- 1State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Plant Protection, Agricultural University of Hebei, Baoding, China
- 3College of Plant Protection, China Agricultural University, Beijing, China
Apolygus lucorum (Hemiptera: Miridae), one of the main insect pests, causes severe damage in cotton and many other economic crops. As is well-known, legs play important roles in the chemoreception of insects. In this study, the putative chemosensory proteins in legs of A. lucorum involved in close or contact chemical communication of adult bugs were investigated using RNA transcriptome sequencing and qPCR methods. Transcriptome data of forelegs, middle legs and hind legs of adult bugs demonstrated that 20 odorant binding protein (OBP) genes, eight chemosensory protein (CSP) genes, one odorant receptor (OR) gene, one ionotropic receptor (IR) gene and one sensory neuron membrane protein (SNMP) gene were identified in legs of A. lucorum. Compared to the previous antennae transcriptome data, five CSPs, IR21a and SNMP2a were newly identified in legs. Results of qPCR analysis indicated that all these putative chemosensory genes were ubiquitously expressed in forelegs, middle legs and hind legs of bugs. Furthermore, four types of sensilla on legs of A. lucorum including sensilla trichodea (subtypes: long straight sensilla trichodea, Str1; long curved sensilla trichodea, Str2), sensilla chaetica (subtypes: sensilla chaetica 1, Sch1; sensilla chaetica 2, Sch2; and sensilla chaetica 3, Sch3), sensilla basiconca (subtypes: medium-long sensilla basiconca, Sba1; short sensilla basiconca, Sba2) and Böhm bristles (BB) were found using scanning electron microscopy. Additionally, the largest number of sensilla was observed on hind legs, while the forelegs had the smallest number of sensilla. Our data provide valuable insights into understanding the chemoreception of legs in A. lucorum.
Introduction
Insects rely on their sensitive olfactory and gustatory organs to find hosts, forage, lay eggs and mate (Romani et al., 2005; Li and Liberles, 2015). Lots of chemosensory sensilla are independently distributed in various organs of insects such as antennae, mouthparts, legs, wings and ovipositors. Insect legs sense chemical signals when the insects land on the host (Klijnstra and Roessingh, 1986; Maher et al., 2006). Legs of Drosophila are involved in making the preliminary contact with food resources and non-volatile pheromones (Frederick et al., 2014; Koh et al., 2014). Likewise, legs of Helicoverpa armigera can sense some salts, sugars and amino acids (Zhang et al., 2010). Generally, chemosensory associated proteins in insect antennae such as odorant binding proteins (OBPs), chemosensory proteins (CSPs), Niemann-Pick type C2 proteins (NPC2s), odorant receptors (ORs), gustatory receptors (GRs), ionotropic receptors (IRs) and sensory neuron membrane proteins (SNMPs) play crucial roles in olfactory and gustatory behaviors of insects (Pelosi et al., 2006; Wynand and Carlson, 2006; Abdel-Latief, 2007; Wanner and Robertson, 2008; Benton et al., 2009; Touhara and Vosshall, 2009; Vogt et al., 2009; Leal, 2013; Adachi et al., 2014). These chemosensory associated proteins are usually also expressed in insect legs. By transcriptome sequencing, OBPs, CSPs, SNMPs, GRs, and ORs were identified in the legs of Apis cerana cerana, Ectropis obliqua, and Adelphocoris lineolatus (Ma et al., 2016; Sun et al., 2017; Du et al., 2019).
OBPs as carrier proteins play an indispensable role in transport of chemical signals through the aqueous sensilla lymph to the olfactory receptor cells in insects (Calvello et al., 2003; Pelosi et al., 2006). OBPs can recognize and distinguish different odorant messages (Schultze et al., 2012). LUSH, one of the Drosophila OBPs, is required for activation of pheromone-sensitive chemosensory neurons (Xu et al., 2005). Additionally, OBPs are essential for mediating olfactory behavioral responses. When the expression of OBPs was suppressed in flies, their behavioral responses to odorants were changed (Swarup et al., 2011). CSPs highly expressed in the sensilla lymph exhibit binding activities to odorants and pheromones, suggesting their olfactory functions (Pelosi et al., 2006; Hua et al., 2013; Sun et al., 2015).
Insect ORs play crucial roles in detecting the volatile molecules, especially in the long distance perception (Missbach et al., 2014). Generally, a typical OR unit functions as a dimer complex with the highly conserved odorant receptor co-receptor (Orco) among insect species (Wicher et al., 2008). In insects, GRs expressed in gustatory neurons acts as taste receptors, which are responsible for feeding behaviors (Dunipace et al., 2001; Scott et al., 2001). As a large and highly divergent family of ionotropic glutamate receptors in Drosophila, IRs are subdivided into “antennal IRs,” which are expressed in antennae specifically and mainly involved in olfactory recognition; and “divergent IRs,” which are found in various tissues taking responsible for sensation of taste (Jaeger et al., 2018; Lee et al., 2018; He et al., 2019; Rimal et al., 2019). Various chemosensory associated proteins in insect play diverse functions, such as those listed in Table 1.
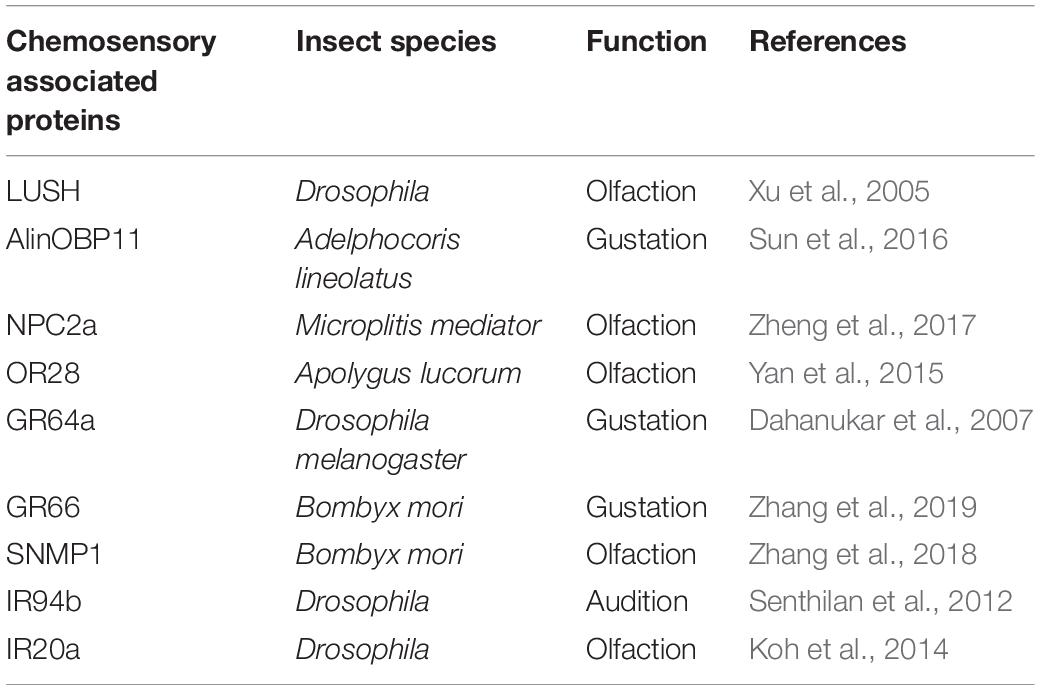
Table 1. Diverse functions of chemosensory associated proteins in insects reported in previous studies.
There are different types of chemosensory sensilla distributed on insect legs. Gustatory sensilla such as contact sensilla chaetica on tarsi enable insects to perceive taste substances on host plant surfaces (Ave et al., 1978). Enormous amount of sensilla were observed on the antennae of Lygus lineolaris and Apolygus lucorum (Chinta et al., 1997; Lu et al., 2007). However, little is known about the sensilla types of legs of mirid bugs so far.
The A. lucorum, one of the dominant mirid bug species, has become a major pest in cotton fields along with the widespread cultivation of Bt transgenic cotton, resulting in substantial economic losses (Lu et al., 2010). Moreover, it is very difficult to control A. lucorum due to their polyphagous host-feeding and host transfer behaviors (Lu and Wu, 2008). In the present study, transcriptome sequencings of forelegs, middle legs and hind legs form adult A. lucorum were performed to identify the candidate chemosensory genes. The tissue- and sex-biased expression patterns of the putative chemosensory genes were assessed by conducting quantitative real-time PCR (qPCR). Moreover, the sensilla types on legs of adult male and female bugs were observed and characterized. Our finds provide valuable insights into understandings the roles of mirid bug legs in olfaction and gustation.
Materials and Methods
Insects Rearing and Tissue Collection
Nymphs and adults of A. lucorum were collected from cotton fields at the Langfang Experimental Station of Chinese Academy of Agricultural Sciences, Hebei Province (39.53° N, 116.70° E), China. Laboratory colonies feeding on green beans (Phaseolus vulgaris L.) were bred in climate chambers under following conditions: 29 ± 1°C, relative humidity (RH) 60 ± 5% and 14: 10 light: dark (L: D) photoperiod (An et al., 2016).
For transcriptome sequencing, 500 forelegs, 500 middle legs and 500 hind legs were collected from 5 days old adult bugs of both sexes. In qPCR measurement, forelegs, middle legs and hind legs samples from A. lucorum of both sexes were separately collected and immediately immersed in liquid nitrogen till to use.
cDNA Library Construction and Transcriptome Sequencing
Total RNAs were isolated from samples using Trizol regent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions. RNA purity was checked using NanoDrop® spectrophotometers (Thermo Fisher, Waltham, MA, United States). RNA integrity was assessed using the RNA Nano 6000 Assay Kit (Agilent Technologies, Santa Clara, CA, United States). The cDNA library construction and transcriptome sequencing were carried out by Sinobiocore Bioinformatics Technology Co. Ltd on an Illumina HiSeq 2500 platform (Beijing, China).
A total amount of 1 μg RNA per sample was used as input material for the library preparation. The sequencing libraries were generated using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, United States). Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was performed using divalent cations under elevated temperature in an Illumina proprietary fragmentation buffer. First strand cDNA was synthesized using random oligonucleotides and SuperScript II (Thermo Fisher, Waltham, MA, United States). Second strand cDNA synthesis was performed using DNA Polymerase I and RNase H. Then, the cDNA fragments were end repaired with the addition of a single ‘A’ base at the 3′-end of each strand, ligated with the special sequencing adapters subsequently. The products were purified and size selected in order to get appropriate size for sequencing. Finally, PCRs were performed and aimed products were purified.
Library concentration was measured using Qubitp® RNA Assay Kit in Qubitp® 3.0 (Thermo Fisher, Waltham, MA, United States) to preliminary quantify. Insert size was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States). When the insert size was in consistent with expectations, qualified insert size was accurately quantified using qPCR by Step One Plus realtime PCR system (Applied Biosystems, Foster City, CA, United States). The clustering of the index-coded samples was performed on a cBot Cluster Generation System (Illumia, San Diego, CA, United States) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform with 150 bp paired-end module.
Transcriptome Assembly and Functional Annotation
De novo transcriptome assembly was performed using Trinity1. A K-mer library was constructed with the filtered reads, and the contigs were formed using Inchworm. Cuffdiff (v2.2.1) was used to calculate FPKMs for coding genes in each sample. Gene FPKMs were computed by summing the FPKMs of the transcripts in each gene group. FPKM stands for “fragments per kilobase of exon per million fragments mapped,” and it is calculated based on the length of the fragments and the reads count mapped to each fragment (Anders et al., 2015). To annotate the unigenes, blastx and blastn searches were performed against the database of Nt, Nr, SwissProt, and eggNOG (e-value < 10–5, bitscore > 60)2. The blast results were then imported into Blast2GO pipeline for Go annotations.
Differential Expression Analysis
Deseq2 provides statistical routines for determining differential expression in digital transcript or gene expression datasets using a model based on a negative binomial distribution. Genes with corrected p-values less than 0.05 and the absolute value of log2 (fold change) < 1.0 were assigned as significantly differentially expressed (Love et al., 2014). Differentially expressed genes (DEGs) were identified by Benjamini and Hochberg FDR according to statistically significant differences with the threshold of false discovery rates (FDR) < 0.05 and Fold Change ≥ 2 (Leng et al., 2013; Nikolayeva and Robinson, 2014).
Identification of Putative Chemosensory Genes
In addition to keywords searching, a FASTA file of unigenes was created from a local nucleotide database file using the BioEdit Sequence Alignment Editor program 7.13, and the local BLASTN program was performed using available A. lucorum antennae chemosensory genes (unpublished data) as the queries. Candidate unigenes encoding putative chemosensory genes were used BLASTX to search in NCBI website (Zhou et al., 2010).
Verification of Candidate Genes
All the sequences of candidate chemosensory genes from transcriptome were further confirmed by gene cloning and sequencing. Gene-specific primers (Supplementary Table S1) amplifying the full length or partial sequences of target genes were designed using Primer Premier 5.0 (PREMIER Biosoft International, Palo Alto, CA, United States). PCR reactions were carried out in a volume of 50 μl with 200 ng cDNA template of each sample and 1 μl TransStartp® FastPfu DNA Polymerase (TransGen Biotech, Beijing, China). The PCR parameters were: 95°C for 1 min, followed by 40 cycles of 95°C for 20 s, 55°C for 20 s, 72°C for 1 min, and a final extension at 72°C for 5 min. PCR products were subsequently gel-purified and cloned into pEASY®-Blunt Cloning vector (TransGen Biotech, Beijing, China) and then sequenced with standard M13 primers.
Phylogenetic Analysis of CSPs
Multiple alignments of the complete CSPs amino acid sequences were performed by ClustalX 2.0 and further edited by GeneDoc 2.7. The phylogenetic tree was constructed by MEGA 7.0 using the Neighbor-joining method with a p-distance model and a pairwise deletion of gaps. Bootstrap support was assessed by a bootstrap procedure based on 1000 replicates (Felsenstein, 1985; Saitou and Nei, 1987; Nei and Kumar, 2000; Kumar et al., 2016). The data sets of CSPs sequences which were chosen from other hemipteran species (Supplementary Table S2).
The qPCR Analysis
The relative expression levels of candidate chemosensory genes in forelegs, middle legs and hind legs of both sexes were examined by qPCR on an ABI Prism 7500 system (Applied Biosystems, Carlsbad, CA, United States). Reaction system contained a mixture of 10 μl 2 × SuperReal PreMix Plus (Tiangen Biotech, Beijing Co., Ltd.), 0.6 μl of each primer (10 μM), 200 ng sample cDNA, 0.4 μl 50 × ROX Reference Dye and proper volume of RNase-free water. PCR cycling parameters were as follows: 95°C for 15 min, followed by 40 cycles of 95°C for 10 s, cooled to 60°C for 32 s. Then, the PCR products were heated to 95°C for 15 s, cooled to 60°C for 1 min, heated to 95°C for 30 s and cooled to 60°C for 15 s to measure the dissociation curves. The GAPDH (GenBank accession No. JX987672) of A. lucorum stably expressed in different tissues (Supplementary Figure S1) was used as a reference gene for normalization (Ji et al., 2013). Primers (Supplementary Table S3) of the target and reference genes were designed using the Beacon Designer 7.9 (PREMIER Biosoft International, Palo Alto, CA, United States). A discrete amplification peak and a subsequent melting curve were evaluated to ensure the primer specificity. Each qPCR reaction for each sample was performed in three technical replicates and three biological replicates. Five serial tenfold dilutions of cDNA from each sample were amplified. For each dilution, amplifications were performed in triplicate using primers for the target gene and GAPDH. As a result, the absolute values of the slopes of all lines from template dilution plots (log cDNA dilution vs. ΔCT) were close to zero, indicating that the amplification efficiencies of the target and reference genes were approximately equal. Then, the comparative 2–Δ Δ CT method (Livak and Schmittgen, 2001) was used to calculate the relative expressions of different tissue samples. The comparative analyses of each target gene among various samples and sexes were determined using a two-way analysis of variance (ANOVA), followed by least significant difference (LSD) post-hoc test, P < 0.05 using the IBM SPSS Statistics 25.0 software (SPSS Inc., Chicago, IL, United States).
Observation of Sensilla on Legs of A. lucorum
The sensilla types on legs of A. lucorum were observed using a scanning electron microscopy (GeminiSEM 500, Zeiss, Germany). Forelegs, middle legs and hind legs were removed from female and male adult A. lucorum, respectively. Leg samples were fixed in 70% ethanol for 3 h, cleaned in an ultrasonic bath (250 W) for 10 s and finally subjected to gradient elution in an ethanol series (70, 80, 90, 95, and 100%). Subsequently, samples were dried in an oven thermostat at 25°C for 10 h. After coated with gold-palladium and mounting on holders, samples were observed under a scanning electron microscopy. Identification of the leg sensilla was mainly based on the description of Chinta et al. (1997) and Lu et al. (2007).
Results
Overview of Transcriptome
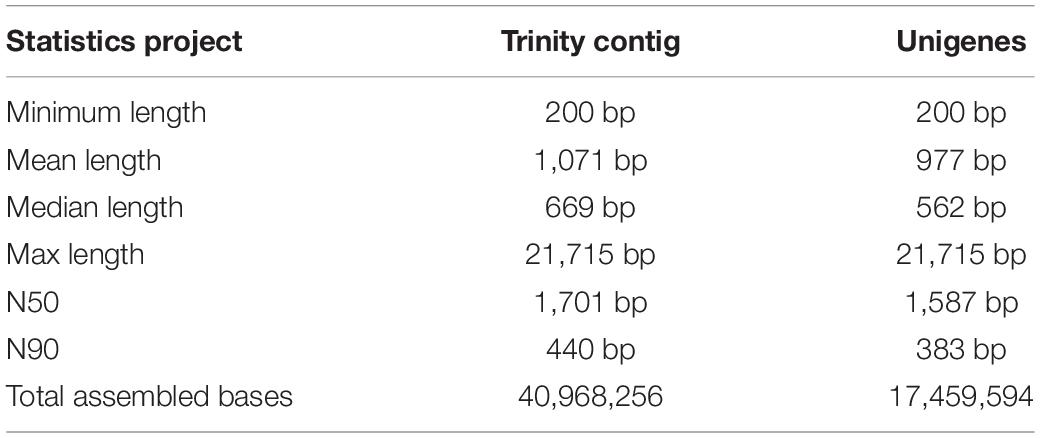
Six transcriptome data from forelegs, middle legs and hind legs of male and female were generated by HiSeq 2500 platform. A total of 106,020,038, 96,631,626, 110,435,520, 104,039,236, 90,768,110, and 106,745,344 raw reads were produced from six female and male leg samples (forelegs, middle legs, and hind legs), respectively. After filtering the low quality and adaptor sequences, we obtained 102,528,364, 93,578,672, 106,681,660, 100,468,622, 87,850,168, and 103,088,688 clean reads, respectively (Supplementary Tables S4, S5). The assembly of all clean reads together led to the generation of 40,968,256 contigs with a mean length of 1,071 bp. After merging and clustering, 17,459,594 unigenes with a mean length of 977 bp and N50 of 1,587 bp were acquired (Table 2). Of the clean reads, the Q30 percentage (proportion of sequences with a sequencing error rate less than 0.1%) for both libraries exceeded 94% (Supplementary Tables S4, S5). The length distributions of the unigenes were listed in Figure 1.
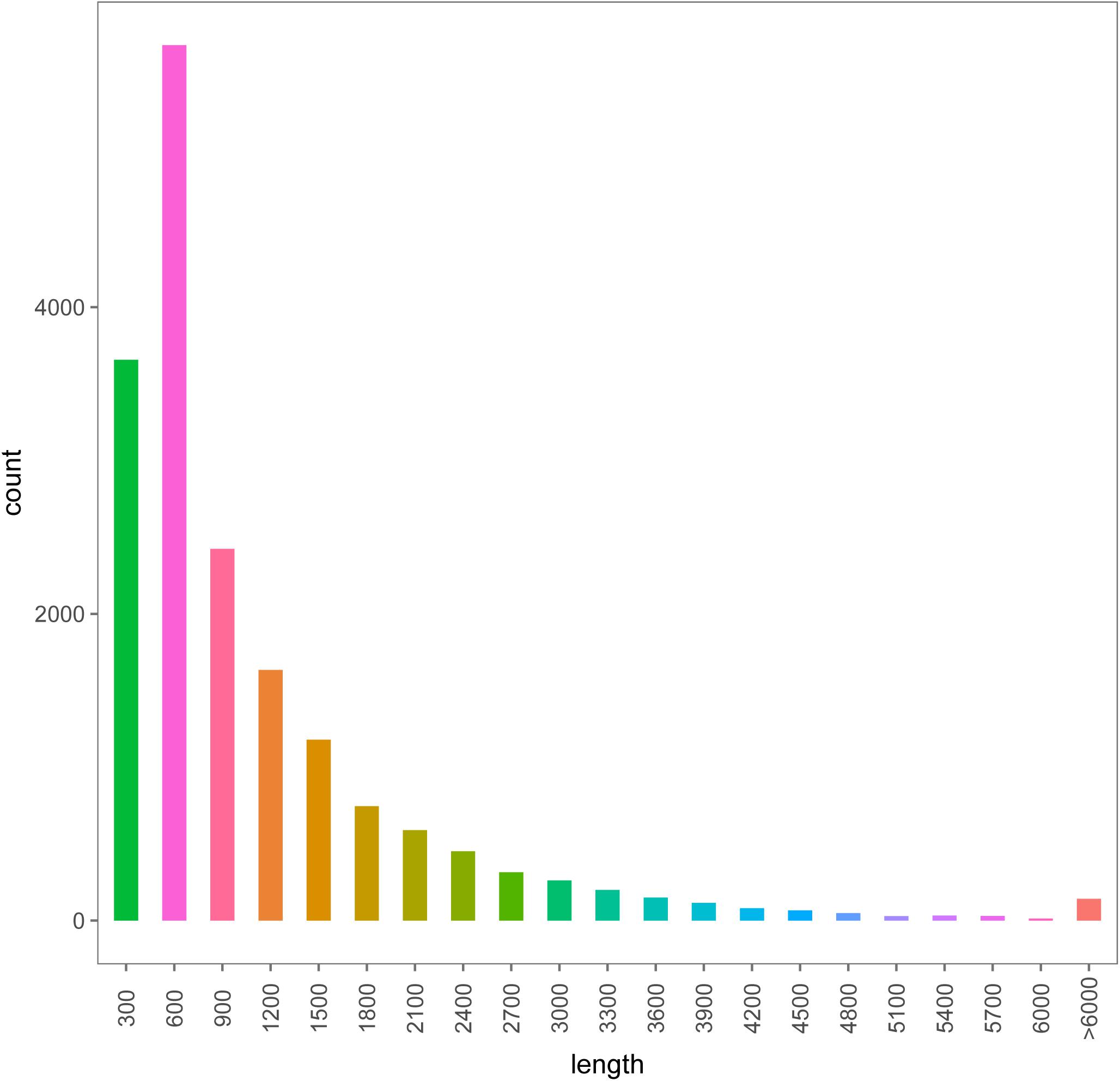
Figure 1. The length distributions of the assembled unigenes from legs transcriptomes of female and male A. lucorum.
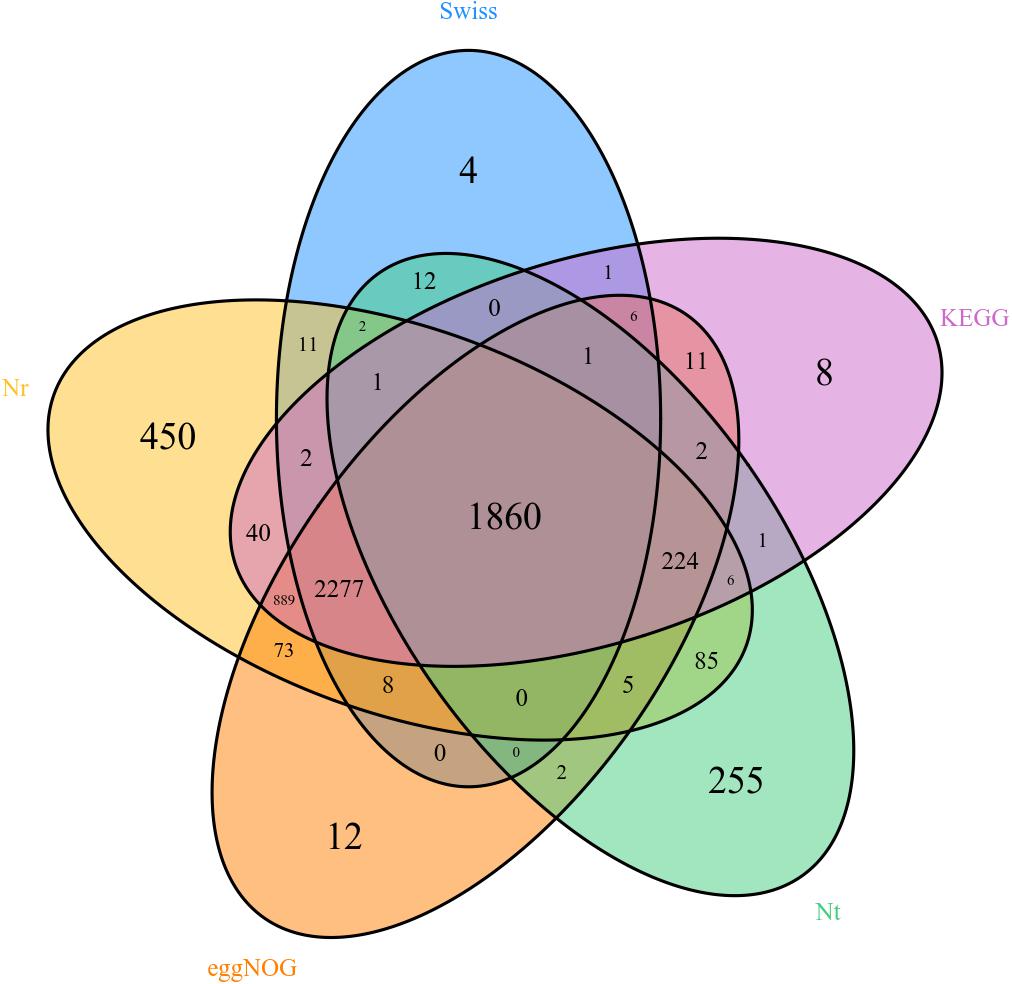
Unigenes were searched with blastx and blastn programs against the sequences in the NCBI GenBank database. The results showed that 5,933 out of the 17,459,594 unigenes had blastx hits in the non-redundant protein (nr) databases, and 2,456 unigenes had blastn hits in the non-redundant nucleotide sequence (nt) databases. Some unigenes were homologous to more than one species, and most of the annotated unigenes had the best hit with hemipteran insect genes (Figure 2). There are 358, 418, 562, 669, 1,347, 1,812, 2,186, 3,141, and 3,519 different genes in female forelegs vs. female middle legs, male middle legs vs. female middle legs, male middle legs vs. male forelegs, female forelegs vs. male forelegs, female hind legs vs. female middle legs, female hind legs vs. female forelegs, female hind legs vs. male hind legs, male middle legs vs. male hind legs, and male hind legs vs. male forelegs, respectively (Supplementary Table S6).
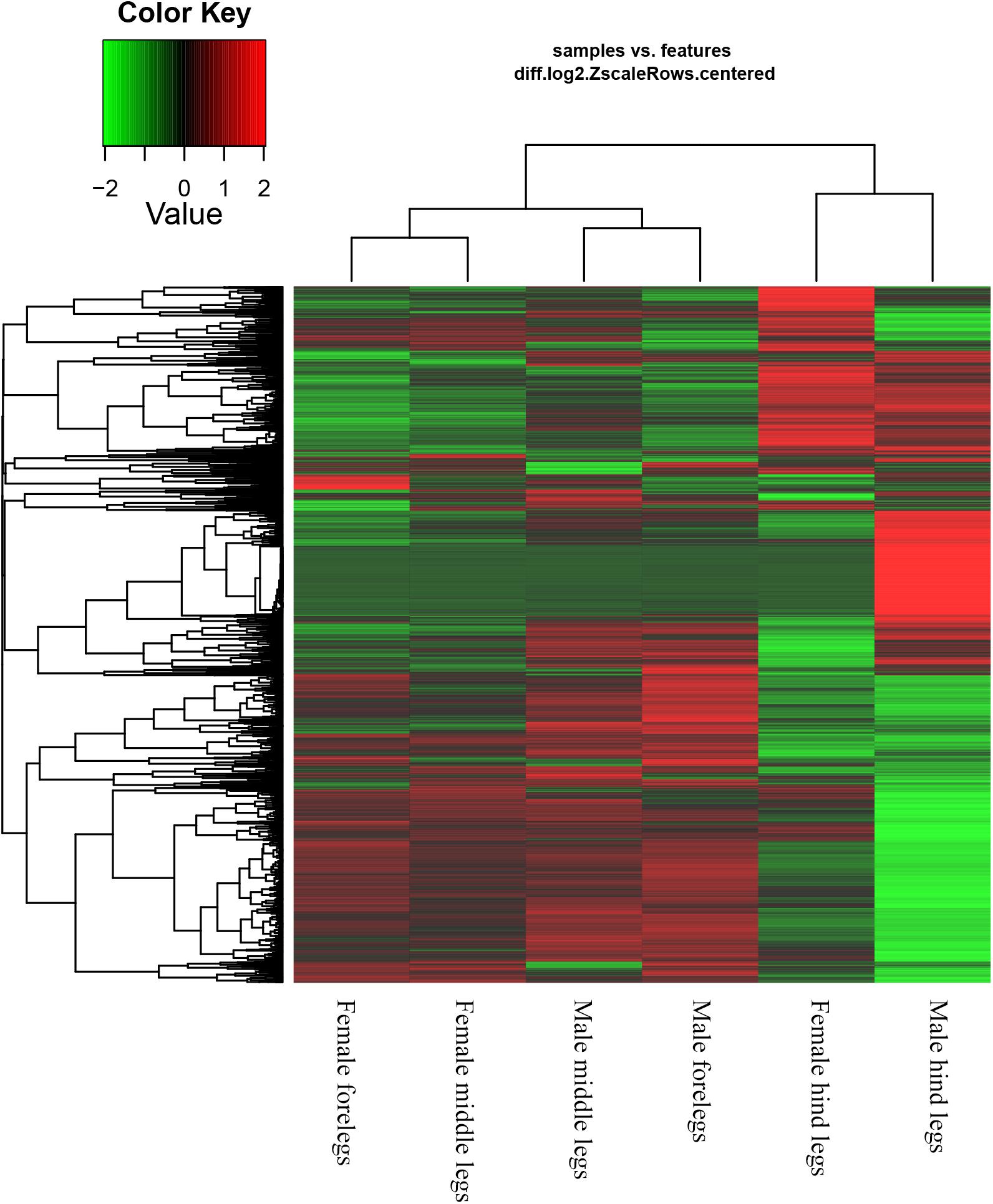
Based on the Gene Ontology (GO) annotations, 4,844 unigenes could be annotated into the following three functional categories: molecular function, cellular components and biological processes. The cellular process (31.92% unigenes) and metabolic process (28.47% unigenes) GO categories were most abundantly represented within the biological process GO. In the cellular components GO, the transcripts were mainly distributed in the membrane part (24.22% unigenes) and cell part (32.23% unigenes). The GO analysis also showed that the unigenes involved in binding (44.51% unigenes) and catalytic activity (44.12% unigenes) were most abundant in the molecular function ontology (Figure 3). The difference clustering results among six samples showed that there was a high correlation between female forelegs and female middle legs, male forelegs and male middle legs, female hind legs and male hind legs, separately (Figure 4).
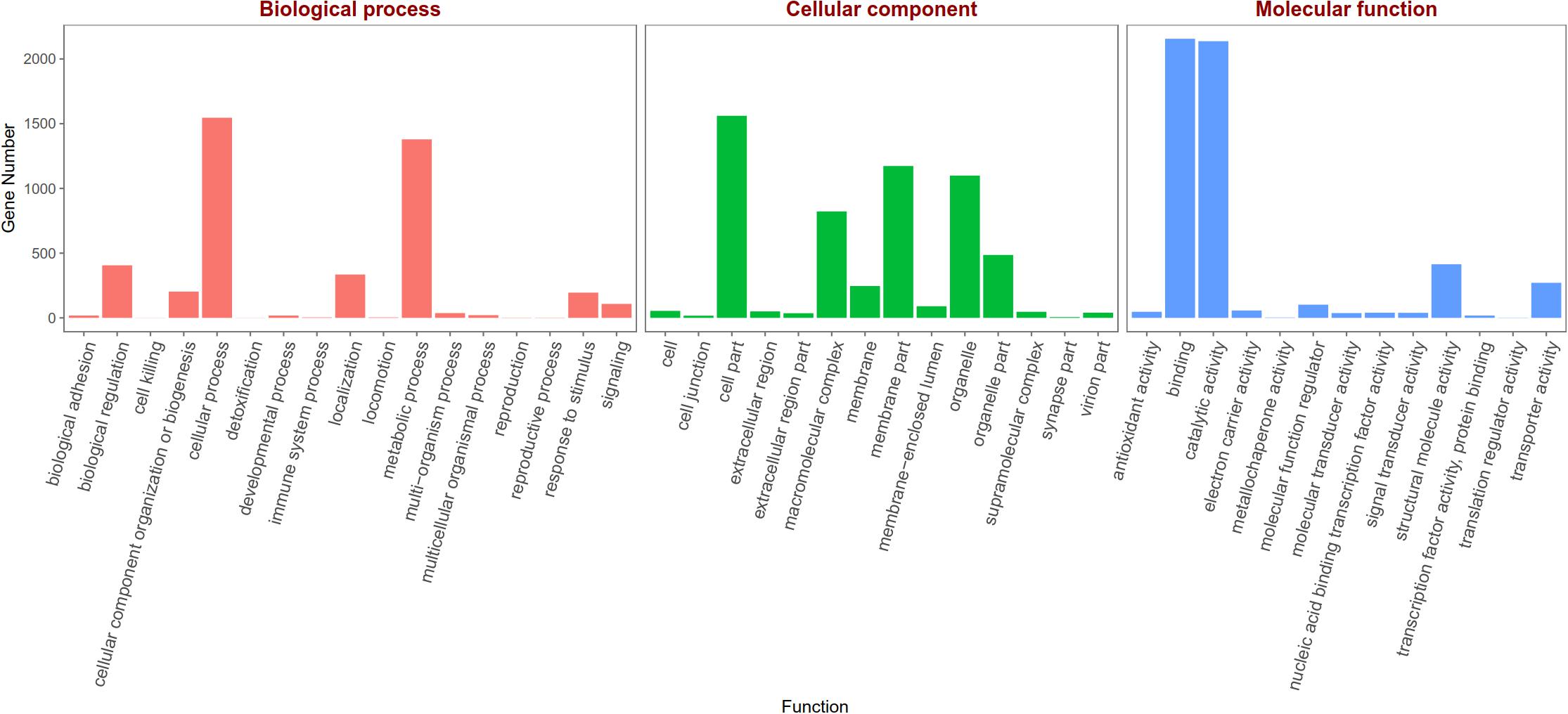
Figure 3. Gene Ontology (GO) classifications of legs transcripts annotated at GO level 2 according to the involvement in biological processes, cellular component and molecular function.
Putative Chemosensory Genes in A. lucorum Legs
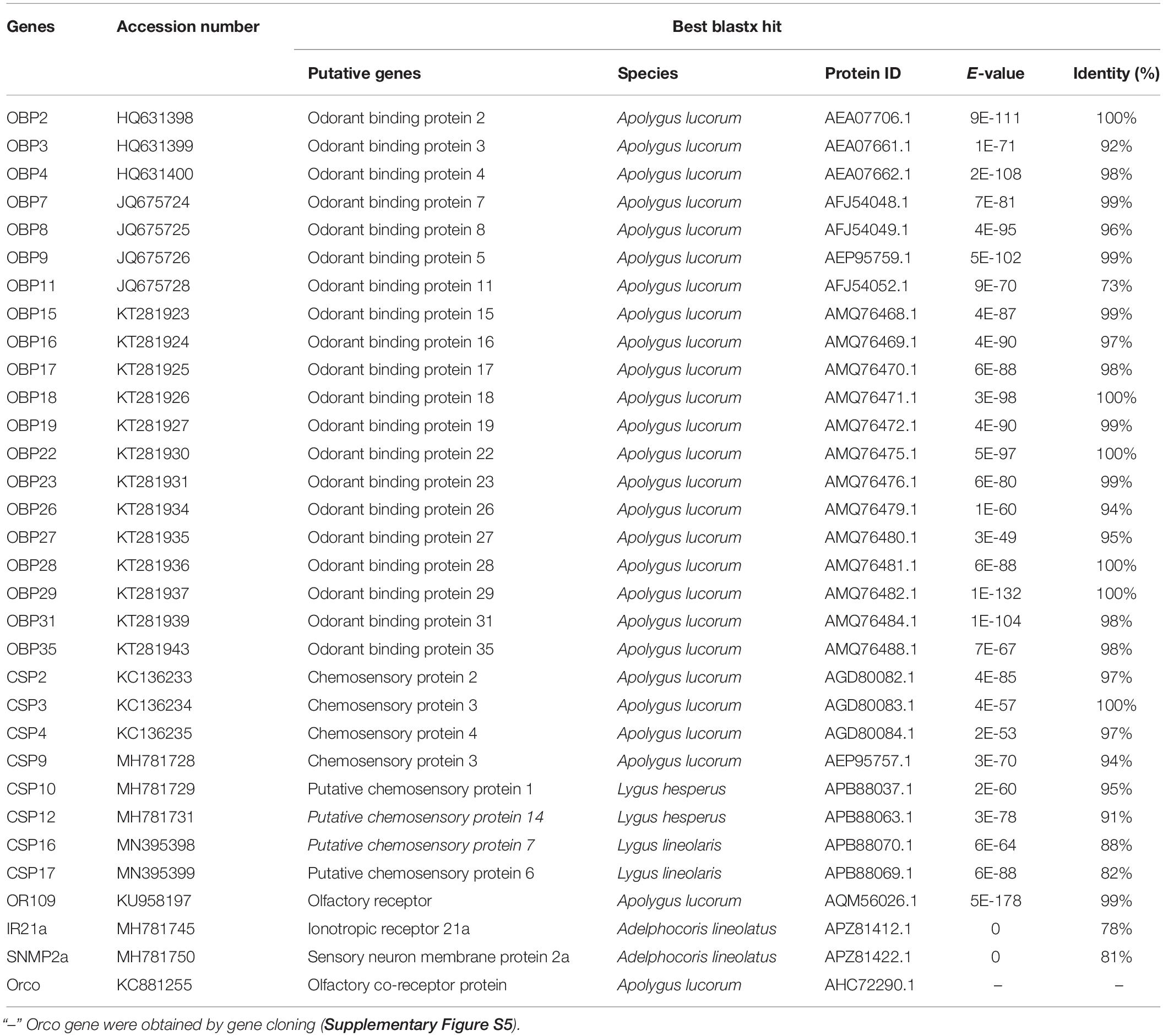
A total of 20 OBP genes, eight CSP genes, one OR gene, one IR gene, and one SNMP gene were identified (Table 3) in cDNA library of bug legs. Among these candidate chemosensory genes, Aluc-OBP2, 3, 4, 7, 8, 9, 11, 15, 16, 17, 18, 19, 22, 23, 26, 27, 28, 29, 31, 35; CSP2, 3, 4 and OR109 were previously reported in antennal transcriptome of A. lucorum (Hua et al., 2013; Yuan et al., 2015; An et al., 2016). However, CSP9, 10, 12, 16, 17, IR21a and SNMP2a (Table 3) were newly found and deposited in GenBank (accession numbers: MH781728, MH781729, MH781731, MN395398, MN395399, MH781745, and MH781750, respectively). All five candidate AlucCSPs represented the typical character of insect CSPs (Figure 5). In phylogenetic tree of 95 CSP sequences from hemipteran species (Supplementary Table S2), most of CSPs from same family were located in the same branch, whereas AlucCSP10, AlucCSP12, AlucCSP13, AlucCSP16, and AlucCSP17 were segregated into different central clusters (Figure 6).

Figure 5. Multiple sequences alignment of CSPs of A. lucorum. Amino acid sequences were aligned by ClustalX 2.1 and edited by GeneDoc 2.7.0 software. Yellow boxes show conserved cysteine residues and other conserved residues. Accession numbers of CSPs of A. lucorum are listed in Table 2.
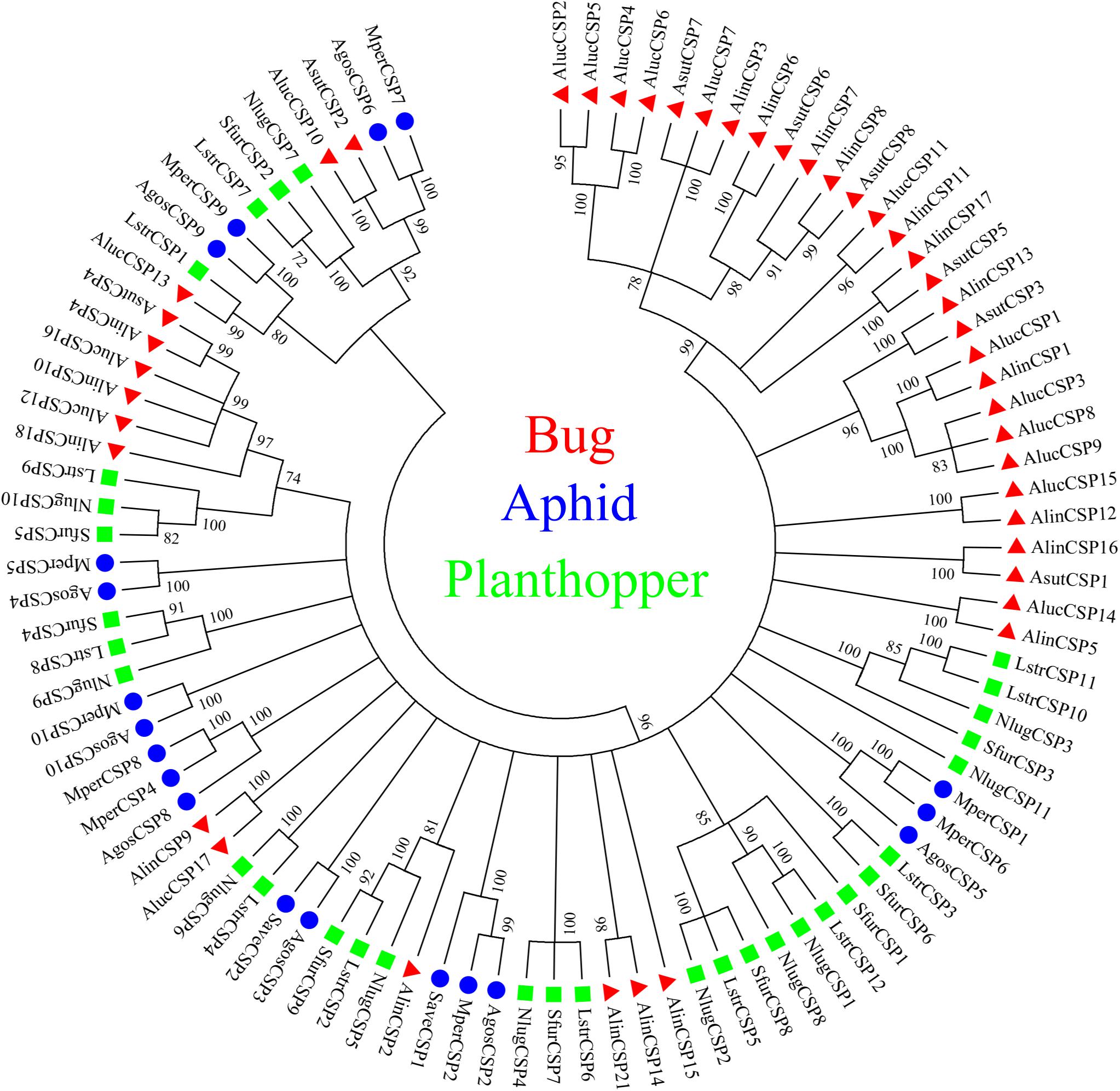
Figure 6. Phylogenetic analysis of CSPs. The CSP sequences used in this analysis are listed in Supplementary Table S4. Species abbreviations: Alin, Adelphocoris lineolatus; Aluc, Apolygus lucorum; Asut, Adelphocoris suturalis; Agos, Aphis gossypii; Mper, Myzus persicae; Save, Sitobion avenae; Sfur, Sogatella furcifera; Nlug, Nilaparvata lugens; Lstr, Laodelphax striatellus.
Expression Profiles of Chemosensory Genes in Legs
FPKM value analysis indicated that the AlucOBP9 was the most abundant (FPKM > 8000 in male forelegs, FPKM > 5000 in male middle legs, FPKM > 300 in male hind legs; FPKM > 5000 in female forelegs, FPKM > 3000 in female middle legs, FPKM > 1000 in female hind legs), followed by AlucOBP19 (FPKM > 800 in male forelegs, FPKM > 600 in male middle legs, FPKM > 25 in male hind legs; FPKM > 600 in female forelegs, FPKM > 400 in female middle legs, FPKM > 100 in female hind legs), AlucOBP3 (FPKM > 800 in male forelegs, FPKM > 600 in male middle legs, FPKM > 30 in male hind legs; FPKM > 500 in female forelegs, FPKM > 300 in female middle legs, FPKM > 90 in female hind legs), and AlucOBP26 (FPKM > 300 in male forelegs, FPKM > 300 in male middle legs, FPKM > 100 in male hind legs; FPKM > 200 in female forelegs, FPKM > 200 in female middle legs, FPKM > 200 in female hind legs). In term of CSPs, AlucCSP9 was the most abundant (FPKM > 2000 in male forelegs, FPKM > 2000 in male middle legs, FPKM > 700 in male hind legs; FPKM > 1900 in female forelegs, FPKM > 1800 in female middle legs, FPKM > 1000 in female hind legs), followed by AlucCSP2 (FPKM > 1400 in male forelegs, FPKM > 1600 in male middle legs, FPKM > 600 in male hind legs; FPKM > 1200 in female forelegs, FPKM > 1300 in female middle legs, FPKM > 1000 in female hind legs). FPKM values of both OR109 and SNMP2a were lower than 10, while FPKM values of IR21a were lower than 150 (Supplementary Table S7).
The qPCR results proved that OBP9, OBP31, and CSP3 were highly expressed in male forelegs, while OBP2, OBP8, OBP17, CSP9, and CSP16 were highly expressed in female forelegs. In details, the expression levels of Aluc-OBP2, 8, 11, 17, CSP3, 9, 10 and CSP16 in female forelegs were significantly higher than that in female middle and hind legs. For males, Aluc-OBP9, 17, 31, and CSP3 genes showed high expression levels in the forelegs of male, and the gene expression levels were significantly different from those in the middle and hind legs of male. The expression levels of AlucOBP8, 17 and CSP16 in female forelegs were significantly higher than those in male forelegs, while the expression levels of AlucOBP9 and CSP3 in male forelegs were significantly higher than those in female forelegs. Moreover, OBP18 was highly expressed in male and female hind legs. Additionally, the expression levels of OBP4, OBP15, OBP26, OBP27, OBP28, OBP29, CSP2, CSP4, CSP17, OR109, IR21a and SNMP2a showed no significant difference among forelegs, middle legs and hind legs of both sexes (Figures 7–9).
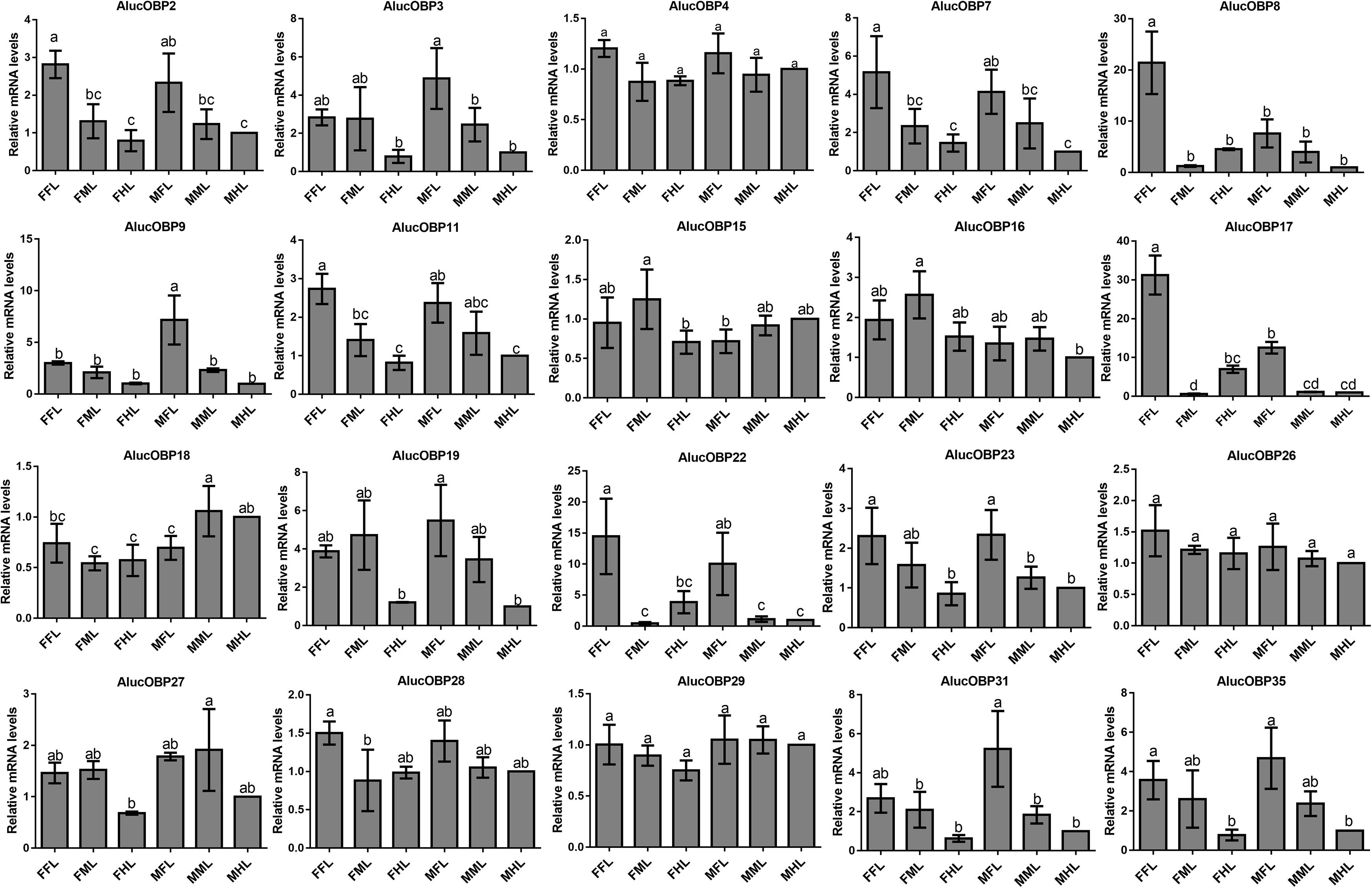
Figure 7. OBP transcript levels in different tissues of A. lucorum assessed by qPCR. FFL, female forelegs; FML, female middle legs; FHL, female hind legs; MFL, male forelegs; MML, male middle legs; MHL, male hind legs. The error bars present the standard error, and the different letters indicate significant differences (P < 0.05).
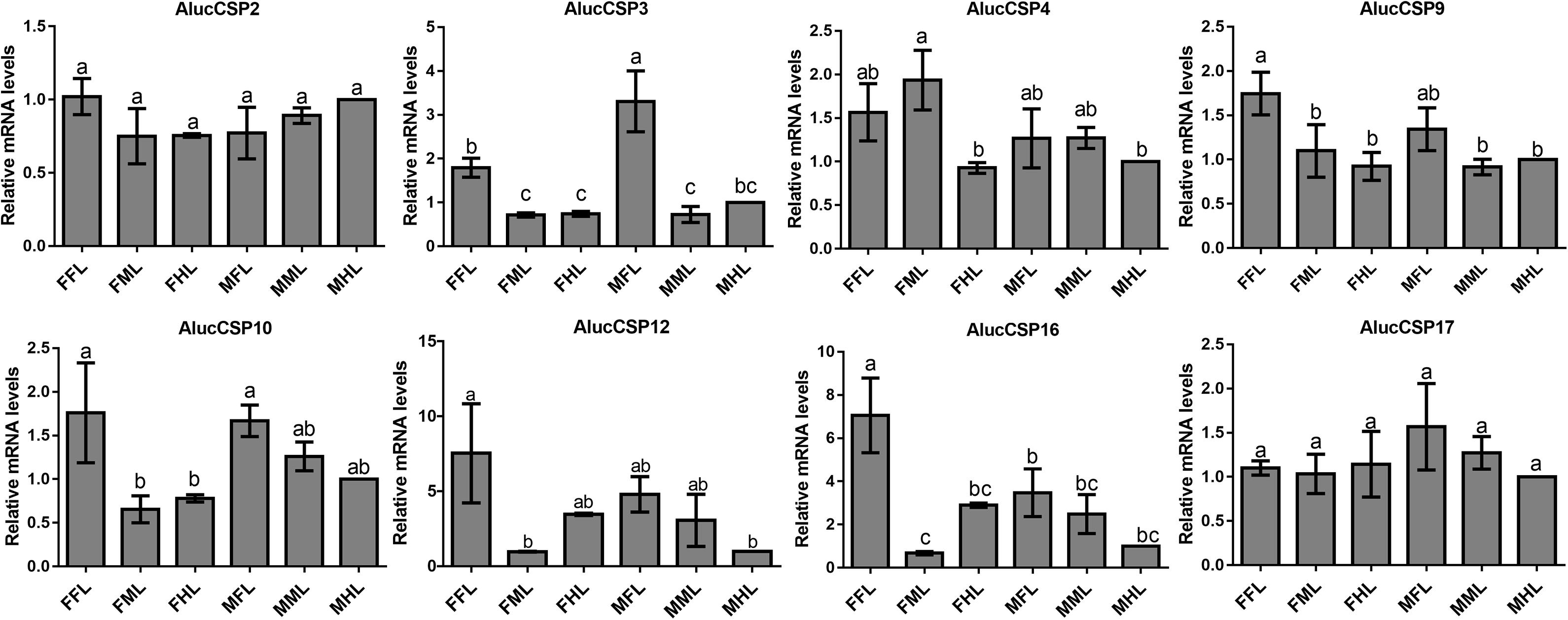
Figure 8. CSP transcript levels in different tissues of A. lucorum assessed by qPCR. FFL, female forelegs; FML, female middle legs; FHL, female hind legs; MFL, male forelegs; MML, male middle legs; MHL, male hind legs. The error bars present the standard error, and the different letters indicate significant differences (P < 0.05).
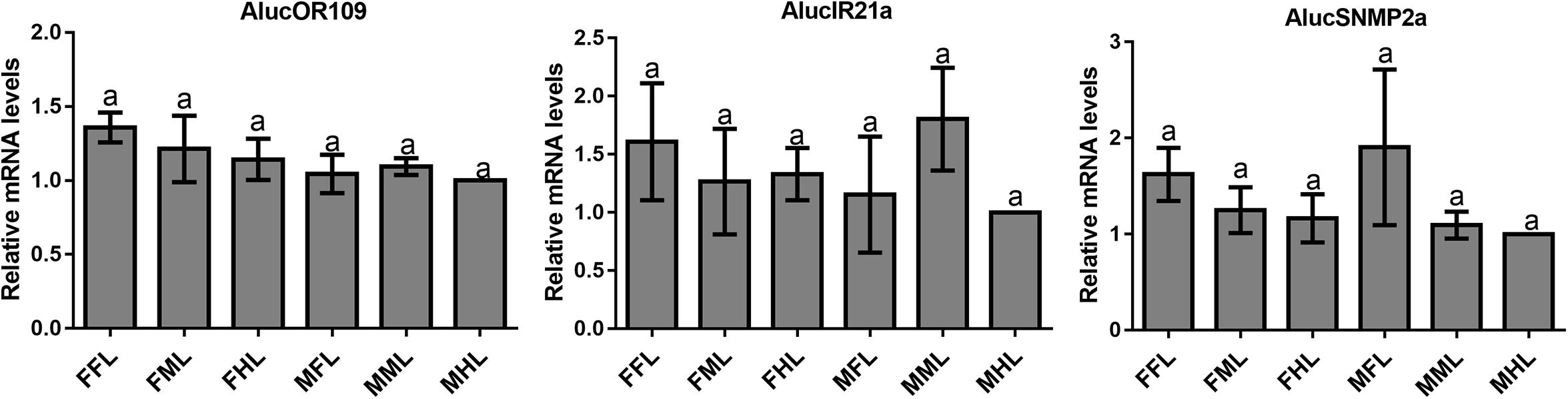
Figure 9. OR, IR and SNMP transcript levels in different tissues of A. lucorum assessed by qPCR. FFL, female forelegs; FML, female middle legs; FHL, female hind legs; MFL, male forelegs; MML, male middle legs; MHL, male hind legs. The error bars present the standard error, and the different letters indicate significant differences (P < 0.05).
Sensilla Types on Legs of A. lucorum
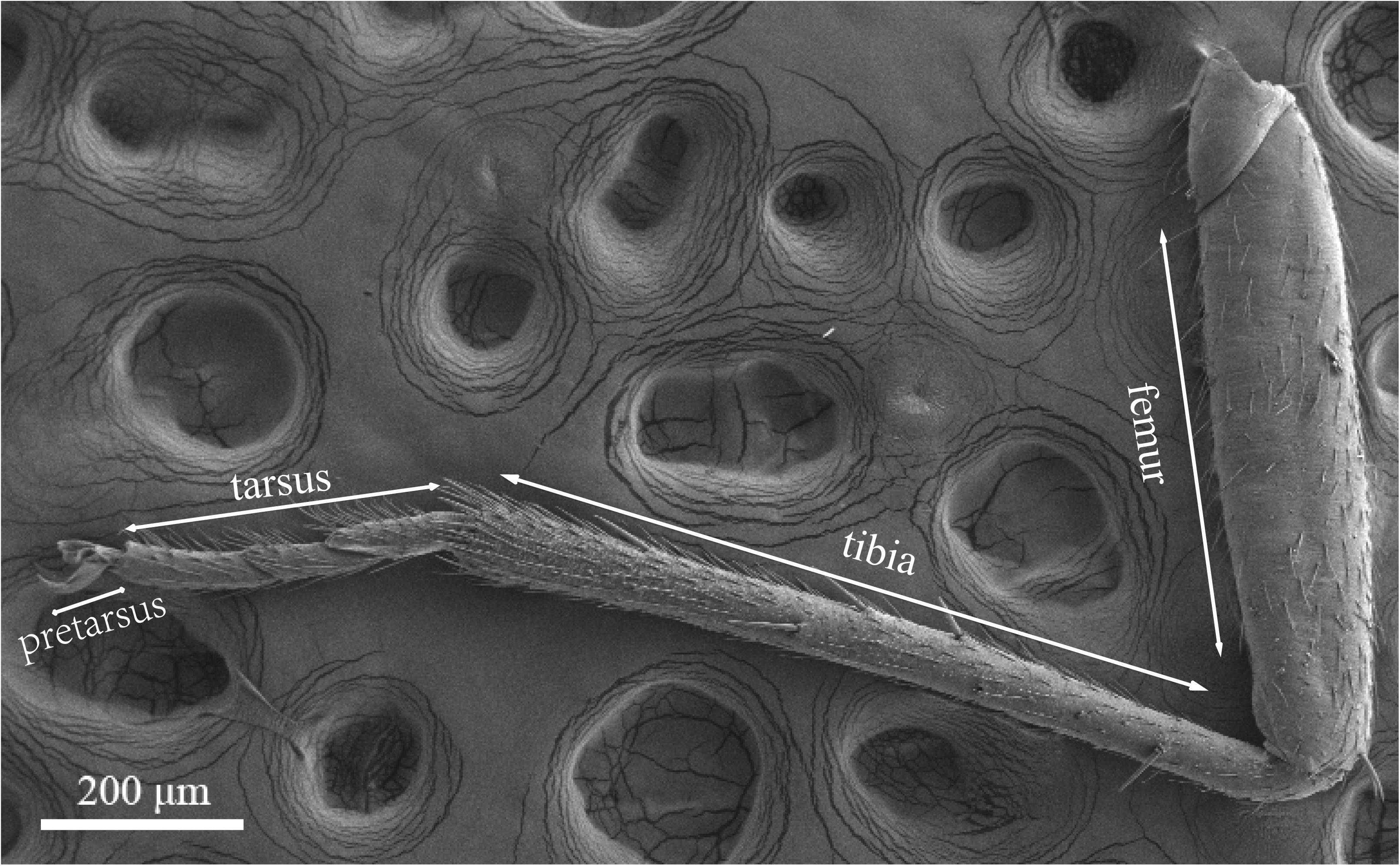
Legs of both males and females are consisted of four components: femur, tibia, tarsus and pretarsus (Figure 10). No sexual dimorphism was observed in the legs sensilla types. Scanning electron microscopy results showed that there were four sensilla types on legs of A. lucorum: sensilla trichodea (subtypes: long straight sensilla trichodea, Str1; long curved sensilla trichodea, Str2), sensilla chaetica (subtypes: sensilla chaetica 1, Sch1; sensilla chaetica 2, Sch2; and sensilla chaetica 3, Sch3), sensilla basiconca (subtypes: medium-long sensilla basiconca, Sba1; short sensilla basiconca, Sba2), and Böhm bristles (BB) (Figures 11–13). The types of sensilla of forelegs, middle legs and hind legs were the same (Supplementary Figures S2–S4). Additionally, the largest number of sensilla was on hind legs whereas the forelegs had the minimum number of sensilla.
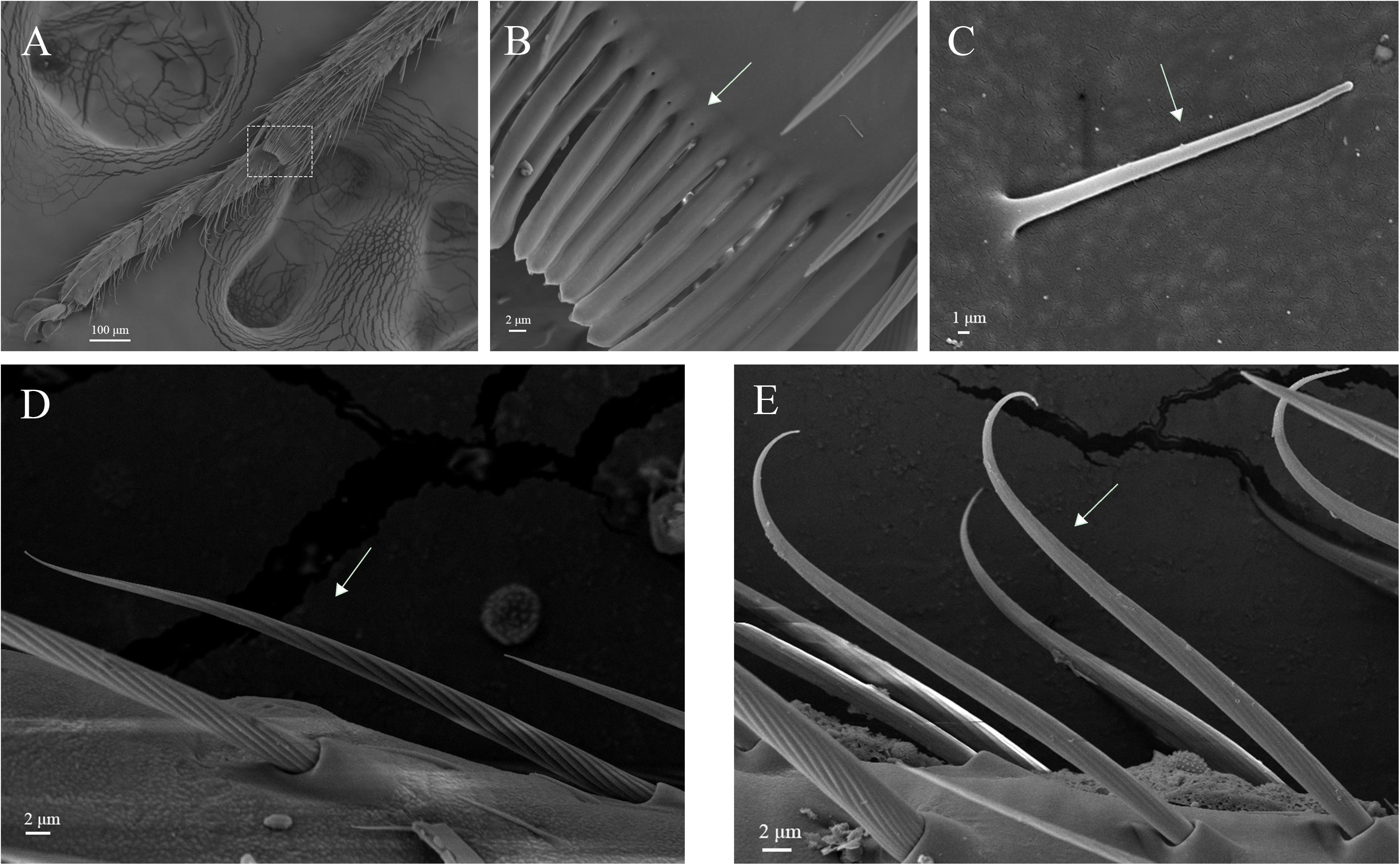
Figure 11. Hair brush, Böhm bristles and sensilla trichodea on legs of A. lucorum. (A) Brush at the junction of the tibia and tarsus. (B) Rows of hair brush with micropores at base. (C) Böhm bristles (BB). (D) Long straight sensilla trichodea (Str1). (E) Long curved sensilla trichodea (Str2).
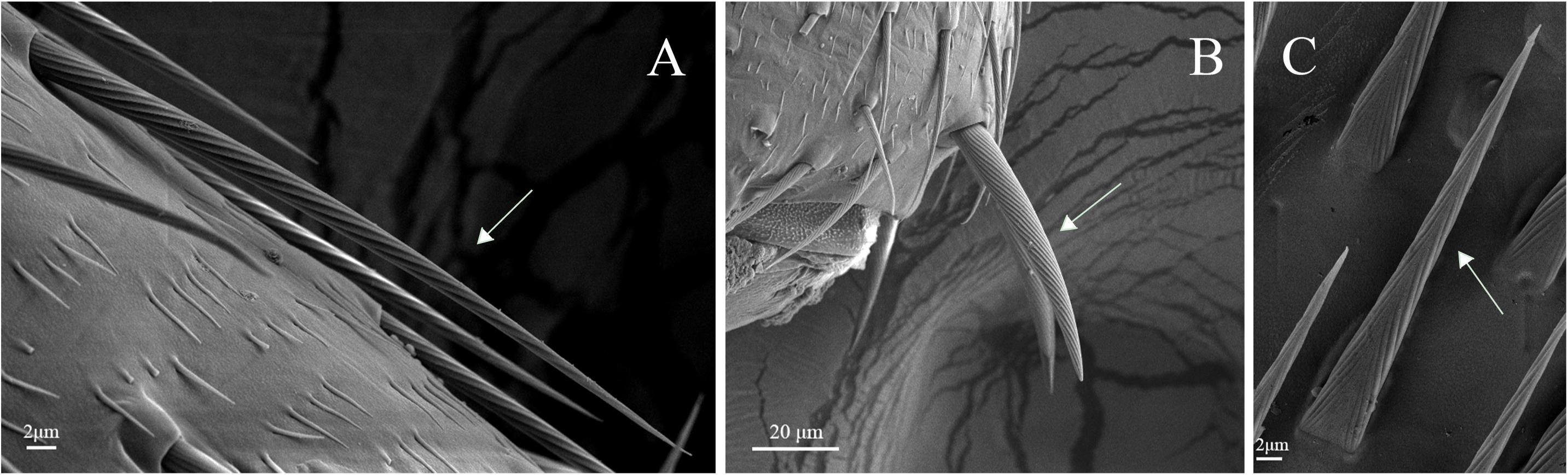
Figure 12. Sensilla chaetica on legs of A. lucorum. (A) Sensilla chaetica 1 (Sch1). (B) Sensilla chaetica 2 (Sch2). (C) Sensilla chaetica 3 (Sch3).
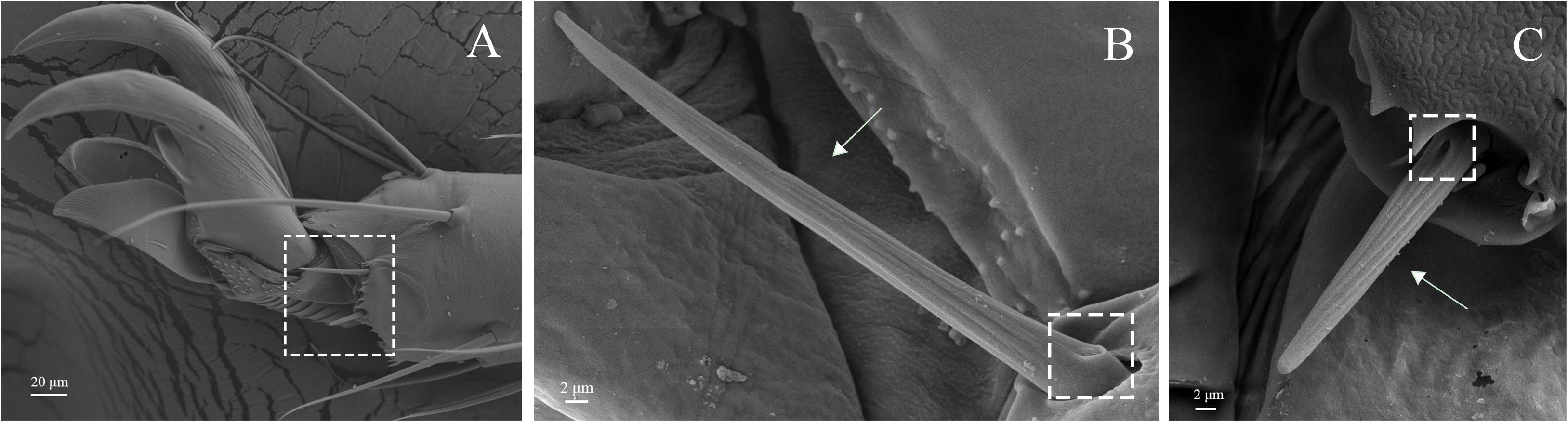
Figure 13. Sensilla basiconca on legs of A. lucorum. (A) Sensilla basiconca on distal of the tarsus. (B) Medium-long sensilla basiconca (Sba1). (C) Short sensilla basiconca (Sba2).
The rows of hair brush with micropores at base were observed in the junction of the tibia and tarsus (Figures 11A,B). The Böhm bristles had smooth surface without pores and base socket, which were distributed in all segments of the legs (Figure 11C).
Sensilla trichodea were divided into two different subtypes and presented in the tarsus of both sexes. The Str1 had a longitudinal grooved surface and softly pointed tip (Figure 11D). The Str2 had a longitudinal grooved surface and gradually curved with blunt tip (Figure 11E). Moreover, two subtypes of sensilla trichodea inserted in the cavities of the legs’ socket (Figures 11D,E). The sensilla chaetica were distributed in the tibia of both sexes. Based on their shapes, sensilla chaetica were further divided into three subtypes: Sch1, Sch2, Sch3 (Figure 12). Each sensillum had a grooved surface and a sharp tip. Sch1 and Sch2 inserted into a socket (Figures 12A,B). However, Sch3 was only distributed in the tibia of hind legs the base of which is directly attached to the cuticle without socket (Figure 12C). The sensilla basiconca were observed in the distal of the tarsus (Figure 13A). Based on their different length, sensilla basiconca were further divided into two subtypes: Sba1 and Sba2 (Figure 13). Each sensillum had a shallow grooved surface and a blunt tip with a pore at base inserting in the cavities of the legs’ socket (Figures 13B,C).
Discussion
In our previous study, up to 38 OBPs and 109 ORs have been found in the antennae of A. lucorum (Yuan et al., 2015; An et al., 2016). Here, we focused on the putative chemosensory genes in legs of both sexes of A. lucorum adults. There were 20 OBP genes, eight CSP genes, one OR gene, one IR gene and one SNMP gene identified in legs of A. lucorum (Table 3). The total number of chemosensory genes identified in legs of A. lucorum was much more than in legs of A. lineolatus (Sun et al., 2017). Moreover, the differences in clustering results among six tissue samples indicted that there was a high correlation between female forelegs and female middle legs, male forelegs and male middle legs, female hind legs and male hind legs, respectively, suggesting the similar chemosensory roles of forelegs and middle legs of both sexes, as well as female hind legs and male hind legs (Figure 4).
All the identified AlucOBPs and AlucOR109 in legs are also presented in antennae of A. lucorum (Yuan et al., 2015; An et al., 2016), indicating their potential chemosensory function in legs. In A. lineolatus, AlinOBP11 was labeled in the tarsal sensilla chaetica and involved in a complicated chemical recognition (Sun et al., 2017). A large number of ORs expressed in insect antennae (Touhara and Vosshall, 2009; Leal, 2013; An et al., 2016). However, we only identified one OR in the legs of A. lucorum. It was reported that HoblOR22 specifically expressed in Holotrichia oblita legs is a receptor of ligands (Li et al., 2017). We proposed that AlucOR109 in legs may be a receptor involved in olfaction or gustation of A. lucorum.
CSPs, another type of odorant carrier proteins, act as chemoreceptors to transport semiochemicals (Wanner et al., 2004). There were five new CSPs identified in legs of A. lucorum adults. In a previous study, AlucCSPs showed higher affinities with the secondary metabolites of cotton plants (Hua et al., 2013), suggesting their taste role in insect herbivores. In phylogenetic tree, although CSPs belonging to the same family (CSPs of bug, aphid and plant hopper) cluster together locally, the CSPs from same family segregate into different central clusters. Additionally, some CSPs from mirid bug species showed a very high protein identity and sat at the same branches as orthologous groups (AlucCSP10 and AsutCSP2; AlucCSP13 and LstrCSP1; AluCSP16 and AlinCSP10), which indicated these CSPs may have the same ancestor and differentiate along sex isolation and speciation (Figure 6).
Insect SNMPs play important roles in signal transduction. MmedSNMP2 from M. mediator is not only expressed in antennae, but also in head, legs and other tissues, suggesting the multiple roles of MmedSNMP2 (Shan et al., 2019). AlucSNMP2a identified in legs of A. lucorum may participate in multiple ecological functions. IR21a and IR25a in Drosophila could mediate cool sensing (Ni et al., 2016). Thus, AlucIR21a expressed in legs of bugs may perceive the changes of temperature.
The qPCR results indicated that all the putative chemosensory genes were ubiquitously expressed in forelegs, middle legs and hind legs of adult bugs. Additionally, most genes expressed in forelegs, middle legs and hind legs showed no significant difference such as Aluc-OBP4, OBP15, OBP26, OBP27, OBP28, OBP29, CSP2, CSP4, CSP17, OR109, IR21a and SNMP2a. The Aluc-OBP2, 8, 11, 17, CSP3, 9, 10 and CSP16 showed high expression levels in forelegs of female, and the gene expressions were significantly different from those in middle and hind legs of female (Figures 7, 8), which illustrate that those genes may play more crucial roles in chemosensory behavior in female forelegs than in female middle and female hind legs. On the other hand, Aluc-OBP9, 17, 31, and CSP3 genes showed high expression levels in the forelegs of male, and the gene expression levels were significantly different from those in the middle and hind legs of male (Figures 7, 8), suggesting that the differentially expressed genes in males may be involved in courtship behavior.
Furthermore, the expression differences of chemosensory genes in forelegs, middle legs and hind legs were compared between males and females. It was found that the expression levels of Aluc-OBP8, 17 and CSP16 in female forelegs were significantly higher than in male forelegs, indicating their more roles in female forelegs. The expression levels of Aluc-OBP9 and CSP3 in male forelegs were significantly higher than those in female forelegs, which suggest that these genes may be involved in male courtship in the near distance. The AlucOBP18 showed significantly higher expressions in male middle and hind legs than in female middle and hind legs, respectively, which implies AlucOBP18 may be responsible for the senses of touch and taste in male bugs. However, the expression levels of chemosensory genes except AlucOBP18 in female and male hind legs had no significant difference, revealing their same roles in both sexes (Figures 7–9). All in all, all the forelegs, middle legs and hind legs of bugs may be involved in close or contact chemical communication. Therefore, chemosensory genes with different expression profiles are associated with different physiological roles in legs of bugs.
Four types of sensilla were observed on the legs of A. lucorum, which is consistent with that of A. lucorum antennae (Lu et al., 2007). The types of sensilla identified on legs of A. lucorum were more than on legs of A. lineolatus. Moreover, the types of sensilla on legs of A. lucorum showed no sexual dimorphism. The similar results also appeared in A. lineolatus and Adelphocoris suturalis (Sun et al., 2017; Lu et al., 2009). In L. lineolaris and A. lineolatus, sensilla trichodea were involved in perception of olfactory stimuli (Chinta et al., 1997; Sun et al., 2013a). In lepidopteran species, sensilla trichodea are reported to respond to female-produced pheromones (Leal, 2005). Additionally, sensilla trichodea in lepidopteran females detect their own sex pheromone and lead to aggregating to increase chance of mating, moving away from calling females, or stimulating oviposition (Birch, 1977; Saad and Scott, 1981). We proposed that sensilla trichodea on legs of A. lucorum may play a vital role in olfactory behavior. Böhm bristles were distributed in all segments of legs of A. lucorum (Figure 11C). However, in antennae of Adelphocoris fasciaticollis, Böhm bristles are found at the scape and pedicel segments, especially abundant in the joints of the antennal segments (Sun et al., 2013b). Three types of sensilla chaetica were observed on legs of A. lucorum (Figure 12). In A. lineolatus, AlinOBP11 located in sensilla chaetica show highly binding abilities to the bitter substances catechin and quercetin (Sun et al., 2017). Thus, sensilla chaetica may be involved in the perception of bitter substances. Additionally, sensilla basiconca enriched in micropores contain abundant nerve cells, suggesting their potential chemoreception roles (Ochieng et al., 2000; Bleeker et al., 2004). In A. lineolatus, AlinOBP1 located in sensilla basiconca could bind the volatile compounds (Gu et al., 2011). Moreover, AfasOBP11 was located in the two types of sensilla basiconca in A. fasciaticollis (Li et al., 2019). In addition, AlinOBP13 expressed specifically in basiconic sensilla had strong binding affinity to terpenoids (Sun et al., 2014). In this work, sensilla basiconca with pores were found on legs of A. lucorum (Figure 13) and may be involved in olfactory perception. To sum up, we proposed that sensilla on legs of A. lucorum could play dual roles in gustation or olfaction.
Data Availability Statement
The datasets generated for this study can be found in the PRJNA590311.
Author Contributions
YJZ, ZL, and SL conceived and designed the experimental plan. ZL, YYZ, and XA performed the experiments. QW and AK analyzed the data. ZL and SG drafted the manuscript. SL and YJZ refined and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31772176, 31972338, 31672038, and 31621064).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00276/full#supplementary-material
Footnotes
- ^ https://github.com/trinityrnaseq/trinityrnaseq/wiki
- ^ ftp://ftp.ncbi.nlm.nih.gov/genomes/Gossypium_hirsutum
References
Abdel-Latief, M. (2007). A family of chemoreceptors in Tribolium castaneum (Tenebrionidae: Coleoptera). PLoS One 2:e1319. doi: 10.1371/journal.pone.0001319
Adachi, T., Ishii, K., Matsumoto, Y., Hayashi, Y., Hamamoto, H., and Sekimizu, K. (2014). Niemann-Pick disease type C2 protein induces triglyceride accumulation in silkworm and mammalian cell lines. Biochem. J. 459, 137–147. doi: 10.1042/BJ20130876
An, X. K., Sun, L., Liu, H. W., Liu, D. F., Ding, Y. X., Li, L. M., et al. (2016). Identification and expression analysis of an olfactory receptor gene family in green plant bug Apolygus lucorum (Meyer-Dür). Sci. Rep. 6:37870. doi: 10.1038/srep37870
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Ave, D., Frazier, J. L., and Hatfield, L. D. (1978). Contact chemoreception in the tarnished plant bug Lygus lineolaris. Entomol. Exp. Appl. 24, 217–227. doi: 10.1111/j.1570-7458.1978.tb02776.x
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Birch, M. C. (1977). Response of both sexes of Trichoplusia ni (Lepidoptera: Noctuidae) to virgin females and to synthetic pheromone. Ecol. Entomol. 2, 99–104. doi: 10.1111/j.1365-2311.1977.tb00870.x
Bleeker, M. A. K., Smid, H. M., van Aelst, A. C., van Loon, J. J. A., and Vet, L. E. M. (2004). Antennal sensilla of two parasitoid wasps: a comparative scanning electron microscopy study. Microsc. Res. Tech. 63, 266–273. doi: 10.1002/jemt.20038
Calvello, M., Guerra, N., Brandazza, A., D’Ambrosio, C., Scaloni, A., Dani, F. R., et al. (2003). Soluble proteins of chemical communication in the social wasp Polistes dominulus. Cell. Mol. Life Sci. 60, 1933–1943. doi: 10.1007/s00018-003-3186-5
Chinta, S., Dickens, J. C., and Baker, G. T. (1997). Morphology and distribution of antennal sensilla of the tarnished plant bug, Lygus lineolaris (Palisot de beauvois) (Hemiptera: Miridae). Int. J. Insect Morphol. 2, 21–26. doi: 10.1016/S0020-7322(96)00022-0
Dahanukar, A., Lei, Y. T., Kwon, J. Y., and Carlson, J. R. (2007). Two Gr genes underlie sugar reception in Drosophila. Neuron 56, 503–516. doi: 10.1016/j.neuron.2007.10.024
Du, Y., Xu, K., Ma, W., Su, W., Tai, M., Zhao, H., et al. (2019). Contact chemosensory genes identified in leg transcriptome of Apis cerana cerana (Hymenoptera: Apidae). J. Econ. Entomol. 112, 2015–2029. doi: 10.1093/jee/toz130
Dunipace, L., Meister, S., McNealy, C., and Amrein, H. (2001). Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr. Biol. 11, 822–835. doi: 10.1016/s0960-9822(01)00258-5
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Frederick, L., Anupama, D., Linnea, A. W., Jae Young, K., and John, R. C. (2014). The molecular and cellular basis of taste coding in the legs of Drosophila. J. Neurosci 34, 7148. doi: 10.1523/JNEUROSCI.0649-14.2014
Gu, S. H., Wang, W. X., Wang, G. R., Zhang, X. Y., Guo, Y. Y., Zhang, Y. J., et al. (2011). Functional characterization and immunolocalization of odorant binding protein 1 in the lucerne plant bug, Adelphocoris lineolatus (GOEZE). Arch. Insect bioche. 2, 81–98. doi: 10.1002/arch.20427
He, Z., Luo, Y., Shang, X., Sun, J. S., and Carlson, J. R. (2019). Chemosensory sensilla of the Drosophila wing express a candidate ionotropic pheromone receptor. PLoS Biol. 17:e2006619. doi: 10.1371/journal.pbio.2006619
Hua, J. F., Zhang, S., Cui, J. J., Wang, D. J., Wang, C. Y., Luo, J. Y., et al. (2013). Functional characterizations of one odorant binding protein and three chemosensory proteins from Apolygus lucorum (Meyer-Dur) (Hemiptera: Miridae) legs. J. Insect Physiol 59, 690–696. doi: 10.1016/j.jinsphys.2013.04.013
Jaeger, A. H., Stanley, M., Weiss, Z. F., Musso, P. Y., Chan, R. C., Zhang, H., et al. (2018). A complex peripheral code for salt taste in Drosophila. eLife 7:e37167. doi: 10.7554/eLife.37167
Ji, P., Gu, S. H., Liu, J. T., Zhu, X. Q., Guo, Y. Y., Zhou, J. J., et al. (2013). Identification and expression profile analysis of odorant-binding protein genes in Apolygus lucorum (Hemiptera: Miridae). Appl. Entomol. Zool. 48, 301–311. doi: 10.1007/s13355-013-0188-0
Klijnstra, J., and Roessingh, P. (1986). Perception of the oviposition deterring pheromone by tarsal and abdominal contact chemoreceptors in Pieris brassicae. Entomol. Exp. Appl. 40, 71–79. doi: 10.1111/j.1570-7458.1986.tb02157.x
Koh, T. W., He, Z., Gorur-Shandilya, S., Menuz, K., Larter, N. K., Stewart, S., et al. (2014). The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865. doi: 10.1016/j.neuron.2014.07.012
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Lee, Y., Poudel, S., Kim, Y., Thakur, D., and Montell, C. (2018). Calcium taste avoidance in Drosophila. Neuron 97, 67–74. doi: 10.1016/j.neuron.2017.11.038
Leng, N., Dawson, J. A., Thomson, J. A., Ruotti, V., Rissman, A. I., Smits, B. M., et al. (2013). EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043. doi: 10.1093/bioinformatics/btt337
Li, K., Wei, H., Shu, C., Zhang, S., Cao, Y., Luo, C., et al. (2017). Identification and comparison of candidate odorant receptor genes in the olfactory and non-olfactory organs of Holotrichia oblita Faldermann by transcriptome analysis. Comp. Biochem. Physiol. Part D. Genomics Proteomics. 24, 1–11.
Li, Q., and Liberles, S. D. (2015). Aversion and attraction through olfaction. Curr. Biol. 25, R120–R129. doi: 10.1016/j.cub.2014.11.044
Li, Z. B., Wei, Y., Sun, L., An, X. K., Dhiloo, H. K., Zhang, Y. J., et al. (2019). Mouthparts enriched odorant binding protein AfasOBP11 plays a role in the gustatory perception of Adelphocoris fasciaticollis. J. Insect Physiol. 117:103915. doi: 10.1016/j.jinsphys.2019.103915
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, C., Zhu, F., Chen, L. Z., Zhou, L. J., and Lei, C. L. (2009). Observation on antennal sensilla of Adelphocoris suturalis with scanning electron microscopy. J. Appl. Entomol. 46, 879–882.
Lu, Y. H., Tong, Y. J., and Wu, K. M. (2007). Scanning electron microscope observation of the antennal sensilla of the A. lucorum. Acta Entomol. Sin. 50, 863–867. doi: 10.3321/j.issn:0454-6296.2007.08.016
Lu, Y. H., and Wu, K. M. (2008). Biology and Control of Cotton Mirids. Beijing: Golden Shield Press.
Lu, Y. H., Wu, K. M., Jiang, Y. Y., Xia, B., Li, P., Guo, Y. Y., et al. (2010). Mirid bug outbreaks in multiple crops correlated with wide-scale adoption of Bt cotton in China. Science 328, 1151–1154. doi: 10.1126/science.1187881
Ma, L., Li, Z. Q., Bian, L., Cai, X. M., Luo, Z. X., Zhang, Y. J., et al. (2016). Identification and comparative study of chemosensory genes related to host selection by legs transcriptome analysis in the tea Geometrid Ectropis obliqua. PLoS One 11:e0149591. doi: 10.1371/journal.pone.0149591
Maher, N., Thiery, D., and Stadler, E. (2006). Oviposition by Lobesia botranais stimulated by sugars detected by contact chemoreceptors. Physiol. Entomol. 31, 14–22. doi: 10.1111/j.1365-3032.2005.00476.x
Missbach, C., Dweck, H. K., Vogel, H., Vilcinskas, A., Stensmyr, M. C., Hansson, B. S., et al. (2014). Evolution of insect olfactory receptors. eLife 3:e02115. doi: 10.7554/eLife.02115
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. New York, NY: Oxford University Press.
Ni, L., Klein, M., Svec, K. V., Budelli, G., Chang, E. C., Ferrer, A. J., et al. (2016). The Ionotropic Receptors IR21a and IR25a mediate cool sensing in Drosophila. eLife 5:e13254. doi: 10.7554/eLife.13254
Nikolayeva, O., and Robinson, M. D. (2014). edgeR for differential RNA-seq and ChIP-seq analysis: an application to stem cell biology. Methods Mol. Biol. 1150, 45–79. doi: 10.1007/978-1-4939-0512-6_3
Ochieng, S. A., Park, K. C., Zhu, J. W., and Baker, T. C. (2000). Functional morphology of antennal chemoreceptors of the parasitoid Microplitis croceipes (Hymenoptera: Braconidae). Arthropod Struct. Dev. 29, 231–240. doi: 10.1016/S1467-8039(01)00008-1
Pelosi, P., Zhou, J. J., Ban, L. P., and Calvello, M. (2006). Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63, 1658–1676. doi: 10.1007/s00018-005-5607-0
Rimal, S., Sang, J., Poudel, S., Thakur, D., Montell, C., and Lee, Y. (2019). Mechanism of acetic acid gustatory repulsion in Drosophila. Cell Rep. 26, 1432–1442. doi: 10.1016/j.celrep.2019.01.042
Romani, R., Salerno, G., Frati, F., Conti, E., Isidoro, N., and Bin, F. (2005). Oviposition behaviour in Lygus rugulipennis: a morpho-functional study. Entomol. Exp. Appl. 115, 17–25. doi: 10.1111/j.1570-7458.2005.00268.x
Saad, A. D., and Scott, D. R. (1981). Repellency of pheromones released by females of Heliothis armigera and H. zea to females of both species. Entomol. Exp. Appl. 30, 123–127. doi: 10.1111/j.1570-7458.1981.tb03085.x
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Schultze, A., Schymura, D., Forstner, M., and Krieger, J. (2012). Expression pattern of a ‘Plus-C’ class odorant binding protein in the antenna of the malaria vector Anopheles gambiae. Insect Mol. Biol. 21, 187–195. doi: 10.1111/j.1365-2583.2011.01125.x
Scott, K., Brady, R., Cravchik, A., Morozov, P., Rzhetsky, A., Zuker, C., et al. (2001). A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104, 661–673. doi: 10.1016/S0092-8674(01)00263-X
Senthilan, P. R., Piepenbrock, D., Ovezmyradov, G., Nadrowski, B., Bechstedt, S., Pauls, S., et al. (2012). Drosophila auditory organ genes and genetic hearing defects. Cell 150, 1042–1054. doi: 10.1016/j.cell.2012.06.043
Shan, S., Wang, S. N., Song, X., Khashaveh, A., Lu, Z. Y., Dhiloo, K. H., et al. (2019). Molecular characterization and expression of sensory neuron membrane proteins in the parasitoid Microplitis mediator (Hymenoptera: Braconidae). Insect. Sci. doi: 10.1111/1744-7917.12667
Sun, L., Gu, S. H., Xiao, H. J., Zhou, J. J., Guo, Y. Y., Liu, Z. W., et al. (2013a). The preferential binding of a sensory organ specific odorant binding protein of the alfalfa plant bug Adelphocoris lineolatus AlinOBP10 to biologically active host plant volatiles. J. Chem. Ecol. 39, 1221–1231. doi: 10.1007/s10886-013-0333-9
Sun, L., Xiao, H. J., Gu, S. H., Guo, Y. Y., Liu, Z. W., and Zhang, Y. J. (2013b). Perception of potential sex pheromones and host-associated volatiles in the cotton plant bug, Adelphocoris fasciaticollis (Hemiptera: Miridae): morphology and electrophysiology. Appl. Entomol. Zool. 49, 43–57. doi: 10.1007/s13355-013-0223-1
Sun, L., Wang, Q., Wang, Q., Dong, K., Xiao, Y., and Zhang, Y. J. (2017). Identification and characterization of odorant binding proteins in the forelegs of Adelphocoris lineolatus (Goeze). Front. Physiol. 8:735. doi: 10.3389/fphys.2017.00735
Sun, L., Wei, Y., Zhang, D. D., Ma, X. Y., Xiao, Y., Zhang, Y. J., et al. (2016). The mouthparts enriched odorant binding protein 11 of the alfalfa plant bug Adelphocoris lineolatus displays a preferential binding behavior to host plant secondary metabolites. Front. Physiol. 7:201. doi: 10.3389/fphys.2016.00201
Sun, L., Xiao, H. J., Gu, S. H., Zhou, J. J., Guo, Y. Y., Liu, Z. W., et al. (2014). The antenna-specific odorant-binding protein AlinOBP13 of the alfalfa plant bug Adelphocoris lineolatusis expressed specifically in basiconic sensilla and has high binding affinity to terpenoids. Insect Mol. Biol. 23, 417–434. doi: 10.1111/imb.12089
Sun, L., Zhou, J. J., Gu, S. H., Xiao, H. J., Guo, Y. Y., Liu, Z. W., et al. (2015). Chemosensillum immunolocalization and ligand specificity of chemosensory proteins in the alfalfa plant bug Adelphocoris lineolatus (Goeze). Sci. Rep. 5:8073. doi: 10.1038/srep08073
Swarup, S., Williams, T. I., and Anholt, R. R. (2011). Functional dissection of odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 10, 648–657. doi: 10.1111/j.1601-183X.2011.00704.x
Touhara, K., and Vosshall, L. B. (2009). Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 71, 307–332. doi: 10.1146/annurev.physiol.010908.163209
Vogt, R. G., Miller, N. E., Litvack, R., Fandino, R. A., Sparks, J., Staples, J., et al. (2009). The insect SNMP gene family. Insect Biochem. Mol. Biol. 39, 448–456. doi: 10.1016/j.ibmb.2009.03.007
Wanner, K. W., and Robertson, H. M. (2008). The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 17, 621–629. doi: 10.1111/j.1365-2583.2008.00836.x
Wanner, K. W., Willis, L. G., Theilmann, D. A., Isman, M. B., Feng, Q. L., and Plettner, E. (2004). Analysis of the insect os-d-like gene family. J. Chem. Ecol. 30, 889–911. doi: 10.1023/b:joec.0000028457.51147.d4
Wicher, D., Schäfer, R., Bauernfeind, R., Stensmyr, M. C., Heller, R., Heinemann, S. H., et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011. doi: 10.1038/nature06861
Wynand, V. D. G. V. N., and Carlson, J. R. (2006). Insects as chemosensors of humans and crops. Nature 444, 302–307. doi: 10.1038/nature.05403
Xu, P., Atkinson, R., Jones, D. N., and Smith, D. P. (2005). Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 193–200. doi: 10.1016/j.neuron.2004.12.031
Yan, S. W., Zhang, J., Liu, Y., Li, G. Q., and Wang, G. R. (2015). An olfactory receptor from Apolygus lucorum (Meyer-Dur) mainly tuned to volatiles from flowering host plants. J. Insect Physiol. 79, 36–41. doi: 10.1016/j.jinsphys.2015.06.002
Yuan, H. B., Ding, Y. X., Gu, S. H., Sun, L., Zhu, X. Q., Liu, H. W., et al. (2015). Molecular characterization and expression profiling of odorant-binding proteins in Apolygus lucorum. PLoS One 10:e0140562. doi: 10.1371/journal.pone.0140562
Zhang, H. J., Xu, W., Chen, Q. M., Sun, L. N., Anderson, A., Xia, Q. Y., et al. (2018). Functional characterization of sensory neuron membrane proteins (SNMPs). BioRxiv. [preprint]. doi: 10.1101/262154
Zhang, Y. F., van Loon, J. J. A., and Wang, C. Z. (2010). Tarsal taste neuron activity and proboscis extension reflex in response to sugars and amino acids in Helicoverpa armigera (Hubner). J. Exp. Biol. 213, 2889–2895. doi: 10.1242/jeb.042705
Zhang, Z. J., Zhang, S. S., Niu, B. L., Palli, S. R., Wang, C. Z., Tan, A. J., et al. (2019). A determining factor for insect feeding preference in the silkworm. Bombyx mori. PLoS Biol. 17:e3000162. doi: 10.1371/journal.pbio.3000162
Zheng, Y., Wang, S. N., Peng, Y., Lu, Z. Y., Shan, S., Zhang, Y. J., et al. (2017). Functional characterization of a Niemann-Pick type C2 protein in the parasitoid wasp Microplitis mediator. Insect Sci. 25, 765–777. doi: 10.1111/1744-7917.12473
Keywords: Apolygus lucorum, legs, transcriptome sequencing, chemosensory genes, expression profiles, sensilla
Citation: Li Z, Zhang Y, An X, Wang Q, Khashaveh A, Gu S, Liu S and Zhang Y (2020) Identification of Leg Chemosensory Genes and Sensilla in the Apolygus lucorum. Front. Physiol. 11:276. doi: 10.3389/fphys.2020.00276
Received: 21 November 2019; Accepted: 11 March 2020;
Published: 15 April 2020.
Edited by:
Graziano Fiorito, Stazione Zoologica Anton Dohrn, ItalyReviewed by:
Giulia Di Cristina, University of Cologne, GermanySergio Angeli, Free University of Bozen-Bolzano, Italy
Copyright © 2020 Li, Zhang, An, Wang, Khashaveh, Gu, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongjun Zhang, eWp6aGFuZ0BpcHBjYWFzLmNu
 Zibo Li
Zibo Li Yaoyao Zhang
Yaoyao Zhang Xingkui An
Xingkui An Qi Wang
Qi Wang Adel Khashaveh
Adel Khashaveh Shaohua Gu
Shaohua Gu Shun Liu
Shun Liu Yongjun Zhang
Yongjun Zhang