- State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Agasicles hygrophila is an effective biological control agent for the invasive weed Alternanthera philoxeroides, and because of this it has been introduced to many parts of the world where A. philoxeroides is a problem. Despite this, there are no reports at present about the reproduction of this important insect. Vitellogenin receptors (VgRs) belong to the superfamily of low-density lipoprotein receptors (LDLRs). One of the roles of VgRs is to regulate the absorption of yolk protein in insects. In this study, the full length vitellogenin receptor gene (AhVgR) from A. hygrophila was sequenced and found to encode a predicted protein of 1,642 amino acids. Sequence analysis of AhVgR revealed that it contains conserved structural motifs common to LDLR family members, and a phylogenetic analysis placed AhVgR as a separate group among the order Coleoptera. AhVgR was found to be specifically expressed in ovarian tissues, and it is first transcribed in the newly-emerged females. The expression patterns are consistent with VgR genes in other insects. RNA interference (RNAi)-mediated suppression of AhVgR gene expression in adult A. hygrophila females inhibited yolk protein deposition in the ovaries, shortened the ovariole, drastically reduced egg production, and ultimately led to a decrease in fecundity. In summary, our work shows that AhVgR is critical for transporting Vg into the oocytes and plays an important role in A. hygrophila reproduction.
Introduction
Alternanthera philoxeroides, (Mart.) Griseb (Amaranthaceae), commonly known as alligatorweed, is a perennial aquatic plant native to South America that has invaded many regions of the world, including North America, Asia, and Australia (Julien et al., 1995; Xu et al., 2003). In China, A. philoxeroides was introduced as a forage crop in the late 1930s, and subsequently spread to the country’s eastern and southern regions (Ye et al., 2010). A. philoxeroides is highly competitive, and causes economic and ecological problems almost everywhere it is found (Maddox and Rhyne, 1975). Agasicles hygrophila (Selman & Vogt) (Coleoptera: Chrysomelidae), the alligatorweed flea beetle, has proven to be an efficient control agent for A. philoxeroides (Buckingham, 1996). The beetle was introduced into China from Florida in 1987, and was first released in Chongqing, Fuzhou, and Changsha in 1988. Agasicles hygrophila became established in the release sites and is now distributed mostly in southern China, where it has controlled A. philoxeroides for over 30 years (Guo et al., 2011).
As the precursor of the major yolk protein vitellin (Vn), vitellogenin (Vg) provides nutrition necessary for egg development, and plays an important role in insect reproduction (Tufail and Takeda, 2009). During reproduction, Vg is primarily synthesized in the fatbody and then secreted into the hemolymph, where it is absorbed by the developing oocytes via the vitellogenin receptor (VgR) (Lu et al., 2015). Thus, VgR plays an essential role in the process of oocyte maturation (Sappington and Raikhel, 1998). In insects, the VgR gene encodes a large ovary-specific protein of 180–214 kDa, which is twice the size of the vertebrate VgR proteins (Sappington and Raikhel, 1998). The amino acid sequences of insect VgRs show that they are members of the low-density lipoprotein receptor (LDLR) family, with two ligand binding domains (LBDs) containing several LDLR-domain class A repeats. The function of VgR is different from than that of Vg, but silencing either of the genes can lead to malformed ovaries and decreased fecundity, as has been shown in Helicoverpa armigera (Zhang et al., 2016), Nilaparvata lugens (Lu et al., 2015), Spodoptera litura (Shu et al., 2011), and Aphis citricidus (Shang et al., 2018).
Although VgRs have been described in other insects, there are no studies that describe the VgR of A. hygrophila. In our study, we characterized the AhVgR gene and explore its expression patterns in A. hygrophila. We cloned the complete sequence of AhVgR, and showed its molecular characteristics, phylogenetic relationships and expression patterns. Furthermore, we verified the function of AhVgR in ovary development and reproduction through RNA interference experiments.
Materials and Methods
Insect Rearing and Sample Collection
Alternanthera philoxeroides plants were collected at the Institute of Plant Protection, Hunan Academy of Agricultural Sciences, and grown in the greenhouse at Langfang Experimental Station, Chinese Academy of Agricultural Sciences. Plants were grown in sterilized soil in plastic boxes (40 cm × 18 cm × 15 cm) and watered every other day. Agasicles hygrophila adults were initially collected from the field in Changsha. The population was reared on A. philoxeroides plants in the laboratory at the Chinese Academy of Agricultural Sciences (Beijing, China) under controlled conditions (28 ± 1°C, 12 h light:dark photoperiod, relative humidity [RH] = 75 ± 5%) (Guo et al., 2011).
To clone the AhVgR gene sequence, the ovaries of A. hygrophila females were dissected in 1X PBS (phosphate-buffered saline, pH 7.4) using an Olympus stereomicroscope (SZX16, Olympus, Tokyo, Japan). For tissue-specific expression of AhVgR, the head, thorax, ovary, fat body, midgut, and wing tissues of 6-day-old beetles were also dissected in PBS using the Olympus SZX16 stereomicroscope. To analyze the expression of the different developmental stages, freshly pupated pupa were collected daily, and female adults were collected every 2 day after emergence. All samples were immediately frozen in liquid nitrogen and stored at −80°C until they were used for RNA isolation.
RNA Isolation and Gene Cloning
Total RNA was extracted from A. hygrophila tissues using TRIzol reagent (Life Technologies, Carlsbad, CA, United States) according to the manufacturer’s protocol. The concentrations of the RNA samples were determined with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States), while degradation and genomic DNA contamination were monitored by electrophoresis through 1% agarose gels. First-strand cDNA from the different RNA samples was synthesized using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen, Beijing, China) following the manufacturer’s protocol. Our lab mined the A. hygrophila transcriptome data to obtain expressed sequence tags (ESTs) that showed similarities to other insect vitellogenin receptor genes. The PCR primers were designed based on the A. hygrophila vitellogenin receptor gene cDNA fragment from the transcriptome (Supplementary Table 1). Positive clones were confirmed by PCR amplification and then sequenced. To obtain full-length cDNA sequence of the AhVgR gene, we used a SMARTTM RACE cDNA amplification kit (Clontech, Mountain View, CA, United States) to amplify the 5′- and 3′-ends. Gene-specific primers were designed (Supplementary Table 1), and the RACE cycling conditions were performed as described in Zhang et al. (2016). The RACE products were then gel-purified and sequenced.
Gene Sequence Analysis and Phylogenetic Relationships
Sequence similarities were analyzed using the BLAST website1. The open reading frames (ORFs) of the AhVgR clones were predicted using the NCBI ORF finder2. The molecular weights and isoelectric points (pIs) of the inferred AhVgR protein sequences were predicted with the ExPASy proteomics server3. The conserved domains and signal peptide were analyzed by the online SMART server4 and the SignalP 4.1 Server5. The VgR amino acid sequences from other insects were downloaded from the GenBank database and used to in the phylogenetic analyses. The VgR sequence alignments were carried out using Clustal X software. A phylogenetic tree was constructed with molecular evolutionary genetics analysis software (MEGA version 5.0), using the neighbour-joining (NJ) method with a bootstrap test of 1,000 replicates (Tamura et al., 2011).
Synthesis of Double-Stranded RNA (dsRNA) and Injection of dsRNA
dsRNA was synthesized from the AhVgR cDNA, and EGFP (Enhanced Green Fluorescent Protein, GenBank Accession No. AIR08541.1) dsRNA was synthesized as a negative control. To ensure the interference effect, we synthesized two dsRNA fragments from AhVgR (dsVgR-A and dsVgR-B) and EGFP (ds EGFP- and, ds EGFP-B). The target gene dsRNAs were synthesized and purified using the HiScribeTM T7 Quick High Yield RNA Synthesis Kit (New England BioLabs, Ipswich, MA, United States #E2050S) as directed by the manufacturer. The purity of the dsRNA was checked by electrophoresis on a 1.0% agarose gel and the concentration of the dsRNA was measured on a NanoDrop ND-2000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE, United States). To ensure the injection volume between the control group and treatment group was consistent, we adjusted the concentration of all synthesized dsRNA to 10,000 ng/μl. Newly-emerged adult females were collected, and the dsRNA was injected into the conjunctivum using a PLI-100 Pico-Injector (Harvard Apparatus, Holliston, MA, United States) with an MP-255 Micromanipulator (Sutter, Novato, CA, United States) under an Olympus stereomicroscope. Each experiment was repeated three times. Each repeat consisted of 60 female individuals for later experimental observation after injection. The injection dose was 0.1 μl of each dsRNA (dsVgR group: dsVgR-A 0.1 μl, dsVgR-B 0.1 μl; dsEGFP group: dsEGFP-A 0.1 μl, dsEGFP-B 0.1 μl). After injection, female adults were kept in separate plastic bottles (8 cm × 10 cm) and paired with freshly emerged wild-male adults. Each bottle was kept moist with a piece of moistened filter paper at the bottom, and fresh A. philoxeroides stems were added.
qPCR Experiments
Total RNA was extracted from the samples with TRIzol reagent (Life Technologies, Carlsbad, CA, United States) following the manufacturer’s instructions. First-strand cDNA was synthesized by following the procedures mentioned above. Quantitative real-time PCR (qPCR) analysis was performed with the TransStart Green qPCR SuperMix Kit (Transgen, Beijing, China) on an ABI Prism 7500 system (Applied Biosystems, Carlsbad, CA, United States). CoxI (GenBank Accession No. FJ977926.1) was used as an internal control gene. Specific primers were designed for qPCR with Beacon Designer 7.9 software (PREMIER Biosoft International, Davis, CA, United States) (Supplementary Table 1). Thermal cycler amplification conditions were as recommended by the manufacturer. The qPCR reactions were performed in 20 μl volumes containing 10 μl of 2X TransStart Green qPCR SuperMix, 0.4 μl of each primer (10 μM), 0.4 μl of 50X Rox Reference Dye, 200 ng of sample cDNA, and 7.8 μl of ddH2O. The qPCR cycling parameters were as follows: 94°C for 30 s, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 10 s, with melt curve stages at 95°C for 15 s, 60°C for 1 min, and 9°C for 15 s. To ensure reliability, each reaction for each sample was performed three times with three technical replicates. Each run included negative controls with no cDNA template. The relative expression of each gene was calculated using the 2–ΔΔCT method of Livak and Schmittgen (2001).
Observation of Ovary Development and Fecundity After RNAi
Each dsRNA-treated A. hygrophila female was observed 4 days after dsRNA injection to examine ovary development. The ovaries were dissected using high-precision tweezers (IDEAL-TEK, Balerna, Switzerland) in 1X PBS under the Olympus SZX16 stereomicroscope. Dissected ovaries were washed three times with 1X PBS and photographed as described by Zhao et al. (2016). Eggs laid by each pair of A. hygrophila adults in the control and treatment groups were collected and counted once. A total of 1,000 eggs were used to calculate the egg hatch rate, and the eggs were assessed every 12 h until they started to decay (Zhao et al., 2016).
Statistical Analysis
Statistical analyses were performed using SPSS 18.0 (SPSS, Inc., Chicago, IL, United States). All the experimental data are shown as means ± SD (standard deviation). Experimental data were checked for normality and homoscedasticity, and if needed, were arcsine square-root or log-transformed before analysis. Egg hatching rates and ovariole lengths were arcsine-transformed. The number of eggs was square root-transformed. We performed the least significant difference (LSD) test after one-way analysis of variance (ANOVA) to analyze differences in AhVgR expression levels between different tissues and developmental stages. AhVgR expression levels after injection, ovariole lengths, fecundity of females, and egg hatch rate were analyzed by Student’s t-test. We considered means significantly different at P < 0.05.
Results
Sequence Analysis and Phylogenetic Analysis of the AhVgR Gene
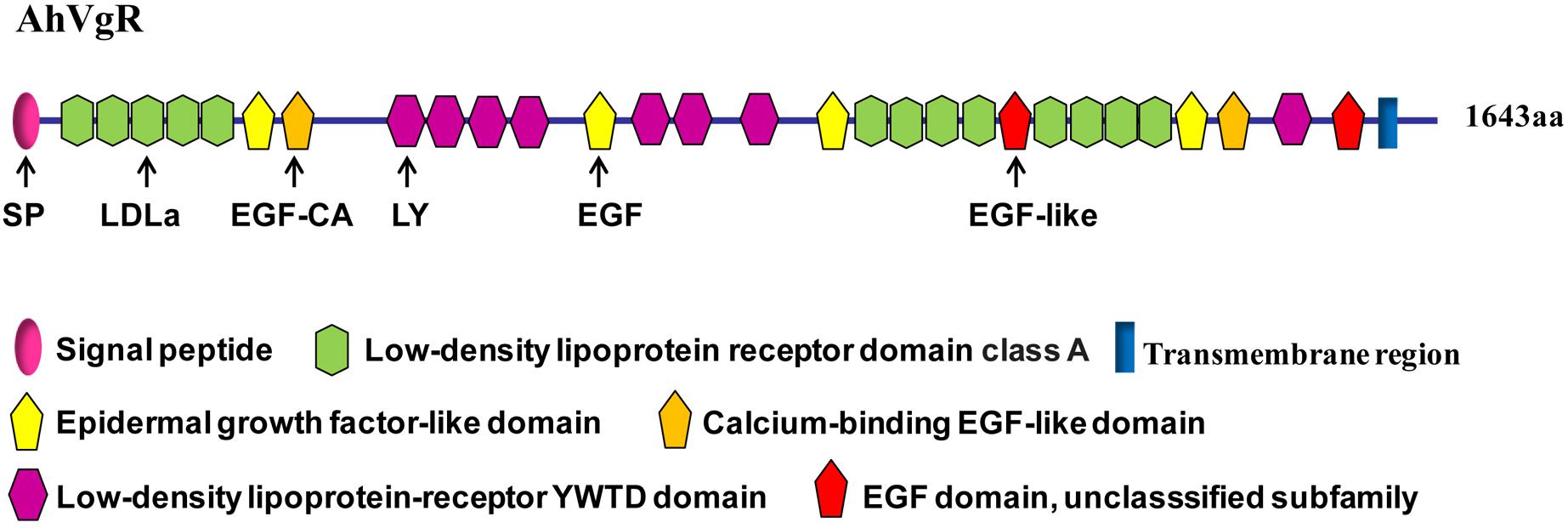
The full-length AhVgR cDNA was found to be 5,386 bp in length, including a 224 bp 5′ untranslated region (UTR), a 4,929 bp ORF encoding a predicted protein of 1,642 amino acids, and a 233 bp 3′ UTR (GenBank Accession No. MH428915). The theoretical molecular weight is 183.32 kDa, and the pI is 5.08. Analysis of the AhVgR amino acid sequence showed that a 16-amino acid signal peptide (MLVLLLLGVVTPSLGF) is located at the N-terminus of AhVgR. Domain architecture analysis showed that AhVgR is a transmembrane protein with several typical domains, including a LDLR domain class A, an epidermal growth factor (EGF)-like domain, a calcium-binding EGF-like domain, a low-density lipoprotein-receptor YWTD domain, and the transmembrane region (Figure 1). The analysis indicated that AhVgR has three LBDs with five class A (LDLa) cysteine-rich repeats in the first domain and eight in the second domain (Figure 1). Each repeat contains six cysteine residues, and each LDLa is followed by an EGF-like domain. The second and the seventh EGF-like domains contain a calcium-binding domain. The second, third, and seventh EGF-like domains are followed by 4, 3, and 1 repeats of the LDLR YWTD motif, respectively (Figure 1).
A phylogenetic analysis of the AhVgR protein showed that VgR sequences from insects are clearly separated from mammalian VgR sequences (Figure 2). In the neighbor-joining tree, VgRs from Coleoptera clustered in one clade, well-separated from VgR proteins from other insect orders, showing that VgRs from Coleoptera all had high amino acid sequence identity and are derived from a common ancestor (Figure 2). The dendrogram also showed that the VgR proteins from A. hygrophila and Leptinotarsa decemlineata are close, suggesting that the VgRs from these two insects share a close evolutionary relationship. The phylogenetic tree also showed that VgRs from the same insect orders clustered into individual clades, suggesting the conservation of insect VgRs and showing the close evolutionary relationships within the same taxonomic groups. However, the VgRs of Coleoptera clustered most closely with the VgRs of Blattaria with high bootstrap support, showing that the VgRs of these two insect orders share a closer evolutionary relationship than they do with VgRs from other insect orders.
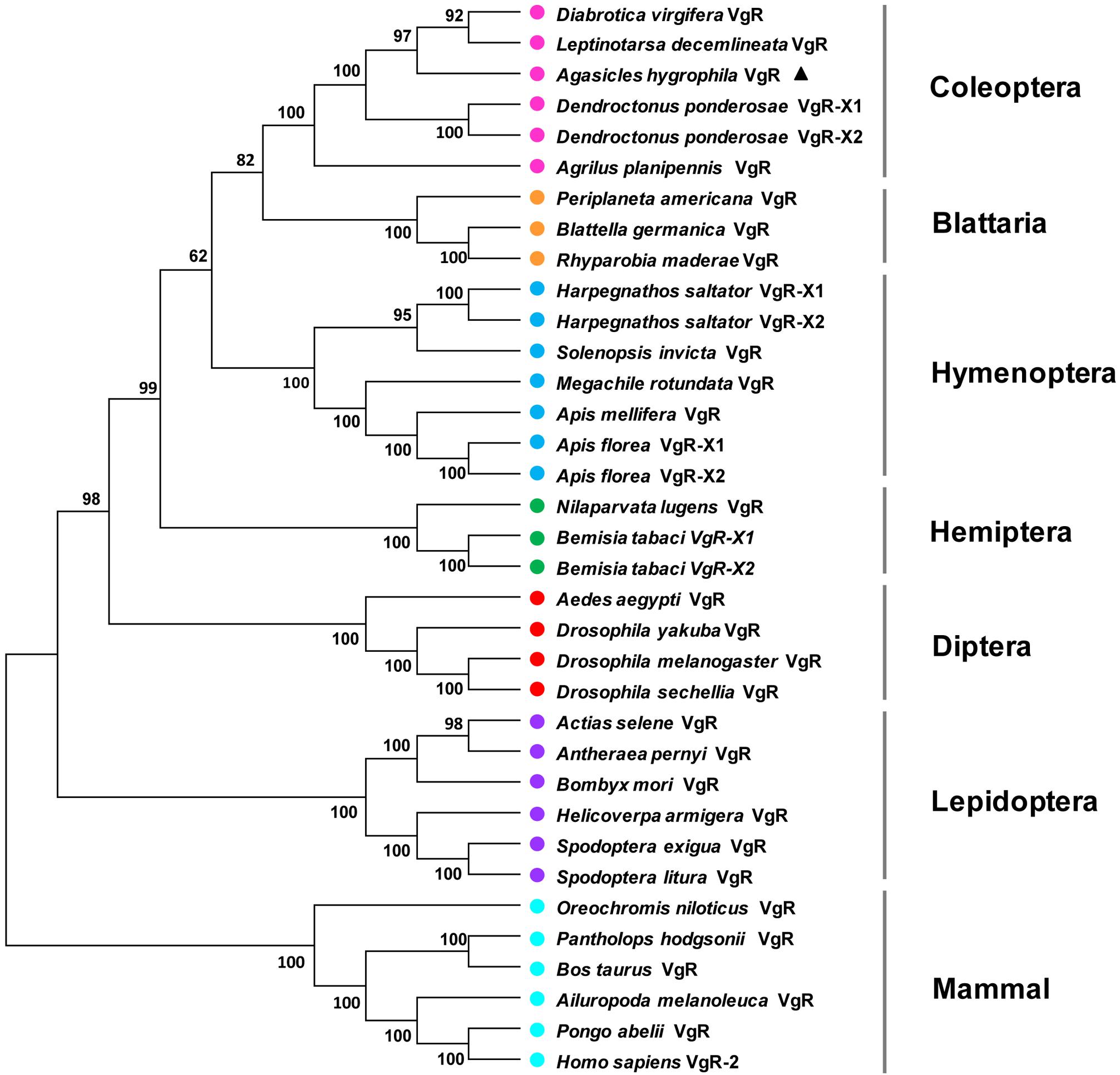
Figure 2. Unrooted consensus neighbor-joining tree showing the evolutionary relationships among vitellogenin receptor proteins. Phylogenetic trees were generated with MEGA 5 using VgR amino acid sequences from insect species in the orders Coleoptera, Hemiptera, Dictyoptera, Lepidoptera, Hymenoptera, and Diptera and five mammal species. The names and accession numbers of the protein sequences used in this analysis are given in Supplementary Table 2.
Tissue-Specific Expression and Temporal Expression Patterns of AhVgR
qPCR analysis shows that AhVgR mRNA is most highly expressed in the ovaries (Figure 3). Analysis of AhVgR mRNA expression in the pupal and adult stages revealed that AhVgR expression was not detectable in the pupal stage, but was detected in newly-emerged females, reaching a maximum level in 11-day-old individuals (Figure 4). Subsequently, expression decreased gradually to a lower level that was maintained in the adults.
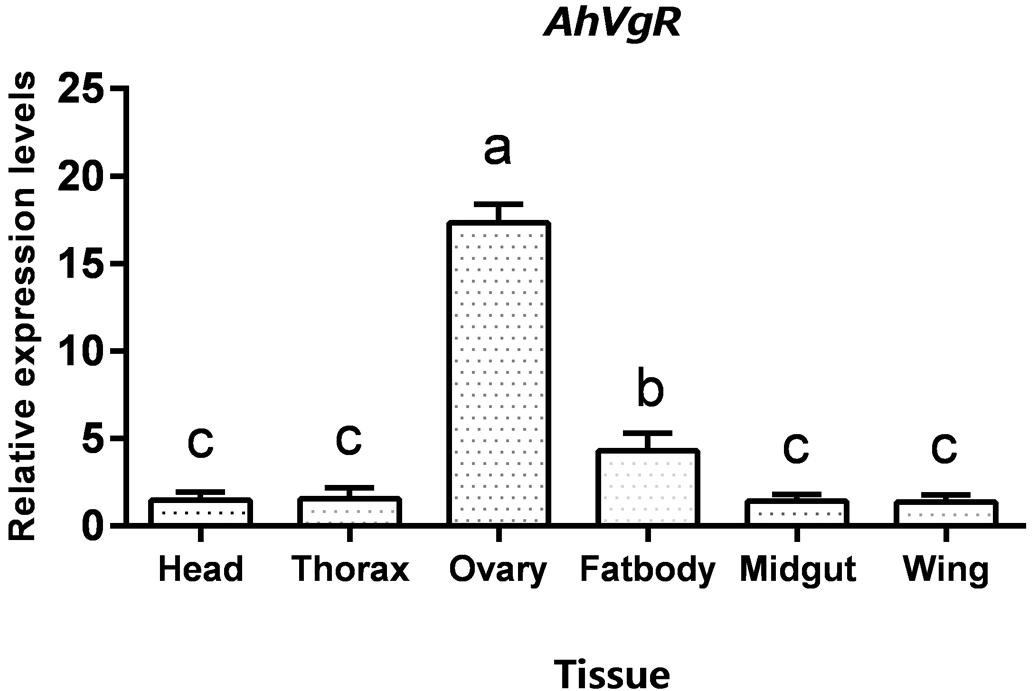
Figure 3. Tissue-specific expression levels of AhVgR. The relative mRNA levels were normalized to the expression of the CoxI gene and analyzed by the 2–ΔΔCT method. The figure shows data from three replicates that were analyzed by one-way analysis of variance (ANOVA) and the least significant difference (LSD) test. All values are shown as the mean ± SD and bars with different letters indicate significant differences (P < 0.05).
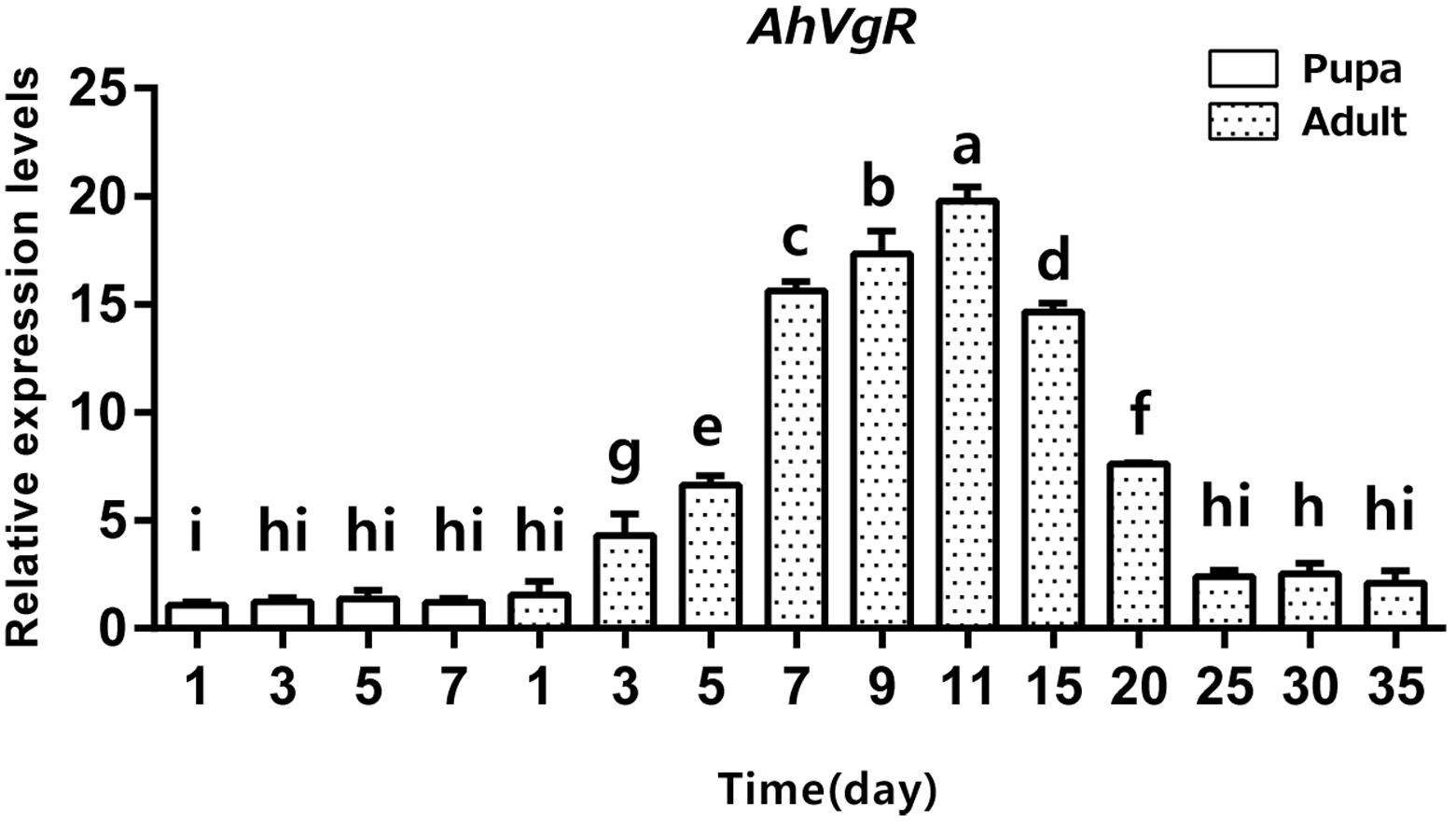
Figure 4. Temporal expression patterns of AhVgR in pupae and adults of A. hygrophila. The relative mRNA levels were normalized to the expression of the CoxI gene and analyzed by the 2–ΔΔCT method. The figure shows data from three replicates that were analyzed by a one-way analysis of variance (ANOVA) and the LSD test. All values are shown as the mean ± SD and bars with different letters indicate significant differences (P < 0.05).
Effects of dsRNA Injection on AhVgR Expression
qPCR assays showed that AhVgR expression in dsVgR-injected females decreased significantly at all sampled time points from 3 to 25 days post-injection (Figure 5). However, AhVgR expression did not show an obvious difference between the dsEGFP-injected group and dsVgR-injected group after 25 days. This may be because of the timeliness of dsRNA. Furthermore, from 3 to 25 days after the injection of dsAhVgR, the transcription levels of AhVgR decreased by 45.6–81.2% (Figure 5) compared to the dsEGFP treatment control group.
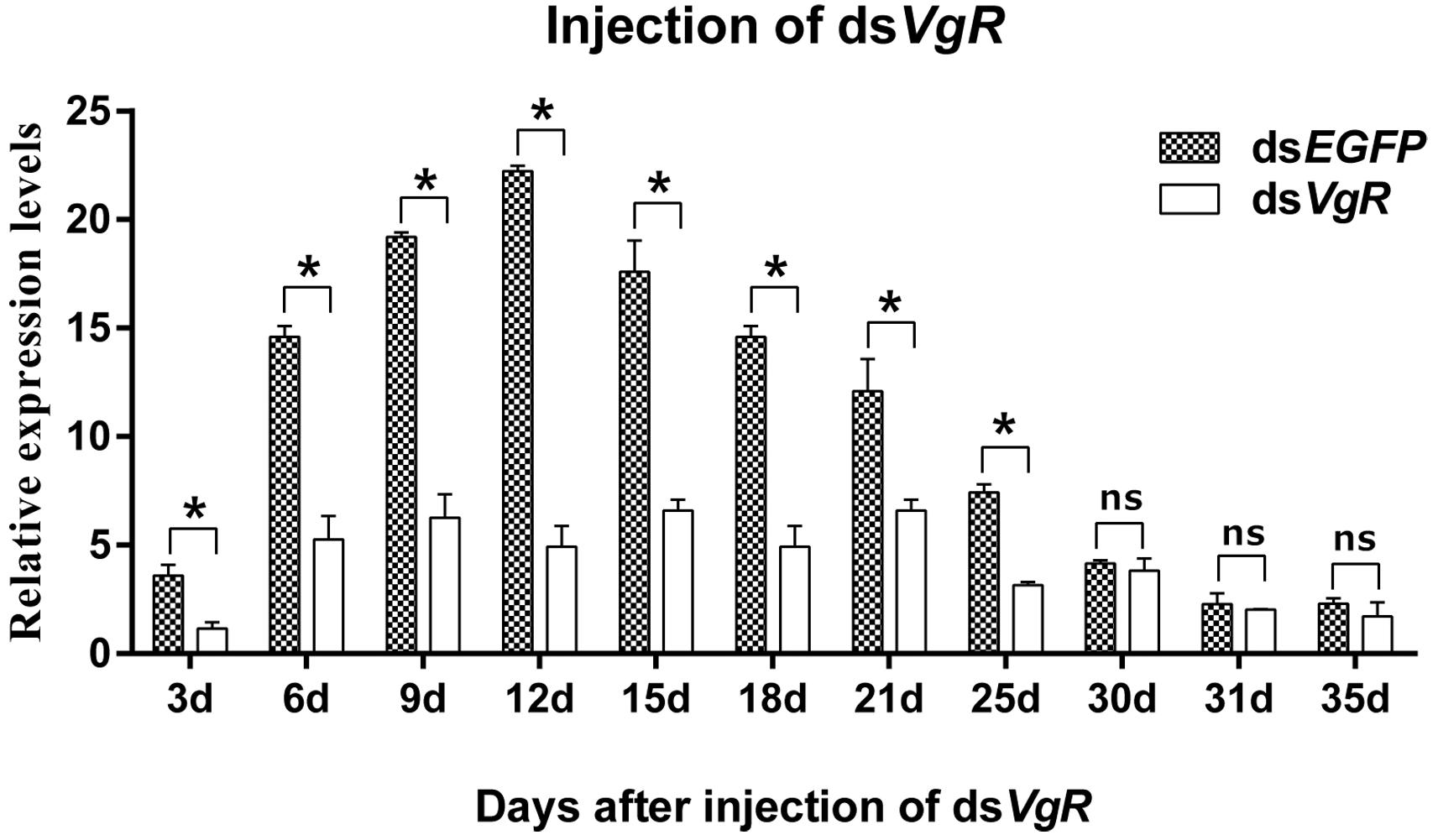
Figure 5. Relative expression of AhVgR after injection of dsVgR RNA into newly emerged A. hygrophila females (n = 3). All values are shown as the mean ± SD. The data were analyzed by Student’s t-test. *P < 0.05. ns, not significant. dsEGFP RNA was used as the control.
dsVgR Injection Inhibits Ovarian Development
After 4 days, the ovaries of the dsVgR-injected group showed a decrease in yolk protein deposition compared to the dsEGFP-injected group (Figure 6). The length of the ovariole in the dsVgR-injected group was shorter than in the dsEGFP-injected group (Figure 7), showing that ovary development in A. hygrophila females was inhibited after injection with dsVgR RNA, and that AhVgR plays an important role in ovary development and yolk protein deposition.
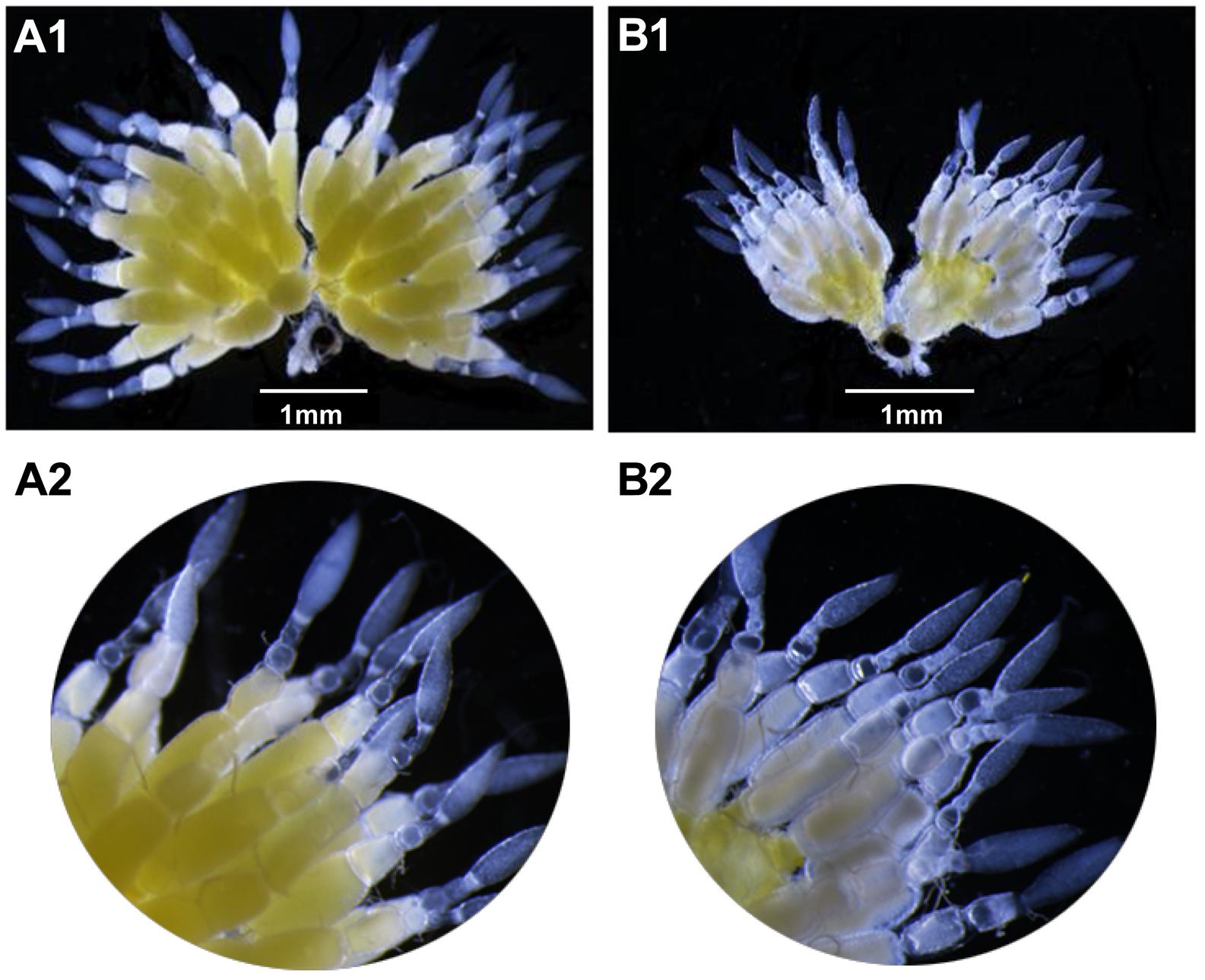
Figure 6. Effect of dsVgR on A. hygrophila ovary development. (A1) It shows an ovary that developed normally. (A2) It is an enlargement of (A1). (B1) Shows an abnormal ovary that changed morphologically after injection of dsVgR. (B2) Is an enlargement of (B1).
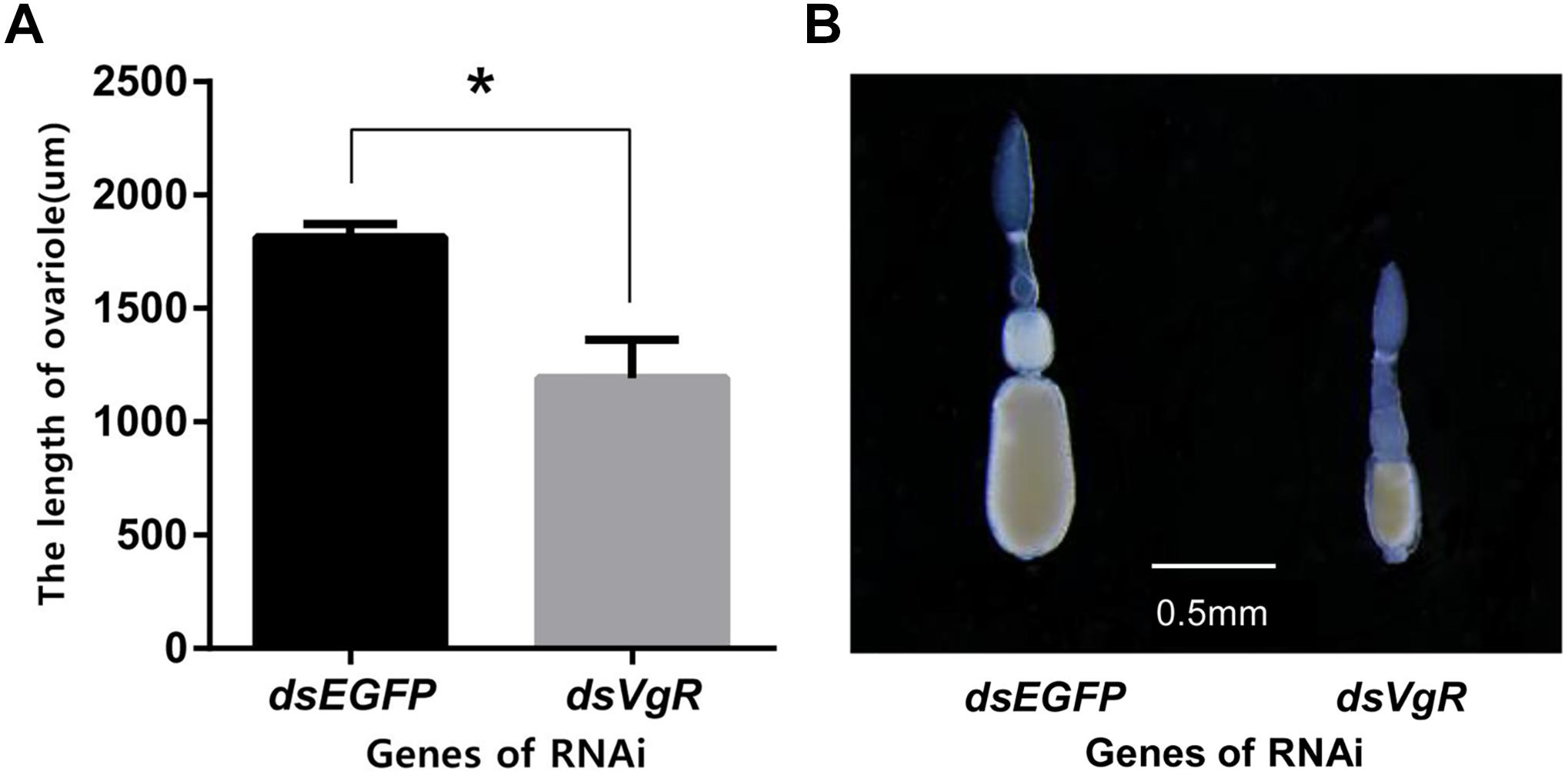
Figure 7. Length of the A. hygrophila ovariole after injection with dsVgR RNA. (A) Ovariole lengths measured from the dsVgR-injected and the dsEGFP-injected group were analyzed by Student’s t-test. *P < 0.05. All values are shown as the mean ± SD. (B) Representative ovarioles dissected from the ovaries from the dsVgR-injected and dsEGFP-injected groups of A. hygrophila females.
Knockdown of AhVgR Affects Fecundity in A. hygrophila Females
After the injection of dsVgR RNA into newly-emerged adult females, they laid significantly fewer eggs than did the dsEGFP-injected group (Figure 8A). Furthermore, when the hatch rates were compared, eggs produced by the dsVgR-injected females had lower hatching success than did eggs from the dsEGFP-injected group (Figure 8B). Results of fecundity and egg hatching rate both confirmed that RNA interference of AhVgR gene expression significantly inhibited reproduction in A. hygrophila.
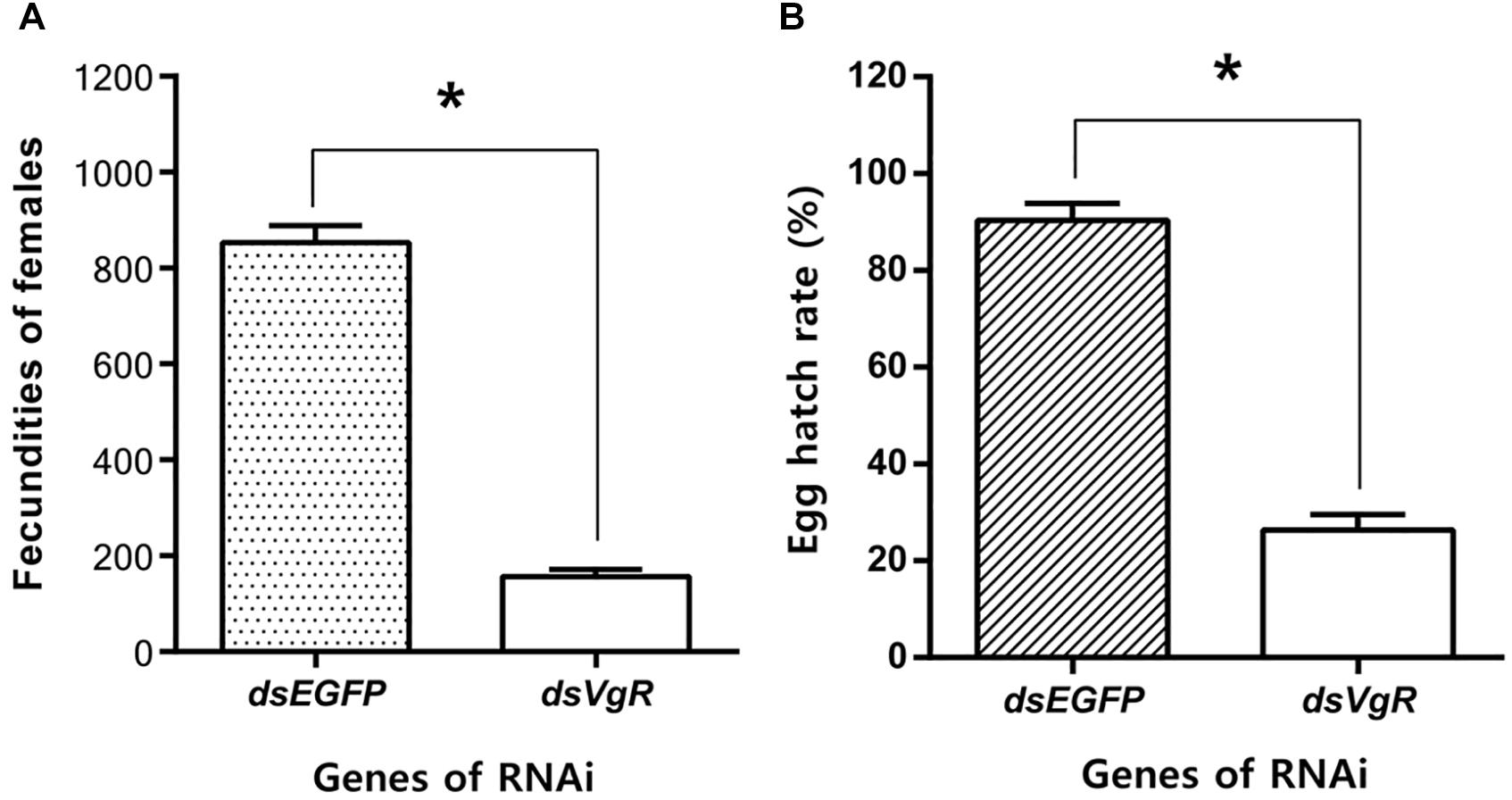
Figure 8. Knockdown of AhVgR gene expression reduces fecundity in A. hygrophila females. (A) Fecundity of females from the dsVgR-injected and dsEGFP-injected groups (n = 20). (B) Egg hatch rate from the dsVgR-injected and dsEGFP-injected groups (n = 10). All values are shown as the mean ± SD. The data were analyzed by Student’s t-test. *P < 0.05.
Discussion
The VgR gene have been studied in many insects, however, this gene has not been characterized in A. hygrophila, and studying it is relevant because that this species is used as a biological control agent for the invasive weed A. philoxeroides. In our experiments, we characterized AhVgR as the LDLR homolog, and analyzed the expression patterns of this gene in different tissues and developmental stages. Our study is the first to use the RNA interference method to explore the function of AhVgR, and the results showed that silencing the AhVgR gene inhibits ovary development and significantly reduces fecundity in A. hygrophila.
VgR proteins belong to the LDLR family, and regulate the process through which Vg enters the oocyte by endocytosis. All oviparous vertebrates and invertebrates have VgR family member proteins (Tufail and Takeda, 2009; Shu et al., 2011). In our study, the full-length cDNA of the AhVgR gene was cloned, and the molecular characteristics of this gene were analyzed. The AhVgR protein shares several typical domains with other insect VgRs, such as a LDLR domain class A, an EGF-like domain, a calcium-binding EGF-like domain, and a low-density lipoprotein-receptor YWTD domain. As the structural arrangement found in most insect VgRs, the AhVgR protein contains five cysteine-rich repeats in the first binding site and eight in the second binding site (Figure 1). However, the structural characteristics in other insect VgRs are different. For example, in Spodoptera litura, SlVgR has four cysteine-rich repeats in the first binding site and seven in the second binding site. Phylogenetic analysis shows that these insect VgRs cluster into six clades (Coleoptera, Blattaria, Hymenoptera, Hemiptera, Diptera, and Lepidoptera). As expected, AhVgR is in the Coleoptera clade, and the phylogenetic tree indicates that AhVgR shares homology with other Coleoptera VgR proteins, such as those from Leptinotarsa decemlineata, Diabrotica virgifera, Dendroctonus ponderosae, and Agrilus planipennis (Figure 2).
In A. hygrophila, expression profiles of AhVgR showed that this gene is expressed at high levels in the ovaries (Figure 3), indicating that it is an ovary-specific gene, consistent with its role in insect reproduction (Ciudad et al., 2010; Lin et al., 2013). However, VgR gene expression has also been detected in other tissues. In Apis mellifera, AmVgR expression was detected in the hypopharyngeal gland and other tissues (Guidugli-Lazzarini et al., 2008). The various expression patterns of VgR genes in different tissues may be related to their ligand Vg, which regulates longevity and the juvenile hormone titer in bees (Corona et al., 2007; Nelson et al., 2013). The temporal expression patterns of AhVgR were found to be closely correlated with the reproductive process, and expression levels significantly increased in the ovaries beginning 3 days after emergence (Figure 4). This pattern is consistent with VgR expression patterns in other insects, such as in Nilaparvata lugens, Thitarodes pui, and Aphis citricidus (Lu et al., 2015; Shang et al., 2018; Wu et al., 2018).
RNA interference is considered to be a specific tool to efficiently determine gene functions (Hannon, 2002) and also to control gene expression at the mRNA level (Fire et al., 1998). We used RNAi to explore the functions of AhVgR in vivo. The results showed that injection with dsVgR RNA displayed high efficiency and caused significant differences in fecundity and ovarian development compared to the control group (Figures 6–8). AhVgR expression decreased after the dsVgR treatment, and the effects of dsVgR RNA lasted for about 25 days after injection (Figure 5). The high gene silencing efficiency of dsRNA has also been verified in other Coleoptera species such as Tribolium castaneum, Leptinotarsa decemlineata, and Diabrotica virgifera (Arakane et al., 2005; Shi et al., 2016; Camargo et al., 2018). After injecting the dsVgR RNA, the ovaries exhibited a decrease in yolk protein deposition compared to the control group (Figure 6), and the lengths of the ovarioles were also reduced (Figure 7). Furthermore, we observed that A. hygrophila fecundity, as measured by the number of eggs laid and their hatch rate, both decreased sharply after injection with dsVgR RNA (Figure 8). Our results are consistent with an earlier study which suggested that the capability of reproduction in insects depends on two primary steps, (1) the process of vitellogenin (Vg) formation and deposition and (2) transport of the Vg to oocytes by the vitellogenin receptor (VgR) (Sappington and Raikhel, 1998). The functions of VgR have been also explored in other insect species in which the functions of Vg have been studied, and the results are in accordance with ours (Shu et al., 2011; Lin et al., 2013; Shang et al., 2018).
Conclusion
In our study, the molecular characterization of AhVgR and the expression patterns of this gene in different tissues and developmental stages were analyzed. Analysis of amino acid sequences of AhVgR suggests AhVgR is a member of the LDLR family and displays high sequence similarity to VgRs from other insects. The tissue and developmental stage-specific mRNA expression patterns of AhVgR were also similar to those in other insects. We found that AhVgR was most highly expressed in the ovary of adult stage. After the basic characterization of the gene, we explored the functions of AhVgR via RNA interference, and the results indicate that the AhVgR protein is necessary to stimulate yolk uptake, as well as being critical for ovary development and egg laying in A. hygrophila.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
HZ and JG conceived and designed the experiments, and wrote the manuscript. HZ performed the experiments. HZ, YL, JJ, and ZZ analyzed the data.
Funding
This work was supported by the National Natural Science Foundation of China (31572068) and the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2015BAD08B03).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00969/full#supplementary-material
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/BLAST/
- ^ https://www.ncbi.nlm.nih.gov/orffinder/
- ^ http://www.expasy.org
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.cbs.dtu.dk/services/SignalP
References
Arakane, Y., Muthukrishnan, S., Kramer, K. J., Specht, C. A., Tomoyasu, Y., Lorenzen, M. D., et al. (2005). The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol. Biol. 14, 453–463. doi: 10.1111/j.1365-2583.2005.00576.x
Buckingham, G. R. (1996). Biological control of alligatorweed, Alternanthera philoxeroides, the world’s first aquatic weed success story. Castanea 61, 232–243.
Camargo, C., Wu, K., Fishilevich, E., Narva, K. E., and Siegfried, B. D. (2018). Knockdown of RNA interference pathway genes in western corn rootworm, Diabrotica virgifera virgifera, identifies no fitness costs associated with argonaute 2 or dicer-2. Pestic. Biochem. Physiol. 148, 103–110. doi: 10.1016/j.pestbp.2018.04.004
Ciudad, L., Piulachs, M. D., and Bellés, X. (2010). Systemic RNAi of the cockroach vitellogenin receptor results in a phenotype similar to that of the Drosophila yolkless mutant. FEBS J. 273, 325–335. doi: 10.1111/j.1742-4658.2005.05066.x
Corona, M., Velarde, R. A., Remolina, S., Moranlauter, A., Wang, Y., Hughes, K. A., et al. (2007). Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc. Natl. Acad. Sci. U.S.A. 104, 7128–7133. doi: 10.1073/pnas.0701909104
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Guidugli-Lazzarini, K. R., do Nascimento, A. M., Tanaka, E. D., Piulachs, M. D., Hartfelder, K., Bitondi, M. G., et al. (2008). Expression analysis of putative vitellogenin and lipophorin receptors in honey bee (Apis mellifera L.) queens and workers. J. Insect Physiol. 54, 1138–1147. doi: 10.1016/j.jinsphys.2008.04.021
Guo, J. Y., Fu, J. W., Xian, X. Q., Ma, M. Y., and Wan, F. H. (2011). Performance of Agasicles hygrophila (Coleoptera: Chrysomelidae), a biological control agent of invasive alligator weed, at low non-freezing temperatures. Biol. Invasions 14, 1597–1608. doi: 10.1007/s10530-010-9932-3
Julien, M. H., Skarratt, B., and Maywald, G. F. (1995). Potential geographical distribution of alligator weed and its biological control by Agasicles hygrophila. J. Aquat. Plant Manage. 33, 55–60.
Lin, Y., Meng, Y., Wang, Y. X., Luo, J., Katsuma, K., Yang, C. W., et al. (2013). Vitellogenin receptor mutation leads to the oogenesis mutant phenotype “scanty vitellin” of the silkworm. Bombyx mori. J. Biol. Chem. 288, 13345–13355. doi: 10.1074/jbc.M113.462556
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, K., Shu, Y., Zhou, J., Zhang, X., Zhang, X., Chen, M., et al. (2015). Molecular characterization and RNA interference analysis of vitellogenin receptor from Nilaparvata lugens (Stal). J. Insect Physiol. 73, 20–29. doi: 10.1016/j.jinsphys.2015.01.007
Maddox, D. M., and Rhyne, M. (1975). Effects of Induced host-plant mineral deficiencies on attraction, feeding, and fecundity of the alligatorweed flea beetle. Enviro. Entomol. 4, 682–686. doi: 10.1093/ee/4.5.682
Nelson, C. M., Ihle, K. E., Fondrk, M. K. Jr., Page, R. E., and Amdam, G. V. (2013). The gene vitellogenin has multiple coordinating effects on social organization. Plos Biol 5:e62. doi: 10.1371/journal.pbio.0050062
Sappington, T. W., and Raikhel, A. S. (1998). Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect Biochem. Mol. Biol. 28, 277–300. doi: 10.1016/s0965-1748(97)00110-0
Shang, F., Niu, J. Z., Ding, B. Y., Zhang, Q., Ye, C., Zhang, W., et al. (2018). Vitellogenin and its receptor play essential roles in the development and reproduction of the brown citrus aphid, Aphis (Toxoptera) citricidus. Insect Mol. Biol. 27, 221–233. doi: 10.1111/imb.12366
Shi, J. F., Mu, L. L., Chen, X., Guo, W. C., and Li, G. Q. (2016). RNA interference of chitin synthase genes inhibits chitin biosynthesis and affects larval performance in Leptinotarsa decemlineata (Say). Int. J. Biol. Sci. 12, 1319–1331. doi: 10.7150/ijbs.14464
Shu, Y. H., Wang, J. W., Lu, K., Zhou, J. L., Zhou, Q., and Zhang, G. R. (2011). The first vitellogenin receptor from a Lepidopteran insect: molecular characterization, expression patterns and RNA interference analysis. Insect Mol. Biol. 20, 61–73. doi: 10.1111/j.1365-2583.2010.01054.x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tufail, M., and Takeda, M. (2009). Insect vitellogenin/lipophorin receptors: molecular structures, role in oogenesis, and regulatory mechanisms. J. Insect Physiol. 55, 88–104. doi: 10.1016/j.jinsphys.2008.11.007
Wu, H., Jiang, F. Z., Guo, J. X., Yi, J. Q., Liu, J. B., Cao, Y. S., et al. (2018). Molecular Characterization and expression of vitellogenin and vitellogenin receptor of Thitarodes pui (Lepidoptera: Hepialidae), an insect on the tibetan plateau. J. Insect Sci. 18:23. doi: 10.1093/jisesa/iey010
Xu, C. Y., Zhang, W. J., Fu, C. Z., and Lu, B. R. (2003). Genetic diversity of alligator weed in China by RAPD analysis. Biodivers. Conserv. 12, 637–645.
Ye, W. H., Li, J., Cao, H. L., and Ge, X. J. (2010). Genetic uniformity of Alternanthera philoxeroides in South China. Weed Res. 43, 297–302. doi: 10.1046/j.1365-3180.2003.00346.x
Zhang, W., Ma, L., Xiao, H., Xie, B., Smagghe, G., Guo, Y., et al. (2016). Molecular characterization and function analysis of the vitellogenin receptor from the cotton bollworm. PloS One 11:e0155785. doi: 10.1371/journal.pone.0155785
Keywords: Agasicles hygrophila, vitellogenin receptor, RNAi, ovarian development, fecundity
Citation: Zhang H, Liu Y, Jin J, Zhou Z and Guo J (2019) Identification and Characterization of the Vitellogenin Receptor Gene and Its Role in Reproduction in the Alligatorweed Flea Beetle, Agasicles hygrophila. Front. Physiol. 10:969. doi: 10.3389/fphys.2019.00969
Received: 08 April 2019; Accepted: 11 July 2019;
Published: 31 July 2019.
Edited by:
Fernando Ariel Genta, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Qi Fang, Zhejiang University, ChinaKatia Calp Gondim, Federal University of Rio de Janeiro, Brazil
Copyright © 2019 Zhang, Liu, Jin, Zhou and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianying Guo, Z3VvamlhbnlpbmdAY2Fhcy5jbg==
 Hong Zhang
Hong Zhang Yiran Liu
Yiran Liu