- Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
The polycomb group proteins (PcGs) are a group of epigenetic factors associated with gene silencing. They are found in several families of multiprotein complexes, including polycomb repressive complex 2 (PRC2). EZH2, EED and SUZ12 form the core components of the PRC2 complex, which is responsible for the mono, di- and trimethylation of lysine 27 of histone 3 (H3K27Me3), the chromatin mark associated with gene silencing. Loss-of-function studies of Ezh2, the catalytic subunit of PRC2, have shown that PRC2 plays a role in regulating developmental transitions of neuronal progenitor cells (NPCs); from self-renewal to differentiation and the neurogenic-to-gliogenic fate switch. To further address the function of EZH2 and H3K27me3 during neuronal development, we generated a conditional mutant in which Ezh2 was removed in the mammalian isthmic (mid-hindbrain) region from E10.5 onward. Loss of Ezh2 changed the molecular coding of the anterior ventral hindbrain leading to a fate switch and the appearance of ectopic dopaminergic (DA) neurons. The correct specification of the isthmic region is dependent on the signaling factors produced by the Isthmic organizer (IsO), located at the border of the mid- and hindbrain. We propose that the change of cellular fate is a result of the presence of Otx2 in the hindbrain of Ezh2 conditional knock-outs (cKOs) and a dysfunctional IsO, as represented by the loss of Fgf8 and Wnt1. Our work implies that next to controlling developmental transitions, EZH2 mediated gene silencing is important for specification of the isthmic region by influencing IsO functioning and repressing Otx2 in the hindbrain.
Introduction
In recent years, it has become apparent that next to transcription factors, gene regulation via epigenetics also plays an important part in the development of a multicellular organism (Bernstein et al., 2006; Mikkelsen et al., 2007; Mohn et al., 2008). Epigenetics can be defined as the biological processes that alter transcriptional activity by influencing the accessibility of the DNA by reorganizing the chromatin structure. Adaptation of chromosomal regions can rely on three distinct processes: (1) Modification of histones; (2) DNA methylation; and (3) non-coding RNAs mediated changes (van Heesbeen et al., 2013). Histones contain numerous sites for different types of modifications, such as methylation, acetylation and phosphorylation (Podobinska et al., 2017), which are highly dynamic and cause readily reversible changes in chromosomal organization (Cedar and Bergman, 2009). Methylation of histones can have different outcomes, while some sites correlate to transcriptional activity, others are associated with gene silencing (Podobinska et al., 2017). The polycomb repressive complex 2 (PRC2) mediates the mono, di and tri-methylation of lysine 27 of histone 3 (H3K27me1, 2 and 3; Shen et al., 2008) and consists of three core subunits: enhancer of zeste homolog 2 (EZH2), embryonic ectoderm development (EED) and suppressor of zeste 12 (SUZ12), that are all crucial for the catalytic activity of the complex (Corley and Kroll, 2015). H3K27me3 is associated with gene silencing and shows a highly dynamic profile during development (Mohn et al., 2008). Together with histone 3 lysine 4 tri-methylation (H3K4me3), H3K27me3 is found on bivalent domains that silence developmental genes during one stage of development but keeps them poised for activation in later stages of development (Bernstein et al., 2006; Mohn et al., 2008). The importance of H3K27 methylation and the PRC2 complex during development were shown in previous loss-of-function studies. The genetic ablation of one of the core subunits of the PRC2 complex led to a global loss of H3K27 di- and tri-methylation and a significant reduction in mono-methylation (Ferrari et al., 2014). In addition, knock-out (KO) embryos for one of the three core subunits of PRC2 fail to complete gastrulation due to defects in morphogenetic movements (Faust et al., 1998; O’Carroll et al., 2001; Cao and Zhang, 2004a). In the later stages of development, it was shown that when Ezh2 was conditionally removed before the onset of neurogenesis in cortical progenitors, the balance between self-renewal and differentiation was shifted toward differentiation (Pereira et al., 2010). In addition, neurogenesis was accelerated and the onset of gliogenesis was earlier (Pereira et al., 2010). However, when Ezh2 or Eed were removed during neurogenesis, the neurogenic phase was prolonged at the expense of the onset of astrogenesis (Hirabayashi et al., 2009). These data suggest that PRC2 plays a role in regulating developmental transitions in cortical progenitor cells; from self-renewal to differentiation and the neurogenic-to-gliogenic switch (Hirabayashi et al., 2009; Pereira et al., 2010). A similar role for Ezh2 was found in neuronal progenitors (NPs) of the dorsal midbrain. Wnt1Cre driven deletion of Ezh2 led to reduced number of NPs in the dorsal midbrain, due to elevated cell cycle exit and differentiation (Zemke et al., 2015). In addition to its role in regulating the transition from proliferation to differentiation, Ezh2 was also found to be required for the maintenance of regional identity by suppressing forebrain traits (Zemke et al., 2015). To further address the function of EZH2 and H3K27me3 during neuronal development we generated a conditional mutant in which Ezh2 was removed in the isthmic (mid-hindbrain) region from E10.5 onward (Sunmonu et al., 2009; Zemke et al., 2015). The mid-hindbrain region gives rise to several essential neurotransmitter systems, including dopaminergic (DA) and serotonergic (5-HT) neurons (Brodski et al., 2003). A critical event in the development of the different neuronal populations found in the mid-hindbrain region is the formation of the Isthmic Organizer (IsO; Crossley et al., 1996; Brodski et al., 2003). The IsO secretes inductive signals that specify the mid-hindbrain region and it has been shown that a disorganized IsO influences the location and size of the DA and 5-HT system (Brodski et al., 2003; Kouwenhoven et al., 2016). The functioning and maintenance of the IsO is dependent on several feedback loops formed by multiple developmental factors, including Fgf8, Wnt1, Lmx1b and En1 (Danielian and McMahon, 1996; Martinez et al., 1999; Adams et al., 2000; Guo et al., 2006; Canning et al., 2007; Kouwenhoven et al., 2016). In this study, we show that the En1Cre driven deletion of Ezh2 and a consequential loss of H3K27me3 leads to a disorganized IsO and ectopic expression of the transcription factor Otx2 in the hindbrain. In accordance with other studies in which Otx2 expression is expanded caudally, DA cells are formed in rhombomere (R) 1 at the expense of R1 born neuronal subtypes, including 5-HT neurons (Brodski et al., 2003; Sherf et al., 2015; Kouwenhoven et al., 2016). Together, our data suggest that next to transcription factors, gene repression via EZH2 and H3K27me3 are required for the repression of Otx2 in the hindbrain, maintenance of the IsO and the correct fate-determination of the isthmic area.
Materials and Methods
Ethics Statement
All animal studies are performed in accordance with local animal welfare regulations, as this project has been approved by the animal experimental committee (Dier ethische commissie, Universiteit van Amsterdam; DEC-UvA), and international guidelines.
Animals
All lines were maintained on a C57BL/6J background (Charles River). Ezh2-floxed animals were generated by S.H. Orkin and a kind gift from F. Zilbermann (Friedrich Miescher Institute, Basel, Switzerland) and have been previously described (Shen et al., 2008). En1Cre animals were generated by A.L Joyner and a kind gift from S. Blaess (Rheinische Friedrich-Wilhelms-Universität, Bonn, Gemany). En1Cre/+ animals were crossed with En1Cre-ERT +/+; R26RYFP/R26RYFP to obtain En1Cre/+; R26RYFP/R26RYFP (Kimmel et al., 2000). Ezh2 L/L animals were crossed with En1Cre/+ or En1Cre/+; R26RYFP/R26RYFP animals to obtain En1Cre/+; Ezh2 L/+ or En1Cre/+; Ezh2 L/+; R26RYFP/R26RYFP animals. For the generation of embryos we crossed En1Cre/+; Ezh2 L/+ animals with Ezh2 L/+ animals and En1Cre/+; Ezh2 L/+ with En1Cre/+; Ezh2 L/+ animals for the generation of En1/Ezh2 double mutant embryos. Embryos were isolated at embryonic day (E) 12.5, E14.5, considering the morning of plug formation as E0.5. Pregnant dams were euthanized by CO2 asphyxiation and embryos were collected in 1× PBS and immediately frozen on dry-ice (fresh frozen) or fixed by immersion in 4% paraformaldehyde (PFA) for 4–5 h at 4°C. After PFA incubation, samples were cryoprotected O/N in 30% sucrose at 4°C. Embryos were frozen on dry-ice and stored at −80°C. Cryosections were slices at 16 μm, mounted at Superfrost plus slides, air-dried and stored at −80°C until further use.
E14.5 Wn1Cre/+; Ezh2 L/L and wildtype littermate embryonic tissue were a kind gift from Prof. Dr. Filippo Rijli of the Friedrich Miescher Institute in Basel, Switzerland.
Genotyping
The genotyping for the Ezh2-flox allele was executed with 50–100 ng of genomic DNA together with forward primer 5′-ACCATGTGCTGAAACCAACAG-3′ and reverse primer 5′-TGACATGGGCCTCATAGTGAC-3′ resulting in a 395 bp product for the wild type allele and a 361 bp product for the floxed allele.
Genotyping of the En1Cre allele was performed with 50–100 ng of genomic DNA together with primer pair En1Cre 5UTR_F3 5′-CTTCGCTGAGGCTTCGCTTT-3′ and En1Cre Cre_R2 5′-AGTTTTTACTGCCAGACCGC-3′ resulting in a product at 240 bp for the Cre-allele.
Genotyping for the R26R-YFP allele was performed using 3′ primers Rosa_mutant primer 5′-AAGACCGCGAAGAGTTTGTC-3′, Rosa_wildtype primer 5′-GGAGCGGGAGAAATGGATATG-3′ and a Rosa_common primer 5′-AAAGTCGCTCTGAGTTGTTAT-3′ with 50–100 ng of genomic DNA. The PCR reaction gave a product at 320 bp for the mutant R26R-YFP allele and a product of 600 bp for the wildtype allele.
In situ Hybridization
In situ hybridization with digoxigenin (DIG)-labeled RNA probes was performed as described (Smits et al., 2003; Smidt et al., 2004). The following DIG-Labeled RNA probes were used: Th (Grima et al., 1985), Pitx3, Nurr1 (Smidt et al., 2000), Ahd2 (Jacobs et al., 2007), En1, Cck (Jacobs et al., 2011), Lmx1a (Hoekstra et al., 2013), Wnt1 (Mesman et al., 2014), Fgf8, Otx2 (Kouwenhoven et al., 2016) and a Nkx6.1 probe containing bp 1182–1713 (532 bp) of the mouse cDNA sequence (NM_144955.1). Experiments we performed in two independent sets of animals to ensure the results. Wnt1Cre driven mutants, En1 mutants and En1; Ezh2 double mutants were performed on one set of embryos.
Fluorescence Immunohistochemistry
Cryosections were blocked with 4% HIFCS in THZT buffer (50 mM Tris-HCL, pH 7.6; 0.5 M NaCl; 0.5% Triton X-100) and incubated with a primary antibody; Th [Rabbit-TH (Pelfreeze 1:1,000), Sheep-TH (Millipore AB1542, 1:750)], EYFP [Chicken-GFP (Abcam, 1:500)], H3K27me3 [rabbit-anti-H3K27me3 (Millipore, 17-622 1:2,000)] or Serotonin [rabbit-anti-Serotonin (Immunostar, 1:500)], in THZT buffer overnight at room temperature. The following day the slides were washed and incubated for 2 h at room temperature with secondary Alexafluor antibody [anti-rabbit, anti-sheep and anti-chicken (Invitrogen, 1:1,000)] in Tris-Buffer saline (TBS). After immunostaining nuclei were stained with DAPI (1:3,000), embedded in Fluorsave (Calbiogen) and analyzed with the use of a fluorescent microscope (Leica). All washes were done in TBS and double staining was performed sequentially. The antibody against H3K27me3 required antigen retrieval, which was executed as follows: slides were incubated 10 min in PFA after which they were submerged in 0.1 M citrate buffer pH 6.0 for 3 min at 1,000 Watts followed by 9 min at 200 Watts. Slides were left to cool down, after which protocol was followed as described above. Per embryonic stage two sets, En1Cre/+; Ezh2 +/+ vs. En1Cre/+; Ezh2 L/L, were analyzed. Wnt1Cre driven mutants, En1 mutants and En1;Ezh2 double mutants were performed on one set of embryos.
Quantitative PCR (qPCR)
RNA was isolated from dissected E12.5 or E14.5 midbrains of En1Cre/+; Ezh2 +/+ and Ezh2 conditional KO (cKO) embryos. RNA was isolated with Trizol (ThermoFisher) according to the manufacturer’s protocol. For E12.5 a single midbrain was used per sample (En1Cre/+; Ezh2 +/+ n = 3 and En1Cre/+; Ezh2 L/L n = 4). For E14.5 two midbrains were pooled for the Ezh2 cKO samples and a single midbrain was used per sample for the wildtype (wildtype n = 4 and En1Cre/+; Ezh2 L/L n = 3). Relative expression levels were determined by using the QuantiTect SYBR green PCR lightCycler kit (Qiagen) according to the manufacturer’s instructions. For each reaction, 10 ng (dissected midbrain) of total RNA was used as input. Primers used for En1 were previously published (Jacobs et al., 2011). Further, primer designs: Fgf8: forward 5′-GCAGAAGACGGAGACCCCT and reverse 5′-CGTTGCTCTTGGCAATTAGCTTCC (product size 136 bp), Wnt1 forward 5′-CAGCAACCACAGTCGTCAGA and reverse 5′-TTCACGATGCCCCACCATC (produce size 170 bp) and Otx2: forward 5′-GGGAAGAGGTGGCACTGAAA and reverse 5′-CTTCTTGGCAGGCCTCACTT (produce size 137 bp). Statistical analysis was performed by a Student-T test and one or two tails is indicated in the figure legend.
Results
En1Cre Driven Deletion of Ezh2 Causes a Widespread Loss of H3K27me3
As described above, EZH2 functions as the methyltransferase of PRC2, which catalyzes the methylation of H3K27 (Cao and Zhang, 2004b; Margueron et al., 2008; Shen et al., 2008). Early deletion of Ezh2 has been shown to cause an overall ablation of H3K27me3 and transitions in developmental phases, like the switch from self-renewal to differentiation, are disrupted (Hirabayashi et al., 2009; Pereira et al., 2010; Zemke et al., 2015). In the mid-hindbrain region several neuronal subtypes arise, including DA and serotonergic (5-HT) neurons (Brodski et al., 2003; Kouwenhoven et al., 2016). An essential event in the emergence of these neuronal populations is the formation of the IsO, which secretes inductive signals that determine the size and location of the different neuronal subtypes originating in the mid-hindbrain region (Crossley et al., 1996; Brodski et al., 2003; Kouwenhoven et al., 2016). In this study, we aim to gain further insight into the role of EZH2 and PRC2 functioning in the development of the mid-hindbrain region by generating a cKO mouse. Ezh2 was removed from the mid-hindbrain region from E10.5 onward by crossing Ezh2-floxed mice (Shen et al., 2008) with En1Cre animals (Kimmel et al., 2000). The CRE recombinant region, as visualized by YFP (R26RYFP, Srinivas et al., 2001), extends rostrally into the ventral region of prosomere 3 and caudally to the presumed R1/R2 border (Figure 1A, arrowheads point to border regions; Kouwenhoven et al., 2016). To determine whether the deletion of Ezh2 in the mid-hindbrain leads to the loss of H3K27me3, an immunohistochemistry for H3K27me3 was performed in combination with DAPI staining (Figure 1B). In En1Cre/+; Ezh2 L/L animals, H3K27me3 could not be observed in the mid-hindbrain region from E12.5 onward [Figure 1B(3), arrowheads], while the mark could be detected in this region in wildtype embryos [Figure 1B(1)]. In addition, H3K27me3 is present in the Ezh2 cKO outside of the CRE recombination region [Figure 1B(4)], indicating that the deletion of Ezh2 is specific for the En1 positive area and that the ablation of Ezh2 alone is sufficient to remove H3K27me3 in this region.
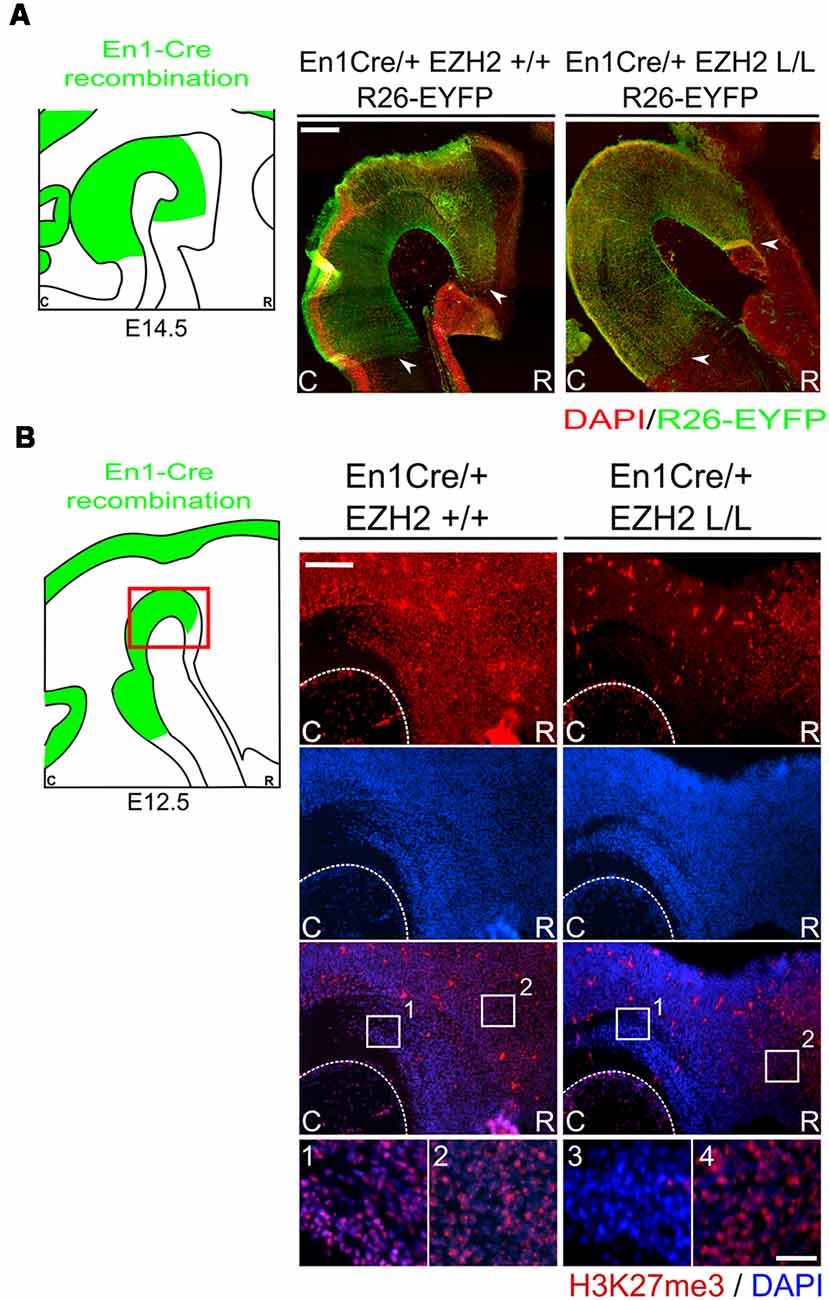
Figure 1. H3K27me3 is lost in the En1 expression domain of En1Cre/+; Ezh2 L/L E12.5 Embryos. (A) Immunohistochemistry of YFP (green; counter stain DAPI in red) was performed in E14.5 wildtypes and Ezh2 conditional knock-out (cKO) embryos to determine the CRE recombinant area. YFP expression is found from the ventral region of P3 and extended caudally up until the R1/R2 border. Border areas are indicated by arrow heads. Scale bar = 225 μM. (B) The presence of H3K27me3 was determined in E12.5 En1Cre/+; Ezh2 +/+ and En1Cre/+; Ezh2 L/L embryos by means of immunohistochemistry. H3K27me3 (red) was lost in the midbrain of Ezh2 cKO embryos (3) but was still present in the dorsal pretectum (4). Nuclear localization of H3K27me3 was verified by co-localization with DAPI staining (blue). Scale bars are 150 μM and 15 μM in the magnifications.
Ectopic Dopaminergic Neurons Arise in the Hindbrain of En1Cre/Ezh2 L/L Animals
One of the essential neuronal subtypes that develop in the Isthmic region are mesodiencephalic DA (mdDA) neurons. MdDA progenitors originate from the floor plate of the ventral midbrain under the influence of intersecting signals of FGF8, OTX2 and WNT1 (Placzek and Briscoe, 2005; Ono et al., 2007; Mesman et al., 2014). During neurogenesis, mdDA progenitors enter the G0 phase of the cells cycle to give rise to post-mitotic mdDA precursors that start migrating towards their final diencephalic (Prosomere 1–2) and midbrain domains (Shults et al., 1990; Kawano et al., 1995; Mesman et al., 2014). During this phase, mdDA precursors differentiate further into mdDA neurons characterized by the presence of tyrosine hydroxylase (TH), the rate-limiting enzyme in dopamine synthesis. Although most TH+ cells are born around E11.0 (Bayer et al., 1995), it is not until E14.5 that they express most genes that define a mature mdDA neuron, like Dat, Ahd2 and Cck (Iversen, 2010; Veenvliet et al., 2013; Arenas et al., 2015). To examine whether the loss of Ezh2 in the mid-hindbrain region affects the DA neuronal population we performed immunohistochemistry for TH at E14.5 (Figure 2). When comparing En1Cre/+; Ezh2 +/+ embryos to En1Cre/+; Ezh2 L/L embryos a caudal expansion of TH+ neurons is observed (Figure 2, arrowheads). The biggest changes in the TH+ domain are observed in the medial sections where a group of TH+ neurons is found in an ectopic location caudal to the midbrain.
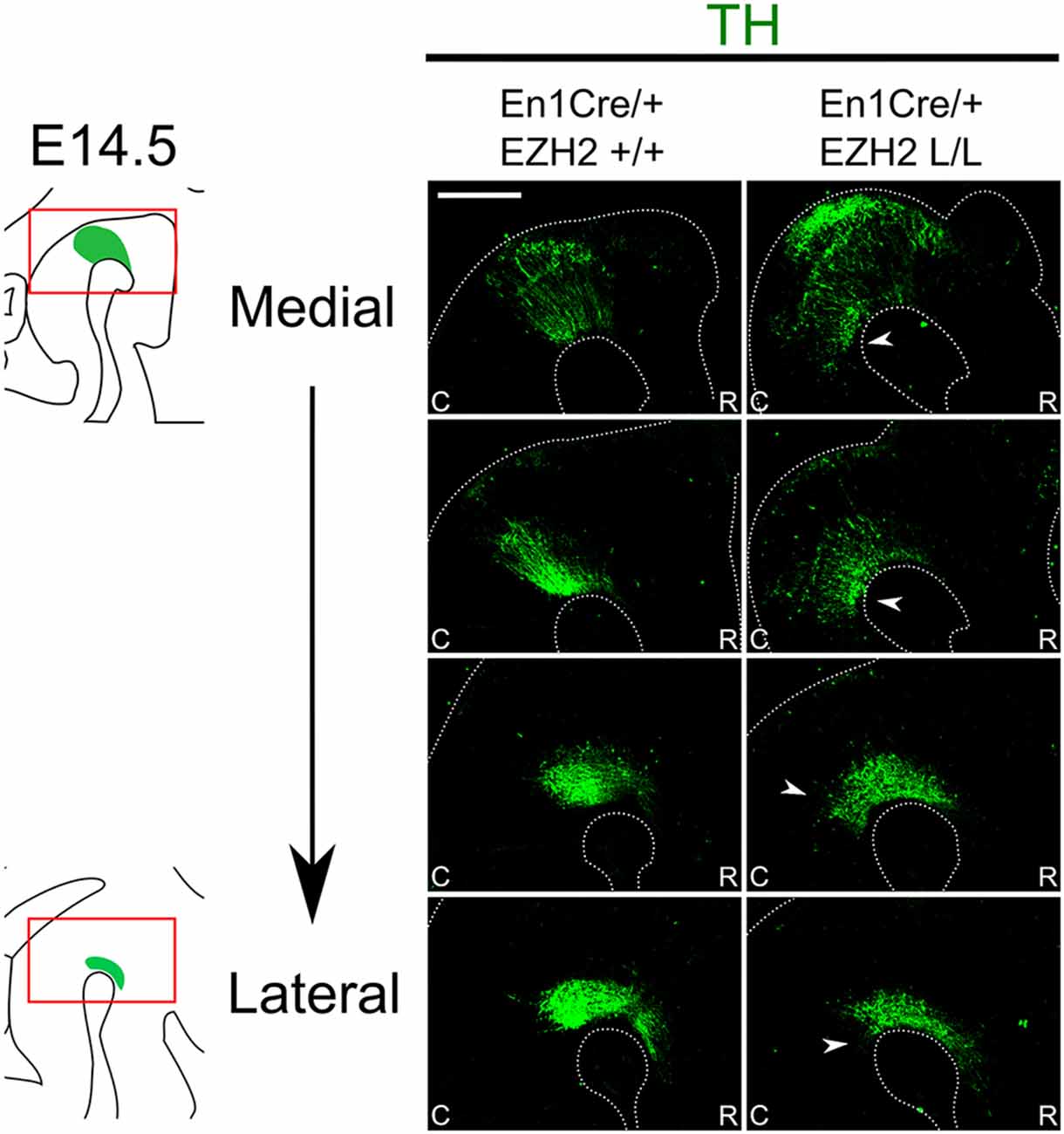
Figure 2. En1Cre driven deletion of Ezh2 leads to a caudal expansion of the TH+ domain. Protein expression of TH (green) was evaluated by means of immunohistochemistry. In E14.5 En1Cre/+; Ezh2 L/L embryos the TH+ domain is shifted caudally in comparison to En1Cre/+; Ezh2 +/+ embryos (arrowheads). The caudal expansion of the TH+ domain is most apparent in medial sections (upper panels). Scale bar = 150 μM.
Next to mdDA neurons, the mid-hindbrain region also gives rise to 5-HT neurons (Brodski et al., 2003; Fox and Deneris, 2012; Alonso et al., 2013; Sherf et al., 2015). 5-HT neurons develop caudally to the IsO and are arranged into three distinct nuclei; the dorsal raphe nucleus (DRN), the median raphe nucleus (MRN) and the prepontine raphe nucleus (PpnR), which are R2-derived (Fox and Deneris, 2012; Alonso et al., 2013). Analysis of serotonin by immunohistochemistry at E14.5 showed that the cytoarchitecture of the 5-HT system is changed in Ezh2 cKOs (Figure 3A). Under normal conditions, no 5-HT+ cells can be found in between the DRN and the median raphe nucleus (MRN) where the decussation of the superior cerebellar peduncle runs, however in En1Cre/+; Ezh2 L/L embryos 5-HT+ neurons are present in this region (Figure 3A, arrowheads). In addition to the disorganized structure, a loss of cells in the DRN is observed in lateral sections of En1Cre/+; Ezh2 L/L embryos when compared to wildtypes (Figure 3A, arrows). Previous studies have already shown that the caudal expansion of the DA domain negatively affects the generation of 5-HT+ neurons in R1 (Brodski et al., 2003; Sherf et al., 2015; Kouwenhoven et al., 2016). So to get a better insight into the effect of the caudal expansion of the TH+ domain into the ventral hindbrain on the generation of the 5-HT+ population in this region, we performed a double immunohistochemistry for 5-HT and TH on adjacent slides (Figure 3B). In line with these studies, no 5-HT+ cells were found dorsally of the ventral ectopic patch of TH+ neurons or directly caudal to the dorsal DA domain in the Ezh2 cKOs [Figure 3B(2), arrowhead]. In wildtypes, the 5-HT+ population of the DRN aligns the dorsal DA domain [Figure 3B(1), arrowhead] and the lack of cells dorsal of the ectopic TH+ patch suggests that the caudal expansion of the TH+ domain is at the expense of the 5-HT+ domain corresponding to the DRN.
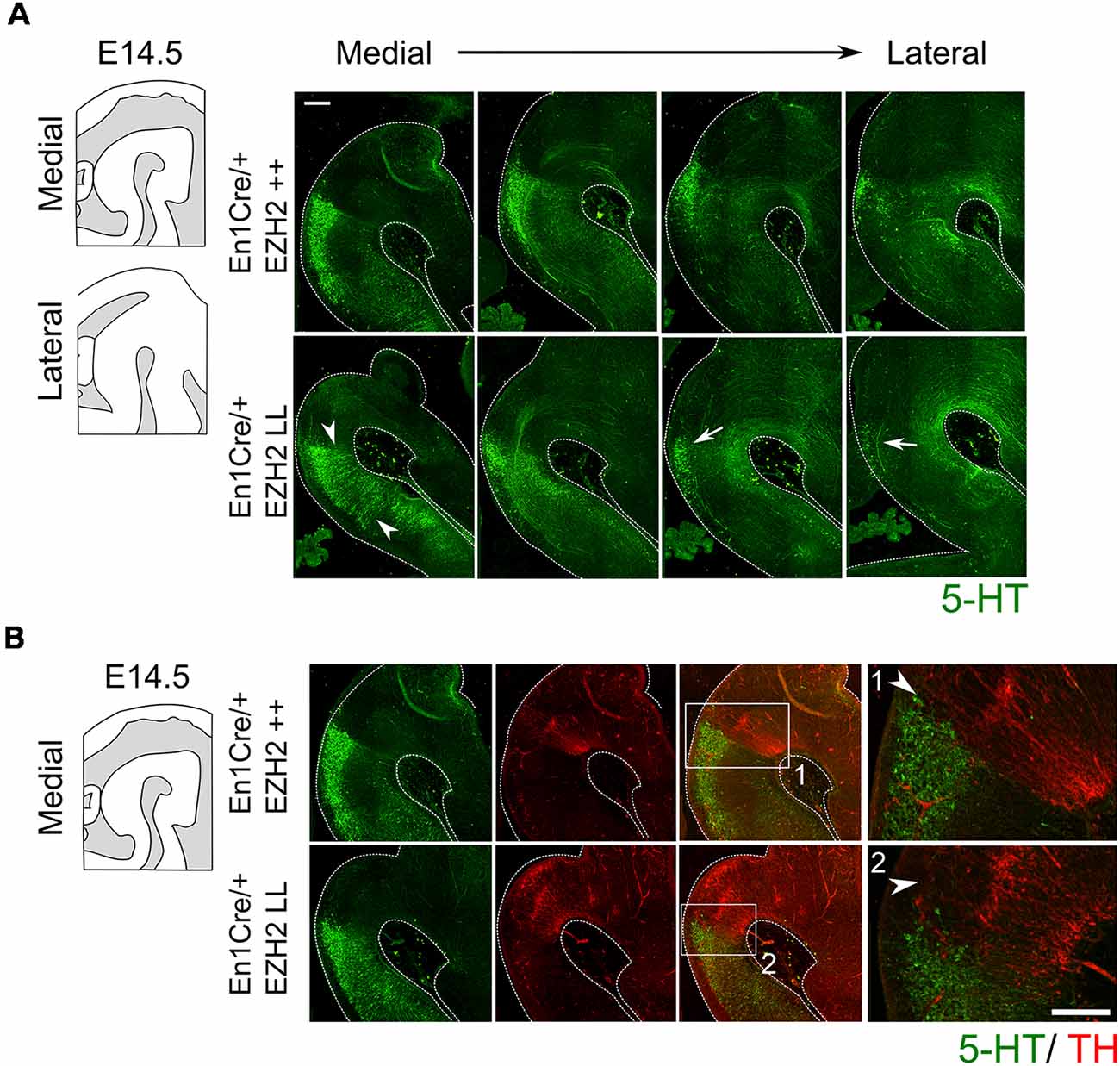
Figure 3. Genetic ablation of Ezh2 affects the cytoarchitecture of the 5-HT population and leads to the loss of neurons of the dorsal raphe nucleus (DRN). (A) The 5-HT positive population was examined by performing a immunohistochemistry for 5-HT (green). The overall cytoarchitecture of the 5-HT population is affected (arrowheads) and cells of the DRN are lost in the lateral sections of En1Cre/+; Ezh2 L/L E14.5 embryos (arrows). Scale bar = 250 μM. (B) A double immunohistochemistry for 5-HT (green) and TH+ (red). (1) In wildtypes 5-HT cells align the dorsal mdDA domain (arrowhead), while in En1Cre driven Ezh2 cKOs (2) TH+ cells are found in the hindbrain and the 5-HT cells of the DRN cells are lost dorsal of the ectopic TH+ patch (arrowhead). Scale bars are 150 μM.
Together our data indicate that En1Cre driven deletion of Ezh2 leads to the appearance of TH positive cells caudal to the midbrain and that this extension of the TH+ domain is at the expense of 5-HT+ neurons.
Conditional Deletion of Ezh2 Changes the Molecular Coding of the Anterior Hindbrain
To further substantiate whether the ectopic TH+ cells are DA-like neurons, we performed in situ hybridization for several DA marks (Figure 4). Lmx1a, Nurr1 and Pitx3 have been shown to play an important role in mdDA development (Saucedo-Cardenas et al., 1998; Jankovic et al., 2005; Deng et al., 2011; Yan et al., 2011). While Lmx1a has been shown to be required for the activation of neurogenesis and for the suppression of alternative cells fates (Andersson et al., 2006; Omodei et al., 2008), Nurr1 induces the DA neurotransmitter phenotype (Saucedo-Cardenas et al., 1998; Smits et al., 2003). Pitx3 expression is initiated in later stages of development and has been shown to be important for the specification of the different mdDA subsets and for the survival of the DA neurons of the Substantia Nigra pars compacta (SNc; Smidt et al., 1997; Hwang et al., 2003; Maxwell et al., 2005; Veenvliet et al., 2013). The spatial expression of Th shows a corresponding expression pattern as was observed with immunohistochemistry, an ectopic patch of expression caudal to the midbrain (arrowhead). Analysis of Lmx1a and Nurr1 expression on adjacent sections demonstrated that they are also expressed in the ectopic Th area caudal to the midbrain in Ezh2 cKO embryos [Figure 4A(*), arrowheads]. In addition, Pitx3 was also found ectopically, caudal to the midbrain in En1Cre/+; Ezh2 L/L embryos [Figure 4A(*), lowest panel, arrowhead], indicating that the ectopic TH+ cells have a DA-like phenotype. Next to DA marks that are expressed by all mdDA neurons, we also examined the expression pattern of two subset marks, Cck and Ahd2 (Figure 4B; Veenvliet et al., 2013). Cck is normally expressed by the caudomedial mdDA neuronal population and when examining En1Cre/+; Ezh2 +/+ and Ezh2 cKO midbrains, expression was also detected caudal to the midbrain in Ezh2 cKO embryos (Figure 4B, left panel, arrowhead). Interestingly, Adh2 expression was also sparsely observed caudal to the midbrain in medial sections of En1Cre/+; Ezh2 L/L embryos (Figure 4B, right panel, arrowhead), while Ahd2 is restricted to the rostrolateral DA population in wildtypes (Veenvliet et al., 2013).
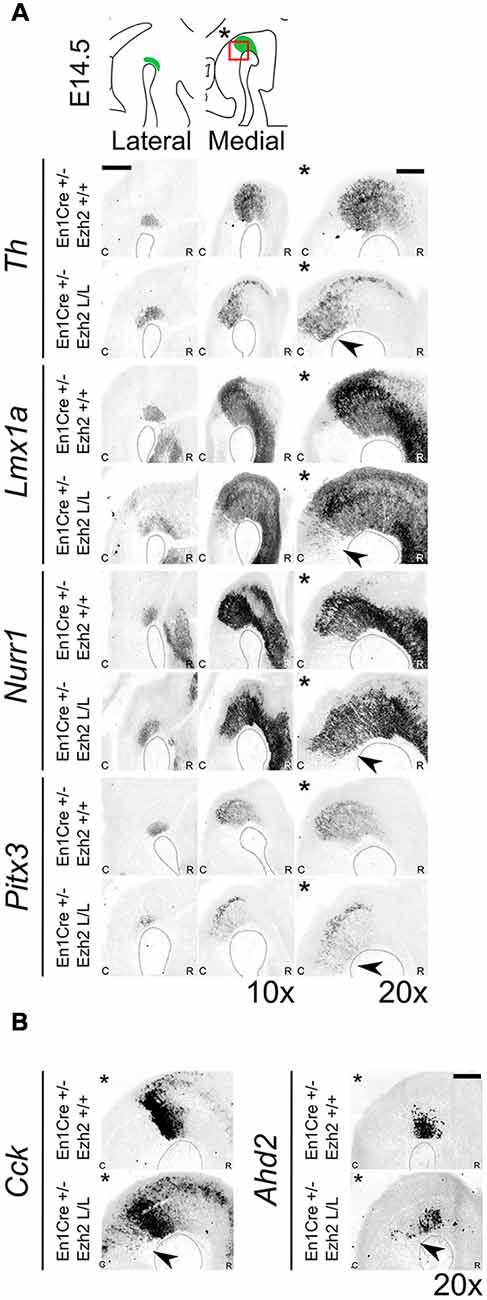
Figure 4. Dopaminergic (DA) marks can be detected more caudally in Ezh2 cKO animals. Analysis of DA marks, Th, Lmx1a, Nurr1, Pitx3 and subset marks Cck and Ahd2 at E14.5 by means of in situ hybridization (A,B). (A) An ectopic patch expressing Th, Lmx1a, Nurr1 and Pitx3 can be observed caudal to the midbrain in the Ezh2 mutant, which is not present in En1Cre/+; Ezh2 +/+ animals (arrowheads). Scale bar = 400 μM. (B) In addition to general marks, the caudomedial subset mark Cck and the rostrolateral mark Ahd2 were also found to be shifted caudally in En1Cre/+; Ezh2 L/L animals compared to wildtypes (arrowheads). Scale bar = 200 μM.
In normal conditions, Nkx6.1 expressing trochlear motor neurons are located in and directly caudal to the IsO (Chandrasekhar, 2004; Prakash et al., 2009). As described above, Lmx1a has been demonstrated to suppress alternative cell fates, including motor neurons by the repression of Nkx6.1 (Andersson et al., 2006). The presence of Lmx1a and other DA-related genes in ventral R1 suggest that the molecular coding in this region might be altered, leading to a DA-like cell fate. To assess whether the deletion of Ezh2 leads to an alternative cell fate and the suppression of Nkx6.1, we assessed the presence of Nkx6.1 in the ventral Isthmic area by means of in situ hybridization (Figure 5). When examining the mid-hindbrain region of En1Cre/+; Ezh2 L/L embryos almost all Nkx6.1 staining was lost in the Isthmic area [Figure 5(2), arrowhead], corresponding to the region where normally the trochlear motor neurons develop [Figure 5(1); Chandrasekhar, 2004]. In contrast, Nkx6.1 expression in other regions, like the red nucleus, were similar to En1Cre/+; Ezh2 +/+ littermates, indicating that the loss of Ezh2 specifically affects Nkx6.1 expression in the anterior hindbrain.
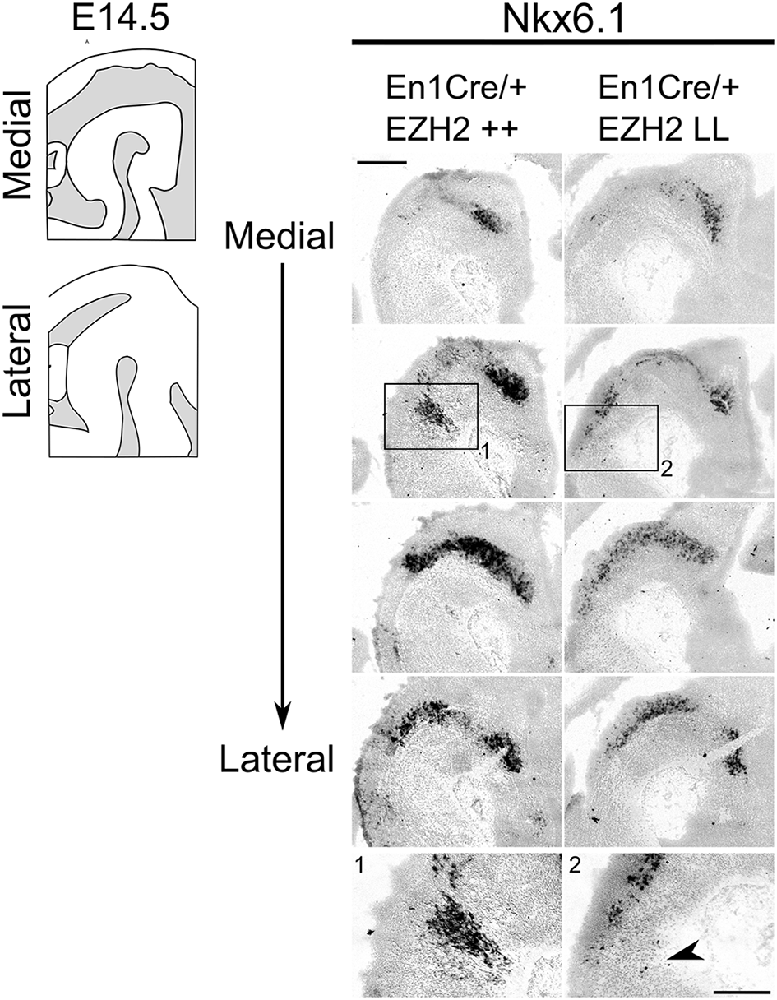
Figure 5. Nkx6.1 expression is lost in the ventral R1 region in Ezh2 cKO E14.5 embryos. In situ hybridization for Nkx6.1 was performed on E14.5 En1Cre/+; Ezh2 +/+ and Ezh2 cKO embryos. Analysis of the spatial expression of Nkx6.1 shows that the expression is lost in the ventral region of R1 in En1Cre/+; Ezh2 L/L embryos (1 and 2, arrowhead), but not in the midbrain. Scale bars are 400 μM and 100 μM in the magnifications.
Together, these results suggest that the loss of Ezh2 changes the molecular coding of the anterior hindbrain, leading to the presence of an ectopic patch of dopamine-like cells caudal to the midbrain and the loss of Nkx6.1 + expression in this region.
En1Cre Driven Ezh2 cKOs Show a Similar Phenotype as En1 Mutants Independent of En1
Previous studies have shown that in the absence of En1, DA-related genes display a caudal expansion of their expression domain in more medial sections. The presence of these ectopic DA cells can be detected as early as E12.5 and they show a similar molecular and electrophysiological profile as mdDA neurons (Veenvliet et al., 2013; Kouwenhoven et al., 2016). The Ezh2 cKO is heterozygous for En1 and to verify that the ectopic presence of DA neurons caudal to the midbrain in the En1Cre/+; Ezh2 L/L is not a consequence of the combined loss of En1 and Ezh2, we verified the levels and spatial expression of En1 via qPCR and in situ hybridization at E14.5 in En1Cre/+; Ezh2 L/L embryos (Supplementary Figures S1A,B) and performed a second set of experiments on Wnt1Cre driven Ezh2 cKOs (Figure 6; Kratochwil et al., 2017). When examining the spatial expression of En1 in En1Cre/+; Ezh2 L/L embryos a similar pattern of expression as in En1Cre/+; Ezh2 +/+ animals is observed (Supplementary Figure S1A). However, the data also suggest that in the medial sections, En1 is ectopically expressed in the ventral R1 region of En1Cre driven Ezh2 cKOs (Supplementary Figure S1A, arrowhead), as was found for Nurr1, Lmx1a and Pitx3 as shown above. QPCR analysis indicated that the level of expression was not significantly changed between En1Cre/+; Ezh2 +/+ and En1Cre/+; Ezh2 L/L animals (Supplementary Figure S1B). Wnt1Cre/+; Ezh2 L/L embryos have been studied before and a previous study on the developing midbrain of these animals showed that Wnt1Cre driven deletion of Ezh2 leads to a loss of H3K27me3 in the Cre-recombinant area from E12.5 onward (Zemke et al., 2015). In addition, they showed full Wnt1Cre-mediated recombination in the caudal midbrain and R1 and while they used the Wnt1Cre; Ezh2 model to study early development of the dorsal midbrain, the presence of CRE-recombinase in the caudal midbrain and R1 makes it possible for us to also use this model to validate our initial findings observed in En1Cre driven Ezh2 cKOs independent of the dosage of En1 (Zemke et al., 2015). We first performed immunohistochemistry for TH at E14.5 (Figure 6A). Similar to the En1Cre/+; Ezh2 L/L phenotype, we observed a group of TH+ cells caudal to the midbrain (Figure 6A, arrowheads). In addition to Th expression, we also verified the presence of several other DA marks by means of in situ hybridization (Figures 6B,C). Next to TH protein, Th, Pitx3 and Cck were found to be expanded caudally in the medial sections of the Wnt1Cre; Ezh2 L/L compared to wildtypes (Figures 6B,C, arrowheads). However, in contrast to En1Cre; Ezh2 L/L animals almost no Ahd2 expression was detected in the Wnt1Cre; Ezh2 L/L embryos in comparison to the Wnt1Cre/+; Ezh2 +/+ embryos (Figure 6C, lower panel, arrows).
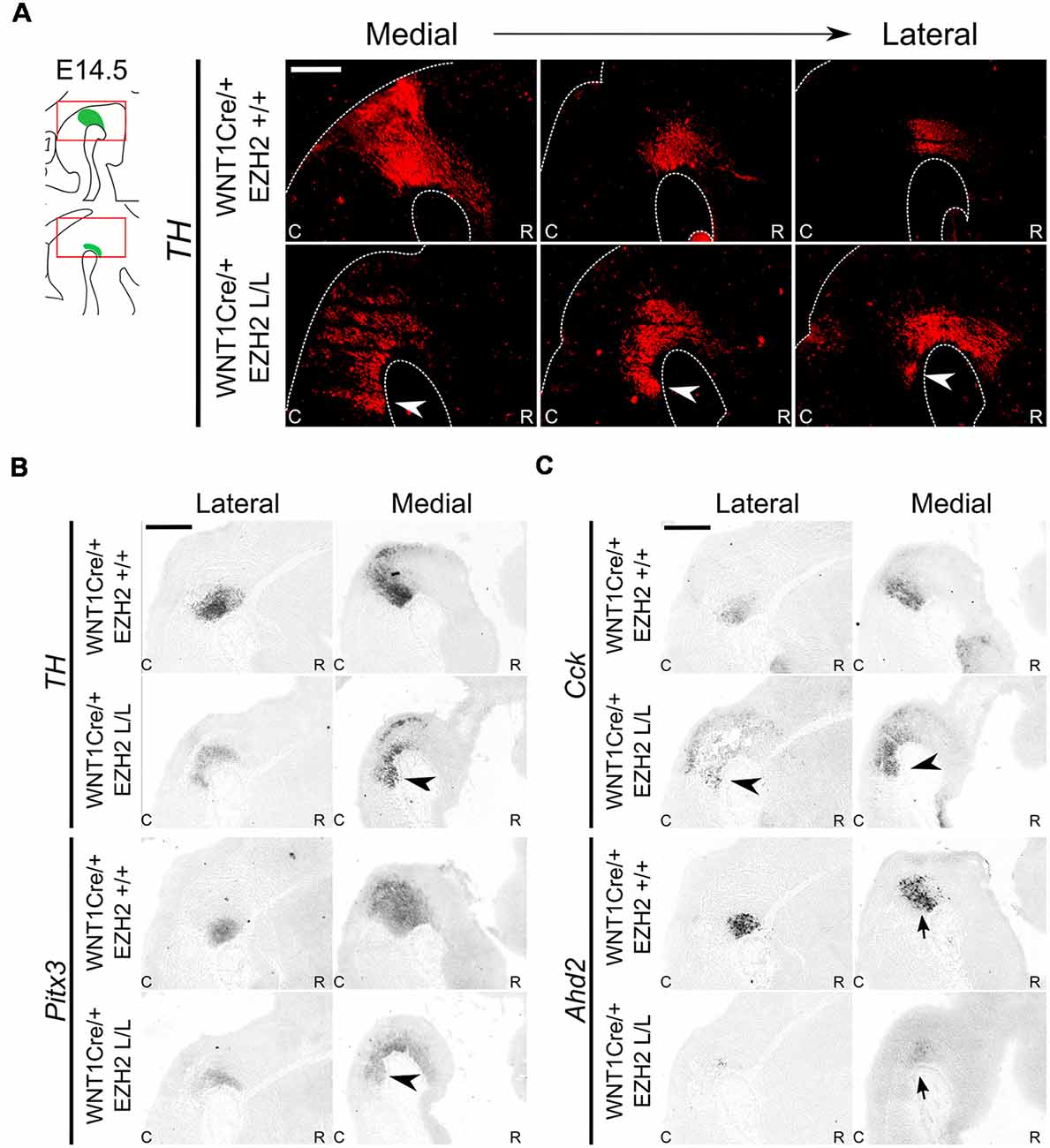
Figure 6. Wnt1Cre driven deletion of Ezh2 results in a comparable presence of ectopic DA cells as observed in the En1Cre/+; Ezh2 L/L. (A) Immunohistochemistry for TH (red) demonstrates that TH+ cells are present caudal to the midbrain in Wnt1Cre/+; Ezh2 L/L embryos at E14.5. The ectopic TH+ neurons can be observed in lateral and medial sections (arrowheads). Scale bar = 250 μM. (B,C) Analysis of spatial expression of Th, Pitx3, Cck and Ahd2 in adjacent E14.5 sections by means of in situ hybridization. Expression of Th, Pitx3 and Cck is found in a similar location caudal to the midbrain (arrowheads). Expression of Ahd2 is reduced in both the lateral and medial sections (arrows). Scale bar = 400 μM.
To further investigate a possible relationship between the phenotypes of En1 ablation (Kouwenhoven et al., 2016) and Ezh2 ablation, we investigated whether genetic removal of En1 influences the presence of H3K27me3 in the mid-hindbrain region. Therefore, an immunohistochemistry experiment was performed in En1 mutant animals (Figure 7A). H3K27me3 was found in the mid-hindbrain region of both the En1 mutants and wildtype controls. In addition, H3K27me3 was also detected in the ectopic DA cells in the En1 mutant [Figure 7A(*), arrowheads], suggesting that Ezh2 and general PRC2 activity is not affected in these cells. Next to analyzing the individual mutants, we also generated double mutants by crossing En1Cre/+; Ezh2 L/+ with En1Cre/+; Ezh2 L/+. Immunohistochemistry on TH was performed to analyze the DA population in the resulting mutants (Figure 7B). In En1CreCre; EZh2 L/L embryos the DA domain expanded even further into the hindbrain than in the En1Cre/+; Ezh2 L/L embryos (Figure 7B, arrowheads) and the overall cytoarchitecture is more disorganized in the double mutant when compared to both the Ezh2 cKO and the wildtype (Figure 7B, arrows), suggesting that En1 and Ezh2 do have a functional convergence although the molecular mechanism towards that convergence is not identical.
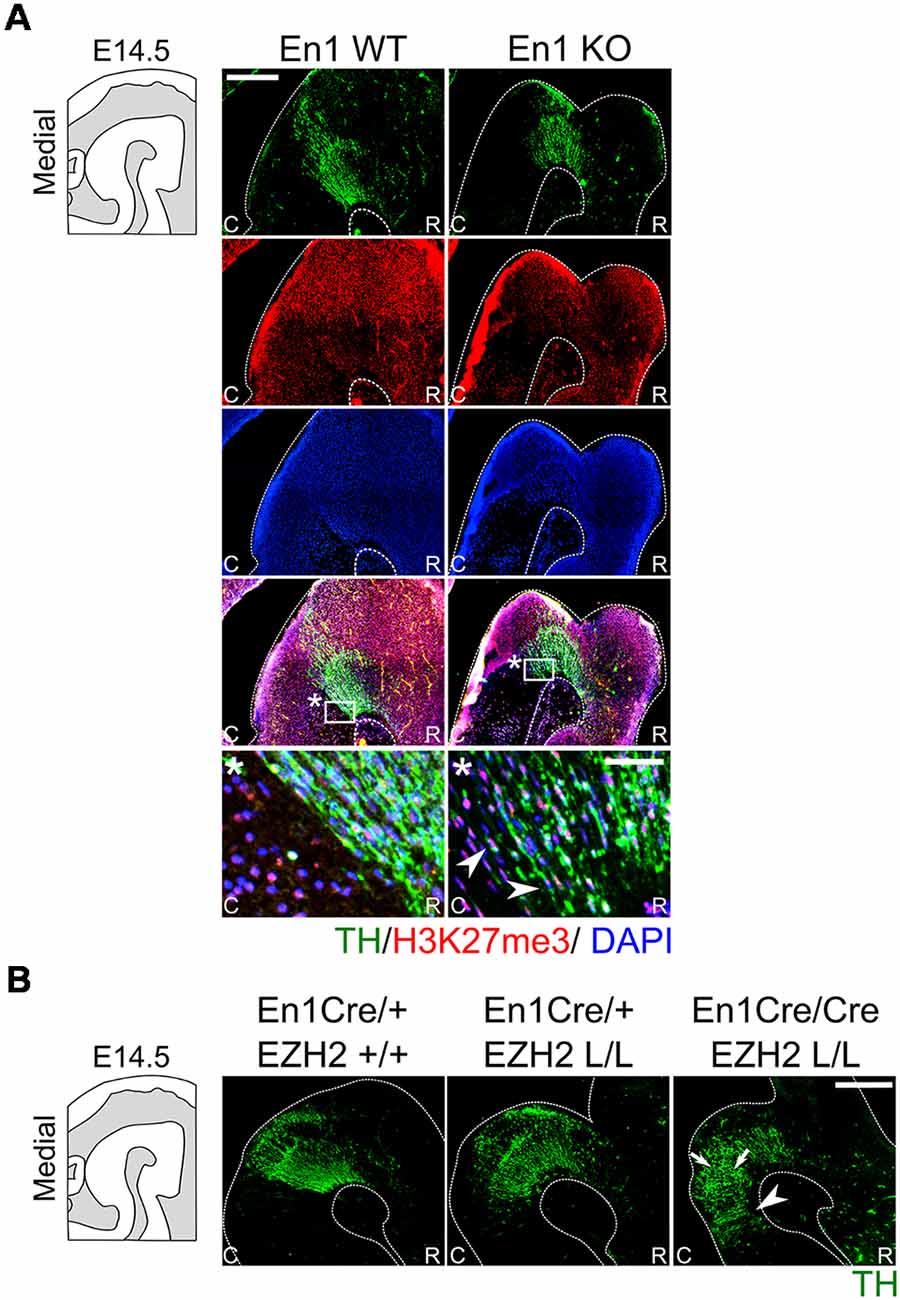
Figure 7. H3K27me3 is present in En1-deficient ectopic DA cells, whereas En1/ Ezh2 double mutants display an aggravated caudal expansion of the DA domain. (A) The presence of H3K27me3 in ectopic DA cells observed in En1 mutants was investigated by means of a double immunohistochemistry for TH (green) and H3K27me3 (red) in wildtypes and En1 −/− littermates. At E14.5 the TH+ cells found ectopically in the hindbrain of En1 mutant are also positive for H3K27me3 (arrowheads). Nuclear localization was confirmed by performing a DAPI staining (blue). Scale bars are 250 μM and 50 μM in the zoom. (B) Immunohistochemistry for TH (green) was performed on wildtypes, Ezh2 cKOs and En1CreCre; Ezh2 L/L double mutant animals. TH+ cells can be found further caudal into the hindbrain in double mutants compared to both the En1Cre/+; Ezh2 L/L and En1Cre/+ Ezh2 +/+ animals (arrowheads). In addition, the overall cytoarchitecture is disorganized in En1CreCre; Ezh2 L/L animals (arrows).
H3K27me3 is present in En1-deficient ectopic DA cells, whereas En1/ Ezh2 double mutants display an aggravated caudal expansion of the DA domain.
In summary, our data demonstrate that the loss of Ezh2 and H3K27me3 in the mid-hindbrain region lead to a caudal expansion independently of En1 expression. In addition, we show that although En1 mutant animals have a similar phenotype, global H3K27me3 is not affected in these mutants, suggesting that overall EZH2 functioning is not affected in En1 mutants in the isthmic area.
Ehz2 Influences IsO Determinants
The IsO is located at the border between the mid- and hindbrain and has been shown to be critical for the specification of both the midbrain as the anterior hindbrain (Crossley et al., 1996; Joyner et al., 2000; Brodski et al., 2003). The location of the IsO is initially determined by the mutual repression of two opposing factors, Otx2 and Gbx2 (Wurst and Bally-Cuif, 2001; Brodski et al., 2003). Loss- and gain-of-function showed that when the IsO is moved over the anterior-posterior axis the size of the two main neurotransmitter systems formed in this region, DA and serotonergic, is affected. When the IsO is moved to a more anterior position, the serotonergic system expands into the midbrain, while a caudal shift causes an expansion of the DA system into the hindbrain (Brodski et al., 2003; Kouwenhoven et al., 2016). A disorganization of the IsO might explain the caudal shift of the DA population observed in En1Cre/+; Ezh2 L/L embryos. We performed in situ hybridization of several genes associated with IsO functioning to determine whether the IsO was affected in Ezh2 cKOs (Figure 8). Fgf8 mediates the inducing capacity of the IsO (Crossley et al., 1996; Martinez et al., 1999; Joyner et al., 2000) and its expression can be observed as a transverse ring that encircles the neural tube in the vicinity of the mid-hindbrain border (Figure 8A, upper panel; Martinez et al., 1999). When comparing En1Cre/+; Ezh2 +/+ to En1Cre/+; Ezh2 L/L mid-hindbrain sections, we observed a partial loss of expression in medial sections of E12.5 En1Cre/+; Ezh2 L/L embryos (Figure 8A, upper panel, arrow), while lateral no clear changes in the expression pattern could be observed. The reduction of the levels of Fgf8 was verified using qPCR (n = 3, P < 0.05, one-tailed; Figure 8B). In addition to Fgf8, we also studied the expression pattern of Wnt1. Wnt1 is essential for the maintenance of several genes associated with IsO functioning, including Fgf8 and En1 (McMahon et al., 1992; Danielian and McMahon, 1996; Canning et al., 2007). Similar to Fgf8, reduced expression of Wnt1 was observed in medial sections of the Ezh2 cKO animals (Figure 8A, middle panel, arrow) and levels of Wnt1 were significantly lower in En1Cre/+; Ezh2 L/L animals compared to wildtypes (n = 3, P < 0.05, one-tailed; Figure 8B). Another important determinant for the IsO is Otx2. As described above, Otx2 plays a role in positioning the IsO, as the caudal limit of the Otx2 expression domain marks the location of the IsO (Broccoli et al., 1999; Wurst and Bally-Cuif, 2001). In contrast to Fgf8 and Wnt1, Otx2 expression was found to be expanded caudally into the hindbrain of Ezh2 cKO embryos (Figures 8A,C, lower panels, arrowheads). Normally Otx2 expression is not found caudal to Fgf8 expression, however in medial sections of En1Cre/+; Ezh2 L/L embryos, we could observe expression of Otx2 caudal to the Fgf8 expression domain (Figure 8C, arrowheads). Even though an expansion of the expression domain of Otx2 is visible, the overall levels of Otx2 were found to be reduced in the Ezh2 mutant (n = 3, P < 0.01, one-tailed; Figure 8B). To further substantiate the caudal Otx2 expression, we also performed in situ hybridization for Otx2 on E14.5 Wnt1Cre/+; Ezh2 L/L embryos (Supplementary Figure S2). Using the Th expression domain from Figure 6B as a reference, a patch of Otx2 expression could be observed caudal to the midbrain in medial sections of Wnt1Cre driven Ezh2 cKOs, which was not present in wildtype littermates (Supplementary Figure S2, arrowhead). Together these data indicate that Ezh2 plays a role in suppressing Otx2 expression in the R1 region, next to influencing expression levels of Fgf8 and Wnt1.
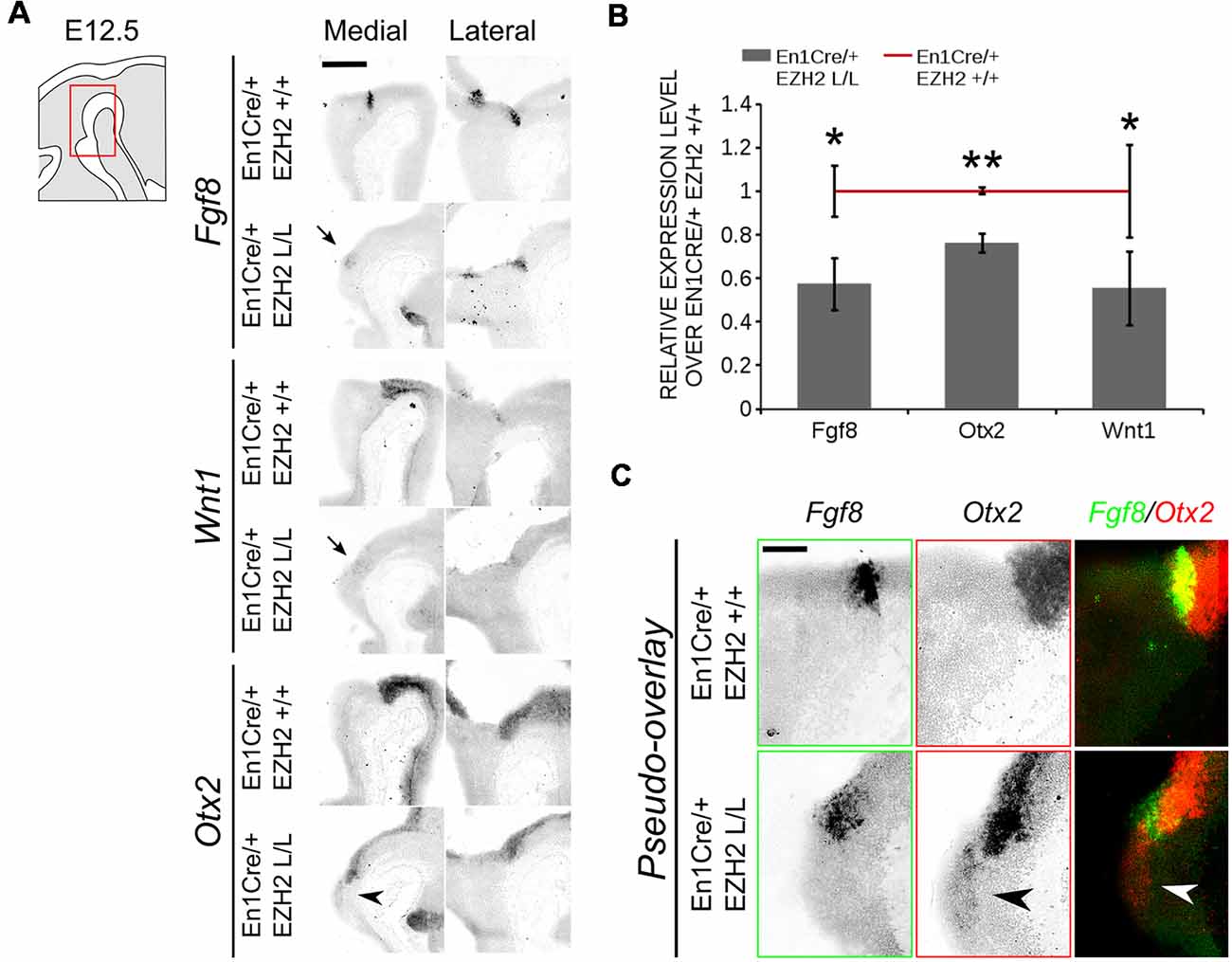
Figure 8. Otx2 expression can be found caudal to the IsO in the hindbrain of En1Cre/+; Ezh2 L/L E12.5 embryos. (A) The expression of Fgf8, Otx2 and Wnt1 was analyzed in E12.5 wildtypes and Ezh2 cKO embryos via in situ hybridization. Otx2 expression was found in the hindbrain of medial sections (arrowhead) while expression of both Fgf8 and Wnt1 was reduced in the medial sections of E12.5 En1Cre/+; Ezh2 L/L embryos (arrows). Scale bar = 300 μM. (B) Quantitative PCR (qPCR) for Fgf8, Otx2 and Wnt1 comparing the wildtype (n = 3, red line) expression levels to the En1Cre/; Ezh2 L/L (n = 4, gray bars) levels. All factors were significantly down-regulated in the Ezh2 cKO, Fgf8 is ~43% reduced (* P < 0.05, one-tailed), Otx2 levels were reduced to ~76% (** P < 0.01, one-tailed) and ~45% of the Wnt1 expression is lost (* P < 0.05, one-tailed). (C) Pseudo-overlay of adjacent slides for Fgf8 and Otx2 demonstrates that Otx2 is found caudal of Fgf8 expression in Ezh2 cKO animals but not in controls. Scale bar = 100 μM.
Discussion
A critical early event in the generation of different neuronal subtypes is the regionalization of the neural tube. This process in the mid-hindbrain region is dependent on the signaling factors produced by the IsO (Crossley et al., 1996; Wurst and Bally-Cuif, 2001). The IsO is formed at the juxtaposition of Otx2/Gbx2 expression around ~E7–8 (Broccoli et al., 1999; Joyner et al., 2000), and depends on several transcription factors for its proper functioning and maintenance (Adams et al., 2000; Li et al., 2002; Guo et al., 2006; Canning et al., 2007). In the current study, we show that next to transcription factors, transcriptional inhibition via PRC2 and H3K27me3 also play a role in maintaining the IsO and determining the fate of cells in the mid-hindbrain region. The loss of Ezh2 and consequently H3K27me3 affected the molecular coding of the anterior R1. DA-related genes were found ectopically in the ventral anterior hindbrain and a caudal expansion of the TH positive population was observed. The emergence of DA neurons in the Isthmic area was at the expanse of 5-HT neurons and Nkx6.1 expression in the trochlear motor neurons located directly caudal to the IsO.
The deletion of Ezh2 mimics the phenotype of the En1 mutant, in which a disorganized IsO causes a posterior expansion of the midbrain a the expanse of the anterior ventral R1 region (Kouwenhoven et al., 2016). Even though both proteins might mechanistically function via the regulation of Otx2, no direct relationship between the two factors was found. The expression of En1 is not affected in En1Cre/+; Ehz2 L/L embryos and the results were verified in a second independent model, Wnt1Cre/Ezh2. In addition, no global changes were observed in H3K27me3 levels in En1 mutants compared to wildtypes, suggesting that general PRC2 functioning is not impaired. However, the targeting of the PRC2 complex might be altered in En1 mutants, leading to local changes in H3K27me3 levels, which are not visualized by overall immunohistochemistry for H3K27me3. Furthermore, analysis of the TH+ domain in En1/ Ezh2 double mutants does imply that EZH2 and EN1 have a phenotypic relationship as the expansion of the TH+ region is aggravated in animals depleted of both En1 and Ezh2.
The induction of DA neurons in R1 of Ezh2 cKOs can be explained by the presence of Otx2 in the hindbrain (Brodski et al., 2003; Sherf et al., 2015). At E12.5 we observed Otx2 expression caudal to the midbrain in En1Cre/+; Ezh2 L/L embryos and Wnt1Cre driven Ezh2 cKOs also displayed a caudal expansion of the Otx2 domain. It has been shown that repression of Otx2 in the hindbrain initially depends on GBX2, however, after E8.5 a GBX2-independent mechanism can repress Otx2 in R1 (Li et al., 2002). At E12.5 the Fgf8 expression domain marks the caudal limit of the Otx2 expression domain and a study in which a FGF8-bead was implanted in p2, showed a regional repression of Otx2, suggesting that FGF8 can compensate for the loss of GBX2 in repressing Otx2 after E8.5 (Li et al., 2002). However, the introduction of FGF8 into this region also led to the induction of several other IsO related genes, including En1, which might also contribute to the repression of Otx2 (Shamim et al., 1999). Although the expansion of Otx2 into the hindbrain might additionally be a consequence of alterations in IsO related genes, we argue that the direct loss of H3K27me3 on the promoter of Otx2 may cause the observed reactivation of the Otx2 gene in our model. Otx2 has already been identified as a direct target of PRC2 in embryonic stem cells (Boyer et al., 2006; Lee et al., 2006; Pasini et al., 2007). Loss of a functional PRC2 complex led to the loss of H3K27me3 on the promoter of Otx2 and an increase in its expression level (Pasini et al., 2007, Supplementary Figure S1). In addition, H3K27me3 is found on the promoter of Otx2 in cell types that do not express Otx2 (Supplementary Figure S3), suggesting that the silencing of Otx2 might be H3K27me3-mediated (Mikkelsen et al., 2007; Mohn et al., 20081; mouse NCBI37/mm9). Interestingly, in contrast to previous studies the altered expression of Otx2 was not mimicked by Wnt1 in En1Cre/+; Ezh2 L/L animals (Broccoli et al., 1999; Kouwenhoven et al., 2016; Broccoli et al., 1999; Kouwenhoven et al., 2016). The levels of Wnt1 were down-regulated in En1Cre driven Ezh2 mutants and reduced spatial expression was observed in medial sections of E12.5 En1Cre/+; Ezh2 L/L embryos. In addition, reduced canonical Wnt signaling was found in Wnt1Cre/+; Ezh2 L/L embryos (Zemke et al., 2015), suggesting that the sole presence of Otx2 in the hindbrain of Ezh2 cKOs is enough to fate switch the hindbrain R1 region towards a midbrain phenotype.
Taken together, our data show that next to transcription factors, transcriptional regulation via epigenetics also plays an important part in the development of the Isthmic area. Deletion of Ezh2 leads to Otx2 expression in the hindbrain and the induction of DA neurons at the expense of 5-HT positive and Nkx6.1 positive neurons. The observed phenotype mimics that of En1 mutant animals and even though Ezh2 and En1 do not appear to have a direct regulatory relationship, we theorize that EN1 might be required for the targeting of EZH2 and PRC2 to the Otx2 locus.
Author Contributions
IW performed the experiments, analyzed the data and wrote the manuscript. CW performed the experiments. MS wrote the manuscript, initiated the study, analyzed data and funded the study.
Funding
This work was sponsored by the NWO-ALW (Nederlandse Organisatie voor Wetenschappelijk Onderzoek-Aard en Levenswetenschappen) VICI grant (865.09.002) awarded to MS The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr Frederic Zilbermann of the Friedrich Miescher Institute for his generous gift of the Ezh2-floxed mouse line and Dr Sandra Blaess for kindly providing us with the En1Cre; R26RYFP mouse strain. In addition, we would like to thank Prof. Dr. Filippo Rijli for sending us E14.5 frozen and PFA/Sucrose Wnt1Cre; Ezh2 embryos. This work is present as preprint at: bioRxiv 442111; doi: 10.1101/442111.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2019.00076/full#supplementary-material
FIGURE S1 | The loss of Ezh2 does not influence En1 expression. The expression of En1 was analyzed with in situ hybridization and qPCR at E14.5 (A, B). (A) En1 shows a similar expression pattern between the En1Cre/+; Ezh2 +/+ and the En1Cre/+; Ezh2 L/L animals. However ectopic expression of En1 might be present in the ventral region of R1 (arrowhead). (B) The levels of En1 are not significantly changed between wildtypes and Ezh2 cKO animals at E14.5. Scale bar = 300 μM.
FIGURE S2 | Otx2 is found caudal to the midbrain in Wnt1Cre/+; Ezh2 L/L embryos. Otx2 expression was assessed by means of in situ hybridization using Th expression as a reference for the location. A pseudo-overlay of adjacent slides demonstrates the presence of Otx2 expression dorsal of the ectopic Th expression in medial sections of Wnt1Cre driven Ezh2 cKOs (arrowhead). Scale bar = 300 μM.
FIGURE S3 | H3K27me3 is present on the promoter of Otx2 in tissue in which Otx2 is not expressed. (A) Visualization of ChIP-sequencing peaks of H3K27me3 and RNA-polymerase II (POLII) at the Otx2 promoter obtained from https://genome.ucsc.edu/index.html using the mouse NCBI37/mm9 assembly. In tissue where almost no RNA-sequencing reads are detected for Otx2, a high peak for H3K27me3 is observed, while the peak for POLII is low. (B) Analysis of ChIP-sequencing data from Mohn et al. (2008). The Otx2 promoter shows a high enrichment for H3K27me3 in Embryonic stem cells (ES), Neuronal progenitors (NP) and Terminal pyramidal glutamatergic neurons (TN), while enrichment for POLII is low, indicating that Otx2 is not activated in these cells.
References
Adams, K. A., Maida, J. M., Golden, J. A., and Riddle, R. D. (2000). The transcription factor Lmx1b maintains Wnt1 expression within the isthmic organizer. Development 127, 1857–1867.
Alonso, A., Merchán, P., Sandoval, J. E., Sánchez-Arrones, L., Garcia-Cazorla, A., Artuch, R., et al. (2013). Development of the serotonergic cells in murine raphe nuclei and their relations with rhombomeric domains. Brain Struct. Funct. 218, 1229–1277. doi: 10.1007/s00429-012-0456-8
Andersson, E., Tryggvason, U., Deng, Q., Friling, S., Alekseenko, Z., Robert, B., et al. (2006). Identification of intrinsic determinants of midbrain dopamine neurons. Cell 124, 393–405. doi: 10.1016/j.cell.2005.10.037
Arenas, E., Denham, M., and Villaescusa, J. C. (2015). How to make a midbrain dopaminergic neuron. Development 142, 1918–1936. doi: 10.1242/dev.097394
Bayer, S. A., Wills, K. V., Triarhou, L. C., and Ghetti, B. (1995). Time of neuron origin and gradients of neurogenesis in midbrain dopaminergic neurons in the mouse. Exp. Brain Res. 105, 191–199. doi: 10.1007/bf00240955
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. doi: 10.1016/j.cell.2006.02.041
Boyer, L. A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L. A., Lee, T. I., et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. doi: 10.1038/nature04733
Broccoli, V., Boncinelli, E., and Wurst, W. (1999). The caudal limit of Otx2 expression positions the isthmic organizer. Nature 401, 164–168. doi: 10.1038/43670
Brodski, C., Weisenhorn, D. M. V., Signore, M., Sillaber, I., Oesterheld, M., Broccoli, V., et al. (2003). Location and size of dopaminergic and serotonergic cell populations are controlled by the position of the midbrain-hindbrain organizer. J. Neurosci. 23, 4199–4207. doi: 10.1523/JNEUROSCI.23-10-04199.2003
Canning, C. A., Lee, L., Irving, C., Mason, I., and Jones, C. M. (2007). Sustained interactive Wnt and FGF signaling is required to maintain isthmic identity. Dev. Biol. 305, 276–286. doi: 10.1016/j.ydbio.2007.02.009
Cao, R., and Zhang, Y. (2004a). SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15, 57–67. doi: 10.1016/j.molcel.2004.06.020
Cao, R., and Zhang, Y. (2004b). The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14, 155–164. doi: 10.1016/j.gde.2004.02.001
Cedar, H., and Bergman, Y. (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304. doi: 10.1038/nrg2540
Chandrasekhar, A. (2004). Turning heads: development of vertebrate branchiomotor neurons. Dev. Dyn. 229, 143–161. doi: 10.1002/dvdy.10444
Corley, M., and Kroll, K. L. (2015). The roles and regulation of polycomb complexes in neural development. Cell Tissue Res. 359, 65–85. doi: 10.1007/s00441-014-2011-9
Crossley, P. H., Martinez, S., and Martin, G. R. (1996). Midbrain development induced by FGF8 in the chick embryo. Nature 380, 66–68. doi: 10.1038/380066a0
Danielian, P. S., and McMahon, A. P. (1996). Engrailed-1 as a target of the Wnt-1 signalling pathway in vertebrate midbrain development. Nature 383, 332–334. doi: 10.1038/383332a0
Deng, Q., Andersson, E., Hedlund, E., Alekseenko, Z., Coppola, E., Panman, L., et al. (2011). Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in ventral midbrain development. Development 138, 3399–3408. doi: 10.1242/dev.065482
Faust, C., Lawson, K. A., Schork, N. J., Thiel, B., and Magnuson, T. (1998). The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development 125, 4495–4506.
Ferrari, K. J., Scelfo, A., Jammula, S., Cuomo, A., Barozzi, I., Stützer, A., et al. (2014). Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 53, 49–62. doi: 10.1016/j.molcel.2013.10.030
Fox, S. R., and Deneris, E. S. (2012). Engrailed is required in maturing serotonin neurons to regulate the cytoarchitecture and survival of the dorsal raphe nucleus. J. Neurosci. 32, 7832–7842. doi: 10.1523/JNEUROSCI.5829-11.2012
Grima, B., Lamouroux, A., Blanot, F., Biguet, N. F., and Mallet, J. (1985). Complete coding sequence of rat tyrosine hydroxylase mRNA. Proc. Natl. Acad. Sci. U S A 82, 617–621. doi: 10.1073/pnas.82.2.617
Guo, C., Qiu, H.-Y., Huang, Y., Chen, H., Yang, R.-Q., Chen, S.-D., et al. (2006). Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development 134, 317–325. doi: 10.1242/dev.02745
Hirabayashi, Y., Suzki, N., Tsuboi, M., Endo, T. A., Toyoda, T., Shinga, J., et al. (2009). Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600–613. doi: 10.1016/j.neuron.2009.08.021
Hoekstra, E. J., von Oerthel, L., van der Heide, L. P., Kouwenhoven, W. M., Veenvliet, J. V., Wever, I., et al. (2013). Lmx1a encodes a rostral set of mesodiencephalic dopaminergic neurons marked by the Wnt/B-catenin signaling activator R-spondin 2. PLoS One 8:e74049. doi: 10.1371/journal.pone.0074049
Hwang, D.-Y., Ardayfio, P., Kang, U. J., Semina, E. V., and Kim, K.-S. (2003). Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Mol. Brain Res. 114, 123–131. doi: 10.1016/s0169-328x(03)00162-1
Jacobs, F. M. J., Smits, S. M., Noorlander, C. W., von Oerthel, L., van der Linden, A. J. A., Burbach, J. P. H., et al. (2007). Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development 134, 2673–2684. doi: 10.1242/dev.02865
Jacobs, F. M. J., Veenvliet, J. V., Almirza, W. H., Hoekstra, E. J., von Oerthel, L., van der Linden, A. J. A., et al. (2011). Retinoic acid-dependent and -independent gene-regulatory pathways of Pitx3 in meso-diencephalic dopaminergic neurons. Development 138, 5213–5222. doi: 10.1242/dev.071704
Jankovic, J., Chen, S., and Le, W. D. (2005). The role of Nurr1 in the development of dopaminergic neurons and Parkinson’s disease. Prog. Neurobiol. 77, 128–138. doi: 10.1016/j.pneurobio.2005.09.001
Joyner, A. L., Liu, A., and Millet, S. (2000). Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr. Opin. Cell Biol. 12, 736–741. doi: 10.1016/s0955-0674(00)00161-7
Kawano, H., Ohyama, K., Kawamura, K., and Nagatsu, I. (1995). Migration of dopaminergic neurons in the embryonic mesencephalon of mice. Dev. Brain Res. 86, 101–113. doi: 10.1016/0165-3806(95)00018-9
Kimmel, R. A., Turnbull, D. H., Blanquet, V., Wurst, W., Loomis, C. A., and Joyner, A. L. (2000). Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 14, 1377–1389. doi: 10.1101/gad.14.11.1377
Kouwenhoven, W. M., Veenvliet, J. V., van Hooft, J. A., van der Heide, L. P., and Smidt, M. P. (2016). Engrailed 1 shapes the dopaminergic and serotonergic landscape through proper isthmic organizer maintenance and function. Biol. Open 5, 279–288. doi: 10.1242/bio.015032
Kratochwil, C. F., Maheshwari, U., and Rijli, F. M. (2017). The long journey of pontine nuclei neurons: from rhombic lip to cortico-ponto-cerebellar circuitry. Front. Neural Circuits 11:33. doi: 10.3389/fncir.2017.00033
Lee, T. I., Jenner, R. G., Boyer, L. A., Guenther, M. G., Levine, S. S., Kumar, R. M., et al. (2006). Control of developmental regulators by polycomb in human embryonic stem cells. Cell 125, 301–313. doi: 10.1016/j.cell.2006.02.043
Li, J. Y. H., Lao, Z., and Joyner, A. L. (2002). Changing requirements for Gbx2 in development of the cerebellum and maintenance of the mid/hindbrain organizer. Neuron 36, 31–43. doi: 10.1016/s0896-6273(02)00935-2
Margueron, R., Li, G., Sarma, K., Blais, A., Zavadil, J., Woodcock, C. L., et al. (2008). Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518. doi: 10.1016/j.molcel.2008.11.004
Martinez, S., Crossley, P. H., Cobos, I., Rubenstein, J. L., and Martin, G. R. (1999). FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development 126, 1189–1200.
Maxwell, S. L., Ho, H.-Y., Kuehner, E., Zhao, S., and Li, M. (2005). Pitx3 regulates tyrosine hydroxylase expression in the substantia nigra and identifies a subgroup of mesencephalic dopaminergic progenitor neurons during mouse development. Dev. Biol. 282, 467–479. doi: 10.1016/j.ydbio.2005.03.028
McMahon, A. P., Joyner, A. L., Bradley, A., and McMahon, J. A. (1992). The midbrain-hindbrain phenotype of Wnt-1− Wnt-1− mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69, 581–595. doi: 10.1016/0092-8674(92)90222-x
Mesman, S., von Oerthel, L., and Smidt, M. P. (2014). Mesodiencephalic dopaminergic neuronal differentiation does not involve GLI2A-mediated SHH-signaling and is under the direct influence of canonical WNT signaling. PLoS One 9:e97926. doi: 10.1371/journal.pone.0097926
Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560. doi: 10.1038/nature06008
Mohn, F., Weber, M., Rebhan, M., Roloff, T. C., Richter, J., Stadler, M. B., et al. (2008). Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755–766. doi: 10.1016/j.molcel.2008.05.007
O’Carroll, D., Erhardt, S., Pagani, M., Barton, S. C., Surani, M. A., and Jenuwein, T. (2001). The polycomb-group GeneEzh2 is required for early mouse development. Mol. Cell. Biol. 21, 4330–4336. doi: 10.1128/mcb.21.13.4330-4336.2001
Omodei, D., Acampora, D., Mancuso, P., Prakash, N., Di Giovannantonio, L. G., Wurst, W., et al. (2008). Anterior-posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development 135, 3459–3470. doi: 10.1242/dev.027003
Ono, Y., Nakatani, T., Sakamoto, Y., Mizuhara, E., Minaki, Y., Kumai, M., et al. (2007). Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development 134, 3213–3225. doi: 10.1242/dev.02879
Pasini, D., Bracken, A. P., Hansen, J. B., Capillo, M., and Helin, K. (2007). The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27, 3769–3779. doi: 10.1128/mcb.01432-06
Pereira, J. D., Sansom, S. N., Smith, J., Dobenecker, M.-W., Tarakhovsky, A., and Livesey, F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U S A 107, 15957–15962. doi: 10.1073/pnas.1002530107
Placzek, M., and Briscoe, J. (2005). The floor plate: multiple cells, multiple signals. Nat. Rev. Neurosci. 6, 230–240. doi: 10.1038/nrn1628
Podobinska, M., Szablowska-Gadomska, I., Augustyniak, J., Sandvig, I., Sandvig, A., and Buzanska, L. (2017). Epigenetic modulation of stem cells in neurodevelopment: the role of methylation and acetylation. Front. Cell. Neurosci. 11:23. doi: 10.3389/fncel.2017.00023
Prakash, N., Puelles, E., Freude, K., Trümbach, D., Omodei, D., Di Salvio, M., et al. (2009). Nkx6–1 controls the identity and fate of red nucleus and oculomotor neurons in the mouse midbrain. Development 136, 2545–2555. doi: 10.1242/dev.031781
Saucedo-Cardenas, O., Quintana-Hau, J. D., Le, W.-D., Smidt, M. P., Cox, J. J., Mayo, F. D., et al. (1998). Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc. Natl. Acad. Sci. U S A 95, 4013–4018. doi: 10.1073/pnas.95.7.4013
Shamim, H., Mahmood, R., Logan, C., Doherty, P., Lumsden, A., and Mason, I. (1999). Sequential roles for Fgf4, En1 and Fgf8 in specification and regionalisation of the midbrain. Development 126, 945–959.
Shen, X., Liu, Y., Hsu, Y.-J., Fujiwara, Y., Kim, J., Mao, X., et al. (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502. doi: 10.1016/j.molcel.2008.10.016
Sherf, O., Nashelsky Zolotov, L., Liser, K., Tilleman, H., Jovanovic, V. M., Zega, K., et al. (2015). Otx2 requires Lmx1b to control the development of mesodiencephalic dopaminergic neurons. PLoS One 10:e0139697. doi: 10.1371/journal.pone.0139697
Shults, C. W., Hashimoto, R., Brady, R. M., and Gage, F. H. (1990). Dopaminergic cells align along radial glia in the developing mesencephalon of the rat. Neuroscience 38, 427–436. doi: 10.1016/0306-4522(90)90039-7
Smidt, M. P., Asbreuk, C. H. J., Cox, J. J., Chen, H., Johnson, R. L., and Burbach, J. P. H. (2000). A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat. Neurosci. 3, 337–341. doi: 10.1038/73902
Smidt, M. P., Smits, S. M., Bouwmeester, H., Hamers, F. P. T., van der Linden, A. J. A., Hellemons, A. J., et al. (2004). Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development 131, 1145–1155. doi: 10.1242/dev.01022
Smidt, M. P., van Schaick, H. S. A., Lanctôt, C., Tremblay, J. J., Cox, J. J., van der Kleij, A. A. M., et al. (1997). A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc. Natl. Acad. Sci. U S A 94, 13305–13310. doi: 10.1073/pnas.94.24.13305
Smits, S. M., Ponnio, T., Conneely, O. M., Burbach, J. P. H., and Smidt, M. P. (2003). Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur. J. Neurosci. 18, 1731–1738. doi: 10.1046/j.1460-9568.2003.02885.x
Srinivas, S., Watanabe, T., Lin, C. S., William, C. M., Tanabe, Y., Jessell, T. M., et al. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4. doi: 10.1186/1471-213X-1-4
Sunmonu, N. A., Chen, L., and Li, J. Y. H. (2009). Misexpression of Gbx2 throughout the mesencephalon by a conditional gain-of-function transgene leads to deletion of the midbrain and cerebellum in mice. Genesis 47, 667–673. doi: 10.1002/dvg.20546
van Heesbeen, H. J., Mesman, S., Veenvliet, J. V., and Smidt, M. P. (2013). Epigenetic mechanisms in the development and maintenance of dopaminergic neurons. Development 140, 1159–1169. doi: 10.1242/dev.089359
Veenvliet, J. V., Dos Santos, M. T., Kouwenhoven, W. M., von Oerthel, L., Lim, J. L., van der Linden, A. J. A., et al. (2013). Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development 140, 4116–4116. doi: 10.1242/dev.094565
Wurst, W., and Bally-Cuif, L. (2001). Neural plate patterning: upstream and downstream of the isthmic organizer. Nat. Rev. Neurosci. 2, 99–108. doi: 10.1038/35053516
Yan, C. H., Levesque, M., Claxton, S., Johnson, R. L., and Ang, S.-L. (2011). Lmx1a and Lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J. Neurosci. 31, 12413–12425. doi: 10.1523/JNEUROSCI.1077-11.2011
Keywords: Ezh2, mid-hindbrain border, dopamine, seretonin, epigenetics
Citation: Wever I, Wagemans CMRJ and Smidt MP (2019) EZH2 Is Essential for Fate Determination in the Mammalian Isthmic Area. Front. Mol. Neurosci. 12:76. doi: 10.3389/fnmol.2019.00076
Received: 04 December 2018; Accepted: 11 March 2019;
Published: 09 April 2019.
Edited by:
Ildikó Rácz, Universitätsklinikum Bonn, GermanyReviewed by:
Anna La Torre, University of California, Davis, United StatesSungkun Chun, Chonbuk National University, South Korea
Copyright © 2019 Wever, Wagemans and Smidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marten P. Smidt, bS5wLnNtaWR0QHV2YS5ubA==
 Iris Wever
Iris Wever Cindy M. R. J. Wagemans
Cindy M. R. J. Wagemans Marten P. Smidt
Marten P. Smidt