, T., and Weiner, J. A. (2011). Direct and indirect regulation of spinal cord Ia afferent terminal formation by the gamma-protocadherins. Front. Mol. Neurosci. 4:54. doi: 10.3389/fnmol.2011.00054
Puller, C., and Haverkamp, S. (2011). Cell-type-specific localization of protocadherin beta16 at AMPA and AMPA/Kainate receptor-containing synapses in the primate retina. J. Comp. Neurol. 519, 467–479.
Rhoades, K. L., Singh, N., Simon, I., Glidden, B., Cedar, H., and Chess, A. (2000). Allele-specific expression patterns of interleukin-2 and Pax-5 revealed by a sensitive single-cell RT-PCR analysis. Curr. Biol. 10, 789–792.
Ribich, S., Tasic, B., and Maniatis, T. (2006). Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. U.S.A. 103, 19719–19724.
Sago, H., Kitagawa, M., Obata, S., Mori, N., Taketani, S., Rochelle, J. M., Seldin, M. F., Davidson, M., St John, T., and Suzuki, S. T. (1995). Cloning, expression, and chromosomal localization of a novel cadherin-related protein, protocadherin-3. Genomics 29, 631–640.
Schippert, R., Schaeffel, F., and Feldkaemper, M. P. (2009). Microarray analysis of retinal gene expression in Egr-1 knockout mice. Mol. Vis. 15, 2720–2739.
Schmucker, D., and Chen, B. (2009). Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 23, 147–156.
Schreiner, D., and Weiner, J. A. (2010). Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc. Natl. Acad. Sci. U.S.A. 107, 14893–14898.
Sugino, H., Hamada, S., Yasuda, R., Tuji, A., Matsuda, Y., Fujita, M., and Yagi, T. (2000). Genomic organization of the family of CNR cadherin genes in mice and humans. Genomics 63, 75–87.
Suzuki, S. C., and Takeichi, M. (2008). Cadherins in neuronal morphogenesis and function. Dev. Growth Differ. 50(Suppl. 1), S119–S130.
Takeichi, M. (2007). The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 8, 11–20.
Tan, Y. P., Li, S., Jiang, X. J., Loh, W., Foo, Y. K., Loh, C. B., Xu, Q., Yuen, W. H., Jones, M., Fu, J., Venkatesh, B., and Yu, W. P. (2010). Regulation of protocadherin gene expression by multiple neuron-restrictive silencer elements scattered in the gene cluster. Nucleic Acids Res. 38, 4985–4997.
Tasic, B., Nabholz, C. E., Baldwin, K. K., Kim, Y., Rueckert, E. H., Ribich, S. A., Cramer, P., Wu, Q., Axel, R., and Maniatis, T. (2002). Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10, 21–33.
Vanhalst, K., Kools, P., Vanden Eynde, E., and van Roy, F. (2001). The human and murine protocadherin-beta one-exon gene families show high evolutionary conservation, despite the difference in gene number. FEBS Lett. 495, 120–125.
Wang, X., Su, H., and Bradley, A. (2002a). Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 16, 1890–1905.
Wang, X., Weiner, J. A., Levi, S., Craig, A. M., Bradley, A., and Sanes, J. R. (2002b). Gamma protocadherins are required for survival of spinal interneurons. Neuron 36, 843–854.
Watakabe, A., Ichinohe, N., Ohsawa, S., Hashikawa, T., Komatsu, Y., Rockland, K. S., and Yamamori, T. (2007). Comparative analysis of layer-specific genes in Mammalian neocortex. Cereb. Cortex 17, 1918–1933.
Watakabe, A., Komatsu, Y., Ohsawa, S., and Yamamori, T. (2010). Fluorescent in situ hybridization technique for cell type identification and characterization in the central nervous system. Methods 52, 367–374.
Wu, Q., and Maniatis, T. (1999). A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97, 779–790.
Wu, Q., Zhang, T., Cheng, J. F., Kim, Y., Grimwood, J., Schmutz, J., Dickson, M., Noonan, J. P., Zhang, M. Q., Myers, R. M., and Maniatis, T. (2001). Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 11, 389–404.
Yagi, T. (2012). Molecular codes for neuronal individuality and cell assembly in the brain. Front. Mol. Neurosci. 5:45. doi: 10.3389/fnmol.2012.00045
Yokota, S., Hirayama, T., Hirano, K., Kaneko, R., Toyoda, S., Kawamura, Y., Hirabayashi, M., Hirabayashi, T., and Yagi, T. (2011). Identification of the cluster control region for the Protocadherin-{beta} genes located beyond the Protocadherin-{gamma} cluster. J. Biol. Chem. 286, 31886–31895.
Keywords: single-neuron diversity, neuronal individuality, protocadherin, Pcdh, monoallelic, combinatorial expression, single-cell 3′-RACE, neural circuit
Citation: Hirano K, Kaneko R, Izawa T, Kawaguchi M, Kitsukawa T and Yagi T (2012) Single-neuron diversity generated by Protocadherin-β cluster in mouse central and peripheral nervous systems. Front. Mol. Neurosci. 5:90. doi: 10.3389/fnmol.2012.00090
Received: 20 July 2012; Paper pending published: 09 August 2012;
Accepted: 14 August 2012; Published online: 31 August 2012.
Edited by:
Alistair N. Garratt, Max Delbrück Center for Molecular Medicine, GermanyReviewed by:
Sheriar Hormuzdi, University of Dundee, UKAlistair N. Garratt, Max Delbrück Center for Molecular Medicine, Germany
Copyright © 2012 Hirano, Kaneko, Izawa, Kawaguchi, Kitsukawa and Yagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Takeshi Yagi, Graduate School of Frontier Biosciences, Osaka University, 1-3 Yamadaoka, Suita, Osaka 565-0871, Japan. e-mail:eWFnaUBmYnMub3Nha2EtdS5hYy5qcA==
†These authors equally contributed to this work.


