- 1Department of Pharmacy, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Institute of Clinical Pharmacy, Central South University, Changsha, China
- 3Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha, China
- 4Hunan Key Laboratory of Pharmacogenetics, Institute of Clinical Pharmacology, Central South University, Changsha, China
- 5Department of Family Medicine, Faculty of Medicine and Dentistry, University of Alberta, Edmonton, AB, Canada
Background and Objective: Apolipoprotein E (APOE) plays important roles in lipoprotein metabolism and cardiovascular disease. Evidence suggests the APOE gene epsilon2/epsilon3/epsilon4 (ε2/ε3/ε4) polymorphisms might be associated with the susceptibility of coronary artery disease (CAD) in patients with type 2 diabetes mellitus (T2DM). However, no clear consensus has yet been established. Therefore, the aim of this meta-analysis is to provide a precise conclusion on the potential association between APOE ε2/ε3/ε4 polymorphisms and the risk of CAD in patients with T2DM based on case-control studies.
Methods: Pubmed, Embase, Chinese National Knowledge Infrastructure (CNKI), and Wanfang databases were searched for all relevant studies prior to August 2017 in English and Chinese language. The pooled odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) were used to assess the strength of the relationships. The between-study heterogeneity was evaluated by Cochran's Q-test and the I2 index to adopt fixed- or random- effect models.
Results: A total of 13 studies were eligible for inclusion. There was evidence for significant associations between APOE ε4 mutation and the risk of CAD in patients with T2DM (for ε3/ε4 vs. ε3/ε3: OR = 1.69, 95% CI = 1.38–2.08, P < 0.001; for ε4/ε4 vs. ε3/ε3: OR = 2.72, 95% CI = 1.61–4.60, P < 0.001; for ε4/ε4+ε3/ε4 vs. ε3/ε3: OR = 1.83, 95% CI = 1.52–2.22, P < 0.001; for ε4 allele vs. ε3 allele: OR = 1.64, 95% CI = 1.40–1.94, P < 0.001). In contrast, no significant associations were found in genetic model of APOE ε2 mutation (for ε2/ε2 vs. ε3/ε3: OR = 1.67, 95% CI = 0.90–3.09, P = 0.104; for ε2/ε3 vs. ε3/ε3: OR = 1.18, 95% CI = 0.93–1.51, P = 0.175; for ε2/ε2+ε2/ε3 vs. ε3/ε3: OR = 1.26, 95% CI = 0.88–1.82, P = 0.212; for ε2 allele vs. ε3 allele: OR = 1.34, 95% CI = 0.98–1.84, P = 0.07).
Conclusions: The APOE gene ε4 mutation is associated with an increased risk of CAD in patients with T2DM, while the ε2 variation has null association with this disease.
Introduction
Type 2 diabetes mellitus (T2DM) is a long-term metabolic disease with a high incidence and prevalence in the world. T2DM is often accompanied by various complications such as hypertension, dyslipidemia and coronary artery disease (CAD) (Naito and Miyauchi, 2017). As the disease progresses, patients with T2DM have a 2 to 4-fold increased risk for developing CAD compared with non-diabetic individuals (Mohan et al., 2001; Emerging Risk Factors et al., 2010). In addition, cardiovascular disease including CAD in patients with T2DM is associated with significant mortality (Zhang et al., 2014b; Freitas Lima et al., 2015). Therefore, early prevention and vigorous control of T2DM and its complications are becoming an ever-increasing global health priority. A better understanding of the etiology of CAD in patients with T2DM will result in better clinical management.
Dyslipidemia, hypertension, obesity, and smoking status are well-established risk factors for T2DM (Paneni et al., 2013; Wang et al., 2015a). Additionally, human genetic association studies have revealed that numerous genetic mutations and polymorphisms also play a critical role (Wei et al., 2014; Raj et al., 2015; Sumi et al., 2017). Among the previous studies, apolipoprotein E (APOE) gene has been regarded as one of the most likely candidate genes which may be associated with CAD in T2DM patients.
APOE is a class of plasma apolipoprotein totaling 299 amino acids, and it is involved in lipoprotein metabolism and the development of cardiovascular diseases (Zheng et al., 1998). The APOE gene is mapped to chromosome 19q13.2 in a cluster with apolipoprotein C1 and C2 gene, and it consists of three introns and four exons. APOE is a polymorphic gene and the most commonly studied alleles/isoforms are: epsilon2 (ε2), epsilon3 (ε3), and epsilon4 (ε4). The differences between the three isoforms are the location of 112 and 158 in the amino acid chain where cysteine or arginine is present. These three APOE alleles are determined by the rs7412 and rs429358 single-nucleotide polymorphisms. The three alleles, APOE-ε2 (cys112 and cys158), APOE-ε3 (cys112 and arg158) and APOE-ε4 (arg112 and arg158), yield six different genotypes for the APOE gene: ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4, and ε4/ε4. Because the ε3 allele or ε3/ε3 genotype is the most common allele or genotype among the population, they are well accepted as the “wild-type” and used as the “reference” in the genetic models (Zhang et al., 2000; Guo et al., 2007; Izar et al., 2009; Chaudhary et al., 2012; Hong et al., 2017).
The role of APOE ε2/ε3/ε4 polymorphisms in the development of CAD in patients with T2DM is widely studied, but the results are still controversial and conflicting. In 1998, Zheng et al. firstly investigated the association between APOE gene polymorphism and T2DM complicated with CAD in the Chinese population. The results showed that APOE-ε4 allele increased the risk of CAD in T2DM (Zheng et al., 1998). Other studies have also confirmed Zheng's findings (Chaaba et al., 2008; Hong et al., 2017). However, APOE-ε2 allele was also found to be associated with the risk of CAD in T2DM (Halim et al., 2012). In addition, no significant association between APOE ε2/ε3/ε4 polymorphisms and the risk of CAD in T2DM was reported in some studies (Zhang et al., 2000; Guo et al., 2007; Izar et al., 2009). To demonstrate with certainty the associations between the APOE ε2/ε3/ε4 polymorphisms and the risk of CAD in patients with T2DM, we conducted a systematic review and meta-analysis on published case-control studies.
Materials and Methods
Literature Search
This study was undertaken according to the methodology of MOOSE (Meta-analysis of Observational Studies in Epidemiology) statement (Stroup et al., 2000). We thoroughly searched all published studies in the Embase, PubMed, China National Knowledge Infrastructure (CNKI) and Wanfang databases up to August 2, 2017. The included articles were limited to Chinese and English language. The following keywords were used for searching: “apolipoprotein E” OR “APOE” AND “polymorphism” OR “single nucleotide polymorphism” OR “SNP” OR “variant” OR “variation” AND “coronary artery disease” OR “coronary heart disease” OR “CAD” OR “CHD” OR “atherosclerosis” OR “myocardial infarction” OR “myocardial infarct” OR “heart attack” OR “MI” AND “type 2 diabetes” OR “non-insulin dependent diabetes mellitus” OR “diabetes mellitus, type 2” OR “diabetes, type 2” OR “diabetes mellitus, non-insulin dependent” The Chinese databases were searched using the equivalent Chinese terms. In addition, hand searches for all related articles were performed. The detailed search strategies are presented in Supplementary Table 1.
Inclusion and Exclusion Criteria
The first two investigators independently accessed the eligibility of the studies by screening the title, abstract and full-text, based on the inclusion and exclusion criteria. The inclusion criteria for all studies were as follows: (1) study on the associations between APOE ε2/ε3/ε4 polymorphisms and CAD in patients with T2DM, regardless of sample size. (2) case-control design. (3) detailed data for the APOE alleles or genotype distribution in case and control groups to estimate odds ratio (OR) with 95% confidence interval (CI). Exclusion criteria: (1) duplication of previous data; (2) review, comment and editorial; (3) no sufficient genotype data. Any dispute about the eligibility of an article was resolved by discussion.
Data Extraction
The data was drawn out based on a standard protocol. The following information was carefully extracted from all eligible publications independently by two authors (JQL and HR) using a standardized form: last name of first author, year of publication, study country, sample size in cases and controls, methods of genotyping, number genotypes and alleles. If similar data sets presented in different articles by the same research group, the data would be adopted only once. The collected data were compared, and possible disagreements were discussed by the authors and resolved with consensus.
Quality Score Assessment
The study quality was independently assessed by two reviewers. Quality assessment of genetic associations between APOE ε2/ε3/ε4 polymorphisms and CAD in patients with T2DM is described in the Supplementary Table 2. The scores were adjusted according to the criteria developed for meta-analysis of molecular association studies by Thakkinstian et al. (2005). The total scores ranged from 0 to 13, with 13 representing the highest quality.
Statistics Analysis
All the statistical analysis in this study was performed using Stata 12.0 (StataCorp, College Station, TX). Hardy-Weinberg equilibrium was performed in control groups using the chi-square test. The combined OR and 95%CI were calculated to evaluate the strength of the association between the APOE ε2/ε3/ε4 polymorphisms and risk of CAD in T2DM subjects. The pooled ORs were, respectively, performed for nine genetic models (ε2/ε2 vs. ε3/ε3; ε2/ε3 vs. ε3/ε3; ε2/ε4 vs. ε3/ε3; ε3/ε4 vs. ε3/ε3; ε4/ε4 vs. ε3/ε3; ε2 allele vs. ε3 allele; ε4 allele vs. ε3 allele; ε2/ε2+ε2/ε3 vs. ε3/ε3; ε4/ε4+ε3/ε4 vs. ε3/ε3). The statistically significant level of the combined OR was determined by the Z-test with P < 0.05. Heterogeneity between studies was calculated by using the Cochran's Q-test and Higgins I2 index. In the absence of between-study heterogeneity (I2 < 50%), the fixed effect model (Mantel–Haenszel method) was chosen to calculate the pooled estimates. Otherwise, random effect model (DerSimonian and Laird method) would be adopted if the I2 > 50% (Higgins et al., 2003). Subgroup analysis was performed according to the source of patients (Chinese and non-Chinese). Galbraith plot analysis and sensitivity analysis were conducted to detect whether there were outliers that could be the potential sources of heterogeneity between studies when heterogeneity was moderately large. Publication bias was evaluated by Begg's funnel plot and Egger's regression test (Begg and Mazumdar, 1994; Egger et al., 1997). If there is evidence of significant publication bias, the trim and fill method was performed to assess the potential influence of publication bias (Duval and Tweedie, 2000).
Results
The Characteristics of the Included Studies
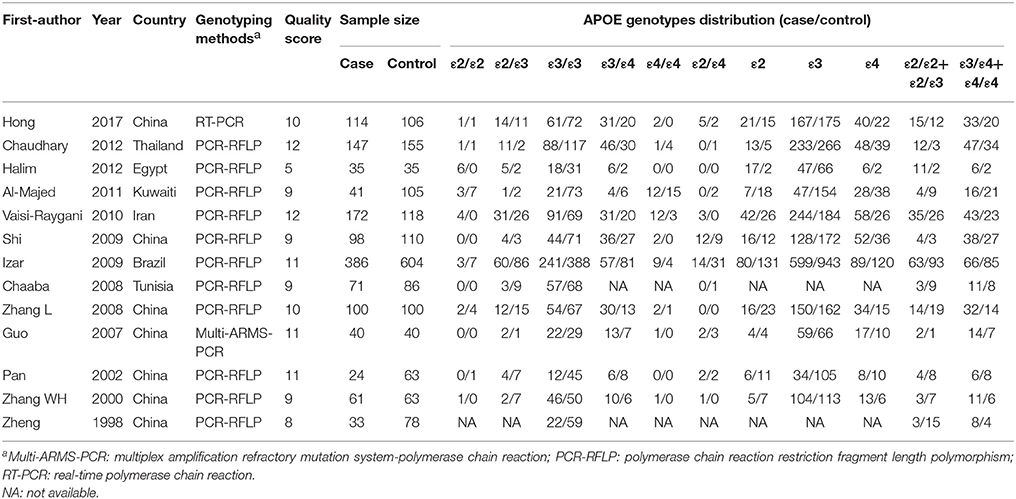
As depicted in Figure 1, a total of 222 articles were obtained by online search, and 2 articles were included by manual search. After removing duplicates, 175 articles were included. After screening title and abstract, 115 articles were excluded. As a result, 13 articles (Zheng et al., 1998; Zhang et al., 2000, 2008; Pan et al., 2002; Guo et al., 2007; Chaaba et al., 2008; Izar et al., 2009; Shi et al., 2009; Vaisi-Raygani et al., 2010; Al-Majed et al., 2011; Chaudhary et al., 2012; Halim et al., 2012; Hong et al., 2017) were eligible for the meta-analysis. The characteristics of the included articles are summarized in Table 1. The included studies were conducted in several countries including China, Brazil, Thailand, Egypt, Iran, Kuwait, and Tunisia. All studies were performed in a case-control design and the sample sizes varied from 70 to 990. The quality score of the included studies ranged from 5 to 12 (mean: 9.69) out of a maximal score of 13.
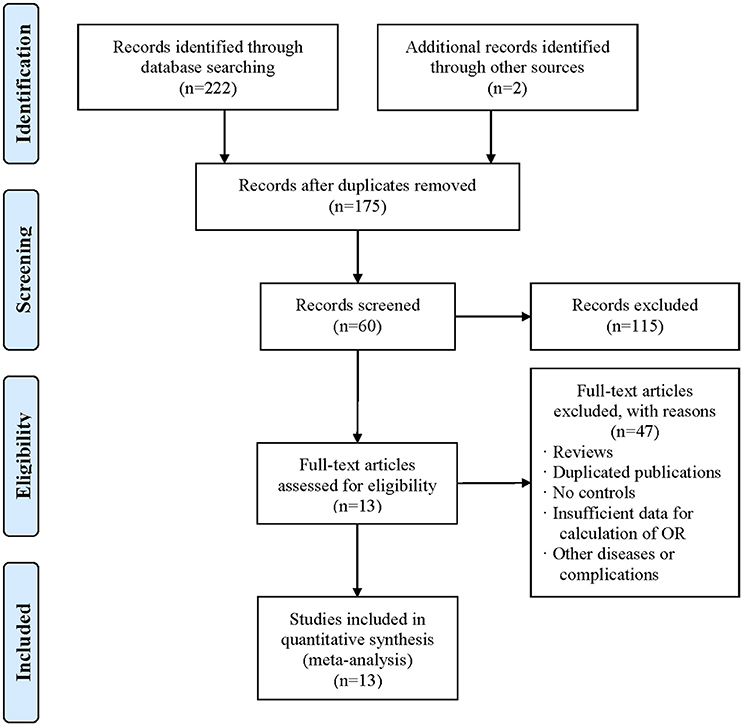
Figure 1. Flow diagram of study selection process. The term “n” in the boxes represens the number of corresponding studies.
Quantitative Synthesis
The main results of this meta-analysis for the association between APOE ε2/ε3/ε4 polymorphisms and the risk of CAD in T2DM patients are presented in Table 2. There was significant association in three genetic models which demonstrate, ε4 mutation contributed to an increased risk of CAD in patients with T2DM (Figure 2). The pooled results for the three genetic models in the overall analysis were as follows: for ε3/ε4 vs. ε3/ε3: OR = 1.69, 95% CI = 1.38–2.08, P < 0.001; for ε4/ε4 vs. ε3/ε3: OR = 2.72, 95% CI = 1.61–4.60, P < 0.001; for ε4/ε4+ε3/ε4 vs. ε3/ε3: OR = 1.83, 95% CI = 1.52–2.22, P < 0.001. In contrast, the ε2 variation had null association with this disease (Figure 3). No significant association in the overall analysis was found in genetic model of ε2/ε2 vs. ε3/ε3 (OR = 1.67, 95% CI = 0.90–3.09, P = 0.104); ε2/ε3 vs. ε3/ε3 (OR = 1.18, 95% CI = 0.93–1.51, P = 0.175); ε2/ε4 vs. ε3/ε3 (OR = 1.20, 95% CI = 0.78–1.84, P = 0.405); ε2/ε2+ε2/ε3 vs. ε3/ε3 (OR = 1.26, 95% CI = 0.88–1.82, P = 0.212). In addition, the genetic models of allele-based contrasts in the overall analysis also revealed a statistically significant pooled estimates for ε4 allele vs. ε3 allele (OR = 1.64, 95% CI = 1.40–1.94, P < 0.001) but not for ε2 allele vs. ε3 allele (OR = 1.34, 95% CI = 0.98–1.84, P = 0.07).
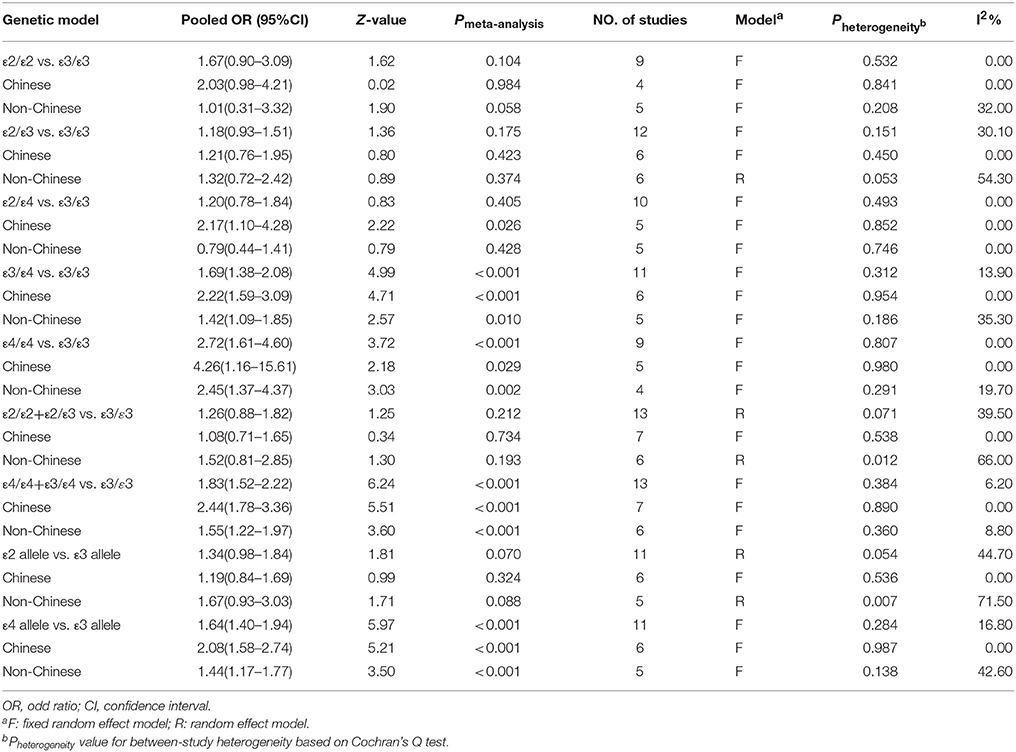
Table 2. Meta-analysis results of the associations between APOE ε2/ε3/ε4 polymorphisms and risk of coronary artery diseases in type 2 diabetes patients.
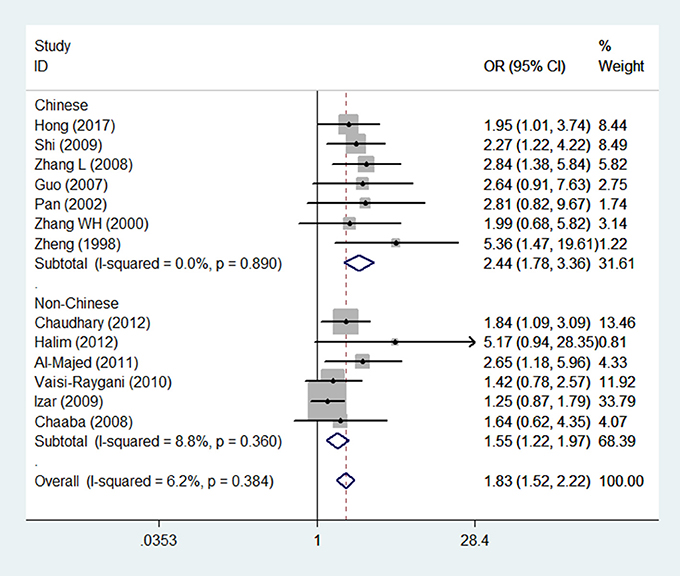
Figure 2. Forest plot for the association between APOE gene polymorphism and the risk of coronary artery diseases in type 2 diabetes patients under the genetic model of ε4/ε4+ε3/ε4 vs. ε3/ε3. The center of each square represents the OR, the area of the square is for the weight of studies, and the horizontal line indicates the 95% CI.
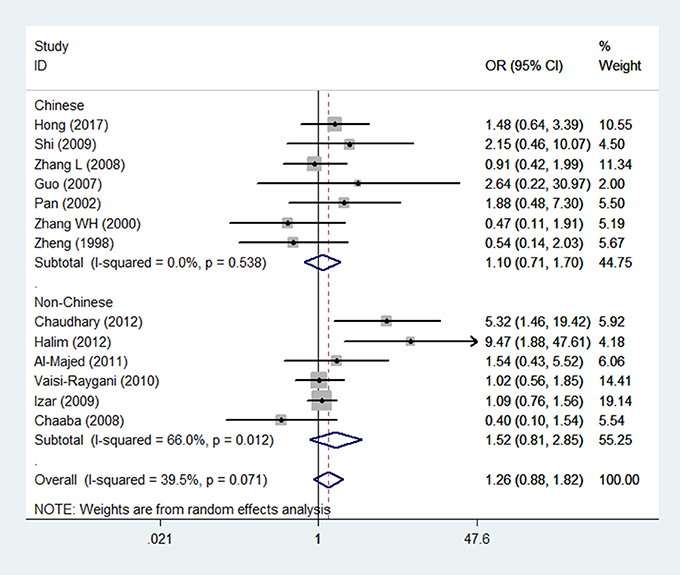
Figure 3. Forest plot for the association between APOE gene polymorphism and the risk of coronary artery diseases in type 2 diabetes patients under the genetic model of ε2/ε2+ε2/ε3 vs. ε3/ε3. The center of each square represents the OR, the area of the square is for the weight of studies, and the horizontal line indicates the 95% CI.
In the subgroup analysis according to the source of patients (Chinese and non-Chinese), the pooled ORs of all genetic models except the ε2/ε4 vs. ε3/ε3 model were consistent with the results in the overall population. In the Chinese population, the ε2/ε4 genotype increased the risk of CAD in patients with T2DM (OR = 2.17, 95% CI = 1.10–4.28, P = 0.026).
Heterogeneity Analysis
As shown in Table 2, there was moderate between-study heterogeneity in the genetic model of ε2 allele vs. ε3 allele (Pheterogeneity = 0.054, I2 = 44.70%) and ε2/ε2+ε2/ε3 vs. ε3/ε3 (Pheterogeneity = 0.071, I2 = 39.50%) in the overall analysis. However, no significant heterogeneity was found in other genetic models (for ε2/ε2 vs. ε3/ε3: Pheterogeneity = 0.532, I2 = 0%; for ε2/ε3 vs. ε3/ε3: Pheterogeneity = 0.151, I2 = 30.10%; for ε2/ε4 vs. ε3/ε3: Pheterogeneity = 0.493, I2 = 0%; for ε3/ε4 vs. ε3/ε3: Pheterogeneity = 0.312, I2 = 13.90%; for ε4/ε4 vs. ε3/ε3: Pheterogeneity = 0.807, I2 = 0%; for ε4 allele vs. ε3 allele: Pheterogeneity = 0.284, I2 = 16.80%; for ε4/ε4+ε3/ε4 vs. ε3/ε3: Pheterogeneity = 0.384, I2 = 6.20%). The heterogeneity analysis results indicated that the pooled results of this meta-analysis in most genetic models were statistically steady and robust. In addition, subgroup analysis indicated that there was no heterogeneity under all nine genetic models in the Chinese population.
Galbraith Plot Analysis and Sensitivity Analysis
There was evidence of moderately large between-study heterogeneity in the genetic model of ε2 allele vs. ε3 allele (Pheterogeneity = 0.054, I2 = 44.70%) and ε2/ε2+ε2/ε3 vs. ε3/ε3 (Pheterogeneity = 0.071, I2 = 39.50%), so Galbraith plot analysis and sensitivity analysis were performed to detect the possible sources of heterogeneity. Under the genetic model of ε2 allele vs. ε3 allele, the Galbraith plot analysis (Figure 4A) showed that the Halim et al. study was the outlier, which is consistent with the results of sensitivity analysis (Figure 4B). No heterogeneity existed after this outlier study was omitted (Pheterogeneity = 0.460, I2 = 0%). Thus, the study by Halim et al. may be the source of heterogeneity in the meta-analysis for the ε2 allele vs. ε3 genetic model.
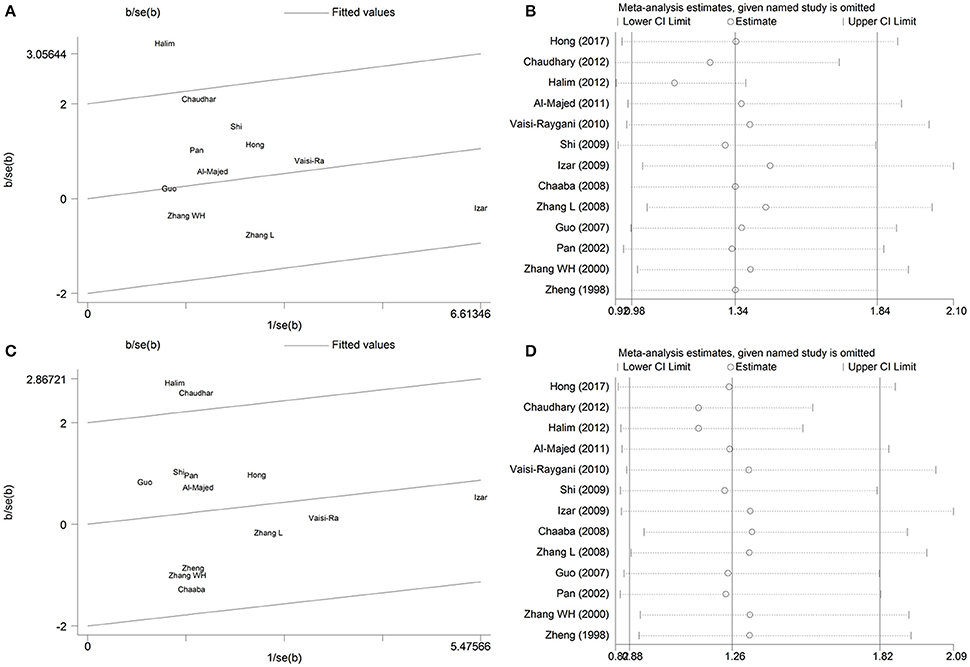
Figure 4. Galbraith plot analysis and sensitivity analysis of the association between APOE gene polymorphism and the risk of coronary artery diseases in type 2 diabetes patients under the genetic model of ε2 allele vs. ε3 allele (A,B) and ε2/ε2+ε2/ε3 vs. ε3/ε3 (C,D). For sensitivity analysis, open circle indicates the pooled ORs, horizontal lines represent the 95% CIs, given named study is omitted.
Similarly, under the genetic model of ε2/ε2+ε2/ε3 vs. ε3/ε3, the Galbraith plot analysis (Figure 4C) and sensitivity analysis (Figure 4D) indicated that Halim and Chaudhary's study were the outliers. When the two outlier studies were omitted, no heterogeneity existed in the remaining studies (Pheterogeneity = 0.681, I2 = 0%). Therefore, the studies of Halim et al. and Chaudhary et al. may be the main contributors to the source of heterogeneity in the meta-analysis for the ε2/ε2+ε2/ε3 vs. ε3/ε3 genetic model.
Publication Bias
No obvious asymmetry was observed in the shape of the funnel plot for the following genetic models: ε2/ε2 vs. ε3/ε3 (Figure 5A); ε2/ε3 vs. ε3/ε3 (Figure 5B); ε2/ε4 vs. ε3/ε3 (Figure 5C); ε4/ε4 vs. ε3/ε3 (Figure 5D); ε2 allele vs. ε3 allele (Figure 5E); ε2/ε2+ε2/ε3 vs. ε3/ε3 (Figure 5F). In addition, the Begg's test and Egger's test also did not show any evidence of publication bias (PBegg = 0.251 and PEgger = 0.08 for ε2/ε2 vs. ε3/ε3, PBegg = 0.373 and PEgger = 0.320 for ε2/ε3 vs. ε3/ε3, PBegg = 0.283 and PEgger = 0.403 for ε2/ε4 vs. ε3/ε3, PBegg = 0.466 and PEgger = 0.988 for ε4/ε4 vs. ε3/ε3, PBegg = 0.119 and PEgger = 0.053 for ε2 allele vs. ε3 allele, PBegg = 0.300 and PEgger = 0.331 for ε2/ε2+ε2/ε3 vs. ε3/ε3).
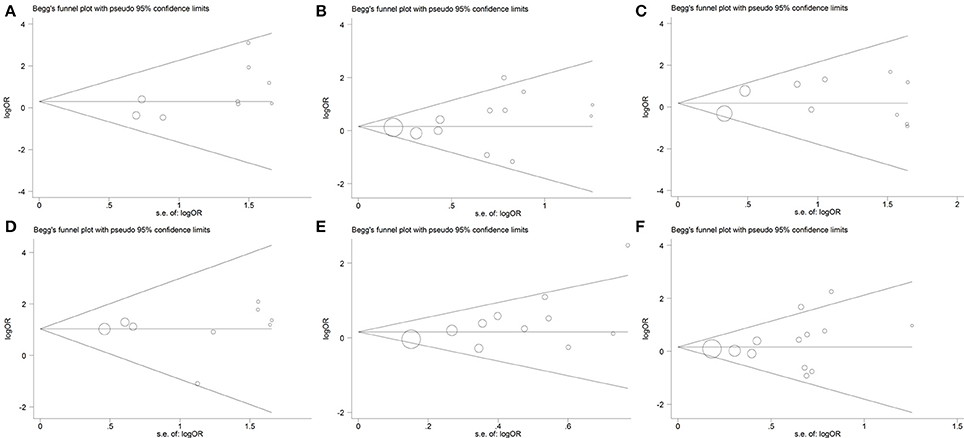
Figure 5. Begg's funnel plot for the association between APOE gene polymorphism and the risk of coronary artery diseases in type 2 diabetes patients under the genetic model of ε2/ε2 vs. ε3/ε3 (A), ε2/ε3 vs. ε3/ε3 (B), ε2/ε4 vs. ε3/ε3 (C), ε4/ε4 vs. ε3/ε3 (D), ε2 allele vs. ε3 allele (E), and ε2/ε2+ε2/ε3 vs. ε3/ε3 (F). Size of the open circles is proportional to the weight of studies.
The results from the following three genetic models ε3/ε4 vs. ε3/ε3; ε3/ε4+ε4/ε4 vs. ε3/ε3 and ε4 allele vs. ε3 allele performed by Begg's test (PBegg = 0.213, PBegg = 0.033, and PBegg = 0.043, respectively) or Egger's test (PEgger = 0.013; PEgger = 0.001 and PEgger = 0.001, respectively) revealed publication bias. Nevertheless, by using the trim and fill method, the recalculated estimates (OR = 1.50, 95%CI = 1.24–1.82; OR = 1.59, 95%CI = 1.34–1.89 and OR = 1.40, 95%CI = 1.22–1.62, respectively) remained statistically significant, which indicated that our meta-analysis results were steady and not influenced by publication bias. Figure 6 shows the funnel plot of trim and fill method in the genetic model of ε3/ε4 vs. ε3/ε3 (Figure 6A), ε3/ε4+ε4/ε4 vs. ε3/ε3 (Figure 6B), ε4 allele vs. ε3 allele (Figure 6C).
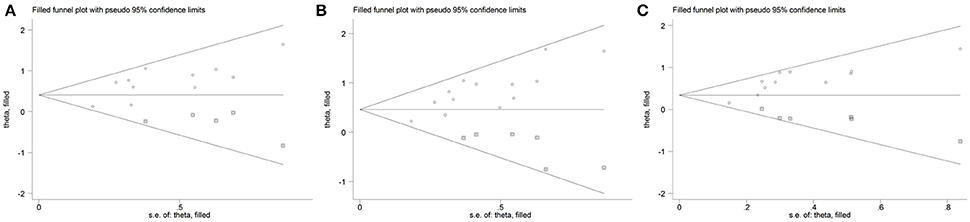
Figure 6. Funnel plot with trim and fill method for the association between APOE gene polymorphism and the risk of coronary artery diseases in type 2 diabetes patients under the genetic model of ε3/ε4 vs. ε3/ε3 (A), ε4/ε4+ε3/ε4 vs. ε3/ε3 (B), and ε4 allele vs. ε3 allele (C). Circle represents the included studies; Square represents the possibly missing studies.
Discussion
T2DM is a well-established risk factor for the development of CAD. The management of CAD in patients with T2DM poses great challenges to the medical profession (Wei et al., 2015). The identification of susceptibility genes would be very helpful for the management of CAD in patients with T2DM. The link between APOE ε2/ε3/ε4 polymorphisms and CAD in diabetic patients has been highlighted in our study. This meta-analysis provides evidence for the significant associations between APOE ε4 mutation (ε3/ε4 vs. ε3/ε3; ε4/ε4 vs. ε3/ε3; ε4/ε4+ε3/ε4 vs. ε3/ε3; ε4 allele vs. ε3 allele) and an elevated risk of CAD in patients with T2DM. In contrast, no significant association was found in genetic model of APOE ε2 variation (ε2/ε2 vs. ε3/ε3; ε2/ε3 vs. ε3/ε3; ε2/ε2+ε2/ε3 vs. ε3/ε3; ε2 allele vs. ε3 allele). However, CAD in patients with T2DM is believed to be multifactorial and involved in many susceptibility genes with small individual effects. Therefore, the integration of information derived from several polymorphisms in multiple susceptibility genes may become clinically useful.
It has been reported that lipoprotein-related mechanisms are associated with the impairment of the cardiovascular system among patients with diabetes (Jenkins et al., 2004). For example, serum low-density lipoprotein cholesterol (LDL-C) level was identified as an independent risk factor for CAD in T2DM patients (Jayashankar et al., 2016). APOE is initially recognized for its important role in plasma lipid metabolism and thus affects the serum lipid profiles in the body. The three APOE alleles (ε2, ε3, ε4) differ from each other by only one or two amino acids at positions 112 and 158, but these slight differences alter the structure and function of APOE. In general, the APOE-ε4 allele is associated with higher and the APOE-ε2 allele with lower total plasma cholesterol and LDL-C concentrations compared with the APOE-ε3 allele (Bennet et al., 2007; Larifla et al., 2017). Therefore, abnormalities of lipoprotein metabolism may explain, at least in part, the associations between APOE ε2/ε3/ε4 polymorphisms and the risk of CAD in patients with T2DM.
Several meta-analysis studies have been conducted to assess the association between APOE ε2/ε3/ε4 polymorphisms and risk of CAD in the general population. In 2004, Song et al firstly found that carriers of the APOE-ε4 allele had a 42% increased risk for CAD (OR = 1.42, 95% CI = 1.26–1.61) compared with the ε3/ε3 genotypes (Song et al., 2004). Xu et al. found similar results which showed that the ε4 allele had a 46% higher risk of CAD (OR = 1.46, 95% CI = 1.28–1.66) (Xu et al., 2016). Similar findings were also observed in other meta-analysis (Yin et al., 2013; Xu et al., 2014, 2016; Zhang et al., 2014a, 2015; Wang et al., 2015b). Interestingly, the role of APOE-ε2 allele in the risk of CAD may be dependent on the patient ethnicity (Xu et al., 2016). In addition, the association between APOE ε2/ε3/ε4 polymorphisms and the risk of T2DM in the general population was also well explored in previous meta-analysis (Anthopoulos et al., 2010; Yin et al., 2014). The results indicated that both APOE ε2 and ε4 alleles were associated with an increased risk of T2DM in the general population. In 2015, Wu et al. performed a meta-analysis on the association between APOE ε2/ε3/ε4 polymorphisms and T2DM patients with CAD among Chinese Han population. They found that APOE-ε4 allele resulted in an increased risk of T2DM patients with CAD in China (Wu et al., 2015). However, only five individual studies were included in their meta-analysis. To our knowledge, our meta-analysis represents the largest study to investigate the association between APOE ε2/ε3/ε4 polymorphisms and risk of CAD in the T2DM patients.
Heterogeneity across studies is common in meta-analysis of genetic association study (Munafo and Flint, 2004). Heterogeneity should be taken into consideration in the interpretation of the meta-analysis results. However, one of the strengths in this meta-analysis was the lack of significant heterogeneity in all genetic models except the genetic model of ε2 allele vs. ε3 allele. Between-study heterogeneity can be attributed to the potential differences such as the definition of disease, ethnicity, genotyping methods and sample size in the included studies. To explore the potential sources of heterogeneity under the genetic model of ε2 allele vs. ε3 allele, Galbraith plot analysis and sensitivity analysis were employed to detect whether there were outliers that could be the potential sources of heterogeneity between studies. The study conducted by Halim et al was considered as the main contributors to between-study heterogeneity. The heterogeneity was effectively decreased after omitting the study. The frequency of APOE-ε3 allele was nearly 95% in Halim's study, whereas lower than 90% in other studies (Zhang et al., 2008; Izar et al., 2009; Chaudhary et al., 2012; Hong et al., 2017). Consequently, the heterogeneity can be due to the distinct frequency of APOE ε2/ε3/ε4 polymorphisms among the included studies. Although Halim's study caused the substantial heterogeneity in the genetic model of ε2 allele vs. ε3 allele, the pooled effect was still insignificant after removing it.
There are several limitations in this meta-analysis that should be noted. First, the included studies were limited to only English or Chinese languages in our research and some eligible studies may be published in other languages, which would cause bias of the results. Second, all the included studies in this meta-analysis were the type of retrospective case-control studies, which may result in some selection bias. Third, publication bias existed in the following three genetic models: ε3/ε4 vs. ε3/ε3; ε3/ε4+ε4/ε4 vs. ε3/ε3; ε4 allele vs. ε3 allele. However, by using the trim and fill method, the recalculated ORs and their 95% CIs did not change, which indicated the stability and robustness of meta-analysis results. Last but not the least, T2DM complicated with CAD is a multifactorial disease caused by both genetic and environmental factors. The APOE-environment interactions should be considered. For example, the study by Talmud et al. has found that the impact of the APOE-ε4 on the risk of CAD appeared to be restricted to smokers (Talmud et al., 2004).
In conclusion, we observed a significant association between the APOE gene ε4 mutation and an increased risk of CAD in patients with T2DM, while the ε2 variation had null association with this disease. Taking into account the above limitations, more studies with larger sample size and incorporated with gene-environment interactions are needed to definitively determine the association between the APOE gene ε2/ε3/ε4 polymorphisms and the risk of CAD in patients with T2DM.
Author Contributions
Conceived and designed the study: J-QL and HR. Performed the search: J-QL, HR, M-ZL. Analyzed the data: J-QL and HR. Contributed reagents/material/analysis tools: J-QL, HR, M-ZL, PX, P-FF, and D-XX. Wrote and review the manuscript: J-QL, HR and HB. Reference collection, data management, statistical analyses, paper writing, and study design: J-QL and HR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (NO. 81703623).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2017.01031/full#supplementary-material
References
Al-Majed, H. T., Qasem, J. A., Al-Sherifi, A. K., Al-Attar, A. A., Qasem, A. A., and Abdullah, S. A. (2011). Association between apolipoprotein E-polymorphism and Ischemic heart disease patients with or without type 2 diabetes mellitus: a preliminary study in Kuwait. Arch. Iran. Med. 14, 385–388.
Anthopoulos, P. G., Hamodrakas, S. J., and Bagos, P. G. (2010). Apolipoprotein E polymorphisms and type 2 diabetes: a meta-analysis of 30 studies including 5423 cases and 8197 controls. Mol. Genet. Metab. 100, 283–291. doi: 10.1016/j.ymgme.2010.03.008
Begg, C. B., and Mazumdar, M. (1994). Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101. doi: 10.2307/2533446
Bennet, A. M., Di Angelantonio, E., Ye, Z., Wensley, F., Dahlin, A., Ahlbom, A., et al. (2007). Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA 298, 1300–1311. doi: 10.1001/jama.298.11.1300
Chaaba, R., Attia, N., Hammami, S., Smaoui, M., Ben Hamda, K., Mahjoub, S., et al. (2008). Association between apolipoprotein E polymorphism, lipids, and coronary artery disease in Tunisian type 2 diabetes. J. Clin. Lipidol. 2, 360–364. doi: 10.1016/j.jacl.2008.08.441
Chaudhary, R., Likidlilid, A., Peerapatdit, T., Tresukosol, D., Srisuma, S., Ratanamaneechat, S., et al. (2012). Apolipoprotein E gene polymorphism: effects on plasma lipids and risk of type 2 diabetes and coronary artery disease. Cardiovasc. Diabetol. 11:36. doi: 10.1186/1475-2840-11-36
Duval, S., and Tweedie, R. (2000). Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 56, 455–463. doi: 10.1111/j.0006-341X.2000.00455.x
Egger, M., Davey Smith, G., Schneider, M., and Minder, C. (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629.
Emerging Risk Factors, C., Sarwar, N., Gao, P., Seshasai, S. R., Gobin, R., Kaptoge, S., et al. (2010). Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet 375, 2215–2222. doi: 10.1016/S0140-6736(10)60484-9
Freitas Lima, L. C., Braga, V. A., do Socorro de França Silva, M., Cruz, J. C., Sousa Santos, S. H., de Oliveira Monteiro, M. M., et al. (2015). Adipokines, diabetes and atherosclerosis: an inflammatory association. Front. Physiol. 6:304. doi: 10.3389/fphys.2015.00304
Guo, J. J., Ju, J., and Xu, X. H. (2007). Association of polymorphisms of apolipoprotein E gene and High-sensitive C-reactive protein with type 2 diabetes with coronary heart disease. Shaanxi Med. J. 36, 1613–1616. doi: 10.3969/j.issn.1000-7377.2007.12.010
Halim, E. F., Reda, A. A., Hendi, A. A., Zaki, S. A., Essa, E. S., and Khalifa, A. S. (2012). Apolipoprotein E gene variants as a risk factor for coronary artery disease in type 2 diabetic Egyptian patients. Egypt. J. Immunol. 19, 1–10.
Higgins, J. P., Thompson, S. G., Deeks, J. J., and Altman, D. G. (2003). Measuring inconsistency in meta-analyses. BMJ 327, 557–560. doi: 10.1136/bmj.327.7414.557
Hong, G. Q., Cai, J. L., and Chen, J. P. (2017). Association of genetic polymorphisms of apolipoprotein E gene and type-2 diabetic mellitus complicated with coronary heart disease in Chaoshan patients. Int. Med. Health Guid. News 23, 792–794. doi: 10.3760/cma.j.issn.1007-1245.2017.06.004
Izar, M. C., Helfenstein, T., Ihara, S. S., Relvas, W. G., Santos, A. O., Fischer, S. C., et al. (2009). Association of lipoprotein lipase D9N polymorphism with myocardial infarction in type 2 diabetes: the genetics, outcomes, and lipids in type 2 diabetes (GOLD) study. Atherosclerosis 204, 165–170. doi: 10.1016/j.atherosclerosis.2008.08.006
Jayashankar, C. A., Andrews, H. P., Vijayasarathi, Pinnelli, V. B., Shashidharan, B., Nithin Kumar, H. N., et al. (2016). Serum uric acid and low-density lipoprotein cholesterol levels are independent predictors of coronary artery disease in Asian Indian patients with type 2 diabetes mellitus. J. Nat. Sci. Biol. Med. 7, 161–165. doi: 10.4103/0976-9668.184703
Jenkins, A. J., Rowley, K. G., Lyons, T. J., Best, J. D., Hill, M. A., and Klein, R. L. (2004). Lipoproteins and diabetic microvascular complications. Curr. Pharm. Des. 10, 3395–3418. doi: 10.2174/1381612043383188
Larifla, L., Armand, C., Bangou, J., Blanchet-Deverly, A., Numeric, P., Fonteau, C., et al. (2017). Association of APOE gene polymorphism with lipid profile and coronary artery disease in Afro-Caribbeans. PLoS ONE 12:e0181620. doi: 10.1371/journal.pone.0181620
Mohan, V., Deepa, R., Rani, S. S., Premalatha, G., and Chennai Urban Population, S. (2001). Prevalence of coronary artery disease and its relationship to lipids in a selected population in south india: the chennai urban population study (CUPS No. 5). J. Am. Coll. Cardiol. 38, 682–687. doi: 10.1016/S0735-1097(01)01415-2
Munafo, M. R., and Flint, J. (2004). Meta-analysis of genetic association studies. Trends Genet. 20, 439–444. doi: 10.1016/j.tig.2004.06.014
Naito, R., and Miyauchi, K. (2017). Coronary artery disease and type 2 diabetes mellitus. Int. Heart J. 58, 475–480. doi: 10.1536/ihj.17-191
Pan, M., Zhu, J. H., Liu, Y., Pan, H. Y., Wang, H. M., Liang, S., et al. (2002). Association of apolipoprotein E with essential hypertension, NIDDM and coronary heart disease in Chinese population. China Synth. Med. 3, 1–3. Available online at: http://www.cqvip.com/QK/85550X/200201/6817449.html
Paneni, F., Beckman, J. A., Creager, M. A., and Cosentino, F. (2013). Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part I. Eur. Heart J. 34, 2436–2443. doi: 10.1093/eurheartj/eht149
Raj, R., Bhatti, J. S., Badada, S. K., and Ramteke, P. W. (2015). Genetic basis of dyslipidemia in disease precipitation of coronary artery disease (CAD) associated type 2 diabetes mellitus (T2DM). Diabetes Metab. Res. Rev. 31, 663–671. doi: 10.1002/dmrr.2630
Shi, Y. R., Zhou, H. Y., and Zhang, Y. (2009). Relationship of gene polymorphisms of fatty acid binding protein-2 and apolipoprotein E with coronary heart disease in type-2 diabetic patients. J. Xi'an Jiaotong Univ. 30, 85–88. Available online at: http://d.wanfangdata.com.cn/Periodical/xaykdxxb200901022
Song, Y., Stampfer, M. J., and Liu, S. (2004). Meta-analysis: apolipoprotein E genotypes and risk for coronary heart disease. Ann. Intern. Med. 141, 137–147. doi: 10.7326/0003-4819-141-2-200407200-00013
Stroup, D. F., Berlin, J. A., Morton, S. C., Olkin, I., Williamson, G. D., Rennie, D., et al. (2000). Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 283, 2008–2012. doi: 10.1001/jama.283.15.2008
Sumi, S., Ramachandran, S., RamanKutty, V., Patel, M. M., Anand, T. N., Mullasari, A. S., et al. (2017). ENPP1 121Q functional variant enhances susceptibility to coronary artery disease in South Indian patients with type 2 diabetes mellitus. Mol. Cell. Biochem. 435, 67–72. doi: 10.1007/s11010-017-3057-2
Talmud, P. J., Lewis, S. J., Hawe, E., Martin, S., Acharya, J., Marmot, M. G., et al. (2004). No APOEepsilon4 effect on coronary heart disease risk in a cohort with low smoking prevalence: the Whitehall II study. Atherosclerosis 177, 105–112. doi: 10.1016/j.atherosclerosis.2004.06.008
Thakkinstian, A., McEvoy, M., Minelli, C., Gibson, P., Hancox, B., Duffy, D., et al. (2005). Systematic review and meta-analysis of the association between β2-adrenoceptor polymorphisms and asthma: a HuGE review. Am. J. Epidemiol. 162, 201–211. doi: 10.1093/aje/kwi184
Vaisi-Raygani, A., Rahimi, Z., Tavilani, H., and Pourmotabbed, T. (2010). Butyrylcholinesterase K variant and the APOE-ε4 allele work in synergy to increase the risk of coronary artery disease especially in diabetic patients. Mol. Biol. Rep. 37, 2083–2091. doi: 10.1007/s11033-009-9666-4
Wang, L., Lin, P., Ma, A., Zheng, H., Wang, K., Li, W., et al. (2015a). C-peptide is independently associated with an increased risk of coronary artery disease in T2DM subjects: a cross-sectional study. PLoS ONE 10:e0127112. doi: 10.1371/journal.pone.0127112
Wang, Y. L., Sun, L. M., Zhang, L., Xu, H. T., Dong, Z., Wang, L. Q., et al. (2015b). Association between Apolipoprotein E polymorphism and myocardial infarction risk: a systematic review and meta-analysis. FEBS Open Bio. 5, 852–858. doi: 10.1016/j.fob.2015.10.006
Wei, X., Ma, X., Lu, R., Bai, G., Zhang, J., Deng, R., et al. (2014). Genetic variants in PCSK1 gene are associated with the risk of coronary artery disease in type 2 diabetes in a Chinese Han population: a case control study. PLoS ONE 9:e87168. doi: 10.1371/journal.pone.0087168
Wei, Y., Guo, H., The, E., Che, W., Shen, J., Hou, L., et al. (2015). Persistent lipid abnormalities in statin-treated coronary artery disease patients with and without diabetes in China. Int. J. Cardiol. 182, 469–475. doi: 10.1016/j.ijcard.2015.01.024
Wu, Z. R., Chen, Z. C., Fu, M. X., Chen, J. Y., Han, L. Y., Zhou, L. H., et al. (2015). Polymorphism of apolipoprotein E gene and type 2 diabetic patients with coronary heart disease among Chinese Han population: a meta-analysis. J. Clin. Exp. Med. 14, 982–985. doi: 10.3969/j.issn.1671-4695.2015.012.008
Xu, H., Li, H., Liu, J., Zhu, D., Wang, Z., Chen, A., et al. (2014). Meta-analysis of apolipoprotein E gene polymorphism and susceptibility of myocardial infarction. PLoS ONE 9:e104608. doi: 10.1371/journal.pone.0104608
Xu, M., Zhao, J., Zhang, Y., Ma, X., Dai, Q., Zhi, H., et al. (2016). Apolipoprotein E gene variants and risk of coronary heart disease: a meta-analysis. Biomed Res. Int. 2016:3912175. doi: 10.1155/2016/3912175
Yin, Y. W., Qiao, L., Sun, Q. Q., Hu, A. M., Liu, H. L., Wang, Q., et al. (2014). Influence of apolipoprotein E gene polymorphism on development of type 2 diabetes mellitus in Chinese Han population: a meta-analysis of 29 studies. Metab. Clin. Exp. 63, 532–541. doi: 10.1016/j.metabol.2013.12.008
Yin, Y. W., Sun, Q. Q., Zhang, B. B., Hu, A. M., Liu, H. L., Wang, Q., et al. (2013). Association between apolipoprotein E gene polymorphism and the risk of coronary artery disease in Chinese population: evidence from a meta-analysis of 40 studies. PLoS ONE 8:e66924. doi: 10.1371/journal.pone.0066924
Zhang, L., Qi, X. Y., Wang, Y., Jia, Z. L., and Xie, Y. (2008). A study on the association between polymorphism of apolipoprotein E gene and type 2 diabetic patients with coronary heart disease. Acta. Acad. Med. CPAF 17, 572–575. Available online at: http://d.g.wanfangdata.com.cn/Periodical_wjyxyxb200807007.aspx
Zhang, M. D., Gu, W., Qiao, S. B., Zhu, E. J., Zhao, Q. M., and Lv, S. Z. (2014a). Apolipoprotein E gene polymorphism and risk for coronary heart disease in the Chinese population: a meta-analysis of 61 studies including 6634 cases and 6393 controls. PLoS ONE 9:e95463. doi: 10.1371/journal.pone.0095463
Zhang, W. H., Zhang, G. W., Zhang, X., Fang, Y. M., Xu, Z. F., and Zhang, A. Z. (2000). Relationship between Apo E gene polymorphism and type 2 diabetes mellitus with its cardiovascular complication in Chinese. Med. J. Chin. Civ. Adm 12, 206–209. Available online at: http://www.cnki.com.cn/Article/CJFDTOTAL-ZMYX200004008.htm
Zhang, Y., Hu, C., Hong, J., Zeng, J., Lai, S., Lv, A., et al. (2014b). Lipid profiling reveals different therapeutic effects of metformin and glipizide in patients with type 2 diabetes and coronary artery disease. Diabetes Care 37, 2804–2812. doi: 10.2337/dc14-0090
Zhang, Y., Tang, H. Q., Peng, W. J., Zhang, B. B., and Liu, M. (2015). Meta-analysis for the association of apolipoprotein E epsilon2/epsilon3/epsilon4 polymorphism with coronary heart disease. Chin. Med. J. 128, 1391–1398. doi: 10.4103/0366-6999.156803
Keywords: coronary artery disease, type 2 diabetes mellitus, apolipoprotein E, epsilon2, epsilon3, epsilon4, genetic polymorphism
Citation: Luo J-Q, Ren H, Banh HL, Liu M-Z, Xu P, Fang P-F and Xiang D-X (2017) The Associations between Apolipoprotein E Gene Epsilon2/Epsilon3/Epsilon4 Polymorphisms and the Risk of Coronary Artery Disease in Patients with Type 2 Diabetes Mellitus. Front. Physiol. 8:1031. doi: 10.3389/fphys.2017.01031
Received: 22 August 2017; Accepted: 28 November 2017;
Published: 12 December 2017.
Edited by:
Gerald A. Meininger, University of Missouri, United StatesReviewed by:
Aaron J. Trask, The Research Institute at Nationwide Children's Hospital, United StatesNaifeng Liu, Southeast University, China
Copyright © 2017 Luo, Ren, Banh, Liu, Xu, Fang and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Quan Luo, bHVvamlhbnF1YW54eUBjc3UuZWR1LmNu
†These authors have contributed equally to this work.
 Jian-Quan Luo
Jian-Quan Luo Huan Ren
Huan Ren Hoan Linh Banh5
Hoan Linh Banh5