- 1State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Agriculture, Ludong University, Yantai, China
- 3College of Plant Protection, Hunan Agricultural University, Changsha, China
The alligatorweed flea beetle Agasicles hygrophila is an insect used for biological control of the aquatic weed Alternanthera philoxeroides (alligatorweed). Because these insects are oviparous, synthesis, and transportation of yolk proteins is integral to reproduction. Vitellin, the chief protein constituent in egg yolk, is mainly synthesized in the fat body and its synthesis is regulated by the transcript levels of Vitellogenin (Vg). In our study, we first cloned and characterized three Vg genes from A. hygrophila and quantified the expression levels of these Vgs in different tissues and developmental stages by RT-qPCR. Analysis of the full-length cDNA sequences of the three A. hygrophila Vg genes revealed that the open reading frames of AhVg1, AhVg2, and AhVg3 were 5175, 5346, and 5385 bp, encoding 1724, 1781, and 1794 amino acids, respectively. RT-qPCR analysis revealed that these three AhVgs have similar expression patterns; expression in the fat body was significantly higher than that in other tissues, and the highest expression was observed in the adult developmental stage. RNA interference was used to explore the functions of the AhVgs. A. hygrophila female adults injected with dsRNA targeting the AhVg genes showed decreased AhVg gene expression. Down regulation of all three AhVgs significantly affected ovary development, reduced egg laying capacity, and reduced the egg hatch rate compared with the control groups. Our findings provide the basis for further study of the functions of Vg genes in other insect species.
Introduction
Vitellogenin (Vg), the precursor protein of egg yolk vitellin (Vn), provides the energy and material for the development of ovaries and embryos in insects (Sappington and Raikhel, 1998; Tufail and Takeda, 2008; Veerana et al., 2015). Vgs are mainly synthesized and processed in the fat body and then secreted into the hemolymph (Raikhel and Dhadialla, 1992) and transported into oocytes by the Vitellogenin receptor (VgR). Vgs are incorporated into oocytes and stored as Vns, a crystalline substance, and reserved for development of the ovary and future embryo (Sappington and Raikhel, 1998; Tufail and Takeda, 2008). In insects, Vg synthesis is regulated by the expression levels of Vg (Raikhel and Dhadialla, 1992), and previous research has shown that ecdysone, juvenile hormone, and neuropeptides each play a role in Vg gene regulation (Engelmann and Friedel, 1974; Wyatt and Davey, 1996).
The first insect Vg was discovered in the cecropia moth (Hyalophora cecropia) as a female-specific protein precursor for Vn or yolk protein (Pan et al., 1969). Insect Vgs were then shown to be multipart oligomeric glycolipophosphoproteins (Anderson, 1974). The molecular weights of insect Vgs vary in size from 200 to 250 kDa, with the large subunit ranging from 150 to 200 kDa and the small subunit from 40 to 65 kDa (Dhadialla and Raikhel, 1990; Kim et al., 2010). Comparison of insect Vg amino acid sequences revealed that they are highly conserved (Chen et al., 1997; Sappington and Raikhel, 1998). The similarity between Vgs is also evident based on their similar antigenicity (Tufail and Takeda, 2008). Vitellogenesis is important for fecundity and ovary development (Boldbaatar et al., 2010). For example, overexpression of Vg resulted in increased fecundity in Tetranychus cinnabarinus (Xing et al., 2016), and Vgs were shown to be crucial for ovary development in Haemaphysalis longicornis (Boldbaatar et al., 2010).
Alternanthera philoxeroides is commonly considered an invasive species and has spread to many countries (Julien et al., 1995; Zhang H. J. et al., 2016). The spread of A. philoxeroides has had a negative impact on the economy, environment, and society (Andres, 1977). Agasicles hygrophila (Selman & Vogt) (Chrysomelidae), the alligatorweed flea beetle, is a classical biological agent that has proven effective in controlling alligatorweed and is presently used in many countries (Napompeth, 1991; Buckingham, 1996). The life cycle of A. hygrophila can be completed in about 25 days in the appropriate habitat and temperature (∼22°C), and it can produce four generations per year (Zhao et al., 2016). A. hygrophila was introduced to the United States to control alligatorweed in the late 1960s (Zhao et al., 2016), and subsequently brought from Florida to China in 1987, where it was released in several provinces, including Jiangsu, Yunnan, Sichuan, and Hunan (Pan et al., 2006). Many studies have focused on the geographical distribution of A. hygrophila and its ability to control alligatorweed; however, studies of the reproductive physiology of A. hygrophila are rare. Knowledge of reproduction-related genes and proteins involved in ovary development and fecundity are necessary for better understanding the molecular mechanisms of reproduction in this species. Because Vg genes are known to be important reproductive genes regulating ovary development and fecundity in other insects, understanding the expression patterns and functions of these genes is essential for revealing the mechanism of reproduction in A. hygrophila.
In our study, we identified Vgs in A. hygrophila, characterizing Vgs at multiple levels with the use of gene cloning and sequence analysis. We established the full length AhVg cDNA sequences and compared the molecular and structural characteristics of these genes to Vgs from other insect species. We further analyzed the expression levels of AhVgs in different tissues and across developmental stages. Finally, we analyzed the role of the AhVgs in A. hygrophila ovary development and fecundity by utilizing RNA interference (RNAi).
Materials and Methods
Host Plants and Experimental Insects
Roots of A. philoxeroides were collected from standing water at the Institute of Plant Protection, Hunan Academy of Agricultural Sciences and planted in sterilized soil in plastic boxes (40 × 18 × 15 cm). Plants were grown in the greenhouse at Langfang Experimental Station, Chinese Academy of Agricultural Sciences (LF, CAAS), and watered every other day. A. philoxeroides were selected for experiments when they reached the four- to six-internodes stage.
Agasicles hygrophila adults were collected from a field in Changsha, Hunan province and were maintained on A. philoxeroides plants in the laboratory at the Chinese Academy of Agricultural Sciences (BJ, CAAS) under controlled conditions: 28 ± 2°C, 75 ± 5% relative humidity (RH), and a 12 h light:12 h dark regime. The insects were cultivated for three generations to eliminate maternal effects.
Sample Collection
For cloning the A. hygrophila Vg genes, the abdomens of A. hygrophila females were dissected in 1× phosphate-buffered saline (PBS) under an Olympus stereomicroscope (SZX16, Olympus, Tokyo, Japan). For analysis of expression during different developmental stages, the freshly pupated pupae were collected daily, and female adults were collected every 2 days after emergence. For analysis of expression in different tissues, head, thorax, ovary, fat body, midgut, and wing tissues were dissected from 6-day-old females under an Olympus stereomicroscope (SZX16, Olympus, Tokyo, Japan) in PBS. All samples were frozen immediately in liquid nitrogen and subsequently kept at −80°C until further experimentation.
RNA Isolation and Gene Cloning
TRIzol reagent (Life Technologies, Carlsbad, CA, United States) was used to extract total RNA from the above samples according to the manufacturer’s protocols. RNA integrity and concentration were assessed as described in An et al. (2016). The cDNA of A. hygrophila Vg genes was synthesized from total RNA isolated from the abdomen. We mined A. hygrophila transcriptome data to obtain three expressed sequence tags (ESTs) showing similarity to other insect Vg proteins. Gene-specific primers were designed based on these A. hygrophila Vg ESTs (Supplementary Table S1). Cloning was performed following the procedure from Ding et al. (2018). In brief, 5′ rapid amplification of cDNA ends (5′-RACE) and 3′ rapid amplification of cDNA ends (3′-RACE) to obtain the full length cDNA sequences of the AhVgs were done using the SMART RACE cDNA amplification kit (Clontech, Mountain View, CA, United States) following the manufacturer’s protocol. PCR amplification was carried out using the Phusion DNA Polymerase mix (New England BioLabs, Ipswich, MA, United States #M0530). The PCR conditions were as follows: 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 65°C for 10 s, and 72°C for 1 min, with a final 5 min extension. The RACE PCR products were analyzed by 1% agarose gel electrophoresis and purified using an AxyPrepTM DNA Gel Extraction Kit (Axygen, West Orange, NJ, United States). Finally, the purified distinct single-band PCR products were cloned into the pEASY-Blunt vector (Transgen, Beijing, China) and sequenced.
Sequence Analysis and Phylogenetic Analysis
Open reading frames (ORFs) of the AhVgs were obtained using the NCBI ORF finder.1 The putative molecular weights, isoelectric points, and signal peptide positions were determined using the Signal IP 4.1 Server. For phylogenetic analysis, the Vg sequences from insects in Coleoptera, Hemiptera, Dictyoptera, Lepidoptera, Hymenoptera, and Diptera were downloaded from the NCBI reference sequences (RefSeq) database (Supplementary Table S2). A. hygrophila Vg sequences were aligned with these insect Vgs using ClustalW and MEGA 5.0 software (Tamura et al., 2011), following the methods of Zhang W. N. et al. (2016).
Double-Stranded RNA Synthesis and RNAi
For each of the three AhVgs, we synthesized two double-stranded RNAs (dsRNAs) using gene-specific primers; the T7 promoter was added to the 5′-end of each primer (Supplementary Table S1). EGFP (GenBank accession number: AIR08541.1) dsRNA (dsEGFP) was synthesized as a negative control. dsRNA was synthesized using the HiScribeTM T7 Quick High YieldRNA Synthesis Kit (New England BioLabs, Ipswich, MA, United States #E2050S) following the manufacturer’s protocol. The concentration of dsRNA was evaluated using a NanoVue spectrophotometer (GE-Healthcare, Germany), and the purity was verified by running the dsRNA on a 1.0% agarose gel (Supplementary Figure S1). In order to ensure that the volume of dsRNA injected into the control group and treatment group was the same, we adjusted the concentration of all synthesized dsRNA to 10,000 ng/μl (Supplementary Table S3).
The freshly emerged adult A. hygrophila females (<12 h following eclosion) were collected for dsRNA injection utilizing a PLI-100 Pico-Injector (Harvard Apparatus, Holliston, MA, United States) with an MP-255 Micromanipulator (Sutter, Novato, CA, United States) under an Olympus stereomicroscope. The dsRNA solution was injected into the conjunctivum on the abdomen of A. hygrophila female adults. The amount of dsRNA injected for different groups is shown in Supplementary Table S4. Each experiment was repeated at three times. Each repeat included 60 female individuals. Based on the results of the preliminary experiment, we injected about 80–90 females for our experiment to ensure the survival of 60 females for later experimental observation after injection. The female adults and freshly emerged wild adult males were kept in plastic bottles (8 × 10 cm) with fresh A. philoxeroides stems and a piece of moistened filter paper at the bottom to maintain a moist environment. Each pair of adults kept in a separate bottle (Guo et al., 2011). Of the 60 adult females, 20 were used for observation of oviposition, 30 were used for expression analysis of the three Vg genes, and 10 were used for observation of ovary development and measuring the lengths of the ovarioles. The plastic bottles with A. hygrophila adults were transferred to a climate chamber maintained at 28 ± 2°C, 75 ± 5% RH, and a 12 h light:12 h dark photoperiod (Guo et al., 2011).
Expression Analysis of AhVgs
Total RNA was extracted with TRIzol reagent (Life Technologies, Carlsbad, CA, United States) following established protocols. qRT-PCR was conducted with the TransStart Green qPCR SuperMix Kit (Transgen, Beijing, China) on an ABI Prism 7500 instrument (Applied Biosystems, Carlsbad, CA, United States). We designed specific primer pairs for AhVgs with Beacon Designer 7.9 software (PREMIER Biosoft International, CA, United States) (Supplementary Table S1). qRT-PCR reactions and cycling were performed according to the instruction manual of the TransStart Tip Green qPCR SuperMix Kit (TransGen, Beijing, China). The qRT-PCR reactions were conducted in a 20 μl mixture containing 10 μl of 2 × TransStart Green qPCR SuperMix, 0.4 μl of each primer (10 μM), 0.4 μl of 50 × Rox Reference Dye, 200 ng of sample cDNA and 7.8 μl of ddH2O. The qRT-PCR cycling parameters were as follows: 94°C for 30 s, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 10 s, with melt curve stages at 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. To ensure reliability, each experiment included three biological and three technical replicates. Relative expression for each gene was calculated using the comparative Ct method (2−ΔΔCt) (Livak and Schmittgen, 2001). CoxI (GenBank accession number: FJ977926.1) was used for normalization (Supplementary Figure S2).
Agasicles hygrophila Ovary Development and Fecundity After RNAi
Ten 4-day-old female adults from each treatment group were randomly selected to observe ovary development. Ovaries were dissected under an Olympus stereomicroscope (SZX16, Olympus, Tokyo, Japan) using high-precision tweezers (IDEAL-TEK, Balerna, Switzerland). The dissected ovaries were washed three times with 1 × PBS. Photography was performed as described in Liu et al. (2017). Observation and measurement of ovarioles was done as described by Zhao et al. (2016). Each pair of A. hygrophila adults was observed and laid eggs were counted and collected once a day. The egg hatch rate was assessed for a total of 1000 eggs every 12 h for 6 days until the unhatched eggs started to rot (Zhao et al., 2016).
Data Analysis
All analyses of experimental data were conducted with SPSS 18.0 (SPSS Inc., Chicago, IL, United States) and are shown as means ± SD (standard deviation). Experimental data were checked for normality and homoscedasticity, and if needed, were arcsine square-root or log-transformed before analysis. Egg hatching rates and ovariole lengths were arcsine-transformed. The number of eggs was square root-transformed. We performed the least significant difference (LSD) test after one-way analysis of variance (ANOVA) to analyze differences in AhVgs expression levels between different tissues and developmental stages, fecundity and egg hatch rate. AhVgs expression levels after injection and ovariole lengths were analyzed by Student’s t-test. P-values of 0.05 or lower were considered significant.
Results
Sequence and Structural Analysis of the AhVgs
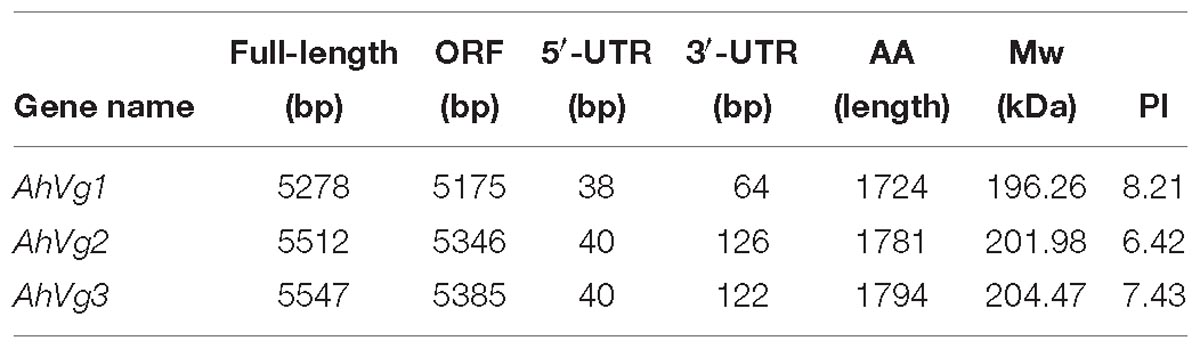
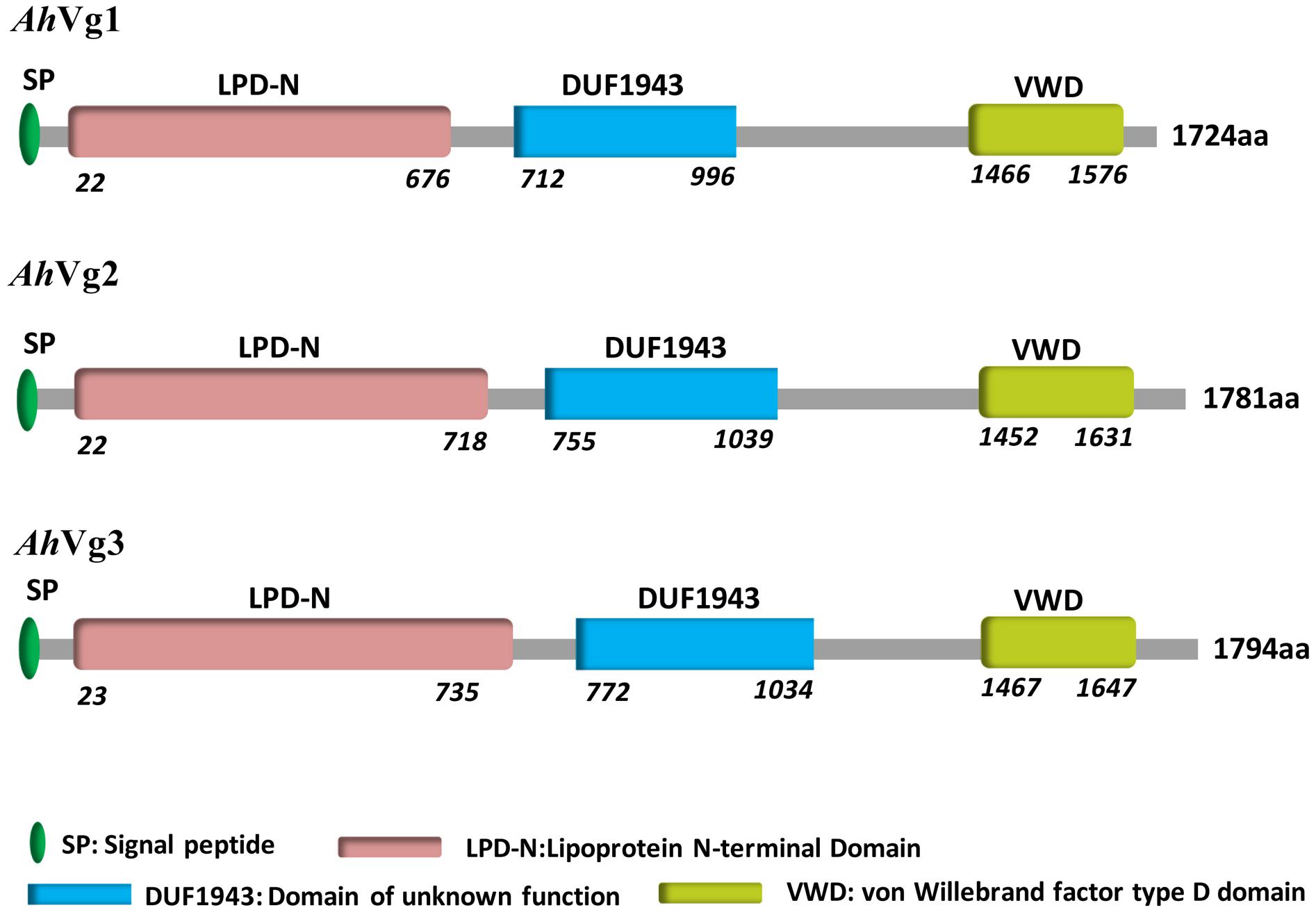
We obtained the full-length cDNAs of the three A. hygrophila genes AhVg1, AhVg2, and AhVg3 by performing RACE-PCR and submitted them to NCBI (GenBank accession numbers: MH423679, MH423680, and MH423681). Information about these full-length AhVg cDNAs including their 5′- and 3′-untranslated regions (5′-UTRs and 3′-UTRs), ORFs, molecular weights and isoelectric points are presented in Table 1. Analyses of the Ahvg1, AhVg2, and AhVg3 protein sequences revealed that all of them have three conserved domains similar to those in other insect Vgs: lipoprotein N-terminal domain (LPD_N), domain of unknown function 1943 (DUF1943), and von-Willebrand factor type D domain (VWD) (Figure 1). Additionally, AhVg1, AhVg2, and AhVg3 all possess a signal peptide, and each protein is predicted to be cleaved between amino acids 16 and 17.
Phylogenetic Analysis of AhVgs Sequences
A phylogenetic tree was constructed to evaluate the phylogenetic relationships of AhVgs with other insect Vgs (Figure 2). Vg sequences from Coleoptera were clustered in one branch, indicating that these Vg sequences had relatively similar amino acid sequences. The clade formed by Coleoptera Vgs was most closely related to those formed by Dictyoptera and Diptera Vgs, suggesting that Vgs in Coleoptera shared a more recent common ancestor with Dictyoptera and Diptera Vgs than with Vgs in other insect species.
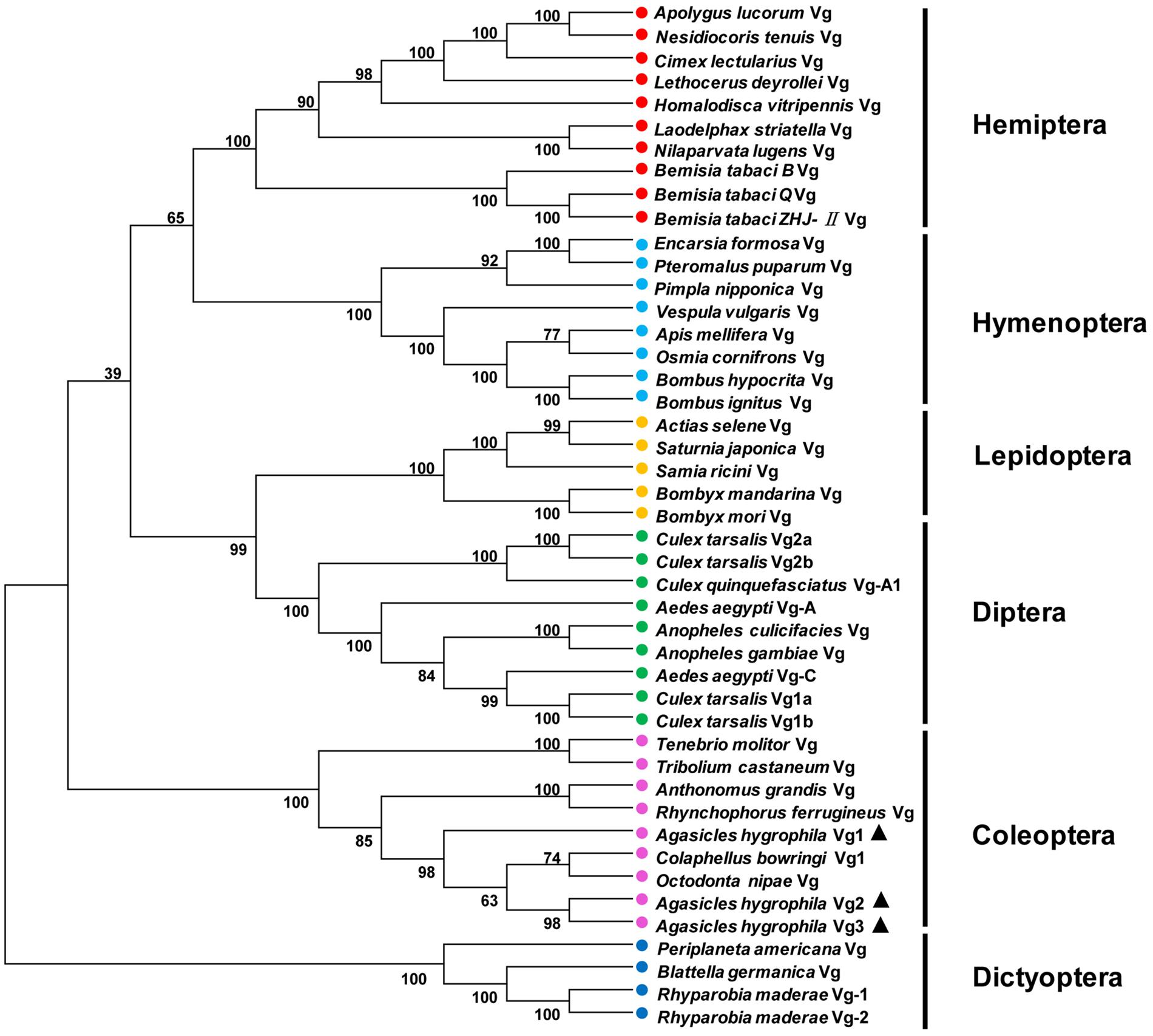
Figure 2. Unrooted consensus neighbor-joining trees for vitellogenin (Vg) proteins. Phylogenetic trees were generated with MEGA 5 using the amino acid sequences of Vgs from insect species from Coleoptera, Hemiptera, Dictyoptera, Lepidoptera, Hymenoptera, and Diptera.
Expression Patterns of AhVgs Across Tissues and Developmental Stages
AhVg1, AhVg2, and AhVg3 mRNA expression levels across different tissues and developmental stages of A. hygrophila were determined by qRT-PCR. AhVg1, AhVg2, and AhVg3 were all mainly expressed in the female fat body (Figure 3); the expression level of each Vg was significantly higher in the fat body than in other tissues. During the pupa and adult stages, expression of AhVgs in the fat body was first detected in the newly emerged 3-day-old females (Figure 3). Additionally, AhVg1, AhVg2, and AhVg3 were each maximally expressed in 9-day-old individuals, after which the expression level decreased significantly.
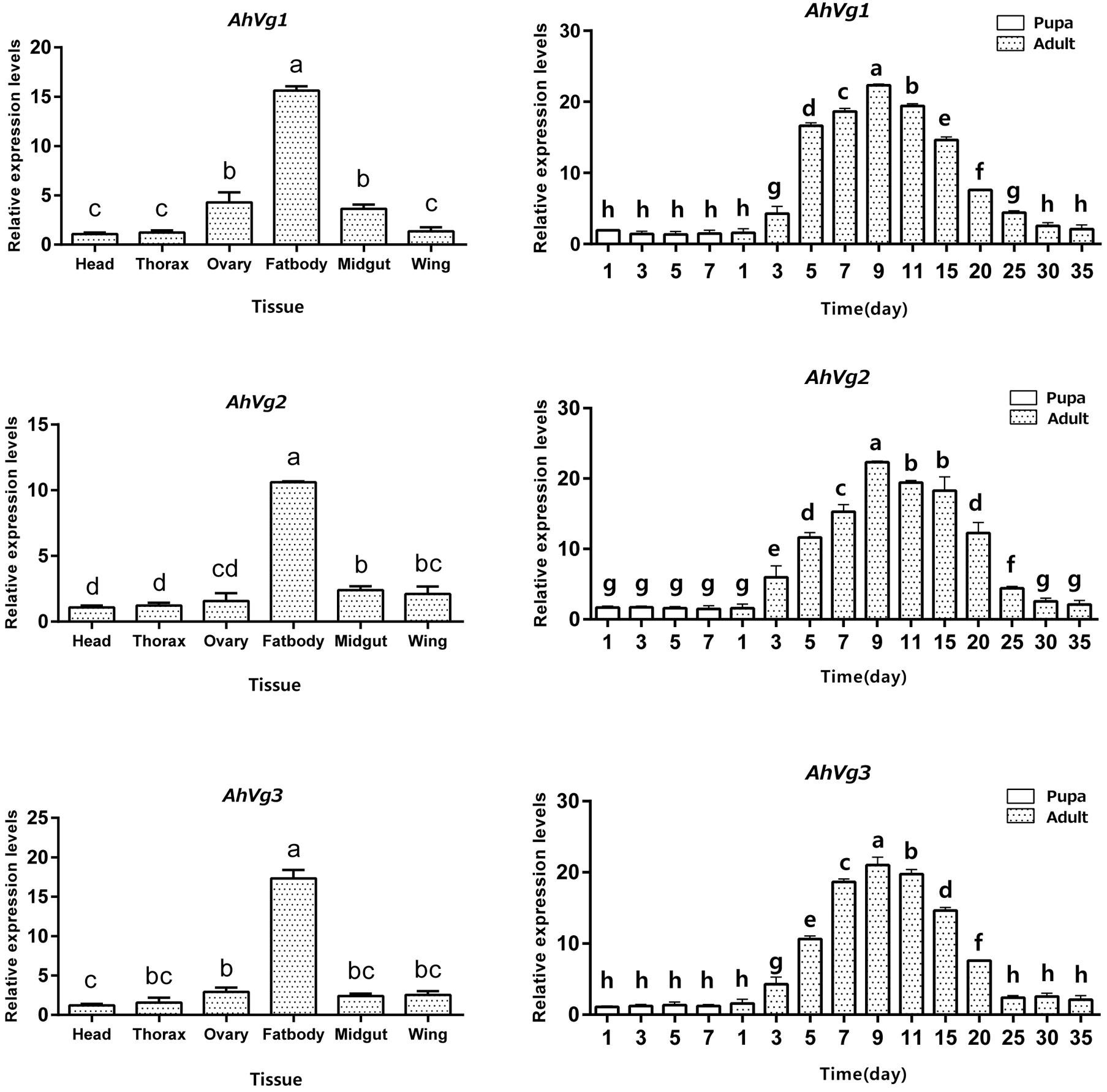
Figure 3. Tissue- and developmental stage-specific expression patterns. The relative mRNA levels were normalized to those of the CoxI gene and analyzed using the 2−ΔΔCT method. All values are shown as the mean ± SD. The data were analyzed by the least significant difference (LSD) test after one-way analysis of variance (ANOVA). Different letters indicate significant differences between means (P < 0.05).
Effects of dsRNA Injection on AhVg Expression
Injection of dsAhVg1, dsAhVg2, and dsAhVg3 into freshly emerged A. hygrophila female adults significantly inhibited endogenous expression of AhVg mRNAs at each time point sampled (3, 6, 9, 12, 15, 20, 25, 30, and 35 days) (Figure 4). From 3 to 20 days after the injection of dsAhVg1, dsAhVg2, and dsAhVg3, the expression levels of these genes decreased significantly by 57.14–78.9% (Figure 4) compared with the dsEGFP control group. However, the decrease in expression levels of AhVg1, AhVg2, and AhVg3 was not significant after 20 days, perhaps because of the timeliness of dsRNA.
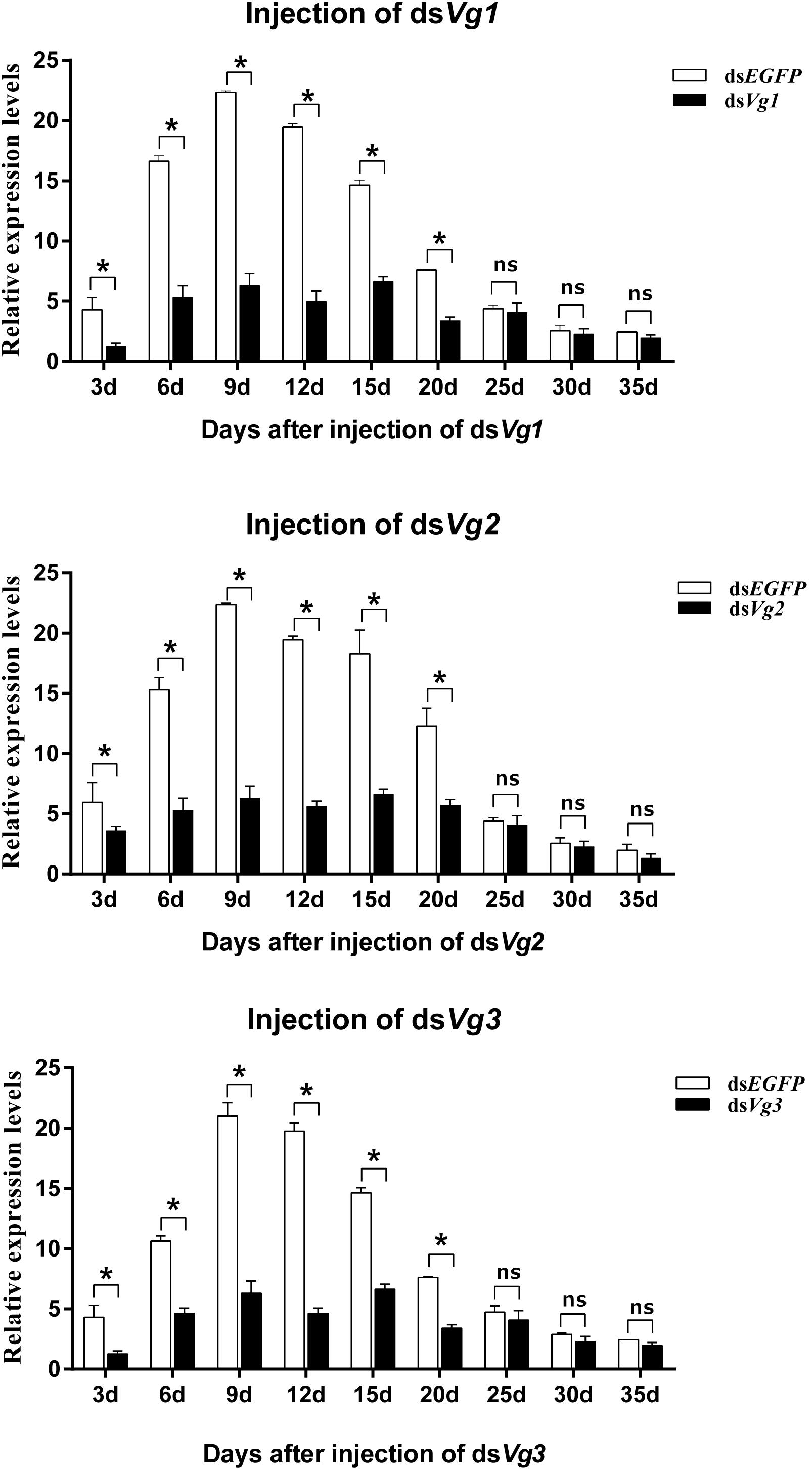
Figure 4. Relative expression levels of AhVg1, AhVg2, and AhVg3 after injection of dsRNA into freshly emerged female Agasicles hygrophila adults. All values are shown as the mean ± SD. The data were analyzed by Student’s t-test. ∗P < 0.05. ns, not significant.
Knockdown of AhVgs Affected Female A. hygrophila Fecundity
After injection of dsAhVg1, dsAhVg2, and dsAhVg3 into the abdomens of freshly emerged female adults, we observed that the number of laid eggs for the dsAhVg1&AhVg2&AhVg3 group was significantly lower than that of the dsEGFP group, while the number of laid eggs for the other injection groups was not significantly different compared with the dsEGFP group (Figure 5). Additionally, the egg hatch rates of the eggs collected from the dsAhVg1&AhVg2&AhVg3 group, but not the other groups, was obviously lower compared with the dsEGFP group (Figure 6). The finding that injection of one or two of the three AhVg genes had no distinct effect on A. hygrophila fecundity indicates that there is strong functional compensation among AhVg1, AhVg2, and AhVg3.
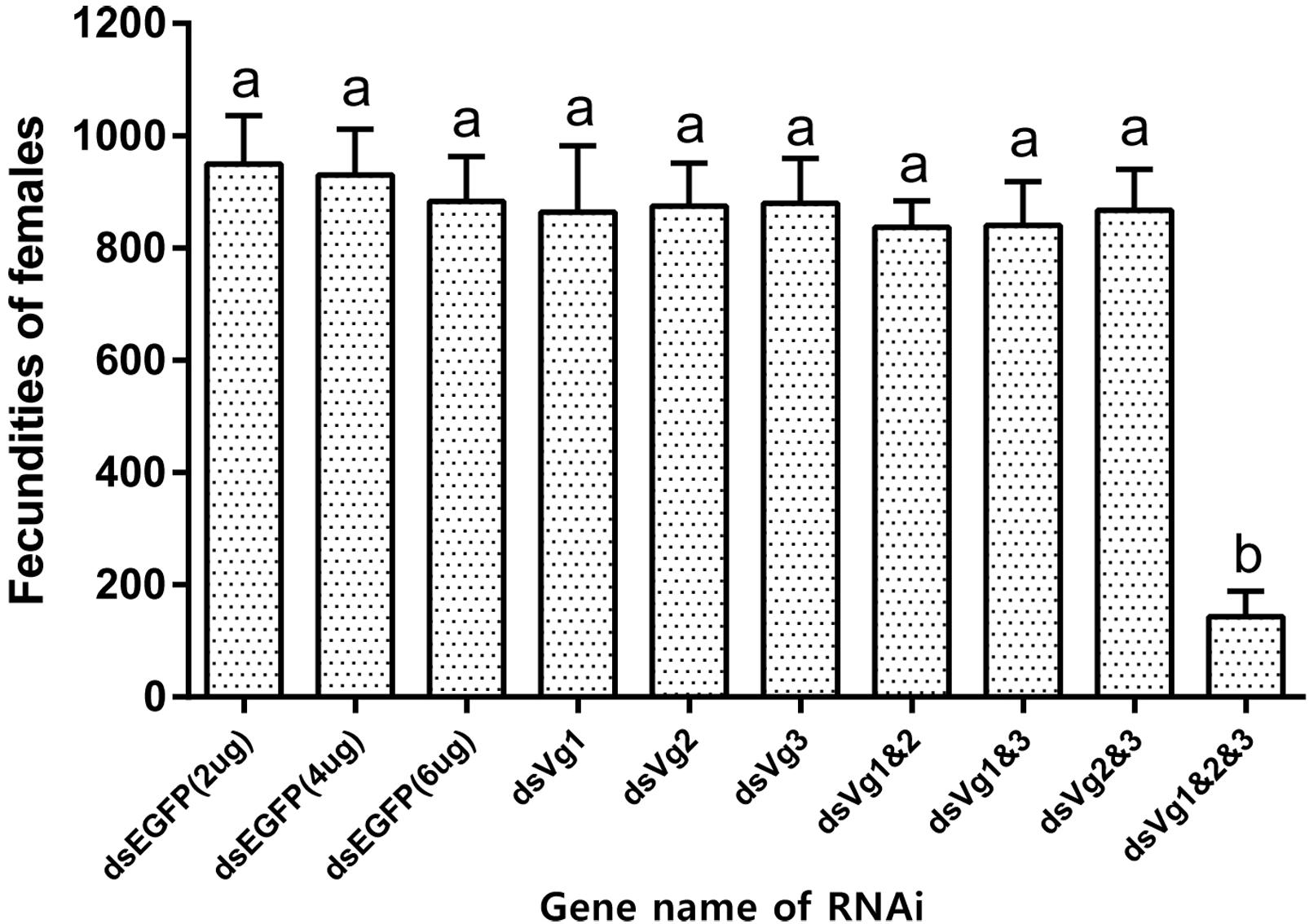
Figure 5. Fecundities of Agasicles hygrophila after injection of dsAhVg1, dsAhVg2, and/or dsAhVg3. Different amounts of dsEGFP were injected as a control. All values are shown as the mean ± SD. The data were analyzed by the least significant difference (LSD) test after one-way analysis of variance (ANOVA). Different letters indicate significant differences between means (P < 0.05).
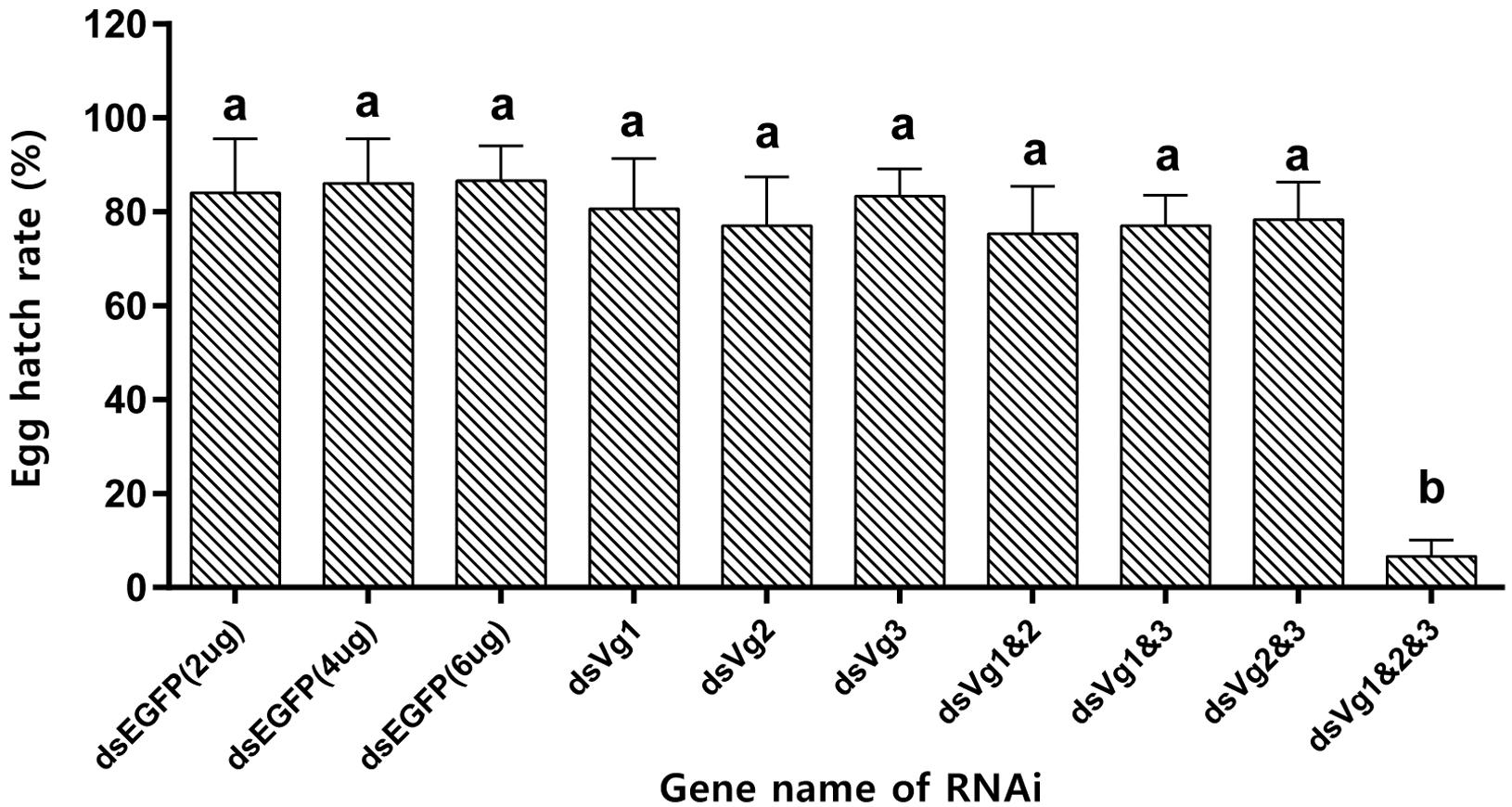
Figure 6. Egg hatch rate of Agasicles hygrophila offspring after injection with dsAhVg1, dsAhVg2, and/or dsAhVg3. Different amounts of dsEGFP were injected as a control. All values are shown as the mean ± SD. The data were analyzed by the least significant difference (LSD) test after one-way analysis of variance (ANOVA). Different letters indicate significant differences between means (P < 0.05).
Knockdown of AhVgs Inhibited Female A. hygrophila Ovarian Development
Because the dsAhVg1&AhVg2&AhVg3 group showed an obvious difference in the number of laid eggs and egg hatch rate compared with the dsEGFP group, we dissected the ovaries of 4-day-old female adults from these two groups to observe ovary development. The ovaries dissected from the triple injection group showed a reduction in deposition of yolk protein and a decrease in oocyte yolk uptake compared with the dsEGFP group (Figure 7). The lengths of the ovarioles of the dsAhVg1&AhVg2&AhVg3 group were significantly shorter than those of the dsEGFP group, while the lengths of the ovarioles among other injection groups were not significantly different from those of the dsEGFP group (Figure 8). These results show that A. hygrophila ovarian development was only inhibited after injection with all three AhVgs.
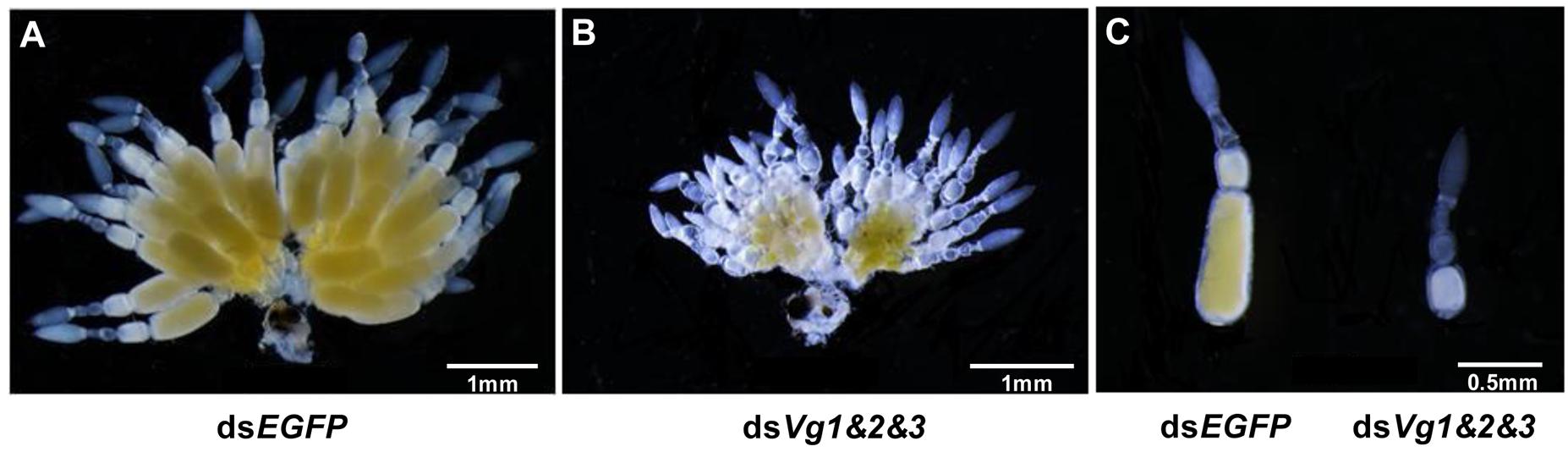
Figure 7. Effect of injection of dsVg1&Vg2&Vg3 on Agasicles hygrophila ovary development. (A) Image of an ovary that developed normally from an adult female injected with dsEGFP. (B) Image of an abnormal ovary from an adult injected with dsVg1&Vg2&Vg3. (C) Image of ovarioles from A, B.
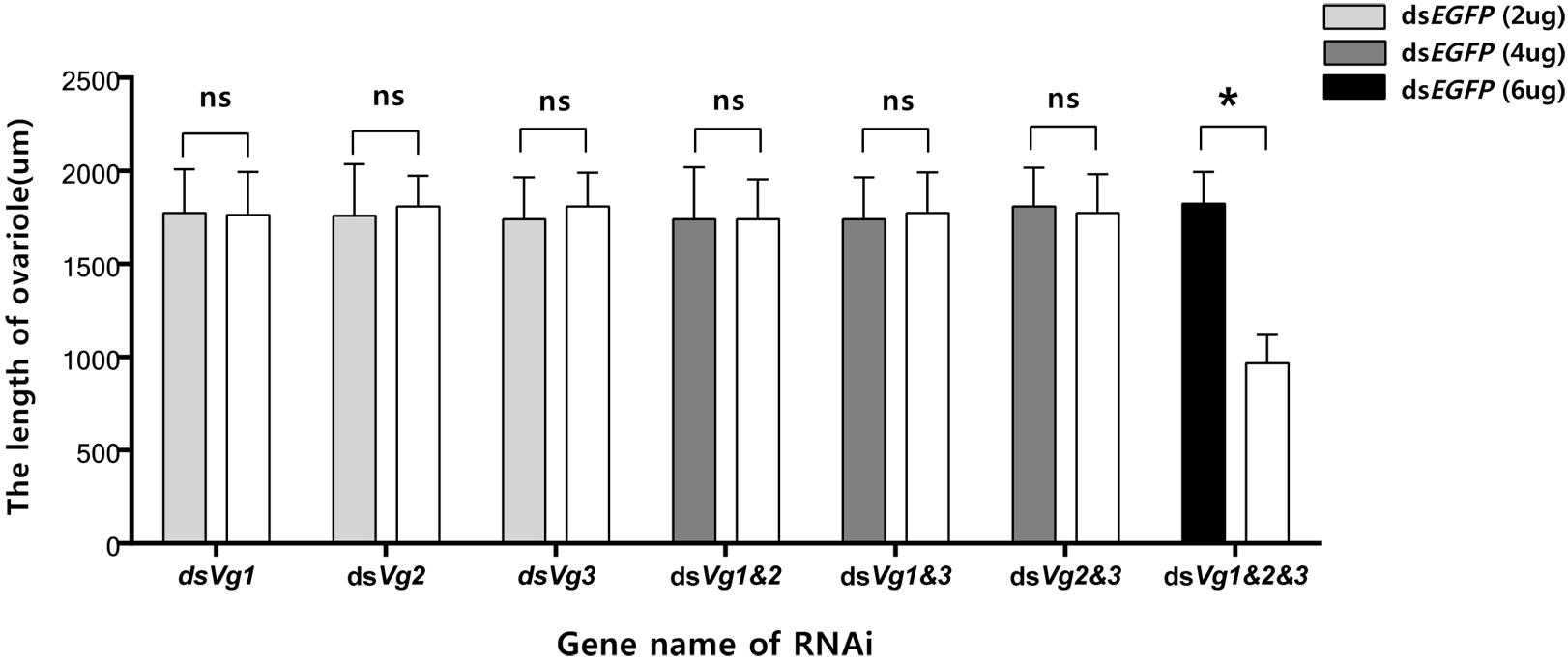
Figure 8. Lengths of the Agasicles hygrophila ovarioles after injection with dsAhVg1, dsAhVg2, and/or dsAhVg3. Different amounts of dsEGFP were injected as a control. All values are shown as the mean ± SD. The data were analyzed by Student’s t-test. ∗P < 0.05. ns, not significant.
Discussion
The hemolymph proteins Vgs are the precursors of egg yolk, and are conserved across many eukaryotes, including insects (Tufail and Takeda, 2008). In insects, these large Vgs (∼200 kDa) are synthesized in the fat body, and the number of cleavage sites in these proteins and their expression patterns can vary (Tufail and Takeda, 2008; Tufail et al., 2014). In our study, we cloned and characterized three Vg genes from the alligatorweed flea beetle, A. hygrophila. This is the first study to investigate the functions of AhVgs in A. hygrophila reproduction. We observed inhibition of ovarian development and reduced fecundity only when silencing of all three AhVgs, which indicates that there is strong functional compensation among the three AhVgs. Our findings provide the basis for studying the functions of Vg genes in other insects.
Previous studies of Vgs have shown that they are members of a multi-gene family, with some insect species possessing more than one representative. For example, Neoseiulus barkeri has three Vg genes (Ding et al., 2018) and Caenorhabditis elegans expresses six Vg genes (Blumenthal et al., 1984). Our study represents the first attempt to clone the three A. hygrophila genes. The ORFs of AhVg1, AhVg2, and AhVg3 were 5175, 5346, and 5385 bp, respectively. Sequence analysis of AhVg1, AhVg2, AhVg3, and Vgs of other insects revealed that all three AhVgs possess domains conserved in Vgs; LPD_N, DUF1943, and VWD. The calculated molecular weights of AhVg1, AhVg2, and AhVg3 are all ∼200 kDa (Table 1), which is similar to the molecular weight of most insect Vgs, such as Thitarodes pui Vg (204.43 kDa; Wu et al., 2018), Spodoptera litura Vg (198.73 kDa; Shu et al., 2009), and Harmonia axyridis Vg (211.88 kDa; Zhang et al., 2017). Phylogenetic analysis indicated that AhVgs is homologous to other Coleoptera Vgs, including those from Tribolium castaneum, Anthonomus grandis, Colaphellus bowringi, and Tenebrio molitor (Figure 2). Additionally, the AhVg2 and AhVg3 sequences were more similar to each other than to AhVg1 (Figure 2).
In our current study, AhVg1, AhVg2, and AhVg3 showed similar expression patterns (Figure 3), with highest expression observed in the fat body, which is consistent with previous studies showing that insects typically synthesize Vg in the fat body (Ding et al., 2018). The VgR takes Vg up from the hemolymph and transports it into developing oocytes (Sappington and Raikhel, 1998). Thus, based on the expression pattern of Vgs in A. hygrophila, we speculate that the specific expression pattern of Vg indicates its function in synthesizing nutrition in the fat body. Although Vg protein synthesis in insects usually takes place in the fat body (Tufail and Takeda, 2008; Tufail et al., 2014), there are some exceptions. For instance, in Rhodnius prolixus, Vg can be synthesized in part in the follicle cells and in part in the fat body, and these both are later incorporated into the oocytes (Melo et al., 2000). After analysis of the expression levels of AhVgs at different stages, we found very high levels of AhVgs mRNA in adult insects but no expression at the pupal stage, so we conclude that AhVg1, AhVg2, and AhVg3 expression is not required during the pupal stage. We propose that AhVg expression is upregulated during sexual maturation in A. hygrophila. Similar expression patterns have also been reported in other insects, such as Bemisia tabaci (Upadhyay et al., 2016) and Actias selene (Qian et al., 2011). However, upregulation of Vg expression in some insects was observed earlier, upregulation beginning during the pupal stage. For example, Vg expression was first detected during the late pupal stage in Spodoptera litura (Shu et al., 2009).
Gene silencing using RNAi is an efficient method to explore gene function (Hannon, 2002), and its use has proven successful in Coleoptera (Arakane et al., 2005; Shi et al., 2016). In our study, the target genes AhVg1, AhVg2, and AhVg3 were silenced with high efficiency in A. hygrophila, but only suppression of all three AhVgs simultaneously negatively affected fecundity and ovary development, causing a significant difference in egg laying, egg hatch rates, and ovary development (Figures 5–8). This phenomenon indicates that AhVg1, AhVg2, and AhVg3 play the same role in reproduction and that there is strong functional compensation among these three AhVgs. Previous studies indicated that successful reproduction in insects depends on two key steps: (1) creation and deposition of Vg and (2) the uptake of Vgs by the VgR into developing oocytes (Sappington and Raikhel, 1998). The silencing of AhVgs significantly affects reproduction in A. hygrophila, ultimately resulting in a decline in egg laying. Silencing of Vgs also results in depressed fecundity and abnormal ovary development in other insects. For instance, suppressing Cimex lectularius Vg caused ovarian tissue atrophy and reduced egg production (Moriyama et al., 2016). Additionally, Shang et al. (2018) demonstrated that Aphis citricidus Vgs are crucial for development during different stages.
Conclusion
We present the first study of three A. hygrophila Vg genes. The amino acid sequences of the three AhVgs contain conserved domains (LPD_N, DUF1943, and VWD) found in Vg sequences from other insects. The tissue- and developmental stage-specific mRNA expression patterns were also similar to those in other insects. We found that all three AhVgs were most highly expressed in the adult stage. Our results provided further information about these highly conserved Vgs in insects. We showed via RNAi bioassays that AhVgs play important roles in A. hygrophila fecundity and ovary development, and that there is strong functional compensation among these three AhVgs. Whether these three AhVg genes have the same function in regulating reproduction in A. hygrophila needs to be further investigated.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
HZ and JG conceived and designed the experiments. HZ, JG, and YW performed the experiments. HZ, YL, JJ, MZ, and ZZ analyzed the data. HZ and JG wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31572068) and the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2015BAD08B03).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a shared affiliation, though no other collaboration, with several of the authors, HZ, YW, YL, ZZ, and JG, at the time of review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00368/full#supplementary-material
Footnotes
References
An, X. K., Sun, L., Liu, H. W., Liu, D. F., Ding, Y. X., Li, L. M., et al. (2016). Identification and expression analysis of an olfactory receptor gene family in green plant bug Apolygus lucorum (Meyer-Dur). Sci. Rep. 6:37870. doi: 10.1038/srep37870
Anderson, E. (1974). Comparative aspects of the ultrastructure of the female gamete. Int. Rev. Cytol. 4, 1–70.
Andres, L. A. (1977). The economics of biological control of weeds. Aquat. Bot. 3, 111–123. doi: 10.1016/0304-3770(77)90011-0
Arakane, Y., Muthukrishnan, S., Kramer, K. J., Specht, C. A., Tomoyasu, Y., Lorenzen, M. D., et al. (2005). The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol. Biol. 14, 453–463. doi: 10.1111/j.1365-2583.2005.00576.x
Blumenthal, T., Squire, M., Kirtland, S., Cane, J., Donegan, M., Spieth, J., et al. (1984). Cloning of a yolk protein gene family from Caenorhabditis elegans. J. Mol. Biol. 174, 1–18. doi: 10.1016/0022-2836(84)90361-9
Boldbaatar, D., Umemiya-Shirafuji, R., Min, L., Tanaka, T., Xuan, X., and Fujisaki, K. (2010). Multiple vitellogenins from the Haemaphysalis longicornis tick are crucial for ovarian development. J. Insect Physiol. 56, 1587–1598. doi: 10.1016/j.jinsphys.2010.05.019
Buckingham, G. R. (1996). Biological control of alligatorweed, Alternanthera philoxeroides, the world’s first aquatic weed success story. Castanea 61, 232–243.
Chen, J. S., Sappington, T. W., and Raikhel, A. S. (1997). Extensive sequence conservation among insect, nematode, and vertebrate vitellogenins reveals ancient common ancestry. J. Mol. Evol. 44, 440–451. doi: 10.1007/PL00006164
Dhadialla, T. S., and Raikhel, A. S. (1990). Biosynthesis of mosquito vitellogenin. J. Biol. Chem. 265, 9924–9933.
Ding, L., Chen, F., Luo, R., Pan, Q., Wang, C., Yu, S., et al. (2018). Gene cloning and difference analysis of vitellogenin in Neoseiulus barkeri (Hughes). B. Entomol. Res. 108, 141–149. doi: 10.1017/S0007485317000591
Engelmann, F., and Friedel, T. (1974). Insect yolk protein precursor, a juvenile hormone induced phosphoprotein. Life Sci. 14, 587–594. doi: 10.1016/0024-3205(74)90373-7
Guo, J. Y., Fu, J. W., Xian, X. Q., Ma, M. Y., and Wan, F. H. (2011). Performance of Agasicles hygrophila (Coleoptera: Chrysomelidae), a biological control agent of invasive alligator weed, at low non-freezing temperatures. Biol. Invasions. 14, 1597–1608. doi: 10.1007/s10530-010-9932-3
Julien, M. H., Skarratt, B., and Maywald, G. F. (1995). Potential geographical distribution of alligator weed and its biological control by Agasicles hygrophila. J. Aquat. Plant Manage. 33, 55–60.
Kim, H. R., Ko, Y. G., and Mayer, R. T. (2010). Purification, characterization, and synthesis of vitellin from the cabbage butterfly Pieris rapae L. Arch. Insect Biochem. 9, 67–79. doi: 10.1002/arch.940090105
Liu, F. Z., Li, K. Y., and Cai, W. (2017). Knockdown of TOR causing ovarian diapause in a genetically stable brachypterous strain of Nilaparvata lugens. Arch. Insect Biochem. 95:e21400. doi: 10.1002/arch.21400
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Melo, A. C., Valle, D., Machado, E. A., Salerno, A. P., Paivasilva, G. O., Nl, C. E. S., et al. (2000). Synthesis of vitellogenin by the follicle cells of Rhodnius prolixus. Insect Biochem. Mol. 30, 549–557. doi: 10.1016/S0965-1748(00)00023-0
Moriyama, M., Hosokawa, T., Tanahashi, M., Nikoh, N., and Fukatsu, T. (2016). Suppression of bedbug’s reproduction by rna interference of vitellogenin. PLoS One 11:e0153984. doi: 10.1371/journal.pone.0153984
Napompeth, B. (1991). “Brief review of biological control activities in Thailand,” in Biological Control in SoutheastAsia, ed. Y. Hirose (Fukuoka: IOBC/SEARS. Kyushu University Press).
Pan, M. L., Bell, W. J., and Telfer, W. H. (1969). Vitellogenic blood protein synthesis by insect fat body. Science 165, 393–394. doi: 10.1126/science.165.3891.393
Pan, X. Y., Geng, Y., Zhang, W. J., Li, B., and Chen, J. K. (2006). The influence of abiotic stress and phenotypic plasticity on the distribution of invasive Alternanthera philoxeroides along a riparian zone. Acta Oecol. 30, 333–341. doi: 10.1016/j.actao.2006.03.003
Qian, C., Liu, C., Zhu, B., Meng, Y., and Wei, G. (2011). Identification and expression analysis of Vitellogenin from Silk-producing Insect, Actias selene Hubner. Afr. J. Biotechnol. 10, 999–1010.
Raikhel, A. S., and Dhadialla, T. S. (1992). Accumulation of yolk proteins in insect oocytes. Annu.Rev. Entomol. 37, 217–251. doi: 10.1146/annurev.en.37.010192.001245
Sappington, T. W., and Raikhel, A. S. (1998). Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect Biochem. Mol. 28, 277–300. doi: 10.1016/S0965-1748(97)00110-0
Shang, F., Niu, J. Z., Ding, B. Y., Zhang, Q., Ye, C., Zhang, W., et al. (2018). Vitellogenin and its receptor play essential roles in the development and reproduction of the brown citrus aphid, Aphis (Toxoptera) citricidus. Insect Mol. Biol. 27, 221–233. doi: 10.1111/imb.12366
Shi, J. F., Mu, L. L., Chen, X., Guo, W. C., and Li, G. Q. (2016). RNA interference of chitin synthase genes inhibits chitin biosynthesis and affects larval performance in Leptinotarsa decemlineata (Say). Int. J. Biol. Sci. 12, 1319–1331. doi: 10.7150/ijbs.14464
Shu, Y. H., Zhou, J. L., Tang, W. C., Lu, K., Zhou, Q. A., and Zhang, G. R. (2009). Molecular characterization and expression pattern of Spodoptera litura (Lepidoptera: Noctuidae) vitellogenin, and its response to lead stress. J. Insect Physiol. 55, 608–616. doi: 10.1016/j.jinsphys.2009.03.005
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tufail, M., Nagaba, Y., Elgendy, A. M., and Takeda, M. (2014). Regulation of vitellogenin genes in insects. Entomol. Sci. 17, 269–282. doi: 10.1111/ens.12086
Tufail, M., and Takeda, M. (2008). Molecular characteristics of insect vitellogenins. J. Insect Physiol. 54, 1447–1458. doi: 10.1016/j.jinsphys.2008.08.007
Upadhyay, S. K., Singh, H., Dixit, S., Mendu, V., and Verma, P. C. (2016). Molecular Characterization of vitellogenin and Vitellogenin Receptor of Bemisia tabaci. PLoS One 11:e0155306. doi: 10.1371/journal.pone.0155306
Veerana, M., Kubera, A., and Ngernsiri, L. (2015). Analysis of the Vitellogenin gene of rice moth, Corcyra cephalonica STAINTON. Arch. Insect Biochem. 87, 126–147. doi: 10.1002/arch.21185
Wu, H., Jiang, F. Z., Guo, J. X., Yi, J. Q., Liu, J. B., Cao, Y. S., et al. (2018). Molecular characterization and expression of vitellogenin and vitellogenin receptor of Thitarodes pui (Lepidoptera: Hepialidae), an insect on the tibetan plateau. J. Insect Sci. 18:23. doi: 10.1093/jisesa/iey010
Wyatt, G. R., and Davey, K. G. (1996). Cellular and molecular actions of juvenile hormone. II. Roles of juvenile hormone in adult insects. Adv. Insect Physiol. 26, 1–155. doi: 10.1016/S0065-2806(08)60030-2
Xing, L., Shen, G., Xu, H., and Lin, H. (2016). The fenpropathrin resistant Tetranychus cinnabarinus showed increased fecundity with high content of vitellogenin and vitellogenin receptor. Pestic. Biochem. Phys. 134:31. doi: 10.1016/j.pestbp.2016.04.010
Zhang, H. J., Liu, F. H., Wang, R. Q., and Liu, J. (2016). Roles of clonal integration in both heterogeneous and homogeneous habitats. Front. Plant Sci. 7:551. doi: 10.3389/fpls.2016.00551
Zhang, W. N., Ma, L., Xiao, H. J., Xie, B. T., Smagghe, G., Guo, Y. Y., et al. (2016). Molecular characterization and function analysis of the vitellogenin receptor from the cotton bollworm, Helicoverpa armigera (Hubner) (Lepidoptera, Noctuidae). PLoS One 11:e0155785. doi: 10.1371/journal.pone.0155785
Zhang, T., Zhang, G., Zeng, F., Mao, J., Liang, H., and Liu, F. (2017). Molecular cloning of the vitellogenin gene and the effects of vitellogenin protein expression on the physiology of Harmonia axyridis (Coleoptera: Coccinellidae). Sci. Rep. 7:13926. doi: 10.1038/s41598-017-14339-3
Keywords: Agasicles hygrophila, vitellogenin, RT-qPCR, RNAi, fecundity, ovary
Citation: Zhang H, Wang Y, Liu Y, Zhao M, Jin J, Zhou Z and Guo J (2019) Identification and Expression Patterns of Three Vitellogenin Genes and Their Roles in Reproduction of the Alligatorweed Flea Beetle Agasicles hygrophila (Coleoptera: Chrysomelidae). Front. Physiol. 10:368. doi: 10.3389/fphys.2019.00368
Received: 28 November 2018; Accepted: 18 March 2019;
Published: 02 April 2019.
Edited by:
Youjun Zhang, Institute of Vegetables and Flowers (CAAS), ChinaCopyright © 2019 Zhang, Wang, Liu, Zhao, Jin, Zhou and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianying Guo, Z3VvamlhbnlpbmdAY2Fhcy5jbg==
 Hong Zhang
Hong Zhang Yao Wang1
Yao Wang1