- 1Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
As the main structural protein of muscle fiber, myosin is essential for multiple cellular processes or functions, especially for muscle composition and development. Although the shrimp possess a well-developed muscular system, the knowledge about the myosin family in shrimp is far from understood. In this study, we performed comprehensive analysis on the myosin genes in the genome of the Pacific white shrimp, Litopenaeus vannamei. A total of 29 myosin genes were identified, which were classified into 14 subfamilies. Among them, Myo2 subfamily was significantly expanded in the penaeid shrimp genome. Most of the Myo2 subfamily genes were primarily expressed in abdominal muscle, which suggested that Myo2 subfamily genes might be responsible for the well-developed muscular system of the penaeid shrimp. In situ hybridization detection showed that the slow-type muscle myosin gene was mainly localized in pleopod muscle and superficial ventral muscle of the shrimp. This study provides valuable insights into the evolutionary and functional characterization of myosin genes in shrimps, which provides clues for us to understand the well-developed muscular system of shrimp.
Introduction
Nearly all eukaryotic cells possess myosin proteins, which bind to filamentous actin and produce physical forces through ATP hydrolysis (Richards and Cavalier-Smith, 2005; Sebé-Pedrós et al., 2014). Myosin plays key roles in muscle composition and various cellular activities, including cytokinesis, organelle transport, cell polarization, intracellular transport and signal transduction (Hofmann et al., 2009; Bloemink and Geeves, 2011; Hartman et al., 2011; Pette and Staron, 2015). The Myo2 subfamily (Myosin 2), also known as myosin heavy chain (MYH or MHC), are the main component of the contractile muscle. They are considered to be conventional myosins, while the other subfamilies are considered to be unconventional myosins. The Myo2 subfamily genes can be classified into three groups, including fast-type, slow-type and non-muscle type. The Myo2 subfamily proteins can form large bipolar filaments through tail-directed homo-oligomerization, while the tails of the unconventional myosins typically direct binding to membrane and other proteins (Woolner and Bement, 2009).
Myosin consists of three domains, including a conserved motor (or head) domain with actin-binding activities, a short neck that serves as a binding site for myosin light chains, and a variable tail that generally mediates interaction with the motor “cargo” to determine the functional specificity of the motor (Berg et al., 2001; Richards and Cavalier-Smith, 2005). In general, it is very difficult to investigate the myosin gene family systemically due to their extensive alternative splicing events, large sizes and high copy numbers. Although crustaceans are a large, diverse group, limited myosin genes have been reported, except that 17 myosin genes in 13 classes have been characterized in Daphnia duplex (Odronitz and Kollmar, 2008). In addition, a few Myo2 sequences have been cloned from Marsupenaeus japonicus, Penaeus monodon, and Litopenaeus vannamei, respectively (Koyama et al., 2013). Hence, studies on the myosin gene family in crustaceans will enhance our understanding on their structure, function and evolution.
The Pacific white shrimp L. vannamei is one of the most economically important marine aquaculture species in the world (FAO, 2014). Based on the whole genome sequences (Zhang et al., 2019) and the transcriptome data, we performed a genome-wide analysis on the myosin gene family of L. vannamei. This study provides valuable resources for the shrimp myosin genes, which will increase our understanding on the crustacean muscular system.
Materials and Methods
Genome and Transcriptome Data
The genomic data of L. vannamei (PRJNA438564), Lepeophtheirus salmonis (GCA_001005205.1), Parhyale hawaiensis (GCA_001587735.1), Eulimnadia texana (GCA_002872375.1), were obtained from NCBI GenBank and Ensemble database. Our previous research conducted RNA-seq on several libraries, (I) five larval stages, including embryo, nauplius, zoea, mysis and post-larvae (Wei et al., 2014); (II) eight molting stages, including the inter-molt (C), pre-molt (D0, D1, D2, D3, D4), and post-molt (P1 and P2) stages (Gao et al., 2015); (III) 16 adult tissues, including antennal gland, brain, hemocyte, epidermis, eyestalk, gill, hepatopancreas, heart, intestine, abdominal muscle, lymphoid organ, ovary, stomach, testis, thoracic ganglion, and abdominal ganglion (Zhang et al., 2018).
Isolation and Annotation of Myosin Genes
The full protein sequences of the L. vannamei genome were searched against the myosin head (motor domain) (PF00063) to find all the candidate myosin genes by using HMMER3.0 (Potter et al., 2018). All possible myosin transcripts were collected from the transcriptome data. Then, the redundant sequences were removed using CAP3 program (Huang and Madan, 1999). The candidate sequences were submitted to SMART (Letunic et al., 2014) and InterPro (Finn et al., 2016) databases to determine the integrity of the motor domain. More myosin sequences used in this study were collected from CyMoBase1 and (Hammesfahr et al., 2010).
Gene Structure and Alternative Splicing
The characteristics of myosin genes, including the location, gene length, open reading frame (ORF), exon number and the number of deduced amino acids were analyzed in detail. To illustrate the structure of myosin genes, all alternative spliced exons were characterized by mapping all variant transcripts to the shrimp genome.
Phylogenetic Analysis
The amino acid sequences of the conserved myosin head domain were aligned by MUSCLE program (version 3.8.31) (Edgar, 2004) with default parameters. A neighbor-joining (NJ) phylogenetic tree with 1000 bootstrap replicates was constructed by MEGA7.0 program (Kumar et al., 2016), and visualized using iTOL (Letunic and Bork, 2007). To investigate the evolution of the myosin gene family of shrimp, a class occurrence tree of 29 arthropods was generated by using the method described by Odronitz et al. (2009).
Transcription Regulatory Element Identification
To investigate whether the expanded muscle-type Myo2 genes are associated with muscle composition or development, transcription regulatory elements of muscle-type Myo2 genes were predicted. In detail, the promoter regions, located at the 2 kb upstream of the transcriptional start site, were first extracted from the shrimp genome. The transcription regulatory elements were then predicted by Signal Scan2 with TRANSFAC database and the PATCH algorithm integrated in the GeneXplain platform3. In order to decrease the false positive rate, we intersected the predicted results of Signal Scan and GeneXplain.
Expression Patterns at Early Development Stages and Different Tissues of Adults
In previous studies, we conducted Digital Gene Expression Profiling (DGE) to sequence 20 larval stages and 16 adult tissues of L. vannamei, and the RPKM (reads per kilobases per million reads) values of all transcripts were calculated (Zhang et al., 2018). The RPKM values of myosin genes were extracted and normalized with log2 conversion. Heat maps were created using TBtools software (Chen et al., 2018).
In situ Hybridization
In this study, LvMYH5 was characterized to be a slow-type muscle Myo2 gene. To further distinguish fiber types and muscle distribution of the abdominal muscle of shrimp, in situ hybridization was performed according to the protocol developed in our laboratory (Zhang et al., 2018). Briefly, primers LvMYH5-pF with T7 promoter sequence and LvMYH5-R (Supplementary Table 1) were designed to amplify the cDNA fragments as the template of LvMYH5 to synthesize sense probe. Primers LvMYH5-pR with T7 promoter sequence and LvMYH5-F were designed as the template to synthesize antisense probe. PCR products were purified using MiniBEST DNA Fragment Purification Kit (Takara, Japan). The Digoxigenin (DIG)-labeled sense and antisense RNA probes were transcribed by TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific, United States) and synthesized using DIG RNA Labeling Mixture (Roche, Germany), respectively, and stored at -80°C.
Transverse sections with 5–7 μm thickness were prepared from polyformaldehyde fixed abdominal muscle of juvenile shrimp (about 2 cm in body length) embedded in paraffin (Sigma, Germany), and hybridization was performed following the general protocol of DIG RNA labeling kit (SP6/T7) (Roche, Germany). The final concentration of both anti-sense and sense RNA probes were 1.5 ng/μl. Subsequently, alkaline phosphatase-conjugated anti-DIG antibody and NBT/BCIP (DIG Nucleic Acid Detection Kit, Roche) were used to detect RNA probes and observed under Nikon Eclipse 80i microscope (Nikon, Japan).
Results
Identification of Myosin Genes
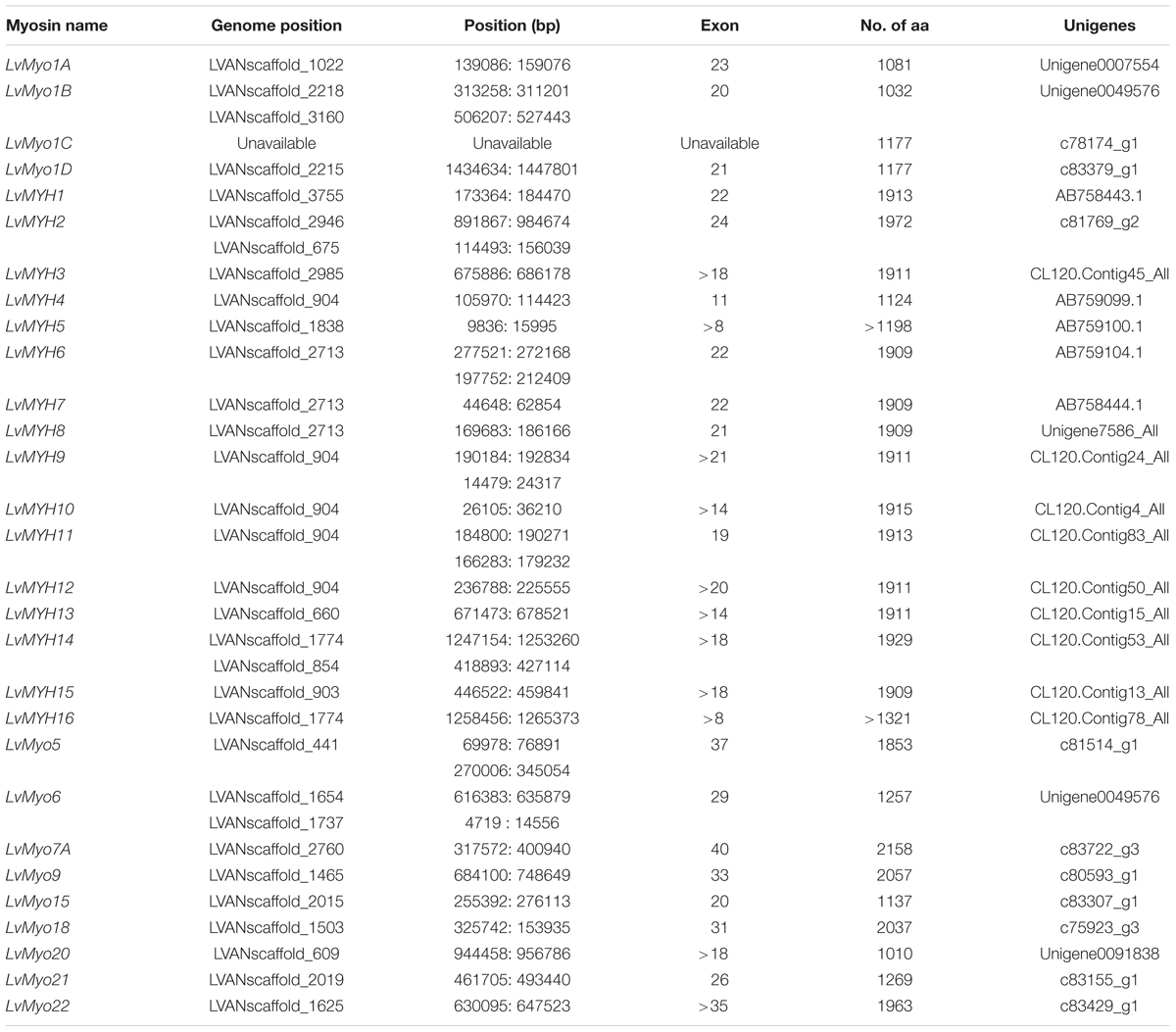
A total of 29 myosin genes were identified in L. vannamei, including 15 muscle-type Myo2 genes, one non-muscle type Myo2 gene and 13 unconventional myosin genes (Table 1). Among them, five muscle-type myosin genes had been reported previously, including LvMHC1, LvMHC2, LvMHC4, LvMHC5, and LvMHC6 (Koyama et al., 2012, 2013). They were designated as LvMYH1, LvMYH7, LvMYH4, LvMYH5, and LvMYH6, respectively, in this study to avoid confusion with major histocompatibility complex genes. The gene structure of these 29 myosin genes showed high complexity with the sizes ranging from 6.159 to 171.807 kb that corresponded to encode proteins of 1,010-2,158 aa (Table 1). Phylogenetic analysis indicated that these myosin genes were clustered into 14 distinct groups, namely 14 subfamilies, Myo1, muscle-type Myo2, non-muscle type Myo2, Myo3, Myo5, Myo6, Myo7, Myo9, Myo15, Myo18, Myo19, Myo20, Myo21, and Myo22. Myo1 can be further divided into four subfamilies, Myo1A, Myo1B, Myo1C, and Myo1D. These myosin genes within each subfamily shared similar protein domains. Furthermore, Myo1 and the muscle-type Myo2 were the two largest clades including 22 and 40 myosin genes in the phylogenetic tree of this study, respectively. However, the smallest clade Myo19 only included 4 myosin genes. All the 29 myosin genes identified in this study fell into 14 distinct clades and were closely related to the homologous proteins from other species (Figure 1). Of these genes, 15 muscle-type Myo2 genes were monophyletic with 27 muscle-type Myo2 genes from other arthropods.
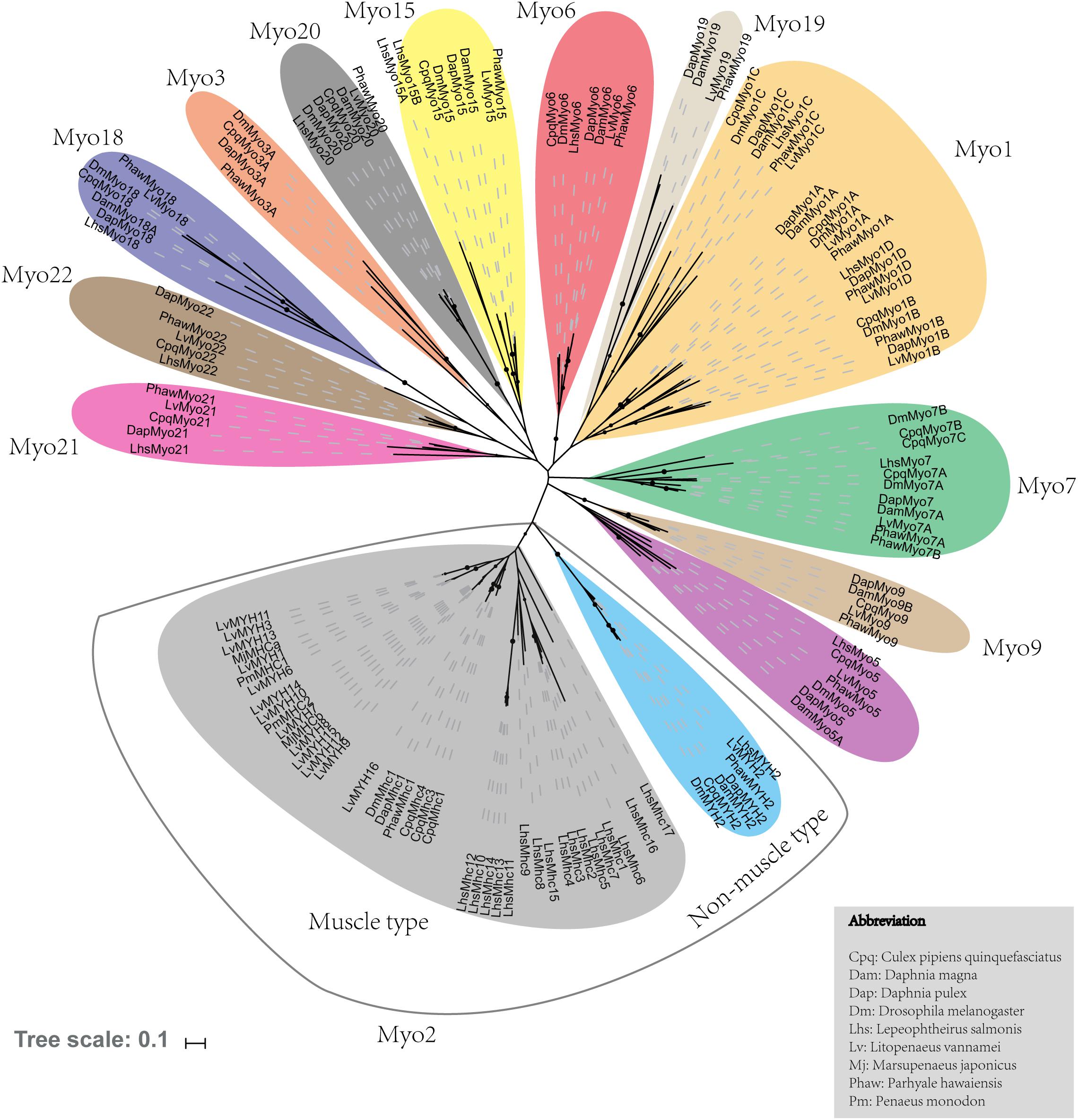
Figure 1. Phylogenetic tree of myosin head domains generated using the neighbor-joining method (MEGA7 software) with bootstrapping replication set at 1000. The myosin groups are identified in roman numerals beside each clade. Myosin genes from the following organisms were used: Cpq, Culex pipiens quinquefasciatus; Dam, Daphnia magna; Dm, Drosophila melanogaster; Dap, Daphnia duplex; Lhs, Lepeophtheirus salmonis; Lv, Litopenaeus. vannamei; Mj, Marsupenaeus japonicus; Phaw, Parhyale hawaiensis; Pm, Penaeus monodon.
Fifteen muscle-type Myo2 genes exhibited conspicuous lineage-specific expansion in the shrimp genome. Among them, eight Myo2 genes were tandemly distributed in two clusters (Figure 2), LvMYH9, LvMYH10, LvMYH4, LvMYH11, and LvMYH12 were located on the Scaffold 904, LvMYH7, LvMYH8, and LvMYH6 were located on the Scaffold 2713. Besides, at least 19 fragmental muscle-type Myo2-like sequences or pseudogenes were located on the Scaffold 903, Scaffold 2713 and Scaffold 3755 (Figure 2). The others scattered in the shrimp genome (Table 1).
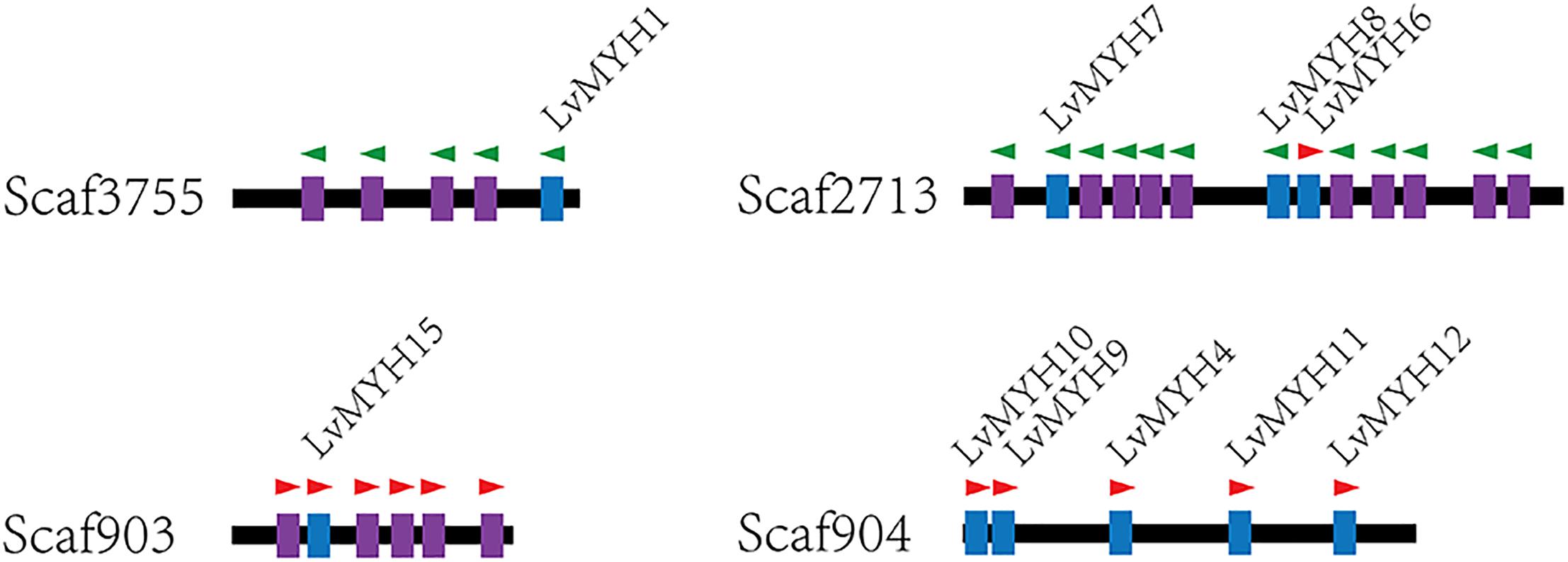
Figure 2. Representation of paralogous muscle-type Myo2 gene clusters arranged on different scaffolds of L. vannamei. Blue: the complete muscle-type MYH gene. Purple: fragmental Myo2-like gene sequences or pseudogenes of MYH. The green and red triangles represent transcription orientation.
Myosin Functional Domain
Generally, homologous myosin genes of different species possess similar domain architecture (Richards and Cavalier-Smith, 2005). All myosin genes identified in this study contained a large ATPase motor domain which hydrolyzes ATP, except for LvMYH4, which encodes a headless myosin. In addition, all the myosin genes contained at least one isoleucine-glutamine (IQ) motif except for LvMyo6 (Figure 3). The myosin genes could be classified into different groups with diverse functions based on the remaining domains and variable number of IQ motifs. LvMyo5 contained six IQ domains and a unique C-terminal DIL domain. LvMyo19, LvMyo20, and LvMyo21 had truncated tail regions. Notably, all muscle-type Myo2 proteins contained an N-terminal SH3-like fold, a large ATPase motor domain, an IQ motif, and a large C-terminal myosin tail. However, the N-terminal SH3-like domain was absent in unconventional myosin genes of L. vannamei. Compared with DamMyo18 and DapMyo18, the upstream PDZ domain was absent in PhawMyo18 and LvMyo18.
Figure 3. Schematic diagram of shrimp unconventional myosin structures. The putative domains or motifs were identified using the Pfam and SMART databases with the default parameters. MYSc, Myosin head, large ATPase; IQ, isoleucine-glutamine motif; DIL, dilute; CBD, cargo binding domain; MyTH4, myosin tail homology 4; B41, band 4.1, ezrin, radixin, and moesin; SH3, src homology 3; RA, Ras association (RalGDS/AF-6) domain; C1, Protein kinase C conserved region 1; RhoGAP, Rho GTPase-activating protein; S_TKC, Serine/Threonine protein kinases, catalytic domain. Myosin_TH1, myosin tail homology 1.
Phylogenetic Analysis
As expected, the crustacea formed a distinct clade from other arthropods, and L. vannamei was most closely related to P. hawaiensis belonging to Malacostraca clades (Figure 4). In addition, arthropods contained 14 classes (groups) of myosin genes including muscle-type Myo2, non-muscle Myo2 (MYH2), Myo5, Myo6, Myo7A, Myo15, Myo18, Myo20, and Myo21 with a few exceptions. For example, non-muscle type Myo2, Myo6, and Myo15 were absent in Ixodes scapularis, and Myo18 and Myo21 were not found in Tetranychus urticae and Strigamia maritima, respectively. Most of these myosin genes were present in crustaceans, so we inferred that the last common crustacean ancestor had already contained these classes of myosin genes.
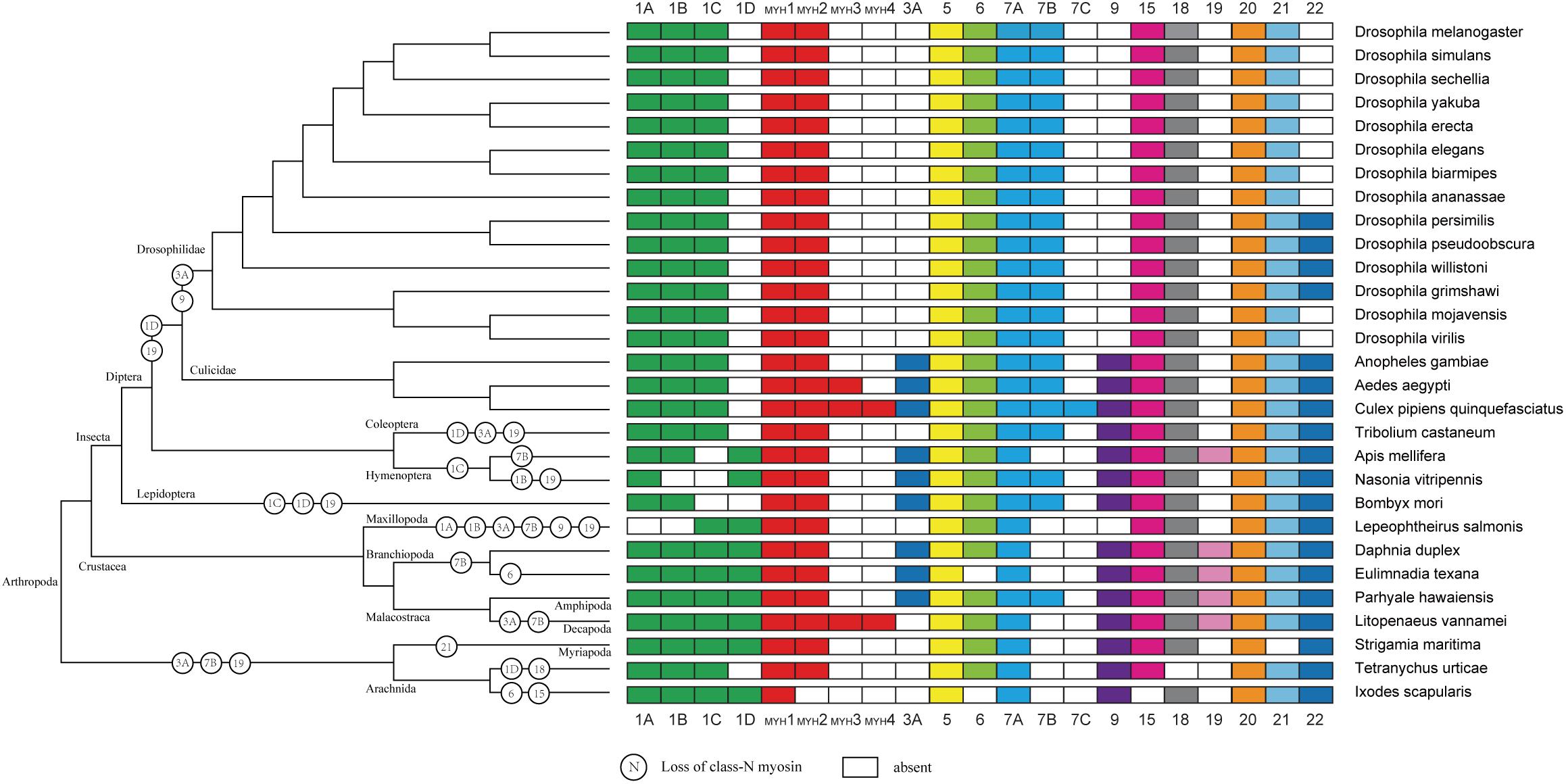
Figure 4. Myosin gain and loss in arthropod species. Tree on the left illustrates the phylogenetic relationships of species used (no scale). Colored boxes indicate the presence of certain myosin subfamily members in each species. Empty boxes mean the potential loss of particular myosin genes.
Alternative Splicing
Among the 29 myosin genes, four genes underwent alternative splicing (Figure 5 and Supplementary Table 2). The unspliced transcript of LvMYH2 encoded 1,972 aa protein, and the corresponding spliced transcripts encoded 1,982 and 1,970 aa when the exon 20 was retained and exon 4 was mutually excluded, respectively. LvMyo5 possessed at least three splicing events, the unspliced transcript encoded 1,853 aa and the corresponding spliced transcript encoded 1,833, 1,804 and 1,846 aa with the exon 7, 24 and 28 were skipped, respectively. In the case of LvMyo6, an exon skipping event occurred in exon 22, and the corresponding spliced transcript encoded 1,238 aa, whereas the unspliced transcript encoded 1,257 aa. As for LvMyo18, the unspliced transcript generated 2,037 aa in length. Exon 8 of LvMyo18-A1 transcript was retained, and the corresponding spliced transcript encoded 2,061 aa. Alternatively, a novel 660 bp intron between exon 28 and exon 29 of LvMyo18-A2 transcript was retained. Furthermore, an alternative acceptor site was found in exon 29 of LvMyo18-A3 transcript, and the corresponding spliced transcript encoded 2,066 aa.
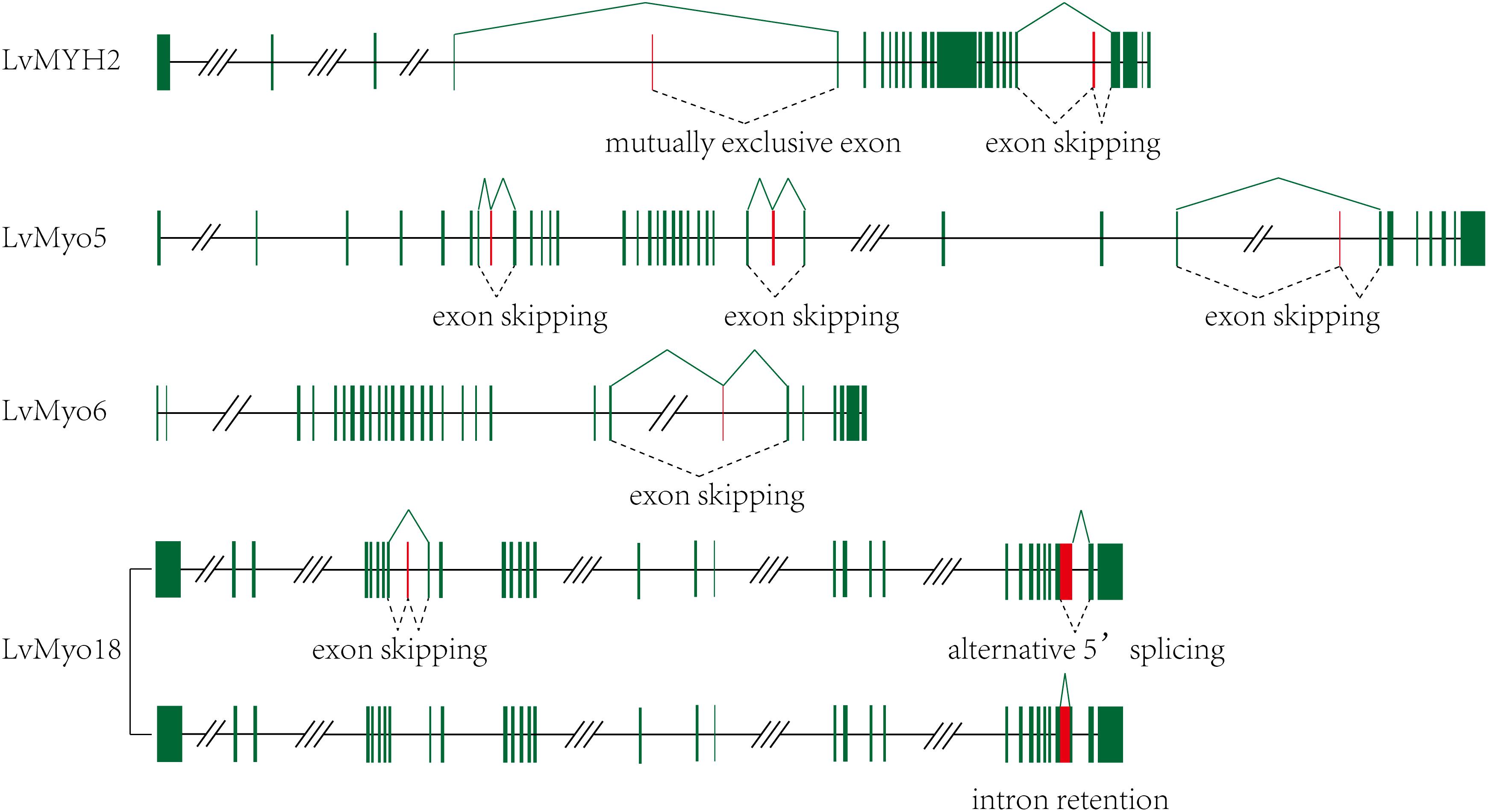
Figure 5. Transcript evidences and splicing patterns for unconventional myosin genes in L. vannamei. Green boxes: exons; red boxes: alternative splicing exons; line, introns. Three-slashes represent large intron, and two-slashes represent genomic gap.
Transcription Regulatory Elements of the Muscle-Type Myo2 Promoters
A large number of muscle development-related transcription factors binding to the upstream promoter sequences of 10 LvMYHs were identified, including TATA-binding protein (TBP), GATA-1, activator protein 1 (AP1), specificity protein 1 (Sp1), serum response factor (SRF), myogenic differentiation 1 (MyoD) and myogenin (Figure 6). LvMYH1, LvMYH2, LvMYH3, LvMYH6, LvMYH8, LvMYH9, LvMYH11, LvMYH12, LvMYH14, and LvMYH15 possessed 11, 5, 8, 13, 12, 11, 14, 18, 14, and 7 binding sites of muscle-related transcription factors, respectively. Among them, LvMYH12 contained the highest number of binding sites of five muscle-related transcription factors. In addition, all analyzed LvMYH genes possessed at least one binding site of MyoD, except for LvMYH2 and LvMYH15. The LvMYH3, LvMYH9, LvMYH11, and LvMYH14, might be activated by myogenin (Figure 6).
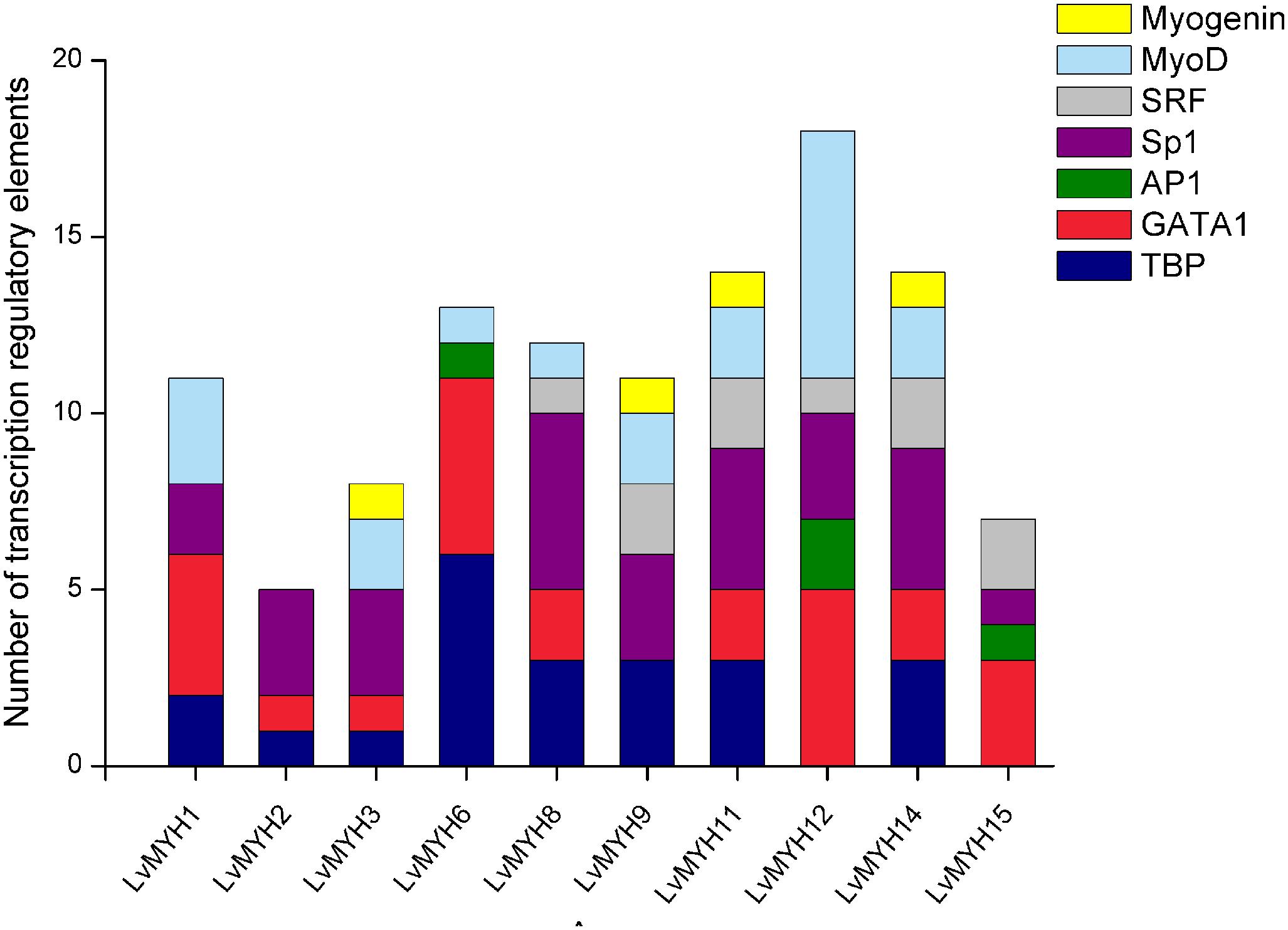
Figure 6. Transcription regulatory elements in the promoters of several muscle-type Myo2 genes that are related to muscle development. SRF, serum response factor; Sp1, specificity protein 1; AP1, activator protein 1; TBP, TATA-binding protein.
Temporal and Spatial Expression Patterns of Myosin Genes
We analyzed the temporal and spatial expression patterns of myosin genes based on the transcriptome of tissues or development stages (Figure 7). The results revealed that seven Myo2 subfamily genes, LvMYH3, LvMYH6, LvMYH8, LvMYH9, LvMYH10, LvMYH13, and LvMYH15, were specifically highly expressed in the abdominal muscle. LvMYH12 was exclusively expressed in stomach, and LvMYH16 was particularly abundant in heart and epidermis. The expression levels of LvMYH11 and LvMYH14 were very low in all tissues. Similarly, the unconventional myosin genes also showed tissue-specific expression patterns; for instance, LvMyo5 was specifically expressed in hemocyte, and LvMyo21 exhibited high expression level in eyestalk.
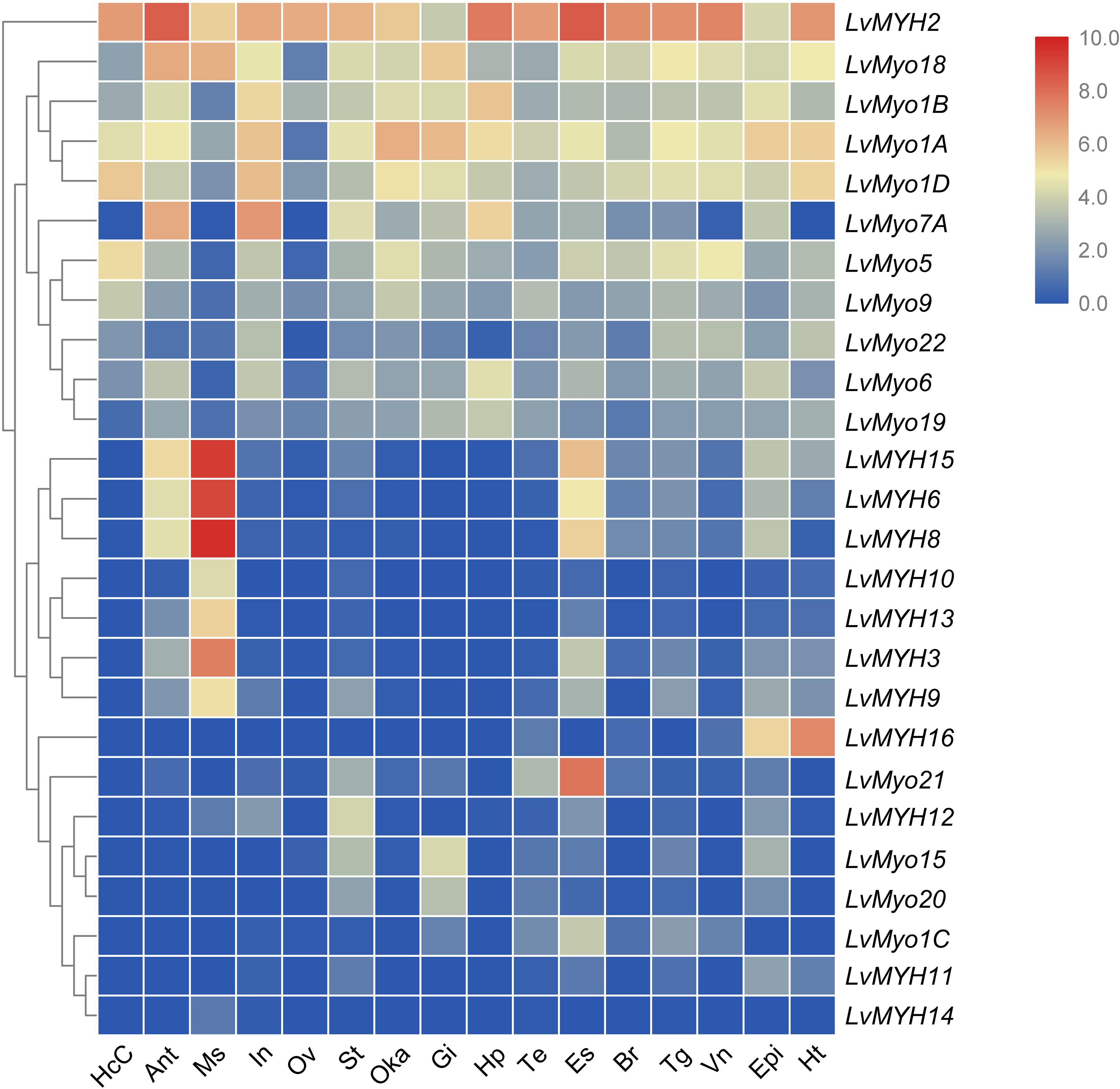
Figure 7. The expression profiles of myosin genes in 16 adult tissues. Log2-transformed expression values were used to create the heat map. The red or blue colors represent the higher or lower relative abundance of each myosin gene, respectively. To the left are myosin genes; the top lists the adult tissues of L. vannamei. (HcC, hemocyte cells; Ant, antennary gland; Ms, abdominal muscle; In, intestine; Ov, ovary; St, stomach; Oka, lymphoid organ; Gi, gill; Hp, hepatopancreas; Te, testis; Es, eyestalk; Br, brain; Tg, thoracic ganglion; Vn, ventral nerve; Epi, epidermis; Ht, heart).
All myosin genes were detectable with dynamic expression patterns during the whole developmental stage. Most of muscle-type Myo2 genes were highly expressed between the larvae in membrane (Lim) stage and the post-larva stage (P1) (Figure 8). For instance, LvMYH2, LvMYH3, and LvMYH10 were expressed from Lim stage to nauplius 6 (N6) stage, and LvMYH6, LvMYH8, LvMYH9, and LvMYH15 were highly expressed from zoea 1 (Z1) to P1 stage. However, unconventional myosin genes exhibited a distinct pattern, LvMyo1A, LvMyo1B, LvMyo1D, LvMyo9, LvMyo15, and LvMyo19 were mainly expressed from zygote to gastrula stage, and LvMyo7A and LvMyo20 were expressed from limb bud embryo 1 (Lbe1) stage to N6 stage.
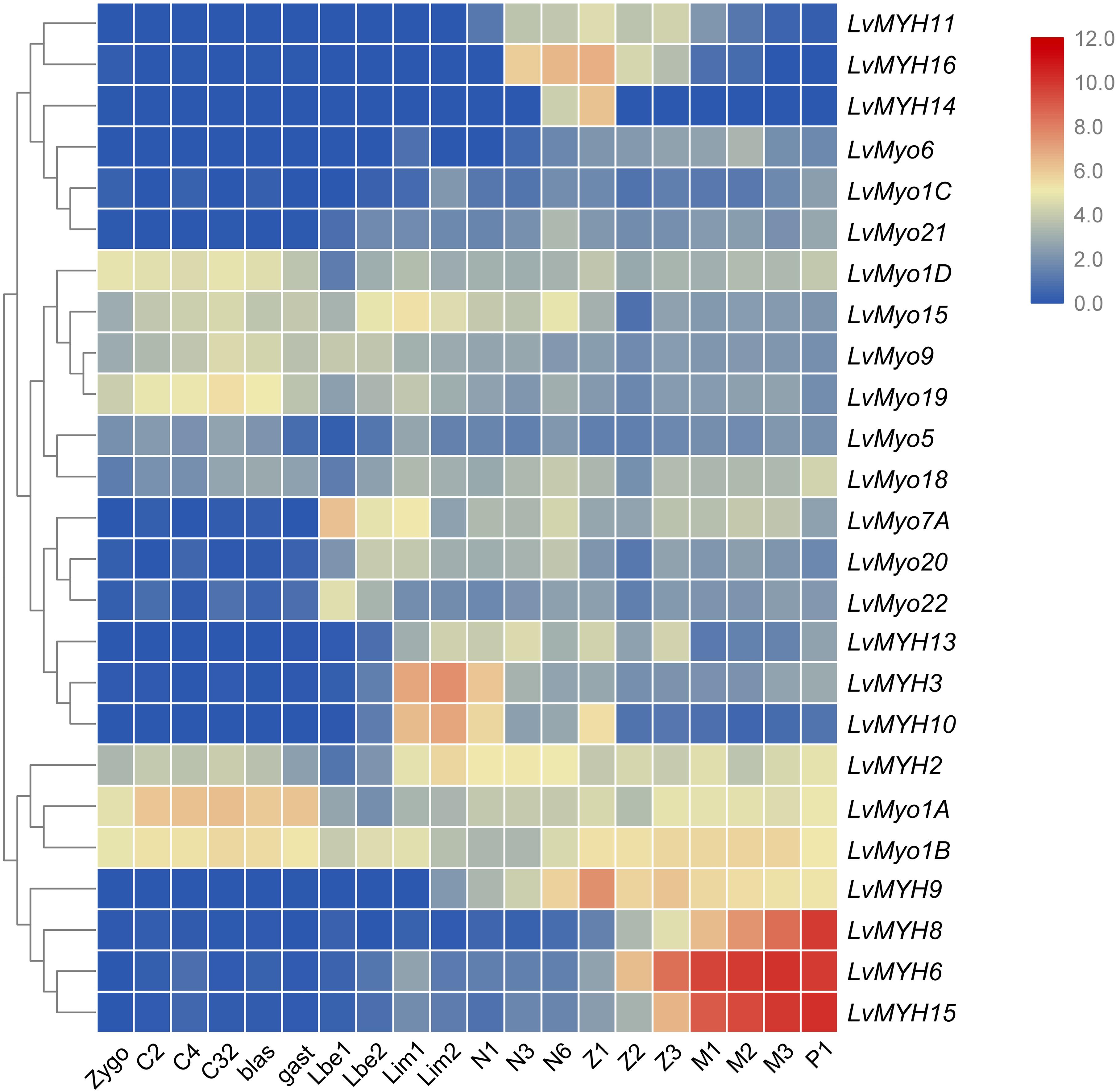
Figure 8. The expression profiles of myosin genes during development of L. vannamei. Log2-transformed expression values were used to create the heat map. The red or blue colors represent the higher or lower relative abundance of each myosin gene, repectively. On the right are myosin genes; the bottom lists different larval stages of L. vannamei, from left to right: zygote, 2 cells (C2), 4 cells (C4), 32 cells (C32), blastula (blas), gastrula (gast), limb bud embryo 1 (Lbe1), limb bud embryo 2 (Lbe2), larva in membrane 1 (Lim1), larva in membrane 2 (Lim2), Nauplius 1 (N1), Nauplius 2 (N2), Nauplius 3 (N3), Nauplius 6 (N6), Zoea 1 (Z1), Zoea 2 (Z2), Zoea 3 (Z3), Mysis 1 (M1), Mysis 2 (M2), Mysis 3 (M3), and Post-larva 1 (P1).
In situ Hybridization
Shrimp abdominal muscle could be divided into four functional muscle groups, the superficial ventral muscle, the lateral muscle, the dorsal muscle and the main ventral muscle. The morphological organization of the skeletal muscle was displayed by the hematoxylin-eosin (H&E) staining (Figure 9A). In situ hybridization detection showed that LvMYH5, characterized as a marker gene of slow-type skeletal muscle Myo2, was mainly expressed in pleopod muscle and superficial ventral muscle (Figure 9C). They were attached between yokes of thin cuticle lying transversely across the posterior portion of each abdominal segment beneath the ventral nerve cord, and presumably function to hold the articular cuticle between the abdominal segments in place. However, no significant signal was observed in flexor and extensor muscle (Figure 9B).
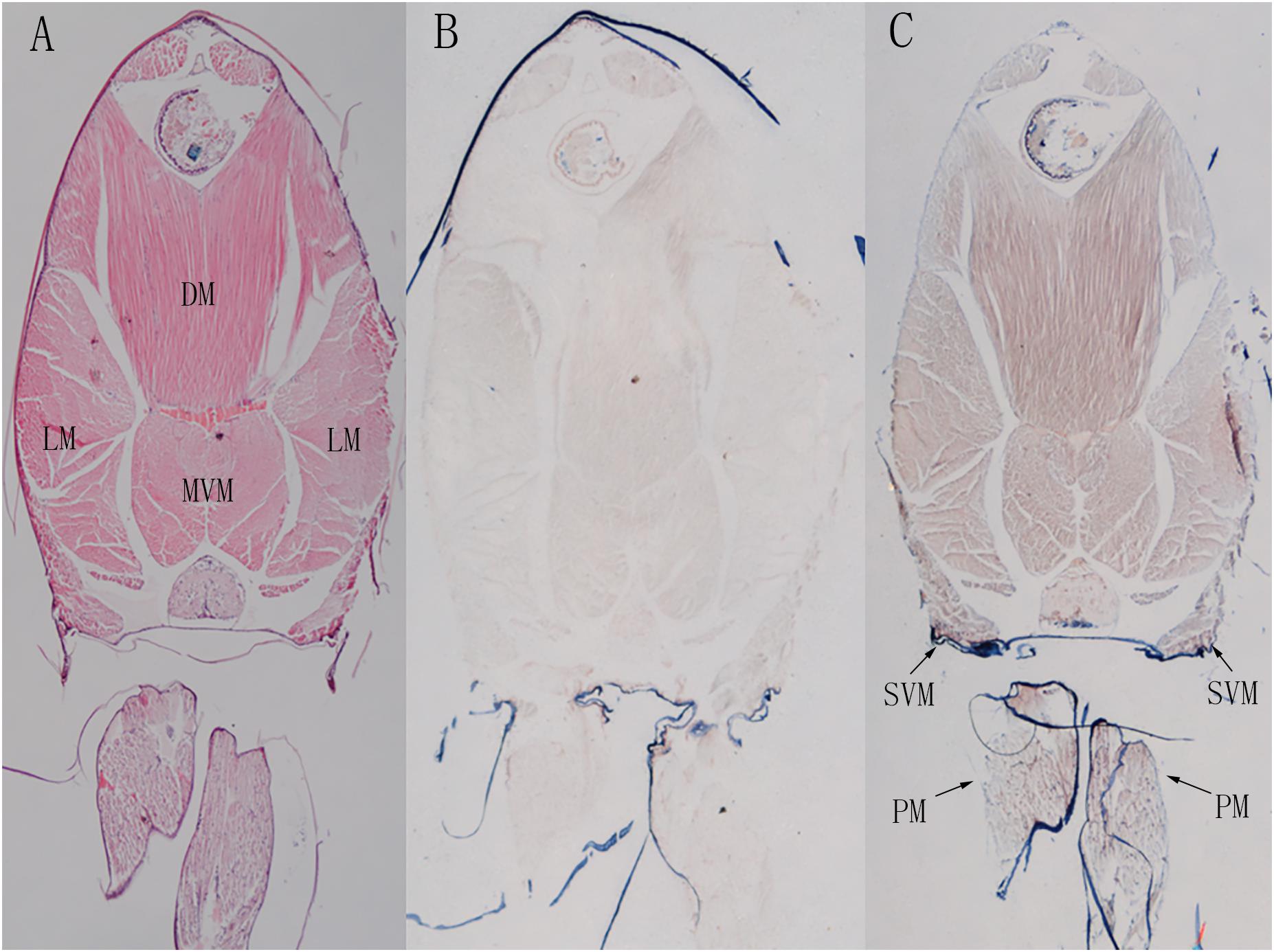
Figure 9. The localization of LvMYH5 in the abdominal muscle and pleopod muscle of L. vannamei. (A) Hematoxylin-eosin (H&E) staining and (B) a sense probe were used as the control for (C) the antisense probe hybridization. LM, the lateral muscle; DM, the dorsal muscle; MVM, the main ventral muscle; PM, the pleopod muscle; SVM, the superficial ventral muscle. The picture of H&E staining was from our previous study (Zhang et al., 2018).
Discussion
The Evolutionary Relationship of Myosin Genes in Arthropods
The evolutionary relationship of myosin family is limited in crustaceans due to the lack of Crustacea genome and difficulties for myosin gene identification. With the decoding of more and more Crustacea genomes, we are now in a position to validate the evolution of myosin genes proposed previously in arthropods and gain new insights in crustaceans. In accordance with previous study (Odronitz et al., 2009), all myosin subfamilies are shared in arthropods, suggesting that the common ancestor of arthropod has already owned all myosin classes (Odronitz and Kollmar, 2007). Meanwhile, the evolution of arthropods is likely accompanied by taxon- or species-specific losses of certain myosin classes. For example, Myriopoda and Arachnida appeared to have lost Myo3A and Myo19 classes, and Drosophilidae have lost Myo1D, Myo3A, Myo9, and Myo19 classes. As Crustacea, Maxillopoda may have lost Myo1A and Myo1B after separating from Branchiopoda and Malacostraca. The potential lack of the Myo3 class in L. vannamei and L. salmonis may be specific characteristic of species. In agreement with the results of several comparative genome analyses (Hill, 2005; Colbourne et al., 2011; Chipman et al., 2014; Kao et al., 2016), the Crustacea clade is more closely related to the Hexapoda clade, but the Myriapoda clade is closer relative to the Arachnida.
Gene Expansion of Muscle-Type Myo2 Subfamilies
Gene family expansion generally results for a strengthened phenotype. In the L. vannamei genome, the muscle-type Myo2 subfamilies showed significant expansion. Eight of 15 muscle-type Myo2 genes are tandemly duplicated. In addition, numerous fragmental muscle-type Myo2-like sequences or pseudogenes near the LvMYH cluster were also detected. Furthermore, most of the expanded muscle-type Myo2 genes possess binding sites of various myogenesis related transcription factors, such as Sp1 (Biesiada et al., 1999), AP-1 (Andreucci et al., 2002), myogenin (Hasty et al., 1993) and MyoD (Tapscott et al., 1988). It was reported that LvMYH1 and LvMYH7 were located in all fibers of abdominal muscles containing extensor and flexor muscles by in situ hybridization (Koyama et al., 2012, 2013). These results indicate that these expanded muscle-type Myo2 genes might be associated with muscle composition or development. In accordance with our previous study (Zhang et al., 2018), this study reveals the pleopod muscle and superficial ventral muscle of L. vannamei contain slow-type oxidative muscle fibers.
The muscle of shrimp accounts for more than half of the total body weight (Chen and Chen, 2001), but the mechanism of its well-developed muscular system is unknown. Our previous study has also indicated large expansion of actin gene family in L. vannamei (Zhang et al., 2018), which encodes structural proteins of muscle fiber together with muscle-type myosin heavy chains. Taken together, we speculate that the well-developed muscular system of L. vannamei might be associated with the massive expansion of muscle-type actin genes and muscle-type Myo2 genes.
Alternative Splicing of Shrimp Myosin Genes
In most arthropods, MYH1 with multiple complex alternative splicing patterns was observed to be involved in muscle composition (Bernstein et al., 1986; Rozek and Davidson, 1986; Nyitray et al., 1994; Odronitz and Kollmar, 2008; Kollmar and Hatje, 2014). For example, the D. duplex MYH1 (Mhc1) contains nine sets of mutually exclusive spliced exons (MXEs), and Drosophila melanogaster MYH1 contains five clusters of MXEs and 480 combinations of alternative splicing patterns are possibly existed (Odronitz and Kollmar, 2008). Consistent with the report that crustacean L. salmonis contains 17 Myo2 genes without MXEs (Kollmar and Hatje, 2014), no MXE has been found in the 15 muscle-type Myo2 genes in this study. This implies that the last common ancestor of crustacean might have developed a MXE-less muscle-type Myo2 gene and followed by extensive gene duplications. Multiple, but not alternatively spliced myosin heavy chain genes are therefore a common characteristic of crustacean (Kollmar and Hatje, 2014). This might be caused by the loss of introns in the ancient muscle-type myosin heavy chain gene during arthropods evolution (Odronitz and Kollmar, 2008). However, unconventional myosin genes exhibited higher diversity due to the exon skipping in L. vannamei. It indicated that alternative splicing could potentially yield unconventional myosin variants with diverse functions.
Special Expression Patterns of Shrimp Myosin Genes
Most of muscle-type Myo2 genes of L. vannamei were primarily expressed during larva in membrane and post-larvae stages. Combined with our previous study on actin gene family (Zhang et al., 2018), the whole early development stage of shrimp can be grouped into three periods. In the beginning of early development period (zygote to gastrula), most muscle-type Myo2 and actin genes are not expressed, then begin to be expressed in the second period (limb embryo and larvae in membrane stage), and their expression levels reach to the peak and remain at high levels in the third period (zoea to post-larvae stage). According to the report by Hertzler and Freas (2009), major ventral pleonal muscle and dorsal pleonal muscle of L. vannamei formed at the zoea stage, while pleopod muscle and major pleonal muscle formed in Mysis stage. Therefore, the high expression of muscle-type Myo2 and actin genes in these stages is compatible with the formation and development of pleonal and pleopod muscle.
The genes regulating muscle development, such as transcription factors, start to be expressed and to activate the co-expression of actin and myosin gene clusters at the nauplius stage. It promotes growth of muscle fibers in the larvae stage. Furthermore, compared with multiple alternative splicing of a single gene, the gene clusters have equivalent effects on improving the expression of transcripts, and strengthening the function of proteins (Talavera et al., 2007). Therefore, it is reasonable to conclude that the gene expansion of muscle-type Myo2 and actin genes may contribute to abundant fast-type muscle fibers and the explosive force of the shrimp abdominal muscles.
Conclusion
In summary, we identified 29 myosin genes in the L. vannamei genome, and classified them into 14 subfamilies. Their genome localization, gene structure, protein domains, trans-acting elements in promoter regions and molecular evolution were comprehensively analyzed. Their temporal and spatial expression profiles provide insights into their important functions in muscle composition and development. Moreover, the expanded muscle-type Myo2 subfamily may explain the well-developed muscular system of L. vannamei. Collectively, this study will provide important clues for future research on the function of myosin genes in shrimp.
Ethics Statement
This study was carried out in accordance with the recommendations of Welfare ethics of experimental animals and safety inspection system of animal experiments, laboratory animal management and ethics Committee of IOCAS. The protocol was approved by the laboratory animal management and ethics Committee of IOCAS.
Author Contributions
JX, FL, and XnZ conceived and designed the study. XiZ and CL collected the data. XiZ conducted the bioinformatics analyses, performed all experiments and wrote the manuscript. JY, XnZ, and FL revised the manuscript.
Funding
This work was financially supported by the National Key R&D Program of China (2018YFD0900103), the National Natural Science Foundation of China (31830100 and 41876167), and China Agriculture Research System-47 (CARS-47).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Qing Guo for her help in performing the in situ hybridization experiment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00610/full#supplementary-material
Footnotes
- ^https://www.cymobase.org/
- ^https://www-bimas.cit.nih.gov/molbio/signal/
- ^http://gene-regulation.com/
References
Andreucci, J. J., Grant, D., Cox, D. M., Tomc, L. K., Prywes, R., Goldhamer, D. J., et al. (2002). Composition and function of AP-1 transcription complexes during muscle cell differentiation. J. Biol. Chem. 277:16426.
Berg, J. S., Powell, B. C., and Cheney, R. E. (2001). A millennial myosin census. Mol. Biol. Cell 12, 780–794.
Bernstein, S. I, Hansen, C. J., Becker, K. D., Roche, E. S., Donady, J. J., and Emerson, C. P. (1986). Alternative RNA splicing generates transcripts encoding a thorax-specific isoform of Drosophila melanogaster myosin heavy chain. Mol. Cell. Biol. 6:2511.
Biesiada, E., Hamamori, Y., Kedes, L., and Sartorelli, V. (1999). Myogenic basic helix-loop-helix proteins and sp1 interact as components of a multiprotein transcriptional complex required for activity of the human cardiac α-actin promoter. Mol. Cell. Biol. 19, 2577–2584.
Bloemink, M. J., and Geeves, M. A. (2011). Shaking the myosin family tree: biochemical kinetics defines four types of myosin motor. Semin. Cell Dev. Biol. 22, 961–967. doi: 10.1016/j.semcdb.2011.09.015
Chen, C., Xia, R., Chen, H., and He, Y. (2018). TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv
Chen, Q., and Chen, X.-H. (2001). Effect of different salinity culture on flesh content and nutrients of Penaeus vannamei. Mar. Sci. 25, 16–28.
Chipman, A. D., Ferrier, D. E. K., Brena, C., Qu, J., Hughes, D. S. T., Schröder, R., et al. (2014). The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12:e1002005. doi: 10.1371/journal.pbio.1002005
Colbourne, J. K., Pfrender, M. E., Gilbert, D., Thomas, W. K., Tucker, A., Oakley, T. H., et al. (2011). The ecoresponsive genome of Daphnia pulex. Science 331:555. doi: 10.1126/science.1197761
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., et al. (2016). InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. doi: 10.1093/nar/gkw1107
Gao, Y., Zhang, X., Wei, J., Sun, X., Yuan, J., Li, F., et al. (2015). Whole transcriptome analysis provides insights into molecular mechanisms for molting in Litopenaeus vannamei. PLoS One 10:e0144350. doi: 10.1371/journal.pone.0144350
Hammesfahr, B., Odronitz, F., and Kollmar, M. (2010). Cymobase – the reference database for cytoskeletal and motor proteins. Biophys. J. 98. doi: 10.1016/j.bpj.2009.12.3036
Hartman, M. A., Finan, D., Sivaramakrishnan, S., and Spudich, J. A. (2011). Principles of unconventional myosin function and targeting. Ann. Rev. Cell Dev. Biol. 27:133. doi: 10.1146/annurev-cellbio-100809-151502
Hasty, P., Bradley, A., Morris, J. H., Edmondson, D. G., Venuti, J. M., Olson, E. N., et al. (1993). Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364:501.
Hertzler, P. L., and Freas, W. R. (2009). Pleonal muscle development in the shrimp Penaeus (Litopenaeus) vannamei (Crustacea: Malacostraca: Decapoda: Dendrobranchiata). Arth. Struct. Dev. 38, 235–246. doi: 10.1016/j.asd.2008.12.003
Hill, C. A. (2005). The Ixodes scapularis genome project: an opportunity for advancing tick research. Trends Parasitol. 21:151.
Hofmann, W. A., Richards, T. A., and De, L. P. (2009). Ancient animal ancestry for nuclear myosin. J. Cell Sci. 122, 636–643. doi: 10.1242/jcs.030205
Kao, D., Lai, A. G., Stamataki, E., Rosic, S., Konstantinides, N., Jarvis, E., et al. (2016). The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion. eLife 5:e20062. doi: 10.7554/eLife.20062
Kollmar, M., and Hatje, K. (2014). Shared gene structures and clusters of mutually exclusive spliced exons within the metazoan muscle myosin heavy chain genes. PLoS One 9:e88111. doi: 10.1371/journal.pone.0088111
Koyama, H., Akolkar, D. B., Piyapattanakorn, S., and Watabe, S. (2012). Cloning, expression, and localization of two types of fast skeletal myosin heavy chain genes from black tiger and Pacific white shrimps. J. Exp. Zool. Part A Ecol. Genet. Physiol. 317, 608–621. doi: 10.1002/jez.1752
Koyama, H., Piyapattanakorn, S., and Watabe, S. (2013). Cloning of skeletal myosin heavy chain gene family from adult pleopod muscle and whole larvae of shrimps. J. Exp. Zool. Part A Ecol. Genet. Physiol. 319, 268–276. doi: 10.1002/jez.1791
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33:1870. doi: 10.1093/molbev/msw054
Letunic, I., and Bork, P. (2007). Interactive Tree Of Life (ITOL): an Online Tool for Phylogenetic Tree Display and Annotation. Oxford: Oxford University Press.
Letunic, I., Doerks, T., and Bork, P. (2014). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Nyitray, L., Jancsó, A., Ochiai, Y., Gráf, L., and Szentgyörgyi, A. G. (1994). Scallop striated and smooth muscle myosin heavy-chain isoforms are produced by alternative RNA splicing from a single gene. Proc. Natl. Acad. Sci. U.S.A. 91:12686.
Odronitz, F., Becker, S., and Kollmar, M. (2009). Reconstructing the phylogeny of 21 completely sequenced arthropod species based on their motor proteins. BMC Genomics 10:173. doi: 10.1186/1471-2164-10-173
Odronitz, F., and Kollmar, M. (2007). Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, 1–23.
Odronitz, F., and Kollmar, M. (2008). Comparative genomic analysis of the arthropod muscle myosin heavy chain genes allows ancestral gene reconstruction and reveals a new type of ‘partially’ processed pseudogene. BMC Mol. Biol. 9:21. doi: 10.1186/1471-2199-9-21
Pette, D., and Staron, R. S. (2015). Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 50, 500–509.
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Richards, T. A., and Cavalier-Smith, T. (2005). Myosin domain evolution and the primary divergence of eukaryotes. Nature 436, 1113–1118.
Rozek, C. E., and Davidson, N. (1986). Differential processing of RNA transcribed from the single-copy Drosophila myosin heavy chain gene produces four mRNAs that encode two polypeptides. Proc. Natl. Acad. Sci. U.S.A. 83, 2128–2132.
Sebé-Pedrós, A., Grau-Bové, X., Richards, T. A., and Ruiz-Trillo, I. (2014). Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol. Evol. 6, 290–305. doi: 10.1093/gbe/evu013
Talavera, D., Vogel, C., Orozco, M., Teichmann, S. A., and Cruz, X. D. L. (2007). The independence of alternative splicing and gene duplication. PLoS Comput. Biol. 3:e33.
Tapscott, S. J., Davis, R. L., Thayer, M. J., Cheng, P. F., Weintraub, H., and Lassar, A. B. (1988). MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405–411.
Wei, J., Zhang, X., Yu, Y., Huang, H., Li, F., and Xiang, J. (2014). comparative transcriptomic characterization of the early development in Pacific White Shrimp Litopenaeus vannamei. PLoS One 9:e106201. doi: 10.1371/journal.pone.0106201
Woolner, S., and Bement, W. M. (2009). Unconventional myosins acting unconventionally. Trends Cell Biol. 19, 245–252. doi: 10.1016/j.tcb.2009.03.003
Zhang, X., Yuan, J., Sun, Y., Li, S., Gao, Y., Yu, Y., et al. (2019). Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 10:356. doi: 10.1038/s41467-018-08197-4
Keywords: myosin genes, muscle development, alternative splicing, penaeid shrimp, gene family
Citation: Zhang X, Yuan J, Zhang X, Liu C, Li F and Xiang J (2019) Genome-Wide Identification and Expression Profiles of Myosin Genes in the Pacific White Shrimp, Litopenaeus vannamei. Front. Physiol. 10:610. doi: 10.3389/fphys.2019.00610
Received: 23 January 2019; Accepted: 29 April 2019;
Published: 21 May 2019.
Edited by:
Carlos Rosas, National Autonomous University of Mexico, MexicoReviewed by:
Theresa Joan Grove, Valdosta State University, United StatesTiziano Verri, University of Salento, Italy
Copyright © 2019 Zhang, Yuan, Zhang, Liu, Li and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Zhang, eGp6aGFuZ0BxZGlvLmFjLmNu Fuhua Li, ZmhsaUBxZGlvLmFjLmNu
 Xiaoxi Zhang
Xiaoxi Zhang Jianbo Yuan
Jianbo Yuan Xiaojun Zhang
Xiaojun Zhang Chengzhang Liu1,2,4
Chengzhang Liu1,2,4 Fuhua Li
Fuhua Li Jianhai Xiang
Jianhai Xiang