- 1Department of Biology, University of Konstanz, Konstanz, Germany
- 2Institute of Biological Sciences, University of Rostock, Rostock, Germany
Diatoms, heterokont microalgae found in all aquatic habitats, can be distinguished by their typical brown colour due to the presence of a characteristic light-harvesting carotenoid: fucoxanthin. The biosynthesis of fucoxanthin involves several intermediates, some of which also play a key role in photoprotection via the xanthophyll cycle, controlling the dissipation of excessively absorbed light energy in the form of Non-Photochemical Quenching (NPQ). The regulation of the fucoxanthin pathway is therefore crucial to direct xanthophyll biosynthesis towards light harvesting or photoprotective functions. Yet, until recent years most of the steps in this key metabolical route remained unknown. Interestingly, diatoms possess multiple homologs of the ancestral genes encoding the two xanthophyll cycle enzymes: Violaxanthin De-Epoxidase (VDE) and Zeaxanthin Epoxidase (ZEP). Here, we review the recent discoveries of the function of most VDE and ZEP isoforms in the fucoxanthin pathway of the model diatom Phaeodactylum tricornutum. Some of these enzymes have a central role in photoprotection, while other have been identified as ideal targets for engineering and industrial applications. We discuss the physiological role of these proteins and address missing links in the pathway and unknown properties of these enzymes. Finally, we argue that the expansion of the VDE and ZEP gene families represented a turning point in the evolution of xanthophyll cycling and fucoxanthin biosynthesis in diatoms.
1 Introduction
Carotenoids are essential molecules for life on Earth, contributing to photosynthesis and several other biological processes (Stange, 2016). Xanthophylls, carotenoids that contain oxygen, stand out for their role in light harvesting and photoprotection: they are structural components of the photosynthetic apparatus, increase the absorption of blue-green light wavelengths and actively protect the photosystems from light-induced damage via constitutive energy dissipation or via the inducible xanthophyll cycle, involved in the Non-Photochemical Quenching (NPQ) of excess light energy (Cazzaniga et al., 2016; Jahns and Holzwarth, 2012). Since the discovery of the violaxanthin cycle in the early 1960s (Yamamoto et al., 1962), xanthophyll cycling and its connection to NPQ have been described numerous times in plants and algae (Arsalane et al., 1994; Demmig et al., 1988; 1987; Niyogi et al., 1998; 1997; Olaizola et al., 1994, to cite the most seminal works). These processes are characterised by the interconversion between two or more xanthophylls, through the epoxidation and de-epoxidation of the ionone rings present at the two ends of the polyene chain of the molecule (Latowski et al., 2011; Stransky and Hager, 1970). In the two main forms of xanthophyll cycling (the violaxanthin and the diadinoxanthin cycle) the enzyme Violaxanthin De-Epoxidase (VDE) catalyses the formation of de-epoxidized xanthophylls (zeaxanthin or diatoxanthin), that trigger the activation of NPQ. The cycle is closed with the counteracting reaction operated by Zeaxanthin Epoxidase (ZEP), that restores the epoxidized xanthophyll (violaxanthin or diadinoxanthin) thus abolishing NPQ (for a comprehensive review we refer to: Fernández-Marín et al., 2021; Goss and Latowski, 2020; Goss and Lepetit, 2015). Beside their role in photoprotection, epoxy xanthophylls are also intermediates in the biosynthesis of other carotenoids and derived compounds (called apocarotenoids), like abscisic acid (DellaPenna and Pogson, 2006; Moreno et al., 2021). Thus, the regulation of xanthophyll cycle enzymes has both a photophysiologic and a metabolic relevance.
Xanthophylls have been gaining scientific and commercial interest in recent years, due to their relevance in sectors like human health or agriculture (Aziz et al., 2020; Beyer et al., 2002; Demmig-Adams et al., 2020; Karniel et al., 2020; Mann et al., 2000; Van Der Straeten et al., 2020). One of the frontiers of this industry is represented by microalgae, regarded as a powerful and sustainable source for xanthophyll production (Smaoui et al., 2021). In this framework, diatoms (brown heterokont microalgae) are emerging as a new platform for the production of xanthophylls of high interest, like fucoxanthin but also diadinoxanthin and diatoxanthin. For this purpose, the pennate diatom Phaeodactylum tricornutum has been identified as an ideal target: several molecular tools are already available for this species, that is also fast and easy to grow compared to other microalgal models (Butler et al., 2020; Morelli et al., 2025; Russo et al., 2023).
Diatoms are a major component of marine and freshwater phytoplankton dominating turbulent waters, nutrient-rich environments characterised by frequent and intense light changes (Falkowski et al., 2004; Lavaud, 2007). In response to this potential source of stress, diatoms display a highly efficient NPQ that heavily relies on the diadinoxanthin cycle and on antenna proteins of the Lhcx family (Buck et al., 2019; Croteau et al., 2025; Lavaud, 2007). The extent of this remarkable photoprotection capacity seems to be habitat dependent, with diatoms thriving in highly dynamic environments (such as coastal or intertidal waters) displaying a higher NPQ capacity compared to populations in the open ocean (Barnett et al., 2015; Lavaud et al., 2007). Furthermore, the xanthophyll cycle carotenoids involved in this photoprotective process are also precursors of the major light harvesting carotenoid fucoxanthin (as illustrated in Figure 1). Thus, while the fine regulation of xanthophyll-mediated NPQ is crucial to respond to dynamic light fluctuations, diatoms must also coordinate this with a complex xanthophyll biosynthesis pathway. Due to these characteristic features, the role of the enzymes regulating this key metabolical route takes on an even more marked importance in this relevant group of microalgae.
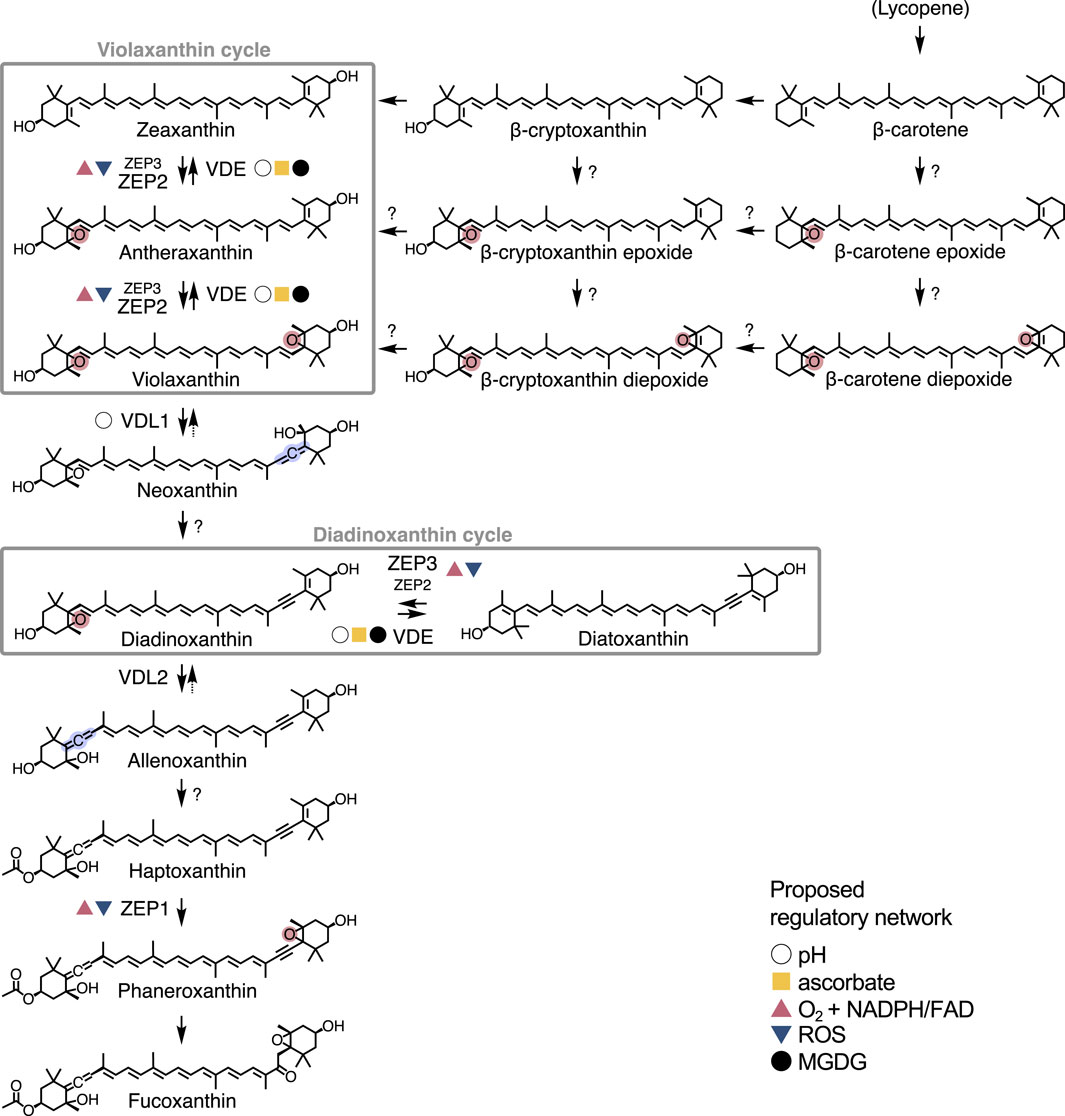
Figure 1. Confirmed roles of VDE and ZEP isoforms in the xanthophyll biosynthesis pathway of Phaeodactylum tricornutum. Reactions catalysed by other enzymes are represented with an unlabelled arrow, while uncharacterised steps are indicated by a question mark. Formation of epoxy and allenic groups are highlighted in red and blue, respectively. Next to each VDE/ZEP isoform, coloured symbols represent the proposed regulatory network (pH, ascorbate, O2 + NADPH/FAD, ROS and MGDG). A complete overview of fucoxanthin biosynthesis is presented in Bai et al. (2022) and Cao et al. (2023).
Diatoms harbor multiple paralogs of the VDE and ZEP enzymes (homologous to those regulating the violaxanthin cycle in land plants) that are essential for the regulation of xanthophyll cycling and fucoxanthin biosynthesis (Bai et al., 2022; Cao et al., 2023; Coesel et al., 2008; Frommolt et al., 2008). In this work, we review the recent advances on the characterisation of these key enzymes in P. tricornutum and address persisting research gaps and future challenges.
2 A quick overview of xanthophyll biosynthesis in diatoms
In all oxygenic photoautotrophs, the biosynthesis of xanthophylls begins from the conversion of active isoprene (the backbone of all carotenoids) into lycopene, the precursor of α- and β-carotene (Roy et al., 2011). Diatoms synthesise only β-carotene and therefore contain solely “β-branch” xanthophylls (Dautermann and Lohr, 2017; Roy et al., 2011). First, zeaxanthin is synthesised, followed by the two epoxy xanthophylls that complete the violaxanthin cycle: antheraxanthin and violaxanthin. Violaxanthin might also be synthetised via alternative routes, involving epoxi- or diepoxidized forms of β-carotene and/or β-cryptoxanthin (Bertrand, 2010; Lohr and Wilhelm, 2001). An allenic group is then added to violaxanthin for the synthesis of its allenic derivatives: first neoxanthin (in its trans- isomer), followed by a yet unknown reaction that leads to the formation of diadinoxanthin (Dautermann et al., 2020). The latter can then be de-epoxidized into diatoxanthin in the diadinoxanthin cycle. Finally, fucoxanthin, the main light harvesting carotenoid of diatoms, is also synthesised from diadinoxanthin, in a pathway that involves a series of allenic intermediates recently described in two seminal papers (Bai et al., 2022; Cao et al., 2023). Several reactions on this pathway involve the removal (de-epoxidation), addition (epoxidation) or rearrangement of the epoxy moiety and are catalysed by different VDE or ZEP isoforms (Figure 1).
3 Functional characterization of VDE and ZEP isoforms in Phaeodactylum tricornutum
VDE and ZEP belong to the lipocalin protein family and are ubiquitous among photosynthetic eukaryotes (Goss and Jakob, 2010). According to the latest genome annotation (Rastogi et al., 2018), P. tricornutum harbours 4 paralogs of VDE (VDE: Phatr3_J51703; VDL1: Phatr3_J36048; VDL2: Phatr3_J45846; VDR: Phatr3_J43240) and 3 of ZEP (ZEP1: Phatr3_J45845; ZEP2: Phatr3_J5928; ZEP3: Phatr3_J10970), annotated based on their similarity with the VDE or ZEP from green algae and land plants (Coesel et al., 2008). Four of these genes are coupled in tandem, with a tail-to-tail orientation: ZEP3-VDE (on chromosome 4), and ZEP1-VDL2 (on chromosome 8). In addition, VDR and ZEP2 are located on the same chromosome (1) but at more distant positions, while VDL1 is located on chromosome 9. This structure, also found in other heterokonts and prasinophytes, indicates that the ancestral VDE-ZEP tandem was subject to several duplication events followed by functional specialisation in these taxa (Coesel et al., 2008; Frommolt et al., 2008). Indeed, recent research has shown that, while some of these paralogs still operate the violaxanthin and diadinoxanthin cycles, others have specialised in different steps of xanthophyll biosynthesis (Figure 1).
3.1 VDE and ZEP3 orchestrate photoprotective xanthophyll cycling
Early physiological studies already proposed that de-epoxidation of both violaxanthin and diadinoxanthin must be operated by the same enzyme in P. tricornutum (Jakob et al., 2001; Lohr and Wilhelm, 1999). Subsequent experiments in recombinant and transient systems (Bojko et al., 2013; Dautermann et al., 2020; Lavaud et al., 2012; Olchawa-Pajor et al., 2019) identified the responsible isoform as VDE, as recently confirmed by the corresponding CRISPR/Cas9 gene knockout (Giossi et al., 2025). Identification of the specific ZEP isoform(s) responsible for the counteracting reactions (i.e., the epoxidation of zeaxanthin and diatoxanthin) has proven more challenging. Experiments on transformed Arabidopsis thaliana initially pointed to ZEP2 as best candidate for diatoxanthin epoxidation, due to its broader substrate specificity (Eilers et al., 2016). However, three independent studies employing knockout mutants later identified ZEP3 as the main epoxidase of the photoprotective diadinoxanthin cycle in P. tricornutum (Giossi et al., 2025; Græsholt et al., 2024; Ware et al., 2024). While most likely both ZEP2 and ZEP3 can accept diatoxanthin, zeaxanthin and antheraxanthin as substrates (Eilers et al., 2016; Giossi et al., 2025), these isoforms clearly developed different functions, with ZEP3 being mostly responsible for the rapid epoxidation of diatoxanthin (and thus for NPQ recovery) after light stress. This might be explained by differences in the light-dependent activation properties, substrate specificity and/or localisation of the two enzymes in the plastid (for a detailed elucidation of these hypotheses and corresponding models we refer to: Giossi et al., 2025).
In most photosynthetic eukaryotes VDE is localised in the thylakoid lumen and is activated by its acidification. In diatoms, it is inhibited at relatively higher pH (7.5) and requires lower amounts of co-factor ascorbate compared to plants (Bratt et al., 1995; Goss et al., 2006; Grouneva et al., 2006; Jakob et al., 2001). VDE is active as a dimer, formed upon acidification of the lumen pH, and requires the presence monogalactosyl diacylglycerols (MGDGs) in the thylakoid membrane (Goss and Latowski, 2020; Grouneva et al., 2006; Yamamoto and Higashi, 1978). ZEPs are instead found at the stromal side of the thylakoids, have a pH optimum slightly above neutral (∼7.5) and require O2, NADPH and FAD as co-substrates for zeaxanthin or diatoxanthin epoxidation (Büch et al., 1995; Goss and Latowski, 2020; Hager, 1975; Siefermann and Yamamoto, 1975). In plants, ZEP activity is inactivated in presence of certain reactive oxygen species (ROS) (Bethmann et al., 2019; Holzmann et al., 2022; Reinhold et al., 2008). In diatoms, epoxidase activity is significantly faster compared to the green lineage counterparts (Giossi et al., 2025; Goss et al., 2006; Lohr and Wilhelm, 1999) and is active at non-saturating irradiances (i.e., is inactive in both high light and darkness) due to a yet unknown mechanism which might be independent of pH (Blommaert et al., 2021). It is also inhibited by cadmium (Bertrand et al., 2001). Due to its clear role in the recovery of the diadinoxanthin cycle, we can now reasonably attribute these properties at least to ZEP3.
3.2 ZEP2: at the interface between photoprotection and de novo xanthophyll biosynthesis
As discussed above, ZEP2 can most likely catalyse the epoxidation of both zeaxanthin/antheraxanthin and diatoxanthin, but showed a broader substrate specificity compared to ZEP3 when transformed into land plants (Eilers et al., 2016; Giossi et al., 2025). Mutants lacking ZEP2 display a wild type-like diatoxanthin epoxidation capacity after a short light stress, but accumulate significantly higher amounts of pigments of the violaxanthin cycle when exposed to extreme high light for several hours (Giossi et al., 2025; Græsholt et al., 2024). This indicates that ZEP2 is mostly employed in de novo xanthophyll biosynthesis, while ZEP3 is likely specialised to strictly control photoprotective xanthophyll cycling in response to light stress (Giossi et al., 2025).
This leads to the hypothesis that ZEP2 is rather constitutively active to support downstream xanthophyll biosynthesis under a broad range of environmental conditions. Such feature could be extremely relevant for diatoms: while plants and green algae can accumulate their main NPQ-inducing pigment, zeaxanthin, directly from β-carotene, diatoms must synthesise violaxanthin to accumulate diadinoxanthin and diatoxanthin (Figure 1). Thus, an alternative ZEP activity (here attributed to ZEP2) would allow the flow of de novo xanthophyll biosynthesis independent of photoprotective ZEP3 regulation, leading to the increase of the diadinoxanthin cycle pool under high light and uncoupling the synthesis of fucoxanthin from photoprotective xanthophyll cycling. If ZEP3 would also be responsible for de novo xanthophyll biosynthesis, no accumulation of the total diadinoxanthin cycle pool would be possible in prolonged high light, as ZEP3 seems to be completely switched off under these conditions to enable photoprotection by diatoxanthin-based NPQ.
Still, further studies are needed to clarify these hypotheses. Transient expression of diatoms ZEPs in plant leaves is a powerful tool for investigating their catalytic activity, but this method presents limitations for studying their substrate specificity or their physiological role as plants lack several potential carotenoid substrates present in diatom plastids (such as diatoxanthin). Further functional studies on ZEP2 and ZEP3 will be needed to assess the relative contributions of these two enzymes in photoprotection and de novo xanthophyll biosynthesis and to examine postulated differences in their light dependency or cellular localisation.
3.3 VDL1, VDL2 and ZEP1 are involved in fucoxanthin biosynthesis
Frommolt et al. (2008) proposed that VDE-like (VDL) proteins, found in chromist algae but absent in the green lineage (Coesel et al., 2008), were unlikely to participate in xanthophyll cycling due to the lack of characteristic amino acids of the catalytic domain of VDE. Indeed, it was recently established that these enzymes catalyse the formation of allenic xanthophylls (Figure 1): VDL1 synthesises neoxanthin from violaxanthin, while VDL2 is responsible for the conversion of diadinoxanthin to allenoxanthin, an intermediate product of fucoxanthin biosynthesis (Bai et al., 2022; Dautermann et al., 2020). Further down the pathway, ZEP1 epoxidizes haptoxanthin to phaneroxanthin, the direct precursor of fucoxanthin (Bai et al., 2022; Cao et al., 2023).
Due to competition for substrates (violaxanthin and diadinoxanthin, as shown in Figure 1), the activity of VDE, VDL1 and VDL2 must be finely regulated to direct carotenoid biosynthesis towards photoprotective xanthophyll cycling or light-harvesting fucoxanthin depending on the physiological needs of the cell (Dautermann et al., 2020). Indeed, in vitro assays indicated that VDL1 activity is modulated solely by pH, in contrast to VDE that has a more complex regulation relying also on the cofactor ascorbate (Dautermann et al., 2020). VDL2 should also be subject to a tight regulation by a yet unknown factor (Bai et al., 2022). Finally, in vitro experiments suggested that ZEP1 is regulated by NADPH, FAD and molecular O2 (Bai et al., 2022), as known for the ZEP from plants (Büch et al., 1995).
Due to their involvement in the regulation of downstream fucoxanthin biosynthesis, VDL1, VDL2 and ZEP1 have a strong influence on the accumulation of light harvesting carotenoids but play no role in xanthophyll cycle-related photoprotection. For instance, Li et al. (2024) reported that natural variations of VDL1 expression in different P. tricornutum strains enhance fucoxanthin accumulation, with none or only minor effects on the diadinoxanthin cycle and on the NPQ capacity. For these reasons, these genes represent potential engineering targets for the industrial production of fucoxanthin.
3.4 VDR: an unknown role in carotenoid biosynthesis
VDE-related (VDR) genes were first discovered in the green alga Chlamydomonas reinhardtii and are found in most photosynthetic eukaryotes, including land plants and diatoms (Coesel et al., 2008). VDR proteins share some similarities with the other VDE isoforms but lack characteristic cysteine residues potentially relevant for their enzymatic activity (Coesel et al., 2008; Olchawa-Pajor et al., 2019; Simionato et al., 2015). The sequences from P. tricornutum and the centric diatom Thalassiosira pseudonana (synonym of Cyclotella nana) showed significant similarity to the respective homologs of C. reinhardtii and land plants (Coesel et al., 2008), suggesting conserved properties albeit their exact function remains unresolved.
In P. tricornutum, overexpression of VDR was linked to increased xanthophyll production (Manfellotto et al., 2020), confirming that this paralog is also involved in carotenoid biosynthesis. However, silencing and knockout of VDE caused severe NPQ-deficiency phenotypes (Giossi et al., 2025; Lavaud et al., 2012), indicating that VDR is not involved in photoprotective xanthophyll cycling, while it may still catalyse some kind of de-epoxidation reaction. It was previously postulated that this isoform might also de-epoxidize a lipid-bound pool of diadinoxanthin under conditions of stronger light intensity and prolonged illumination (Lavaud et al., 2012). However, due to the complete absence of diatoxanthin in VDE knockout cultures exposed to extreme high light for several hours (Giossi et al., 2025) we can now reasonably exclude that VDR catalyses the de-epoxidation of diadinoxanthin to diatoxanthin.
4 Conclusion
Xanthophyll biosynthesis in diatoms has been gaining high interest due to the commercial relevance of pigments like fucoxanthin and diatoxanthin. In this context, the tuning of selected VDE and ZEP paralogs already proved successful for increasing carotenoid production and may represent an ideal target for future industrial purposes (Græsholt et al., 2024; Li et al., 2024; Manfellotto et al., 2020). The synthesis of diadinoxanthin is another essential step in fucoxanthin biosynthesis and represents a central hub for regulating the balance between photoprotective and light harvesting pigment pools (Bai et al., 2022). The discovery of the so far unknown diadinoxanthin synthase and of the putative acetyl transferase catalyzing the formation of haptoxanthin from allenoxanthin (Bai et al., 2022; Cao et al., 2023) would represent not only an interesting target for biocatalytic purposes in diatoms, but would also allow the engineering of fucoxanthin biosynthesis into other taxa for both industrial and research purposes.
The gene expansion of the ZEP-VDE cluster in diatoms opened the door to the development of new pathways but also increased the complexity of the regulation of xanthophyll biosynthesis. Indeed, compared to plants that employ VDE and ZEP only in the violaxanthin cycle, the activity of the diatom paralogs must be finely tuned to direct the pathway in response to specific needs (i.e., photoprotection or light harvesting). How this regulation is achieved is still poorly understood. Gene expression studies have highlighted the co-expression of some ZEP-VDE clusters under specific light treatments (Coesel et al., 2008; Nymark et al., 2009). This is not surprising, as light dynamics play a key role in the regulation of photosynthetic and photoprotective genes in diatoms (Madhuri et al., 2024; Manzotti et al., 2025; Zhang et al., 2024). Now that the pathway of fucoxanthin biosynthesis has been almost entirely elucidated, further studies focusing on the involved enzymes might provide new answers to questions related to their regulation.
To summarise, all VDE and ZEP isoforms evolved from the duplication of the ancestral genes devoted to the violaxanthin cycle in the common ancestor of Chromista, red and green algae, in a sequence of gene repurposing (Bai et al., 2022; Coesel et al., 2008; Dautermann and Lohr, 2017; Frommolt et al., 2008). This trend was essential for the development of new pigments like the major light harvesting xanthophyll fucoxanthin, driving the evolution of algae utilising new photosynthetic strategies that now dominate aquatic environments (Bai et al., 2022). This reservoir of adaptations and genetic diversity not only represents an exciting playground for industrial applications but might have also contributed to the widespread ecological success of diatoms.
Author contributions
CG: Conceptualization, Writing – original draft, Writing – review and editing. PK: Writing – review and editing. BL: Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. BL received funding from DFG (German Research Fundation, Bonn, Germany; LE 3358/3-2 and Heisenberg program). PK acknowledges funding from the University of Konstanz (Germany). CG received doctoral scholarship funding from Studienstiftung des Deutschen Volkes (Bonn, Germany) and the LGFG (Act on Graduate Funding of the State of Baden-Württemberg, University of Konstanz, Germany). Publication fees were covered by the Open-Access Funding initiative of the University of Konstanz (Germany).
Acknowledgments
We sincerely thank Martin Lohr for constructive criticism of the manuscript. This review forms part of a Ph.D. thesis submitted at the University of Konstanz (Germany) by CG.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arsalane, W., Rousseau, B., and Duval, J. (1994). Influence of the pool size of the xanthophyll cycle on the effects of light stress in a diatom: competition between photoprotection and photoinhibition. Photochem. Photobiol. 60, 237–243. doi:10.1111/j.1751-1097.1994.tb05097.x
Aziz, E., Batool, R., Akhtar, W., Rehman, S., Shahzad, T., Malik, A., et al. (2020). Xanthophyll: health benefits and therapeutic insights. Life Sci. 240, 117104. doi:10.1016/j.lfs.2019.117104
Bai, Y., Cao, T., Dautermann, O., Buschbeck, P., Cantrell, M. B., Chen, Y., et al. (2022). Green diatom mutants reveal an intricate biosynthetic pathway of fucoxanthin. Proc. Natl. Acad. Sci. 119, e2203708119. doi:10.1073/pnas.2203708119
Barnett, A., Méléder, V., Blommaert, L., Lepetit, B., Gaudin, P., Vyverman, W., et al. (2015). Growth form defines physiological photoprotective capacity in intertidal benthic diatoms. ISME J. 9, 32–45. doi:10.1038/ismej.2014.105
Bertrand, M. (2010). Carotenoid biosynthesis in diatoms. Photosynth. Res. 106, 89–102. doi:10.1007/s11120-010-9589-x
Bertrand, M., Schoefs, B., Siffel, P., Rohacek, K., and Molnar, I. (2001). Cadmium inhibits epoxidation of diatoxanthin to diadinoxanthin in the xanthophyll cycle of the marine diatom Phaeodactylum tricornutum. FEBS Lett. 508, 153–156. doi:10.1016/s0014-5793(01)03050-2
Bethmann, S., Melzer, M., Schwarz, N., and Jahns, P. (2019). The zeaxanthin epoxidase is degraded along with the D1 protein during photoinhibition of photosystem II. Plant Direct 3, e00185. doi:10.1002/pld3.185
Beyer, P., Al-Babili, S., Ye, X., Lucca, P., Schaub, P., Welsch, R., et al. (2002). Golden rice: introducing the β-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency. J. Nutr. 132, 506S-510S–510S. doi:10.1093/jn/132.3.506S
Blommaert, L., Chafai, L., and Bailleul, B. (2021). The fine-tuning of NPQ in diatoms relies on the regulation of both xanthophyll cycle enzymes. Sci. Rep. 11, 12750–16. doi:10.1038/s41598-021-91483-x
Bojko, M., Olchawa-Pajor, M., Tuleja, U., Kuczyńska, P., Strzałka, W., Latowski, D., et al. (2013). Expression of three diadinoxanthin de-epoxidase genes of Phaeodacylum tricornutum in Escherichia coli Origami b and BL21 strain. Acta Biochim. Pol. 60, 857–860. doi:10.18388/abp.2013_2072
Bratt, C. E., Arvidsson, P.-O., Carlsson, M., and Åkerlund, H.-E. (1995). Regulation of violaxanthin de-epoxidase activity by pH and ascorbate concentration. Photosynth. Res. 45, 169–175. doi:10.1007/BF00032588
Büch, K., Stransky, H., and Hager, A. (1995). FAD is a further essential cofactor of the NAD(P)H and O2 -dependent zeaxanthin-epoxidase. FEBS Lett. 376, 45–48. doi:10.1016/0014-5793(95)01243-9
Buck, J. M., Sherman, J., Bártulos, C. R., Serif, M., Halder, M., Henkel, J., et al. (2019). Lhcx proteins provide photoprotection via thermal dissipation of absorbed light in the diatom Phaeodactylum tricornutum. Nat. Commun. 10, 4167. doi:10.1038/s41467-019-12043-6
Butler, T., Kapoore, R. V., and Vaidyanathan, S. (2020). Phaeodactylum tricornutum: a diatom cell factory. Trends Biotechnol. 38, 606–622. doi:10.1016/j.tibtech.2019.12.023
Cao, T., Bai, Y., Buschbeck, P., Tan, Q., Cantrell, M. B., Chen, Y., et al. (2023). An unexpected hydratase synthesizes the green light-absorbing pigment fucoxanthin. Plant Cell 35, 3053–3072. doi:10.1093/plcell/koad116
Cazzaniga, S., Bressan, M., Carbonera, D., Agostini, A., and Dall’osto, L. (2016). Differential roles of carotenes and xanthophylls in photosystem i photoprotection. Biochemistry 55, 3636–3649. doi:10.1021/acs.biochem.6b00425
Coesel, S., Oborník, M., Varela, J., Falciatore, A., and Bowler, C. (2008). Evolutionary origins and functions of the carotenoid biosynthetic pathway in marine diatoms. PLoS ONE 3, e2896. doi:10.1371/journal.pone.0002896
Croteau, D., Jaubert, M., Falciatore, A., and Bailleul, B. (2025). Pennate diatoms make non-photochemical quenching as simple as possible but not simpler. Nat. Commun. 16, 2385. doi:10.1038/s41467-025-57298-4
Dautermann, O., and Lohr, M. (2017). A functional zeaxanthin epoxidase from red algae shedding light on the evolution of light-harvesting carotenoids and the xanthophyll cycle in photosynthetic eukaryotes. Plant J. 92, 879–891. doi:10.1111/tpj.13725
Dautermann, O., Lyska, D., Andersen-Ranberg, J., Becker, M., Fröhlich-Nowoisky, J., Gartmann, H., et al. (2020). An algal enzyme required for biosynthesis of the most abundant marine carotenoids. Sci. Adv. 6, eaaw9183. doi:10.1126/sciadv.aaw9183
DellaPenna, D., and Pogson, B. J. (2006). Vitamin synthesis in plants: tocopherols and carotenoids. Annu. Rev. Plant Biol. 57, 711–738. doi:10.1146/annurev.arplant.56.032604.144301
Demmig, B., Winter, K., Krüger, A., and Czygan, F. C. (1987). Photoinhibition and zeaxanthin formation in intact leaves: a possible role of the xanthophyll cycle in the dissipation of excess light energy. Plant Physiol. 84, 218–224. doi:10.1104/pp.84.2.218
Demmig, B., Winter, K., Krüger, A., and Czygan, F.-C. (1988). Zeaxanthin and the heat dissipation of excess light energy in Nerium oleander exposed to a combination of high light and water stress. Plant Physiol. 87, 17–24. doi:10.1104/pp.87.1.17
Demmig-Adams, B., López-Pozo, M., Stewart, J. J., and Adams, W. W. (2020). Zeaxanthin and lutein: photoprotectors, anti-inflammatories, and brain food. Molecules 25, 3607. doi:10.3390/molecules25163607
Eilers, U., Dietzel, L., Breitenbach, J., Büchel, C., and Sandmann, G. (2016). Identification of genes coding for functional zeaxanthin epoxidases in the diatom Phaeodactylum tricornutum. J. Plant Physiol. 192, 64–70. doi:10.1016/j.jplph.2016.01.006
Falkowski, P. G., Katz, M. E., Knoll, A. H., Quigg, A., Raven, J. A., Schofield, O., et al. (2004). The evolution of modern eukaryotic phytoplankton. Science 305, 354–360. doi:10.1126/science.1095964
Fernández-Marín, B., Roach, T., Verhoeven, A., and García-Plazaola, J. I. (2021). Shedding light on the dark side of xanthophyll cycles. New Phytol. 230, 1336–1344. doi:10.1111/nph.17191
Frommolt, R., Werner, S., Paulsen, H., Goss, R., Wilhelm, C., Zauner, S., et al. (2008). Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol. Biol. Evol. 25, 2653–2667. doi:10.1093/molbev/msn206
Giossi, C. E., Wünsch, M. A., Dautermann, O., Schober, A. F., Buck, J. M., Kroth, P. G., et al. (2025). Both major xanthophyll cycles present in nature promote non-photochemical quenching in a model diatom. Plant Physiol. kiaf371 199, kiaf371. doi:10.1093/plphys/kiaf371
Goss, R., and Jakob, T. (2010). Regulation and function of xanthophyll cycle-dependent photoprotection in algae. Photosynth. Res. 106, 103–122. doi:10.1007/s11120-010-9536-x
Goss, R., and Latowski, D. (2020). Lipid dependence of xanthophyll cycling in higher plants and algae. Front. Plant Sci. 11, 455–22. doi:10.3389/fpls.2020.00455
Goss, R., and Lepetit, B. (2015). Biodiversity of NPQ. J. Plant Physiol. 172, 13–32. doi:10.1016/j.jplph.2014.03.004
Goss, R., Ann Pinto, E., Wilhelm, C., and Richter, M. (2006). The importance of a highly active and DeltapH-regulated diatoxanthin epoxidase for the regulation of the PS II antenna function in diadinoxanthin cycle containing algae. J. Plant Physiol. 163, 1008–1021. doi:10.1016/j.jplph.2005.09.008
Græsholt, C., Brembu, T., Volpe, C., Bartosova, Z., Serif, M., Winge, P., et al. (2024). Zeaxanthin epoxidase 3 knockout mutants of the model diatom Phaeodactylum tricornutum enable commercial production of the bioactive carotenoid diatoxanthin. Mar. Drugs 22, 185. doi:10.3390/md22040185
Grouneva, I., Jakob, T., Wilhelm, C., and Goss, R. (2006). Influence of ascorbate and pH on the activity of the diatom xanthophyll cycle-enzyme diadinoxanthin de-epoxidase. Physiol. Plant. 126, 205–211. doi:10.1111/j.1399-3054.2006.00613.x
Hager, A. (1975). Die reversiblen, lichtabhängigen Xanthophyllumwandlungen im Chloroplasten. Berichte Dtsch. Bot. Ges. 88, 27–44. doi:10.1111/j.1438-8677.1975.tb02448.x
Holzmann, D., Bethmann, S., and Jahns, P. (2022). Zeaxanthin epoxidase activity is downregulated by hydrogen peroxide. Plant Cell Physiol. 63, 1091–1100. doi:10.1093/pcp/pcac081
Jahns, P., and Holzwarth, A. R. (2012). The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim. Biophys. Acta BBA - Bioenerg. 1817, 182–193. doi:10.1016/j.bbabio.2011.04.012
Jakob, T., Goss, R., and Wilhelm, C. (2001). Unusual pH-dependence of diadinoxanthin de-epoxidase activation causes chlororespiratory induced accumulation of diatoxanthin in the diatom Phaeodactylum tricornutum. J. Plant Physiol. 158, 383–390. doi:10.1078/0176-1617-00288
Karniel, U., Koch, A., Zamir, D., and Hirschberg, J. (2020). Development of zeaxanthin-rich tomato fruit through genetic manipulations of carotenoid biosynthesis. Plant Biotechnol. J. 18, 2292–2303. doi:10.1111/pbi.13387
Latowski, D., Kuczyńska, P., and Strzałka, K. (2011). Xanthophyll cycle – a mechanism protecting plants against oxidative stress. Redox Rep. 16, 78–90. doi:10.1179/174329211X13020951739938
Lavaud, J. (2007). Fast regulation of photosynthesis in diatoms: mechanisms, evolution and ecophysiology. Funct. Plant Sci. Biotechonology 1, 267–287.
Lavaud, J., Strzepek, R. F., and Kroth, P. G. (2007). Photoprotection capacity differs among diatoms: possible consequences on the spatial distribution of diatoms related to fluctuations in the underwater light climate. Limnol. Oceanogr. 52, 1188–1194. doi:10.4319/lo.2007.52.3.1188
Lavaud, J., Materna, A. C., Sturm, S., Vugrinec, S., and Kroth, P. G. (2012). Silencing of the violaxanthin de-epoxidase gene in the diatom Phaeodactylum tricornutum reduces diatoxanthin synthesis and non-photochemical quenching. PLoS ONE 7, e36806. doi:10.1371/journal.pone.0036806
Li, C., Pan, Y., Yin, W., Liu, J., and Hu, H. (2024). A key gene, violaxanthin de-epoxidase-like 1, enhances fucoxanthin accumulation in Phaeodactylum tricornutum. Biotechnol. Biofuels Bioprod. 17, 49. doi:10.1186/s13068-024-02496-3
Lohr, M., and Wilhelm, C. (1999). Algae displaying the diadinoxanthin cycle also possess the violaxanthin cycle. Proc. Natl. Acad. Sci. U. S. A. 96, 8784–8789. doi:10.1073/pnas.96.15.8784
Lohr, M., and Wilhelm, C. (2001). Xanthophyll synthesis in diatoms: quantification of putative intermediates and comparison of pigment conversion kinetics with rate constants derived from a model. Planta 212, 382–391. doi:10.1007/s004250000403
Madhuri, S., Lepetit, B., Fürst, A. H., and Kroth, P. G. (2024). A knockout of the photoreceptor PtAureo1a results in altered diel expression of diatom clock components. Plants Basel Switz. 13, 1465. doi:10.3390/plants13111465
Manfellotto, F., Stella, G. R., Falciatore, A., Brunet, C., and Ferrante, M. I. (2020). Engineering the unicellular alga Phaeodactylum tricornutum for enhancing carotenoid production. Antioxidants 9, 757. doi:10.3390/antiox9080757
Mann, V., Harker, M., Pecker, I., and Hirschberg, J. (2000). Metabolic engineering of astaxanthin production in tobacco flowers. Nat. Biotechnol. 18, 888–892. doi:10.1038/78515
Manzotti, A., Monteil, R., Cheminant Navarro, S., Croteau, D., Charreton, L., Hoguin, A., et al. (2025). Circadian regulation of key physiological processes by the RITMO 1 clock protein in the marine diatom Phaeodactylum tricornutum. New Phytol. 246, 1724–1739. doi:10.1111/nph.70099
Morelli, L., Patwari, P., Pruckner, F., Bastide, M., and Fabris, M. (2025). Specific light-regime adaptations, terpenoid profiles and engineering potential in ecologically diverse Phaeodactylum tricornutum strains. Algal Res. 86, 103920. doi:10.1016/j.algal.2025.103920
Moreno, J. C., Mi, J., Alagoz, Y., and Al-Babili, S. (2021). Plant apocarotenoids: from retrograde signaling to interspecific communication. Plant J. 105, 351–375. doi:10.1111/tpj.15102
Niyogi, K. K., Bjorkman, O., and Grossman, A. R. (1997). Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9, 1369–1380. doi:10.1105/tpc.9.8.1369
Niyogi, K. K., Grossman, A. R., and Björkman, O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10, 1121–1134. doi:10.1105/tpc.10.7.1121
Nymark, M., Valle, K. C., Brembu, T., Hancke, K., Winge, P., Andresen, K., et al. (2009). An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 4, e7743. doi:10.1371/journal.pone.0007743
Olaizola, M., La Roche, J., Kolber, Z., and Falkowski, P. G. (1994). Non-photochemical fluorescence quenching and the diadinoxanthin cycle in a marine diatom. Photosynth. Res. 41, 357–370. doi:10.1007/BF00019413
Olchawa-Pajor, M., Bojko, M., Strzalka, W., Strzalka, K., and Latowski, D. (2019). Violaxanthin conversion by recombinant diatom and plant de-epoxidases, expressed in Escherichia coli-a comparative analysis. Acta Biochim. Pol. 66, 249–255. doi:10.18388/abp.2019_2831
Rastogi, A., Maheswari, U., Dorrell, R. G., Vieira, F. R. J., Maumus, F., Kustka, A., et al. (2018). Integrative analysis of large scale transcriptome data draws a comprehensive landscape of Phaeodactylum tricornutum genome and evolutionary origin of diatoms. Sci. Rep. 8, 4834. doi:10.1038/s41598-018-23106-x
Reinhold, C., Niczyporuk, S., Beran, K. C., and Jahns, P. (2008). Short-term down-regulation of zeaxanthin epoxidation in Arabidopsis thaliana in response to photo-oxidative stress conditions. Biochim. Biophys. Acta BBA - Bioenerg. 1777, 462–469. doi:10.1016/j.bbabio.2008.03.002
Roy, S., Llewellyn, C., Egeland, E. S., and Johnsen, G. (2011). “Phytoplanton pigments - characterization,” in Chemotaxonomy and applications in oceanography pigments. Cambridge University Press.
Russo, M. T., Rogato, A., Jaubert, M., Karas, B. J., and Falciatore, A. (2023). Phaeodactylum tricornutum: an established model species for diatom molecular research and an emerging chassis for algal synthetic biology. J. Phycol. 59, 1114–1122. doi:10.1111/jpy.13400
Siefermann, D., and Yamamoto, H. Y. (1975). Properties of NADPH and oxygen-dependent zeaxanthin epoxidation in isolated chloroplasts. A transmembrane model for the violaxanthin cycle. Arch. Biochem. Biophys. 171, 70–77. doi:10.1016/0003-9861(75)90008-9
Simionato, D., Basso, S., Zaffagnini, M., Lana, T., Marzotto, F., Trost, P., et al. (2015). Protein redox regulation in the thylakoid lumen: the importance of disulfide bonds for violaxanthin de-epoxidase. FEBS Lett. 589, 919–923. doi:10.1016/j.febslet.2015.02.033
Smaoui, S., Barkallah, M., Ben Hlima, H., Fendri, I., Mousavi Khaneghah, A., Michaud, P., et al. (2021). Microalgae xanthophylls: from biosynthesis pathway and production techniques to encapsulation development. Foods 10, 2835. doi:10.3390/foods10112835
C. Stange (2016). Carotenoids in nature: biosynthesis, regulation and function, subcellular biochemistry (Cham: Springer International Publishing). doi:10.1007/978-3-319-39126-7
Stransky, H., and Hager, A. (1970). Das Carotinoidmuster und die Verbreitung des lichtinduzierten Xanthophyllcyclus in verschiedenen Algenklassen - VI. Chemosyst. Betracht. Arch. Für Mikrobiol. 73, 315–323. doi:10.1007/BF00412298
Van Der Straeten, D., Bhullar, N. K., De Steur, H., Gruissem, W., MacKenzie, D., Pfeiffer, W., et al. (2020). Multiplying the efficiency and impact of biofortification through metabolic engineering. Nat. Commun. 11, 5203. doi:10.1038/s41467-020-19020-4
Ware, M. A., Paton, A. J., Bai, Y., Kassaw, T., Lohr, M., and Peers, G. (2024). Identifying the gene responsible for non-photochemical quenching reversal in Phaeodactylum tricornutum. Plant J. 120, 2113–2126. doi:10.1111/tpj.17104
Yamamoto, H. Y., and Higashi, R. M. (1978). Violaxanthin de-epoxidase. Lipid composition and substrate specificity. Arch. Biochem. Biophys. 190, 514–522. doi:10.1016/0003-9861(78)90305-3
Yamamoto, H. Y., Nakayama, T. O. M., and Chichester, C. O. (1962). Studies on the light and dark interconversions of leaf xanthophylls. Arch. Biochem. Biophys. 97, 168–173. doi:10.1016/0003-9861(62)90060-7
Keywords: diatoms, carotenoid biosynthesis, violaxanthin de-epoxidase (VDE), xanthophyll cycle, zeaxanthin epoxidase (ZEP)
Citation: Giossi CE, Kroth PG and Lepetit B (2025) Xanthophyll cycling and fucoxanthin biosynthesis in the model diatom Phaeodactylum tricornutum: recent advances and new gene functions. Front. Photobiol. 3:1680034. doi: 10.3389/fphbi.2025.1680034
Received: 05 August 2025; Accepted: 17 September 2025;
Published: 01 October 2025.
Edited by:
Alessandra Rogato, National Research Council (CNR), ItalyReviewed by:
Maxwell A. Ware, Free University of Berlin, GermanyCopyright © 2025 Giossi, Kroth and Lepetit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiara E. Giossi, Y2hpYXJhLmdpb3NzaUBoaHUuZGU=
†Present address: Chiara E. Giossi, Photosynthesis and Stress Physiology of Plants, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
 Chiara E. Giossi
Chiara E. Giossi Peter G. Kroth
Peter G. Kroth Bernard Lepetit
Bernard Lepetit