Abstract
As an effective method of transgenesis, the plasmid of PiggyBac transposon containing GFP (PiggyBac) transposon system has been widely used in various organisms but not yet in mollusks. In this work, piggyBac containing green fluorescent protein (GFP) was transferred into the Pacific oyster (Crassostrea gigas) by sperm-mediated gene transfer with or without electroporation. Fluorescent larvae were then observed and isolated under an inverted fluorescence microscope, and insertion of piggyBac was tested by polymerase chain reaction (PCR) using genomic DNA as template. Oyster larvae with green fluorescence were observed after transgenesis with or without electroporation, but electroporation increased the efficiency of sperm-mediated transgenesis. Subsequently, the recombinant piggyBac plasmid containing gGH (piggyBac-gGH) containing GFP and a growth hormone gene from orange-spotted grouper (gGH) was transferred into oysters using sperm mediation with electroporation, and fluorescent larvae were observed and isolated. The insertion of piggyBac-gGH was tested by PCR and genome walking analysis. PCR analysis indicated that piggyBac-gGH was transferred into oyster larvae; genome walking analysis further showed the detailed location where piggyBac-gGH was inserted in the oyster genome. This is the first time that piggyBac transposon-mediated transgenesis has been applied in mollusks.
Introduction
Many methods can be employed to achieve genetic transformation, including viral vectors, non-viral vectors and transposons. Among these relatively effective methods of transgenesis, transposons have the ability to integrate into chromosomes, and gene expression from transposons is permanent (Ivics and Izsvak, 2006). Transposons are mobile elements that can transpose between vectors and a target genome. The original piggyBac transposon was first identified and isolated from the genome of the cabbage looper moth, Trichoplusia ni, in the late 1980s (Cary et al., 1989). Later,
other piggyBac-like transposons were identified in different insect species (Wu et al., 2011; Depra et al., 2012; Sormacheva et al., 2012), frogs (Lam et al., 1996; Leaver, 2001), fish (Cocca et al., 2011; Costa et al., 2013), and mammals (Jurka, 2000; Sarkar et al., 2003). The original piggyBac element is approximately 2.4 kb, with identical 13 bp inverted terminal repeats and additional asymmetric 19 bp internal repeats (Elick et al., 1997; Li et al., 2001, 2005). Previous data have shown that the original piggyBac transposon can effectively transfer up to 9.1 kb of transgene sequence into chromosomes with the help of the piggyBac transposase enzyme from a separate plasmid (Ding et al., 2005). PiggyBac allows “cut and paste” transfer of genes of interest between inverted terminal repeats and internal repeats into a host genome at TTAA nucleotide elements (Fraser et al., 1996; Bauser et al., 1999; O’Brochta et al., 2003; Wu and Burgess, 2004). PiggyBac transposon can insert into gene regions and cause mutagenesis, which contributes to infer function of the inserted gene (Ding et al., 2005). Furthermore, PiggyBac transposon can introduce exogenous genes into the genome for constructing transgenic organisms or testing gene function (Yusa, 2015). Thus, piggyBac transposons have been successfully used as a promising tool for basic genetic studies (Wang et al., 2008; Kaji et al., 2009; Woltjen et al., 2009; Yusa et al., 2009), gene therapies (Carlson et al., 2005) and construction of transgenic animals (Ding et al., 2005). Studies have also shown that the original piggyBac transposon has a broad host spectrum ranging from yeast to mammals, and this mobile element has been widely used for a variety of applications in a diverse range of organisms. However, the piggyBac transposition system has not yet been utilized or characterized in mollusks.
It has been reported that exogenous DNA can stick to spermatozoa and then be carried into an egg via fertilization. This method of sperm mediation is widely used for gene transfer (Parrington et al., 2011). Previous research has also investigated approaches for increasing the proportion of spermatozoa carrying exogenous DNA to improve the efficiency of transgenesis. One effective method is electroporation of spermatozoa during transformation, which increases the capacity of DNA bound by sperm in fish (Patil and Khoo, 1996) and mollusks (Tsai, 2000).
Pacific oyster is one of the most economically important cultivated oysters in the world. Genome sequencing of Pacific oyster has been completed (Zhang et al., 2012). However, the functions of most Pacific oyster genes are unknown due to a lack of effective research methods, such as transgenesis technology. In this work, we demonstrate that the piggyBac transposon system is suitable for Pacific oyster transgenesis.
Materials and Methods
Plasmid Construction
The donor plasmid piggyBac Dual Promoter and helper plasmid HA-mPB were constructed by Applied Biosystems. PiggyBac Dual Promoter carries the CMV promoter upstream of a MCS. Downstream of the expression cassette is an EF1alpha promoter driving the expression of a GFP+Puro marker. The entire cassette is flanked by genomic insulator elements to stabilize expression and piggyBac ITR for mobilization and integration. The helper plasmid HA-mPB contains the mPB transposase coding sequence in a pcDNA3-Kz-HA backbone, and it features a 5′ HS4 insulator to support robust transcription from the rPolr2A promoter.
Sequence encoding the mature GH peptide of orange-spotted grouper was amplified by primers GH-F and GH-R, with EcoRI at the 5′ end and BamHI at the 3′ end, using grouper pituitary cDNA as template. Then, the resultant GH fusion gene was digested with EcoRI and BamHI, purified and ligated into the EcoRI and BamHI sites of plasmid piggyBac to construct piggyBac-gGH.
Transfection of piggyBac Into Pacific Oysters
Five hundred μL of semen sampled from mature male Pacific oysters was collected into conical polystyrene sample cups, and then diluted to a concentration of 105 sperm/mL. Then, a combination of “helper-independent” transposase (15 μg) and transposon (30 μg) were added to the semen, and the mixture was incubated for 5 min at 20°C. For sperm-mediated gene transfer, approximately 104 eggs collected from mature female Pacific oysters were added to the semen-plasmid mixture, and then the mixture was incubated for 5 min. Finally, seawater was added into the mixture to activate fertilization.
For electroporation-mediated gene transfer, the semen-plasmid mixture was transferred into an electric shock cup, and then treated five times with an electroporation apparatus (Scientz, Wuhan, China) with the following parameters: resistance, 100 Ω; voltage, 300 V; capacitance, 100 μF. And each electric pulse duration was 20 ms, and the interval between two pulses was 100 ms. Then, fertilization was carried out as described above. Finally, fertilized ova were cultured in a 10-L tank at 20°C. During transgenesis, the same treatment was performed without plasmid as a negative control. All treatments were repeated three times, and three individual experiments were performed. Additionally, Schematic diagram of procedures for transfection of piggyBac into Pacific Oysters was shown in Figure 1.
FIGURE 1
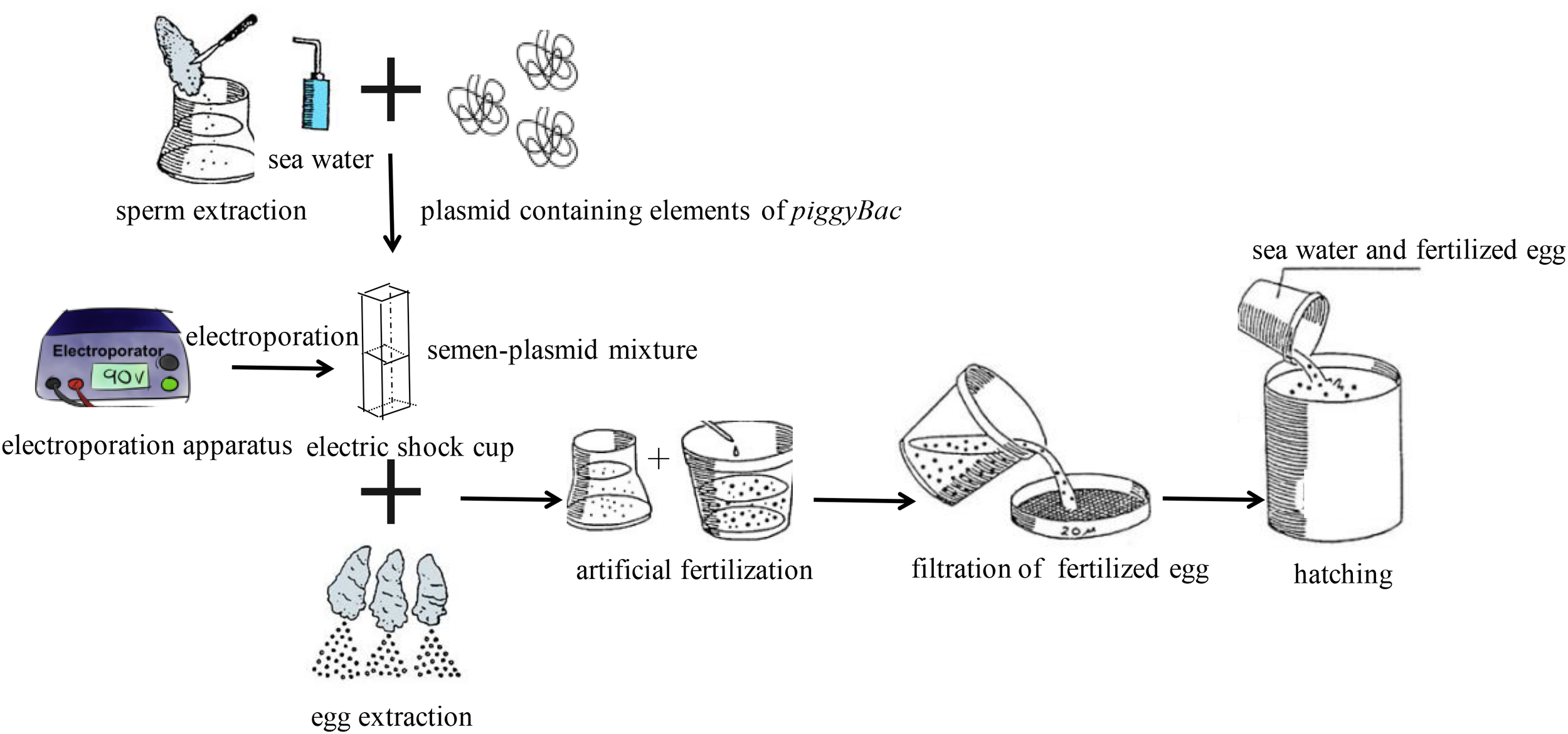
Schematic diagram of procedures for transfection of piggyBac into Pacific oyster.
Microscopic Observation of Fluorescent Oyster Larvae and Calculation of Transfection Efficiency and Relative Transfection Efficiency
Survival rates of oyster larvae were initially estimated by counting the density of oyster larvae using an optical microscope 0 and 72 h after fertilization. Oyster larvae transfected with both donor and helper plasmids were observed at 72 h post-transfection using a fluorescence microscope (IX71, Olympus, Japan), under which the expressed fluorescence appeared green. Fluorescent oyster larvae were photographed, counted and isolated.
Next, the efficiency of transfection (%) and relative efficiency of transfection (%) were calculated as below. Efficiency of transfection (%) = Number of larvae expressing fluorescence/Total number of fertilized eggs; relative efficiency of transfection (%) = Number of larvae expressing fluorescence/Total number of larvae at 72 h.
PCR and Sequence Analysis
QIAamp® DNA Micro Kit (Qiagen, United States) was used for trace DNA extraction from oyster larvae. Primers specific for GFP, GH, GH-GFP and the sequence downstream of GFP were used to confirm that piggyBac-GFP and piggyBac-gGH were successfully inserted and to identify the insertion site within the Pacific oyster genome (primers are shown in Table 1). Briefly, PCR amplification for GFP, GH, or GH-GFP sequences was conducted with 35 cycles of 95°C for 20 s, 60°C/60°C/56°C for 20 s, and 72°C for 50 s/1 min/1 min 20 s. PCR products were analyzed by agarose gel electrophoresis and sequencing. Additionally, the oyster genomic insertion site was analyzed using a genome walking kit (Takara, Japan). All primers are shown in Table 1.
Table 1
| Primer | Sequence (5′→3′) | Purpose |
|---|---|---|
| GFP-F | TGACCAACAAGATGAAGAGCACC | Cloningof GFP gene |
| GFP-R | TCCACGTCACCGCATGTTAGAAGA | Cloning of GFP gene |
| GH-F | CGGAATTCATGGACCGA GTCGTCCTCCTG | Cloning of GH gene |
| GH-R | CGGGATCCCTACAGGGTA CAGTTGGCCTC | Cloning of GH gene |
| GH-GFP-F | AACTGCTGGCTTGTTTCA | Cloning of GH-GFP gene |
| GH-GFP-R | CGATGCGGGTGTTGGTGT | Cloning of GH-GFP gene |
| SP1 | ATTCCACACAACATACGAGCCG | Genome walking |
| SP2 | CTGGGGTGCCTAATGAGTGAGC | Genome walking |
| SP3 | GGGAGAGGCGGTTTGCGTATTG | Genome walking |
Primers used in this study.
The base sequences of restriction enzyme cutting sites are underlined.
Statistical Analysis
All data were analyzed using SPSS 20.0 software (Chicago, IL, United States) with one-way variance (ANOVA) followed by Student–Newman–Keuls multiple comparisons test. Differences were considered statistically significant when p < 0.05.
Results
Construction of piggyBac-gGH
As shown in Figure 2, a fusion gene encoding the mature GH peptide of orange-spotted grouper was subcloned upstream of GFP in the donor plasmid PiggyBac, producing the recombinant plasmid piggyBac-gGH.
FIGURE 2
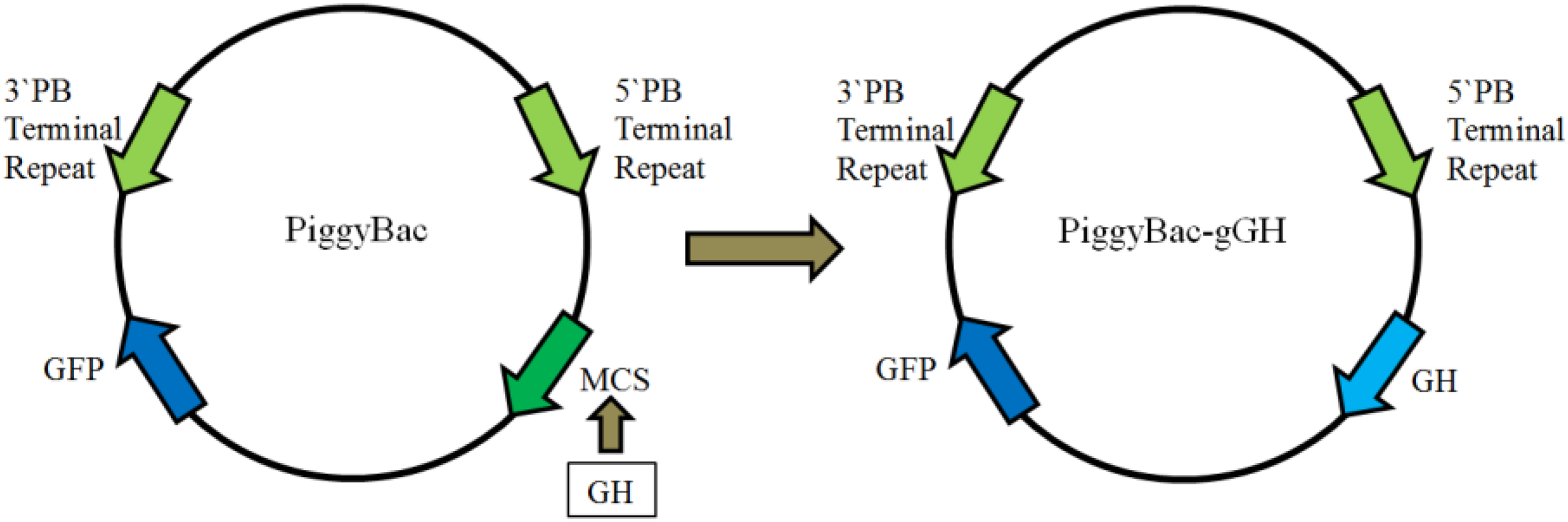
Construction of plasmid piggyBac-gGH. The left is the plasmid PiggyBac, the right the recombinant plasmid piggyBac-gGH. PB, piggyBac; GFP, green fluorescent protein; MCS, multiple clone sites; gGH, growth hormone gene of orange-spotted grouper.
Observation of Fluorescent Larvae and Efficiency of Transfection
Fluorescent oyster larvae transfected with helper and donor plasmids were observed and counted using a confocal microscope, and then were isolated 72 h after transfection. As shown in Figure 3, Pacific oyster larvae with green fluorescence were observed after transformation with piggyBac without electroporation (Figure 3B), with electroporation (Figure 3F) or with piggyBac-gGH with electroporation (Figure 3J), while green fluorescence was not observed in the control group (Figures 3D,H,L).
FIGURE 3
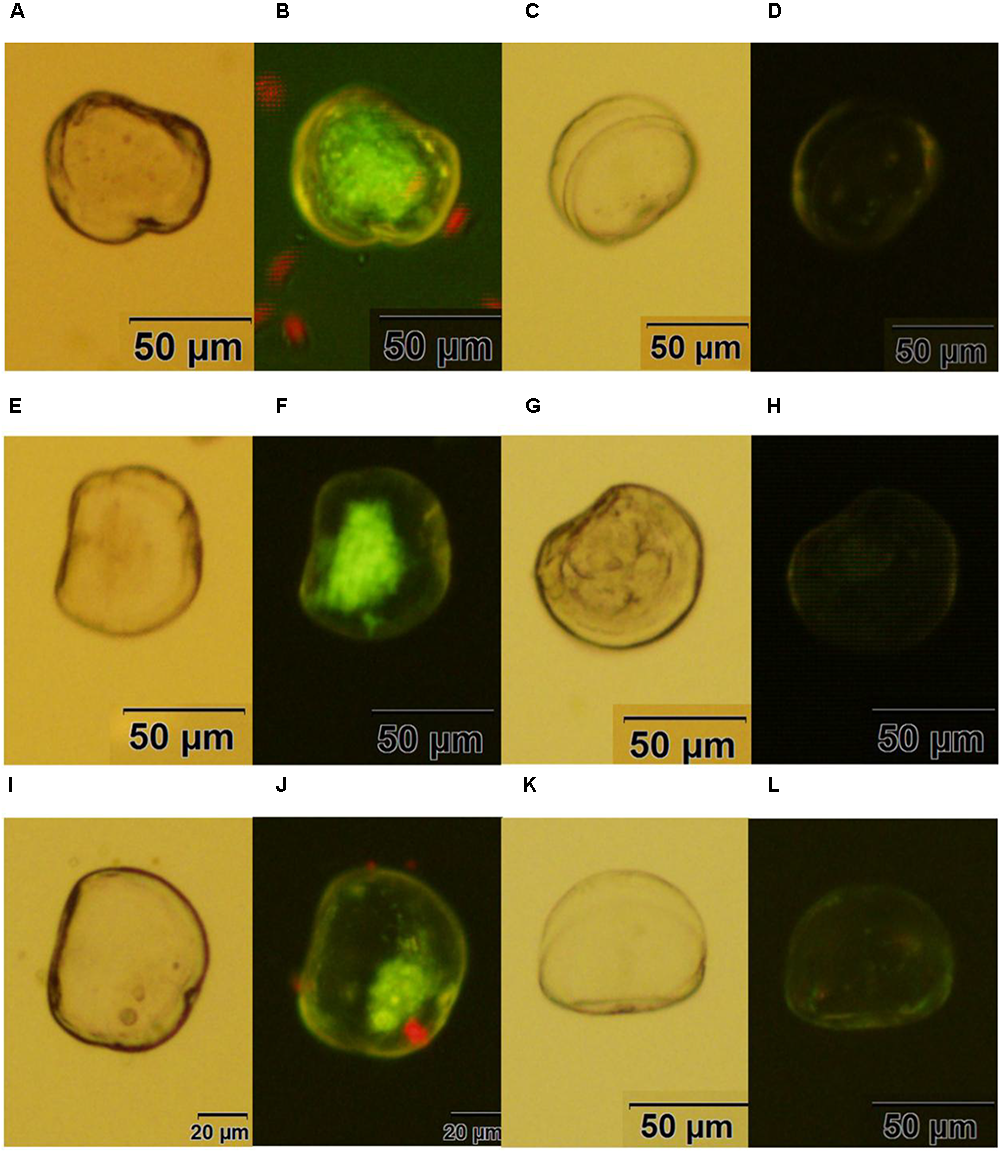
Transgenic oyster larvae and controls. (A,B) Oyster larvae transfected with piggyBac using sperm mediation without electroporation under white light or green fluorescence; (C,D) non-transfected oyster larvae as a control for (A,B); (E,F) oyster larvae transfected with piggyBac using sperm mediation with electroporation under white light or green fluorescence; (G,H) non-transfected oyster larvae as a control for (E,F); (I,J) oyster larvae transfected with piggyBac-gGH using sperm mediation with electroporation under white light or green fluorescence; (K,L) non-transfected oyster larvae as a control for (I,J) Red spots are the algae Isochrysis galbana used as food for oyster larvae.
As shown in Figure 4A, the survival rates of oyster larvae were approximately 10 and 1% after gene transfer of piggyBac without or with electroporation, respectively. The survival rate of the electroporation group was significantly lower than that of the control group or the group without electroporation (p < 0.01). However, the results (Figure 4B) showed that the efficiency of piggyBac transfection with electroporation (0.4%) was significantly higher than without electroporation (0.03%). In addition, the relative efficiency of piggyBac transfection with electroporation was dramatically higher than transfection without electroporation (40 vs. 0.3%, Figure 4C).
FIGURE 4
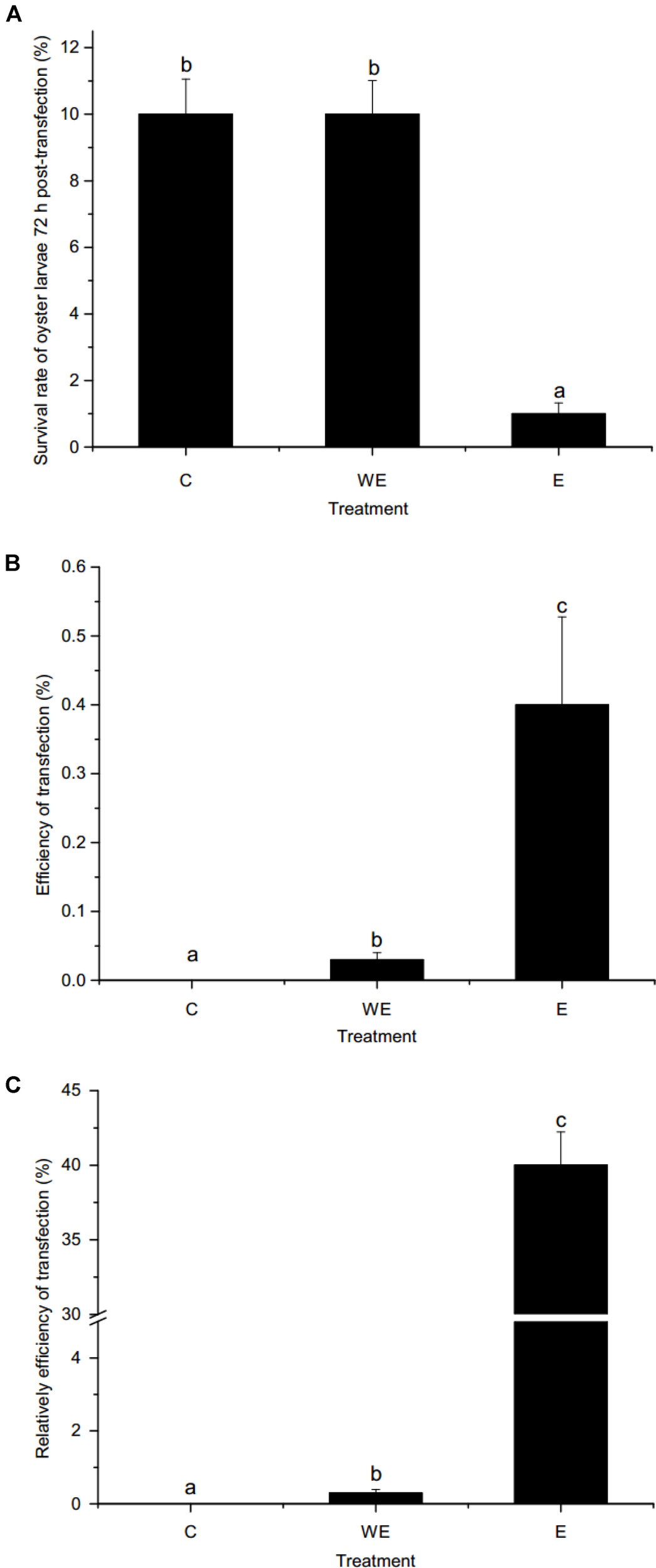
Survival rate and transfection efficiency using piggyBac system in oyster larvae. (A) Survival rate of oyster larvae 72 h post-transfection; (B) efficiency of transfection; (C) relative efficiency of transfection. C, non-transfected control group. WE, sperm-mediated transfection without electroporation. E, sperm-mediated transfection with electroporation. Data are shown as the means ± SEM (standard error of mean, SEM), n = 3. Different letters indicate statistically significant differences (p < 0.05).
Characterization of Exogenous Gene Insertion
Polymerase chain reaction and agarose gel electrophoresis detected the target gene segment in fluorescent oyster larvae, but not in non-fluorescent oyster larvae. As shown in Figure 5A, the GFP gene was detected in oyster larvae transfected with piggyBac. Both the GFP and GH genes were amplified from genomic DNA of oyster larvae transfected with piggyBac-gGH (Figure 5B), and sequencing results showed that their sequences matched the appropriate gene sequences in piggyBac-gGH (Supplementary Figures S1, S2). Meanwhile, segments covering partial sequences of GFP and GH were amplified using GH-GFP-F and GH-GFP-R as primers, which indicated that the GFP and GH genes were inserted into the oyster genome together (Figure 5C and Supplementary Figure S3). Additionally, the genomic insertion site of piggyBac-gGH was identified using the method of genome walking, and as shown in Figure 6, GFP-GH had been inserted into Scaffold200 of Pacific oyster genome at the sequence “AAGA.”
FIGURE 5
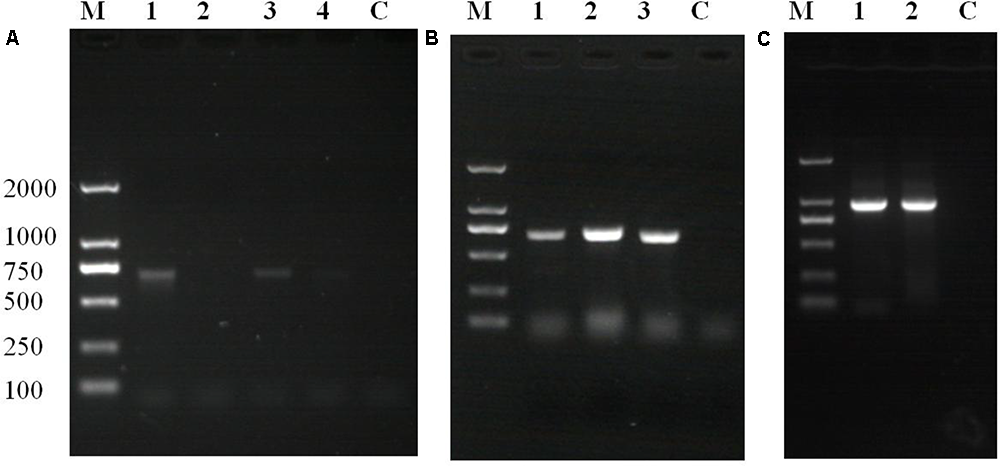
Polymerase chain reaction (PCR) amplification of target genes using the transgenic oyster genome as template. (A) GFP gene products amplified from oyster larvae transformed with piggyBac: lane M, marker; lane 1, amplification of GFP gene using fluorescent larvae from transgenesis experiment without electroporation; lane 2, amplification of GFP gene using non-fluorescent larvae from transgenesis experiment without electroporation; lane 3, amplification of GFP gene using fluorescent oyster larvae in transgenesis experiment with electroporation; lane 4, amplification of GFP gene using non-fluorescent larvae from transgenesis experiment with electroporation; lane C, control. (B) Gene products amplified from oyster larvae transformed with piggyBac or piggyBac-gGH with electroporation: lane M, marker; lane 1, amplification of GFP gene using fluorescent larvae transfected with piggyBac; lane 2, amplification of GFP gene using fluorescent larvae transfected with piggyBac-gGH; lane 3, amplification of GH gene using fluorescent larvae transfected with piggyBac-gGH; lane C, control. (C) Gene products covering partial sequences of GFP and GH genes amplified from oyster larvae transformed with piggyBac-gGH with electroporation: lane M, marker; lanes 1 and 2, amplification of segments covering GH and GFP genes using genome walking; lane C, control.
FIGURE 6

Sequence obtained by genome walking using genomic DNA from transgenic oyster larvae as template. The sequence in red corresponds to partial vector sequence downstream of the GFP gene in piggyBac-gGH, and sequence in blue aligns with the scaffold200 in Pacific oyster genome (http://www.oysterdb.com/FrontToolsAction.do?method=blastInit).
Discussion
Sperm-mediated transformation in mollusks has been reported in previous studies with or without electroporation (Tsai et al., 1997; Guerra et al., 2005; Guerra and Esponda, 2006), and CRISPR/Cas9-mediated transformation has also been utilized (Perry and Henry, 2015). In this work, our transgenesis of Pacific oysters with the piggyBac transposon system is the first time this system has been used in mollusks.
Our study showed that transgenic fluorescent oyster larvae can be acquired with the piggyBac transposon system, and PCR amplification of target genes from the genomes of transgenic fluorescent oyster larvae indicated that exogenous genes were integrated into Pacific oyster genomes. Genome walking is commonly used to identify regions upstream and downstream from known DNA sequences (Rishi et al., 2004; Leoni et al., 2008, 2011; Shapter and Waters, 2014). In this study, the DNA sequence of unknown genomic regions flanking the piggyBac-gGH insertion were determined by genome walking, then these sequences were aligned with the oyster genome sequence to determine the insertion site. These results further showed that piggyBac transposons can mediate integration of exogenous genes into the oyster genome. However, the insertion site of piggyBac-gGH in this study was different from previous studies, which reported that piggyBac insertions showed a high preference for genomic TTAA nucleotide elements (Fraser et al., 1995; Balu et al., 2005; Ding et al., 2005; Su et al., 2012; Yusa, 2015).
In this study, the efficiency of transfection using electroporation in Pacific oysters was approximately 0.4%, which is similar to reported rates of 0.1–10% in insects (Fraser, 2012); electroporation of the mixture of spermatozoa and transposase–transposon vectors resulted in about 10-fold higher efficiency of transgenesis than without electroporation, in agreement with earlier research (Patil and Khoo, 1996; Tsai, 2000). These results indicated that electroporation of the mixture of spermatozoa and vectors increased the efficiency of transgenesis. However, data in this study revealed that survival rate of Pacific oysters was decreased by electroporation prior to gene transfer, maybe because spermatozoa were damaged by the electrical impulse.
In addition, the present work would support a new method, piggyBac transposon system, for the research fields of genome and gene function in oyster. Firstly, some genes related with economic importance, such as resistance gene, growth hormone gene, or glycogenesis gene, could be introduced into oyster genome by piggyBac transposon system to improve the quality of oyster. Moreover, the genes with unknown function also could be transferred into oyster genome and their physiological functions could be deduced according to the phenotype of the transgenic oysters.
Conclusion
This study demonstrated that the piggyBac transposon system can be used for transgenesis of Pacific oysters. This work provides a new chance for studying the gene function of mollusks. And our further research will focus on enhancing the transfection efficiency of Pacific oysters.
Statements
Author contributions
XW and BZ conceived and designed the experiments. JC, CW, ZC, LW, and ZL performed the experiments. GL, TG, and YL analyzed the data. WG contributed reagents, materials, analysis tools. XW, JC, and CW wrote the paper. All authors reviewed the manuscript.
Funding
This research was supported by the Shandong Provincial Natural Science Foundation, China (Grant Nos. ZR2013CM026, ZR2014CP009, and ZR2017BC058), the Modern Agricultural Industry Technology System of Shandong Province, China (Grant No. SDAIT-14-03), Key R&D program of Shandong Province, China (Grant No. 2018GHY115027), the Key R&D Program of Yantai City, China (Grant No. 2017ZH054), the Innovation Plan for Marine/Fishery Science and Technology of Shandong Province, China (Grant No. 2017YY03), and A Project of Shandong Province Higher Educational Science and Technology Program (Grant No. J17KA129).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00811/full#supplementary-material
FIGURE S1Sequence alignment between piggyBac sequence and GFP segment amplified using the genome of the oyster transfected with piggyBac as template (PCR amplification using primers GFP-F and GFP-R).
FIGURE S2Sequence alignment between piggyBac-gGH sequence and GH segment amplified using the genome of the oyster transfected with piggyBac-gGH as template (PCR amplification using primers GH-F and GH-R).
FIGURE S3Sequence alignment between piggyBac-gGH sequence and GH-GFP segment amplified using the genome of the oyster transfected with piggyBac-gGH as template (PCR amplification using primers GH-GFP-F and GH-GFP-R). The sequence at upstream of the first red arrow is GH segment, that between the two red arrows is the partial plasmid sequence, and that at downstream of the second red arrow is GFP segment.
Abbreviations
- CMV
human cytomegalovirus
- GFP
green fluorescent protein
- gGH
a growth hormone gene from orange-spotted grouper
- MCS
multiple cloning site
- PCR
polymerase chain reaction
- piggyBac
the plasmid of PiggyBac transposon containing GFP
- piggyBac-gGH
the recombinant piggyBac plasmid containing gGH
References
1
Balu B. Shoue D. A. Fraser M. J. Jr. Adams J. H. (2005). High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac.Proc. Natl. Acad. Sci. U.S.A.10216391–16396. 10.1073/pnas.0504679102
2
Bauser C. A. Elick T. A. Fraser M. J. (1999). Proteins from nuclear extracts of two lepidopteran cell lines recognize the ends of TTAA-specific transposons piggyBac and tagalong.Insect. Mol. Biol.8223–230. 10.1046/j.1365-2583.1999.820223.x
3
Carlson C. M. Frandsen J. L. Kirchhof N. Mcivor R. S. Largaespada D. A. (2005). Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse.Proc. Natl. Acad. Sci. U.S.A.10217059–17064. 10.1073/pnas.0502974102
4
Cary L. C. Goebel M. Corsaro B. G. Wang H. G. Rosen E. Fraser M. J. (1989). Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of Nuclear polyhedrosis viruses.Virology172156–169. 10.1016/0042-6822(89)90117-7
5
Cocca E. De Iorio S. Capriglione T. (2011). Identification of a novel helitron transposon in the genome of Antarctic fish.Mol. Phylogenet. Evol.58439–446. 10.1016/j.ympev.2010.12.020
6
Costa G. W. Cioffi M. B. Bertollo L. A. Molina W. F. (2013). Transposable elements in fish chromosomes: a study in the marine cobia species.Cytogenet. Genome Res.141126–132. 10.1159/000354309
7
Depra M. Ludwig A. Valente V. L. Loreto E. L. (2012). Mar, a MITE family of hAT transposons in Drosophila.Mob. DNA3:13. 10.1186/1759-8753-3-13
8
Ding S. Wu X. Li G. Han M. Zhuang Y. Xu T. (2005). Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice.Cell122473–483. 10.1016/j.cell.2005.07.013
9
Elick T. A. Lobo N. Fraser M. J. Jr. (1997). Analysis of the cis-acting DNA elements required for piggyBac transposable element excision.Mol. Gen. Genet.255605–610. 10.1007/s004380050534
10
Fraser M. J. Jr. (2012). Insect transgenesis: current applications and future prospects.Annu. Rev. Entomol.57267–289. 10.1146/annurev.ento.54.110807.090545
11
Fraser M. J. Cary L. Boonvisudhi K. Wang H. G. (1995). Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA.Virology211397–407. 10.1006/viro.1995.1422
12
Fraser M. J. Ciszczon T. Elick T. Bauser C. (1996). Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera.Insect. Mol. Biol.5141–151. 10.1111/j.1365-2583.1996.tb00048.x
13
Guerra R. Carballada R. Esponda P. (2005). Transfection of spermatozoa in bivalve molluscs using naked DNA.Cell Biol. Int.29159–164. 10.1016/j.cellbi.2004.11.018
14
Guerra R. Esponda P. (2006). Transfection of eggs in the bivalve mollusc Chamelea gallina (Bivalvia, Veneridae).J. Submicrosc. Cytol. Pathol.385–10.
15
Ivics Z. Izsvak Z. (2006). Transposons for gene therapy!Curr. Gene Ther.6593–607.
16
Jurka J. (2000). Repbase update: a database and an electronic journal of repetitive elements.Trends Genet.16418–420. 10.1016/S0168-9525(00)02093-X
17
Kaji K. Norrby K. Paca A. Mileikovsky M. Mohseni P. Woltjen K. (2009). Virus-free induction of pluripotency and subsequent excision of reprogramming factors.Nature458771–775. 10.1038/nature07864
18
Lam W. L. Seo P. Robison K. Virk S. Gilbert W. (1996). Discovery of amphibian Tc1-like transposon families.J. Mol. Biol.257359–366. 10.1006/jmbi.1996.0168
19
Leaver M. J. (2001). A family of Tc1-like transposons from the genomes of fishes and frogs: evidence for horizontal transmission.Gene271203–214.10.1016/S0378-1119(01)00530-3
20
Leoni C. Gallerani R. Ceci L. R. (2008). A genome walking strategy for the identification of eukaryotic nucleotide sequences adjacent to known regions.Biotechniques44229232–235. 10.2144/000112680
21
Leoni C. Volpicella M. De Leo F. Gallerani R. Ceci L. R. (2011). Genome walking in eukaryotes.FEBS J.2783953–3977. 10.1111/j.1742-4658.2011.08307.x
22
Li X. Harrell R. A. Handler A. M. Beam T. Hennessy K. (2005). piggyBac internal sequences are necessary for efficient transformation of target genomes.Insect. Mol. Biol.1417–30. 10.1111/j.1365-2583.2004.00525.x
23
Li X. Lobo N. Bauser C. A. Fraser M. J. Jr. (2001). The minimum internal and external sequence requirements for transposition of the eukaryotic transformation vector piggyBac.Mol. Genet. Genomics266190–198.10.1007/s004380100525
24
O’Brochta D. A. Sethuraman N. Wilson R. Hice R. H. Pinkerton A. C. Levesque C. S. et al (2003). Gene vector and transposable element behavior in mosquitoes.J. Exp. Biol.2063823–3834. 10.1242/jeb.00638
25
Parrington J. Coward K. Gadea J. (2011). Sperm and testis mediated DNA transfer as a means of gene therapy.Syst. Biol. Reprod. Med.5735–42.10.3109/19396368.2010.514022
26
Patil J. G. Khoo H. W. (1996). Nuclear internalization of foreign DNA by zebrafish spermatozoa and its enhancement by electroporation.J. Exp. Zool.274121–129. 10.1002/(SICI)1097-010X(19960201)274:2<121::AID-JEZ5>3.0.CO;2-R
27
Perry K. J. Henry J. Q. (2015). CRISPR/Cas9-mediated genome modification in the mollusc, Crepidula fornicata.Genesis53237–244. 10.1002/dvg.22843
28
Rishi A. S. Nelson N. D. Goyal A. (2004). Genome walking of large fragments: an improved method.J. Biotechnol.1119–15. 10.1016/j.jbiotec.2004.03.008
29
Sarkar A. Sim C. Hong Y. S. Hogan J. R. Fraser M. J. Robertson H. M. et al (2003). Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences.Mol. Genet. Genomics270173–180. 10.1007/s00438-003-0909-0
30
Shapter F. M. Waters D. L. (2014). Genome walking.Methods Mol. Biol.1099133–146. 10.1007/978-1-62703-715-0_12
31
Sormacheva I. Smyshlyaev G. Mayorov V. Blinov A. Novikov A. Novikova O. (2012). Vertical evolution and horizontal transfer of CR1 non-LTR retrotransposons and Tc1/mariner DNA transposons in Lepidoptera species.Mol. Biol. Evol.293685–3702. 10.1093/molbev/mss181
32
Su H. Liu X. Yan W. Shi T. Zhao X. Blake D. P. et al (2012). piggyBac transposon-mediated transgenesis in the apicomplexan parasite Eimeria tenella.PLoS One7:e40075. 10.1371/journal.pone.0040075
33
Tsai H. J. (2000). Electroporated sperm mediation of a gene transfer system for finfish and shellfish.Mol. Reprod. Dev.56281–284. 10.1002/(SICI)1098-2795(200006)56:2+<281::AID-MRD15>3.0.CO;2-B
34
Tsai H. J. Lai C. H. Yang H. S. (1997). Sperm as a carrier to introduce an exogenous DNA fragment into the oocyte of Japanese abalone (Haliotis divorsicolor suportexta).Transgenic Res.685–95. 10.1023/A:1018413318223
35
Wang W. Lin C. Lu D. Ning Z. Cox T. Melvin D. et al (2008). Chromosomal transposition of piggyBac in mouse embryonic stem cells.Proc. Natl. Acad. Sci. U.S.A.1059290–9295. 10.1073/pnas.0801017105
36
Woltjen K. Michael I. P. Mohseni P. Desai R. Mileikovsky M. Hamalainen R. et al (2009). piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells.Nature458766–770. 10.1038/nature07863
37
Wu M. Sun Z. Luo G. Hu C. Zhang W. Han Z. (2011). Cloning and characterization of piggyBac-like elements in lepidopteran insects.Genetica139149–154. 10.1007/s10709-010-9542-0
38
Wu X. Burgess S. M. (2004). Integration target site selection for retroviruses and transposable elements.Cell Mol. Life Sci.612588–2596. 10.1007/s00018-004-4206-9
39
Yusa K. (2015). piggyBac Transposon. Microbiol. Spectr.3MDNA3–MDNA0028. 10.1128/microbiolspec.MDNA3-0028-2014
40
Yusa K. Rad R. Takeda J. Bradley A. (2009). Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon.Nat. Methods6363–369. 10.1038/nmeth.1323
41
Zhang G. Fang X. Guo X. Li L. Luo R. Xu F. et al (2012). The oyster genome reveals stress adaptation and complexity of shell formation.Nature49049–54. 10.1038/nature11413
Summary
Keywords
Pacific oyster, transgenesis, piggyBac transposons, sperm-mediated gene transfer, electroporation
Citation
Chen J, Wu C, Zhang B, Cai Z, Wei L, Li Z, Li G, Guo T, Li Y, Guo W and Wang X (2018) PiggyBac Transposon-Mediated Transgenesis in the Pacific Oyster (Crassostrea gigas) – First Time in Mollusks. Front. Physiol. 9:811. doi: 10.3389/fphys.2018.00811
Received
09 April 2018
Accepted
08 June 2018
Published
16 July 2018
Volume
9 - 2018
Edited by
Menghong Hu, Shanghai Ocean University, China
Reviewed by
Weiwei You, Xiamen University, China; Vengatesen Thiyagarajan, University of Hong Kong, Hong Kong
Updates
Copyright
© 2018 Chen, Wu, Zhang, Cai, Wei, Li, Li, Guo, Li, Guo and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaotong Wang, wangxiaotong999@163.com
†These authors have contributed equally to this work.
This article was submitted to Aquatic Physiology, a section of the journal Frontiers in Physiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.