- 1Functional Genomics, Council of Scientific and Industrial Research (CSIR), Institute of Genomics and Integrative Biology (IGIB), New Delhi, India
- 2Academy of Scientific and Innovative Research (AcSIR), New Delhi, India
Kalirin, a key player in axonal development, nerve growth and synaptic re-modeling, is implicated in many pathological conditions like schizophrenia and autism-spectrum disorders. Alternative promoters and splicing lead to functionally distinct isoforms, but the post-transcriptional regulation of Kalirin has not been studied. Here, we report a novel non-coding RNA, which we name durga, arising from the first exon of kalirin a (kalrna) in the antisense orientation in zebrafish. The kalrna and durga transcripts are barely detectable during early development, but steadily increase by 24 hours post-fertilization (hpf) as the brain develops. Over-expression of durga in the zebrafish embryo led to an increase in kalrna expression. The morphology of the neurons cultured from durga injected embryos had significantly fewer and shorter dendrites. Although durga has no apparent sequence homolog in mammals, based on gene synteny, we found a non-coding RNA arising from the 5′ end of the human Kalrn gene and expressed in the human neuronal cell line, SH-SY5Y. We propose that the zebrafish lncRNA durga maintains dendritic length and density through regulation of kalrna expression and this may have further implications in mammalian systems.
Introduction
Development of the brain involves a fine balance between excitatory and inhibitory signaling at synapses, ensuring plasticity while avoiding excitotoxicity. Kalirin, originally reported as a protein that interacts with Huntington associated protein (HAP), is known to be involved in active remodeling of synapses (Colomer et al., 1997). Kalirin has been implicated in a variety of neuropsychiatric and neurodegenerative diseases like schizophrenia, depression and Alzheimer’s disease (Hill et al., 2006; Mandela and Ma, 2012; Remmers et al., 2014; Makrythanasis et al., 2016). At the molecular level, this RhoGEF kinase is expressed highly at excitatory synapses and reduced at inhibitory synapses. Its binding to receptors leads to RhoGTPase mediated signaling and regulation of actin cytoskeletal organization. This intra-cellular signaling pathway leads to synaptic remodeling through altered dendrite numbers and dendritic morphology (Penzes et al., 2001). The mammalian Kalirin (Kalrn) gene locus gives rise to several isoforms which have diverse and sometimes antagonistic effects on dendritogenesis. The Kalrn gene can code for a protein with a lipid binding motif, several tandem spectrin homology domains and additional protein-protein interaction domains like SH3 domain commonly found in members of signaling pathways (McPherson et al., 2002, 2004; Vishwanatha et al., 2012; Miller et al., 2017). The functional differences between isoforms are thought to arise from the combinations of protein domains retained in the splice isoforms. The effects of alternative isoforms of Kalrn on dendritic morphology have been studied extensively (Penzes et al., 2001; McPherson et al., 2002, 2004; Vishwanatha et al., 2012). However, the regulation of Kalrn transcription during development has not been explored extensively.
The human Kalrn gene is located on chromosome 3 and spans a huge 0.63 Mb region with a complex gene structure encompassing 17 alternatively spliced transcripts. The largest transcript of 19.9 kb contains 60 exons (McPherson et al., 2002). The shorter isoforms of human Kalrn fall into two broad categories, the ones containing combinations of the 30 exons from the 5′ end or 20 exons from the 3′ end. Johnson et al. (2000) and McPherson et al. (2002) reported the domain architecture of Kalirin protein isoforms and their relation to the alternative spliced transcript isoforms. Kalrn7, the major brain-specific isoform, and the closely related Kalrn8 isoform consist of the core Sec14p-like putative lipid-binding motif, nine spectrin-like repeats and a Dbl homology/pleckstrin homology (DH-PH) domain but differs at the amino terminus and have a distinct PDZ binding motif or an SH3 motif at the -COOH end respectively. Kalrn9 and Kalrn12 have additional C-terminal domains and show distinct sub-cellular localization in cortical neurons while Kalrn7 shows characteristic punctate localization in the neuronal processes. The transcript isoforms also have distinct spatio-temporal expression patterns with Kalrn7 being expressed in the adult brain, while Kalrn9 and Kalrn12 are expressed in the developing brain and yet another isoform Trio is expressed in many tissues (Hansel et al., 2001).
Zebrafish is emerged as a convenient model to explore axonal growth (Chen et al., 2013), signaling by neurotropic factors during development and in human diseases. Thus, we studied the kalrna locus in detail on chromosome 9 of the zebrafish genome (Zv10). The 2.9 kb predicted transcript from the longest isoform (ENSDART00000164543.1) consists of the expected core domains (spectrin repeats, DH-PH domains). The kalrna locus has been extensively re-annotated during the transition from zebrafish genome version 9 (Zv9) to version 10 (Zv10). A notable change is the extension of the 5′ end to include an exon positioned 95 kb upstream to the previously reported kalrna gene. We noticed that in Zv9, this region was inverted and hence the link with the kalrna gene was not evident. Large scale studies, most notably, the ENCODE project, have shown that extended 5′ terminal exons can play an important role in the regulation of the gene. On closer inspection, we found that the region also corresponds to an lncRNA reported by Kaushik et al. (2013) from an RNA-seq based developmental profiling of non-coding RNAs.
Here, we show that this lncRNA is transcribed in anti-sense to the largest kalrna transcript in zebrafish. The multi-domain Kalirin was named after the multi-armed goddess Kali from Indian mythology (Alam et al., 1997), and hence, we propose to name this lncRNA that is anti-sense to kalrna, durga- the mythological alter-ego of Kali. The lncRNA is expressed in the oocyte and as embryo is developing, it gets gradually restricted to the brain. The presence of this lncRNA seems to be critical for appropriate temporal expression of kalrna. The durga transcripts present in the cytoplasm of the fertilized embryo, subsequently increases and localize to the developing zebrafish brain. The rise in durga expression is accompanied by an increase in the level of kalrna mRNA and coincides with the developmental time when the zebrafish brain is being formed and neurites are growing as rapidly as 20 microns/h (Chen et al., 2013). Ectopic injection of the durga RNA results in further increase in the expression of kalrna. Primary zebrafish neurons with elevated durga and kalrna showed striking morphological changes in dendrites. Using a transgenic fish with neurons marked with kaede, we found that the ectopically injected lncRNA triggered a profound loss of dendrites, perhaps corresponding to the increased kalrna expression. Thus, transcription from the first exon of the kalrna gene, located about 95 kb upstream to the next exon in zebrafish, seems to be regulated by lncRNA durga.
Materials and Methods
Animal Husbandry
Zebrafish (Danio rerio) stocks were maintained according to standard procedures (Westerfield, 2000) with 13/11 h light/dark cycle at the zebrafish facility at Institute of Genomics and Integrative Biology (IGIB). Zebrafish experiments were performed in strict accordance with the recommendations and guidelines of the CSIR-Institute of Genomics and Integrative Biology, India. The protocol was approved by the Institutional Animal Ethics Committee (IAEC) of the CSIR-Institute of Genomics and Integrative Biology, India. Embryos from casper (White et al., 2008) and [Tg(HuC: Kaede)] (Sato et al., 2006) strains were used for all experiments. Fish were paired in mating cages a night before obtaining embryos. Embryos were collected and harvested at different stages of development and eggs were obtained by squeezing abdomen of gravid females.
LncRNA durga Over-Expression by Microinjections
LncRNA durga was over-expressed at one-cell stage by injecting 5 ng of in vitro synthesized (MEGAshortscript transcription kit, Thermo Fisher Scientific) RNA. The embryos were monitored constantly and fixed at 1cell, high (3 hpf), shield (6 hpf), 2Somites (11 hpf) and 30Somites (24 hpf) for in situ hybridization and RNA isolation.
RNA Extraction, Reverse Transcriptase and Polymerase Chain Reaction
Total RNA was extracted using TRIzol (Invitrogen) as per manufacturer’s instructions from mentioned time points. cDNA was synthesized using 500 ng total RNA with 5000 pg spiked in exogenous RNA with gene specific reverse primer or random hexamer primers and M-MLV reverse transcriptase at 42°C for 1 h. Standard polymerase chain reaction (PCR) was performed to amplify durga and kalrna with intron spanning primers.
Real Time Polymerase Chain Reaction
Real-time PCR (RT-PCR) for durga and kalrna was performed using spiked in exogenous RNA as internal control. SYBR green master mix (Takara) was used as per manufacturer’s protocol on Roche LightCycler480. Data was extracted and analyzed manually using excel.
RNA Detection by In Situ Hybridization
lncRNA durga and kalrna specific sequence was amplified using specific primers from adult brain cDNA and cloned in TOPO2 dual promoter vector (Invitrogen). Plasmids were linearized using SpeI and ECoRV restriction enzymes and probes were in vitro synthesized using T7 and SP6 RNA polymerase and digoxigenin labeled UTPs. Embryos were fixed using 4%w/v PFA(Sigma) made in 1× PBS, pH 7.4. Embryos were washed with 1× PBS 0.1%, tween and stored at −20°C in 100% methanol. Embryos were hybridized with lncRNA durga and kalrna antisense probes in hybridization buffer at 65°C overnight, thereafter given stringency washes with 25%, 50%, 75% and 100% 2× SSC. Embryo were then washed with maleic acid buffer with 0.1% tween. Embryos were then incubated with digoxigenin-alkaline phosphatase Fab fragments (Roche) in 1:1000 dilutions overnight at 4°C. Post antibody incubation, stainings were developed using Nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3′-indolyphosphate (BCIP) substrate (Roche). After completion of staining, reaction was terminated by fixing embryos with PFA. Embryos were then taken serially through 25%, 50% and 75% of glycerol and imaged in 2.5% methyl cellulose.
Northern Blotting
Total RNA was extracted from zebrafish adult brain and 30 μg of total RNA was run on denaturing agarose gel. RNA was then transferred to Amersham hybond N+ membrane by capillary action and then crosslinked to the membrane using stratalinker. Probes for lncRNA and kalrna were made using 50 μCi αP32 dATP and Klenow polymerase (M0210S, NEB) incubating reaction mix at 37°C for 1 h. Blot was prehybridized in Church buffer for 3 h at 65°C and incubated in hybridization buffer overnight. Blot was then washed with 2× SSC + 0.1% SDS, 1× SSC + 0.5% SDS and 0.1× SSC + 0.5% SDS serially for 1 h each at 65°C with agitation. Blot was then exposed to the film for 45 h and film was scanned in phosphoimager to obtain an image.
Zebrafish Primary Neuron Culture
Control or durga over-expressed 48 hpf embryos were decorionated and deyolked under microscope. Then they were washed with sterile cold PBS in cell culture hood. Embryos were disintegrated by adding Trypsin and incubating at 37°C for 20 min, with intermittent mixing. Trypsin was removed by centrifugation. Fresh Neurobasal-A media was added to trypsinized embryos and triturated slowly and carefully with pipette to get single cell suspension (Westerfield, 2000). Cells were passed through 70 μm mesh to get rid of clumps and plated in poly-D lysine coated plates or coverslip in Neurobasal-A medium with B27, glutamax and primocin. Cultured cells were incubated at 29°C in the CO2 incubator for 24 h, then collected for RNA isolation or fixed for imaging.
Imaging Acquisition and Analysis
Images of in situ hybridized embryos were captured using NIKON SMZ800N microscope at 6× magnification. Fluorescence images of primary cultured neurons were captured using LeicaTCSSP8 microscope at 100× magnification using 488 laser in randomly selected fields. Dendritic density and length was measured using NeuronJ plug-in of the ImageJ tool in the images of control or durga over-expressed neurons (Meijering et al., 2004).
Results
The advent of next generation sequencing technologies has led to the rapid, high-throughput identification of thousands of putative non-coding RNAs. The zebrafish genome (latest version Zv10) has been recently re-annotated to incorporate modifications at many loci. We set out to study a putative lncRNA locus that was re-annotated in the recent version of the zebrafish genome.
Genomic Organization of Zebrafish kalrna in Zv9 and Zv10 Genome Assembly
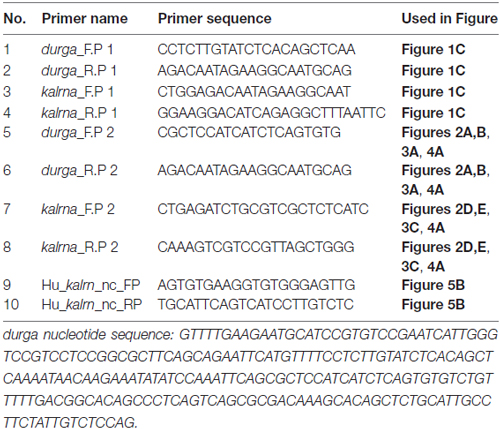
In the Zv9 version of the zebrafish genome, mylk1 and kalrna were assembled in a convergent orientation on the same strand with ~20 kb intergenic region. However in Zv10, this was revised to include a genomic inversion, which places mylk1 and kalrna in the divergent orientation on opposite strand, separated by ~11 kb intergenic region (Kent et al., 2002; Figure 1A). Apart from changes in the orientation, we also noticed the addition of a poorly conserved 5′ exon (denoted as E0 in Figures 1A,B), ~95 kb upstream to the previously reported kalrna transcript which confers a 22 amino acids N-terminal extension. Interestingly, this 5′ exon also includes a 240 nucleotide long 5′UTR, which was not annotated in Zv9. Such extended or alternative 5′ terminal exon can play an important role in the regulation of the mRNA stability and translation by interacting with translational machinery, microRNAs or lncRNAs. Regulation of kalrna by lncRNA is not known. Thus, we checked for presence of any lncRNA near the 5′ locus of kalrna in the zebrafish lncRNA database zflncRNApedia (Dhiman et al., 2015). We found one such lncRNA ZF_LNC002146 in the database; although its location is ~72 kb away from kalrna according to Zv9 annotation, overlaps with extended 5′ exon of kalrna in Zv10 (Figure 1A). This lncRNA was first reported as lncBR47, to be highly enriched in the brain (Kaushik et al., 2013). However, in this study, lncRNAs were detected using a RNA-seq data generated by non-directional RNA-seq method, suggesting that the lncBR47 (named durga hereafter) may correspond to the kalrna first exon. Therefore, it was not clear if the reported lncRNA is in fact part of an extended isoform of kalrna or a distinct lncRNA antisense to kalrna.
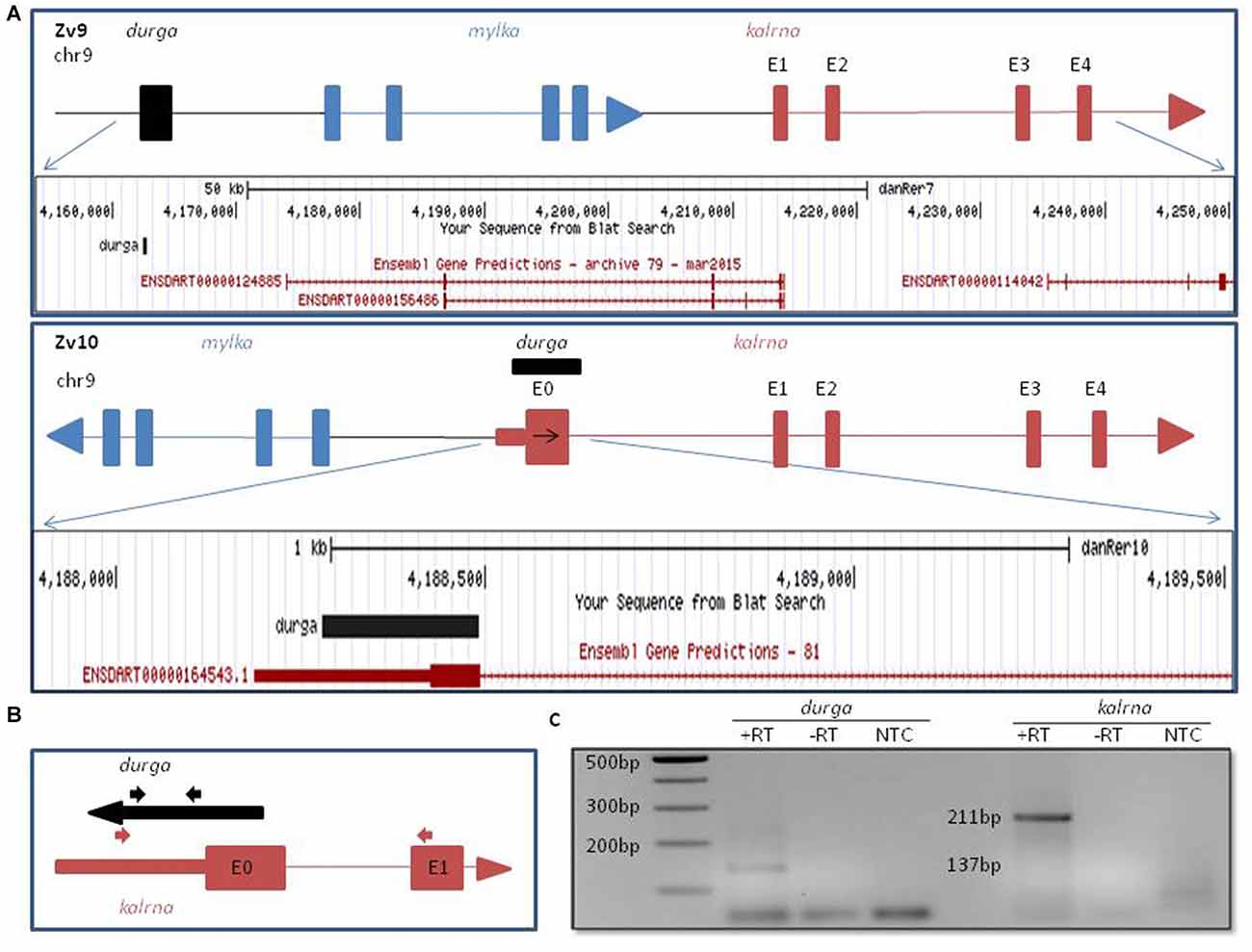
Figure 1. Genomic organization of zebrafish kalrn in zebrafish genome version 9 (Zv9) and zebrafish genome version 10 (Zv10) genome assembly. (A) Pictorial representation and UCSC genome browser tracks of gene re-arrangement of kalrna, mylka and durga in the Zv9 and Zv10 genome assembly. (B) Primers are shown with their position and orientation. Representation not to scale. (C) Expression of durga (137 bp) and novel first exon of kalrna (211 bp) was confirmed by polymerase chain reaction (PCR) with cDNA synthesized from strand specific primer and zebrafish brain RNA.
To verify which of the two strands are transcribed at the kalrna locus, we used gene specific reverse primers to synthesize cDNA from zebrafish brain RNA (Figure 1B; Table 1). Both the cDNA samples gave rise to expected size PCR products suggesting that the locus is transcribed bi-directionally (Figure 1C). We further confirmed the expression of both transcripts by northern blotting and oligodT primed RT-PCR (Supplementary Figure S1).The positive strand would produce a kalrna isoform while the negative strand would give rise to durga.
Developmental Expression Profile of durga and kalrna in Zebrafish
To study the spatio-temporal expression pattern of durga, we used gene-specific reverse primers for cDNA synthesis and primer pairs were designed to produce PCR products of different sizes corresponding to kalrna and durga (Table 1). The reverse transcriptase-PCR analysis showed that durga (Figures 2A,B; 96 bp) is expressed at low levels during the early stages of development but steadily increases by 24 hpf (30Somites). We also used riboprobes from the sense (with respect to kalrna) and anti-sense strand in in situ hybridizations to confirm that both strands produce transcripts in vivo. In situ hybridization confirms the presence of durga transcripts in the egg (Supplementary Figure S2) and a clear expression throughout the developing embryo upto 11 hpf (2Somites stage). Subsequently as the brain develops, the expression of the lncRNA becomes localized to the head region of the embryo (Figure 2C). The kalrna starts expressing after zygotic genome activation and becomes apparent in anterior region of the embryo as brain starts developing at 2Somites stage (Figures 2D–F; 156 bp). By 24 hpf, the kalrna expression is almost entirely in the anterior region (Figure 2F).
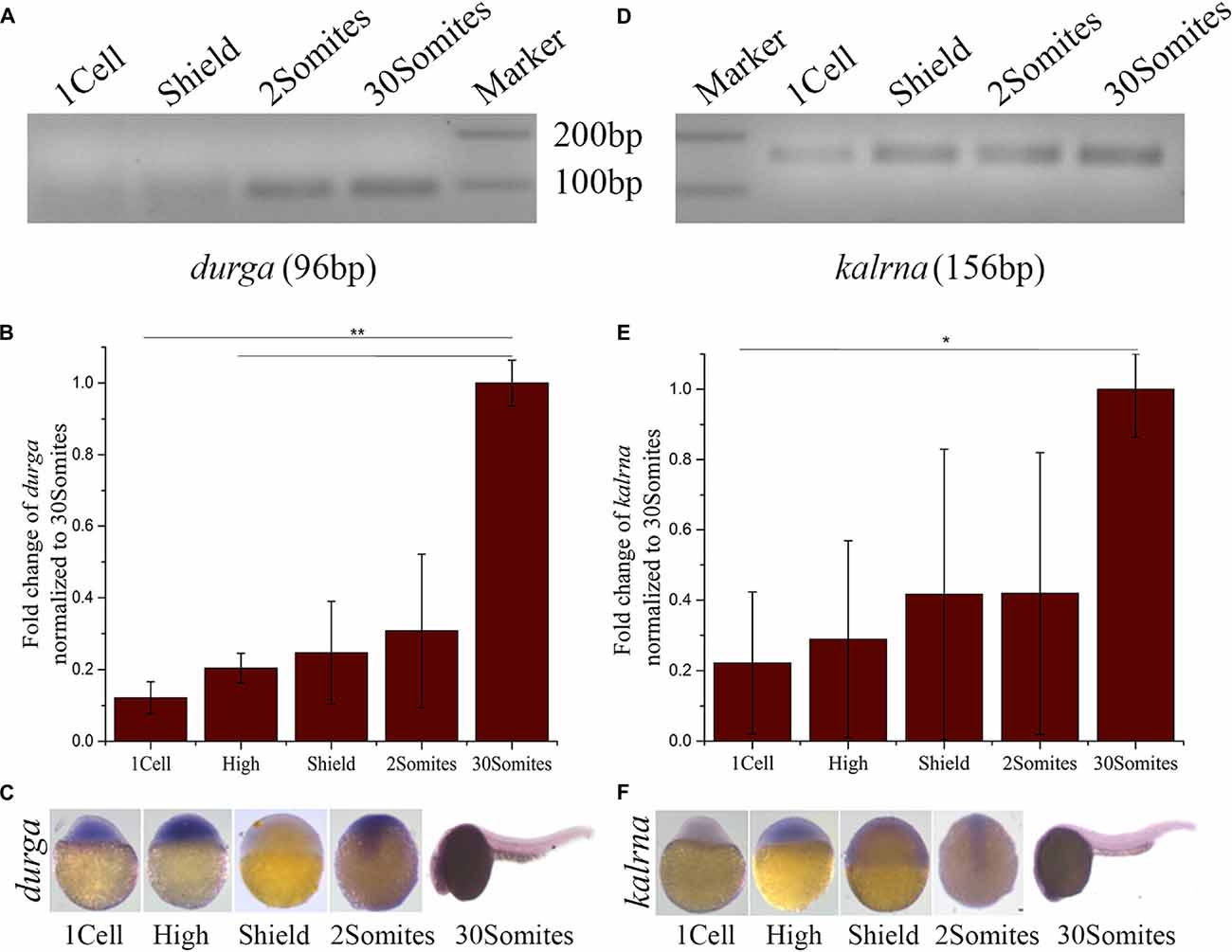
Figure 2. Developmental expression profile of durga and kalrna in zebrafish. (A) Expression of durga and kalrna was checked by semi-quantitative PCR (A,D respectively), qPCR (B,E respectively) and in situ hybridization (C,F respectively) during 1cell, High, Shield, 2Somites, 30Somites stages. Data shown represent more than three independent experiments. p-values were calculated using Student’s t-test. **p ≤ 0.01, *p ≤ 0.05.
LncRNA durga Modulates the Expression Profile of kalrna
Like kalrna and durga many lncRNAs are found in close proximity of the genes they regulate and share spatio-temporal expression patterns with their targets (Yap et al., 2010; Stojic et al., 2016; Tran et al., 2016). To find if durga could affect the expression profile of kalrna, we injected in vitro transcribed durga RNA into the embryo at the 1-cell stage. The injected RNA was stable upto the 30Somites stage and localized to the anterior region, matching the spatial expression pattern of the endogenous durga transcripts (Figures 3A,B). In these embryos, kalrna showed a two-fold upregulation in quantitative RT-PCR (Figure 3C). A strong upregulation of kalrna was also evident in the brain region in in situ hybridization experiments (Figure 3D). The induction of kalrna is more evident in in situ hybridization perhaps because of the dilution due to homogenization of the whole embryo in the quantitative RT-PCR.
Figure 3. Over-expression of durga enhances kalrna expression. In vitro transcribed durga was injected in one cell stage of zebrafish embryo and over-expression was confirmed by qPCR (A) and in situ hybridization (B) at 2Somites and 30Somites stages. Increase in kalrna expression was seen in qPCR (C) and in situ hybridization (D) at 2Somites and 30Somites stages. Data represent three independent experiments. p-value was calculated using Student’s t-test. *p ≤ 0.05. OE denotes overexpression. N is number of biological replicates.
LncRNA durga Alters Dendritic Morphology in Primary Culture of Zebrafish Neurons
kalrna is a guanine nucleotide exchange factor (GEF) that affects cytoskeletal arrangement in neurons, resulting in changes in dendritic spine morphology. During development, spine dynamics plays an important role in the proper formation of neuronal circuits, while in adults spine morphogenesis is linked to synaptic plasticity, learning and cognition. By virtue of its role in neuritogenesis and dendritic spine morphology, kalrna is implicated in several neuro-psychiatric diseases like schizophrenia, Alzheimer’s disease and depression. Therefore, we were interested in testing the effect of the lncRNA durga on neuronal cell morphology. We used a double transgenic zebrafish line with radial glial cells marked with mCherry driven by a Her4.1 promoter (Yeo et al., 2007) and neuronal cells marked by the kaede reporter under the regulation of the HuC promoter (Sato et al., 2006). After 48 hpf, durga injected and control embryos were triturated and plated in Neurobasal-A medium to study the morphology of individual neurons. In these cells, we confirmed that the over-expression of durga and up-regulation of kalrna were consistent with the effects we found in vivo (Figure 4A). A closer examination of the morphology of the neurons showed that cells from durga injected cells had significantly fewer dendrites (Figure 4B). As shown in Figure 4C, 60% of the control cells showed more than five dendrites per cell while 75% of the durga—kalrna over-expressing cells had less than five dendrites per cell (Figure 4C). The average length of dendrites also reduced significantly in the durga—kalrna over-expressing cells (Figure 4D).
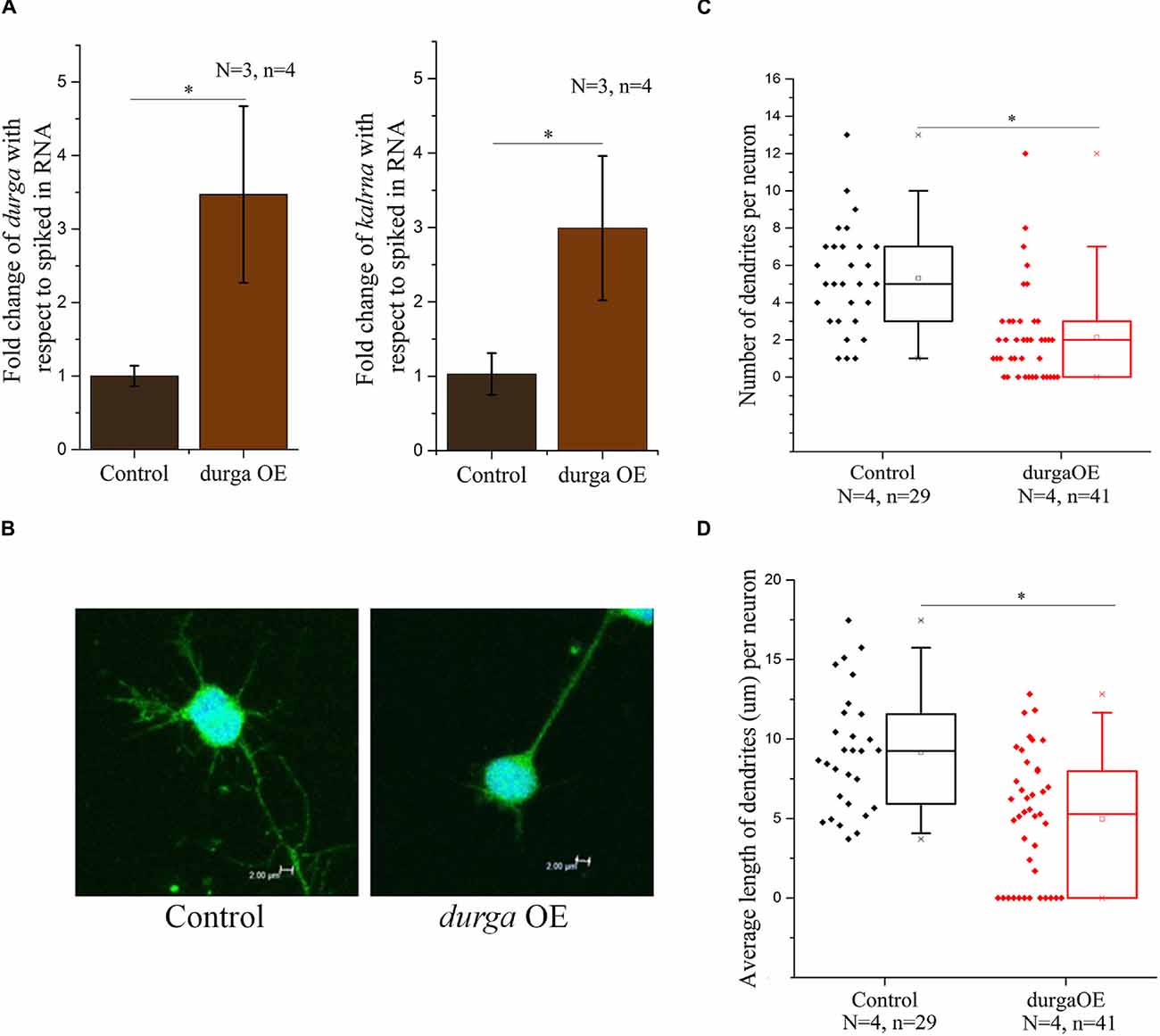
Figure 4. Over-expression of durga in zebrafish embryos alters dendritic morphology in primary culture of neurons. Over-expression of durga and kalrna was confirmed by qPCR in primary culture of neurons (A). Confocal image of durga over-expressed neuron showed reduced average dendrite number (B,C) and dendrite length (B,D). Data shown represent minimum three independent experiments. p-values were calculated using Student’s t-test. *p < 0.05. OE denotes overexpression.
Discussion
Kalirin acts a pivotal point in the regulation of cytoskeletal organization and thus neuronal morphology because of integration of diverse, excitatory as well as inhibitory signals. Human Kalrn gene is a large gene that shows extensive alternative splicing leading to multi-domain protein isoforms whose functionality is determined by the combination of exons included in each transcript type. The alternative transcripts are of two broad categories, the ones that have predominantly 5′ exons, for instance, Kalrn-7, 9, 12 and others that have predominantly 3′ exons like Duo and Trio. These transcript isoforms also differs in the spatio-temporal expression pattern as seen in Kalrn7 found in the adult brain and Kalrn9 and 12 predominantly expressed during development. Kalirin protein isoforms seem to respond to a variety of signals including Nerve Growth Factor signaling through the TrkB receptor, BDNF signaling through Rac1 (Chakrabarti et al., 2005; Yan et al., 2016). It also interacts with many PDZ domain containing proteins such as PSD95 (Penzes et al., 2001). Kalirin proteins can integrate these signals to modify actin cytoskeletal dynamics eventually modifying neurite morphology and axonal growth of neurons.
We noted a putative, brain enriched lncRNA that directly overlaps with the extended exon of the kalrna gene in the Zv10 annotation of the zebrafish genome. Using a combination of semi-quantitative RT-PCR with directional primers and in situ hybridization, we established that the locus gives rise to an lncRNA durga and the transcript of kalrna gene. Both durga and kalrna showed increasing expression during development, finally localizing to the brain region by 24 hpf. Further, over-expression of the lncRNA resulted in an increase in the expression of kalrna and decrease in the number of dendrites in zebrafish neurons. Currently, no detailed information on the transcript isoforms of kalrn in zebrafish is known. However in future, it will be interesting to explore the isoform-specific effects of the durga in neurodevelopment and neurodegenerative disorders. Besides durga, additional factors are likely to be involved in the regulation of zebrafish kalrna.
Depletion of the endogenous durga lncRNA is technically challenging due to the overlap with the kalrna gene. Standard tools like CRISPR or morpholinos are not readily applicable. CRISPR based editing would disrupt the kalrna gene on the opposite strand. Lack of information on exon-intron structure in durga gene prevented use of morpholinos to knock-down the expression of the lncRNA. In future, it will be interesting to study the mechanism of kalrna regulation by durga.
The Kalrn over-expression, especially in the form of the transcript isoform Kalrn7 is usually associated with increased dendritic length while Kalrn9 and 12 isoforms seem to have age dependent, contrasting effects on dendritic length. In early development, Kalrn9 and 12 isoforms induce dendritic elongation while in mature neurons Kalrn9 over-expression reduces dendritic length (Grubisha et al., 2016). The dual role of Kalrn9 and 12 isoforms to induce or retract dendrites comes from their two GEF domains. The first GEF domain activates RhoG/Rac1, which promotes the formation and elongation of dendrites. The second GEF domain interacts with RhoA, which induces spine elimination (Tolias et al., 2011; Yan et al., 2015). The Kalrn7 isoform does not have a second GEF domain and thus over expression of Kalrn9 and 12 isoforms but not Kalrn7 reduces dendritic length (Deo et al., 2012; Grubisha et al., 2016). Interestingly Kalrn9 expression has been observed to be increased during normal human aging in the cortex and also in schizophrenia subjects (Deo et al., 2012; Grubisha et al., 2016).
Reduction in dendritic length, pruning of synaptic connections and decrease in brain size are common pathological symptoms of neurodevelopmental, neuro-degenerative and neuro-psychiatric disorders like autism, schizophrenia, Alzheimer’s (Remmers et al., 2014). On the other hand, controlled synaptic pruning and maintenance of dendritic arborization are also essential to maintain structural plasticity of brain throughout life. lncRNAs have been reported to be involved in almost every aspect of brain development with diverse mechanisms of action. For example, lncRNA POU3F2, Gomafu, HAR1F are involved in neural stem cell proliferation while lncRNA Pnky, NEAT1, TUG1, EVF2 regulates neuronal differentiation (Iyengar et al., 2014). Besides, the role of lncRNAs in regulation of neuronal architecture and synaptic plasticity is beginning to be understood. Here we showed that lncRNA durga shortens the dendrites and reduces dendritic arborization in zebrafish, which may have higher level implications during neurodevelopment, neuropathologies and aging.
lncRNAs are poorly conserved at nucleotide levels, but criteria of sequence level conservation is too narrow for lncRNAs. Large number of lncRNAs shows structural, functional conservation and expression from syntenic loci despite lack of sequence homology (Diederichs, 2014; Johnsson et al., 2014). We found that the 5′ end of human and mouse Kalrn locus gives rise to non-coding RNA. Although the sequence of durga, human and mouse lncRNA is not conserved, the gene arrangement is similar (Figure 5A). Putative human counterpart of durga has three exons and its first exon has overlap with first exon of Kalrn. Further expression of human Kalrn non-coding RNA was checked by PCR using cDNA prepared from RNA of SH-SY5Y cell line. We found both spliced (239 bp) as well as unspliced (500 bp) form of human Kalrn non-coding RNA (Figure 5B).
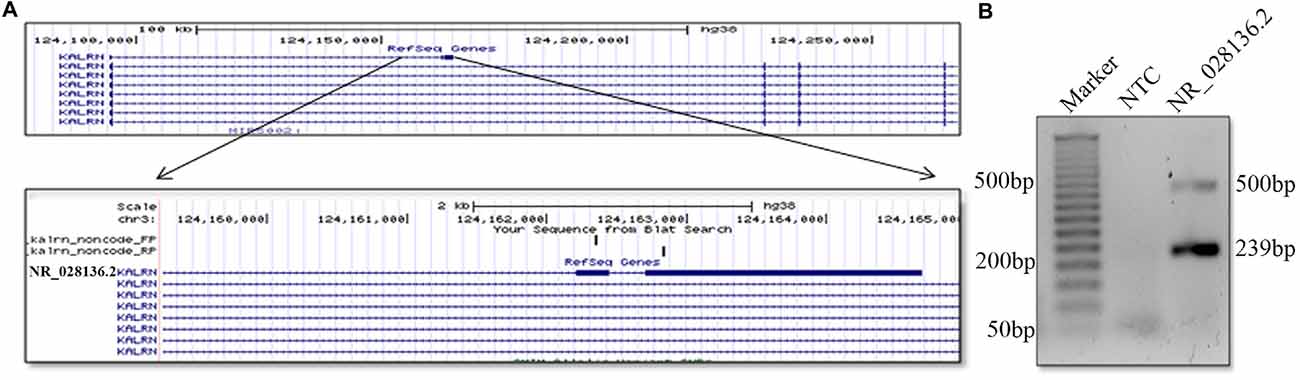
Figure 5. Presence of non-coding RNA at 5′ end of human Karln. Pictorial representation and UCSC genome browser tracks of non-coding (NR_028136.2) RNA located at 5′ end of human Kalrn. Representation is not to scale (A). Expression of non-coding RNA (NR_028136.2) located at 5′ end of human Kalrn was confirmed by PCR in SH-SY5Y cell line (B).
In both the mammalian genomes, the lncRNA is annotated in the same orientation as the Kalrn gene, while in zebrafish it is anti-sense and overlapping. Further functional studies and evaluation of durga in mouse model of neurodegenerative diseases may pave a way for better understanding of Kalrn regulation during development and in various neuropathologies.
Conclusion
Kalirin regulates higher level brain functions by modulating dendritic spine density and morphology. We find an antisense, promoter-proximal lncRNA co-expressed with Kalirin that affects dendrite length and density in zebrafish.
Author Contributions
BP conceived the idea. BP and MAS designed the experiments. DC and MKM collected zebrafish samples. MKM and SR maintained zebrafish stocks. MAS and DC performed PCR, qPCR, primary cell culture experiments and data analysis. DC did Northern blotting experiments. AB and DC performed the in situ hybridization experiments. MK captured the confocal images. BP, MAS and DC co-wrote the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge CSIR INDIA, project code BSC0123 for funding this work. Zebrafish facility and core imaging facility (funded through BSC0403) of CSIR-IGIB are also acknowledged. MAS acknowledges University Grant Commission, New Delhi for fellowship.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fnmol.2017.00095/full#supplementary-material
FIGURE S1 | Northern blot representing transcript corresponding kalrna and lncRNA durga. Northern blotting experiment shows the presence of transcripts corresponding kalrna and durga above 5 KB and ~400 bp respectively (A). Polymerase chain reaction (PCR) detection of durga transcript using anchored oligodT reverse primer for cDNA synthesis and amplified using durga forward primer (B).
FIGURE S2 | In situ hybridization of kalrna and durga in the zebrafish egg. In situ hybridization shows kalrna and durga transcript expression in zebrafish egg.
References
Alam, M. R., Johnson, R. C., Darlington, D. N., Hand, T. A., Mains, R. E., and Eipper, B. A. (1997). Kalirin, a cytosolic protein with spectrin-like and GDP/GTP exchange factor-like domains that interacts with peptidylglycine α-amidating monooxygenase, an integral membrane peptide-processing enzyme. J. Biol. Chem. 272, 12667–12675. doi: 10.1074/jbc.272.19.12667
Chakrabarti, K., Lin, R., Schiller, N. I., Wang, Y., Koubi, D., Fan, Y. X., et al. (2005). Critical role for Kalirin in nerve growth factor signaling through TrkA. Mol. Cell. Biol. 25, 5106–5118. doi: 10.1128/mcb.25.12.5106-5118.2005
Chen, Z., Lee, H., Henle, S. J., Cheever, T. R., Ekker, S. C., and Henley, J. R. (2013). Primary neuron culture for nerve growth and axon guidance studies in zebrafish (Danio rerio). PLoS One 8:e57539. doi: 10.1371/journal.pone.0057539
Colomer, V., Engelender, S., Sharp, A. H., Duan, K., Cooper, J. K., Lanahan, A., et al. (1997). Huntingtin-associated protein 1 (HAP1) binds to a Trio-like polypeptide, with a rac1 guanine nucleotide exchange factor domain. Hum. Mol. Genet. 6, 1519–1525. doi: 10.1093/hmg/6.9.1519
Deo, A. J., Cahill, M. E., Li, S., Goldszer, I., Henteleff, R., Vanleeuwen, J. E., et al. (2012). Increased expression of Kalirin-9 in the auditory cortex of schizophrenia subjects: its role in dendritic pathology. Neurobiol. Dis. 45, 796–803. doi: 10.1016/j.nbd.2011.11.003
Dhiman, H., Kapoor, S., Sivadas, A., Sivasubbu, S., and Scaria, V. (2015). zflncRNApedia: a comprehensive online resource for zebrafish long non-coding RNAs. PLoS One 10:e0129997. doi: 10.1371/journal.pone.0129997
Diederichs, S. (2014). The four dimensions of noncoding RNA conservation. Trends Genet. 30, 121–123. doi: 10.1016/j.tig.2014.01.004
Grubisha, M. J., Lin, C.-W., Tseng, G. C., Penzes, P., Sibille, E., and Sweet, R. A. (2016). Age-dependent increase in Kalirin-9 and Kalirin-12 transcripts in human orbitofrontal cortex. Eur. J. Neurosci. 44, 2483–2492. doi: 10.1111/ejn.13351
Hansel, D. E., Quinones, M. E., Ronnett, G. V., and Eipper, B. A. (2001). Kalirin, a GDP/GTP exchange factor of the Dbl family, is localized to nerve, muscle, and endocrine tissue during embryonic rat development. J. Histochem. Cytochem. 49, 833–844. doi: 10.1177/002215540104900704
Hill, J. J., Hashimoto, T., and Lewis, D. A. (2006). Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol. Psychiatry 11, 557–566. doi: 10.1038/sj.mp.4001792
Iyengar, B. R., Choudhary, A., Sarangdhar, M. A., Venkatesh, K. V., Gadgil, C. J., and Pillai, B. (2014). Non-coding RNA interact to regulate neuronal development and function. Front. Cell. Neurosci. 8:47. doi: 10.3389/fncel.2014.00047
Johnson, R. C., Penzes, P., Eipper, B. A., and Mains, R. E. (2000). Isoforms of kalirin, a neuronal Dbl family member, generated through use of different 5′- and 3′- ends along with an internal translational initiation site. J. Biol. Chem. 275, 19324–19333. doi: 10.1074/jbc.m000676200
Johnsson, P., Lipovich, L., Grandér, D., and Morris, K. V. (2014). Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 1840, 1063–1071. doi: 10.1016/j.bbagen.2013.10.035
Kaushik, K., Leonard, V. E., Kv, S., Lalwani, M. K., Jalali, S., Patowary, A., et al. (2013). Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS One 8:e83616. doi: 10.1371/journal.pone.0083616
Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M., et al. (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. doi: 10.1101/gr.229102.
Makrythanasis, P., Guipponi, M., Santoni, F. A., Zaki, M., Issa, M. Y., Ansar, M., et al. (2016). Exome sequencing discloses KALRN homozygous variant as likely cause of intellectual disability and short stature in a consanguineous pedigree. Hum. Genomics 10:26. doi: 10.1186/s40246-016-0082-2
Mandela, P., and Ma, X. M. (2012). Kalirin, a key player in synapse formation, is implicated in human diseases. Neural Plast. 2012:728161. doi: 10.1155/2012/728161
McPherson, C. E., Eipper, B. A., and Mains, R. E. (2002). Genomic organization and differential expression of Kalirin isoforms. Gene 284, 41–51. doi: 10.1016/s0378-1119(02)00386-4
McPherson, C. E., Eipper, B. A., and Mains, R. E. (2004). Kalirin expression is regulated by multiple promoters. J. Mol. Neurosci. 22, 51–62. doi: 10.1385/jmn:22:1-2:51
Meijering, E., Jacob, M., Sarria, J. C., Steiner, P., Hirling, H., and Unser, M. (2004). Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58, 167–176. doi: 10.1002/cyto.a.20022
Miller, M. B., Yan, Y., Wu, Y., Hao, B., Mains, R. E., and Eipper, B. A. (2017). Alternate promoter usage generates two subpopulations of the neuronal RhoGEF Kalirin-7. J. Neurochem. 140, 889–902. doi: 10.1111/jnc.13749
Penzes, P., Johnson, R. C., Kambampati, V., Mains, R. E., and Eipper, B. A. (2001). Distinct roles for the two Rho GDP/GTP exchange factor domains of kalirin in regulation of neurite growth and neuronal morphology. J. Neurosci. 21, 8426–8434.
Remmers, C., Sweet, R. A., and Penzes, P. (2014). Abnormal kalirin signaling in neuropsychiatric disorders. Brain Res. Bull. 103, 29–38. doi: 10.1016/j.brainresbull.2013.12.006
Sato, T., Takahoko, M., and Okamoto, H. (2006). HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis 44, 136–142. doi: 10.1002/gene.20196
Stojic, L., Niemczyk, M., Orjalo, A., Ito, Y., Ruijter, A. E., Uribe-Lewis, S., et al. (2016). Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 7:10406. doi: 10.1038/ncomms10406
Tolias, K. F., Duman, J. G., and Um, K. (2011). Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog. Neurobiol. 94, 133–148. doi: 10.1016/j.pneurobio.2011.04.011
Tran, N. T., Su, H., Khodadadi-Jamayran, A., Lin, S., Zhang, L., Zhou, D., et al. (2016). The AS-RBM15 lncRNA enhances RBM15 protein translation during megakaryocyte differentiation. EMBO Rep. 17, 887–900. doi: 10.15252/embr.201541970
Vishwanatha, K. S., Wang, Y. P., Keutmann, H. T., Mains, R. E., and Eipper, B. A. (2012). Structural organization of the nine spectrin repeats of Kalirin. Biochemistry 51, 5663–5673. doi: 10.1021/bi300583s
Westerfield, M. (2000). The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th Edn. Eugene: University of Oregon Press.
White, R. M., Sessa, A., Burke, C., Bowman, T., LeBlanc, J., Ceol, C., et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189. doi: 10.1016/j.stem.2007.11.002
Yan, Y., Eipper, B. A., and Mains, R. E. (2015). Kalirin-9 and Kalirin-12 play essential roles in dendritic outgrowth and branching. Cereb. Cortex 25, 3487–3501. doi: 10.1093/cercor/bhu182
Yan, Y., Eipper, B. A., and Mains, R. E. (2016). Kalirin is required for BDNF-TrkB stimulated neurite outgrowth and branching. Neuropharmacology 107, 227–238. doi: 10.1016/j.neuropharm.2016.03.050
Yap, K. L., Li, S., Muñoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S., et al. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. cell 38, 662–674. doi: 10.1016/j.molcel.2010.03.021
Keywords: Kalirin, long non-coding RNA, dendritic morphology, zebrafish, primary neuron culture
Citation: Sarangdhar MA, Chaubey D, Bhatt A, KM M, Kumar M, Ranjan S and Pillai B (2017) A Novel Long Non-coding RNA, durga Modulates Dendrite Density and Expression of kalirin in Zebrafish. Front. Mol. Neurosci. 10:95. doi: 10.3389/fnmol.2017.00095
Received: 21 November 2016; Accepted: 21 March 2017;
Published: 10 April 2017.
Edited by:
Philip Washbourne, University of Oregon, USAReviewed by:
Christian Gonzalez-Billault, University of Chile, ChileVidisha Tripathi, National Centre for Cell Science, India
Copyright © 2017 Sarangdhar, Chaubey, Bhatt, KM, Kumar, Ranjan and Pillai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beena Pillai, YmVlbmFAaWdpYi5pbg==
 Mayuresh A. Sarangdhar
Mayuresh A. Sarangdhar Divya Chaubey1
Divya Chaubey1 Beena Pillai
Beena Pillai