- 1Key Laboratory of Tropical Biological Resources of Ministry of Education, Hainan University, Haikou, China
- 2Liaoning Key Laboratory of Marine Animal Immunology and Disease Control, Dalian Ocean University, Dalian, China
- 3Department of Marine Sciences, University of Connecticut, Groton, CT, United States
Vibrio coralliilyticus is known to cause coral diseases, especially under environmental perturbation, but its impact on coral physiology and underpinning mechanism is poorly understood. In the present study, we investigated cytological, immunological, and metatranscriptomic responses of the scleractinian coral Pocillopora damicornis to V. coralliilyticus infection. The density and chlorophyll content of symbiotic zooxanthellae decreased significantly at 12 and 24 h after Vibrio challenge. The activities of antioxidant enzymes such as superoxide dismutase and catalase, nitric oxide synthase, phenoloxidase (PO), and the activation level of caspase3 all rose significantly in P. damicornis after Vibrio challenge. In the metatranscriptomic analysis, we found 10 significantly upregulated genes in the symbionts at 24 h after the challenge, which were mostly involved in the metabolism of nucleic acid and polysaccharide, and 133 significantly down-regulated symbiont genes, which were mainly related to amino acid catabolism and transport. Meanwhile, 1432 significantly upregulated coral genes were revealed, highly overrepresented in GO terms that are mostly related to the regulation of immune response, the regulation of cytokine production, and innate immune response. Furthermore, at 24 h after Vibrio challenge, 890 coral genes were significantly downregulated, highly overrepresented in four GO terms implicated in defense response. These results in concert suggest that V. coralliilyticus infection triggered the innate immune response including the redox, PO, and apoptosis systems, but repressed the response of the complement system in the scleractinian coral P. damicornis, accompanied by symbiont density decrease and symbiosis collapse through disordering the metabolism of the symbionts. These findings shed light on the molecular regulatory processes underlying bleaching and degradation of P. damicornis resulting from the infection of V. coralliilyticus.
Introduction
Reef-building scleractinian corals are the pillar of the biologically and economically important coral reef ecosystems. Scleractinian corals flourish in shallow tropical waters with low and often undetectable concentrations of dissolved inorganic nutrients, depending on the elaborate coral-Symbiodinium endosymbiosis (Fabina et al., 2013). The unicellular symbiont lives in the intracellular vacuoles of host gastrodermal cells, and supplies the host coral with indispensable organic nutrients and oxygen through photosynthesis (Loram et al., 2007). In return the host coral provides inorganic nutrients and carbon dioxide for symbionts. However, due to environmental changes and anthropogenic perturbations, scleractinian corals have been subjected to impacts of frequent and severe diseases over the past decades (Rosenberg and Ben-Haim, 2002).
Bacterial pathogens are one of main etiologies of scleractinian coral diseases, and pathogenic Vibrio has been found in several scleractinian corals. For example, V. alginolyticus has been identified to be the pathogenic agent of Montastraea spp. yellow band disease in Caribbean and Indo-Pacific and Porites andrewsi white syndrome in the South China Sea (Cervino et al., 2008; Zhenyu et al., 2013). V. coralliilyticus is a coral bleaching pathogen, which was characterized firstly as temperature-dependent pathogen (strain YB1) of scleractinian coral Pocillopora damicornis (Ben-Haim et al., 2003a), and therefore has received attention. It has been reported that V. coralliilyticus used dimethylsulfoniopropionate as a chemotaxis and chemokinesis to target the mucus of the coral host (Garren et al., 2014), and resulted in coral tissue lysis and symbiont density decrease through extracellular proteases (Ben-Haim et al., 2003b). Furthermore, it has been observed recently that non-virulent V. coralliilyticus induced an “efficient” immune response, whereas virulent one caused immuno-suppression in the coral host (Vidal-Dupiol et al., 2014). However, the molecular mechanisms underlying the pathogenesis process of V. coralliilyticus to scleractinian corals, especially for symbiosis collapse, remain relatively poorly understood and underexplored particularly by high-throughput transcriptome sequencing technology.
Scleractinian corals possess a suite of innate immune defense mechanisms that enable them to maintain a close association with bacterial communities and eliminate potentially harmful microbes (van de Water et al., 2015). Pattern recognition receptors such as lectin, Toll/NOD-like receptor, and scavenger receptor identified in scleractinian corals can recognize the potential bacterial pathogen (Kvennefors et al., 2008; Hamada et al., 2013; Neubauer et al., 2016). The components of Toll-like receptor-to-NF-κB signaling pathway have been identified, and their immune-related functions have also been confirmed (Libro et al., 2013; Poole and Weis, 2014; Williams et al., 2018). The redox system, phenoloxidase (PO) system, complement system, antimicrobial peptide, apoptosis, and autophagy of scleractinian coral all have been thought to be involved in the killing and elimination of pathogenic microbes (Palmer et al., 2011; Vidal-Dupiol et al., 2011a; Brown et al., 2013; Fuess et al., 2017). Furthermore, innate immunity in scleractinian corals is implicated not only in the defense against invasive pathogenic microbes but also in the maintenance of coral-Symbiodinium symbiosis (Kvennefors et al., 2010; Zhou et al., 2017).
Pocillopora damicornis is a scleractinian coral in the family Pocilloporidae, and is widely distributed in the tropical and subtropical regions of the Indian and Pacific Oceans. It has been used as a model to address effects of Vibrio infection (Vidal-Dupiol et al., 2011b) and interactive effects of thermal stress and Vibrio infection (Garren et al., 2014). To elaborate the immune defense mechanism of scleractinian coral to pathogenic Vibrio as well as its effect on coral-Symbiodinium symbiosis, the symbiont density and chlorophyll content, the crucial immune parameters, and the metatranscriptome of coral P. damicornis were investigated after the challenge of pathogen V. coralliilyticus. The results provide insights for further understanding the potent functions and potential mechanisms of innate immune response in pathogen elimination and symbiosis maintenance in the scleractinian coral.
Materials and Methods
Coral
Colonies of the scleractinian coral P. damicornis were collected from a coral reef in Wenchang, Hainan Province, China, and transported to Hainan University where it was cultured in flow-through aquaria (ca. 500 L) filled with natural seawater (26°C). Cultures were illuminated with LED light bulbs in a 12 h/12 h light–dark cycle (light intensity 518 μmol photons m-2 s-1) for 1 month to acclimatize in laboratory conditions.
Vibrio Challenge Experiment
The Gram-negative bacterium Vibrio coralliilyticus SCSIO 43001 was kindly provided by Dr. Xiaoxue Wang and grown in marine broth 2216E at 28°C. Live V. coralliilyticus were added into the natural seawater with a final concentration of 108 CFU ml-1, and meanwhile a total of 60 coral nubbins were prepared for the Vibrio challenge experiment. Then, 25 of the coral nubbins were transferred into the Vibrio-containing seawater, hereafter referred to as the Vibrio challenge group, while another 25 were used as the control group, which were incubated only in the natural seawater, and the remaining 10 were the blank group. Coral nubbins were randomly sampled in the Vibrio challenge and control groups after 6, 12, and 24 h incubation, while the blank group was randomly sampled at 0 h. The three nubbins from each of the Vibrio challenge and control groups after 24 h exposure were immediately stored in liquid nitrogen for subsequent RNA extraction and transcriptome sequencing. Meanwhile, six nubbins were prepared from each group at each time point and used for evaluating the density and chlorophyll concentration of symbiotic zooxanthellae, and crucial immune parameters of the coral host.
The Density Determination of Symbiotic Zooxanthellae
The variation of symbiont density after Vibrio challenge was determined by the method described by Higuchi et al. (2008, 2015) with few modifications. Briefly, tissue homogenates were prepared using Waterpik water jet to strip the tissue from the coral skeleton into approximately 10 ml filtered seawater. Two milliliter homogenates were then centrifuged at 5000 rpm, 4°C for 15 min, and the supernatants were used to determine coral enzyme activities. The symbionts were resuspended in filtered seawater, and counted using a Neubauer hemocytometer (QIUJING, China). The surface area of the nubbins was determined by the aluminum foil method (Johannes et al., 1970). The density of symbiotic zooxanthellae was defined as the ratio of the symbiont number to the surface area of coral nubbin (cell cm-2).
The Measurement of Chlorophyll Content
The chlorophyll content of symbiotic zooxanthellae was determined after Vibrio challenge using the method described by Hedouin et al. (2016). Briefly, 2 ml homogenates were centrifuged at 5000 rpm, 4°C for 10 min to obtain symbionts. The harvested symbionts were resuspended in filtered seawater, and then centrifuged at 12,000 rpm, 4°C for 30 s. Chlorophyll a and c were extracted for 24 h at 4°C in 100% acetone, following by a centrifugation at 12,000 rpm, 4°C for 30 s. The absorbance values of the extracts were measured at 630 and 663 nm. The total content of chlorophyll a+c2 was computed according to the equations of Jeffrey and Humphrey (1975). The chlorophyll content was the ratio of the total content of chlorophyll a+c2 to the number of symbionts, and expressed as pg cell-1.
Activity Assay of Antioxidases and Nitric Oxide Synthetase
Total activities of superoxide dismutase (SOD), catalase (CAT), and nitric oxide synthetase (NOS) in the supernatants were measured using commercial kits (JIANCHENG, A001, A007, and A014), following the manufacturer’s recommendations. Total SOD activity was determined by the hydroxylamine method, where 1 SOD activity unit was defined as the enzyme amount causing 50% inhibition in 1 ml reaction solution. Total CAT activity was determined using spectrophotometry to measure the yellowish complex compound generated after the reaction between hydrogen peroxide and ammonium molybdate. Here, 1 CAT activity unit referred to the amount of enzyme needed to degrade 1 mmol hydrogen peroxide per second. Spectrophotometry was also used to determine total NOS activity by measuring the production of NO synthesized by NOS using L-arginine as the substrate. For this, 1 NOS activity unit represented the amount of enzyme catalyzing the production of 1 nmol NO per min. After the concentration of total protein in the supernatant was quantified by BCA method (Dodd and Drickamer, 2001), the measured enzyme activity was divided by the total protein to yield specific activity expressed as U mg-1 protein.
Activity Assay of Phenoloxidase (PO)
Phenoloxidase activity was measured by the photocolorimetric method described by Palmer et al. (2011) with slight modifications. L-DOPA (Sigma-Aldrich) was used as substrates, and the formation of o-quinones was recorded spectrophotometrically. Briefly, 20 μl supernatant was mixed with 65 μl PBS in the well of microplate (Costar). After the addition of 30 μl of 10 mmol l-1 L-DOPA (in PBS), the absorbance at 490 nm in each reaction well was measured every 5 min for 45 min using a microplate spectrophotometer (TECAN, infinite F50). The PO activity was defined as the change of absorbance (∆OD490) catalyzed by 1 mg supernatant protein in 1 min for the linear portion of the reaction curve, and expressed as ∆OD490 mg-1 min-1.
Caspase3 Assay
The caspase3 activity in the supernatant was measured by Caspase-3 Colorimetric Assay Kit (KeyGEN BioTECH) according to the instruction. Briefly, the supernatants of all samples were diluted firstly to the same protein concentration. Then, 50 μl supernatants were added in the reaction mixture containing 50 μl reaction buffer and 5 μl substrate. After an incubation in the dark at 37°C for 4 h, the color change was detected spectrophotometrically at the wavelength of 405 nm. The activity of caspase3 was defined as the absorbance of the reaction solution at 405 nm (OD405), and the activation level of caspase3 in the scleractinian coral was defined as the ratio of OD405 in samples to that of the blank group.
Deep Sequencing of P. damicornis Metatranscriptome
The coral nubbins with symbionts were ground in liquid nitrogen and mixed with 1 ml Trizol reagent (Invitrogen). The mixture was transferred into a 1.5 ml centrifuge tube and centrifuged at 5000 rpm, 4°C for 5 min to discard calcium carbonate skeleton. Subsequently, total RNA was isolated according to the protocol described by Stefanik et al. (2013). The extracted total RNA was quantified by Nanodrop 2000 (Thermo Scientific) at 260/280 nm (ratio > 2.0) and its integrity was checked with Angilent 2100 Bioanalyzer (Agilent Technologies). After the total RNA extraction and DNase I treatment, magnetic beads with Oligo(dT) are used to isolate mRNA. Then, the paired-end fragment library (2 × 150 bp) was constructed and sequenced on the Illumina HiseqX platform according to the manufacturer’s instructions (BGI, Shenzhen, China). The generated raw sequencing reads were deposited at the NCBI Short Read Archive (SRA) under Accession No. SRP133521.
Reads Mapping and Identification of Differentially Expressed Genes
Raw reads obtained from Illumina sequencing were processed by the Fastx-toolkit pipeline1 to summarize data production, evaluate sequencing quality, and remove low quality reads and adaptor sequences. Quality filtration removed sequences in which 80% of the base pairs had a Phred score of less than 20, while adaptor sequences were removed using the trimmer package. The assembled coral and Symbiodinium transcripts in our previous study served as reference sequences for reads mapping, and the mapping of paired-end reads was performed using the HISAT2 software (Pertea et al., 2016). After reads mapping, the resultant BAM files were used by StringTie and DESeq2 software to estimate transcript abundances and identify differentially expressed genes between the Vibrio challenge and control groups.
GO Enrichment of Differentially Expressed Genes
After identification of differentially expressed genes, the lists of significantly upregulated and downregulated genes were generated for both the scleractinian coral and symbionts. GO enrichment analysis was implemented via the hypergeometric test with filter value of 0.01. The significantly upregulated or downregulated genes were selected as test set, while all assembled coral or symbiont genes were used as the reference set. The BiNGO tool was employed to calculate the overrepresented GO terms in the network and display them as a network of significant GO terms.
Statistical Analysis
All data were presented as means ± standard deviation (SD). All of the data were subjected to two-way analysis of variance (two-way ANOVA) followed by multiple comparison (SNK) to determine significant differences among the treatments and controls. Differences were considered significant at p < 0.05.
Results
Density and Chlorophyll Content of Symbiotic Zooxanthellae After the Challenge of V. coralliilyticus
The density of symbiotic zooxanthellae and their chlorophyll content both decreased significantly in P. damicornis after Vibrio challenge. The symbiont density in the Vibrio challenge group was significantly lower (1.92 ± 0.83 × 105 cell cm-2, p < 0.05) after 12 h of pathogen exposure, in comparison with those in the blank (3.96 ± 1.19 × 105 cell cm-2) and control groups (4.05 ± 1.16 × 105 cell cm-2) (Figure 1A). The chlorophyll content of symbiotic zooxanthellae began to decrease at 12 h, and reached the lowest level (18.43 ± 2.71 pg cell-1, p < 0.05) at 24 h after Vibrio challenge (Figure 1B). There was no significant difference in both symbiont density and chlorophyll content between the control and blank groups for the entire duration of the experiment.
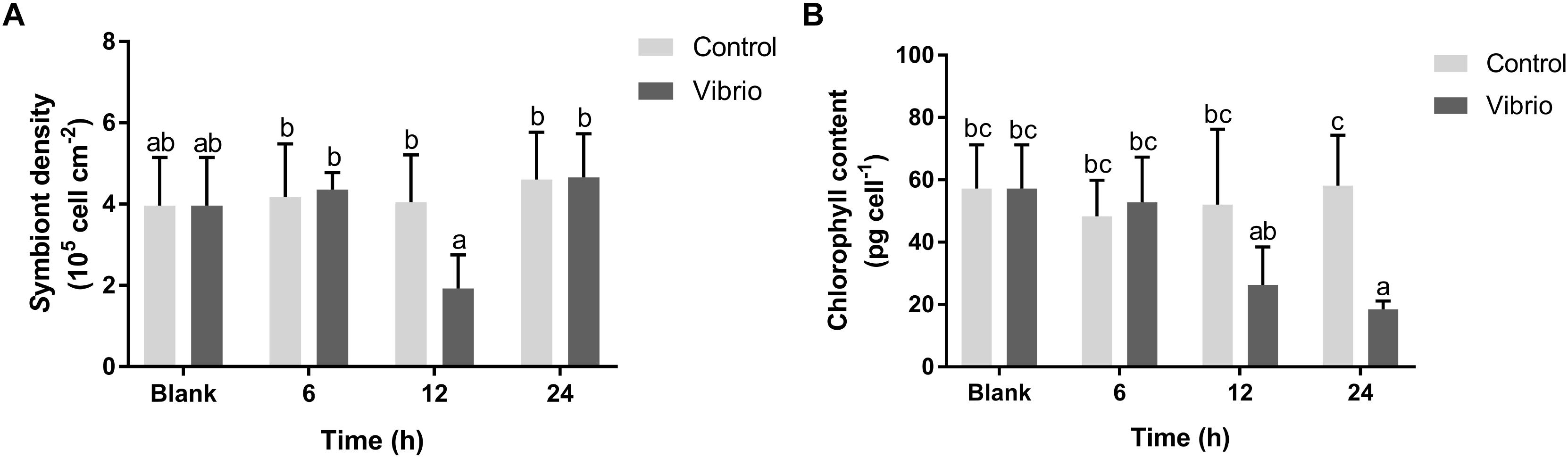
Figure 1. Density (A) and chlorophyll (B) variation of the symbionts in the scleractinian coral Pocillopora damicornis after Vibrio challenge. Vertical bars represent the mean ± SD (N = 6), bars with different letters were significantly different (p < 0.05).
Effects of Vibrio Challenge on the SOD, CAT, and NOS Activities
The activities of SOD, CAT, and NOS increased significantly in P. damicornis after Vibrio challenge. For SOD activity, the significant increase only occurred at 6 h (668.20 ± 138.97 U mg-1, p < 0.05) after Vibrio challenge (Figure 2A). CAT activity increased significantly (268.09 ± 43.58 U mg-1, p < 0.05) at 24 h after Vibrio challenge, which was significantly higher than those in the blank and control groups (Figure 2B). The NOS activities at 6 h (1.23 ± 0.21 U mg-1, p < 0.05) after Vibrio challenge were higher than those in the control group (Figure 2C). There was no significant difference in the activities of SOD, CAT, and NOS between the control and blank groups for the entire duration of the experiment.
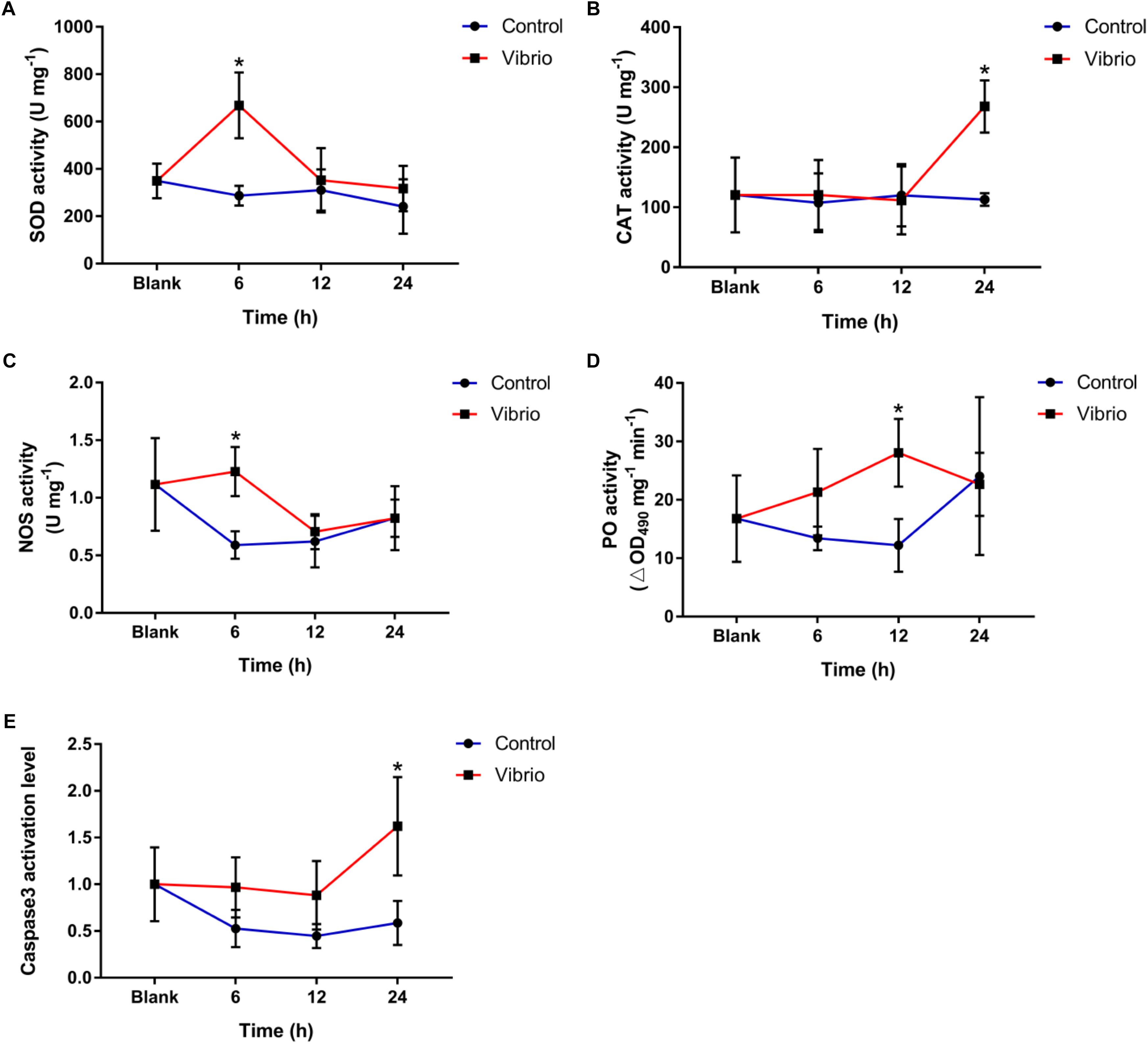
Figure 2. Temporal changes of superoxide dismutase (SOD, A), catalase (CAT, B), nitric oxide synthetase (NOS, C), and phenoloxidase (PO, D) and caspase3 activation level (E) in the scleractinian coral Pocillopora damicornis after Vibrio challenge. Vertical bars represent the mean ± SD (N = 6), and bars with asterisks are significantly different (p < 0.05).
Effect of Vibrio Challenge on the PO Activities and Caspase3 Activation Levels
The PO activity reached the peak level at 12 h of Vibrio challenge (28.05 ± 5.80 ∆OD490 mg-1 min-1, p < 0.05), which was significantly higher than that in the control group (Figure 2D), while no significant difference was observed at the other time points. The caspase3 activation level increased significantly in the scleractinian coral P. damicornis after Vibrio challenge, and it reached the peak (1.62 ± 0.53-fold, p < 0.05) at 24 h after Vibrio challenge (Figure 2E). There was no significant difference in the caspase3 activation level between the control and blank groups for the entire duration of the experiment.
Construction and Sequencing of Transcriptome Libraries
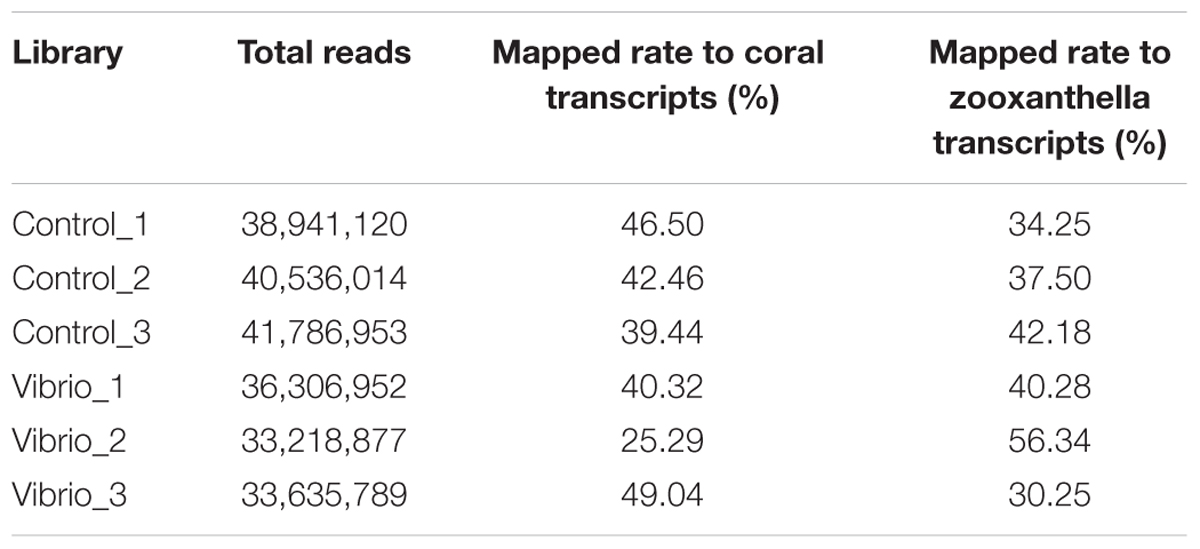
A total of six paired-end transcriptome libraries were constructed, including three libraries in the control group and three libraries in the Vibrio challenge group. To compare the abundance and difference of gene expression, these six libraries were sequenced to saturated level. After filtration of low-quality sequence and adaptor sequence, a total of 224,425,705 paired-end reads with a length of 2 × 150 bp were obtained, including 38,941,120, 40,536,014, and 41,786,953 reads from three control libraries, and 36,306,952, 33,218,877, and 33,635,789 reads from three Vibrio challenge libraries, respectively (Table 1).
Gene Expression
The high-quality clean paired-end reads obtained from all six transcriptome libraries were processed in HISAT2 software to map to the reference sequences built from the assembled coral and Symbiodinium transcripts in our previous study. The mapping rates of the six libraries ranged from 25.29 to 49.04% for coral and from 30.25 to 56.34% for zooxanthella (Table 1). The gene expression level was analyzed using StringTie software, and the counts of mapped reads for each coral and symbiont gene were further fed into the DESeq2 software to identify differentially expressed genes (Supplementary Tables S1, S2). Out of the 38,951 genomically predicted coral genes, 36,534 and 36,306 genes (more than three reads) were detectable in the control and Vibrio challenge groups, respectively. Meanwhile, we detected 45,357 and 45,207 genes of the genomically predicted 489,305 symbiont genes in the control and Vibrio challenge groups, respectively.
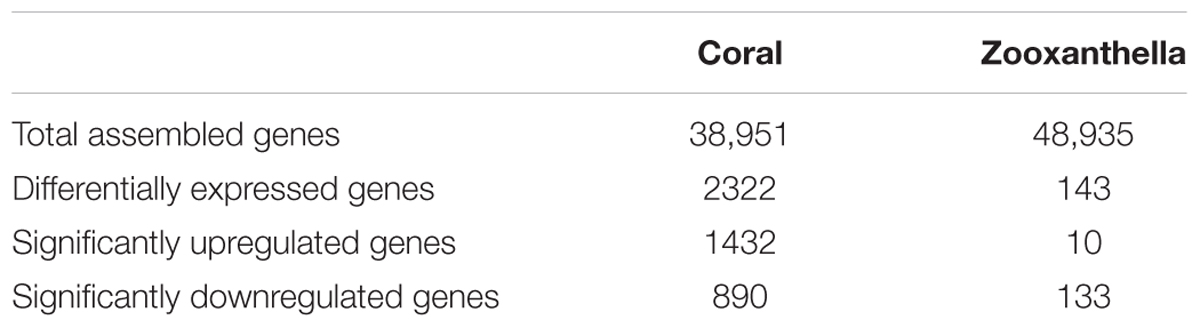
After library calibration, the expression levels of these genes were compared between the control and Vibrio challenge groups. The result revealed 2322 differentially expressed coral and only 143 zooxanthella genes (Table 2), which accounted for 5.96 and 0.03% of total number of coral and zooxanthella genes, respectively. There were 1432 significantly upregulated and 890 significantly downregulated genes for the scleractinian coral, while 10 and 133 for the symbionts (Supplementary Tables S3, S4).
Functional Annotation of the Differentially Expressed Genes in the Symbiotic Zooxanthellae
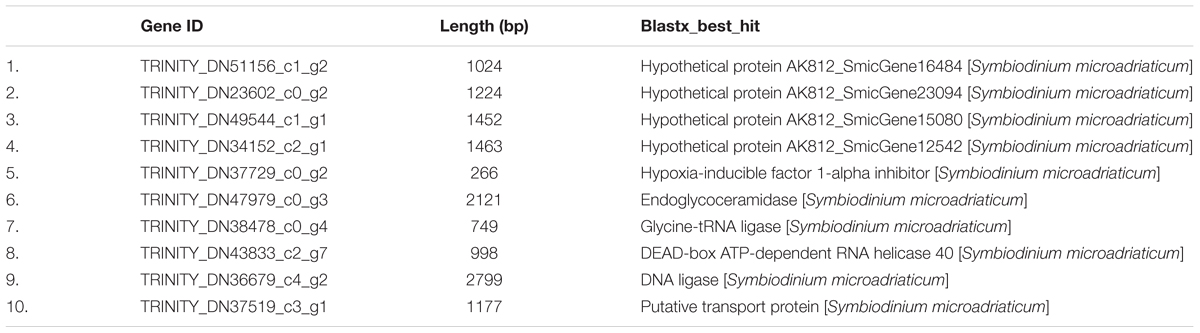
Ten significantly upregulated genes and 133 significantly downregulated genes were observed in the symbiont at 24 h after Vibrio challenge. Blastx searches revealed these significantly upregulated genes including four hypothetical protein genes, one hypoxia-inducible factor 1-alpha inhibitor, one endoglycoceramidase, one glycine–tRNA ligase, one DEAD-box ATP-dependent RNA helicase, one DNA ligase, and one putative transport protein (Table 3).
For the 133 significantly downregulated genes, their GO overrepresentation was analyzed at multiple GO levels in the Biological Process category. A total of seven major GO terms were overrepresented, which were mainly related to the regulation of cytokine production and the catabolic processes of small molecules, organic acids, cellular amino acids, carboxylic acid, and amine (Figure 3 and Supplementary Table S5). Furthermore, three transporter-related genes were observed through Blastx searches, including intraflagellar transport protein (TRINITY_DN54551_c5_g3), sodium-coupled neutral amino acid transporter (TRINITY_DN48832_c0_g1), and sodium-dependent dicarboxylate transporter (TRINITY_DN51317_c0_g2) (Supplementary Table S6).
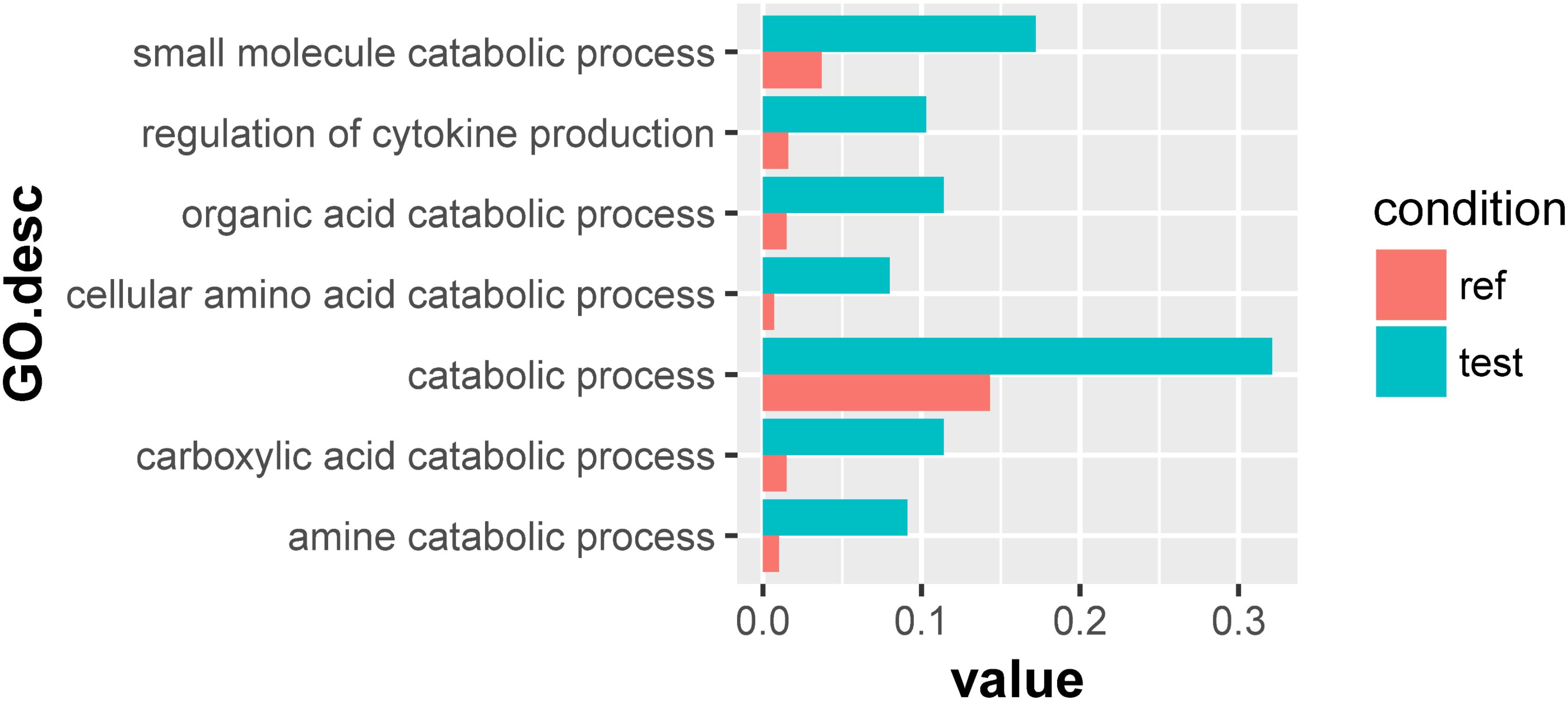
Figure 3. Overrepresented GO terms of the 133 significantly downregulated genes in the symbionts of the scleractinian coral P. damicornis at 24 h after Vibrio challenge. A total of seven GO terms were overrepresented for these significantly downregulated genes. “Ref” referred to the proportion of genes annotated to a GO term in all reference gene of symbiotic zooxanthellae, while “test” referred to the proportion of genes annotated to that GO term in all significantly downregulated genes.
Functional Annotation of the Differentially Expressed Genes in the Scleractinian Coral
The expression levels of 1432 coral genes in the Vibrio challenge group were higher than those in the control group, while the expression levels of 890 genes were significantly lower. The GO overrepresentation of these differentially expressed genes was further analyzed at multiple GO levels in the Biological Process category.
For the 1432 significantly upregulated genes, there were 26 major overrepresented GO terms (Figure 4 and Supplementary Table S7). These overrepresented GO terms could be divided into three groups, including the regulation of immune response, the regulation of cytokine production, and innate immune response (Figure 5). The first group contained the activation and positive regulation of immune response, and immune signaling pathway. Furthermore, a total of four overrepresented GO terms were observed for the 890 significantly downregulated genes (Figure 6 and Supplementary Table S8), which were mainly related to defense response. Of these two GO terms were overrepresented for both significantly upregulated genes and significantly downregulated genes, including GO:0045087 innate immune response and GO:0006952 defense response.
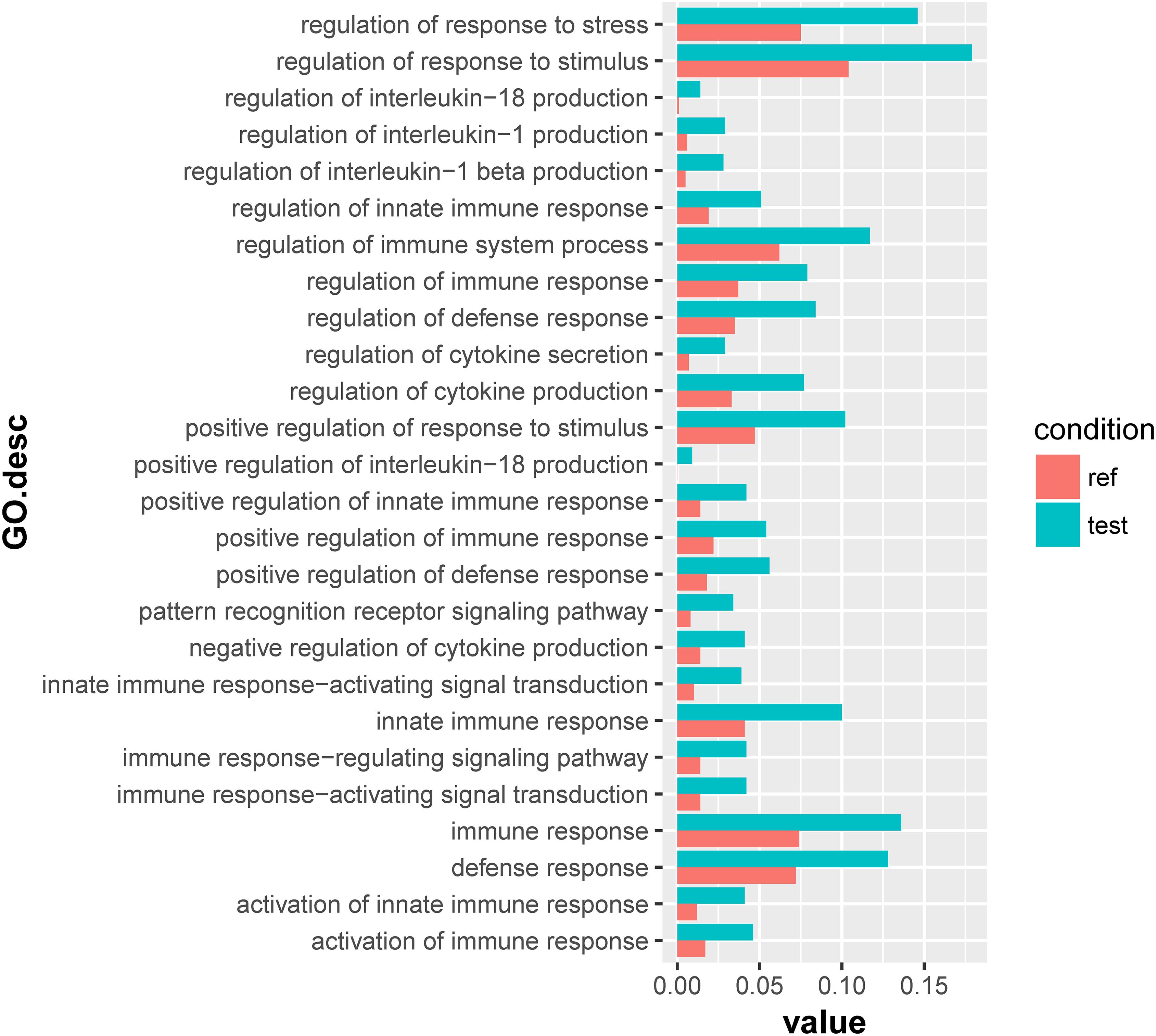
Figure 4. Overrepresented GO terms of the 1432 significantly upregulated genes in the scleractinian coral P. damicornis at 24 h after Vibrio challenge. A total of 26 GO terms were overrepresented for these significantly upregulated genes. “Ref” referred to the proportion of genes annotated to a GO term in all reference gene of the scleractinian coral P. damicornis, while “test” referred to the proportion of genes annotated to that GO term in all significantly upregulated genes.
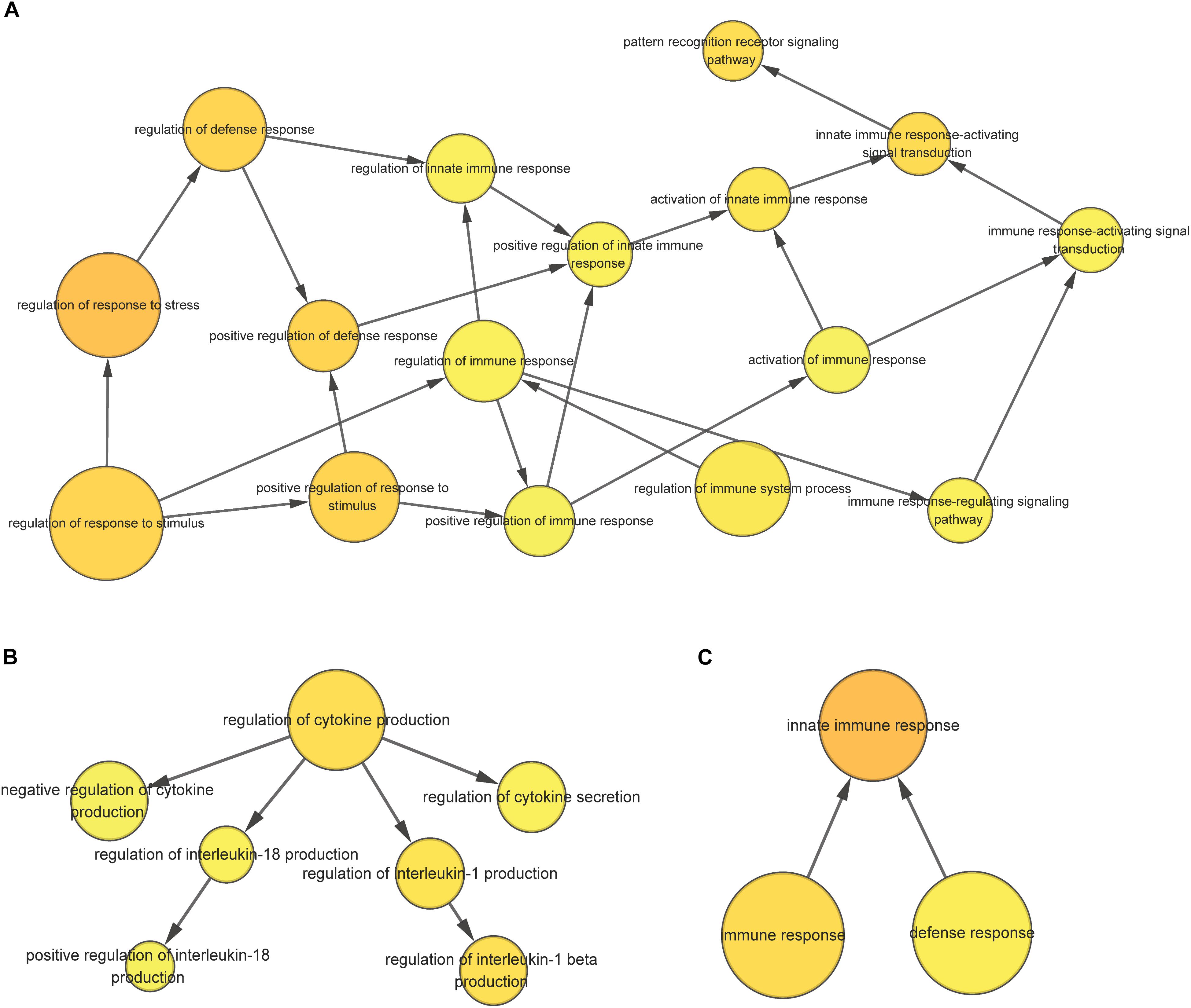
Figure 5. The overrepresented GO term group correlated with the regulation of immune response (A), the regulation of cytokine production (B), and innate immune response (C) for significantly upregulated genes in the scleractinian coral P. damicornis at 24 h after Vibrio challenge.
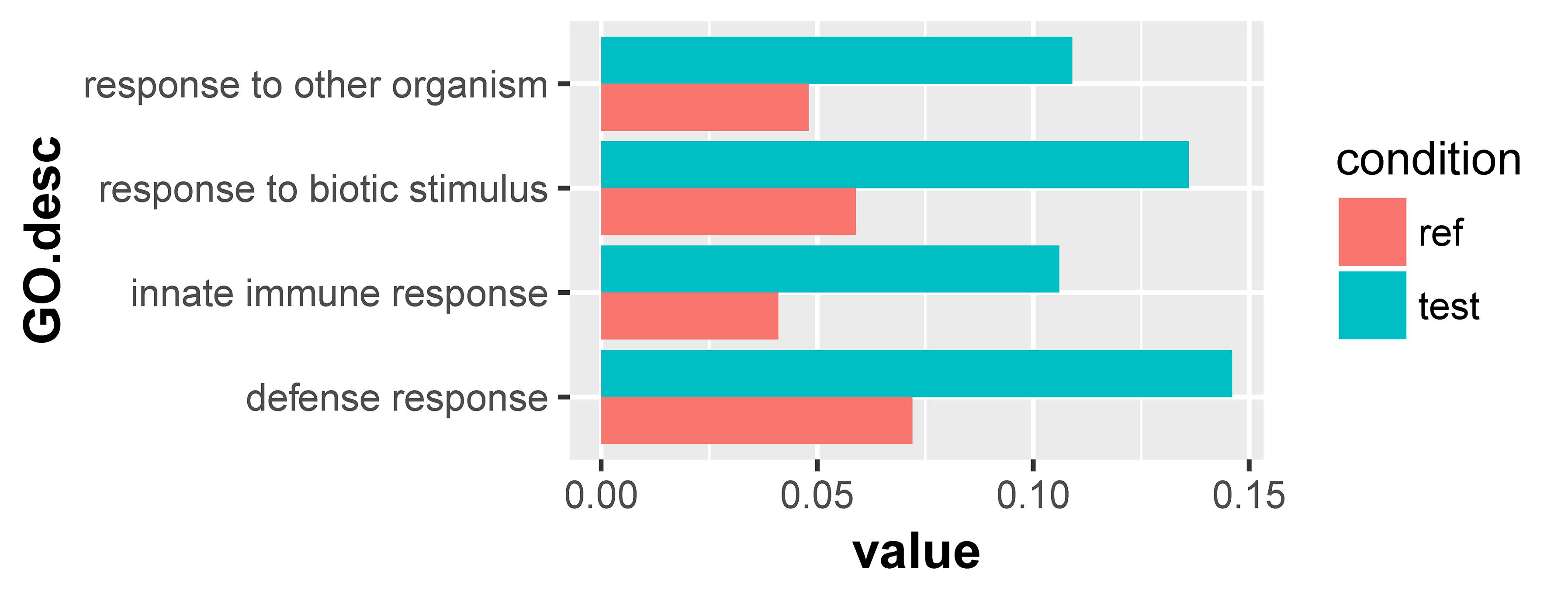
Figure 6. Overrepresented GO terms of the 890 significantly downregulated genes in the scleractinian coral P. damicornis at 24 h after Vibrio challenge. A total of four GO terms were overrepresented for these significantly downregulated genes. “Ref” referred to the proportion of genes annotated to a GO term in all reference gene of the scleractinian coral P. damicornis, while “test” referred to the proportion of genes annotated to that GO term in all significantly downregulated genes.
Functional Annotation of the Differentially Expressed Genes Under Common Overrepresented GO Terms
Differentially expressed genes contained in two common overrepresented GO terms (GO:0045087, GO:0006952) mentioned above were annotated through NCBI blastx, and the genes under GO:0045087 was a part of those under GO:0006952 (Supplementary Tables S9–S12). For GO:0006952, more upregulated genes were related to positive regulation of immune response than downregulated genes, such as macrophage mannose receptor, Toll-like receptor, NACHT, LRR, and PYD domains-containing protein and TNF receptor-associated factor. Furthermore, the genes of complement components, including complement C3 and CUB, were exclusively downregulated genes (Table 4).
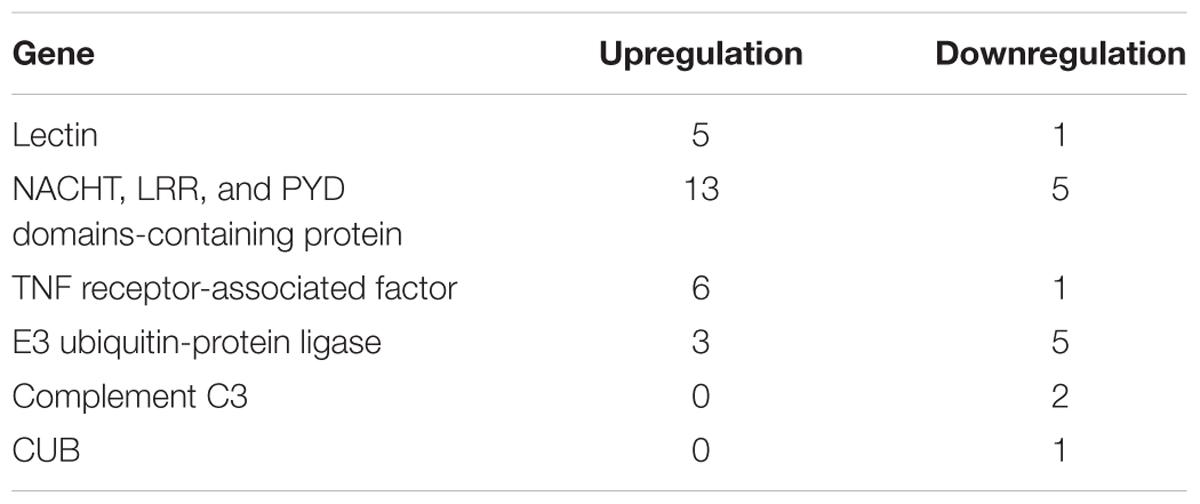
Table 4. The number of crucial immune genes significantly upregulated and downregulated in P. damicornis at 24 h after Vibrio challenge.
Discussion
The survival and reproduction of scleractinian coral is increasingly threatened by a wide variety of environmental insults, such as increased seawater temperature, ocean acidification, and pathogenic microorganisms. The Gram-negative bacterium V. coralliilyticus has been identified as a coral bleaching pathogen in the scleractinian coral P. damicornis and pathogenic agent of other corals; however, its molecular mechanism underlying the effect on coral physiology is still not well understood (Ben-Haim et al., 2003a,b; Kimes et al., 2012). Through physiological, biochemical, and transcriptomic approaches, we have investigated effects of V. coralliilyticus challenge on P. damicornis and its symbiotic zooxanthellae and found some interesting responses.
The Disruption of the Coral-Symbiodinium Symbiosis After Vibrio Challenge
Vibrio coralliilyticus SCSIO 43001 challenge led to significant decreases in the density and chlorophyll content of symbiotic zooxanthellae in P. damicornis, demonstrating that the symbiosis could be disrupted by V. coralliilyticus infection. This is consistent with a previous report that the infection of V. coralliilyticus YB1 caused the decline of density (cell g-1 of tissue) and chlorophyll content (chlorophyll a, μg g-1 of tissue) of symbionts in P. damicornis, eventually leading to coral bleaching and lysis (Ben-Haim et al., 2003b). The similar results suggest that the challenge of V. coralliilyticus SCSIO 43001 could also potentially result in symbiosis collapse and coral bleaching through depressing the symbiont density and chlorophyll content. The innate immune response of P. damicornis could be triggered by the infection with pathogen V. coralliilyticus, as previously shown to manifest in the induced significant change of the expression level of immune-related coral genes such as lectin, metal-binding protein, and cysteine protease inhibitor (Vidal-Dupiol et al., 2011b). Both the innate immune response and symbiosis collapse in P. damicornis as a result of Vibrio challenge suggest a cascade of pathogenic effects from eliciting immune response, decreasing symbiont abundance, and photosynthetic capacity (chlorophyll content), to causing coral bleaching or even degradation. Whether V. coralliilyticus can infect both the coral and Symbiodinium or the effect on Symbiodinium was a secondary impact stemming from the perturbed coral host remains to be further studied in the future.
Immune Response of the Scleractinian Coral to Vibrio Challenge at the Biochemical Level
Scleractinian corals employ the innate immune response as the sole defense mechanism against pathogen infection, such as the redox, PO, and apoptosis systems (Palmer et al., 2010, 2011; Fuess et al., 2017). To understand the immune response of scleractinian coral against the challenge of V. coralliilyticus, the activities of crucial enzymes in the redox, PO, and apoptosis systems were monitored in P. damicornis. Elevated activities were observed for SOD and NOS at 6 h, PO at 12 h, CAT and caspase3 at 24 h in the Vibrio challenge group relative to the control group, showing that the redox, PO, and apoptosis systems in this scleractinian coral were activated by Vibrio challenge. It also demonstrates that the scleractinian coral could launch full innate immune response against Vibrio infection. Substantial ROS and RNS is produced generally by invertebrate hosts to kill the invaded pathogens once recognized (Nyholm and Graf, 2012), the production of ROS or RNS in the coral after challenge was not determined in the present study owing to sample operation difficulty. However, we observed the significant rise of the antioxidase and NOS activities, which might further reflect the production of more ROS and RNS in P. damicornis after Vibrio challenge. Furthermore, the induced PO would catalyze the production of insoluble melanin to aid in the encapsulation of invaded pathogenic vibrios, and more caspase3 was activated to trigger apoptosis to eliminate injured coral cells containing vibrios and symbionts. Taken together, these results suggest that the infection of V. coralliilyticus could activate the redox, PO, and apoptosis systems in P. damicornis to initiate the innate immune defense against Vibrio infection.
The inductions of these immune components have also been observed in the scleractinian coral under environmental stresses, especially heat stress that causes bleaching (Downs et al., 2002; Mydlarz et al., 2009; Palmer et al., 2011; Tchernov et al., 2011; Yu et al., 2017). This suggests that the triggered immune response in P. damicornis by Vibrio challenge might be responsible for the decrease in the symbiont density and disruption of the symbiosis between Symbiodinium and the coral. On the other hand, coral immunity is crucial to the metabolism and health of the scleractinian coral, because it can regulate and control bacterial community in coral biofilms (Cardenas et al., 2012; Kvennefors et al., 2012), and the bacterial community can provide necessary nitrogen and sulfur resources for the growth and reproduction of the coral (Raina et al., 2009; Lema et al., 2012). Therefore, it was speculated that there was a trade-off between the elimination of a potential pathogen and the maintenance of coral-Symbiodinium symbiosis and beneficial bacteria in the scleractinian coral, and the needed balance can be sensitive to environmental changes. This is supported by the observation that the scleractinian coral was more susceptible to bleaching, accompanied by an improved immune response, when it suffered from heat stress (Mydlarz et al., 2009; Palmer et al., 2010; Kimes et al., 2012).
Repressed Complement System of the Scleractinian Coral at the Transcriptional Level After Vibrio Challenge
To understand the molecular mechanisms underlying the response of the coral-Symbiodinium association to pathogenic infection, we sequenced the metatranscriptome of P. damicornis at 24 h after the challenge of V. coralliilyticus. Analysis of differentially expressed genes and corresponding enriched GO terms revealed that significantly upregulated genes were mainly related to the immune response, immune regulation, and cytokine production regulation, and the immune regulation was comprised of the activation of immune response, pattern recognition receptor-activation signal transduction, and positive regulation of immune response. The result was consistent with the increase of immune-related enzyme activities, and agreeable to a previous report on the scleractinian coral challenged by V. coralliilyticus YB1 (Vidal-Dupiol et al., 2014). Furthermore, it could be supported by the binding of lectin protein in the scleractinian coral to LPS or Gram-negative bacteria (Kvennefors et al., 2008; Zhou et al., 2017).
However, among the four overrepresented GO terms for the 890 significantly downregulated genes, two terms (innate immune response and defense response) were also observed in the analysis of the significantly upregulated genes. There were more upregulated genes for immune response, including macrophage mannose receptor, Toll-like receptor, NACHT, LRR, and PYD domains-containing protein and the TNF receptor-associated factor. This indicates that immunity machinery was remolded at the transcriptional level in the scleractinian coral after Vibrio challenge. As a result, the genes of complement components such as C3 and CUB were significantly downregulated. The complement system is an important part of the immune system that enhances the ability of phagocytic cells to clear microbes and attack the pathogen’s cell membrane, and multi-components in the complement system had been identified in cnidaria (Kimura et al., 2009). The expression level of complement C3 as the core component has been shown to respond to bacterial challenge in the scleractinian coral Acropora millepora (Brown et al., 2013). This demonstrates that the immune ability of the complement system was suppressed in P. damicornis after Vibrio challenge. These results together suggest that the V. coralliilyticus infection can reconfigure the immune landscape, triggering the innate immune response (including the redox, PO, and apoptosis systems), but repressing the response of the complement system in P. damicornis.
Disordered Metabolism of the Symbionts During Host Immune Response
To have a deeper insight into the effect of Vibrio challenge on the coral-Symbiodinium symbiosis, we further examined the functions of differentially expressed genes in the symbionts. We only identified 10 significantly upregulated genes, which were mainly related with nucleic acid and polysaccharide metabolisms. Among the 133 downregulated genes identified, major enriched GO terms were relevant to amino acid catabolism. These altered metabolic pathways were all essential for the maintenance of the symbiont and its symbiosis with the coral host. Similar metabolic alteration of the symbionts has been reported in the other scleractinian corals under heat stress and depth change, which was accompanied by the shift of Symbiodinium genotype (Cooper et al., 2011; Jones and Berkelmans, 2011). The metabolic changes in the present study might have resulted from the interaction of the two symbiotic partners in response to Vibrio infection. The scleractinian coral needed more organic nutrition from the symbiont to meet the massive demand of immune energy after Vibrio challenge, and the symbiont could not photosynthesize at full capacity because of the excessive oxidative stress and chlorophyll content decline. The odds between the scleractinian coral and Symbiodinium might eventually lead to the collapse of the symbiosis.
Data Availability
The datasets generated for this study can be found in SRA, SRP133521.
Ethics Statement
All animal-involving experiments of this study were approved by the Ethics Committee of Hainan University and local government.
Author Contributions
SZ and ZZ conceived and designed the experiments. SZ, JT, YW, and ZZ performed the experiments. SZ, ZL, and ZZ analyzed the data. JT and YW contributed reagents, materials, and analysis tools. SL and ZZ contributed to the discussion and wrote the manuscript. All the authors read and approved the final manuscript.
Funding
ZZ gratefully acknowledges the financial support from China Scholarship Council (201708460042) to enable his visit to the University of Connecticut where part of the work was performed. This research was supported by grant (31772460 and 31661143029) from National Science Foundation of China and the Scientific Research Foundation (kyqd1554) of Hainan University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors were grateful to all of the laboratory members for their continuous technical advice and helpful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00366/full#supplementary-material
Footnotes
References
Ben-Haim, Y., Thompson, F. L., Thompson, C. C., Cnockaert, M. C., Hoste, B., Swings, J., et al. (2003a). Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 53(Pt 1), 309–315. doi: 10.1099/ijs.0.02402-0
Ben-Haim, Y., Zicherman-Keren, M., and Rosenberg, E. (2003b). Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242.
Brown, T., Bourne, D., and Rodriguez-Lanetty, M. (2013). Transcriptional activation of c3 and hsp70 as part of the immune response of Acropora millepora to bacterial challenges. PLoS One 8:e67246. doi: 10.1371/journal.pone.0067246
Cardenas, A., Rodriguez, R. L., Pizarro, V., Cadavid, L. F., and Arevalo-Ferro, C. (2012). Shifts in bacterial communities of two Caribbean reef-building coral species affected by white plague disease. ISME J. 6, 502–512. doi: 10.1038/ismej.2011.123
Cervino, J. M., Thompson, F. L., Gomez-Gil, B., Lorence, E. A., Goreau, T. J., Hayes, R. L., et al. (2008). The Vibrio core group induces yellow band disease in caribbean and indo-pacific reef-building corals. J. Appl. Microbiol. 105, 1658–1671. doi: 10.1111/j.1365-2672.2008.03871.x
Cooper, T. F., Ulstrup, K. E., Dandan, S. S., Heyward, A. J., Kuhl, M., Muirhead, A., et al. (2011). Niche specialization of reef-building corals in the mesophotic zone: metabolic trade-offs between divergent Symbiodinium types. Proc. Biol. Sci. 278, 1840–1850. doi: 10.1098/rspb.2010.2321
Dodd, R. B., and Drickamer, K. (2001). Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology 11, 71R–79R. doi: 10.1093/glycob/11.5.71R
Downs, C. A., Fauth, J. E., Halas, J. C., Dustan, P., Bemiss, J., and Woodley, C. M. (2002). Oxidative stress and seasonal coral bleaching. Free Radic. Biol. Med. 33, 533–543. doi: 10.1016/S0891-5849(02)00907-3
Fabina, N. S., Putnam, H. M., Franklin, E. C., Stat, M., and Gates, R. D. (2013). Symbiotic specificity, association patterns, and function determine community responses to global changes: defining critical research areas for coral-Symbiodinium symbioses. Glob. Chang. Biol. 19, 3306–3316. doi: 10.1111/gcb.12320
Fuess, L. E., Pinzon, C. J., Weil, E., Grinshpon, R. D., and Mydlarz, L. D. (2017). Life or death: disease-tolerant coral species activate autophagy following immune challenge. Proc. Biol. Sci. 284:1856. doi: 10.1098/rspb.2017.0771
Garren, M., Son, K., Raina, J. B., Rusconi, R., Menolascina, F., Shapiro, O. H., et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007. doi: 10.1038/ismej.2013.210
Hamada, M., Shoguchi, E., Shinzato, C., Kawashima, T., Miller, D. J., Satoh, N., et al. (2013). The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Mol. Biol. Evol. 30, 167–176. doi: 10.1093/molbev/mss213
Hedouin, L. S., Wolf, R. E., Phillips, J., and Gates, R. D. (2016). Improving the ecological relevance of toxicity tests on scleractinian corals: influence of season, life stage, and seawater temperature. Environ. Pollut. 213, 240–253. doi: 10.1016/j.envpol.2016.01.086
Higuchi, T., Fujimura, H., Arakaki, T., and Oomori, T. (2008). “Activities of antioxidant enzymes (SOD and CAT) in the coral Galaxea fascicularis against increased hydrogen peroxide concentrations in seawater,” in Proceeding of the 11th International Coral Reef Symposium, Fort Lauderdale.
Higuchi, T., Yuyama, I., and Nakamura, T. (2015). The combined effects of nitrate with high temperature and high light intensity on coral bleaching and antioxidant enzyme activities. Reg. Stud. Mar. Sci. 2, 27–31. doi: 10.1016/j.rsma.2015.08.012
Jeffrey, S. W., and Humphrey, G. F. (1975). New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. 167, 191–194. doi: 10.1016/s0015-3796(17)30778-3
Johannes, R., Coles, S. L., and Kuenzel, N. T. (1970). The role of zooplankton in the nutrition of some scleractinian corals. Limnol. Oceanogr. 15, 579–586. doi: 10.4319/lo.1970.15.4.0579
Jones, A. M., and Berkelmans, R. (2011). Tradeoffs to thermal acclimation: energetics and reproduction of a reef coral with heat tolerantsymbiodiniumtype-D. J. Mar. Biol. 2011, 1–12. doi: 10.1155/2011/185890
Kimes, N. E., Grim, C. J., Johnson, W. R., Hasan, N. A., Tall, B. D., Kothary, M. H., et al. (2012). Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 6, 835–846. doi: 10.1038/ismej.2011.154
Kimura, A., Sakaguchi, E., and Nonaka, M. (2009). Multi-component complement system of Cnidaria: C3, Bf, and MASP genes expressed in the endodermal tissues of a sea anemone, Nematostella vectensis. Immunobiology 214, 165–178. doi: 10.1016/j.imbio.2009.01.003
Kvennefors, E. C., Leggat, W., Hoegh-Guldberg, O., Degnan, B. M., and Barnes, A. C. (2008). An ancient and variable mannose-binding lectin from the coral Acropora millepora binds both pathogens and symbionts. Dev. Comp. Immunol. 32, 1582–1592. doi: 10.1016/j.dci.2008.05.010
Kvennefors, E. C., Leggat, W., Kerr, C. C., Ainsworth, T. D., Hoegh-Guldberg, O., Barnes, A. C., et al. (2010). Analysis of evolutionarily conserved innate immune components in coral links immunity and symbiosis. Dev. Comp. Immunol. 34, 1219–1229. doi: 10.1016/j.dci.2010.06.016
Kvennefors, E. C., Sampayo, E., Kerr, C., Vieira, G., Roff, G., Barnes, A. C., et al. (2012). Regulation of bacterial communities through antimicrobial activity by the coral holobiont. Microb. Ecol. 63, 605–618. doi: 10.1007/s00248-011-9946-0
Lema, K. A., Willis, B. L., and Bourne, D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78, 3136–3144. doi: 10.1128/AEM.07800-11
Libro, S., Kaluziak, S. T., and Vollmer, S. V. (2013). RNA-seq profiles of immune related genes in the staghorn coral Acropora cervicornis infected with white band disease. PLoS One 8:e81821. doi: 10.1371/journal.pone.0081821
Loram, J. E., Trapido-Rosenthal, H. G., and Douglas, A. E. (2007). Functional significance of genetically different symbiotic algae Symbiodinium in a coral reef symbiosis. Mol. Ecol. 16, 4849–4857. doi: 10.1111/j.1365-294X.2007.03491.x
Mydlarz, L. D., Couch, C. S., Weil, E., Smith, G., and Harvell, C. D. (2009). Immune defenses of healthy, bleached and diseased Montastraea faveolata during a natural bleaching event. Dis. Aquat. Organ. 87, 67–78. doi: 10.3354/dao02088
Neubauer, E. F., Poole, A. Z., Weis, V. M., and Davy, S. K. (2016). The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian-dinoflagellate symbiosis. PeerJ 4:e2692. doi: 10.7717/peerj.2692
Nyholm, S. V., and Graf, J. (2012). Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 10, 815–827. doi: 10.1038/nrmicro2894
Palmer, C. V., Bythell, J. C., and Willis, B. L. (2010). Levels of immunity parameters underpin bleaching and disease susceptibility of reef corals. FASEB J. 24, 1935–1946. doi: 10.1096/fj.09-152447
Palmer, C. V., Bythell, J. C., and Willis, B. L. (2011). A comparative study of phenoloxidase activity in diseased and bleached colonies of the coral Acropora millepora. Dev. Comp. Immunol. 35, 1098–1101. doi: 10.1016/j.dci.2011.04.001
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, stringTie and ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Poole, A. Z., and Weis, V. M. (2014). TIR-domain-containing protein repertoire of nine anthozoan species reveals coral-specific expansions and uncharacterized proteins. Dev. Comp. Immunol. 46, 480–488. doi: 10.1016/j.dci.2014.06.002
Raina, J. B., Tapiolas, D., Willis, B. L., and Bourne, D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501. doi: 10.1128/AEM.02567-08
Rosenberg, E., and Ben-Haim, Y. (2002). Microbial diseases of corals and global warming. Environ. Microbiol. 4, 318–326. doi: 10.1046/j.1462-2920.2002.00302.x
Stefanik, D. J., Wolenski, F. S., Friedman, L. E., Gilmore, T. D., and Finnerty, J. R. (2013). Isolation of DNA, RNA and protein from the starlet sea anemone Nematostella vectensis. Nat. Protoc. 8, 892–899. doi: 10.1038/nprot.2012.151
Tchernov, D., Kvitt, H., Haramaty, L., Bibby, T. S., Gorbunov, M. Y., Rosenfeld, H., et al. (2011). Apoptosis and the selective survival of host animals following thermal bleaching in zooxanthellate corals. Proc. Natl. Acad. Sci. U.S.A. 108, 9905–9909. doi: 10.1073/pnas.1106924108
van de Water, J. A., Ainsworth, T. D., Leggat, W., Bourne, D. G., Willis, B. L., and van Oppen, M. J. (2015). The coral immune response facilitates protection against microbes during tissue regeneration. Mol. Ecol. 24, 3390–3404. doi: 10.1111/mec.13257
Vidal-Dupiol, J., Dheilly, N. M., Rondon, R., Grunau, C., Cosseau, C., Smith, K. M., et al. (2014). Thermal stress triggers broad Pocillopora damicornis transcriptomic remodeling, while Vibrio coralliilyticus infection induces a more targeted immuno-suppression response. PLoS One 9:e107672. doi: 10.1371/journal.pone.0107672
Vidal-Dupiol, J., Ladriere, O., Destoumieux-Garzon, D., Sautiere, P. E., Meistertzheim, A. L., Tambutte, E., et al. (2011a). Innate immune responses of a scleractinian coral to vibriosis. J. Biol. Chem. 286, 22688–22698. doi: 10.1074/jbc.M110.216358
Vidal-Dupiol, J., Ladriere, O., Meistertzheim, A. L., Foure, L., Adjeroud, M., Mitta, G., et al. (2011b). Physiological responses of the scleractinian coral Pocillopora damicornis to bacterial stress from Vibrio coralliilyticus. J. Exp. Biol. 214(Pt 9), 1533–1545. doi: 10.1242/jeb.053165
Williams, L. M., Fuess, L. E., Brennan, J. J., Mansfield, K. M., Salas-Rodriguez, E., Welsh, J., et al. (2018). A conserved Toll-like receptor-to-NF-kappaB signaling pathway in the endangered coral Orbicella faveolata. Dev. Comp. Immunol. 79, 128–136. doi: 10.1016/j.dci.2017.10.016
Yu, X., Huang, B., Zhou, Z., Tang, J., and Yu, Y. (2017). Involvement of caspase3 in the acute stress response to high temperature and elevated ammonium in stony coral Pocillopora damicornis. Gene 637, 108–114. doi: 10.1016/j.gene.2017.09.040
Zhenyu, X., Shaowen, K., Chaoqun, H., Zhixiong, Z., Shifeng, W., Yongcan, Z., et al. (2013). First characterization of bacterial pathogen, Vibrio alginolyticus, for Porites andrewsi white syndrome in the South China Sea. PLoS One 8:e75425. doi: 10.1371/journal.pone.0075425
Keywords: coral bleaching, pathogen, immunol response, symbiosis, scleractinian coral
Citation: Zhou Z, Zhao S, Tang J, Liu Z, Wu Y, Wang Y and Lin S (2019) Altered Immune Landscape and Disrupted Coral-Symbiodinium Symbiosis in the Scleractinian Coral Pocillopora damicornis by Vibrio coralliilyticus Challenge. Front. Physiol. 10:366. doi: 10.3389/fphys.2019.00366
Received: 10 October 2018; Accepted: 15 March 2019;
Published: 02 April 2019.
Edited by:
Francesca Carella, University of Naples Federico II, ItalyReviewed by:
Susana Puntarulo, Universidad de Buenos Aires, ArgentinaKarl Blyth Andree, IRTA Institute of Agrifood Research and Technology, Spain
Copyright © 2019 Zhou, Zhao, Tang, Liu, Wu, Wang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Zhou, emhvdXpoaUBoYWluYW51LmVkdS5jbg== Senjie Lin, c2VuamllLmxpbkB1Y29ubi5lZHU=
 Zhi Zhou
Zhi Zhou Shuimiao Zhao
Shuimiao Zhao Jia Tang1
Jia Tang1 Yibo Wu
Yibo Wu Yan Wang
Yan Wang Senjie Lin
Senjie Lin