- 1Department of Plant Protection, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Department of Entomology, University of Kentucky, Lexington, KY, United States
The intersex (ix) gene acts in concert with doublesex (dsx) at the end of the sex determination hierarchy to control somatic sexual differentiation in Drosophila melanogaster. Here, we report the Drosophila ix homolog in Bemisia tabaci (Btix) with differential splicing events. Four isoforms were found in B. tabaci adults, including two sex-specific transcripts (BtixF and BtixM). Knockdown of Btix had no measurable effects on female morphological phenotypes but reduced the expression of the vitellogenin gene and resulted in the production of significantly fewer eggs, a lower eclosion rate, and a shorter body size of female progeny in comparison with control females. These results increase our understanding of the genes underlying sex determination in B. tabaci and reveal a potential target for RNA interference-based pest management.
Introduction
Insect sex determination, which affects the development and reproduction of insects, is the result of long-term natural selection. Through long-term natural selection, insects have developed a great variety of sex determination mechanisms. Even in closely related species, the primary signals are different, for example, dosage of the X-signaling element (XSE) in Drosophila melanogaster (Erickson and Quintero, 2007), maternal input of transformer (tra) messenger RNA (mRNA) in Ceratitis capitata (Pane et al., 2002), and also in Musca domestica (Hediger et al., 2010) and Nasonia vitripennis (Verhulst et al., 2010), heterozygosity of the complementary sex determiner (csd) locus in Apis mellifera (Beye et al., 2003), female-specific piRNA in Bombyx mori (Kiuchi et al., 2014), a dominant male-determining factor (M factor), the Nix gene in Aedes aegypti (Hall et al., 2015), the Guy1 gene in Anopheles stephensi (Criscione et al., 2016), the Yob gene in Anopheles gambiae (Krzywinska et al., 2016), the Mdmd gene in M. domestica (Sharma et al., 2017), the MoY gene in Ceratitis capitate, and the MoY orthologs in Bactrocera oleae and Bactrocera dorsalis (Meccariello et al., 2019). Nevertheless, recent studies have indicated that some of the downstream sex determination genes are generally conserved, such as doublesex (dsx) and intersex (ix) (Sánchez, 2008).
Dsx, a downstream effector of the sex determination cascade, produces male- or female-specific proteins via sex-specific splicing, controlling sexually dimorphic characters in different insect species (Shukla and Nagaraju, 2010). Ix, a gene required for female sexual development in Drosophila, acts together with dsx to regulate terminal differentiation. The gene ix has been characterized in Drosophila virilis, Megaselia scalaris, B. mori, Oncopeltus fasciatus, and Cyclommatus metallifer (Garrett-Engele et al., 2002; Sánchez, 2008; Aspiras et al., 2011; Gotoh et al., 2016). In D. melanogaster, ix is intronless and produce only one mRNA in all tissues of both males and females (Garrett-Engele et al., 2002). However, in Maruca vitrata and B. mori, ix is a multiexon gene with alternative splicing (AS) (Cavaliere et al., 2009; Arunkumar and Nagaraju, 2011). Dmix Bmix and Mvix mRNAs encode highly related and similarly sized proteins. Basic Local Alignment Search Tool (BLAST) analysis revealed 71–74% similarity among these three proteins. The IX proteins exhibit conserved organization at the amino (N)-terminal region similar to known transcriptional activation domains (Garrett-Engele et al., 2002). Functional conservation of ix has been demonstrated through heterologous expression of other Diptera and Lepidoptera ix homologs in transgenic D. melanogaster (Siegal and Baker, 2005; Cavaliere et al., 2009). RNA interference (RNAi) experiments with O. fasciatus (Hemiptera) and C. metallifer (Coleoptera) also indicated that ix has a conserved role in the sex determination pathway (Aspiras et al., 2011; Gotoh et al., 2016).
RNAi is a posttranscriptional gene-silencing mechanism triggered by the introduction of double-stranded RNAs (dsRNAs) and is ubiquitous in many eukaryotes, including insects (Wilson and Doudna, 2013). RNAi has become an effective technique for studying gene function in insects and provides a new method for insect pest control. The RNAi-mediated insect pest management strategy is environment friendly and highly specific. It has great application potential in both species-specific biopesticides and transgenic plants (Burand and Hunter, 2013).
The whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) is a worldwide agricultural pest with a haplodiploid reproductive system (Blackman and Cahill, 1998). Our laboratory previously identified 26 genes putatively associated with sex determination in B. tabaci and found that da, mle, and PSI undergo sex-specific splicing in B. tabaci, while Imp, dsx, and tra2 do not (Liu et al., 2016, 2019; Guo et al., 2018). Using RNAi experiments, we also found that Btdsx and Bttra2 affect each other and are important for male genitalia formation (Guo et al., 2018). Btdsx regulates the expression of Vg in B. tabaci females (Guo et al., 2018). Bttra directly regulates the expression of Btfru and Btdsx. Bttra and Btdsx affect each other, and autoregulation of tra may exist in B. tabaci (unpublished). In this study, we cloned and characterized the ix gene in B. tabaci, explored the AS events of Btix in adult insects, and investigated the functions in regulating Vg expression and reducing female fecundity. The results suggest the potential of Btix as a target for RNAi-based pest management.
Experimental Procedures
Insect Strains
The Q B. tabaci (Mediterranean, MED) population was originally collected from the fields in Beijing, China, in 2009, and was reared on cotton plants (Gossypium herbaceum L. cv. Zhongmian 49) in a glasshouse under natural light after isolation. The purity of the strain was monitored every two to three generations using a mitochondrial cytochrome oxidase I (mtCOI) marker (Chu et al., 2005). Samples of newly emerged adult females and males (within 1 h) were separately collected using a glass tube (5.0 × 0.5 cm), and the sex of each individual was determined with a stereomicroscope.
Gene Cloning and Identification of Alternative Splicing Variants
The annotated sequences obtained from our previous study (Liu et al., 2019) were used to design full-length primers to amplify the Btix gene (Table 1). The PCRs were performed using Es-Taq Master Mix (CWBIO, Beijing, China) under the following conditions: initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. The products were purified by a Monarch DNA Gel Extraction Kit (NEB, Beijing, China), cloned into the pEASY-T1 vector (TransGen, Beijing, China), and sequenced. For AS analysis, complementary DNA (cDNA) from newly emerged female or male samples was used. The products were purified, cloned, and sequenced as described previously.
Phylogenetic Analysis and Sequence Alignment
Sequences were aligned using the program ClustalW with the following default gap penalty parameters: a gap opening of 10 and an extension of 0.2. A neighbor-joining (NJ) tree was constructed using the program MEGA 6.0 with a p-distance model and pairwise deletion of gaps (Tamura et al., 2013). The bootstrap support of tree branches was assessed by resampling amino acid positions 1,000 times.
The open reading frame (ORF) of the target nucleotide sequence was predicted by the ORF Finder tool on the National Center for Biotechnology Information (NCBI) website. The deduced protein domains were determined using the NCBI Conserved Domain Search Service. Molecular weights (MWs) and isoelectric points were analyzed via the ExPASy ProtParam tool. Sequence-similarity analyses were performed with DNAMAN 6.0. The amino acid sequences of IX were aligned using the ClustalW program and then shaded by the ExPASy BOXSHADE tool.
RNA Isolation
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. RNA was quantified using a NanoDrop 2000 (Thermo Scientific, Wilmington, DE, United States), and integrity was checked with 1% Tris/borate/ethylenediaminetetraacetic acid (EDTA) (TBE) agarose gel electrophoresis. For gene cloning and quantitative PCR (qPCR) analysis, first-strand cDNA was synthesized using 1 μg of total RNA with the PrimeScript® RT reagent kit (TaKaRa Biotech, Kyoto, Japan) following the manufacturer’s recommendations. The synthesized first-strand cDNA was used immediately or stored at −20°C for later use.
Real-Time Quantitative PCR
Quantitative PCR was performed using SuperReal PreMix Plus (SYBR Green) (Tiangen, Beijing, China) according to the manufacturer’s instructions. Each reaction contained 1 μl of cDNA template, 10 μl of SuperReal PreMix Plus, 0.4 μl of ROX reference dye, and 0.5 μl of specific primers in a total volume of 20 μl. PCR was performed under the following conditions: denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. The specific primers are shown in Table 1. Three independent biological replicates were included for each treatment. The results were standardized to the expression level of the B. tabaci SDHA gene (Li et al., 2013), and the relative differences in transcript levels were analyzed by the 2–△△Ct method (Livak and Schmittgen, 2001).
RNA Interference
Primers (Table 1) containing a T7 promoter sequence were designed to amplify dsRNA targeting Btix. The dsRNA for enhanced green fluorescent protein (EGFP) was used as the negative control. The primers are listed in Table 1. All dsRNAs were prepared using the T7 RibomaxTM Express RNAi system (Promega, Madison, WI, United States). RNAi was achieved by directly feeding dsRNA to B. tabaci adults in a feeding chamber. A 0.20-ml drop of diet solution [5% yeast extract and 30% sucrose (wt/vol)] containing 0.5 μg/μl dsRNA was placed in the chamber (Upadhyay et al., 2011; Yang et al., 2017). Approximately 40 newly emerged females were introduced into an environmental chamber at 25°C under a photoperiod of L14/D10 and a relative humidity (RH) of 70%. Each sample was represented by three technical replicates. After 2 days of feeding, mortality was recorded, and B. tabaci specimens were collected. To determine the effects of silencing Btix on Vg expression and female development, we observed the RNAi females for 3 days. The phenotype of B. tabaci was observed by a stereomicroscope (Leica, M205C, Germany), and the expression levels of targeted genes were analyzed by real-time qPCR (RT-qPCR). The detail was as follows: first, ∼1,000 newly emerged B. tabaci females were observed to make sure that all individuals were wild type. Second, ∼500 newly emerged females were fed on dsBtix, and the others were fed on dsEGFP. Third, after 2 days of feeding, females were observed immediately or at 24, 48, 72 h with a stereomicroscope, and then were collected for qPCR analysis. Each replicate contained 40 females and 3 replicates per treatment. One-way ANOVA with Tukey’s test (P < 0.05) were used to evaluate differences among treatments.
Fecundity of B. tabaci
To determine the effects of silencing Btix on B. tabaci reproduction, after 2 days of dsRNA feeding, females were placed in a plastic bottle (5.5 cm diameter, 15 cm height) with a cotton seedling (Supplementary Figure S1). Each bottle contained five RNAi females and five newly emerged males (20 replicates per treatment). After 3 days of oviposition, the number of B. tabaci individuals in each bottle was determined, and their eggs were counted using a stereomicroscope. B. tabaci reproduction was reported as the mean number of eggs per surviving whitefly over 3 days (7 days of oviposition was also measured). The eclosion rates were calculated as the total number of emerging adults divided by the number of eggs in each bottle. Newly emerged progenies (females and males) from each treatment were randomly selected and frozen (approximately 90 females and 90 males per treatment). Thereafter, the randomly selected adults were laid on their back, and their length was measured from the top of their head to the end of their abdomen with the aid of a stereomicroscope (Leica, M205C, Germany). Student’s t-test (P < 0.05) were used to evaluate differences among treatments. SPSS version 19.0 (SPSS Inc., Chicago, IL, United States) was used for statistical analyses.
Results
Cloning and Characterization of Btix
Based on our previously annotated sequence from the Q/MED B. tabaci genome database (Xie et al., 2017; Liu et al., 2019), the full-length cDNA sequence of the Btix gene was obtained from PCR amplification, using adult cDNA as a template. The ORF encoded a protein of 181 amino acids with a predicted MW of ∼19.85 kDa and an isoelectric point (pI) of 5.65. In contrast to ix in D. melanogaster and hymenopterans, which is a single-exon gene, Btix possesses four exons (Figure 1A and Supplementary Figure S2).
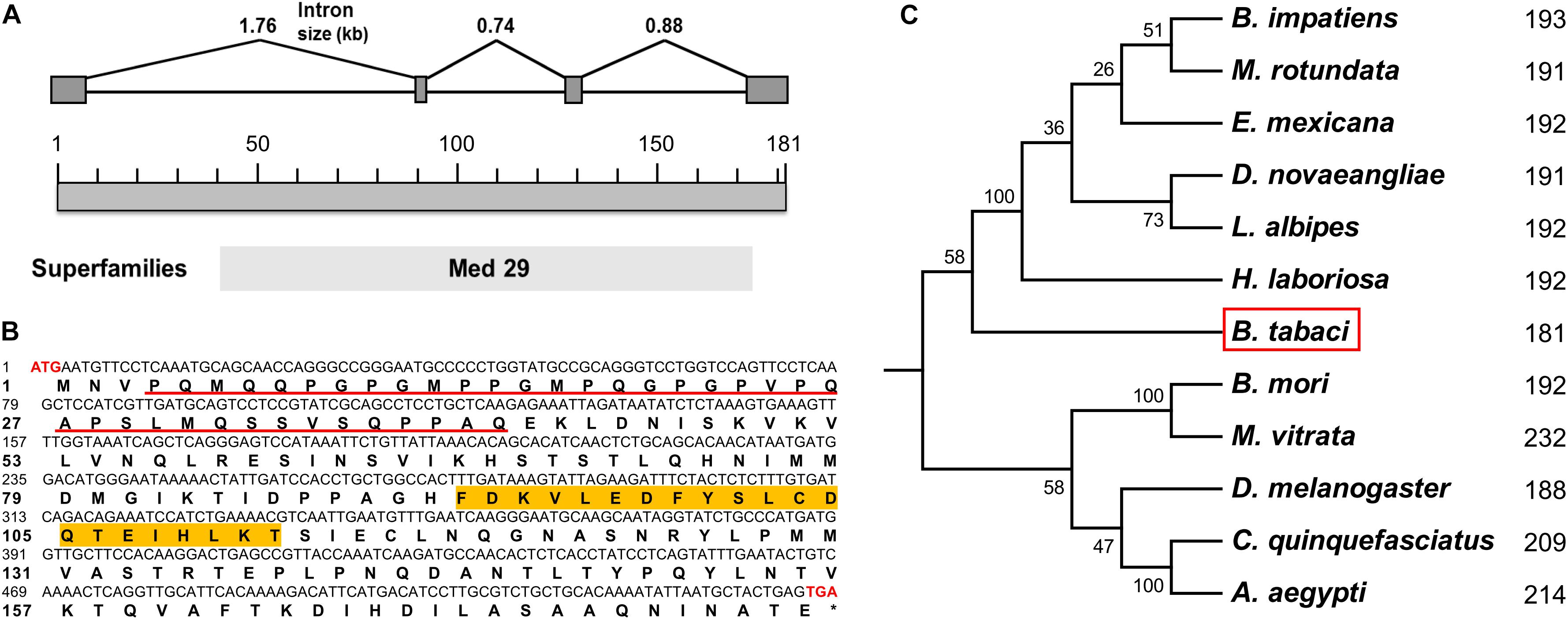
Figure 1. Characteristics of Btix. (A) Genomic structure of the Btix gene in B. tabaci. Gray boxes and the spaces between boxes indicate the exons and introns, respectively. The size of the putative exons and introns is drawn to scale. (B) Complementary DNA (cDNA) nucleotides and deduced amino acid sequence of the Btix gene. The start (ATG) and stop (TGA) codons are highlighted with red bold characters. The glutamine-, glycine-, proline-, and serine-rich regions are underlined, and the DSXF binding region is highlighted in orange. (C) Phylogenetic tree of intersex proteins among several insects. Phylogenetic tree of known IX proteins constructed by the neighbor-joining (NJ) method. Each number corresponds to the length of IX for each species.
BtIX contains glutamine-, glycine-, proline-, and serine-rich regions in the N-terminus (corresponding to positions 4–41), and in the C-terminus, there are several stretches of high amino acid identity (with the most striking such block corresponding to residues 92–112), which is a conserved feature of the IX protein (Figure 1B and Supplementary Figure S3).
Phylogenetic Analysis of Btix
We identified putative homologs of IX in three dipteran, two lepidopteran, and six hymenopteran species by searching the BeeBase (Elsik et al., 2016) or NCBI website. Protein sequences were aligned, and an NJ phylogenetic tree was constructed. The resulting tree clustered the IX proteins within each insect order, and BtIX was closely related to the IX from Hymenoptera (Figure 1C).
Splice Variation of Btix
To analyze splice variations of Btix, we amplified mRNA products from newly emerged female and male adults using full-length primers (Table 1). A total of four isoforms were found in B. tabaci adults: two non-sex-specific isoforms (BtixC), one female-specific isoform (BtixF), and one male-specific isoform (BtixM) (Figure 2). The four isoforms encoded four proteins with different sizes, and among them, only BtixM encoded a truncated non-functional protein (Supplementary Figure S4).
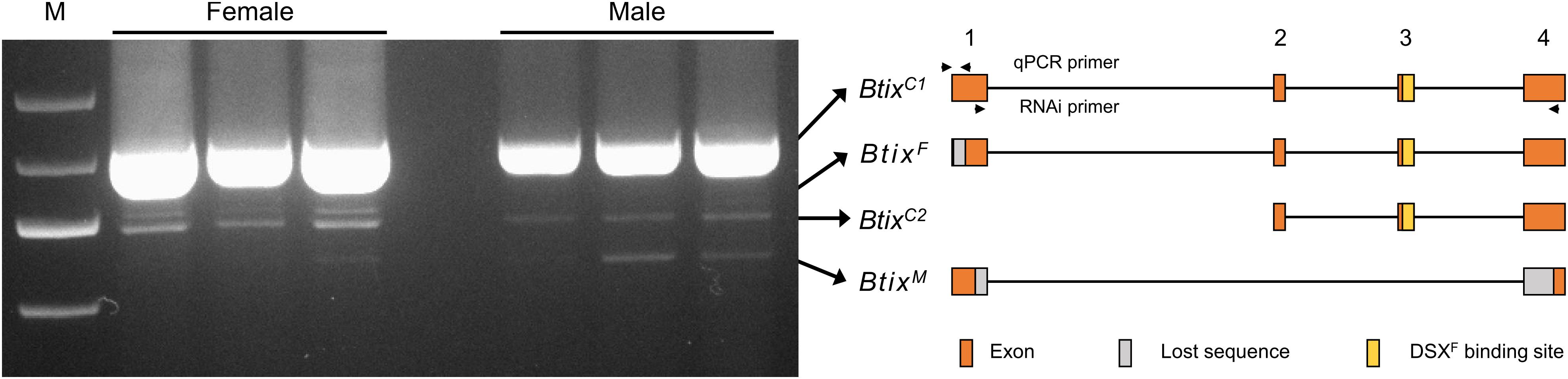
Figure 2. Alternative splicing (AS) analysis of Btix. Photograph of an electrophoretic gel showing real-time PCR (RT-PCR) amplification of Btix of adult male and female total messenger RNA (mRNA) samples. Structural diagrams of the four Btix isoforms found in B. tabaci adults are shown on the right. BtixC, non-sex-specific isoform; BtixF, female-specific isoform; BtixM, male-specific isoform. Orange boxes: exons; gray boxes: lost sequences; yellow boxes: DSXF binding sites.
Btix RNA Interference
To determine the function of Btix, we used RNAi technology to knock down the expression of Btix in the newly emerged female adults of B. tabaci. For dsRNA feeding, RT-qPCR results showed that Btix expression was significantly decreased at 2 days of feeding (77.2%, P < 0.01; Figure 3A), and the survival rate was more than 95% (Supplementary Figure S5). Furthermore, over the next 3 days, the Vg expression in the dsRNA-fed group decreased to varying degrees, particularly on the first (P < 0.01) and 3rd days (P = 0.018) after RNAi (Figure 3B). After 3 days of observation, no phenotypic (body color and morphological defects of genitalia) changes were observed compared with control females (Supplementary Figure S6).
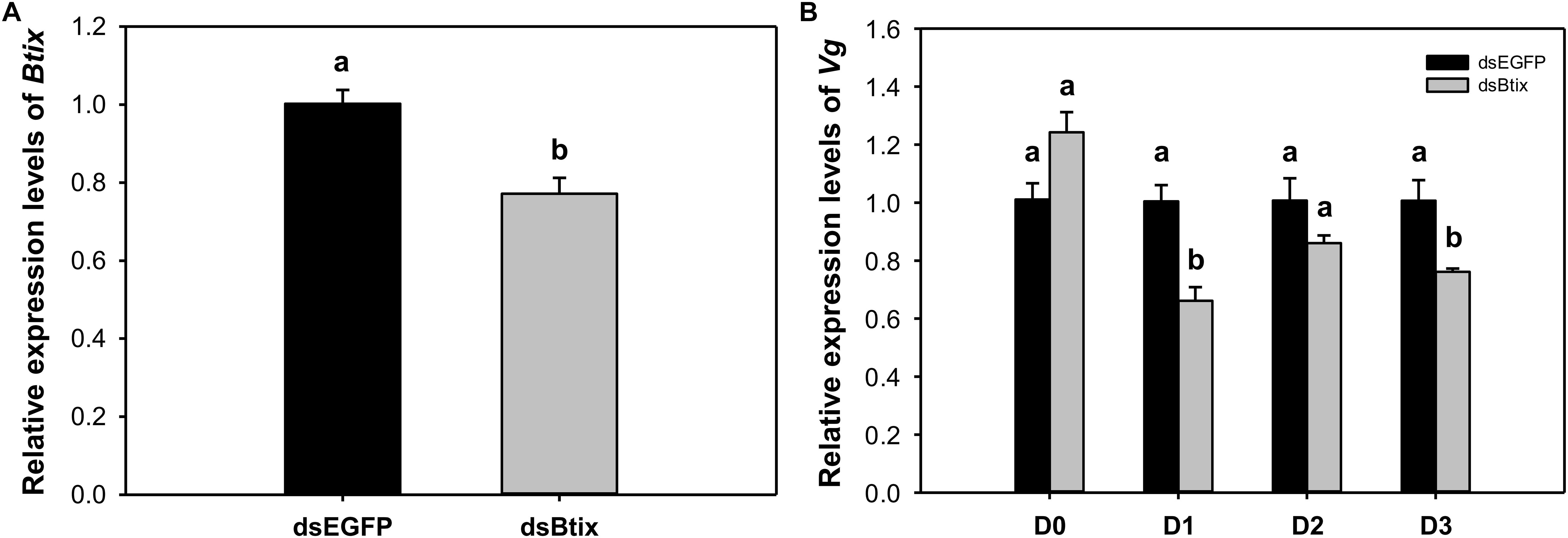
Figure 3. (A) The efficiency of RNA interference (RNAi) of Btix on newly emerged female adults. (B) Relative transcript levels of Vg in control and Btix double-stranded RNA (dsRNA)-fed females. Bars with the same letter are not significantly different from each other at P < 0.05 based on Tukey’s test. Each point represents the mean ± SE from three independent experiments.
Function of Btix in Female Reproduction
We found that knocking down the expression of the Btix gene affected female reproduction in B. tabaci. In Btix dsRNA-fed females, fecundity was significantly reduced by 19% (P < 0.01; Figures 4A,B), and the eclosion rate of progeny was reduced by 16% (P = 0.025; Figure 4C). The average body length of female progeny was also markedly shorter than that of control females (shortened by 7%) (P < 0.01; Figures 4D, 5).
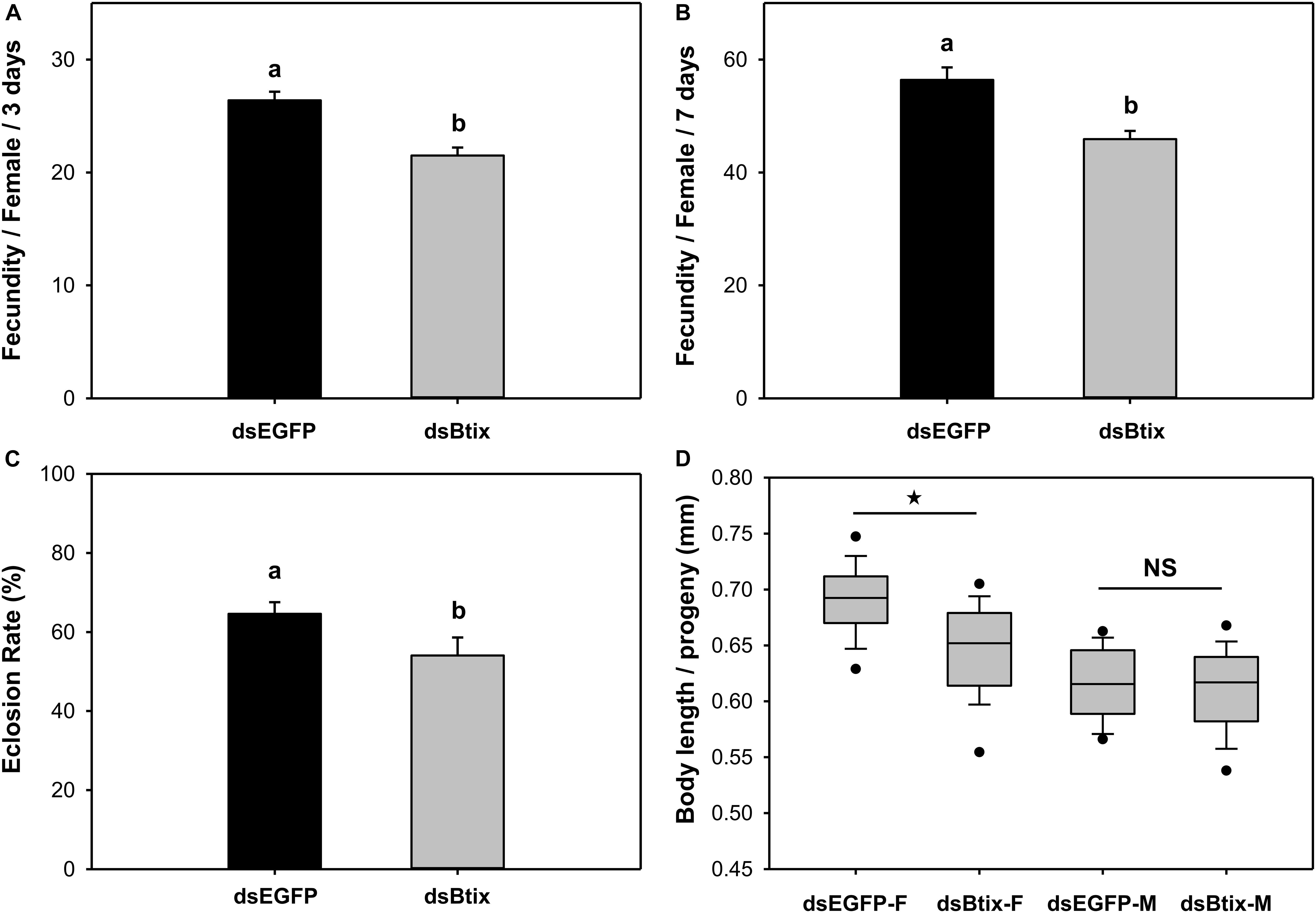
Figure 4. Effect of RNA interference (RNAi) of Btix on female reproductive physiology, including fecundity (A,B), eclosion rate of progeny (C), and the body length of progeny (D). Values shown in the figure are means and standard errors. Different letters indicate significant differences between treatments (P < 0.05; Student’s t-test). The box shows the 25th to the 75th quartiles, and the dark line in the box represents the median value; circles show outliers (5th/95th percentile).
Discussion
In this study, we cloned and characterized the ix gene in B. tabaci (Btix; Figures 1A,B) and found that Btix is structurally conserved. First, BtIX shared several modular elements with other insect IX proteins, including the amino acid composition of the N-terminus and high amino acid identity in the C-terminus (Figure 1B and Supplementary Figure S3). The former region resembles known transcriptional activation domains (Garrett-Engele et al., 2002), and the latter is considered an important region for DSXF binding (Yang et al., 2008). Second, the IX homologs were conserved in their short length. BtIX is only 181 amino acids long, and the D. melanogaster IX is 188 amino acids in length. The M. vitrata IX presents an additional region of 38 amino acids in the C-terminus (Cavaliere et al., 2009) and is the longest homolog, yet it is only 232 amino acids in length (Figure 1C).
ix is a single-exon gene and not sex-specifically spliced in Drosophila (Garrett-Engele et al., 2002). However, in M. vitrata and B. mori, the ix orthologs present, respectively, five and six exons (Cavaliere et al., 2009). A female-specific and pupae-stage specific ix transcript is found in M. vitrata, and a testis-specific splice form of ix is found in B. mori (Cavaliere et al., 2009; Arunkumar and Nagaraju, 2011). In B. tabaci, the Btix gene contains four exons; a female-specific isoform (BtixF) and a male-specific isoform (BtixM) were found in B. tabaci adults (Figure 2). However, we did not find any sex-specific exons, so we were unable to design specific qPCR primers to reveal the developmental and temporal expression patterns of the four isoforms. In addition, since BtixM only has 186 bp, we did not find a suitable RNAi fragment. Therefore, we only investigated the function of BtixF mainly in this paper.
The gene ix is a conserved downstream gene at the end of the sex determination cascade, second to dsx. In D. melanogaster, IX specifically interacts with DSXF, forming a DNA-binding complex to activate Yolk protein (Yp) gene expression and control female sexual differentiation (Garrett-Engele et al., 2002). Female ix mutants of D. melanogaster display characteristic morphological defects that are highly similar to those in dsx mutants (Chase and Baker, 1995; Garrett-Engele et al., 2002). Ectopic expression of D. virilis and M. scalaris ix cDNA fully rescues the D. melanogaster female ix mutants, and expression of B. mori and M. vitrata cDNA partially restores the mutants (Siegal and Baker, 2005; Cavaliere et al., 2009). Knockdown of ix in O. fasciatus produced defects in both sexes, reducing the size of both male and female genitalia (Aspiras et al., 2011). Depletion of ix in C. metallifer affected sex-specific morphological characters such as mandible size and body color in females only (Gotoh et al., 2016). These results highlight the conserved function of ix in insect sex determination. However, silencing Btix did not affect the morphological changes in B. tabaci females (Supplementary Figure S6). This may be because we treated the adults. Our lab is working on a new RNAi method for B. tabaci nymph and egg stage. When the method is mature, the role of Btix in sex determination will be better studied.
In this paper, the expression of Vg decreased significantly on the 1st and 3rd day after silencing Btix (Figure 3B). This result indicates that Btix plays an important role in regulating Vg expression. However, the detection of Vg expression immediately after RNAi had no effect. This may be because Vg is not a target gene for Btix. In D. melanogaster, yolk protein genes (Yp1 and Yp2) are the best characterized target genes of sex-specific DSX proteins and are affected by any change in the sex-determination pathway (Shukla and Nagaraju, 2010). DSXF regulates Yp expression and requires IX; similarly, the female-specific function of IX is mediated by DSXF (Waterbury et al., 1999; Garrett-Engele et al., 2002). To understand the interaction between Btix and Btdsx, we silenced Btdsx and Btix, respectively, to detect the expression of these two genes. The results showed that silencing Btdsx did not affect the expression of Btix and vice versa (Supplementary Figure S7). Since ix functions as a complex with DSX in Drosophila, the RNAi results can only indicate that there is no cascade control between Btix and Btdsx. Whether Btix combined with Btdsx participate in sex determination remain further explained by subsequent experiments.
The Vg gene plays a vital role in oocyte and embryo development in insects. Silencing of vitellogenin or vitellogenin receptor genes resulted in significant mortality and reduced fecundity in B. tabaci adults (Upadhyay et al., 2016; Grover et al., 2018). Therefore, it is reasonable to believe that Btix may affect reproduction in B. tabaci. In the present study, disruption of the expression of Btix in female adults reduced fecundity, eclosion rate of progeny, and shortened the body length of female progeny (Figure 4). These results suggest that Btix does have an effect on B. tabaci reproduction. In previous transgenic D. melanogaster tests, female ix mutants were sterile; despite expression of the B. mori ix, fertility was severely reduced (Siegal and Baker, 2005). These results showed that ix has an evolutionarily conserved function in the control of female fertility.
RNAi technology is promising for the control of insect pests; its main challenge is to find suitable target genes. In general, functional genes encoding essential proteins can be suitable RNAi targets. Downregulation of the expression of these genes can reduce the fitness or survival rate of insect pests (Katoch et al., 2013). For example, RNAi knockdown of the P450 CYP6CM1, GST, or HOT gene increased mortality in a B. tabaci-resistant strain (Grover et al., 2018). Knockdown of SexiCSP3 in Spodoptera exigua caused high mortality, low hatching rate, and reduced oviposition (Gong et al., 2012). Knockdown of genes in the ecdysone pathway reduced the survival and delayed the development of B. tabaci during nymphal stages (Luan et al., 2013). In this paper, we confirmed the important role of Btix in B. tabaci reproduction, indicating that Btix can be used as a potential target for RNAi-based pest management.
Conclusion
In the present study, we cloned and characterized the Btix gene from B. tabaci and provided evidence for the presence of alternatively spliced forms of ix in B. tabaci. Furthermore, silencing Btix downregulated Vg expression and had some negative effects on female reproduction in B. tabaci. Our findings indicate that ix is functionally conserved in the control of female fertility. In addition to expanding our understanding of the genes that may determine sex in B. tabaci, these results will help us develop new methods for the effective control of B. tabaci. The functional relationships of Btix and Btdsx, as well as the specific contribution of Btix in regulating the terminal differentiation of B. tabaci, remain to be further studied.
Data Availability Statement
The datasets generated for this study can be found in the GenBank: QAB02863.1.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This research was supported by the National Natural Science Foundation of China (31420103919, 31871970, and 31672032), the Beijing Nova Program (Z171100001117039), the China Postdoctoral Science Foundation (2018M641559), the China’s Agricultural Research System (CARS-24-C-02), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS), and the Beijing Key Laboratory for Pest Control and Sustainable Cultivation of Vegetables.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00055/full#supplementary-material
References
Arunkumar, K. P., and Nagaraju, J. (2011). Drosophila intersex, orthologue in the silkworm, Bombyx mori, and related species. Genetica 139, 141–147. doi: 10.1007/s10709-010-9529-x
Aspiras, A. C., Smith, F. W., and Angelini, D. R. (2011). Sex-specific gene interactions in the patterning of insect genitalia. Dev. Biol. 360, 369–380. doi: 10.1016/j.ydbio.2011.09.026
Beye, M., Hasselmann, M., Fondrk, K. M., Page, E. R., and Omholt, W. S. (2003). The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114, 419–429. doi: 10.1016/s0092-8674(03)00606-8
Blackman, R., and Cahill, M. (1998). The karyotype of Bemisia tabaci (Hemiptera: Aleyrodidae). B Entomol. Res. 88, 213–215. doi: 10.1017/S0007485300025785
Burand, J. P., and Hunter, W. B. (2013). RNAi: future in insect management. J. Invertebr. Pathol. 112, S68–S74. doi: 10.1016/j.jip.2012.07.012
Cavaliere, D., Cara, F. D., Polito, L. C., and Digilio, F. A. (2009). Cloning and functional characterization of the intersex homologous gene in the pest lepidopteron Maruca vitrata. Int. J. Dev. Biol. 53, 1057–1062. doi: 10.1387/ijdb.082840dc
Chase, B. A., and Baker, B. S. (1995). A genetic analysis of intersex, a gene regulating sexual differentiation in Drosophila melanogaster females. Genetics 139, 1649–1661.
Chu, D., Zhang, Y. J., Cong, B., Xu, B. Y., Wu, Q. J., and Zhu, G. R. (2005). Sequences analysis of mtDNA COI gene and molecular phylogeny of different geographical populations of Bemisia tabaci (Gennadius). Agr. Sci. China 4, 533–541.
Criscione, F., Qi, Y., and Tu, Z. (2016). GUY1 confers complete female lethality and is a strong candidate for a male-determining factor in Anopheles stephensi. eLife 5:e19281. doi: 10.7554/eLife.19281
Elsik, C. G., Tayal, A., Diesh, C. M., Unni, D. R., Emery, M. L., Nguyen, H. N., et al. (2016). Hymenoptera genome database: integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 44, 793–800. doi: 10.1093/nar/gkv1208
Erickson, J. W., and Quintero, J. J. (2007). Indirect effects of ploidy suggest X chromosome dose, not the X: a ratio, signals sex in Drosophila. PLoS Biol. 5:e332. doi: 10.1371/journal.pbio.0050332
Garrett-Engele, C. M., Siegal, M. L., Manoli, D. S., Williams, B. C., Li, H., and Baker, B. S. (2002). intersex, a gene required for female sexual development in Drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development 129, 4661–4675.
Gong, L., Luo, Q., Rizwan-Ul-Haq, M., and Hu, M. Y. (2012). Cloning and characterization of three chemosensory proteins from spodoptera exigua and effects of gene silencing on female survival and reproduction. B Entomol Res 102, 600–609. doi: 10.1017/S0007485312000168
Gotoh, H., Zinna, R. A., Warren, I., Denieu, M., Niimi, T., Dworkin, I., et al. (2016). Identification and functional analyses of sex determination genes in the sexually dimorphic stag beetle Cyclommatus metallifer. BMC Genomics 17:250. doi: 10.1186/s12864-016-2522-8
Grover, S., Jindal, V., Banta, G., Taning, C. N. T., Smagghe, G., and Christiaens, O. (2018). Potential of RNA interference in the study and management of the whitefly, Bemisia tabaci. Arch. Insect. Biochem. Physiol. 100:e21522. doi: 10.1002/arch.21522
Guo, L., Xie, W., Liu, Y., Yang, Z., Yang, X., Xia, J., et al. (2018). Identification and characterization of doublesex in Bemisia tabaci. Insect. Mol. Biol. 27, 620–632. doi: 10.1111/imb.12494
Hall, A. B., Basu, S., Jiang, X., Qi, Y., Timoshevskiy, V. A., Biedler, J. K., et al. (2015). A male-determining factor in the mosquito Aedes aegypti. Science 348, 1268–1270. doi: 10.1126/science.aaa2850
Hediger, M., Henggeler, C., Meier, N., Perez, R., Saccone, G., and Bopp, D. (2010). Molecular characterization of the key switch f provides a basis for understanding the rapid divergence of the sex-determining pathway in the housefly. Genetics 184, 155–170. doi: 10.1534/genetics.109.109249
Katoch, R., Sethi, A., Thakur, N., and Murdock, L. L. (2013). RNAi for insect control: current perspective and future challenges. Appl. Biochem. Biotechnol. 171, 847–873. doi: 10.1007/s12010-013-0399-4
Kiuchi, T., Koga, H., Kawamoto, M., Shoji, K., Sakai, H., Arai, Y., et al. (2014). A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509:633. doi: 10.1038/nature13315
Krzywinska, E., Dennison, N. J., Lycett, G. J., and Krzywinski, J. (2016). A maleness gene in the malaria mosquito Anopheles gambiae. Science 353, 67–69. doi: 10.1126/science.aaf5605
Li, R., Xie, W., Wang, S., Wu, Q., Yang, N., Yang, X., et al. (2013). Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS One 8:e53006. doi: 10.1371/journal.pone.0053006
Liu, Y., Xie, W., Guo, L., Yang, X., Yang, J., Wang, S., et al. (2019). Genome-wide dissection of sex determination genes in the highly invasive whitefly species Bemisia tabaci Q/MED. Insect. Mol. Biol 28, 509–519. doi: 10.1111/imb.12568
Liu, Y., Xie, W., Yang, X., Guo, L., Wang, S., Wu, Q., et al. (2016). Molecular cloning of the sex-related gene PSI in Bemisia tabaci and its alternative splicing properties. Gene 580, 104–110. doi: 10.1016/j.gene.2016.01.005
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luan, J. B., Ghanim, M., Liu, S. S., and Czosnek, H. (2013). Silencing the ecdysone synthesis and signaling pathway genes disrupts nymphal development in the whitefly. Insect Biochem. Mol. Biol. 43, 740–746. doi: 10.1016/j.ibmb.2013.05.012
Meccariello, A., Salvemini, M., Primo, P., Hall, B., Koskinioti, P., Dalikova, M., et al. (2019). Maleness-on-the-Y (MoY) orchestrates male sex determination in major agricultural fruit fly pests. Science 365, 1457–1460. doi: 10.1101/533646
Pane, A., Salvemini, M., Bovi, P. D., Polito, L. C., and Saccone, G. (2002). The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129, 3715–3725. doi: 10.1101/gad.1008902
Sánchez, L. (2008). Sex-determining mechanisms in insects. Int. J. Dev. Biol. 52, 837–856. doi: 10.1387/ijdb.072396ls
Sharma, A., Heinze, S. D., Wu, Y., Kohlbrenner, T., and Bopp, D. (2017). Male sex in houseflies is determined by Mdmd, a paralog of the generic splice factor gene CWC22. Science 356, 642–645. doi: 10.1126/science.aam5498
Shukla, J. N., and Nagaraju, J. (2010). Doublesex: a conserved downstream gene controlled by diverse upstream regulators. J. Genet. 89, 341–356. doi: 10.1007/s12041-010-0046-6
Siegal, M. L., and Baker, B. S. (2005). Functional conservation and divergence of intersex, a gene required for female differentiation in Drosophila melanogaster. Dev. Genes Evol. 215, 1–12. doi: 10.1007/s00427-004-0445-x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Upadhyay, S. K., Chandrashekar, K., Thakur, N., Verma, P. C., Borgio, J. F., Singh, P. K., et al. (2011). RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosciences 36, 153–161. doi: 10.1007/s12038-011-9009-1
Upadhyay, S. K., Singh, H., Dixit, S., Mendu, V., and Verma, P. C. (2016). Molecular characterization of vitellogenin and vitellogenin receptor of Bemisia tabaci. PLoS One 11:e0155306. doi: 10.1371/journal.pone.0155306
Verhulst, E. C., Beukeboom, L. W., and van de Zande, L. (2010). Maternal control of haplodiploid sex determination in the wasp Nasonia. Science 328, 620–623. doi: 10.1126/science.1185805
Waterbury, J. A., Jackson, L. L., and Schedl, P. (1999). Analysis of the doublesex female protein in Drosophila melanogaster. Role in sexual differentiation and behavior and dependence on Intersex. Genetics 152, 1653–1667.
Wilson, R. C., and Doudna, J. A. (2013). Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 42, 217–239. doi: 10.1146/annurev-biophys-083012-130404
Xie, W., Chen, C., Yang, Z., Guo, L., Yang, X., Wang, D., et al. (2017). Genome sequencing of the sweetpotato whitefly Bemisia tabaci MED/Q. GigaScience 6, 1–7. doi: 10.1093/gigascience/gix018
Yang, X., Xie, W., Li, R. M., Zhou, X. M., Wang, S. L., Wu, Q. J., et al. (2017). RNA interference-mediated knockdown of the hydroxyacid-oxoacid transhydrogenase gene decreases thiamethoxam resistance in adults of the whitefly Bemisia tabaci. Sci. Rep. 7:41201. doi: 10.1038/srep41201
Keywords: Bemisia tabaci, intersex, RNA interference, sex determination, pest control
Citation: Liu Y, Yang J, Huo Z, Wang S, Wu Q, Zhou X, Xie W and Zhang Y (2020) Characteristic and Functional Study of Intersex, a Gene Related to Female Fertility in Bemisia tabaci. Front. Physiol. 11:55. doi: 10.3389/fphys.2020.00055
Received: 30 October 2019; Accepted: 21 January 2020;
Published: 25 February 2020.
Edited by:
Peng He, Guizhou University, ChinaReviewed by:
Xiaoling Tan, Chinese Academy of Agricultural Sciences, ChinaHongbo Jiang, Southwest University, China
Giuseppe Saccone, University of Naples Federico II, Italy
Copyright © 2020 Liu, Yang, Huo, Wang, Wu, Zhou, Xie and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Xie, eGlld2VuQGNhYXMuY24=; Youjun Zhang, emhhbmd5b3VqdW5AY2Fhcy5jbg==
†These authors have contributed equally to this work
 Yating Liu1†
Yating Liu1† Shaoli Wang
Shaoli Wang Qingjun Wu
Qingjun Wu Xuguo Zhou
Xuguo Zhou Wen Xie
Wen Xie Youjun Zhang
Youjun Zhang