- 1Key Laboratory of Freshwater Animal Breeding, Ministry of Agriculture, Fishery College, Huazhong Agricultural University, Wuhan, China
- 2Collaborative Innovation Center for Efficient and Health Production of Fisheries in Hunan Province, Changde, China
SUMOylation is a kind of important post-translational modification. In the present study, we identified 10 key genes involved in SUMOylation and deSUMOylation (sumo1, sumo2, sumo3, sae1, uba2, ubc9, pias1, senp1, senp2, and senp3) in yellow catfish Pelteobagrus fulvidraco, investigated their tissue expression patterns and transcriptional responses to carbohydrate addition both in vivo and in vitro. All of these members shared similar domains to their orthologous genes of other vertebrates. Their mRNAs were widely expressed in all the tested tissues, but at variable levels. Dietary carbohydrate levels differentially influenced the mRNA levels of these genes in liver, muscle, testis, and ovary of yellow catfish. Their mRNA levels in primary hepatocytes were differentially responsive to glucose addition. Our study would contribute to our understanding into the molecular basis of SUMOylation modification and into the potential SUMOylation function in the carbohydrate utilization in fish.
Introduction
SUMOylation is a kind of important post-translational modification. It is characterized by SUMO proteins which covalently binds to the ε-amino group of lysine residues in intracellular target proteins in an ATP-dependent process (Gareau and Lima, 2010). To date, four SUMO proteins have been identified and characterized in humans: SUMO 1, SUMO 2, SUMO 3, and SUMO 4 (Pseudogene) (Krumova and Weishaupt, 2013). During the SUMOylation modification, a series of enzymatic reactions mediate the process. These enzymes include the heteromeric activating enzyme E1 (Uba2), the conjugation enzyme E2 (Ubc9), and SUMO E3 ligases represented by PIAS family (Johnson, 2004; Hay, 2005). SUMOylation plays a central role in cell homeostasis by regulating a large number of key biological processes such as signal transduction, nucleoplasm transport, transcription regulation, DNA repair, and cell cycle (Johnson, 2004; Hay, 2005; Yang and Paschen, 2009). In addition, SUMOylation is a dynamic process which can be reversed by a family of Sentrin/SUMO-specific protease (Bailey and O’Hare, 2004; Hay, 2005). SENPs act to deSUMOylate the proteins by cleaving the covalent conjugation between SUMO and its target (Wilkinson and Henley, 2010).
To date, most studies involved in SUMOylation and deSUMOylation mainly focuse on mammals. The data is still very scarce in aquatic organisms, including fish. To our best knowledge, only partial genes in the process of SUMOylation and deSUMOylation have been identified in very limited fish, such as in grouper [sumo1 and sumo2 (Xu et al., 2016)], medaka fish [senp1 (Obata et al., 2013)], zebrafish [ubc9 (Yuan et al., 2010)], channel catfish [ubc9 (Chen et al., 2010)], yellow croaker [ubc9 (Zhou et al., 2009)], half-smooth tongue sole [ubc9 (Hu and Chen, 2013)], grass carp [pias1 (Wong, 2013)], and rainbow trout [sumo1, sumo2, sumo3, sae1, sae2, ubc9, and pias1 (Zang, 2013)]. Very limited studies also explored their mRNA tissue expression profiles of genes involving SUMOylation and deSUMOylation processes (Hu and Chen, 2013; Xu et al., 2016).
SUMOylation modification of proteins plays important roles in the function, compartmentalization and stability of target proteins, contributing to the regulation of diverse processes (Bailey and O’Hare, 2004). The exploration of the emerging roles of SUMOylation modification in nutrient metabolism is a growing and fascinating field. In mammals, Huang et al. (2013) suggested that SUMOylation mediated the high carbohydrate (glucose)-induced process of NF-κB signaling activation. Generally speaking, compared to mammals, fish show relatively low capacity of utilization for dietary carbohydrate. However, compared to dietary lipid and proteins, carbohydrates are relatively inexpensive and a readily available source of energy, which makes their inclusion in the diet attractive. Previously, Ye et al. (2009) found that carbohydrate could cause protein-sparing effect in the diets for juvenile yellow catfish Pelteobagrus fulvidraco, a widely cultured freshwater teleost in China and other Asian countries. Since SUMOylation and deSUMOylation mediate the regulation of many transcriptional factors involved in nutrient metabolism, we are eager to know the effects of dietary carbohydrate levels on transcriptional responses of these genes. To this end, the present study identified the cDNA sequences of 10 key genes in SUMO- and deSUMOylation (sumo1, sumo2, sumo3, sae1, uba2, ubc9, pias1, senp1, senp2, and senp3) in yellow catfish, and examine their transcriptional responses in several tissues of yellow catfish fed different levels of dietary carbohydrate. Taken together, this study provided the basis for further exploring the biological characteristics of SUMO- and deSUMOylation and their potential functions for the utilization of dietary carbohydrate.
Materials and Methods
The experiment is divided into three parts. In Experiment 1, we cloned sumoylation-related genes and explored their tissue distribution of gene expression. Experiment 2 was conducted to determinate their transcriptional responses to different levels of dietary carbohydrate. Experiment 3 was taken to determine the effect of glucose incubation on mRNA expression of these genes in primary hepatocytes from yellow catfish. The experimental protocol was approved by the Committee of Huazhong Agricultural University on the Ethics of Laboratory Animal Experiments.
Experiment 1: Cloning and Tissue Distribution of Gene Expression
RNA Isolation and cDNA Cloning
The experimental protocols were similar to those described in the study by Wei et al. (2017). Yellow catfish (23.5 ± 3.3 g, mean ± SEM) were obtained from a local commercial farm. After 2-week acclimation, fish were fasted for 24 h and then euthanized with MS-222 (100 mg/L). Heart, liver, muscle, brain, kidney, intestine, spleen, mesenteric fat, testis, gill, and ovary were removed and stored at -80°C for subsequent tissue expression analysis. Total RNA was extracted using TRIzol RNA reagent (Invitrogen, United States) based on the acid guanidinium thiocyanate–phenol–chloroform extraction method. The quality of total RNA was check by agarose gel electrophoresis, and determined by Nanodrop ND-2000 spectrophotometer (1.8 < OD260/OD280 < 2.0, OD260/OD230 > 1.5). The nested 3′ and 5′ RACE PCR were performed to obtain the 3′ and 5′ end sequences with a SMART RACE cDNA Amplification Kit (Clontech, United States). The primers were shown in Supplementary Table 1.
Sequence Analysis
The full-length cDNA sequences of sumo1-3, sae1/uba2, ubc9, pias1, senp2/senp3, and partial sequences of senp1 were edited and analyzed using the program EDITSEQ (DNAstar) to search for the open reading frame (ORF). They were then translated into amino acid sequences using standard genetic codes. Sequence alignments and percentage of amino acid conservation were assessed by Clustal-W multiple alignment algorithm. Domains were analyzed with the online CDD tool at NCBI1 and the SMART program2. For phylogenetic analysis, multiple sequence alignments were made with MAFFT using an amino acid model at the GUIDANCE web-server3, which pruned unreliably aligned regions by rejecting columns with confidence scores below 0.93. The phylogenetic trees were generated through neighbor-joining (NJ) method with MEGA 6.0 (Tamura et al., 2013) based on the JTT + G model (Jones et al., 1992), and the best-fit model of sequence evolution was obtained by ML model selection. Bootstrap sampling was reiterated 1000 times.
Experiment 2: Effects of Dietary Carbohydrate Levels on mRNA Expression of Genes
Three experimental diets were formulated with dietary carbohydrate levels at 17.2% (low), 22.8% (moderate), and 30.2% (high), based on our published studies (Ye et al., 2009). Starch was used as dietary carbohydrate sources. Yellow catfish were obtained from a local fish farm (Wuhan, China). After 2-week acclimation, 270 uniformly size fish (4.1 ± 0.01 g, mean ± SEM) were randomly stocked to 9 circular fiberglass tanks, 30 fish for each tank. Each diet was assigned to three tanks in a completely randomized manner. The feeding protocols were similar to those in our recent study (Wei et al., 2017, 2018). The experiment lasted for 10 weeks. During the experiment, water temperature ranged from 25.7 to 28.6°C. Dissolved oxygen and NH4-N were 5.87–6.41 and 0.107–0.142 mg/L, respectively.
Sampling
At the end of the experiment, all fish were fasted for 24 h. Fish were killed (MS222 at a dose of 100 mg/L). Then, fish were dissected on ice to obtain liver, muscle, testis, and ovary. All samples were quickly frozen in liquid nitrogen, and reserved at -80°C for the subsequent real-time quantitative PCR (qPCR) analysis.
Quantitative PCR
Quantitative PCR (qPCR) was carried out according to the methods described in our previous studies (Cheng et al., 2017; Wei et al., 2017, 2018). The primer sequences of each gene used in this analysis are given in Supplementary Table 2. A set of 10 housekeeping genes (18S rRNA, β-actin, rpl7, tuba, b2m, elfa, gapdh, tbp, hprt, and ubce) were selected in order to test their transcriptional stability. The relative expression of genes was calculated by 2-ΔΔCt method and GeNorm was used to normalize the geometric mean of two best combination genes under different experimental conditions.
Experiment 3: Effect of Glucose Incubation on mRNA Expression of Genes in Primary Hepatocytes From Yellow Catfish
Primary Hepatocyte Isolation and Glucose Incubation
Primary hepatocytes were isolated from yellow catfish and cultured according to the methods described in our previous studies (Zhuo et al., 2014; Wei et al., 2018). Three glucose concentration, control (5.41 mmol/L glucose), 10 mM (9.75 mmol/L glucose), and 20 mM (20.38 mmol/L glucose), were used to incubate the isolated hepatocytes, respectively. Hepatocyte cell suspension (CS) was plated onto 25 cm2 flasks at the density of 1 × 106 cells per mL, and three different levels of glucose were added and incubated at 28°C in humidified air containing 5% CO2. Sampling occurred at 12, 24, and 48 h, respectively. Each treatment was performed in triplicate.
Cell Livability Assay
The MTT assay was used to test the cell viability according to Zhuo et al. (2014). The wells containing the medium without any cells were used as the positive control. The results are presented as percentage cell viability, which was calculated as the ratio of absorbance A in the experimental well to A in the positive control well.
mRNA Preparation and qPCR Analysis
Hepatocytes were collected from 25 cm2 flasks for the total RNAs extracted by TRIzol regent (TaKaRa, Japan). Reverse transcriptions were performed with equal quantities of each total RNA as template using Quantitect Reverse Transcription Kit (TaKaRa, Japan) for real-time PCR according to the manufacturer’s protocol. qPCR was carried out according to the methods described in our previous studies (Cheng et al., 2017; Wei et al., 2018). The primer sequences of each gene used in this analysis are the same as Supplementary Table 2. A set of 10 housekeeping genes (18S rRNA, β-actin, rpl7, tuba, b2m, elfa, gapdh, tbp, hprt, and ubce) were selected in order to test their transcriptional stability. The relative expression of genes was calculated by 2-ΔΔCt method and GeNorm was used to normalize the geometric mean of two best combination genes under different experimental conditions.
Statistical Analysis
Results are presented as means with their standard errors (mean ± SEM). Before the statistical analysis, all data were tested for normality of distribution using the Kolmogornov–Smirnov test. Data were evaluated using one-way ANOVA, Homoscedasticity analysis and Duncan’s multiple range tests. The analysis was carried out using the SPSS 19.0 for Windows (SPSS, Chicago, IL, United States), and the minimum significant level was set at 0.05.
Results
Molecular Characterization
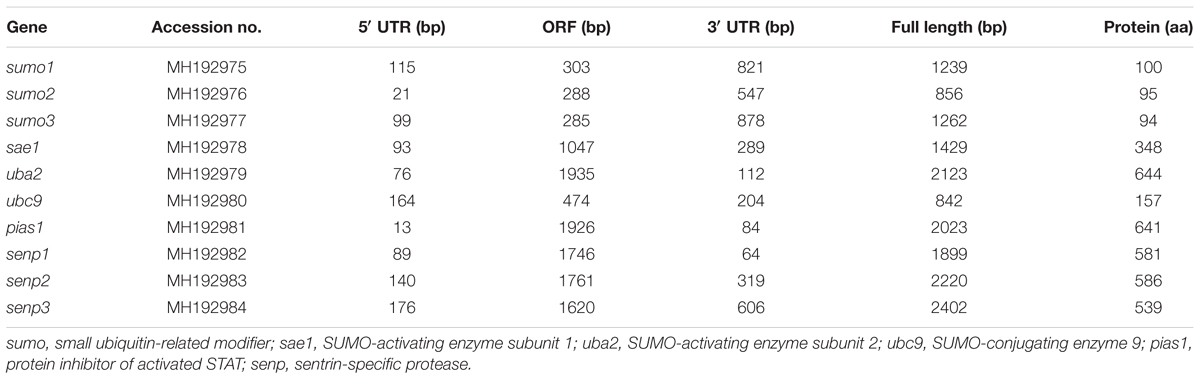
In the present study, the full-length cDNA sequences of nine SUMOylation-related genes (sumo1, sumo2, sumo3, sae1, uba2, ubc9, pias1, senp2, and senp3) were successfully obtained by RT-PCR and RACE methods from P. fulvidraco. Their sequences were 1239, 856, 1262, 1429, 2123, 842, 2023, 2220, and 2402 bp, respectively (Table 1). We also cloned and obtained the partial cDNA sequence of senp1, which was 1899 bp. We have made exhaustive efforts to clone the full-length cDNA sequence of senp1 but met failure. The pair-wise amino acid sequence comparison of 10 sumoylation-related genes between different species were shown in Table 2. The amino acid identities of P. fulvidraco sumo1, sumo2, sumo3, sae1, uba2, ubc9, pias1, senp1, senp2, and senp3 were similar to those of orthologous from other species, exhibiting 28.12–100% amino acid sequence identities among different species.
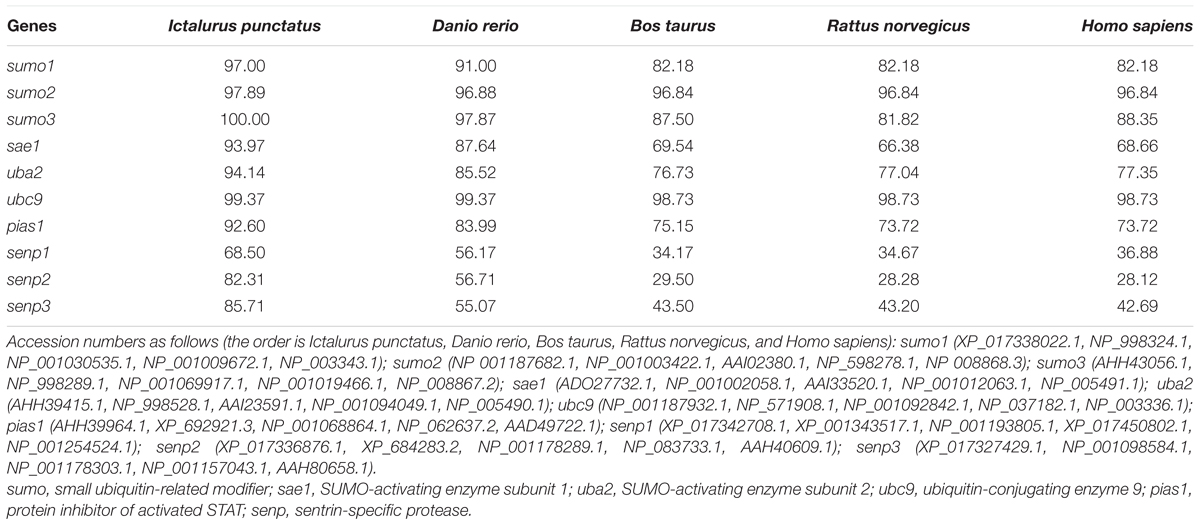
TABLE 2. Amino acid sequence identities of SUMOylation-related genes between P. fulvidraco and other species (%).
The protein sequence of P. fulvidraco sumo1, sumo2, and sumo3 revealed similar domains with mammals, including the UBQ (ubiquitin-like proteins) domain, the hydrophobic surface, the Ulp1-Smt3 interaction sites, a VKTE motif and the C-terminal Gly residues (Supplementary Figure 1). The P. fulvidraco sae1 consisted of three conserved regions, such as the E1 enzyme family domain, the ThiF domain and the dimer interaction sites (Supplementary Figure 2). The P. fulvidraco uba2 comprised E1 enzyme family domain, the Ubiquitin/SUMO-activating enzyme ubiquitin-like domain (UAE-Ubl), and the SUMO-activating enzyme subunit 2 C-terminus (UBA2-C). The E1 enzyme domain of P. fulvidraco uba2 included several specific hits, such as the putative ATP binding sites, putative substrate interface, the catalytic residue and the putative zinc binding sites (Supplementary Figure 3). The protein sequence of P. fulvidraco ubc9 contained the UBCc domain, E3 interaction residues, active cysteine residue and Ub thioester intermediate interaction residues (Supplementary Figure 4). The protein sequence of P. fulvidraco pias1 revealed all the characteristic features of the PIAS protein family, including the SAP (Saf-A/B, Acinus, and Pias) domain, the PINIT domain, a DWNN domain and the MIZ/SP-RING zinc finger. The MIZ/SP-RING zinc finger contained SP-RING finger motif and Zn binding sites (Supplementary Figure 5). As the member of SUMO-specific proteases, senp1, senp2, and senp3 from P. fulvidraco possessed three distinct functional domains, such as the ubiquitin and ubiquitin-like protein 1 domain (Ulp1), the PLN03189 domain (protease specific for SUMO) and the C-terminal catalytic domain (Supplementary Figures 6–8).
Phylogenetic tree was performed with putative amino acid sequences, and further revealed the inferred evolutionary relationship. For sumo, P. fulvidraco sumo2, sumo3, and sumo1 were separated into two clades. sumo2 and sumo3 were in a clade but sumo1 clustered separately (Supplementary Figure 9). For E1 activated enzymes, sae1 and uba2 clustered close with Ictalurus punctatus but far from Poecilia reticulata, Poecilia formosa, and Poecilia latipinna. The P. fulvidraco ubc9 was closer to teleost and more distant from insecta and crustacea, in agreement with established taxonomic relationship. pias1 from P. fulvidraco clustered with Ictalurus punctatus but appeared far from the cluster of amphibians, aves and mammals. According to the amino acid sequences of P. fulvidraco sae1, uba2, ubc9 and pias1, all teleost formed an independent cluster, while mammals formed another cluster (Supplementary Figures 10–13). Similar to sumo, senp1, senp2, and senp3 were clearly divided into three clades, and senp2 was closer to senp1 (Supplementary Figure 14). Moreover, phylogenetic tree indicated that these amino acid sequences from P. fulvidraco were closer to the corresponding members of fish than those of amphibians and mammals, suggesting that the names of the genes were correct.
Tissue Distribution of Gene Expression
The mRNA levels of 10 SUMOylation-related genes were detected in all the tested tissues, but at variable levels (Figure 1). Generally speaking, these genes showed relatively high mRNA levels in ovary, gill and testis, and lowest mRNA levels in liver, muscle, intestine, and mesenteric fat.
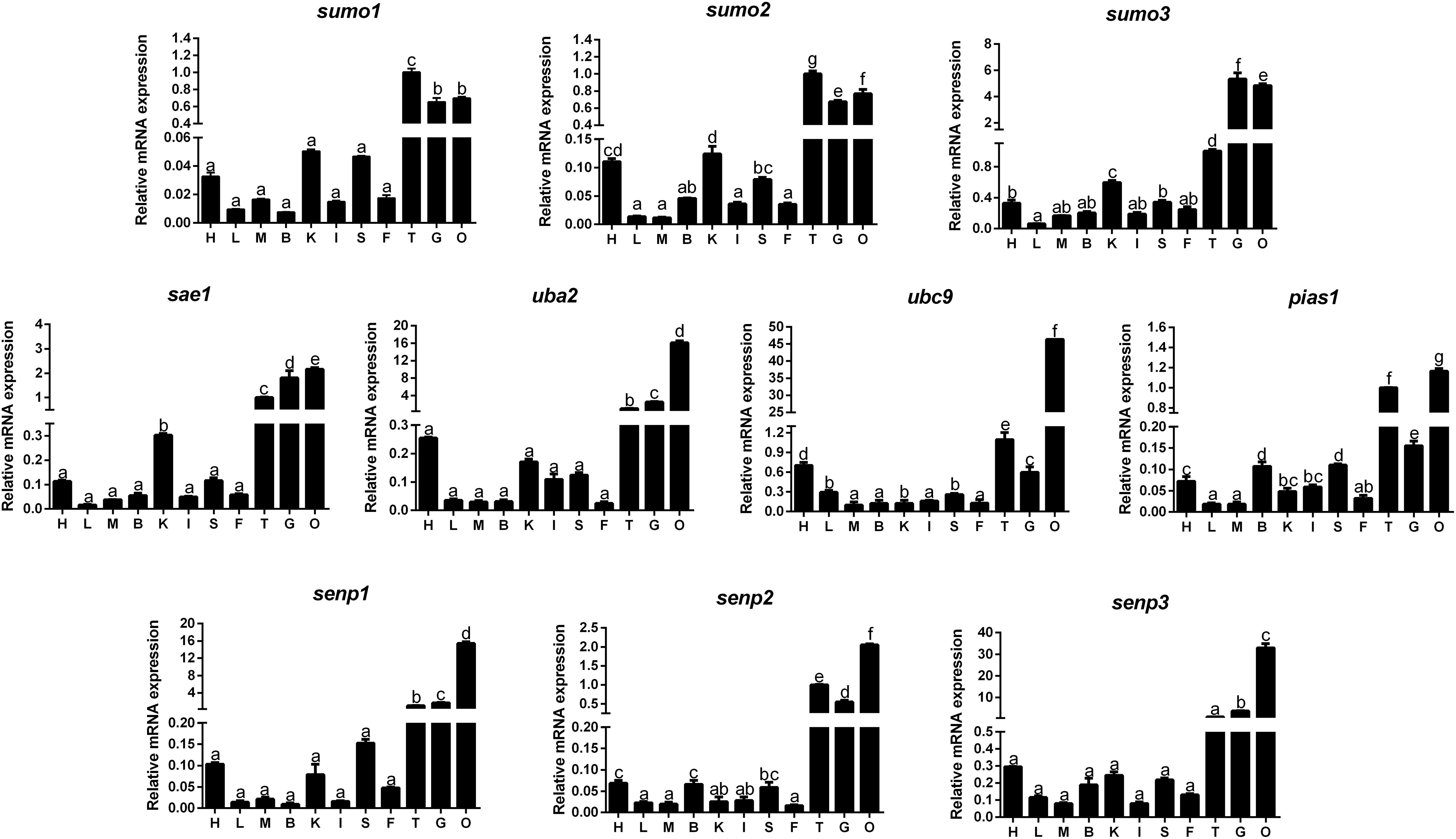
FIGURE 1. Quantitative PCR (Q-PCR) analysis for tissue distribution of gene expression across heart (H), liver (L), muscle (M), brain (B), kidney (K), intestine (I), spleen (S), mesenteric fat (F), testis (T), gill (G) and ovary (O) of Pelteobagrus fulvidraco. Data (mean ± SEM, n = 3) were expressed relative to expression of housekeeping gene [β-actin and elfa (M = 0.6245)]. Bars that share different letters indicate significant differences among groups (P < 0.05).
Transcriptional Responses of SUMOylation-Related Genes to Different Dietary Carbohydrate in Distinct Tissues
The mRNA abundances of SUMOylation-related genes in liver, muscle, testis, and ovary were responsive to three dietary carbohydrate levels. In liver (Figure 2), the mRNA levels of sumo1, sae1, ubc9, and pias1 were the highest for fish fed the middle carbohydrate diets and showed no significant differences between other two groups. mRNA levels of sumo2 and uba2 were the highest for fish fed the middle carbohydrate diet and lowest for fish fed the control. mRNA levels of sumo3 and senp2 were lowest for fish fed the control and showed no significant differences between other two groups. senp3 mRNA levels were the highest fish fed the highest carbohydrate diets and showed no significant differences between other two groups. senp1 mRNA levels showed no significant differences among three groups.
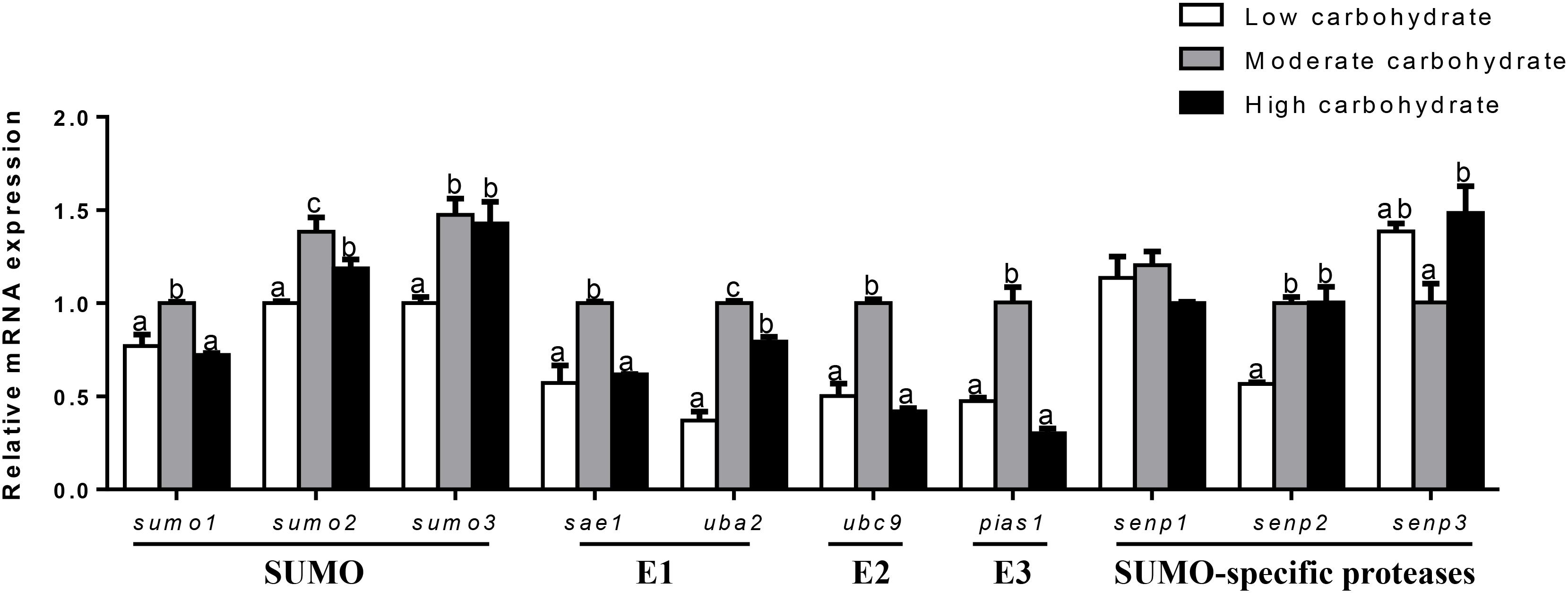
FIGURE 2. Effect of different dietary carbohydrate levels on hepatic mRNA expression of SUMOylation related genes in P. fulvidraco. a,b,cValues are mean ± SEM, n = 3 (replicates of three fish) and data were expressed relative to expression of housekeeping gene [β-actin and hprt (M = 0.2627)]; bars that share different letters within the same gene indicate significant differences among three groups (P < 0.05).
In muscle (Figure 3), sumo1 mRNA levels were the highest for fish fed the control and lowest for fish fed middle carbohydrate diet. mRNA levels of sumo2, sumo3, ubc9, and senp2 were highest for fish fed the control and showed no significant differences between other two groups. uba2 mRNA levels were the highest for fish fed the highest carbohydrate diets and showed no significant differences between other two groups. pias1 mRNA levels were the lowest for fish fed the middle carbohydrate diets and showed no significant differences between other two groups. senp3 mRNA levels declined with increasing dietary carbohydrate levels. mRNA levels of sae1 and senp1 showed no significant differences among three groups.
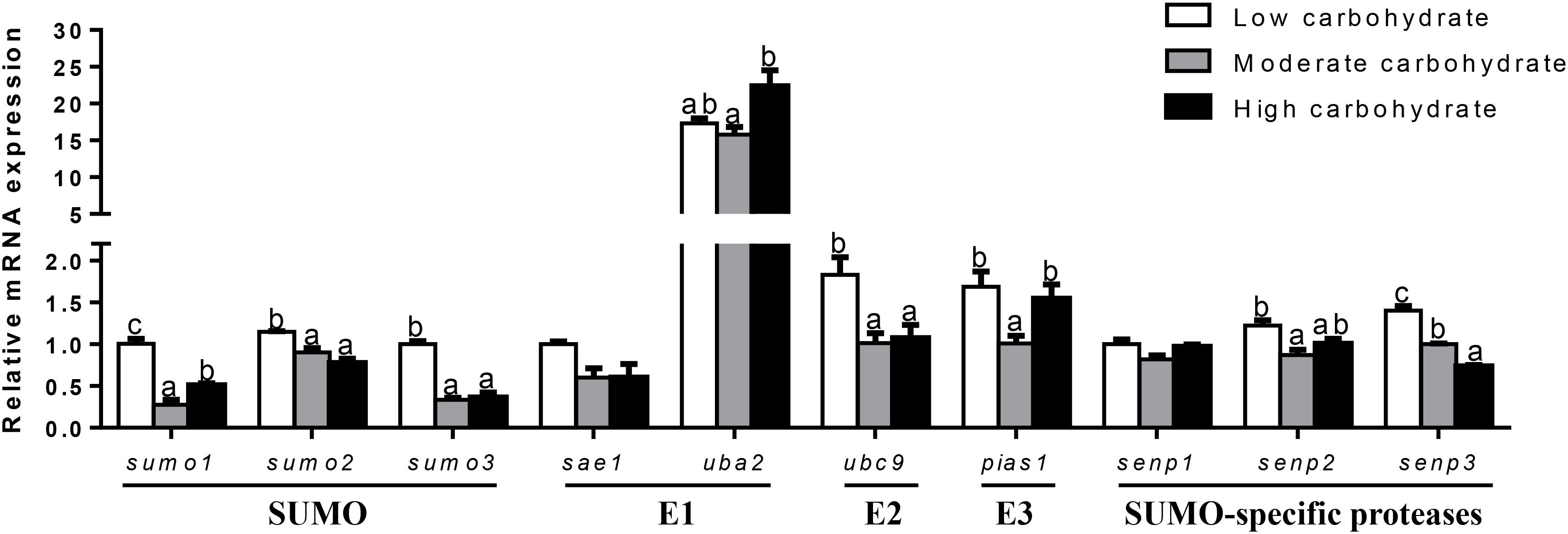
FIGURE 3. Effect of different dietary carbohydrate levels on muscle mRNA expression of SUMOylation related genes in P. fulvidraco. a,b,cValues are mean ± SEM, n = 3 (replicates of three fish) and data were expressed relative to expression of housekeeping gene [elfa and tbp (M = 0.0484)]; bars that share different letters within the same gene indicate significant differences among groups (P < 0.05). The absence of different letters indicates no significant differences.
In the testis (Figure 4), sumo1 mRNA levels were the highest for fish fed the highest carbohydrate diet and lowest for fish fed the middle carbohydrate diet. mRNA levels of sumo2, uba2, senp1, and senp2 increased with increasing dietary carbohydrate levels. mRNA expression of sumo3, sae1, and ubc9 was the highest for fish the highest carbohydrate diets and showed no significant differences between other two groups. pias1 mRNA expression was the highest for fish fed the middle carbohydrate diets and showed no significant differences between other two groups. senp3 mRNA expression showed no significant differences among three groups.
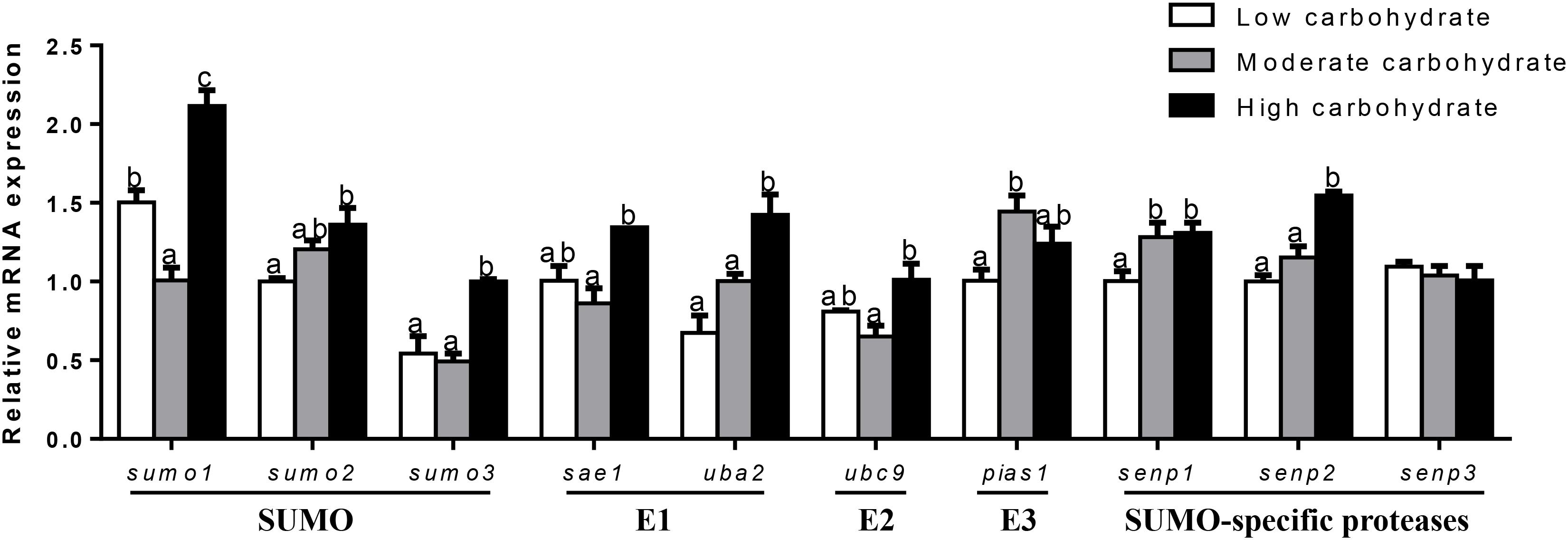
FIGURE 4. Effect of different dietary carbohydrate levels on testis mRNA expression of SUMOylation related genes in P. fulvidraco. a,b,cValues are mean ± SEM, n = 3 (replicates of three fish) and data were expressed relative to expression of housekeeping gene [rpl7 and ubce (M = 0.1046)]; bars that share different letters within the same gene indicate significant differences among groups (P < 0.05). The absence of different letters indicates no significant differences.
In ovary (Figure 5), mRNA expression of sumo1, sumo2, and sae1 showed no significant differences among three groups. mRNA expression of sumo3 and ubc9 declined with increasing dietary carbohydrate levels. uba2 mRNA abundance was the highest for fish fed the middle carbohydrate diets and lowest for fish the control. mRNA expression of pias1 and senp2 were the lowest for fish fed the control and showed no significant differences between other two groups. senp3 mRNA expression was the lowest for fish fed highest carbohydrate diet and showed no significant differences between other two groups. senp1 mRNA levels were the highest for fish fed the middle carbohydrate diet and showed no significant differences between other two groups.
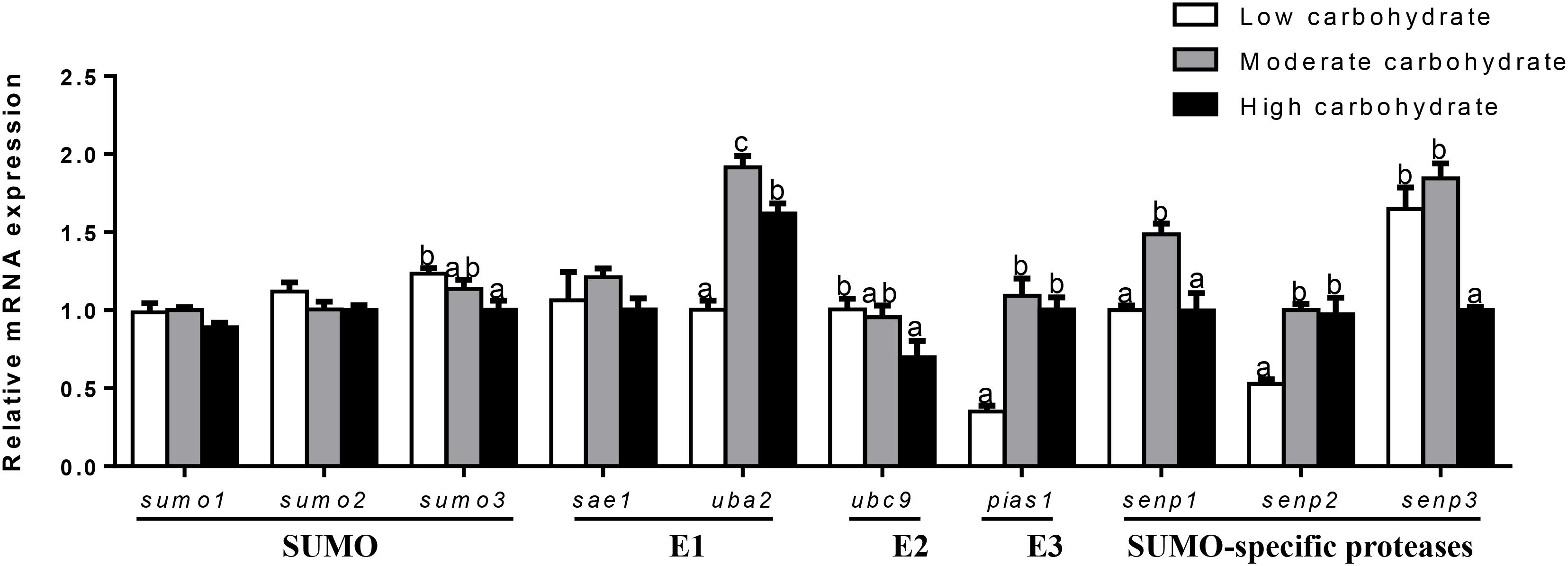
FIGURE 5. Effect of different dietary carbohydrate levels on ovarian mRNA expression of SUMOylation related genes in P. fulvidraco. a,b,cValues are mean ± SEM, n = 3 (replicates of three fish) and data were expressed relative to expression of housekeeping gene [β-actin and tbp (M = 0.1636)]; bars that share different letters within the same gene indicate significant differences among groups (P < 0.05). The absence of different letters indicates no significant differences.
Effect of Glucose Incubation on Cell Viability, mRNA Expressions of Sumoylation Related Genes in Primary Hepatocytes From Yellow Catfish
Glucose treatment showed no significant effect on cell viability among the three treatments (Figure 6).
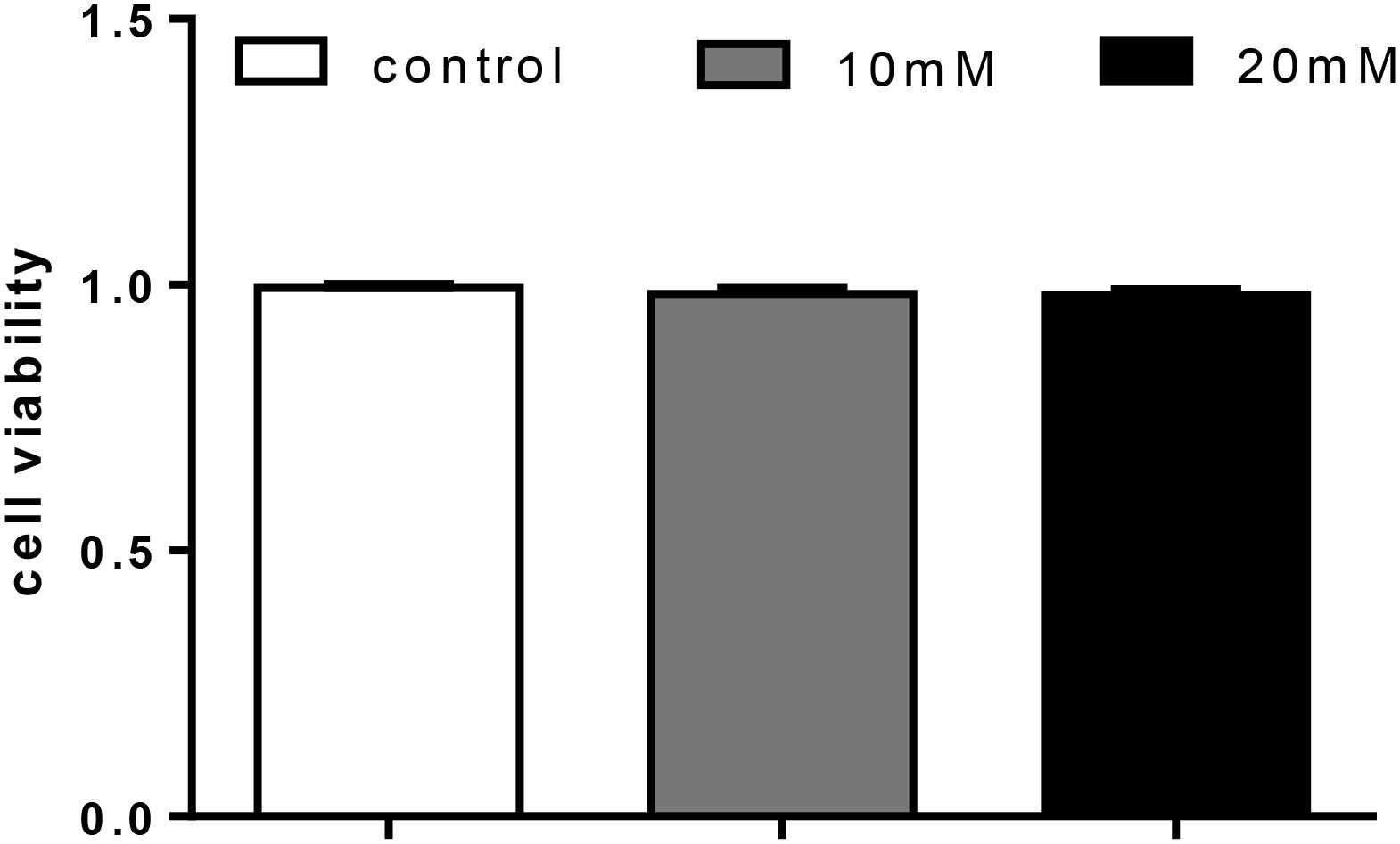
FIGURE 6. Effect of glucose [control (5.41 mmol/L), 10 mM (9.75 mmol/L), 20 mM (20.38 mmol/L)] on cell livability in primary hepatocytes from yellow catfish, and incubation lasted for 48 h. Values are mean ± SEM, n = 3 (replicates of three biological experiments); Bars that did not mark different letters indicate no significant differences among groups (P < 0.05).
At 12 h (Figure 7), mRNA expression of senp3 showed no significant differences among three groups. sumo1 and sumo2 mRNA abundances were the highest for 20 mM glucose and the lowest for 10 mM glucose. mRNA expressions of sumo3, sae1, and senp2 were the lowest for 10 mM glucose and showed no significant differences between other two groups. uba2 and ubc9 mRNA abundances were the highest for control group, senp1 mRNA levels were the highest for 20 mM glucose and showed no significant differences between other two groups. pias1 mRNA abundance was the highest for control group and the lowest for 10 mM glucose.
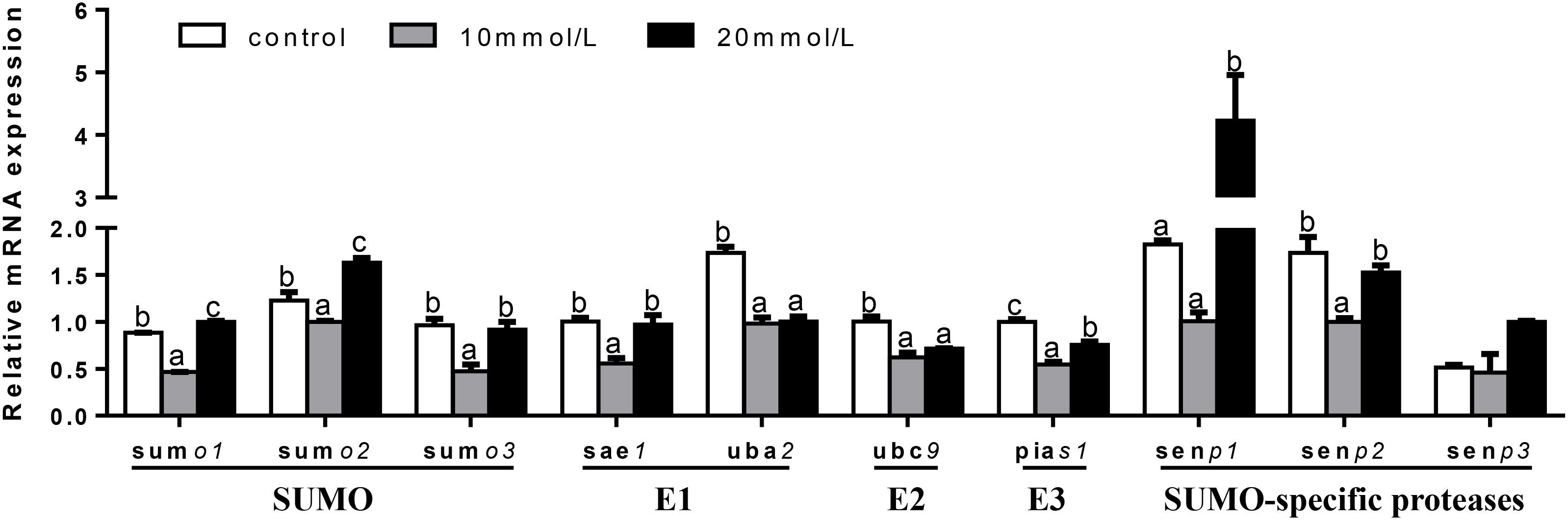
FIGURE 7. Effects of glucose (control, 10 and 20 mM) incubation on mRNA expressions of SUMOylation related genes in primary hepatocytes from yellow catfish, and incubation lasted for 12 h. a,b,cValues are mean ± SEM, n = 3 (replicates of three biological experiments) and data were expressed relative to expression of housekeeping gene [b2m and rpl7 (M = 0.1078)]; bars that share different letters within the same gene indicate significant differences among groups (P < 0.05).
At 24 h (Figure 8), mRNA expression of sumo2 and ubc9 showed no significant differences among three groups. sumo1 and senp3 mRNA abundances were the lowest for control group and showed no significant differences between other two groups. sumo3, pias1, and senp2 mRNA abundances were the highest for 10 mM glucose and showed no significant differences between other two groups. sae1 mRNA expression was the highest for 20 mM glucose and showed no significant differences between other two groups. mRNA expressions of uba2 and senp1 were the highest for 10 mM glucose and the lowest for control group.
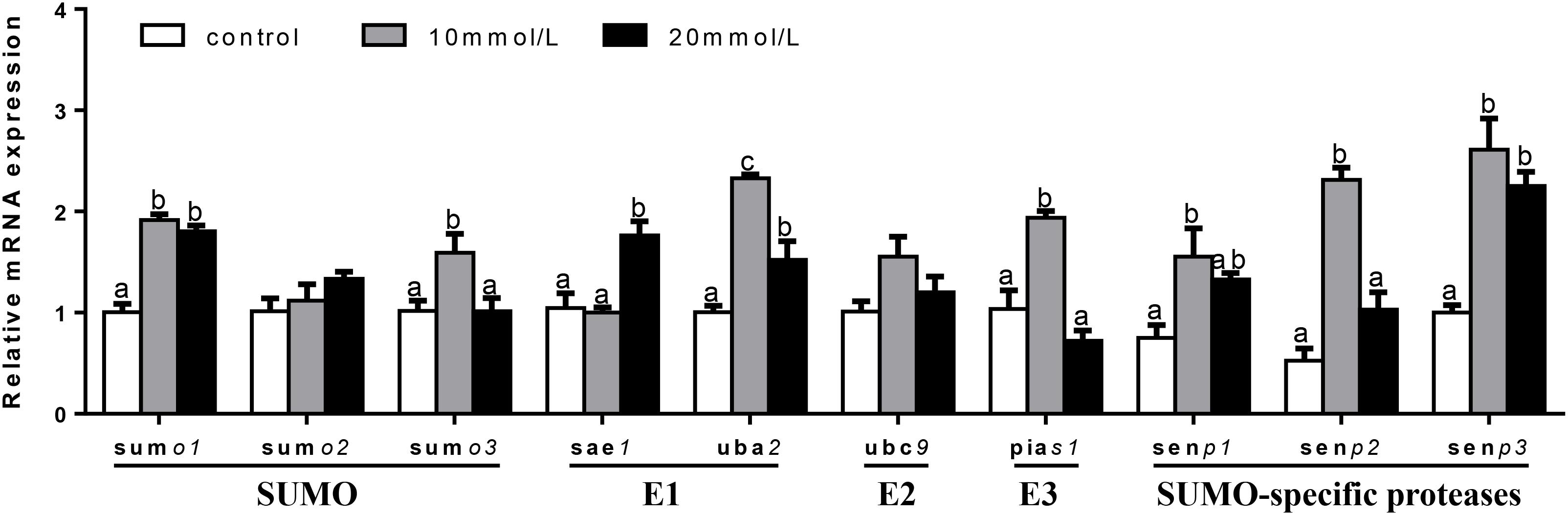
FIGURE 8. Effects of glucose (control, 10 and 20 mM) incubation on mRNA expressions of SUMOylation related genes in primary hepatocytes from yellow catfish, and incubation lasted for 24 h. a,b,cValues are mean ± SEM, n = 3 (replicates of three biological experiments) and data were expressed relative to expression of housekeeping gene [hprt and rpl7 (M = 0.0808)]; bars that share different letters within the same gene indicate significant differences among groups (P < 0.05).
At 48 h (Figure 9), mRNA expression of ubc9 and senp3 showed no significant differences among three groups. sumo1, sumo2, sumo3, and sae1 mRNA levels were the highest for 20 mM glucose and the lowest for 10 mM glucose. uba2 and senp2 mRNA abundances were the highest for 20 mM glucose and showed no significant differences between other two groups. pias1 mRNA expression was the highest for 10 mM glucose and the lowest for 20 mM glucose. mRNA levels of senp1 increased with increasing glucose concentrations.
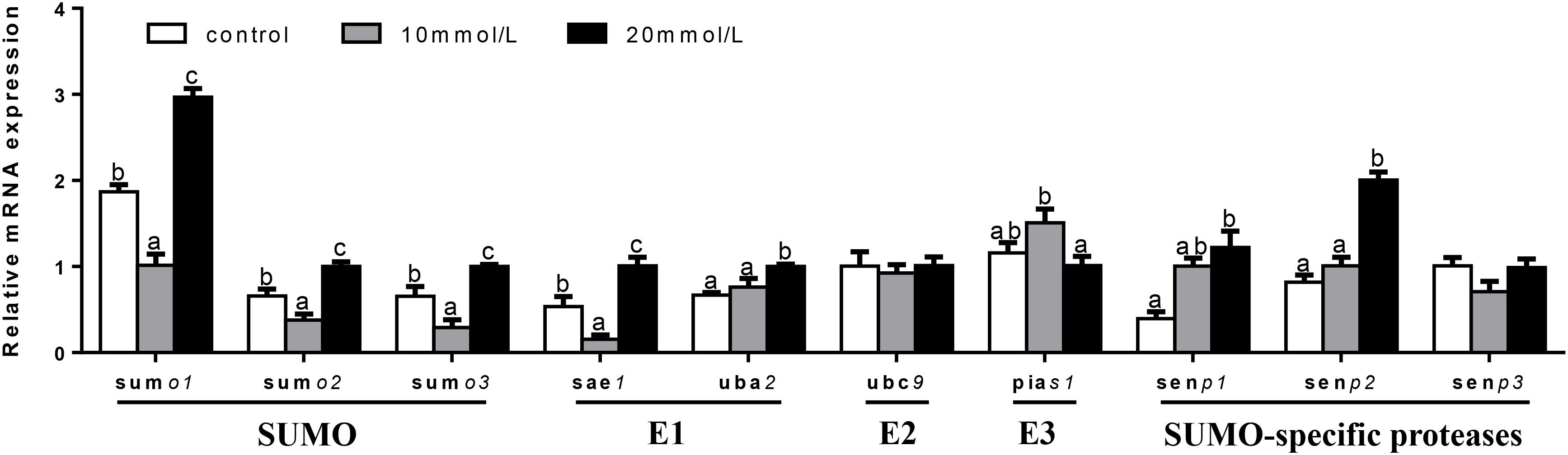
FIGURE 9. Effects of glucose (control, 10 and 20 mM) incubation on mRNA expressions of SUMOylation related genes in primary hepatocytes from yellow catfish, and incubation lasted for 48 h. a,b,cValues are mean ± SEM, n = 3 (replicates of three biological experiments) and data were expressed relative to expression of housekeeping gene [ubce and rpl7 (M = 0.0994)]; bars that share different letters within the same gene indicate significant differences among groups (P < 0.05).
Discussion
SUMOylation plays many important roles in signaling pathway, transcriptional regulation, and cell cycle progression. In the present study, for the first time, we identified the cDNA sequences of 10 SUMOylation genes, explored their mRNA tissue expression profiles and transcriptional responses to carbohydrate levels both in vivo and in vitro, which establish a basis for further investigation on their functions.
In the present study, the protein sequences of P. fulvidraco sumo1, sumo2, and sumo3 revealed similar domains with mammals, indicating that the protein’s structure and function of SUMO were highly evolutionarily conserved, as reported by Yuan et al. (2010). sumo1, sumo2, and sumo3 in yellow catfish had the conserved Gly–Gly motifs in their C-terminus, which are the critical region for their maturation and conjugation (Bayer et al., 1998; Xu et al., 2016). Comparing to sumo1, sumo2/sumo3 of yellow catfish possess a VKTE motif, consistent with the SUMOylation consensus YKXE (Y represents a hydrophobic amino acid, X means any amino acid), and this consensus sequence is functional for possible polymerization (Tatham et al., 2001). In contrast, in mud crab, Dai et al. (2012) reported that SUMO1 also contained an AKPE motive site.
The present study also indicated that the P. fulvidraco sae1 consisted of the conserved ThiF domain. The ThiF domain in sae1 was related to the nucleotide-binding function (Dou et al., 2005). The P. fulvidraco uba2 comprised E1 enzyme family domain, the Ubiquitin/SUMO-activating enzyme ubiquitin-like domain (UAE-Ubl), and the SUMO-activating enzyme subunit 2 C-terminus (UBA2-C). These conserved domains of uba2 were similar to these in mammals (Schwartz and Hochstrasser, 2003; Huang et al., 2004). In the present study, the protein sequence of P. fulvidraco ubc9 contained the UBCc domain, E3 interaction residues, active cysteine residue and Ub thioester intermediate interaction residues. Similar domains of ubc9 have been reported by Hu and Chen (2013). The PIAS protein family acts as an E3 ubiquitin ligase in the SUMOylation pathway. Our study indicated that the protein sequence of P. fulvidraco pias1 revealed all the characteristic features of the PIAS protein family, including the SAP (after SAF-A/B, Acinus, and PIAS) domain, the Pro-Ile-Asn-Ile-Thr (PINIT) domain, a DWNN domain and the MIZ/SP-RING zinc finger, in agreement with other reports (Duval et al., 2003; Huang et al., 2015). A RING finger signature confers an E3-SUMO ligase function (Palvimo, 2007).
SENP1, SENP2, and SENP3 all belong to SUMO-specific proteases. As an unique crucial deSUMOylation-specific proteinase, SENPs play an important role to determine the intracellular SUMOylation level (Xia et al., 2016). In the present study, senp1, senp2, and senp3 from P. fulvidraco possessed three distinct functional domains, such as the ubiquitin and ubiquitin-like protein 1 domain (Ulp1), the PLN03189 domain and the C-terminal catalytic domain, in agreement with other reports (Drag and Salvesen, 2008). The C-terminal catalytic domain of ubiquitin-like 1 domain contains the catalytic triad Cys-His-Asn and appears to function with sSUMOylation and deSUMOylation (Mossessova and Lima, 2000).
In this work, we examined the tissue distribution of these identified genes as a preliminary step to shed some light on their physiological roles. The present study demonstrated that the mRNA levels of 10 SUMOylation-related genes were detected in all the tested tissues, suggesting that they participated in general SUMOylation reactions occurring at basal levels in all tissues. In fish, to our best knowledge, mRNA tissue expression profiles of SUMOylation-relevant genes have been reported only in ubc9 (Hu and Chen, 2013), sumo1 and sumo2 (Xu et al., 2016) and pias1 (Wong, 2013) and these studies indicated that they were widely distributed. Moreover, our study indicated that these genes showed relatively high mRNA levels in ovary, gill and testis, and lowest mRNA levels in liver, muscle, intestine, and mesenteric fat. Similarly, several studies have demonstrated that sumo1 gene was highly expressed in the ovary (Shen et al., 1996; Kamitani et al., 1998; Dai et al., 2012) and testis (Shen et al., 1996). Huang et al. (2015) found the higher expression level of Pias1 in the gonad of S. paramamosain, and implied that pias1 may exert an important function in the maintenance of spermatogenesis. High expression of these genes in the ovary implies that the SUMOylation and deSUMOylation were highly active in the regulation of gonad development (Hu and Chen, 2013). In mouse, Ihara et al. (2008) also reported that Ubc9 had an important role in the development of oocytes. High mRNA expression of sumo1 and sumo2 in gills also had been reported in grouper (Xu et al., 2016). The gills were primary sites where fish encounter environmental stresses. Therefore, we can reasonably infer that the high expression level of these genes in the gills might be a sensor for environmental stress in fish that stimulates the SUMOylation and deSUMOylation responses.
In mammals, several studies indicated that SUMOylation modification of proteins were influenced by carbohydrate (or glucose) levels (Huang et al., 2013; Jung et al., 2016), but no such studies were conducted in fish. For the first time, our study explored the effects of dietary carbohydrate levels on mRNA expressions of the 10 genes in four tissues in fish, and detected the effect of glucose incubation on mRNA expressions of SUMOylation related genes in primary hepatocytes from yellow catfish. Our study indicated that dietary carbohydrate levels differentially influenced their mRNA expression in a dose- and tissue-dependent manner, and glucose incubation differentially influence their mRNA expression in dose- and time-dependent ways. At present, we did not know the exact reason and further mechanism for carbohydrate (glucose)-induced changes of their mRNA expression levels. However, since these genes and proteins mediated the regulation of many transcriptional factors and signaling pathways related to nutrient metabolism (Sentis et al., 2005; Chung et al., 2011; Koo et al., 2015), it is understandable that the carbohydrate-induced changes of these gene expression will in turn influence the utilization and metabolism of carbohydrate in fish. Similarly, Huang et al. (2013) reported that high glucose increased the expression of sumo1 and sumo2/3 in glomerular mesangial cells of the rats. Jung et al. (2016) pointed out that different stimulations were required for inducing different SENPs according to cell types, and concluded that chronic high glucose was responsible for inducing Senp2 expression in insulin-producing cells.
Conclusion
In summary, for the first time, we characterized the cDNA sequences of 10 SUMOylation related genes from yellow catfish, determined their transcriptional responses to different levels of dietary carbohydrate, and detected the effect of different levels glucose on mRNA expressions of SUMOylation related genes in primary hepatocytes, which would contribute to our understanding of the molecular basis of SUMOylation and the potential mechanisms of SUMOylation modifications in influencing carbohydrate metabolism in fish. Further studies should be directed to analyze the functional roles of these identified genes and proteins, and clarify the mechanism of dietary carbohydrate levels influencing their transcriptional responses.
Author Contributions
S-BY and ZL designed the experiments. S-BY conducted the experiments, with the help of X-YT, D-GZ, and JC. S-BY, ZL, and X-YT analyzed the data. S-BY drafted the manuscript. ZL revised the manuscript. All the authors read and approved the manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities, China (Grant No. 2662018PY089).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the staff of Aquatic Animal Nutrition and Feed Laboratory of Huazhong Agricultural University for their assistance with excellent sample analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01544/full#supplementary-material
Abbreviations
DMSO, dimethyl sulfoxide; FBS, fetal bovine serum; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide; ORF, open reading fame; PBS, phosphate buffered saline; PIAS1, protein inhibitor of activated STAT1; RACE, rapid-amplification of cDNA ends; RT-PCR, reverse transcription-PCR; Sae1, SUMO-activating enzyme subunit 1; SAP, Saf-A/B, Acinus and Pias; SEM, standard error of mean; SENP, sentrin-specific protease; Smt3, yeast ubiquitin-like protein; SUMO, small ubiquitin-related modifier; UAE-Ubl, ubiquitin/SUMO-activating enzyme ubiquitin-like protease; Uba2, SUMO-activating enzyme subunit 2; UBA2-C, SUMO-activating enzyme subunit 2 C-terminus; Ubc9, ubiquitin-conjugating enzyme 9; UBCc, ubiquitin-conjugating enzyme E2 catalytic; UBQ, ubiquitin and ubiquitin-like protein; Ulp1, ubiquitin-like protease 1.
Footnotes
- ^http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^http://smart.embl-heidelberg.de/
- ^http://guidance.tau.ac.il/
References
Bailey, D., and O’Hare, P. (2004). Characterization of the localization and proteolytic activity of the SUMO-specific protease. SENP1. J. Biol. Chem. 279, 692–703. doi: 10.1074/jbc.M306195200
Bayer, P., Arndt, A., Metzger, S., Mahajan, R., Melchior, F., Jaenicke, R., et al. (1998). Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280, 275–286. doi: 10.1006/jmbi.1998.1839
Chen, F., Lee, Y., Jiang, Y., Wang, S., Peatman, E., Abernathy, J., et al. (2010). Identification and characterization of full-length cDNAs in channel catfish (Ictalurus punctatus) and blue catfish (Ictalurus furcatus). PLoS One 5:e11546. doi: 10.1371/journal.pone.0011546
Cheng, J., Luo, Z., Chen, G. H., Wei, C. C., and Zhuo, M. Q. (2017). Identification of eight copper (Cu) uptake related genes from yellow catfish Pelteobagrus fulvidraco, and their tissue expression and transcriptional responses to dietborne Cu exposure. J. Trace Elem. Med. Biol. 44, 256–265. doi: 10.1016/j.jtemb.2017.09.004
Chung, S. S., Ahn, B. Y., Kim, M., Kho, J. H., Jung, H. S., and Park, K. S. (2011). SUMO modification selectively regulates transcriptional activity of peroxisome-proliferator-activated receptor gamma in C2C12 myotubes. Biochem. J. 433, 155–161. doi: 10.1042/BJ20100749
Dai, Y., Han, K., Zou, Z., Yan, S., Wang, Y., and Zhang, Z. (2012). SUMO-1 of mud crab (Scylla paramamosain) in gametogenesis. Gene 503, 260–268. doi: 10.1016/j.gene.2012.04.056
Dou, T., Gu, S., Liu, J., Chen, F., Zeng, L., Guo, L., et al. (2005). Isolation and characterization of ubiquitin-activating enzyme E1-domain containing 1, UBE1DC1. Mol. Biol. Rep. 32, 265–271. doi: 10.1007/s11033-005-4822-y
Drag, M., and Salvesen, G. S. (2008). DeSUMOylating enzymes-SENPs. IUBMB Life 60, 734–742. doi: 10.1002/iub.113
Duval, D., Duval, G., Kedinger, C., Poch, O., and Boeuf, H. (2003). The ’PINIT’ motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 554, 111–118. doi: 10.1016/S0014-5793(03)01116-5
Gareau, J. R., and Lima, C. D. (2010). The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871. doi: 10.1038/nrm3011
Hay, R. T. (2005). SUMO: a history of modification. Mol. Cell 18, 1–12. doi: 10.1016/j.molcel.2005.03.012
Hu, Q., and Chen, S. (2013). Cloning, genomic structure and expression analysis of ubc9 in the course of development in the half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 165, 181–188. doi: 10.1016/j.cbpb.2013.03.007
Huang, A. M., Geng, Y., Fang, W. H., Wang, K. Y., Chen, D. F., Huang, X. L., et al. (2015). cDNA cloning and expression pattern analysis of protein inhibitor of activated STAT (PIAS) of the mud crab. Scylla paramamosain. Aquaculture 444, 21–27. doi: 10.1016/j.aquaculture.2015.03.023
Huang, W., Xu, L., Zhou, X., Gao, C., Yang, M., Chen, G., et al. (2013). High glucose induces activation of NF-κB inflammatory signaling through IκBα sumoylation in rat mesangial cells. Biochem. Biophys. Res. Commun. 438, 568–574. doi: 10.1016/j.bbrc.2013.07.065
Huang, W. C., Ko, T. P., Li, S. S., and Wang, A. H. (2004). Crystal structures of the human SUMO-2 protein at 1.6 A and 1.2 A resolution: implication on the functional differences of SUMO proteins. Eur. J. Biochem. 271, 4114–4122. doi: 10.1111/j.1432-1033.2004.04349.x
Ihara, M., Stein, P., and Schultz, R. M. (2008). UBE2I (ubc9), a SUMO-conjugating enzyme, localizes to nuclear speckles and stimulates transcription in mouse oocytes. Biol. Reprod. 79, 906–913. doi: 10.1095/biolreprod.108.070474
Johnson, E. S. (2004). Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382. doi: 10.1146/annurev.biochem.73.011303.074118
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi: 10.1093/bioinformatics.8.3.275
Jung, H. S., Kang, Y. M., Park, H. S., Ahn, B. Y., Lee, H., Kim, M. J., et al. (2016). Senp2 expression was induced by chronic glucose stimulation in INS1 cells, and it was required for the associated induction of Ccnd1 and Mafa. Islets 8, 207–216. doi: 10.1080/19382014.2016.1235677
Kamitani, T., Kito, K., Nguyen, H. P., Fukuda-Kamitani, T., and Yeh, E. T. (1998). Characterization of a second member of the sentrin family of ubiquitin-like proteins. J. Biol. Chem. 273, 11349–11353. doi: 10.1074/jbc.273.18.11349
Koo, Y. D., Choi, J. W., Kim, M., Chae, S., Ahn, B. Y., Kim, M., et al. (2015). SUMO-specific protease 2 (SENP2) is an important regulator of fatty acid metabolism in skeletal muscle. Diabetes Metab. Res. Rev. 64, 2420–2431. doi: 10.2337/db15-0115
Krumova, P., and Weishaupt, J. H. (2013). Sumoylation in neurodegenerative diseases. Cell. Mol. Life Sci. 70, 2123–2138. doi: 10.1007/s00018-012-1158-3
Mossessova, E., and Lima, C. D. (2000). Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876. doi: 10.1016/S1097-2765(00)80326-3
Obata, S., Yuasa, E., Seki, D., Kitano, T., and Saitoh, H. (2013). Molecular cloning and bacterial expression of the catalytic domain of the SENP1 gene of Oryzias latipes. Biosci. Biotechnol. Biochem. 77, 1788–1791. doi: 10.1271/bbb.130357
Palvimo, J. J. (2007). PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem. Soc. Trans. 35, 1405–1408. doi: 10.1042/BST0351405
Schwartz, D. C., and Hochstrasser, M. (2003). A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends. Biochem. Sci. 28, 321–328. doi: 10.1016/S0968-0004(03)00113-0
Sentis, S., Le Romancer, M., Bianchin, C., Rostan, M. C., and Corbo, L. (2005). Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol. Endocrinol. 19, 2671–2684. doi: 10.1210/me.2005-0042
Shen, Z., Pardington-Purtymun, P. E., Comeaux, J. C., Moyzis, R. K., and Chen, D. J. (1996). UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics 36, 271–279. doi: 10.1006/geno.1996.0462
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tatham, M. H., Jaffray, E., Vaughan, O. A., Desterro, J. M., Botting, C. H., Naismith, J. H., et al. (2001). Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374. doi: 10.1074/jbc.M104214200
Wei, C. C., Luo, Z., Song, Y. F., Pan, Y. X., Wu, K., and You, W. J. (2017). Identification of autophagy related genes LC3 and ATG4 from yellow catfish Pelteobagrus fulvidraco and their transcriptional responses to waterborne and dietborne zinc exposure. Chemosphere 175, 228–238. doi: 10.1016/j.chemosphere.2017.02.042
Wei, C. C., Luo, Z., Song, Y. F., Pan, Y. X., and Zhuo, M. Q. (2018). Characterization of twelve autophagy-related genes from yellow catfish Pelteobagrus fulvidraco and their transcriptional responses to waterborne zinc exposure. Ecol. Indicat. 93, 677–686. doi: 10.1016/j.ecolind.2018.05.068
Wilkinson, K. A., and Henley, J. M. (2010). Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428, 133–145. doi: 10.1042/BJ20100158
Wong, W. W. (2013). Novel Functions of PIAS1 and PIAS4 as Intracellular Feedback Repressors of Growth Hormone Signaling in Grass Carp. Hong Kong: The University of Hong Kong.
Xia, W., Tian, H., Cai, X., Kong, H., Fu, W., Xing, W., et al. (2016). Inhibition of SUMO-specific protease 1 induces apoptosis of astroglioma cells by regulating NF-κB/Akt pathways. Gene 595, 175–179. doi: 10.1016/j.gene.2016.09.040
Xu, M., Wei, J. G., Chen, X. L., Gao, P., Zhou, Y. C., and Qin, Q. W. (2016). Molecular cloning and expression analysis of small ubiquitin-like modifier (SUMO) genes from grouper (Epinephelus coioides). Fish Shellfish Immunol. 48, 119–127. doi: 10.1016/j.fsi.2015.11.022
Yang, W., and Paschen, W. (2009). Gene expression and cell growth are modified by silencing SUMO2 and SUMO3 expression. Biochem. Biophys. Res. Commun. 382, 215–218. doi: 10.1016/j.bbrc.2009.03.013
Ye, W. J., Tan, X. Y., Chen, Y. D., and Luo, Z. (2009). Effects of dietary protein to carbohydrate ratios on growth and body composition of juvenile yellow catfish, Pelteobagrus fulvidraco (Siluriformes, Bagridae, Pelteobagrus). Aquacult. Res. 40, 1410–1418. doi: 10.1111/j.1365-2109.2009.02239.x
Yuan, H., Zhou, J., Deng, M., Liu, X., Bras, L. M., de The, H., et al. (2010). Small ubiquitin-related modifier paralogs are indispensable but functionally redundant during early development of zebrafish. Cell Res. 20, 185–196. doi: 10.1038/cr.2009.101
Zang, X. W. (2013). Characterization of Genes Involved in SUMOylation During Embryogenesis in Rainbow Trout (Oncorhynchus mykiss). Dissertations: West Virginia University.
Zhou, P., Zhang, Z. P., Wang, Y. L., Xie, F. J., and Zou, Z. H. (2009). Cloning and characterization of Ubc9 gene of large yellow croaker Larimichthys crocea and its expression pattern. Biotechnol. Bull. 8, 76–82.
Keywords: SUMOylation, molecular characterization, transcriptional responses, carbohydrate, fish
Citation: Yang S-B, Tan X-Y, Zhang D-G, Cheng J and Luo Z (2018) Identification of 10 SUMOylation-Related Genes From Yellow Catfish Pelteobagrus fulvidraco, and Their Transcriptional Responses to Carbohydrate Addition in vivo and in vitro. Front. Physiol. 9:1544. doi: 10.3389/fphys.2018.01544
Received: 26 June 2018; Accepted: 15 October 2018;
Published: 08 November 2018.
Edited by:
José Luis Soengas, University of Vigo, SpainReviewed by:
Luis Vargas-Chacoff, Universidad Austral de Chile, ChileBeiping Tan, Guangdong Ocean University, China
Copyright © 2018 Yang, Tan, Zhang, Cheng and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Luo, bHVvemhpOTlAbWFpbC5oemF1LmVkdS5jbg==; bHVvemhpOTlAYWxpeXVuLmNvbQ==
 Shui-Bo Yang1
Shui-Bo Yang1 Zhi Luo
Zhi Luo