- 1National Xenopus Resource, Marine Biological Laboratory, Woods Hole, MA, United States
- 2European Xenopus Resource Centre, Portsmouth, United Kingdom
- 3School of Biological Sciences, King Henry Building, Portsmouth, United Kingdom
- 4Amphibian Research Center, Hiroshima University, Higashihiroshima, Japan
- 5Centre de Ressources Biologiques Xenopes, CNRS, Inserm, BIOSIT – UMS 3480, Université de Rennes 1, Rennes, France
- 6Department of Microbiology and Immunology, University of Rochester Medical Center, Rochester, NY, United States
- 7Xenbase, Division of Developmental Biology, Cincinnati Children’s Research Foundation, Cincinnati, OH, United States
Two species of the clawed frog family, Xenopus laevis and X. tropicalis, are widely used as tools to investigate both normal and disease-state biochemistry, genetics, cell biology, and developmental biology. To support both frog specialist and non-specialist scientists needing access to these models for their research, a number of centralized resources exist around the world. These include centers that hold live and frozen stocks of transgenic, inbred and mutant animals and centers that hold molecular resources. This infrastructure is supported by a model organism database. Here, we describe much of this infrastructure and encourage the community to make the best use of it and to guide the resource centers in developing new lines and libraries.
Introduction
Xenopus laevis was first described in 1802 in Daudin’s “Histoire naturelle des rainettes, des grenouilles, et des crapauds” as Bufo laevis. For the next century it was used for comparative anatomical studies [reviewed in Gurdon and Hopwood (2000)] but then became widely distributed as a bioassay for human pregnancy (Hogben, 1939). The availability of Xenopus underpinned their adoption for the study of biochemical mechanisms driving development and cell physiology (Gurdon and Hopwood, 2000). Xenopus has an extraordinary track record as a model organism, playing key roles in discoveries as disparate as isolation of the first eukaryotic gene and transcription factor, the first demonstration of nuclear reprogramming and the discovery of the mesoderm inducing and Spemann organizer molecules, as reviewed in Harland and Grainger (2011) and Tandon et al. (2017). More recently, the size and number of its synchronous embryos, its blastomere size, its well-defined fate map (Shindo et al., 1987) and accurate genome sequences (Hellsten et al., 2010; Session et al., 2016) have enabled the allotetraploid X. laevis and X. tropicalis, its diploid relative, to be used very successfully for transcriptome (e.g., Briggs et al., 2018), proteome (e.g., Baxi et al., 2018), and metabolome studies. Together with the genetic resources described below, and the high efficiency of gene editing in these species, which allows very rapid studies of human genetic disease variants, these techniques continue to demonstrate the value of Xenopus as key model organisms. Here, we review the various resources that are in place to underpin and support research using the Xenopus models.
Transgenic Resources
Transgenesis, or the ability to transfer DNA from one genome to another, is a powerful tool which can be used in established model systems for the study of regulatory and coding DNA in normal and disease-associated processes as well as changes in gene function and control that occur during evolution. The origins of transgenesis can be traced back to the discovery of bacterial restriction enzymes and their use to generate recombinant DNA plasmids in the early 1970s (Cohen et al., 1973). In Xenopus, a number of reports in the 1980s demonstrated that simply microinjecting exogenous plasmid DNA into fertilized eggs can result in its successful integration into the frog genome (Rusconi and Schaffner, 1981; Etkin and Roberts, 1983; Andres et al., 1984; Bendig and Williams, 1984; Etkin et al., 1984), although showing mosaic distribution in the adult tissues. Etkin and Pearman (1987) demonstrated that exogenous, linearized plasmid DNA can be successfully transmitted through the germline of the transformed male parent to following generations; however, this approach proved to be highly inefficient since only ∼1% of the injected individuals demonstrated mosaic integration of the DNA into their germline (Yergeau et al., 2010). It took another decade for the idea of using Xenopus in transgenic studies to become a practicable endeavor with the development of much more efficient methods of transgenesis, initially via restriction enzyme mediated integration (REMI) (Kroll and Amaya, 1996) and then through the use of phiC31 integrase (Allen and Weeks, 2005), I-SceI meganuclease (Ogino et al., 2006; Pan et al., 2006), and various transposable element-based approaches (Yergeau et al., 2007). The use of I-SceI meganuclease has been shown to be especially effective with the reported ratios of non-mosaic integration in the F0 generation and germline transmission as high as 30% in X. tropicalis and 20% in X. laevis (Ogino et al., 2006). These transgenesis methods result in random integration of the exogenous DNA and, although approaches for targeted, precise integration using gene editing have recently been described (Aslan et al., 2017), their use in the Xenopus community is at an early stage (see below).
Several practical aspects make Xenopus an enticing model to use in transgenic studies. These include the ability of a single female to produce as many as 4000 eggs per spawning (Wlizla et al., 2017), thus providing a large batch of sibling embryos that are synchronous and develop externally. Furthermore, embryonic development is relatively rapid, with most major organs formed within 5 days following fertilization, and is easily observable since the tissues surrounding major viscera are transparent during the same time frame (Nieuwkoop and Faber, 1994; Khokha et al., 2002). However, the model is somewhat limited by the generation time with the two most commonly used species, X. laevis and X. tropicalis taking approximately 6–12 and 5–8 months, respectively, to reach sexual maturity, with males maturing slightly faster than females. Due to this limitation, most individual labs tend not to spend their time and resources to generate true breeding transgenic animal lines and instead focus on experiments that take advantage of the rapid early development. The disadvantage of a long generation time is to some extent balanced by the long period of fertility in the animals with X. laevis as old as 15 years producing viable offspring (Tinsley and Kobel, 1996); careful breeding strategies can thus be used to avoid significant genetic drift in this species.
The Xenopus resource centers including the National BioResource Project (NBRP) in Japan, the European Xenopus Resource Centre (EXRC) in Europe, and the National Xenopus Resource (NXR) in the United States of America were established, in part, to serve as centralized repositories with sufficient infrastructure to allow for maintenance of the extant Xenopus transgenic lines at capacities allowing for their distribution to individual labs on an as needed basis (Pearl et al., 2012). The stock centers also have expertise in generating new lines which can then be grown and made available to the research community. This has effectively eliminated the need for labs to contribute crucial resources into generation of novel transgenic lines since these are readily available for distribution as adults, tadpoles, embryos, isolated testes, or cryopreserved sperm (Pearl et al., 2017). Currently, the stock centers hold over 130 different transgenic lines, a number that is continuously increasing, and which can be grouped into four different categories: (1) reporter expression lines, (2) inducible lines for disruption and regulation of signaling pathway activity, (3) GAL4 and Cre driver lines, and (4) single landing site lines (Supplementary Table S1).
Reporter expression lines form by far the largest category of transgenic Xenopus lines available and can be further subdivided into several groups. First are the tissue/region specific lines which typically contain a fluorescent protein driven by a specific promoter to mark a particular tissue, region, or organ in the developing embryo (Supplementary Table S1A). Besides being useful for observation of normal development, these lines are highly amenable to investigations of abnormal development following disruption of gene activity, as demonstrated by a recent study from the Miller lab using a line marking the developing kidney, Xla.Tg (Dre.cdh17:eGFP)NXR (RRID:NXR_0102), to investigate the disruption in kidney development following morpholino induced knockdown of the Planar Cell Polarity pathway component, Daam1 (Corkins et al., 2018). Studies using a similar design can easily be performed in lines marking other parts of the developing embryo taking advantage of the other tissue/region specific lines available. Certain experimental conditions, in particular ones relying on the use of formalin-fixed, paraffin-embedded tissues, result in a considerable loss of true fluorescent protein signal, and an increase in tissue autofluorescence (Jiang et al., 2005; Swenson et al., 2007). In cases where such histological preparations are necessary, transgenic lines that mark tissues/regions through colorimetric methods can be used in place of fluorescent reporters. An example of such a line is Xtr.Tg (tubb2b:Has.ALPP;cryga:dsRed)Amaya (RRID:EXRC_3003, NXR_1099), which allows for rapid detection of axonal projections in situ by a simple alkaline phosphatase reaction at any stage of development in whole embryos, which can then be further processed for histology (Huang et al., 2007). This reporter group also includes lines driven by ubiquitous promoters like CMV and human ubiquitin C. These are particularly useful for cut-and-paste, transplantation-based experiments to label and fate map regions of host embryos.
The second group of reporter expression lines includes transgenics marking subcellular organelles (Supplementary Table S1B). These are highly useful for the study of molecular processes involved in cell function and are especially effective when utilized in the context of Xenopus egg extracts, the only cell-free system that permits full investigation of all DNA transactions related to cell cycle progression and DNA damage repair (Cross and Powers, 2009; Hoogenboom et al., 2017). Many of these lines have been generated by the Ueno lab and mark a diverse range of organelles including plasma membrane, Xla.Tg (CMV:RFP-CAAX)Ueno (RRID:EXRC_0075, NXR_0115), golgi bodies, Xla.Tg (CMV:Has.B4GALT1-eGFP)Ueno (RRID:EXRC_0077, NXR_0110), and microtubule plus ends, Xla.Tg (CMV:Has.MAPRE3-eGFP)Ueno (RRID:EXRC_0078, NXR_0109) (Takagi et al., 2013). Lines generated by other labs, as well as by the stock centers themselves, that label other organelles are also available.
The third group of reporter lines are those that serve as indicators of signaling activity (Supplementary Table S1C). These include Wnt signaling reporter lines generated by the Vleminckx lab in both X. laevis and X. tropicalis (Tran et al., 2010), an apoptosis sensor (Kominami et al., 2006), a histone H3 lysine 9 acetylation sensor for in vivo epigenetic analysis (Sato et al., 2013; Suzuki et al., 2016), a neural tissue specific calcium signaling sensor (Chen et al., 2013), and two distinct lines for detection of oxidative stress response (Love et al., 2013).
The fourth group of reporter lines are transposable element enhancer trap lines, all generated by the Mead lab (Supplementary Table S1D; Yergeau and Mead, 2009; Yergeau et al., 2010). Although the integration sites for some of these lines have been identified, this is not the case for all of them. These lines are of use in studies where the particular promoter driving expression is not important and it is the labeled region of the embryo that matters. They can also be used for further study of transposable element activity and additional discovery of regulatory regions via enhancer trap approaches and include lines which result in remobilization of the transposons in the offspring produced.
The fifth group currently contains a single line generated in the Buchholz lab allowing for temporal regulation of fluorescent marker expression via treatment with doxycycline (Supplementary Table S1E; Rankin et al., 2009). Other lines where temporal transgene expression can be regulated through use of simple small molecule treatments or heat shock will likely be added to this group in the future.
The final group includes reporter lines that are especially suitable in fate mapping studies via temporal or regional switch in fluorescence (Supplementary Table S1F). These lines work best when used in crosses with transgenic driver lines designed to regulate the switch in expression pattern (Supplementary Tables S1, S3). Among these reporters are two Brainbow lines generated at the NXR as well as a number of lines made in the Ryffel lab (Waldner et al., 2006; Livet et al., 2007). The initial fluorescence is ubiquitous; however, these lines contain loxP or FRT sites, and the initial fluorescence can be altered through crosses with driver lines that express Cre or FLP recombinase, respectively. The change in fluorescence can be spatially or temporally regulated through use of region specific, or inducible promoters or can alternatively be induced by targeted microinjection of Cre or FLP mRNA. This group of reporters also includes two types of lines which rely on a binary effector-transactivator design to function, the GAL4-UAS system and the tet-on system (Gossen and Bujard, 1992; Chae et al., 2002; Hartley et al., 2002).
In the GAL4-UAS system, the reporter gene is regulated by a UAS effector and will only be expressed when crossed to a driver line expressing GAL4. This allows for spatial or temporal control of reporter expression dependent on the promoter used to control expression of GAL4. Similarly, in the tet-on, the reporter is regulated by a TRE effector and will only be expressed if crossed to a driver line containing an rtTA transactivator. However, besides the promoter used to control the rtTA expression, an additional level of temporal control is provided since rtTA only works in the presence of doxycycline.
The remaining three main categories are somewhat less varied than the reporter lines but have important functional applications and include: inducible lines for disruption and regulation of signaling pathway activity (Supplementary Tables S1, S2), GAL4 and Cre driver lines (Supplementary Tables S1, S3), and single landing site lines (Supplementary Tables S1, S4). Disrupting signaling pathways can be achieved by microinjection of constitutively active or dominant negative pathway components into early embryos, however, many of the transgenic lines available are designed to allow for precise temporal control thus permitting the study of signaling pathway roles at much later stages of development than microinjection allows.
There are two ways these transgenes are regulated. First is through use of a heat shock promoter as in the Xla.Tg (hsp70:nog;cryga:GFP)Jmws (RRID:EXRC_0018 NXR_0020), which was used to study temporal requirement for BMP signaling in haematopoiesis (Kirmizitas et al., 2017), or the Xla.Tg (hsp70:Xtr.dkk1;cryga:GFP)Jmws (RRID:NXR_0021), which could be used in a similar manner to interfere with Wnt signaling. The second way the expression of these transgenes can be regulated is through use of binary control systems like CRE-Lox, FLP-FRT, GAL4-UAS, and tet-on. These lines may require the use of driver transgenic lines to regulate their activity, but not always, as in the case of Xla.Tg (Mmu.col1a2:rtTA;TRE:DNthra-GFP)Brown (RRID:EXRC_0026) which has both components of the tet-on system and allows, in the presence of for expression of a GFP tagged dominant negative form of the thyroid hormone receptor (DNthra) specifically in cartilage, and which was used in the study of thyroid hormone function in limb development (Brown et al., 2005).
The currently available driver lines allow for use of CRE or GAL4 in a manner described above in order to control changes in fluorescence patterns of reporter lines or activities of signaling. In X. laevis the CRE drivers are mainly designed to allow for temporal control through the use of a heat shock promoter or doxycycline treatments as part of a tet-on system, although a transgenic line from the Ryffel lab that allows for CRE expression specifically in muscle cells is also available (Waldner et al., 2006; Roose et al., 2009; Kerney et al., 2012). In X. tropicalis there is a GAL4 driver line which allows for both temporal and spatial control, Xtr.Tg (tubb2b:PR-Gal4;cryga:CFP)Zimml (RRID:EXRC_3033, NXR_1109). The tubb2b promoter, used here, drives expression specifically in neural cells, however, the GAL4 DNA binding domain is fused to the ligand binding domain of the progesterone receptor and thus, to function, requires the progesterone antagonist RU-482. This allows for full temporal regulation (Waldner et al., 2006). When used together with some of the available UAS effector lines temporal disruption of Wnt or Hedgehog signaling can be induced specifically in the nervous system. Additional driver lines that allow expression in other tissues and the use of FLP-FRT or the tet-on systems will very likely become available in the future.
Finally, there is the landing site category which currently contains a single line, Xla.Tg (CFP-ATTP)Ryff (RRID:EXRC_0058). This line contains an attP docking site within a functional blue fluorescent protein coding sequence. Using phiC31 mediated integration, exogenous proteins can be introduced into this line, and loss of blue fluorescence can be used to screen for effective integration. Transgenic animals generated this way are likely to show very similar levels of the exogenous proteins expressed, due to local transcriptional control elements and chromatin environment being the same.
The transgenic lines available from the stock centers can be used in versatile ways to investigate many questions that are essentially low hanging fruit, ready to be plucked. The whole list may be a bit overwhelming at first glance, but the staff at the centers is there to provide advice and guidance regarding the best lines available for use in the study of particular questions. Individual investigators are encouraged to take advantage of this expertise and the lines available to move their research forward. Furthermore, in particular the EXRC and the NXR have expertise in generating novel transgenic lines and growing them to adulthood. If a line of interest is not available, we encourage investigators to take the initiative and request it to be made or provide guidance to aid the stock centers in determining which lines should be generated as a priority.
Gene Editing Resources
Xenopus have proven to be excellent organisms in which to perform gene editing experiments due to their external fertilization and efficiency of RNA and protein microinjection into synchronous embryos to deliver ZFNs (Nakajima et al., 2012; Young and Harland, 2012), TALENS (Lei et al., 2013; Miyamoto et al., 2015), or CRISPR/Cas9 (Blitz et al., 2013; Nakayama et al., 2013). Numerous labs have shown that production of simple insertions and deletions (indels) is very straightforward, fast, and efficient (McQueen and Pownall, 2017; Tandon et al., 2017). Studies using CRISPR/Cas9 also allow loss of function experiments to take place much later than those previously possible in Xenopus, which relied on injection of mRNAs expressing dominant negative proteins (Amaya et al., 1991) or antisense morpholino oligonucleotides that have recently been shown to be able to cause off-target, generic, phenotypic effects (Gentsch et al., 2018). The main drawback to gene editing in Xenopus is that, in F0 animals, it produces mosaicism (Ratzan et al., 2017); this is mainly due to the rapid cell divisions that occur every 30 min and the low temperature at which Xenopus are raised. Frogs made this way have nonetheless been used to address various biomedical topics, including cancer, immunology, neurobiology, cell biology, and other developmental biology questions (Naert et al., 2016; Banach et al., 2017; Hassnain Waqas et al., 2017; Delay et al., 2018) as well as to provide embryos that can be used as tools to understand human genetic diseases (Feehan et al., 2017; Deniz et al., 2018; Naert and Vleminckx, 2018a; Sega et al., 2018). Although mosaic, mutations in F0 animals are useful due to the high level of penetrance, often exceeding 90% in X. tropicalis when measured using TIDE (Brinkman et al., 2018; Naert and Vleminckx, 2018b).
In addition to indel-based knockout F0 animals, the resource centers are also producing knockout lines both for the community and to order. Staff in the centers work closely with individual researchers to create mutants in specific target genes and then breed the animals to determine germline transmission. In addition, a large program targeting 200 developmentally important genes prioritized by the research community is being undertaken at the NXR, although it is funded separately from the resource center itself. The first of these lines are now being inbred to produce F2 homozygous mutant animals and knockout phenotypes are being assayed; phenotypic analysis is then done in collaboration with individual researchers. These mutants are being made available and cataloged on Xenbase as soon as they are initially characterized. Making locus-specific gene knock-ins in embryos has, in all vertebrates, proven a significant challenge. Although this is possible in Xenopus by using injection of fertilized eggs and screening large numbers of animals (for example in the runx1 locus shown in Figure 1), it is inefficient and there are no reports of precise DNA construct integration in the germline by egg injection. Recent work shows, however, that being able to access the Xenopus oocyte to apply gene editing techniques for insertion largely overcomes this challenge (Aslan et al., 2017), most likely because the levels of the homology directed repair (HDR) machinery are much higher in oocytes than eggs (Hagmann et al., 1996). To prevent mosaicism, oocytes can be cultured long enough (3 days) for injected sgRNA and Cas9 to decay before fertilization and treatment with SCR-7, a DNA ligase IV inhibitor, and increases the likelihood that the genetic lesion is repaired via HDR mechanisms instead of the double-strand break repair pathway (DSBR) (Aslan et al., 2017). There are other approaches being taken in several organisms to improve HDR-mediated integration using CRISPR/Cas9 and it should become clear in the near future which becomes the dominant technology in Xenopus.
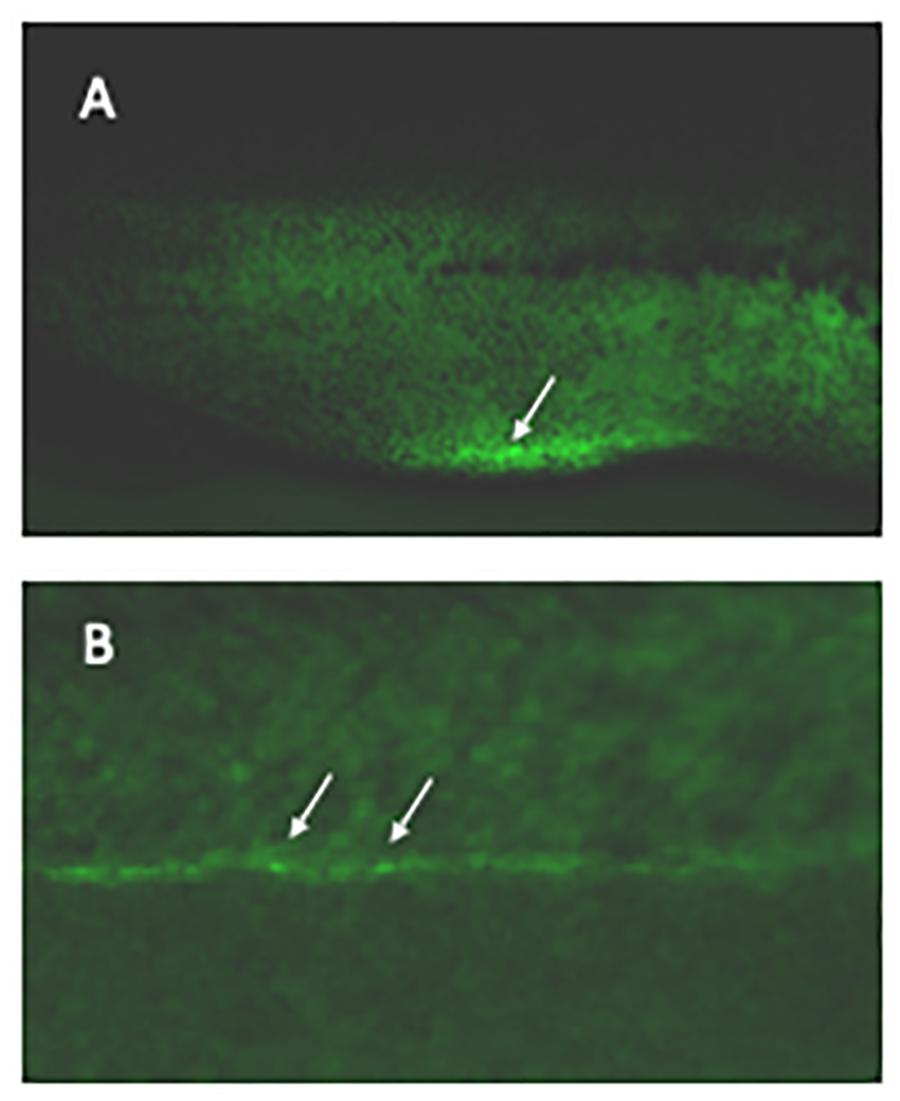
Figure 1. Labeling haematopoietic stem cells in live embryos. Xenopus laevis eggs were microinjected with CRISPR/Cas9 targeting the 3′UTR of runx1 together with a DNA construct containing 400 bp homology arms from either side of the cut site flanking an IRES controlling eGFP expression. Founder embryos were screened for mosaic expression of GFP in the correct regions and 24 grown to adulthood. Their offspring were then screened for germline transmission; two sets of offspring showed strong expression in the vbi (A, arrowed) and vasculature (B, examples arrowed) as expected for runx1 at this stage. The transmission rate was 46 and 52% in the 2 sets. An F2 embryo is shown.
Robust Inbred Strains of X. tropicalis
There are two major wild type strains of X. tropicalis: “Ivory Coast” and “Nigerian” that were originally collected from different localities (Tymowska and Fischberg, 1973; Grainger, 2012). After their introduction to the community in 1990 they were bred in several different labs, resulting in a number of strains that can be distinguished by mitochondrial haplotype and genotypic data on SSLP markers. The Ivory Coast and Nigerian strains are evolutionally diverged, and some Ivory Coast strains have diverged from one another while Nigerian strains share a unique mitochondrial haplotype (Kashiwagi et al., 2010; Igawa et al., 2015). Some of these strains are particularly suited to genetic studies, for example those with a short generation times such as Golden and Superman.
When using reverse genetic tools such as CRISPR/Cas9, that rely on a lack of polymorphisms in target regions for cutting, sequence insertion and analysis, inbred strains greatly increase the efficiency of experiments. Four strains in the NBRP X. tropicalis have been successfully maintained to achieve inbred status (Igawa et al., 2015). Heterozygosity value (HE) of SSLP markers decreased following a theoretical reduction curve due to brother-sister mating in every generation (Figure 2). The offspring are vigorous in viability, comparable to other non-inbred strains, and thus these strains have stabilized past the stage of inbreeding depression. The inbred strains have been tested successfully for transgenesis and gene editing (Sakane et al., 2018) and should begin to become available to the community late in 2019.
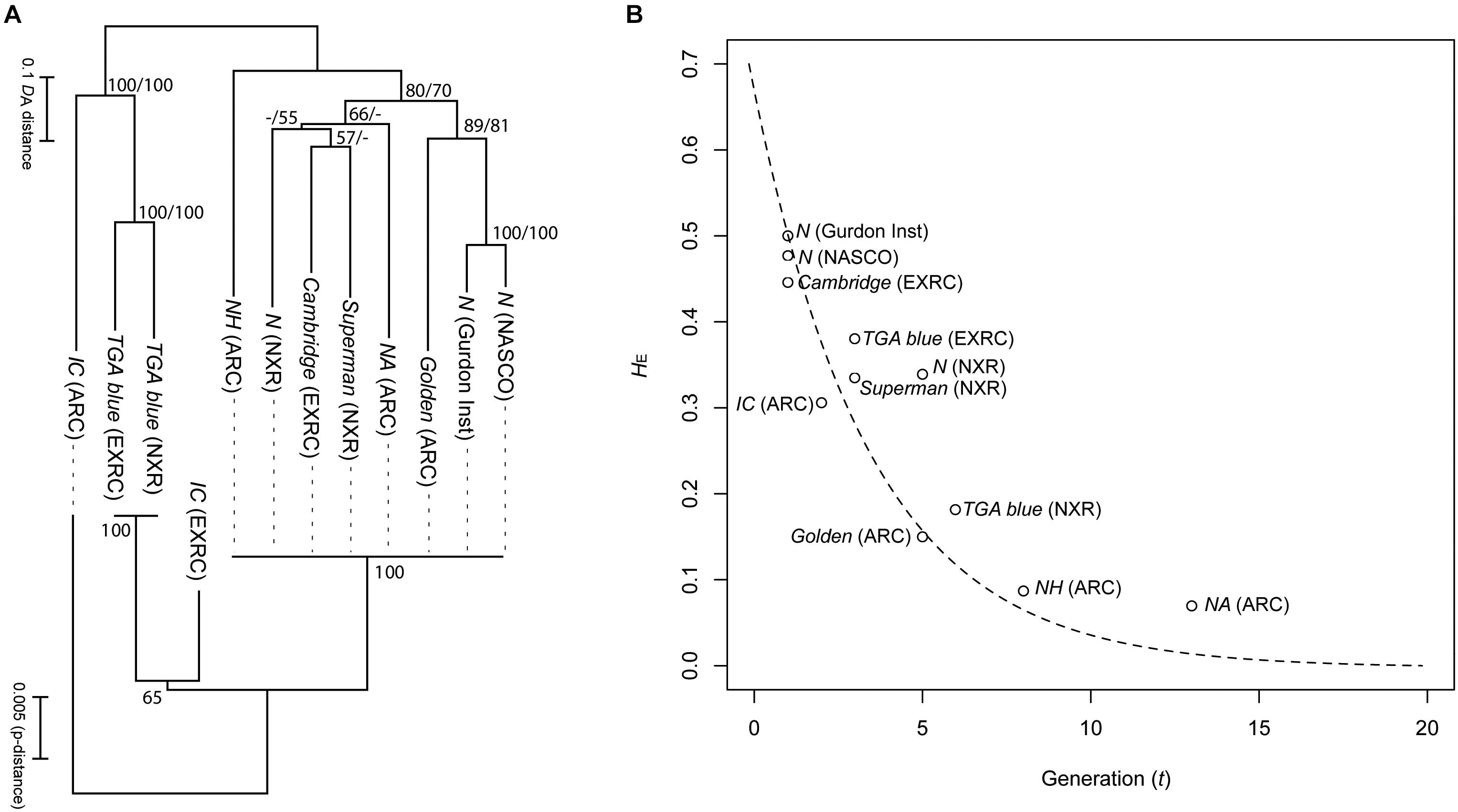
Figure 2. Genetic relationships and heterozygosity reduction curve in the strains of X. tropicalis. (A) Neighbor joining trees of the strains based on genetic distances (DA) of 60 Simple Sequence Length Polymorphism (SSLP) markers (upper) and p-distance of mitochondrial haplotypes (2,328 bp; lower). Numbers on branches indicate percent bootstrap probability (only indicated > 50%). (B) Plot of expected heterozygosity (HE) against generation number (t). The curved dotted strain indicates theoretical reduction of heterozygosity via single brother-sister matings for every generation, calculated from the following equation: HE = H0.
Training Resources
As described above, new genetic and biochemical techniques are being developed for Xenopus research, and the application and usability of Xenopus in basic and biomedical sciences are expanding into a wide range of research areas such as human disease modeling in CRISPR/Cas9 induced mutants. To meet this increasing demand, the resource centers are not only distributing materials but also providing a variety of advanced training courses and research opportunities for users who are not familiar with basic or advanced methods. Each resource center organizes a unique combination of educational opportunities and updates the content of courses annually based on requests from users.
In 2013, the NBRP X. tropicalis started running annual technical courses in Hiroshima, to teach several basic techniques such as artificial fertilization of eggs, microinjection of synthetic mRNAs/oligonucleotides, and husbandry of tadpoles and frogs. Trainees include faculty members, post-doctoral researchers, and graduate and undergraduate students from across Japan. The training course has been continually updated, and now includes transgenesis, genome editing and bioinformatics. Moreover, a user survey resulted in further development of the course in 2018. To allow more time for genome editing techniques, the NBRP technical courses are now held in both summer and winter annually. The technical courses represent a good opportunity not only for providing training, but also for distributing and testing genetic tools – more than 200 plasmid samples from the X. tropicalis cDNA plasmid collection and transgenesis plasmids have been distributed during the past six courses. In addition to its technical courses, the NBRP X. tropicalis has been organizing a joint technical course with the NBRP Medaka in Okazaki, Japan. This joint course is designed for overseas trainees from around the world and aims to facilitate the beneficial use of two different aquatic model organisms. Xenopus has become a popular animal for education in schools, members of the NBRP X. tropicalis have been providing opportunities to introduce Xenopus research to medical university and high-school students across Japan and to students from other Asian countries. To reach a wider range of researchers, the NBRP X. tropicalis is also running XenoBiores1, which facilitates the exchange of questions and troubleshooting among users and provides the latest information on a variety of topics including conferences reports, technical courses and genetic/live-animal resources.
The NXR at the Marine Biological Laboratory (MBL) in Woods Hole, has a range of advanced training courses, which are run annually and are aimed at advanced and beginner Xenopus researchers. These courses bring experts in the fields of bioinformatics, specialized imaging, and gene editing to teach cutting edge techniques to the participants. The Bioinformatics Workshop is designed for wet lab biologists who want to extend their understanding of computational tools and methods and may also want to acquire grounded computational skills to enable them to work independently. The imaging workshop provides hands on training in imaging Xenopus embryos, including live imaging of embryos and explants and image processing, analysis and quantitation. The genome editing workshop focuses on providing theoretical and practical aspects of genome editing in Xenopus, allowing participants to generate mutants during the workshop. Husbandry training is also offered at the NXR to promote harmonization among Xenopus researchers and improve yields of healthy and productive frogs throughout the community. Furthermore, the NXR offers a research facility service, which allows visiting scientists to integrate themselves in the NXR facility and MBL. This allows full access to all available Xenopus lines, equipment, reagents, and expertise in gene editing. Uniquely, the NXR allows for a rich collaborative environment established by the diverse group of scientists from different disciplines who come every year to do their research at the MBL.
The EXRC at Portsmouth does not run specific training courses, but within its remit is a “research hotel” service that is becoming heavily used. Researchers, who may be very familiar with Xenopus or used to other models, come to the center and work alongside staff to learn new techniques and carry out their own experiments, using all of the resources at cost. Often, they make new lines to be grown up in the center then return to analyze them.
Specialized Xenopus Resources
Also situated in Europe and regularly providing training is the French Xenopus Biological Resource Center (CRB2), which has been located in Rennes since 2008. Training is provided by members of the CRB in the fields of Xenopus breeding techniques, embryo manipulation and, like at the EXRC, how to care for Xenopus as required by EU laws. Research-wise, the CRB has special expertise in projects centered on the screening of biologically active compounds in Xenopus oocytes with electrophysiological assays; the center is equipped to perform this type of analysis at high throughput using robotic approaches. The CRB can also offer transgenesis, CRISPR/Cas9 for targeted knockouts and screening of the mutations. Many of the wild-type X. laevis and X. tropicalis used in Europe are provided by the CRB and it is closely involved with projects to improve the health of aquatic laboratory animals. Shortly the CRB will relocate to the biomedical campus in Rennes. Excitingly, this relocation should involve the development of a state-of-the-art facility for the development and housing of transgenic animals.
The X. laevis research resource for immunobiology (XLRRI3) at the University of Rochester Medical Center, is a comprehensive research resource specializing in the use of X. laevis as a multi-faceted experimental platform for immunological research. The XLRRI maintains and provides to the research community MHC-defined inbred strains and clones of frogs, as well as tools such as lymphoid tumor cell lines, MHC-defined fibroblast cell lines, monoclonal antibodies, MHC tetramers and batteries of validated PCR primers for immune-relevant genes. An important effort of the XLRRI is the development of X. laevis transgenic lines with fluorescent, traceable immune cells and specific immune deficiency, including the establishment of a reliable CRISPR/Cas9-mediated genome editing platform focused on immunity.
The XLRRI plays also plays a key role in understanding infectious diseases that plague amphibians worldwide and contribute to their decline. Notably, the XLRRI includes satellite facilities devoted to studying pathogenesis and immunity to ranaviruses and mycobacteria. Importantly, the XLRRI provides protocols, technical assistance and training to new as well as established investigators and students from a wide area of scientific disciplines from comparative, developmental and evolutionary immunology to field and conservation biology. Training and assistance cover general Xenopus breeding and husbandry, transgenesis and reverse genetics as well as Xenopus-specific in vivo and in vitro immunological methodologies.
Integration and Support on Xenbase
Xenopus research around the world is supported by Xenbase4 the Xenopus model organism database. Xenbase maintains, curates, and freely disseminates all of the diverse genomic, genetic, expression, and functional data for Xenopus and interrelates these data to human and other model organisms (Karimi et al., 2018). In addition, Xenbase allows researchers to identify Xenopus resources and reagents, including the transgenic and mutant lines (as detailed in this paper), as well as clones (ORFeome, plasmid, fosmid, and BAC) which are supplied by the EXRC.
Xenbase links to all the major Xenopus stock center websites (EXRC, NXR, XLRRI, NBRP, and CRB) from the Xenbase home page and from resource pages where appropriate. A fully searchable “Lines and Strains” module includes all of the mutant and transgenic lines and wild-type strains that are, or have been, available from NXR, EXRC and NBRP. Each Xenopus line has a dedicated page which summarizes targeted gene (s) or transgenic construct details, phenotype, genetic background and provenance, and provides direct links to the relevant stock centre (s) from which the line may be ordered. In the database, wild-type strains are also given “XB-LINE” identifiers so that they can be assigned as background to transgenic and mutant lines. Stable research identifiers (RRIDs) are recorded for all Xenopus stocks, a measure taken to promote reproducibility, rigor and transparency. Lines and strains shared between stock centers have been given unique RRIDs, as these colonies are now genetically isolated, and may be affected by genetic drift.
Importantly, Xenbase staff developed the transgenic nomenclature guidelines in consultation with the Xenopus stock centers and following best practice used by all other model organism databases. Using standard nomenclature is essential to ensure Xenopus research is accessible to the broadest possible scientific community, and improves rigor and reproducibility while also establishing provenance. These naming guidelines are posted on Xenbase5. Help naming maintained lines is always available via emailing the Xenbase help desk (xenbase@ucalgary.ca) or by contacting stock center staff.
The Xenopus ORFeome project, funded by NICHD (R01 HD069352), was developed as a molecular toolkit to probe the cellular and genetic mechanisms underlying many human diseases using Xenopus. It is another resource that is fully integrated, cataloged and searchable on Xenbase6. The ORFeome project produced two sets of full-length, validated, open reading frame clones, one for X. laevis (representing ∼ 10,250 genes, ∼7,700 with human orthologs; Grant et al., 2015), and one for X. tropicalis (representing ∼3,970 genes, ∼3,800 with human orthologs). Each ORFeome clone has a dedicated Xenbase page with details including gene symbol and name, full sequence, translation, availability, and links to the gene page and BLAST tool, and to the EXRC which supplies small numbers of ORFeome clones (James-Zorn et al., 2018). Similarly, all IMAGE Consortium Xenopus Gene Collection (XGC) clones (Morin et al., 2006) have dedicated pages with supporting sequence and other data, and these clones can be searched, filtering for only those supplied by the EXRC.
Xenbase enables efficient access to the physical resources for scientists wanting to use the Xenopus models; these resources are already extensive and growing rapidly as can be seen from the review above. As we write, there is still capacity in the resource centers to hold and develop new transgenic and knockout lines and we encourage all Xenopus users to take the opportunity to enhance their research programs by using these community facilities and taking advantage of the knowledge of their staff.
Member Of The Centre De Ressource Biologique Xenope Team In France And Team Xenopus tropicalis Nbrp In Japan
The Centre de Ressource Biologique Xenope (CRBX) team in France is Morgane Nicolas, Thomas Lafond, Daniel Boujard, Yann Audic, and Brigitte Guillet. Team Xenopus tropicalis NBRP in Japan is Hajime Ogino, Akihiko Kashiwagi, Takeshi Igawa, Keiko Kashiwagi, Nanoka Suzuki, Atsushi Suzuki, Ichiro Tazawa, Haruki Ochi, Nobuaki Furuno, Minoru Takase, Keisuke Nakajima, Hideki Hanada, Ikuo Miura, Atsushi Kurabayashi, Takashi Kato, Kei Sato, Kimiko Takebayashi-Suzuki, and Hitoshi Yoshida.
Author Contributions
All of the authors except DG wrote and reviewed the text. DG and TI produced the data shown. MW, AA-D, SM, and CJ-Z produced the table of transgenic animals.
Funding
The European Xenopus Resource Centre was funded by the Wellcome Trust (212942/Z/18/Z) and BBSRC (BB/R014841/1). The National Xenopus Resource was funded by the NIH (ORIP/NICHD) (Grant P40 OD010997). Major funding for Xenbase was provided by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (Grant P41 HD064556). XLRRI was funded by the NIH/NIAID (R24-AI-059830 and R21AI139718). CRBX was funded by GIS-IBISA 2014 and Fondation Maladies Rares 2016. The National BioResource Project (NBRP) for Xenopus tropicalis was funded by Japan Agency for Medical Research and Development (AMED).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The staff of all of the research centers are very grateful to those scientists who have donated lines, plasmids, and other reagents for the benefit of the community. The CRBX would like to acknowledge Christophe Heligon for his past involvement in the CRBX.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00387/full#supplementary-material
Footnotes
- ^ https://home.hiroshima-u.ac.jp/amphibia/xenobiores/forum/
- ^ https://xenopus.univ-rennes1.fr
- ^ https://www.urmc.rochester.edu/microbiology-immunology/xenopus-laevis.aspx
- ^ www.xenbase.org
- ^ www.xenbase.org/gene/static/tgNomenclature.jsp
- ^ www.xenbase.org/reagents/orf.do
References
Allen, B. G., and Weeks, D. L. (2005). Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat. Methods 2, 975–979. doi: 10.1038/nmeth814
Amaya, E., Musci, T. J., and Kirschner, M. W. (1991). Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in xenopus embryos. Cell 66, 257–270. doi: 10.1016/0092-8674(91)90616-7
Andres, A. C., Muellener, D. B., and Ryffel, G. U. (1984). Persistence, methylation and expression of vitellogenin gene derivatives after injection into fertilized eggs of Xenopus laevis. Nucleic Acids Res. 12, 2283–2302. doi: 10.1093/nar/12.5.2283
Aslan, Y., Tadjuidje, E., Zorn, A. M., and Cha, S.-W. (2017). High-efficiency non-mosaic CRISPR-mediated knock-in and indel mutation in F0 Xenopus. Development 144, 2852–2858. doi: 10.1242/dev.152967
Banach, M., Edholm, E. S., and Robert, J. (2017). Exploring the functions of nonclassical MHC class Ib genes in Xenopus laevis by the CRISPR/Cas9 system. Dev. Biol. 426, 261–269. doi: 10.1016/j.ydbio.2016.05.023
Baxi, A. B., Lombard-Banek, C., Moody, S. A., and Nemes, P. (2018). Proteomic characterization of the neural ectoderm fated cell clones in the Xenopus laevis embryo by high-resolution mass spectrometry. ACS Chem. Neurosci. 9,2064–2073. doi: 10.1021/acschemneuro.7b00525
Bendig, M. M., and Williams, J. G. (1984). Differential expression of the Xenopus laevis tadpole and adult beta-globin genes when injected into fertilized Xenopus laevis eggs. Mol. Cell. Biol. 4,567–570. doi: 10.1128/MCB.4.3.567
Blitz, I. L., Biesinger, J., Xie, X., and Cho, K. W. Y. (2013). Biallelic genome modification in F0 Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis 51, 827–834. doi: 10.1002/dvg.22719
Briggs, J. A., Weinreb, C., Wagner, D. E., Megason, S., Peshkin, L., Kirschner, M. W., et al. (2018). The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360:eaar5780. doi: 10.1126/science.aar5780
Brinkman, E. K., Kousholt, A. N., Harmsen, T., Leemans, C., Chen, T., Jonkers, J., et al. (2018). Easy quantification of template-directed CRISPR/Cas9 editing. Nucleic Acids Res. 46:e58. doi: 10.1093/nar/gky164
Brown, D. D., Cai, L., Das, B., Marsh-Armstrong, N., Schreiber, A. M., and Juste, R. (2005). Thyroid hormone controls multiple independent programs required for limb development in Xenopus laevis metamorphosis. Proc. Natl. Acad. Sci. 102, 12455–12458. doi: 10.1073/pnas.0505989102
Chae, J., Zimmerman, L. B., and Grainger, R. M. (2002). Inducible control of tissue-specific transgene expression in Xenopus tropicalis transgenic lines. Mech. Dev. 117, 235–241. doi: 10.1016/S0925-4773(02)00219-8
Chen, T.-W., Wardill, T. J., Sun, Y., Pulver, S. R., Renninger, S. L., Baohan, A., et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. doi: 10.1038/nature12354
Cohen, S. N., Chang, A. C. Y., Boyer, H. W., and Helling, R. B. (1973). Construction of biologically functional bacterial plasmids in vitro (R factor/restriction enzyme/transformation/endonuclease/antibiotic resistance). PNAS 70,3240–3244. doi: 10.1073/PNAS.70.11.3240
Corkins, M. E., Hanania, H. L., Krneta-Stankic, V., DeLay, B. D., Pearl, E. J., Lee, M., et al. (2018). Transgenic Xenopus laevis line for in vivo labeling of nephrons within the kidney. Genes 9:197. doi: 10.3390/genes9040197
Cross, M. K., and Powers, M. A. (2009). Learning about cancer from frogs: analysis of mitotic spindles in Xenopus egg extracts. Dis. Model. Mech. 2, 541–547. doi: 10.1242/dmm.002022
Delay, B. D., Corkins, M. E., Hanania, H. L., Salanga, M., Deng, J. M., Sudou, N., et al. (2018). Tissue-specific gene inactivation in Xenopus laevis: knockout of lhx1 in the kidney with CRISPR/Cas9. Genetics 208, 673–686. doi: 10.1534/genetics.117.300468
Deniz, E., Mis, E. K., Lane, M., and Khokha, M. K. (2018). CRISPR/Cas9 F0 screening of congenital heart disease genes in Xenopus tropicalis. Methods Mol. Biol. 1865, 163–174. doi: 10.1007/978-1-4939-8784-9_12
Etkin, L. D., and Pearman, B. (1987). Distribution, expression and germ line transmission of exogenous DNA sequences following microinjection into Xenopus laevis eggs. Development 99, 15–23.
Etkin, L. D., Pearman, B., Roberts, M., and Bektesh, S. L. (1984). Replication, integration and expression of exogenous DNA injected into fertilized eggs of Xenopus laevis. Differentiation 26, 194–202. doi: 10.1111/j.1432-0436.1984.tb01395.x
Etkin, L. D., and Roberts, M. (1983). Transmission of integrated sea urchin histone genes by nuclear transplantation in Xenopus laevis. Science 221, 67–69. doi: 10.1126/SCIENCE.6857265
Feehan, J. M., Chiu, C. N., Stanar, P., Tam, B. M., Ahmed, S. N., and Moritz, O. L. (2017). Modeling dominant and recessive forms of retinitis pigmentosa by editing three rhodopsin-encoding genes in Xenopus laevis using crispr/Cas9. Sci. Rep. 7:6920. doi: 10.1038/s41598-017-07153-4
Gentsch, G. E., Spruce, T., Monteiro, R. S., Owens, N. D. L., Martin, S. R., and Smith, J. C. (2018). Innate immune response and off-target mis-splicing are common morpholino-induced side effects in Xenopus. Dev. Cell 44,597.e10–610.e10. doi: 10.1016/j.devcel.2018.01.022
Gossen, M., and Bujard, H. (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 89, 5547–5551. doi: 10.1073/pnas.89.12.5547
Grainger, R. M. (2012). Xenopus tropicalis as a model organism for genetics and genomics: past, present, and future. Methods Mol. Biol. 917, 3–15. doi: 10.1007/978-1-61779-992-1_1
Grant, I. M., Balcha, D., Hao, T., Shen, Y., Trivedi, P., Patrushev, I., et al. (2015). The Xenopus ORFeome: a resource that enables functional genomics. Dev. Biol. 408, 345–357. doi: 10.1016/j.ydbio.2015.09.004
Gurdon, J. B., and Hopwood, N. (2000). The introduction of Xenopus laevis into developmental biology: of empire, pregnancy testing and ribosomal genes. Int. J. Dev. Biol. 44, 43–50. doi: 10.1387/ijdb.10761846
Hagmann, M., Adlkofer, K., Pfeiffer, P., Bruggmann, R., Georgiev, O., Rungger, D., et al. (1996). Dramatic changes in the ratio of homologous recombination to nonhomologous DNA-end joining in oocytes and early embryos of Xenopus laevis. Biol. Chem. Hoppe Seyler 377, 239–250. doi: 10.1515/bchm3.1996.377.4.239
Harland, R. M., and Grainger, R. M. (2011). Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 27, 507–515. doi: 10.1016/j.tig.2011.08.003
Hartley, K. O., Nutt, S. L., and Amaya, E. (2002). Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc. Natl. Acad. Sci. 99, 1377–1382. doi: 10.1073/pnas.022646899
Hassnain Waqas, S. F., Noble, A., Hoang, A. C., Ampem, G., Popp, M., Strauß, S., et al. (2017). Adipose tissue macrophages develop from bone marrow–independent progenitors in Xenopus laevis and mouse. J. Leukoc. Biol. 102, 845–855. doi: 10.1189/jlb.1A0317-082RR
Hellsten, U., Harland, R. M., Gilchrist, M. J., Hendrix, D., Jurka, J., Kapitonov, V., et al. (2010). The genome of the western clawed frog Xenopus tropicalis. Science 328, 633–636. doi: 10.1126/science.1183670
Hoogenboom, W. S., Klein Douwel, D., and Knipscheer, P. (2017). Xenopus egg extract: a powerful tool to study genome maintenance mechanisms. Dev. Biol. 428, 300–309. doi: 10.1016/j.ydbio.2017.03.033
Huang, J. K., Dorey, K., Ishibashi, S., and Amaya, E. (2007). BDNF promotes target innervation of Xenopus mandibular trigeminal axons in vivo. BMC Dev. Biol. 7:59. doi: 10.1186/1471-213X-7-59
Igawa, T., Watanabe, A., Suzuki, A., Kashiwagi, A., Kashiwagi, K., Noble, A., et al. (2015). Inbreeding ratio and genetic relationships among strains of the western clawed frog, Xenopus tropicalis. PLoS One 10:e0133963. doi: 10.1371/journal.pone.0133963
James-Zorn, C., Ponferrada, V., Fisher, M. E., Burns, K., Fortriede, J., Segerdell, E., et al. (2018). Navigating xenbase: an integrated Xenopus genomics and gene expression database. Methods Mol. Biol. 1757, 251–305. doi: 10.1007/978-1-4939-7737-6_10
Jiang, X., Kalajzic, Z., Maye, P., Braut, A., Bellizzi, J., Mina, M., et al. (2005). Histological analysis of GFP expression in murine bone. J. Histochem. Cytochem. 53, 593–602. doi: 10.1369/jhc.4A6401.2005
Karimi, K., Fortriede, J. D., Lotay, V. S., Burns, K. A., Wang, D. Z., Fisher, M. E., et al. (2018). Xenbase: a genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 46, D861–D868. doi: 10.1093/nar/gkx936
Kashiwagi, K., Kashiwagi, A., Kurabayashi, A., Hanada, H., Nakajima, K., Okada, M., et al. (2010). Xenopus tropicalisx: an ideal experimental animal in amphibia. Exp. Anim. 59, 395–405. doi: 10.1538/expanim.59.395
Kerney, R. R., Brittain, A. L., Hall, B. K., and Buchholz, D. R. (2012). Cartilage on the move: cartilage lineage tracing during tadpole metamorphosis. Dev. Growth Differ. 54, 739–752. doi: 10.1111/dgd.12002
Khokha, M. K., Chung, C., Bustamante, E. L., Gaw, L. W. K., Trott, K. A., Yeh, J., et al. (2002). Techniques and probes for the study of Xenopus tropicalis development. Dev. Dyn. 225, 499–510. doi: 10.1002/dvdy.10184
Kirmizitas, A., Meiklejohn, S., Ciau-Uitz, A., Stephenson, R., and Patient, R. (2017). Dissecting BMP signaling input into the gene regulatory networks driving specification of the blood stem cell lineage. Proc. Natl. Acad. Sci. U.S.A. 114, 5814–5821. doi: 10.1073/pnas.1610615114
Kominami, K., Takagi, C., Kurata, T., Kitayama, A., Nozaki, M., Sawasaki, T., et al. (2006). The initiator caspase, caspase-10β, and the BH-3-only molecule, Bid, demonstrate evolutionary conservation in Xenopus of their pro-apoptotic activities in the extrinsic and intrinsic pathways. Genes Cells 11, 701–717. doi: 10.1111/j.1365-2443.2006.00983.x
Kroll, K. L., and Amaya, E. (1996). Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122, 3173–3183.
Lei, Y., Guo, X., Deng, Y., Chen, Y., and Zhao, H. (2013). Generation of gene disruptions by transcription activator-like effector nucleases (TALENs) in Xenopus tropicalis embryos. Cell Biosci. 3:21. doi: 10.1186/2045-3701-3-21
Livet, J., Weissman, T. A., Kang, H., Draft, R. W., Lu, J., Bennis, R. A., et al. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62. doi: 10.1038/nature06293
Love, N. R., Chen, Y., Ishibashi, S., Kritsiligkou, P., Lea, R., Koh, Y., et al. (2013). Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat. Cell Biol. 15, 222–228. doi: 10.1038/ncb2659
McQueen, C., and Pownall, M. E. (2017). An analysis of MyoD-dependent transcription using CRISPR/Cas9 gene targeting in Xenopus tropicalis embryos. Mech. Dev. 146, 1–9. doi: 10.1016/j.mod.2017.05.002
Miyamoto, K., Suzuki, K. T., Suzuki, M., Sakane, Y., Sakuma, T., Herberg, S., et al. (2015). The expression of TALEN before fertilization provides a rapid knock-out phenotype in Xenopus laevis founder embryos. PLoS One 10:e0142946. doi: 10.1371/journal.pone.0142946
Morin, R. D., Chang, E., Petrescu, A., Liao, N., Griffith, M., Chow, W., et al. (2006). Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 16, 796–803. doi: 10.1101/gr.4871006
Naert, T., Colpaert, R., Van Nieuwenhuysen, T., Dimitrakopoulou, D., Leoen, J., Haustraete, J., et al. (2016). CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci. Rep. 6:35264. doi: 10.1038/srep35264
Naert, T., and Vleminckx, K. (2018a). CRISPR/Cas9 disease models in zebrafish and Xenopus: the genetic renaissance of fish and frogs. Drug Discov. Today Technol. 28, 41–52. doi: 10.1016/j.ddtec.2018.07.001
Naert, T., and Vleminckx, K. (2018b). Genotyping of CRISPR/Cas9 genome edited Xenopus tropicalis. Methods Mol. Biol. 1865, 67–82. doi: 10.1007/978-1-4939-8784-9_5
Nakajima, K., Nakajima, T., Takase, M., and Yaoita, Y. (2012). Generation of albino Xenopus tropicalis using zinc-finger nucleases. Dev. Growth Differ. 54, 777–784. doi: 10.1111/dgd.12006
Nakayama, T., Fish, M. B., Fisher, M., Oomen-Hajagos, J., Thomsen, G. H., and Grainger, R. M. (2013). Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis 51, 835–843. doi: 10.1002/dvg.22720
Nieuwkoop, P. D., and Faber, J. (1994). Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development From the Fertilized Egg Till the End of Metamorphosis. Spokane, WA: Garland Pub & Grill.
Ogino, H., McConnell, W. B., and Grainger, R. M. (2006). High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat. Protoc. 1, 1703–1710. doi: 10.1038/nprot.2006.208
Pan, F. C., Chen, Y., Loeber, J., Henningfeld, K., and Pieler, T. (2006). I-SceI meganuclease-mediated transgenesis in Xenopus. Dev. Dyn. 235, 247–252. doi: 10.1002/dvdy.20608
Pearl, E., Morrow, S., Noble, A., Lerebours, A., Horb, M., and Guille, M. (2017). An optimized method for cryogenic storage of Xenopus sperm to maximise the effectiveness of research using genetically altered frogs. Theriogenology 92, 149–155. doi: 10.1016/j.theriogenology.2017.01.007
Pearl, E. J., Grainger, R. M., Guille, M., and Horb, M. E. (2012). Development of Xenopus resource centers: the national Xenopus resource and the European Xenopus resource center. Genesis 50, 155–163. doi: 10.1002/dvg.22013
Rankin, S. A., Hasebe, T., Zorn, A. M., and Buchholz, D. R. (2009). Improved Cre reporter transgenic Xenopus. Dev. Dyn. 238, 2401–2408. doi: 10.1002/dvdy.22043
Ratzan, W., Falco, R., Salanga, C., Salanga, M., and Horb, M. E. (2017). Generation of a Xenopus laevis F1 albino J strain by genome editing and oocyte host-transfer. Dev. Biol. 426, 188–193. doi: 10.1016/j.ydbio.2016.03.006
Roose, M., Sauert, K., Turan, G., Solomentsew, N., Werdien, D., Pramanik, K., et al. (2009). Heat-shock inducible cre strains to study organogenesis in transgenic Xenopus laevis. Transgenic Res. 18, 595–605. doi: 10.1007/s11248-009-9253-4
Rusconi, S., and Schaffner, W. (1981). Transformation of frog embryos with a rabbit beta-globin gene. Proc. Natl. Acad. Sci. U.S.A. 78, 5051–5055. doi: 10.1073/pnas.78.8.5051
Sakane, Y., Iida, M., Hasebe, T., Fujii, S., Buchholz, D. R., Ishizuya-Oka, A., et al. (2018). Functional analysis of thyroid hormone receptor beta in Xenopus tropicalis founders using CRISPR-Cas. Biol. Open 7:bio030338. doi: 10.1242/bio.030338
Sato, Y., Mukai, M., Ueda, J., Muraki, M., Stasevich, T. J., Horikoshi, N., et al. (2013). Genetically encoded system to track histone modification in vivo. Sci. Rep. 3:2436. doi: 10.1038/srep02436
Sega, A. G., Mis, E. K., Lindstrom, K., Mercimek-Andrews, S., Ji, W., Cho, M. T., et al. (2018). De novo pathogenic variants in neuronal differentiation factor 2 (NEUROD2) cause a form of early infantile epileptic encephalopathy. J. Med. Genet. 56, 113–122. doi: 10.1136/jmedgenet-2018-105322
Session, A. M., Uno, Y., Kwon, T., Chapman, J. A., Toyoda, A., Takahashi, S., et al. (2016). Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343. doi: 10.1038/nature19840
Shindo, A., Wallingford, J. B., Dale, L., and Slack, J. M. (1987). Fate map for the 32-cell stage of Xenopus laevis. Development 99, 527–551.
Suzuki, M., Takagi, C., Miura, S., Sakane, Y., Suzuki, M., Sakuma, T., et al. (2016). In vivo tracking of histone H3 lysine 9 acetylation in Xenopus laevis during tail regeneration. Genes Cells 21, 358–369. doi: 10.1111/gtc.12349
Swenson, E. S., Price, J. G., Brazelton, T., and Krause, D. S. (2007). Limitations of green fluorescent protein as a cell lineage marker. Stem Cells 25, 2593–2600. doi: 10.1634/stemcells.2007-0241
Takagi, C., Sakamaki, K., Morita, H., Hara, Y., Suzuki, M., Kinoshita, N., et al. (2013). Transgenic Xenopus laevis for live imaging in cell and developmental biology. Dev. Growth Differ. 55, 422–433. doi: 10.1111/dgd.12042
Tandon, P., Conlon, F., Furlow, J. D., and Horb, M. E. (2017). Expanding the genetic toolkit in Xenopus: approaches and opportunities for human disease modeling. Dev. Biol. 426, 325–335. doi: 10.1016/j.ydbio.2016.04.009
Tinsley, R. C., and Kobel, H. R. (1996). The Biology of Xenopus. London: Zoological Society of London.
Tran, H. T., Sekkali, B., Van Imschoot, G., Janssens, S., and Vleminckx, K. (2010). Wnt/β-catenin signaling is involved in the induction and maintenance of primitive hematopoiesis in the vertebrate embryo. Proc. Natl. Acad. Sci. 107, 16160–16165. doi: 10.1073/pnas.1007725107
Tymowska, J., and Fischberg, M. (1973). Chromosome complements of the genus Xenopus. Chromosoma 44, 335–342. doi: 10.1007/BF00291027
Waldner, C., Sakamaki, K., Ueno, N., Turan, G., and Ryffel, G. U. (2006). Transgenic Xenopus laevis strain expressing Cre recombinase in muscle cells. Dev. Dyn. 235, 2220–2228. doi: 10.1002/dvdy.20880
Wlizla, M., Falco, R., Peshkin, L., Parlow, A. F., and Horb, M. E. (2017). Luteinizing Hormone is an effective replacement for hCG to induce ovulation in Xenopus. Dev. Biol. 426, 442–448. doi: 10.1016/j.ydbio.2016.05.028
Yergeau, D. A., Kelley, C. M., Zhu, H., Kuliyev, E., and Mead, P. E. (2010). Transposon transgenesis in Xenopus. Methods 51, 92–100. doi: 10.1016/j.ymeth.2010.03.001
Yergeau, D. A., Kuliyev, E., and Mead, P. E. (2007). Injection-mediated transposon transgenesis in Xenopus tropicalis and the identification of integration sites by modified extension primer tag selection (Epts) linker-mediated pcr. Nat. Protoc. 2, 2975–2986. doi: 10.1038/nprot.2007.428
Yergeau, D. A., and Mead, P. E. (2009). Transposon-mediated transgenesis in the frog: new tools for biomedical and developmental studies. Front. Biosci. 14, 225–236. doi: 10.2741/3242
Keywords: Xenopus laevis, Xenopus tropicalis, transgenesis, gene editing, inbred strains, ORFeome, model organism database, resource centers
Citation: Horb M, Wlizla M, Abu-Daya A, McNamara S, Gajdasik D, Igawa T, Suzuki A, Ogino H, Noble A, Centre de Ressource Biologique Xenope Team in France, Robert J, James-Zorn C and Guille M (2019) Xenopus Resources: Transgenic, Inbred and Mutant Animals, Training Opportunities, and Web- Based Support. Front. Physiol. 10:387. doi: 10.3389/fphys.2019.00387
Received: 31 January 2019; Accepted: 21 March 2019;
Published: 25 April 2019.
Edited by:
Emily Sempou, Yale University, United StatesReviewed by:
Caroline Beck, University of Otago, New ZealandAnna Philpott, University of Cambridge, United Kingdom
Copyright © 2019 Horb, Wlizla, Abu-Daya, McNamara, Gajdasik, Igawa, Suzuki, Ogino, Noble, Centre de Ressource Biologique Xenope Team in France, Robert, James-Zorn and Guille. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew Guille, bWF0dGhldy5ndWlsbGVAcG9ydC5hYy51aw==
†On behalf of Team Xenopus tropicalis NBRP in Japan
 Marko Horb
Marko Horb Marcin Wlizla
Marcin Wlizla Anita Abu-Daya
Anita Abu-Daya Sean McNamara
Sean McNamara Dominika Gajdasik3
Dominika Gajdasik3 Anna Noble
Anna Noble Jacques Robert
Jacques Robert Christina James-Zorn
Christina James-Zorn Matthew Guille
Matthew Guille