- 1Institute of Human Genetics, University Medical Center Mainz, Mainz, Germany
- 2Focus Program of Translational Neurosciences, University Medical Center Mainz, Mainz, Germany
- 3First Department of Internal Medicine, University Medical Center Mainz, Mainz, Germany
- 4German Resilience Centre, University Medical Center Mainz, Mainz, Germany
The RNA-binding protein RBFOX1 is an important regulator of neuron development and neuronal excitability. Rbfox1 is a dosage-sensitive gene and in both mice and humans, decreased expression of Rbfox1 has been linked to neurodevelopmental disorders. Alternative promoters drive expression of Rbfox1 transcript isoforms that encode an identical protein. The tissue- and developmental stage-specific expression of these isoforms, as well as the underlying regulatory mechanisms, are, however, unclear. Here, we set out to capture all of the Rbfox1 transcript isoforms and identify transcriptional mechanisms that regulate brain-specific Rbfox1 expression. Isoform sequencing identified multiple alternative Rbfox1 transcript variants in the mouse cerebral cortex, including transcripts with novel first exons, alternatively spliced exons and 3′-truncations. Quantitative RT-PCR determined the expression of the alternative first exons in the developing cerebral cortex and different subregions of the juvenile brain. Alternative first exons were found to be highly stage- and subregion specific in their expression patterns suggesting that they fulfill specific functions during cortex development and in different brain regions. Using reporter assays we found that the promoter regions of the two first exons E1B and E1C/E1C.1 contain several functional E-boxes. Together, we provide an extensive picture of Rbfox1 isoform expression. We further identified important regulatory mechanisms that drive neuron-specific Rbfox1 expression. Thus, our study forms the basis for further research into the mechanisms that ensure physiological Rbfox1 expression in the brain. It also helps to understand why, in patients with neurodevelopmental disorders deletion of individual RBFOX1 transcript isoforms could affect brain function.
Introduction
The RNA Binding Fox-1 Homolog 1 (Rbfox1) gene is one of the longest genes in the genome with its remarkable size being caused by an extended 5′-noncoding region containing several alternative 5′UTR exons, transcription of which is controlled by alternative promoter usage (Kuroyanagi, 2009; Damianov and Black, 2010; Conboy, 2017). An important yet unanswered question is why so many alternative Rbfox1 transcripts exist that encode an identical protein. These transcripts could theoretically encode RBFOX1 proteins associated with specific functions. Alternatively, tissue-specific transcription factors might bind the alternative Rbfox1 promoters and fail-safe Rbfox1 expression levels in a tissue-specific manner.
RBFOX1 is a member of the RBFOX family of RNA binding proteins that also includes RBFOX2 and RBFOX3. Rbfox1 expression is restricted to neurons, heart, and muscle (Shibata et al., 2000; Jin et al., 2003; Underwood et al., 2005). In the cell, RBFOX1 shuttles between the nucleus and cytoplasm, an event which is regulated by alternative splicing of the exon A53. Splicing of exon A53 into the Rbfox1 mRNA causes cytoplasmic localization of RBFOX1 by introducing a frame-shift and an alternative C-terminus lacking the nuclear localization signal (NLS; Underwood et al., 2005; Lee et al., 2009). In the nucleus, RBFOX proteins regulate alternative pre-mRNA splicing by binding the consensus sequence (U)GCAUG in introns flanking alternative exons. Cytoplasmic RBFOX1 influences gene expression, by binding the 3′-untranslated regions (3′UTRs) of its target genes (Dredge and Jensen, 2011; Lee et al., 2016; Rajman et al., 2017).
RBFOX1 is linked to the etiology of neurodevelopmental disorders. Thus, studies in knockout mice have shown that in the brain, RBFOX1 controls splicing and expression of transcripts involved in neuronal transmission and excitability (Gehman et al., 2011; Vuong et al., 2018; Wamsley et al., 2018). Subsequent studies identified multiple additional RBFOX1 splicing targets in differentiating human fetal neural progenitor cells and in mouse brain (Fogel et al., 2012). These targets included many genes related to autism spectrum disorders (ASD). For example, the vSNARE protein VAMP1 was identified as a major RBFOX1 target in inhibitory neurons of the hippocampus (Vuong et al., 2018). Re-expression of Vamp1 rescued the electrophysiological abnormalities seen in the Rbfox1 knockout mice. Furthermore, RBFOX1 was identified as a hub gene in ASD transcriptional networks. The downregulation of RBFOX1 observed in autistic brains was reflected by RBFOX1-dependent splicing changes (Parikshak et al., 2013).
Copy number variations (CNVs) in the RBFOX1 gene are associated with various neurodevelopmental disorders such as epilepsy, intellectual disability (ID) and ASD (Bhalla et al., 2004; Martin et al., 2007; Lal et al., 2013; Zhao, 2013). A characteristic feature of these CNVs is their location in the RBFOX1 gene, which is typically restricted to the long 5′-noncoding part of the gene. Each of the CNVs described so far in patients with neurodevelopmental disorders affect some, but not all, of the alternative RBFOX1 transcripts. Since it is unknown at which developmental stages and in which tissues every single alternative transcript is expressed and how each transcript contributes to the overall RBFOX1 expression levels it remains unclear to what extent these CNVs change overall RBFOX1 expression levels in the brain.
Deeper insight into the expression patterns of the alternative Rbfox1 transcripts and their transcriptional regulation through alternative promoter usage is key for the understanding of how the RBFOX1 function is controlled in the healthy and diseased brain. In this study, by performing Isoform sequencing (Iso-Seq) and RT-qPCR we have characterized in detail which alternative Rbfox1 transcripts are expressed in the embryonic and postnatal cerebral cortex and the juvenile mouse brain. Besides Rbfox1 transcript variants already known before, we have identified several novel transcripts some of which contain novel coding exons. These exons are located even further upstream of the known Rbfox1 5′ exons and encode a unique N terminus with a predicted classic NLS. We could confirm the protein expression of these isoforms and found that they localize to the cell nucleus even if they lacked the C-terminal NLS. To investigate the transcriptional regulation of Rbfox1 we have searched for transcription factor binding sites in the alternative Rbfox1 promoters. We have found that two of the alternative Rbfox1 promoters contain functional E-boxes that promote Rbfox1 expression in primary neurons.
Materials and Methods
Ethics Approval
Sacrificing mice to obtain brain tissue was performed according to European (EU directive 86/609/EEC), national (TierSchG), and institutional guidelines.
Nomenclature of Gene and Protein Names
Throughout the manuscript, we followed the rules of the Human Genome Organization Nomenclature Committee1 and the Mouse Genome Informatics Nomenclature Committee2.
Mice
NMRI (12 weeks old for neuron culture, 6 weeks old for isolation of brain subregions) mice from Janvier labs (Saint Berthevin, France) were sacrificed by cervical dislocation upon arrival. For the collection of brain subregions from juvenile mice, brains were removed and incubated in RNAlater® (Sigma, Munich, Germany) for 2 days at 4°C. After dehydration, brain regions were dissected and further processed. Three male mice were used for expression analyses. For neuron culture, cortical neurons were isolated from E14.5 embryos from pregnant female NMRI mice.
Plasmids, in vitro Mutagenesis
Rbfox1 1B (positions −2,147 to +1,039), 1C (positions −989 to +470) and 1D (positions −2,302 to +56) promoter fragments were PCR amplified from mouse DNA and cloned into the multiple cloning site of pGL4.10 firefly luciferase reporter vector using KpnI and HindIII (Rbfox1 1B) or XhoI and BglII (Rbfox1 1C and 1D). Site-directed mutagenesis was performed using the QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies, Waldbronn, Germany) and following the manufacturer’s recommendations. Primer sequences are given in Supplementary Table S1.
Rbfox1 isoforms starting from E1A with exon A53 skipped or included were PCR amplified from total RNA of P0 mouse cerebral cortex with primers containing XmaI and EcoRI overhangs and cloned into the multiple cloning sites of the pCAGGs-IRES-GFP vector. The accuracy of cloned constructs was verified by Sanger sequencing. Primer sequences are given in Supplementary Table S1.
Cell Culture, Reporter Assays and Immunofluorescence Experiments
P19 cells were cultured in DMEM high glucose supplemented with 10% FBS, 1% NEAA and 100 μg/ml Penicillin/Streptomycin. N2A cells were cultured in DMEM high glucose supplemented with 10% FBS and 100 μg/ml Penicillin/Streptomycin. Primary cortical neurons, prepared from E14.5 mouse embryos, were cultured in neurobasal medium containing 2% B27 supplement (Gibco Life Technologies, Carlsbad, CA, USA) and 500 μM Glutamax (Gibco Life Technologies, Carlsbad, CA, USA). See Supplementary Methods for a detailed description of luciferase assays and immunofluorescence experiments.
RT-qPCR
Total RNA was extracted from mouse brain tissue using TRIzol (Thermo Fisher Scientific, Rockford, IL, USA) and from primary neurons and P19 cells using the High Pure RNA Isolation kit (Roche, Basel, Switzerland). Reverse transcription was performed with the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Rockford, IL, USA). Quantitative PCR was performed on an ABI StepOnePlusTM Real-Time PCR System using Sybr Green or Rox Mix (Applied Biosystems, Carlsbad, CA, USA). The Universal Probe Library Technology (UPL; Roche, Basel, Switzerland) was used to directly compare the expression of different transcripts. Primer efficiencies were calculated with the software LinRegPCR (Ruijter, 2004). Relative expression was calculated using the equation published by Pfaffl (2001) which adjusts for the primer efficiency. Primer sequences are given in Supplementary Table S1.
Iso-Seq Sequencing and Analysis
For isoform sequencing total RNA from the cerebral cortex of a mouse at stage P0 with a RIN value >9 was sent to the Vienna Biocenter Core Facilities (VBCF) of the Vienna Biocenter. The single-molecule real-time (SMRT) library preparation and sequencing was performed at the VBCF NGS Unit3. See Supplementary Methods for a detailed description.
Data Availability
The binary Alignment/Map files (BAM) from the combined sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under the BioProject accession number PRJNA587931.
In situ Hybridization
Two riboprobes, one corresponding to exon 1B and the other one to a part of the coding sequence of Rbfox1 were generated and hybridized on E15.5 mouse brain sections (ZYAGEN, San Diego, CA, USA). In situ hybridizations were analyzed with an EVOS XL Core Cell Imaging System (Thermo Fisher Scientific, Rockford, IL, USA). See Supplementary Methods for a detailed protocol.
Design and Statistical Analyses
All statistical analyses were done with GraphPad Prism 5 or Microsoft Office Excel. Data are shown as mean + standard error of the mean. Statistical analyses were done using student’s t-test. p values < 0.05 were considered statistically significant.
Results
Identification of Alternative Rbfox1 Transcripts by Isoform Sequencing
Rbfox1 is one of the longest genes in the genome with an extensive 5′-region. It is transcribed from several alternative promoters. The complexity of Rbfox1 transcripts further increases by alternative splicing and alternative polyadenylation events. It is, however, unclear how many different alternative Rbfox1 transcript isoforms exist and whether there is any preference in alternative exon combinations. To identify all existing full-length Rbfox1 transcript isoforms in the mouse cerebral cortex, we performed Iso-Seq. Iso-Seq is a relatively recently developed sequencing method that creates long reads and therefore provides sequences of full-length transcripts. Iso-Seq of total RNA from mouse postnatal day (P) 0 cerebral cortex revealed a very complex picture of Rbfox1 isoforms with multiple transcripts containing alternative first, internal or last exons or alternative polyadenylation sites (PAS; Figures 1A–C, Supplementary Figure S1). Interestingly, in addition to the already known brain-specific alternative first exons E1B, E1C, E1C.1 and E1D (Figure 1A) we identified several novel first exons that are, as the already known alternative 5′-terminal exons, likely to be transcribed from their promoters (Figure 1B, Supplementary Figure S1). All of these additional first exons are spliced to the known Rbfox1 internal exons and are therefore expected to encode RBFOX1 protein variants. One of these first exons is located 578 kb upstream of E1B and was therefore named E1A. It is spliced to two additional novel exons upstream of E1B, which we named exons 2 and 3 followed by the already known internal Rbfox1 exons. While it was originally thought that most of the alternative Rbfox1 first exons are noncoding, the novel exons E1A, 2 and 3 are coding exons and, when integrated into the Rbfox1 open reading frame, add 92 amino acids to the RBFOX1 N-terminus. The splice sites of these novel exons are conserved in mammals. Their overall conservation at the protein level is, however, low (59.3% identity) between mouse and human. The first exon E1A encodes a predicted cNLS which is conserved in humans (Figure 1D). RBFOX1 isoforms containing E1A are therefore supposed to localize to the nucleus and participate in the alternative splicing process.
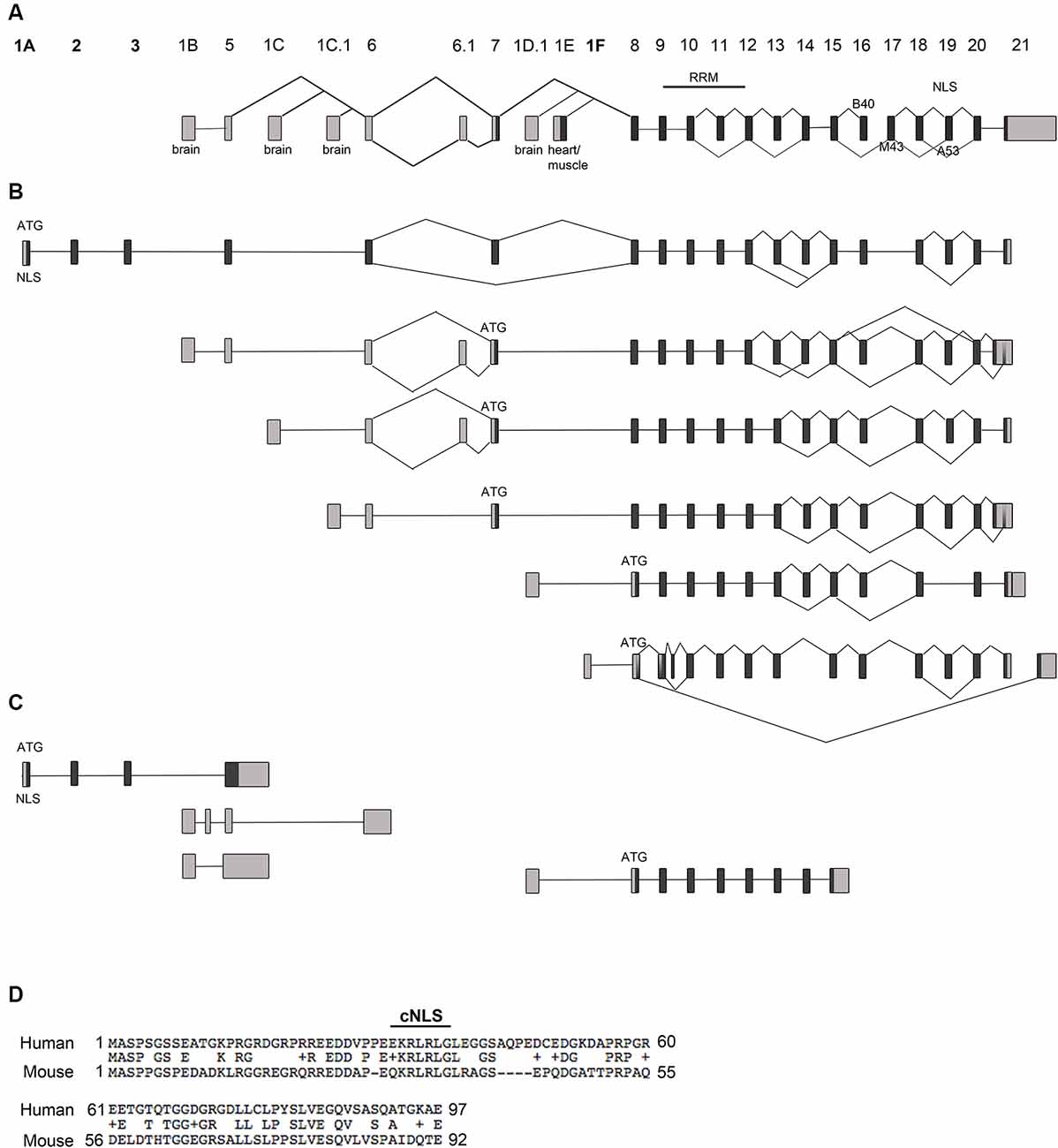
Figure 1. Rbfox1 transcript identification in the cerebral cortex. (A) Scheme of the Rbfox1 gene with the known exon annotation (Damianov and Black, 2010). (B,C) A summary of the alternative Rbfox1 transcripts expressed in the perinatal cerebral cortex. Total RNA was isolated from P0 mouse cortices and used for Iso-Seq. (B) Full-length Rbfox1 transcripts. (C) Three prime truncated Rbfox1 transcripts. Boxes represent exons. Dark gray boxes indicate coding exons and light gray boxes non-coding exons. Novel exons are named 1A, 2, 3, and 1F. Alternative exons B40, M43, and A53 are shown. RRM, RNA recognition motif; NLS, nuclear localization signal. (D) Conservation of the novel exons 1A, 2 and 3 between human and mouse. cNLS, predicted classic nuclear localization signal.
Several other novel first exons (E1F, E1F.1, E1F.2) were identified downstream of the heart and muscle-specific first exon E1E (Figures 1A,B; Supplementary Figure S1). As for transcripts starting in E1E, their translational start codon is in exon 8.
The Rbfox1 gene contains a long 3′-untranslated region (3′UTR) of 3.7 kb with several alternative PAS. Almost all Rbfox1 transcript isoforms identified here were polyadenylated at an alternative PAS located 150 bp downstream of the translation termination codon, which suggests that Rbfox1 transcripts in the P0 cerebral cortex typically contain a short 3′UTR.
In addition to alternative 5′ and 3′ splicing, alternative splicing of internal exons were observed in the Iso-Seq data. Inclusion of the alternative Rbfox1 exon A53 introduces a frame-shift in the C-terminus and causes a lack of the C-terminal NLS with the consensus sequence FAPY. These protein isoforms are therefore expected to be mostly cytoplasmic. Transcription of a given alternative first exon was generally not associated with specific splicing events. Alternative splicing of exon A53, for example, was a frequent event in the P0 cerebral cortex and occurred in transcript isoforms containing the alternative first exons E1A, E1B, E1C, E1C.1 and E1F (Figure 1B). Likewise, alternative splicing of the brain-specific exon B40 and Rbfox1 exons 13 and 14 was frequently observed. Similar to the inclusion of exon A53 the skipping of exon B40 introduces a frame-shift at the C-terminus.
In addition to the full-length transcripts, we identified several 3′-truncated transcripts (Figure 1C). Most of them do not contain an open reading frame of substantial length.
Rbfox1 Isoforms Carrying the Alternative First Exon 1A Are Translated Into Protein and Localize to the Nucleus
Nuclear localization of Rbfox1 is regulated by alternative splicing of exon A53. Inclusion of this exon causes a frame-shift and a lack of the C-terminal NLS and has been shown to guide Rbfox1 to the cytoplasm (Lee et al., 2009). Rbfox1 isoforms starting from the novel exon E1A contain a predicted cNLS which theoretically might ensure nuclear localization of Rbfox1 isoforms lacking the C-terminal NLS.
To test for protein expression and subcellular localization of Rbfox1 isoforms starting from E1A we generated two different constructs, the first encoding untagged Rbfox1 isoform 1A without exon A53 and the second untagged Rbfox1 isoform 1A with exon A53 included. Both constructs also contained an IRES GFP downstream of the Rbfox1 coding sequence to detect transfected cells. We chose untagged Rbfox1 constructs because a tag at the N-terminus might bias expression of the Rbfox1 open reading frame while a tag at the C-terminus might hinder the NLS which is encoded by the last four amino acids. After transfecting these constructs into Neuro2A (N2A) cells and primary cortical neurons we performed immunofluorescent stainings with Rbfox1 and GFP specific antibodies. We could not detect the expression of endogenous Rbfox1 in N2A cells suggesting that expression levels of Rbfox1 are either absent or too low for antibody detection in this cell line. Although we could detect endogenous Rbfox1 expression in primary cortical neurons using the Rbfox1 specific antibody the signal was rather weak and could be clearly distinguished from the Rbfox1 signal of the overexpressing neurons (Figures 2A,B). The immunofluorescence stainings revealed nuclear localization of Rbfox1 regardless of whether exon A53 was included in the respective constructs or not (Figures 2A,B).
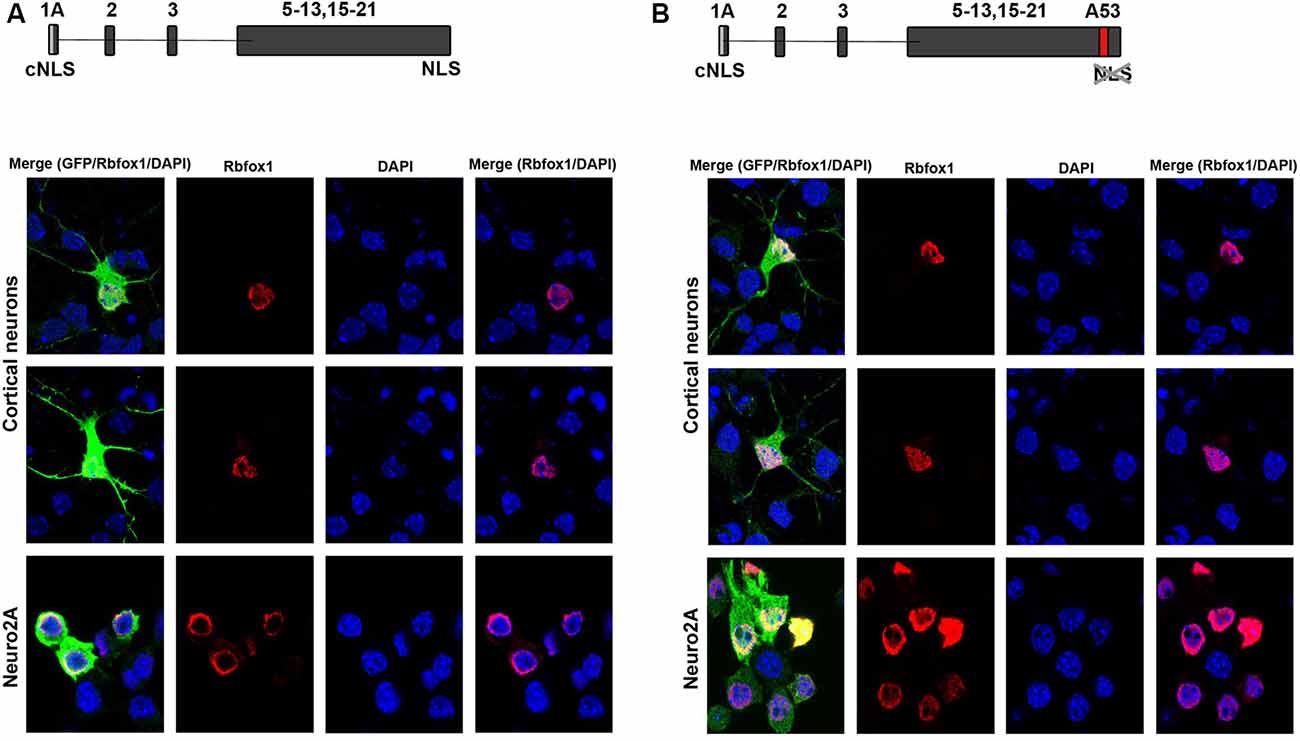
Figure 2. Expression of novel Rbfox1 isoforms starting from exon 1A in N2A cells and primary neurons. (A,B) Immunofluorescence analysis of the subcellular localization of the two Rbfox1 isoforms starting from exon 1A. N2A cells or primary cortical neurons were transfected with constructs encoding the untagged Rbfox1 isoforms followed by an IRES GFP. Twenty-four hours later cell were fixed and immunofluorescence stainings were performed with antibodies specific for Rbfox1 and GFP. (A) Construct encoding Rbfox1 isoform starting from exon 1A and lacking exon A53. (B) Construct encoding Rbfox1 isoform starting from exon 1A and containing exon A53.
Expression of Rbfox1 Isoforms Carrying Alternative First Exons in the Pre- and Post-natal Cerebral Cortex and in the Juvenile Brain
To further study the expression of the Rbfox1 alternative first exon transcripts during cortical development we performed RT-qPCR experiments with RNA from the cerebral cortex of mice at embryonic days (E) 13.5, 15.5, 17.5, P0, P7, and at 3 and 6 weeks of age. To cover the expression of the majority of Rbfox1 transcripts within a single RT-qPCR reaction we generated primers located in the Rbfox1 exons 20 and 21. This primer combination amplifies all Rbfox1 transcripts except the 3′-truncated transcripts and weakly expressed Rbfox1 transcripts that are terminated at an alternative PAS upstream of exon 21 (Supplementary Figures S1, S2). Using these primers, we observed a 7.8-fold increase in Rbfox1 expression from E13.5 until 3 weeks of age (Figure 3A) whereafter Rbfox1 expression decreased strongly (3.7-fold from 3 weeks to 6 weeks of age). To analyze the expression levels of the alternative first exons and their contribution to overall Rbfox1 expression levels we then generated primers corresponding to sequences in these exons. Expression of most of these alternative first exons also increased from E13.5 until perinatal or postnatal stages and declined thereafter. We observed an increase in expression until P0 for E1B until P7 for E1C and until 3 weeks of age for E1A (Figures 3B–D). After reaching these peaks expression of these first exons declined and at 6 weeks of age reached levels similar to or slightly above the levels observed at E13.5. E1C.1 expression increased from E13.5 to P0, decreased from P0 to P7 and increased again thereafter (Figure 3E). By contrast, Rbfox1 1D expression was stable from E13.5 to P0, undetectable at P7 and 3 weeks of age and increased again at 6 weeks of age (Figure 3F).
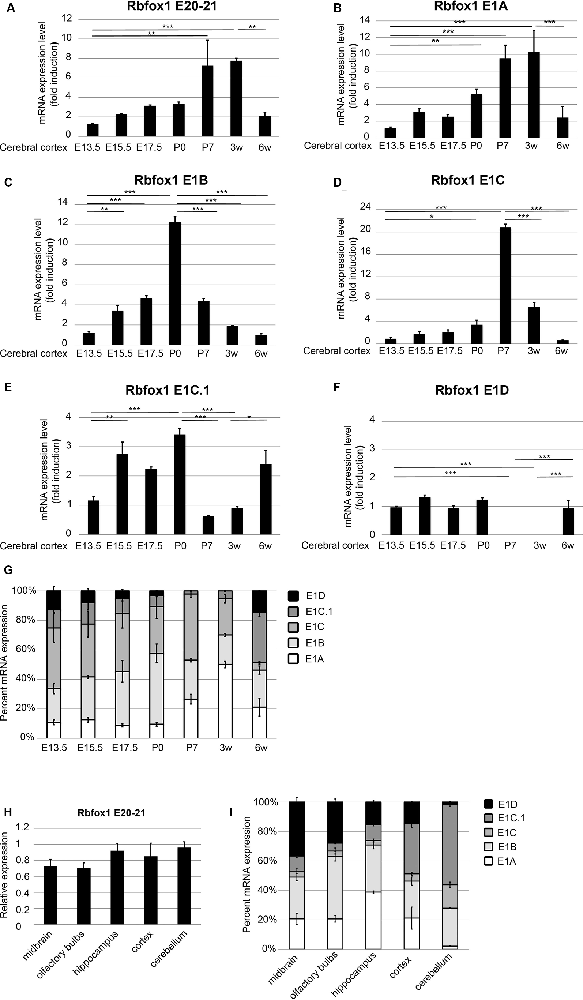
Figure 3. Expression of the Rbfox1 alternative first exons during embryonic and postnatal development of the cerebral cortex and in different brain subregions. (A–F,H) RT-qPCR with primers corresponding to Rbfox1 exons 20/21 or the alternative first exons. Total RNA was isolated from the cerebral cortex at different embryonic/postnatal stages (A–G) and different brain subregions of 6-weeks old mice (H,I), and cDNA synthesis and qPCR were performed; mRNA expression was normalized to Gapdh. (G,I) Direct comparison of the expression levels of the alternative Rbfox1 first exons. The sum of the expression levels of all alternative first exons was set to 100%. Data represent the average of three (juvenile brain regions) to four (embryonic/postnatal stages) biological replicates. The standard error of the mean is indicated by black bars. Statistical analyses were done using one-way-ANOVA with Bonferroni’s multiple comparison test. *P < 0.05; **P < 0.01; ***P < 0.001.
Since, we used probe-based qPCR assays we were able to compare the transcript levels of the different Rbfox1 isoforms directly after correcting for differences in primer efficiencies (see “Materials and Methods” section). A direct comparison of the expression levels of the different Rbfox1 first exons showed that E1B and E1C/E1C.1 together accounted for approximately 80% of the total amount of all measured Rbfox1 first exon variants in the E13.5 to P0 cerebral cortex (Figure 3G). In situ hybridization experiments confirmed the expression of Rbfox1 E1B in the cortical plate of E15.5 embryos (Supplementary Figure S3). From P0 to P3 weeks of age the percentage of E1A in the total amount of expressed first exons increased from 9.5% to 50% and declined to 22% at 6 weeks of age.
To test how Rbfox1 alternative first exon isoforms contribute to total Rbfox1 expression levels in juvenile brain regions we carried out RT-qPCR with RNA from the midbrain, olfactory bulbs, hippocampus, cerebral cortex, and cerebellum of 6-weeks old mice. We found that while the total Rbfox1 levels (detected with primer combination in exons 20/21) are relatively constant throughout different brain regions (Figure 3H) some of the alternative first exons are expressed in a brain region-specific manner (Figure 3I; Supplementary Figure S4). In the hippocampus, E1A was the predominant first exon variant and accounted for 40% of the total amount of all measured Rbfox1 first exon variants. In the cerebellum, the expression of E1A was very low. E1B expression levels were relatively constant in all brain regions examined with a 2-fold higher expression in olfactory bulbs and hippocampus compared to other brain regions. Rbfox1 E1C/E1C.1 was expressed most strongly in the cerebellum (71% of all measured Rbfox1 first exon variants) whereas Rbfox1 E1D was highly expressed in the midbrain (37% of all measured Rbfox1 first exon variants).
Several E-Boxes Regulate Neuronal Expression of Rbfox1 Isoforms 1B and 1C/1C.1
The first exon variants E1B and E1C/E1C.1 are the most strongly expressed variants in the developing cerebral cortex (Figures 3C–E,G). This suggests that the promoter regions driving expression of E1B and E1C/E1C.1 are highly active in this region. To further confirm this we cultured primary cortical neurons from E14.5 mouse embryos and measured expression of E1B, E1C/E1C.1 and for comparison E1D. As expected, these neurons expressed Rbfox1 exons E1B and E1C/E1C.1 strongly and exon E1D only weakly (Figure 4A). To assess whether the regions upstream of exons E1B, E1C/E1C.1 and E1D contain neuron-specific promoters we cloned these regions into the pGL4.10 luciferase vector and carried out luciferase experiments in cortical neurons (Figures 4B,C). All three regions produced significantly more luciferase activity than the empty vector (Figure 4C). Also, the regions upstream of E1B and E1C/E1C.1 produced significantly more luciferase activity in primary neurons than in undifferentiated P19 cells which do not express endogenous Rbfox1. They also produced three times more luciferase activity than the region upstream of E1D suggesting that these regions are more active in cortical neurons derived from embryonic tissue. We therefore concentrated on these regions in subsequent experiments.
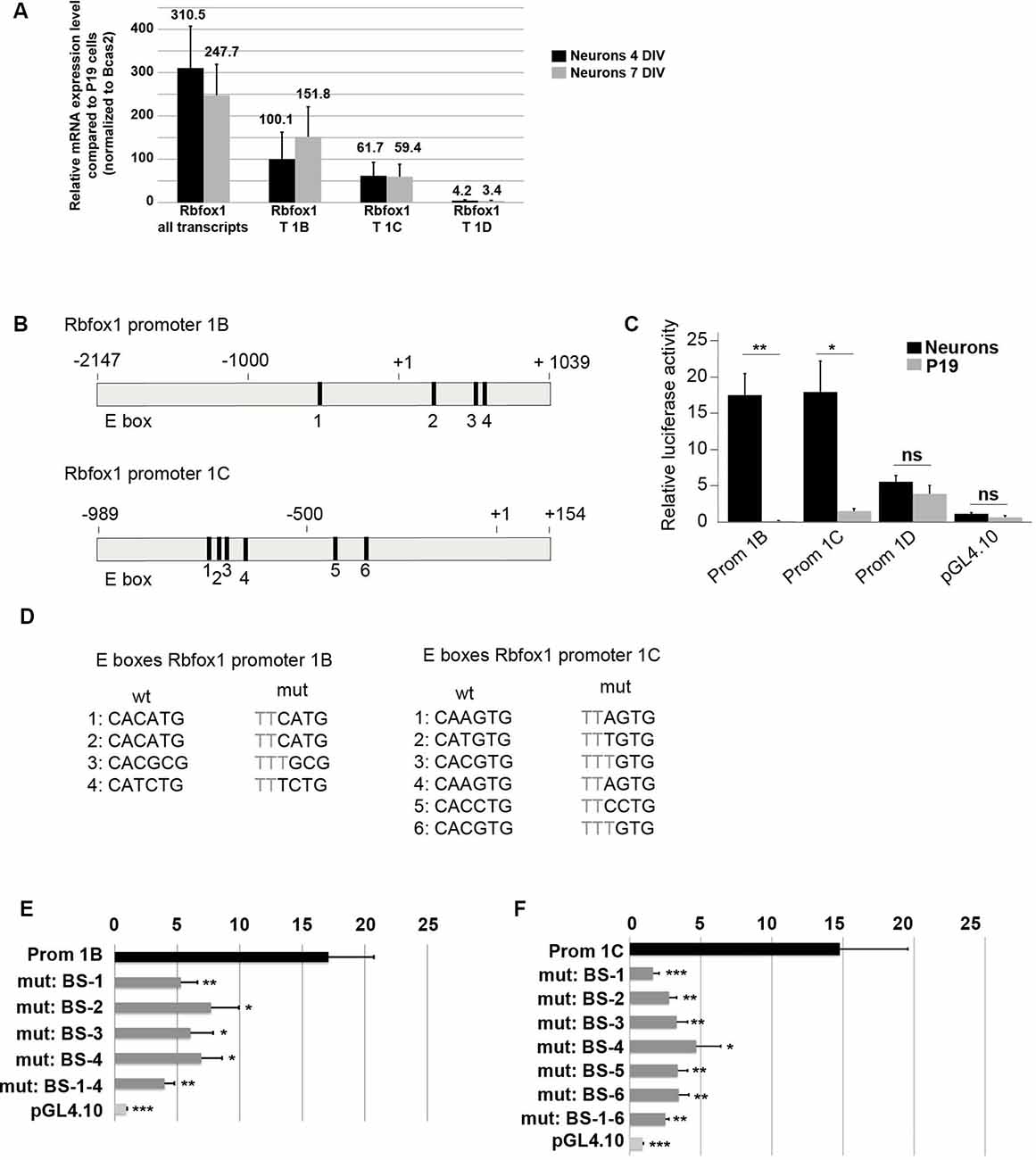
Figure 4. E-boxes promote expression of Rbfox1 1B and 1C/1C.1 in neurons. (A) Expression of Rbfox1 alternative first exons in primary cortical neurons. Total RNA was isolated from primary cortical neurons that had been in culture for 4 or 7 days, and cDNA synthesis and qPCR experiments with primers corresponding to Rbfox1 exons 20/21, 1B, 1C or 1D were performed (n = 4). (B) The scheme of the Rbfox1 1B and 1C/1C.1 upstream regulatory regions that were cloned into the pGL4.10 vector. Black boxes represent evolutionary conserved E-boxes. Numbers indicate the nucleotide position relative to the transcriptional start sites. (C) Luciferase assays were performed in primary cortical neurons and undifferentiated P19 cells to determine the promoter activities of the Rbfox1 1B, 1C/1C.1 and 1D upstream regulatory regions (n = 8). Statistical analyses were done using student’s t-test. ns, not significant; *P < 0.05; **P < 0.01. (D) Sequences of the wildtype and mutated E-boxes as depicted in (B). (E,F) Luciferase assays were performed to determine the activity of the Rbfox1 1B and 1C/1C.1 upstream regulatory regions with or without mutations in all E-boxes or every single E-box (n = 6). The standard error of the mean is indicated by black bars. Statistical analyses were done using one-way-ANOVA with Bonferroni’s multiple comparison test. *P < 0.05; **P < 0.01; ***P < 0.001.
A visual inspection of the cloned regions upstream of E1B and E1C revealed several evolutionarily conserved E-boxes (Figures 4B,D); four in the region upstream of E1B and six in the region upstream of E1C. We could not detect conserved E-boxes in the upstream regions of any of the other Rbfox1 first exon variants. E-boxes are well described binding sites for bHLH transcription factors. To test whether the E-boxes upstream of E1B and E1C were functional, we mutated each of these binding sites individually. Also, we generated constructs carrying mutations in all of these binding sites. Mutating the E boxes upstream of E1B and E1C caused a significant drop in luciferase activity for each of the single mutations (Figures 4E,F). This suggests that the E boxes upstream of E1B and E1C are all functional and might act together to drive transcription of E1B and E1C.
Discussion
Haploinsufficiency of the Rbfox1 gene causes neuronal hyperexcitability and seizures in Rbfox1 knockout mice (Gehman et al., 2011). In humans, heterozygous CNVs in the 5’-noncoding part of the RBFOX1 gene that typically only affect some but not all RBFOX1 transcript isoforms are associated with a range of neurodevelopmental disorders (Bhalla et al., 2004; Martin et al., 2007; Zhao, 2013). While this indicates dosage sensitivity for Rbfox1, regulatory mechanisms that balance Rbfox1 expression levels are unclear. One reason for this might be the size and complexity of the Rbfox1 gene which suggests complex modes of regulation. In this study, we identified all Rbfox1 transcript variants detectable by Iso-Seq in the mouse cerebral cortex at P0, and characterized the alternative Rbfox1 promoters.
Here, we show that transcripts starting in the novel first exon E1A encode a unique N-terminus containing a predicted cNLS, which is conserved in humans. RBFOX1 contains a C-terminal NLS of the sequence FAPY, which is typical for nuclear RNA binding proteins (Sun et al., 2012). Skipping of exon B40 or inclusion of exon A53, however, leads to a frame-shift, an alternative C-terminus lacking the FAPY NLS and cytoplasmic localization of the encoded RBFOX1 protein isoforms (Lee et al., 2009). RBFOX1 protein isoforms containing the N-terminal NLS brought in by exon E1A however localized to the nucleus even if exon A53 was included in the respective Rbfox1 transcripts. Strong expression of E1A as seen in the cerebral cortex at 3 weeks of age and in the hippocampus at 6 weeks of age might be a guarantee for correct splicing patterns through a high concentration of RBFOX1 in the nucleus. We have recently found a similar phenomenon in the Rbfox2 gene, which contains an N-terminal cNLS in its isoform 1A (Wenzel et al., 2016). Rbfox2 protein isoforms with this cNLS and lacking the FAPY NLS in the C-terminus are indeed nuclear.
Skipping of exon 11 produces a dominant-negative RBFOX1 isoform, which has been found in muscle and heart but not in the brain (Damianov and Black, 2010). In agreement, we did not detect Rbfox1 transcripts lacking exon 11 in P0 cerebral cortex. This suggests that skipping of exon 11 is a heart and muscle-specific event and does not play a role in the brain, at least not in the regions and stages tested here. Rather, we found frequent skipping of exon 14 in the P0 cerebral cortex, a splicing event, which to our knowledge has not been described before. Skipping of exon 13 was detected as well although in fewer transcripts. Skipping of exons 13 and 14 leads to in-frame deletions of 18 and 27 amino acids, respectively, in the RBFOX1 protein. Both exons encode parts of the C-terminus of RBFOX1, which interacts with multiple binding partners including several RNA binding proteins and is crucial for exon activation and repression (Sun et al., 2012). Skipping of exons 13 and 14 could therefore theoretically change the binding of RBFOX1 to its interactors or influence the ability of RBFOX1 to activate and/or repress target exons.
In mammals, and eukaryotes in general, splicing is coupled to transcription (Kornblihtt, 2015). Not only the speed of transcription can pre-determine alternative splicing events but also the expression from different promoters (Tasic et al., 2002). In our study, we could not observe that expression from alternative Rbfox1 promoters was linked to skipping or inclusion of alternative internal exons. For example, we could detect both skipping and inclusion of exon A53 in Rbfox1 transcripts expressed from almost all of the alternative promoters. This suggests that alternative promoters in the Rbfox1 gene do not regulate alternative splicing choices. However, Iso-Seq is not a quantitative measure of transcript abundance and it might be that the ratio between skipped and included alternative exons varies in transcripts generated from the different promoters. Despite we cannot exclude this possibility our results rather suggest that the many different promoters in the Rbfox1 gene are needed to contribute to total Rbfox1 levels in different developmental stages and brain subregions. This is supported by the observation that expression levels of total Rbfox1 were relatively constant between different brain subregions whereas those of the alternative transcripts were stage- and region specific. It must, however, be mentioned that the analysis of brain regions with high cellular heterogeneity is a limitation of this study.
The usage of alternative promoters may theoretically be linked to specific RBFOX1 functions as has been observed for other proteins (Bae et al., 2014; Zhu et al., 2014; Philips et al., 2015; Perera and Kim, 2016). A similar mechanism has been described for the BDNF gene, which contains nine alternative promoters driving the expression of identical BDNF proteins (Maynard et al., 2016). Knockout studies in which BDNF production from each one of the four major promoters was disrupted provided evidence that BDNF transcripts produced from different promoters mediate different aspects of behavior. Whether this is the case for the alternative Rbfox1 transcripts as well is not clear. In humans, CNVs that are located in different parts of the 5’-region of the RBFOX1 gene have been found in patients with ID, ASD, ADHD, epilepsy, and schizophrenia (Bhalla et al., 2004; Martin et al., 2007; Elia et al., 2010; Lal et al., 2013; Zhao, 2013). These CNVs span 5′-UTR exons of unique RBFOX1 transcript isoforms. They are therefore predicted to interfere with the expression of some but not all RBFOX1 transcripts. If the location of these CNVs was correlated with specific phenotypes in the patients this might be an indication of transcript specific functions. This is, however, not the case. Rather, CNVs spanning overlapping regions have been observed in patients with variable phenotypes and patients with similar phenotypes carry CNVs spanning distinct regions in the RBFOX1 gene (personal observation). Approximately 50% of the genes that are linked to neurological disorders are transcribed from alternative promoters, and specific transcript variants are involved in these diseases. It may, therefore, be important to study the expression and functions of these gene isoforms instead of concentrating on the whole gene’s function (Pal et al., 2011). For Rbfox1, future studies should aim to generate transcript specific knockouts in mice or human cell models.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
Ethical review and approval was not required for the animal study because the study does not include any animal experiments requiring approval. To carry out this study mice were killed for organ removal. In Germany, this procedure is notifiable but does not require approval by an ethics committee.
Author Contributions
SC and LS designed and performed experiments and analyzed the data. SM, LZ, and MS performed experiments and analyzed the data. SD, DS, and SS analyzed the data. JW planned experiments, analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
SC and LS were supported by stipends from the Focus Program of Translational Neurosciences of the Johannes Gutenberg-University Mainz. This work was supported by the Deutsche Forschungsgemeinschaft (CRC 1193 and WI 3837/10-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Susann Schweiger for critical reading of the manuscript.
Footnotes
- ^ https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/HGNC:18222
- ^ www.informatics.jax.org/marker/MGI:1926224
- ^ www.vbcf.ac.at
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2020.00066/full#supplementary-material.
FIGURE S1 | Rbfox1 transcript identification in the cerebral cortex. All spliced Rbfox1 transcript variants that were detected by Iso-Seq in the P0 cerebral cortex are shown. At the top of each page is a schematic of the known Rbfox1 exons (18). RRM, RNA recognition motif; NLS, nuclear localization signal; pA, polyadenylation site; ATG, translational start codon.
FIGURE S2 | Expression of the alternative 3′end of the Rbfox1 gene during cortex development. Total RNA was isolated from the cerebral cortex of different embryonic and perinatal stages and subjected to RT-qPCR with primers corresponding to Rbfox1 exons 20/21 or the alternative 3′ end (n = 3); mRNA expression was normalized to Gapdh.
FIGURE S3 | Rbfox1 first exon 1B is expressed in the cortical plate of the mouse embryo. (A) In situ hybridization of Rbfox1 (genepaint.org) showing Rbfox1 expression in the cortical plate, and several other brain regions. (B,C) In situ hybridization carried out with probes corresponding to total Rbfox1 (B) or Rbfox1 1B (C). A high expression level of Rbfox1 1B was detected in the cortical plate.
FIGURE S4 | Expression of the Rbfox1 alternative first exons in different subregions of the juvenile brain. RT-qPCR with primers corresponding to Rbfox1 exons 20/21 or the alternative first exons. Total RNA was isolated from different brain subregions of 6-weeks old mice, and cDNA synthesis and qPCR were performed; mRNA expression was normalized to Gapdh.
TABLE S1 | Primer sequences.
References
Bae, B. I., Tietjen, I., Atabay, K. D., Evrony, G. D., Johnson, M. B., Asare, E., et al. (2014). Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science 343, 764–768. doi: 10.1126/science.1244392
Bhalla, K., Phillips, H. A., Crawford, J., Mckenzie, O. L., Mulley, J. C., Eyre, H., et al. (2004). The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J. Hum. Genet. 49, 308–311. doi: 10.1007/s10038-004-0145-4
Conboy, J. G. (2017). Developmental regulation of RNA processing by Rbfox proteins. Wiley Interdiscip. Rev. RNA 8:2. doi: 10.1002/wrna.1398
Damianov, A., and Black, D. L. (2010). Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA 16, 405–416. doi: 10.1261/rna.1838210
Dredge, B. K., and Jensen, K. B. (2011). NeuN/Rbfox3 nuclear and cytoplasmic isoforms differentially regulate alternative splicing and nonsense-mediated decay of Rbfox2. PLoS One 6:e21585. doi: 10.1371/journal.pone.0021585
Elia, J., Gai, X., Xie, H. M., Perin, J. C., Geiger, E., Glessner, J. T., et al. (2010). Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry 15, 637–646. doi: 10.1038/mp.2009.57
Fogel, B. L., Wexler, E., Wahnich, A., Friedrich, T., Vijayendran, C., Gao, F., et al. (2012). RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum. Mol. Genet. 21, 4171–4186. doi: 10.1093/hmg/dds240
Gehman, L. T., Stoilov, P., Maguire, J., Damianov, A., Lin, C. H., Shiue, L., et al. (2011). The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet. 43, 706–711. doi: 10.1038/ng.841
Jin, Y., Suzuki, H., Maegawa, S., Endo, H., Sugano, S., Hashimoto, K., et al. (2003). A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22, 905–912. doi: 10.1093/emboj/cdg089
Kornblihtt, A. R. (2015). Transcriptional control of alternative splicing along time: ideas change, experiments remain. RNA 21, 670–672. doi: 10.1261/rna.051151.115
Kuroyanagi, H. (2009). Fox-1 family of RNA-binding proteins. Cell. Mol. Life Sci. 66, 3895–3907. doi: 10.1007/s00018-009-0120-5
Lal, D., Reinthaler, E. M., Altmüller, J., Toliat, M. R., Thiele, H., Nürnberg, P., et al. (2013). RBFOX1 and RBFOX3 mutations in rolandic epilepsy. PLoS One 8:e73323. doi: 10.1371/journal.pone.0073323
Lee, J. A., Damianov, A., Lin, C. H., Fontes, M., Parikshak, N. N., Anderson, E. S., et al. (2016). Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron 89, 113–128. doi: 10.1016/j.neuron.2015.11.025
Lee, J. A., Tang, Z. Z., and Black, D. L. (2009). An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 23, 2284–2293. doi: 10.1101/gad.1837009
Martin, C. L., Duvall, J. A., Ilkin, Y., Simon, J. S., Arreaza, M. G., Wilkes, K., et al. (2007). Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144B, 869–876. doi: 10.1002/ajmg.b.30530
Maynard, K. R., Hill, J. L., Calcaterra, N. E., Palko, M. E., Kardian, A., Paredes, D., et al. (2016). Functional role of BDNF production from unique promoters in aggression and serotonin signaling. Neuropsychopharmacology 41, 1943–1955. doi: 10.1038/npp.2015.349
Pal, S., Gupta, R., Kim, H., Wickramasinghe, P., Baubet, V., Showe, L. C., et al. (2011). Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 21, 1260–1272. doi: 10.1101/gr.120535.111
Parikshak, N. N., Luo, R., Zhang, A., Won, H., Lowe, J. K., Chandran, V., et al. (2013). Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021. doi: 10.1016/j.cell.2013.10.031
Perera, B. P., and Kim, J. (2016). Alternative promoters of Peg3 with maternal specificity. Sci. Rep. 6:24438. doi: 10.1038/srep24438
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Philips, M. A., Lillevali, K., Heinla, I., Luuk, H., Hundahl, C. A., Kongi, K., et al. (2015). Lsamp is implicated in the regulation of emotional and social behavior by use of alternative promoters in the brain. Brain Struct. Funct. 220, 1381–1393. doi: 10.1007/s00429-014-0732-x
Rajman, M., Metge, F., Fiore, R., Khudayberdiev, S., Aksoy-Aksel, A., Bicker, S., et al. (2017). A microRNA-129–5p/Rbfox crosstalk coordinates homeostatic downscaling of excitatory synapses. EMBO J. 36, 1770–1787. doi: 10.15252/embj.201695748
Ruijter, J. (2004). LinRegPCR: Analysis of Real-Time PCR Data [Computer Pro-gram]. Version 7.5. Amsterdam: Department of Anatomyand Embryology, Academic Medical Centre.
Shibata, H., Huynh, D. P., and Pulst, S. M. (2000). A novel protein with RNA-binding motifs interacts with ataxin-2. Hum. Mol. Genet. 9, 1303–1313. doi: 10.1093/hmg/9.9.1303
Sun, S., Zhang, Z., Fregoso, O., and Krainer, A. R. (2012). Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA 18, 274–283. doi: 10.1261/rna.030486.111
Tasic, B., Nabholz, C. E., Baldwin, K. K., Kim, Y., Rueckert, E. H., Ribich, S. A., et al. (2002). Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10, 21–33. doi: 10.1016/s1097-2765(02)00578-6
Underwood, J. G., Boutz, P. L., Dougherty, J. D., Stoilov, P., and Black, D. L. (2005). Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 25, 10005–10016. doi: 10.1128/mcb.25.22.10005-10016.2005
Vuong, C. K., Wei, W., Lee, J. A., Lin, C. H., Damianov, A., De La Torre-Ubieta, L., et al. (2018). Rbfox1 regulates synaptic transmission through the inhibitory neuron-specific vSNARE Vamp1. Neuron 98, 127–141.e7. doi: 10.1016/j.neuron.2018.03.008
Wamsley, B., Jaglin, X. H., Favuzzi, E., Quattracolo, G., Nigro, M. J., Yusef, N., et al. (2018). Rbfox1 mediates cell-type-specific splicing in cortical interneurons. Neuron 100, 846.e7–859.e7. doi: 10.1016/j.neuron.2018.09.026
Wenzel, M., Schule, M., Casanovas, S., Strand, D., Strand, S., and Winter, J. (2016). Identification of a classic nuclear localization signal at the N terminus that regulates the subcellular localization of Rbfox2 isoforms during differentiation of NMuMG and P19 cells. FEBS Lett. 590, 4453–4460. doi: 10.1002/1873-3468.12492
Zhao, W. W. (2013). Intragenic deletion of RBFOX1 associated with neurodevelopmental/neuropsychiatric disorders and possibly other clinical presentations. Mol. Cytogenet. 6:26. doi: 10.1186/1755-8166-6-26
Keywords: Rbfox1, alternative splicing, neurodevelopmental disorders, autism, alternative promoters
Citation: Casanovas S, Schlichtholz L, Mühlbauer S, Dewi S, Schüle M, Strand D, Strand S, Zografidou L and Winter J (2020) Rbfox1 Is Expressed in the Mouse Brain in the Form of Multiple Transcript Variants and Contains Functional E Boxes in Its Alternative Promoters. Front. Mol. Neurosci. 13:66. doi: 10.3389/fnmol.2020.00066
Received: 27 September 2019; Accepted: 06 April 2020;
Published: 05 May 2020.
Edited by:
David Murphy, University of Bristol, United KingdomReviewed by:
Paul Bishop, University of Bristol, United KingdomCharles Colin Thomas Hindmarch, Queen’s University—Kingston, Canada
Copyright © 2020 Casanovas, Schlichtholz, Mühlbauer, Dewi, Schüle, Strand, Strand, Zografidou and Winter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer Winter, amV3aW50ZXJAdW5pLW1haW56LmRl
† These authors have contributed equally to this work
 Sonia Casanovas1,2†
Sonia Casanovas1,2† Laura Schlichtholz
Laura Schlichtholz Sophia Mühlbauer
Sophia Mühlbauer Sri Dewi
Sri Dewi Martin Schüle
Martin Schüle Lea Zografidou
Lea Zografidou Jennifer Winter
Jennifer Winter